Abstract
Background
The prognostic value of polybromo 1 (PBRM1) gene mutations in clear cell renal carcinoma (CCRCC) with anti-programmed death-ligand 1 (PD-L1) therapy remains controversial, and few studies have reported the impact of PBRM1 mutations in other cancer types.
Methods
The patient information was obtained from cBioPortal and the Tumor Immune Estimation Resource (TIMER) databases. Mann-Whitney U test were used for correlation analysis. For survival analyses, Kaplan-Meier survival curves were used and compared using the log-rank test. Cox’s regression model was used to perform univariable and multivariable analyses
Results
Our study, for the first time, performed comprehensive analyses of PBRM1 mutation frequency, PBRM1 expression, relationship of PBRM1 mutations with clinical benefit from immunotherapy, and PBRM1 expression with immune infiltrates in diverse cancer types. The results showed that the expression of PBRM1 was significantly lower in diverse cancer types compared with normal tissues. Based on multivariable analysis, PBRM1 mutations trended towards worse clinical outcomes from anti-PD-L1 in CCRCC, lung adenocarcinoma (LUAD), bladder urothelial carcinoma (BLCA), and skin cutaneous melanoma (SKCM), and a significant association was observed in LUAD and BLCA. PBRM1 mutations were associated with higher TMB in diverse cancer types and significant associations were observed in LUAD and BLCA. The expression of PBRM1 was found to positively correlate with immune infiltrates in diverse cancer types.
Conclusions
Our findings suggested caution in starting immunotherapy alone in PBRM1 mutant patients. Further studies are needed to improve treatment for PBRM1 mutant patients.
Keywords: PBRM1 mutations, PBRM1 expression, immunotherapy, immune infiltrates, multiple cancer types
Introduction
Immune checkpoint inhibitor (ICI) drugs have revolutionized the treatment landscapes in multiple cancer types (1,2). The use of ICIs against cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed death-1 (PD-1), and programmed death-ligand 1 (PD-L1) has been approved for treating a variety of malignancies (3-5). Patients with biomarkers such as PD-L1, tumor mutational burden (TMB), and high microsatellite instability (MSI-H), may have a survival advantage with the use of ICIs (6-9). Nevertheless, these biomarkers not enough for clinicians to precisely distinguish responders to immunotherapy. Patient intrinsic factors, tumor intrinsic factors, and environmental factors may impact the efficacy of ICIs (10,11). There is an urgent need to identify specific predictive molecular biomarkers for immunotherapy to facilitate precision of treatment.
The PBRM1 gene encodes the bromodomain-containing protein BAF180, which is a subtype of the switch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex and the second most commonly mutated gene in clear cell renal carcinomas (CCRCC) after VHL (Vov Hippel-Lindau) (12-15). Approximately 80% of PBRM1 somatic mutations may result in loss of function of the protein (16). Mutations in PBRM1 have also been found in other cancer types including pancreatic, gastric, renal, and biliary cancers (17,18). Decreased expression of PBRM1 has been reported to correlate with poor prognosis and advanced clinicopathological features in CCRCC (19-21).
Different studies have tried to analyze the impact of PBRM1 status on response to immunotherapy in CCRCC but the results have seemed controversial. Miao et al. reported that in patients with metastatic CCRCC who received prior treatment (largely with inhibitors of vascular endothelial growth factor (VEGF), PBRM1 mutations were associated with increased progression free survival (PFS) with anti-PD-L1 therapy, but the association was not observed in patients who underwent first-line anti-PD-(L)-1 therapy (22). A further study validated the relationship between PBRM1 truncating mutations and improved response to nivolumab (anti-PD-1) in participants who received prior antiangiogenic therapy (23). In treatment-naive metastatic RCC, PBRM1 mutant patients had a trend towards better PFS in the sunitinib (anti-VEGF) arm vs. both atezolizumab (anti-PD-L1) and atezolizumab + bevacizumab (anti-VEGF) treatment arms (both HR <1) (24). In brief, no evidence has demonstrated PBRM1 mutant patients have better clinical outcomes with first-line immunotherapy. Some other studies have suggested that PBRM1 mutations may benefit from antiangiogenic therapy in CCRCC (25,26). Furthermore, a comprehensive analysis of PBRM1 frequency and PBRM1 expression, as well as their predictive value for ICIs on clinical outcome in other cancer types has not yet been reported.
