Abstract
Background
High data quality is of crucial importance to the integrity of research projects. In the conduct of multi-center observational cohort studies with increasing types and quantities of data, maintaining data quality is challenging, with few published guidelines.
Methods
The Cure Glomerulonephropathy (CureGN) Network has established numerous quality control procedures to manage the 70 participating sites in the United States, Canada, and Europe. This effort is supported and guided by the activities of several committees, including Data Quality, Recruitment and Retention, and Central Review, that work in tandem with the Data Coordinating Center to monitor the study. We have implemented coordinator training and feedback channels, data queries of questionable or missing data, and developed performance metrics for recruitment, retention, visit completion, data entry, recording of patient-reported outcomes, collection, shipping and accessing of biological samples and pathology materials, and processing, cataloging and accessing genetic data and materials.
Results
We describe the development of data queries and site Report Cards, and their use in monitoring and encouraging excellence in site performance. We demonstrate improvements in data quality and completeness over 4 years after implementing these activities. We describe quality initiatives addressing specific challenges in collecting and cataloging whole slide images and other kidney pathology data, and novel methods of data quality assessment.
Conclusions
This paper reports the CureGN experience in optimizing data quality and underscores the importance of general and study-specific data quality initiatives to maintain excellence in the research measures of a multi-center observational study.
Keywords: Data quality, Quality metrics, Site performance, Observational studies, Kidney disease
1. Introduction
In the conduct of multi-center observational cohort studies, strategies to maintain high data quality (DQ) are critical. This is particularly important with multiple data types, such as clinical, laboratory, patient-reported, biomarker, pathology, proteomic and genetic data. Inaccurate or missing data compromise statistical accuracy and power and can lead to wrong conclusions. In clinical trials, high-quality data have been a priority. The Child and Adolescent Trial for Cardio-vascular Health (CATCH) incorporated an extensive quality control (QC) system into the trial design [1]. The Diabetes Control and Complications Trial described their 30-year experience using a DQ assurance committee [2]; other studies have described QC oversight [[3], [4], [5], [6], [7], [8], [9], [10]].
Observational studies vary widely in data quality. In a liver transplant study, difficulties in collecting quality data were illustrated by modest concordance of data elements from two independent extractions from medical records for the same 785 transplant candidates and 386 donors [11]. Other studies have made special efforts to improve DQ. A prospective, population-based study of maternal and newborn health in low resource settings describe their data quality metrics and reporting process implemented to identify cluster- and site-level quality issues that could be tracked over time. This study illustrates how the sites increasingly achieved acceptable values of performance [12]. In a large observational study of atrial fibrillation, source data verification was key. Sites were ranked based on four performance indicators, e.g., number of missing critical variables, which facilitated identification of sites with suboptimal data quality. Corrective actions improved performance at the next monitoring phase [13]. To improve quality of data in an international HIV research network, audit codes identified error rates >10% in key study variables. Laboratory data, weight measurements, and medication start and stop dates were prone to error. Verifying key data elements against source documents through audits improved the quality of the research databases [14]. Another HIV study [15] found a discrepancy rate of 17% between pre- and post-audit data of 250 participants, and then investigated the effect of the poor data quality on epidemiologic inference. In 2020, Guidelines for Data Acquisition, Quality and Curation for Observational Research Designs (DAQCORD) were published [16]. Based on a modified Delphi process, data curation indicators were developed that could be used in the design and reporting of large observational studies. Although developed around neuroscience studies, DAQCORD guidelines were designed to be relevant and generalizable, in whole or in part, to other fields of health research.
The Cure Glomerulonephropathy (CureGN) Network is an ongoing multi-center, prospective observational cohort study of children and adults with one of four biopsy-proven glomerular kidney diseases: minimal change disease (MCD), focal segmental glomerulosclerosis (FSGS), membranous nephropathy (MN), and immunoglobulin A (IgA) nephropathy (IgAN). The objectives of CureGN are to support a broad range of scientific approaches to identify mechanistically distinct subgroups, identify biomarkers of disease activity, identify genetic associations with disease pathways, delineate disease-specific treatment targets, and inform future therapeutic trials [17].
Given the complexity of CureGN, with 70 sites across North America and Europe, over 2400 patients enrolled, long-term follow-up, wide scientific scope and many types of data collected, maintaining high DQ is a major effort. The goals of this paper are to: (1) describe the DQ initiatives of CureGN, including coordinator training, data queries, site Report Cards, and QC of biospecimen, pathology and genetic materials; (2) document improvements in DQ and completeness over time; (3) present novel methods for quality assessment of pathology images and proteinuria data; and (4) demonstrate the importance of DQ to statistical accuracy, power, and quality of research products.
2. Methods
2.1. CureGN study design
Enrollment began in 2014 with a target of 2400 participants, and over time has included 67 to 70 clinical sites managed by four Participating Clinical Center (PCC) networks. Now in its second funding cycle (CureGN-2) the scope of data and sample collection is comprehensive, and includes hundreds of clinical data values, including demographics, medical history, medications, family history, and laboratory data. Patient or parent-proxy reported Outcomes (PROs) applied selected PRO Measurement Information System measures as well as newly developed content, and were administered during in-person visits, twice in year one and annually thereafter. Study coordinators enter the data for all of these measures using a flexible in-house data entry platform (GN-Link) that includes real-time reports, task and completeness reminders, and alerts for lab data outside the expected range, using logic that reduces coordinator burden while ensuring that relevant questions are asked. Entry within 30 days of a patient visit or phone call is expected.
Biospecimens (serum, plasma, urine, Deoxyribonucleic Acid [DNA], and Ribonucleic Acid [RNA]) are collected and shipped for storage in the National Institutes of Health (NIH) central biorepository. Additional genetic data (peripheral blood leukocyte gene expression and blood whole genome sequencing [WGS] data sets) are currently generated and will be submitted to National Center for Biotechnology Information (NCBI) data repository for access by the research community after completion of quality control measures. Pathology materials (scans of biopsy reports, glass slides, and electron microscopy [EM] and immunofluorescence [IF] jpeg images) are sent to the NIH Image Coordinating Center, where glass slides are scanned into whole slide images (WSI). All digital materials are uploaded and stored in the CureGN digital pathology repository (DPR). All patients provided written informed consent to participate; the study was carried out in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of each site and the Data Coordinating Center.
Patient follow-up in CureGN-1 included two in-person study visits in the first year and one in-person and two additional visits (by phone, email, or in-person) each subsequent year, during a disease flare or relapse when possible. Subjects are followed for kidney disease activity, progression, and non-renal complications of disease or treatment, including infection, malignancy, and cardiovascular and thromboembolic events. Patients in CureGN-1 could continue into CureGN-2 with re-consent. CureGN-2 includes one in-person study visit each year, and two additional visits (by phone, email, or other modality) for those in the study for years one to three. Starting in year four, one in-person and one additional visit are completed each year.
