Abstract
Japanese quail is a low-fat, meat-bird species exhibiting high disease resistance. Cathelicidins (CATHs) are host defense peptides conserved across numerous vertebrate species that play an important role in innate immunity. The activity of host defense peptides can be affected by amino acid substitutions. However, no polymorphisms in avian CATH genes have been reported to date. The aim of this study was to clarify the polymorphisms in CATHs in Japanese quail. DNA for genomic analyses was extracted from the peripheral blood of 99 randomly selected quail from 6 inbred lines. A total of 6, 4, 6, and 4 CjCATH1, -2, -3, and -B1 alleles were identified, respectively. Nine haplotypes, including 4 strain-specific haplotypes, were identified by combining alleles at the CjCATH1, -2, -3, and -B1 loci. In addition, 2 and 1 amino acid substitutions (I145F, Q148H, and P245H) predicted by PROVEAN and PolyPhen-2 to have deleterious effects were detected in CjCATH2 and -B1, respectively. Synthetic CjCATH2 and -B1 peptides exhibited greater antibacterial activity against Escherichia coli than chicken CATH2 and -B1, respectively. Furthermore, the CjCATHB1∗04 peptide exhibited less potent antimicrobial activity than other CjCATHB1 peptides examined. This is the first report of amino acid substitutions accompanied by changes in antibacterial activity in avian CATHs. These findings could be employed as indicators of improvements in innate immune response in poultry.
Key words: amino acid substitution, antimicrobial activity, cathelicidin, host defense peptide, SNP
Introduction
Host defense peptides (HDPs) are small cationic peptides conserved among all vertebrate species (Zasloff, 2002) that function as important mediators of innate immune responses (Yang et al., 2004). Cathelicidins (CATHs) are short (<40 amino acid residues), cationic, amphipathic peptides that have been identified in a variety of vertebrate species (Zanetti, 2004). CATHs are composed of a cathelin-like domain in the N-terminal region of the precursor. Although the CATH signal peptide and cathelin-like domain are highly conserved across species, the sequence of the C-terminal mature peptide varies significantly (Zhang and Sunkara, 2014).
CATH genes have been identified in 15 avian species (Feng et al., 2011; Zhang and Sunkara, 2014; Cheng et al., 2015; Ishige et al., 2017). The 4 chicken CATH genes (GgCATH1, -2, -3, and -B1) are tightly clustered in a 7.5-kb region at the proximal end of chromosome 2 (Xiao et al., 2006). Avian CATHs exhibit antimicrobial activity against gram-positive bacteria such as Listeria monocytogenes and Staphylococcus (Xiao et al., 2006; Bommineni et al., 2007).
In the genome of Japanese quail, we identified a cluster of 4 chicken orthologous CATH genes (CjCATH1, -2, -3, and -B1) located within approximately 13 kb of one another. This cluster was highly conserved in synteny with chickens and quail (Ishige et al., 2017). Therefore, investigations of polymorphisms and the function of quail CATHs may contribute to improving the anti-disease properties of chickens and other pheasants.
The identification of amino acid substitutions in mediators of innate immune responses can help elucidate the relationship between host genomic variations and protection against invading pathogens (Hasenstein and Lamont, 2007; Pankratz et al., 2010; Barreiro et al., 2012). SNPs and other mutations in HDP genes can affect the function of HDPs and cause differential susceptibility to infections (Matsushita et al., 2002; Hasenstein and Lamont, 2007; Rivas-Santiago et al., 2009; Hellgren et al., 2010).
CATHs are polymorphic in nature, with several known sequence variations in frogs (Yu et al., 2013), buffalos (Brahma et al., 2015), and humans (López et al., 2014). Furthermore, copy number variations have been reported in CATH genes in cattle (Chen et al., 2017). A substitution from Phe to Trp in an N-terminal segment of GgCATH2 resulted in variants with improved antimicrobial activity against Bacillus anthracis and Yersinia pestis (Molhoek et al., 2010). Inactive truncated GgCATH2-derived peptides containing a Phe to Trp substitution exhibited endotoxin neutralization capacity (van Dijk et al., 2016). In contrast, a Phe to Tyr amino acid substitution abrogated the endotoxin neutralization activity of GgCATH2 analogues (van Dijk et al., 2016).
In the present study, we identified the haplotypes and amino acid substitutions in the CATH gene cluster of Japanese quail (CjCATH) and characterized the effects of these amino acid substitutions on antimicrobial activity.
Materials and methods
Animal Care
Management of Japanese quail and all procedures involving animals in the present study were performed in accordance with the Animal Experimental Guidelines of Tokyo University of Agriculture.
Japanese Quail
A total of 99 adult quail representing 6 strains (18 A [high IgG], 15 B [low IgG], 15 K [dark], 19 ND [neuron disease], 16 P [panda], and 16 Y [yellow]) were maintained in the Laboratory of Animal Physiology at the Tokyo University of Agriculture, Kanagawa, Japan (Ishige et al., 2020).
Nucleic Acid Isolation
Peripheral blood samples were collected from the jugular vein. Genomic DNA was isolated from peripheral red blood cells using a DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer's protocol.
