Abstract
Commensal microorganisms present at mucosal surfaces play a vital role in protecting the host organism from bacterial infection. There are multiple factors that contribute to selecting for the microbiome, including host genetics. Flavobacterium psychrophilum, the causative agent of Bacterial Cold Water Disease in salmonids, accounts for acute losses in wild and farmed rainbow trout (Oncorhynchus mykiss). The U.S. National Center for Cool and Cold Water Aquaculture has used family-based selective breeding to generate a line of rainbow trout with enhanced resistance to F. psychrophilum. The goal of this study is to determine whether selective breeding impacts the gut and gill microbiome of the F. psychrophilum-resistant as compared to a background matched susceptible trout line. Mid-gut and gill samples were collected from juvenile fish maintained at high or low stocking densities and microbial diversity assessed by 16S rDNA amplicon sequencing. Results indicate that alpha diversity was significantly higher in the mid-gut of the susceptible line compared to the resistant line, while no significant differences in alpha diversity were observed in the gills. Mycoplasma sp. was the dominant taxon in the mid-gut of both groups, although it was present at a decreased abundance in the susceptible line. We also observed an increased abundance of the potential opportunistic pathogen Brevinema andersonii in the susceptible line. Within the gills, both lines exhibited similar microbial profiles, with Candidatus Branchiomonas being the dominant taxon. Together, these results suggest that selectively bred F. psychrophilum-resistant trout may harness a more resilient gut microbiome, attributing to the disease resistant phenotype. Importantly, interactions between host genetics and environmental factors such as stocking density have a significant impact in shaping trout microbial communities.
Keywords: Rainbow trout, Aquaculture, Flavobacterium psychrophilum, Bacterial cold water disease, ARS-Fp-R, ARS-Fp-S, Microbiome, Disease resistance
1. Introduction
The microbiome has well established roles in pathogen exclusion and host immunity, including systemic and mucosal innate and adaptive immune responses and development of the immune system [1-3]. Across species, the intestinal microbiome is established at nascent developmental stages upon exposure to external environments. Under homeostatic conditions, primordial commensal microbes colonizing host mucosal surfaces must outcompete any other microorganisms present in the environment. Resident microbes possess an advantage in resource acquisition driving their evolution to better adapt in a specific host microenvironment, which in some cases provides benefits to the host organism. In response to these phenomena, species can adapt to select for those microorganisms that are most beneficial, resulting in microbial assemblies that are to a large degree unique to each species. Studies in teleost fish have supported this by identifying a 'core' gut microbiota in zebrafish (Danio rerio) [4], Atlantic salmon (Salmo salar) [5], and rainbow trout (O. mykiss) cultured in water recirculation systems [6].
Host genetics have been proposed to play a supporting role in the selection of the gut microbiome in humans and other mammals [7-11], while environmental factors have also been shown to largely contribute to host microbiome assembly [12-15]. There have been some efforts to investigate factors that intrinsically influence microbiome assembly in fish, providing support for both host-associated and environmental factors. Host genetic factors that contribute to microbiome assembly have been well characterized in stickleback, as differences in MHC genotype have been shown to affect microbiome composition [16]. Additionally, a longitudinal microbiome analysis conducted on channel catfish (Ictalurus punctatus) characterized how the intestinal microbiota shifts during ontogeny and how diet and environmental microbes influence microbiota in this species [17]. Further work by this group showed that host genetic factors had a minimal impact on microbial composition [18]. Additionally, a study conducted on hatchery-reared and wild caught Atlantic salmon across various regions highlights diet and genetic factors as major contributors to microbiome assembly, particularly in the gut [19]. To further disentangle the contribution of host genetics and environmental factors shaping the fish microbiome, here we utilize a rainbow trout model in which two genetic lines of rainbow trout have been established by selective breeding that differ in susceptibility to a common environmental gram-negative pathogen, Flavobacterium psychrophilum.
F. psychrophilum is the causative agent of Bacterial Cold Water Disease (BCWD), which is a major concern in the United States aquaculture industry affecting a range of cold-water fish species, including the commercially relevant rainbow trout (O. mykiss). Outbreaks of this pathogen have been reported across all areas of the world that contribute to salmonid aquaculture, posing a substantial threat to the future of this industry [20]. F. psychrophilum is a mucosal pathogen that typically infects the skin and gills of fish but also has the ability to adhere to and damage the intestinal epithelium [21,22]. Additionally, supplementing rainbow trout with probiotic bacteria that are able to colonize the gastrointestinal tract has been shown to decrease F. psychrophilum induced mortality [23]. Symptoms of BCWD in developed fish include necrosis of the caudal region, skin lesions, eroded fin tips, and loss of appetite. F. psychrophilum has a more pronounced effect on young fry, a condition referred to as rainbow trout fry syndrome. Rainbow trout fry syndrome is responsible for acute losses in trout farms worldwide, as the associated mortality rate is reported to be greater than 50% [20]. BCWD is becoming an increasingly difficult disease to treat, as F. psychrophilum strains have developed resistance to several commonly used antibiotics [24-26], and there is currently no commercially available licensed vaccine.
The National Center for Cool and Cold Water Aquaculture (NCCCWA) utilized family-based selective breeding to develop two distinctive genetic lines of rainbow trout that confer enhanced resistance (ARS-Fp-R), or susceptibility (ARS-Fp-S) to the pathogen F. psychrophilum [27]. Enhanced resistance to F. psychrophilum-induced mortality in the ARS-Fp-R line has been described, both in the laboratory setting and on trout farms [28,29]. Previous studies have begun to investigate possible host mechanisms that attribute to enhanced resistance. For instance, a strong correlation between resistance to F. psychrophilum and increased spleen size has been described, although this relation does not appear to translate to other common fish pathogens, such as Yersinia ruckeri [30]. Additionally, whole-body transcriptome analysis has identified numerous acute phase proteins and inflammatory cytokines that are differentially expressed in each line following challenge with F. psychrophilum [31]. Further work is needed to better characterize the mechanism(s) by which enhanced resistance is achieved in the ARS-Fp-R line.
In this paper, we begin to investigate whether ARS-Fp-R and ARS-Fp-S trout lines have different microbial assemblages associated with the gut and gills. Using 16S rDNA amplicon sequencing, we evaluate the effect of host genetics (F. psychrophilum resistance or susceptibility) as well as the effect of different tank conditions on trout microbiome assembly.
2. Materials and methods
2.1. Animals and sampling
The Institutional Animal Care and Use Committee (Leetown, WV) reviewed and approved all animal husbandry practices and disease challenge protocols per standards set forth in the USDA, ARS Policies and Procedures 130.4.V.3 titled ‘Institutional Animal Care and Use 84 Committee’. Fish used in these experiments were from the 2017 Year Class and maintained as specific pathogen free as determined by biannual testing as previously described [28]. A total of 33 single siredam matings contributed to the pool of ARS-Fp-R line fish and 31 matings contributed to the pool of ARS-Fp-S line.
