Abstract
Reactive oxygen species (ROS) are a common product of active mitochondrial respiration carried in mitochondrial cristae, but whether cristae shape influences ROS levels is unclear. Here we report that the mitochondrial fusion and cristae shape protein Opa1 requires mitochondrial ATP synthase oligomers to reduce ROS accumulation. In cells fueled with galactose to force ATP production by mitochondria, cristae are enlarged, ATP synthase oligomers destabilized, and ROS accumulate. Opa1 prevents both cristae remodeling and ROS generation, without impinging on levels of mitochondrial antioxidant defense enzymes that are unaffected by Opa1 overexpression. Genetic and pharmacologic experiments indicate that Opa1 requires ATP synthase oligomerization and activity to reduce ROS levels upon a blockage of the electron transport chain. Our results indicate that the converging effect of Opa1 and mitochondrial ATP synthase on mitochondrial ultrastructure regulate ROS abundance to sustain cell viability.
Keywords: Mitochondrial cristae, Ultrastructure, ROS, Opa1, F1FO-ATP synthase, Bioenergetics
Graphical abstract

Highlights
-
•
Mitochondrial ROS levels mirror changes in cristae shape.
-
•
F1FO-ATP synthase oligomerization and reversal link mitochondrial ultrastructure to ROS generation.
-
•
Disrupted F1FO-ATP synthase dimerization abolishes Opa1 ability to curtail mitochondrial ROS accumulation.
1. Introduction
One of the key roles of mitochondria is to supply ATP, synthesized by the F1FO-ATP synthase [1,2]. The F1FO-ATP synthase uses the proton electrochemical gradient generated by the proton pumping of the respiratory chain complexes (RCC) [1,3], located in the cristae folding of the inner mitochondrial membrane (IMM) [[4], [5], [6]]. Given that these complexes are all located at the IMM, cristae morphology was proposed to influence mitochondrial respiration and energy conversion efficiency [7]. Genetic and apoptotic manipulations proved indeed that cristae morphology favors assembly of RCCs into quaternary respiratory supercomplexes (RCS). The existence of respiratory supercomplexes (RCS) is becoming unambiguously accepted, reinforced by the resolution of their structure by cryo-electron microscopy that provides a basis for their functional role [[15], [16], [17]]. Notwithstanding, controversies on supercomplexes function still exist. This may be due to technical issues along with coexistence of RCS different conformations [14]. Controversies particularly concern RCS engagement in electron transfer, stability of complexes, substrate channeling, ROS generation, protein nucleation, and pathological consequences [[13], [14], [15]]. Recent cryo-EM structural data and functional experiments of assembly factors manipulation suggest that RCS facilitate cytochrome c shuttle between CIII and CIV and hence maximal respiration [12]. Structural data also suggest that transfer of electrons can be facilitated the in the “respirasome” RCS (Letts et al., 2016).
Both cristae shape and RCS stability depend on the formation of cristae junctions (CJ), narrow tubular structures connecting the cristae to the inner boundary membrane [20,21]. CJ fulfill a variety of biological functions: they needed to individualize cristae as independent, dynamic bioenergetic units [22]; CJ are also central to keep the bulk of cytochrome c inside the cristae [23,24] and pro-apoptotic BH3-only BCL-2 family members such as BID, BIM-S or BNIP3 induce cytochrome c mobilization to the IMS by causing CJ enlargement and inversion of the cristae curvature [23,[25], [26], [27]]. From a bioenergetics point of view, the aberrant ultrastructure caused by apoptotic signals destabilizes RCS composition. Conversely, cristae preservation favors RCS stability and mitochondrial-dependent cell growth [8].
The IMM dynamin-related protein optic atrophy 1 (OPA1) is central for the processes described above: its oligomers maintain CJ tight to prevent cytochrome c mobilization and therefore control the mitochondrial arm of apoptosis [24,26]. By narrowing cristae independently of its mitochondrial pro-fusion role, OPA1 also stabilizes RCS and regulates respiratory efficiency [[8], [9], [10]]. OPA1 favors also the supramolecular assembly of the F1Fo-ATP (ATP) synthase into oligomers [[28], [29], [30]]. This configuration sustains mitochondrial ATP levels and homeostasis during starvation [31], limited respiratory substrate availability [30] or compromised electron transfer [29]. In this latter condition, improved bioenergetics and electron transfer by RCS may prevent both reactive oxygen species (ROS) generation [18,19,[32], [33], [34]], and their effects [[35], [36], [37], [38]]. However, the role of mitochondrial ultrastructure in controlling ROS levels remains unclear [39]. We therefore dissected whether and how OPA1 can influence mitochondrial ROS accumulation. Stabilization of cristae by OPA1 preserves ATP synthase dimerization and reversal activity, required to sustain the electrochemical gradient, and reduces mitochondrial ROS levels. We illustrate how Opa1 limits ROS accumulation via ATP synthase dimerization and activity.
2. Results
2.1. OPA1 overexpression prevents mitochondrial ROS accumulation
The master regulator of cristae shape Opa1 [24,[40], [41], [42]] protects from compromised mitochondrial function and respiratory chain blockage by sustaining F1FO-ATP synthase (ATP synthase) reversal activity [29]. It is however unclear whether Opa1 also impacts ROS accumulation, a known product of oxidative phosphorylation (OXPHOS) and a player in cell death caused by respiratory dysfunction. To address this question, we first confirmed that viability of Opa1Tg mouse adult fibroblasts (MAFs) where Opa1 is mildly upregulated (Fig. S1a), was sustained when they were cultured in medium supplemented with galactose to halt glycolysis and force ATP biosynthesis by mitochondria [18,43,44] (Fig. 1a). Reciprocally, deletion of Opa1 in MAFs from Opa1flx/flx mice (Fig. S1b) doubled cell death in galactose-containing media (Fig. 1b). We then profiled mitochondrial function in cell lines cultured in glucose vs. galactose. We did not measure any difference in the basal levels of the potentiometric dye tetra methyl rhodamine methyl ester (TMRM) in glucose or galactose containing media, between Wt and Opa1Tg cells (Fig. S2a). In glucose-containing media, TMRM response to the ATP synthase inhibitor oligomycin, a reliable assay to unveil latent mitochondrial dysfunction [45] was also similar between Wt and Opa1Tg cells (Fig. S3a). We obtained similar results when we analyzed the chemical component of the proton electrochemical gradient (ΔμH+) using mtSypHer, a genetically encoded pH ratiometric sensor targeted to the mitochondrial matrix [46]. In galactose-containing medium, basal mtSypHer ratios were slightly lower, irrespective of the genetic background of the cells or even the absence of Opa1 (Fig. 1c, d, S3b). This Opa1-independent matrix acidification likely reflects the activation of mitochondrial ATP synthase when glycolysis is marginal and mitochondrial OXPHOS is engaged to produce ATP. We therefore compared resting matrix ATP levels measured using the genetically encoded sensor ATeam1.03 [47]. Interestingly, we did not see differences in mitochondrial ATP levels between WT and Opa1Tg MAFs, irrespective of the monosaccharide supplemented to the culture media. ATP levels were lower in mitochondria of both genotypes when cells were cultured in galactose-containing medium (Fig. 1e and f). Altogether, these experiments indicate that in glucose- or galactose-containing media, the mitochondrial electrochemical gradient and steady state mitochondrial ATP levels are similar in WT and Opa1tg MAFs. Nevertheless, when cells rely on mitochondrial respiration to provide ATP, Opa1 overexpression preserves cell viability, independently from changes in ΔμH+ or mitochondrial ATP levels. We therefore evaluated ROS levels in mitochondria in the two cell types using mt-roGFP1, a genetically encoded ROS sensor targeted to the mitochondrial matrix, responsive to 1 mM H2O2 (Fig. S4). In glucose containing media steady state mt-roGFP1 fluorescence ratio was slightly but not significantly lower in Opa1tg vs. WT MAFs. When cells were switched to galactose media, mt-roGFP1 fluorescence ratio significantly increased in WT but not in Opa1tg cells (Fig. 1g). These data indicate that when mitochondrial respiration is stimulated, ROS accumulation is lowered by Opa1 overexpression.
