Abstract
Background
Laryngeal cancer (LC) is a common malignant tumor of the head and neck. As circular RNAs (circRNAs) and other non-coding RNAs are involved in various malignant processes, we analyzed circRNAs to better understand LC and explored specific tumor markers.
Methods
High-throughput sequence was performed to analyze the differential circular RNAs in four coupled laryngeal cancers and para-cancerous tissues. The differential expression of selected circ-RANBP9 in laryngeal cancer tissues and cells was verified by RT-qPCR assay. CCK8, EDU, Transwell and wound healing assays were used to confirm the biological function of circ-RANBP9 in laryngeal cancer. Western blot assay was performed to identify the effects of circ-RANBP9 having on the epithelial to mesenchymal transition process. One-way AN0VA was used to analyze the correlation between the expression of circ-RANBP9 and clinicopathological parameters of the included patients. Kaplan-Meier analysis was used to investigate whether the expression level of circ-RANBP9 correlated with survival in LC patients. Bioinformatic analyses were also conducted to predict the functions and possible signaling pathways of the targeted mRNAs of circ-RANBP9 via co-expression and competing endogenous RNA network.
Results
We found a transcript from RNA sequence data, termed hsa_circ_0001578, which is a circRNA spliced from RANBP9. Circ-RANBP9 was downregulated in the LC cell lines tissues, relating to a better prognosis. Circ-RANBP9 was found to inhibit the proliferation, migration, and invasion ability of LC, exerting a suppressive role in the epithelial to mesenchymal transition process as well. For the diagnostic value of circ-RANBP9, the sensitivity and the specificity were 0.979 and 0.553, respectively. Circ-RANBP9 downregulation was significantly correlated with differentiation (P=0.031), T-stage (P=0.018), lymphatic metastasis (P=0.046), and clinical stage (P=0.003). Circ-RANBP9 was involved in insulin-like growth factor receptor binding, cell polarity, focal adhesion, and MAPK signaling pathways. CeRNA analysis identified the possible involvement of circ-RANBP9 in the ECM-receptor interaction, cAMP, calcium, and Wnt signaling pathways by harboring miRNA genes.
Conclusions
Circ-RANBP9 was confirmed to play important roles in inhibiting laryngeal cancers. Circ-RANBP9 was also validated to be associated with the clinicopathological parameters and diagnostic value, suggesting that circ-RANBP9 is a promising biomarker for LC prognosis and early diagnosis.
Keywords: Circular RNAs (circRNAs), laryngeal neoplasms, biomarkers
Introduction
Laryngeal cancer (LC) is the second most common malignancy of the head and neck, accounting for 30–50% of these tumors (1). Laryngeal squamous cell carcinoma accounts for 85–90% of all LC cases (2). The initiation and progression of LC is a complicated process driven by various pathogenetic factors and dysregulation of multiple genes (3). Smoking and heavy alcohol consumption have been deemed to be the most significant risk factors for LC, and HPV infection, air pollution and sex hormone levels have also been considered as the risk factors for LC (4). With respect to dysregulated genes, accumulating studies have shown that the dysregulation of multiple genes contribute to the malignant progression of various tumors, especially LC (5). For instance, hsa_circ_0057481 was upregulated in LC, and promoted LC proliferation and migration by modulating the miR-200c/ZEB1 axis (6). Currently, surgery and radiotherapy are the main treatment methods for LC, but the results of these treatments are frequently unsatisfactory. Due to the difficulty in early diagnosis, a large number of patients are diagnosed with an advanced stage or lymph node metastasis. Furthermore, the 5-year survival rate for patients in advanced stages is less than 50% (7). Therefore, it is particularly important to explore new biomarkers for the early diagnosis of LC.
Due to the rapid development of high-throughput sequencing, a multitude of non-coding RNAs have been proven to participate in the tumorigenesis and progression of malignant tumors (8). Circular RNAs (circRNAs), which are a new type of non-coding RNA with strong stability, high tissue specificity, and high expression abundance, have attracted more and more attention recently. The majority of previous studies have shown that circRNAs act as microRNA sponges, which regulate tumorigenesis though ceRNA network (9). Additionally, circRNAs were also found to play important roles in gene splicing, transcription and translation (10,11). For example, circITGA7 was found to promote the transcription of its parent gene integrin alpha 7 (ITGA7) through suppressing a transcription factor RAS responsive element-binding protein 1 (RREB1) (10). Moreover, circ-UBR5 was observed to be involved in RNA splicing regulatory process by interacting with splicing regulatory factor QKI, NOVA alternative splicing regulator 1 (NOVA1) and U1 small nuclear RNA (snRNA) in the NSCLC cells (11). Interestingly, accumulating studies have focus on the RBP mechanism of RNA interacting with protein and on how circular RNA regulate the RNA binding protein (12,13). For instance, circACC1 binds to β and γ subunits of AMP activated protein kinase (AMPK) to assemble a large complex, thus stabilizing and enhancing the enzymatic activity of AMPK holoenzyme (13). So far, there have been many studies on molecular mechanisms investigations of LC (14). And a few studies have been proven that circular RNAs play a very important role in the pathogenesis of LC (15,16). However, different from those studies on LC, our study proposed a novel circular RNA, analyzing its prognostic value and diagnostic value, and made a comprehensive analysis for its downstream signaling pathway. Therefore, in this study, we aimed to analyze circRNAs to better understand the pathogenesis and progression of LC, and explored specific tumor biomarkers that can help facilitate the early diagnosis of LC. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-567).
Methods
Patients and specimens
A total of 47 LC samples and matched paracancerous tissues were obtained from the Department of Otorhinolaryngology of the First Affiliated Hospital of China Medical University (Shenyang, China) during 2016–2017. The patients who were included in the study underwent total or partial laryngectomy and did not receive any treatment before diagnosis. The diagnosis of LC was based on postoperative pathology. Inclusion criteria: (I) clear pathological diagnosis of laryngeal cancer; (II) new cases of LC; (III) the patient did not receive any radiotherapy, chemotherapy or other anticancer treatments before surgery. Exclusion criteria: (I) the diagnosis was not clear, such as atypical hyperplasia, laryngeal keratosis and other non-cancerous lesions; (II) recurrent cases; (III) cases receiving treatment other than surgery. LC was classified according to the American Joint Committee on Cancer Classification criteria. The study was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University (number[2016]2016-30-2), and informed consent was obtained from all patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
The human LC cell lines LCC, and LLN were purchased from the Institute of Basic Medical Sciences (Beijing, China). The human LC cell lines Tu212, and the normal cell line Hacat were obtained from Laryngeal cancer Research Office (the First Affiliated Hospital of China Medical University, Shenyang, China). TU212 and Hacat were cultured with RPMI 1640 (Gibco, MA, USA) containing 10% fetal bovine serum (Bovogen, Australia). LCC and LLN were cultivated in RPMI 1640 (Gibco, MA, USA) containing 15% fetal bovine serum (Hyclone, MA, USA). All cells were incubated in an incubator at 37 °C, under 5% CO2. All cell lines were authenticated by STR DNA profiling and tested for Mycoplasma (GENEWIZ Co, Ltd, Suzhou, China) within 6 months.