Initial coordinator training is provided by the Data Coordinating Center (DCC) at the beginning of the study phase via webinars. To improve uniformity across sites, quarterly Town Halls are held to continue coordinators’ education in CureGN. Topics addressed during Town Halls include updates to the GN-Link database, changes in processes, and special guest presenters providing information and tips regarding topics relevant to coordinators day-to-day activities. The trainings and Town Halls are recorded and posted on the study website for coordinators to access. Also included on the study website are the Manual of Operations, GN-Link User Manual, and other reference documents. The PCC lead coordinators use the videos and documents as training aids for new coordinators and as refresher trainings for established coordinators. We note that each PCC lead coordinator is funded by the grant to manage their own site as well as to assist their participating site coordinators. Effort for the participating site coordinators is covered in the per-visit reimbursement schedule, which is limited by NIDDK budget constraints.
2.2. The CureGN DQ committee (DQC)
Although CureGN had a DCC data management team from study start, the DQC was established in 2015 with the charge to ensure a high level of DQ for the CureGN study and to develop and implement strategies to improve DQ as needed. The DQC met via teleconference twice a month for a year, followed by monthly and then quarterly meetings. The DQC includes at least one clinician (nephrologist) and one study coordinator from each of the four PCCs, and members of the DCC, including biostatisticians, analysts, programmers, study monitors, and pathologists. The activities of the DQC are integrated with other consortium committees (Table 1).
Table 1.
CureGN Committees with data quality as part of their mission.
| Committee | Mission |
|---|---|
| Data Quality | Develop and implement strategies to improve data quality, oversee development and modification of the study Site Report Card, provide data to the Central Review Committee on site metrics, and work with Data Coordinating Center staff on new data queries. |
| Recruitment and Retention | Monitor recruitment and loss to follow-up, identify challenges and mitigation strategies, generate newsletters and other patient support materials, and guide metric development for recruitment and retention reports, as well as relevant metrics for the site Report Cards. |
| Central Review | Work with Participating Clinical Center (PCC) coordinators who have sites with quality metrics that are below a defined level to: develop a remediation plan to improve site performance; use ideas from all sites to achieve the best possible outcomes; allow flexibility in approaches to sites that have differing problems or issues; and develop metrics that may indicate a need for formal review by the study leadership for retention versus removal of a site from the consortium. |
| Ancillary Studies | Review ancillary study proposals for feasibility and scientific merit, with input from the Data Coordinating Center regarding sufficient biospecimen sample size, data availability and quality to answer the research question. |
| Biospecimens | Provide expert input on collection, storage, and optimal use of the non-renewable biosample resource: meet study aims while shepherding this resource for the community; review the current biosample inventory; provide input for proposed ancillary studies requesting CureGN biosamples; review study biospecimen performance; and recommend protocol and procedure changes related to biospecimens. |
| Pathology | Provide morphology-based inclusion and exclusion criteria, set guidelines for monitoring completeness and de-identification of pathology reports, and develop a protocol defining core scoring elements and establishing a process for core scoring. |
| Digital Pathology Repository (DPR) | Provide monitoring and metrics on proper shipment of pathology material to and from the Image Coordinating Center at the National Institutes of Health (NIH); evaluate completeness and de-identification of the pathology material, image quality, and adequacy of metadata. |
2.3. Queries methodology
As part of the CureGN DQ effort, queries are generated when important data fields are missing, deemed physiologically implausible, or inconsistent with data in related fields (e.g., medication stop date prior to start date). Queries, with explanation, are sent to each site quarterly (except medications, queried every 6 months). Coordinators can make changes directly in GN-Link, and/or report details in their query spreadsheet. Queries that are not resolved in the same query cycle are re-sent in the next cycle, along with new queries. This process is managed by study monitors and programmers at the DCC who interact regularly with staff at the participating sites. The number of variables queried, currently 382, has increased during the study and includes variables related to screening/termination (n = 33), demographics (n = 24), other clinical study participation (n = 7), unique events such as pregnancy and hospitalization (n = 43), laboratory measures (n = 32), medications (n = 64), comorbidities (n = 91), patient birth history (n = 6), family history (n = 23), physical exam (n = 21), clinical disease course (n = 25), and outcomes (n = 13). Non-statistical methods for detecting outliers and missing values could include real-time alerts during the data entry process, similar to the current alerts for suspected out-of-range lab values. Outlier alerts for important variables such as eGFR and proteinuria relapse/remission status could reduce the need for queries on these key variables.
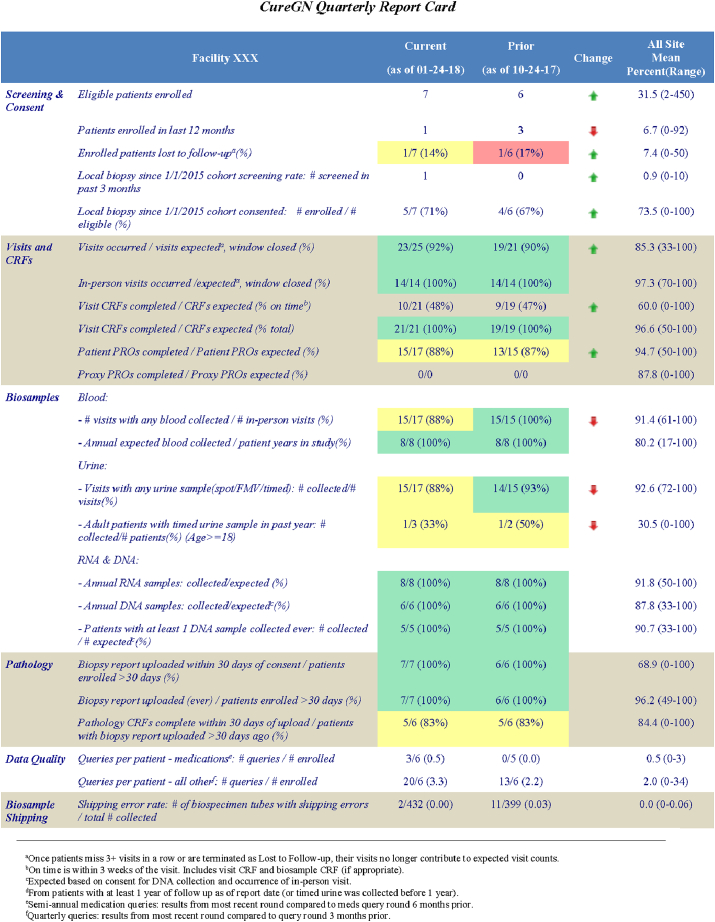
2.4. Site report card development
The Report Cards were developed to assist study coordinators and Principal Investigators (PIs) in monitoring site compliance for major study components on a regular basis (Table 2; enlarged version in Supplemental Table S1). The Report Card is divided into six sections: screening and consent, visits and case report forms (CRFs), biosample collection, pathology, data quality as reflected in query volume, and biosample shipping. Metrics may be patient-level (e.g., percent of patients with any DNA collected) or visit-level (e.g., percent of visits that occurred divided by visits expected). Sites are rated on their percent compliance with each metric. To identify areas needing improvement, the DQC established cut-points for excellent (≥90%), acceptable (70–89%), and poor (<70%) compliance, using a color-coded system of green, yellow, and red, respectively. The Report Card column labeled 'Change' shows arrows pointing up (green) or down (red) to indicate respective trends of improved or worse performance compared with the previous report. The Report Card includes a study-wide summary to enable each site to compare their performance with the mean and range of all consortium sites. In CureGN-1, Report Cards remained largely unchanged over time. CureGN-2 has added greater focus on subject retention as well as new DPR metrics.