Amplification of Genomic Fragments of 4 CjCATH Genes
Four fragments (approximately 325–517 bp) were amplified using 4 primer pairs (Supplementary Table 1) designed based on each of the 4 CjCATH genes (GenBank accession no. LC136907). Each 50-μL PCR reaction mixture contained 50 ng of quail genomic DNA, 1.25 U of PrimeSTAR GXL DNA polymerase (Takara, Kyoto, Japan), 1 × PrimeSTAR GXL buffer (Mg2+ concentration 1 mmol/L), 0.2 mmol/L of each dNTP, and 0.2 μmol/L of each primer. Cycling parameters were as follows: initial denaturation at 98°C for 1 min, followed by 35 cycles of denaturation at 98°C for 10 s, annealing at 58°C for 15 s, and extension at 68°C for 30 s. A 2-μL aliquot of each PCR reaction was then analyzed by electrophoresis on a 1.0% agarose gel. PCR products were purified using a QIAquick PCR Purification Kit (QIAGEN) and then subjected to direct sequencing using an ABI Capillary System (Macrogen Research, Macrogen Inc., Seoul, Korea).
Amplicon Sequencing Using MiSeq
The partial nucleotide sequences of CjCATH1, -2, -3, and -B1, including insertions or deletions, were determined using MiSeq (Illumina Inc., San Diego, CA). The forward and reverse primers included the Illumina adapter overhang nucleotide sequences (Supplementary Table 1). Each 50-μL PCR reaction mixture contained 50 ng of quail genomic DNA, 25 μL of KAPA HiFi HotStart ReadyMix (KAPA, Biosystems, Wilmington, WA), and 1 μmol each of forward and reverse primers. PCR products were purified using AMPure XP (Beckman Coulter, Brea, CA). The index PCR was conducted using a 50-μL mixture consisting of 25 μL of KAPA HiFi HotStart ReadyMix, 5 μL of Nextera XT Index Primer 1 (Illumina Inc.), 5 μL of Nextera XT Index Primer 2 (Illumina Inc.), 10 μL of sterile H2O, and 5 μL of the first-PCR product. PCR products were purified using AMPure XP. Library quality was assessed using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA). The libraries were sequenced as paired-end, 300-bp reads using a MiSeq (Illumina Inc.) platform according to the manufacturer's instructions. All sequence data associated with this project were submitted to the DNA Data Bank of Japan Sequence Reads Archive (DRA006654).
Amplicon Sequencing Analysis
The overall quality of the MiSeq reads was evaluated using FastQC. The reads were trimmed 1 bp from both the site and low-quality ends (Phred score <30) using the FASTX ToolKit (http://hannonlab.cshl.edu/fastx_toolkit/), and unpaired reads were removed. The MiSeq reads were joined using fastq-join (Esposito et al., 2015) with the option –p 5 –m 40 and adapter trimming was performed using cutadapt 1.2.1 (Martin, 2011) with the option –anywhere = CTGTCTCTTATACACATCT, -o 9, -e 0.2, -m 250, and -M 600. The joined reads were mapped using Burrows-Wheeler Aligner (Li and Durbin, 2009) with default parameters against the PhiX sequence (GenBank accession no. NC_001422), creating unmapped reads for further analysis. Reads containing complete primer sequences were recovered from among the unmapped reads, and primer sequences were trimmed. Trimmed primer reads with an average quality score of >30 and without unknown nucleotides were retained for further analysis. High-quality reads were converted to a FASTA file, and identical sequences were collapsed using the FASTX Toolkit. The BLAST database was generated from each CATH using the makeblastdb program included with the BLAST 2.5.0+ package. BLAST searches (BLASTN algorithm) were used to identify the most-similar CATH sequences. In the analysis of CATH1, -2, -3, and -B1, identification as a heterozygote or homozygote was based on whether the second highest number of reads was more than one-third of the highest read number.
Comparison of Nucleotide Sequences
The length of PCR products differed between Sanger sequencing and next-generation sequencing. The sequences were therefore aligned using ClustalW (DNA Data Bank of Japan; https://clustalw.ddbj.nig.ac.jp/) in order to ensure each gene had the same length and the alleles were then identified. Genotypes and haplotypes were estimated using PHASE software, version 2 (http://stephenslab.uchicago.edu/phase/download.html) (Stephens and Scheet, 2005).
Comparison of Nucleotide and Amino Acid Sequences
The net charge of each peptide was calculated using Innovagen's Peptide Property Calculator (https://pepcalc.com/;Innovagen, Lund, Sweden). Hydropathy indexes used the value reported by Kyte and Doolittle (1982). The impact of all non-synonymous SNPs on the function of each CATH peptide was assessed using the PROVEAN (http://provean.jcvi.org/index.php) (Choi et al., 2012) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) (Adzhubei et al., 2010) software packages. A resulting PROVEAN score of less than −2.500 indicated a deleterious variant, whereas a score of greater than −2.500 indicated a neutral variant.
Antimicrobial Region Peptide Synthesis
GgCATH peptides were designed based on the mature peptide region (Figure 1). CjCATH peptides were designed from 6 peptides isolated from the mature peptide region of CjCATHs examined in this experiment based on amino acid substitutions (Figure 1). All 10 peptides were synthesized by Funakoshi Corporation (Tokyo, Japan), and all were >95% pure. Lyophilized peptides were stored desiccated at −20°C and dissolved in dimethyl sulfoxide and diluted in 10 mmol/L phosphate buffer (pH 7.2) without sodium chloride and serum.
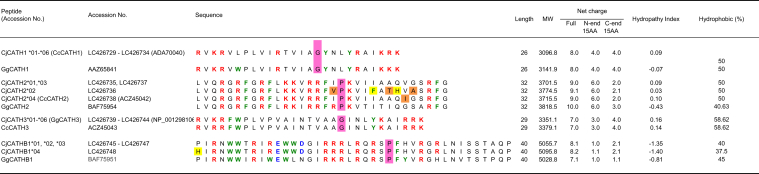
Figure 1.
