Abstract
Genetic testing is an integral component in the workup of male infertility as genetic conditions may be responsible for up to 15% of all cases. Currently, three genetic tests are commonly performed and recommended by major urologic associations: karyotype analysis (KA), Y-chromosome microdeletion testing, and CFTR mutation testing. Despite widespread adoption of these tests, an etiology for infertility remains elusive in up to 80% of cases. Recent work has identified intriguing new targets for genetic testing which may soon see clinical relevance. This review will discuss the indications and techniques for currently offered genetic tests and briefly explore ongoing research directions within this field.
Keywords: Genetic techniques, Klinefelter syndrome, male infertility, Y chromosome microdeletion, next-generation sequencing (NGS)
Introduction
Infertility, commonly defined as inability to achieve pregnancy despite 1 year of unprotected intercourse, affects nearly 15% of couples (1), and a male factor contributes in up to 50% of these couples (2). The etiology of male infertility can be broadly divided into four major categories: hypothalamic-pituitary axis dysfunction, quantitative spermatogenic defects, qualitative spermatogenic defects, and ductal obstruction or dysfunction (3). Known genetic anomalies underlie ~15% of male infertility cases (4) and can cause abnormalities within any of these four etiologic categories. With significant advancement in our understanding of genetics came a concordant rise in the medical applications of genetic testing. Many of these tests now comprise essential components of the male infertility workup. This review will provide an overview of common genetic tests used for male infertility and the indications for each of these tests.
Initial workup of infertility
The American Urologic Association (AUA), European Association of Urology (EAU), and American Society of Reproductive Medicine (ASRM) agree that the initial workup of infertility should include at least a comprehensive medical history, physical examination with focus on male genitalia, and at least one (although some committees recommend two) semen analysis (5-7). If any part of this workup is abnormal, the man should be referred to a urologist or male reproductive specialist for a more thorough evaluation. In particular, the examiner should note the location of urethral meatus, size and consistency of testes, presence or absence of both vasa and epididymides, and secondary sex characteristics. Digital rectal examination may also be performed if there is concern for prostatic or seminal vesicle anomalies (7). Completion of this evaluation, in addition to serum FSH and testosterone levels, may identify the etiology of infertility in up to 70% of men (8).
Genetic testing is not indicated for all infertile men. Those men who have had previous fertility, known prior gonadotoxic (e.g., chemotherapeutic) exposure, or have an exam highly suspicious for ejaculatory duct dysfunction do not warrant a full genetic workup. Rather, genetic testing should be considered in men with nonobstructive azoospermia, severe oligospermia, or nonpalpable vasa (9). Genetic testing serves two major purposes in the setting of infertility: determination of heritable conditions that may be passed to offspring and evaluation for conditions which may impact the success of assisted reproductive techniques (10). Diagnosis of genetic problems causing infertility may also have implications for management of a patient’s overall health (e.g., consideration of testosterone therapy for skeletal and mood benefits in men with Klinefelter syndrome).
The most common genetic tests for male infertility used in clinical practice today are karyotyping, Y-chromosome microdeletion screening, and CFTR gene mutation testing. The AUA, EAU, and ASRM all have guidelines outlining the appropriate uses for each of these diagnostic techniques (Table 1). This review will primarily discuss the indications, rationale, and methodology of these three genetic tests. Other rarer genetic tests used for specific populations will also be described.
Table 1. Guidelines for genetic testing in male infertility (5-7).
| Genetic test | Society | ||
|---|---|---|---|
| AUA | EAU | ASRM | |
| Karyotyping | NOA or <5 million/mL | Sperm conc <10 million/mL | NOA or <5 million/mL |
| YCMD | NOA or <5 million/mL | Sperm conc <5 million/mL | NOA or <5 million/mL |
| CFTR | CBAVD | CBAVD or CUAVD without renal abnormalities | CBAVD, CUAVD without renal abnormalities, or bilateral epididymal obstruction |
| Female partners should also be tested | Female partners should also be tested | Female partners should also be tested | |
Karyotype analysis (KA)
KA is a cytogenetic technique in which human chromosomes are visualized using light microscopy and subsequently analyzed for abnormalities in number or structure. To produce a karyotype, lymphocytes obtained from peripheral blood cultures are chemically arrested in metaphase with colcemid, a drug which depolymerizes microtubules and inhibits spindle formation (11). Conventional G-banded karyotype analysis, in which Giemsa stain is added to these arrested lymphocytes, produces characteristic banding patterns on each chromosome and allows for detection of abnormalities greater than 5 megabase (Mb) in size. Other staining patterns such as C (centromere)-banding and T (telomere)-banding exist but are less commonly used in the evaluation of infertility. Due to the relatively low resolution of KA, only large chromosomal abnormalities such as aneuploidies, Robertsonian and balanced translocations, and inversions can be detected (12).
With over 2,000 genes thought to play a role in proper spermatogenesis (13), countless chromosomal anomalies could theoretically cause infertility. However, the most commonly detected etiologies on KA are Klinefelter syndrome, structural chromosomal aberrations, and 46, XX male syndrome. Klinefelter syndrome (47, XXY) and its variants (such as mosaic 47, XXY/46, XY) affect one in 660 men and are the most frequent genetic causes of nonobstructive azoospermia (13,14). Structural chromosome aberrations such as translocations and inversions are found in up to 10% of infertile men and are the most frequent causes of oligospermia (4,13). 46, XX male syndrome, also known as de la Chappelle syndrome, is a rare condition with a prevalence of one in 20,000 males and occurs when Y chromosomal material including the SRY gene is translocated onto another, usually autosomal, chromosome during paternal meiosis (15,16). Successful sperm retrieval rates vary widely among these populations, ranging from 0% in 46, XX males (due to an absent AZF region) to ~30% in males with mosaic Klinefelter syndrome (10).