Table 2.
CureGN Quarterly Report Cards. Designed to monitor site compliance for major study elements, with consortium averages for comparison. Cells are color-coded based on compliance, defined as excellent (≥90%, green), acceptable (70%–89%, yellow), and poor (<70%, red). Arrows indicate change in performance compared with the previous quarter (better = green up-arrow; worse = red down-arrow). For details, see Supplemental Table S1.
Report Cards are sent quarterly to provide time between cycles for site personnel to make changes and improve performance. The first Report Card was distributed as a 'soft release' with no operational consequences. Subsequent releases included letters to sites requesting a remediation plan if the site had more than two measures with <70% compliance, or the same measure with <70% compliance in two consecutive quarters without improvement.
A Central Review Committee (CRC) was established in 2018 to provide a study-wide approach to assist PCC coordinators and P.I.s in working with sites that have poor compliance in crucial study components, such as enrollment, retention, and bio-sample collection. CRC members include clinician scientists and study coordinators. They work with the PCCs to develop site remediation plans for areas needing improvement, evaluate the success of prior plans, and modify the plans as needed. The CRC follows a collegial approach to remediate problems, with the goal of retaining sites and participants in the study. In the rare event that sites are unable to make progress, the CRC works with the PCC and study leadership to consider the site's termination and transfer of patients to other sites when feasible.
2.5. Data reports
All reports based on data are generated using SAS software, and are disseminated as Word or Excel documents or Adobe Portable Document Formats (PDFs). Report Cards are distributed quarterly, by e-mail for confidentiality; other reports are posted on Basecamp (a web-based tool for group communication). ‘Enrollment & Retention’ and ‘Eligibility by Facility’ reports are generated weekly. ‘Recruitment & Retention,’ ‘Quality Improvement,’ ‘Pathology Core Scoring,’ and ‘Digital Pathology Repository’ reports are generated monthly. ‘Observational Study Monitoring Board’ (OSMB) and ‘Research Performance Progress Report’ (RPPR, an NIH-mandated report showing enrollment by sex, race and ethnicity) are annual.
2.6. Other data issues
There are scant data on the proportion of research subjects whose genetic samples carry the wrong subject ID. Subject labeling (“sample swap” errors) can occur at several ‘touch points’ that individually can introduce errors. Front-end prevention could require at least two methods of patient identification on each sample. A back-end check could involve comparing self-reported race with race assessed by genetic profile, or options such as the KING [18] or HYSYS [19] software, which estimate sample relatedness based on the concordance between samples of homozygous SNPs (single-nucleotide polymorphisms). CureGN is exploring this important issue.
2.7. Statistical methods
The detection of data outliers, implausible data, missing data, and crossed data (wrong patient, wrong blood sample, wrong date) require constant vigilance. Laboratory data outliers are flagged at data entry; implausible values are challenged, but not rejected if the coordinator confirms the value. Further checks are made during the quarterly data queries process. Although we query approximately 382 different data elements, the process requires regular monitoring. For example, although we have queried medication data for several years, it was primarily to detect missing data; current CureGN manuscripts request detailed medication data, which require further cleaning. In another example, eGFR data may be incorrect due to data entry errors in different components of the eGFR calculation, such as height errors (in children) and date errors (e.g., recording the date as several years prior).
Data query and Report Card metric summaries are described using boxplots and line graphs to show variability by site and changes over time.
3. Results
3.1. Data queries
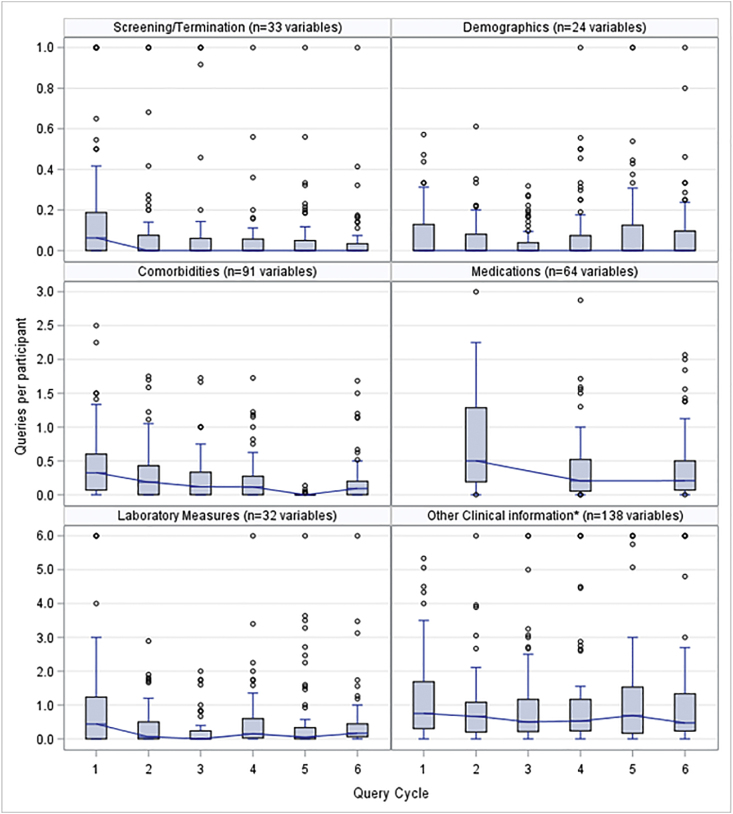
Boxplot distributions of the average number of queries per subject at each site are depicted in Fig. 1 by quarterly cycle and query type (e.g., laboratory values, medications). In almost every query category, there is a substantial drop in the number of queries from the first to the second cycle, but a fairly consistent level of queries per patient from the second cycle on. In every cycle, there are a few 'outlier sites' with relatively high numbers of queries per subject, partly explained by the addition of new queries and new sites. The site query volume is monitored as a Report Card metric.
Fig. 1.
Data quality improvement over time with quarterly queries. Boxplots show the distribution over six query cycles (quarters) of data query volumes (average number of queries per subject), shown for six categories of variables queried over 66 study sites. The number of variables queried has increased during the study and was 382 at cycle 6. Most sites had joined the study by cycle 1. Values were capped at the maxima shown in each graph. Median values in each cycle are shown as a horizontal line across each box. Each box spans the 25th to 75th percentiles. Medication queries were sent every other quarter. *Other Clinical Information includes family and birth history, physical exam (e.g., blood pressure, height, weight), and disease course (proteinuria relapse or remission).