Fig. 1.
OPA1 prevents mitochondrial dysfunction and cell death under bioenergetic stress.
a, b) Cell death of MAFs of the indicated genotype, incubated for the indicated time in medium supplemented with the indicated monosaccharide. Data are mean ± SEM of four independent experiments.
c) Gem-color coded images of mtSypHer fluorescence ratios (em: 535 nm) after alternate excitation (exc: 500/430 nm) of WT or Opa1tg MAFs incubated for 24 h in complete DMEM supplemented with the indicated monosaccharide. The gem color scale is shown. Scale bar, 20 μm
d) Quantitative analysis of experiments performed as in c. mtSypHer ratios are shown as mean excitation 500/430 nm ratios.
e) Representative pseudocolor-coded FRET ratio images of mtATeam matrix ATP reporter in cells incubated for 24 h in the indicated medium. The pseudocolor scale is shown. Scale bar, 20 μm.
f) FRET fluorescence ratios of WT or Opa1tg MAFs, as in (e). Results show the average emission ratio (525/475 nm).
g) Mitochondrial redox levels quantified from ratiometric excitation ratio (400/480 nm) image analysis mt-roGFP1 fluorescence, in cells grown for 24 h in medium supplemented with either glucose or galactose. In box plots, boxes represent SEM, middle line mean, whiskers 5th-95th percentile, dots the individual values of the independent experiments. *p < 0.05 in a two-way ANOVA versus control.
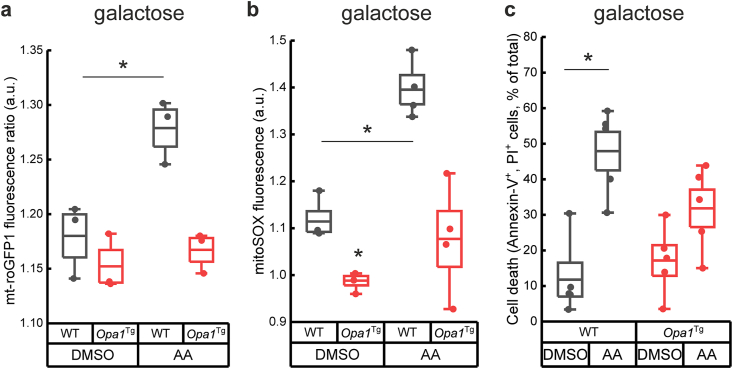
We next addressed if Opa1 overexpression could also prevent ROS production when mitochondrial respiratory activity was inhibited. To this end, we treated cells with antimycin A (AA) which stimulates ROS production by interfering with the Q cycle in complex III. Two independent sensors of mitochondrial ROS production, the fluorescent dye MitoSOX and the ratiometric probe mt-roGFP1 concurrently reported that mitochondrial ROS accumulate in WT but not Opa1tg MAFs treated with AA (Fig. 2a and b). In Opa1tg cells, TMRM decayed in response to the concomitant inhibition of complex III using AA and ATP synthase using oligomycin slower than in WT cells (t1/2: 20 min in WT cells, 24 min in Opa1tg, Fig. S3c). Accordingly, Opa1tg MAFs grown in galactose were protected from cell death induced by AA (Fig. 2c), as we previously reported [29]. Altogether, these data indicate that Opa1 overexpression protects from ROS accumulation produced by forced respiration and complex III inhibition.
Fig. 2.
Opa1 prevents ROS accumulation in cells relying on mitochondrial metabolism.
a) Mitochondrial redox levels quantified from ratiometric excitation ratio (400/480 nm) image analysis mt-roGFP1 fluorescence in WT or Opa1tg MAFs challenged for 2 h with vehicle or 5 μM AA before recordings. Results are expressed as mean ± SEM of three independent experiments. *p < 0.05 in an unpaired two-sample Student's t-test.
b) Fold increase in MitoSOX fluorescence after treatment with AA (10 μM, 15 min) in cells of the indicated genotype grown for 24 h in the indicated media. Data are mean ± SEM of four independent experiments. *p < 0.05 in a two-way ANOVA versus control (a, b) or paired two-sample Student's t-test versus control (c)
c) Cell death determined by flow cytometry as the percentage of annexin-V, PI double positive events from WT and Opa1tg MAFs MAFs, incubated for 48 h in the presence of the indicated monosaccharides and exposed to 5 μM AA for 4 h. In box plots, boxes represent SEM, middle line mean, whiskers 5th-95th percentile, dots the individual values of the independent experiments.
2.2. OPA1 requires ATP synthase activity to reduce mitochondrial ROS accumulation
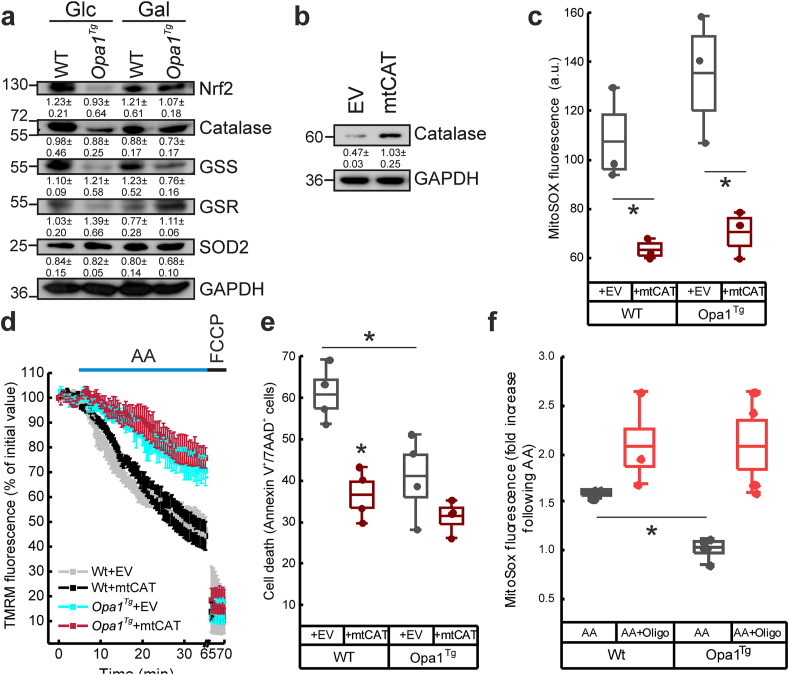
We wished to understand how Opa1 overexpression reduced ROS accumulation. We therefore first measured levels of endogenous antioxidant enzymes. Immunoblotting showed no difference in the levels of superoxide dismutase 2 (SOD2), glutathione synthetase (GSS) or glutathione reductase (GSR) as well as of the master transcriptional regulator of the antioxidant response NRF2 in Opa1tg vs. WT MAFs, irrespective of whether they were grown in galactose or glucose containing media (Fig. 3a). These results suggested that the reduced ROS accumulation in Opa1tg MAFs was not due to an increase in the antioxidant enzymatic defenses. We next asked whether expression of catalase, an antioxidant enzyme genetically modified to be targeted to the mitochondrial matrix (mtCAT) [35] could enhance the protection afforded by Opa1 overexpression. Moderate mtCAT expression (Fig. 3b) reduced superoxide accumulation in both WT and Opa1tg MAFs grown in galactose (Fig. 3c), without altering basal mitochondrial membrane potential (Fig. S1b). Judging from experiments of real time mitochondrial membrane potential measurements, mtCAT expression did not protect from acute AA-induced mitochondrial depolarization in cells of either genotype, indicating that mtCAT does not directly interfere with loss of membrane potential upon acute AA exposure in our setting (Fig. 3d). However, mtCAT prevented the spontaneous cell death observed in WT MAFs grown in galactose, whereas it did not increase the protection afforded by Opa1 overexpression (Fig. 3e), indicating that this mitochondrially targeted catalase protects WT cells from ROS accumulation and cell death but is not additive to Opa1 overexpression. Altogether, these experiments point to the existence of a mechanism different than induction of antioxidant defense that explains the resistance of Opa1tg cells to ROS. We therefore wondered whether the reduced ROS accumulation observed in Opa1tg MAFs involved the known stimulatory effect of Opa1 on the activity of the ATP synthase [29]. Indeed, the ATP synthase inhibitor oligomycin abolished the protective effect of Opa1 overexpression on AA-induced ROS accumulation (Fig. 3f).
