Abstract
Vascular calcification is the ectopic deposition of calcium hydroxyapatite minerals in arterial wall, which involves the transdifferentiation of vascular smooth muscle cells (VSMCs) toward an osteogenic phenotype. However, the underlying molecular mechanisms regulating the VSMC osteogenic switch remain incompletely understood. In this study, we examined the roles of microRNAs (miRNAs) in vascular calcification. miRNA-seq transcriptome analysis identified miR-223-3p as a candidate miRNA in calcified mouse aortas. MiR-223-3p knockout aggravated calcification in both medial and atherosclerotic vascular calcification models. Further, RNA-seq transcriptome analysis verified JAK-STAT and PPAR signaling pathways were upregulated in both medial and atherosclerotic calcified aortas. Overlapping genes in these signaling pathways with predicted target genes of miR-223-3p derived from miRNA databases, we identified signal transducer and activator of transcription 3 (STAT3) as a potential target gene of miR-223-3p in vascular calcification. In vitro experiments showed that miR-223-3p blocked interleukin-6 (IL-6)/STAT3 signaling, thereby preventing the osteogenic switch and calcification of VSMCs. In contrast, overexpression of STAT3 diminished the effect of miR-223-3p. Taken together, the results indicate a protective role of miR-223-3p that inhibits both medial and atherosclerotic vascular calcification by regulating IL-6/STAT3 signaling-mediated VSMC transdifferentiation.
Keywords: vascular smooth muscle cell, interleukin-6 (IL-6), signal transducer and activator of transcription 3 (STAT3), osteogenic phenotype, vascular calcification, miR-223-3p, epigenetics
Abbreviations: BUN, blood urea nitrogen; CKD, chronic kidney disease; DKO, double knockout; DMEM, Dulbecco’s modified Eagle’s medium; FBS, fetal bovine serum; IL-6, interleukin-6; JAK, Janus kinase; KEGG, Kyoto Encyclopedia of Genes and Genomes; KO, knockout; OIM, osteoblast-induced medium; PPAR, peroxisome proliferator-activated receptor; STAT3, signal transducer and activator of transcription 3; VSMC, vascular smooth muscle cell
Vascular calcification is a characteristic pathological change in atherosclerosis, hypertension, diabetes and is also an independent risk factor for all-cause mortality in chronic kidney disease (CKD) (1). Vascular calcification mainly manifests as increased blood vessel wall stiffness and decreased compliance, which in turn lead to cardiovascular events (2). With the increasing prevalence of coronary heart disease, diabetes, and CKD in the aging population, the harmfulness of vascular calcification has become increasingly obvious (3). Unfortunately, the pathological mechanism of vascular calcification is currently poorly understood, and there is a lack of effective clinical treatments. Therefore, further investigation of the molecular mechanisms regulating vascular calcification could have great significance, enabling the discovery of novel therapeutic targets for effective clinical prevention and treatment strategies.
Vascular calcification is a regulated osteogenic process in arterial tissue, which is similar to bone development (4). Although different mechanisms are involved in medial and atherosclerotic calcification, the osteogenic transformation of vascular smooth muscle cells (VSMCs) is a common characteristic pathological process (5, 6). In response to cytokines, oxidized lipids, or high phosphorus levels, VSMCs express osteoblast-specific transcription factors (e.g., Runx2, Osterix) (7, 8), and differentiation markers (e.g., Osteopontin, Osteocalcin and Alp) (8). Osteogenic switch of VSMC accelerates calcium- and phosphorus-rich matrix vesicles releasing and apoptotic cells (9) depositing in the remodeled extracellular matrix, leading to the formation of vascular calcifications. However, the mechanism regulating the process of osteogenic switch remains to be elucidated.
MicroRNAs (miRNAs) are short (20–25 nucleotides) single-stranded RNAs that regulate the translational processing of their target mRNAs. MiRNAs regulate several key checkpoints in the cellular processes involved in vascular calcification, such as osteoblast differentiation (miR-125b (10), miR-205 (11), miR-223-3p (12)) and regulation of calcium and phosphate homeostasis (miR-221 (13), miR-9 (14)). However, our understanding of the roles of miRNAs in vascular calcification remains incomplete.
In this study, miRNA transcriptome analysis of CKD-induced calcified mouse aortas revealed that miR-223-3p is among the most upregulated microRNAs. It’s reported that miR-223-3p is downregulated in patients with CKD stage 4 and 5 (15), and miR-223-3p regulates osteogenic differentiation (12) and osteoclastogenesis (16). However, the role and mechanism of miR-223-3p in vascular calcification are unknown. Our study showed thatmiR-223-3p knockout (KO) accelerated calcification in mouse models of both medial and endothelial vascular calcification. Mechanistically, we found that the effects of miR-223-3p were linked to its target gene, gene signal transducer, and activator of transcription 3 (STAT3). MiR-223-3p inhibited interleukin-6 (IL-6)/STAT3 signaling, which is required for the induction of VSMC osteoblastic differentiation and vascular calcification.
Results
MiR-223-3p is upregulated in CKD-induced vascular calcification
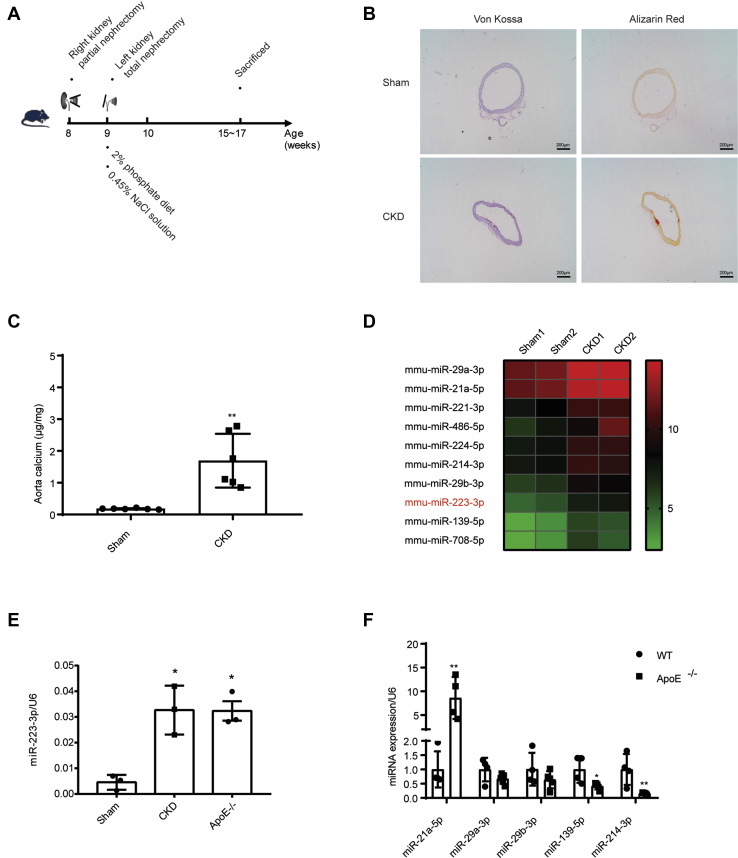
To identify novel miRNAs involved in vascular calcification, mice were subjected to a modified two-step surgical model of subtotal nephrectomy, involving the removal of 5/6 of the kidney tissue (17, 18, 19, 20), to induce medial vascular calcification (Fig. 1A). Aortic calcification (Fig. 1B) and aortic calcium levels (Fig. 1C) were detected 6 weeks after nephrectomy, confirming formed vascular calcification. MiRNA levels in calcified aortas from CKD group and control aortas from sham group were analyzed by miRNA sequencing. When sorted based on their fold change (FC) values, the top ten upregulated miRNAs include several miRNAs that have been reported to be involved in vascular calcification, such as miR-29a/b (21), miR-214 (22), miR-139 (23), and miR-223-3p (24) (Fig. 1D, Table S1). Among them, miR-223-3p drew our attention, as it is also significantly increased in the plasma of patients with coronary artery calcifications (25). RT-PCR analysis confirmed the upregulation of miR-223-3p in medial calcification (Fig. 1E). Meanwhile, miR-223-3p was also upregulated in the aortas of aged ApoE KO mice, which displayed significant atherosclerotic calcification (Figs. 1E and S1). These results suggest a correlation between miR-223-3p expression and vascular calcification. We further analyzed other up-regulated miRNA in medial calcification, and found miR-21a-5p increased while other miRNA decreased or showed no difference between WT and aged ApoE−/− mice. These data suggest medial calcification and atherosclerotic calcification may have different miRNA expression profile.
