Abstract
Sialic acid (Sia)-binding immunoglobulin-like lectin 7 (Siglec-7) is an inhibitory receptor primarily expressed on natural killer (NK) cells and monocytes. Siglec-7 is known to negatively regulate the innate immune system through Sia binding to distinguish self and nonself; however, a counter-receptor bearing its natural ligand remains largely unclear. Here, we identified a counter-receptor of Siglec-7 using K562 hematopoietic carcinoma cells presenting cell surface ligands for Siglec-7. We affinity-purified the ligands using Fc-ligated recombinant Siglec-7 and diSia-dextran polymer, a strong inhibitor for Siglec-7. We then confirmed the counter-receptor for Siglec-7 as leukosialin (CD43) through mass spectrometry, immunoprecipitation, and proximity labeling. Additionally, we demonstrated that the cytotoxicity of NK cells toward K562 cells was suppressed by overexpression of leukosialin in a Siglec-7-dependent manner. Taken together, our data suggest that leukosialin on K562 is a counter-receptor for Siglec-7 on NK cells and that a cluster of the Sia-containing glycan epitope on leukosialin is key as trans-ligand for unmasking the cis-ligand.
Keywords: Siglec-7, ligand, counter-receptor, CD43, leukosialin, NK cell
Abbreviations: NCBI, National Center for Biotechnology Information; NK, natural killer; Sia, sialic acid; TBS, Tris-buffered saline
Siglecs are sialic-acid-binding immunoglobulin-like lectins that are mainly expressed on immune cells in a cell-type-specific manner (1). Siglecs are type I membrane immunoreceptors containing a Sia-binding N-terminal V-set domain and several C2-type Ig-like domains that act as regulators of oligomerization or as spacers in projecting the ligand-binding site away from the membrane surface. The majority of Siglecs are involved in the innate immune system by distinguishing molecules as self or nonself; Sia is considered a self-molecule (1). Siglecs regulate immune cell functions by binding to Sia residues. Disrupting these interactions causes changes in the states of infection, inflammation, and tumorigenesis. It is known that enhanced sialylation on the surface of cancer cells is relevant in their escape mechanism from the host immune system (2); therefore, Siglecs are used as targeting therapeutic molecules toward cancer cells for drug delivery (3, 4, 5, 6).
Siglec-7 (CD328), a member of the human CD33-related Siglecs, is localized predominantly on human NK cells and monocytes. Siglec-7 consists of an extracellular N-terminal V-set domain, two C2-type Ig-like domains, and an immunoreceptor tyrosine-based inhibition motif in the cytosolic region (7, 8). It acts as an inhibitory receptor in a Sia-binding-dependent manner. The recognition of Siglec ligands is usually modulated by the endogenous ligands at the cellular surface via cis-interaction, which is known as masking (3, 9, 10). In NK cells, Siglec-7 is considered to be masked by the endogenous cis-ligand (10). The masking effect was clearly shown for CD22, which specifically binds to α2,6-linked Sia on B cells (9). Unmasking was achieved by exogenous addition of sialidase or cellular activation (11, 12, 13). It has also been shown that microglia activation by pathogens or inflammatory cytokines causes glycan-related degrading enzymes, such as the sialidase Neu1, to reduce Sia on the cell surface (14). However, the natural unmasking mechanism of NK cells, which is involved in the surveillance system, and the identity of the trans-ligand on the cancer cells that escape from NK cell remain unclear. Therefore, it is necessary to confirm the natural trans-ligand for Siglec-7 for further understanding of the cis-ligand unmasking mechanism.
The glycan structure of the ligand for Siglec-7 has been studied in part. Siglec-7 binds to the Neu5Acα2,8Neu5Acα2- (diSia) sequence present within the GD3 structure and the branched α2,6-sialyl residues present in DSGb5 and DSLc4 (15, 16). The clustering of diSia epitopes is also important for enhancing the affinity of molecular units for the binding to Siglec-7 (8). Complex ligand specificity has also been shown by several sets of glycan array experiments [http://www.functionalglycomics.org/glycomics/publicdata/primaryscreen.jsp]. Although it is still unknown why such complex specificity occurs in the region of the Siglec-7 V-set domain containing residue R124, which was proven by X-ray crystallography as necessary for ligand binding (17, 18), the existence of a new Sia-binding region (site 2 containing R67) was recently demonstrated based on in silico docking simulation and subsequent mutation experiments (19).
Functional analysis of Siglec-7 in sialidase-treated NK cells, with the ganglioside ligands GD3 and DSGb5 was shown to inhibit NK cell cytotoxicity toward GD3-or DSGb5-expressing cancer cells (10, 16). However, the counter-receptor for Siglec-7 in cancer cells and its involvement in NK cell functions are still unknown. Recently, we demonstrated the presence of the counter-receptor for Siglec-7 in K562 cancer cells, often used as the target cells for NK cytotoxicity assays (20). In this study, we identified the counter-receptor for Siglec-7 using diSia-dextran (diSia-Dex), a strong inhibitor of Siglec-7-ligand binding, and demonstrated its involvement in cytotoxicity.
Results
Observation of the cell surface ligands for Siglec-7
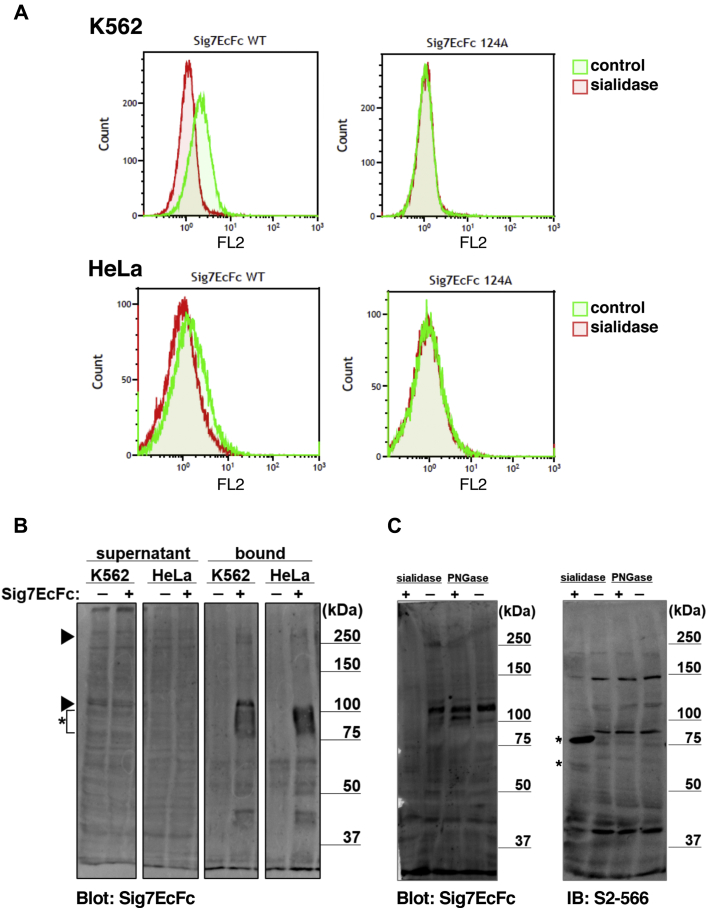
Recently, we identified the presence of the counter-receptor for Siglec-7 on the K562 cell surface (20). To confirm this, we investigated whether K562 and HeLa cells express natural ligands for Siglec-7 on their surfaces (21). Both K562 and HeLa cells had ligands for Siglec-7 on the cell surface, as demonstrated by flow cytometry (Fig. 1A), and the binding was Sia-dependent because Siglec-7 binding disappeared after sialidase treatment (Fig. 1A). In addition, the Siglec-7 R124A mutant, in which the Arg residue essential for Sia binding is replaced by Ala (18, 19, 20), showed no binding, indicating that binding occurred with the V-set domain of Siglec-7. The binding signal of Siglec-7 to K562 was stronger than that of HeLa cells, indicating that K562 expressed the ligands at a higher level than HeLa cells. To confirm the presence of the counter-receptor for Siglec-7, pull-down assays were performed using Siglec-7EcFc on K562 and HeLa cell homogenates. As shown in Figure 1B, a 110kDa-gp was observed in K562, and a 270kDa-gp was observed in both K562 and HeLa. The results obtained in K562 cells were consistent with those of our previous study (20). To analyze the glycan structures of the Siglec-7 counter-receptor, enzyme digestion with sialidase and peptide-N-glycosidase F (PNGase F) was performed on K562 homogenates. Removal of N-linked glycans by PNGase F did not decrease the Siglec-7 staining; however, sialidase treatment markedly abolished this signal (Fig. 1C). It is noteworthy that a weakly stained band appeared at a molecular weight of 105 kDa after PNGase F digestion. Using an anti-diSiaα2,3Gal antibody, S2-566 (22), the 110kDa-gp was observed at a lower level than the other stained bands in a Sia-dependent manner but not in an N-glycan dependent manner.
Figure 1.
Binding of Siglec-7 to the ligands in K562 and HeLa cells.A, the binding of Siglec-7EcFc WT (Sig7EcFc WT) and R124A (Sig7EcFc R124A) to the surface of K562 and HeLa cells was analyzed by flow cytometry. Cells treated with sialidase (red) were compared with those untreated (green). B, ligand pull-down of Siglec-7. The K562 or HeLa cell lysate (90 μg) was mixed with (+) or without (−) Siglec-7EcFc (1 μg), and Siglec-7 ligands were pulled down by Protein A-Sepharose (10 μl). Proteins bound to the beads (bound) and supernatant were analyzed by western blotting with Siglec-7EcFc WT (Sig7EcFc). The exogenous Siglec-7EcFc was observed as smear band at 80 to 100 kDa and marked by asterisk. The precipitated 110 kDa and 270 kDa proteins are indicated by arrowheads. C, sialidase and PNGase F treatment for ligands in K562 cells. The K562 cell lysate was treated with (+) or without (−) sialidase (A. ureafaciens sialidase 2.5 mU, V. cholerae sialidase 6.7 mU, pH 5.5) or with (+) or without (−) PNGase (20 U, pH 7.5) and analyzed by western blotting using Siglec-7EcFc WT (Sig7EcFc) or anti-diSiaα2,3-Gal monoclonal antibody (S2-566). The bands derived from sialidases are marked by asterisks.