3.2. Report card metrics
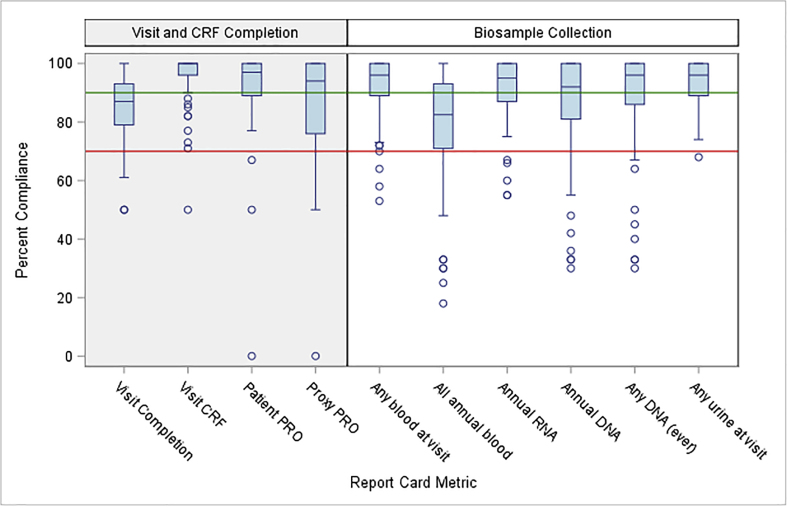
For each of 10 Report Card metrics, there is considerable variability among sites in percent compliance (Fig. 2; data are shown for cycle 5). A site below the 70% reference line would be labeled ‘poor compliance’ for the given metric. For most metrics, the majority of sites have excellent or acceptable compliance, as seen for all four Visit and CRF Completion metrics, and for all biosample metrics except 'All annual blood.' For all metrics, however, the distribution has a long tail of poorly performing sites. Poor compliance was seen at approximately 25% of sites, primarily pediatric, for 'All annual blood'. This was anticipated due to competition for blood samples between clinical and research needs, parental concern, and regulatory limits regarding the total volume that can be obtained in children. While there is high compliance for RNA and DNA samples at most sites, a few sites had persistent difficulty in this area. Comparison across the metrics enables the DQC to assess needs and recommend priorities. An example was a DQC project to increase collection of DNA samples and reduce the number of blood tubes required to obtain all study materials, particularly important for children.
Fig. 2.
Variability across study sites (boxplots) for 10 Report Card metrics at cycle 5. Sites below the reference line at 70% have 'poor compliance’ for the given metric. Comparison across the metrics allows the Data Quality Committee (DQC) to assess needs and recommend priorities within the network. CRF=Case Report Form; PRO=Patient-Reported Outcome. Medians = horizontal line across each box; 25th to 75th percentiles = bottom to top of each box. Metrics are defined as follows: Visit Completion (#visits occurred/#visits expected where visit window has closed); Visit CRF (#visit CRFs completed/#visit CRFs expected); Patient or Proxy PRO (#PROs completed/#PROs expected); Any blood at visit (#visits with any blood collected/#in-person visits); All annual blood (#tubes annual expected blood collected/#patient-years in study); Annual RNA or DNA (#annual samples collected/#expected); Any DNA (ever) (#consenting patients with at least one DNA sample collected ever/# expected); Any urine at visit (#visits with any urine sample collected [spot, First Morning Void, timed]/#visits).
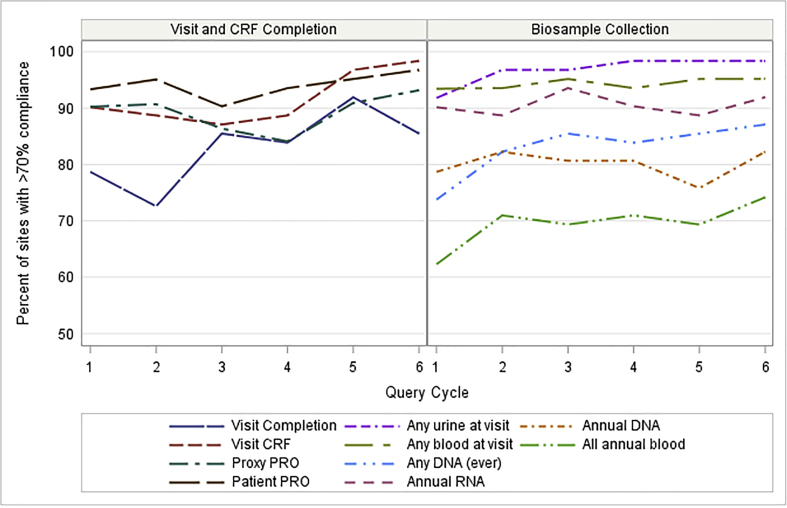
For the same 10 metrics, compliance trends over six cycles of Report Cards are shown in Fig. 3. The line for each metric shows the percent of sites at each cycle achieving over 70% compliance. Increases in performance over time are seen for most metrics. In the left panel showing Visit and CRF Completion, >70% compliance was seen for ~85% of sites across all but the first two cycles, and Visit Completion had substantial improvement over time. The right panel showing Biosample Collection demonstrates favorable trends with persistent room for improvement.
Fig. 3.
Percent of study sites with acceptable or better (>70%) achievement for 10 Report Card metrics over 6 query cycles, for sites with all six Report Card cycles (n = 62). Most metrics show increases in achievement over time. Network-wide efforts addressed metrics where larger numbers of sites underperformed. See Fig. 2 caption for metric definitions.
Study coordinators were informally asked for their perspectives on the CureGN QC systems. Their responses included: "I think the Report Cards are really useful. They do give some pressure, but a sort of 'positive pressure' that is key to identifying weaknesses and developing strategies to improve at the site/PCC level." "High performing sites that consistently receive green flags for biosample metrics can share tips with underperforming sites to develop best practices. In this way, open dialogue between the lead coordinators and the DQC can be instrumental." “The workflow to send pathology material to the DPR is quite demanding, and adjusting to the procedural changes over time has been difficult. However, the goal of a central pathology repository is of such great value for research in glomerular diseases that we are all making every effort to accomplish it.”
Three examples below show how CureGN addressed specific DQ issues.
3.3. Special initiative I: Biosample shipping
Biosample Shipping. In CureGN-1, after discovering substantial problems with labeling and shipping of biosamples to the NIH biorepository, the DCC provided additional training on biosample shipping to PCC coordinators on monthly conference calls and to all coordinators on quarterly “town hall” conference calls. Helpful strategies included sharing of experiences among coordinators, providing quantitative feedback on performance, and setting clear expectations. These initiatives led to major improvements across the consortium.
3.4. Special initiative II: The digital pathology repository (DPR)
The DPR became a special initiative because of its unique requirements, the complexity of coordination, and the importance of pathology to the CureGN research effort. The following passage illustrates the problems exposed and solutions found. The DPR, housed at the NIH, began in 2017 as a cloud-based image repository of the enrolled patients' biopsy reports, WSI, and EM and IF images obtained as part of routine care. The materials are obtained from the enrolling institution's pathology archives or from non-CureGN institutions where the biopsy was performed or sent for interpretation and/or storage. These irreplaceable slides are used in clinical care, and sites often have legitimate concerns about the risks involved in shipping to and from the NIH. Requests have been made to ship the bare minimum of slides, with a limit on time away from the site. Use of standardized shipping materials has been advocated to ensure that slides are not damaged in transit. These concerns may require negotiations and coordination between the DCC, the NIH Image Coordinating Center, and each site.