There is robust evidence that karyotypic abnormalities are much more prevalent in oligospermic and azoospermic men compared to fertile controls (seen in ~0.4% of general population, ~3.6% of oligospermic men, and up to 15% of azoospermic men) (17-19). Overall, approximately 5% of male infertility cases can be attributed to chromosomal abnormalities identified on KA (20). Accordingly, both AUA and EAU guidelines recommend karyotype testing in all nonobstructive azoospermic or severely oligospermic men (AUA <5 million/mL, EAU <10 million/mL) (5,7). EAU guidelines additionally recommend karyotype analysis for infertile couples with family history of recurrent spontaneous abortion, malformation, or mental retardation (5).
Although commonly used, KA does have some important limitations. It has comparatively poor resolution relative to other genetic tests so point mutations, frameshift mutations, or microdeletions cannot be detected. Additionally, even in men with sperm concentration <10 million/mL, KA often does not produce a definitive diagnosis and may indeed find clinically insignificant chromosomal abnormalities. Ventimiglia et al. recognized this deficiency within the EAU guidelines and proposed a nomogram with parameters of sperm concentration, mean testis volume, and luteinizing hormone level to predict which infertile men would benefit from KA. Using this nomogram resulted in 94% sensitivity in detecting karyotype abnormalities compared to 80% when using EAU criteria, but there was no difference in the specificity (i.e., overtesting) (21). As with many laboratory tests for infertility, cost can be a limiting factor. Insurance coverage of KA varies widely based upon individual plans, and out-of-pocket expenses for these tests range from $700 to $1,200 (22,23) in the United States. Despite these drawbacks, KA remains an integral diagnostic test for the initial workup of many cases of infertility.
Fluorescence in-situ hybridization (FISH), a related microscopic cytogenetic technique in which specific DNA sequences are hybridized to collected samples and visualized under fluorescence microscopy, has a comparatively increased resolution of about 1 Mb (24) and is primarily used to analyze specific genomic regions. This technique is not typically used for routine KA but rather as an adjunct to further characterize specific cytogenetic anomalies, such as determining whether the sex-determining region of the Y chromosome (SRY) is present in patients with XX karyotype. Other important roles for FISH include analysis of spermatic chromosomal integrity and preimplantation genetic diagnosis (25). For those patients with an abnormal karyotype who are candidates for IVF/ICSI, FISH in retrieved sperm can be performed to assess for spermatic aneuploidy or structural defects. Males with Klinefelter syndrome and other sex chromosome aneuploidies such as 47, XYY have an increased incidence of aneuploid (e.g., hyperhaploid or diploid) sperm production, and men with other karyotype anomalies have similarly higher rates of spermatic chromosomal rearrangements (26). For this reason, it has been suggested that FISH analysis of spermatozoa may be a useful supplement to KA, as it may provide better prognostic data prior to assisted reproductive technology (27).
Y-chromosome microdeletion testing
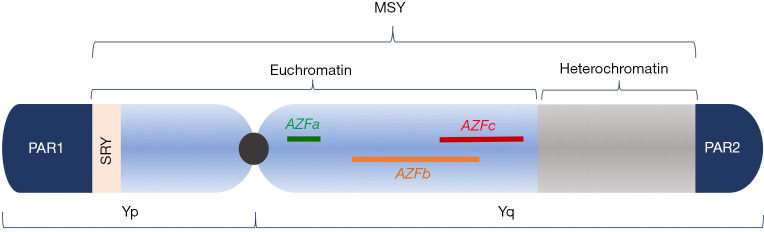
The Y chromosome (Figure 1) is an acrocentric chromosome containing 60 Mb on a short arm (Yp) and long arm (Yq) separated by a centromere (28). Nearly 95% of its length is comprised of the male-specific region of the Y chromosome (MSY), a collection of ~80 genes which play an important role in male sex development and spermatogenesis (29). The MSY, unlike large segments of autosomes, does not recombine during meiosis as there is no homologous region to pair with on the X chromosome (30). Rather, the MSY contains eight repetitive, palindromic, and redundant segments which are highly susceptible to intra-chromosomal rearrangement during meiosis—a process known as non-allelic homologous recombination (31). This process is relevant because ectopic reinsertion of these segments can result in deletions, duplications, or inversions within the Y chromosome, all of which can affect genes necessary for fertility (32-34).
Figure 1.
Schematic representation of the Y chromosome. The human X and Y chromosomes share short homologous regions known as pseudoautosomal regions (PAR) 1 and 2, colored in dark blue. The male-specific region (MSY) spans the remainder of the chromosome and includes regions of euchromatin and genetically inert heterochromatin. Within the euchromatin exists testis-determining factor (SRY, light pink) and the azoospermia factor regions. AZFb and AZFc overlap by 1.5 Mb.
Of particular clinical importance is the azoospermia factor region (AZF) found on Yq which has been rigorously studied due to its richness of genes implicated in spermatogenesis (35). First described in the 1970’s (36) and further characterized in the mid-1990’s to early 2000’s (29,37), AZF is subdivided into three sub-regions—AZFa, AZFb, and AZFc—with AZFb and AZFc overlapping by 1.5 Mb (32,35). Deletions in each of these regions vary in their clinical phenotypes. Deletions of the entire AZFa region are associated with Sertoli cell only syndrome, a histological diagnosis of germ cell aplasia in tissue obtained from testis biopsy that invariably results in nonobstructive azoospermia without the possibility of ART (38). Spermatocytes are also not present in patients with complete AZFb deletions due to absence of essential maturation factors (39) and consequently, ART is not offered in these men. Complete AZFc or partial AZFb+c deletions, however, do not preclude the presence of normal spermatozoa. Accordingly, normal sperm can be retrieved in up to 70% of cases (39,40) and assisted reproductive technology (ART) remains a viable option in this population.