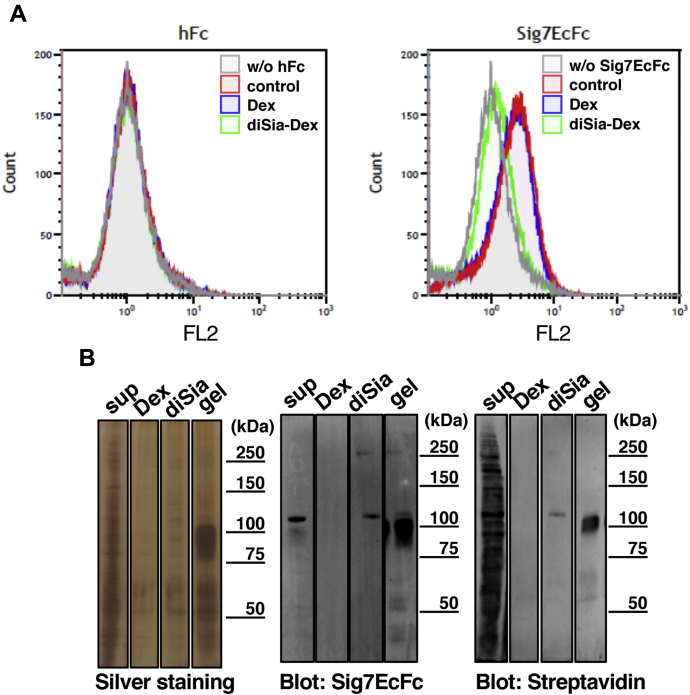
Next, to develop a method for purification of the counter-receptor, we used diSia-Dex, which has been demonstrated to inhibit the binding of Siglec-7 to the diSia-containing ganglioside, GD3 (23). As shown in Figure 2A, the binding to K562 was strongly inhibited by diSia-Dex, but not by Dex alone, indicating that Siglec-7 binding was competitive with diSia epitopes. To determine the ability of diSia-Dex to release the counter-receptor from Siglec-7, we performed a pull-down assay for K562 cell homogenates and attempted to elute the ligand using diSia-Dex. As shown in Figure 2B, diSia-Dex successfully released 110kDa-gp and 270kDa-gp, which were confirmed to be present on the cell surface via cell surface biotin labeling. The coprecipitated proteins (gel) contained these two components. DiSia-Dex-eluted components of the same size were also detected by Siglec-7 blotting, suggesting that they are counter-receptors for Siglec-7. Upon silver staining, several components were visualized in both the diSia-Dex-eluted and gel fractions. These data suggest that the 110kDa- and 270kDa-gps are surface counter-receptors for Siglec-7 and that several other proteins are also associated via indirect binding to Siglec-7.
Figure 2.
Effects of diSia-Dex on the binding of Siglec-7 to the ligands of K562 cells.A, the inhibitory effects of diSia-Dex on Siglec-7EcFc binding to K562 cell surfaces were analyzed using flow cytometry. The reaction solution of 10 μg/ml Siglec-7EcFc and anti-hIgG+M+A antibody was mixed with 100 nM Dex or diSia-Dex and subsequently anti-goat IgG-Alexa Fluor 555; then, the fluorescence intensity was measured using flow cytometry. Human Fc was used as a negative control (hFc). Gray: cells incubated without Siglec-7EcFc (w/o Sig7EcFc), red: cells incubated with Siglec-7EcFc without inhibitor (control), blue: cells incubated with Siglec-7EcFc and Dex (Dex), green: cells incubated with Siglec-7EcFc and diSia-Dex (diSia-Dex). B, cell surface biotin labeling and pull-down experiments with Siglec-7. The cell surfaces of K562 were randomly biotin-labeled using Sulfo-NHS-LC-Biotin. Siglec-7 ligands on the cell surface were pulled down from the lysates (400 μg) using Siglec-7EcFc (6 μg) and eluted with 100 nM Dex (Dex) and diSia-Dex (diSia); beads after elution (gel) and the supernatant (sup) were also analyzed. Samples were denatured, and each sample was separated using SDS-PAGE and analyzed using silver staining and western blotting. Ligands were detected using a complex of 0.5 μg/ml Siglec-7EcFc and anti-human IgG+A+M-HRP (Sig7EcFc) or 5000-fold diluted streptavidin-HRP (Streptavidin).
Identification of the cell surface counter-receptor for Siglec-7
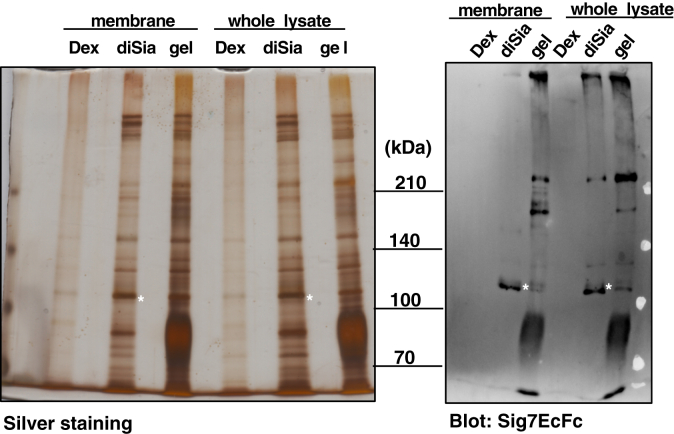
To identify the cell surface counter-receptor for Siglec-7, it was purified by affinity chromatography using a Siglec-7 column. Proteins captured by Siglec-7 from the membrane fraction and the whole cell lysate were eluted with Dex for nonspecific release or with diSia-Dex for specific release. As shown in Figure 3, the 110kDa-gp could be purified from the membrane fraction and the whole cell lysate by diSia-Dex, and the 270kDa-gp could be purified from the whole cell lysate (Fig. 3). To identify the 110kDa-gp and 270kDa-gp observed by Siglec-7 blotting, we excised the bands at approximately 110 kDa and 270 kDa on the silver stained gel, treated them with trypsin, and performed LC-mass spectrometry (MS)/MS analysis. Based on several trials of MS and ProteinPilot software analyses (Table S1), we identified leukosialin (CD43/sialophorin: gene name SPN), nucleolin (gene name: NCL), and heterogeneous nuclear ribonucleoprotein U (gene name: HNRNPU) as potential counter-receptor candidates for Siglec-7 with an approximate molecular weight of 110 kDa. MS analyses for 270kDa-gp were not successful due to the small amount of purified proteins.
Figure 3.
Purification of Siglec-7 Ligands for MS Analysis. Ligands were purified from the crude membrane fraction (2 mg) and the whole cell lysate (6 mg) of K562 using Siglec-7EcFc (125 μg). Ligands were competitively eluted using 50 nM Dex (Dex) and diSia-Dex (diSia) three times and concentrated by ultrafiltration. Each sample and the eluted beads (gel) were heat denatured in sample buffer and separated using SDS-PAGE. The proteins were visualized using silver staining for MS analysis (Silver staining); western blotting was also performed to confirm the molecular weight of the ligands using Siglec-7EcFc (Sig7EcFc). Bands shown as asterisks at 110 kDa were excised from the gel and used for further sample preparation for MS.
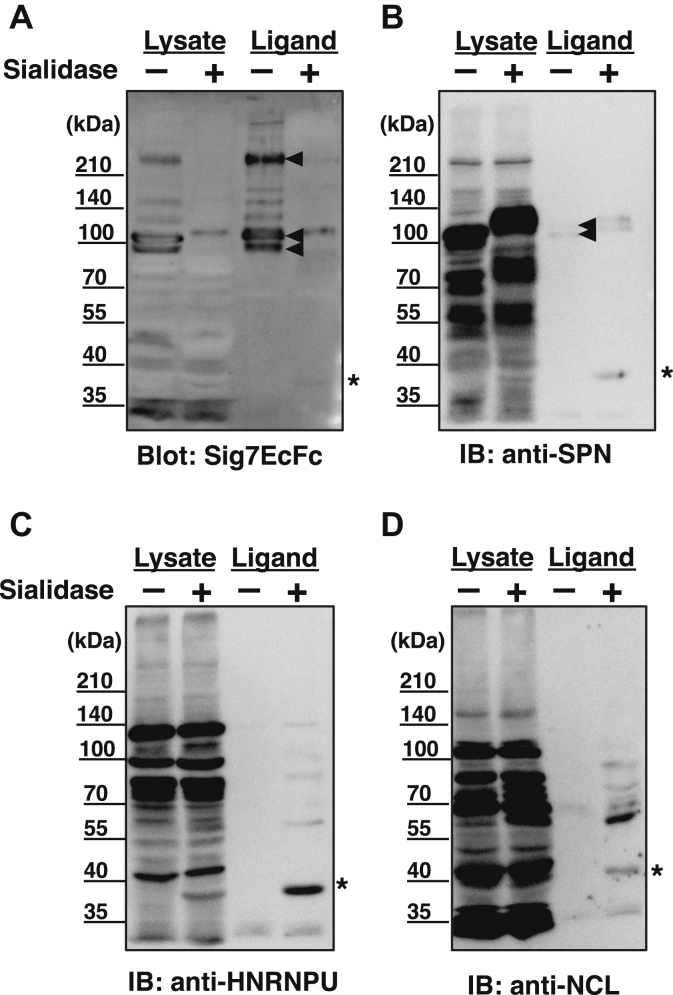
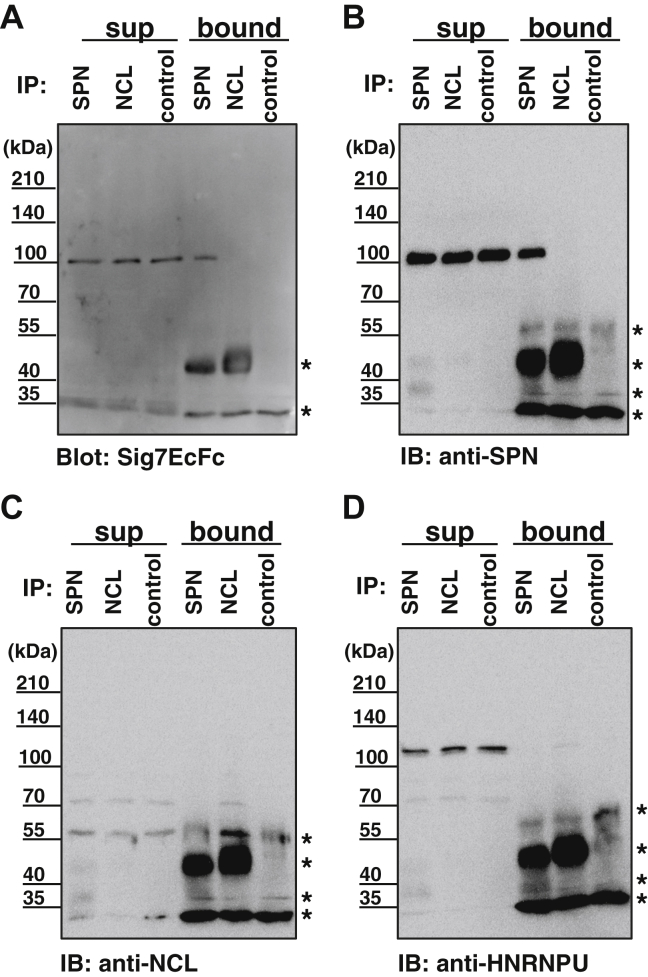
To confirm the MS results, we analyzed the purified fraction by western blotting with specific antibodies against these proteins. As shown in Figure 4, SPN protein bands were clearly observed at approximately 110 kDa by Siglec-7 blotting (Fig. 4A) using both the cell lysate and the Siglec-7 purified fraction. After sialidase treatment, the band shifted but did not disappear, indicating that the SPN was sialylated (Fig. 4B). Migration also decreased, indicating the presence of heavy sialylation on SPN. The 270kDa-gp observed by Siglec-7 blotting was not observed using the anti-SPN antibody, indicating that this was not an oligomeric form of the SPN protein. SPN staining was most intense in the cell lysate, but the staining was faint in the Siglec-7 purified fraction (Fig. 4B), indicating that the glycan epitope recognized by Siglec-7 is, in part, on SPN. Using anti-NCL and -HNRNPU antibodies, we found bands around the 110kDa-gp band (Fig. 4, C and D). The HNRNPU and NCL bands were observed only after sialidase treatment in the Siglec-7 purified proteins (Fig. 4, C and D). To confirm this, we performed co-immunoprecipitation analysis. We immunoprecipitated an identified counter receptor for Siglec-7, SPN, and NCL and immunoblotted them with Siglec-7EcFc, anti-SPN, anti-NCL, and anti-HNRNPU antibodies. As shown in Figure 5, SPN-immunoprecipitated 110kDa-gp was detected with Siglec-7EcFc and anti-SPN. However, the immunoprecipitated NCL protein was not detected using either Siglec-7EcFc or anti-SPN but was observed using an anti-NCL antibody. In addition, the anti-NCL antibody immunoprecipitated a 120 kDa band that was observed with the anti-HNRNPU antibody, indicating that the complex of HNRNPU and NCL was precipitated using the anti-NCL antibody.