Fish that were used in this study were reared under two different tank conditions per line. Each line was separated into a low density tank (60 fish/151L) with a turnover rate of 7.5 turnovers/hour, and a high-density tank (800 fish/800L) with a turnover rate of 4.2 turnovers/ hour, as described in Supplemental data file 1. High-density tanks are designated as HD, low-density tanks are designated as LD. Ten healthy fish per tank system were sampled for microbiome analysis at time points 252 days post-hatch for the low-density tanks and 256 days post-hatch for the high-density tanks (Supplemental data file 1). Fish were euthanized using 200 mg L−1 MS222 for 5 min. Each fish was photographed, weighted and gill and mid-gut tissue samples were collected. Samples were placed in SLB [32] on ice and then moved to storage at −80 °C. Instruments were cleaned between each fish and gloves changed between tank groups. Three control tubes containing SLB alone were included as negative controls. The spleen of each fish was dissected and weighed within 30 min of euthanasia.
Additionally, the disease resistant phenotype of each genetic line was evaluated at two time-points, 75 and 276 days post-hatch. Select fish were moved into challenge tanks and injected with F. psychrophilum strain CSF259-93. At the first time point, small size fish were injected intraperitoneally to increase injection delivery accuracy, while at the second time point, larger size fish were injected intramuscularly, as this is a more lethal challenge route for fish of this size. We have found that the relative resistance/susceptibility phenotypes are present when challenged by either route (Wiens et al., unpublished data). Survival was recorded over 21 days as previously described [30]. Mean fish body weight at the first evaluation was 1.9 g and a total of 120 ARS-Fp-R and 119 ARS-Fp-S fish were challenged by intraperitoneal injection with a dose of 1.4E+07 CFU g−1 in a total volume of 50 μL using a 26 g needle fitted onto an Eppendorf repeating pipette. Mean body weight at the second evaluation was 194 g and a total of 70 ARS-Fp-R and 70 ARS-Fp-S fish were challenged by intramuscular injection with a dose of 3.5E + 06 CFU g−1 body weight in a total volume of 50 μL using a 26 g needle fitted onto an Eppendorf repeating pipette. The fish utilized in the second experiment were part of a larger study evaluating experimental vaccination and these fish had been sham vaccinated with PBS 35 days prior to challenge. In both challenges, F. psychrophilum was isolated from mortalities and confirmed by PCR amplification of a species-specific, 1089 bp F. psychrophilum 16S rRNA gene fragment using previously described primers PSY1 and PSY2 [33].
2.2. DNA extraction, 16S rDNA PCR amplification, and sequencing
Whole genomic DNA was extracted from skin and gill samples by first lysing tissue samples using sterile 3mm tungsten beads (Qiagen) and Qiagen TissueLyser II. Next, using the cetyltrimethylammonium bromide method as previously described [32], DNA was isolated and suspended in 50 μL RNase and DNase free molecular biology grade water. DNA concentration and purity was then assessed using a Nanodrop ND 1000 (Thermo Scientific).
Bacterial DNA was then replicated by PCR using Illumina adapter fused primers targeting the V1-V3 region of the prokaryotic 16S rDNA gene. The primer sequences were as follows: 28F 5′-GAGTTTGATCNTGGCTCAG-3′ and 519R 5’GTNTTACNGCGGCKGCTG-3’ (where N=any nucleotide, and K=T or G). DNA samples were diluted 1:10 or 1:100, and Quantabio 5PRIME HotMasterMix was used. 16S amplicons were generated using the following conditions: 94 °C for 90s; 33 cycles of 94 °C for 30s, 52 °C for 30s, 72 °C for 90s; and a final extension of 72 °C for 7 min. A positive control of a verified 16S V1-V3 amplicon, and a negative control of molecular biology grade water was included in every PCR reaction. In addition, we included a mock community positive control with each sequencing run, which consisted of equal DNA amounts of 7 different bacterial isolates previously cloned and a negative control that consisted of SLB handled in the same way as the rest of the tubes during sampling to which no tissue was added. Amplicons were purified using the Axygen AxyPrep Mag PCR Clean-up Kit (Thermo Scientific), and eluted into 30 μL molecular biology grade water. Unique oligonucleotide barcodes were ligated to the 5′ and 3′ ends of each sample, as well as the Nextera adaptor sequences, using the Nextera XT Index Kit v2 set A (Illumina). DNA concentrations were quantified using a Qubit, and normalized to a concentration of 200 ng μL−1 for DNA library pooling. Pooled samples were cleaned once more using the Axygen PCR clean-up kit before being sent off for sequencing. Paired-end sequencing was performed on the Illumina Miseq platform using the MiSeq Reagent Kit v3 (600 cycle) at the Clinical and Translational Sciences Center at the University of New Mexico Health Sciences Center, generating forward and reverse reads of length 300 bp.
2.3. Data analysis and statistics
Initial sequence data was analyzed using the latest version of Quantitative Insights Into Microbial Ecology 2 (Qiime2 v2018.8) [34]. Demultiplexed paired-end sequence reads were preprocessed using DADA2, a package integrated into Qiime2 that accounts for quality filtering, denoising, joining paired ends, and removing chimeric sequences [35]. The first 35 bp were trimmed from forward and reverse reads before merging to remove adaptors. Samples were then rarefied to a sampling depth of 12,603 sequences per sample for mid-gut reads, and 2020 sequences per sample for gill reads. Amplicon sequence variants (ASVs) were picked by aligning to the latest Silva 16S database (version 132). Core diversity metrics were analyzed, including number of ASVs, Shannon's diversity index, and Chao1 index for alpha diversity, and PERMANOVA for beta diversity. Nonmetric multidimensional scaling and generation of heat maps were performed in RStudio [36] using the phyloseq package [37]. Differential abundance testing was performed in Qiiime2 using ANCOM [38], as well as in RStudio using the DESeq2 package [39]. For all statistical analyses, fish were split into groups based on 'Treatment' (ARS-Fp-R, ARS-Fp-S) or 'Tank Density'.
Furthermore, differences in survival between genetic lines were determined using the product limit method of Kaplan and Meier and calculations were performed using GraphPad v4.0 software. Log-rank (Mantel-Cox) test was used to compare survival curves.
3. Results
3.1. Phenotype confirmation of disease resistance/susceptibility
The relative phenotype of the two genetic lines was evaluated at time-points either before or after microbiome sampling. At both time points, the survival of the ARS-Fp-R genetic line was significantly higher (P < 0.001) than the ARS-Fp-S line and consistent with estimated mid-point breeding values. In the first evaluation, a total of 3/120 (3%) ARS-Fp-R line fish died compared to 82/119 (69%) ARS-Fp-S line fish. In the second evaluation, 2/70 (3%) ARS-Fp-R fish died, while 58/70 (83%) ARS-Fp-S line fish died within the 21-day challenge period. These results validate the relative resistant and susceptible phenotypes of the two genetic lines from the 2017 year class. Fish sampled for microbiome analyses were obtained from these same pools of families that were not challenged.
