Abstract
Aim
Cetuximab is a standard-of-care treatment for KRAS wild-type metastatic colorectal cancer (mCRC), but it may also be effective in a subgroup of KRAS mutant patients by its immunomodulatory activity. Here, we explore if KIR (killer cell immunoglobulin-like receptor) genotyping can provide a significant added value in the clinical outcome of patients with KRAS mutant mCRC based on cetuximab treatment.
Methods
We included 69 patients with histologically confirmed mCRC and KRAS mutation, positive EGFR expression, and Eastern Cooperative Oncology Group performance status ≤2. Based on KIR gene content, haplotype (A or B) was defined and genotypes (AA or Bx) were grouped for each patient.
Results
We demonstrated with new evidence the immunomodulatory activity of cetuximab in patients with KRAS mutant mCRC. Patients with homozygous genotypes (AA or BB) showed shorter 12-month progression-free survival (PFS12) and poorer overall survival (OS) than those with heterozygotes (AB). Moreover, multivariate analysis confirmed stratification of patients based on genotype was an independent marker of PFS12 (HR 2.16) and the centromeric and telomeric distribution of KIRs was an independent predictor of both PFS12 (HR 2.26) and OS (HR 1.93) in patients with mCRC with KRAS mutation treated with cetuximab.
Conclusions
Selection of patients with mCRC based on their KIR genotypes opens a therapeutic opportunity for patients with KRAS mutation, and it should be tested in clinical trials in comparison with other alternatives with scarce benefit.
Trial registration number
NCT01450319, EudraCT 2010-023580-18.
Keywords: biomarkers, tumor, natural killer t-cells, translational medical research, antibodies, neoplasm
Introduction
In 2018, almost 2 million new cases of colorectal cancer (CRC) and nearly 900,000 deaths were estimated. Overall, this cancer ranks third in terms of incidence but second in terms of mortality.1 At the time of diagnosis, about 75% of patients with CRC show metastases and surgery is not possible in most cases.2
In these patients, monoclonal antibodies (mAbs) are the most widely used targeted therapies, such as cetuximab, which inactivates Epidermal Growth Factor Receptor (EGFR) signaling. Cetuximab acts as a functional antagonist by blocking ligand binding to EGFR and therefore inhibits EGFR activation and downstream signaling in tumor cells, preventing cell proliferation, angiogenesis and metastatic tendencies.3 4 Unfortunately, it is not effective in all patients due to resistance to this therapy.5–9
Among the major downstream signaling activated by EGFR, the RAS-RAF-MAPK, PI3K-PTEN-AKT, and JAK/STAT pathways have been described as resistance mechanisms to antibody-mediated EGFR blocking. Any change in these elements, such as KRAS, NRAS, BRAF, and PIK3CA gene mutations, can lead to constitutive activation of EGFR, causing drug resistance.10 Therefore, the status of KRAS in the tumor affects response to cetuximab. It is demonstrated that this therapy is effective in patients with wild-type KRAS CRC; however, some studies have determined that, although the majority of cases of KRAS mutant CRC do not respond to cetuximab, some subjects could be sensitive under certain circumstances,11–13 suggesting that other mechanisms of action apart from EGFR blockage could be involved.14 The identification of this subgroup of patients is very relevant since 36%–46% of patients with metastatic colorectal cancer (mCRC), depending on tumor sidedness, have mutations in KRAS.15
Natural killer (NK) cells are an important subset of lymphocytes in the defense of organisms against viral infection and the development of tumors. They express many activating and inhibiting receptors that regulate their function by an equilibrium between incoming activating and inhibitory signals.16 When NK cells encounter a target cell, these signals are integrated and a response is formed immediately.17
As an IgG1 isotype mAb, cetuximab has two functional motifs: one is that it is able to bind to the extracellular domain of the EGFR and the other is that it can induce immune functions such as antibody-dependent cell-mediated cytotoxicity (ADCC), involving CD16 receptors present in NK cells.3
The antitumor activity of NK cells is not only mediated by CD16, but also by other receptors such as the KIR (killer cell immunoglobulin-like receptor) encoded by different genes (2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, 3DL2, 3DL3 and 3DS1) and pseudogenes (2DP1 and 3DP1). They can also be either activating or inhibitory and thereby are critical in the determination of NK cell activation.18 Therefore, the NK activity in patients with cancer treated with cetuximab is dependent not only on CD16, but also on the balance of signals from activating and inhibitory receptors on the NK cell surface such as the KIR.
The KIR region shows high genomic diversity mainly due to the fact that each person can have a variable number of the genes aforementioned, and it allows us to describe different haplotypes, semihaplotypes and genotypes.19 20 In each of them we can define subtype A or subtype B if the inhibitor or activator genes predominate, respectively. The goal of this variability is probably the diversification of the immune response in the context of a rapidly changing environment. Therefore the identification of KIR genotypes will help to elucidate the influence of these genes on different disease states such as cancer.21
The role of KIR haplotypes and genotypes in patients with KRAS mutant mCRC receiving cetuximab has not been studied. However, it is known that the interaction of KIRs with their human leukocyte antigen (HLA) ligands and CD16 through its fragment crystallizable (Fc) portion of IgG1 isotype antibodies can activate NK effector cells. Thus, the combined action of both may be translated into a superior antitumor effect of cetuximab.22 23 In this context, it seems reasonable that depending on the KIR genotype the response to cetuximab can be modified, since the final result of the NK response depends on the balance of activating and inhibitory signals.
We have previously demonstrated that patients with mCRC harboring FcγRII H131 and non-functional variant of KIR2DS4 showed a significant benefit in terms of prolonging time to progression and overall survival (OS).24 25 In this research, we explore if the use of the KIR genotypes and semihaplotypes could improve the identification of patients with KRAS mutant mCRC that would derive a benefit from cetuximab therapy.
