Abstract
Background and aim
The year 2020 begins with the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that cause the disease COVID-19, and continue till today. As of March 23, 2021, the outbreak has infected 124,313,054 worldwide with a total death of 2,735,707. The use of traditional medicines as an adjuvant therapy with western drugs can lower the fatality rate due to the COVID-19. Therefore, in silico molecular docking study was performed to search potential phytochemicals and drugs that can block the entry of SARS-CoV-2 into host cells by inhibiting the proteolytic cleavage activity of furin and TMPRSS2.
Experimental procedure
The protein-protein docking of the host proteases furin and TMPRSS2 was carried out with the virus spike (S) protein to examine the conformational details and residues involved in the complex formation. Subsequently, a library of 163 ligands containing phytochemicals and drugs was virtually screened to propose potential hits that can inhibit the proteolytic cleavage activity of furin and TMPRSS2.
Results and conclusion
The phytochemicals like limonin, gedunin, eribulin, pedunculagin, limonin glycoside and betunilic acid bind at the active site of both furin and TMPRSS2. Limonin and gedunin found mainly in the citrus fruits and neem showed the highest binding energy at the active site of furin and TMPRSS2, respectively. The polyphenols found in green tea can also be useful in suppressing the furin activity. Among the drugs, the drug nafamostat may be more beneficial than the camostat in suppressing the activity of TMPRSS2.
Keywords: COVID-19, SARS-CoV-2, Protein-protein docking, Molecular docking, Phytochemicals, Drugs
Graphical abstract
Highlights of the findings and novelties
-
•
Protein-protein docking of the host proteases furin and TMPRSS2 with virus spike protein was performed.
-
•
A library of 163 phytochemicals and drugs was virtually screened to propose potential hits against SARS-CoV-2.
-
•
Limonin, gedunin, eribulin, pedunculagin, limonin glycoside and betunilic acid bind at the active site.
-
•
Drug nafamostat may be more beneficial than the camostat.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the seventh human coronavirus is highly contagious that caused the disease COVID-19.1 After the first infected person identified from Wuhan, China in December 2019, the virus SARS-CoV-2 outbreak worldwide and infected 124,313,054 with a total death of 2,735,707 as of March 23rd, 2021. The viral strain of SARS-CoV-2 belongs to beta coronavirus and suspected of zoonotic transfer via bat to human beings. The research study revealed that the pangolin-CoV and batCoV RaTG13 are closely related to SARS-CoV-2, and expected to be the proximal source of SARS-CoV-2.2 It is important to mention here that there are no recommended therapeutics available against the known human coronaviruses, i.e., the two highly pathogenicity coronaviruses (SARS-CoV, MERS-CoV outbreak in 2002 and 2012 respectively), and the other four pathogenic coronaviruses (HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 outbreak in 1966, 1967, 2004 and 2005, respectively).3 Also, there was no recommended therapeutics available at the beginning of the spread of SARS-CoV-2, which triggered exponential growth in research from the beginning of the year 2020 to understand the morphology, genomic sequence and virus-host cell interactions of SARS-CoV-2 to formulate suitable therapeutic approaches to fight against COVID-19.4, 5, 6
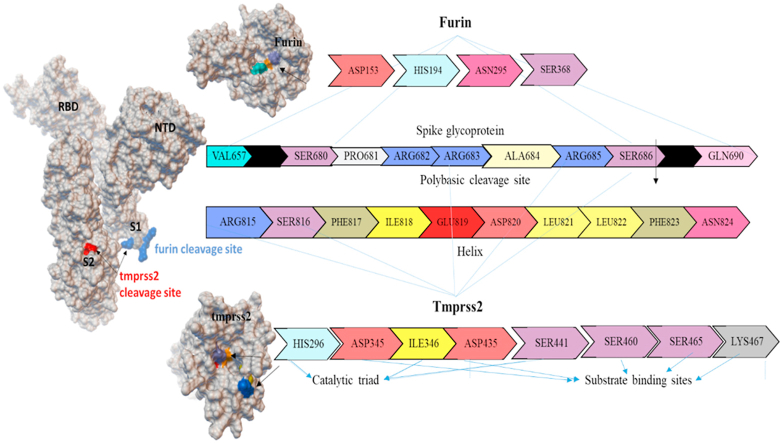
SARS-CoV-2, the enveloped and spherical-shaped virion with an approximate diameter of 120 nm contains a positive-sense ssRNA genome of about 30000 bp. The genome encodes with four important structural proteins (nucleocapsid, membrane, envelop and spike), around sixteen non-structural proteins (NSP1-16) and some accessory proteins.7 The trimeric spike (S) glycoprotein protruding from the SARS-CoV-2 envelope plays the key role in viral infection, where the S1 subunit initiates the virus-receptor binding by interacting with the human host cell receptor angiotensin-converting enzyme 2 (ACE2), and the S2 subunit play the role in viral fusion into the target cell. The S1 subunit possesses two key domains, i.e., N-terminal domain (NTD) and C-terminal domain (CTD). The CTD of SARS-CoV-2 contains the receptor-binding domain (RBD) that interacts with human ACE2 (Fig. 1). Following the interaction of SARS-CoV-2 virus with the human ACE2 receptor, the host proteases like furin and TMPRSS2 play the pivotal role to activate the S protein that allows the fusion of viral genome into the target cell.