Figure 4.
Detection of candidate Siglec-7 ligand molecules by specific antibodies. K562 lysates (20 μg) and Siglec-7 ligand fractions purified from the lysates were treated with (+) or without (−) with sialidase (Sphingobacterium multivorum sialidase 12 mU, pH 7.4) and western blotting analyses were performed. The probes used were Siglec-7EcFc (Sig7EcFc) (A), anti-leukosialin (anti-SPN) (B), anti-nucleolin (anti-NCL) (C), and anti-heterogeneous nuclear ribonucleooprotein U (anti-HNRNPU) (D). Sialidase-derived staining is indicated by asterisks. The major sialidase sensitive specific bands are indicated by arrowheads.
Figure 5.
Immunoprecipitation of candidate molecules for Siglec-7 by specific antibodies. Leukosialin and nucleolin were immunoprecipitated with antileukosialin or antinucleolin antibodies (2 μl) from K562 lysate (200 μg). Controls were prepared without antibodies (control) via Protein A-Sepharose. Supernatant (sup) and precipitated proteins (bound) were denatured and analyzed by SDS-PAGE and western blotting. A, Siglec-7EcFc. B, anti-leukosialin. C, anti-nucleolin. D, anti-HNRNPU. Antibody and Protein A-derived staining are indicated by asterisks.
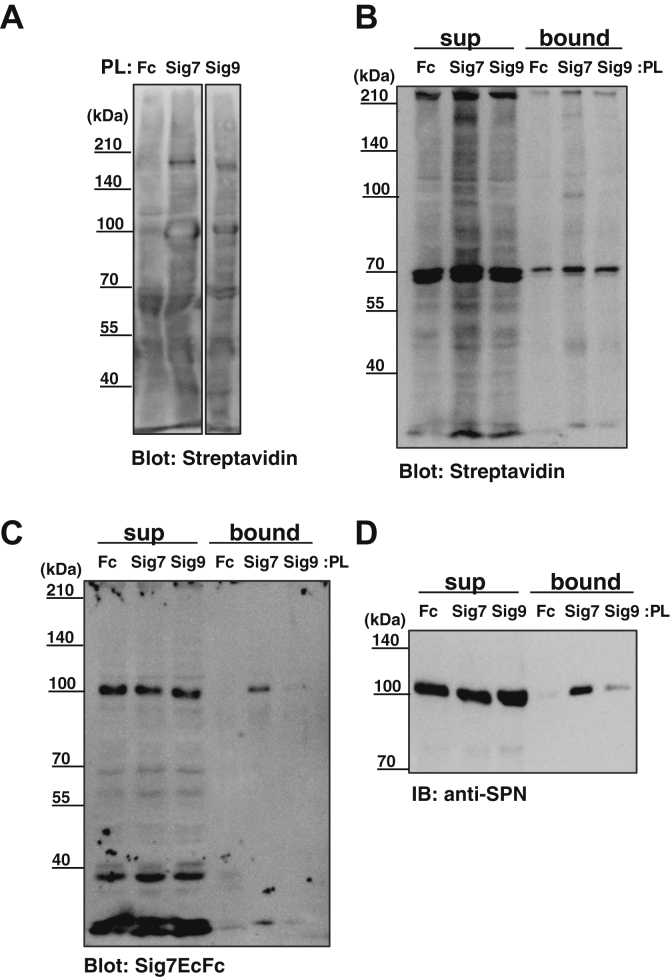
To confirm the binding of Siglec-7 to SPN on the cell surface, we performed proximity labeling (PL) (24). Siglec-9, which favors monoSia ligands and shows binding activity with K562, was used for comparison, and Fc was used as a negative control. After binding with Siglec-7-EcFc, Siglec-9EcFc, and Fc to the surface of K562 cells, we labeled the proteins present near Siglec-7, Siglec-9, and Fc using H2O2 and biotinyl-tyramide. We then analyzed the biotin-labeled lysates via streptavidin and found that the 110kDa-gp and a 170kDa-gp were labeled with Siglec-7 and Siglec-9, but Siglec-7 labeling was more intense (Fig. 6A). Then, biotinylated molecules were purified using streptavidin beads and blotted using streptavidin and Siglec-7EcFc. As shown in Figure 6C, the 110kDa-gp was clearly stained using Siglec-7 and faint using Siglec-9 but not using the negative control of Fc. Streptavidin blot analysis also showed similar results for 110kDa-gp (Fig. 6B). We then analyzed with anti-SPN antibody and found that the 110kDa-gp, which was precipitated from lysates proximity-labeled using Siglec-7 and Siglec-9, could also be detected (Fig. 6D). Taken together, these results indicate that SPN on the surface of K562 cells was a counter-receptor for Siglec-7 and Siglec-9, although the SPN association with Siglec-7 was more pronounced.
Figure 6.
Confirmation of the candidate ligand molecule for Siglec-7 using proximity labeling (PL). K562 cells were incubated with complexes of recombinant Fc fusion proteins (Fc, Sig7EcFc, Sig9EcFc) and anti-human IgG+M+A-HRP. The proteins adjacent to HRP molecules were labeled with hydrogen peroxide and biotinyl-tyramide as substrates. A, lysates were prepared, and total biotin-modified proteins were detected using western blotting with Streptavidin-HRP. Pull-down experiments of the proximity labeled lysates (200 μg) with Fc, Sig7EcFc, and Sig9EcFc were performed using streptavidin-Sepharose. The precipitates were detected with (B) streptavidin, (C) Sig7EcFc, or (D) anti-leukosialin antibody.
Significance of binding of SPN on K562 and Siglec-7 on NK cells
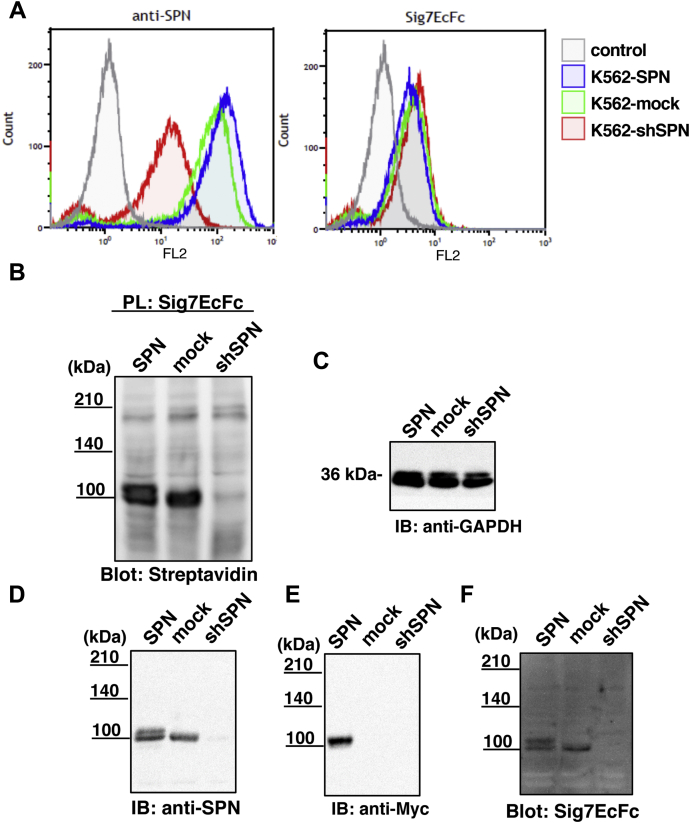
To understand the biological significance of the interaction between SPN and Siglec-7, we established K562 cell lines in which SPN was either stably overexpressed or knocked down by RNA interference. We named the cells K562-SPN and K562-shSPN, respectively. First, the cells were validated by flow cytometry using anti-SPN antibody and Siglec-7. As shown in Figure 7A, K562-SPN expressed SPN at a higher level and K562-shSPN expressed SPN at a lower level on the cell surface than K562-mock. Unexpectedly, Siglec-7 binding was not correlated with the expression levels of SPN, but rather inversely correlated to a small extent. We then analyzed the Siglec-7-labeled cell surface proteins by the PL method. As shown in Figure 7B, the 110kDa-gp was observed using the streptavidin probe, although K562-SPN showed doublet bands. The expression levels of endogenous and exogenous SPN were confirmed using anti-SPN and anti-Myc antibodies (Fig. 7, D and E). The loading control is shown in Figure 7C. The anti-SPN antibody could recognize the endogenous and exogenous SPN, because the doublet bands around 110 kDa in Figure 7B were also observed in the anti-SPN blot (Fig. 7D). K562-shSPN showed a tiny amount of SPN protein. Exogenous SPN, which was Myc-tagged, could be detected using an anti-Myc antibody (Fig. 7E) and corresponded to the upper band. Both SPN proteins were recognized by Siglec-7 (Fig. 7F).
Figure 7.
Validation of the established K562 cell lines. SPN-overexpressing K562 cells (K562-SPN) and SPN-knockdown cells (K562-shSPN) were established. A, the established transfectants (K562-SPN, K562-mock, K562-shSPN) were analyzed using anti-SPN antibody and Sig7EcFc by flow cytometry. Gray: control untreated with probes, blue: K562-SPN, green: K562-mock, red: K562-shSPN. The transfectants were examined using the PL method via reaction with hydrogen peroxide and biotinyl-tyramide as substrates after treatment with a complex of Sig7EcFc and anti-human IgG+M+A-HRP. B, lysates were prepared and biotin-modified proteins were detected with streptavidin-HRP. C, anti-GAPDH antibody was used for the loading control for the proteins. D, anti-leukosialin antibody was used for the detection of both endogenous and exogenous leukosialin proteins. E, anti-Myc antibody was used for the exogenous leukosialin proteins and (F) Sig7EcFc for Siglec-7 ligands.