Materials and methods
Study cohort
The design and the methods of this multicenter, phase II clinical trial have been previously reported,25 and a summary of the methods follows. Clinical data and biospecimens were collected from patients with mCRC enrolled between September 2011 and December 2013. Written informed consent was obtained from all patients before enrollment. Eligibility included patients 18 years or older with mCRC with verified KRAS mutation (all patients, except one, showed mutation in KRAS G12), positive EGFR (immunohistochemistry determination), carrier of polymorphism H131 allele in FcγRIIa and Eastern Cooperative Oncology Group (ECOG) performance status of 0–2. In our study, analyses were restricted to patients who had informative KIR genes to perform haplotype analyses (N=69). Details of patient recruitment are shown in online supplemental figure 1.
jitc-2020-001705supp001.pdf (42.1KB, pdf)
Baseline measurements
Several prognostic factors have been described in CRC, such as blood levels of carcinoembryonic antigen (CEA), lactate dehydrogenase (LDH) and β2 microglobulin.26–28 Therefore, they were evaluated before initiation of cetuximab therapy. Patients were grouped according to the upper limit of the normal range for each measure (>5 μg/L for CEA, 333 UI/L for LDH and 3 mg/L for β2 microglobulin).
KIR genotyping
To perform this analysis we used the previously determined presence or absence of KIR genes.25 Briefly, KIR Genotyping SSP Kit (Applied Biosystems) was used to determine the 17 KIR genes. Genotype-specific PCR products were amplified and later resolved using agarose gels, and the interpretation of the results was performed following the manufacturer’s instructions.
Based on gene content, haplotype (A or B) was defined and genotypes (AA or Bx, where X can be A or B) were grouped for each patient. Genotype AA is homozygous for A inhibitory haplotype (formed by 3DL3, 2DL3, 2DP1, 2DL1, 3DP1, 2DL4, 3DL1, 2DS4 and 3DL2). Bx genotype could be heterozygous (A and B haplotypes) or homozygous for B activator haplotype (constituted by 3DL3, 2DS2, 2DL2, 2DL5B, 2DS3, 2DP1, 2DL1, 3DP1, 2DL4, 3DS1, 2DL5A, 2DS3/2DS5, 2DS1 and 3DL2).
KIR haplotypes consist of two regions, centromeric (CEN) and telomeric (TEL), with different gene content. According to the presence or absence of one or more B haplotype-defining KIR genes, the genotypes for the CEN and TEL parts were defined. Thus, cenA (cA) is the CEN motif and telA (tA) is the TEL motif of the A haplotype; cenB (cB) and telB (tB) are the CEN and TEL motifs of the B haplotype, respectively.29 cenA is defined by centromeric 2DL3, cenB is composed of centromeric 2DS2, 2DL2, 2DL5B and 2DS3, telA is determined by telomeric 3DL1 and 2DS4, and telB is defined by telomeric 3DS1, 2DL5A, 2DS5 and 2DS1.
KIR B score
KIR B-content score is defined as the number of CEN and TEL gene content motifs having B haplotype-defining genes. It is calculated by adding the number of cenB and/or telB motifs in each genotype. Permissible values for the KIR B-content score are 0, 1, 2, 3, and 4.29 Additionally, a classification of KIR B status was performed: best (score ≥2, cenB/B and any tel), better (score ≥2, cenA/x and telB/x) and neutral (score 0–1).29
Statistical methods
Survival studies were carried out by Kaplan-Meier curves as well as Cox regression analysis. Twelve-month progression-free survival (PFS12) was defined as the time of survival until progression or death from any cause within 12 months after initial therapy. The OS period (months) was calculated from patient recruitment date until death due to any cause or last follow-up, when appropriate. Patients alive at last follow-up or those who started a new therapeutic regimen (differing from cetuximab) were censored. In such case, those patients were censored at the date of beginning a new drug scheme.
Kaplan-Meier curves as well as univariate and multivariate Cox regression study were developed to evaluate the impact of KIR genotypes, haplotypes and semihaplotypes on survival. Age and gender were included in the multivariate analysis as confounding variables. Data analysis was carried out with SPSS statistics software (V.20.0).
Results
Patient characteristics and clinical outcome
The demographic and disease characteristics of the cohort are shown in table 1. The median time of follow-up was 6.4 months (IQR 3.7–10.1 months). Fifty-four (78.3%) patients died during the study period, with a median OS of 6.30 (95% CI 3.4 to 8.2).
Table 1.
Baseline characteristics of patients included in the study
| Variable | Patients (N=69) |
| Age at diagnosis, years, median (IQR) | 64 (5.5–72.5) |
| Gender, n (%) | |
| Male | 35 (50.7) |
| Female | 34 (49.3) |
| Primary site, n (%) | |
| Colon | 51 (73.9) |
| Rectum | 18 (26.1) |
| Laterality, n (%) | |
| Right-sided | 16 (23.2) |
| Left-sided | 53 (76.8) |
| Number of metastatic sites, n (%) | |
| 1 | 26 (37.7) |
| 2 | 27 (39.1) |
| 3 or more | 16 (23.2) |
| ECOG performance status, n (%) | |
| 0 | 12 (17.4) |
| 1 | 51 (73.9) |
| 2 | 6 (8.6) |
| CEA basal, n (%) | |
| ≤ULN | 7 (10.1) |
| ≥ULN | 60 (87) |
| N/A | 2 (2.9) |
| LDH basal, n (%) | |
| ≤ULN | 34 (49.3) |
| ≥ULN | 30 (43.5) |
| N/A | 5 (7.2) |
| β2 microglobulin basal, n (%) | |
| ≤ULN | 52 (75.4) |
| ≥ULN | 8 (11.6) |
| N/A | 9 (13) |
CEA, carcinoembryonic antigen; ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase; N/A, not available; ULN, upper limit of the normal range.
KIR haplotype frequencies
Tables 2–4 show the genotype, semihaplotype and KIR B-content frequencies from the series of this study. All of them were included in the survival analysis.
Table 2.
Genotypes based on A and B haplotypes
| Genotype | Frequency (%) |
| AA | 16 (23.2) |
| AB | 33 (47.8) |
| BB | 20 (29) |
Table 3.
Distribution of variables in KIR centromere–telomere genotype
| Centromere–telomere | Frequency (%) |
| Centromere | |
| cAcA | 30 (43.5) |
| cBx | 37 (53.6) |
| N/A | 2 (2.9) |
| Telomere | |
| tAtA | 30 (43.5) |
| tBx | 37 (53.6) |
| N/A | 2 (2.9) |
| Centromere–telomere | |
| cAcA-tAtA | 16 (23.2) |
| cAcA-tBx | 15 (21.7) |
| cBx-tAtA | 14 (20.3) |
| cBx-tBx | 22 (31.9) |
| N/A | 2 (2.9) |
Bx, X can be A or B haplotype; cA, centromeric motif A haplotype; cB, centromeric motif B haplotype; KIR, killer cell immunoglobulin-like receptor; N/A, not available; tA, telomeric motif A haplotype; tB, telomeric motif of B haplotype.