Fig. 1.
Active sites of furin and TMPRSS2 involved in S1/S2 site cleavage activity of spike protein. The furin catalytic residues are ASP153, HIS194, ASN295 and SER368. For TMPRSS2, the catalytic sites are HIS296, ILE346 and SER441, whereas the substrate binding sites are ASP345, ASP435, SER460, SER465 and LYS467.
The human furin enzyme cleaves a specific section to convert the synthesized proteins to biologically active form. It is also called as PACE (paired basic amino acid cleaving enzyme) and subtilisin-like peptidase.8 It is a calcium dependent serine endoprotease that cleaves the processing sites of the precursor protein. Furin cleaves polyprotein precursor HIV envelope such as gp41 and gp20 involved in virus host entry.9 Similarly, it also involved in some viral proteins preprocessing such as influenza, Ebola, dengue fever, Marburg virus, Pseudomonas exotoxin, papillomavirus, anthrax toxin and SARS-CoV-2 etc. Furin cleaves S1 (SPRRAR↓ S) site of SARS-CoV-2, direct emergence in spike glycoprotein activation for viral entry into the host system. It is a potent cleavage site known as the polybasic cleavage site or multibasic site of SARS-CoV-2. In addition, the spike glycoprotein S1/S2 site of SARS-CoV-2 is cleaved by the transmembrane protease serine 2 (TMPRSS2), which played a predominant role in enhancing the spike glycoprotein activity.10, 11, 12, 13 TMPRSS2 activates the SARS-CoV and SARS-CoV-2 spike glycoprotein though N-terminal and C-terminal cleavage sites, and priming the S protein for virus endocytosis. TMPRSS2 also activates human metapneumovirus and influenza virus hemagglutinin entry into the host cells.14 TMPRSS2 cleaves the S protein in the lungs of SARS-CoV-2 infected person and promotes the pathogenicity. Therefore, the TMPRSS2 serine protease opted as an important therapeutic target. In short, SARS-CoV-2 S protein S1 site is cleaved by furin, and subsequently, TMPRSS2 mediates the S2 site cleavage and activation of S protein upon interactions with S1/S2 site residues ARG683, ARG685, SER686, ARG815 and SER816 (Fig. 1).15,16
In this manuscript, the protein-protein docking studies were performed to investigate the binding and conformational details of the complexes formed between the proteases furin and TMPRSS2 with the SARS-CoV-2 S protein. In addition, the human proteases furin and TMPRSS2 active sites were targeted to identify potential ligands through molecular docking analyses that can inhibit the functions of furin and TMPRSS2 during the disease establishment. Molecular docking is a useful computational approach to screen potential ligands from various databases/libraries, which saves both the experimental time and cost in the field of drug discovery. Molecular docking not only predicts the binding affinity or strength of association of the ligand at the active site of target protein but also the mode of interactions.17 As a part of our ongoing in silico computational research to search potential inhibitors for SARS-CoV-2, a library of 163 ligands was created by collecting the best phytochemicals from triterpenoids and polyphenols that cleared the limitations of ADMET/drug-likeliness from our recently reported work,18,19 the terpenoids based synthetic compounds/phytochemicals available in the drugbank, and also the drugs that are known for their activity against furin and TMPRSS2.20, 21, 22, 23 The library also consists of several important triterpenoids and polyphenols known for their antiviral activity against SARS-CoV whose protein content is almost 80% similar to SARS-CoV-2.24 These ligands were docked with the proteases (furin and TMPRSS2), and the protein-ligand interaction study was performed to identify the potential ligands that bind at the active sites with high binding energy.