Fig. 3.
Opa1 requires ATP synthase activity to prevent mitochondrial ROS accumulation.
a) Protein lysates from MAFs of the indicated genotype and incubated for 24 h in medium supplemented with the indicated monosaccharide, were separated by SDS-PAGE and immunoblotted with the indicated antibodies.
b) WT cells transfected with empty (EV) or mitochondrial-targeted catalase (mtCAT) were lysed after 24 h equal amounts of protein were separated by SDS-PAGE and immunoblotted using the indicated antibodies.
c) MitoSOX fluorescence values of WT and Opa1Tg cells transfected as indicated and incubated in galactose medium for 24 h; a.u.: arbitrary units.
d) Analysis of TMRM fluorescence in cells transfected as indicated, incubated in galactose medium for 24 h and treated where indicated with 10 μM AA and 2 μM FCCP. Data are mean ± SEM of four independent experiments.
e) Cell death after 48 h incubation in galactose medium in cells transfected with the indicated plasmids and treated with AA (5 μM, 4 h).
f) MitoSOX mean fold changes from four experiments performed with cells grown in galactose supplemented medium for 24 h and challenged with AA (10 μM, 15 min). Where indicated, cells were pre-incubated with oligomycin (1 μM, 5 min). In box plots, boxes represent SEM, middle line mean, whiskers 5th-95th percentile, dots the individual values of the independent experiments. *p < 0.05 in a two-way ANOVA.
2.3. OPA1 stabilizes ATP synthase monomers and oligomers under restricted glycolysis
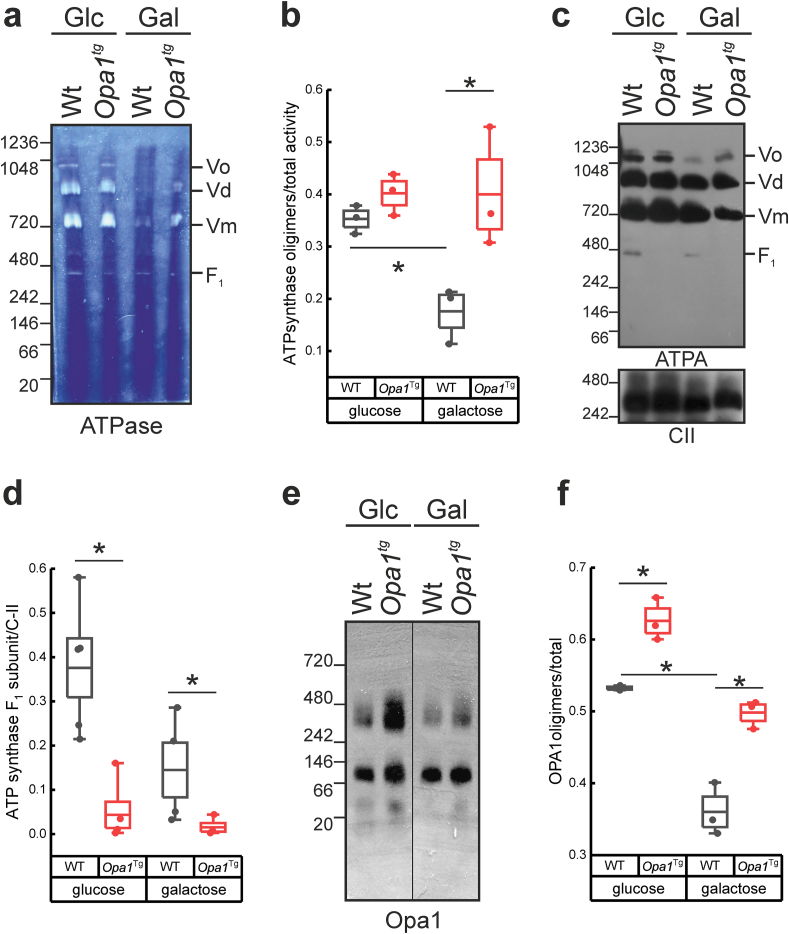
Because our results pointed to a role for ATP synthase in the reduced ROS accumulation observed in Opa1 overexpressing cells, we wished to characterize the relationship between Opa1 and ATP synthase in ROS accumulation. We first compared ATP synthase oligomerization in cells cultured in glucose and in galactose, a condition where ROS accumulate. We measured in blue-native gel electrophoresis (BNGE) gels the assembly ATP synthase in mitochondria from WT MEFs cultured in glucose and galactose containing media. In-gel activity (as well as levels) of ATP synthase oligomeric forms were lower in galactose than in glucose-based media (Fig. 4a and b). However, we noticed that in galactose ATP synthase monomers were more efficiently assembled, as we could not detect any free F1 subunit that was conversely visible in BNGE from cells cultured in glucose (Fig. 4c and d). Importantly, oligomycin inhibited in gel activity of monomeric ATP synthase, but not of free F1, indicating that OSCP does not copurify with F1 and that under our experimental conditions this band represents non assembled F1 (Fig. S5). These results indicate that in galactose containing media ATP synthase monomer assembly is favored, whereas superassembly is impaired. This decrease in ATP synthase oligomers observed in galactose was accompanied by the destabilization of Opa1 oligomers (Fig. 4e and f). We therefore tested whether Opa1 overexpression prevented the destabilization of ATP synthase oligomers in galactose, and the accumulation of free F1 in glucose-rich media. In Opa1Tg cells cultured in both glucose and galactose oligomeric OPA1 was increased, as expected (Fig. 4e and f). In glucose, levels of oligomeric ATP synthase were slightly but not significantly increased, but we could not detect any free F1. Oligomers of ATP synthase were significantly more stable in Opa1Tg than in WT cells cultured in galactose (Fig. 4b, d). Genetic Opa1 deletion yielded the opposite results of Opa1 overexpression: ATP synthase oligomeric levels and activity were reduced (Fig. S6a-d). Free F1 levels were greatly reduced also in Opa1 knockout cells when cultured in galactose containing media (Fig. S6d). Taken together, these results indicate that OPA1 abolishes the reduction in ATP synthase dimers observed in cells cultured in galactose.
Fig. 4.
