Abstract
Human sirtuins (SIRT1–7) regulate not only deacetylation but also deacylation of fatty acid-derived acyl moieties (defatty-acylation) at the ε-amino group of lysine residues. SIRT-subtype-specific defatty-acylase activity modulators are needed for detailed investigation of the biological roles of these enzymes, and to find suitable small molecules, we require appropriate screening systems. Here, we designed and synthesized a set of SIRT defatty-acylase activity probes with various quencher moieties and peptide sequences based on our previously developed one-step FRET-based SIRT probe SFP3, using improved methodology. Scanning of this set of probes with SIRT isozymes revealed that certain probe/isozyme combinations showed especially high responses. To illustrate the utility of the combinations thus identified, we applied compound 18/SIRT2 for inhibitor screening of a large chemical library. This enabled us to discover a new small molecule SIRT2-specific defatty-acylase inhibitor.
Keywords: sirtuin, fluorescence probes, defatty-acylase activity, screening, inhibitors
Human sirtuins (SIRT1–7) are a family of NAD+-dependent histone deacetylases.1 SIRTs are involved in metabolic regulation, stabilization of genomic DNA, stress responses, and even aging,2−6 and SIRT modulators are considered to have potential therapeutic value.7−10 For a long time, it was thought that SIRTs only catalyze deacetylation reactions of histones and the adenine diphosphate (ADP)-ribosylation reaction. However, more recently, it was found that SIRTs can remove various acyl groups, including propionyl, butyryl, crotonyl, succinyl, hexanoyl, octanoyl, decanoyl, dodecanoyl, myristoyl, palmitoyl, lipoyl, and benzoyl groups,11,12 from histones and many other protein substrates.13,14 SIRT1–3, which have relatively potent deacetylase activity, can also remove long-chain fatty acyl groups at the ε-amino group of lysine residues in vitro.11,12
Protein fatty-acylation is crucial for anchoring proteins to the cell membrane and plays important roles in cell signaling and protein–protein interactions. Early studies of protein fatty acylation were focused on N-terminal glycine myristoylation and cysteine palmitoylation.15,16 Among human SIRTs, SIRT6 is a prominent defatty-acylase, which regulates secretion of tumor necrosis factor α (TNF-α) and exosomes.17−19 Recent work suggests that the defatty-acylase activity of SIRT2 toward the ε-amino group of lysine residues regulates the localization of K-Ras4a oncoprotein20 and promotes cellular transformation via interaction with A-Raf.20 In addition to K-Ras4a, the defatty-acylase activity of SIRT2 regulates the activity of RalB and cell migration.21 Therefore, SIRT2 defatty-acylase activity plays an important role in cancer proliferation. However, so far, there has been no report on the physiological roles of the defatty-acylase activities of SIRT1 and SIRT3. Therefore, SIRT-subtype-specific modulators are required both as tools for basic research to understand the physiological functions of SIRTs and as candidate therapeutic agents.
So far, SIRT fluorescent probes employing various detection methods have been developed. Generally, they involve trypsin digestion of a C-terminal lysine residue after the SIRT enzymatic reaction.22,23 However, this requirement is problematic for screening purposes because trypsin inhibitors generate false-positive signals. Therefore, a one-step procedure for detection of SIRT activity would be preferable. Some one-step probes employing an intramolecular reaction mechanism, which affords a fluorescence increase after deacylation of a lysine residue, have been reported,24−26 but unfortunately, the background fluorescence signal gradually increases even in the absence of enzymes. Dai and coworkers27 developed a Förster resonance energy transfer (FRET)-based fluorescence probe containing a 2-aminobenzoylamide group at the acyl side chain as a fluorophore. Its fluorescence is activated by SIRT-mediated hydrolytic release of the fluorophore moiety, but the absorption and fluorescence wavelengths of the fluorophore, 2-aminobenzoylamide, are quite short and may overlap with those of chemical compounds to be screened. Therefore, there is still a need for a stable, one-step fluorescence probe with a longer fluorescence wavelength for chemical screening purposes.
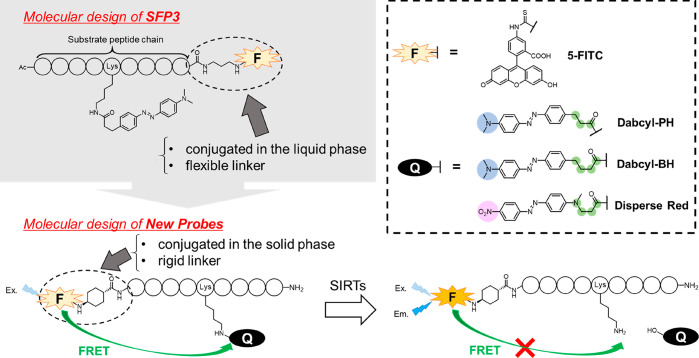
We have already reported a one-step chemical probe for SIRT activity, SFP3, which can evaluate the defatty-acylase activity of SIRT1–3 and SIRT6 (Figure S1).28 SFP3 is a FRET-based fluorescence probe containing a 4-(4-dimethylaminophenylazo)benzoyl (Dabcyl) group as a quencher, and its fluorescence is activated by SIRT-mediated hydrolytic release of the Dabcyl moiety, as shown in Figure 1. The long structure of the Dabcyl group appears to mimic a long-chain fatty acyl group because the Km values of the Dabcyl quencher and the myristoyl group are similar.28,29 However, for purposes such as high-throughput chemical screening and subtype-specific imaging of cellular SIRT, it would be useful to have a set of probes with different characteristics. Therefore, in this work, we set out to build a set of probes incorporating various peptide sequences and quenchers using improved synthetic methodology (Figure 1). We evaluated the kinetic parameters of the obtained probes toward SIRT1–7 and compared them with the corresponding parameters of SFP3. We found that certain combinations of probes and isozymes showed especially high responses, supporting the value of our approach for developing SIRT-subtype-specific probes. We confirmed its practical utility by applying one of the new probes, 18, which showed especially high sensitivity to SIRT2, for SIRT2 inhibitor screening. This enabled us to discover a new small molecule SIRT2 defatty-acylase inhibitor.