Sequencing analysis
Five out of the 47 pairs of specimens were selected for whole-transcriptome sequencing using the Illumina HiSeq X Ten instrument. CIRI (17) and find_circ (18) were used to predict circRNAs. Differentially expressed transcripts and genes were identified by the criteria of |log2(fold-change)| ≥1 and a P value ≤0.05 between the paired samples.
Plasmids, siRNAs and transfection
A cDNA fragment that contained the sequence of circ-RANBP9 was cloned into the PLCDH vector (GenePharma) to overexpress circ-RANBP9. The siRNA of circ-RANBP9 was synthesized by GenePharma (Suzhou, China). The plasmids and circ-RANBP9 siRNA was transiently transfected into cells using the X-tremeGENE siRNA or DNA Transfection Reagent (Roche, Basel, Switzerland) according to the manufacturer’s instructions. The sequences of plasmids, siRNAs are shown in Table S1.
Real-time polymerase chain reaction
A Trizolkit (Thermo Fisher, Waltham, MA, USA) was used to extract total RNA. Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, Waltham, MA, USA). The expression of circ-RANBP9 was determined by quantitative real-time polymerase chain reaction (RT-PCR). The primer sequences of circ-RANBP9 were 5'-ATCCACAATTTAGCATCAGGCAAAG-3' (forward) and 3'-ACTTCTCCTGGTGTTTGAAGCC-5' (reverse). The primer sequences of the internal reference gene β-actin were 5'-GTGGCCGAGGACTTTGATTG-3' (forward) and 3'-CCTGTAACAACGCATCTCATATT-5' (reverse). The primers in our study were synthesized by Shanghai Shenggong Company (Shanghai, China). The PCR program was set at 95 °C for 5 min, then 95 °C (5 sec) and 60 °C (34 sec) in a total of 40 cycles. The 2−ΔΔCt method was used to analyze RNA expression levels (19).
Western blotting
Cells were lysed with radioimmunoprecipitation buffer (Beyotime, Beijing, China) on ice for 30 min. The protein concentrations were quantified using bicinchoninic acid (Beyotime, Beijing, China). Lysates were resolved by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). The membranes were blocked and incubated with primary antibodies against E-cadherin (ab212059, 1:1,000, Abcam, Cambridge, MA, USA), N-cadherin (ab76011, 1:1,000, Abcam, Cambridge, MA, USA), snail (ab216347, 1:1,000, Abcam, Cambridge, MA, USA) and β-catenin (ab223075, 1:1,000, Abcam, Cambridge, MA, USA) overnight at 4 °C and then incubated with HRP-conjugated secondary monoclonal antibody (ab205718, 1:5,000, Abcam, Cambridge, MA, USA) for 1 h at room temperature. The gels were detected using a chemiluminescence system (Bio-Rad) and the protein was quantified.
RNA fluorescence in situ hybridization (RNA-FISH)
Cells were fixed on slides with 4% paraformaldehyde for 20 minutes.
Then, the circ-RANBP9 probe was hybridized overnight at a concentration of 6 ng/µL. The hsa_circ_0001578 probe sequence for FISH was 5'-GCTTCCTTTGCCTGCTGAAAAATGTG-3'. Images were captured using a fluorescence microscope (Eclipse CI; Nikon Corporation).
Cell counting Kit-8 (CCK-8) assay
Twenty-four hours after transfection, the transfected cells were seeded into 96-well plates at a density of 2×103 cells per well. Then, cell proliferation ability was measured at 0, 24, 48, 72, 96 h using the CCK-8 kit (Dojindo, Tokyo, Japan), according to the manufacturer’s protocol. Absorbance was measured at 450 nm using a microplate reader for CCK-8 assay (Bio-Rad, Hercules, CA, USA).
EdU assay
Twenty-four hours after transfection, the transfected cells were seeded into 12-well plates at a density of 1×104 cells per well, cultrred with EDU for 2 h, and then stained with the Cell Light EdU Apollo488 In Vitro Imaging Kit (RiboBio). The percentage of EdU positive cells were defined as proliferation rate in EdU assay. The images were obtained using the Olympus microscope (Olympus, Tokyo, Japan). Independent proliferation assays were conducted three times.
Cell migration and invasion assay
Transwell chambers containing a polycarbonate membrane with 8.0 µm pores (Corning, NY, USA) were placed in a 24-well plate. For the migration assays, transfected cells were digested and diluted to a density of 5×104 cells per 200 µL and then added to upper chamber, medium with 20% FBS was added to the lower chamber. After incubation for 24–48 h, the cells were fixed with 4% formaldehyde, stained with 0.1% crystal violet. For invasion assays, the polycarbonate membrane was coated with Matrigel matrix (BD Bioscience). For quantification, cells were counted in five randomly selected fields using a microscope (IX51, Olympus, Japan). Independent transwell assays were performed three times. The data was shown as mean ± standard deviation (SD).
Wound healing assay
Cells were seeded in a 6-well plate and transfected when cell confluence reached 60–70%. When the cells reached confluency, the well was scratched with a 200 µL pipette tip, three lines at least acrossing each well. The cells were incubated with 1% FBS. The images were obtained at 0 and 48 h after healing using a microscope (IX51, Olympus, Japan).
Bioinformatics analysis
The miRNAs harbored by circ-RANBP9 and the corresponding mRNAs were predicted by TargetScan (http://www.targetscan.org/), miRWalk3.0 (http://129.206.7.150/), and mirDB (http://www.mirdb.org/). Pearson’s correlation coefficient was used to assess the correlation between circ-RANBP9 and differentially expressed genes. The ceRNA network was constructed using Cytoscape 3.6 (http://www.cytoscape.org). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted for the prediction of functions and pathways of differentially expressed genes. The possible target protein of circ-RANBP9 was predicted using the circular RNA interactome (https://circinteractome.nia.nih.gov/).
Statistical analysis
Statistical significance was determined using Student’s t-test, with SPSS 22.0 (IBM, Armonk, NY,USA). MeV4 (http://mev.tm4.org) and GraphPad Prism 6 (GraphPad, San Diego, USA) were used to generate figures. P<0.05 was considered to indicate statistical significance, and P<0.01 was considered to indicate high significance.