Sequence diversity and properties of mature quail and chicken CATH peptides synthesized for antibacterial activity testing. Amino acid sequences of mature peptides were aligned by MEGA6. “G” and “P” with pink highlight indicate glycine and proline residues necessary for the hinge structure. Yellow highlight indicates amino acid substitution that were predicted to have possibly damaging effects on protein function. Orange highlight indicates amino acid substitutions that were predicted to have no effects on protein function. Red, blue, and green bold letters indicate cationic amino acids (K and R), acidic amino acids (D and E), and aromatic amino acid (F, Y, and W), respectively. Length: number of amino acid residues. Net charge at pH7 calculated by PepCalc.com-Peptide property (https://pepcalc.com/calculator). Hydropathy index (Kyte and Doolittle, 1982). Abbreviations: CATH, cathelicidin; MW, molecular weight.
Antimicrobial Activity Assay
Escherichia coli (NRIC 1023) was provided by the Nodai Culture Collection Center (Tokyo, Japan) and used to assess the antimicrobial activity of the 10 synthesized peptides (Figure 1). Following overnight incubation at 37°C, E. coli cells were subcultured for an additional 2 h at 37°C to reach the mid-logarithmic phase. The cells were then washed once with 10 mmol/L phosphate buffer (pH 7.2) and suspended to a concentration of 1 × 104 cfu/mL in the same buffer. Ninety microliters of the E. coli suspension were transferred into 0.2-mL tubes, followed by the addition of 10 μL of serially diluted peptide in triplicate. After 3-hour incubation at 37°C, the cultures were plated onto Lennox L Broth agar (pH 7.0, 0.086 M NaCl) and incubated overnight at 37°C, after which the number of surviving bacterial colonies was determined.
Statistical Analyses
Differences in antimicrobial activity were analyzed using one-way ANOVA, followed by the Holm-Bonferroni test for unpaired data. Differences with a P-value of <0.05 were considered statistically significant.
Results
Sequence Diversity
To assess allelic diversity among the 4 CjCATH mature peptide–encoding regions, PCR products were amplified from 99 quail specimens representing 6 strains. All PCR products were sequenced using a capillary sequencer. However, a portion of the CATH1, -2, and -B1 alleles had deletions or insertions. In contrast, the CATH3 primer amplified both CATH1 and -3; therefore, alleles in these samples were identified using MiSeq (Supplementary Table 2).
A total of 7 SNPs were detected in the CATH1 gene (517 bp), with 5 in intron3 and 2 in exon4 (Supplementary Figure 1). One SNP with a synonymous substitution was detected in the mature peptide–encoding region. Six alleles (CjCATH1∗01–∗06: GenBank accession nos. LC426729–LC426734) were identified based on these 7 SNPs in CjCATH1 (Supplementary Figure 1), including 3 major alleles (CjCATH1∗01 [36.4%], CjCATH1∗02 [25.8%], and CjCATH1∗03 [18.7%]) and 3 minor alleles (CjCATH1∗04 [9.1%], CjCATH1∗05 [6.1%], and CjCATH1∗06 [4.0%]). CjCATH1∗04 and CjCATH1∗06 were specifically detected in strains K and B, respectively (Table 1).
Table 1.
CjCATH haplotype frequencies in 6 Japanese quail strains.
| Haplotype | Locus |
Strain (number) |
Total |
|||||
|---|---|---|---|---|---|---|---|---|
| CATH3-2-B1-1 | A |
B |
K |
ND |
P |
Y |
||
| (18) | (15) | (15) | (19) | (16) | (16) | (99) | ||
| HT1 | ∗01-∗01-∗01-∗01 | 0.36 | - | 0.07 | 0.841 | 0.561 | 0.19 | 0.36 |
| HT2 | ∗02-∗02-∗02-∗02 | 0.19 | 0.731 | 0.10 | - | 0.44 | 0.16 | 0.26 |
| HT3 | ∗02-∗01-∗01-∗01 | - | - | - | 0.032 | - | - | 0.01 |
| HT4 | ∗02-∗01-∗03-∗03 | - | - | - | 0.032 | - | - | 0.01 |
| HT5 | ∗03-∗01-∗03-∗03 | 0.39 | - | - | - | - | 0.19 | 0.10 |
| HT6 | ∗03-∗01-∗03-∗05 | 0.06 | - | 0.23 | 0.03 | - | 0.06 | 0.06 |
| HT7 | ∗04-∗04-∗04-∗04 | - | - | 0.601,2 | - | - | - | 0.09 |
| HT8 | ∗05-∗01-∗03-∗03 | - | - | - | 0.08 | - | 0.41 | 0.08 |
| HT9 | ∗06-∗03-∗03-∗06 | - | 0.272 | - | - | - | - | 0.04 |
- Not observed.
Majority frequency (>50%) of strain.
Strain specific.
A total of 11 SNPs were detected in the CjCATH2 gene (464 bp) with 1 in intron3 and 10 in exon4 (Supplementary Figure 2). Four alleles (CjCATH2∗01–∗04: GenBank accession nos. LC426735–LC426738) were identified based on these 11 SNPs in CjCATH2 (Supplementary Figure 2). Six of the 10 SNPs identified in the region encoding the mature peptide were non-synonymous. This resulted in the detection of 6 amino acid substitutions: Ile140Val (I140 V), Ile145Phe (I145F), Ala147Thr (A147T), Gln148His (Q148H), Ile149Val (I149V), and Gly150Ala (G150A) (Figure 1). These 4 alleles included 2 major alleles (CjCATH2∗01 [61.1%] and CjCATH2∗02 [25.8%]) and 2 minor alleles (CjCATH2∗03 [4.0%] and CjCATH2∗04 [9.1%]). CjCATH2∗03 and CjCATH2∗04 were specifically detected in strains B and K, respectively (Table 1).