Figure 1.
Molecular design strategy and function of newly designed one-step SIRT activity probes employing the FRET mechanism.
Results and Discussion
Molecular Design Strategy for New SIRT Activity Probes
For the development of the new fluorescence probes, we modified the original design and synthetic scheme used for our previously reported probe, SFP3,28,29 as shown in Figure 1. In SFP3, the linker and fluorophore were conjugated at the C-terminus of the H3K9 peptide, which was obtained by solid-phase synthesis (SPS). However, the linker and fluorophore moieties were subsequently conjugated in the liquid phase, which requires an increased number of synthetic steps, resulting in a low yield and many byproducts. In our new design, the linker and fluorophore are conjugated at the N-terminus of the peptide chain, and the scheme was modified to allow all the components to be incorporated by means of SPS. In addition, SFP3 showed unwanted interactions (e.g., intramolecular stacking interactions) between the fluorophore and the quencher group due to its highly flexible propyl linker. In our new design, the linker structure was changed to a more rigid cyclohexane ring with the aim of preventing such stacking interactions and increasing the reactivity with SIRTs.
According to the above strategy, we designed 13 with a 9-residue H4K16 peptide as a substrate peptide and Dabcyl-PH as a quencher, followed by 14 and 15, which have a 7- and an 11-residue H4K16 peptide, respectively. We next designed 16 and 17, which contain the “RIKRY” sequence of peptide-type SIRT2-selective inhibitors (S2iL8, S2iD7),30 anticipating that these sequences would increase the binding affinity to SIRT2. Our previous findings indicated that a longer linker structure of the Dabcyl group increases the reactivity with SIRT.28 Thus, we next designed 18 with Dabcyl-BH as a quencher, which is longer by one carbon atom than Dabcyl-PH. We then considered the influence of the electron density of the quencher group. All the above probes have Dabcyl-type quenchers with an electron-donating dimethylamino group at the end of the structure. We thus designed 19, which contains an H4K16 peptide and a Disperse Red quencher moiety bearing an electron-withdrawing nitro group at the end of the structure. We also designed 20–23 containing peptide sequences RIKRY, RalB, H2BK12, and H3K9 with a Disperse Red quencher. The characteristics of these probes are summarized in Table 1.
Table 1. Structures of Quenchers and Peptides Used in the New Probes.
| quencher | peptide sequence | length | |
|---|---|---|---|
| SFP3 | Dabcyl-PH | H3K9 (QTARKSTGG) | 9 residues |
| 13 | Dabcyl-PH | H4K16 (KGGAKRHRK) | 9 residues |
| 14 | Dabcyl-PH | H4K16 (GGAKRHR) | 7 residues |
| 15 | Dabcyl-PH | H4K16 (GKGGAKRHRKV) | 11 residues |
| 16 | Dabcyl-PH | S2iL8 (NFRIKRYSN) | 9 residues |
| 17 | Dabcyl-PH | S2iD7 (DYRIKRYHT) | 9 residues |
| 18 | Dabcyl-BH | H4K16 (KGGAKRHRK) | 9 residues |
| 19 | Disperse Red | H4K16 (KGGAKRHRK) | 9 residues |
| 20 | Disperse Red | S2iD7 (DYRIKRYHT) | 9 residues |
| 21 | Disperse Red | RalB (KKSFKERSS) | 9 residues |
| 22 | Disperse Red | H2BK12 (APAPKKGSK) | 9 residues |
| 23 | Disperse Red | H3K9 (QTARKSTGG) | 9 residues |
Synthesis of FRET-Based SIRT Activity Probes 13–23
13–23 were synthesized as shown in Scheme S1. Dabcyl-PH (3), Dabcyl-BH (6), and Disperse Red (10) were synthesized and conjugated with Fmoc-Lys-OH in 4, 3, and 4 steps, respectively. Finally, each probe was obtained by Fmoc solid-phase synthesis on Sieber amide resin, cleaved, and purified by preparative reversed-phase HPLC (purity: >95%). The structures were confirmed by HRMS.
Photochemical Properties of 13–23
We measured the absorption and fluorescence spectra and calculated the fluorescence quantum yields of all the synthesized probes (Figure 2, Table 2). The absorption spectrum of each probe showed overlapping absorbance of FITC and the quencher dye, and the fluorescence spectra indicated that the fluorescence of each probe was sufficiently quenched by the FRET mechanism compared to the reference, 5-FITC (Figure 2B).
Figure 2.
