Abstract
Rationale & Objective
Pathogenic variants in type IV collagen have been reported to account for a significant proportion of chronic kidney disease. Accordingly, genetic testing is increasingly used to diagnose kidney diseases, but testing also may reveal rare missense variants that are of uncertain clinical significance. To aid in interpretation, computational prediction (called in silico) programs may be used to predict whether a variant is clinically important. We evaluate the performance of in silico programs for COL4A3/A4/A5 variants.
Study Design, Setting, & Participants
Rare missense variants in COL4A3/A4/A5 were identified in disease cohorts, including a local focal segmental glomerulosclerosis (FSGS) cohort and publicly available disease databases, in which they are categorized as pathogenic or benign based on clinical criteria.
Tests Compared & Outcomes
All rare missense variants identified in the 4 disease cohorts were subjected to in silico predictions using 12 different programs. Comparisons between the predictions were compared with: (1) variant classification (pathogenic or benign) in the cohorts and (2) functional characterization in a randomly selected smaller number (17) of pathogenic or uncertain significance variants obtained from the local FSGS cohort.
Results
In silico predictions correctly classified 75% to 97% of pathogenic and 57% to 100% of benign COL4A3/A4/A5 variants in public disease databases. The congruency of in silico predictions was similar for variants categorized as pathogenic and benign, with the exception of benign COL4A5 variants, in which disease effects were overestimated. By contrast, in silico predictions and functional characterization classified all 9 pathogenic COL4A3/A4/A5 variants correctly that were obtained from a local FSGS cohort. However, these programs also overestimated the effects of genomic variants of uncertain significance when compared with functional characterization. Each of the 12 in silico programs used yielded similar results.
Limitations
Overestimation of in silico program sensitivity given that they may have been used in the categorization of variants labeled as pathogenic in disease repositories.
Conclusions
Our results suggest that in silico predictions are sensitive but not specific to assign COL4A3/A4/A5 variant pathogenicity, with misclassification of benign variants and variants of uncertain significance. Thus, we do not recommend in silico programs but instead recommend pursuing more objective levels of evidence suggested by medical genetics guidelines.
Index Words: Alport syndrome, FSGS, type IV collagen variants, genomics, in silico predictions, computational predictions, ClinVar, ARUP, LOVD, gnomAD
Plain-Language Summary.
Type IV collagen mutations have been reported to account for a significant proportion of chronic kidney disease. As a result, genetic testing is increasingly being used for diagnosis but can uncover DNA changes that are of uncertain clinical significance. To determine whether causative for disease (called pathogenic), DNA changes can be tested with cell and animal models, an approach that is limited by the absence of well-established models for most genes, expense, and time-consuming nature. Alternatively, computational programs can be used to make predictions for pathogenicity. In this report, we begin to define the test characteristics for these computational predictions using bioinformatic and experimental approaches, with results suggesting that programs tend to overestimate the effects of DNA changes.
Chronic kidney disease (CKD) represents a heterogeneous group of disorders that result in irreversible fibrosis over time. Current diagnostic methods often fail to distinguish molecular mechanisms or predict disease course. CKD affects more than 750 million people globally and results in more than 1 million deaths annually. As such, kidney disease is a major health burden with substantive costs.1,2
Genomics is emerging as one tool to identify mechanistically relevant CKD subtypes. Using whole-exome sequencing, we have recently reported that pathogenic variants in the COL4A3/A4/A5 genes are the leading single gene causes (∼5%) of focal and segmental glomerulosclerosis (FSGS), a histopathologic entity representing diverse causes.3 Similarly, pathogenic variants in the COL4A3/A4/A5 genes have also been reported to account for a significant proportion of CKD.4 Pathogenic variants in type IV collagen are well known to cause Alport syndrome.5, 6, 7, 8, 9, 10
The human genome has tremendous sequence variation and the effect of rare nonsynonymous single-nucleotide variants (SNVs) in a disease-associated gene can be unclear. The American College of Medical Genetics (ACMG) has standards based on expert consensus for declaring the pathogenicity of rare variants that are organized into supporting, moderate, strong, and very strong levels of evidence.11 Some of these criteria include assessment of frequency in population data, type of variant change (eg, null variant), identification of familial cosegregation, presence in clinically ascertained mutation databases, bioinformatics, and functional data.12, Well-established functional studies that show a deleterious effect are considered strong levels of evidence.13
Computational predictions, also known as in silico programs, are one part of clinical variant classification in the diagnostic setting but are considered supportive compared with stronger lines of evidence. These programs have been developed to predict the functional effects of rare missense variants. Broadly, the algorithms use different types of variant information, including sequence conservation, protein structure analysis, and meta prediction (using results from multiple programs) for predictions.14, 15, 16, 17, 18
The predictive performance of in silico programs has been evaluated with computational methods against data sets that contain pathogenic and benign variants obtained from public resources (eg, Universal Protein Resource [Uniprot]), literature, and curated disease databases in which variants in kidney disease genes are not highly represented.19, 20, 21, 22, 23, 24, 25, 26 We evaluate the predictive performance of in silico programs for COL4A3/A4/A5 missense variants by first comparing with clinically categorized variants deposited in 3 public disease databases and a local FSGS cohort. As a second approach, in silico predictions are compared with functionally characterized missense variants identified in the local FSGS cohort.
Methods
Whole-Exome Sequencing Analysis
Details on how patients were recruited and exome data analyzed have been previously described.3 Study participants gave their written informed consent and the study protocol was approved by the Toronto General Hospital’s committee on human research (98-UO13). Whole-exome sequencing and data processing were performed by The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada. Exomic capture was achieved with Agilent SureSelect Human All Exon V5. Reads were mapped to the hg19 reference sequence.