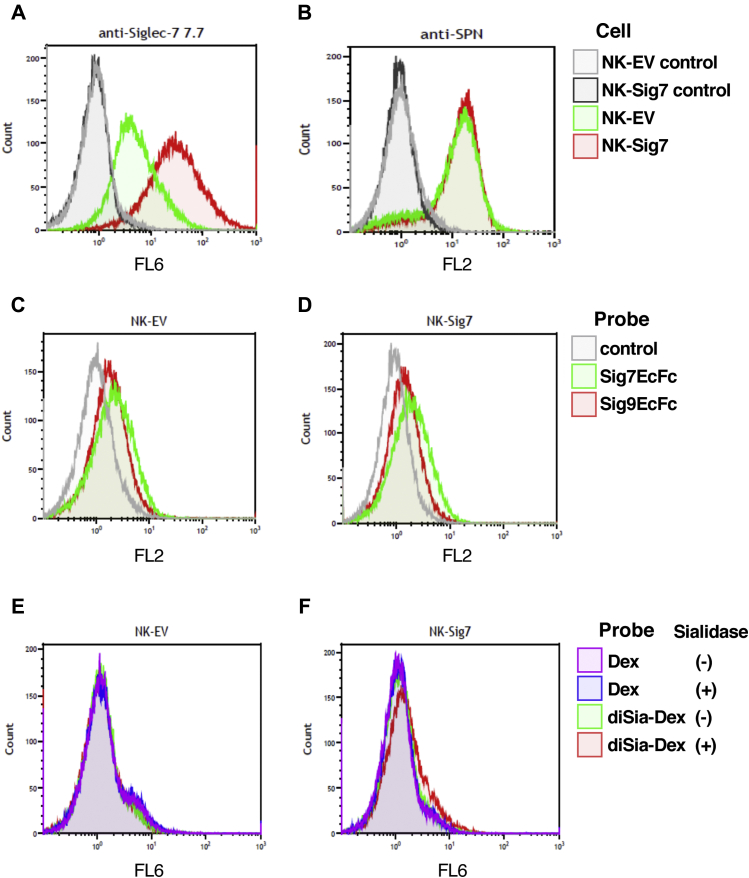
To understand the involvement of Siglec-7 in the cytotoxicity of NK cells, we used Siglec-7-overexpressed (NK-Sig7) and empty vector-containing NK92-MI cells (NK-EV) (25). First, we analyzed the surface of the NK cells and confirmed cell surface expression of Siglec-7 (Fig. 8A) on NK-Sig7 cells compared with that on NK-EV as a negative control. Both NK-EV and NK-Sig7 cells expressed similar levels of SNP on the cell surface (Fig. 8B). When we analyzed Siglec-7 or Siglec-9 binding to the NK cells, we found that the ligands of Siglec-7 and Siglec-9 were present at low levels in NK-EV and NK-Sig7 cells (Fig. 8, C and D). When we added diSia-Dex to the NK cells, the binding was almost the same as with Dex (Fig. 8, E and F), indicating that, in both cases, NK cells showed no diSia-Dex binding and that Siglec-7 on NK cells was completely masked. After treating the NK cells with sialidase, binding was observed only when using diSia-Dex for NK-Sig7 cells (Fig. 8, E and F), indicating that the cis-ligands for Siglec-7 can be released by sialidase treatment but not effectively.
Figure 8.
Characterization of NK92-MI transgenic cell lines. The expression levels of (A) Siglec-7 and (B) leukosialin of cells transfected with empty vector (NK-EV) and Siglec-7 expressing vector (NK-Sig7) were analyzed using flow cytometry. The expression of Siglec-7 was assessed using specific antibody 7.7-containing medium (1/20) and secondary anti-mouse IgG+M-Alexa Fluor 647 (1/600). Leukosialin was analyzed using the anti-SPN antibody conjugated with PE (1/100). Cells without antibody reaction (gray: NK-EV, black: NK-Sig7) and cells with antibody reaction (green: NK-EV, red: NK-Sig7) are shown. The binding of 10 μg/ml Sig7EcFc and Sig9EcFc to the cell surface of (C) NK-EV and (D) NK-Sig7 was also analyzed. Gray: cells not reacted with SigEcFc, green: cells reacted with Sig7EcFc, red: cells reacted with Sig9EcFc. The binding of 50 nM diSia-Dex or Dex to the cell surface of (E) NK-EV and (F) NK-Sig7 before and after sialidase treatment was analyzed by measuring the fluorescence of Cyanine 5 conjugated with Dex derivatives. The reactivity of cells not treated with sialidase (purple: Dex, green: diSia-Dex) and cells treated with sialidase (blue: Dex, red: diSia-Dex) against NK-EV or NK-Sig7 is shown.
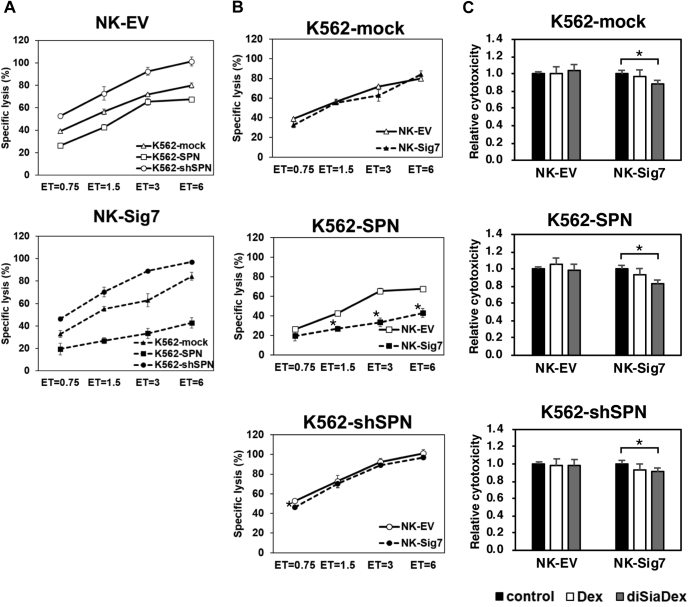
Finally, using K562-SPN or K562-shSPN, we analyzed the influence of Siglec-7 binding to SPN on NK cell cytotoxicity toward K562 cells. As shown in Figure 9A, there was a clear relationship between the expression level of SPN and the cytotoxicity of NK cells. The cytotoxicity of NK-EV and NK-Sig7 cells toward K562-mock cells or K562-shSPN cells was similar (Fig. 9B). In contrast, the cytotoxicity of NK-Sig7 was significantly inhibited compared with that of NK-EV toward K562-SPN cells. To confirm the involvement of the diSia epitope on the function of NK cells, we used diSia-Dex as an inhibitor of Siglec-7 to regulate cytotoxicity. As shown in Figure 9C, significant inhibition was observed using NK-Sig7 cells as effector cells, and this effect was not observed using NK-EV. DiSia-Dex bound Siglec-7 on NK-Sig7 and suppressed NK cell cytotoxicity, regardless of whether the target cells expressed SPN at a high level, indicating that SPN on K562 and diSia-Dex independently inhibited the cytotoxicity of NK cells via Siglec-7.
Figure 9.
Cytotoxicity assay for K562 cell lines with NK cells.A, cytotoxicity assay for NK-EV and NK-Sig7 cells. NK cells were harvested, counted, and then incubated with K562 cells (K562-Mock, K562-SPN and K562-shSPN) stained with Calcein-AM at cell ratios (ET ratio, effector cells: target cells) of 0.75 to 6:1. Specific lysis was calculated by measuring the fluorescence (ex. 485 nm, em. 535 nm) in supernatant. Experiments were performed in triplicate, and error bars indicate standard deviation. B, cytotoxicity of NK cells for K562 cell lines. The Student's t-test was performed for each ET ratio. ∗p < 0.05. C, inhibition of Siglec-7 by the diSia epitope of cytotoxicity in NK cells. Calcein-AM prestained K562 transfectants (K562-Mock, K562-SPN, and K562-shSPN) were seeded onto plates. NK cells were prereacted with 50 nM Dex or diSia-Dex and added to plates with K562 cells to an ET ratio of 2:1 and reacted for 2 h. The fluorescence of the culture supernatant was measured. Values were standardized with no additives as a control. Experiments were performed in triplicate. The Student's t-test was used to determine the differences between cells treated with additives and control. Error bars indicate standard deviation. ∗p < 0.05.
Discussion
NK cells are innate immune cells involved in the elimination of host cells infected by viruses and cancer cells. During NK cell surveillance, the balance of signals from activating and inhibitory receptors is thought to determine the NK cell decision to attack “unhealthy” cells. Siglec-7 is well known as an inhibitory lectin-type receptor on NK cells and is involved in inhibition of cytotoxicity toward cancer cells via Sia-containing ligands, such as GD3 or DSGb5 (10, 16). In this study, the K562 cell line (a human chronic myeloid leukemia carcinoma cell line), frequently used as a target cell in NK cytotoxicity assays (10), was used to identify the counter-receptor for Siglec-7. It has been reported that NK cells, in the presence of a neutralizing antibody against Siglec-7, enhanced cancer immunity (5, 21); however, a counter-receptor for K562 has not been identified. We first identified leukosialin (SPN) as a counter-receptor on K562 cells by MS analysis and specific antibodies. To date, the known natural trans-ligands for Siglec-7 are gangliosides such as GD3 and DSGb5. However, using the PL method with K562 cells showed the ligand to be a glycoprotein. We also analyzed the possibility of a ganglioside ligand on K562 cells; however, there were almost no diSia-containing gangliosides, and Siglec-7 binding to GM3 was very faint (20). Glycodelin A (Progestagen-associated endometrial protein) and MUC16 (CA125) have been reported as counter-receptors for Siglec-7 (26, 27). Using K562, we adopted a new approach to identify the counter-receptor for Siglec-7. We used a strong inhibitor of Siglec-7, diSia-Dex, to elute ligands from a Siglec-7 column. Therefore, ligand elution was achieved by competition between the polyvalent diSia structure on diSia-Dex and the ligands for Siglec-7. Using this method, we could purify the counter-receptor for Siglec-7 and some other proteins that were also included in the Siglec-7-containing complex (Fig. 3). We also purified the Siglec-7 interacting molecules by direct analysis of Siglec-7 binding beads for MS; however, this produced different results. Using diSia-Dex and the membrane fraction, we identified leukosialin as a counter-receptor for Siglec-7. We also identified several molecules localized in the nucleolus and cytoskeleton (Table S1). As false-positive proteins can be identified because of the large amounts of cytosolic proteins, the direct interaction between Siglec-7 and leukosialin on the cell surface was confirmed using the PL method (Figs. 6 and 7). Using this method, leukosialin was shown to interact with Siglec-7 on the K562 cell surface, indicating that leukosialin is a counter-receptor. In this study, we also identified NCL, which is usually localized in the nucleolus, although there are several reports indicating that NCL is translocated to the cell surface and modified with glycans (28, 29, 30, 31). Pre-B acute lymphoblastic leukemia has 9-O-acetyl diSia-containing NCL on the cell surface (32). It has also been reported that NCL on the surface of macrophages binds to poly-N-acetyllactosamine on leukosialin in Jurkat cells and functions as a scavenger receptor (33). Unfortunately, we did not observe a direct interaction between Siglec-7 and NCL using the pull-down assay (Fig. 5); however, a complex containing Siglec-7, NCL, and other molecules might exist. In this regard, it is worth noting that HNRNPU was present in the Siglec-7-containing complex by MS analysis (Table S1). NCL has been shown to interact with HNRNPU during mitosis (34). We also observed an interaction between NCL and HNRNPU (Fig. 5D). It may be interesting to study the complex formation between cytosolic proteins and Siglecs.