Table 4.
KIR B-content score and KIR B status based on score and centromeric/telomeric localization of activator KIR genes
| B-content | Frequency (%) |
| Score | |
| 0 | 16 (23.2) |
| 1 | 23 (33.3) |
| 2 | 25 (36.2) |
| 3 | 4 (5.8) |
| 4 | 1 (1.4) |
| Status | |
| Neutral | 39 (56.5) |
| Better | 23 (33.3) |
| Best | 7 (10.1) |
KIR, killer cell immunoglobulin-like receptor.
The genotype distribution in the study population was 33 (47.8%) AB heterozygotes, 20 (29%) homozygotes for the B haplotype, and 16 (23.2%) homozygotes for the A haplotype (table 2). Since the gene content is different in the CEN and TEL regions, genotypes AA and Bx were examined based on their distribution. Four different combinations were found. The cAcA-tAtA was found in 23% of patients, cAcA-tBx and cBx-tAtA were observed in 22% and 20%, respectively, and cBx-tBx was found in 32% of patients; 2 (2.9%) patients were not informative (table 3). Regarding KIR B-content score, 39 patients showed scores of 0–1, 25 showed a score of 2, and 5 patients showed scores of 3–4 (table 4).
Survival analysis
Of the clinical and biochemical parameters included in the study, the number of metastatic sites was significantly associated with increased risk of death by univariate analysis (table 5).
Table 5.
Univariate analysis of clinical variables for 12-month progression-free survival and overall survival
| Univariate PFS12 | Univariate OS | |||
| Variable | HR (95% CI) | P value | HR (95% CI) | P value |
| Age (continuous) | 1.016 (0.991 to 1.042) | 0.20 | 1.010 (0.98 to 1.04) | 0.47 |
| Gender | ||||
| Male | 1 | 1 | ||
| Female | 1.074 (0.663 to 1.739) | 0.77 | 1.105 (0.65 to 1.88) | 0.71 |
| Primary site | ||||
| Rectum | 1 | 1 | ||
| Colon | 1.567 (0.901 to 2.727) | 0.11 | 1.193 (0.65 to 2.17) | 0.56 |
| Laterality | ||||
| Right-sided | 1 | 1 | ||
| Left-sided | 1.114 (0.632 to 1.965) | 0.71 | 1.373 (0.65 to 2.17) | 0.34 |
| Number of metastatic sites | ||||
| 1 | 1 | 1 | ||
| 2 | 1.277 (0.736 to 2.217) | 0.38 | 1.265 (0.67 to 2.40) | 0.47 |
| 3 or more | 1.750 (0.910 to 3.367) | 0.09 | 2.942 (1.463 to 5.915) | 0.002 |
| ECOG | ||||
| 0 | 1 | 1 | ||
| 1 | 1.037 (0.550 to 1.955) | 0.91 | 1.178 (0.57 to 2.43) | 0.66 |
| 2 | 1.154 (0.418 to 3.187) | 0.78 | 1.100 (0.36 to 3.30) | 0.87 |
| CEA basal | ||||
| ≤ULN | 1 | 1 | ||
| ≥ULN | 1.064 (0.482 to 2.351) | 0.87 | 1.151 (0.49 to 2.70) | 0.75 |
| LDH basal | ||||
| ≤ULN | 1 | 1 | ||
| ≥ULN | 1.173 (0.709 to 1.940) | 0.53 | 1.157 (0.66 to 2.02) | 0.61 |
| β2 microglobulin basal | ||||
| ≥ULN | 1 | 1 | ||
| ≤ULN | 1.281 (0.603 to 2.722) | 0.52 | 1.241 (0.58 to 2.67) | 0.58 |
P-values numbers marked in bold indicate those that are lower than 0.05.
CEA, carcinoembryonic antigen; ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase; OS, overall survival; PFS12, 12-month progression-free survival; p-values in bold, indicate those that are lower than 0.05; ULN, upper limit of the normal range.
The prognostic impact of specific known KRAS codon 12 mutations (G12C, G12V and G12D) on survival in patients with CRC has been previously described.30 31 Therefore, we assessed the impact of mutations at KRAS codon 12 on PFS12 and OS in our cohort. Significant differences in PFS12 or OS were not found between patients carrying p.G12C, p.G12V or p.G12D point mutations and those who carried any of the rest of the mutations, nor was any significant difference observed when mutations were stratified by transitions (p.G12D, p.G12R and p.G12S) and transversions (p.G12A, p.G12C and p.G12V) (online supplemental figure 2).
jitc-2020-001705supp002.pdf (988.4KB, pdf)
Regarding KIR genotypes, Cox regression analysis showed significant associations with PFS12 and OS (tables 6 and 7).
Table 6.
Univariate and multivariate analyses for 12-month progression-free survival
| Univariate | Multivariate | |||
| HR (95% CI) | P value | HR (95% CI) | P value* | |
| Genotype | ||||
| AB | 1 | 1 | ||
| AA | 2.27 (1.21 to 4.25) | 0.01 | 2.58 (1.31 to 5.10) | 0.006 |
| BB | 1.99 (1.12 to 3.55) | 0.02 | 1.93 (1.04 to 3.58) | 0.03 |
| Genotype | ||||
| AB | 1 | 1 | ||
| AA or BB | 2.10 (1.28 to 3.47) | 0.004 | 2.16 (1.26 to 3.78) | 0.005 |
| Centromere–telomere genotype | ||||
| cA/tB or cB/tA | 1 | 1 | ||
| cA/tA or cB/tB | 2.18 (1.29 to 3.67) | 0.003 | 2.26 (1.29 to 3.97) | 0.005 |
| Centromere genotype | ||||
| Bx/cenA | 1 | |||
| AA | 2.31 (1.08 to 4.95) | 0.03 | 2.50 (1.02 to 6.14) | 0.04 |
*Multivariate analysis includes age, gender and number of metastases as covariates for 12-month progression-free survival; p≤0.05 denotes statistical differences.