Opa1 promotes ATP synthase monomers assembly and oligomers stability.
a) Complexes from 40 μg mitochondrial extracts from cells of the indicated genotypes, incubated for 24 h in the indicated media, were separated by Blue-native PAGE (BNGE) and analyzed for ATP synthase in-gel activity.
b) Densitometric analysis of experiments as in (a).
c, d) BNGE (c) and densitometric analysis (d) of immunoblotted mitochondrial extracts from WT or Opa1tg cells incubated for 24 h as indicated from at least four independent experiments as in a). Ratios between the bands corresponding to oligomeric (oligomers, Vo + dimers, Vd) and the sum of all conformations (Vo + Vd + Vm + F1) dimeric are shown.
e, f) BNGE blot (e) and densitometric analysis (f) of OPA1 High Molecular Weight (HMW) complexes of protein extracts (40 μg) from MAFs of the indicated genotype incubated for 24 h in the presence of the referenced media. In box plots, boxes represent SEM, middle line mean, whiskers 5th-95th percentile, dots the individual values of the independent experiments. *p < 0.05 in a two-way ANOVA.
2.4. Opa1 requires ATP synthase dimerization to prevent ROS accumulation
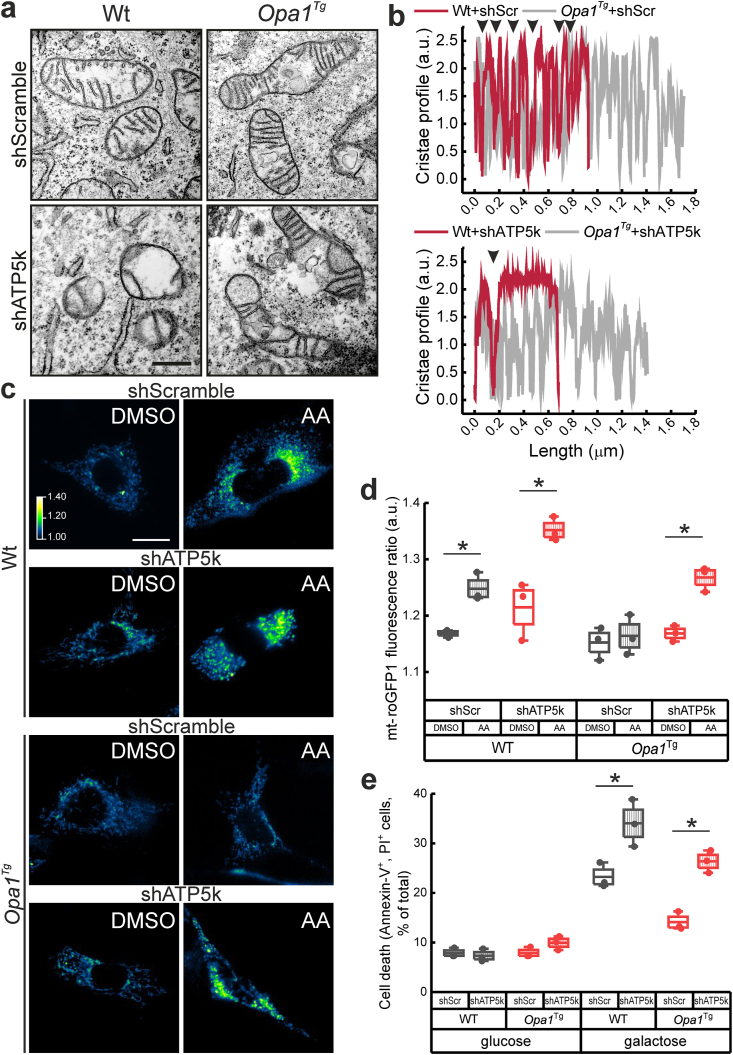
Because Opa1 protects from mitochondrial dysfunction by sustaining cristae shape and ATP synthase supramolecular assembly [29], we wished to address the role of cristae shape and ATP synthase oligomerization in the observed reduced ROS accumulation. We therefore deleted the supernumerary ATP synthase subunit e (ATP5k) that is critical to form dimers at cristae tips [[48], [49], [50], [51]]. Importantly, ATP5k deletion and subsequent impairment of dimerization does not significantly impact resting ATP, mitochondrial content and mtDNA [29,[52], [53], [54], [55], [56]], although it may compromise OXPHOS [56] and ATPase activity that is favored when ATP synthase is in its oligomeric forms [29].
We first addressed whether cristae shape changed depending on the monosaccharide supplied to the cells. Switching cells from glucose to galactose resulted in a reduction in the total number of cristae per mitochondrion (Fig. S6a-c) but more importantly in a reduction in cristae width (Fig. S6a,b,d). These data suggest that cristae dynamics is modulated by the fuel utilized for mitochondrial respiration and confirm that Opa1 overexpression reduces cristae width, irrespective of the fuel burned. We next silenced ATP5k in cells grown in glucose. Cristae were altered morphologically and reduced in number in WT but not in Opa1Tg cells (Fig. 5a and b) [29,56,57]. Silencing of subunit e on the other hand blunted the protective effect of Opa1 on accumulation of mitochondrial ROS induced by AA (Fig. 5c). Similarly, it abolished the ability of Opa1 to prevent spontaneous cell death of MAFs grown in galactose containing medium (Fig. 5d). In conclusion, Opa1 requires F1FO-ATP synthase dimerization to block ROS production and cell death caused by mitochondrial bioenergetic stress.
Fig. 5.
Opa1 requires the ATP synthase dimerization subunit e to prevent ROS accumulation.
a, b) Representative electronic micrographs (a) and cristae profile (b) of mitochondria from MAFs of the indicated genotype transfected with the indicated shRNA for 72 h. The cristae profiles illustrate cristae and their width along the overall mitochondrial length (μm). Arrowheads indicate intercristal matrix space.
c) Analysis of mitochondrial matrix redox status in cells co-transfected with mt-roGFP1 and the indicated shRNAs. Data represent mean ± SEM of 400/492 nm fluorescence ratio. Where indicated, cells were treated for 2 h with 5 μM AA.
d) Spontaneous death of cells of the indicated genotype transfected with the indicated shRNA after 72 h incubation in glucose or galactose medium. In box plots, boxes represent SEM, middle line mean, whiskers 5th-95th percentile, dots the individual values of the independent experiments. *p < 0.05 in a two-way ANOVA.
3. Discussion
Accumulation of ROS is a common consequence of mitochondrial dysfunction. Whether shaping of cristae by Opa1 that protects mitochondrial function also inhibits mitochondrial ROS accumulation was unclear. Here we show that Opa1 requires ATP synthase activity and dimerization to reduce ROS accumulation when mitochondrial respiration is enforced by culturing cells in galactose, or when the respiratory chain is inhibited with Antimycin A.
Opa1 is a central modulator of cristae shape and because of its ability to reduce cytochrome c release and to engage the reversal activity of the ATP synthase it protects mitochondria from dysfunction downstream a plethora of different stimuli. This protective effect can be harnessed therapeutically to ameliorate induced and inherited mitochondrial dysfunction. However, whether and how Opa1 affects mitochondrial ROS generation or accumulation, a well-known consequence of mitochondrial dysfunction is unclear. Our results indicate that Opa1 overexpression did not affect levels of endogenous antioxidant enzymatic defenses, but it nevertheless blunted ROS accumulation recorded when mitochondrial respiration was forced in galactose supplemented medium, where glycolysis is residual and ROS arise as products of forced respiration [9,18,43,44], as well as when we inhibited complex III with Antimycin A to induce ROS generation [18,19]. In principle, Opa1 could lower ROS production by favoring RCS formation and respiratory efficiency [8,9]. However, we found that Opa1 required ATP synthase activity to reduce ROS generation. This result was interesting because it indicated that ATP synthase reversal in antimycin A treated mitochondria not only sustains membrane potential and hence ion homeostasis across the inner membrane [29], but also reduces ROS production.