All pathology materials, including glass slides, are de-identified by the study coordinators at the enrolling institutions and CureGN identifiers are added prior to shipping to the NIH Image Coordinating Center. De-identification requires careful checking, as identifiers used for clinical care are not labeled consistently across sites, or even within sites. Repeated requests by the DCC for full de-identification were insufficient, as the identifiers could lurk in unexpected places. The proposed solution going forward will require sites send a photocopy of slides to the DCC for evaluation. Only upon DCC approval can materials be shipped; disapproval requires additional de-identification. Biopsy reports and EM images will no longer be shipped by CDs; instead, these digital materials will be uploaded digitally to the DCC where another quality control check will ensure only de-identified EM images are subsequently uploaded to the DPR. This solution is expected to largely eliminate the de-identification problem.
At NIH, the glass slides are scanned into WSI and uploaded to the DPR; other pathology materials undergo quality assessment, as recorded in the DPR CRF (Supplemental Table S2), prior to uploading to the DPR. Materials are organized in patient folders by disease. The material is then made available for review and core scoring by Pathology Committee members and for future use by the CureGN consortium and ancillary studies. Populating the DPR is an ongoing coordinated effort between enrollment sites, the PCCs, the DCC, and the NIH Image Coordinating Center. The new initiatives are underway; we anticipate that the plans described above will make an important difference in DPR quality and use.
3.5. Special initiative III: Data cleaning for proteinuria relapse and remission
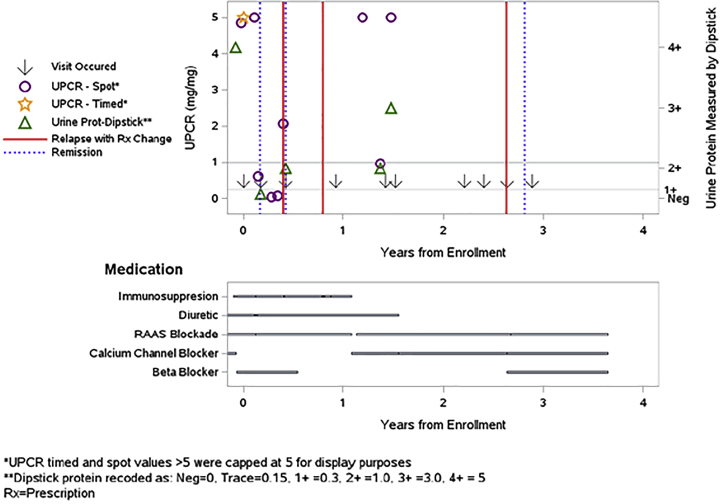
Proteinuria is a primary study outcome; accurate information is crucial, but the accuracy of isolated proteinuria values is difficult to assess. We first developed automated queries to identify cases such as proteinuria events reported without supporting laboratory values, or "steroid-resistant" disease without record of steroid use. These queries handled simple discrepancies but did not address more complicated issues. We then developed a patient-specific plot that simultaneously shows laboratory values, self-reports of relapse or remission, and immunosuppression and other medications (Fig. 4). Although still in the early stage of use, coordinators find these plots helpful for solving issues of incongruent data patterns when several factors are involved.
Fig. 4.
To query or not to query: Logic checking for internal consistency of proteinuria data, reported relapse/remission events, and medication changes for patients with FSGS, MCD, or MN. Patient-specific plots show lab values (in mg urine protein per mg urine creatinine, UPCR; or Dipstick levels), self-reports of relapse with medication change (red lines) or remission (blue dotted lines). Relapse is defined, when possible, as UPCR≥3.0 or Dipstick≥3+, and remission is defined as UPCR<0.3 or Dipstick = Negative. The plot above shows data for a patient with MCD who has several inconsistent or potentially missing data elements: Of the three self-reported relapses, none were confirmed by UPCR≥3.0 or Dipstick≥3+ in the data entered. The first two occurred while on immunosuppression treatment (although possibly during steroid tapering); the last relapse was associated with starting a beta-blocker but not any immunosuppression medication. Nonetheless, the patient achieved self-reported remission thereafter. There were two UPCR-confirmed relapses between days 365 and 730, after immunosuppression had been stopped, with no subsequent data on remission or further immunosuppression medication. Finally, there were three self-reported remissions, only one of which was confirmed as UPCR<0.3. Queries to the site coordinator may resolve some of these issues. FSGS = focal segmental glomerulosclerosis; MCD = minimal change disease; MN = membranous nephropathy; UPCR = urinary protein/creatinine ratio. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Statistical estimates of increased power with complete data
Because the primary goal of data cleaning is to improve statistical accuracy and power, we provide a hypothetical example of power with and without DQ checks for missing data, comparing the percent with edema between two disease groups. We consider complete data and 10%, 20%, and 30% missing or implausible data. We assume a maximum sample size of 600 (the CureGN target enrollment for each of four diseases), and test for differences between subgroups of 300 each. We assume two-sided testing and significance level 0.05 for all tests. Missing data would result in total sample sizes of 540 (10% missing), 480 (20% missing), and 420 (30% missing). With a full sample size of 600, a chi-square test has power = 0.84 to detect a difference in percent with edema of 50% versus 62%; the power drops to 0.80 with 10% missing, to 0.75 with 20% missing, and to 0.69 with 30% missing. Data entry errors, misclassification, or bias in missingness may further erode the statistical power.
4. Discussion
4.1. Guidance for data quality (DQ) initiatives in multi-center observational studies
DQ is motivated by the goal of assuring informational accuracy and statistical power to answer research questions. High DQ also demonstrates respect for the contributions of study participants by responsible stewardship of their data. Ensuring DQ begins with acknowledging the difficult and multi-faceted aspects of data collection performed primarily by study coordinators along with the site PI. The DQC, with members from the DCC, site coordinators, and clinicians, is charged with developing quality guidelines with clear expectations. Standardization of QC processes and reports provides an established work environment that has been vital in creating and sustaining quality.
In CureGN, support from all members has been key to building a collegial team dedicated to collecting accurate and complete data. Early on, the CureGN PIs and PCC coordinators took steps to develop DQ efforts to ensure the value of this unique prospective cohort study of children and adults with primary glomerular disease. The ample use of data queries has been fundamental to maintaining DQ and completeness for individual data elements. Beginning the data query process early in the study established a standard for DQ that has led to a significant reduction in queries over time (Fig. 1). The development of the site Report Card has enhanced both DQ and study performance. Given the generally high level of performance of study sites, Report Cards have been most useful in identifying and working with the minority of underperforming sites. The DQC has created transparent metrics and straightforward management plans to help the CRC facilitate remediation planning to promote improved performance. In a new consortium, an early step would be to identify the most important metrics for study management, keeping the list as concise as possible.
4.2. Incorporating plans for DQ early in study planning and implementation
DQ effors should be interwoven into early study planning and implementation, including in the development of the case report forms and other study tools. For example, in considering a data entry platform, decisions will include whether range checks can be implemented in real time, if over-rides can be allowed, and if data queries can be made after cross-checking data in different fields. Study coordinators should play a key role in this effort. The benefit of robust research coordinator training at the start of study involvement, as well as ongoing refresher training, is well worth the effort. These initiatives can make subsequent DQ efforts a natural outgrowth of data collection and set the expectations for all study staff with respect to DQ. In CureGN, we were not quite so proactive. Enrollment began in December 2014, and the query effort started approximately 6 months later. The DQ committee was established in September 2015, and the first cycle of Report Cards was generated in July 2016. In an ideal world, the DQC could be formed, CRFs could be reviewed, queries could be planned, the Report Card could be developed, and the CRC put into place, all prior to study start. Our Report Card in Table 2 could be modified by future studies to streamline this process.