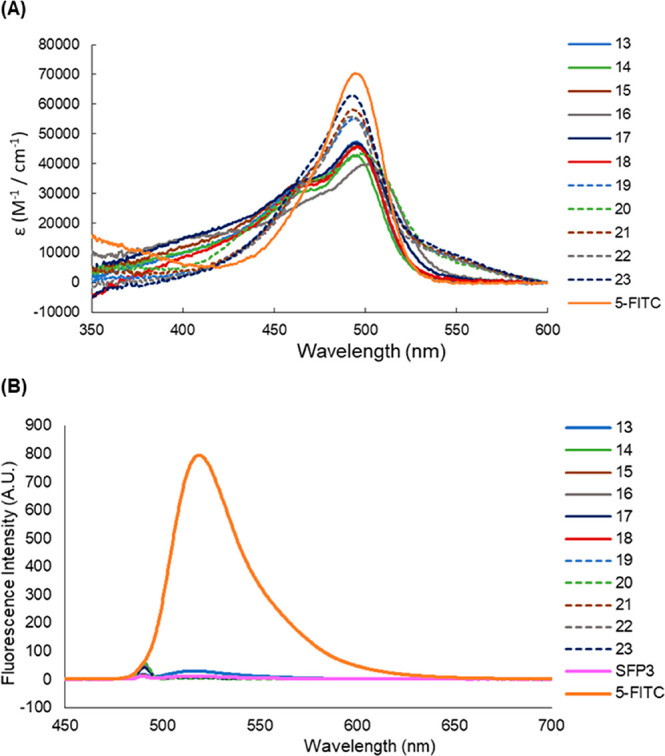
(A) Absorption and (B) fluorescence spectra of 1 μM 5-FITC and 13–23. Data were measured in Tris-buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl), containing 0.01% DMSO as a cosolvent.
Table 2. Photophysical Properties of SFP3, 13–23,a and 5-FITC.
| λmax (nm) | ϵ (104 M–1 cm–1) | λem (nm) | ΦFLb | |
|---|---|---|---|---|
| SFP3 | 496 | 8.91 | 517 | 0.008 |
| 13 | 496 | 4.75 | 516 | 0.053 |
| 14 | 494 | 4.26 | 516 | 0.014 |
| 15 | 494 | 5.52 | 517 | 0.007 |
| 16 | 499 | 3.96 | 516 | 0.021 |
| 17 | 496 | 4.69 | 516 | 0.018 |
| 18 | 496 | 4.58 | 517 | 0.012 |
| 19 | 496 | 4.59 | 517 | 0.015 |
| 20 | 497 | 4.34 | 513 | 0.005 |
| 21 | 493 | 5.82 | 515 | 0.008 |
| 22 | 493 | 5.59 | 515 | 0.010 |
| 23 | 493 | 6.31 | 516 | 0.006 |
| 5-FITC | 494 | 7.04 | 518 | 0.696 |
Data were measured in Tris-buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl).
For determination of ΦFL, fluorescein in 0.1 N NaOH (ΦFL = 0.85) was used as a fluorescence standard.
Reactivity of 13–23 with SIRT1–7
Next, we examined the reactivity of each probe with SIRT1–7 by means of enzymatic assay in 96-well half area plates at 37 °C for 1 h. The values of fluorescence enhancement (fold change) after 1 h are shown in Table 3 and Figure S2.
Table 3. Fluorescence Enhancement (SIRT(+)/SIRT(−)) of 13–23 and SFP3 with SIRT1–7 after Enzymatic Reaction for 1 h.
| SIRT1 | SIRT2 | SIRT3 | SIRT4 | SIRT5 | SIRT6 | SIRT7 | |
|---|---|---|---|---|---|---|---|
| SFP3 | 13.84 | 5.89 | 2.20 | 1.03 | 1.08 | 3.60 | 1.04 |
| 13 | 11.99 | 5.45 | 4.00 | 0.93 | 1.07 | 1.33 | 1.03 |
| 14 | 10.90 | 4.72 | 5.45 | 0.92 | 1.03 | 1.05 | 1.01 |
| 15 | 19.74 | 6.68 | 6.55 | 1.12 | 1.02 | 1.04 | 1.07 |
| 16 | 4.79 | 1.85 | 1.92 | 1.14 | 1.11 | 1.07 | 1.04 |
| 17 | 4.76 | 1.92 | 1.96 | 1.11 | 1.09 | 1.05 | 1.01 |
| 18 | 14.23 | 12.04 | 12.48 | 1.11 | 1.06 | 2.21 | 1.00 |
| 19 | 14.74 | 10.34 | 27.23 | 1.04 | 1.07 | 2.55 | 1.09 |
| 20 | 5.85 | 6.97 | 26.71 | 1.25 | 1.15 | 1.19 | 1.05 |
| 21 | 8.86 | 5.64 | 26.18 | 1.20 | 1.05 | 3.36 | 0.99 |
| 22 | 5.00 | 3.83 | 12.31 | 1.18 | 0.99 | 3.51 | 1.01 |
| 23 | 6.06 | 4.06 | 14.07 | 1.20 | 1.07 | 9.38 | 1.04 |
Most of the probes reacted with SIRT1, SIRT2, and SIRT3, and some showed a large fluorescence enhancement (>10-fold) after 1 h enzymatic reaction. In contrast, none of the probes reacted significantly with SIRT4, 5, or 7, suggesting that our quencher-hydrolyzing strategy might not be suitable for detection of their activities. In the case of SIRT6, several probes, including 22 and 23, showed moderate to high reactivity compared to SFP3. This result suggests that the peptide sequence, especially H3K9, may be critical for improving the binding affinity with SIRT6, and the Disperse Red quencher may be more suitable than the Dabcyl quencher in these cases, although the overall reactivity with SIRT6 was lower than those with SIRT1–3.
