Abstract
Increasing understanding of metabolic and regulatory networks underlying microbial physiology has enabled creation of progressively more complex synthetic biological systems for biochemical, biomedical, agricultural, and environmental applications. However, despite best efforts, confounding phenotypes still emerge from unforeseen interplay between biological parts, and the design of robust and modular biological systems remains elusive. Such interactions are difficult to predict when designing synthetic systems and may manifest during experimental testing as inefficiencies that need to be overcome. Transforming organisms such as Escherichia coli into microbial factories is achieved via several engineering strategies, used individually or in combination, with the goal of maximizing the production of chosen target compounds. One technique relies on suppressing or overexpressing selected genes; another involves introducing heterologous enzymes into a microbial host. These modifications steer mass flux towards the set of desired metabolites but may create unexpected interactions. In this work, we develop a computational method, termed Metabolic Disruption Workflow (MDFlow), for discovering interactions and network disruptions arising from enzyme promiscuity – the ability of enzymes to act on a wide range of molecules that are structurally similar to their native substrates. We apply MDFlow to two experimentally verified cases where strains with essential genes knocked out are rescued by interactions resulting from overexpression of one or more other genes. We demonstrate how enzyme promiscuity may aid cells in adapting to disruptions of essential metabolic functions. We then apply MDFlow to predict and evaluate a number of putative promiscuous reactions that can interfere with two heterologous pathways designed for 3-hydroxypropionic acid (3-HP) production. Using MDFlow, we can identify putative enzyme promiscuity and the subsequent formation of unintended and undesirable byproducts that are not only disruptive to the host metabolism but also to the intended end-objective of high biosynthetic productivity and yield. As we demonstrate, MDFlow provides an innovative workflow to systematically identify incompatibilities between the native metabolism of the host and its engineered modifications due to enzyme promiscuity.
Keywords: Metabolic disruption, Enzyme promiscuity, Metabolic models, Metabolic engineering, Synthetic biology, Bio design automation
Highlights
-
•
Engineering modifications to cellular hosts result in undesirable byproducts.
-
•
Metabolic Disruption: changes in engineered host due to enzyme promiscuity.
-
•
Metabolic Disruption Workflow (MDFlow) uncovers metabolic disruption.
-
•
MDFlow corroborates previously experimentally verified promiscuous interactions.
-
•
MDFlow compares disruption due to heterologous pathways targeting 3-HP production.
1. Introduction
Integrating heterologous synthesis pathways within microbial hosts has been instrumental in the biomanufacturing of industrial products such as biofuels, polymers, pharmaceuticals, therapeutics, flavors and chemical commodities (Lee et al., 2008; Madison and Huisman, 1999; Nakamura and Whited, 2003; Trantas et al., 2015; George et al., 2015). One strategy to improve yield is to use well-established metabolic engineering techniques such as gene deletion, promoter engineering, media optimization, etc. (Lee et al., 2009). Another strategy is to directly engineer enzymatic properties such as activity, selectivity, inhibition-resistance, and solubility (Yoshikuni et al., 2008). Using one or more of these strategies has proven effective in the development of strains with desired target yields, productivity, and titers.
Often, such metabolic engineering strategies yield unexpected enzyme-compound interactions. Some interactions can be beneficial for the survival of the host. For instance, Patrick et al. documented 41 rescue instances where the lethality of an essential protein deletion was suppressed by overexpression of one noncognate E. coli gene, attributing some of them to catalytic promiscuity and substrate ambiguity (Patrick et al., 2007). In other cases, beneficial interactions can come at a cost. The overexpression or knockout of enzymes can result in interactions that are disruptive for growth and maintenance by siphoning off key metabolic intermediates like pyruvate, acetyl-CoA, and NADH. For example, while seeking to suppress lethality of inactivating the pyridoxal-5-phosphate (PLP) cofactor synthesis pathway, Kim et al. experimentally identified a four-step serendipitous pathway in E. coli that restored the strain’s ability to grow on glucose at the expense of consuming essential intermediates in the native serine biosynthetic pathway (Kim et al., 2010), producing toxic byproducts (Kim and Copley, 2012).
The presence of high concentrations of heterologous enzymes and metabolites within microbial cells causes unexpected promiscuous interactions with host enzymes and metabolites. For example, the short-chain dehydrogenase YMR226C used to produce 3-hydroxypropionic acid (3-HP) in yeast is associated with 15 known substrates (Fujisawa et al., 2003; Jessen et al., 2015). In E. coli strains featuring the malonyl-CoA pathway for 3-HP synthesis, significant quantities of lactate and acetate are produced, even after lactate dehydrogenase (ldh) and pta were knocked out (Rathnasingh et al., 2012). Another instance of promiscuous activity can be observed for pathways intended for butanol production: the promiscuity of the bifunctional butyryl-CoA dehydrogenase (AdhE2) enzyme with substrate acetyl-CoA often results in concomitant synthesis of ethanol with butanol (Inui et al., 2008; Atsumi et al., 2008a; Nielsen et al., 2009). In yet another example, Liao and colleagues leveraged promiscuity of ketoacid decarboxylase (KVD) and alcohol dehydrogenase (ADH) enzymes to synthesize a spectrum of alcohols from branched-chain amino acid metabolic intermediates (Atsumi et al., 2008b). Yet, a caveat of this promiscuous activity is that no single alcohol can be made alone. That is, pyruvate itself is a ketoacid, which can be converted to ethanol by the same promiscuous activity of KVD and ADH. Thus, isobutanol synthesis is also coupled to ethanol synthesis due to enzyme promiscuity (Trinh et al., 2011).
While ubiquitous (D’Ari and Casadesús, 1998; Nobeli et al., 2009; Khersonsky et al., 2006; Tawfik and D. S, 2010) and often observed, effects of enzyme promiscuity on the host metabolic network are often ignored during design and only identified during experimental studies. Predicting such interactions early in the design cycle could yield improved design outcomes and reduce experimental efforts. The prediction of enzymatic products due to substrate promiscuity has mainly relied on hand-curated rules that capture well-known enzymatic transformations. For example, a list of 50 reaction rules, each associated with one or more reaction, was previously defined to explore novel synthesis pathways (Cho et al., 2010; Campodonico et al., 2014). Another set of rules was applied repetitively to generate novel synthesis (Li et al., 2004) or degradation pathways (Finley et al., 2009). Further use of such rules allowed the compilation of over 130,000 hypothetical enzymatic reactions that connect two or more KEGG metabolites (Hadadi et al., 2016), and the compilation of predicted metabolic products into databases such as MINEs (Jeffryes et al., 2015). MyCompoundID (Li et al., 2013) utilizes a similar paradigm and generates products by the repeated application of addition or subtraction of common functional groups. BioTransformer (Djoumbou-Feunang et al., 2019) predicts derivatives by utilizing five separate prediction modules in concert with machine learning and a rule-based knowledge base. PROXIMAL (Yousofshahi et al., 2015) utilizes enzyme-specific reactant–product transformation patterns from the KEGG database (Moriya et al., 2010) as a lookup table to predict products for query molecules. The PROXIMAL algorithm was utilized to create organism-specific Extended Metabolic Models (EMMs) that extend reference metabolic models catalogued in databases to include putative products due to promiscuous native enzymatic activities on native metabolites (Amin et al., 2019; Hassanpour et al., 2020). Despite advances in predicting promiscuous products, however, these efforts have not been put forward in a systematic way to analyze metabolic network disruption in engineered microbial hosts due to enzyme promiscuity.