The CureGN DPR and other international DPRs [20] were built on the framework of the Nephrotic Syndrome Study Network (NEPTUNE) DPR; however, the complexity of the CureGN DPR required substantial modification and enhancement of data tracking. CureGN biopsies occurred prior to enrollment and were located at multiple CureGN and non-CureGN institutions; study coordinators spent extensive effort obtaining the pathology materials. The innovative contribution of CureGN was to develop tools to: a) monitor the flow of renal biopsies into the DPR; b) track shipping, receipt, and return of renal biopsy components from multiple locations to prevent loss of materials; and c) monitor quality, completeness, and de-identification of the pathology material through the DPR CRFs (Supplemental Table S2) to assure high DQ and standardization of the process.
4.3. Appropriate funding is required to support DQ efforts
Funding is required for DCC monitors to manage the query process, programmers and statisticians to implement the query and Report Card process, and study coordinators and site investigators to respond to queries and implement DQ efforts. In complex multi-center studies like CureGN, the scope and cost of this effort is considerable. Use of electronic health records may reduce cost and create opportunities to systematize data collection but will require novel approaches to data cleaning. Funding bodies should recognize the need for adequate funding of DQ efforts to maximize the validity of study results. The benefits of DQ efforts that increase statistical power and accuracy of conclusions are well worth the cost to answer important scientific questions.
4.4. Current limitations and future possibilities
Comparing the efficacy of our DQ procedures and remedial interventions with those of other groups is not common practice, underscoring the lack of published analyses similar to ours in other long-term cohort studies. We speculate that unlike interventional studies in which data accuracy is closely monitored to enable assessment of safety and efficacy of standard and test therapies, less attention has been paid to the completeness and validity of information collected in non-treatment trials. Second, although access to downloadable medical record data would be ideal, the different data platforms across our ~70 centers have so far precluded this possibility. Finally, we note that missing data can arise for a variety of reasons, from data missing completely at random [MCAR] to deliberate omissions (data missing not at random [NMAR]). Considering such possible mechanisms has not yet been implemented in CureGN but is recommended for variables with high missingness.
4.5. In conclusion, achieving high DQ in observational studies represents an ongoing, multi-faceted challenge that requires constant dedication and sufficient funding
The CureGN DQ effort has addressed issues common to most observational studies as well as those unique to nephrology. Collecting correct and complete data and samples from study participants upholds ethical standards of data stewardship and makes the most of study funding to answer important scientific questions.
Authors’ contributions
Each author has participated sufficiently in the work to take public responsibility for the content. This participation includes: Conception or design, or analysis and interpretation of data, or both: BWG, LPL, DZ, RL, MM, SEW, SV, CP, LB, JZ, MH, FL, MK, PHC, SMH, LHM, WES, LAG, DSG, BMR, AGG, LMG-W, HT; drafting the article or revising it: BWG, LPL, DZ, RL, MM, SEW, SV, CP, LB, JZ, MH, FL, MK, PHC, SMH, LHM, WES, LAG, DSG, BMR, AGG, LMG-W, HT; providing intellectual content of critical importance to the work described: BWG, LPL, DZ, RL, MM, SEW, SV, CP, LB, JZ, MH, FL, MK, PHC, SMH, LHM, WES, LAG, DSG, BMR, AGG, LMG-W, HT; final approval of the version to be published: All authors contributed to review of the manuscript, and all authors have read and approved the final submitted version of the manuscript.
Funding
This work was supported by the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Deseases (NIDDK, United States) [grant numbers: UM1DK100845, UM1DK100846, UM1DK100876, UM1DK100866, and UM1DK100867]. Patient recruitment was supported by NephCure Kidney International (United States). Laura Mariani is funded by the Integrative Molecular Epidemiology Approach to Identify Nephrotic Syndrome Subgroups project, funded by the NIDDK, United States [grant number K08 DK115891-01]. Dates of funding for CureGN-1 are 9/16/2013-5/31/2019, and for CureGN-2 are 8/16/2019-5/31/2024.
Collaborators
*Consortium Collaborators (to be listed in PubMed): The CureGN Consortium members listed below, from within the four Participating Clinical Center networks and Data Coordinating Center, are acknowledged by the authors as Collaborators (not coauthors) on this manuscript and must be indexed in PubMed as “Collaborators”. CureGN Principal Investigators are noted (**).
Columbia University: Wooin Ahn, Columbia; Gerald B. Appel, Columbia; Revekka Babayev, Columbia; Ibrahim Batal, Columbia; Andrew S. Bomback, Columbia; Eric Brown, Columbia; Eric S. Campenot, Columbia; Pietro Canetta, Columbia; Brenda Chan, Columbia; Debanjana Chatterjee, Columbia; Vivette D. D'Agati, Columbia; Elisa Delbarba, Columbia; Hilda Fernandez, Columbia; Bartosz Foroncewicz, University of Warsaw, Poland; Ali G. Gharavi, Columbia**; Gian Marco Ghiggeri, Gaslini Children's Hospital, Italy; William H. Hines, Columbia; Namrata G. Jain, Columbia; Byum Hee Kil, Columbia; Krzysztof Kiryluk, Columbia; Wai L. Lau, Columbia; Fangming Lin, Columbia; Francesca Lugani, Gaslini Children's Hospital, Italy; Maddalena Marasa, Columbia; Glen Markowitz, Columbia; Sumit Mohan, Columbia; Xueru Mu, Columbia; Krzysztof Mucha, University of Warsaw, Poland; Thomas L. Nickolas, Columbia; Stacy Piva, Columbia; Jai Radhakrishnan, Columbia; Maya K. Rao, Columbia; Simone Sanna-Cherchi, Columbia; Dominick Santoriello, Columbia; Michael B. Stokes, Columbia; Natalie Yu, Columbia; Anthony M. Valeri, Columbia; Ronald Zviti, Columbia.