As Y chromosome microdeletions (YCMDs) are too small to be detected with karyotype, polymerase chain reaction (PCR) amplification must be used. Primers specific to unique sequence-tagged sites (STS)—short DNA sequences with a single occurrence within the genome—on the MSY are then used to initiate PCR. Many STS primers have been described for use in the clinical setting but variability among these primers and protocols used by laboratories complicates the interpretation of test results and may reduce diagnostic accuracy (41). To remedy this, the European Academy of Andrology and the European Molecular Genetics Quality Network have released guidelines which detail a “basic” set of six STS primers, two on each of AZFa, AZFb, and AZFc, which have given robust and reproducible results across several laboratories and quality control trials (42). The ideal number of STS to maximize efficacy of PCR has not yet been elucidated (7).
Nearly 7.5% of all oligo- and azoospermic men are believed to harbor YCMDs, although this prevalence varies worldwide (35). In patients with nonobstructive azoospermia or severe oligospermia this prevalence rises to 10–15% (20). Due to the relatively high frequency of YCMD in oligo- and azoospermic patients, both the EAU and ASRM recommend offering YCMD testing to all men with sperm counts lower than 5 million/mL (5,6). Although this threshold has high sensitivity, multiple large retrospective studies have suggested that YCMDs are exceedingly rare in men with sperm counts greater than 1–2 million/mL (43-45). Kohn et al. recently reviewed 37 studies of oligospermic men (N=12,492) that identified 261 men with YCMD (46). Of these men with YCMD, 93% had sperm counts <1 million/mL and only 5% had sperm counts >1–5 million/mL. Consequently, they proposed lowering the testing threshold to from 5 to 1 million/mL given the rarity of YCMDs in men with sperm counts above this value.
Genetic counseling must also be considered for men who are found to have YCMD, as these deletions are necessarily transmitted to all male offspring (5). Although men with AZFc deletions have fathered sons via ICSI, the spermatogenic and reproductive capabilities of these children are currently unknown; however, there is thought to be a spectrum of infertility ranging from complete sterility to spermatogenic potential. Pan et al. described three options for fully informed couples who wished to proceed with ART: proceeding with ICSI and conceiving infertile sons, not using the retrieved sperm to conceive, or performing preimplantation genetic analysis and selecting only for 46, XX embryos (47). All couples should be aware of the risks and benefits for each of these options prior to make a decision.
CFTR gene testing
Congenital absence of the vas deferens (CAVD) can affect one (congenital unilateral absence of vas deferens, CUAVD) or both vasa deferentia (congenital bilateral absence of vasa deferentia, CBAVD). Bilateral absence occurs in 2–10% of all infertile men but accounts for up to 40% of obstructive azoospermia cases (48,49). Over 90% of men with CBAVD additionally have morphologically abnormal (absent, atrophic, hypotrophic, or cystic) seminal vesicles (50). Physiologically, this results in abnormal semen parameters characterized by low semen volume, acidic pH, and absence of spermatozoa. Nearly 80% of men with CBAVD have mutations within the cystic fibrosis transmembrane conductor (CFTR) gene (51,52), a finding which has prompted widespread adoption of CFTR genetic testing in infertile men with the aforementioned physical exam findings.
CFTR is a 250 Kb gene on chromosome 7 which encodes an ATP-dependent chloride channel found primarily in epithelial cells (53,54). To date, over 2,000 mutations have been described with varying phenotypic consequences from mild to severe (55). The prevalence of these mutations differs among different ethnicities and geographic regions (52,56). Cystic fibrosis (CF), an autosomal recessive disease which causes progressive respiratory and pancreatic failure, occurs when an individual has two severe mutations of CFTR, one on each homologous chromosome. Overall prevalence of CF in the United States ranges from 1 in 2,500 in Caucasians to 1 in 35,000 for Asian-Americans (57). Up to 80% of these cases are due to a deleterious mutation within the CFTR gene termed DeltaF508 which ultimately results in abnormal protein folding (58). Observed CFTR mutation carrier frequency is 1 in 38 Americans with significant variability based on race (59). While CF leads to pancreatic and pulmonary dysfunction, more mild mutations of the CFTR gene (or simply being a carrier) can result in either CBAVD or CUAVD as the sole clinical manifestation of the mutation (10). The precise mechanism for why CFTR mutations cause vasal agenesis is currently unknown; however, it has been proposed that epithelial secretory defects caused by these mutations disrupt proper Wolffian duct development in utero (60,61). Of note, men with CFTR mutations have normal renal anatomy because this disruption of Wolffian duct development occurs after kidney formation (60). The prevalence of unilateral renal agenesis has been reported as 25–85% in men with CUAVD and 10-15% in men with CBAVD (60,62-64). This abnormality is not associated with CFTR mutations, but rather may be explained by different genetic mutations which leads to anomalies within Wolffian duct structures (65).
Detection of CFTR mutations is usually accomplished using direct gene analysis techniques. As no standard protocol for gene analysis exists, these techniques are laboratory dependent. It is neither cost-effective nor practical to directly assess for all known CFTR mutations, so many commercial assays are available which test for ~20 to ~60 of the most common mutations (66). Different assays with alternate mutations can be tailored to the patient’s ethnicity and/or geographic location. Whole-gene sequencing, while the most thorough diagnostic method, was previously reserved for those patients with signs of CFTR dysfunction with normal mutation panels given the high cost. Recent advances in next-generation sequencing (NGS), however, have significantly improved diagnostic accuracy and reduced the cost of whole-gene sequencing which may make NGS a viable-first line test in the future (67,68).
Men with CBAVD typically have normal spermatogenic potential and are therefore excellent candidates for ART (69,70). Both the AUA and EAU recommend evaluating for CFTR mutations in men with CBAVD (5,7). Men with CUAVD should instead undergo renal imaging due to the high incidence of non-CFTR-associated ipsilateral renal agenesis (60). Partners of men with CBAVD should additionally be screened prior to attempts at ART in order to evaluate CF risk in offspring. If the female partner is also a CFTR mutation carrier, the risk of having a child with CF can rise to 50% depending on parental mutations. In these cases, pre-implantation diagnosis should be considered.