Figure 1.
MiR-223-3p is upregulated in the calcified aortas.A, experimental scheme of CKD-induced medial calcification. B, representative Alizarin Red S and Von Kossa staining of aortas in sham and CKD groups 6 weeks after nephrectomy. C, aortic calcium levels after induction of medial calcification. The calcium content was normalized to the weight of the aorta (n = 6–7 per group). D, heat map of the ten most upregulated miRNAs in the aortas of CKD mice. Data points represent the log2 TPM (Transcripts Per Million) mapped values of the indicated genes. E, MiR-223-3p expression in the aortas of sham, CKD and aged ApoE knockout mice (n = 3 per group). F, expression of miR-21a-5p, miR-29a-3p, miR-29b-3p, miR-139-5p and miR-214-3p in the aortas of aged ApoE knockout mice (n = 3 per group). Data are expressed as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01 by unpaired two-tailed Student's t-test.
MiR-223-3p deficiency aggravates both medial and atherosclerotic calcification
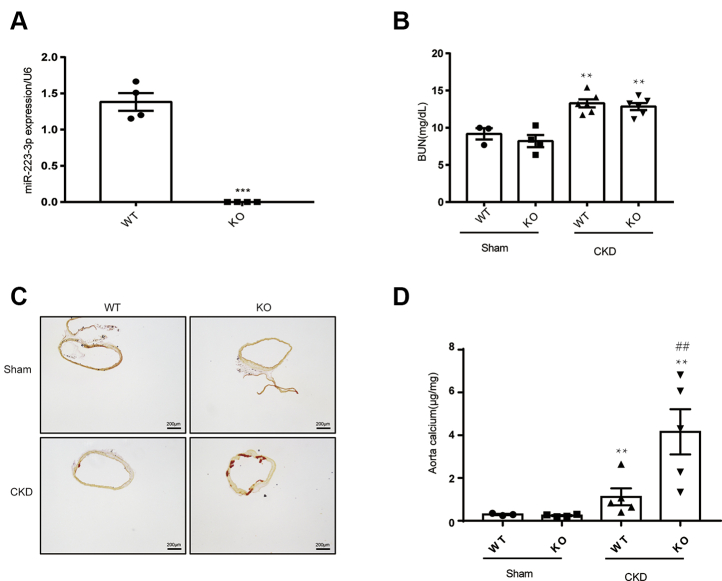
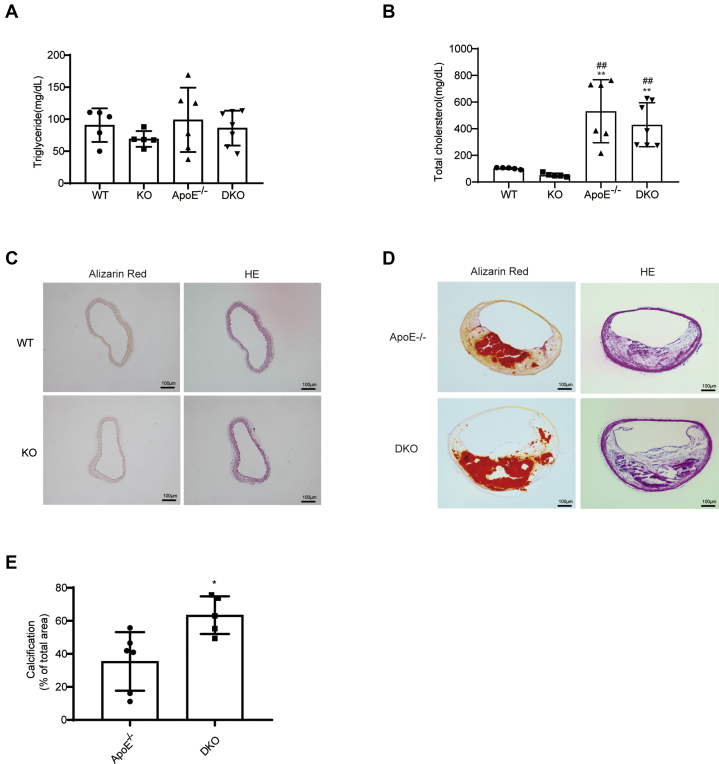
We next sought to determine whether miR-223-3p plays a role in the development of vascular calcification. In this study, miR-223-3p knockout mice which have been published before in our research group (26) were used. MiR-223-3p deficiency in miR-223-3p KO mice was confirmed by RT-PCR analysis of spleen tissue, which should contain high levels of miR-223-3p (Fig. 2A). Increased blood urea nitrogen (BUN) concentration was detected after CKD surgery, while there was no difference between WT and KO mice (Fig. 2B). Six weeks after nephrectomy, KO mice had increased calcium deposition compared with WT mice, shown by both Alizarin Red S staining (Fig. 2C) and aortic calcium content analysis (Fig. 2D). Next, to clarify the role of miR-223-3p in atherosclerotic calcification, we crossbred ApoE KO (ApoE−/−) mice with miR-223-3p KO mice to generate miR-223-3p/ApoE double knockout mice (DKO). ApoE−/− and DKO mice were fed a chow diet for up to 16 months to generate aging-induced atherosclerotic calcification. WT and miR-223 KO mice in chow diet were used as control. MiR-223-3p deficiency did not affect the plasma triglyceride (Fig. 3A) or cholesterol (Fig. 3B) levels. However, DKO mice displayed aggravated atherosclerosis (Fig. S2) and calcification (Fig. 3, D and E) in the brachiocephalic artery (BCA) compared with ApoE−/− mice. No calcification was detected in WT or KO mice (Fig. 3C). These results indicate that the loss of miR-223-3p increases both medial and atherosclerotic calcification.
Figure 2.
MiR-223-3p KO promotes CKD-induced medial calcification.A, RT-PCR verification of miR-223-3p KO in the spleens of WT and KO mice (n = 4 per group). B, BUN concentrations in WT and KO mice with or without CKD (n = 3–6 per group). C, representative Alizarin Red S staining of the thoracic aortas of WT and KO mice after 6 weeks with or without CKD. D, calcium content of the thoracic aortas of WT and KO mice after 8 weeks with or without CKD (n = 3–5 per group). ∗p <0.05 by unpaired two-tailed Student’s t-test.
Figure 3.