In this study, we investigated PBRM1 mutation frequency and PBRM1 expression across different cancer types. The correlation between PBRM1 mutations and clinical outcomes from anti-PD-L1 treatment and TMB was analyzed in CCRCC, lung adenocarcinoma (LUAD), bladder urothelial carcinoma (BLCA), and skin cutaneous melanoma (SKCM). We further evaluated the association of PBRM1 expression with immune infiltrates in a total of 32 cancer types.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-289).
Methods
Participants
The publicly available databases CbioPortal (https://www.cbioportal.org/) and Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) were used in this study. All PBRM1 genetic mutations and related clinical data were downloaded from three datasets in the cBioPortal database. A dataset with 10,945 samples was used to analyze the frequency of PBRM1 mutations across different cancer types (27). A dataset containing 1,661 patients was used to analyze the association of PBRM1 mutations with the overall survival (OS) in CCRCC, LUAD, BLCA, and SKCM with immunotherapy (28). A dataset containing 240 non-small cell lung cancer (NSCLC) patients was used to analyze the association of PBRM1 mutations with PFS and durable clinical benefit (DCB) in LUAD with immunotherapy (29). The TIMER database that includes 10,897 samples across 32 cancer types from The Cancer Genome Atlas (TCGA) was used to analyze the expression of PBRM1 and its relationship with immune infiltration levels (30).
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
For survival analyses, Kaplan-Meier survival curves were used and compared using the log-rank test. Cox’s regression model was used to perform univariable and multivariable analyses. For testing the association of TMB with PBRM1 mutation, the Mann-Whitney U test was used. The association between PBRM1 expression and immune infiltrates was analyzed via the TIMER database. We analyzed the PBRM1 expression in 32 cancer types via the “DiffExp” module, and the correlation of PBRM1 expression with the abundance of immune infiltrates, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, via the “gene” module. All reported P values are 2-sided.
Results
PBRM1 mutation frequency and PBRM1 expression in different cancer types
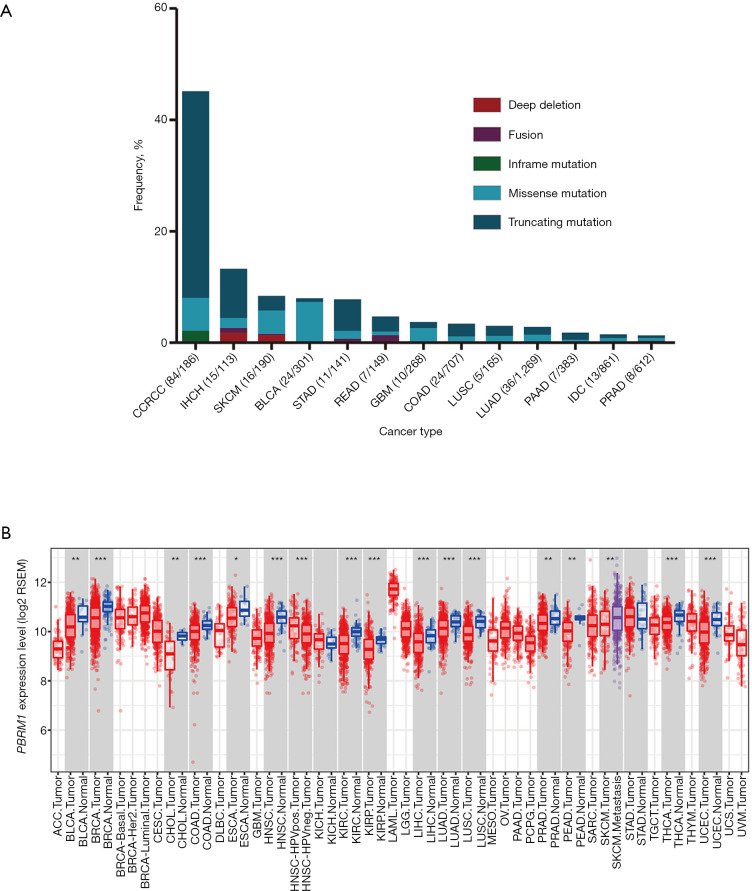
We assessed the frequency of PBRM1 gene alterations in a cBioPortal dataset of 10,336 patients with different cancer types (27). The frequency of PBRM1 mutations was 3.8% across all cancer types. Truncating mutations were the most common type of mutation. We further analyzed the PBRM1 mutation frequency in detailed cancer types, and the cancer types with a sample size less than 100 patients or PBRM1 mutant patients less than 5 were filtered out. The highest level of PBRM1 mutations was seen in CCRCC, with a frequency of 45%. The results showed 13 cancer types with a PBRM1 mutation frequency of more than 1.3% (Figure 1A).