We develop in this paper a computational method, Metabolic Disruption Workflow (MDFlow), to analyze the disruptive impact of enzyme promiscuity on engineered microbial hosts in a systematic manner. We define metabolic disruption as off-target changes in host metabolism that arise because of enzyme-substrate interactions upon gene or pathway overexpression, where such interactions neither exist in the wild-type chassis organism nor are they expected due to presence of recombinant enzyme(s). Accordingly, this definition encompasses all enzyme promiscuity that arises because of adding heterologous enzymes and their chemical products to a host microbe. Therefore, MDFlow is designed to consider two different disruption scenarios. In “Scenario 1”, promiscuous activity is predicted in the context of overexpressed enzymes, whether heterologous or native, acting promiscuously on native host metabolites. Meanwhile, in “Scenario 2,” predictions are made by assuming that native enzymes exhibit promiscuous interactions with synthesis pathway metabolites introduced with engineering changes. Of course, in a biological system, both scenarios would occur simultaneously to some degree, and even higher-order interactions would be possible (e.g., subsequent use of promiscuous reaction products as substrates for additional transformations). Using PROXIMAL (Yousofshahi et al., 2015) and flux analysis (Orth et al., 2010; Segrè et al., 2002), MDFlow models and evaluates such promiscuous scenarios. We demonstrate the use of MDFlow to evaluate how engineered microbial hosts are impacted by enzyme promiscuity under three engineering strategies. First, MDFlow is used to explain how essential gene deletion can be suppressed by overexpression of another native gene. Next, MDFlow is used to identify multi-step interactions that may compensate for essential gene knockouts. Third, MDFlow is used to evaluate the potential disruption when a heterologous pathway is added to a microbial host. The first two cases represent Scenario 1 and are evaluated against experimentally verified data. The last case represents a simultaneous application of both Scenarios 1 and 2 and demonstrates that the choice of synthesis pathway can impact metabolic disruption scenarios.
This work is novel as it is the first to systematically investigate effects of heterologous and native enzyme promiscuity on host metabolism and its consequences on biocatalysts. Our method serves as the first computational tool that can assist metabolic engineers in (1) identifying sources of unexpected byproducts, (2) assessing the consequences of metabolic engineering on the host, and (3) quantifying pathway-host incompatibility using metabolic network disruption. Outcomes from this work will aid in future studies to design robust systems with more predictable behaviors and improved desired product yield.
2. Methods
MDFlow (Fig. 1) is an integrated method that combines several techniques to analyze cellular metabolic disruption that may occur due to enzyme promiscuity. To predict such putative interactions, MDFlow relies on PROXIMAL (Yousofshahi et al., 2015) to predict byproducts resulting from promiscuous activities under Scenarios 1 and 2. Stoichiometrically balanced reactions are then derived based on substrate, product, and the reaction associated with the promiscuous enzymatic transformation pattern. To assess the disruption impact in a systematic fashion, the host metabolic network model is incrementally modified and evaluated after each change – first to set a baseline, then augmented with the engineering strategy of interest. Quantitative flux analysis (Flux Balance Analysis (FBA) (Orth et al., 2010) and/or Minimization of Metabolic Adjustment (MOMA) (Segrè et al., 2002)) was then used to evaluate the impact of such changes on biomass growth rates or product yield. We provide a detailed overview of each of these steps.
Fig. 1.
An overview of the four-step process used by MDFlow to identify and evaluate byproducts formed due to enzymes promiscuity for Scenarios 1 and 2. The original host metabolic model is progressively augmented with engineered modifications and predicted interactions. The updated models are evaluated using FBA and/or MOMA at different stages.
2.1. Step 1 – establish the baseline growth rate and/or target yield using FBA
To model the E. coli metabolic network, we built upon the iML1515 model published by Monk et al. (Monk et al., 2017). When evaluating gene knockout modifications, we used a derivative of that model, iML1428, that offers improved accuracy of lethality predictions in such experiments. iML1428 achieves this improvement by removing isozymes that are minimally expressed in glucose M9 media, thus preventing them from incorrectly compensating for the removal of essential genes. We used the conditions suggested by the authors of iML1515 for constraint-based modeling – the lower bounds of all exchange reactions were set to zero, except for glucose, oxygen, and all inorganic ions: the lower bound for each of those reactions was set to –10, –20, and –1000 mmol gDW–1 h–1, respectively. With the constraints configured for aerobic growth on glucose, we evaluated the baseline growth rate, and target metabolite yield (if applicable), of the host using FBA. Then, we set the lower bound on biomass growth to be equal to 10 % of the baseline growth rate to prioritize minimum growth of the strain required to ensure its long-term survival. This lower bound may be increased or decreased as appropriate for the application.
2.2. Step 2 – evaluate direct impact of the engineering change using flux analysis
We implemented intended changes for metabolic engineering in the context of addition or removal of reactions and metabolites in the network. To construct a synthesis pathway for transforming a metabolite within a host into a target compound, we added each synthesis step to the stochiometric matrix (S-matrix) of the metabolic model as a new reaction, along with a demand reaction for the target metabolite. To model a gene knockout, we enacted the effect of deleting the gene from the model by setting flux to zero for all inactive reactions, identified based on their individual gene-protein-reaction (GPR) rules (Schellenberger et al., 2011). The new “engineered” model was then evaluated using FBA to demonstrate the impact of introduced changes. At this stage in the workflow, it is important to verify that the model’s prediction reflects expectations or experimental data.
2.3. Step 3 – predict promiscuous interactions using PROXIMAL
The modified model from the previous step was used to predict promiscuous byproducts for Scenarios 1 and 2. Each added reaction was assumed reversible unless indicated otherwise. For each scenario, PROXIMAL first created a lookup table of all known biotransformation operators based on catalogued reactions within the model. These operators encode molecular transformation patterns associated with an enzyme and the reactions it catalyzes. Given a query molecule, PROXIMAL first identified operators in the lookup tables that can act on the molecule. Then, the query molecule was mapped to possible byproducts. An operator acts on a query molecule if its reaction center and its nearest neighbor atom(s) exactly match those of the native substrate, as encoded in the lookup tables. Each potential byproduct was reported in the form of a mol file, which was then used to identify if the potential byproduct is a known metabolite. The mol file was matched to either a metabolite in the model, a KEGG ID, or a PubChem ID using InChIKeys (Heller et al., 2015) generated by an open-source chemical toolbox RDKit (RDKit).
For Scenario 1, biotransformation operators were derived from the overexpressed enzymes along the synthesis pathway and applied to all native metabolites in the model. For Scenario 2, biotransformations were derived from native host enzymes within the model and applied to pathway metabolites in the engineered pathway. Applying PROXIMAL operators resulted in a list of byproducts for each scenario. For each predicted byproduct, we developed a new balanced enzymatic reaction based on the catalyzing enzyme’s reaction pattern. A reaction template with suitable cofactors was obtained from reaction(s) associated with each enzymatic biotransformation. A reaction is balanced when the number of atoms for each element on the reactant side matches those on the product side. Reactions were verified to be balanced using ChemPy (Dahlgren, 2018). If a reaction was not balanced, it was discarded and not considered for further analysis as it violated the assumptions of FBA.
2.4. Step 4 – evaluate network disruption using flux analysis
To predict the effect of promiscuity, the engineered model network was augmented with balanced promiscuous reactions. The metabolites in these reactions were either mapped to existing metabolites in the model or added to the model along with the corresponding exchange reactions allowing unlimited export (but not import) of a metabolite from the host. Each predicted reaction was first added to the engineered model separately. The flux range of each promiscuous reaction was calculated by minimizing (“min”) and maximizing (“max”) its flux. Since the new reactions did not have any particular direction associated with them, and the most disruptive one was not yet known at this stage, flux ranges calculations were performed twice: once assuming a forward (“fwd”) flux direction and another assuming the reverse (“rev”) flux. As a result, four flux values were estimated for each reaction: vfwd_min, vfwd_max, vrev_min, and vrev_max. The maximum reverse flux (vrev_max) and minimum forward flux (vfwd_min) may be trivial solutions; however, they could be non-zero for reactions that were required for the model to maintain the minimum growth rate. The added reactions were either new to the model or formed biotransformation routes that were catalyzed differently than those already in the model. This updated network represented the “disrupted” model.