Midwest Pediatric Nephrology Consortium (MWPNC): Larry A. Greenbaum**, Emory University; William E. Smoyer**, Nationwide Children's; Josephine Abruzs, Arkana; Amira Al-Uzri Oregon Health & Science University; Isa Ashoor, Louisiana State University Health Sciences Center; Diego Aviles, Louisiana State University Health Sciences Center; Rossana Baracco, Children's Hospital of Michigan; John Barcia, University of Virginia; Sharon Bartosh, University of Wisconsin; Craig Belsha, Saint Louis University/Cardinal Glennon; Corinna Bowers, Nationwide Children's Hospital; Michael C Braun, Baylor College of Medicine/Texas Children's Hospital; Aftab Chishti, University of Kentucky; Donna Claes, Cincinnati Children's Hospital; Carl Cramer, Mayo Clinic; Keefe Davis, Washington University in St. Louis; Elif Erkan, Cincinnati Children's Hospital Medical Center; Daniel Feig, University of Alabama, Birmingham; Michael Freundlich, University of Miami/Holtz Children's Hospital; Joseph Gaut, Washington University in St. Louis; Rasheed Gbadegesin, Duke University Medical Center; Melisha Hanna, Children's Colorado/University of Colorado; Guillermo Hidalgo, East Carolina University; Tracy E. Hunley, Monroe Carell Jr Children's Hospital at Vanderbilt University Medical Center; Amrish Jain, Children's Hospital of Michigan; Mahmoud Kallash MD, Nationwide Children's Hospital; Myda Khalid, JW Riley Hospital for Children, Indiana University School of Medicine, Indianapolis IN; Jon B. Klein, The University of Louisville School of Medicine; Jerome C. Lane, Feinberg School of Medicine, Northwestern University; Helen Liapis, Arkana; John Mahan, Nationwide Children's; Nisha Mathews, University of Oklahoma Health Sciences Center; Carla Nester, University of Iowa Stead Family Children's Hospital; Cynthia Pan, Medical College of Wisconsin; Larry Patterson, Children's National Health System; Hiren Patel, Nationwide Children's Hospital; Adelaide Revell, Nationwide Children's Hospital; Michelle N. Rheault, University of Minnesota Masonic Children's Hospital; Cynthia Silva, Connecticut Children's Medical Center; Rajasree Sreedharan, Medical College of Wisconsin; Tarak Srivastava, Children's Mercy Hospital; Julia Steinke, Helen DeVos Children's Hospital; Katherine Twombley, Medical University of South Carolina; Scott E. Wenderfer, Baylor College of Medicine/Texas Children's Hospital; Tetyana L. Vasylyeva, Texas Tech University Health Sciences Center; Donald J. Weaver, Levine Children's Hospital at Carolinas Medical Center; Craig S. Wong, University of New Mexico Health Sciences Center Hong Yin, Emory University.
The University of North Carolina (UNC): Anand Achanti, Medical University of South Carolina (MUSC); Salem Almaani, The Ohio State University (OSU); Isabelle Ayoub,OSU; Milos Budisavljevic, MUSC; Anthony Chang, University of Chicago; Vimal Derebail, UNC; Huma Fatima, The University of Alabama at Birmingham (UAB); Ronald Falk**, UNC; Huma Fatima, University of Alabama; Agnes Fogo, Vanderbilt; Todd Gehr, Virginia Commonwealth University (VCU); Keisha Gibson, UNC; Dorey Glenn, UNC; Raymond Harris, Vanderbilt; Susan Hogan, UNC; Koyal Jain, UNC; J. Charles Jennette, UNC; Bruce Julian, UAB; Jason Kidd, VCU; Louis-Philippe Laurin, Hôpital Maisonneuve-Rosemont (HMR) Montreal, H. Davis Massey, VCU; Amy Mottl, UNC; Patrick Nachman, UNC; Tibor Nadasdy, MUSC; Jan Novak, UAB; Samir Parikh, OSU; Vincent Pichette, HMR Montreal; Caroline Poulton, UNC; Thomas Brian Powell, Columbia Nephrology Associates; Matthew Renfrow, UAB; Dana Rizk, UNC, Brad Rovin, OSU; Virginie Royal, HMR Montreal; Manish Saha, UNC; Neil Sanghani, Vanderbilt, Sally Self, MUSC.
University of Pennsylvania (UPENN): Sharon G. Adler, Los Angeles Biomedical Research Institute at Harbor, University of California Los Angeles (UCLA); Carmen Avila-Casado, University Health Network, University of Toronto; Serena Bagnasco, Johns Hopkins; Raed Bou Matar, Cleveland Clinic; Elizabeth Brown, University of Texas (UT) Southwestern Medical Center; Daniel Cattran, University of Toronto; Michael Choi, Johns Hopkins; Katherine M. Dell, Case Western/Cleveland Clinic; Ram Dukkipati, Los Angeles Biomedical Research Institute at Harbor UCLA; Fernando C. Fervenza, Mayo Clinic; Alessia Fornoni, University of Miami; Crystal Gadegbeku, Temple University; Patrick Gipson, University of Michigan; Leah Hasely, University of Washington; Sangeeta Hingorani, Seattle Children's Hospital; Michelle Hladunewich, University of Toronto/Sunnybrook; Jonathan Hogan, University of Pennsylvania; Jean Hou, Cedars-Sinai Medical Center; Lawrence B. Holzman**, University of Pennsylvania; J. Ashley Jefferson, University of Washington; Kenar Jhaveri, North Shore University Hospital; Duncan B. Johnstone, Temple University; Frederick Kaskel, Montefiore Medical Center; Amy Kogan, CHOP; Jeffrey Kopp, NIDDK Intramural Research Program; Richard Lafayette, Stanford; Kevin V. Lemley, Children's Hospital of Los Angeles; Laura Malaga- Dieguez, NYU; Kevin Meyers, Children's Hospital of Pennsylvania; Alicia Neu, Johns Hopkins; Michelle Marie O'Shaughnessy, Stanford; John F. O'Toole, Case Western/Cleveland Clinic; Matthew Palmer, University of Pennsylvania; Rulan Parekh, University Health Network, Hospital for Sick Children; Heather Reich, University Health Network, University of Toronto, Toronto, CAN; Kimberly Reidy, Montefiore Medical Center; Helbert Rondon, UPMC; Kamalanathan K. Sambandam, UT Southwestern; John R. Sedor, Case Western/Cleveland Clinic; David T. Selewski, University of Michigan; Christine B. Sethna, Cohen Children's Medical Center-North Shore Long Island Jewish (LIJ) Health System; Jeffrey Schelling, Case Western; John C. Sperati, Johns Hopkins; Agnes Swiatecka-Urban, Children's Hospital of Pittsburgh; Howard Trachtman, New York University; Katherine R. Tuttle, Spokane Providence Medical Center; Joseph Weisstuch, New York University; Suzanne Vento, New York University Langone Medical Center; Olga Zhdanova, New York University.
Data Coordinating Center (DCC): Laura Barisoni, Duke University, Durham, NC; Brenda W. Gillespie**, University of Michigan; Debbie S. Gipson**, University of Michigan; Peg Hill-Callahan, Arbor Research Collaborative for Health; Margaret Helmuth, Arbor Research Collaborative for Health; Emily Herreshoff, University of Michigan; Matthias Kretzler**, University of Michigan; Chrysta Lienczewski, University of Michigan; Sarah Mansfield, Arbor Research Collaborative for Health; Laura Mariani, University of Michigan; Cynthia C. Nast, Cedars-Sinai Medical Center; Bruce M. Robinson**, Arbor Research Collaborative for Health; Jonathan Troost, University of Michigan; Matthew Wladkowski, Arbor Research Collaborative for Health; Jarcy Zee, Arbor Research Collaborative for Health; Dawn Zinsser, Arbor Research Collaborative for Health.
Steering Committee Chair: Lisa M. Guay-Woodford, Center for Translational Research, Children's National Hospital, Washington DC.