MiR-223-3p KO promotes atherosclerotic calcification.A, plasma triglyceride and B, total cholesterol concentrations of WT, KO, aged ApoE KO and aged DKO mice (n = 6–7 per group). C, representative Alizarin Red S staining and HE staining in the brachiocephalic artery (BCA) of WT and KO groups. D and E, representative Alizarin Red S staining, HE staining (D) and quantification (E) in the BCA of aged ApoE KO and DKO mice (n = 6–7 per group). Data are expressed as the mean ± SD. ∗p <0.05 by unpaired two-tailed Student’s t-test.
The Janus kinase (JAK)/STAT and peroxisome proliferator-activated receptor (PPAR) signaling pathways are upregulated in both medial and atherosclerotic calcification
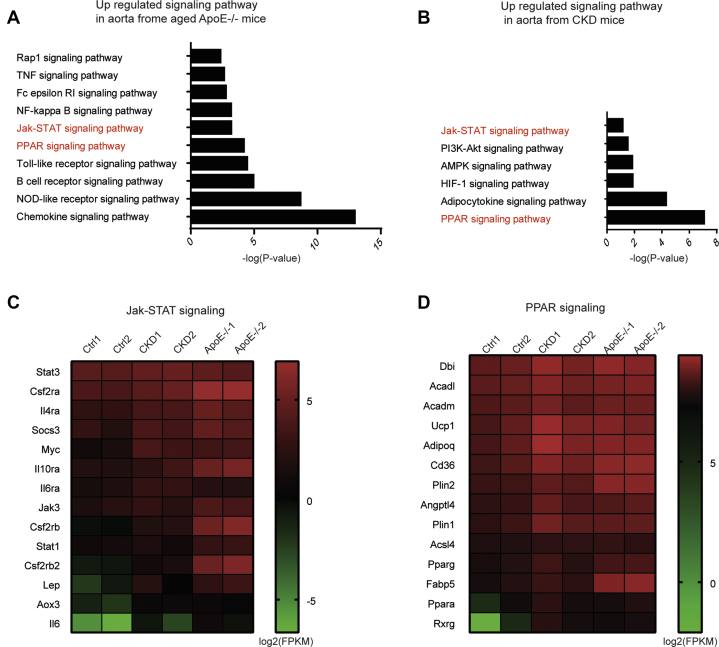
As miR-223-3p deficiency aggravated both medial and atherosclerotic calcification, we hypothesized that miR-223-3p interrupts a signaling pathway common to both forms of calcification. RNA-Seq was applied to identify differentially regulated signaling pathways in the aortas of CKD, aged ApoE−/−, and control mice. We performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and ranked the upregulated signaling pathways in CKD (Fig. 4A) and aged ApoE−/− (Fig. 4B) mice compared with control mice. The JAK/STAT and PPAR signaling pathways were upregulated and overlapped in medial and atherosclerotic calcification (Fig. 4, C and D). Both signaling pathways have been reported relating to vascular calcification. N-3 fatty acids directly inhibit vascular calcification via the p38 mitogen-activated protein kinase and PPARγ pathways (27), and PPARγ counteracts vascular calcification in VSMCs by inhibiting Wnt5a signaling (28). IL-6, a cytokine that activates JAK/STAT signaling, is identified as a new susceptibility gene underlying calcific aortic valve stenosis (29). What’s more, IL-6 induces the STAT3-dependent differentiation of human VSMCs into osteoblast-like cells (30). While which signaling is involved in miR-223-3p mediated protection effect in vascular calcification is unknown.
Figure 4.
The JAK/STAT and PPAR signaling pathways are upregulated in both medial and atherosclerotic calcification.A and B, upregulated KEGG signaling pathways in CKD (A) and aged ApoE KO (B) mice. C and D, heat maps of selected upregulated genes in the JAK/STAT and PPAR signaling pathways in (C) CKD and (D) aged ApoE KO mice. Each point represents the log2 FPKM (fragments per kilobase per million) mapped fragments value of the indicated gene.
MiR-223-3p deficiency doesn’t affect IL-6 level in plasma and aorta
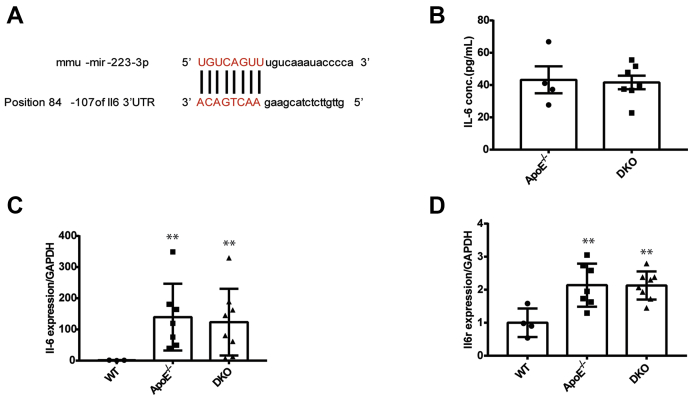
We next screened all upregulated genes involved in the PPAR and JAK/STAT signaling pathways using the MiRTarBase and TargetScan databases. In the JAK/STAT pathway, miR-223-3p-binding sites were predicted in the 3′ UTRs of IL-6 and STAT3 (Figs. 5A and 6A), and no gene in the PPAR pathway was predicted. IL-6-induced inflammation initiates and increases vascular calcification, and previous studies have reported that miR-223-3p inhibits proinflammatory responses in the liver and injured skeletal muscle by directly targeting IL-6 (26, 31). So we detected plasma IL-6 in aged ApoE−/− and DKO mice by ELISA. However, no differences in plasma IL-6 levels were detected between aged ApoE−/− and DKO mice (Fig. 5B). RT-PCR also showed no differences in IL-6 (Fig. 5C) or IL-6 receptor (IL-6R; Fig. 5D) expression between these two groups. These results suggest that IL-6 is probably not the target gene of miR-223-3p during vascular calcification progression.
Figure 5.
MiR-223-3p does not affect IL-6 expression.A, MiR-223-3p target sites (in red letters) in the IL-6 3′ UTRs. B, plasma IL-6 concentrations in aged ApoE−/− and DKO mice were analyzed by ELISA (n = 4–7 per group). C and D, IL-6 (C) and IL-6R (D) expression in the aortas of WT, ApoE KO, and DKO mice (n = 3–8 per group). Data are expressed as the mean ± SD. ∗p <0.05 by unpaired two-tailed Student’s t-test or one-way analysis of variance.
Figure 6.