To model promiscuous activity, we assumed a non-zero flux through the added reaction (vfwd_added, vrev_added) based on a pre-defined percentage p, referred to as a coupling percentage, of the calculated maximum flux. As in case of all reactions when performing FBA, the absolute maximum for any reaction was set at ±1,000 mmol/h/gDW. The constraints imposed on the reaction can be described by the following inequalities – the first one pertains to the forward flux through the reaction, while the second describes the reverse flux:
| vfwd_min + p · (vfwd_max – vfwd_min) ≤ vfwd_added ≤ 1000 |
| –1000 ≤ vrev_added ≤ vrev_max + p · (vrev_min – vrev_max) |
Once constraints were set, the metabolic network disruption caused by the developed reactions could be evaluated. The disrupted yield value was calculated and compared with the value of the yield of the undisrupted engineered model. The direction of each reaction was then fixed to the one that caused maximum disruption. For the purposes of subsequent analysis, reactions were considered irreversible and their individual flux was constrained by only one of the two inequalities. Disruption in yield was then evaluated under various coupling percentages, with a random subset of developed reactions added to the undisrupted model. The results were then placed on a scatter plot relating the extent of metabolic disruption to the intensity of promiscuous activity for visualization purposes.
2.5. Implementation
The workflow was written in Python, targeting versions 3.7 and up, and is available on GitHub (https://github.com/HassounLab/MDFlow). Instructions for running the workflow are provided on the project’s GitHub page and in the README file accompanying the source code. Promiscuous reactions predicted by PROXIMAL (Yousofshahi et al., 2015) (https://hassounlab.cs.tufts.edu/proximal/) are provided in the repository. The FBA implementation and model handling logic were provided by the COBRApy package (Ebrahim et al., 2013), and all visualizations were built using matplotlib (Hunter, 2007), seaborn (Waskom, 2020), and RDKit (RDKit). All experiments were conducted on an Intel® Core™ i7-2600 machine with 8 GB of RAM, though the workflow does not require the use of any specific hardware platform.
3. Results
3.1. MDFlow explains how suppressors can rescue growth of essential gene deletions
We first demonstrated that MDFlow can be used to predict how enzymatic promiscuity suppresses the effect of gene deletions. We validated MDFlow against experimental data suggested by Patrick et al. (Patrick et al., 2007). The authors set out to identify and categorize multifunctional genes that enabled cells’ adaptability to genetic lesions. Within the Keio collection (Baba et al., 2006), the authors identified 107 single-gene knockout strains that were unable to grow on the M9-glucose medium. Patrick et al. found 21 strains that could be rescued via overexpression of one non-cognate gene, for a total of 41 unique suppression examples. Comparing structural superimpositions of deleted proteins and their suppressors, Patrick et al. observed significant structural homology between a handful of enzyme pairs and attributed several examples to substrate ambiguity and catalytic promiscuity (Patrick et al., 2007). The fact that an essential gene deletion was suppressed by overexpression of another native gene and that enzyme/substrate non-specificity played a role in the rescue suggests that the responsible mechanism in many instances could have been a single enzymatic reaction mediated by promiscuous activity of overexpressed enzymes – that is, activity following Scenario 1. In this test case, we applied MDFlow to each of the 41 unique knockout-suppressor pairs and evaluated its effectiveness in explaining the suppression of gene deletion via overexpression of another native gene. The observations made by Patrick et al. were thus used to both guide the workflow and validate it against experimental data.
To establish the baseline for validating rescue, we first determined if the deletion of each of the 20 essential genes responsible for the 41 pairs arrests the growth of the strain. To this end, we performed gene knockouts in iML1428 and measured the biomass growth rate reported by FBA. Out of the 20 lethal knockouts reported by the authors, we were able to confirm lack of growth for 9 of them, accounting for 15 out of the 41 unique knockout-suppressor pairs. Since we cannot computationally validate rescue without first establishing the lethality of the genetic lesion, we only focused on those 15 cases where the deletion led to no biomass growth. For each of the suppressor genes, we used PROXIMAL to generate promiscuous reactions due to Scenario 1 type interactions (e.g., overexpressed gene acting on native metabolites) and then applied FBA to determine their individual effect on the biomass, resulting in the complete recovery of 6 strains (ΔilvE, ΔglnA, ΔilvD, ΔhisH, ΔpabA, and ΔilvA). In the majority of the recovered cases, a promiscuous reaction replicated the deleted essential reaction either exactly (e.g., ΔilvA/tdcB, ΔpabA/pabB, and ΔhisH/hisF) or with different cofactors (e.g., ΔilvE/avtA and ΔglnA/asnB). Both pabA/pabB and hisH/hisF form well-characterized heterodimeric enzymes consisting of a larger subunit (PabB and HisF) and a smaller one (PabA and HisH). In both cases, the larger subunit has shown activity in the absence of the smaller subunit (Ye et al., 1990; Klem and Davisson, 1993). Therefore, overexpressing PabB and HisF could be reasonably expected to compensate for the deletions in the ΔpabA and ΔhisH strains. Meanwhile, in the ΔilvA/tdcB case, the two proteins are known to be isozymes of one another, thus having the same function (Patrick et al., 2007). Therefore, there is biological justification for the rescuer enzyme’s potential ability to exactly replicate the reactions lost by the deletion. From the perspective of our method, both the deleted gene and its suppressor had the same Enzyme Commission (EC) numbers, which made them interchangeable for the purposes of transformation pattern extraction. In the other 3 case sets, however, the outcome appears to be due to enzyme promiscuity – although the first two or three EC groups matched in two instances, the deleted protein and its replacement were not identical enzymes in all three. For ΔglnA/asnB, the promiscuous activity was responsible for creating a reaction edge between L-glutamate and L-glutamine, which appears to be the mechanism of recovery. For ΔilvE/avtA, the promiscuous reaction allowed the production of L-isoleucine, which is essential for survival as double mutants (ΔilvE ΔavtA) are known to require supplementation of both, isoleucine and valine, for growth (Whalen and Berg, 1982). Notably, the deletion of ilvE also caused the loss of an L-valine-producing reaction in the model, however, its essentiality was not confirmed through FBA. For ΔilvD/avtA, no single predicted reaction was responsible for growth recovery, however, including various combinations of two predicted reactions demonstrated rescuing effect. When using MOMA, the lethality and rescuing effects of knockouts and multicopy suppressors were identical as predicted when using FBA. A summary of multicopy suppression results can be found in Table 1 along with example compensating reactions predicted for each case. A complete set of predicted reactions is provided in Supplementary File 1.
Table 1.