A total of 10 SNPs were detected in the CjCATH3 gene (325 bp), with 5 in intron3 and 5 in exon4 (Supplementary Figure 3). However, no SNPs were found in the mature peptide–encoding region. Six alleles (CjCATH3∗01–∗06: GenBank accession nos. LC426739–LC426744) were identified based on these 10 SNPs in CjCATH3 (Supplementary Figure 3), including 3 major alleles (CjCATH3∗01 [35.9%], CjCATH3∗02 [26.8%], and CjCATH3∗03 [16.2%]) and 3 minor alleles (CjCATH1∗04 [9.1%], CjCATH3∗05 [8.1%], and CjCATH3∗06 [4.0%]). CjCATH3∗04 and CjCATH3∗06 were specifically detected in strains K and B, respectively (Table 1).
A total of 5 SNPs were detected in the CjCATHB1 gene (509 bp), with 3 in intron3 and 2 in exon4 (Supplementary Figure 4). This resulted in the detection of 1 amino acid substitution: P246H (Figure 1). Four alleles (CjCATHB1∗01–∗04: GenBank accession nos. LC426745–LC426748) were identified based on these 5 SNPs of CjCATHB1 (Supplementary Figure 4). Two SNPs in the mature peptide–encoding region were identified, including 1 that was non-synonymous. These 4 alleles included 3 major alleles (CjCATHB1∗01 [34.3%], CjCATHB1∗02 [25.8%], and CjCATHB1∗03 [30.8%]) and 1 minor allele (CjCATHB1∗04 [9.1%]). CjCATHB1∗04 was specifically detected in strain K (Table 1).
A comparison of the CATH1, -2, -3, and -B1 genes in the 99 quail specimens revealed 6, 4, 4, and 6 alleles, respectively. We identified 9 haplotypes (HT1–HT9) based on combinations of the CATH1, -2, -3, and -B1 alleles using PHASE software (Table 1). HT1 was the major haplotype in strains ND and P, whereas HT2 was the major haplotype in strain B. Haplotypes HT3 and HT4 were detected only in strain ND, and HT9 was found only in strain B. HT7 was a major haplotype specific to strain K.
Among the 26 amino acid residues of the sequence of the mature CjCATH1 peptide, 2 differed from the sequence of GgCATH1 (L128W and R147K). The sequence of the mature CjCATH1 peptide perfectly matched that of CcCATH1. However, no significant differences in net charge or hydropathy index were observed between CjCATH1 and GgCATH1.
The 29 amino acid residues of the mature CjCATH3 peptide perfectly matched that of GgCATH3. By comparison, 1 of the 29 amino acid residues of the sequence of the mature CjCATH3 peptide differed from the sequence of CcCATH3 (K125 R).
Nine of the 32 amino acid residues of the sequence of the mature CjCATH2 peptides differed from the sequence of GgCATH2 (K134R, V136I, I/V140R, I144T, I/F146I, A147T, A/T148I, V/I150G, G/A151S, and S152A). The amino acid sequence of the mature CjCATH2∗04 peptide perfect matched that of CcCATH2. The net charge of the 15 C-terminal amino acid residues of both CjCATH2 and CcCATH2 was lower than that of GgCATH2. Conversely, the hydropathy index of both CjCATH2s and CcCATH2 was higher than that of GgCATH2. No significant inter-allelic differences were found in terms of net charge or hydropathy index among the 4 alleles of CjCATH2. The PROVEAN and PolyPhen-2 software programs were used to evaluate the possibility that 6 amino acid substitutions among the 4 alleles of CjCATH2 constituted missense mutations that could adversely affect the function of the peptide. These analyses suggested that the I145F and Q148H substitutions detected in CjCATH2∗02 would have an adverse effect on the function of the peptide (Supplementary Table 3).
Nine of the 40 amino acid residues of the mature CjCATHB1 peptide differed from the sequence of GgCATHB1 (P/H246P, T252I, R255W, W258L, D259N, R263K, H272Y, R276H, I279V, and S280T). Although no significant difference in net charge was detected between the 15 N-terminal amino acid residues of both the CjCATHB1s and GgCATHB1, the N-terminal hydropathy index of the 15 CjCATHB1s was lower than that of GgCATHB1. The net charge of the 15 C-terminal AA of CjCATHB1s was higher than that of the 15 C-terminal amino acid residues of GgCATHB1, but there was no significant difference in the C-terminal hydropathy index between the CjCATHB1s and GgCATHB1. No significant inter-allelic differences in terms of net charge and hydropathy index of the mature peptides were found among the 4 alleles of CjCATHB1. However, the PROVEAN and PolyPhen-2 analyses suggested that an amino acid substitution detected in CjCATHB1∗04 (Pro245His [P245H]) could affect the function of the CjCATHB1 peptide (Supplementary Table 3).
Antimicrobial Activity
Synthetic peptides representing all quail and chicken CATHs exhibited concentration-dependent antibacterial activity against E. coli. The amino acid sequence of the antimicrobial region of both chicken and Japanese quail CATH3 was the same. However, the amino acid sequences of CATH1, -2, and -B1 differed between quail and chicken (Figure 1).