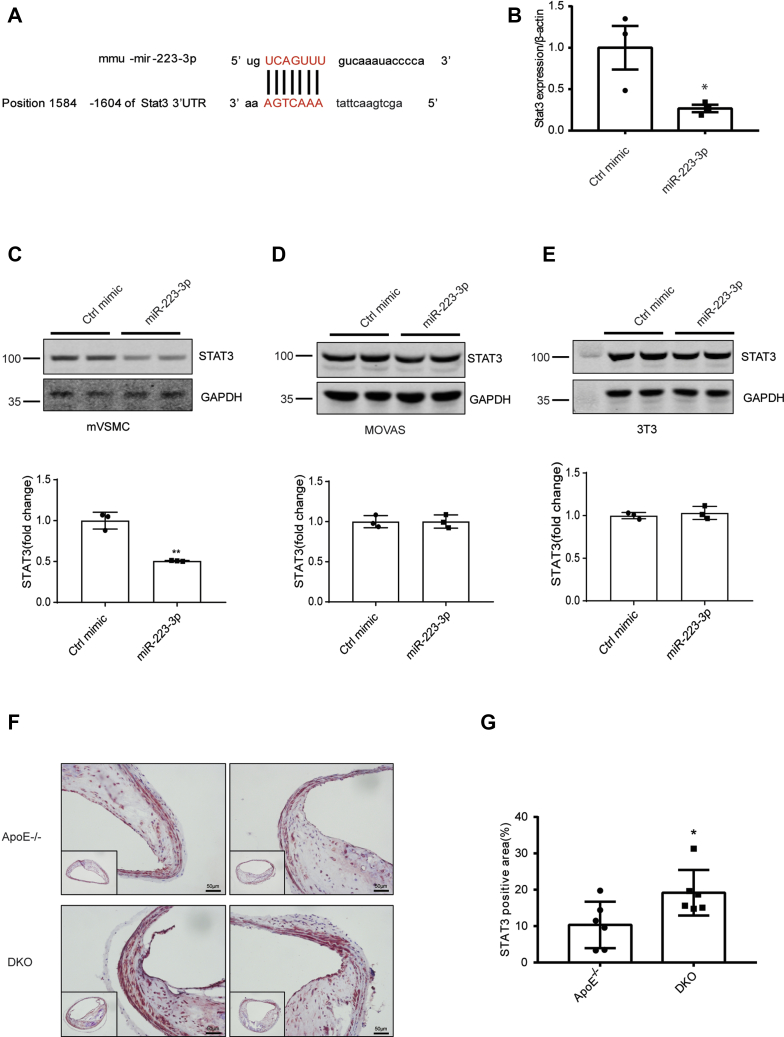
STAT3 is a potential miR-223-3p target in vascular calcification.A, MiR-223-3p target sites (in red letters) in the STAT3 3′ UTRs. B, STAT3 expression in primary vascular smooth muscle cells (mVSMCs) transfected with miR-223-3p or a control mimic (n = 3 per group). C–E, representative western blot and quantification of STAT3 expression in mVSMCs (C), MOVAS (D), and 3T3 cells (E) transfected with miR-223-3p or a control mimic (n = 3 per group). F–G, Representative images (E) and quantification (F) of STAT3 IHC in the brachiocephalic trunks of aged ApoE KO and DKO mice (n = 6 per group). Data are expressed as the mean ± SD. ∗p <0.05 by unpaired two-tailed Student’s t-test.
STAT3 is a target gene of miR-223-3p during vascular calcification
We next focused on STAT3, another known target gene of miR-223-3p (Fig. 6A). By directly targeting STAT3, miR-223-3p regulates the number and function of myeloid-derived suppressor cells in multiple sclerosis (32). In addition, IL-6 and soluble (s)IL-6R induce the STAT3-dependent differentiation of human VSMCs into osteoblast-like cells (30). As there was no difference in IL-6 expression between ApoE−/− and DKO mice, we next hypothesized that miR-223-3p inhibited vascular calcification by blocking IL-6-induced STAT3 signaling in VSMCs. VSMCs transfected with miR-223-3p mimics showed decreased level of STAT3 compared with VSMCs transfected with negative control mimics (Figs. 6B, and S3A). Western blotting confirmed that STAT3 was downregulated in VSMCs transfected with miR-223-3p compared with those transfected with control mimics; while miR-223-3p did not affect STAT3 expression in MOVAS (mouse aortic smooth muscle cells) or 3T3 fibroblasts (Figs. 6, C and D, S3, B and C), suggesting a cellular specificity of miR-223-3p. To determine whether STAT3 was involved in the aggravated vascular calcification observed in DKO mice, immunohistochemistry (IHC) was performed. Compared with ApoE−/− group, more STAT3 were detected in BCAs from DKO group (Fig. 6, F and G). Taken together, these results suggest that STAT3 is a target gene of miR-223-3p in VSMCs during vascular calcification.
MiR-223-3p blocks IL-6/sIL-6R-induced STAT3 and VSMC calcification
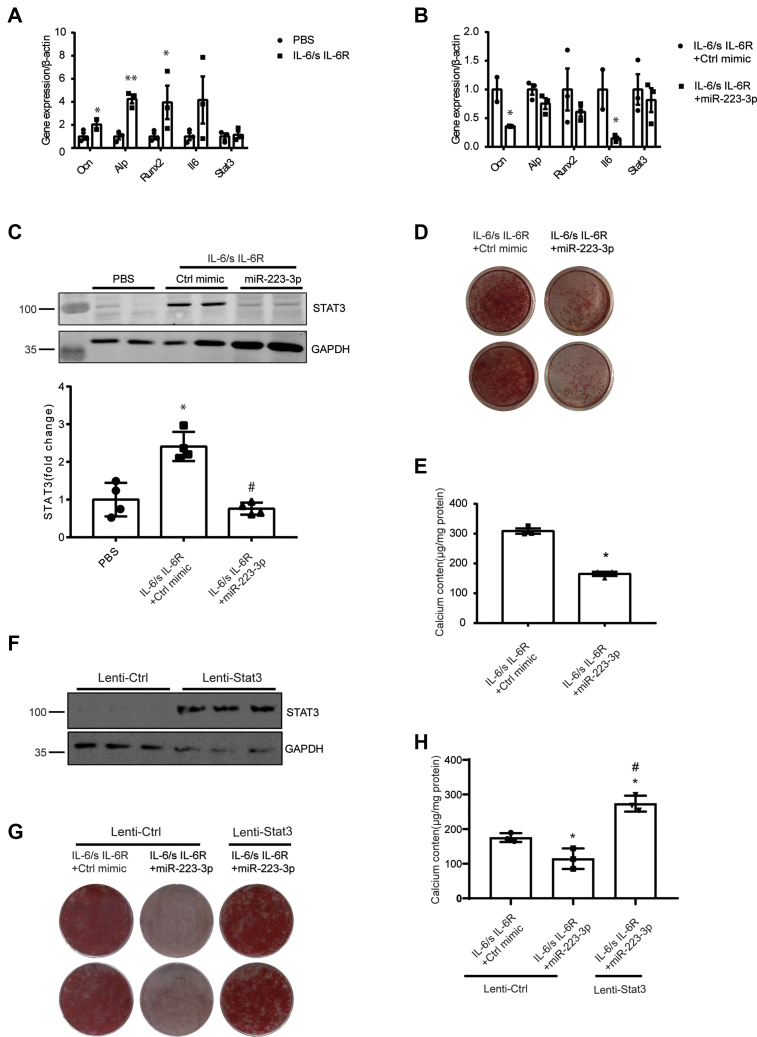
To identify whether miR-223-3p inhibited calcification by targeting STAT3, we used IL-6/sIL-6R to induce STAT3 expression in VSMCs cultured with osteoblast-induced medium (OIM). Consistent with previous studies, IL-6/sIL-6R enhanced the mRNA expression of alkaline phosphatase (Alp), osteocalcin (Ocn), and Runx2, which are osteoblast-specific genes in VSMCs (Fig. 7A). Overexpression of miR-223-3p in VSMCs inhibited IL-6/sIL-6R complex-induced osteogenic gene expression (Fig. 7B). Accordingly, the STAT3 protein level significantly increased when VSMCs were cultured in OIM, and this was blocked by miR-223-3p mimics (Fig. 7C). Accordingly, the calcium deposition and cellular calcium content were reduced in miR-223-3p-transfected VSMCs (Fig. 7, D and E). Knockdown STAT3 using siRNA also significantly decreased calcium accumulation in IL-6/s IL6R-induced VSCM calcification (Fig. S4). While overexpression of STAT3 (Fig. 7F) eliminated the effect of miR-223-3p during VSMC calcification (Fig. 7, G and H). These results show that miR-223-3p inhibits IL-6/STAT3-induced VSMC calcification by blocking STAT3 expression.