Confirmed-lethal single-gene deletion strains from Patrick et al. (Patrick et al., 2007) with the corresponding multicopy suppressors (MS) and representative subsets of predicted compensating reactions. Biomass growth rate in the disrupted network computed using FBA is presented as a percentage of the wild type strain growth rate.
| Deletion | Deficient Reactions | MS | MS Biomass | Compensating Reactions |
|---|---|---|---|---|
| ΔilvA (4.3.1.19) | L-threonine → 2-oxobutanoate + NH4+ | tdcB (4.3.1.17, 4.3.1.19) | 100.00% | L-threonine → 2-oxobutanoate + NH4+ |
| 100.47% | L-allo-threonine → 2-oxobutanoate + NH4+ | |||
| ΔilvE (2.6.1.42) | (S)-3-methyl-2-oxopentanoate + L-glutamate → 2-oxoglutarate + L-isoleucine 3-methyl-2-oxobutanoate + L-glutamate → 2-oxoglutarate + L-valine |
avtA (2.6.1.66) | 100.00% | (S)-3-methyl-2-oxopentanoate + L-alanine → L-isoleucine + pyruvate |
| 100.79% | 3 3-methyl-2-oxobutanoate + 2 L-alanine → 2 L-isoleucine + 3 pyruvate | |||
| ΔglnA (6.3.1.2) | ATP + L-glutamate + NH4+ → ADP + L-glutamine + H+ + PO43- | asnB (6.3.5.4) | 100.94% | AMP + L-asparagine + L-glutamate + P2O74- → L-aspartate + ATP + L-glutamine + H2O |
| 101.26% | L-glutamate + NH4+ → L-glutamine + H2O | |||
| 99.38% | AMP + L-asparagine + D-glucose 1-phosphate + L-glutamate → ADP-glucose + L-aspartate + L-glutamine + H2O | |||
| (and others) | ||||
| ΔilvD (4.2.1.9) | (R)-2,3-dihydroxy-3-methylbutanoate → 3-methyl-2-oxobutanoate + H2O (R)-2,3-dihydroxy-3-methylpentanoate → (S)-3-methyl-2-oxopentanoate + H2O |
avtA (2.6.1.66) | 100.23% | 2 Pyruvate + 3 L-valine → 2 (S)-3-methyl-2-oxopentanoate + 3 L-alanine 2 4-methyl-2-oxopentanoate + 3 L-alanine → 2 pyruvate + 3 L-valine |
| 100.23% | 3 3-methyl-2-oxobutanoate + 2 L-alanine → 2 L-isoleucine + 3 pyruvate 2 L-leucine + 3 Pyruvate → 3 3-methyl-2-oxobutanoate + 2 L-alanine |
|||
| (other combinations may be possible) | ||||
| ΔhisH (4.3.2.10, 3.5.1.2) | L-glutamine + phosphoribulosylformimino-AICAR-phosphate → AICAR + erythro-imidazole-glycerol-phosphate + L-glutamate + H+ | hisF (4.3.2.10) | 100.02% | L-glutamine + phosphoribulosylformimino-AICAR-phosphate → AICAR + erythro-imidazole-glycerol-phosphate + L-glutamate |
| 100.02% | L-glutamine + phosphoribosylformiminoaicar-phosphate → AICAR + erythro-imidazole-glycerol-phosphate + L-glutamate | |||
| 100.05% | L-glutamine + 2 phosphoribulosylformimino-AICAR-phosphate → (S)-2-hydroxyglutarate + 2 AICAR + 2 erythro-imidazole-glycerol-phosphate | |||
| (and others) | ||||
| ΔpabA (2.6.1.85) | chorismate + L-glutamine → 4-amino-4-deoxychorismate + L-glutamate | pabB (2.6.1.85) | 100.00% | chorismate + L-glutamine → 4-amino-4-deoxychorismate + L-glutamate |
| 100.00% | 2 chorismate + L-glutamine → 2 4-amino-4-deoxychorismate + (S)-2-hydroxyglutarate | |||
| 100.00% | chorismate + L-glutamine → 4-amino-4-deoxychorismate + O-acetyl-L-serine | |||
| 100.00% | L-asparagine + chorismate → 4-amino-4-deoxychorismate + L-aspartate | |||
| 100.00% | L-glutamine + isochorismate → 4-amino-4-deoxychorismate + L-glutamate | |||
| 100.00% | L-glutamine + prephenate → 4-amino-4-deoxychorismate + L-glutamate | |||
3.2. MDFlow can identify multi-step bypasses to essential gene functions
Because MDFlow anticipates interactions between native metabolites and enzymes due to multiple genes that are simultaneously overexpressed, MDFlow can be used to identify multi-step interactions where a native pathway contributes metabolites to an unexpected, serendipitous process. We validated MDFlow for predicting promiscuous interactions in the serendipitous pathway identified by Kim et al. (Kim et al., 2010). The authors identified a four-step chain of interactions that compensated for an essential gene knockout. The pdxB gene deletion disrupted the PLP synthesis pathway, resulting in its inability to grow on M9-glucose at 37 °C. Simultaneous overexpression of seven other genes, however, rescued the strain. Using genetic complementation experiments, the authors were able to separate those genes into groups and describe one of the rescuing pathways in detail. The serendipitous pathway, shown in Fig. 2, was found to bypass the knocked-out enzyme in the PLP pathway by diverting flux from serine biosynthesis. The interactions comprising this pathway were catalyzed by three enzymes – two of which exhibited either promiscuity (ThrB) or broad specificity (LtaE). The function of the third enzyme (YeaB) was unknown, and one of the interactions appeared to be non-enzymatic. Therefore, at least part of the pathway could have emerged due to promiscuous activity classified as Scenario 1 type interactions by our method.
Fig. 2.
Serendipitous pathway discovered by Kim et al. that bypasses the deletion of an intermediate gene pdxB in the native PLP pathway by siphoning off material from the serine biosynthesis pathway. Circles highlight reaction steps that were predicted – or not predicted – by PROXIMAL. Pathway layout for the drawing was adapted from the authors’ original paper.
The new pathway bypasses the lesion by diverting a metabolite (3-phosphohydroxypyruvate, 3-PHP) from the serine production pathway and converting it to a metabolite (L-4-phosphohydroxythreonine, 4-PHT) downstream the PLP synthesis pathway via a series of 4 ad-hoc reactions (Kim et al., 2010). Assuming that the reactions comprising the pathway are due to promiscuity, discovering each step of the pathway entails predicting interactions between the candidate enzymes – that is, all native enzymes except the knocked out PdxB – and all available metabolites in the model – which includes the native metabolites as well as metabolites generated by promiscuous activity in previous steps. Repeating this process up to four times, in principle, would allow the extraction of the entire pathway. Unfortunately, this approach would be computationally intensive as it amounts to an exhaustive enumeration of all possible promiscuous reaction sequences. However, because the steps comprising the serendipitous PLP pathway are already known, verifying if the exhaustive search would eventually find the pathway can be done efficiently. At each step, instead of predicting interactions for all available metabolites, we only considered the small subset that overlaps with the ΔpdxB bypass pathway, significantly reducing the search space. Using MDFlow configured in this way, we were able to rediscover 3 out of the 4 reactions along this novel pathway. The elusive step, the LtaE-catalyzed transformation of glycolaldehyde to L-4-hydroxythreonine (4-HT), was not predicted due to the lack of an appropriate reaction pattern in PROXIMAL’s operator lookup table. This observation highlights an important shortcoming of rule-based methods such as BioTransformer (Djoumbou-Feunang et al., 2019) and PROXIMAL (Yousofshahi et al., 2015): their metabolism predictions are limited to the finite set of biotransformation rules encoded in their knowledgebase. Nonetheless, the results for this pathway and multicopy suppressors from (Patrick et al., 2007) show that our technique can be used to discover experimentally validated unexpected interactions that lead to the survival of the host.
To further illustrate the utility of MDFlow in predicting multi-step pathways, we used MDFlow to discover two-step promiscuous pathways in iML1515 that may compensate for the deletion of pdxB. Such a two-step pathway transforms a metabolite within iML1515 to one of the following metabolites that are downstream from 4-phosphoerythronate: 2-oxo-3-hydroxy-4-phosphobutanoate, 4-hydroxy-L-threonine, pyridoxal 5-phosphate. Intermediate metabolites at each step were predicted by PROXIMAL using biotransformations specific to E. coli as listed in the KEGG database. Then, we retained pathways that only had intermediates with known PubChem IDs. We further filtered these results based on thermodynamic feasibility of the pathways as estimated by eQuilibrator (Flamholz et al., 2011; Noor et al., 2012; Noor et al., 2013; Noor et al., 2014). There were 21 such feasible two-step pathways. There were 13 and 8 pathways that terminated on pyridoxal 5-phosphate and 2-oxo-3-hydroxy-4-phosphobutanoate, respectively, whereas none that terminated on 4-hydroxy-L-threonine. As the iML1428 model does not capture the lethality of knocking out pdxB, we were unable to evaluate the impact of these two-step pathways on growth rates using FBA. A complete set of predicted two-step pathways is provided in Supplementary File 2.