18 and 19, whose quenchers are Dabcyl-BH and Disperse Red, respectively, showed large fluorescence enhancements with SIRT1–3. Because the linker structures of Dabcyl-BH and Disperse Red are longer by one carbon atom than that of Dabcyl-PH, the difference of reactivity of 18 and 19 compared with 13 appears to be due to the difference of linker length in the quencher moieties because all three probes have the same peptide chain.
In the case of SIRT1, SFP3 and 13–18 showed large fluorescence enhancements regardless of the peptide sequence, suggesting that SIRT1 favors the Dabcyl structure having a dimethylamino group over the Disperse Red structure having a nitro group, except for 16 and 17, which have peptide sequences optimized for SIRT2. In the case of SIRT2, 18 and 19 showed the greatest fluorescence enhancement, suggesting that SIRT2 precisely recognizes the peptide sequence H4K16. The linker length of the quencher moiety appears to be a key factor for high reactivity. As for SIRT3, interestingly, the probes with the Disperse Red quencher (19–23) showed significantly higher fluorescence enhancement than those with the Dabcyl quencher. Notably, among these probes, 20 and 21 selectively reacted with SIRT3 over SIRT1 and SIRT2.
Overall, these results indicate that our FRET-based fluorescence probes having a quencher dye as a SIRT-hydrolyzable motif are suitable for detecting SIRT1–3 and 6. Moreover, the factors influencing recognition of the fluorescence probes as a substrate by each SIRT subtype are slightly different, suggesting that it is possible to design SIRT-subtype-specific fluorescence probes by precisely optimizing their peptide sequences, quencher structures, and linker lengths.
Determination of the Kinetic Constants of 13–23 with SIRT1–3
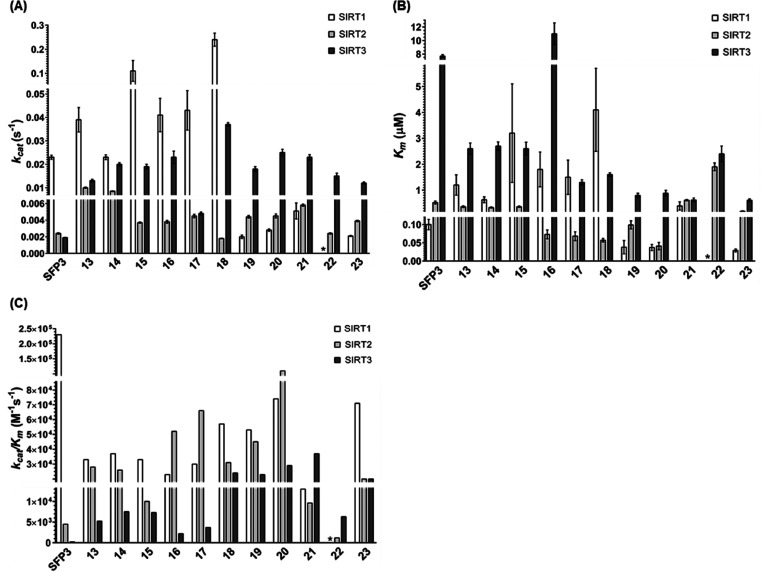
To quantitatively evaluate the reactivity of each probe with SIRT1–3, the Michaelis constant (Km), the catalytic rate constant (kcat), and the specificity constant (kcat/Km) were determined from Michaelis–Menten plots (Figure 3, Figure S3A–C, Table S1A–C).
Figure 3.
Kinetic parameters of the enzymatic reactions of SIRT1–3 with SFP3 and 13–23. (A) kcat values of each probe. (B) Km values of each probe. (C) kcat/Km values of each probe. Asterisks indicate the kinetic parameters of 22 for SIRT1 were not determined. Data for SFP3 were taken from ref (28).
For SIRT1, the kcat and Km values of Dabcyl-type probes (SFP3 and 13–18) are larger than those of Disperse Red-type probes (19–23), suggesting that Dabcyl-type probes are more efficiently hydrolyzed by SIRT1 than Disperse Red-type probes, though SIRT1 has a higher affinity for the Disperse Red quencher than for the Dabcyl quencher. As a result, the difference of kcat/Km values for the two quenchers is small. Indeed, all the newly developed probes have kcat/Km values of the order of 104.
For SIRT2, the Km values of 16 and 17, which contain peptide sequences derived from SIRT2-selective inhibitors, are smaller than those of the other probes with H4K16 sequences. At the same time, the Km values of 18 and 19 are smaller than that of 13, suggesting that the longer linker length on the quencher is crucial for higher affinity with SIRT2. 20 showed the highest affinity with SIRT2. However, the kcat values tend to be larger for probes with larger Km values, and as a result, there is no significant difference in kcat/Km values among all the probes.
For SIRT3, the kcat/Km values are of the order of 104 for most of the Disperse Red-type probes, i.e., about 10 times larger values than those of the Dabcyl-type probes. This is consistent with the results shown in Table 3, and the higher reactivity of the Disperse Red-type probes with SIRT3 might be attributed to their higher affinity for SIRT3 compared with Dabcyl-type probes. It is noteworthy that the kcat/Km value of 22 was smaller than those of other Disperse Red-type probes, suggesting that the H2BK12 sequence may not be favorable and that the peptide sequence can influence SIRT3 substrate recognition.