3.2. High throughput sequencing analysis
A total number of 3,598,038 raw reads were obtained from all mid-gut samples. After merging paired ends, quality filtering, and removal of chimeric reads with DADA2, as well as filtering out non-specific trout genomic reads, a total of 1,413,104 reads remained, with a mean of 35,328 reads per sample. Samples were rarified to a sample depth of 12,031, which excluded two ARS-Fp-R samples and two ARS-Fp-S samples. The sample size after rarefaction was n=18 for both the ARS-Fp-R and ARS-Fp-S lines.
Gill sample sequencing produced a total of 4,646,971 raw reads. Quality filtering in DADA2 and removal of non-specific reads retained 333,281 reads, with a mean of 13,331 reads per sample. Samples were rarified to 2,010 reads. The sample size after rarefaction was n=11 for ARS-Fp-R and n=14 for ARS-Fp-S.
3.3. Resistant and susceptible lines display significant differences in alpha diversity in the mid-gut, but not the gills
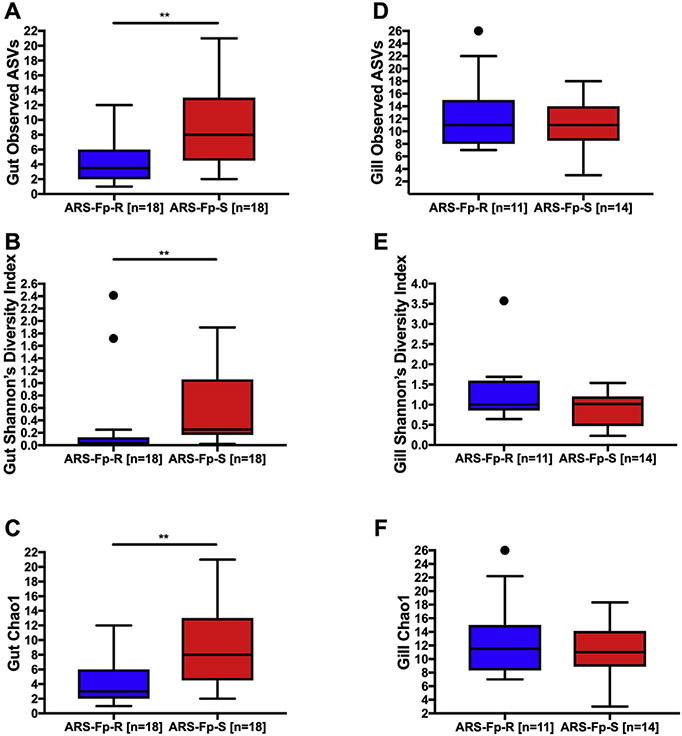
Comparison of alpha diversity metrics obtained from the mid-gut showed significantly lower measures of gut microbial community richness (Observed ASVs), diversity (Shannon's diversity index), and abundance (Chao1) in the ARS-Fp-R line compared to the ARS-Fp-S line (Fig. 1A-C). There were a total of 15 ASVs reported in the mid-gut of the resistant line, and 29 in the mid-gut of the susceptible line. In the gills, there were no significant differences in alpha diversity between lines with a total of 57 ASVs found in the gills of the resistant line, and 50 in the susceptible line (Fig. 1D-F).
Fig. 1. Comparison of alpha diversity metrics for the mid-gut and gill microbiome of ARS-Fp-R and ARS-Fp-S trout.
(A) Total number of observed ASVs in the mid-gut. (B) Shannon's diversity index in the mid-gut. (C) Chao1 index in the mid-gut. (D) Total number of observed ASVs in the gills. (E) Shannon's diversity index in the gills. (F) Chao1 index in the gills. ** indicates statistically significant differences p < 0.01, · indicates outliers.
3.4. Beta diversity analyses: impact of “treatment” and “tank density”
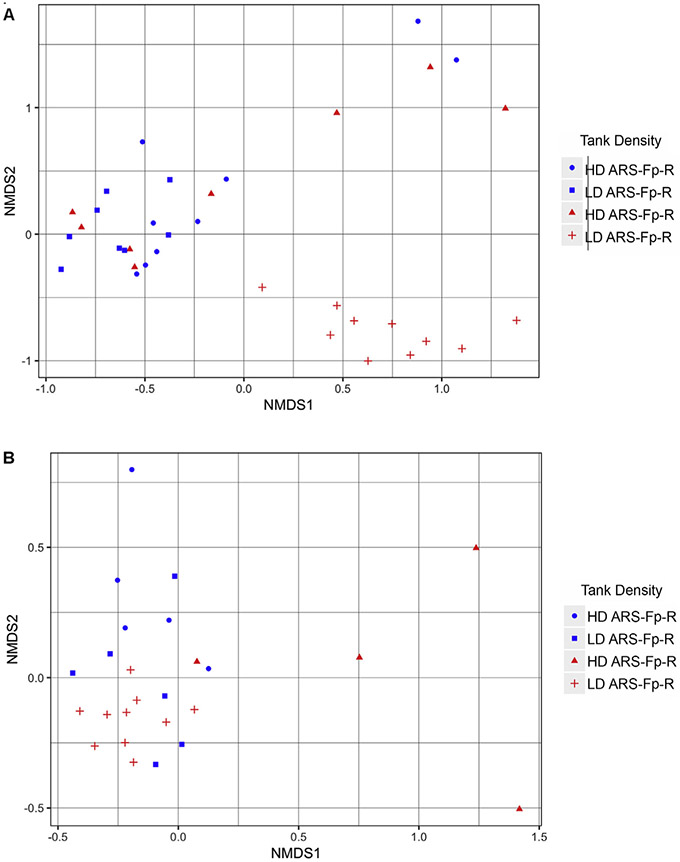
We assessed the microbial diversity between different treatments, as well as between tanks by performing Nonmetric Multidimensional Scaling (NMDS) using the Bray Curtis distance metric. This ordination showed a discrete grouping of the low-density ARS-Fp-S tank. Five of the eight samples taken from the high-density ARS-Fp-S tank clustered tightly together, while the remaining three had a wider spread. Tank density did not have a dramatic effect on ARS-Fp-R samples, as nearly all samples were clustered together. (Fig. 2A). PERMANOVA analysis [40] identified “Treatment” (P value=0.02) and “Tank Density” (P value=0.048 for ARS-Fp-R, P value=0.001 for ARS-Fp-S) as significant determinants of the mid-gut microbial community composition. In the gills, NMDS ordination showed a similar pattern to that found in the gut, where fish from the low-density ARS-Fp-S tank clustered tightly together while fish from the high-density ARS-Fp-S tank showed greater variability. Meanwhile, there was no clear separation between individuals held in separate tanks of the resistant line (Fig. 2B). PERMANOVA identified only tank density within the ARS-Fp-S line (P value=0.004) as a significant factor in determining gill microbial communities.
Fig. 2. NMDS ordination plots.
NMDS ordination performed using Bray Curtis distance matrix of the (A) mid-gut and (B) gill microbiome of ARS-Fp-R and ARS-Fp-S trout.