At concentrations ranging from 0.1 to 1 μmol, the antimicrobial activity of the CjCATH2s (CjCATH2∗01∗03, CjCATH2∗02, and CjCATH2∗04) against E. coli was significantly more potent than that of GgCATH2. At concentrations of 0.5 and 1 μmol, the antimicrobial activity of CjCATH2∗01∗03 and CjCATH2∗02 against E. coli tended to be higher than that of CjCATH2∗04. However, there were no significant differences in the antimicrobial activity of the CjCATH2s and GgCATH2 against E. coli at concentrations of 10 and 100 μmol (Table 2). At a concentration of 0.5 μM, the antimicrobial activity of CjCATHB1∗01∗02∗03 against E. coli was significantly more potent than that of CjCATHB1∗04 or GgCATHB1. The antimicrobial activity of CjCATHB1∗01∗02∗03 and CjCATHB1∗04 at a concentration of 1 μmol was significantly more potent than that of GgCATHB1 (Table 2). However, there was no significant difference in the antimicrobial activity of CjCATHB1s and GgCATHB1 against E. coli at concentrations of 0.1, 10, and 100 μmol (Table 2). In addition, there were no significant differences in the antimicrobial activity of CjCATH1 and GgCATH1 against E. coli at any concentration examined (Table 2).
Table 2.
Effect of synthetic CATH-derived peptides on viability of the gram-negative bacterium Escherichia coli.
| Locus peptide (see Figure 1) | Peptide concentration (μmol) |
||||
|---|---|---|---|---|---|
| 0.1 | 0.5 | 1 | 10 | 100 | |
| CATH1 | |||||
| CjCATH1 (CcCATH1) | 48.0 ± 7.8 | 32.8 ± 7.8 | 8.6 ± 1.5 | 2.9 ± 1.1 | 0 |
| GgCATH1 | 54.4 ± 9.4 | 27.4 ± 9.9 | 13.8 ± 2.1 | 5.4 ± 1.6 | 0 |
| CATH2 | |||||
| CjCATH2∗01∗03 | 66.9 ± 2.7a | 17.4 ± 5.6a | 6.4 ± 1.5a | 4.7 ± 0.7 | 0 |
| CjCATH2∗02 | 76.7 ± 4.3a | 18.7 ± 3.5a | 6.1 ± 0.7a | 4.8 ± 1.0 | 0 |
| CjCATH2∗04 (CcCATH2) | 79.3 ± 6.6a | 24.6 ± 4.8a | 10.5 ± 2.0a | 6.2 ± 1.8 | 0 |
| GgCATH2 | 97.8 ± 5.4b | 82.9 ± 1.8b | 34.7 ± 7.1b | 11.0 ± 4.4 | 0 |
| CATH3 | |||||
| CjCATH3 (GgCATH3) | 42.9 ± 1.7 | 22.4 ± 7.4 | 13.1 ± 0.5 | 6.3 ± 0.9 | 0 |
| CATHB1 | |||||
| CjCATHB1∗01∗02∗03 | 92.7 ± 6.9 | 43.0 ± 0.7a | 31.4 ± 3.0a | 14.2 ± 2.2 | 0 |
| CjCATHB1∗04 | 94.4 ± 0.5 | 69.9 ± 10.9b | 31.6 ± 4.3a | 12.3 ± 3.1 | 0 |
| GgCATHB1 | 97.5 ± 4.8 | 83.6 ± 3.1b | 62.6 ± 9.1b | 8.2 ± 1.1 | 0 |
a,bMeans followed by the same small letter in the same row are not significantly different (P < 0.05).
Results are presented as means ± SEM (n = 3). Concentration-dependent differences in each CATH: a > b.
Abbreviation: CATH, cathelicidin.
Discussion
In nature, CATHs are polymorphic, exhibiting several known sequence variants in frogs (Yu et al., 2013), buffalo (Brahma et al., 2015), and humans (López et al., 2014). In addition, copy number variations have been reported in CATH genes in cattle (Chen et al., 2017). The amino acid sequences of the mature CjCATH3 and GgCATH3 peptides are identical, and the similarity between CjCATH1 and GgCATH1 is >90%. Furthermore, CjCATH2/GgCATH2 and CjCATHB1/GgCATHB1 exhibit 83.1 and 68.7% similarity, respectively (Ishige et al., 2017). The amino acid substitutions identified in CATH2 and CATHB1 may be associated with enhanced antigen sensitivity. The observed less potent antimicrobial activity of CATH2 and CATHB1 against E. coli compared with CATH3 and CATH1 supports this hypothesis. Moreover, amino acid substitutions in both CjCATH2 and CjCATHB1 were shown to affect the antimicrobial activity of these peptides against E. coli. These results were supported by analyses using 2 different software programs that predicted that amino acid substitutions in CjCATH2 and CjCATHB1 would adversely affect peptide function (PROVEAN and PolyPhen-2).