Overall, the effects of quencher structure and peptide sequence on the reaction rate appear to be relatively large, whereas the effect of the peptide length is smaller. Although the newly developed probes have lower reactivity toward SIRT1 than SFP3, they have similar or higher reactivity toward SIRT2 and SIRT3. In particular, 20 and 21 exhibited significantly higher reactivity than SFP3 with SIRT2 (>24-fold) and SIRT3 (>154-fold).
Next, we conducted molecular docking simulations of Dabcyl-PH, Dabcyl-BH, and Disperse Red with the SIRT2 catalytic site (PDB ID: 4X3P) using the CDOCKER algorithm in Discovery Studio Client v17.2.0.16349 (BIOVIA Inc.) (Figure S4). A comparison of the CDOCKER interaction energies of the three quenchers suggests that the binding affinities of Dabcyl-BH and Disperse Red are larger than that of Dabcyl-PH, which is consistent with the Km values of 13, 18, and 19. Interestingly, all the quenchers share some interactions (e.g., hydrogen bonding with Val233, and π–π stacking interaction with Phe190), but only Disperse Red can also form a π-cation interaction between its nitro group and Tyr139 (Figure S5A-C).
Validation of an HTS System for SIRT2 Inhibitors Using 18
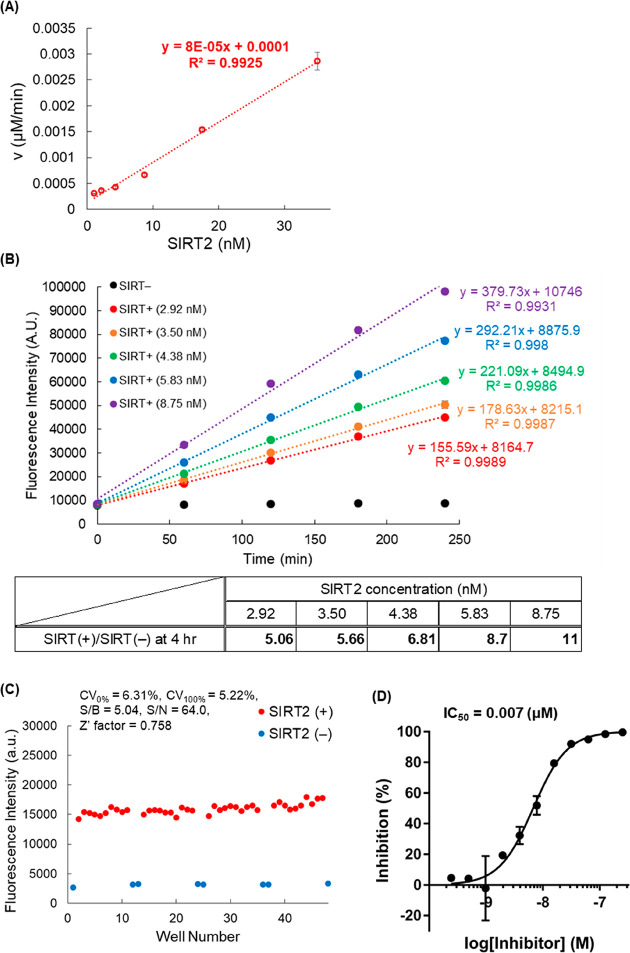
To illustrate the utility of our newly synthesized probes, we conducted a chemical screening assay to find SIRT2 inhibitors because these would be candidate anticancer agents.20 Based on the results shown in Table 3, we chose 18 as the most suitable probe for SIRT2 defatty-acylase modulator screening. Initially, we confirmed that the reaction rate of 18 increased linearly with increasing concentration of SIRT (Figure 4A). We next evaluated the time dependence of the enzymatic reaction of 18 with various concentrations of SIRT2 (Figure 4B) and found that the fluorescence increase was linear for at least 4 h. A low concentration of SIRT2 (2.92 nM) was sufficient for SIRT2 activity detection, reflecting the high reactivity of 18. We further validated the reliability of our screening system with 18 (Figure 4C) by using 48 wells of a 96-well half area plate (SIRT2 (+) in 40 wells and SIRT2 (−) in 8 wells). The CV100% value was less than 10%, the S/B ratio was more than 3, and the Z′ factor was more than 0.5, confirming the reliability of our screening system even with as low a concentration of SIRT2 as 2.92 nM.
Figure 4.
(A) Dependency of the enzymatic reaction rate of 18 on SIRT2 concentration. Results are mean ± SD (n = 3). (B) Time dependence of the enzymatic reaction of 18 with various concentrations of SIRT2 in the range of 2.92–8.75 nM. Results are mean ± SD (n = 3). (C) Reliability of the optimized screening conditions, evaluated in 48 wells of a 96-well half area microplate. CV (coefficient of variation) = SD/Av ≤ 10%; S/B = signal/background ≥3.0; S/N = signal/noise; Z′ factor = 1 – (3 × SD100% + 3 × SD0%)/(Av100%–Av0%) ≥ 0.5 for reliable screening. SD = standard deviation, Av = average. (D) Inhibition curve of S2DMi-6 toward SIRT2 obtained with 18. Results are mean ± SD (n = 3).