Other genetic tests for infertility
A comprehensive discussion of all genetic causes of male infertility is beyond the scope of this review, but it is worthwhile to note many of these disorders have known (or suspected) causative genetic alterations which can be detected using modern-day targeted tests (71,72). As a result, patients with clinical features suspicious for these disorders can undergo confirmatory testing to better predict their reproductive potential. For example, men with hypogonadotropic hypogonadism and anosmia suggestive of Kallmann syndrome can be screened for mutations in one of the known causative genes (e.g., KAL1, FGFR1, or FGF8) using a specialized assay (73), but given its rarity, routinely testing infertile men for this condition would have limited diagnostic utility. Accordingly, these tests should be reserved only for infertile men with symptoms consistent with a known syndrome associated with infertility.
Novel techniques
Despite significant advances in our understanding of clinical genetics, a diagnosis remains elusive in up to 80% of men with infertility (10), indicating that continued research must be performed. Unfortunately, due to the complexity of spermatogenesis and variable phenotypes of infertile men, it has been difficult to elucidate many treatable genetic targets. However, newer technologies and techniques have shown some promise in improving diagnosis, and possible treatment, of infertility.
Epigenetics
Epigenetics is the study of changes in gene regulation and function caused by mechanisms other than alterations in DNA sequence. The most common epigenetic regulators include DNA methylation, histone modification, and presence of various RNA transcripts (74). Multiple studies have demonstrated an association between abnormal DNA methylation and infertility, but whether this association is causative has not yet been determined (75-77). Histone modifications appear to play an important role in germ cell development, and abnormalities in histone function may decrease number of spermatids and alter sperm motility and morphology (78-80). Differences in spermatic micro RNA (miRNA) profiles have been demonstrated in oligo- and azoospermic men compared to fertile controls (81,82). One of the more intriguing applications of sperm epigenetics is discovery of environmental factors which cause infertility (83) and recent studies have linked diet, smoking, and stress to altered spermatic epigenetic regulation (84-87). Although epigenetic testing has not yet been validated in a clinical setting, it may help identify personalized modifiable risk factors for infertile men in the future.
Genomic microarrays and NGS
With improvements in genetic testing, new population-based experimental approaches have emerged which have furthered our understanding of the genetic basis of disease. Successful sequencing of the human genome in the early 2000s enabled the creation of large databases of common sequence variants known as single nucleotide polymorphisms (SNPs). Over the past 15 years, these databases have been used to conduct large genome-wide association studies (GWAS), observational studies which aim to detect genomic differences between populations (88). Candidate genes can then be selected from this data and further studied. Methods for characterizing the genome can be broadly classified into microarray and NGS approaches.
Genomic microarrays are tools used to detect the expression of many genes simultaneously by analyzing binding patterns of complimentary DNA strands from cells of interest to probes on the array. Both custom and commercially-made assays are available which can genotype thousands to millions of genomic regions simultaneously with high accuracy (89). These assays have already determined potentially causative genes for some etiologies of infertility. Globozoospermia, for one, was found to be associated with mutations in a gene called DPY19L2 using a 250 k SNP array (90) and this discovery prompted research into the role of this protein in sperm function (91).
Comparative genomic hybridization (CGH) is another research technique which utilizes fluorescently labeled DNA to study copy number variations (CNVs), segments of DNA with varying number of repeats among individuals. DNA samples from a case and control are hybridized with different colored labels then subsequently applied to a microarray. By visualizing the color of each probe in the array, it is possible to determine the relative quantity of complimentary DNA between the two subjects. CGH-based CNV has shown utility not only for discovering new causes of genetic conditions but also diagnosis of known etiologies. In 2014, Yuen et al. developed a custom CGH microarray which could reliably identify YCMD at higher resolutions than PCR but at nearly double the cost (92).
Next-generation sequencing (NGS) technology allows for accurate and rapid sequencing of large portions of the human genome at a cost far lower than prior techniques. The first NGS sequencing platform in the mid-2000s yielded a 50,000-fold decrease in cost compared to technologies used previously and efficiency continues to rise (93). Three applications of NGS are currently employed: targeted sequencing (TS), whole exome sequencing (WES), and whole genome sequencing (WGS). TS utilizes disease-specific gene panels to simultaneously sequence genes of interest. As these panels can assess for chromosomal abnormalities, microdeletions, and single-gene mutations, they have been a subject of particular interest in infertility. Recently, a gene panel was developed for diagnosing infertile men and women which had a >90% sensitivity for detecting sex chromosome aneuploidies and YCMDs at nearly one-fifth the cost of traditional testing ($599 vs. ~$3,300) (94). WES and WGS have also seen some success in identifying candidate genes such as CATSPER2, MNS1, CFAP65, and FANCM using GWAS (95-98). Over the past four years, these techniques have helped characterize seven novel genes accounting for nearly half of cases of multiple morphological abnormalities of the sperm flagella (MMAF) in a 78-patient cohort (99).
Both microarrays and NGS have enormous clinical potential to find possible causative genetic variants without prior hypotheses of genomic location (100). Unfortunately, these findings have not yet translated into meaningful clinical applications. Many of these studies are limited by small population sizes, unknown clinical relevance of identified genes, and inconsistent results in validation studies. In a large systematic review and clinical validity assessment of 521 gene-disease relationships by Oud et al., only 92 had at least moderate evidence for a role in male infertility, which highlights the need for confirmatory studies when new genes are identified (101). Nonetheless, further advances in genomics and bioinformatics will improve our ability to interpret future genetic research.