Figure 1.
PBRM1 mutation frequency and PBRM1 expression pattern in different cancer types. (A) Frequency of PBRM1 mutations across different cancer types; (B) PBRM1 expression levels in diverse cancer types determined by TIMER. *, P<0.05; **, P<0.01; ***, P<0.001. PBRM1, polybromo 1; TIMER, Tumor Immune Estimation Resource.
The expression of PBRM1 was examined using the RNA-seq data of multiple cancer types in the TIMER database (Figure 1B). It is worth noting that PBRM1 expression was significantly lower in almost all cancer types that had matched normal tissues, except kidney chromophobe (KICH) and stomach adenocarcinoma (STAD).
Association of PBRM1 mutations with OS in CCRCC, LUAD, BLCA, and SKCM treated with anti-PD-L1
To investigate the association between PBRM1 mutations and OS in cancers with anti-PD-L1 treatment, the dataset containing 1,661 advanced cancer patients with ICI treatment from the cBioPortal was used (28). The OS was defined as the time of the first ICI treatment to the time of death or most recent follow-up. In this dataset, 139 patients had PBRM1 mutations, 55 in CCRCC, 16 in SKCM, 14 in LUAD, 6 in BLCA, and 48 in other cancer types. The study included patients who received PD-1 or PD-L1 therapy. Given the varying PBRM1 mutation frequency and clinical outcomes of immunotherapy across cancer types, we performed analysis of the association of PBRM1 mutations with OS in patients with CCRCC, SKCM, LUAD, and BLCA treated with anti-PD-L1, respectively.
As shown in Figure 2, patients with PBRM1 mutations showed a shorter median OS (mOS) in all four cancer types. The OS was significantly worse in PBRM1-mutant BLCA versus PBRM1-wildtype BLCA treated with ICIs.
Figure 2.
Association between PBRM1 mutations and OS in 4 cancer types treated with anti-PD-(L)-1. Kaplan-Meier plots of OS in PBRM1 mutant vs. non-mutant patients with (A) CCRCC, (B) LUAD, (C) BLCA and (D) SKCM. Censored data are indicated by vertical tick marks. P values of log-rank test are indicated. Median survival time in each group is indicated. PBRM1, polybromo 1; OS, overall survival; CCRCC, renal clear cell carcinoma; LUAD, lung adenocarcinoma; BLCA, bladder urothelial carcinoma; SKCM, skin cutaneous melanoma.