Bx, X can be A or B haplotype; cA, centromeric motif A haplotype; cB, centromeric motif B haplotype; p-values in bold, indicate those that are lower than 0.05; tA, telomeric motif A haplotype; tB, telomeric motif of B haplotype.
Table 7.
Univariate and multivariate analyses for overall survival
| Univariate | Multivariate | |||
| HR (95% CI) | P value | HR (95% CI) | P value* | |
| Genotype | ||||
| AB | 1 | |||
| AA | 2.08 (1.04 to 4.15) | 0.04 | 2.02 (0.97 to 4.2) | 0.06 |
| BB | 1.89 (1 to 3.55) | 0.05 | 1.58 (0.79 to 3.11) | 0.19 |
| Genotype | ||||
| AB | 1 | |||
| AA or BB | 1.96 (1.13 to 3.41) | 0.017 | 1.74 (0.97 to 3.15) | 0.06 |
| Centromere–telomere genotype | ||||
| cA/tB or cB/tA | 1 | 1 | ||
| cA/tA or cB/tB | 2.20 (1.23 to 3.93) | 0.007 | 1.93 (1.02 to 3.65) | 0.044 |
| Centromere genotype | ||||
| Bx/cenA | 1 | |||
| AA | 2.31 (0.99 to 5.37) | 0.05 | 1.67 (0.61 to 4.56) | 0.31 |
*Multivariate analysis includes age, gender and number of metastases as covariates for overall survival; p≤0.05 denotes statistical differences.
Bx, X can be A or B haplotype; cA, centromeric motif A haplotype; cB, centromeric motif B haplotype; p-values in bold, indicates those that are lower than 0.05; tA, telomeric motif A haplotype; tB, telomeric motif of B haplotype.
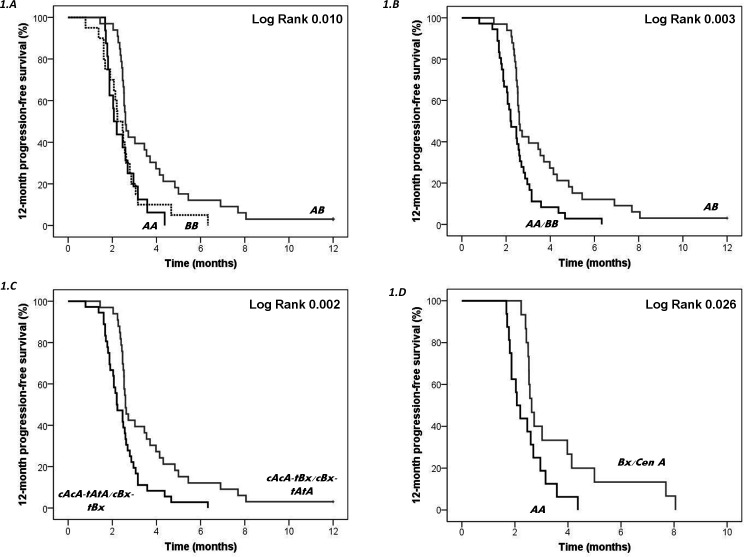
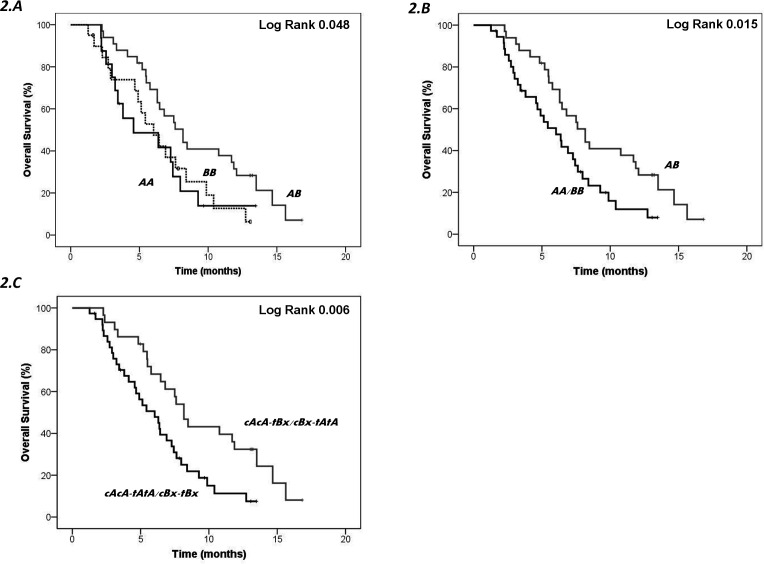
Homozygous patients carrying genotype AA or BB showed significantly lower PFS12 (p=0.01, log-rank test) (figure 1A) and OS (p=0.048, log-rank test) (figure 2A) than heterozygotes AB. When homozygous patients were grouped (AA+BB) and compared with heterozygotes (AB), the difference in Kaplan-Meier curves was more significant for both PFS12 (p=0.003, log-rank test) (figure 1B) and OS (p=0.015, log-rank test) (figure 2B), supporting homozygotes have poorer survival than heterozygotes. Genotypes AA and Bx were analyzed based on the CEN and TEL regions; cAcA-tAtA or cBx-tBx carriers had lower PFS12 (p=0.002, log-rank test) (figure 1C) and OS (p=0.006, log-rank test) (figure 2C) than cAcA-tBx or cBx-tAtA carriers. Moreover, genotype AA showed lower PFS12 than genotype Bx/cenA (p=0.026, log-rank test) (figure 1D).
Figure 1.
Kaplan-Meier curve for 12-month progression-free survival (PFS12) according to genotypes or centromeric and telomeric KIR content. Heterozygous individuals (AB) were compared with homozygotes (AA or BB) separately (A) or in combination (AA and BB) (B). The median PFS12 among homozygotes (AA+BB) was 2.21 months and among heterozygous patients 2.59 months (p=0.003, log-rank test). Genotypes (AA and Bx) were assessed based on the centromeric and telomeric regions (C) or only based on the centromeric region (D). The median PFS12 among homozygotes (cAcA-tAtA or cBx-tBx) was 2.28 months, whereas among heterozygotes (cAcA-tBx or cBx-tAtA) was 2.63 months (p=0.002, log-rank test). The median PFS12 among homozygotes (AA) was 2.13 months, whereas among heterozygotes (Bx/cenA) was 2.63 months (p=0.026, log-rank test). cenA (cA) and telA (tA), centromeric and telomeric motifs of the A haplotype; cenB (cB), centromeric motif of the B haplotype; KIR, killer cell immunoglobulin-like receptor; telB (tB), telomeric motif of the B haplotype.