2. Computational methods
2.1. Proteins and ligand preparations
The proteins 3D structures were retrieved from the PDB database (www.rcsb.org). The crystallography structures of the spike protein of SARS-CoV-2 (PDB ID: 6ZGH), ACE2 (PDB ID: 6M17), furin (PDB ID: 5JXH) and TMPRSS2 (PBD ID: 1Z8G, 6K5D, 3T2N and 5CE1) were modelled using Modeller version 10.0. The modelling of TMPRSS2 was performed with five template structures obtained from PBD ID: 1Z8G, 6K5D, 3T2N and 5CE1, and the best model having the least discrete optimized protein energy (DOPE) score −29028.181641 was selected for further in silico computational studies. The missing residues and atoms of the protein were reconstructed using SWISS-PdBViewer 4.1.0. The modelled proteins were validated using PDBsum online tool. The structures of the 163 ligands were retrieved from EMBL-EBI (www.ebi.ac.uk/chebi/advancedSearchFT.do), PUBCHEM (https://pubchem.ncbi.nlm.nih.gov/) or drugbank (https://www.drugbank.ca/). The collected structures of the ligands were optimized further by semi-empirical PM6 method coded in the computational program Gaussian 09W in the gas phase.25
2.2. Protein-protein docking
The protein-protein molecular docking was performed through ClusPro online docking server (https://cluspro.bu.edu/queue.php).26 This server generates 25 protein-protein docking models binding with multiple conformations and calculates the weighted energies with the scoring function [E = Eattr + w1Erep + w2Eelec + w3Epair] for every binding conformation,27 where Eattr and Erep denote the attractive and repulsive contributions to the van der Waals interaction energy Evdw, Eelec is an electrostatic energy term, and Epair represents the desolvation contributions. Epair has been parameterized on a set of complexes that included a substantial number of enzyme inhibitor pairs and multi-subunit proteins, and therefore the resulting potential assumes good shape and electrostatic complementarity. The coefficients w1, w2, and w3 specify the weights of the corresponding terms, and are optimally selected for different types of docking problems. The protein-protein interactions were visualized by using the BioVia discovery studio.28