To further test the independent prognostic value in terms of OS within each cancer type, univariable and multivariable analyses based on the Cox proportional hazards regression model were conducted (Table 1). Univariable analysis showed that only in LUAD, high TMB was positively correlated with OS with immunotherapy, while in CCRCC and SKCM, high TMB tended to respond poorly to immunotherapy [hazard ratio (HR) >1]. The impact of PBRM1 mutations on OS did not reach statistical significance in any of the 4 cancer types, but the numerical trend of poor OS (HR >1) was observed in the univariable analysis. Multivariable analysis with adjustment for age, gender, PBRM1 status, and TMB in the four cancer types, respectively, indicated that PBRM1 mutations were an independent biomarker for poor prognosis in LUAD and BLCA, while TMB in these two cancer types was an independently improved prognostic biomarker for ICIs therapy. In multivariable analysis of CCRCC and SKCM patients, no factors were found to be significantly correlated with OS, but the trends of PBRM1-mutant patients towards a worse survival (HR >1) and high TMB towards clinical benefit from immunotherapy in CCRCC and SKCM (HR <1) were observed.
Table 1. Univariate and multivariate analysis of factors associated with OS in CCRCC, LUAD, BLCA, and SKCM.
| Category | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| CCRCC | |||||
| Age (>60 vs. ≤60) | 1.782 (0.897–3.541) | 0.099 | 1.744 (0.862–3.528) | 0.122 | |
| Gender (male vs. female) | 0.85 (0.407–1.777) | 0.667 | 0.903 (0.428–1.903) | 0.789 | |
| PBRM1 (Mut vs. Wt) | 1.287 (0.661–2.506) | 0.458 | 1.229 (0.61–2.476) | 0.563 | |
| TMB (continuous) | 1.455 (0.746–2.836) | 0.271 | 0.971 (0.858–1.098) | 0.634 | |
| LUAD | |||||
| Age (>60 vs. ≤60) | 1.167 (0.828–1.645) | 0.378 | 1.075 (0.76–1.52) | 0.683 | |
| Gender (male vs. female) | 1.188 (0.87–1.622) | 0.278 | 1.208 (0.88–1.658) | 0.242 | |
| PBRM1 (Mut vs. Wt) | 1.736 (0.938–3.213) | 0.079 | 2.369 (1.243–4.517) | 0.009 | |
| TMB (continuous) | 0.967 (0.948–0.987) | 0.001 | 0.962 (0.942–0.982) | 0.000 | |
| BLCA | |||||
| Age (>60 vs. ≤60) | 0.929 (0.532–1.622) | 0.796 | 1.086 (0.617–1.912) | 0.776 | |
| Gender (male vs. female) | 0.986 (0.501–1.943) | 0.968 | 1.009 (0.511–1.993) | 0.980 | |
| PBRM1 (Mut vs. Wt) | 2.41 (0.964–6.03) | 0.060 | 3.877 (1.462–10.283) | 0.006 | |
| TMB (continuous) | 0.793 (0.464–1.354) | 0.395 | 0.972 (0.947–0.997) | 0.030 | |
| SKCM | |||||
| Age (>60 vs. ≤60) | 2.721 (0.922–8.026) | 0.070 | 2.894 (0.928–9.029) | 0.067 | |
| Gender (male vs. female) | 1.394 (0.516–3.761) | 0.512 | 1.601 (0.584–4.389) | 0.360 | |
| PBRM1 (Mut vs. Wt) | 1.886 (0.638–5.579) | 0.251 | 1.561 (0.507–4.806) | 0.437 | |
| TMB (continuous) | 1.004 (0.991–1.017) | 0.550 | 0.998 (0.983–1.012) | 0.753 | |
OS, overall survival; CCRCC, renal clear cell carcinoma; LUAD, lung adenocarcinoma; BLCA, bladder urothelial carcinoma; SKCM, skin cutaneous melanoma; HR, hazard ratio; CI, confidence interval; TMB, tumor mutational burden; Mut, mutation type; Wt, wild type.