Results
Differentially expressed circRNAs
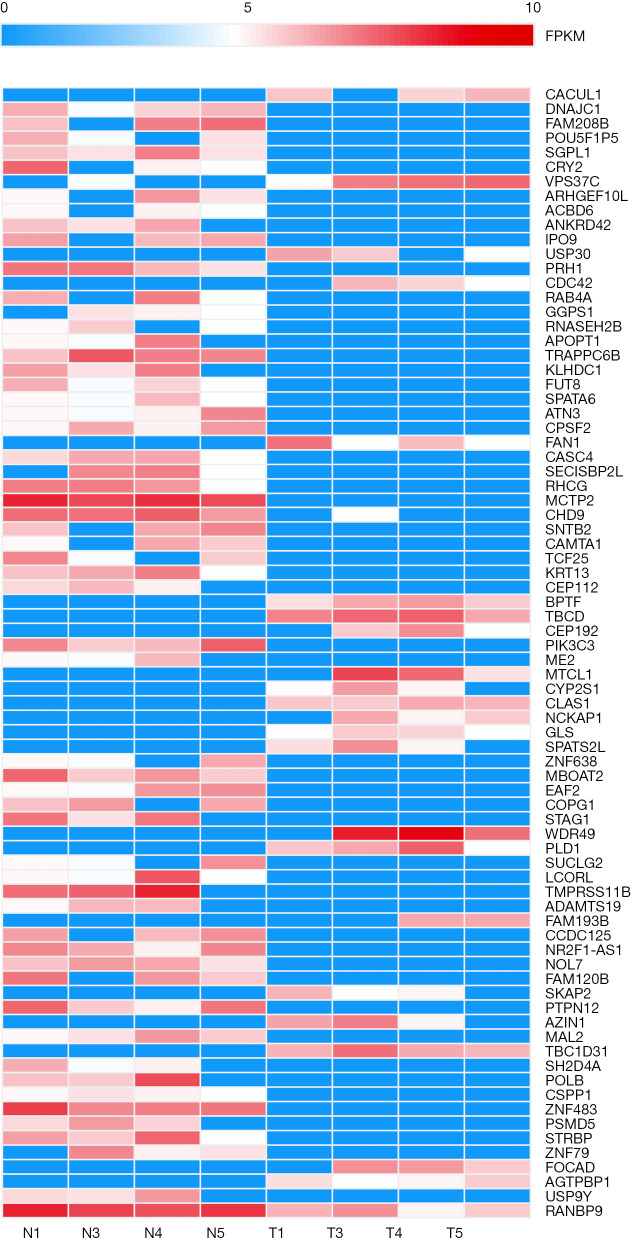
We conducted high-throughput transcriptome sequencing on 5 pairs of LC tissues. According to the criteria of fold-change ≥2.0 and P<0.05, 77 differentially expressed circRNAs were found in the LC samples. Compared with adjacent normal tissues, 22 circRNAs were upregulated and 55 circRNAs were downregulated in LC tissues. The significantly differentially expressed circRNAs are shown in the heatmap in Figure 1.
Figure 1.
Heatmap of significantly differentially expressed circRNAs. Blue color represents low expression and red color represents high expression.
Downregulation of circ-RANBP9
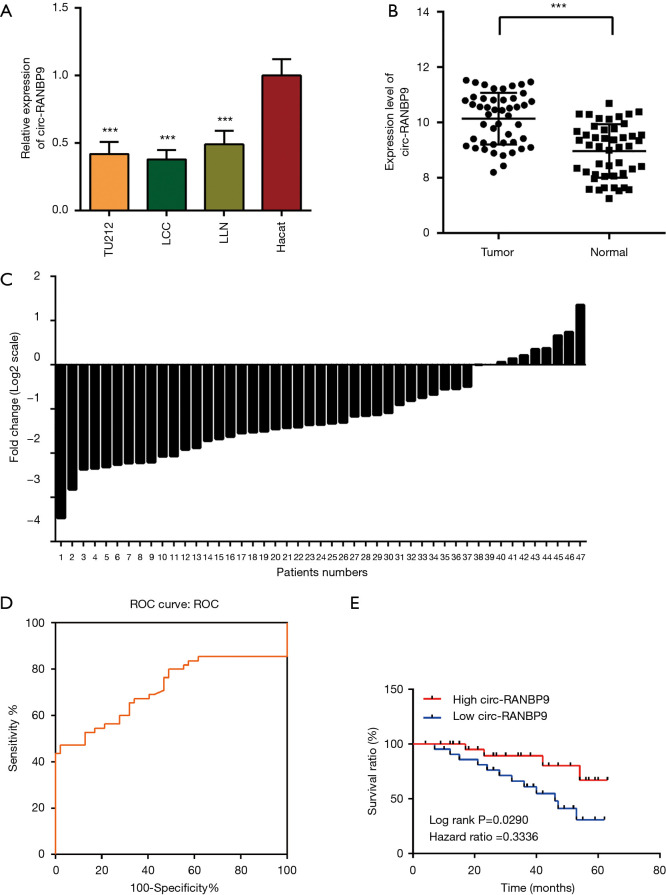
Our sequencing data demonstrated that expression of circ-RANBP9 was low in LC (Table 1). We used RT-qPCR to assess the differential expression of circ-RANBP9 in the cell lines TU212, LCC, LLN, and HaCat. Our results revealed that circ-RANBP9 expression was downregulated in all 3 LC cell lines by 3.2-fold compared with that of HaCat cells (Figure 2A). We also validated the differential expression of circ-RANBP9 in 47 pairs of cancer and non-cancer tissue samples, and observed low circ-RANBP9 expression in most LC tissues (Figure 2B,C). Furthermore, we constructed a receiver operating characteristic curve to examine the diagnostic value of circ-RANBP9. The cutoff value was 0.0007 (Figure 2D), the area under the curve was 0.716, and the sensitivity and the specificity were 0.979 and 0.553, respectively. Importantly, we found that higher expression of circ-RANBP9 was related to the higher survival rates (hazard ratio =0.3336, P=0.0290) (Figure 2E).
Table 1. The expression levels of circ-RANBP9 in sequencing data.
| Sample | Tumor (FPKM) | Control (FPKM) | log2 (fold_change) |
|---|---|---|---|
| 1 | 218.23267 | 641.45009 | −1.555469915 |
| 2 | 264.89598 | 537.12866 | −1.019841718 |
| 3 | 70.14053 | 407.19011 | −2.537382146 |
| 4 | 126.74575 | 546.58409 | −2.108506087 |
Figure 2.
The expression level of circ-RANBP9 in laryngeal cancer tissues and cell lines and its diagnostic and prognosis value.(A) Relative expression of circ-RANBP9 in laryngeal cancer cell lines (TU212, LCC, and LLN) and the normal control cell line HaCat. Data are shown as mean ± SD, and asterisks indicate significant P values (*** indicate P<0.001). (B) Circ-RANBP9 expression levels in 47 pairs of laryngeal cancer and paracancerous tissues (***, P<0.001). (C) Circ-RANBP9 was significantly downregulated in 80.9% (38/47) of laryngeal cancer tissues compared with normal control tissues. (D) The receiver operating characteristic (ROC) curve indicates the diagnostic characteristics of circ-RANBP9. (E) Kaplan-Meier curves indicated that the higher expression of circ-RANBP9 was correlated with the higher survival rates. P<0.05. Data indicate mean ± SD, n=3; ***, P<0.001.