Previous studies focusing on ostrich defensins and avian NK-lysin identified an association between membrane leakage/microbicidal activity and peptide charge (Sugiarto and Yu, 2006; Lee et al., 2012). However, this association has not been verified with Japanese quail and chicken CATH2. CjCATH2 has a lower net charge than GgCATH2 but exhibited more potent antimicrobial activity at concentrations of 0.1, 0.5, and 1 μmol. Hydrophobicity is thought to be an important mediator of the interaction of CATHs with bacterial membranes (Oren and Shai, 1998; Nicolas, 2009). Loss of the first tryptophan residue in GgCATH1 diminishes the peptide's antimicrobial activity, indicating the importance of this hydrophobic residue for the activity of the peptide (Bommineni et al., 2007). Loss of the more hydrophobic C-terminal residue in GgCATH2 analogues also results in diminished bactericidal activity (Xiao et al., 2009), although C-terminal truncation of a GgCATH2 analogue (C1–15 mature peptides) enhanced its antibacterial activity (van Dijk et al., 2009). Interestingly, N-terminal truncation leaving only the hydrophobic C-terminal alpha-helix resulted in almost complete loss of antibacterial activity, probably because the first interaction with bacterial cells involves the polar portions of the N- and C-termini (van Dijk et al., 2009; Xiao et al., 2009). Substitution of phenylalanine with the more hydrophobic tryptophan in the C1 to 15 mature peptides resulted in enhanced bactericidal activity and better stability in the presence of high salt concentrations (Molhoek et al., 2010). Additionally, substitution of tyrosine with an alanine in a cecropin A–magainin-2 fusion peptide resulted in markedly reduced antibacterial activity (Dathe et al., 2001; Nan et al., 2012). These results support the hypothesis that this hydrophobic residue is important for the antimicrobial activity of avian CATH2. Although CjCATH2 synthetic peptides exhibited similar hydrophobicity indexes and antimicrobial activity against E. coli, the antimicrobial activity against E. coli of the CjCATH2∗02 synthetic peptide did not differ from that of CjCATH2∗01∗03. This suggests that 5 amino acid substitutions (including I145F and Q148H) detected in CjCATH2∗02 do not affect the antimicrobial activity against E. coli. However, the antimicrobial activity against E. coli of the CjCATH2∗04 synthetic peptide tended to be slightly lower than that of both CjCATH2∗01∗03 and CjCATH∗02. Compared with synthetic CjCATH2∗01∗03 and CjCATH2∗02, synthetic CjCATH2∗04 has only 1 amino acid substitution (V149I). These results suggest that the amino acid substitution (V149I) in CjCATH2 may affect the peptide's antimicrobial activity against E. coli. The effect of amino acid substitutions on the antimicrobial activity of CjCATH2 will be investigated in a future study.
Synthetic GgCATH1, -2, and -B1 peptides exhibit antimicrobial activity against most strains, including gram-positive and gram-negative bacteria (Xiao et al., 2006; Goitsuka et al., 2007). By contrast, synthetic CcCATH2, -3, and common pheasant CATH1 (PcCATH1) peptides exhibit antimicrobial activity against not only gram-positive and gram-negative bacteria, but also fungi (Feng et al., 2011; Wang et al., 2011). In the present study, all CjCATH peptides exhibited antimicrobial activity against E. coli. The amino acid sequences of the mature CjCATH2∗04 and CcCATH2, CjCATHB1∗01, ∗02, ∗03, and GgCATHB1 peptides are the same. These data suggest that these peptides should exhibit antimicrobial activity against gram-positive and gram-negative bacteria, as well as fungi. Among the 26 amino acids of the sequence of mature CjCATH1, 2 differed from the sequence of GgCATH1 (L128W and R147K). However, no significant differences in net charge or hydropathy index were observed between CjCATH1 and GgCATH1. Furthermore, these amino acid substitutions were determined to have no effect on the function of the peptide according to both PROVEAN (variant: score [prediction], L128W: 10.625 [neutral], and R147K: 2.25 [neutral]) and PolyPhen-2 (L128W: 0.000 [benign], and R147K: 0.000 [benign]) analyses. These results suggest that both CjCATH1 and GgCATH1 should exhibit the same antimicrobial activity against gram-positive and gram-negative bacteria. The 29-residues amino acid sequence of the mature CjCATH3 peptide perfectly matched that of GgCATH3, whereas the sequence of the mature CjCATH3 peptide differed from CcCATH3 by only 1 residue (K125R). However, no significant differences in net charge or hydropathy index were observed between CjCATH3 and CcCATH3. Furthermore, this amino acid substitution was determined to have no effect on function according to both PROVEAN (neutral: −2.0) and PolyPhen-2 (possibly damaging: 0.659) analyses. These results suggest that both CjCATH3 and CcCATH3 would exhibit different antimicrobial activity against gram-positive and gram-negative bacteria, and fungi.
Japanese quail have higher disease resistance than other poultry species (Vali, 2008). Modifications of the sequence of natural peptides can increase the hydrophobic moment and net charge while reducing hydrophobicity, all factors that are reported as crucial for cationic antimicrobial peptides, including cecropins, magainins, melittin, and the CATHs (Oliveira et al., 2020). GgCATH1, -2, and -3, and CcCATH1, -2, and -3 reportedly exhibit antimicrobial activity against Salmonella typhimurium, Salmonella maltophilia, and E. coli (Xiao et al., 2006; Bommineni et al., 2007; Feng et al., 2011; Veldhuizen et al., 2013). In this study, the antimicrobial activity of CjCATH2 and -B1 was more potent than that of GgCATH2 and -B1. The net charge of synthetic CjCATH2 was lower than that of GgCATH2. However, the percent hydrophobicity and hydropathy index values of synthetic CjCATH2 peptides were higher than those of GgCATH2. These results suggest that the antimicrobial activity of CATH2 peptides is affected by hydrophobicity. In contrast, the net charges of synthetic CjCATHB1 peptides were higher than that of GgCATHB1. However, the percent hydrophobicity and hydropathy index values of synthetic CjCATHB1 peptides were higher than those of GgCATHB1. Furthermore, comparisons of CjCATHB1∗01∗02∗03 and CjCATHB1∗04 revealed difference in antimicrobial activity at 0.5 μmol and differences in net charge, percent hydrophobicity, and hydropathy index values. As these results suggest that the antimicrobial activity of CATHB1 peptides is affected by net charge, the antimicrobial activity of CjCATHB1 could be affected by the P/H246P substitutions. The differences in antimicrobial activity observed in this study between Japanese quail and chicken HDPs could explain why Japanese quail exhibits greater disease resistance than other poultry species.