Figure 7.
IL-6-induced VSMC calcification is blocked by miR-223-3p.A, RT-PCR analysis of osteogenic genes in VSMCs. VSMCs were cultured in osteogenic media (OIM) with 50 ng/ml mIL-6/sIL-6R complex or PBS for 10 days (n = 3). B, effects of miR-223-3p on osteogenic gene expression in VSMCs. VSMCs were transfected with 100 nM miR-223-3p or a control mimic and cultured in osteogenic medium (n = 3). C, representative western blot and quantification of STAT3 expression in VSMCs cultured as in B for 14 days (n = 4 per group). D, representative images of Alizarin Red S staining of VSMCs cultured as in B for 21 days. E, total calcium content in VSMCs cultured as in B for 14 days (n = 3). F, representative western blot of STAT3 in VSMCs transfected with vector (Lenti-Ctrl) or lentivirus packaged with STAT3(Lenti-Stat3). G, representative images of ARS staining of VSMCs. VSMCs transfected with Lenti-Ctrl or Lenti-Stat3 were cultured in osteogenic media with 50 ng/ml mIL-6/sIL-6R complex for 21 days, and miR-223-3p or a control mimic(100 nM) was transfected in indicated wells. H, total calcium content in VSMCs cultured as in G for 14 days (n = 3). Data are expressed as the mean ± SD. ∗p <0.05, ∗∗p <0.01 compared with the first group, #p <0.05 compared with the second group. Two groups were assessed by unpaired two-tailed Student's t-test. For comparisons of more than two groups, one-way analysis of variance was used.
Taken together, our data suggest that upregulated expression of miR-223-3p during medial and atherosclerotic calcification negatively regulates VSMC calcification by targeting STAT3.
Discussion
Our data demonstrate that miR-223-3p plays an important role in regulating vascular calcification. MiR-223-3p deficiency aggravates both medial and atherosclerotic calcification. Mechanistically, miR-223-3p negatively regulates calcification by targeting IL-6/STAT3 signaling in VSMCs.
MiR-223-3p is mainly derived from myeloid cells (33) and regulates hematopoietic differentiation (34), human embryonic stem cell differentiation (35). MiR-223-3p is delivered by exosomes to recipient cells such as endothelial cells (36) and VSMCs (37), where it participates in cardiovascular diseases such as atherosclerosis (38). Our data show that miR-223-3p expression increases in medial and atherosclerotic calcified aortas. Consistently, previous studies have reported increased miR-223-3p levels in atherosclerosis (37) and advanced stages of CKD (39). Since inflammation is prevalent in the pathophysiology of atherosclerosis and CKD (40), increased miR-223 may due to accumulated immune cells.
VMSC osteogenic switch occurs in both medial and atherosclerotic vascular calcification (41). RNA-Seq analysis revealed that the JAK/STAT and PPAR signaling pathways are involved in both medial and atherosclerotic calcification. This suggests that targeting these pathways may inhibit both calcification types. In the JAK/STAT pathway, IL-6 and STAT3 are reported to be direct targets of miR-223-3p. IL-6/STAT3 signaling plays important roles in the regulation of inflammation, differentiation, and immunity (42). Previous studies have shown that miR-223-3p binding to the 3′ UTR of IL-6 can inhibit inflammation during alcoholic liver injury (31), pulmonary tuberculosis (43), and skeletal muscle regeneration (26). However, we did not detect any differences of IL-6 from plasma and the aortas of ApoE−/− and DKO mice. These results suggest that IL-6 may not be involved in miR-223-3p-induced vascular calcification.
The downstream transcriptional factor of IL-6, STAT3, is also a direct target of miR-223-3p (32). A previous study showed that IL-6 and sIL-6R induce the STAT3-dependent differentiation of human VSMCs into osteoblast-like cells, and knockdown STAT3 blocks IL-6/s IL-6R-induced calcification (30). Accordingly, our data also showed that knockdown STAT3 by siRNA decreased IL-6/s IL6R-induced mouse VSMCs calcification, suggesting that IL-6/STAT3 pathway plays an important role in VSMC calcification. IHC staining of calcified plaques revealed upregulation of STAT3 in DKO mice, and in vitro experiments confirmed that miR-223-3p downregulated STAT3 expression in VSMCs. Additionally, miR-223-3p significantly inhibited VSMC calcification induced by the IL-6/sIL-6R complex, while overexpression of STAT3 diminished the protection of miR-223-3p in VSMC calcification. Consistently, previous studies have shown that miR-223-3p inhibits osteogenic differentiation in human bone-marrow-derived mesenchymal stem cells by targeting dehydrogenase/reductase 3, another target gene of miR-223-3p. These data suggest that miR-223-3p could negatively regulate vascular calcification by blocking VMSC osteogenic differentiation.
In summary, we found that miR-223-3p inhibits vascular calcification by targeting STAT3, and blocking IL-6/STAT3 signaling inhibits the osteogenic differentiation of VMSCs. These results contribute to our understanding of the mechanism by which miR-223-3p inhibits vascular calcification and demonstrate that miR-223-3p has protective effects on both medial and atherosclerotic calcification.
Experimental procedures
Animals
As published previously by our research group, we purchased miR-223-3p KO mice in the C57BL/6 background from the Jackson Laboratory (stock no. 013198; Bar Harbor, ME, USA). MiR-223-3p KO mice are viable and fertile, with body weights similar to their WT control littermates. ApoE KO (ApoE−/−) mice were purchased from HFK Bioscience (Beijing, China). To generate miR-223-3p/ApoE DKO mice, we bred miR-223-3p KO mice with ApoE−/− mice to generate miR-223-3p ± ApoE ± mice (F1). Interbreeding the F1 mice yielded miR-223-3p ± ApoE−/− (F2) mice, among other genotypes. Interbreeding the F2 mice yielded miR-223-3p/ApoE DKO, ApoE−/−, and heterozygous mice. Mice were bred in the animal facility of Beijing Anzhen Hospital, where they were fed a standard diet and housed in a specific-pathogen-free environment with a 12-h/12-h light/dark cycle. The Animal Subjects Committee of Capital Medical University approved all animal housing and experimental protocols.
Vascular calcification model
Subtotal nephrectomy was performed using a two-step method as previously described (18). Mice (8–10 weeks old) were anesthetized with 1% pentobarbital sodium (100 mg/kg) and two poles of the left kidney were removed. Mice were allowed to recover for 1 week, then anesthetized for removal of the right kidney. Mice were fed a diet containing 2% phosphate and water containing 0.45% NaCl for 4 to 8 weeks after 5/6 nephrectomy. In the sham group, the kidneys were exposed with a flank incision before suturing. Sham mice received a chow diet. BUN was detected 2 weeks after nephrectomy. MiR-223-3p/ApoE DKO and ApoE−/− mice were fed a chow diet for 16 months to form atherosclerotic calcifications. WT and KO mice in chow diet were used as control (8 months).
Tissue harvesting, processing, and analysis
Mice were anesthetized with 1% pentobarbital sodium (100 mg/kg) and subsequently perfused with PBS via the left ventricle. The aorta was used for total calcium measurements, mRNA expression, and transcriptome sequencing. Spleen was harvested for miR-223-3p detection. Tissue-Tek OCT Compound (Sakura, Finetek)-embedded aortic arches, aortic roots, and brachiocephalic artery (BCA) were cryosectioned into 7-μm sections for further experiments.