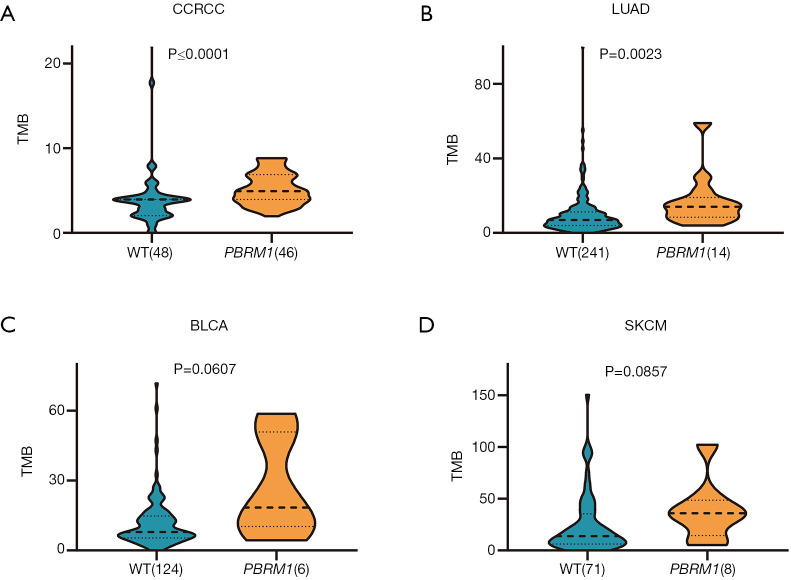
Association of PBRM1 mutations with TMB in CCRCC, LUAD, BLCA, and SKCM
We assessed the association between PBRM1 mutation and TMB in the above four cancer types. The results indicated a trend of PBRM1 mutants towards higher TMB in all the four cancer types (Figure 3). In LUAD and BLCA, PBRM1 mutations were significantly associated with higher TMB (P<0.0001 and P<0.0023, respectively). The effect in BLCA and SKCM did not reach statistical significance, which may have been due to the small sample size.
Figure 3.
Association between PBRM1 mutations and TMB in CCRCC, LUAD, BLCA, and SKCM. TMB in PBRM1 mutant vs. non-mutant patients with (A) CCRCC, (B) LUAD, (C) BLCA and (D) SKCM. P values of Mann-Whitney U test are indicated. PBRM1, polybromo 1; TMB, tumor mutational burden; CCRCC, renal clear cell carcinoma; LUAD, lung adenocarcinoma; BLCA, bladder urothelial carcinoma; SKCM, skin cutaneous melanoma.
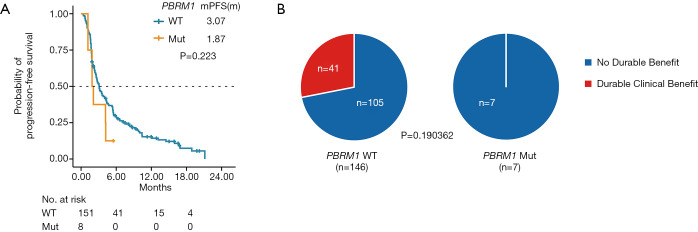
Association of PBRM1 mutations with PFS and DCB in LUAD treated with anti-PD-L1
In cBioPortal, we also identified another dataset comprising 240 advanced NSCLC patients. Most sample IDs in this dataset were included in a dataset of 1,661 patients. The PFS and DCB of patients were available in this dataset (29). We analyzed the association between PBRM1 mutations, PFS, and DCB. A total of 159 LUAD patients treated with anti-PD-(L)-1 monotherapy in the dataset were included in our study. Although not statistically significant, PBRM1 mutant LUAD tended to have a worse PFS (HR: 1.601; 95% CI: 0.743 to 3.450) (Figure 4). None of the 7 PBRM1 mutant patients had DCB from ICI, while 41 out of 146 PBRM1 wild-type patients had DCB. The trends of PFS and DCB in this dataset were consistent with OS in the dataset with 1,661 patients.
Figure 4.
Association between PBRM1 mutations, PFS, and DCB in LUAD treated with anti-PD-(L)-1. (A) Kaplan-Meier plots of PFS in PBRM1 mutant vs. non-mutant patients with LUAD. Censored data are indicated by vertical tick marks. P value of log-rank test is indicated. Median survival time in each group is indicated. (B) Pie charts of the proportion of patients with durable clinical benefits with or without PBRM1 mutation in LUAD. P value of Fisher’s exact test is indicated. PBRM1, polybromo 1; PFS, progression-free survival; DCB, durable clinical benefit; LUAD, lung adenocarcinoma.