Leukosialin (CD43 or sialophorin) is a transmembrane-type sialomucin expressed in all types of immune cells, except for resting B cells (35, 36). Leukosialin is a negatively charged type I glycoprotein, and its extracellular part is thought to extend from the cell surface and form a rod-shaped structure (37). The sequence is rich in Ser/Thr residues, and about 80 residues are modified with O-linked glycans (35). The highly charged nature of sialylated glycans contributes to the antiadhesive effect of leukosialin. The glycans show cell-type specificity depending on the set of biosynthesizing enzymes for O-linked glycans (38). Previously, leukosialin was reported as a counter-receptor for Siglec-1 in T cells (39). The known glycan structures on leukosialin are interesting, as Siglec-7 and Siglec-1 have different binding preferences. The sialic acid ligand for Siglec-7 remained mostly unchanged after PNGase F treatment, indicating that the ligands were modified on O-linked glycans (Fig. 1C). The 105kDa-gp band in the Siglec-7 blots against cell lysate after PNGase F treatment might be derived from leukosialin at 110 kDa, as there is only one potential N-linked glycan site in leukosialin. Siglec-7 binding to overexpressed leukosialin on K562 was not observed in the flow cytometry assay (Fig. 7A) but was observed using the PL method and western blotting (Fig. 7, B and F). Leukosialin has antiadhesive properties, and its high expression could inhibit Siglec-7 binding to surface molecules when leukosialin is not sufficiently modified with glycan epitopes recognized by Siglec-7 due to the restricted action of glycosyltransferases. In this respect, the biosynthetic enzymes for the glycan structures on leukosialin are key. Recent cell-based studies suggested that Siglec-7 binding to some cancer cells is dependent on mucin-type proteins (40) and that O-linked glycans are required for Siglec-7 binding (41). Based on the result that the diSiaα2,3Gal glycan epitope, a well-known GD3 component, was hardly detected on SPN from K562 cells (Fig. 1C), it can be hypothesized that the epitope diSiaα2,6Gal/GalNAc is present on K562 cells because diSia is one of the glycan ligands for Siglec-7. Among α2,8-sialyltransferases (ST8SIAs), ST8SIA2, -3, -4, and -6 are strong candidates for the synthesis of diSia structures on glycoproteins (42, 43). Siglec-7 has been shown to bind to branched α2,6-sialyl residues (15), α2,3-/α2,6-disialyl residues (44), disialyl Lewisa, and sialyl 6-sulfo Lewisx (45), which can be modified on O-linked glycans other than diSia. A precise understanding of the specificity of Siglec-7 for glycan ligands is required. Attempts to identify enzymes and structures are underway.
Recently, we demonstrated the presence of a secondary Siglec-7 binding site (site 2) in addition to the previously known binding site (site 1) (19). It has been suggested that Siglec-7 has several Sia-binding sites and that its counter-receptor for Siglec-7 has polyvalent glycans (e.g., clustered O-linked glycans); further, the binding mechanism may not be explained by a simple 1:1 model. Usually, the lectin–glycan interaction is weaker than the protein–protein interaction. We recently showed that the interaction between a cluster of ligands and Siglec-7 is stronger than the mono-ligand and Siglec-7 alone (23). Therefore, the identified counter-receptor for Siglec-7, leukosialin, with many O-linked glycans, is likely able to interact with Siglec-7 under physiological conditions. In this study, we specifically used diSia-Dex for elution, which strongly indicates that the binding affinity between Siglec-7 and leukosialin is lower than that between Siglec-7 and diSia-Dex. The amount of diSia, which regulates the interaction with Siglec-7, might be achieved by the enzymatic activity of ST8SIAs. Interestingly, Siglec-7 on NK cells cannot bind to the diSia-Dex until sialidase treatment, strongly indicating that diSia-Dex cannot be used for unmasking because of the strong binding between the cis-ligand and Siglec-7. This is consistent with previous reports (10). It is interesting that sialidase treatment induced diSia-Dex binding to Siglec-7 in NK cells (Fig. 8F). At this point, the sialidase Neu1 is already shown to be involved in the cleavage of sialic acid on the cell surface after activation by LPS (14, 46). Sialidase might be a key regulator of Siglec unmasking.
Cancer cells expressing Siglec-7 ligands can escape NK cell immunosurveillance and suppress immune reactions (21). To assay the cytotoxicity of NK cells, K562 cells are often used. To understand the biological significance of the interaction of Siglec-7 with its ligand, we prepared K562-SPN, -mock, and -shSPN cells and analyzed NK cell cytotoxicity. Surprisingly, Siglec-7 exerted no effects on the cytotoxicity toward K562-mock and K562-shSPN cells (Fig. 9B), although K562 itself had Siglec-7 ligands other than SPN. In contrast, the cytotoxicity toward K562-SPN was significantly inhibited in a Siglec-7-dependent manner. It is suggested that the total amount of ligand is crucial for Siglec-7 binding, and this may be a regulator of NK cell cytotoxicity. K562-SPN might display a sufficient number of ligands to release the cis-ligand from Siglec-7 on NK cells. In addition, cytotoxicity activity is determined based on the balance of activating and inhibitory signals from many receptors on NK cells. The cytotoxicity toward K562-SPN was lower than that toward K562-mock and -shSPN (Fig. 9A), and the inhibitory signal of Siglec-7 on NK cells decreased the cytotoxicity toward K562-SPN. Siglec-7 binding to K562-SPN was not high compared with that to K562-mock or -shSPN (Fig. 7A). This means that the binding levels analyzed by flow cytometry might not directly correlate with the inhibition of NK cells. Cell-to-cell contact forms an optimal molecular platform (e.g., immunological synapse), leading to enhancement of the receptor–ligand interaction. In this context, the interaction between Siglec-7 and SPN should be considered in spatiotemporal terms, as SPN is a large, highly charged molecule with antiadhesive properties. Interestingly, in neutrophils, the extracellular domain of leukosialin is shed by cathepsin G (47). In HEK cells expressing Siglec-11, cleavage of the extracellular domain of Siglec-11 occurred (48). The same shedding mechanism might occur in K562 cells or/and NK cells, and shed leukosialin or/and Siglec-7 might influence the cytotoxicity. The diSia mimic diSia-Dex showed inhibitory effects on the cytotoxicity of NK-Sig7 cells (Fig. 9C), indicating that the diSia cluster is one of the regulators of Siglec-7. DiSia-Dex functioned as an agonistic molecule for Siglec-7, as cytotoxicity was inhibited regardless of the expression level of SPN. Identification of the antagonists of Siglec-7 is required to further study the regulatory mechanism of cytotoxicity via SPN.
When this paper was under review, the supportive evidence of our work happened to be reported (49).
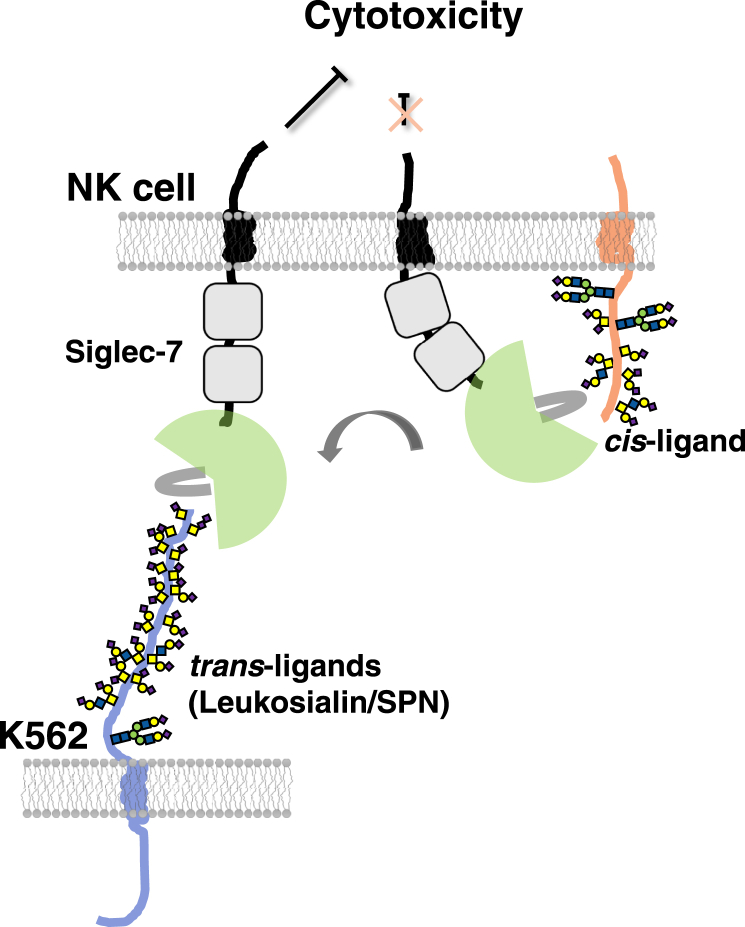
In summary, as shown in Figure 10, we focused on the counter-receptor for Siglec-7 in K562 cells, which have Siglec-7 ligands, and identified leukosialin. For Siglec-7 binding, the polyvalent diSia epitopes on diSia-Dex and O-linked glycans on leukosialin expressed in K562 were effective ligands. In addition, leukosialin on K562 and diSia-Dex showed inhibitory effects on NK cell cytotoxicity via Siglec-7. As Siglec-7 and leukosialin are enriched in immune cells, it is necessary to consider the complex interaction between cis- and trans-interactions in the future.
Figure 10.
Predicted model of counter-receptor on K562 cells for Siglec-7 on NK cells. Siglec-7 on NK cells is normally masked via cis-ligands even after Siglec-7 overexpression. Some of these are released by sialidase treatment but not addition to diSia-Dex. Leukosialin on K562 inhibited the cytotoxicity of NK cells by binding to Siglec-7 and that inhibition was enhanced by addition of diSia-Dex, indicating that the diSia cluster is the modulator of Siglec-7 function.
Experimental procedures
Materials
DNA polymerase KOD plus Neo was purchased from Toyobo. Protein A-Sepharose and enhanced chemiluminescence western blotting detection reagent were purchased from GE Healthcare. Sialidase from Vibrio cholerae, bovine serum albumin (BSA), and biotinyl-tyramide were purchased from Merck. Sialidase from Arthrobacter ureafaciens was obtained from Nacalai. The polyvinylidene difluoride (PVDF) membrane Immobilon P was purchased from Millipore. The In-Fusion HD Cloning Kit and pBApo plasmid were purchased from Takara Bio. Calcein-AM was purchased from Dojindo. Minimum Essential Medium Eagle Alpha (MEMα), Roswell Park Memorial Institute (RPMI)-1640, Dulbecco’s Modified Eagles Medium (DMEM), BSA, and puromycin dihydrochloride were purchased from Fujifilm. Serum-free medium COSMEDIUM 005 and the PEI “Max” transfection reagent were purchased from Cosmobio. Fetal bovine serum, Alexa Fluor 555-labeled anti-goat IgG antibody, horseradish peroxidase (HRP)-conjugated mouse anti-human IgG1 Fc antibody, and EZ-Link Sulfo-NHS-LC-Biotin were purchased from Thermo Fisher Scientific. HRP-conjugated anti-human IgG+M+A (anti-hIgG+M+A-HRP) was purchased from American Qualex. Vivaspin 6 concentrators were purchased from Sartorius. Anti-SPN antibody, anti-NCL antibody, and anti-HNRNPU antibody were purchased from ABclonal. Alexa Fluor 647-conjugated anti-mouse IgG+M was purchased from Jackson ImmunoResearch. The mouse monoclonal antibody (7.7) toward Siglec-7 was gifted by Dr Paul Crocker (College of Life Sciences, University of Dundee, UK). Siglec-7EcFc chimeric proteins were produced and purified as described previously (23). Sphingobacterium multivorum–derived sialidase was prepared as described previously (50).