3.5. Gut microbial community composition
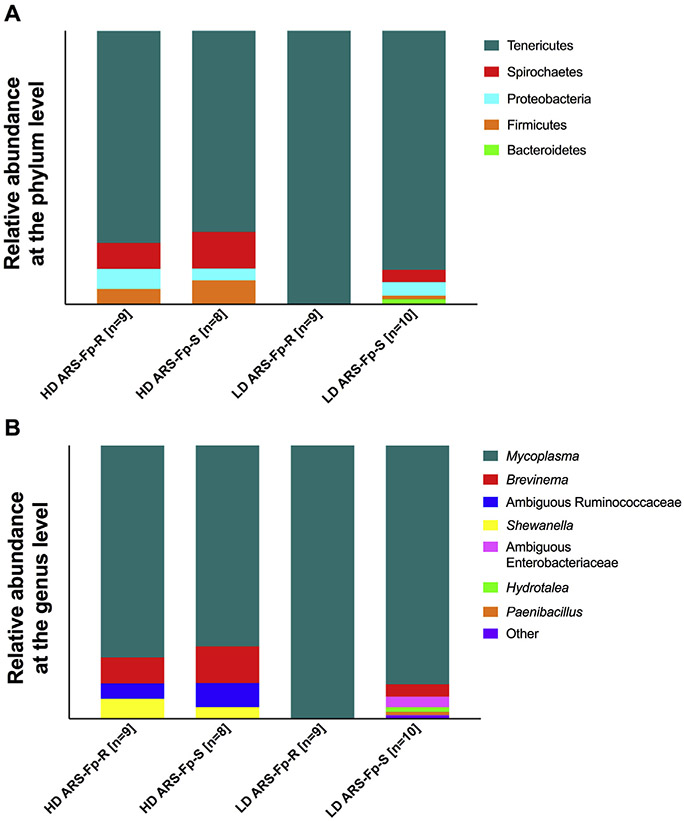
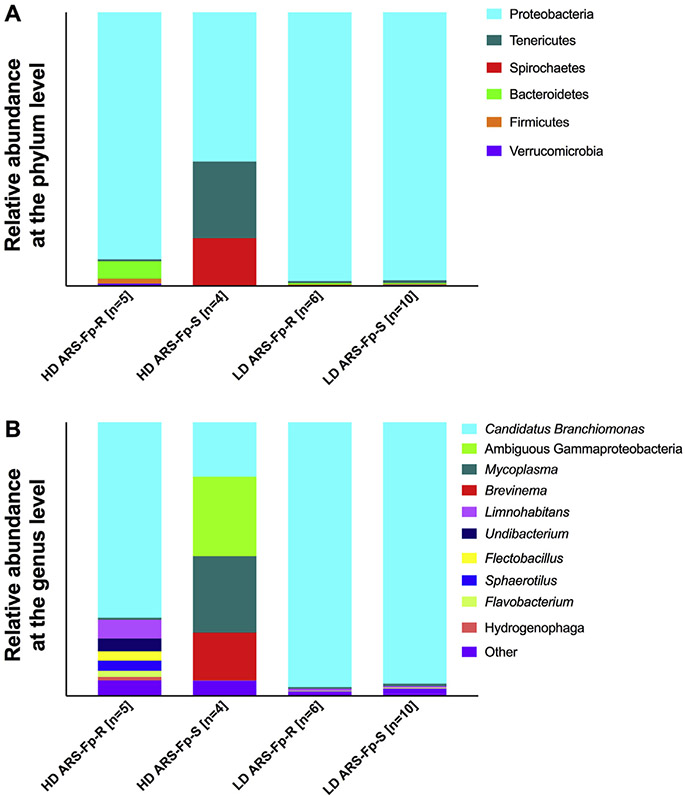
A total of eight different phyla were identified in the mid-gut across both lines, although only four of these were represented over 1%, including Tenericutes, Spirochaetes, Proteobacteria, and Firmicutes. Tenericutes composed the vast majority of the mid-gut microbiome of both lines, although it was more abundant in the low-density tanks. This phylum constituted 99.83% of the total microbial diversity in the low-density ARS-Fp-R tank and 87.35% in the low-density ARS-Fp-S tank. In the high-density tanks, Tenericutes accounted for 77.46% of microbial diversity in the ARS-Fp-R line and 73.48% in the ARS-Fp-S line (Fig. 3A).
Fig. 3. Relative microbial composition in the gut of each line.
(A) Relative abundance of ASVs at the phylum level for each tank. (B) Relative abundance of ASVs at the genus level for each tank.
At the genus level, all Tenericutes reads were identified as Mycoplasma sp. We identified three taxa that were unique to the high-density tanks. These included ambiguous Ruminococcae family members, which were present at 5.62% in the ARS-Fp-R line and 8.80% in the ARS-Fp-S line, as well as Shewanella sp., which was present at 7.04% in the ARS-Fp-R line and 4.11% in the ARS-Fp-S line. In addition, Brevinema sp. was present at 9.64% in the high-density ARS-Fp-R line and 13.55% in the high-density ARS-Fp-S line. This taxon was also present in the low-density ARS-Fp-S line at 4.62% but was not identified in the low-density ARS-Fp-R line. Differential abundance testing with ANCOM revealed three genera that were differentially abundant across tanks in the mid-gut of both trout lines; including Hydrotalea sp., Paenibacillus sp., and Variovorax sp. These three taxa are unique to the low-density ARS-Fp-S tank. This was replicated by differential abundance testing using DESeq2, an R package originally developed for differential expression analysis in RNA-seq data, but has been shown to be effective in analyzing 16S microbiome datasets as well [41,42].