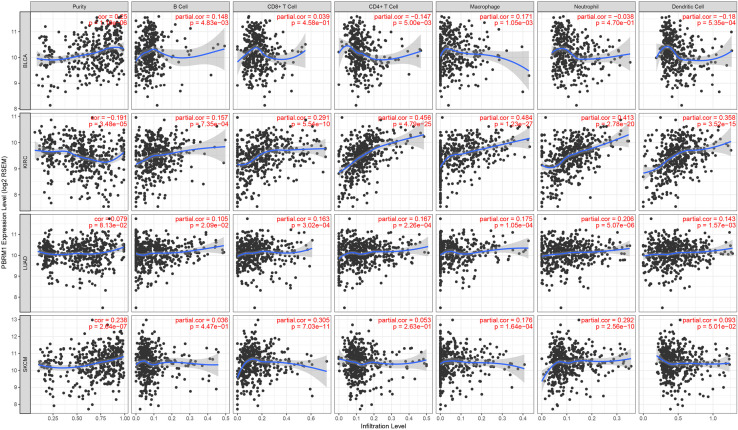
Association of PBRM1 expression with immune infiltrates
We then attempted to assess if PBRM1 expression correlated with immune infiltrates in the 32 cancer types via the TIMER database (Figure S1). A trend of PBRM1 expression towards higher immune infiltrates was observed in many cancer types, including breast (BRCA), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSC), KICH, kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), low-grade glioma (LGG), liver hepatocellular carcinoma (LIHC), LUAD, lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), SKCM, and thymoma (THYM). The results of relationship of PBRM1 expression with immune infiltrates in the four cancer types is shown in Figure 5. In KIRC and LUAD, the expression of PBRM1 was positively correlated with infiltration of B cells, CD8+ cells, CD4+ cells, macrophages, neutrophils, and dendritic cells. In BLCA, PBRM1 expression was positively correlated with B cells and macrophages, negatively correlated with CD4+ T cells and dendritic cells, and no significance was observed with CD8+ cells and neutrophils. In SKCM, there was a positive association of PBRM1 expression with infiltration levels of CD8+ T cells, macrophages, and neutrophils, and no significant correlation with B cells, CD4+ T cells, and dendritic cells was observed.
Figure 5.
Association between PBRM1 expression and immune infiltrates in KIRC, LUAD, BLCA, and SKCM. “Gene” module in TIMER was used to determine the association. PBRM1, polybromo 1; KIRC, kidney renal clear cell carcinoma; LUAD, lung adenocarcinoma; BLCA, bladder urothelial carcinoma; SKCM, skin cutaneous melanoma; TIMER, Tumor Immune Estimation Resource.
Discussion
Previous studies have revealed that the mutation frequency of SWI/SNF complexes in all human tumors was about 20%, similar to that of TP53, KRAS, and PTEN (31,32). Our work showed that PBRM1 mutation frequency was 3.8% across all cancer types and that PBRM1 expression was significantly decreased in most cancer types. This may imply that PRBM1 plays an important role in tumorigenesis in many cancer types.
To our knowledge, studies about the predictive value of PBRM1 were mainly reported in CCRCC. Our study did not observe a positive correlation between PBRM1 mutations and clinical benefit from anti-PD-L1 therapy. The PBRM1 mutant patients tended to respond poorly to the therapy in CCRCC, LUAD, BLCA, and SKCM based on multivariable analysis, especially so in LUAD and BLCA. These findings are consistent with the results from IMmotion150 (24).