A surprising result was that in galactose containing media where respiration is stimulated, F1Fo-ATP synthase dimers were less abundant. Conversely, monomers were fully assembled as we could not detect any free F1 subunit. Monomers display a limited ATP hydrolysis capacity [29,31], but they can still support cell growth [[52], [53], [54], [55]] and do not affect mitochondrial ATP levels [29]. We noticed an interesting parallelism between the scenario observed in cells grown in galactose and Opa1 overexpressing cells where we silenced the e subunit of the ATP synthase, required for its dimerization. In both conditions, ATP synthase dimerization was impaired, cristae were tightened, and ROS accumulated. It is tempting to speculate that the increased ROS accumulation observed in cells grown in galactose might be explained by the reduced levels of ATP synthase dimers.
Another perhaps surprising result was that in cells cultured in galactose measured mitochondrial matrix ATP levels were lower. This might reflect be the consequence of less abundant F1Fo-ATP synthase dimers, or of the rapid export of neosynthesized ATP from mitochondria to match cytosolic demands. Alternatively, ATeam1.03 fluorescence could be reduced by the higher basal ROS levels and by the matrix acidification observed in galactose (Imamura et al., 2009). However, in galactose the reduction in matrix ATP levels were comparable between WT and Opa1Tg cells where matrix acidification and ROS accumulation are conversely different, thus reducing the likelihood of this explanation.
A yet unresolved issue was how dimerization and crista shape impact on ROS production [6,39]. Manipulations of cristae morphology based on Opa1 [8,24,40] alter the stability of F1Fo-ATP synthase dimers [29] that accommodate at the edge of cristae and maintain their orthodox structure [11,49,50,58]. Capitalizing on Opa1 ability to prevent the aberrant ultrastructure that follows subunit e (ATP5k) deletion, which impairs ATP synthase dimerization [29], we could use it as a strategy to dissect that F1Fo-ATP synthase acts downstream of cristae remodeling in mitochondrial ROS production. Functionally, Opa1 is unable to lower ROS generation and cell death in galactose medium unless F1Fo-ATP synthase dimers are preserved, indicating that these two players define not only cristae shape and bioenergetics, but also ROS accumulation. A potential caveat of our finding is that ATP synthase subunit e may affect other ATP synthase complex functions or mitochondrial bioenergetics (Everard-Gigot et al., 2005; Hahn et al., 2016). While ATP5k downregulation does not alter the activity of monomeric ATP synthase in yeast (Arnold et al., 1998; Bornhövd et al., 2006; Boyle et al., 1999; Paumard, 2002), its downregulation in human cells alters mitochondrial ultrastructure and reduces respiratory complex stability, ultimately compromising OXPHOS [56]. The reduction in respiratory complex stability might be a consequence of the altered mitochondrial ultrastructure as also observed in models of Opa1 acute deletion [8]. However, in our system resting matrix ATP levels are not affected by e subunit deletion, suggesting that OXPHOS and ATP synthase activity are not impaired (Quintana-Cabrera et al., 2018).
Our study did not address the role of other modulators of cristae shape such as MICOS components [11,[59], [60], [61]], prohibitins [62], lipid fluidity [12] or cardiolipin composition [63,64] in the regulation of mitochondrial dysfunction and ROS production. Whether they participate in pathways like the one described here remains to be ascertained. Nevertheless, Opa1 emerges as an interesting tool to protect mitochondria from ROS accumulation, a common issue for example in neurodegeneration [65] or ischemia-reperfusion injury [66], two of the many disease conditions characterized by compromised bioenergetics and excessive ROS production.
4. Conclusions
Our work sheds light on a still elusive mechanism by which mitochondrial ultrastructure sets ROS abundance. We demonstrate that changes in cristae shape are exerted by Opa1 to stabilize ATPase oligomers and reversal activity, that in turn avoids the rise of ROS that follows a blockage of complex III. While providing a so far missing mechanism for the ability of the shaping protein Opa1 to blunt ROS production, our results may be extended to other cristae modulators, that could play a similar role to preserve mitochondrial function from the deleterious consequences of excess ROS generation.
4.1. Materials and methods
4.1.1. Cell culture
WT, Opa1flx/flx, Opa1tg MAFs SV40 transformed cell lines were generated from the diaphragm of the respective 7-week-old mouse as described [8]. Cells were grown in Dulbecco's Modified Eagle Medium (DMEM, Invitrogen) containing 4.5 mg/ml glucose and supplemented with 10% fetal bovine serum (FBS, Invitrogen), 2 mM l-glutamine, 100 μM non-essential amino acids, 50 U/ml penicillin and 50 μg/ml streptomycin (Invitrogen) at 37 °C in a 5% CO2 atmosphere. Unless otherwise stated, glucose was substituted with 0.9 mg/ml galactose [18,43,44].
4.1.2. Transfection
Scramble or ATP5k (F1Fo-ATP synthase sub. e; NM_007507.2) shRNA encoding SureSilencing® plasmids (Qiagen, KM31364H, plasmid #3) were transiently transfected using Transfectin (Biorad). Briefly, a 4:3 (μg/μL) DNA:Transfectin ratio was mixed in Optimem® (Invitrogen) medium and added to cultures in penicillin/streptomycin and FBS-free, glucose-supplemented DMEM. After 4–6 h incubations, transfection medium was replaced with complete DMEM and incubated for the indicated time points. Following overnight transduction, the rate of GFP expression was typically around 60–70%, as determined by flow cytometry. Transfections were made with the following plasmids: SureSilencing® shRNA, pEGFP-N1-mt-roGFP1 (Qiagen), mt-roGFP1 [67] or wt (AT1.03) or inactive (AT1.03R122K/R126K) ATeam cloned into a pRSET-B vector (Invitrogen) [47]. OPA1 ablation in Opa1flx/flx MAFs was performed acutely at least 48 h after co-delivery of the Cre-recombinase controlled by a PGK promoter (pPGK-Puro, Addgene).
Transfection with empty pIRES or pIRES-mtCAT vectors [35] was performed Lipofectamine LTX-PLUS Reagent (Life Technologies) following the manufacture's instructions.
4.1.3. Imaging
For live imaging of mitochondrial probes, cells were seeded onto 24-mm round glass coverslips and mounted on the stage of an Olympus IX81 inverted microscope (Melville, NY) equipped with a CellR imaging system and a beam-splitter optical device (Multispec Microimager; Optical Insights). Images were acquired at the indicated time points and excitation/emission ratios using a × 40, 1.4 NA objective (Olympus) and the CellR software. Mitochondrial toxins were added at the indicated concentrations and time points in Ca2+/Mg2+ supplemented HEPES buffer (HBSS, Invitrogen), without further washings between additions. Analysis of fluorescence variance was performed over regions of interests (ROIs) including clusters of mitochondria, using the multi-measure plug-in of Image J software (NIH) following background subtraction. Representative micrographs of experiments are represented, using red hot (TMRM), gem (mtSypHer) or pseudocolor (ATeam) ImageJ filters for color coded images.