Variant Calling From FSGS Whole-Exome Sequencing Data
Variants were identified using GATK (version 4.0.5.1).27 Gene-based annotation features of ANNOVAR were applied (access date, April 16, 2018).28 The frequency of variants was determined using Genome Aggregation Database (gnomAD; version 2.1.1; access date, March 18, 2019).26,29, 30, 31 Variants in COL4A3/A4/A5 were categorized as rare if having a minor allele frequency ≤ 0.005 in the ethnically matched population within gnomAD. This cutoff was selected in consideration of the low prevalence of FSGS, estimated at 7 per million for the general population, 20 per million for Africans, and 5 per million for Europeans.32,33 It was also selected in consideration of inheritance patterns: COL4A3/A4/A5 is associated with autosomal recessive, dominant, or X-linked recessive disease. Rare missense variants in COL4A3/A4/A5 were designated as pathogenic if reported in other cases of kidney disease after searching the literature and disease databases ClinVar, ARUP, and LOVD.3,34
In Silico Predictions Programs
Rare COL4A3/A4/A5 missense variants from our FSGS whole-exome sequencing data and disease databases ClinVar, ARUP, and LOVD (accessed October 22, 2019, September 13, 2019, and August 28, 2019, respectively) were identified. All rare SNVs reported in these sources have already been categorized. We in turn submitted the missense variants to 12 in silico programs for predictions (Table S1).15,24,35, 36, 37, 38, 39,40, 41, 42, 43, 44, 45,46 A variant was categorized as pathogenic if the majority, selected as 10 or more of 12 programs, categorized the variant as pathogenic using the program’s recommended scoring cutoffs.
COL4A Split Luciferase Assay
From our FSGS cohort with whole-exome sequencing data, 9 pathogenic variants and 8 variants of uncertain significance in COL4A3 and COL4A5 were randomly selected. We defined pathogenic variants as rare (minor allele frequency < 0.005) and reported in other cases with kidney disease, whereas variants of uncertain significance were defined as any other rare missense variant. To assess heterotrimer formation ability of these missense variants, we used the split complementation Nano-luciferase assay system that we have previously developed.47 Tagged plasmid constructs of COL4A4-FLAG, wild-type or mutant COL4A3-SmBiT, and COL4A5-LgBiT were generated as described previously.47 Corresponding SmBiT and LgBiT tags were attached at either the N-terminal or C-terminal of COL4A3 and COL4A5. After mutagenesis, sequences were verified. The COL4A3/A4/A5 tagged constructs were subsequently cotransfected into human embryonic kidney 293 (HEK293T) cells. Twenty-four hours posttransfection, cells were replated in LumiNunc 96-well white plates (Thermo Fisher Scientific) and cultured in phenol red-free Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum, 100 U of penicillin and streptomycin, 2 mmol/L of glutamine, and 200 μmol/L of l-ascorbic acid 2-phosphate trisodium salt. After 24 hours, cells (intracellular heterotrimer) and media (secreted heterotrimer) were assayed using Nano-Glo Live Cell Assay reagent and GloMax Navigator system (Promega).
Results
Computational Validation
We identified 70 SNVs in COL4A3 (29), COL4A4 (26), and COL4A5 (15) across 186 adults with FSGS with whole-exome sequencing. Of these, 31 were rare (minor allele frequency < 0.005), of which 30 were missense (14 in COL4A3, 10 in COL4A4, and 6 in COL4A5) and 1 was a stop-gain in COL4A4. Characteristics of the sequenced cohort have been published previously.3
In parallel, 2,803 nonsynonymous COL4A3/A4/A5 variants were identified in 125,748 unscreened participants with whole-exome data in gnomAD, a public database of genomic variation. Of these, 2,307 were rare and 2,279 were missense variants.
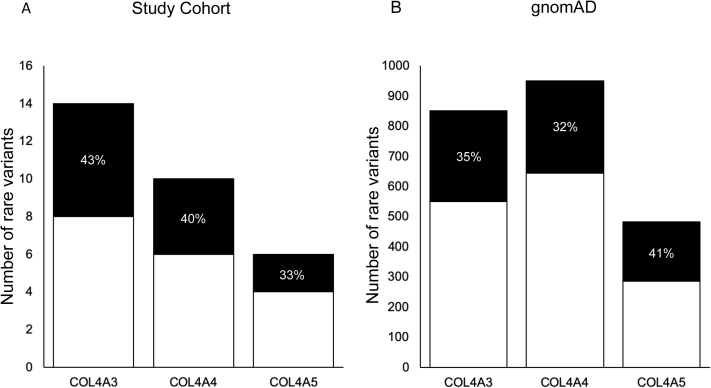
Rare missense variants in our local FSGS cohort, gnomAD, and Alport databases were each interrogated using 12 in silico programs for predictions (Table S1; Item S1). In the FSGS cohort, for rare missense variants in COL4A3, COL4A4, and COL4A5, 43% (6/14), 40% (4/10), and 33% (2/6) were predicted to be deleterious by at least 10 of 12 programs, respectively (Fig 1; Item S1).
Figure 1.
Number of rare missense variants predicted to be pathogenic in: (A) the focal segmental glomerulosclerosis (FSGS) study cohort and (B) Genome Aggregation Database (gnomAD). For rare missense COL4A3, COL4A4, and COL4A5 variants in our FSGS cohort, 43% (6/14), 40% (4/10), and 33% (2/6) were predicted to be deleterious by at least 10 of 12 programs, respectively. For rare missense COL4A3, COL4A4, and COL4A5 variants identified in gnomAD, 35% (301/851), 32% (306/949), and 41% (197/483) were predicted to be deleterious by at least 10 of 12 programs, respectively.