2.3. Protein-ligand docking
The protein-ligand molecular docking was performed by Autodock Vina 1.1.2 software29 to estimate the binding energies of the ligands towards the protein targets furin and TMPRSS2. The protein and the ligand structures were prepared using MGL 1.5.6, and the grid files were generated that covers all the substrate binding sites and active sites. The molecular docking were performed by applying the Lamarckian genetic algorithm (LGA). The docking output files were analyzed by using the BioVia discovery studio and Ligplotplus.30,31
3. Results and discussion
3.1. Protein-protein docking
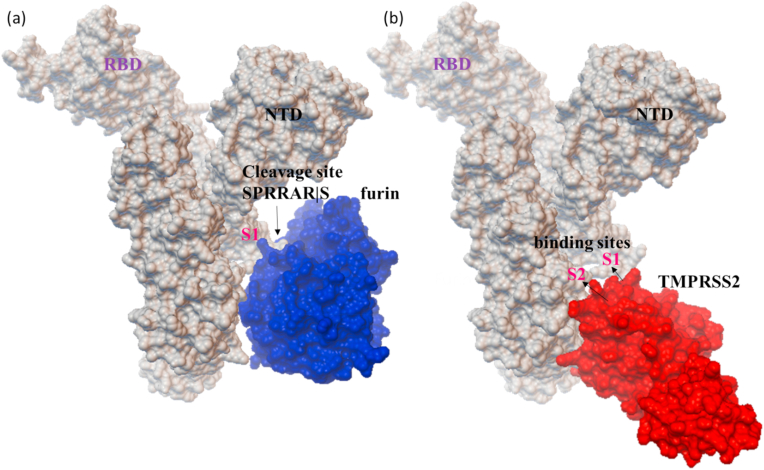
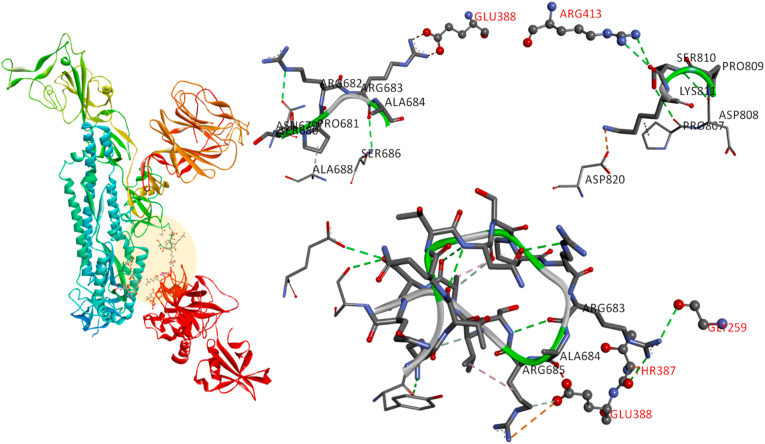
The interaction of the spike glycoprotein of SARS-CoV-2 with human ACE2 receptor initiates the virus-receptor binding followed by the host proteases (furin and TMPRSS2) activate the spike protein that allows the fusion of viral genome into the target cell. The protein-protein docking between spike glycoprotein with ACE2 in ClusPro lower the energy by −908.4 kJ/mol due to multiple non-covalent interactions between the two proteins (Fig. S1). The interaction of spike glycoprotein with ACE2 is studied, and their active sites are targeted extensively to propose potential phytochemicals and drugs compared to the proteases furin and TMPRSS2.32, 33, 34, 35 Therefore, the binding and conformational details of the proteases furin and TMPRSS2 were examined by performing the in silico protein-protein docking studies. Among the 25 generated models, the best pose of the proteases furin and TMPRSS2 binding at the active sites (substrate binding site and/or catalytic site) of S protein is shown in Fig. 2. The furin binds firmly with S protein PRRAR|S site, whereas the TMPRSS2 at the active residues present in both S1 and S2 domains.
Fig. 2.
Protein-protein docked structure of (a) furin and (b) TMPRSS2 with S protein demonstrates the primary contacts at the proteolytic sites of S protein.