Plasmid construction
The plasmid encoding the extracellular domain of Siglec-7 (amino acid #1–353) fused to the human IgG1 Fc region and multiple peptide tags (PA-3×FLAG-Strep (II)-SBP) was prepared using the In-Fusion kit. The Siglec-7 sequence was confirmed using the deoxynucleotide chain termination-based sequencing method and was found to be the same as the National Center for Biotechnology Information (NCBI) reference sequence: NP_055200.1. The Siglec-7 mutant plasmid (R124A) was prepared using PCR-based mutagenesis. pBApo-SPN-Myc (C-terminal tag) was prepared using the PCR amplicon of SPN from K562 cDNA. The SPN sequence was confirmed to be the same as the NCBI reference sequence: NP_003114.1. To construct the knockdown of SPN, we prepared the plasmid by changing the CAG promoter region of pEBMulti-Puro (FUJFILM Wako Pure Chemical) to the H1 promoter from pSUPER.neo (Oligoengine) and inserting the 21-bp target sequence, 5-GATGTACACCACTTCAATAAC-3 into the cloning site downstream of the H1 promoter according to manufacturer’s instructions.
Cell culture
K562 cells and HeLa cells were purchased from the Riken Cell Bank. All media used in this study were purchased from Wako and contained 10% FBS, 100 U/ml penicillin G, and 100 μg/ml streptomycin. The K562 cell line was cultured in RPMI-1640, and HeLa cells were cultured in H-DMEM. NK92-MI cells expressing Siglec-7 (NK-Sig7) or the mock transfectant (NK-EV) were maintained in MEMα containing 12.5% horse serum, 12.5% FBS, 0.2 mM inositol, 0.1 mM β-mercaptoethanol, and 0.02 mM folic acid. All cells were maintained in a humidified atmosphere of 5% CO2 and 95% air at 37 °C.
Transfection and establishment of the cells
K562 cells (106 cells) were plated onto a 6-well plate and incubated overnight at 37 °C in 5% CO2. After 50% to 80% confluency was achieved, the cells were transfected with the plasmid (3 μg) premixed with PEI “MAX” (6 μg) in 200 μl of PBS and incubated for 24 h. New medium with 4 μg/ml of puromycin was added for selection, and the cells were maintained for 20 days.
As NK-92MI does not express Siglec-7 (25), Siglec-7 was expressed by lentiviral transduction. In brief, cDNA for Siglec-7 was subcloned into the pCDH-CMV-MCS-EF1-copGFP transfer vector (System Biosciences) and packaged into lentiviral particles using HEK293T cells by cotransfection with pCMVΔR8.91 and pMD.G plasmids (both from RNA Technology Platform and Gene Manipulation Core, National Biotechnology Research Park and Academia Sinica, Taipei, Taiwan). A control virus containing an empty vector was also prepared in parallel. The supernatant from the transfected cells was cleared of cell debris by centrifugation and used for the transduction of NK-92MI cells. GFP+ cells were sorted twice at the Flow Cytometric Cell Sorting Facility (Institute of Biomedical Sciences, Academia Sinica) to obtain NK-Sig7 cells that express Siglec-7 and control NK-EV cells. These cells were used without further cloning.
Flow cytometric analysis
Cells were washed with PBS and fixed with 4% paraformaldehyde at 25 °C for 12 min. After washing, the cells were blocked with FCM buffer (0.5% BSA and 5 mM ethylenediaminetetraacetic acid, EDTA) in PBS. Sialidase treatment was performed using a mixture of A. ureafaciens neuraminidase (10 mU/ml) and V. cholerae neuraminidase (10 mU/ml) at room temperature for 1 h. Sig7EcFc (10 μg/ml) was precomplexed with anti-hIgG+M+A antibody (10 μg/ml) in FCM buffer on ice for 30 min. The cells were incubated with Sig7EcFc complexes on ice for 30 min. When Dex derivative sialopolymers (42) were used as inhibitors against Siglec-7, they were mixed with the cell suspension to a final concentration of 100 nM. After washing, the cells were stained using the secondary Alexa Fluor 555-labeled anti-goat IgG antibody (4 μg/ml) in FCM buffer. Cell surface staining was measured using a flow cytometer (Gallios, Beckman Coulter), and the collected data were analyzed using the Kaluza software (Beckman Coulter).
Pull-down assay for Siglec-7 ligands and western blotting
Lysates from K562 or HeLa cells were prepared using lysis buffer (15 mM Tris/HCl at pH 7.8, 0.15 M NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail at 1/500). Sig7EcFc (1 μg) was mixed with the lysate (90 μg of protein), and the mixture was rotated at 4 °C overnight. Protein A-sepharose (10 μl of beads) was added to the mixture and rotated at 4 °C for 30 min. The supernatant was collected, and the beads were washed three times with lysis buffer and heat denatured with SDS sample buffer (SB) (6×SB: 2% SDS; 125 mM Tris/HCl at pH 6.8, 10% 2-mercaptoethanol, 10% glycerol, and 0.01% bromophenol blue). The samples were then subjected to western blotting, as described below.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the denatured samples. After the separation gel was transferred to a PVDF membrane, the membrane was blocked using 1% BSA or 0.5% powder milk in PBS containing 0.05% Tween-20 for 1 h at room temperature during Siglec-7 blotting or antibody blotting, respectively. When detecting Siglec-7 ligands, Sig7EcFc (5 μg/ml) and goat anti-human IgG+M+A-HRP (5 μg/ml) were precomplexed in 1% BSA at room temperature for 30 min. The Sig7EcFc complex was diluted tenfold with PBS containing 0.05% Tween-20 and shaken with the membrane for 2 h at room temperature. The antibodies were diluted to 1 μg/ml or 1000-fold with the blocking solution and shaken with the membrane at 4 °C overnight. The HRP-labeled secondary antibody (1/5000) was then added and shaken with the membrane at room temperature for 1 h. The membrane was reacted with ECL solution, and chemiluminescence was measured using a WSE-6200H LuminoGraphII (Atto).
Surface biotinylation and ligand purification using diSia-Dex
To confirm that the Siglec-7 ligand was present on the cell surface, random biotinylation of the cell surface was performed, followed by ligand purification. After washing the K562 cells (108 cells), they were suspended in 900 μl of PBS, and 1 mg of EZ-Link Sulfo-NHS-LC-Biotin was dissolved into the solution. The reaction was carried out at 4 °C for 30 min, and 100 μl of 1 M Tris/HCl (pH 7.8) was added to quench the reaction. The cells were washed twice with PBS, and the lysates were prepared. The lysate (400 μg of protein) was reacted with Sig7EcFc (6 μg) at 4 °C overnight. Protein A-sepharose (20 μl of beads) was reacted with the mixture at 4 °C for 30 min. The supernatant was collected, and the bound proteins were eluted using Dex derivatives. First, Dex (100 nM) as a control compound could react with the proteins at 4 °C for 20 min to perform nonspecific elution. Subsequently, diSia-Dex (100 nM) was used to elute the bound ligands. Each eluted fraction and the beads were heat denatured with SB, and the supernatants were analyzed using silver staining and western blotting.
Siglec-7 ligand purification for MS
Ligand purification for MS was performed using the prepared crude membrane fraction and whole lysate of K562 cells. The crude membrane fraction was prepared by the ultracentrifugation method. After washing the cells twice with PBS, cells were suspended in a hypotonic solution (10 mM Tris/HCl at pH 7.8, 30 mM NaCl, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail at 1/500) and sonicated. The suspension was centrifuged at 600g for 10 min to precipitate the nuclei and intact living cells, and the supernatant was subjected to ultracentrifugation. The membrane fraction was precipitated by centrifugation at 100,000g for 70 min. The precipitate was dissolved in lysis buffer to obtain a crude membrane fraction. Purified Sig7EcFc (125 μg) was added to the K562 crude membrane fraction (2 mg of protein) or whole lysate (6 mg of protein), and the mixture was rotated overnight at 4 °C. Protein A-Sepharose (150 μl of beads) blocked overnight with a bead blocking solution (5% polyvinylpyrrolidone K 30, 50 mM Tris/HCl at pH 8.0) was placed on the column. The mixed sample was passed through the column three times, and the column was washed four times. Subsequently, nonspecific proteins were eluted three times with 50 nM Dex, and the ligands were eluted using 50 nM diSia-Dex three times. Finally, the bound proteins were directly heat denatured and released from the beads. For the Dex and diSia-Dex fractions, the solutions were concentrated about sixfold using the Vaivacon 500 (10 kDa MWCO). Each sample was subjected to SDS-PAGE, silver staining for MS, and western blotting to confirm the molecular weight of the binding partners.
In-gel digestion and MS analysis
Samples for MS were prepared by in-gel trypsin digestion. The target 110 kDa area on the silver stained gel was cut out and finely chopped on the dice. A decolorizing solution (15 mM potassium ferricyanide, 50 mM sodium thiosulfate) was added to the gel pieces, and the mixture was shaken for 1 min at room temperature. The gel pieces were washed three times with water and then twice with a washing solution (30% acetonitrile, 100 mM ammonium bicarbonate), dried in a vacuum Concentrator Plus (Eppendorf) for 15 min, incubated with a reducing solution (10 mM dithiothreitol) at 37 °C for 30 min, shaken with an alkylating solution (1% acrylamide) at room temperature for 15 min, washed once with the washing solution, shaken with 100% acetonitrile for 5 min, and dried again using a vacuum concentrator. Trypsin/Lys-C Mix (Promega) was diluted to 5 ng/μl with a detergent solution (0.01% ProteaseMAX surfactant (Promega), 50 mM ammonium bicarbonate) and absorbed as much as possible into the gel pieces. Fifty microliters of the detergent solution was added and reacted at 50 °C for 1 h. The supernatant containing the digested peptides was collected and 15 μl of 10% formic acid was added to the gel pieces to extract the remaining peptides. The decomposed surfactant was precipitated by centrifuging (20,000g, 10 min, 4 °C), and the combined solution and supernatant were passed through a MonoSpin C18 column (GL Sciences) to purify the peptides. The eluted solution (60% acetonitrile, 0.1% formic acid) containing the peptides was dried using a vacuum concentrator, and the dissolved solution (2% acetonitrile, 0.1% formic acid) was loaded onto an ekspert microLC 200 (Eksigent) directly equipped with an ESI-triple quadrupole-linear ion trap system (Triple Quad 5500+LC-MS/MS system, SCIEX). Liquid chromatography was performed with a C18 column (phase: HALO C18, 2.7 μm, 90Å, size: 0.5 × 100 mm, Eksigent) across a 45 min gradient from 2% to 60% acetonitrile using two mobile phases (water/0.1% formic acid and acetonitrile/0.1% formic acid) at 0.5 μl/min flowrate. The column was washed with 95% acetonitrile for 7 min and then equilibrated with 2% acetonitrile for 8 min. Precursor ion scanning was set from 400 to 1000 m/z with a charge state between 2 and 4 using Information Dependent Acquisition criteria. The scans of Enhanced Product Ions were obtained at a 10,000 Da/s scan rate over a mass range of 100 to 1000 Da. The total 60 min survey included 2840 cycles (1.2674 s/cycle). The collected peptide fragmentation spectra were analyzed using Analyst Ver1.6.1 (SCIEX) and stored in.wiff format. The obtained data were analyzed using ProteinPilot software 5.0 (SCIEX). The Swiss-Prot database (obtained on November 2018) containing 20,411 human sequences was used for protein validation. The following settings were selected in ProteinPilot: Sample Type; Identification, Cys Alkylation: Acrylamide; Digestion: Trypsin+Lys-C; Instrument: 5500 QTRAP ESI; Species: Homo sapiens; ID Focus: Biological modifications; Search Effort: Thorough ID; Results Quality: Detected Protein Threshold [Unused ProtScore (Conf)]>: 0.47 (66.0%) without false discovery rate.