In the present study, we examined the antimicrobial activity of Japanese quail HDPs using only E. coli NRIC 1023 because we verified that all CATHs exhibit antimicrobial activity against bacteria. In a future study, we will investigate the antimicrobial activity against gram-positive and gram-negative bacteria as well as fungi using all synthetic CjCATH mature peptides.
In this study, several SNPs in the mature peptide–encoding regions of CjCATH genes were identified. CjCATH2 contains 6 non-synonymous SNPs, whereas CjCATHB1 contains 1. Three and 2 different peptides, respectively, were synthesized based on each allele. Various amino acid substitutions were identified in CATH2 and -B1 could affect the immunomodulatory and antimicrobial activity of the peptides. Thus, the genetic diversity exhibited in HDPs could affect the resistance of Japanese quail to pathogen infection.
Acknowledgments
This research was supported in part by the Advanced Research Project Type A, Tokyo University of Agriculture (no. 02, 2006-2008) and by the MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2013-2017 (S1311017).
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2021.101046.
Disclosures
The authors declare that they have no conflict of interest.
Supplementary data
References
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L.B., Tailleux L., Pai A.A., Gicquel B., Marioni J.C., Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommineni Y.R., Dai H., Gong Y.X., Soulages J.L., Fernando S.C., Desilva U., Prakash O., Zhang G. Fowlicidin-3 is an α-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities. FEBS J. 2007;274:418–428. doi: 10.1111/j.1742-4658.2006.05589.x. [DOI] [PubMed] [Google Scholar]
- Brahma B., Patra M.C., Karri S., Chopra M., Mishra P., De B.C., Kumar S., Mahanty S., Thakur K., Poluri K.M., Datta T.K., De S. Diversity, antimicrobial action and structure-activity relationship of buffalo Cathelicidins. PLoS One. 2015;19:e0144741. doi: 10.1371/journal.pone.0144741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chamberlain A.J., Reich C.M., Daetwyler H.D., Hayes B.J. Detection and validation of structural variations in bovine whole-genome sequence data. Genet. Sel. Evol. 2017;49:13. doi: 10.1186/s12711-017-0286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Prickett M.D., Gutowska W., Kuo R., Belov K., Burt D.W. Evolution of the avian β-defensin and cathelicidin genes. BMC Evol. Biol. 2015;15:188. doi: 10.1186/s12862-015-0465-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P., A P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe M., Nikolenko H., Meyer J., Beyermann M., Bienert M. Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 2001;501:146–150. doi: 10.1016/s0014-5793(01)02648-5. [DOI] [PubMed] [Google Scholar]
- Esposito A., Ahmed E., Ciccazzo S., Sikorski J., Overmann J., Holmström S.J., Brusetti L. Comparison of rock varnish bacterial communities with surrounding non-varnished rock surfaces: taxon-specific analysis and morphological description. Microb. Ecol. 2015;70:741–750. doi: 10.1007/s00248-015-0617-4. [DOI] [PubMed] [Google Scholar]
- Feng F., Chen C., Zhu W., He W., Guang H., Li Z., Wang D., Liu J., Chen M., Wang Y., Yu H. Gene cloning, expression and characterization of avian cathelicidin orthologs, Cc-CATHs, from Coturnix. FEBS J. 2011;278:1573–1584. doi: 10.1111/j.1742-4658.2011.08080.x. [DOI] [PubMed] [Google Scholar]
- Goitsuka R., Chen C.L., Benyon L., Asano Y., Kitamura D., Cooper M.D. Chicken cathelicidin-B1, an antimicrobial guardian at the mucosal M cell gateway. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15063–15068. doi: 10.1073/pnas.0707037104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenstein J.R., Lamont S.J. Chicken gallinacin gene cluster associated with Salmonella response in advanced intercross line. Avian Dis. 2007;51:561–567. doi: 10.1637/0005-2086(2007)51[561:CGGCAW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Hellgren O., Sheldon B.C., Buckling A. In vitro tests of natural allelic variation of innate immune genes (avian β-defensins) reveal functional differences in microbial inhibition. J. Evol. Biol. 2010;23:2726–2730. doi: 10.1111/j.1420-9101.2010.02115.x. [DOI] [PubMed] [Google Scholar]
- Ishige T., Hara H., Hirano T., Kono T., Hanzawa K. Characterization of the cathelicidin cluster in the Japanese quail (Coturnix japonica) Anim. Sci. J. 2017;88:1249–1257. doi: 10.1111/asj.12752. [DOI] [PubMed] [Google Scholar]
- Ishige T., Hara H., Hirano T., Kono T., Hanzawa K. Analysis of the diversity of the AvBD gene region in Japanese quail. J. Hered. 2020;111:436–443. doi: 10.1093/jhered/esaa035. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee M.O., Kim E.H., Jang H.J., Park M.N., Woo H.J., Han J.Y., Womack J.E. Effects of a single nucleotide polymorphism in the chicken NK-lysin gene on antimicrobial activity and cytotoxicity of cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12087–12092. doi: 10.1073/pnas.1209161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Campos G.N., Velarde Félix J.S., Sandoval Ramírez L., Cazares Salazar S., Corona Nakamura A.L., Amaya Tapia G., Prado Montes de Oca E. Polymorphism in cathelicidin gene (CAMP) that alters Hypoxia-inducible factor (HIF-1α:: ARNT) binding is not associated with tuberculosis. Int. J. Immunogenet. 2014;41:54–62. doi: 10.1111/iji.12080. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17:10–12. [Google Scholar]