The main characteristics of circ-RANBP9
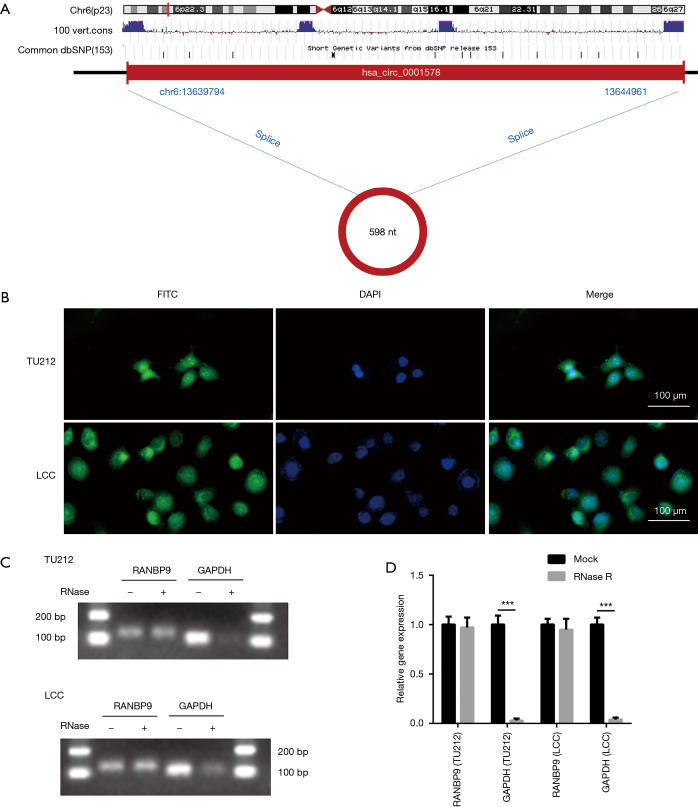
Next, we confirmed the characteristics of circ-RANBP9. According to circBase, circ-RANBP9 is spliced from the RANBP9 gene, and is rooted in chr6:13639794|13644961 with a length of 598 nt (Figure 3A). Localization of circ-RANBP9 was validated by FISH analysis, which indicated that it was localized both in the nucleus and cytoplasm of TU212 and LCC cells (Figure 3B). Circ-RANBP9 was resistant to RNase R digestion, demonstrating the cyclic structure by RT-qPCR (Figure 3C,D).
Figure 3.
The characteristics of circ-RANBP9. (A) The ideogram of genomic location and splicing mode of circ-RANBP9. (B) Fluorescence in situ hybridization (FISH) analysis for circ-RANBP9 was performed in TU212 and LCC cells. Scale bar: 100 µm. (C,D) The RNA enzyme digestion test was conducted to confirm the stability of circ-RANBP9. Data indicate mean ± SD, n=3; ***, P<0.001.
The anticancer role of circ-RANBP9
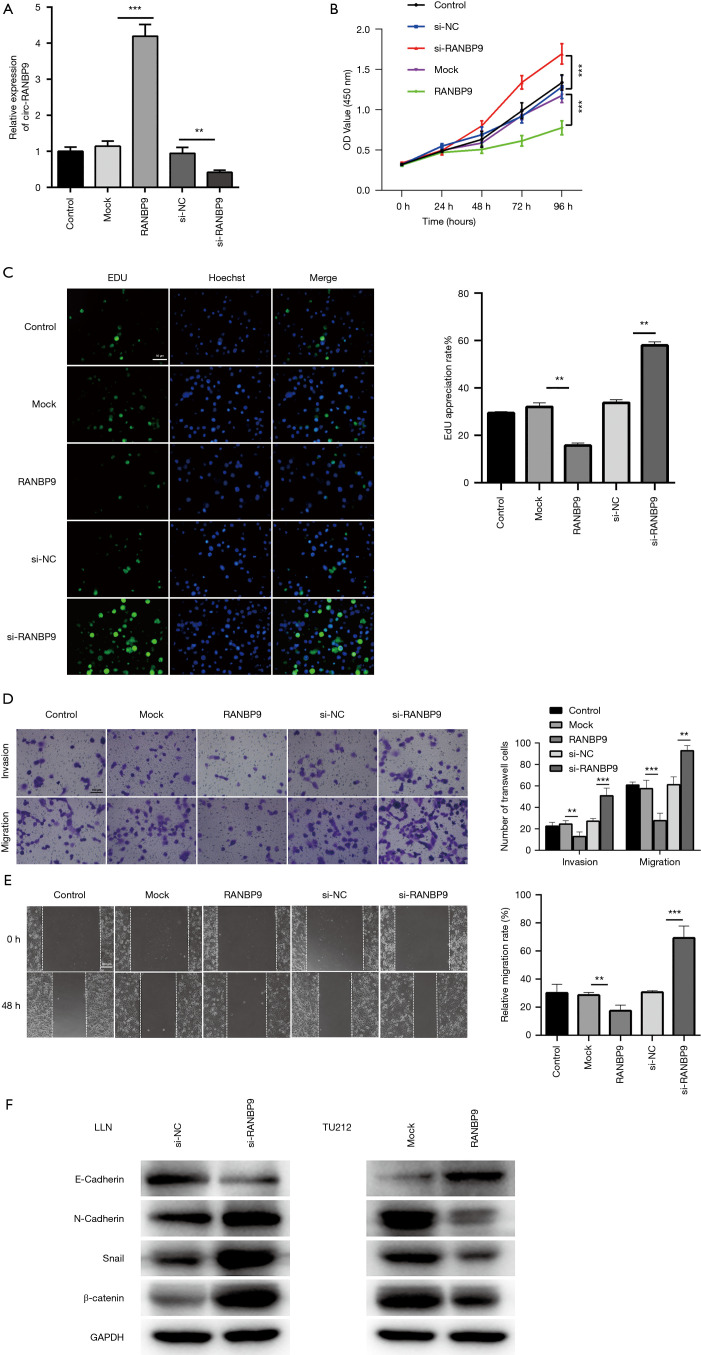
To validate the functional role of circ-RANBP9 in LC, we used silenced or overexpressed circ-RANBP9 by transfection with si-RNA or RANBP9, respectively. Transfection efficiency was determined using RT-qPCR (Figure 4A). Subsequently, the effects of circ-RANBP9 on LC cell proliferation, migration, and invasion were assessed. Overexpression of RANBP9 markedly inhibited cell proliferation, whereas knockdown of circ-RANBP9 significantly enhanced cell viability (Figure 4B). The EDU assay was performed to observe the effects of circ-RANBP9 on LC cells, and revealed that higher circ-RANBP9 inhibited cell proliferation, whereas cell viability was enhanced by knockdown of circ-RANBP9 (Figure 4C). Additionally, the transwell assay was conducted in LCC cells transfected with siRNA or RANBP9, and showed that the overexpression of circ-RANBP9 significantly inhibited the invasiveness and mobility of LC cells, whereas knockdown of circ-RANBP9 had the opposite effect (Figure 4D). Also, overexpression significantly increased the wound healing ability of cells, whereas downregulation of circ-RANBP9 significantly inhibited cell motility (Figure 4E). Our results confirm the role of circ-RANBP9 in inhibiting LC cell growth. Furthermore, the western bolt assay was used to observe the effects of circ-RANBP9 on the epithelial to mesenchymal transition process, and demonstrated that higher circ-RANBP9 inhibited the epithelial to mesenchymal transition process, whereas lower circ-RANBP9 promoted the epithelial to mesenchymal transition process (Figure 4F). The results indicated that circ-RANBP9 inhibited the metastasis of laryngeal cancer.