By comparison, for rare missense variants in COL4A3, COL4A4, and COL4A5 identified in gnomAD, 35% (301/851), 32% (306/949), and 41% (197/483) were predicted to be deleterious by at least 10 of 12 programs, respectively (Fig 1). gnomAD is a database in which some rare diseases would be even less represented than population estimates given that severe pediatric cases are not included.48 However, the lack of clinical data to correlate rare variants in gnomAD controls does not enable us to draw conclusions as to the accuracy of these predictions.
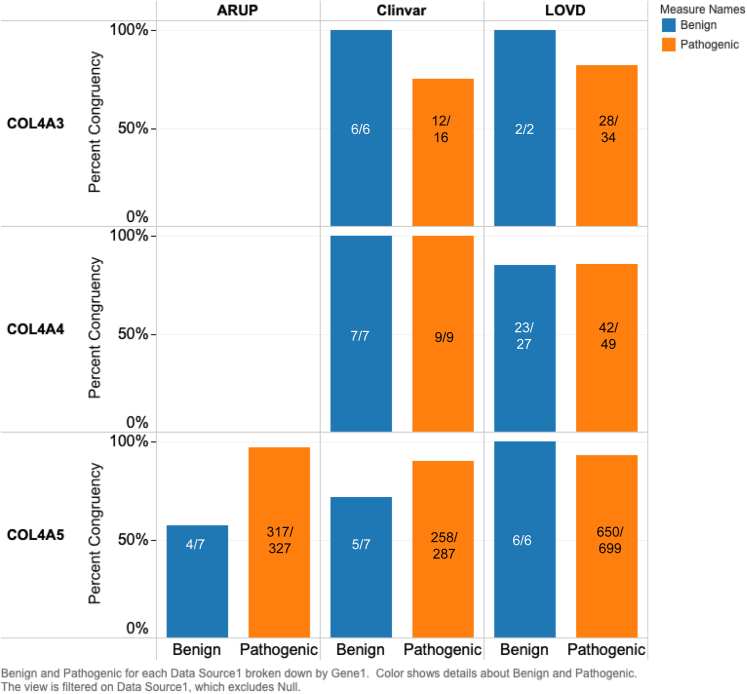
We also accessed disease databases in which COL4A3, COL4A4, and COL4A5 variants would be deposited, which included ARUP, ClinVar, and LOVD (accessed October 22, 2019, September 13, 2019, and August 28, 2019, respectively; Fig 2). ARUP documented 346 SNVs in COL4A5. Three hundred twenty-seven were categorized as pathogenic, with 97% (317/327) concordance to in silico predictions. ARUP does not document SNVs in COL4A3 or COL4A4. In ClinVar, 120 COL4A3 SNVs were documented. Sixteen were categorized as pathogenic and 75% were assigned (12/16) correctly by in silico programs. For COL4A4, 55 SNVs were reported. Nine were classified as pathogenic, with 100% assigned correctly by in silico programs. For COL4A5, there were 367 SNVs. Two hundred eighty-seven were categorized as pathogenic, with 90% (258/287) concordance to in silico predictions. In LOVD, 412 COL4A3 SNVs were catalogued. Of these, 34 were pathogenic and 82% (28/34) were predicted accurately. For COL4A4, there were 306 SNVs, of which 49 were classified as pathogenic, with 86% (42/49) concordance to in silico predictions. For COL4A5, there were 987 SNVs. Six hundred ninety-nine were classified as pathogenic, with 94% (650/699) concordant in silico predictions. In silico program sensitivity could be overestimated given that they may have been used in the categorization of variants labeled as pathogenic in these disease databases.
Figure 2.
Comparison of COL4A3, COL4A4, and COL4A5 in silico predictions with disease database categorization. For ARUP COL4A5 pathogenic variants, there was 97% (317/327) concordance with in silico predictions. For ClinVar COL4A3/A4/A5 pathogenic variants, there was 75% (12/16), 100% (9/9), and 89% (258/287) concordance with in silico predictions, respectively. For LOVD COL4A3/A4/A5 pathogenic variants, there was 82% (28/34), 86% (42/49), and 94% (650/699) concordance. Congruency of in silico predictions was similar for variants categorized as benign, with the exception of COL4A5 variants documented in ARUP and ClinVar, in which the effects were overestimated by in silico programs, though there were fewer variants to interrogate. In ARUP, 57% (4/7) of COL4A5 variants were classified correctly by in silico predictions. In ClinVar, 100% (6/6), 100% (9/9), and 71% (5/7) of COL4A3/A4/A5 variants, respectively, were correctly assigned. Finally, for LOVD, 100% (2/2), 85% (23/37), and 100% (6/6) of COL4A3/A4/A5 variants were correctly classified.
The congruency of in silico predictions was similar for variants categorized as benign, with the exception of COL4A5 variants documented in ARUP and ClinVar, in which the effects were overestimated by in silico programs, though there were fewer variants to interrogate (Fig 2). In ARUP, 7 COL4A5 variants were classified as benign, 57% (4/7) of which were assigned as such by in silico predictions. In ClinVar, 6 COL4A3, 7 COL4A4, and 7 COL4A5 variants were classified as benign, with 100%, 100%, and 71% (5/7) concordance with predictions, respectively. In LOVD, 2 COL4A3 and 6 COL4A5 variants were classified as benign, with 100% concordance for both. For COL4A4, there were 27 benign variants, with 85% (23/27) concordance with predictions.