3.3. MDFlow predicts metabolic disruption during 3-hydroxypropionic acid (3-HP) biosynthesis adversely affects yield
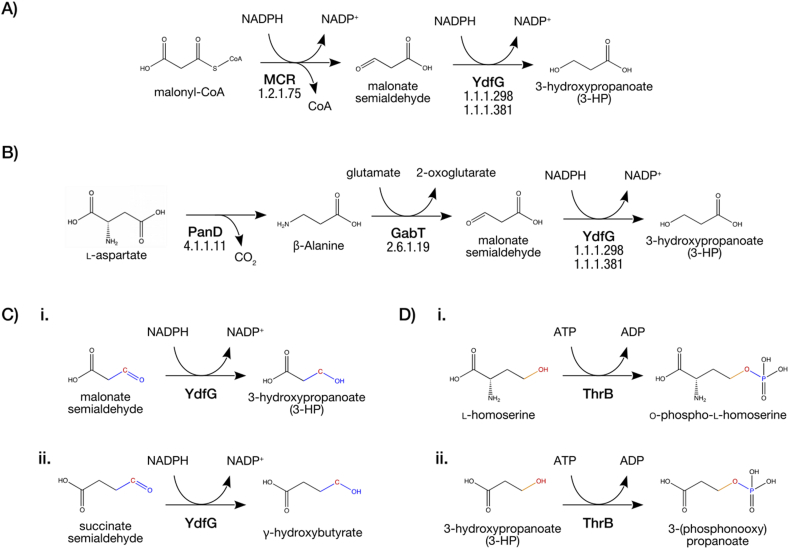
We used our workflow to analyze metabolic disruption through Scenarios 1 and 2 for two different synthetic 3-HP pathways, an important precursor metabolite to useful derivatives such as acrylic acid, 1,3-propanediol, and malonic acid (Chen and Nielsen, 2013; Della Pina et al., 2011; Kumar et al., 2013). Several groups have reported production of 3-HP in various organisms (Jiang et al., 2009; Huang et al., 2013), including E. coli (Raj et al., 2008; Kim et al., 2014). Using E. coli as a host, we first added a two-step 3-HP synthesis pathway (Rathnasingh et al., 2012) (Fig. 3A) catalyzed by malonyl-CoA reductase (MCR) (Cheng et al., 2016) and 3-hydroxyacid dehydrogenase (YdfG). Then, separately, we added a three-step 3-HP pathway (Wang et al., 2014) (Fig. 3B) comprising of panD, gabT, and ydfG. The 3-HP yield of the baseline unmodified model was estimated to be 0.90 mol/mol of glucose, and the addition of the new pathways increased that to 1.62 mol/mol of glucose for the first pathway and 1.66 mol/mol for the second. These figures correspond to the maximum theoretical yield of 3-HP assuming no disruption. When using MOMA instead of FBA to evaluate the 3-HP yield, we found that the added reactions do not increase the yield of 3-HP over the baseline. We believe this is a direct consequence of the MOMA approach: when the engineering change is the addition of reactions, the problem of minimizing flux redistribution has a trivial solution that allows no flux through the new reactions. As a result, we do not believe MOMA is applicable to this particular case. Thus, we conducted all subsequent analysis using FBA only. After the addition of a synthesis pathway to E. coli, our workflow utilized PROXIMAL to predict derivatives for both Scenarios 1 and 2. For both Scenarios, identities of predicted derivatives were looked up in either iML1515, KEGG, or PubChem. As not all predicted derivatives were identifiable through this method, we considered only derivatives that are documented in at least one of the three databases. Example reactions for each Scenarios 1 and 2 are shown in Fig. 3, panels C and D, respectively. Other predicted reactions are given in Supplementary File 3.
Fig. 3.
Overview of the two heterologous 3-HP pathways integrated into the E. coli model and the method used to construct putative promiscuous reactions for each scenario. (A) Pathway 1, which converts malonyl-CoA into 3-HP via two reactions catalyzed by malonyl-CoA reductase (MCR) and YdfG. (B) Pathway 2 that produces 3-HP from L-aspartate using PanD and GabT in addition to YdfG. Developed reactions examples of Scenario 1 (C) and Scenario 2 (D). Both panels (C) and (D) are divided in three sections: i. the native reaction catalyzed by the potentially promiscuous enzyme, ii. the RDM pattern showing the rction center in red where the biotransformation occurs, and iii. the developed balanced reaction indicating the reactants, products, and the promiscuous enzyme. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Balanced predicted reactions due to each scenario were first evaluated using FBA to determine the range of fluxes they can potentially sustain in the context of the E. coli metabolic model. The model was then augmented with all predicted reactions, along with the 3-HP synthesis pathway. We then assumed that each of the predicted reactions exhibits a certain minimum activity, measured as a fraction of its maximum possible flux. We refer to this fraction as the coupling percentage of a given reaction.
As not all enzymes and metabolites in E. coli are present in high concentrations, not all predicted promiscuous reactions occur at appreciable levels to disrupt the metabolic network. In addition, the level of promiscuous activity is not constant across the network and is likely to vary from reaction to reaction. To address these issues, we performed a two-stage randomized probabilistic analysis. For each scenario, we assumed that only a portion of the predicted promiscuous reactions act on their target molecules. First, a random mean coupling percentage pmean was sampled from a normal distribution with μ = 1 % and σ = μ/3. The standard deviation was chosen in such way to place 99.7 % of the samples within a range strictly above 0 %, and in the remaining 0.3 % of the cases, the coupling percentage was resampled repeatedly to obtain a value above 0 %, still. Then, we randomly selected 10 % of the developed reactions to exhibit promiscuous activity. For each of the selected reactions, a coupling percentage was chosen randomly from another normal distribution with μ = pmean and σ = μ/3. The selected coupling percentage was then used to set the minimum required flux for the reaction according to one of the inequalities presented earlier in the methods section, while the lower and upper bounds of all other (non-selected) developed reactions were set to zero. Sampling the mean coupling percentage thus allowed us to vary the extent of the overall promiscuous activity, while sampling individual reaction-specific coupling percentages allowed each reaction to behave independently from any other. 10,000 FBA runs were performed, each time selecting a different set of promiscuous reactions and re-sampling the mean and individual reaction coupling percentages. In each run, the FBA objective was to maximize the yield of 3-HP while maintaining biomass growth of at least 10 % of wildtype. The analysis was also repeated assuming 25 % and 50 % of the developed reactions to have promiscuous activity. Additionally, another run was performed, where the total number of active reactions was fixed at 10 and each Scenario was either allocated the entire set (10) or half of it (5). This was done to account for the fact that Scenario 2 reactions were much more plentiful than Scenario 1 reactions. Avoiding overrepresentation of the former reactions enabled qualitative comparison of the two scenarios.