Figure 4.
The anticancer role of circ-RANBP9. (A) qRT-PCR was used to measure the expression level of circ-RANBP9 in LC cells transfected with si-circ-RANBP9 si-NC, mock, or circ-RANBP9. (B) The CCK-8 assay demonstrated increased or decreased viability of circ-RANBP9 knockdown or overexpressed LC cells, respectively. (C) Proliferation was validated using the EDU assay. Scale bar: 50 µm. (D) Cell motility was measured by cell invasion or migration assays using a transwell chamber with or without matrigel in cells transfected with the indicated vectors. Cells were dyed by 0.1% crystal violet. Scale bar: 100 µm. (E) Cell motility was measured in LC cells transfected with si-RANBP9 or RANBP9 using the wound healing assay. Scale bar: 200 µm. (F) The effects of circ-RANBP9 on the epithelial to mesenchymal transition process was measured in LC cells transfected with circ-RANBP9 overexpressed vector using the western blot assay. Data indicate mean ± SD, n=3; **, P<0.01; ***, P<0.001.
Correlation between circ-RANBP9 level and clinical parameters
We investigated the relationship between circ-RANBP9 expression and the clinicopathological parameters of the included patients (Table 2). Downregulation of circ-RANBP9 was significantly associated with differentiation (P=0.031), T-stage (P=0.018), lymphatic metastasis (P=0.046), and clinical stage (P=0.003), but not with age, gender, tumor size, or tumor location.
Table 2. The association between circ-RANBP9 expression levels and clinicopathological characteristics of 47 patients with laryngeal cancer (LC).
| Characteristics | No. of cases | Mean ± SD* | P value |
|---|---|---|---|
| Age (years) | 0.615 | ||
| ≥60 | 29 | 1.23±0.98 | |
| <60 | 18 | 1.07±1.12 | |
| Gender | 0.502 | ||
| Male | 42 | 1.20±1.03 | |
| Female | 5 | 0.87±1.07 | |
| Size (cm) | 0.424 | ||
| ≥3 | 21 | 1.03±1.12 | |
| <3 | 26 | 1.28±0.96 | |
| Location | 0.197 | ||
| Supraglottic | 15 | 1.02±0.99 | |
| Glottic | 28 | 1.12±0.99 | |
| Subglottic | 4 | 2.04±0.48 | |
| Differentiation | 0.031 | ||
| Well | 17 | 1.07±1.12 | |
| Moderate | 24 | 0.99±1.03 | |
| Poor | 6 | 2.18±0.39 | |
| T stage | 0.018 | ||
| T1 | 3 | 0.16±0.67 | |
| T2 | 9 | 0.46±1.05 | |
| T3 | 15 | 1.49±1.02 | |
| T4 | 20 | 1.40±0.87 | |
| Lymphatic metastasis | 0.046 | ||
| N0 | 33 | 1.36±0.98 | |
| N+ | 14 | 1.02±0.27 | |
| Clinical stage | 0.003 | ||
| I | 2 | −0.21±0.22 | |
| II | 6 | 0.62±1.12 | |
| III | 14 | 1.88±0.60 | |
| IV | 25 | 1.17±0.63 | |
Italic P values indicate statistical significance. One-way ANOVA was used to analyze the correlation between the expression level of circ-RANBP9 and clinicopathological parameters of the patients. *, P<0.05.
Bioinformatics analysis
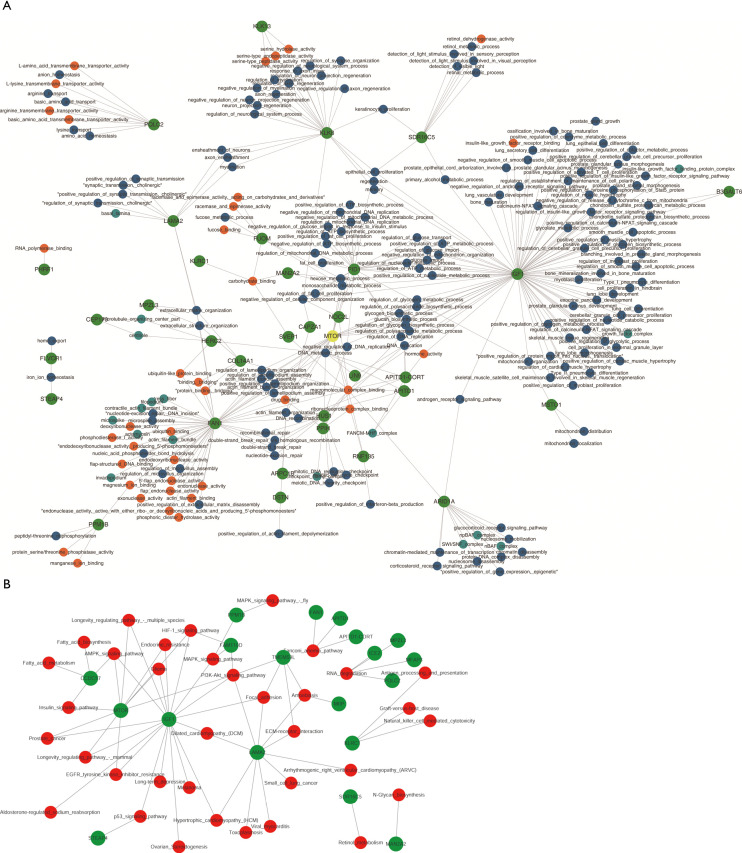
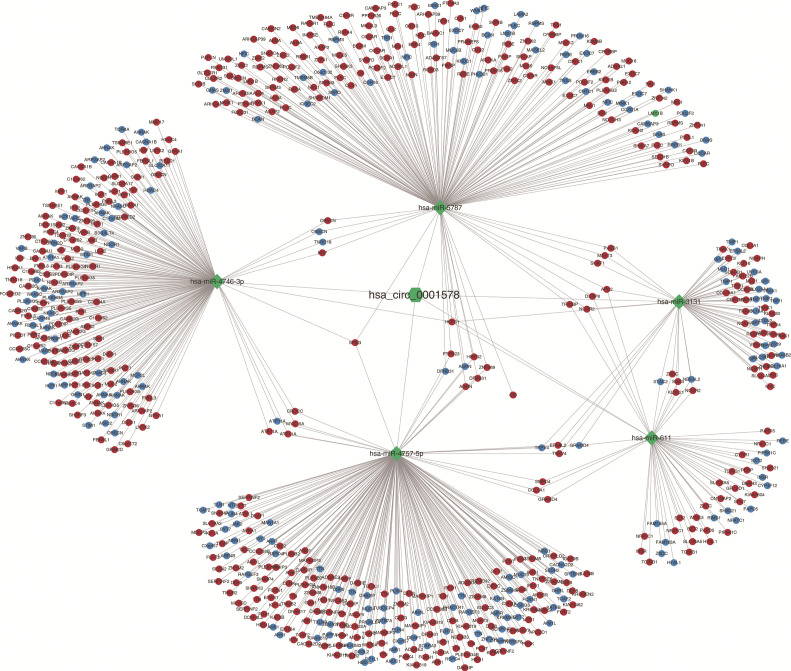
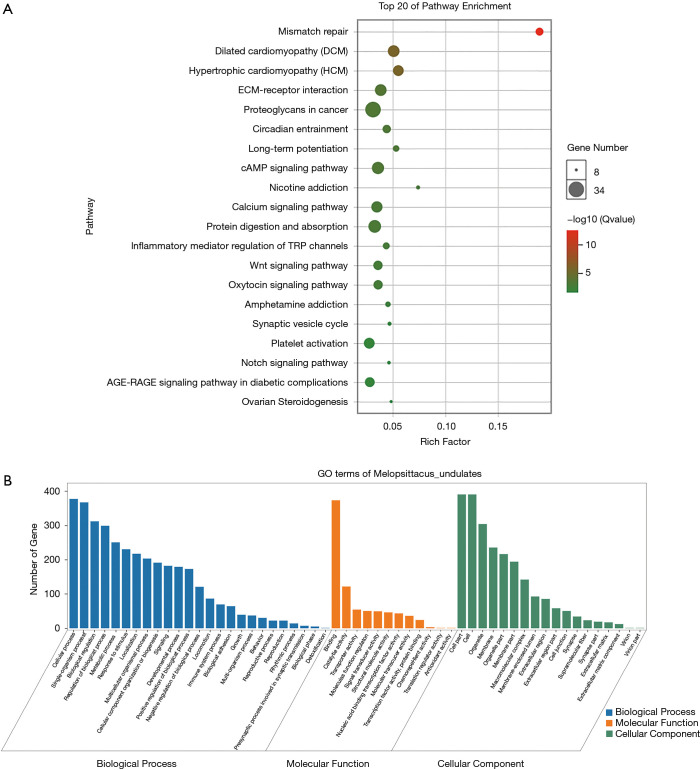
Pearson’s correlation coefficient was calculated to assess the correlation between circ-RANBP9 and differentially expressed mRNAs, and transcripts were identified according to the criteria of P>0.05 or P<−0.05. GO (Figure 5A) and KEGG (Figure 5B) analyses from the co-expressed mRNAs were performed to predict possible functions and pathways. Insulin-like growth factor receptor binding and regulation of establishment or maintenance of cell polarity were identified as significant GO items. Focal adhesion and MAPK pathways were considered to be the primary enrichment signaling pathways. The ceRNA network was also constructed to predict miRNAs and the corresponding targeted mRNAs