A report suggests that one in silico classifier called M-CAP outperforms popular scores such as SIFT, PolyPhen-2, and CADD in its ability to separate pathogenic from benign variants.45 As a result, analysis of variants from disease databases was performed using M-CAP only. None of the benign variants in the disease databases were correctly classified, either as a result of incorrect categorization as pathogenic or by not generating an output (Fig S1). The accuracy for classification of pathogenic variants was much better, ranging from 89% to 96%. Additionally, a receiver operating curve for each of the 12 in silico programs was generated using the disease database type IV collagen variants and their in silico scores (Fig S2). When each curve is examined, we find that the score cutoff that maximizes the true-positive rate while minimizing the false-positive rate does not coincide with the in silico programs’ recommendations. For instance, we find that the cutoff for SIFT should be approximately less than 0.004, whereas the recommended cutoff is less than 0.05 (Fig S2). As a result, variants with scores between 0.004 and 0.05 are being predicted as pathogenic, leading to false positives.
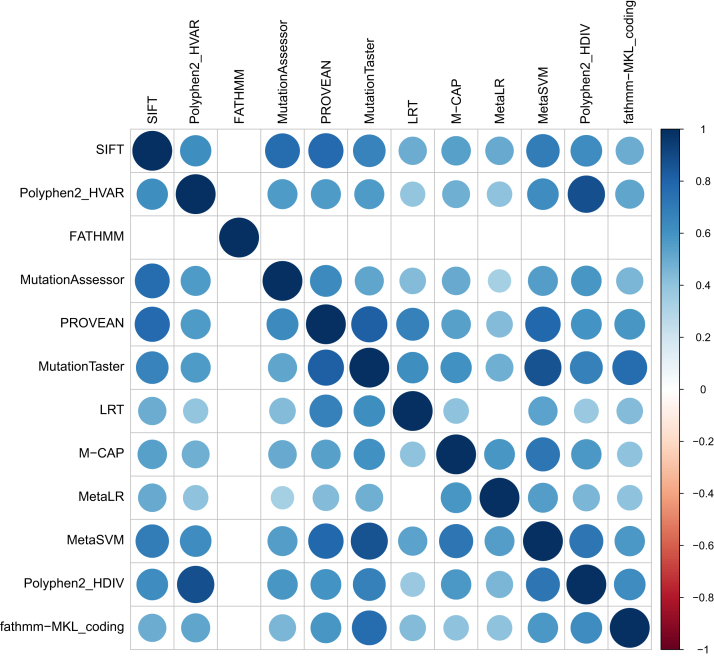
Congruency in classification between in silico programs was also explored (Fig 3). Most programs had similar prediction scores when comparing with each other except for FATHMM and M-CAP.
Figure 3.
Spearman correlation coefficient heatmap comparing results of various prediction models. Most programs had similar prediction scores when comparing with each other except for FATHMM and M-CAP. Dark blue signifies a strong direct correlation while dark red signifies a strong indirect correlation. Squares that are lighter in color signify a weak correlation between the results of the 2 prediction models. Figure created using the corrplot package available in Rstudio.
Functional Validation
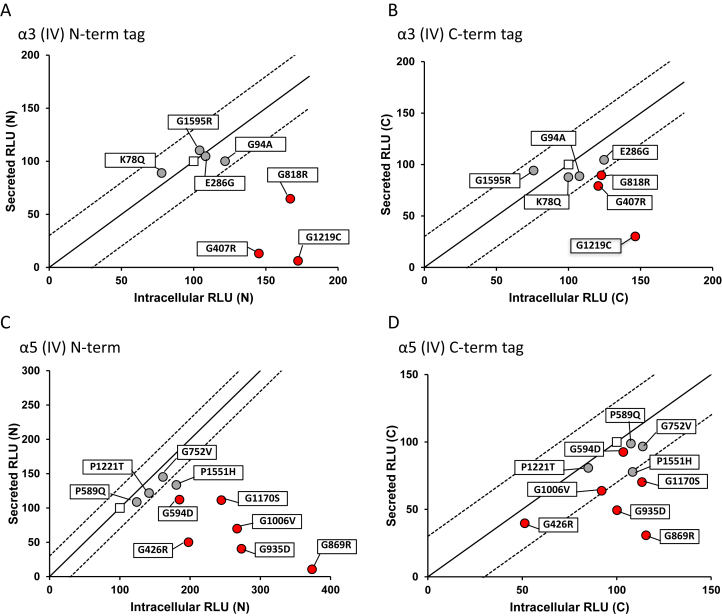
We evaluated 9 pathogenic missense variants in COL4A3 and the X-linked COL4A5 identified in the local FSGS cohort. For COL4A3 and COL4A4, the mode of inheritance has traditionally been reported as recessive, but next-generation sequencing studies have reported about 20% to 30% of patients with dominant disease.49, 50, 51, 52 Of 3 pathogenic heterozygous missense variants in COL4A3 (ie, rare variant reported in other individuals with kidney disease), all were predicted to be deleterious by at least 10 of 12 in silico programs (Table 1; Item S2). Of 6 pathogenic variants in COL4A5, all were predicted to be deleterious by at least 10 of 12 in silico programs (Table 1). Under normal conditions, COL4A3, COL4A4, and COL4A5 each encodes a protein that heterotrimerizes and is secreted into the glomerular basement membrane. To determine the secretory behavior of the COL4A3 and COL4A5 mutants, we used an assay system that quantified the intracellular trimerization and trimer secretion of COL4A3/4/5.47 Using this split luciferase complementation assay, all COL4A3 and COL4A5 pathogenic variants were found to have secretory defect with the N-terminal tagged versions of COL4A3 and COL4A5 (Fig 4A and C). The pathogenic variants could form trimers intracellularly but could not be efficiently secreted. By contrast, this was not always observed for C-terminal tagged versions of COL4A3 and COL4A5 (Fig 4B and D). We speculate that this could be due to heterotrimer formation being initiated at the noncollagenous (NC1) domain of the C-terminal region of collagen and terminates at the N-terminal region. The fusion of the monomers initially at the C-terminal region brings the reporter tags closer together to produce luminesce regardless of whether the trimer is completely formed.