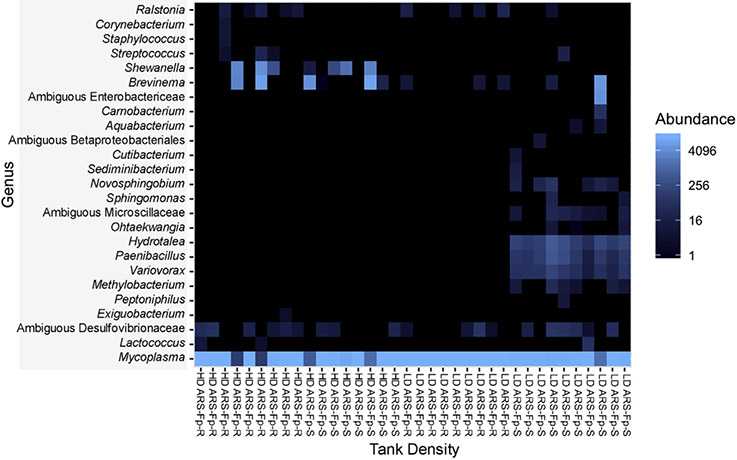
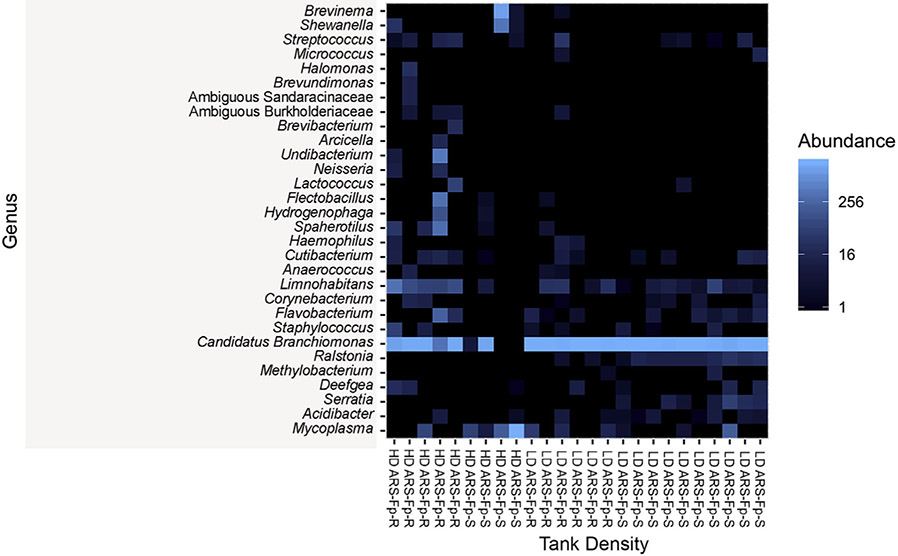
This trend is further shown in a heatmap representing the top 25 ASVs observed in each sample (Fig. 4). The high-density tanks displayed similar microbial profiles, regardless of the holding trout lines. This was not the case for the ARS Fp-S low-density tanks, which displayed a microbial profile different from all other tanks with ASVs representative of Enterobacterieaceae, Hydrotalea sp., Paenibacillus sp, and Variovorax sp.
Fig. 4. Heatmap representing the top 25 ASVs present in the mid-gut of ARS-Fp-R and ARS-Fp-S trout.
Each column represents one individual. Each row represents one ASV.
3.6. Gill microbial community composition
A total of nine different phyla were present in the trout gills across both lines. Five of these were represented at abundance greater than 1%, including Proteobacteria, Tenericutes, Spirochaetes, Bacteroidetes, and Firmicutes. Proteobacteria was the most abundant phylum in the gill microbiome of each line, though it was more represented in the low-density tanks. Proteobacteria made up 96.71% and 95.49% of the gill microbial community in low-density ARS-Fp-R and ARS-Fp-S tanks, respectively. In the high-density tanks, this phylum represented 71.4% of all the gill diversity of the ARS-Fp-R tank and 19.95% of all the gill diversity the ARS-Fp-S tank. (Fig. 5A). At the genus level, most Proteobacteria reads were identified as Candidatus Branchiomonas. Distribution of Candidatus Branchiomonas among tanks was as follows: 96.71% in low-density ARS-Fp-R, 95.49% in low-density ARS-Fp-S, 71.40% in high-density ARS-Fp-R, and 19.95% in high-density ARS-Fp-S We identified trace amounts of Flavobacterium sp. in both lines, as this taxon constituted 0.16% of the ARS-Fp-S line and 1.1% of the ARS-Fp-R line (Fig. 5B). No ASVs were differentially abundant in the gill microbial community of both groups through ANCOM or DEseq2 analyses. A heatmap of the top 30 ASVs in each sample (Fig. 6) shows signatures of Brevinema sp. in fish from the low-density ARS-Fp-S tank, as well as a reduced abundance of Candidatus Branchiomonas in this tank compared to all others.
Fig. 5. Microbial community composition of the gills of ARS-Fp-R and ARS-Fp-S trout.
(A) Relative abundance of ASVs at the phylum level for each tank. (B) Relative abundance of ASVs at the genus level for each tank.
Fig. 6. Heatmap representing the top 30 ASVs represented in the gill of ARS-Fp-R and ARS-Fp-S trout.
Each column represents one individual fish. Each row represents one ASV.
4. Discussion
Commensal microbes have co-evolved with their eukaryotic counterparts, forming an intricate relationship that benefits both parties involved. Several studies have revealed that host genetics influences gut microbiota composition in a variety of species, including humans and rodents [7,8], chickens [43], and Drosophila [44]. However, other factors such as host diet and environmental conditions are also deeply intertwined and are crucial in determining host microbial communities [12-14].
Teleost fish live in symbiosis with complex microbial communities that inhabit every mucosal barrier (gut, gills, skin and nose) [45]. Fish microbial community composition is influenced by age [46], tissue site [47], diet [48-50], stress [51] and pathogen infection [52]. However, few studies have investigated the impact host genetics have on shaping teleost microbiomes. A study on brook charr identified three quantitative trait loci associated with abundance of commensal strains in the skin [53]. Another study in Atlantic salmon found significant differences in the skin and gut microbial composition amongst distinct wild populations that are not likely attributed to environmental conditions alone [54]. These observations suggest that host genetics play an important role in teleost microbiome assembly, although more research is needed to better understand this relation.
Farmed fish are susceptible to many pathogens that threaten the sustainability of the finfish farming industry. Among the most prominent bacterial diseases, BCWD is particularly problematic in rainbow trout. The development of two genetic trout lines with different susceptibilities to the BCWD agent, F. psychrophilum, offers an excellent platform to understand how host genetics may shape fish microbial communities. The current study suggests that host genetics influence the microbial community composition in the mid-gut, since the mid-gut bacterial community of the susceptible line was significantly more diverse than that of the resistant line. In agreement with previous studies, the mid-gut communities of both lines were dominated by Mycoplasma sp. This taxon appears to be highly abundant in the gastrointestinal microbiome of many salmonid species studied, including Atlantic salmon [55-57], rainbow trout [47,58] and Chinook salmon [59]. Interestingly, we identified a lower abundance of Mycoplasma sp. in the susceptible line under both stocking density conditions, suggesting that disease susceptibility may be associated with decreased Mycoplasma sp. levels in the gut. In support, a recent study investigated the intestinal microbiome of offshore farmed Chinook salmon [59], finding that abundance of potential pathogenic Vibrio sp. appeared to be inversely correlated with the presence of Mycoplasma sp. Mycoplasma sp. are characterized by their uniquely small genome and lack of a cell wall, which makes culture-based approaches to studying this bacterium difficult to achieve. Considering the widespread presence and abundance of Mycoplasma sp. in salmonid gastrointestinal tract across a wide range of geographical locations, including both lab-reared fish and wild-caught, it appears as if there have been strong evolutionary forces that have enabled Mycoplasma sp. to thrive in this microenvironment. Future studies are needed to determine the nature of this relationship and the ability of Mycoplasma sp. to prevent pathogen colonization in the gastrointestinal tract of salmonids.