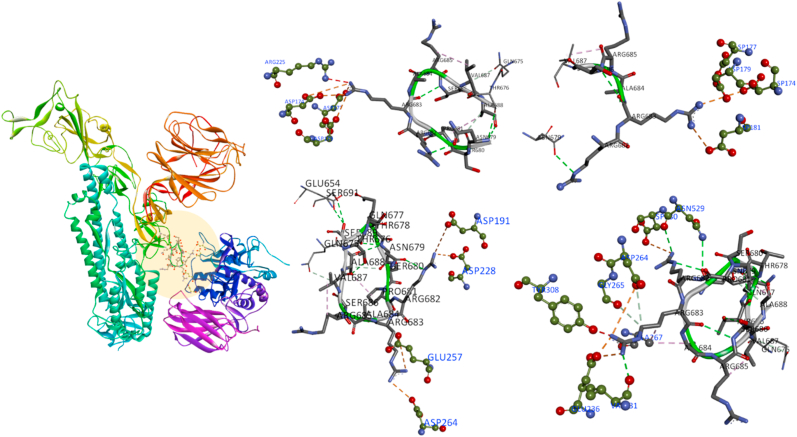
The protein-protein docking of furin and S protein from Cluspro server resulting the binding of furin at the cleavable site with the lowest energy −1026.8 kJ/mol. Furin binding to SARS-CoV-2 S1 site and cleavable PRRAR|S residues with the amino acid residues ALA267, PRO266, LYS261, ASP264, GLY265, TYR308, PRO256, GLY255, GLU236, VAL231, SER253, MET226, LEU227, ASP153, ASP154, ASP228, HIS194, ARG185, ASN192 and ASP191. The residues ARG683 and ARG682 are popped out of the closed state of S protein to form multiple non-covalent interactions with the furin (Fig. 3). The deep cavity of catalytic site in furin buried the residues ASP153 and ASN295 play a critical role in cleaving PRRAR|S site. ARG683, forming a salt bridge between ASP174, ASP177, TYR308, charge-charge interaction with ASP179, ASP181, ARG225, GLU257, ASP264 residues, carbon-hydrogen bond with GLY265, ASP264 residues and conventional hydrogen bond with VAL231. ARG682 forming conventional hydrogen bonds with ASN529 and ASP530 residues, and also forming a salt bridge with the residues ASP191 and ASP228.
Fig. 3.
Representing the furin interactions with S protein cleavage site, with possible binding conformations of furin residues VAL231, GLY265, TYR308 binding affinity to S1 ARG683, ARG682 sites to cleave the PRRAR|S residues.
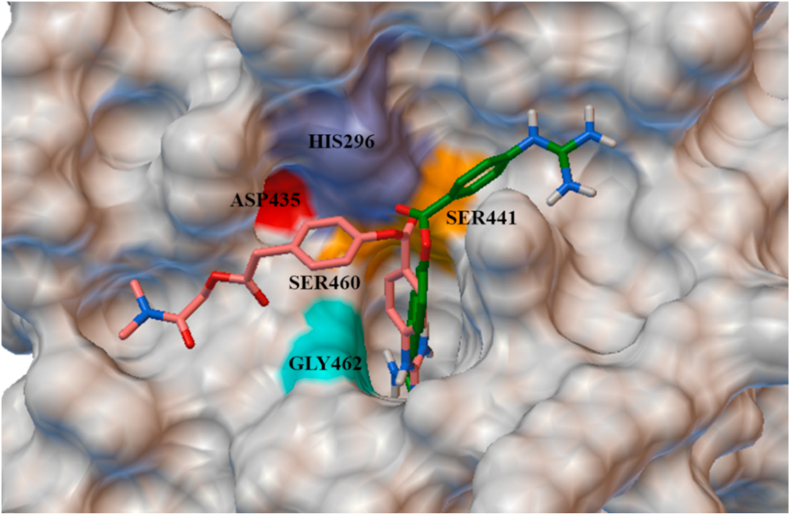
TMPRSS2 docked with S protein predicted 25 binding conformations, and the model binding at the active site of S1/S2 was selected that showed the lowest energy −886.7 kJ/mol (Fig. 4). TMPRSS2 cleaved the spike protein at two different sites, i.e. ARG815/SER816 and ARG685/SER686. Accordingly, the protein-protein docked structure was analyzed and the important non-covalent interactions of TMPRSS2 at the cleavage of S protein is shown in Fig. 4. The S2 domain residue SER810 forming conventional hydrogen bond with ARG413. The residues GLY259 and THR387 pose conventional hydrogen bonds with ARG683 of PRRAR site. GLU388 forming charge-charge and carbon hydrogen bond with ARG685, and also pose a bump with the ALA684 residue of the S protein. The residue ARG683 of S protein is interacting with GLY259, THR387 by conventional hydrogen bond, whereas ARG685 posing carbon hydrogen bond and charge-charge interaction with GLU388. The substrate binding and the catalytic sites residues such as HIS296, ILE346, SER441, ASP435, SER460, GLY462 LYS467 and ASN465 plays a critical role in proteolytic cleavage, and therefore, these possible binding conformations between TMPRSS2 and S protein at S1/S2 sites leads to cleavage activity.