Aortic calcium analysis
Calcium deposits in aortic sections were stained by Alizarin Red S (Sigma Aldrich, USA) or Von Kossa staining (Sigma Aldrich, USA). Descending aortas were decalcified with 0.5 mmol/L hydrochloric acid overnight. Calcium released from the aortic tissues was determined colorimetrically using a calcium diagnostic kit (BIOSINO, China). The amount of vascular calcium was normalized to the weight of the tissues and expressed as μg/mg.
MiRNA sequencing
MiRNA sequencing and analysis were performed on the Illumina platform at Annoroad Gene Technology Corporation (Beijing, China). In brief, after the quality and integrity of total RNA were assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, USA), small RNAs (18–30-nucleotide segments) were separated from the total RNA by PAGE and processed for library construction and sequencing. High-quality clean read sequences were screened by alignment with Bowtie1 to build the reference genome index. Mapping reads to mature miRNA and hairpin that recorded in miRbase (release 21) to identify known-miRNA. The p-value could be assigned to each gene and adjusted by the Benjamini and Hochberg’s approach for controlling the false discovery rate. MiRNA with p < 0.01 and fold change≥1.5 are identified as differentially expressed miRNA.
RNA-Seq analysis
RNA sequencing and analysis were performed on the Illumina platform at Annoroad Gene Technology Corporation (Beijing, China). Two samples per group were used for RNA-Seq analysis. The RNA samples were used to generate individual cDNA libraries and sequenced on an Illumina platform and 150 bp paired-end reads. The p-value could be assigned to each gene and adjusted by the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with q ≤ 0.05 and |log2_ratio|≥1 are identified as differentially expressed genes (DEGs). DEGs were included in KEGG pathway analysis.
Cell culture
Thoracic aortas were isolated, and the adventitia and intima were stripped away. Then, the aortic tissues were vertically sliced and incubated with digestion buffer (1 mg/ml type I collagenase) for 15 min. Then, the aortas were cut into 1-mm2 pieces and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 20% fetal bovine serum (FBS) until smooth muscle cells sprouted from the tissue. Cells in passages 3–8 were used for further experiments.
3T3 cells and MOVAS were purchased from the American-Type Culture Collection (Manassas, USA) and cultured in DMEM containing 10% FBS.
In vitro VSMC calcification
VSMCs were cultured in osteoblastOIM containing 0.25 mmol/L L-ascorbic acid, 10 mmol/L β-glycerophosphate, and 10–8 M dexamethasone. Fresh medium was changed every 3 days. Cells were harvested after 10 days for RNA extraction and RT-PCR. Proteins were extracted after 3 weeks for western blotting. Calcium levels were visualized by Alizarin Red S staining and quantified by measuring the total calcium in the cell lysates.
In vitro miRNA mimic transfection
Primary mouse VSMCs, MOVAS, or 3T3 cells were transfected with 100 nM mmu-miR-223-3p or negative control mimic (RiboBio, China) using Lipofectamine RNAiMAX (Invitrogen, USA) according to the manufacturer’s guidelines.
Inhibition of STAT3 expression by siRNA
The siRNA against mouse STAT3 and scrambled siRNA were purchased from Santa Cruz Biotechnology, Inc (TX, USA). Primary moused VSMCs were transfected with 50 nmol/L siRNA using Lipofectamin RNAiMAX Transfection Reagent (Invitrogen, USA). After 48 h of transfection, cells were harvested for determination of protein expression or switched to OIM to induce calcification
Overexpression of STAT3 in VSMCs
Lentiviral particles carrying mouse STAT3 or control plasmid were packaged by GeneChem (Shanghai, China). Lentiviral transduction was performed by incubating VSMCs with lentiviruses in growth media, and stable transfectants were selected with puromycin (2 μg/ml) for 1 week.
RNA extraction and RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, USA). For miR223-3p analysis, 2 μg RNA was reverse transcribed to cDNA using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Lithuania) and primers targeting miR-223-3p and U6 small nuclear RNA (snRNA; TaqMan MicroRNA assay, Applied Biosystems). MiR223-3p expression levels were measured by RT-PCR using TaqMan Universal Master Mix II (Applied Biosystems, CA, USA) and probes targeting miR-223-3p and U6 (TaqMan MicroRNA Assay, Applied Biosystems) in a CFX Connect real-time PCR detection system (Bio-Rad, USA). For other miRNA analysis, cDNA was generated from 2 μg total RNA using the reverse transcription kit (RiboBio, China). MiRNA expression levels were measured by qPCR kit (RiboBio, China) targeting miRNA and U6 (RiboBio, China). For mRNA analysis, cDNA was generated from 2 μg total RNA using the reverse transcription kit (Promega, USA). The sequences of all cDNA primers are listed in Table S2. Transcript levels were measured by RT-PCR using SYBR Master Mix II (Takara, Japan).
Western blot analysis
Cells were lysed in protein extraction buffer (Thermo Fisher, USA) on ice for 30 min. After centrifugation at 12,000 rpm for 15 min at 4 °C, the supernatants were collected. Protein concentrations were determined using a Pierce BCA Protein Assay Kit (Thermo Fisher, USA). For each sample, 20 μg total protein was resolved by SDS-PAGE and transferred to membranes to detect STAT3 (Cell Signaling Technologies, USA) and GAPDH (Cell Signaling Technologies, USA). After blocking in 5% milk powder, membranes were incubated with the primary antibodies at 4 °C overnight, washed, and then incubated with IRD800-conjugated secondary antibodies (Li-COR Biosciences, USA) at room temperature for 1 h in the dark. The membranes were then washed and analyzed using the Odyssey software system (Li-COR). STAT3 levels were normalized to GAPDH.
Histology and IHC
Serial transverse cryosections (7-μm thick) were stored at –80 °C until their use. Sections were used for hematoxylin and eosin (H&E) staining and calcification staining. To detect STAT3 expression, sections were blocked with serum, incubated with anti-STAT3 antibodies (Cell Signaling Technologies, USA) at 4 °C overnight, then secondary antibodies(ZSGB-BIO, China) at room temperature for 30 min, and detected with 3,3ʹ-diaminobenzidine (DAB). Images were obtained from each section on an ECLIPSE 90i microscope (Nikon, Japan) and analyzed using ImageJ software (National Institutes of Health, USA).
Statistical analysis
Values are reported as the mean ± standard deviation (SD). Statistical differences between two groups were assessed by unpaired two-tailed Student's t-test. For comparisons of more than two groups, one-way analysis of variance was used. For all statistical tests, p < 0.05 was considered statistically significant.
Data availability
miRNA sequencing and mRNA sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE159833. All other data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Author contributions
Y. H., J. D. conceptualization; Y. H., J. Z. data curation; Y. H., J. Z., S. H. software; Y. H., J. Z., N. C. formal analysis; Y. H., J. Z., S. H., N. C. investigation; Y. H., J. Z., S. H., X. W. methodology; Y. H. writing—original draft; Y. H., J. D. writing—review and editing; Y. L., J. D. project administration; Y. L., C. Z. supervision; J. D., J. L., B. Y. funding acquisition; J. D. validation; Y. H., J. D. visualization.
Funding and additional information
This work was supported by the National Natural Science Foundation of China (grant no. 81861128025, 81790622 to J. D., 8187020148 to B. Y., 81970291 to J. L).