Conclusions
Genetic testing has an important and necessary clinical role in the diagnosis and treatment of infertility. Some genetic tests, such as karyotyping, YCMD screening, and CFTR sequencing, clearly benefit a subpopulation of infertile men by providing clearer prognoses and treatment options for future fertility. Still, many men have no known cause of their fertility. Recent advances in our ability to sequence the genome harbor the potential to improve diagnostic capabilities and develop novel treatments aimed at newly discovered genetic targets associated with male infertility.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of this work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Keith Jarvi and Jared Bieniek) for the series “Genetic Causes and Management of Male Infertility” published in Translational Andrology and Urology. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-19-725). The series “Genetic Causes and Management of Male Infertility” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
References
- 1.Thoma ME, McLain AC, Louis JF, et al. Prevalence of infertility in the United States as estimated by the current duration approach and a traditional constructed approach. Fertil Steril 2013;99:1324-31.e1. 10.1016/j.fertnstert.2012.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Kretser DM. Male infertility. Lancet 1997;349:787-90. 10.1016/S0140-6736(96)08341-9 [DOI] [PubMed] [Google Scholar]
- 3.Tournaye H, Krausz C, Oates RD. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol 2017;5:544-53. 10.1016/S2213-8587(16)30040-7 [DOI] [PubMed] [Google Scholar]
- 4.Ferlin A, Raicu F, Gatta V, et al. Male infertility: role of genetic background. Reprod Biomed Online 2007;14:734-45. 10.1016/S1472-6483(10)60677-3 [DOI] [PubMed] [Google Scholar]
- 5.Jungwirth A, Diemer T, Dohle GR, et al. European Association of Urology guidelines on Male Infertility: the 2016 update. Available online: https://uroweb.org/guideline/male-infertility/ [DOI] [PubMed]
- 6.Practice Committee of the American Society for Reproductive Medicine . Diagnostic evaluation of the infertile male: a committee opinion. Fertil Steril 2015;103:e18-25. 10.1016/j.fertnstert.2014.12.103 [DOI] [PubMed] [Google Scholar]
- 7.American Urological Association Education and Research. The Optimal Evaluation of the Infertile Male: AUA Best Practice Statement. 2010.
- 8.Bhasin S, de Kretser DM, Baker HW. Clinical review 64: Pathophysiology and natural history of male infertility. J Clin Endocrinol Metab 1994;79:1525-9. [DOI] [PubMed] [Google Scholar]
- 9.Wosnitzer MS. Genetic evaluation of male infertility. Transl Androl Urol 2014;3:17-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hotaling J, Carrell DT. Clinical genetic testing for male factor infertility: current applications and future directions. Andrology 2014;2:339-50. 10.1111/j.2047-2927.2014.00200.x [DOI] [PubMed] [Google Scholar]
- 11.Bates SE. Classical Cytogenetics: Karyotyping Techniques. In: Schwartz PH, Wesselschmidt RL, editors. Human Pluripotent Stem Cells. Methods in Molecular Biology (Methods and Protocols), vol 767. Totowa, NJ: Humana Press, 2011:177-90. [DOI] [PubMed] [Google Scholar]
- 12.Van Assche E, Bonduelle M, Tournaye H, et al. Cytogenetics of infertile men. Hum Reprod 1996;11 Suppl 4:1-24; discussion 25-6. 10.1093/humrep/11.suppl_4.1 [DOI] [PubMed] [Google Scholar]
- 13.Krausz C, Riera-Escamilla A. Genetics of male infertility. Nature Reviews Urology 2018;15:369-84. 10.1038/s41585-018-0003-3 [DOI] [PubMed] [Google Scholar]
- 14.Groth KA, Skakkebæk A, Høst C, et al. Clinical review: Klinefelter syndrome--a clinical update. J Clin Endocrinol Metab 2013;98:20-30. 10.1210/jc.2012-2382 [DOI] [PubMed] [Google Scholar]
- 15.Vorona E, Zitzmann M, Gromoll J, et al. Clinical, endocrinological, and epigenetic features of the 46,XX male syndrome, compared with 47,XXY Klinefelter patients. J Clin Endocrinol Metab 2007;92:3458-65. 10.1210/jc.2007-0447 [DOI] [PubMed] [Google Scholar]
- 16.de la Chapelle A. The etiology of maleness in XX men. Hum Genet 1981;58:105-16. 10.1007/BF00284157 [DOI] [PubMed] [Google Scholar]
- 17.Chandley AC. Chromosome anomalies and Y chromosome microdeletions as causal factors in male infertility. Hum Reprod 1998;13 Suppl 1:45-50. 10.1093/humrep/13.suppl_1.45 [DOI] [PubMed] [Google Scholar]
- 18.Pandiyan N, Jequier AM. Mitotic chromosomal anomalies among 1210 infertile men. Hum Reprod 1996;11:2604-8. 10.1093/oxfordjournals.humrep.a019178 [DOI] [PubMed] [Google Scholar]
- 19.Ravel C, Berthaut I, Bresson JL, et al. Prevalence of chromosomal abnormalities in phenotypically normal and fertile adult males: large-scale survey of over 10,000 sperm donor karyotypes. Hum Reprod 2006;21:1484-9. 10.1093/humrep/del024 [DOI] [PubMed] [Google Scholar]
- 20.O’Flynn O’Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: A review. Fertility and Sterility 2010;93:1-12. 10.1016/j.fertnstert.2009.10.045 [DOI] [PubMed] [Google Scholar]
- 21.Ventimiglia E, Capogrosso P, Boeri L, et al. When to Perform Karyotype Analysis in Infertile Men? Validation of the European Association of Urology Guidelines with the Proposal of a New Predictive Model. Eur Urol 2016;70:920-3. 10.1016/j.eururo.2016.06.015 [DOI] [PubMed] [Google Scholar]
- 22.Geddes GC, Butterly M, Sajan I. FISH for 22q11.2 deletion not cost-effective for infants with congenital heart disease with microarray. Pediatr Cardiol 2015;36:531-6. 10.1007/s00246-014-1045-9 [DOI] [PubMed] [Google Scholar]