Table 1.
Comparison of Functional Annotation With In Silico Predictions for Pathogenic COL4A3 and COL4A5 Variants Identified in the FSGS Cohort
| Gene | No. of Pathogenic Variants | No. Predicted Deleterious by 10/12 Programs | No. of Variants With Secretory Defect | Congruence |
|---|---|---|---|---|
| COL4A3 | 3 | 3 | 3 | 100% |
| COL4A5 | 6 | 6 | 6 | 100% |
Note: All pathogenic COL4A3 and COL4A5 variants were categorized as such as a result of being identified in other kidney disease cases. All pathogenic variants demonstrated a secretory defect with functional characterization and were correctly assigned by in silico predictions.
Abbreviation: FSGS, focal segmental glomerulosclerosis.
Figure 4.
Functional characterization of COL4A3 and COL4A5 using the split-luciferase assay. Scatterplots of the intracellular/secreted relative light unit (RLU) ratio from human embryonic kidney 293 (HEK293T) cells expressing (A, B) mutant α3 chain or (C, D) mutant α5 chain compared to wild type (WT) using N-terminal and C-terminal split-luciferase tagged constructs. Pathogenic α3 and α5 chain mutants showed clearer secretory defect with N-terminal tagged constructs. Solid line: Y = X, dotted line: Y = X + 50, Y = X – 50. Square, WT; red circles are pathogenic variants and grey circles are variants of uncertain significance. Any data point under the −50 line was considered as a significant secretory defect. Experiments were performed in triplicate. Data presented are representative of 2 independent experiments.
Similarly, 8 variants of uncertain significance in COL4A3 and COL4A5 identified in our FSGS cohort were selected, comparing in silico predictions with functional characterization. Of 4 variants of uncertain significance in COL4A3, 3 were predicted to be deleterious by at least 10 of 12 in silico programs (Table 1). Of 4 variants of uncertain significance in COL4A5, 2 were predicted to be deleterious by at least 10 of 12 in silico programs (Table 1). Using the split luciferase complementation assay, only 1 variant of uncertain significance in COL4A5 was found to have a secretory defect using N-terminal tagged proteins, though not to the degree observed for the definitely pathogenic variants (Fig 4C). Any data point under the −50 line was considered as a significant secretory defect. Thus, there was poor concordance between in silico predictions and functional characterization, with the former potentially overestimating the functional characteristics of missense variants (Table 2).
Table 2.
Comparison of Functional Annotation With In Silico Predictions for Variants of Uncertain Significance in COL4A3 and COL4A5 Identified in the FSGS Cohort
| Gene | No. of Variants of Uncertain Significance | No. Predicted Deleterious by 10/12 Programs | No. of Variants With Secretory Defect | Congruence |
|---|---|---|---|---|
| COL4A3 | 4 | 3 | 0 | 0% |
| COL4A5 | 4 | 2 | 1 | 50% |
Note: Only 1 variant of uncertain significance in COL4A5 was found to have a secretory defect, which was accurately predicted by in silico predictions. However, 1 variant of uncertain significance without evidence of a secretory defect was also predicted to be deleterious.
Abbreviation: FSGS, focal segmental glomerulosclerosis.
To further determine the functional nature of 8 variants of uncertain significance in COL4A3 and COL4A5, clinical characteristics for these patients were obtained (Tables S2 and S3). Many individuals with Alport syndrome have hematuria and basement membrane abnormalities. In our cohort, microscopic hematuria data were reported for 9 patients with pathogenic variants and 8 patients with a variant of uncertain significance in COL4A3 and COL4A5. Microscopic hematuria was observed in 4 of 9 patients with pathogenic variants and 2 of 8 patients with a variant of uncertain significance (Tables S2 and S3). For the 2 patients with variants of uncertain significance (COL4A3 p.G1595R and COL4A5 p.P589Q) and hematuria, neither variant was characterized as functionally deleterious (Fig 4).
Discussion
Our results demonstrate that in silico predictions correctly classified most pathogenic COL4A3/A4/A5 variants catalogued in ClinVar, ARUP, and LOVD. In silico predictions performed similarly for benign variants with the exception of COL4A5 (concordance in ARUP and ClinVar with predictions and classification was 57% [4/7] and 71% [5/7], respectively) but there were also far fewer benign variants to interrogate in these disease databases. Our second approach of correlating in silico predictions with functional testing showed that both accurately classified all pathogenic COL4A3/A4/A5 missense variants in the FSGS cohort. These variants were labeled as pathogenic because they are rare and already reported as disease-causing in other individuals with kidney disease, which are considered strong lines of evidence (ACMG criteria PS1 and PS4; Item S2).11 By contrast, in silico predictions overestimated the effects of COL4A3/A5 variants of uncertain significance when compared with functional characterization. A variant of uncertain significance was defined as a rare variant that did not satisfy ACMG criteria for definite pathogenicity. Interestingly, interrogation of COL4A3/A4/A5 variants found in gnomAD predicted a high percentage to be deleterious, but the lack of clinical data for correlation prevents us from making any conclusion with these data.