Edited by Ronald Wek
Supporting information
References
- 1.Rogers M.A., Aikawa E. Cardiovascular calcification: Artificial intelligence and big data accelerate mechanistic discovery. Nat. Rev. Cardiol. 2019;16:261–274. doi: 10.1038/s41569-018-0123-8. [DOI] [PubMed] [Google Scholar]
- 2.Lanzer P., Boehm M., Sorribas V., Thiriet M., Janzen J., Zeller T., St Hilaire C., Shanahan C. Medial vascular calcification revisited: Review and perspectives. Eur. Heart J. 2014;35:1515–1525. doi: 10.1093/eurheartj/ehu163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bostrom K.I. Where do we stand on vascular calcification? Vasc. Pharmacol. 2016;84:8–14. doi: 10.1016/j.vph.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Zhao X., Wu H. Arterial stiffness: A Focus on vascular calcification and its Link to bone Mineralization. Arterioscler. Thromb. Vasc. Biol. 2020;40:1078–1093. doi: 10.1161/ATVBAHA.120.313131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanahan C.M., Crouthamel M.H., Kapustin A., Giachelli C.M. Arterial calcification in chronic kidney disease: Key roles for calcium and phosphate. Circ. Res. 2011;109:697–711. doi: 10.1161/CIRCRESAHA.110.234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacolley P., Regnault V., Segers P., Laurent S. Vascular smooth muscle cells and arterial stiffening: Relevance in development, aging, and disease. Physiol. Rev. 2017;97:1555–1617. doi: 10.1152/physrev.00003.2017. [DOI] [PubMed] [Google Scholar]
- 7.Raaz U., Schellinger I.N., Chernogubova E., Warnecke C., Kayama Y., Penov K., Hennigs J.K., Salomons F., Eken S., Emrich F.C., Zheng W.H., Adam M., Jagger A., Nakagami F., Toh R. Transcription factor Runx2 promotes aortic Fibrosis and stiffness in type 2 diabetes Mellitus. Circ. Res. 2015;117:513–524. doi: 10.1161/CIRCRESAHA.115.306341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y., Byon C.H., Yuan K., Chen J., Mao X., Heath J.M., Javed A., Zhang K., Anderson P.G., Chen Y. Smooth muscle cell-specific runx2 deficiency inhibits vascular calcification. Circ. Res. 2012;111:543–552. doi: 10.1161/CIRCRESAHA.112.267237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapustin A.N., Chatrou M.L.L., Drozdov I., Zheng Y., Davidson S.M., Soong D., Furmanik M., Sanchis P., De Rosales R.T.M., Alvarez-Hernandez D., Shroff R., Yin X., Muller K., Skepper J.N., Mayr M. Vascular smooth muscle cell calcification is mediated by regulated exosome secretion. Circ. Res. 2015;116:1312–1323. doi: 10.1161/CIRCRESAHA.116.305012. [DOI] [PubMed] [Google Scholar]
- 10.Goettsch C., Rauner M., Pacyna N., Hempel U., Bornstein S.R., Hofbauer L.C. miR-125b regulates calcification of vascular smooth muscle cells. Am. J. Pathol. 2011;179:1594–1600. doi: 10.1016/j.ajpath.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiao W., Chen L., Zhang M. MicroRNA-205 regulates the calcification and osteoblastic differentiation of vascular smooth muscle cells. Cell Physiol. Biochem. 2014;33:1945–1953. doi: 10.1159/000362971. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S., Liu Y., Zheng Z., Zeng X., Liu D., Wang C., Ting K. MicroRNA-223 suppresses osteoblast differentiation by inhibiting DHRS3. Cell Physiol. Biochem. 2018;47:667–679. doi: 10.1159/000490021. [DOI] [PubMed] [Google Scholar]
- 13.Mackenzie N.C.W., Staines K.A., Zhu D., Genever P., Macrae V.E. miRNA-221 and miRNA-222 synergistically function to promote vascular calcification. Cell Biochem. Funct. 2014;32:209–216. doi: 10.1002/cbf.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clement T., Salone V., Charpentier B., Jouzeau J.Y., Bianchi A. Identification of new microRNAs targeting genes regulating the Pi/PPi balance in chondrocytes. Biomed. Mater. Eng. 2014;24 doi: 10.3233/BME-140969. [DOI] [PubMed] [Google Scholar]
- 15.Ulbing M., Kirsch A.H., Leber B., Lemesch S., Münzker J., Schweighofer N., Hofer D., Trummer O., Rosenkranz A.R., Müller H., Eller K., Stadlbauer V., Obermayer-Pietsch B. MicroRNAs 223-3p and 93-5p in patients with chronic kidney disease before and after renal transplantation. Bone. 2017;95:115–123. doi: 10.1016/j.bone.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J., Xing G., Zhang L., Shang J., Li Y., Li C., Tian F., Yang X. Satb1 promotes osteoclastogenesis by recruiting CBP to upregulate miR-223 expression in chronic kidney disease-mineral and bone disorder. Pharmazie. 2017;72:680–686. doi: 10.1691/ph.2017.7606. [DOI] [PubMed] [Google Scholar]
- 17.Lau W.L., Leaf E.M., Hu M.C., Takeno M.M., Kuro-o M., Moe O.W., Giachelli C.M. Vitamin D receptor agonists increase klotho and osteopontin while decreasing aortic calcification in mice with chronic kidney disease fed a high phosphate diet. Kidney Int. 2012;82:1261–1270. doi: 10.1038/ki.2012.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L., Rajan V., Lin E., Hu Z., Han H.Q., Zhou X., Song Y., Min H., Wang X., Du J., Mitch W.E. Pharmacological inhibition of myostatin suppresses systemic inflammation and muscle atrophy in mice with chronic kidney disease. FASEB J. 2011;25:1653–1663. doi: 10.1096/fj.10-176917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramann R., Goettsch C., Wongboonsin J., Iwata H., Schneider R.K., Kuppe C., Kaesler N., Chang-Panesso M., Machado F.G., Gratwohl S., Madhurima K., Hutcheson J.D., Jain S., Aikawa E., Humphreys B.D. Adventitial MSC-like cells are Progenitors of vascular smooth muscle cells and drive vascular calcification in chronic kidney disease. Cell Stem Cell. 2016;19:628–642. doi: 10.1016/j.stem.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aikawa E., Aikawa M., Libby P., Figueiredo J.L., Rusanescu G., Iwamoto Y., Fukuda D., Kohler R.H., Shi G.P., Jaffer F.A., Weissleder R. Arterial and aortic valve calcification abolished by elastolytic cathepsin S deficiency in chronic renal disease. Circulation. 2009;119:1785–1794. doi: 10.1161/CIRCULATIONAHA.108.827972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Y., Gao C., Liu Z., Wang L., Liu B., He F., Zhang T., Wang Y., Wang X., Xu M., Luo G.-Z., Zhu Y., Xu Q., Wang X., Kong W. Upregulation of a disintegrin and metalloproteinase with thrombospondin motifs-7 by miR-29 repression mediates vascular smooth muscle calcification. Arterioscler. Thromb. Vasc. Biol. 2012;32:2580–2588. doi: 10.1161/ATVBAHA.112.300206. [DOI] [PubMed] [Google Scholar]
- 22.Gupta S.K., Kumari S., Singh S., Barthwal M.K., Singh S.K., Thum T. Non-coding RNAs: Regulators of valvular calcification. J. Mol. Cell. Cardiol. 2020;142:14–23. doi: 10.1016/j.yjmcc.2020.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Long H., Sun B., Cheng L., Zhao S., Zhu Y., Zhao R., Zhu J. miR-139-5p Represses BMSC Osteogenesis via targeting Wnt/β-Catenin signaling pathway. DNA Cell Biol. 2017;36:715–724. doi: 10.1089/dna.2017.3657. [DOI] [PubMed] [Google Scholar]