Declaration of competing interest
HT is a consultant to Otsuka and Chemocentryx (Data Monitoring Committees) and receives support from Travere Therapeutic and Goldfinch BioPharma Inc. through separate grants to New York University. In addition, he has received research support from Natera Inc.
LAG receives research support from Vertex Pharmaceuticals and Reata Pharmaceuticals.
SEW has a consulting agreement with Bristol-Myers Squibb.
The remaining authors declare no perceived or actual COIs.
Acknowledgements
Heather Van Doren, Senior Medical Editor with Arbor Research Collaborative for Health, provided editorial assistance on this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.conctc.2021.100749.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Stone E.J., Osganian S.K., McKinlay S.M., Wu M.C., Webber L.S., Luepker R.V., Perry C.L., Parcel G.S., Elder J.P. Operational design and quality control in the CATCH multicenter Trial. Prev. Med. 1996;25(4):384–399. doi: 10.1006/pmed.1996.0071. [DOI] [PubMed] [Google Scholar]
- 2.Lorenzi G.M., Braffett B.H., Arends V.L., Danis R.P., Diminick L., Klumpp K.A., Morrison A.D., Soliman E.Z., Steffes M.W., Cleary P.A. DCCT/EDIC research group. Quality control measures over 30 Years in a multicenter clinical study: results from the Diabetes control and complications trial/epidemiology of Diabetes interventions and complications (DCCT/EDIC) study. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0141286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams G.W. The other side of clinical trial monitoring; assuring data quality and procedural adherence. Clin. Trials. 2006;3(6):530–537. doi: 10.1177/1740774506073104. [DOI] [PubMed] [Google Scholar]
- 4.Tolmie E.P., Dinnett E.M., Ronald E.S., Gaw A. AURORA Clinical Endpoints Committee. Clinical Trials: minimising source data queries to streamline endpoint adjudication in a large multi-national trial. Trials. 2011;12:112. doi: 10.1186/1745-6215-12-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward L., Steel J., Le Compte A., Evans A., Tan C.S., Penning S., Shaw G.M., Desaive T., Chase J.G. Data entry errors and design for model-based tight glycemic control in critical care. J. Diabetes Sci. Technol. 2012;6(1):135–143. doi: 10.1177/193229681200600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao H., Shang H., Mu W., Zhai J., Hou Y., Wang H. Internal challenge to clinical trial project management: strategies for managing investigator compliance. J. Evid. Base Med. 2013;6(3):157–166. doi: 10.1111/jebm.12053. [DOI] [PubMed] [Google Scholar]
- 7.Cagnon C.H., Cody D.D., McNitt-Gray M.F., Seibert J.A., Judy P.F., Aberle D.R. Description and implementation of a quality control program in an imaging-based clinical trial. Acad. Radiol. 2006;13(11):1431–1441. doi: 10.1016/j.acra.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Nakajima K., Inomata M., Akagi T., Etoh T., Sugihara K., Watanabe M., Yamamoto S., Katayama H., Moriya Y., Kitano S. Quality control by photo documentation for evaluation of laparoscopic and open colectomy with D3 resection for stage II/III colorectal cancer: Japan Clinical Oncology Group Study JCOG 0404. Jpn. J. Clin. Oncol. 2014;44(9):799–806. doi: 10.1093/jjco/hyu083. [DOI] [PubMed] [Google Scholar]
- 9.Boellaard R., Rausch I., Beyer T., Delso G., Yaqub M., Quick H.H., Sattler B. Quality control for quantitative multicenter whole-body PET/MR studies: a NEMA image quality phantom study with three current PET/MR systems. Med. Phys. 2015;42(10):5961–5969. doi: 10.1118/1.4930962. [DOI] [PubMed] [Google Scholar]
- 10.Yan G., Greene T. Statistical analysis and design for estimating accuracy in clinical-center classification of cause-specific clinical events in clinical trials. Clin. Trials. 2011;8(5):571–580. doi: 10.1177/1740774511411320. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie B.W., Merion R.M., Ortiz-Rios E., Tong L., Shaked A., Brown R.S., Ojo A.O., Hayashi P.H., Berg C.L., Abecassis M.M., Ashworth A.S., Friese C.E., Hong J.C., Trotter J.T., Everhart J.E., the A2ALL Study Group Database comparison of the adult-to-adult living donor liver transplantation cohort study (A2ALL) and the SRTR U.S. Transplant registry. Am. J. Transplant. 2010;10(7):1621–1633. doi: 10.1111/j.1600-6143.2010.03039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goudar S.S., Stolka K.B., Koso-Thomas M., Honnungar N.V., Mastiholi S.C., Ramadurg U.Y., Dhaded S.M., Pasha O., Patel A., Esamai F., Chomba E., Garces A., Althabe F., Carlo W.A., Goldenberg R.L., Hibberd P.L., Liechty E.A., Krebs N.F., Hambidge M.K., Moore J.L., Wallace D.D., Derman R.J., Bhalachandra K.S., Bose C.L. Data quality monitoring and performance metrics of a prospective, population-based observational study of maternal and newborn health in low resource settings. Reprod. Health. 2015;12(Suppl 2):S2. doi: 10.1186/1742-4755-12-S2-S2. Epub 2015 Jun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiele F. Quality of data in observational studies: separating the wheat from the chaff. Eur. Heart J. Qual. Care Clin. Outcomes. 2017 Apr 1;3(2):99–100. doi: 10.1093/ehjqcco/qcx003. [DOI] [PubMed] [Google Scholar]
- 14.Duda S.N., Shepherd B.E., Gadd C.S., Masys D.R., McGowan C.C. Measuring the quality of observational study data in an international HIV research network. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0033908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giganti M.J., Shepherd B.E., Caro-Vega Y., Luz P.M., Rebeiro P.F., Maia M., Julmiste G., Cortes C., McGowan C.C., Duda S.N. The impact of data quality and source data verification on epidemiologic inference: a practical application using HIV observational data. BMC Publ. Health. 2019;19:1748. doi: 10.1186/s12889-019-8105-2. PMID: 31888571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ercole A., Brinck V., George P., Hicks R., Huijben J., Jarrett M., Vassar M., Wilson L. DAQCORD collaborators. Guidelines for data acquisition, quality and curation for observational research designs (DAQCORD) J. Clin. Transl. Sci. 2020;4:354–359. doi: 10.1017/cts.2020.24. PMID: 33244417, PMCID: PMC7681114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariani L.H., Bomback A.S., Canetta P.A., Flessner M.F., Helmuth M., Hladunewich M.A. CureGN study rationale, design, and methods: establishing a large prospective observational study of glomerular disease. Am. J. Kidney Dis. 2019;73(2):218–229. doi: 10.1053/j.ajkd.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schröder J., Corbin V., Papenfuss A.T. HYSYS: have you swapped your samples? Bioinformatics. 2017 Feb 15;33(4):596–598. doi: 10.1093/bioinformatics/btw685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barisoni L., Gimpel C., Kain R., Laurinavicius A., BuenoG G., Zeng C. Digital pathology imaging as a novel platform for standardization and globalization of quantitative nephropathology. Clin. Kidney J. 2017;10(2):176–187. doi: 10.1093/ckj/sfw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.