related to circ-RANBP9. As shown in Figure 6, we found that circ-RANBP9 could target JAK3, FOXN1, MYC, and APC2 by harboring miR-4746-3p, miR-4757-5p, miR-3131, and miR-611. Subsequently, we conducted GO and KEGG analyses based on the mRNAs in the ceRNA network (Figure 7A,B) and found that circ-RANBP9 exerted anticancer activity through the extracellular matrix (ECM)-receptor interaction, cAMP, calcium, and Wnt signaling pathways. Four proteins were found to be the possible target RNA-binding proteins of circ-RANBP9:AGO2, EIF4A3, HuR, and U2AF65 (Table 3).
Figure 5.
GO and KEGG analysis of co-expressed mRNAs of circ-RANBP9. (A) Gene Ontology (GO) analysis for co-expressed mRNAs of circ-RANBP9 with a standard of correlation >0.95 or <−0.95; (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis for co-expressed mRNAs of circ-RANBP9, kappa scores of enriched terms are shown using bubble scale.
Figure 6.
The ceRNA analysis for circ-RANBP9. Red nodes represent upregulated mRNAs and blue nodes represent downregulated mRNAs.
Figure 7.
GO and KEGG analysis of mRNAs targeted by circ-RANBP9 in the ceRNA network. (A) Gene Ontology (GO) analysis for mRNAs targeted by circ-RANBP9 in the ceRNA network; (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis for mRNAs targeted by circ-RANBP9 in the ceRNA network. The enrichment factor represents the ratio between the differentially expressed genes and all annotated genes enriched in the pathway. Bubble scale represents the number of different genes; depth of bubble color represents the P value.
Table 3. The possible RNA-binding proteins predicted by the circular RNA interactome database.
| RNA-binding protein | RNA-binding protein sites matching to circRNAs | ||
|---|---|---|---|
| Tag name | % Identity | Alignment length | |
| AGO2 | HHFKP_121760_cluster-19652_14_28_41 | 100.00 | 41 |
| EIF4A3 | (I) HHLE1_296880_eIF4AIII_rep1_296880_25_188 | 100.00 | 187 |
| (II) HHLE2_1012767_eIF4AIII_rep2_1012767_1_29 | 100.00 | 229 | |
| HuR | None | None | None |
| U2AF65 | HIUUS_877725_U2AF65_ctrl_plus_HNRNPC_kd_877725_26 | 100.00 | 26 |
Discussion
A multitude of non-coding RNAs have been found, which can be attributed to the rapid development of high-throughput sequencing (20). Although non-coding RNAs were once considered as meaningless transcriptional noise, relevant studies have confirmed their involvement in disease progression (8,21). CircRNAs, a new type of non-coding RNA, have closed-loop strings formed by 3'-5' terminal reverse splicing (18). CircRNAs can act as sponges for miRNAs and participate in the mechanisms of ceRNAs (22). They also encode proteins at internal ribosome entry sites and regulate transcription factors by binding to proteins. CircRNA dysregulation has been observed in tumors and is involved in tumor pathogenesis. For example, circAGFG1 expression is upregulated in breast cancer tissues and is related to both clinical stage and poor prognosis (23). However, the mechanism of action of most circRNAs is not well known. In our sequencing results, we found that circ-RANBP9 expression was low in cancer tissue samples compared with that in normal tissue samples. Furthermore, qRT-PCR was used to assess the differential expression of circ-RANBP9 in LC cell lines and tissue samples. Circ-RANBP9 expression was significantly lower in the TU212, LCC, and LLN cell lines and in most of the LC tissues. Therefore, circ-RANBP9 is likely to play an anticancer role in the pathogenesis and development of LC.