- 24.M'Baya-Moutoula E., Louvet L., Metzinger-Le Meuth V., Massy Z.A., Metzinger L. High inorganic phosphate concentration inhibits osteoclastogenesis by modulating miR-223. Biochim. Biophys. Acta. 2015;1852:2202–2212. doi: 10.1016/j.bbadis.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Liu W., Ling S., Sun W., Liu T., Li Y., Zhong G., Zhao D., Zhang P., Song J., Jin X., Xu Z., Song H., Li Q., Liu S., Chai M. Circulating microRNAs correlated with the level of coronary artery calcification in symptomatic patients. Sci. Rep. 2015;5:16099. doi: 10.1038/srep16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng N., Liu C., Li Y., Gao S., Han Y.-C., Wang X., Du J., Zhang C. MicroRNA-223-3p promotes skeletal muscle regeneration by regulating inflammation in mice. J. Biol. Chem. 2020;295:10212–10223. doi: 10.1074/jbc.RA119.012263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abedin M., Lim J., Tang T.B., Park D., Demer L.L., Tintut Y. N-3 fatty acids inhibit vascular calcification via the p38-mitogen-activated protein kinase and peroxisome proliferator-activated receptor-gamma pathways. Circ. Res. 2006;98:727–729. doi: 10.1161/01.RES.0000216009.68958.e6. [DOI] [PubMed] [Google Scholar]
- 28.Woldt E., Terrand J., Mlih M., Matz R.L., Bruban V., Coudane F., Foppolo S., El Asmar Z., Chollet M.E., Ninio E., Bednarczyk A., Thiersé D., Schaeffer C., Van Dorsselaer A., Boudier C. The nuclear hormone receptor PPARγ counteracts vascular calcification by inhibiting Wnt5a signalling in vascular smooth muscle cells. Nat. Commun. 2012;3:1077. doi: 10.1038/ncomms2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thériault S., Dina C., Messika-Zeitoun D., Le Scouarnec S., Capoulade R., Gaudreault N., Rigade S., Li Z., Simonet F., Lamontagne M., Clavel M.-A., Arsenault B.J., Boureau A.-S., Lecointe S., Baron E. Genetic Association Analyses Highlight , , and as 3 new susceptibility genes underlying calcific aortic valve stenosis. Circ. Genomic Precision Med. 2019;12:e002617. doi: 10.1161/CIRCGEN.119.002617. [DOI] [PubMed] [Google Scholar]
- 30.Kurozumi A., Nakano K., Yamagata K., Okada Y., Nakayamada S., Tanaka Y. IL-6 and sIL-6R induces STAT3-dependent differentiation of human VSMCs into osteoblast-like cells through JMJD2B-mediated histone demethylation of RUNX2. Bone. 2019;124:53–61. doi: 10.1016/j.bone.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Li M., He Y., Zhou Z., Ramirez T., Gao Y., Gao Y., Ross R.A., Cao H., Cai Y., Xu M., Feng D., Zhang P., Liangpunsakul S., Gao B. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47-oxidative stress pathway in neutrophils. Gut. 2017;66:705–715. doi: 10.1136/gutjnl-2016-311861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cantoni C., Cignarella F., Ghezzi L., Mikesell B., Bollman B., Berrien-Elliott M.M., Ireland A.R., Fehniger T.A., Wu G.F., Piccio L. Mir-223 regulates the number and function of myeloid-derived suppressor cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Acta Neuropathol. 2017;133:61–77. doi: 10.1007/s00401-016-1621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taibi F., Metzinger-Le Meuth V., Massy Z.A., Metzinger L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochim. Biophys. Acta. 2014;1842:1001–1009. doi: 10.1016/j.bbadis.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Yuan X., Berg N., Lee J.W., Le T.-T., Neudecker V., Jing N., Eltzschig H. MicroRNA miR-223 as regulator of innate immunity. J. Leukoc. Biol. 2018;104:515–524. doi: 10.1002/JLB.3MR0218-079R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y.-H., Zhang L., Wu D.-S., Zhang Z., Huang F.-F., Zhang J., Chen X.-P., Liang D.-S., Zeng H., Chen F.-P. MiR-223 regulates human embryonic stem cell differentiation by targeting the IGF-1R/Akt signaling pathway. PLoS ONE. 2013;8:e78769. doi: 10.1371/journal.pone.0078769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabet F., Vickers K.C., Cuesta Torres L.F., Wiese C.B., Shoucri B.M., Lambert G., Catherinet C., Prado-Lourenco L., Levin M.G., Thacker S., Sethupathy P., Barter P.J., Remaley A.T., Rye K.-A. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan Z., Qin S., Li W., Wu W., Yang J., Chu M., Li X., Huo Y., Schaer G.L., Wang S., Zhang C. An Endocrine Genetic signal between blood cells and vascular smooth muscle cells: Role of MicroRNA-223 in smooth muscle function and Atherogenesis. J. Am. Coll. Cardiol. 2015;65:2526–2537. doi: 10.1016/j.jacc.2015.03.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhen S., Shanshan Q., Wen L., Weibin W., Jian Y., Maoping C., Xiaokun L., Yuqing H., Gary L.S., Shenming W., Chunxiang Z. An Endocrine Genetic signal between blood cells and vascular smooth muscle cells: Role of MicroRNA-223 in smooth muscle function and Atherogenesis. J. Am. Coll. Cardiol. 2015;65:2526–2537. doi: 10.1016/j.jacc.2015.03.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taïbi F., Metzinger-Le Meuth V., M'Baya-Moutoula E., Djelouat M.s. e.I., Louvet L., Bugnicourt J.-M., Poirot S., Bengrine A., Chillon J.-M., Massy Z.A., Metzinger L. Possible involvement of microRNAs in vascular damage in experimental chronic kidney disease. Biochim. Biophys. Acta. 2014;1842:88–98. doi: 10.1016/j.bbadis.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz-Ortega M., Rayego-Mateos S., Lamas S., Ortiz A., Rodrigues-Diez R.R. Targeting the progression of chronic kidney disease. Nat. Rev. Nephrol. 2020;16:269–288. doi: 10.1038/s41581-019-0248-y. [DOI] [PubMed] [Google Scholar]
- 41.Furmanik M., Chatrou M., van Gorp R.H., Akbulut A., Willems B., Schmidt H.H., van Eys G., Bochaton-Piallat M.-L., Proudfoot D., Biessen E.A., Hedin U., Matic L., Mees B., Shanahan C.M., Reutelingsperger C. Reactive Oxygen-Forming Nox5 Links vascular smooth muscle cell phenotypic switching and extracellular Vesicle-mediated vascular calcification. Circ. Res. 2020;127:911–927. doi: 10.1161/CIRCRESAHA.119.316159. [DOI] [PubMed] [Google Scholar]
- 42.Yang J., Stark G.R. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18:443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 43.Dorhoi A., Iannaccone M., Farinacci M., Faé K.C., Schreiber J., Moura-Alves P., Nouailles G., Mollenkopf H.-J., Oberbeck-Müller D., Jörg S., Heinemann E., Hahnke K., Löwe D., Del Nonno F., Goletti D. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Invest. 2013;123:4836–4848. doi: 10.1172/JCI67604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
miRNA sequencing and mRNA sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE159833. All other data are contained within the article.