4.1.4. Analysis of mitochondrial matrix pH
Matrix ΔpH analyses were performed 24 h after transfection with the pSypHer-dMito [46]. Sequential images of the pSypHer-dMito 535-emission fluorescence were acquired every 10 s for 30 min after alternate excitation for 100 ms at 430 and 500 nm. Mean fluorescence ratios of selected ROIs matching mitochondria were estimated in at least three experiments and expressed as mtSypHer (430/500 nm) ratios.
4.1.5. Analysis of mitochondrial membrane potential
For imaging of Δᴪm, MAFs grown on coverslips were loaded with 10 nM TMRM (Molecular Probes) in the presence of 1 μM cyclosporine H, a P-glycoprotein inhibitor (30 min at 37 °C in a 5% CO2 atmosphere). Sequential images of TMRM fluorescence were acquired at every 30 s as described [24] upon the addition of the indicated inhibitors. The mitochondrial uncoupler carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP, 2 μM, Sigma) was added at the end of every experiment as a control for mitochondrial depolarization.
4.1.6. Analysis of mitochondrial ATP levels
Mitochondrial ATP content was determined by FRET image analysis of cells transfected with the plasmid pRSET-B-ATeam1.03, encoding for the ε subunit of the B. subtilis F1Fo-ATP synthase with humanized codons sandwiched between the mseCFPΔC11 (cyan) and cp173-mVenus (yellow) fluorescent proteins [47]. Sequential alternate images of the 525 nm and 475 nm fluorescence emission after excitation at 435 nm for 100 ms were acquired every 30 s. As negative control, cells were transfected with a pRSET-B-mtATeam1.03R122K//R126K plasmid expressing an inactive, ATP-insensitive form of the enzyme.
4.1.7. Determination of ROS production
Mitochondrial ROS levels were analyzed in cells incubated for 24 h in DMEM supplemented with glucose or galactose, as indicated. The mitochondrial matrix redox potential was addressed by analyzing fluorescence changes of the ratiometric, genetically encoded reporter pEGFP-N1-mt-roGFP1 [67]. To this end, images of the 535 nm fluorescence emission were acquired every 30 s for 5 min, following sequential excitation for 100 ms at 400/480 nm, averaged and expressed as 400/480 nm excitation ratios.
Mitochondrial O2−· production was determined as previously described [68]. Briefly, cells were stained with 2 μM MitoSOX (Invitrogen) in Ca2+/Mg2+ HBSS for 30 min at 37 °C in a 5% CO2 atmosphere. After rinsing with PBS, each sample was split into two tubes and treated with either vehicle or 10 μM antimycin A for 20 min at RT; where indicated, 1 μM oligomycin was added 5 min before antimycin A. The MitoSOX fluorescence was assessed by flow cytometry using the FL1 channel of a FACSCalibur (BD Pharmingen) cytometer and expressed as fold increase over vehicle treated cells.
4.1.8. Transmission electron microscopy
For morphometric analysis of cristae structure, MEFs of the specified genotypes and treated as indicated were fixed with 1.25% (v:v) glutaraldehyde in 0.1 M sodium cacodylate at pH 7.2 for 1 h at 37 °C in a 5% CO2 atmosphere. Samples were washed with PBS, fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.4 for 1 h at 37 °C and left at 4 °C in 0.1 M sodium cacodylate buffer until postfixation, performed with a mixture of 1% OsO4, 1.5% K4Fe(CN)6 in 0.1 M sodium cacodylate pH 7.4 for 1 h at 4° and overnight incubation in 0.25% uranyl acetate at 4 °C. After three water washes, samples were dehydrated in series of 15 min steps in 25%, 50%, 75%, 95% and 100% (v/v) ethanol and embedded in an epoxy resin (Sigma-Aldrich). Ultrathin sections (60–70 nm) were obtained with an Ultrotome V (LKB) ultramicrotome and counterstained with 1% uranyl acetate for 15 min and 1% lead citrate for 6 min. In cell populations co-transfected with the pIRES2-eGFP and SureSilencing® shRNA plasmids, GFP+ cells were sorted with a FACS Aria sorter (BD Pharmingen) and fixed 24 h after re-seeding in complete glucose-supplemented DMEM. Electron microscopy (EM) imaging of cells was performed as described [8] and thin sections were imaged using a Tecnai G2 (FEI) transmission electron microscope operating at 100 kV. Images were captured with a Veleta (Olympus Soft Imaging System) digital camera. For morphometric analysis of mitochondrial cristae in randomly selected cells, maximal cristae width was measured on at least five mitochondria/cell from six randomly selected cells (n = 3 independent experiments) using the ImageJ Multimeasure plug-in as previously described [8]. To further quantify ultrastructure changes, the number of horizontal cristae and cristae junctions were manually quantified. Cristae profiles were obtained with the plot profile plugin of ImageJ software from the line transversally crossing individual cristae along the major axis of mitochondria.
4.1.9. Isolated mitochondrial assays
Mitochondria were individualized in Isolation Buffer (IB: 10 mM Tris-MOPS, 1 mM EGTA/Tris, 200 mM Sucrose) as described [69] from cells plated in 150 cm2 plates and incubated for 48 h in complete glucose supplemented DMEM. After isolation, mitochondrial protein concentration was determined by the Bradford assay (BioRad).
4.1.10. Blue-native polyacrylamide gel electrophoresis
Isolated mitochondria were resuspended to 0.5 mg/ml protein in NativePAGE Sample buffer (Invitrogen) containing 1.1% (w/v) digitonin (Sigma) and protease-inhibitor cocktail (Sigma). After 5 min on ice, the lysate was spun at 20,000×g for 30 min at 4 °C. G250 (5%, 1 μl/100 μg protein, Invitrogen) was added to the supernatant at 1 μl/100 μg protein and 40 μg of protein were loaded onto a 3–12% native precasted gels (Invitrogen). Running was performed at 4 °C for 30 min at 150 V in cathode buffer 1 (1X Native Page buffer, 1X Native Blue buffer; Invitrogen) and further 90 min at 250 V in cathode buffer 2 (1X Native Page buffer, 0.1X Native Blue buffer; Invitrogen). Transfer of native gels was carried out overnight at constant 35 V onto PVDF membranes, fixed for 10 min in 8% (v/v) acetic acid and activated for 5 min in absolute methanol before blocking and probing with antibodies in 5% (w/v) skinned milk.
4.1.11. Immunoblotting
Immunoblotting in SDS-PAGE experiments with samples collected in cells transfected and incubated for 24 h in either glucose or galactose medium, was performed with (50 μg) loaded protein immunodecorated with mouse monoclonal anti-GSS (1:1000) (Sc-166882, Santa Cruz Biotechnologies, Heidelberg, Germany), mouse monoclonal anti-GSR (Sc-133159, Santa Cruz Biotechnologies), mouse monoclonal anti-SOD2 (1:2000) (ab16,956, Abcam), rabbit anti-catalase (1:1000) (PA5-23246, Thermo Scientific), anti-Nrf2 (sc30915, Santa Cruz Biotechnologies) or mouse monoclonal anti-GAPDH (1:40,000) (glyceraldehyde-3-phosphate dehydrogenase; 4300 Ambion, Cambridge, UK) antibodies.
4.1.12. In-gel ATPase activity assay
ATPase in-gel activity was measured as previously described [31,70,71]. After running native gels, ATPase activity was assessed in gels by incubating them overnight in ATPase activity buffer (35 mM Tris, 270 mM glycine, 14 mM MgSO4, 0.2% Pb(NO3)2 and 8 mM ATP, pH 7.8) at room temperature. Gels were washed in water to stop the reaction ATP measurement and scanned for densitometric analysis. All uncropped gels and immunoblots are shown in Fig. S8.