We used the established conditions to examine whether 18 can be applied to determine the half-inhibitory activity (IC50) of the reported SIRT defatty-acylase inhibitor S2DMi-6 (Figure 4D, Figure S6A).29 The measured IC50 value was 0.007 μM, which is close to the value given in our previous report (IC50 = 0.019 μM, determined by SFP3).29 In addition, we determined the IC50 value of S2DMi-9 toward SIRT6 by using 23 (Figure S6). The measured IC50 value was 0.10 μM, which is close to the value given in our previous report (IC50 = 0.25 μM, determined with SFP3).29
Chemical Screening of SIRT2 Defatty-Acylase Inhibitors Using 18
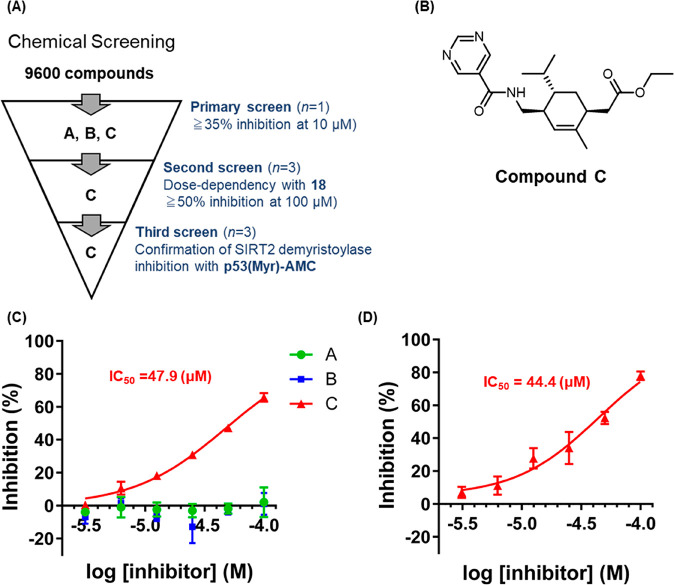
Following the HTS validation of 18, we applied it to screen 9600 compounds from a large chemical library for nonpeptide novel SIRT2 defatty-acylase inhibitors (Figure 5A).
Figure 5.
(A) Three-stage screening. (B) Chemical structure of C. (C) Inhibition curves of A, B, and C toward SIRT2 obtained with 18. The results are mean ± SD (n = 3). (D) Inhibition curve of A toward SIRT2 obtained with p53(Myr)-AMC. The results are mean ± SD (n = 3).
Primary screening was performed in 384-well plates, and all compounds were tested at 10 μM in the presence of 0.5% DMSO. This screen yielded three candidate compounds, A, B, and C, which showed more than 35% inhibition at 10 μM (Figure 5B, Figure S7A). We next conducted a second screening with 18 to confirm dose-dependency and to eliminate false-positive compounds (Figure 5C) and found that compound C reproducibly inhibited SIRT2 with an IC50 value of 47.9 μM. We then conducted a third screening to confirm the SIRT2 defatty-acylase inhibitory activity of the three hit compounds by using our previously developed two-step defatty-acylase probe, p53(Myr)-AMC (Figure S8).29 Among the three candidates, compound C dose-dependently inhibited SIRT2 demyristoylase activity determined with p53(Myr)-AMC, and its IC50 value was calculated to be 44.4 μM, in good accordance with the value obtained using 18 (Figure 5D, Figure S7B).
Further evaluations of compound C were conducted to confirm its selectivity for SIRT2 defatty-acylase activity. We found that compound C selectively inhibits defatty-acylase activity of SIRT2 over SIRT1, 3, and 6 (Figure S9A). However, compound C also inhibited SIRT2 deacetylase activity with an IC50 value of 5.09 μM, indicating that it is a dual inhibitor of SIRT2 deacetylase and defatty-acylase activities (Figure S9B).
In addition, we determined the IC50 values of known SIRT2 inhibitors, nicotinamide,31 AGK2,32 SirReal2,33 and TM,34 by using 18 and compared them with those of compound C (Figure S10). SirReal2 and TM are much more SIRT2 deacetylase-selective than the classical inhibitors nicotinamide and AGK2. It was found that compound C inhibits both SIRT2 deacetylase and defatty-acylase activities, indicating that its defatty-acylase selectivity is similar to those of nicotinamide and AGK2.
Thus, the screening assay with our newly developed, highly sensitive fluorescence probe, 18, enabled us to identify compound C as a nonpeptide, small-molecular SIRT2 defatty-acylase inhibitor.
Conclusion
In this research, we designed and synthesized a set of 11 new one-step fluorescence probes with various peptide sequences and quenchers using an improved synthetic scheme that does not involve liquid-phase steps. Evaluation of these probes with SIRT isozymes indicated first that the length of the quencher structure and the peptide sequence are more important factors than peptide sequence length for determining the reaction rate with SIRT1–3, and second, that the Disperse Red quencher is selectively recognized by SIRT3.