Despite the fact that non-coding RNAs have no protein-coding functions, their functions can, nevertheless, be predicted through ceRNA mechanisms or RNA-binding proteins. According to the parent gene theory (24), the co-expression network was used to predict circRNA function by means of co-expressed mRNAs. KEGG analysis of co-expressed mRNAs indicated that the predominantly enriched signaling pathways were the MAPK and focal adhesion pathways. The MAPK signaling pathway is linked to cell growth, differentiation, apoptosis, and cancer progression (25). Four cascades exist in the MAPK signaling pathway: ERK1/2, p38 MAPK, c-Jun N-terminal kinase (JNK), and ERK5 (26). It has been shown that p38is involved in the invasion and metastasis of breast cancer (27), while ERK5 is involved in both cell proliferation in prostate cancer (28) and cell proliferation, migration, and invasion in bladder cancer (29). The focal adhesion signaling pathway has also been found to be related to the tumorigenesis and progression of various cancers (30). Our study showed that circ-RANBP9 inhibited the proliferation, invasion, and migration of LC cells. Combined with the analysis above, we speculated that circ-RANBP9 might have anticancer effects on LC through the MAPK signaling pathway or the focal adhesion signaling pathway.
CeRNA analysis demonstrated that circ-RANBP9 could harbor miR-4746-3p, miR-4757-5p, miR-3131, and miR-611.The KEGG analysis of the ceRNA network then revealed enrichment in the ECM-receptor interaction, cAMP, calcium, and Wnt signaling pathways. The pathways mentioned above have been shown to be dysregulated in multiple malignant tumors. For example, the cAMP/PKA pathway is abnormally activated and involved in both angiogenesis and vasculogenic mimicry in colorectal cancer (31). The Wnt/β-catenin signaling pathway is associated with a variety of human tumors. β-catenin is a primary component of the Wnt/β-catenin pathway and regulates downstream target genes through their translocation from the cytoplasm to the nucleus (32). Approximately 40–70% of hepatocellular carcinomas show β-catenin nuclear accumulation and abnormal Wnt/β-catenin activation (33,34). Our results showed that circ-RANBP9 expression was lower in LC and that it exerted anticancer activity. These results suggest that circ-RANBP9 may participate in the signaling pathways mentioned above. The ECM-receptor interaction pathway is a common signaling pathway that was found in both the KEGG analysis and the ceRNA network. The shared mRNA was LAMA2, which also showed lower expression in the sequencing data. Therefore, we suspect that circ-RANBP9 may regulate the pathogenesis of LC by harboring hsa-miR-5787 to suppress the expression of LAMA2.
In addition, the circular RNA interactome was used to predict the possible RNA-binding proteins of circ-RANBP9. Among these proteins, HuR has been shown to bind to the long non-coding RNA (lncRNA) OIP5-AS1 and is associated with the proliferation of cervical carcinoma cells (35). U2AF65 was also found to be an RNA-binding protein of lncRNA NEAT1and is related to the proliferation and invasion of hepatocellular carcinoma cells (36). Thus, we speculate that circ-RANBP9 regulates LC through the RNA-binding proteins that were predicted using the circular RNA interactome.
Recently, a few studies have investigated the molecular mechanisms affecting laryngeal cancer. For example, circ_0004507 was found to contribute to laryngeal cancer progression and cisplatin resistance by sponging miR-873 to upregulate multidrug resistance 1 and multidrug resistance protein 1 (37). Similarly, knockdown of hsa_circ_0023028 inhibits cell proliferation, migration, and invasion in laryngeal cancer by sponging miR-194-5p (38). However, the precise pathogenesis underlying LC remains largely unknown. Its frequent recurrence and high mortality may be attributed to resistance to both chemotherapy and radiotherapy. Early diagnosis can lead to an improved quality of life and better outcomes for patients with LC (39). The diagnosis of LC depends largely on its signs and symptoms, along with auxiliary examinations. The symptoms are frequently nonspecific, which may result in misdiagnosis. Despite its traumatic nature, conventional biopsy of the tumor is the best method for the accurate diagnosis of LC. Clinical examinations such as computed tomography, magnetic resonance imaging, or positron emission tomography are not sufficient for an accurate diagnosis (40,41). Thus far, no ideal biomarker has been found to replace these clinical examinations for LC and improve diagnostic accuracy. The results of our study suggest that circ-RANBP9 could have 97.9% accuracy and 55.3% specificity for the diagnosis of LC, with an area under the curve of 0.716. These results indicate that circ-RANBP9 could be useful for the diagnosis of LC. Our results showed that higher expression of circ-RANBP9 was correlated with a better outcome. Specifically, considering that circ-RANBP9 inhibited the invasion and migration capacity through functional assays, combing with the fact that circ-RANBP9 also inhibited epithelial to mesenchymal transition process, we believe that circ-RANBP9 can inhibit the metastasis of laryngeal cancer. Furthermore, through analysis of clinicopathological characteristics, we uncovered that the lower expression of circ-RANBP9 was correlated with differentiation level, T stage, lymphatic metastasis and clinical stage, indicating that circ-RANBP9 may be considered a valuable biomarker for the prediction of clinical outcomes. Furthermore, we can investigate novel medicine targeting the molecular axis of circ-RANBP9, hsa-miR-5787, and LAMA2 for precision treatment. Then the expression level of circ-RANBP9 should be considered as prognostic index for predicting outcome. In addition, as accumulating studies showing that dysregulated genes were related to risk stratification and local regional metastasis in tumors (42), combining with our results that circ-RANBP9 was related to the clinicopathological characteristics and involved in the metastasis of LC, we should speculate that circ-RANBP9 may be used as a potential biomarker for risk stratification and local regional metastasis in patients with laryngeal cancer. Certainly, it needs further identification. Our findings may therefore improve the early diagnosis, early treatment, and overall prognosis for LC.
Conclusions
Our results are the first to indicate that circ-RANBP9 plays a crucial role in the pathogenesis and progression of LC and has the potential to be used as a valuable biomarker for early diagnosis and prognosis. Furthermore, circ-RANBP9 may be a useful target for the development of future therapies.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the Leading Talent of the “Hundred-Thousand-Ten Thousand Project” in Liaoning under grant No. XLYC1905004, and Shenyang High-level Innovation Talents Plan under grant No. RC190403.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All study participants provided informed consent, and the study was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University (number[2016]2016-30-2). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-567
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-567
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-567). The authors have no conflicts of interest to declare.