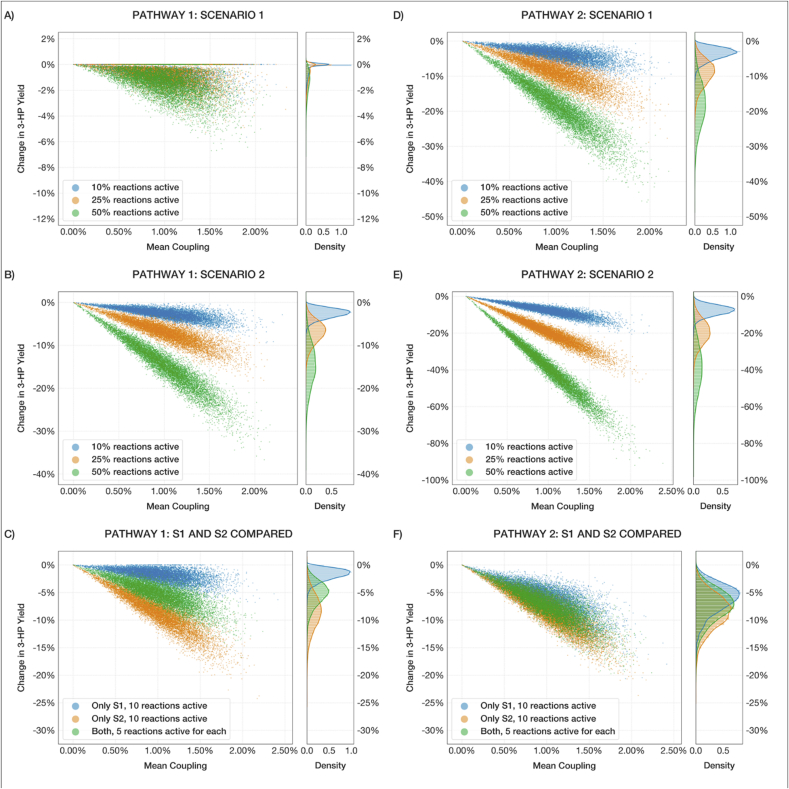
The results in Fig. 4 (A, B, C, and D) for both pathways and scenarios exhibit similar trends. Higher disruption tends to be correlated with more active reactions and with higher coupling percentages. As the mean coupling percentage increases, so does the disruption, and the relationship is almost linear in all but Pathway 1, Scenario 1 (Fig. 3A). The mean coupling percentage was sampled from a normal distribution and the resulting distribution of disruption values followed a very similar bell curve. The same effect occurs with an increase in the number of active reactions – note the mean of the distribution of disrupted yields shifts towards increased disruption with greater reaction activities. In fact, Scenario 2 has between 1.5- and 2-times the number of reactions compared to Scenario 1 (e.g., 34 vs. 17 for pathway 1 and 91 vs. 60 for pathway 2), which leads to a proportional difference in magnitude of disruption observed in the experiments as well, and when the total number of reactions is set to a fixed value as opposed to a percentage, the difference between the two Scenarios almost vanishes (Fig. 4F).
Fig. 4.
Comparison of simulation runs for the two pathways and both Scenarios, demonstrating the effect of mean coupling percentage and fraction of active promiscuous reactions (10, 25, 50 %) on the yield disruption of 3-HP. (A, D) Pathways 1 and 2 with only Scenario 1-type promiscuous interactions incorporated. (B, E) Pathways 1 and 2 with Scenario 2 interactions. (C, F) Comparison of Scenario 1 (S1) and 2 (S2) interactions for each pathway given a fixed total number of promiscuous reactions. The scatter plots document the results of each individual experiment, while the distribution plots on the right represent the probability density of obtaining a given disruption under the specified conditions.
Scenario 1 for Pathway 1 was an exception to this trend, however. In that instance, only ~17 % of predicted reactions had any effect on 3-HP yield disruption when measured individually – suggesting that only a small fraction of sampled reactions would have a disruptive impact at any given instant under those conditions. As a result, there is a disproportionately large number of instances where promiscuous reactions led to a low magnitude of disruption, particularly for low coupling percentages (Fig. 4A). The comparison of the two Scenarios for that Pathway further demonstrates the difference in behavior, with Scenario 1 being significantly less disruptive than Scenario 2 on average (Fig. 4C). This outcome highlights the downside of considering Scenario 1 alone: when few enzymes are being evaluated, the results are highly sensitive to the set of transformation patterns that can be derived from them – subject to knowledge limitations and enzymatic behaviors. Under more realistic biological circumstances, a more diverse range of enzymes participates in metabolism, allowing for more representative results. These observations imply that as promiscuous activity is intensified, it directly competes with 3-HP synthesis pathway flux causing significant disruption. However, such high activity of promiscuous reactions is unlikely under physiological settings, so these exercises likely overestimate the actual extent of disruption.
Overall, Pathway 1 was found to experience comparatively less disruption compared to pathway 2 under the same circumstances, possibly due to a lower number of enzymes, fewer intermediates, and/or lower promiscuous activity of MCR relative to PanD and GabT. In light of our analysis and considering the similar yield of Pathways 1 and 2, the design of Pathway 1 may be the preferred design option.
4. Discussion
In metabolic engineering, a number of strategies are employed to produce a target metabolite of interest, including the introduction of heterologous enzymes and selective overexpression and deletion of certain genes. After such interventions or combinations thereof, it is not uncommon for unexpected and/or undesirable metabolite product profiles to arise from various interactions between the introduced and native machinery in the host. Using MDFlow, it is possible to predict such interactions in the form of single- and multi-step pathways enabled by promiscuous enzymatic activity. The method utilizes PROXIMAL to construct reactions arising from promiscuity and relies on FBA to assess their impact on yield or biomass growth rate in pre- and post-modification hosts. The results for the single-gene deletions that were rescued via single- and multi-step enzymatic pathways, which were experimentally validated in prior studies, provide evidence of the ability of MDFlow in predicting metabolic network disruption. The results for the 3-HP case illustrate how MDFlow can help identify promiscuous interactions early in the design cycle. Further, our sampling-based FBA analysis shows that promiscuity can cause unexpected byproducts and results in yield disruption. Importantly, MDFlow can be used to explain byproducts often observed but not well explained in the literature.
Our Scenario 1 and 2 disruption classification has direct correspondence to the network inference classification proposed by Kim et al. (Kim and Copley, 2012). Interference is classified into three groups: those due to (i) heterologous metabolites in new pathways interfering with native metabolism, (ii) native metabolites interfering with a heterologous pathway, and (iii) heterologous pathway intermediates being diverted by promiscuous activity of native enzymes. MDFlow identifies the same interactions as long as they’re caused by enzyme substrate promiscuity – with groups (i) and (iii) corresponding to Scenario 2 predictions and group (ii) represented by Scenario 1-type interactions.
The computational methodology in MDFlow can be further enhanced. It currently uses PROXIMAL to predict promiscuous byproducts; it is possible to use alternatives for promiscuous product prediction. We selected PROXIMAL because we have established confidence in its capabilities in predicting organism-specific enzymatic transformations. Our earlier study of promiscuity using PROXIMAL on non-engineered E. coli allowed the discovery of 17 putative enzymatic reactions that explained metabolomics measurement (Yousofshahi et al., 2015). Regardless of the tool, however, there always remains the issue of false positives. In our work with PROXIMAL, we discarded byproducts that were not documented prior in PubChem, KEGG, or iML1515/1428. Using machine-learning tools that evaluate the likelihood of compound-enzyme interactions, such as EPP-HMCNF (Visani et al., 2020), might provide further confidence in such predictions. Importantly, better prediction of enzymatic products and their likelihood can improve the workflow’s ability to uncover unexpected interactions and evaluate their impact on the engineered host.
Metabolism simulation aspect of MDFlow also offers opportunities for improvement. In certain applications, MOMA can be an attractive alternative to FBA. While FBA utilizes linear programming to maximize an objective function, typically yield or biomass, MOMA approaches cell modeling from the perspective of minimizing redistribution of metabolic fluxes compared to the wildtype conditions. MOMA can provide improved correlation of predictions with experimental flux data over the steady-state modeling provided by FBA (Segrè et al., 2002). Additionally, the model itself can be tailored to the specific circumstances of an experiment. For example, isozymes that are minimally expressed in glucose M9 media were removed from the iML1515 model to create iML1428. As a result of this reduction in degrees of freedom, the derivative context-specific model tends to offer more accurate lethality predictions than iML1515 in gene knockout experiments (Monk et al., 2017). Such adjustments can be informed by manual investigation of the host’s metabolic network, by leveraging detailed kinetic models (Khodayari and Maranas, 2016), or via techniques such as 13C Metabolic Flux Analysis (Long and Antoniewicz, 2019). Of course, depending on the application, it may be helpful to consider using more complex techniques that can capture translation and regulation aspects of underground metabolism, though we believe – in the three cases we have considered – the overexpressed genes are the dominating factor. Knowing more about the biological sample, such as the concentrations of metabolites and enzymes can shed further light on the amount of disruption.