Probe-scanning with SIRT isozymes revealed that different combinations of probes and isozymes showed different responses. To illustrate the value of our probe design strategy, we selected compound 18 for HTS application to discover SIRT2 defatty-acylase modulators in a one-step manner. Even with a low concentration of SIRT2, we identified compound C as a new, small molecule SIRT2 defatty-acylase inhibitor. Our one-step detection method has the following advantages: (i) low background fluorescence due to the probe’s stability, (ii) longer wavelength fluorescence, minimizing possible overlap, (iii) simple manipulation for chemical screening, and (iv) biocompatibility for cellular SIRT activity imaging. A possible drawback is that the substrate recognition sites are different from those of the natural substrates (quenchers vs long-chain fatty acids). However, in our previous paper, we established that IC50 values evaluated with our one-step probe are well-correlated with those obtained using a two-step probe, p53(Myr)-AMC, which has a myristoyl group as a substrate recognition site.29 Therefore, we believe our probe set will be useful to screen chemical libraries for modulators of not only SIRT2 but also other SIRTs, including SIRT1, 3, and 6. Such modulators would be useful tools to decipher the physiological and pathological roles of the defatty-acylase activities of SIRTs as well as to discover new SIRT inhibitors as candidate anticancer therapeutic agents.
Acknowledgments
The authors thank all the members of H.N.’s laboratory for fruitful discussions and also thank Drs. Hirotatsu Kojima, Takayoshi Okabe, and Tetsuo Nagano at the Drug Discovery Initiative, the University of Tokyo for the assistance of chemical screening. This work was supported in part by JSPS KAKENHI Grants 26893223, 15K18899, and 18K14358 (M.K.) and 16H05103, 19H03354, and 19KK0197 (H.N.) as well as by the Hori Sciences and Arts Foundation (M.K.), a Grant-in-Aid for Scientific Research on Innovative Areas from MEXT (26111012, H.N.) and a grant from Daiichi Sankyo Foundation of Life Science (H.N.). This research was partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research from AMED under Grant JP19am0101086.
Glossary
Abbreviations
- SIRT
sirtuin
- FRET
Förster resonance energy transfer
- NAD
nicotinamide adenine dinucleotide
- ADP
adenine diphosphate
- TNF-α
tumor necrosis factor α
- Dabcyl
4-(4-(dimethylamino)phenylazo)benzoic acid
- SPS
solid-phase synthesis
- HPLC
high performance liquid chromatography
- HTS
high-throughput screening
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.1c00010.
Structure of SFP3, reactivity of SFP3 and 13–23 with SIRT1–7, Michaelis–Menten plots for SFP3 and 13–23 with SIRT1–3, structure of hit compounds A and B and their SIRT2 defatty-acylase inhibitory activity determined by p53(Myr)-AMC, docking simulation of quenchers with SIRT2, selectivity of compound C over SIRT1, 3, and 6 and deacetylase activity of SIRT2, inhibitory activities of known SIRT2 inhibitors evaluated with 18, synthesis of 13–23 and in vitro experimental procedures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Imai S.; Armstrong C. M.; Kaeberlein M.; Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Haigis M. C.; Guarente L. P. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006, 20, 2913–2921. 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Longo V. D.; Kennedy B. K. Sirtuins in aging and age-related disease. Cell 2006, 126, 257–268. 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Sauve A. A.; Wolberger C.; Schramm V. L.; Boeke J. D. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006, 75, 435–465. 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Finkel T.; Deng C. X.; Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature 2009, 460, 587–591. 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozako T.; Suzuki T.; Yoshimitsu M.; Arima N.; Honda S.; Soeda S. Anticancer agents targeted to sirtuins. Molecules 2014, 19, 20295–20313. 10.3390/molecules191220295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavu S.; Boss O.; Elliott P. J.; Lambert P. D. Sirtuins--novel therapeutic targets to treat age-associated diseases. Nat. Rev. Drug Discovery 2008, 7, 841–853. 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- Milne J. C.; Lambert P. D.; Schenk S.; Carney D. P.; Smith J. J.; Gagne D. J.; Jin L.; Boss O.; Perni R. B.; Vu C. B.; Bemis J. E.; Xie R.; Disch J. S.; Ng P. Y.; Nunes J. J.; Lynch A. V.; Yang H.; Galonek H.; Israelian K.; Choy W.; Iffland A.; Lavu S.; Medvedik O.; Sinclair D. A.; Olefsky J. M.; Jirousek M. R.; Elliott P. J.; Westphal C. H. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 2007, 450, 712–716. 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N. Engl. J. Med. 2011, 364, 2235–2244. 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- Hu J.; Jing H.; Lin H. Sirtuin inhibitors as anticancer agents. Future Med. Chem. 2014, 6, 945–966. 10.4155/fmc.14.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman J. L.; Baeza J.; Denu J. M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.; Zhang D.; Wang Y.; Perez-Neut M.; Han Z.; Zheng Y. G.; Hao Q.; Zhao Y. Lysine benzoylation is a histone mark regulated by SIRT2. Nat. Commun. 2018, 9, 3374. 10.1038/s41467-018-05567-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M.; Chen W. Y. Sorting out functions of sirtuins in cancer. Oncogene 2014, 33, 1609–1620. 