- 23.Foo YL, Chow JC, Lai MC, et al. Genetic Evaluation of Children with Global Developmental Delay--Current Status of Network Systems in Taiwan. Pediatr Neonatol 2015;56:213-9. 10.1016/j.pedneo.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 24.Turnpenny PD, Ellard S. Emery’s elements of medical genetics. Edition 15. Philadelphia, PA: Elsevier, 2017. [Google Scholar]
- 25.Hwang K, Weedin JW, Lamb DJ. The use of fluorescent in situ hybridization in male infertility. Ther Adv Urol 2010;2:157-69. 10.1177/1756287210373758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egozcue S, Blanco J, Vendrell JM, et al. Human male infertility: chromosome anomalies, meiotic disorders, abnormal spermatozoa and recurrent abortion. Hum Reprod Update 2000;6:93-105. 10.1093/humupd/6.1.93 [DOI] [PubMed] [Google Scholar]
- 27.Hwang K, Lipshultz LI, Lamb DJ. Use of diagnostic testing to detect infertility. Curr Urol Rep 2011;12:68-76. 10.1007/s11934-010-0154-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilford CA, Kuroda-Kawaguchi T, Skaletsky H, et al. A physical map of the human Y chromosome. Nature 2001;409:943-5. 10.1038/35057170 [DOI] [PubMed] [Google Scholar]
- 29.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003;423:825-37. 10.1038/nature01722 [DOI] [PubMed] [Google Scholar]
- 30.Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 2003;4:598-612. 10.1038/nrg1124 [DOI] [PubMed] [Google Scholar]
- 31.Lange J, Noordam MJ, van Daalen SKM, et al. Intrachromosomal homologous recombination between inverted amplicons on opposing Y-chromosome arms. Genomics 2013;102:257-64. 10.1016/j.ygeno.2013.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Repping S, Skaletsky H, Lange J, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet 2002;71:906-22. 10.1086/342928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamp C, Hirschmann P, Voss H, et al. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet 2000;9:2563-72. 10.1093/hmg/9.17.2563 [DOI] [PubMed] [Google Scholar]
- 34.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 2001;29:279-86. 10.1038/ng757 [DOI] [PubMed] [Google Scholar]
- 35.Colaco S, Modi D. Genetics of the human Y chromosome and its association with male infertility. Reprod Biol Endocrinol 2018;16:14. 10.1186/s12958-018-0330-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet 1976;34:119-24. 10.1007/BF00278879 [DOI] [PubMed] [Google Scholar]
- 37.Vogt PH. AZF deletions and Y chromosomal haplogroups: history and update based on sequence. Hum Reprod Update 2005;11:319-36. 10.1093/humupd/dmi017 [DOI] [PubMed] [Google Scholar]
- 38.Kamp C, Huellen K, Fernandes S, et al. High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndrome. Mol Hum Reprod 2001;7:987-94. 10.1093/molehr/7.10.987 [DOI] [PubMed] [Google Scholar]
- 39.Kleiman SE, Yogev L, Lehavi O, et al. The likelihood of finding mature sperm cells in men with AZFb or AZFb-c deletions: six new cases and a review of the literature (1994-2010). Fertil Steril 2011;95:2005-2012.e20124. 10.1016/j.fertnstert.2011.01.162 [DOI] [PubMed] [Google Scholar]
- 40.Oates RD, Silber S, Brown LG, et al. Clinical characterization of 42 oligospermic or azoospermic men with microdeletion of the AZFc region of the Y chromosome, and of 18 children conceived via ICSI. Hum Reprod 2002;17:2813-24. 10.1093/humrep/17.11.2813 [DOI] [PubMed] [Google Scholar]
- 41.Simoni M. Molecular diagnosis of Y chromosome microdeletions in Europe: state-of-the-art and quality control. Hum Reprod 2001;16:402-9. 10.1093/humrep/16.3.402 [DOI] [PubMed] [Google Scholar]
- 42.Krausz C, Hoefsloot L, Simoni M, et al. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology 2014;2:5-19. 10.1111/j.2047-2927.2013.00173.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simoni M, Tüttelmann F, Gromoll J, et al. Clinical consequences of microdeletions of the Y chromosome: the extended Münster experience. Reprod Biomed Online 2008;16:289-303. 10.1016/S1472-6483(10)60588-3 [DOI] [PubMed] [Google Scholar]
- 44.Johnson M, Raheem A, De Luca F, et al. An analysis of the frequency of Y-chromosome microdeletions and the determination of a threshold sperm concentration for genetic testing in infertile men. BJU Int 2019;123:367-72. 10.1111/bju.14521 [DOI] [PubMed] [Google Scholar]
- 45.Ferlin A, Arredi B, Speltra E, et al. Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab 2007;92:762-70. 10.1210/jc.2006-1981 [DOI] [PubMed] [Google Scholar]
- 46.Kohn TP, Kohn JR, Owen RC, et al. The Prevalence of Y-chromosome Microdeletions in Oligozoospermic Men: A Systematic Review and Meta-analysis of European and North American Studies. Eur Urol 2019;76:626-36. 10.1016/j.eururo.2019.07.033 [DOI] [PubMed] [Google Scholar]
- 47.Pan S, Bearelly P, Oates RD. Fertility in men with Klinefelter syndrome and Y chromosome microdeletions: an update. Curr Opin Endocr Metab Res 2019;6:21-8. 10.1016/j.coemr.2019.04.005 [DOI] [Google Scholar]
- 48.Donat R, McNeill AS, Fitzpatrick DR, et al. The incidence of cystic fibrosis gene mutations in patients with congenital bilateral absence of the vas deferens in Scotland. Br J Urol 1997;79:74-7. 10.1046/j.1464-410X.1997.30816.x [DOI] [PubMed] [Google Scholar]
- 49.Gudeman SR, Townsend B, Fischer K, et al. Etiology of azoospermia in a military population. J Urol 2015;193:1318-21. 10.1016/j.juro.2014.10.048 [DOI] [PubMed] [Google Scholar]
- 50.Daudin M, Bieth E, Bujan L, et al. Congenital bilateral absence of the vas deferens: clinical characteristics, biological parameters, cystic fibrosis transmembrane conductance regulator gene mutations, and implications for genetic counseling. Fertil Steril 2000;74:1164-74. 10.1016/S0015-0282(00)01625-3 [DOI] [PubMed] [Google Scholar]