Another area of improvement is the approach used to determine the direction and maximum flux limits of the predicted reactions. We rely on the preset 1% average mean coupling percentage to estimate the limits of all reactions, which may not be representative of the higher or lower actual mean coupling percentage under the conditions of a given experiment. Future studies may re-evaluate directions and flux limits in the context of each experiment individually. Despite these limitations, the presented results are promising and call for further design exploration of the impact of enzyme promiscuity on engineered microorganisms.
5. Conclusion
We presented MDFlow, a method for quantitatively evaluating the side effects of engineered modifications on the host metabolic network resulting from enzyme promiscuity. Without mitigation, such side effects may lead to either unexpected behaviors during experiments or failure to take advantage of potentially beneficial interactions. By combining PROXIMAL and flux analysis in a streamlined workflow, MDFlow is capable of both discovering new interactions and evaluating their effects without the need for costly, time-consuming studies of in vivo experiments. MDFlow can be used at all stages of the metabolic engineering process. Prior to any design work, the method can be applied to expose native background promiscuous activity, revealing potentially interfering enzymes and metabolites that may be present but not documented in the model, resulting in building Extended Metabolic Models (EMM), as we demonstrated in prior works (Amin et al., 2019; Hassanpour et al., 2020). The workflow can then actively guide the pathway construction process by providing feedback on engineering decisions as they are being made, thus helping to identify modifications that create more robust strains. And finally, MDFlow may be leveraged to compare a fully designed pathway to other candidate pathways for the same target compound: when implemented, a pathway with less predicted metabolic disruption may have a higher yield. MDFlow is a first systematic automated analysis step towards the evaluation of underground metabolism and its interaction with engineered cellular machinery.
Author statement
Conceptualization: NUN, SH.
Methodology: NUN, SH, VP, SA, VEG, TN.
Software: VP, SA.
Investigation: VP, SA.
Data Curation: VP, SA, VEG, TN.
Funding acquisition: SH, NUN.
Writing - Original Draft: VP, SA.
Writing - Review & Editing: VP, SH, NUN, TN.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank the funding provided by NIH grant #DP2HD91798 (to N.U.N) and NSF grant #1909536 (to S.H. and N.U.N.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2021.e00170.
Contributor Information
Vladimir Porokhin, Email: vladimir.porokhin@tufts.edu.
Sara A. Amin, Email: sara.amin@tufts.edu, saraaamin@gmail.com.
Trevor B. Nicks, Email: trevor.nicks@tufts.edu.
Venkatesh Endalur Gopinarayanan, Email: venkatesh.endalur_gopinarayanan@tufts.edu.
Nikhil U. Nair, Email: nikhil.nair@tufts.edu.
Soha Hassoun, Email: soha.hassoun@tufts.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Summary of predicted promiscuous reactions for the multicopy suppressor study done by Patrick et al., categorized by the 8 rescue mechanisms proposed by the authors. For each gene knockout-suppressor pair, we provide the sets of reactions blocked by the deletion and reactions created by the overexpression of the suppressor gene, as well as all relevant growth rates.
Predicted two-step serendipitous pathways that may compensate for the deletion of pdxB in E. coli. Each pathway is listed in a separate numbered section, with illustrations of all involved metabolites. To view a specific step of a pathway, click on the “Step 1″ or “Step 2″ link under the section number. To look up a metabolite, template reaction, or an enzyme, click on the corresponding identifier. To view the eQuilibrator query used to compute thermodynamic feasibility for a given pathway, click on the ΔrG’° symbol.
Summary of predicted promiscuous reactions for two 3-HP synthesis pathways under Scenario 1 and Scenario 2. For each prediction, we list the enzyme and KEGG template reaction used to derive its biotransformation operator. All metabolites in the model are referred to by their iML1515 identifiers; external metabolites not considered to be a part of normal host metabolism are referenced by their KEGG ids or PubChem compound identifiers.
References
- Amin S.A. Towards creating an extended metabolic model (EMM) for E. coli using enzyme promiscuity prediction and metabolomics data. Microb. Cell Factories. 2019;18(1):109. doi: 10.1186/s12934-019-1156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 2008;10(6):305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Atsumi S., Hanai T., Liao J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Baba T. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campodonico M.A. Generation of an atlas for commodity chemical production in Escherichia coli and a novel pathway prediction algorithm, GEM-Path. Metab. Eng. 2014;25:140–158. doi: 10.1016/j.ymben.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Chen Y., Nielsen J. Advances in metabolic pathway and strain engineering paving the way for sustainable production of chemical building blocks. Curr. Opin. Biotechnol. 2013;24(6):965–972. doi: 10.1016/j.copbio.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Cheng Z. Enhanced production of 3-hydroxypropionic acid from glucose via malonyl-CoA pathway by engineered Escherichia coli. Bioresour. Technol. 2016;200:897–904. doi: 10.1016/j.biortech.2015.10.107. [DOI] [PubMed] [Google Scholar]
- Cho A. Prediction of novel synthetic pathways for the production of desired chemicals. BMC Syst. Biol. 2010;4(1):35. doi: 10.1186/1752-0509-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ari R., Casadesús J. Underground metabolism. Bioessays. 1998;20(2):181–186. doi: 10.1002/(SICI)1521-1878(199802)20:2<181::AID-BIES10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Dahlgren B. ChemPy: a package useful for chemistry written in Python. J. Open Source Software. 2018;3(24):565. [Google Scholar]
- Della Pina C., Falletta E., Rossi M. A green approach to chemical building blocks. The case of 3-hydroxypropanoic acid. Green Chem. 2011;13(7):1624. [Google Scholar]
- Djoumbou-Feunang Y. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J. Cheminf. 2019;11(1):2. doi: 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim A. COBRApy: COnstraints-based reconstruction and analysis for Python. BMC Syst. Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley S.D., Broadbelt L.J., Hatzimanikatis V. Computational framework for predictive biodegradation. Biotechnol. Bioeng. 2009;104(6):1086–1097. doi: 10.1002/bit.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamholz A. eQuilibrator--the biochemical thermodynamics calculator. Nucleic Acids Res. 2011;(D1):D770–D775. doi: 10.1093/nar/gkr874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H., Nagata S., Misono H. Characterization of short-chain dehydrogenase/reductase homologues of Escherichia coli (YdfG) and Saccharomyces cerevisiae (YMR226C) Biochim. Biophys. Acta Protein Proteonomics. 2003;1645(1):89–94. doi: 10.1016/s1570-9639(02)00533-2. [DOI] [PubMed] [Google Scholar]
- George K.W. Biotechnology of Isoprenoids. Springer; 2015. Isoprenoid drugs, biofuels, and chemicals—artemisinin, farnesene, and beyond; pp. 355–389. [DOI] [PubMed] [Google Scholar]
- Hadadi N. ATLAS of biochemistry: a repository of all possible biochemical reactions for synthetic biology and metabolic engineering studies. ACS Synth. Biol. 2016;5(10):1155–1166. doi: 10.1021/acssynbio.6b00054. [DOI] [PubMed] [Google Scholar]
- Hassanpour N. Biological filtering and substrate promiscuity prediction for annotating untargeted metabolomics. Metabolites. 2020;10(4):160. doi: 10.3390/metabo10040160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller S.R. InChI, the IUPAC international chemical identifier. J. Cheminf. 2015;7:23. doi: 10.1186/s13321-015-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Co-production of 3-hydroxypropionic acid and 1, 3-propanediol by Klebseilla pneumoniae expressing aldH under microaerobic conditions. Bioresour. Technol. 2013;128:505–512. doi: 10.1016/j.biortech.2012.10.143. [DOI] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9(3):90–95. [Google Scholar]