Sialidase or PNGase F treatment
S. multivorum–derived sialidase (12 mU), with an optimum pH near neutral, was added to the K562 lysate (20 μg of protein) or the purified ligand fraction, and the mixture was incubated overnight at 37 °C.
PNGase F (NEB Japan) treatment was performed according to the manufacturer’s instructions. K562 lysate (20 μg of protein) was heat denatured using the denaturing buffer, mixed with the reaction buffer and PNGase F (20 U), and incubated overnight at 37 °C.
Immunoprecipitation using antibodies against candidate Siglec-7 counter-receptor molecules
Immunoprecipitation (IP) was performed using antibodies against molecules that were predicted to be the counter-receptor candidates for Siglec-7 based on the MS results. Anti-SPN or anti-NCL antibody (2 μl) was mixed with K562 lysate (200 μg of protein), and the mixture was rotated overnight at 4 °C. Protein A-Sepharose (15 μl of beads) blocked with the bead blocking solution was added and reacted at 4 °C for 1 h. The supernatant was collected, and the beads were washed three times and heat denatured. The samples were analyzed by western blotting using Sig7EcFc and the antibodies.
Biotinylation of cell surface ligands using proximity labeling
PL was performed following a previously reported method (24). Briefly, harvested K562 cells were washed with Tris-buffered saline (TBS) and then incubated with Sig7EcFc or Sig9EcFc and anti-hIgG+M+A-HRP complexes on ice for 1 h. After washing, the cells were incubated in TBS containing 20 μM biotinyl-tyramide and 10 mM H2O2 at 25 °C for 10 min, followed by lysis. The lysates were analyzed by western blotting with HRP-labeled streptavidin.
Biotinylated proteins were purified by a pull-down experiment to confirm the binding of Siglec-7 and leukosialin on the cell surface. Streptavidin-Sepharose (20 μl of beads) was reacted with the PL-treated K562 lysates (200 μg of protein) at 4 °C for 30 min. The supernatant was collected, and the beads were washed and denatured. The supernatant and bead-binding proteins were analyzed by western blotting.
Cytotoxicity assay of Siglec-7 stably expressing NK cells
After collecting K562 cells (107 cells), the cells were washed twice with RPMI-1640 without FBS and phenol red. The cells were resuspended in RPMI-1640 containing 10 μM Calcein-AM and incubated in a CO2 incubator at 37 °C for 20 min. The stained K562 cells were washed three times with RPMI-1640 containing 5% FBS (RPMI (5)). After counting the cells, they were seeded at 2 × 105 cells/100 μl in a round-bottomed 96-well plate. NK-EV or NK-Sig7 was washed once with RPMI (5) to prepare a suspension of 1.5 to 12 × 105 cells/100 μl. NK cell suspensions were added so that the ET ratio (Effector cell, NK92-MI: Target cell, K562) equaled 0.75 to 6: 1. The plate was centrifuged (1200 rpm, 1 min) and incubated at 37 °C for 2 h. At 1.5 h after the start of incubation, RPMI (5) was added as a spontaneous control, and 1% Triton X-100 was added as a maximum lysis control. The plate was centrifuged (1800 rpm, 3 min), and 120 μl of the supernatant was transferred to a black plate for fluorescence measurement (Sumitomo Bakelite). The solution fluorescence was measured using a plate reader (EnSpire, PerkinElmer Japan) at an excitation wavelength of 485 nm and an emission wavelength of 535 nm. The specific lysis rate was calculated based on the following formula using the fluorescence intensity (FI):
When binding inhibitors were used, Dex or diSia-Dex was mixed with the suspension of NK92-MI to a final concentration of 50 nM. The cells were seeded with K562 (2 × 105 cells/100 μl) and NK92-MI (4 × 105 cells/100 μl) on a plate to ensure the ET ratio was 2. Experiments were performed in triplicate. Values were standardized with no additives as controls. Student's t-test was used for each ET ratio or the cells treated with additives and control.
Data availability
The MS raw data were deposited in the Figshare repository with DOI (https://doi.org/10.6084/m9.figshare.13602785.v1).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no conflicts of interest regarding this article.
Acknowledgments
We thank RNA Technology Platform and Gene Manipulation Core (National Biotechnology Research Park and Academia Sinica, Taipei, Taiwan) and Flow Cytometric Cell Sorting Facility (Institute of Biomedical Sciences, Academia Sinica) for their technical support. We thank Dr Shunsuke Nishio (Karolinska Institutet, Sweden) and Prof. Hitoshi Mori (Nagoya University, Nagoya, Japan) for technical advice for MS operation.
Author contributions
Conceptualization: A. Y. and C. S.; Data curation: A. Y., K. K., and C. S.; Formal analysis: A. Y. and C. S.; Funding acquisition: C. S.; Investigation: A. Y., K. K., and C. S.; Methodology: A. Y., L. C., T. A., K. K., and C. S.; Project administration: C. S.; Resources for Siglec: A. Y., Y. A., H. T., K. K., and C. S.; Resources for NK-Sig7 and NK-EV cell lines: L. C. and T. A.; Software: A. Y.; Supervision: C. S.; Validation: A. Y., K. K., and C. S.; Visualization: A. Y., K. K., and C. S.; Writing—original draft: A. Y. and C. S.; Writing—review and editing: A. Y., K. K., and C. S.
Funding and additional information
This work was supported by grants from the Mizutani Foundation (C. S.), Daiko Foundation (C. S.), and AMED (20ae0101069h0005) (C. S.). This work was also supported by Academia Sinica Thematic Project Grant (AS-TP-108-ML06) (T. A.).
Edited by Gerald Hart
Supporting information
References
- 1.Crocker P.R., Paulson J.C., Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 2.Macauley M.S., Crocker P.R., Paulson J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angata T., Nycholat C.M., Macauley M.S. Therapeutic targeting of Siglecs using antibody- and glycan-based approaches. Trends Pharmacol. Sci. 2015;36:645–660. doi: 10.1016/j.tips.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daly J., Carlsten M., O'Dwyer M. Sugar free: Novel immunotherapeutic approaches targeting Siglecs and sialic acids to enhance natural killer cell cytotoxicity against cancer. Front. Immunol. 2019;10:1047. doi: 10.3389/fimmu.2019.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudak J.E., Canham S.M., Bertozzi C.R. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol. 2014;10:69–75. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao H., Woods E.C., Vukojicic P., Bertozzi C.R. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc. Natl. Acad. Sci. U. S. A. 2016;113:10304–10309. doi: 10.1073/pnas.1608069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicoll G., Ni J., Liu D., Klenerman P., Munday J., Dubock S., Mattei M.G., Crocker P.R. Identification and characterization of a novel Siglec, Siglec-7, expressed by human natural killer cells and monocytes. J. Biol. Chem. 1999;274:34089–34095. doi: 10.1074/jbc.274.48.34089. [DOI] [PubMed] [Google Scholar]
- 8.Angata T., Varki A. Siglec-7: A sialic acid-binding lectin of the immunoglobulin superfamily. Glycobiology. 2000;10:431–438. doi: 10.1093/glycob/10.4.431. [DOI] [PubMed] [Google Scholar]
- 9.Razi N., Varki A. Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7469–7474. doi: 10.1073/pnas.95.13.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicoll G., Avril T., Lock K., Furukawa K., Bovin N., Crocker P. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur. J. Immunol. 2003;33:1642–1648. doi: 10.1002/eji.200323693. [DOI] [PubMed] [Google Scholar]
- 11.Freeman S.D., Kelm S., Barber E.K., Crocker P.R. Characterization of CD33 as a new member of the sialoadhesin family of cellular interaction molecules. Blood. 1995;85:2005–2012. [PubMed] [Google Scholar]
- 12.Cornish A.L., Freeman S., Forbes G., Ni J., Zhang M., Cepeda M., Gentz R., Augustus M., Carter K.C., Crocker P.R. Characterization of Siglec-5, a novel glycoprotein expressed on myeloid cells related to CD33. Blood. 1998;92:2123–2132. [PubMed] [Google Scholar]
- 13.Munday J., Kerr S., Ni J., Cornish A.L., Zhang J.Q., Nicoll G., Floyd H., Mattei M.G., Moore P., Liu D., Crocker P.R. Identification, characterization and leucocyte expression of Siglec-10, a novel human sialic acid-binding receptor. Biochem. J. 2001;355:489–497. doi: 10.1042/0264-6021:3550489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumida M., Hane M., Yabe U., Shimoda Y., Pearce O.M.T., Kiso M., Miyagi T., Sawada M., Varki A., Kitajima K., Sato C. Rapid trimming of cell surface polysialic acid (PolySia) by exovesicular sialidase triggers release of preexisting surface neurotrophin. J. Biol. Chem. 2015;290:13202–13214. doi: 10.1074/jbc.M115.638759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaji T., Teranishi T., Alphey M.S., Crocker P.R., Hashimoto Y. A small region of the natural killer cell receptor, Siglec-7, is responsible for its preferred binding to alpha 2,8-disialyl and branched alpha 2,6-sialyl residues. A comparison with Siglec-9. J. Biol. Chem. 2002;277:6324–6332. doi: 10.1074/jbc.M110146200. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki Y., Ito A., Withers D.A., Taima T., Kakoi N., Saito S., Arai Y. Ganglioside DSGb5, preferred ligand for Siglec-7, inhibits NK cell cytotoxicity against renal cell carcinoma cells. Glycobiology. 2010;20:1373–1379. doi: 10.1093/glycob/cwq116. [DOI] [PubMed] [Google Scholar]
- 17.Alphey M.S., Attrill H., Crocker P.R., van Aalten D.M. High resolution crystal structures of Siglec-7. Insights into ligand specificity in the Siglec family. J. Biol. Chem. 2003;278:3372–3377. doi: 10.1074/jbc.M210602200. [DOI] [PubMed] [Google Scholar]
- 18.Attrill H., Imamura A., Sharma R.S., Kiso M., Crocker P.R., van Aalten D.M. Siglec-7 undergoes a major conformational change when complexed with the alpha(2,8)-disialylganglioside GT1b. J. Biol. Chem. 2006;281:32774–32783. doi: 10.1074/jbc.M601714200. [DOI] [PubMed] [Google Scholar]