- 51.Chillón M, Casals T, Mercier B, et al. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N Engl J Med 1995;332:1475-80. 10.1056/NEJM199506013322204 [DOI] [PubMed] [Google Scholar]
- 52.Yu J, Chen Z, Ni Y, et al. CFTR mutations in men with congenital bilateral absence of the vas deferens (CBAVD): a systemic review and meta-analysis. Hum Reprod 2012;27:25-35. 10.1093/humrep/der377 [DOI] [PubMed] [Google Scholar]
- 53.Riordan JR, Rommens JM, Kerem B, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 1989;245:1066-73. 10.1126/science.2475911 [DOI] [PubMed] [Google Scholar]
- 54.Zielenski J, Rozmahel R, Bozon D, et al. Genomic DNA sequence of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics 1991;10:214-28. 10.1016/0888-7543(91)90503-7 [DOI] [PubMed] [Google Scholar]
- 55.Sosnay PR, Siklosi KR, Van Goor F, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet 2013;45:1160-7. 10.1038/ng.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Estivill X, Bancells C, Ramos C. Geographic distribution and regional origin of 272 cystic fibrosis mutations in European populations. The Biomed CF Mutation Analysis Consortium. Hum Mutat 1997;10:135-54. [DOI] [PubMed] [Google Scholar]
- 57.Palomaki GE, FitzSimmons SC, Haddow JE. Clinical sensitivity of prenatal screening for cystic fibrosis via CFTR carrier testing in a United States panethnic population. Genet Med 2004;6:405-14. 10.1097/01.GIM.0000139505.06194.39 [DOI] [PubMed] [Google Scholar]
- 58.De Boeck K, Zolin A, Cuppens H, et al. The relative frequency of CFTR mutation classes in European patients with cystic fibrosis. J Cyst Fibros 2014;13:403-9. 10.1016/j.jcf.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 59.Rohlfs EM, Zhou Z, Heim RA, et al. Cystic fibrosis carrier testing in an ethnically diverse US population. Clin Chem 2011;57:841-8. 10.1373/clinchem.2010.159285 [DOI] [PubMed] [Google Scholar]
- 60.Schlegel PN, Shin D, Goldstein M. Urogenital anomalies in men with congenital absence of the vas deferens. J Urol 1996;155:1644-8. 10.1016/S0022-5347(01)66152-4 [DOI] [PubMed] [Google Scholar]
- 61.Radpour R, Gourabi H, Dizaj AV, et al. Genetic investigations of CFTR mutations in congenital absence of vas deferens, uterus, and vagina as a cause of infertility. J Androl 2008;29:506-13. 10.2164/jandrol.108.005074 [DOI] [PubMed] [Google Scholar]
- 62.Kolettis PN, Sandlow JI. Clinical and genetic features of patients with congenital unilateral absence of the vas deferens. Urology 2002;60:1073-6. 10.1016/S0090-4295(02)01973-8 [DOI] [PubMed] [Google Scholar]
- 63.Donohue RE, Fauver HE. Unilateral absence of the vas deferens. A useful clinical sign. JAMA 1989;261:1180-2. 10.1001/jama.1989.03420080100041 [DOI] [PubMed] [Google Scholar]
- 64.Weiske WH, Sälzler N, Schroeder-Printzen I, et al. Clinical findings in congenital absence of the vasa deferentia. Andrologia 2000;32:13-8. 10.1111/j.1439-0272.2000.tb02859.x [DOI] [PubMed] [Google Scholar]
- 65.McCallum T, Milunsky J, Munarriz R, et al. Unilateral renal agenesis associated with congenital bilateral absence of the vas deferens: phenotypic findings and genetic considerations. Hum Reprod 2001;16:282-8. 10.1093/humrep/16.2.282 [DOI] [PubMed] [Google Scholar]
- 66.Dequeker E, Stuhrmann M, Morris MA, et al. Best practice guidelines for molecular genetic diagnosis of cystic fibrosis and CFTR-related disorders--updated European recommendations. Eur J Hum Genet 2009;17:51-65. 10.1038/ejhg.2008.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams DR, Eng CM. Next-Generation Sequencing to Diagnose Suspected Genetic Disorders. N Engl J Med 2018;379:1353-62. 10.1056/NEJMra1711801 [DOI] [PubMed] [Google Scholar]
- 68.Straniero L, Soldà G, Costantino L, et al. Whole-gene CFTR sequencing combined with digital RT-PCR improves genetic diagnosis of cystic fibrosis. J Hum Genet 2016;61:977-84. 10.1038/jhg.2016.101 [DOI] [PubMed] [Google Scholar]
- 69.Esteves SC, Lee W, Benjamin DJ, et al. Reproductive potential of men with obstructive azoospermia undergoing percutaneous sperm retrieval and intracytoplasmic sperm injection according to the cause of obstruction. J Urol 2013;189:232-7. 10.1016/j.juro.2012.08.084 [DOI] [PubMed] [Google Scholar]
- 70.Kamal A, Fahmy I, Mansour R, et al. Does the outcome of ICSI in cases of obstructive azoospermia depend on the origin of the retrieved spermatozoa or the cause of obstruction? A comparative analysis. Fertil Steril 2010;94:2135-40. 10.1016/j.fertnstert.2010.01.041 [DOI] [PubMed] [Google Scholar]
- 71.Cariati F, D’Argenio V, Tomaiuolo R. The evolving role of genetic tests in reproductive medicine. J Transl Med 2019;17:267. 10.1186/s12967-019-2019-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nuti F, Krausz C. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online 2008;16:504-13. 10.1016/S1472-6483(10)60457-9 [DOI] [PubMed] [Google Scholar]
- 73.Dodé C, Hardelin JP. Kallmann syndrome. Eur J Hum Genet 2009;17:139-46. 10.1038/ejhg.2008.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.James E, Jenkins TG. Epigenetics, infertility, and cancer: future directions. Fertil Steril 2018;109:27-32. 10.1016/j.fertnstert.2017.11.006 [DOI] [PubMed] [Google Scholar]
- 75.Aston KI, Uren PJ, Jenkins TG, et al. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril 2015;104:1388-97.e975. 10.1016/j.fertnstert.2015.08.019 [DOI] [PubMed] [Google Scholar]
- 76.Urdinguio RG, Bayón GF, Dmitrijeva M, et al. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum Reprod 2015;30:1014-28. 10.1093/humrep/dev053 [DOI] [PubMed] [Google Scholar]