4.1.13. Cell death assays
For cell death analysis, 3.0 × 103 cells/cm2 of the indicated genotype were seeded and, after 24 h, co-transfected with SureSilencing® shRNA and pIRES2-eGFP plasmids (3:1 ratio) and further incubating for 72 h in galactose-supplemented DMEM. To analyze cell death, after transfection cells were incubated for 24 h in the indicated medium, treated with 5 μM antimycin A for 6 h and subsequently were trypsinized for analysis. Cell death was assessed by flow cytometry detection (FACSCalibur) of double Annexin-V-APC/PI or Annexin-V/7-AAD positive events from the transfected GFP+ cell population when cells were transfected.
4.1.14. Statistical analysis
All measurements in cell culture were carried out, at least, in triplicate, and the results are expressed as the mean ± SEM values of the indicated number (n) of independent experiments. Statistical significance was determined by ANOVA between the indicated samples with Bonferroni post hoc analysis. P values are indicated in the legends (P < 0.05 was considered to indicate a significant difference).
Author contributions
R.Q.C, I.M.R., C.V.G. and M.C. performed research and analyzed data, J.P.B. provided reagents, conceptualized experiments and edited the manuscript. R.Q.C. and L.S. conceived the project, acquired funds, designed and supervised research, analyzed data and wrote the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Graphical abstract was designed using templates from Servier Medical Art (SMART). We thank Drs. F. Caicci and F. Boldrin (Department of Biology, University of Padova) for EM sample preparation. We also thank Drs. S. J. Remington (Univ. of Oregon, U.S.), N. Demaurex (Univ. of Geneva, Switzerland) and H. Imamura and H. Koji (University of Osaka, Japan) for kindly sharing genetic probes for mitochondrial measurements. R.Q.-C was supported by an AIRC and a Fondazione Veronesi fellowship and by a H2020 Marie Sklodowska-Curie IEF GA793987. J.P.B. is funded by Agencia Estatal de Investigación (PID2019-105699RB-I00/AEI/10.13039/501100011033 and RED2018‐102576‐T), Instituto de Salud Carlos III (CB16/10/00282), Escalera de Excelencia CLU-2017-03, Ayudas Equipos Investigación Biomedicina 2017 Fundación BBVA and Fundación Ramón Areces. This work was supported by AIRC IG19991, Italian Ministry of Education, University and Research PRIN 2017BF3PXZ, Fondation Leducq TNE15004, Muscular Dystrophy Association (MDA USA) RG 603731 and Cariparo Foundation SIGMI (to L.S.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101944.
Contributor Information
Rubén Quintana-Cabrera, Email: ruben310@usal.es.
Luca Scorrano, Email: luca.scorrano@unipd.it.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Walker J.E. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 2.Kucharczyk R., Zick M., Bietenhader M., Rak M., Couplan E., Blondel M., Caubet S.D., di Rago J.P. Mitochondrial ATP synthase disorders: Molecular mechanisms and the quest for curative therapeutic approaches. Biochim. Biophys. Acta Mol. Cell Res. 2009;1793:186–199. doi: 10.1016/j.bbamcr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Reid R.A., Moyle J., Mitchell P. Synthesis of adenosine triphosphate by a protonmotive force in rat liver mitochondria. Nature. 1966;212:257–258. doi: 10.1038/212257a0. [DOI] [PubMed] [Google Scholar]
- 4.Gilkerson R.W., Schon E.A., Hernandez E., Davidson M.M. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J. Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel F., Bornhövd C., Neupert W., Reichert A.S. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogliati S., Enriquez J.A., Scorrano L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci. 2016;41:261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Hackenbrock C.R. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J. Cell Biol. 1966;30:269–297. doi: 10.1083/jcb.30.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogliati S., Frezza C., Soriano M.E. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varanita T., Soriano M.E., Romanello V. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metabol. 2015;21:834–844. doi: 10.1016/j.cmet.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Civiletto G., Varanita T., Cerutti R., Gorletta T., Barbaro S., Marchet S., Lamperti C., Viscomi C., Scorrano L., Zeviani M. Opa1 overexpression ameliorates the phenotype of two mitochondrial disease mouse models. Cell Metabol. 2015;21:845–854. doi: 10.1016/j.cmet.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephan T., Brüser C., Deckers M. MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 2020 doi: 10.15252/embj.2019104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernansanz-Agustín P., Choya-Foces C., Carregal-Romero S. Na+ controls hypoxic signalling by the mitochondrial respiratory chain. Nature. 2020:1–5. doi: 10.1038/s41586-020-2551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milenkovic D., Blaza J.N., Larsson N.G., Hirst J. The enigma of the respiratory chain supercomplex. Cell Metabol. 2017;25:765–776. doi: 10.1016/j.cmet.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Javadov S., Jang S., Chapa-Dubocq X.R., Khuchua Z., Camara A.K. Mitochondrial respiratory supercomplexes in mammalian cells: structural versus functional role. J. Mol. Med. 2021;99:57–73. doi: 10.1007/s00109-020-02004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brave F., Becker T. Supercomplex formation boosts respiration. EMBO Rep. 2020;21 doi: 10.15252/embr.202051830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudkina N.V., Eubel H., Keegstra W., Boekema E.J., Braun H.P. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3225–3229. doi: 10.1073/pnas.0408870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lapuente-Brun E., Moreno-Loshuertos R., Aciń-Pérez R. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. 80. [DOI] [PubMed] [Google Scholar]
- 18.Acín-Pérez R., Bayona-Bafaluy M.P., Fernández-Silva P., Moreno-Loshuertos R., Pérez-Martos A., Bruno C., Moraes C.T., Enríquez J.A. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maranzana E., Barbero G., Falasca A.I., Lenaz G., Genova M.L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex i. Antioxidants Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frey T.G., Mannella C.A. The internal structure of mitochondria. Trends Biochem. Sci. 2000;25:319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 21.Quintana-Cabrera R., Mehrotra A., Rigoni G., Soriano M.E. Who and how in the regulation of mitochondrial cristae shape and function. Biochem. Biophys. Res. Commun. 2018;500:94–101. doi: 10.1016/j.bbrc.2017.04.088. [DOI] [PubMed] [Google Scholar]
- 22.Wolf D.M., Segawa M., Kondadi A.K., Anand R., Bailey S.T., Reichert A.S., Bliek A.M., Shackelford D.B., Liesa M., Shirihai O.S. Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J. 2019 doi: 10.15252/embj.2018101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scorrano L., Ashiya M., Buttle K., Weiler S., Oakes S.A., Mannella C.A., Korsmeyer S.J. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 24.Frezza C., Cipolat S., Martins de Brito O. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi R., Lartigue L., Perkins G., Scott R.T., Dixit A., Kushnareva Y., Kuwana T., Ellisman M.H., Newmeyer D.D. Opa1-Mediated cristae opening is bax/bak and BH3 dependent, required for apoptosis, and independent of bak oligomerization. Mol. Cell. 2008;31:557–569. doi: 10.1016/j.molcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cipolat S., Rudka T., Hartmann D. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 27.Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 28.Amutha B., Gordon D.M., Gu Y., Pain D. A novel role of Mgm1p, a dynamin-related GTPase, in ATP synthase assembly and cristae formation/maintenance. Biochem. J. 2004;381:19–23. doi: 10.1042/BJ20040566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quintana-Cabrera R., Quirin C., Glytsou C. The cristae modulator Optic atrophy 1 requires mitochondrial ATP synthase oligomers to safeguard mitochondrial function. Nat. Commun. 