- Matsushita I., Hasegawa K., Nakata K., Yasuda K., Tokunaga K., Keicho N. Genetic variants of human beta-defensin-1 and chronic obstructive pulmonary disease. Biochem. Biophys. Res. Commun. 2002;291:17–22. doi: 10.1006/bbrc.2002.6395. [DOI] [PubMed] [Google Scholar]
- Molhoek E.M., van Dijk A., Veldhuizen E.J., Dijk-Knijnenburg H., Mars-Groenendijk R.H., Boele L.C., Kaman-van Zanten W.E., Haagsman H.P., Bikker F.J. Chicken cathelicidin-2-derived peptides with enhanced immunomodulatory and antibacterial activities against biological warfare agents. Int. J. Antimicrob. Agents. 2010;36:271–274. doi: 10.1016/j.ijantimicag.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Nan Y.H., Bang J.K., Jacob B., Park I.S., Shin S.Y. Prokaryotic selectivity, anti-endotoxic activity and protease stability of diastereomeric and enantiomeric analogs of human antimicrobial peptide LL-37. Bull. Korean Chem. Soc. 2012;33:2883–2889. [Google Scholar]
- Nicolas P. Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J. 2009;276:6483–6496. doi: 10.1111/j.1742-4658.2009.07359.x. [DOI] [PubMed] [Google Scholar]
- Oliveira G.J.N., Cardoso H.M., Velikova N., Giesbers, Wells M.J., Rezende M.B.T., Vries de.R., Octávio L., Fran L.O. Physicochemical-guided design of cathelicidin-derived peptides generates membrane active variants with therapeutic potential. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-66164-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren Z., Shai Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Pankratz V.S., Vierkant R.A., O’Byrne M.M., Ovsyannikova I.G., Poland G.A. Associations between SNPs in candidate immune-relevant genes and rubella antibody levels: a multigenic assessment. BMC Immunol. 2010;11:48. doi: 10.1186/1471-2172-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Santiago B., Serrano C.J., Enciso-Moreno J.A. Susceptibility to infectious diseases based on antimicrobial peptide production. Infect. Immun. 2009;77:4690–4695. doi: 10.1128/IAI.01515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiarto H., Yu P.L. Identification of three novel ostricacins: an update on the phylogenetic perspective of β-defensins. Int. J. Antimicrob. Agents. 2006;27:229–235. doi: 10.1016/j.ijantimicag.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Vali N. The Japanese quail: a review. Int. J. Poult. Sci. 2008;7:925–931. [Google Scholar]
- van Dijk A., Molhoek E.M., Veldhuizen E.J., Bokhoven J.L., Wagendorp E., Bikker F., Haagsman H.P. Identification of chicken cathelicidin-2 core elements involved in antibacterial and immunomodulatory activities. Mol. Immunol. 2009;46:2465–2473. doi: 10.1016/j.molimm.2009.05.019. [DOI] [PubMed] [Google Scholar]
- van Dijk A., van Eldik M., Veldhuizen E.J., Tjeerdsma-van Bokhoven H.L., de Zoete M.R., Bikker F.J., Haagsman H.P. Immunomodulatory and anti-inflammatory activities of chicken cathelicidin-2 derived peptides. PLoS One. 2016;11:e0147919. doi: 10.1371/journal.pone.0147919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuizen E.J., Brouwer E.C., Schneider V.A., Fluit A.C. Chicken cathelicidins display antimicrobial activity against multiresistant bacteria without inducing strong resistance. PLoS One. 2013;8:e61964. doi: 10.1371/journal.pone.0061964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lu Z., Feng F., Zhu W., Guang H., Liu J., He W., Chi L., Li Z., Yu H. Molecular cloning and characterization of novel cathelicidin-derived myeloid antimicrobial peptide from Phasianus colchicus. Dev. Comp. Immunol. 2011;35:314–322. doi: 10.1016/j.dci.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Cai Y., Bommineni Y.R., Fernando S.C., Prakash O., Gilliland S.E., Zhang G. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 2006;281:2858–2867. doi: 10.1074/jbc.M507180200. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Dai H., Bommineni Y.R., Soulages J.L., Gong Y.X., Prakash O., Zhang G. The central kink region of fowlicidin-2, an α-helical host defense peptide, is critically involved in bacterial killing and endotoxin neutralization. J. Innate Immun. 2009;1:268–280. doi: 10.1159/000174822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Biragyn A., Hoover D.M., Lubkowski J., Oppenheim J.J. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu. Rev. Immunol. 2004;22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]
- Yu H., Cai S., Gao J., Zhang S., Lu Y., Qiao X., Yang H., Wang Y. Identification and polymorphism discovery of the cathelicidins, Lf-CATHs in ranid amphibian (Limnonectes fragilis) FEBS J. 2013;280:6022–6032. doi: 10.1111/febs.12521. [DOI] [PubMed] [Google Scholar]
- Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 2004;75:39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Zhang G., Sunkara L.T. Avian antimicrobial host defense peptides: from biology to therapeutic applications. Pharmaceuticals (Basel) 2014;7:220–247. doi: 10.3390/ph7030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.