Tank density had a notable impact on shaping gut microbial community composition. Furthermore, differences between lines reared under the same tank conditions were also observed. For example, in the high-density tanks, we observed an expansion of Ambiguous Ruminococcaceae in the ARS-Fp-S line. This taxon has previously been described as a core member of the Atlantic salmon gut microbiome [60], although deeper taxonomic classification has not been achieved. It is worth noting that this taxon was absent in both of the low-density tanks. Shewanella sp. was also present at a greater abundance in the high-density ARS-Fp-R tank, and was not observed in either of the low-density tanks. Species level taxonomic resolution was not achieved by sequence search using BLAST. Finally, we also observed an expansion of Brevinema andersonii in the high-density ARS-Fp-S tank. This taxon is known to be pathogenic in rodents [61] but has not been described in teleosts. Brevinema andersonii was also identified in the low-density ARS-Fp-S line, suggesting a moderate expansion in potential opportunistic pathogens could be associated with susceptibility to BCWD. Exansions of opportunistic taxa have been previously described in other fish microbiome studies including in the skin of Atlantic salmon experimentally infected with salmonid alphavirus (Reid, 2017) and in Atlantic salmon experimentally infected with the parasite Lepeophtheirus salmonis [52].
It is worth noting that microbial diversity in the trout gut was low across all tanks, a common feature observed in most salmonid species [5,47,60]. However, there were substantial differences in the gut microbial communities between lines in the low-density tanks. The low-density ARS-Fp-R line was nearly solely composed of Mycoplasma sp., while the low-density ARS-Fp-S line was more diverse. Three taxa were unique to the low-density ARS-Fp-S line, including Hydrotalea sp., Paembacdlus sp., and Variovorax sp. These results suggest that environmental factors and the interaction between environmental factors and host genetics also contribute to gut microbiome assembly.
Differences in the microbial community composition between both trout lines were less pronounced in the gills compared to the gut, although it should be noted that the sample size was smaller in this tissue due to difficulties amplifying the 16S rDNA region in certain samples. Nonetheless, alpha diversity metrics were similar in each line. Interestingly, Candidatus Branchiomonas was the dominant taxon in both lines. This is a known pathogen that has been shown to cause gill epitheliocystis in Atlantic salmon [62], although it has not been previously described in rainbow trout. Fish from both lines were visually healthy, suggesting that Candidatus Branchiomonas may be a common member of the trout gill microbiome in certain environments. Further studies should evaluate which factors favor the colonization of Candidatns Branchiomonas in trout gills. Flavobacterinm sp. was identified in the gills of both trout lines, but sequence search using BLAST did not yield species level taxonomic resolution. Although this taxon was present at low abundance in both lines, it was surprising that we detected higher Flavobacterinm sp. abundance in the gill microbiome of the resistant line compared to the susceptible line. Considering that the resistant and susceptible lines exhibited the respective phenotypes at the time of sampling, our results may suggest that susceptibility to F. psychrophilum infection may not be due to increased abundance of this pathogen as a member of the indigenous microbial community. Perhaps susceptibility is due to a greater ability of this pathogen to displace the microbial communities in the susceptible line compared to the resistant line.
Interestingly, and similar to the gut data, Brevinema andersonii was also observed in the gills of the high-density ARS-Fp-S tank, but not the high-density ARS-Fp-R tank, further supporting an expansion of potential opportunistic pathogens in the susceptible line. Diversity was lower in both of the low-density tanks, although not significant. Unlike the gut, the gills are a tissue site that typically exhibits substantial diversity. Thus, our findings suggest that low-density rearing conditions result in decreased complexity in the gill microbial assembly of trout.
One of the interesting aspects of the present study was the identification of differences in the microbial composition between fish of the same genetic lines reared under different tank conditions. All tanks were supplied water from the same source, and proper water quality and temperature were maintained throughout the experiment. This tank effect complicates the ability to discern between genetic lines, although there still appears to be notable differences when comparing each line, particularly in the gut as discussed earlier. Work in zebrafish has shown that interhost dispersal can actually outweigh genetic factors that contribute to microbiome assembly [63]. This is evident in our study, as fish from low-density tanks exhibited a microbiome that was distinctly different from those from high-density tanks. This finding highlights the importance of adequate experimental design in fish microbiome studies as well as the fact that different factors differentially shape microbial assembly at different tissue sites (i.e. gut versus gills).
5. Conclusions
In conclusion, the present study reveals the impact of host genetics and environmental factors on the microbial community composition of rainbow trout, Host genetics shaped the microbial composition of the gut but not the gills of two rainbow trout lines with differential susceptibility to F. psychrophilum infection. Disease susceptibility was associated with a more diverse gut microbiome and the presence of potentially pathogenic taxa although important stocking density effects were also detected. Thus, selective breeding programs and stocking conditions may not only select for host genetic factors but also, as a consequence, unique microbial assemblies, which in turn, may render the host more or less resilient to pathogen invasion or infection.
Supplementary Material
Acknowledgements
We thank Kurt Schwalm and Dr. Darrell Dinwiddie for handling the Illumina MiSeq runs. We thank Dr. Timothy Leeds for breeding the ARS-Fp-R and ARS-Fp-S genetic lines and Travis Moreland for fish rearing and Jeremy Everson for assistance with fish sampling. This work was supported in part by the U.S Agricultural Research Service Project 1930-32000-006. Ryan Brown received support from the University of New Mexico PREP program. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S Department of Agriculture. USDA is an equal opportunity employer.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fsi.2018.11.079.
Data availability
All datasets obtained have been deposited at NCBI BioProject and are publicly available under BioProject ID number PRJNA488363.