Genomics facilitates clinically meaningful classification of CKD but sequencing can reveal rare SNVs for which the relationship to disease is unclear. The ACMG has standards based on expert consensus for declaring pathogenicity wherein in silico predictions are considered only supporting compared with higher levels of evidence that are deemed moderate, strong, or very strong.11 Well-established functional studies that show deleterious effect is an example of one criterion considered strong level of evidence. Against this background, we provide an assessment of in silico programs using both computational and experimental approaches.
Using the Nano-luciferase complementation system, we have recently quantified trimerization of 9 typical glycine substitutions in COL4A5 that differ in disease progression, finding a correlation between in vitro results and phenotype.53 In the data presented here, we observe that the pattern of heterotrimer formation and secretion for pathogenic mutants differed slightly between N-terminal and C-terminal tagged constructs. The N-terminal tagged pathogenic mutants showed clearer secretory defect. We postulate that this could be as a result of heterotrimer formation initiating at the NC1 domain at the C-terminal region of collagen and terminates at the N-terminal region. The fusion of the monomers initially at the C-terminal region could bring the reporter tags closer together to produce luminesce regardless of whether the heterotrimer is completely formed. Therefore, the luciferase reporter attached at the N-terminal region, that is, the N-tagged constructs, may better reflect the state of trimer folding.
The 12 prediction models used in this study can be categorized as solely conservation based: (SIFT Polyphen2-HVAR, Polyphen2-HDIV, MutationAssessor, PROVEAN, and LRT) and multifeatured algorithms (FATHMM, M-CAP, MetaLR, MetaSVM, FATHMM-MKL, MutationTaster; Table S1). Conservation-based models select homologous sequences to create multiple sequence alignments across species (MSA) and use the sequence and predicted structure-based features of the MSA to predict pathogenicity with variants in more conserved areas predicted to be deleterious. The multifeatured algorithms integrate other information, such as epigenomic signals (FATHMM- MKL and MutationTaster), allele frequencies (FATHMM, MetaLR, and MetaSVM), or the results of other prediction algorithms (M-CAP, MetaLR, and MetaSVM). Eleven of the 12 prediction models are trained using databases including UniProt54 (PolyPhen2-HDIV, PolyPhen2-HVAR, FATHMM, PROVEAN, MetaLR, and MetaSVM), Human Gene Mutation Database55 (FATHMM, FATHMM-MKL, MutationTaster, and M-CAP), ExAC56 (M-CAP), Ensembl57 (LRT), 1000 Genomes Project58 (MutationTaster and FATHMM-MKL), and COSMIC59 (MutationAssessor). SIFT was trained using known variants of the E coli LacI gene that have been individually mutated and functionally tested.60,61
M-CAP has been previously reported to outperform popular pathogenicity classifiers but our results demonstrate that it unreliably categorizes the small number of benign type IV collagen variants in disease databases by incorrectly assigning pathogenicity or not generating an output. M-CAP already uses 9 established pathogenicity likelihood scores included in our scoring system: SIFT13, PolyPhen-2, CADD15, MutationTaster20, MutationAssessor21, FATHMM22, LRT23, MetaLR16, and MetaSVM16.45 It incorporates 7 established measures of base pair, amino acid, genomic region, and gene conservation: RVIS24, PhyloP25, PhastCons26, PAM250, BLOSUM62, SIPHY28, and GERP29. Additionally, M-CAP introduces 298 new features derived from multiple-sequence alignment of 99 primate, mammalian, and vertebrate genomes to the human genome30. However, previous reports seeking to demonstrate superiority of one classifier over others are all limited by the veracity of variant assignment in test databases and in which kidney gene variants contribute a small proportion.
Our study highlights several limitations and opportunities for future investigation. Estimating in silico program accuracy using disease databases relies on robust categorization and underscores a need for consistency in variant annotation. The sensitivity of in silico programs could be overestimated given that they may have been used in the categorization of variants labeled as pathogenic. In disease databases, there were far fewer variants classified as benign compared with pathogenic. However, to address these limitations, we have pursued more laborious functional characterization on randomly selected type IV collagen variants from the FSGS cohort as an additional line of evidence.
With respect to functional characterization, we include data to support our conclusions, but only a small number of missense variants were characterized. We use the arbitrary cutoff of ±50 from wild-type data, but characterizing more pathogenic and benign variants would better define a threshold. As per standard convention throughout the literature, we characterize the effects of single variants on the reference haplotype, but there are several common haplotypes documented in the 1000 Genomes Project (Table S4). Studying the effects of single variants on different haplotype backgrounds could provide important information regarding interaction effect between haplotype and mutation. Second, our assay will identify mutations that are associated with secretory defects, but this is a simplification of disease pathogenesis that does not account for the complexities involving extracellular type IV collagen network formation. For instance, a previous report suggests that ∼20% of COL4A5 mutations have detectable heterotrimers in the glomerular basement membrane, suggesting alternate disease mechanisms.62
Recent reports demonstrate that pathogenic variants in COL4A3/A4/A5 account for a significant and unappreciated proportion of patients with Alport syndrome in CKD.3,5, 6, 7, 8, 9, 10,63 Sequencing is increasingly being used to obtain mechanistically relevant diagnoses but often generates rare missense variants that remain of uncertain clinical significance. In silico predictions have been developed to aid in categorizing variants. We show here that computational approaches including M-CAP, which was reported to outperform other classifiers, are sensitive but not sufficiently specific to confidently assign COL4A3/A4/A5 variant pathogenicity. Thus, we do not recommend any in silico program in the consideration of type IV collagen variant categorization, but instead pursuing more objective levels of evidence suggested by medical genetic guidelines.