Figure 2.
Kaplan-Meier curve for overall survival according to genotypes or centromeric and telomeric KIR content. Heterozygous individuals (AB) were compared with homozygotes (AA or BB) separately (A) or in combination (AA and BB) (B). The median overall survival among homozygotes (AA+BB) was 5.28 months and among heterozygous patients 7.60 months (p=0.015, log-rank test). Genotypes (AA and Bx) were assessed based on the centromeric and telomeric regions (C). The median overall survival among homozygotes (cAcA-tAtA or cBx-tBx) was 5.28 months, whereas among heterozygotes (cAcA-tBx or cBx-tAtA) was 8.17 months (p=0.006, log-rank test). cenA (cA) and telA (tA), centromeric and telomeric motifs of the A haplotype; cenB (cB), centromeric motif B haplotype; KIR, killer cell immunoglobulin-like receptor; telB (tB), telomeric motif of the B haplotype.
Multivariate logistic regression model, adjusted for number of metastatic sites and confounding variables (age and gender), showed patients with AA or BB genotypes had a twofold increased risk of progression (HR 2.16, 95% CI 1.26 to 3.78, p=0.005) compared with AB heterozygous (table 6) and confirmed this variable as an independent marker. Moreover, stratification of patients based on CEN and TEL distribution also showed that both CEN genotype and CEN–TEL genotype were independent predictors of PFS12 (table 6), with the latter also an independent predictor of OS in patients with KRAS mutated mCRC receiving cetuximab (table 7). cAcA-tAtA or cBx-tBx carriers showed a twofold increased risk of progression (HR 2.26, 95% CI 1.29 to 3.97, p=0.005) and death (HR 1.93, 95% CI 1.02 to 3.65, p=0.044) than cAcA-tBx or cBx-tAtA carriers.
Discussion
We have previously described higher OS in patients with KRAS mutant mCRC carrying the non-functional receptor KIR2DS4,25 and we also identified that polymorphisms in Fc gamma receptors (FcγRs) contribute to differences in the immune response to cetuximab.24 Now, this study provides new evidence that, in patients with KRAS mutant mCRC, cetuximab plays a primary role as an immunomodulatory treatment. Our results show how stratification of patients based on KIR genotypes (AA or BB vs AB) or based on CEN and TEL KIR distribution was an independent predictor of progression-free survival, with the latter also an independent biomarker of OS, with twofold increased risk of death for cAcA-tAtA or cBx-tBx carriers.
The immune-modulating effects of KIRs are regulated by the balance among inhibitors or activator signals, which depend on the number and type of receptors present in each individual.32 33 In this regard, KIR genes and their combination define two main groups of haplotypes (A and B). The A haplotype predominates genes encoding inhibitory receptors, while B haplotypes are more heterogeneous in their composition, presenting a greater number of activating receptors.34 Due to the evident strength of immunomodulatory activity of cetuximab, we hypothesized that KIR receptors and their genotype may influence the response of this treatment in patients with KRAS mutant mCRC.
Despite the essential role of individual KIR genes in modulating NK cell function, as proven by our group,35 the impact of the KIR genotypes on the outcome of patients with mCRC has not been sufficiently explored.
The association of different KIR genes and the susceptibility to develop CRC has been studied. In contrast to hematological malignancies, CRC seems to be affected by the hyperactivity of the NK cells.36 37
Even more interesting appears to be those studies that evaluate the effect of KIRs in different treatments. The anticancer treatments modify the tumor environment, the expression of HLA, and the release of tumor neoantigens, decreasing the tumor mass furthering as a whole that the immune system can be effective.38 39
The immunomodulatory effect of cetuximab has been studied in different types of cancers, including mCRC. Most of these studies have focused on the analysis of specific polymorphisms and their influence on ADCC response.40–42 However, the interaction between NK cells and target cells is more complex, involving many more molecules, among which KIRs play an important role. The intrinsic mechanism by which KIRs modulate the anticancer effects in patients with KRAS mutant mCRC receiving cetuximab is unknown. Activation signals mediated by both KIR and CD16 could be necessary for NK activity. As NK cells require a positive balance of activating signals, the joint action of KIR and CD16 could enhance cytotoxic activity. Veluchamy et al43 found that cetuximab enhanced NK cell cytotoxic activity in a CD16-dependent manner in EGFR-positive tumor cells regardless of the presence of RAS or BRAF mutations. These results support the importance of NK cell immunotherapy in combination with cetuximab for patients with RAS or BRAF mutant mCRC. Recently, the results obtained by Faden et al44 strongly suggest that HLA-C–KIR interaction is indeed important for NK cell activation in patients treated with cetuximab, and alterations in HLA-C may be involved in the immune evasion mechanism by affecting this activation.
However, none of these studies evaluated the contribution of KIR genotypes, as has been assessed in this work. In this study, we have analyzed how complete KIR genotypes can influence the outcome of patients with KRAS mutant mCRC treated with cetuximab. This aspect may be of great interest since the final effect mediated by the KIRs depends mainly on the balance of inhibitory and activating signals, as Braun et al45 highlighted in their study where ethnicity and tuberculosis status were analyzed. They found that the KIR profile and haplotype were more predictive than the presence or absence of individual genes.
Similarly, Siebert et al46 found that patients with neuroblastoma with B haplotype showed a higher level of ADCC and superior event-free survival (EFS) than those with A inhibitory haplotype. In bone marrow transplant, patients homozygous for A haplotype had an improved OS, higher EFS and non-relapse mortality when donors expressed at least one KIR B haplotype.47 It has also been described that allogeneic transplant donors carrying a KIR B haplotype and lacking a recipient HLA-C epitope protect against relapse from acute myeloid leukemia.48 Apart from these studies, there are few research analyzing the influence of specific combinations of KIR genotypes in the treatment response of solid cancers.