The TMB was demonstrated as a predictor of superior OS with ICI treatment (33). However, some patients do not show DCB from ICIs even with high TMB (28,34). It is worth noting that in our study LUAD or BLCA patients with PBRM1 mutations tended to have higher TMB, yet these people responded poorly to anti-PD-L1 based on the log-rank test. Moreover, univariable analysis revealed that TMB tended to correlate with poor response to immunotherapy in CCRCC and SKCM (HR >1). After adjusting for PBRM1 mutations, age, and gender, the trend of TMB towards clinical benefit was similar across the four cancer types. These findings further suggested that TMB was not sufficient for predicting clinical benefit from immunotherapy response. The identification biomarker is needed as a complement to the existing methods.
Immune cell infiltrations have been suggested as a critical factor for ICIs treatment in recent studies (34-38). Our study revealed that PBRM1 expression correlated with immune infiltrates in many cancer types. Miao et al. reported tumors harboring PBRM1 mutations showed a lower expression of immune inhibitory ligands than those with intact PBRM1 (22). Kamal et al. reported inactivating mutations in PBRM1 was independently associated with reduced senescence enrichment in CCRCC, while high tumor senescence activity associates with clinical benefit from checkpoint blockade therapy (39). These mechanisms may contribute to poor clinical outcomes from immunotherapy in patients with immunotherapy.
The PBRM1 mutant CCRCC patients were reported to have high angiogenesis and respond well to anti-angiogenic therapy (24). The Food and Drug Administration (FDA) has approved anti-angiogenic drugs, such as bevacizumab, sorafenib, and sunitinib, for the treatment of several solid tumors (40). Anti-angiogenic inhibitors were believed to be important players not only in tumor angiogenesis but also in promoting immune cell infiltration (41-43). Further research is required to establish whether PBRM1 mutant patients with LUAD or BLCA or other cancers can get benefit from anti-angiogenic therapy or not.
This study was limited by the sample size and medical history of the patients. Further studies are needed to verify our findings. Our results may provide an impetus for studies and prospective clinical trials based on PBRM1 mutations.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-289
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-289). The authors have no conflicts of interest to declare.
(English Language Editor: J. Jones)
References
- 1.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133-50. 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darvin P, Toor SM, Sasidharan Nair V, et al. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 2018;50:1-11. 10.1038/s12276-018-0191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishino M, Ramaiya NH, Hatabu H, et al. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol 2017;14:655-68. 10.1038/nrclinonc.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong J, Chehrazi-Raffle A, Reddi S, et al. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer 2018;6:8. 10.1186/s40425-018-0316-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotte A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J Exp Clin Cancer Res 2019;38:255. 10.1186/s13046-019-1259-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther 2015;14:847-56. 10.1158/1535-7163.MCT-14-0983 [DOI] [PubMed] [Google Scholar]
- 8.Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. 10.1158/1535-7163.MCT-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao P, Li L, Jiang X, et al. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol 2019;12:54. 10.1186/s13045-019-0738-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway JR, Kofman E, Mo SS, et al. Genomics of response to immune checkpoint therapies for cancer: implications for precision medicine. Genome Med 2018;10:93. 10.1186/s13073-018-0605-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol 2020;20:25-39. 10.1038/s41577-019-0218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue Y, Canman JC, Lee CS, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A 2000;97:13015-20. 10.1073/pnas.240208597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nargund AM, Pham CG, Dong Y, et al. The SWI/SNF Protein PBRM1 Restrains VHL-Loss-Driven Clear Cell Renal Cell Carcinoma. Cell Rep 2017;18:2893-906. 10.1016/j.celrep.2017.02.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawłowski R, Muhl SM, Sulser T, et al. Loss of PBRM1 expression is associated with renal cell carcinoma progression. Int J Cancer 2013;132:E11-17. 10.1002/ijc.27822 [DOI] [PubMed] [Google Scholar]