Article Information
Authors’ Full Names and Academic Degrees
Cole Shulman, Emerald Liang, BSc, Misato Kamura, BSc, Khalil Udwan, MD, PhD, Tony Yao, BSc, Daniel Cattran, MD, Heather Reich, MD, PhD, Michelle Hladunewich, MD, MSc, York Pei, MD, MSc, Judy Savige, MB, PhD, Andrew D. Paterson, MD, PhD, Mary Ann Suico, PhD, Hirofumi Kai, PhD, and Moumita Barua, MD.
Authors’ Contributions
Research idea and study design: CS, EL MK, ADP, HK, MB; patient ascertainment and DNA archiving: KU, DC, HR, MH, YP; acquired LOVD variant data: JS; computational analysis: CS, EL MB, TY; prepared figures: CS, EL, MB; collection and analysis of functional data: MK, MS; supervision/mentorship: HK, MB (equal contribution). CS, EL, and MK contributed equally to this work. Each author contributed important intellectual content during manuscript drafting or revision and accepts accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved.
Support
Dr Barua received a NephCure Kidney International-Neptune Ancillary Studies Grant in 2016, Health Research Grant (14-04) from Physician’s Services Inc in 2015, a McLaughlin Accelerator Award in 2019, and support from the Can-SOLVE CKD Network (https://www.cansolveckd.ca/) and the Toronto General Hospital Foundation.
Financial Disclosure
The authors declare that they have no relevant financial interests.
Acknowledgements
We thank the study participants.
Peer Review
Received September 24, 2020. Evaluated by 2 external peer reviewers, with direct editorial input by the Editor-in-Chief. Accepted in revised form December 13, 2020.
Footnotes
Complete author and article information provided before references.
Figure S1: Comparison of COL4A3, COL4A4, and COL4A5 in silico predictions by M-CAP with disease database categorization.
Figure S2: Receiver operator curves for the 12 in silico programs using scores generated from type IV collagen variants obtained from disease databases.
Table S1: Twelve In Silico Predictions Programs Used.
Table S2: Clinical Characteristics of FSGS Patients With COL4A3 Variants.
Table S3: Clinical Characteristics of FSGS Patients With COL4A5 Variants.
Table S4: Common COL4A3 Haplotypes Identified in Europeans in the 1000 Genomes Project.
Item S1: COL4A3/A4/A5 Variants Identified in FSGS Cohort and Predictions by In Silico Programs.
Item S2: Variants Selected for Functional Testing.
Supplementary Material
References
- 1.Klarenbach S.W., Tonelli M., Chui B., Manns B.J. Economic evaluation of dialysis therapies. Nat Rev Nephrol. 2014;10(11):644–652. doi: 10.1038/nrneph.2014.145. [DOI] [PubMed] [Google Scholar]
- 2.Canadian Institute for Health Information . Canadian Institute for Health Information; Ottawa, ON: 2015. National Health Expenditure Trends, 1975 to 2015. [Google Scholar]
- 3.Yao T., Udwan K., John R. Integration of genetic testing and pathology for the diagnosis of adults with FSGS. Clin J Am Soc Nephrol. 2019;14:213–223. doi: 10.2215/CJN.08750718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groopman E.E., Marasa M., Cameron-Christie S. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380(2):142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashtan C.E. Alport syndrome. An inherited disorder of renal, ocular, and cochlear basement membranes. Medicine (Baltimore) 1999;78(5):338–360. doi: 10.1097/00005792-199909000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Voskarides K., Pierides A., Deltas C. COL4A3/COL4A4 mutations link familial hematuria and focal segmental glomerulosclerosis. Glomerular epithelium destruction via basement membrane thinning? Connect Tissue Res. 2008;49(3):283–288. doi: 10.1080/03008200802148280. [DOI] [PubMed] [Google Scholar]
- 7.Papazachariou L., Papagregoriou G., Hadjipanagi D. Frequent COL4 mutations in familial microhematuria accompanied by later-onset Alport nephropathy due to focal segmental glomerulosclerosis. Clin Genet. 2017;92(5):517–527. doi: 10.1111/cge.13077. [DOI] [PubMed] [Google Scholar]
- 8.Xie J., Wu X., Ren H. COL4A3 mutations cause focal segmental glomerulosclerosis. J Mol Cell Biol. 2014;6(6):498–505. doi: 10.1093/jmcb/mju040. [DOI] [PubMed] [Google Scholar]
- 9.Malone A.F., Phelan P.J., Hall G. Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int. 2014;86(6):1253–1259. doi: 10.1038/ki.2014.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierides A., Voskarides K., Athanasiou Y. Clinico-pathological correlations in 127 patients in 11 large pedigrees, segregating one of three heterozygous mutations in the COL4A3/ COL4A4 genes associated with familial haematuria and significant late progression to proteinuria and chronic kidney disease from focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2009;24(9):2721–2729. doi: 10.1093/ndt/gfp158. [DOI] [PubMed] [Google Scholar]
- 11.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.N. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starita L.M., Ahituv N., Dunham M.J. Variant interpretation: functional assays to the rescue. Am J Hum Genet. 2017;101(3):315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christenhusz G.M., Devriendt K., Dierickx K. Disclosing incidental findings in genetics contexts: a review of the empirical ethical research. Eur J Med Genet. 2013;56:529–540. doi: 10.1016/j.ejmg.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng P.C., Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 17.Thusberg J., Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum Mutat. 2009;30(5):703–714. doi: 10.1002/humu.20938. [DOI] [PubMed] [Google Scholar]
- 18.Cooper G.M., Goode D.L., Ng S.B. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat Methods. 2010;7(4):250–251. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thusberg J., Olatubosun A., Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32(4):358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 20.The UniProt Consortium Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39(database issue):D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C.H., Apweiler R., Bairoch A. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34(database issue):D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasidharan Nair P., Vihinen M. VariBench: a benchmark database for variations. Hum Mutat. 2013;34(1):42–49. doi: 10.1002/humu.22204. [DOI] [PubMed] [Google Scholar]