(English Language Editor: C. Betlazar-Maseh)
References
- 1.Iizuka Y, Yoshimura M, Inokuchi H, et al. Recurrence patterns after postoperative radiotherapy for squamous cell carcinoma of the pharynx and larynx. Acta Otolaryngol 2015;135:96-102. 10.3109/00016489.2014.949848 [DOI] [PubMed] [Google Scholar]
- 2.Li P, Liu H, Wang Z, et al. MicroRNAs in laryngeal cancer: implications for diagnosis, prognosis and therapy. Am J Transl Res 2016;8:1935-44. [PMC free article] [PubMed] [Google Scholar]
- 3.Nadal A, Cardesa A. Molecular biology of laryngeal squamous cell carcinoma. Virchows Arch 2003;442:1-7. 10.1007/s00428-002-0726-6 [DOI] [PubMed] [Google Scholar]
- 4.Yang D, Shi Y, Tang Y, et al. Effect of HPV Infection on the Occurrence and Development of Laryngeal Cancer: A Review. J Cancer 2019;10:4455-62. 10.7150/jca.34016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y, Hou J, Wang X, et al. Circ_0000218 plays a carcinogenic role in laryngeal cancer through regulating microRNA-139-3p/Smad3 axis. Pathol Res Pract 2020;216:153103. 10.1016/j.prp.2020.153103 [DOI] [PubMed] [Google Scholar]
- 6.Fu D, Huang Y, Gao M. Hsa_circ_0057481 promotes laryngeal cancer proliferation and migration by modulating the miR-200c/ZEB1 axis. Int J Clin Exp Pathol 2019;12:4066-76. [PMC free article] [PubMed] [Google Scholar]
- 7.He FY, Liu HJ, Guo Q, et al. Reduced miR-300 expression predicts poor prognosis in patients with laryngeal squamous cell carcinoma. Eur Rev Med Pharmacol Sci 2017;21:760-4. [PubMed] [Google Scholar]
- 8.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16:94. 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Z, Rong Z, Zhang J, et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol Cancer 2020;19:86. 10.1186/s12943-020-01203-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol 2018;246:166-79. 10.1002/path.5125 [DOI] [PubMed] [Google Scholar]
- 11.Qin M, Wei G, Sun X. Circ-UBR5: An exonic circular RNA and novel small nuclear RNA involved in RNA splicing. Biochem Biophys Res Commun 2018;503:1027-34. 10.1016/j.bbrc.2018.06.112 [DOI] [PubMed] [Google Scholar]
- 12.Lou J, Hao Y, Lin K, et al. Circular RNACDR1asdisrupts the p53/MDM2 complex to inhibit Gliomagenesis. Molecular Cancer 2020;19:138. 10.1186/s12943-020-01253-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Wang Y, Wu S, et al. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab 2019;30:157-173.e7. 10.1016/j.cmet.2019.05.009 [DOI] [PubMed] [Google Scholar]
- 14.Yue PJ, Sun YY, Li YH, et al. MYCT1 inhibits the EMT and migration of laryngeal cancer cells via the SP1/miR-629-3p/ESRP2 pathway. Cell Signal 2020;74:109709. 10.1016/j.cellsig.2020.109709 [DOI] [PubMed] [Google Scholar]
- 15.Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 16.Xuan L, Qu L, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res 2016;8:932-9. [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015;16:4. 10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 20.Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids 2019;16:118-29. 10.1016/j.omtn.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arun G, Spector DL. MALAT1 long non-coding RNA and breast cancer. RNA Biol 2019;16:860-3. 10.1080/15476286.2019.1592072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Yang M, Zhou B, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond) 2019;133:1487-503. 10.1042/CS20190394 [DOI] [PubMed] [Google Scholar]
- 23.Yang R, Xing L, Zheng X, et al. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer 2019;18:4. 10.1186/s12943-018-0933-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell 2019;176:869-881.e13. 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rezatabar S, Karimian A, Rameshknia V, et al. RAS/MAPK signaling functions in oxidative stress, DNA damage response and cancer progression. J Cell Physiol 2019. [Epub ahead of print]. doi: . 10.1002/jcp.28334 [DOI] [PubMed] [Google Scholar]
- 26.Guo YJ, Pan WW, Liu SB, et al. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med 2020;19:1997-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen S, Hou Y, Fu L, et al. Cancer-associated fibroblast (CAF)-derived IL32 promotes breast cancer cell invasion and metastasis via integrin β3-p38 MAPK signalling. Cancer Lett 2019;442:320-32. 10.1016/j.canlet.2018.10.015 [DOI] [PubMed] [Google Scholar]
- 28.Zhu M, Huang C, Ma X, et al. Phthalates promote prostate cancer cell proliferation through activation of ERK5 and p38. Environ Toxicol Pharmacol 2018;63:29-33. 10.1016/j.etap.2018.08.007 [DOI] [PubMed] [Google Scholar]
- 29.Yao J, Zhang Z, Li S, et al. Melittin inhibits proliferation, migration and invasion of bladder cancer cells by regulating key genes based on bioinformatics and experimental assays. J Cell Mol Med 2020;24:655-70. 10.1111/jcmm.14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee BY, Timpson P, Horvath LG, et al. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther 2015;146:132-49. 10.1016/j.pharmthera.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 31.Wang S, Zhang Z, Qian W, et al. Angiogenesis and vasculogenic mimicry are inhibited by 8-Br-cAMP through activation of the cAMP/PKA pathway in colorectal cancer. Onco Targets Ther 2018;11:3765-74. 10.2147/OTT.S164982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Smits R. Wnt/β-Catenin signaling in liver cancers. Cancers (Basel) 2019;11:926. 10.3390/cancers11070926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lachenmayer A, Alsinet C, Savic R, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res 2012;18:4997-5007. 10.1158/1078-0432.CCR-11-2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong CM, Fan ST, Ng IO. beta-Catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer 2001;92:136-45. [DOI] [PubMed] [Google Scholar]
- 35.Kim J, Abdelmohsen K, Yang X, et al. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res 2016;44:2378-92. 10.1093/nar/gkw017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mang Y, Li L, Ran J, et al. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Onco Targets Ther 2017;10:1003-16. 10.2147/OTT.S116319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi X, Chen W, Li C, et al. Circular RNA circ_0004507 contributes to laryngeal cancer progression and cisplatin resistance by sponging miR-873 to upregulate multidrug resistance 1 and multidrug resistance protein 1. Head Neck 2021;43:928-41. 10.1002/hed.26549 [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Su X. Knockdown of hsa_circ_0023028 inhibits cell proliferation, migration, and invasion in laryngeal cancer by sponging miR-194-5p. Biosci Rep 2019;39:BSR20190177. 10.1042/BSR20190177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson SM, Yeo JC, Sabey L, et al. Effects of tumor staging and treatment modality on functional outcome and quality of life after treatment for laryngeal cancer. Head Neck 2013;35:1759-63. 10.1002/hed.23230 [DOI] [PubMed] [Google Scholar]
- 40.Becker M, Zaidi H. Imaging in head and neck squamous cell carcinoma: the potential role of PET/MRI. Br J Radiol 2014;87:20130677. 10.1259/bjr.20130677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JT, Branstetter BF, 4th. PET/CT in head and neck oncology: State-of-the-art 2013. Laryngoscope 2014;124:913-5. 10.1002/lary.23942 [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Jiang W, Zhang T, et al. Increased CYFRA 21-1, CEA and NSE are Prognostic of Poor Outcome for Locally Advanced Squamous Cell Carcinoma in Lung: A Nomogram and Recursive Partitioning Risk Stratification Analysis. Transl Oncol 2018;11:999-1006. 10.1016/j.tranon.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as