References
- [1].Thaiss CA, Zmora N, Levy M, Elinav E, The microbiome and innate immunity, Nature 535 (2016) 65–74, 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- [2].Shi N, Li N, Duan X, Niu H, Interaction between the gut microbiome and mucosal immune system, Mil. Med. Res 4 (2017), 10.1186/s40779-017-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Buffie CG, Pamer EG, Microbiota-mediated colonization resistance against intestinal pathogens, Nat. Rev. Immunol 13 (2013) 790–801, 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF, Evidence for a core gut microbiota in the zebrafish, ISME J. (2011), 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gajardo K, Rodiles A, Kortner TM, Krogdahl Å, Bakke AM, Merrifield DL, Sørum H, A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): a basis for comparative gut microbial research, Sci. Rep 6 (2016) 30893 10.1038/srep30893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wong S, Waldrop T, Summerfelt S, Davidson J, Barrows F, Kenney BB, Welch T, Wiens GD, Snekvi K, Rawls JF, Good C, Aquacultured rainbow trout (Oncorhynchus mykiss) possess a large core intestinal microbiota that is resistant to variation in diet and rearing density, Appl. Environ. Microbiol 79 (2013) 4974–4984, 10.1128/AEM.00924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, Zhernakova DV, Jankipersadsing SA, Jaeger M, Oosting M, Cenit MC, Masclee AAM, Swertz MA, Li Y, Kumar V, Joosten L, Harmsen H, Weersma RK, Franke L, Hofker MH, Xavier RJ, Jonkers D, Netea MG, Wijmenga C, Fu J, Zhernakova A, The effect of host genetics on the gut microbiome, Nat. Genet 48 (2016) 1407–1412, 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- [8].Spor A, Koren O, Ley R, Unravelling the effects of the environment and host genotype on the gut microbiome, Nat. Rev. Microbiol 9 (2011) 279–290, 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- [9].Fulde M, Sommer F, Chassaing B, van Vorst K, Dupont A, Hensel M, Basic M, Klopfleisch R, Rosenstiel P, Bleich A, Bäckhed F, Gewirtz AT, Hornef MW, Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition, Nature 560 (2018) 489–493, 10.1038/s41586-018-0395-5. [DOI] [PubMed] [Google Scholar]
- [10].Dabrowska K, Witkiewicz W, Correlations of host genetics and gut microbiome composition, Front. Microbiol (2016), 10.3389/fmicb.2016.01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, De Visser JAGM, De Vos WM, The host genotype affects the bacterial community in the human gastrointestinal tract, Microb. Ecol. Health Dis (2001), 10.1080/089106001750462669. [DOI] [Google Scholar]
- [12].David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ, Diet rapidly and reproducibly alters the human gut microbiome, Nature 505 (2014) 559–563, 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E, Environment dominates over host genetics in shaping human gut microbiota, Nature 555 (2018) 210–215, 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- [14].Singh RK, Chang H-W, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W, Influence of diet on the gut microbiome and implications for human health, J. Transl. Med 15 (2017) 73, 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schreiner VBY, Andrew B, Kao John Y., The gut microbiome in health and in disease, Curr. Opin. Gastroenterol 31 (2015) 69–75, 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bolnick DI, Snowberg LK, Caporaso JG, Lauber C, Knight R, Stutz WE, Major Histocompatibility Complex class IIb polymorphism influences gut microbiota composition and diversity, Mol. Ecol (2014), 10.1111/mec.12846. [DOI] [PubMed] [Google Scholar]
- [17].Bledsoe JW, Peterson BC, Swanson KS, Small BC, Ontogenetic characterization of the intestinal microbiota of channel catfish through 16S rRNA gene sequencing reveals insights on temporal shifts and the influence of environmental microbes, PloS One (2016), 10.1371/journal.pone.0166379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bledsoe JW, Waldbieser GC, Swanson KS, Peterson BC, Small BC, Comparison of channel catfish and blue catfish gut microbiota assemblages shows minimal effects of host genetics on microbial structure and inferred function, Front. Microbiol (2018), 10.3389/fmicb.2018.01073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Webster TMU, Consuegra S, Hitchings M, de Leaniz CG, Interpopulation variation in the Atlantic salmon microbiome reflects environmental and genetic diversity, Appl. Environ. Microbiol (2018), 10.1128/AEM.000691-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Starliper CE, Bacterial coldwater disease of fishes caused by Flavobacterium psychrophilum, J. Adv. Res 2 (2011) 97–108, 10.1016/j.jare.2010.04.001. [DOI] [Google Scholar]
- [21].Nematollahi A, Decostere A, Pasmans F, Haesebrouck F, Flavobacterium psychrophilum infections in salmonid fish, J. Fish. Dis 26 (2003) 563–574, 10.1046/j.1365-2761.2003.00488.x. [DOI] [PubMed] [Google Scholar]
- [22].Nematollahi A, Pasmans F, Van Den Broeck W, Ducatelle R, Haesebrouck F, Decostere A, Association of Flavobacterium psychrophilum strains with intestinal explants of rainbow trout Oncorhynchus mykiss, Dis. Aquat. Org (2005), 10.3354/dao067067. [DOI] [PubMed] [Google Scholar]
- [23].Burbank DR, Shah DH, LaPatra SE, Fornshell G, Cain KD, Enhanced resistance to coldwater disease following feeding of probiotic bacterial strains to rainbow trout (Oncorhynchus mykiss), Aquaculture (2011), 10.1016/j.aquaculture.2011.09.004. [DOI] [Google Scholar]
- [24].Bruun MS, Schmidt AS, Madsen L, Dalsgaard I, Antimicrobial resistance patterns in Danish isolates of Flavobacterium psychrophilum, Aquaculture 187 (2000) 201–212, 10.1016/S0044-8486(00)00310-0. [DOI] [Google Scholar]
- [25].Bruun MS, Madsen L, Dalsgaard I, Efficiency of oxytetracycline treatment in rainbow trout experimentally infected with Flavobacterium psychrophilum strains having different in vitro antibiotic susceptibilities, Aquaculture 215 (2003) 11–20, 10.1016/S0044-8486(01)00897-3. [DOI] [Google Scholar]
- [26].Kum C, Kirkan S, Sekkin S, Akar F, Boyacioglu M, Comparison of in vitro antimicrobial susceptibility in Flavobacterium psychrophilum isolated from rainbow trout fry, J. Aquat. Anim. Health 20 (2008) 245–251, 10.1577/H07-040.1. [DOI] [PubMed] [Google Scholar]
- [27].Silverstein JT, Vallejo RL, Palti Y, Leeds TD, Rexroad CE, Welch TJ, Wiens GD, Ducrocq V, Rainbow trout resistance to bacterial cold-water disease is moderately heritable and is not adversely correlated with growth, J. Anim. Sci 87 (2009) 860–867, 10.2527/jas.2008-1157. [DOI] [PubMed] [Google Scholar]
- [28].Wiens GD, LaPatra SE, Welch TJ, Evenhuis JP, Rexroad CE, Leeds TD, Onfarm performance of rainbow trout (Oncorhynchus mykiss) selectively bred for resistance to bacterial cold water disease: effect of rearing environment on survival phenotype, Aquaculture 388–391 (2013) 128–136, 10.1016/j.aquaculture.2013.01.018. [DOI] [Google Scholar]
- [29].Wiens GD, Palti Y, Leeds TD, Three generations of selective breeding improved rainbow trout (Oncorhynchus mykiss) disease resistance against natural challenge with Flavobacterium psychrophilum during early life-stage rearing, Aquaculture 497 (2018) 414–421. [Google Scholar]
- [30].Hadidi S, Glenney GW, Welch TJ, Silverstein JT, Wiens GD, Spleen size predicts resistance of rainbow trout to Flavobacterium psychrophilum challenge, J. Immunol 180 (2008) 4156–4165, 10.4049/jimmunol.180.6.4156. [DOI] [PubMed] [Google Scholar]
- [31].Marancik D, Gao G, Paneru B, Ma H, Hernandez AG, Salem M, Yao J, Palti Y, Wiens D, Whole-body transcriptome of selectively bred, resistant-, control-, and susceptible-line rainbow trout following experimental challenge with Flavobacterium psychrophilum, Front. Genet 5 (2015), 10.3389/fgene.2014.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mitchell KR, Takacs-Vesbach CD, A comparison of methods for total community DNA preservation and extraction from various thermal environments, J. Ind. Microbiol. Biotechnol 35 (2008) 1139–1147, 10.1007/s10295-008-0393-y. [DOI] [PubMed] [Google Scholar]
- [33].Wiklund T, Madsen L, Bruun MS, Dalsgaard I, Detection of Flavobacterium psychrophilum from fish tissue and water samples by PCR amplification, J. Appl. Microbiol (2000), 10.1046/j.1365-2672.2000.00959.x. [DOI] [PubMed] [Google Scholar]
- [34].Bolyen E CJ, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, Alexander E.J. Aim, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope E, Da Silva R, Dorrestei, QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science, PeerJ Prepr. 6 (2018) e27295v1. [Google Scholar]
- [35].Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP, DADA2: high-resolution sample inference from Illumina amplicon data, Nat. Methods 13 (2016) 581–583, 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].R.C. Team, R: a Language and Environment for Statistical Computing, (2018) https://www.r-project.org/.