- 23.Morrison A.C., Voorman A., Johnson A.D. Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat Genet. 2013;45(8):899–901. doi: 10.1038/ng.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong C., Wei P., Jian X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24(8):2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassan M.S., Shaalan A.A., Dessouky M.I., Abdelnaiem A.E., ElHefnawi M. Evaluation of computational techniques for predicting non-synonymous single nucleotide variants pathogenicity. Genomics. 2019;111(4):869–882. doi: 10.1016/j.ygeno.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 26.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37(3):235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Auwera G.A., Carneiro M.O., Hartl C. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protocols Bioinforms. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11.10.1-11.10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karczewski K.J., Francioli L.C., Tiao G. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. Nature. 2019 https://doi.org/10.1038/s41586-020-2308-7 [Google Scholar]
- 30.Landrum M.J., Lee J.M., Benson M. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitiyakara C., Kopp J.B., Eggers P. Trends in the epidemiology of focal segmental glomerulosclerosis. Semin Nephrol. 2003;23(2):172–182. doi: 10.1053/snep.2003.50025. [DOI] [PubMed] [Google Scholar]
- 33.Kitiyakara C., Eggers P., Kopp J.B. Twenty-one-year trend in ESRD due to focal segmental glomerulosclerosis in the United States. Am J Kidney Dis. 2004;44(5):815–825. [PubMed] [Google Scholar]
- 34.Crockett D.K., Pont-Kingdon G., Gedge F., Sumner K., Seamons R., Lyon E. The Alport syndrome COL4A5 variant database. Hum Mutat. 2010;31(8):E1652–E1657. doi: 10.1002/humu.21312. [DOI] [PubMed] [Google Scholar]
- 35.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei I.A., Schmidt S., Peshkin L. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramensky V. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shihab H.A., Gough J., Cooper D.N. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shihab H.A., Rogers M.F., Gough J. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reva B., Antipin Y., Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8:R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. Mutationtaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 42.Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jagadeesh K.A., Wenger A.M., Berger M.J. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 46.Kim S., Jhong J.H., Lee J., Koo J.Y. Meta-analytic support vector machine for integrating multiple omics data. BioData Mining. 2017;10:2. doi: 10.1186/s13040-017-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omachi K., Kamura M., Teramoto K. A split-luciferase-based trimer formation assay as a high-throughput screening platform for therapeutics in Alport syndrome. Cell Chem Biol. 2018;25(5):634–643 e634. doi: 10.1016/j.chembiol.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Song W., Gardner S.A., Hovhannisyan H. Exploring the landscape of pathogenic genetic variation in the ExAC population database: insights of relevance to variant classification. Genet Med. 2016;18(8):850–854. doi: 10.1038/gim.2015.180. [DOI] [PubMed] [Google Scholar]
- 49.Fallerini C., Dosa L., Tita R. Unbiased next generation sequencing analysis confirms the existence of autosomal dominant Alport syndrome in a relevant fraction of cases. Clin Genet. 2014;86(3):252–257. doi: 10.1111/cge.12258. [DOI] [PubMed] [Google Scholar]
- 50.Moriniere V., Dahan K., Hilbert P. Improving mutation screening in familial hematuric nephropathies through next generation sequencing. J Am Soc Nephrol. 2014;25(12):2740–2751. doi: 10.1681/ASN.2013080912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Loop F.T., Heidet L., Timmer E.D. Autosomal dominant Alport syndrome caused by a COL4A3 splice site mutation. Kidney Int. 2000;58(5):1870–1875. doi: 10.1111/j.1523-1755.2000.00358.x. [DOI] [PubMed] [Google Scholar]
- 52.Pescucci C., Mari F., Longo I. Autosomal-dominant Alport syndrome: natural history of a disease due to COL4A3 or COL4A4 gene. Kidney Int. 2004;65(5):1598–1603. doi: 10.1111/j.1523-1755.2004.00560.x. [DOI] [PubMed] [Google Scholar]
- 53.Kamura M., Yamamura T., Omachiet K. Trimerization and genotype–phenotype correlation of COL4A5 Mutants in Alport SYNDROME. Kidney Int Rep. 2020;5(5):718–726. doi: 10.1016/j.ekir.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bateman A. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stenson P.D., Ball E.V., Mort M. Human Gene Mutation Database (HGMD®): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 56.Lek M., Karczewski K.J., Minikel E.V. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zerbino D.R., Achuthan P., Akanni W. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Auton A., Abecasis G.R., Altshuler D.M. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tate J.G., Bamford S., Jubb H.C. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Markiewicz P., Kleina L.G., Cruz C., Ehret S., Miller J.H. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. J Mol Biol. 1994;240(5):421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 62.Kashtan C.E., Kleppel M.M., Gubler M.C. Immunohistologic findings in Alport syndrome. Contrib Nephrol. 1996;117:142–153. doi: 10.1159/000424811. [DOI] [PubMed] [Google Scholar]
- 63.Groopman E., Goldstein D., Gharavi A. Diagnostic utility of exome sequencing for kidney disease. Reply. N Engl J Med. 2019;380(21):2080–2081. doi: 10.1056/NEJMc1903250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.