Our study indicates that patients with both an inhibitor haplotype and an activator (genotype AB) have longer progression-free survival and OS than homozygous AA or BB. We could explain these data because both activating and inhibitory KIRs can cause NK cell-mediated lysis, mediated since the NK cell kills tumor cells both when the ligand of an inhibitory KIR is missing and when an activating KIR binds to its ligand. Both circumstances can simultaneously occur in solid tumors treated with cetuximab. Loss of HLA ligands would facilitate cell death mediated by inhibitory KIRs, and the appearance of tumor neoantigens would facilitate tumor lysis by activating KIRs. Therefore, the AB genotype (with sufficient number of inhibitory and activating KIRs) would allow patients to be more sensitive to cetuximab, as found in our work where better outcome for patients with AB genotype was observed. Another fact that demonstrates that inhibitory and activating KIRs are interrelated and improve the clinical outcome is the biological interaction between HLA class I ligand and inhibitory KIR that determines NK-cell licensing and subsequent potential to respond to activating signals.49
The majority of these studies included patients with bone marrow tumors. Very few have been performed in solid tumors and most of them analyzed cancer risk rather than prognostic impact. Hernandez et al50 showed that Bx genotype, Bx centromere-Bx telomere, cA01|cB03, and tB01|tB01 were associated with risk of developing gastric cancer, and in a different study KIR CEN B haplotype was also associated with increased risk of multiple basal cell carcinoma.51
Different authors have tried to demonstrate whether the CEN and TEL semihaplotypes influence the course of treatments differently. Analysis comparing the effect of both halves and scales based on the number of activating semihaplotypes have been performed in different studies.29 Recently, Ureshino et al52 found an association between different alleles of KIR2DL4, 3DL1 and 2DS4 (present in telA) and treatment response. Other data seem to contradict the previous results and prioritize the role of 2DS1 and 3DS1 (present in telB).53 The presence of CEN and the absence of TEL KIR B haplotypes were associated with reduced relapse risk of leukemia after (hematopoietic stem-cell transplantation) HSCT for childhood acute lymphoblastic leukaemia (ALL).54 Cooley et al55 showed the presence of KIR B versus KIR A haplotypes makes better the clinical outcome of patients with acute myelogenous leukemia by decreasing the frequency of leukemic relapse and enhancing leukemia-free survival. Although CEN and TEL KIR B genes contributed to this effect, the CEN genes were dominant.
In our study, greater progression-free survival and OS were observed in patients with one activating (+) and one inhibitory (−) semihaplotype. Specifically, cAcA-tAtA (−, −) or cBx-tBx (+, +) carriers showed twofold increased risk of progression and death compared with cAcA-tBx (−, +) or cBx-tAtA (+, −) carriers. Again, the advantage of presenting enough number of inhibitory and activating KIRs is observed. This is similar to the study carried out by Mancusi et al53, who observed that donor activating KIRs had no effects on outcomes when donor-versus-recipient NK cell (inhibitory KIR) alloreactivity is lacking. We observed that both the CEN and the TEL semihaplotype appear to have the same power of action, unlike that observed in hematopoietic progenitor transplantation, although in that case the coexistence of KIR and different HLA ligands in donors and recipients may influence the observed results.
No difference was found when patients were classified according to the number of activating semihaplotypes (0–4) or according to the scale of Cooley et al (best, better or neutral).29 These data support the evidence that the presence of both activating and inhibitory KIRs is necessary and that the results are not proportional to the number of activators presented by each subject.
In our work, the role of KIR genotypes in patients with KRAS mutant mCRC treated with cetuximab has been evaluated, determining those that are associated with a better clinical outcome. However, our study has some limitations that should be considered. The study has been performed in a limited cohort and therefore should be validated in large-scale prospective studies to confirm whether the biological significance of KIR expression profile could be a fitting prognostic marker for patients with KRAS mutant mCRC treated with cetuximab.
Conclusions
We have demonstrated that a more precise selection of patients with mCRC including KIR genotypes can clearly provide a much higher clinical benefit. From a clinical point of view these new data in this subgroup of patients with KRAS mutant mCRC open a therapeutic opportunity to be considered and tested in clinical trials in comparison with other alternatives with scarce benefit.
Acknowledgments
The authors would like to thank all patients who took part in this clinical trial. They also want to thank all medical oncologists who carried out the recruitment and follow-up of patients and the scientific people involved in the laboratory work of this study.
Footnotes
BM-M and ACA contributed equally.
Contributors: Conception and design: JG-F and EA. Data curation: BM-M. Statistical analysis: LP-N and SS. Data analysis and interpretation: ACA, RG and MAG-E. Manuscript writing: BM-M, ACA, LP-N and RG. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding: This study was funded by Merck SL, an affiliate of Merck KGaA, Darmstadt, Germany (research project number 2010-023580-18, date: June 5, 2014). BM-M is a recipient of a contract from Programa Operativo Fondo Social Europeo 2014-2020 (D1113102S1) and Consejería de Salud y Familias Junta de Andalucía (HR-0060-2020).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. Data may be obtained from a third party and are not publicly available. Data are available from Translational Oncology Unit, Department of Oncology, Hospital Fundación Jiménez Díaz (principal investigator: JG-F, jesús.garciafoncillas@oncohealth.eu).
Ethics statements
Patient consent for publication
Not required.