- 15.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011;469:539-42. 10.1038/nature09639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carril-Ajuria L, Santos M, Roldan-Romero JM, et al. Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma. Cancers (Basel) 2019;12:16. 10.3390/cancers12010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 2013;8:e55119. 10.1371/journal.pone.0055119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45:1470-3. 10.1038/ng.2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Peng S, Guo L, et al. Prognostic and clinicopathological value of PBRM1 expression in renal cell carcinoma. Clin Chim Acta 2018;486 9-17. 10.1016/j.cca.2018.07.014 [DOI] [PubMed] [Google Scholar]
- 20.da Costa WH, da Cunha IW, Fares AF, et al. Prognostic impact of concomitant loss of PBRM1 and BAP1 protein expression in early stages of clear cell renal cell carcinoma. Urol Oncol 2018;36: 243.e1-243.e8. 10.1016/j.urolonc.2018.01.002 [DOI] [PubMed] [Google Scholar]
- 21.Gu YF, Cohn S, Christie A, et al. Modeling Renal Cell Carcinoma in Mice: Bap1 and Pbrm1 Inactivation Drive Tumor Grade. Cancer Discov 2017;7:900-17. 10.1158/2159-8290.CD-17-0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801-6. 10.1126/science.aan5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun DA, Ishii Y, Walsh AM, et al. Clinical Validation of PBRM1 Alterations as a Marker of Immune Checkpoint Inhibitor Response in Renal Cell Carcinoma. JAMA Oncol 2019;5:1631-3. 10.1001/jamaoncol.2019.3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDermott DF, Huseni MA, Atkins MB, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 2018;24:749-57. 10.1038/s41591-018-0053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang JC, Haworth L, Sherry RM, et al. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N Engl J Med 2003;349:427-34. 10.1056/NEJMoa021491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh JJ, Chen D, Wang PI, et al. Genomic Biomarkers of a Randomized Trial Comparing First-line Everolimus and Sunitinib in Patients with Metastatic Renal Cell Carcinoma. Eur Urol 2017;71:405-14. 10.1016/j.eururo.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zehir A, Benayed R, Shah RH, S, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703-13. 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. 10.1200/JCO.2017.75.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-e110. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013;45:592-601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masliah-Planchon J, Bieche I, Guinebretiere JM, et al. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol 2015;10:145-71. 10.1146/annurev-pathol-012414-040445 [DOI] [PubMed] [Google Scholar]
- 33.Yarchoan M, Albacker LA, Hopkins AC, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight 2019;4:e126908. 10.1172/jci.insight.126908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng W, Chen JQ, Liu C, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov 2016;6:202-16. 10.1158/2159-8290.CD-15-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodor JN, Boumber Y, Borghaei H. Biomarkers for immune checkpoint inhibition in non-small cell lung cancer (NSCLC). Cancer 2020;126:260-70. 10.1002/cncr.32468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen PL, Roh W, Reuben A, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov 2016;6:827-37. 10.1158/2159-8290.CD-15-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji RR, Chasalow SD, Wang L, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother 2012;61:1019-31. 10.1007/s00262-011-1172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568-71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamal Y, Cheng C, Frost HR, et al. Predictors of disease aggressiveness influence outcome from immunotherapy treatment in renal clear cell carcinoma. Oncoimmunology 2018;8:e1500106. 10.1080/2162402X.2018.1500106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Abd AM, Alamoudi AJ, Abdel-Naim AB, et al. Anti-angiogenic agents for the treatment of solid tumors: Potential pathways, therapy and current strategies - A review. J ADV RES 2017;8:591-605. 10.1016/j.jare.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elamin YY, Rafee S, Toomey S, et al. Immune effects of bevacizumab: killing two birds with one stone. Cancer Microenviron 2015;8:15-21. 10.1007/s12307-014-0160-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusmartsev S, Eruslanov E, Kubler H, et al. Oxidative stress regulates expression of VEGFR1 in myeloid cells: link to tumor-induced immune suppression in renal cell carcinoma. J Immunol 2008;181:346-53. 10.4049/jimmunol.181.1.346 [DOI] [PubMed] [Google Scholar]
- 43.Roland CL, Lynn KD, Toombs JE, et al. Cytokine levels correlate with immune cell infiltration after anti-VEGF therapy in preclinical mouse models of breast cancer. PLoS One 2009;4:e7669. 10.1371/journal.pone.0007669 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as