- 77.Hammoud SS, Nix DA, Hammoud AO, et al. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod 2011;26:2558-69. 10.1093/humrep/der192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gou LT, Kang JY, Dai P, et al. Ubiquitination-Deficient Mutations in Human Piwi Cause Male Infertility by Impairing Histone-to-Protamine Exchange during Spermiogenesis. Cell 2017;169:1090-104.e13. 10.1016/j.cell.2017.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schon SB, Luense LJ, Wang X, et al. Histone modification signatures in human sperm distinguish clinical abnormalities. J Assist Reprod Genet 2019;36:267-75. 10.1007/s10815-018-1354-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steilmann C, Cavalcanti MCO, Bartkuhn M, et al. The interaction of modified histones with the bromodomain testis-specific (BRDT) gene and its mRNA level in sperm of fertile donors and subfertile men. Reproduction 2010;140:435-43. 10.1530/REP-10-0139 [DOI] [PubMed] [Google Scholar]
- 81.Wu W, Hu Z, Qin Y, et al. Seminal plasma microRNAs: potential biomarkers for spermatogenesis status. Mol Hum Reprod 2012;18:489-97. 10.1093/molehr/gas022 [DOI] [PubMed] [Google Scholar]
- 82.Mokánszki A, Molnár Z, Varga Tóthné E, et al. Altered microRNAs expression levels of sperm and seminal plasma in patients with infertile ejaculates compared with normozoospermic males. Hum Fertil (Camb) 2020;23:246-55. 10.1080/14647273.2018.1562241 [DOI] [PubMed] [Google Scholar]
- 83.Cortessis VK, Thomas DC, Levine AJ, et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet 2012;131:1565-89. 10.1007/s00439-012-1189-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schagdarsurengin U, Steger K. Epigenetics in male reproduction: effect of paternal diet on sperm quality and offspring health. Nat Rev Urol 2016;13:584-95. 10.1038/nrurol.2016.157 [DOI] [PubMed] [Google Scholar]
- 85.Donkin I, Versteyhe S, Ingerslev LR, et al. Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metab 2016;23:369-78. 10.1016/j.cmet.2015.11.004 [DOI] [PubMed] [Google Scholar]
- 86.Dickson DA, Paulus JK, Mensah V, et al. Reduced levels of miRNAs 449 and 34 in sperm of mice and men exposed to early life stress. Transl Psychiatry 2018;8:101. 10.1038/s41398-018-0146-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jenkins TG, James ER, Alonso DF, et al. Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology 2017;5:1089-99. 10.1111/andr.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ikegawa S. A short history of the genome-wide association study: where we were and where we are going. Genomics Inform 2012;10:220-5. 10.5808/GI.2012.10.4.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ha NT, Freytag S, Bickeboeller H. Coverage and efficiency in current SNP chips. Eur J Hum Genet 2014;22:1124-30. 10.1038/ejhg.2013.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koscinski I, Elinati E, Fossard C, et al. DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet 2011;88:344-50. 10.1016/j.ajhg.2011.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abdelhedi F, Chalas C, Petit J-M, et al. Altered three-dimensional organization of sperm genome in DPY19L2-deficient globozoospermic patients. J Assist Reprod Genet 2019;36:69-77. 10.1007/s10815-018-1342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yuen RK, Merkoulovitch A, MacDonald JR, et al. Development of a high-resolution Y-chromosome microarray for improved male infertility diagnosis. Fertil Steril 2014;101:1079-85.e3. 10.1016/j.fertnstert.2013.12.027 [DOI] [PubMed] [Google Scholar]
- 93.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17:333-51. 10.1038/nrg.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patel B, Parets S, Akana M, et al. Comprehensive genetic testing for female and male infertility using next-generation sequencing. J Assist Reprod Genet 2018;35:1489-96. 10.1007/s10815-018-1204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luo T, Chen H-Y, Zou Q-X, et al. A novel copy number variation in CATSPER2 causes idiopathic male infertility with normal semen parameters. Hum Reprod 2019;34:414-23. 10.1093/humrep/dey377 [DOI] [PubMed] [Google Scholar]
- 96.Leslie JS, Rawlins LE, Chioza BA, et al. MNS1 variant associated with situs inversus and male infertility. Eur J Hum Genet 2020;28:50-5. 10.1038/s41431-019-0489-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang W, Tu C, Nie H, et al. Biallelic mutations in CFAP65 lead to severe asthenoteratospermia due to acrosome hypoplasia and flagellum malformations. J Med Genet 2019;56:750-7. 10.1136/jmedgenet-2019-106031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yin H, Ma H, Hussain S, et al. A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet Med 2019;21:62-70. 10.1038/s41436-018-0015-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nsota Mbango J-F, Coutton C, Arnoult C, et al. Genetic causes of male infertility: snapshot on morphological abnormalities of the sperm flagellum. Basic Clin Androl 2019;29:2. 10.1186/s12610-019-0083-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aston KI. Genetic susceptibility to male infertility: news from genome-wide association studies. Andrology 2014;2:315-21. 10.1111/j.2047-2927.2014.00188.x [DOI] [PubMed] [Google Scholar]
- 101.Oud MS, Volozonoka L, Smits RM, et al. A systematic review and standardized clinical validity assessment of male infertility genes. Hum Reprod 2019;34:932-41. 10.1093/humrep/dez022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as