- Inui M. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl. Microbiol. Biotechnol. 2008;77(6):1305–1316. doi: 10.1007/s00253-007-1257-5. [DOI] [PubMed] [Google Scholar]
- Jeffryes J.G. MINEs: open access databases of computationally predicted enzyme promiscuity products for untargeted metabolomics. J. Cheminf. 2015;7 doi: 10.1186/s13321-015-0087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen H. Google Patents; 2015. Compositions and Methods for 3-hydroxypropionic Acid Production. [Google Scholar]
- Jiang X., Meng X., Xian M. Biosynthetic pathways for 3-hydroxypropionic acid production. Appl. Microbiol. Biotechnol. 2009;82(6):995–1003. doi: 10.1007/s00253-009-1898-7. [DOI] [PubMed] [Google Scholar]
- Khersonsky O., Roodveldt C., Tawfik D.S. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 2006;10(5):498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Khodayari A., Maranas C.D. A genome-scale Escherichia coli kinetic metabolic model k-ecoli457 satisfying flux data for multiple mutant strains. Nat. Commun. 2016;7(1) doi: 10.1038/ncomms13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Copley S.D. Inhibitory cross-talk upon introduction of a new metabolic pathway into an existing metabolic network. Proc. Natl. Acad. Sci. Unit. States Am. 2012;109(42):E2856–E2864. doi: 10.1073/pnas.1208509109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5’-phosphate synthesis. Mol. Syst. Biol. 2010;6:436. doi: 10.1038/msb.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Enhanced production of 3-hydroxypropionic acid from glycerol by modulation of glycerol metabolism in recombinant Escherichia coli. Bioresour. Technol. 2014;156:170–175. doi: 10.1016/j.biortech.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Klem T.J., Davisson V.J. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry. 1993;32(19):5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- Kumar V., Ashok S., Park S. Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol. Adv. 2013;31(6):945–961. doi: 10.1016/j.biotechadv.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Lee S.K. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 2008;19(6):556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Lee S.Y. Metabolic engineering of microorganisms: general strategies and drug production. Drug Discov. Today. 2009;14(1–2):78–88. doi: 10.1016/j.drudis.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Li C. Computational discovery of biochemical routes to specialty chemicals. Chem. Eng. Sci. 2004;59(22–23):5051–5060. [Google Scholar]
- Li L. MyCompoundID: using an evidence-based metabolome library for metabolite identification. Anal. Chem. 2013;85(6):3401–3408. doi: 10.1021/ac400099b. [DOI] [PubMed] [Google Scholar]
- Long C.P., Antoniewicz M.R. High-resolution 13C metabolic flux analysis. Nat. Protoc. 2019;14(10):2856–2877. doi: 10.1038/s41596-019-0204-0. [DOI] [PubMed] [Google Scholar]
- Madison L.L., Huisman G.W. Metabolic engineering of poly (3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 1999;63(1):21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk J.M. iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 2017;35:904–908. doi: 10.1038/nbt.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y. PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 2010;38(Suppl 2):W138–W143. doi: 10.1093/nar/gkq318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura C.E., Whited G.M. Metabolic engineering for the microbial production of 1, 3-propanediol. Curr. Opin. Biotechnol. 2003;14(5):454–459. doi: 10.1016/j.copbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Nielsen D.R. Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 2009;11(4–5):262–273. doi: 10.1016/j.ymben.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Nobeli I., Favia A.D., Thornton J.M. Protein promiscuity and its implications for biotechnology. Nat. Biotechnol. 2009;27(2):157. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- Noor E. An integrated open framework for thermodynamics of reactions that combines accuracy and coverage. Bioinformatics. 2012;28(15):2037–2044. doi: 10.1093/bioinformatics/bts317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor E. Consistent estimation of Gibbs energy using component contributions. PLoS Comput. Biol. 2013;9(7) doi: 10.1371/journal.pcbi.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor E. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput. Biol. 2014;10(2) doi: 10.1371/journal.pcbi.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick W.M. Multicopy suppression underpins metabolic evolvability. Mol. Biol. Evol. 2007;24(12):2716–2722. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj S.M. Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 strain. Process Biochem. 2008;43(12):1440–1446. [Google Scholar]
- Rathnasingh C. Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J. Biotechnol. 2012;157(4):633–640. doi: 10.1016/j.jbiotec.2011.06.008. [DOI] [PubMed] [Google Scholar]
- RDKit Open-source cheminformatics. http://www.rdkit.org Available from:
- Schellenberger J. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 2011;6(9):1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrè D., Vitkup D., Church G.M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. U. S. A. 2002;99(23):15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik O.K., Dan S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Trantas E.A. When plants produce not enough or at all: metabolic engineering of flavonoids in microbial hosts. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh C.T. Redesigning Escherichia coli metabolism for anaerobic production of isobutanol. Appl. Environ. Microbiol. 2011;77(14):4894–4904. doi: 10.1128/AEM.00382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visani G.M., Hughes M.C., Hassoun S. 2020. Hierarchical Classification of Enzyme Promiscuity Using Positive, Unlabeled, and Hard Negative Examples. arXiv: 2002.07327 [q-bio.CB] [Google Scholar]
- Wang Q. Metabolic engineering of Escherichia coli for poly(3-hydroxypropionate) production from glycerol and glucose. Biotechnol. Lett. 2014;36(11):2257–2262. doi: 10.1007/s10529-014-1600-8. [DOI] [PubMed] [Google Scholar]
- Waskom M.e. 2020. A. Seaborn: Statistical Data Visualization. [DOI] [Google Scholar]
- Whalen W.A., Berg C.M. Analysis of an avtA::Mu d1(Ap lac) mutant: metabolic role of transaminase C. J. Bacteriol. 1982;150(2):739–746. doi: 10.1128/jb.150.2.739-746.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q.Z., Liu J., Walsh C.T. p-Aminobenzoate synthesis in Escherichia coli: purification and characterization of PabB as aminodeoxychorismate synthase and enzyme X as aminodeoxychorismate lyase. Proc. Natl. Acad. Sci. U. S. A. 1990;87(23):9391–9395. doi: 10.1073/pnas.87.23.9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikuni Y. Redesigning enzymes based on adaptive evolution for optimal function in synthetic metabolic pathways. Chem. Biol. 2008;15(6):607–618. doi: 10.1016/j.chembiol.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousofshahi M. PROXIMAL: a method for prediction of xenobiotic metabolism. BMC Syst. Biol. 2015;9(1):94. doi: 10.1186/s12918-015-0241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of predicted promiscuous reactions for the multicopy suppressor study done by Patrick et al., categorized by the 8 rescue mechanisms proposed by the authors. For each gene knockout-suppressor pair, we provide the sets of reactions blocked by the deletion and reactions created by the overexpression of the suppressor gene, as well as all relevant growth rates.
Predicted two-step serendipitous pathways that may compensate for the deletion of pdxB in E. coli. Each pathway is listed in a separate numbered section, with illustrations of all involved metabolites. To view a specific step of a pathway, click on the “Step 1″ or “Step 2″ link under the section number. To look up a metabolite, template reaction, or an enzyme, click on the corresponding identifier. To view the eQuilibrator query used to compute thermodynamic feasibility for a given pathway, click on the ΔrG’° symbol.
Summary of predicted promiscuous reactions for two 3-HP synthesis pathways under Scenario 1 and Scenario 2. For each prediction, we list the enzyme and KEGG template reaction used to derive its biotransformation operator. All metabolites in the model are referred to by their iML1515 identifiers; external metabolites not considered to be a part of normal host metabolism are referenced by their KEGG ids or PubChem compound identifiers.