Ethics approval
This study was approved by the Institutional Ethical Committee at the University Hospital Fundacion Jimenez Diaz (authorization number EC 02-12 IIS-FJD) and accepted by the other participating hospitals located across Spain. This study was performed in accordance with the Declaration of Helsinki. All methods were carried out in accordance with the approved guidelines and regulations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Cervantes A, Nordlinger B, et al. Metastatic colorectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25 Suppl 3:iii1–9. 10.1093/annonc/mdu260 [DOI] [PubMed] [Google Scholar]
- 3.García-Foncillas J, Sunakawa Y, Aderka D, et al. Distinguishing features of cetuximab and panitumumab in colorectal cancer and other solid tumors. Front Oncol 2019;9:849. 10.3389/fonc.2019.00849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shim H. One target, different effects: a comparison of distinct therapeutic antibodies against the same targets. Exp Mol Med 2011;43:539–49. 10.3858/emm.2011.43.10.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandhu J, Lavingia V, Fakih M. Systemic treatment for metastatic colorectal cancer in the era of precision medicine. J Surg Oncol 2019;119:564–82. 10.1002/jso.25421 [DOI] [PubMed] [Google Scholar]
- 6.Rachiglio AM, Lambiase M, Fenizia F, et al. Genomic profiling of KRAS/NRAS/BRAF/PIK3CA wild-type metastatic colorectal cancer patients reveals novel mutations in genes potentially associated with resistance to anti-EGFR Agents. Cancers 2019;11. 10.3390/cancers11060859. [Epub ahead of print: 20 06 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cremolini C, Benelli M, Fontana E, et al. Benefit from anti-EGFRs in RAS and BRAF wild-type metastatic transverse colon cancer: a clinical and molecular proof of concept study. ESMO Open 2019;4:e000489. 10.1136/esmoopen-2019-000489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montagut C, Tsui DW, Diaz LA. Detection of somatic Ras mutations in circulating tumor DNA from metastatic colorectal cancer patients: are we ready for clinical use? Ann Oncol 2018;29:1083–4. 10.1093/annonc/mdy091 [DOI] [PubMed] [Google Scholar]
- 9.Mauri G, Pizzutilo EG, Amatu A, et al. Retreatment with anti-EGFR monoclonal antibodies in metastatic colorectal cancer: systematic review of different strategies. Cancer Treat Rev 2019;73:41–53. 10.1016/j.ctrv.2018.12.006 [DOI] [PubMed] [Google Scholar]
- 10.Zhao B, Wang L, Qiu H, et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget 2017;8:3980–4000. 10.18632/oncotarget.14012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Cutsem E, Peeters M, Siena S, et al. Open-Label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 2007;25:1658–64. 10.1200/JCO.2006.08.1620 [DOI] [PubMed] [Google Scholar]
- 12.Dunn EF, Iida M, Myers RA, et al. Dasatinib sensitizes KRAS mutant colorectal tumors to cetuximab. Oncogene 2011;30:561–74. 10.1038/onc.2010.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13., et alGiordano G, Remo A, Porras A . Immune resistance and EGFR antagonists in colorectal cancer. Cancers 2019;11:1089. 10.3390/cancers11081089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misale S, Di Nicolantonio F, Sartore-Bianchi A, et al. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov 2014;4:1269–80. 10.1158/2159-8290.CD-14-0462 [DOI] [PubMed] [Google Scholar]
- 15.Bylsma LC, Gillezeau C, Garawin TA, et al. Prevalence of Ras and BRAF mutations in metastatic colorectal cancer patients by tumor sidedness: a systematic review and meta-analysis. Cancer Med 2020;9:1044–57. 10.1002/cam4.2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitale M, Cantoni C, Della Chiesa M, et al. An historical overview: the discovery of how NK cells can kill enemies, recruit defense troops, and more. Front Immunol 2019;10:1415. 10.3389/fimmu.2019.01415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long EO, Kim HS, Liu D, et al. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol 2013;31:227–58. 10.1146/annurev-immunol-020711-075005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bontadini A, Testi M, Cuccia MC, et al. Distribution of killer cell immunoglobulin-like receptors genes in the Italian Caucasian population. J Transl Med 2006;4:44. 10.1186/1479-5876-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin MP, Carrington M. Kir locus polymorphisms: genotyping and disease association analysis. Methods Mol Biol 2008;415:49–64. 10.1007/978-1-59745-570-1_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Middleton D, Gonzelez F. The extensive polymorphism of Kir genes. Immunology 2010;129:8–19. 10.1111/j.1365-2567.2009.03208.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Augusto DG. The impact of Kir polymorphism on the risk of developing cancer: not as strong as imagined? Front Genet 2016;7:121. 10.3389/fgene.2016.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binyamin L, Alpaugh RK, Hughes TL, et al. Blocking NK cell inhibitory self-recognition promotes antibody-dependent cellular cytotoxicity in a model of anti-lymphoma therapy. J Immunol 2008;180:6392–401. 10.4049/jimmunol.180.9.6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Erbe AK, Hank JA, et al. Nk cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol 2015;6:368. 10.3389/fimmu.2015.00368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez J, Zarate R, Bandres E, et al. Fc gamma receptor polymorphisms as predictive markers of cetuximab efficacy in epidermal growth factor receptor downstream-mutated metastatic colorectal cancer. Eur J Cancer 2012;48:1774–80. 10.1016/j.ejca.2012.01.007 [DOI] [PubMed] [Google Scholar]
- 25.Borrero-Palacios A, Cebrián A, Gómez Del Pulgar MT, et al. Combination of KIR2DS4 and FcγRIIa polymorphisms predicts the response to cetuximab in KRAS mutant metastatic colorectal cancer. Sci Rep 2019;9:2589. 10.1038/s41598-019-39291-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giessen C, Fischer von Weikersthal L, Laubender RP, et al. Evaluation of prognostic factors in liver-limited metastatic colorectal cancer: a preplanned analysis of the FIRE-1 trial. Br J Cancer 2013;109:1428–36. 10.1038/bjc.2013.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eker B, Ozaslan E, Karaca H, et al. Factors affecting prognosis in metastatic colorectal cancer patients. Asian Pac J Cancer Prev 2015;16:3015–21. 10.7314/APJCP.2015.16.7.3015 [DOI] [PubMed] [Google Scholar]
- 28.Shrout J, Yousefzadeh M, Dodd A, et al. beta(2)microglobulin mRNA expression levels are prognostic for lymph node metastasis in colorectal cancer patients. Br J Cancer 2008;98:1999–2005. 10.1038/sj.bjc.6604399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 2010;116:2411–9. 10.1182/blood-2010-05-283051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayama T, Hashiguchi Y, Okamoto K, et al. G12V and G12C mutations in the gene KRAS are associated with a poorer prognosis in primary colorectal cancer. Int J Colorectal Dis 2019;34:1491–6. 10.1007/s00384-019-03344-9 [DOI] [PubMed] [Google Scholar]