- [37].McMurdie PJ, Holmes S, Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data, PloS One 8 (2013), 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD, Analysis of composition of microbiomes: a novel method for studying microbial composition, Microb. Ecol. Health Dis 26 (2015) 27663, 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15 (2014), 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Anderson MJ, PERMANOVA Permutational multivariate analysis of variance, Austral Ecol. (2005) 1–24, 10.1139/cjfas-58-3-626. [DOI] [Google Scholar]
- [41].McMurdie PJ, Holmes S, Waste not, want not: why rarefying microbiome data is inadmissible, PLoS comput, Biol 10 (2014), 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Väzquez-Baeza Y, Birmingham A, Hyde ER, Knight R, Normalization and microbial differential abundance strategies depend upon data characteristics, Microbiome 5 (2017), 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhao L, Wang G, Siegel P, He C, Wang H, Zhao W, Zhai Z, Tian F, Zhao J, Zhang H, Sun Z, Chen W, Zhang Y, Meng H, Quantitative genetic background of the host influences gut microbiomes in chickens, Sci. Rep 3 (2013), 10.1038/srep01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chaston JM, Dobson AJ, Newell PD, Douglas AE, Host genetic control of the microbiota mediates the Drosophila nutritional phenotype, Appl. Environ. Microbiol 82 (2016) 671–679, 10.1128/AEM.03301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kelly C, Salinas I, Under pressure: interactions between commensal microbiota and the teleost immune system, Front. Immunol 8 (2017), 10.3389/fimmu.2017.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M, Valenzano DR, Regulation of life span by the gut microbiota in the short-lived african turquoise killifish, Elife 6 (2017), 10.7554/eLife.27014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lowrey L, Woodhams DC, Tacchi L, Salinas I, Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin, Appl. Environ. Microbiol 81 (2015) 6915–6925, 10.1128/AEM.01826-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Reid HI, Treasurer JW, Adam B, Birkbeck TH, Analysis of bacterial populations in the gut of developing cod larvae and identification of Vibrio logei, Vibrio anguillarum and Vibrio splendidus as pathogens of cod larvae, Aquaculture 288 (2009) 36–43, 10.1016/j.aquaculture.2008.11.022. [DOI] [Google Scholar]
- [49].Miyake S, Ngugi DK, Stingl U, Diet strongly influences the gut microbiota of surgeonfishes, Mol. Ecol 24 (2015) 656–672, 10.1111/mec.13050. [DOI] [PubMed] [Google Scholar]
- [50].Neuman C, Hatje E, Zarkasi KZ, Smullen R, Bowman JP, Katouli M, The effect of diet and environmental temperature on the faecal microbiota of farmed Tasmanian Atlantic Salmon (Salmo salar L.), Aquacult. Res 47 (2016) 660–672, 10.1111/are.12522. [DOI] [Google Scholar]
- [51].Tacchi L, Lowrey L, Musharrafieh R, Crossey K, Larragoite ET, Salinas I, Effects of transportation stress and addition of salt to transport water on the skin mucosal homeostasis of rainbow trout (Oncorhynchus mykiss), Aquaculture 435 (2015) 120–127, 10.1016/j.aquaculture.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Llewellyn MS, Leadbeater S, Garcia C, Sylvain FE, Custodio M, Ang KP, Powell F, Carvalho GR, Creer S, Elliot J, Derome N, Parasitism perturbs the mucosal microbiome of Atlantic Salmon, Sci. Rep 7 (2017), 10.1038/srep43465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Boutin S, Sauvage C, Bernatchez L, Audet C, Derome N, Inter individual variations of the fish skin microbiota: host genetics basis of mutualism? PloS One 9 (2014), 10.1371/journal.pone.0102649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Uren Webster TM, Consuegra S, Hitchings M, Garcia de Leaniz C, Inter-population variation in the Atlantic salmon microbiome reflects environmental and genetic diversity, Appl. Environ. Microbiol (2018), 10.1128/AEM.00691-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dehler CE, Secombes CJ, Martin SAM, Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.), Sci. Rep 7 (2017), 10.1038/s41598-017-13249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Llewellyn MS, McGinnity P, Dionne M, Letourneau J, Thonier F, Carvalho GR, Creer S, Derome N, The biogeography of the atlantic salmon (Salmo salar) gut microbiome, ISME J. 10 (2015) 1–5, 10.1038/ismej.2015.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Holben WE, Williams P, Saarinen M, Särkilahti LK, Apajalahti JHA, Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon, Microb. Ecol 44 (2002) 175–185, 10.1007/S00248-002-1011-6. [DOI] [PubMed] [Google Scholar]
- [58].Lyons PP, Turnbull JF, Dawson KA, Crumlish M, Phylogenetic and functional characterization of the distal intestinal microbiome of rainbow trout Oncorhynchus mykiss from both farm and aquarium settings, J. Appl. Microbiol 122 (2017) 347–363, 10.1111/jam.13347. [DOI] [PubMed] [Google Scholar]
- [59].Ciric M, Waite D, Draper J, Jones JB, Characterisation of gut microbiota of farmed Chinook salmon using metabarcoding, BioRxiv (2018) 288761, 10.1101/288761. [DOI] [Google Scholar]
- [60].Dehler CE, Secombes CJ, Martin SAM, Environmental and Physiological Factors Shape the Gut Microbiota of Atlantic Salmon Parr, Salmo salar L.), Aquaculture, 2017, 10.1016/j.aquaculture.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Defosse DL, Johnson RC, Paster BJ, Dewhirst FE, Fraser GJ, Brevinema andersonii gen. nov., sp. nov., an infectious spirochete isolated from the short-tailed shrew (Blarina brevicauda) and the white-footed mouse (Peromyscus leucopus), Int. J.Syst. Bacteriol 45 (1995) 78–84, 10.1099/00207713-45-1-78. [DOI] [PubMed] [Google Scholar]
- [62].Toenshoff ER, Kvellestad A, Mitchell SO, Steinum T, Falk K, Colquhoun DJ, Horn M, A novel betaproteobacterial agent of gill epitheliocystis in seawater farmed Atlantic salmon (Salmo salar), PloS One 7 (2012), 10.1371/journal.pone.0032696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Burns AR, Miller E, Agarwal M, Rolig AS, Milligan-Myhre K, Seredick S, Guillemin K, Bohannan BJM, Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model, Proc. Natl. Acad. Sci (2017), 10.1073/pnas.1702511114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets obtained have been deposited at NCBI BioProject and are publicly available under BioProject ID number PRJNA488363.