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T.; Hiratsuka M.; Osaki M.; Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle 2007, 6, 1011–1018. 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- Lanyon-Hogg T.; Faronato M.; Serwa R. A.; Tate E. W. Dynamic protein acylation: new substrates, mechanisms, and drug targets. Trends Biochem. Sci. 2017, 42, 566–581. 10.1016/j.tibs.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Tate E. W.; Kalesh K. A.; Lanyon-Hogg T.; Storck E. M.; Thinon E. Global profiling of protein lipidation using chemical proteomic technologies. Curr. Opin. Chem. Biol. 2015, 24, 48–57. 10.1016/j.cbpa.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.; Khan S.; Wang Y.; Charron G.; He B.; Sebastian C.; Du J.; Kim R.; Ge E.; Mostoslavsky R.; Hang H. C.; Hao Q.; Lin H. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110–113. 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.; Zhang X.; Lin H. Lysine fatty acylation promotes lysosomal targeting of TNF-α. Sci. Rep. 2016, 6, 24371. 10.1038/srep24371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Khan S.; Jiang H.; Antonyak M. A.; Chen X.; Spiegelman N. A.; Shrimp J. H.; Cerione R. A.; Lin H. Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat. Chem. Biol. 2016, 12, 614–620. 10.1038/nchembio.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H.; Zhang X.; Wisner S. A.; Chen X.; Spiegelman N. A.; Linder M. E.; Lin H. SIRT2 and lysine fatty acylation regulate the transforming activity of K-Ras4a. eLife 2017, 6, e32436 10.7554/eLife.32436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman N. A.; Zhang X.; Jing H.; Cao J.; Kotliar I. B.; Aramsangtienchai P.; Wang M.; Tong Z.; Rosch K. M.; Lin H. SIRT2 and lysine fatty acylation regulate the activity of RalB and cell migration. ACS Chem. Biol. 2019, 14, 2014–2023. 10.1021/acschembio.9b00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Liu T.; Liao S.; Li Y.; Lan Y.; Wang A.; Wang Y.; He B. Biochem. Biophys. Res. Commun. 2015, 467, 459–466. 10.1016/j.bbrc.2015.09.172. [DOI] [PubMed] [Google Scholar]
- Yang L.-L.; Wang H.-L.; Yan Y.-H.; Liu S.; Yu Z.-J.; Huang M.-Y.; Luo Y.; Zheng X.; Yu Y.; Li G.-B. Eur. J. Med. Chem. 2020, 192, 112201. 10.1016/j.ejmech.2020.112201. [DOI] [PubMed] [Google Scholar]
- Baba R.; Hori Y.; Mizukami S.; Kikuchi K. Development of a fluorogenic probe with a transesterification switch for detection of histone deacetylase activity. J. Am. Chem. Soc. 2012, 134, 14310–14313. 10.1021/ja306045j. [DOI] [PubMed] [Google Scholar]
- Baba R.; Hori S.; Kikuchi K. Intramolecular long-distance nucleophilic reactions as a rapid fluorogenic switch applicable to the detection of enzymatic activity. Chem. - Eur. J. 2015, 21, 4695–4702. 10.1002/chem.201406093. [DOI] [PubMed] [Google Scholar]
- Xie Y.; Yang L.; Chen Q.; Zhang J.; Feng L.; Chen J. L.; Hao Q.; Zhang L.; Sun H. Single-stepfluorescent probes to detect decrotonylation activity of HDACs through intramolecular reactions. Eur. J. Med. Chem. 2021, 212, 113120. 10.1016/j.ejmech.2020.113120. [DOI] [PubMed] [Google Scholar]
- Dai Q.; Zheng Z.; Xia F.; Liu P.; Li M. A one-step specific assay for continuous detection of sirtuin 2 activity. Acta Pharm. Sin. B 2019, 9, 1183–1192. 10.1016/j.apsb.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M.; Ikegawa S.; Ieda N.; Nakagawa H. A fluorescent probe for imaging sirtuin activity in living cells, based on one-step cleavage of the Dabcyl quencher. ChemBioChem 2016, 17, 1961–1967. 10.1002/cbic.201600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M.; Ieda N.; Nakagawa H. Development of peptide-based sirtuin defatty-acylase inhibitors identified by the fluorescence probe, SFP3, that can efficiently measure defatty-acylase activity of sirtuin. J. Med. Chem. 2019, 62, 5434–5452. 10.1021/acs.jmedchem.9b00315. [DOI] [PubMed] [Google Scholar]
- Morimoto J.; Hayashi Y.; Suga H. Discovery of macrocyclic peptides armed with a mechanism-based warhead: isoform-selective inhibition of human deacetylase SIRT2. Angew. Chem., Int. Ed. 2012, 51, 3423–3427. 10.1002/anie.201108118. [DOI] [PubMed] [Google Scholar]
- Anthony A.; Schramm V. L. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Bio Chemistry. 2003, 42, 9249–9256. [DOI] [PubMed] [Google Scholar]
- Outeiro T. F.; Kontopoulos E.; Altmann S. M.; Kufareva I.; Strathearn K. E.; Amore A. M.; Volk C. B.; Maxwell M. M.; Rochet J.-C.; McLean P. J.; Young A. B.; Abagyan R.; Feany M. B.; Hyman B. T.; Kazantsev A. G. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 2007, 317, 516–519. 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- Rumpf T.; Schiedel M.; Karaman B.; Roessler C.; North B. J.; Lehotzky A.; Oláh J.; Ladwein K. I.; Schmidtkunz K.; Gajer M.; Pannek M.; Steegborn C.; Sinclair D. A.; Gerhardt S.; Ovádi J.; Schutkowski M.; Sippl W.; Einsle O.; Jung M. Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat. Commun. 2015, 6, 6263. 10.1038/ncomms7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H.; Hu J.; He B.; Negron Abril Y. L.; Stupinski J.; Weiser K.; Carbonaro M.; Chiang Y.-L.; Southard T.; Giannakakou P.; Weiss R. S.; Lin H. A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell 2016, 29, 767–768. 10.1016/j.ccell.2016.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.