- 31.Li W, Liu Y, Cai S, et al. Not all mutations of KRAS predict poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol 2019;12:957–67. [PMC free article] [PubMed] [Google Scholar]
- 32.Bakker AB, Phillips JH, Figdor CG, et al. Killer cell inhibitory receptors for MHC class I molecules regulate lysis of melanoma cells mediated by NK cells, gamma delta T cells, and antigen-specific CTL. J Immunol 1998;160:5239–45. [PubMed] [Google Scholar]
- 33.Shilling HG, Guethlein LA, Cheng NW, et al. Allelic polymorphism synergizes with variable gene content to individualize human Kir genotype. J Immunol 2002;168:2307–15. 10.4049/jimmunol.168.5.2307 [DOI] [PubMed] [Google Scholar]
- 34.Martin AM, Freitas EM, Witt CS, et al. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (Kir) gene cluster. Immunogenetics 2000;51:268–80. 10.1007/s002510050620 [DOI] [PubMed] [Google Scholar]
- 35.Morales-Estevez C, De la Haba-Rodriguez J, Manzanares-Martin B, et al. Kir genes and their ligands predict the response to anti-EGFR monoclonal antibodies in solid tumors. Front Immunol 2016;7:561. 10.3389/fimmu.2016.00561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz-Peña R, Mondelo-Macía P, Molina de la Torre AJ, et al. Analysis of killer immunoglobulin-like receptor genes in colorectal cancer. Cells 2020;9:514. 10.3390/cells9020514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghanadi K, Shayanrad B, Ahmadi SAY, et al. Colorectal cancer and the KIR genes in the human genome: A meta-analysis. Genom Data 2016;10:118–26. 10.1016/j.gdata.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang T, Shi T, Zhang H, et al. Tumor neoantigens: from basic research to clinical applications. J Hematol Oncol 2019;12:93. 10.1186/s13045-019-0787-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Re V, Caggiari L, De Zorzi M, et al. Genetic diversity of the KIR/HLA system and outcome of patients with metastatic colorectal cancer treated with chemotherapy. PLoS One 2014;9:e84940. 10.1371/journal.pone.0084940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo Nigro C, Ricci V, Vivenza D, et al. Evaluation of antibody-dependent cell-mediated cytotoxicity activity and cetuximab response in KRAS wild-type metastatic colorectal cancer patients. World J Gastrointest Oncol 2016;8:222–30. 10.4251/wjgo.v8.i2.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trotta AM, Ottaiano A, Romano C, et al. Prospective evaluation of Cetuximab-Mediated antibody-dependent cell cytotoxicity in metastatic colorectal cancer patients predicts treatment efficacy. Cancer Immunol Res 2016;4:366–74. 10.1158/2326-6066.CIR-15-0184 [DOI] [PubMed] [Google Scholar]
- 42.Eriksson M, Leitz G, Fällman E, et al. Inhibitory receptors alter natural killer cell interactions with target cells yet allow simultaneous killing of susceptible targets. J Exp Med 1999;190:1005–12. 10.1084/jem.190.7.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veluchamy JP, Spanholtz J, Tordoir M, et al. Combination of NK cells and cetuximab to enhance anti-tumor responses in Ras mutant metastatic colorectal cancer. PLoS One 2016;11:e0157830. 10.1371/journal.pone.0157830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faden DL, Concha-Benavente F, Chakka AB, et al. Immunogenomic correlates of response to cetuximab monotherapy in head and neck squamous cell carcinoma. Head Neck 2019;41:2591–601. 10.1002/hed.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braun K, Larcombe L, Orr P, et al. Killer immunoglobulin-like receptor (Kir) centromeric-AA haplotype is associated with ethnicity and tuberculosis disease in a Canadian first nations cohort. PLoS One 2013;8:e67842. 10.1371/journal.pone.0067842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siebert N, Jensen C, Troschke-Meurer S, et al. Neuroblastoma patients with high-affinity FCGR2A, -3A and stimulatory KIR 2DS2 treated by long-term infusion of anti-GD2 antibody ch14.18/CHO show higher ADCC levels and improved event-free survival. Oncoimmunology 2016;5:e1235108. 10.1080/2162402X.2016.1235108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Symons HJ, Leffell MS, Rossiter ND, et al. Improved survival with inhibitory killer immunoglobulin receptor (Kir) gene mismatches and Kir haplotype B donors after nonmyeloablative, HLA-haploidentical bone marrow transplantation. Biol Blood Marrow Transplant 2010;16:533–42. 10.1016/j.bbmt.2009.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooley S, Parham P, Miller JS. Strategies to activate NK cells to prevent relapse and induce remission following hematopoietic stem cell transplantation. Blood 2018;131:1053–62. 10.1182/blood-2017-08-752170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boudreau JE, Hsu KC. Natural killer cell education and the response to infection and cancer therapy: stay tuned. Trends Immunol 2018;39:222–39. 10.1016/j.it.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernandez EG, Partida-Rodriguez O, Camorlinga-Ponce M, et al. Genotype B of killer cell immunoglobulin-like receptor is related with gastric cancer lesions. Sci Rep 2018;8:6104. 10.1038/s41598-018-24464-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vineretsky KA, Karagas MR, Christensen BC, et al. Skin cancer risk is modified by KIR/HLA interactions that influence the activation of natural killer immune cells. Cancer Res 2016;76:370–6. 10.1158/0008-5472.CAN-15-0547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ureshino H, Shindo T, Kojima H, et al. Allelic Polymorphisms of KIRs and HLAs Predict Favorable Responses to Tyrosine Kinase Inhibitors in CML. Cancer Immunol Res 2018;6:745–54. 10.1158/2326-6066.CIR-17-0462 [DOI] [PubMed] [Google Scholar]
- 53.Mancusi A, Ruggeri L, Urbani E, et al. Haploidentical hematopoietic transplantation from Kir ligand-mismatched donors with activating KIRs reduces nonrelapse mortality. Blood 2015;125:3173–82. 10.1182/blood-2014-09-599993 [DOI] [PubMed] [Google Scholar]
- 54.Babor F, Peters C, Manser AR, et al. Presence of centromeric but absence of telomeric group B Kir haplotypes in stem cell donors improve leukaemia control after HSCT for childhood all. Bone Marrow Transplant 2019;54:1847–58. 10.1038/s41409-019-0543-z [DOI] [PubMed] [Google Scholar]
- 55.Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol 2014;192:4592–600. 10.4049/jimmunol.1302517 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001705supp001.pdf (42.1KB, pdf)
jitc-2020-001705supp002.pdf (988.4KB, pdf)
Data Availability Statement
Data are available upon reasonable request. Data may be obtained from a third party and are not publicly available. Data are available from Translational Oncology Unit, Department of Oncology, Hospital Fundación Jiménez Díaz (principal investigator: JG-F, jesús.garciafoncillas@oncohealth.eu).