2018;9:3399. doi: 10.1038/s41467-018-05655-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patten D.A., Wong J., Khacho M. OPA1‐dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 2014;33:2676–2691. doi: 10.15252/embj.201488349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes L.C., Benedetto G Di, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Fabuel I., Le Douce J., Logan A., James A.M., Bonvento G., Murphy M.P., Almeida A., Bolaños J.P. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. U. S. A. 2016;113:13063–13068. doi: 10.1073/pnas.1613701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acin-Perez R., Enriquez J.A. The function of the respiratory supercomplexes: the plasticity model. Biochim. Biophys. Acta Bioenerg. 2014;1837:444–450. doi: 10.1016/j.bbabio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Ghelli A., Tropeano C.V., Calvaruso M.A. The cytochrome b p.278Y>C mutation causative of a multisystem disorder enhances superoxide production and alters supramolecular interactions of respiratory chain complexes. Hum. Mol. Genet. 2013;22:2141–2151. doi: 10.1093/hmg/ddt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vicente-Gutierrez C., Bonora N., Bobo-Jimenez V. Astrocytic mitochondrial ROS modulate brain metabolism and mouse behaviour. Nat. Metabol. 2019;1:201–211. doi: 10.1038/s42255-018-0031-6. [DOI] [PubMed] [Google Scholar]
- 36.Jimenez-Blasco D., Busquets-Garcia A., Hebert-Chatelain E. Glucose metabolism links astroglial mitochondria to cannabinoid effects. Nature. 2020;583:603–608. doi: 10.1038/s41586-020-2470-y. [DOI] [PubMed] [Google Scholar]
- 37.Madreiter-Sokolowski C.T., Thomas C., Ristow M. Interrelation between ROS and Ca2+ in aging and age-related diseases. Redox Biol. 2020:101678. doi: 10.1016/j.redox.2020.101678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 39.Plecitá-Hlavatá L., Ježek P. Integration of superoxide formation and cristae morphology for mitochondrial redox signaling. Int. J. Biochem. Cell Biol. 2016;80:31–50. doi: 10.1016/j.biocel.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Glytsou C., Calvo E., Cogliati S. Optic atrophy 1 is epistatic to the core MICOS component MIC60 in mitochondrial cristae shape control. Cell Rep. 2016;17:3024–3034. doi: 10.1016/j.celrep.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pernas L., Scorrano L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 2016;78:505–531. doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]
- 42.Giacomello M., Pyakurel A., Glytsou C., Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020;21:204–224. doi: 10.1038/s41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 43.Gohil V.M., Sheth S.A., Nilsson R. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat. Biotechnol. 2010;28:249–255. doi: 10.1038/nbt.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S.J., Capaldi R.A. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Canc. Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 45.Irwin W.A., Bergamin N., Sabatelli P. Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat. Genet. 2003;35:367–371. doi: 10.1038/ng1270. [DOI] [PubMed] [Google Scholar]
- 46.Santo-Domingo J., Giacomello M., Poburko D., Scorrano L., Demaurex N. OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange. EMBO J. 2013;32:1927–1940. doi: 10.1038/emboj.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imamura H., Huynh Nhat K.P., Togawa H., Saito K., Iino R., Kato-Yamada Y., Nagai T., Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahn A., Parey K., Bublitz M., Mills D.J., Zickermann V., Vonck J., Kühlbrandt W., Meier T. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol. Cell. 2016;63:445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blum T.B., Hahn A., Meier T., Davies K.M., Kühlbrandt W. Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc. Natl. Acad. Sci. U. S. A. 2019;116:4250–4255. doi: 10.1073/pnas.1816556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davies K.M., Anselmi C., Wittig I., Faraldo-Gómez J.D., Kühlbrandt W. Structure of the yeast F 1F o-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss M., Hofhaus G., Schröder R.R., Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold I., Pfeiffer K., Neupert W., Stuart R.A., Schägger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paumard P. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyle G.M., Roucou X., Nagley P., Devenish R.J., Prescott M. Identification of subunit g of yeast mitochondrial F1F0-ATP synthase, a protein required for maximal activity of cytochrome c oxidase. Eur. J. Biochem. 1999;262:315–323. doi: 10.1046/j.1432-1327.1999.00345.x. [DOI] [PubMed] [Google Scholar]
- 55.Bornhövd C., Vogel F., Neupert W., Reichert A.S. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J. Biol. Chem. 2006;281:13990–13998. doi: 10.1074/jbc.M512334200. [DOI] [PubMed] [Google Scholar]
- 56.Habersetzer J., Larrieu I., Priault M., Salin B., Rossignol R., Brèthes D., Paumard P. Human F1F0 ATP synthase, mitochondrial ultrastructure and OXPHOS impairment: a (Super-)Complex matter? PloS One. 2013;8 doi: 10.1371/journal.pone.0075429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stephan T., Roesch A., Riedel D., Jakobs S. Live-cell STED nanoscopy of mitochondrial cristae. Sci. Rep. 2019;9:1–6. doi: 10.1038/s41598-019-48838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daum B., Walter A., Horst A., Osiewacz H.D., Kühlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rabl R., Soubannier V., Scholz R. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anand R., Strecker V., Urbach J., Wittig I., Reichert A.S. Mic13 is essential for formation of crista junctions in mammalian cells. PloS One. 2016;11 doi: 10.1371/journal.pone.0160258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kondadi A.K., Anand R., Hänsch S. Cristae undergo continuous cycles of membrane remodelling in a MICOS ‐dependent manner. EMBO Rep. 2020 doi: 10.15252/embr.201949776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F.T., Von Kleist-Retzow J.C., Waisman A., Westermann B., Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22:476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friedman J.R., Mourier A., Yamada J., McCaffery J.M., Nunnari J. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. Elife. 2015 doi: 10.7554/eLife.07739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Acehan D., Malhotra A., Xu Y., Ren M., Stokes D.L., Schlame M. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys. J. 2011;100:2184–2192. doi: 10.1016/j.bpj.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Formentini L., Pereira M.P., Sánchez-Cenizo L., Santacatterina F., Lucas J.J., Navarro C., Martínez-Serrano A., Cuezva J.M. In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 2014;33:762–778. doi: 10.1002/embj.201386392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M.R., Abramov A.Y., Tinker A., Duchen M.R. Regulation of mitochondrial structure and function by the F1fo-ATPase inhibitor protein, IF1. Cell Metabol. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Hanson G.T., Aggeler R., Oglesbee D., Cannon M., Capaldi R.A., Tsien R.Y., Remington S.J. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 68.Quintana-Cabrera R., Fernandez-Fernandez S., Bobo-Jimenez V., Escobar J., Sastre J., Almeida A., Bolaños J.P. γ-Glutamylcysteine detoxifies reactive oxygen species by acting as glutathione peroxidase-1 cofactor. Nat. Commun. 2012;3:718. doi: 10.1038/ncomms1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frezza C., Cipolat S., Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 70.Bisetto E., Di Pancrazio F., Simula M.P., Mavelli I., Lippe G. Mammalian ATPsynthase monomer versus dimer profiled by blue native PAGE and activity stain. Electrophoresis. 2007;28:3178–3185. doi: 10.1002/elps.200700066. [DOI] [PubMed] [Google Scholar]
- 71.Alirol E., James D., Huber D., Marchetto A., Vergani L., Martinou J.-C., Scorrano L. The mitochondrial fission protein hFis1 requires the endoplasmic reticulum gateway to induce apoptosis. Mol. Biol. Cell. 2006;17:4593–4605. doi: 10.1091/mbc.E06-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.