- 19.Yamakawa N., Yasuda Y., Yoshimura A., Goshima A., Crocker P.R., Vergoten G., Nishiura Y., Takahashi T., Hanashima S., Matsumoto K., Yamaguchi Y., Tanaka H., Kitajima K., Sato C. Discovery of a new sialic acid binding region that regulates Siglec-7. Sci. Rep. 2020;10:8647. doi: 10.1038/s41598-020-64887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimura A., Hatanaka R., Tanaka H., Kitajima K., Sato C. The conserved arginine residue in all Siglecs is essential for Siglec-7 binding to sialic acid. Biochem. Biophys. Res. Commun. 2020;534:1069–1075. doi: 10.1016/j.bbrc.2020.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Jandus C., Boligan K.F., Chijioke O., Liu H., Dahlhaus M., Démoulins T., Schneider C., Wehrli M., Hunger R.E., Baerlocher G.M., Simon H.U., Romero P., Münz C., von Gunten S. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Invest. 2014;124:1810–1820. doi: 10.1172/JCI65899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato C., Fukuoka H., Ohta K., Matsuda T., Koshino R., Kobayashi K., Troy F.A., Kitajima K. Frequent occurrence of pre-existing alpha 2-->8-linked disialic and oligosialic acids with chain lengths up to 7 Sia residues in mammalian brain glycoproteins. Prevalence revealed by highly sensitive chemical methods and anti-di-, oligo-, and poly-Sia antibodies specific for defined chain lengths. J. Biol. Chem. 2000;275:15422–15431. doi: 10.1074/jbc.275.20.15422. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi S., Yoshimura A., Yasuda Y., Mori A., Tanaka H., Takahashi T., Kitajima K., Sato C. Chemical synthesis and evaluation of a disialic acid-containing dextran polymer as an inhibitor for the interaction between Siglec 7 and its ligand. Chembiochem. 2017;18:1194–1203. doi: 10.1002/cbic.201600694. [DOI] [PubMed] [Google Scholar]
- 24.Chang L., Chen Y.J., Fan C.Y., Tang C.J., Chen Y.H., Low P.Y., Ventura A., Lin C.C., Angata T. Identification of Siglec ligands using a proximity labeling method. J. Proteome Res. 2017;16:3929–3941. doi: 10.1021/acs.jproteome.7b00625. [DOI] [PubMed] [Google Scholar]
- 25.Huang C.H., Liao Y.J., Fan T.H., Chiou T.J., Lin Y.H., Twu Y.C. A developed NK-92MI cell line with Siglec-7neg phenotype exhibits high and sustainable cytotoxicity against leukemia cells. Int. J. Mol. Sci. 2018;19:1073. doi: 10.3390/ijms19041073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijayan M., Lee C.L., Wong V.H.H., Wang X., Bai K., Wu J., Koistinen H., Seppälä M., Lee K.F., Yeung W.S.B., Ng E.H.Y., Chiu P.C.N. Decidual glycodelin-A polarizes human monocytes into a decidual macrophage-like phenotype through Siglec-7. J. Cell Sci. 2020;133 doi: 10.1242/jcs.244400. [DOI] [PubMed] [Google Scholar]
- 27.Mitic N., Milutinovic B., Jankovic M. Assessment of sialic acid diversity in cancer- and non-cancer related CA125 antigen using sialic acid-binding Ig-like lectins (Siglecs) Dis. Markers. 2012;32:187–194. doi: 10.3233/DMA-2011-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandes E., Freitas R., Ferreira D., Soares J., Azevedo R., Gaiteiro C., Peixoto A., Oliveira S., Cotton S., Relvas-Santos M., Afonso L.P., Palmeira C., Oliveira M.J., Ferreira R., Silva A.M.N. Nucleolin-Sle A glycoforms as E-selectin ligands and potentially targetable biomarkers at the cell surface of gastric cancer cells. Cancers (Basel) 2020;12:861. doi: 10.3390/cancers12040861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoja-Łukowicz D., Kedracka-Krok S., Duda W., Lityńska A. The lectin-binding pattern of nucleolin and its interaction with endogenous galectin-3. Cell Mol. Biol. Lett. 2014;19:461–482. doi: 10.2478/s11658-014-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galzio R., Rosati F., Benedetti E., Cristiano L., Aldi S., Mei S., D'Angelo B., Gentile R., Laurenti G., Cifone M.G., Giordano A., Cimini A. Glycosilated nucleolin as marker for human gliomas. J. Cell Biochem. 2012;113:571–579. doi: 10.1002/jcb.23381. [DOI] [PubMed] [Google Scholar]
- 31.Carpentier M., Morelle W., Coddeville B., Pons A., Masson M., Mazurier J., Legrand D. Nucleolin undergoes partial N- and O-glycosylations in the extranuclear cell compartment. Biochemistry. 2005;44:5804–5815. doi: 10.1021/bi047831s. [DOI] [PubMed] [Google Scholar]
- 32.Joo E.J., Wasik B.R., Parrish C., Paz H., Mϋhlenhoff M., Abdel-Azim H., Groffen J., Heisterkamp N. Pre-B acute lymphoblastic leukemia expresses cell surface nucleolin as a 9-O-acetylated sialoglycoprotein. Sci. Rep. 2018;8:17174. doi: 10.1038/s41598-018-33873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirano K., Miki Y., Hirai Y., Sato R., Itoh T., Hayashi A., Yamanaka M., Eda S., Beppu M. A multifunctional shuttling protein nucleolin is a macrophage receptor for apoptotic cells. J. Biol. Chem. 2005;280:39284–39293. doi: 10.1074/jbc.M505275200. [DOI] [PubMed] [Google Scholar]
- 34.Ma N., Matsunaga S., Morimoto A., Sakashita G., Urano T., Uchiyama S., Fukui K. The nuclear scaffold protein SAF-A is required for kinetochore-microtubule attachment and contributes to the targeting of aurora-A to mitotic spindles. J. Cell Sci. 2011;124:394–404. doi: 10.1242/jcs.063347. [DOI] [PubMed] [Google Scholar]
- 35.Fukuda M. Leukosialin, a major O-glycan-containing sialoglycoprotein defining leukocyte differentiation and malignancy. Glycobiology. 1991;1:347–356. doi: 10.1093/glycob/1.4.347. [DOI] [PubMed] [Google Scholar]
- 36.Dragone L.L., Barth R.K., Sitar K.L., Disbrow G.L., Frelinger J.G. Disregulation of leukosialin (CD43, Ly48, sialophorin) expression in the B-cell lineage of transgenic mice increases splenic B-cell number and survival. Proc. Natl. Acad. Sci. U. S. A. 1995;92:626–630. doi: 10.1073/pnas.92.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cyster J.G., Shotton D.M., Williams A.F. The dimensions of the T lymphocyte glycoprotein leukosialin and identification of linear protein epitopes that can be modified by glycosylation. EMBO J. 1991;10:893–902. doi: 10.1002/j.1460-2075.1991.tb08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuccillo F.M., de Laurentiis A., Palmieri C., Fiume G., Bonelli P., Borrelli A., Tassone P., Scala I., Buonaguro F.M., Quinto I., Scala G. Aberrant glycosylation as biomarker for cancer: Focus on CD43. Biomed. Res. Int. 2014;2014:742831. doi: 10.1155/2014/742831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Berg T.K., Nath D., Ziltener H.J., Vestweber D., Fukuda M., van Die I., Crocker P.R. Cutting edge: CD43 functions as a T cell counterreceptor for the macrophage adhesion receptor sialoadhesin (Siglec-1) J. Immunol. 2001;166:3637–3640. doi: 10.4049/jimmunol.166.6.3637. [DOI] [PubMed] [Google Scholar]
- 40.Malaker S.A., Pedram K., Ferracane M.J., Bensing B.A., Krishnan V., Pett C., Yu J., Woods E.C., Kramer J.R., Westerlind U., Dorigo O., Bertozzi C.R. The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7278–7287. doi: 10.1073/pnas.1813020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narimatsu Y., Joshi H.J., Nason R., Van Coillie J., Karlsson R., Sun L., Ye Z., Chen Y.H., Schjoldager K.T., Steentoft C., Furukawa S., Bensing B.A., Sullam P.M., Thompson A.J., Paulson J.C. An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol. Cell. 2019;75:394–407.e395. doi: 10.1016/j.molcel.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato C., Kitajima K. Disialic, oligosialic and polysialic acids: Distribution, functions and related disease. J. Biochem. 2013;154:115–136. doi: 10.1093/jb/mvt057. [DOI] [PubMed] [Google Scholar]
- 43.Sato C. Chain length diversity of sialic acids and its biological significance. Trends Glycosci. Glycotech. 2004;14:331–344. [Google Scholar]
- 44.Wu H.R., Anwar M.T., Fan C.Y., Low P.Y., Angata T., Lin C.C. Expedient assembly of Oligo-LacNAcs by a sugar nucleotide regeneration system: Finding the role of tandem LacNAc and sialic acid position towards Siglec binding. Eur. J. Med. Chem. 2019;180:627–636. doi: 10.1016/j.ejmech.2019.07.046. [DOI] [PubMed] [Google Scholar]
- 45.Miyazaki K., Sakuma K., Kawamura Y.I., Izawa M., Ohmori K., Mitsuki M., Yamaji T., Hashimoto Y., Suzuki A., Saito Y., Dohi T., Kannagi R. Colonic epithelial cells express specific ligands for mucosal macrophage immunosuppressive receptors Siglec-7 and -9. J. Immunol. 2012;188:4690–4700. doi: 10.4049/jimmunol.1100605. [DOI] [PubMed] [Google Scholar]
- 46.Amith S.R., Jayanth P., Franchuk S., Finlay T., Seyrantepe V., Beyaert R., Pshezhetsky A.V., Szewczuk M.R. Neu1 desialylation of sialyl alpha-2,3-linked beta-galactosyl residues of TOLL-like receptor 4 is essential for receptor activation and cellular signaling. Cell. Signal. 2010;22:314–324. doi: 10.1016/j.cellsig.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 47.Mambole A., Baruch D., Nusbaum P., Bigot S., Suzuki M., Lesavre P., Fukuda M., Halbwachs-Mecarelli L. The cleavage of neutrophil leukosialin (CD43) by cathepsin G releases its extracellular domain and triggers its intramembrane proteolysis by presenilin/gamma-secretase. J. Biol. Chem. 2008;283:23627–23635. doi: 10.1074/jbc.M710286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hane M., Chen D.Y., Varki A. Human-specific microglial Siglec-11 transcript variant has the potential to affect polysialic acid-mediated brain functions at a distance. Glycobiology. 2020 doi: 10.1093/glycob/cwaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wisnovsky S., Möckl L., Malaker S.A., Pedram K., Hess G.T., Riley N.M., Gray M.A., Smith B.A.H., Bassik M.C., Moerner W.E., Bertozzi C.R. Genome-wide CRISPR screens reveal a specific ligand for the glycan-binding immune checkpoint receptor Siglec-7. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2015024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwaki Y., Matsunaga E., Takegawa K., Sato C., Kitajima K. Identification and characterization of a novel, versatile sialidase from a Sphingobacterium that can hydrolyze the glycosides of any sialic acid species at neutral pH. Biochem. Biophys. Res. Commun. 2020;523:487–492. doi: 10.1016/j.bbrc.2019.12.079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS raw data were deposited in the Figshare repository with DOI (https://doi.org/10.6084/m9.figshare.13602785.v1).