Fig. 4.
Representing the TMPRSS2 interactions with S protein cleavage site with possible binding conformations at S1/S2 site ARG683, ALA684, ARG685 and SER810.
3.2. Docking with furin
The protease furin is a proprotein that activates protein functions and converts into a biologically active form by cleaving some part of the protein. It also cleaves the S protein of the SARS-CoV-2 virus, and promotes the S protein to bind with TMPRSS2 that activates the process of endocytosis. The S protein residues ASN657 and GLN690 are the prime interacting amino acids with 105 residues of the furin protease pose multiple non-covalent interactions to cleave the S protein residues, and concomitantly activates S protein to bind with TMPRSS2. During the cleavage of S protein, the residues ASP153, HIS194, ASN295 and SER368 of furin play the important role in catalytic activity. Therefore, the molecular docking of 163 ligands were performed by adjusting the grid dimensions localizing the active site at the centers x: 34.6056, y: 37.5109, z: 3.3422. After docking, the protein-ligand interaction study was performed to identify the ligands that bind with the catalytically active residues of furin. Total 23 ligands bind with the catalytic residues of furin at the active site with the dock score in the range −8.7 to −7.0 kcal/mol (Table S1). The dock score supported the effective binding of the ligands at the active site of furin, and also comparably higher than the well-known furin inhibitor decanoyl-RVKR-chloromethylketone (CMK) binding with dock score −6.1 kcal/mol (Fig. S2).19 Based on the reported medicinal properties and dock score at the active site of furin, twelve best ligands (Fig. S3) were identified and summarized in Table 1.
Table 1.
Compounds pose at the furin active site with their binding energy (B.E., kcal/mol) and medicinal properties.
| Compounds | B.E. | Residues interacting at active site | Medicinal importance | Ref. |
|---|---|---|---|---|
| Limonin | −8.7 | ASN295, ARG298, GLU299, THR365, ASP259, ASP258, TRP328, GLY296. | Anti-viral properties, Inhibiting HIV-I, HTLV-I replication, and inhibiting proliferation of colon cancer cell. | 36,37 |
| Eribulin | −8.6 | ASN295, ASP191, ASN190, ARG193, ASP259, HIS194, SER368, ARG197 | Anticancer agent, treats breast cancer and liposarcoma. | 38 |
| Pedunculagin | −8.5 | ASN295, HIS194, ASP258, GLU299, GLY296, TRP328, SER368, ARG193 | Carbonic anhydrase inhibitor, anti-human myelogenous leukemia. | 39,40 |
| Maytansine | −8.4 | HIS194, ASN295, TRP328, THR365, HIS364, ARG193, ASP191 | Cytotoxic agent, microtubules inhibitor. | 41 |
| Bevirimat | −8.1 | HIS194, ASN295, TRP328, THR365, HIS364, ARG193 | HIV gap protein inhibitor in development of maturation, protease inhibitor. | 42 |
| Ellagic Acid | −8.0 | HIS194, SER368, ASN295, THR365, HIS364, ARG197, ARG193, ASP191. | Antioxidant agent. | 43 |
| Glycyrrhetinic acid | −8.0 | HIS194, SER368, THR365, HIS364, ASN295, TRP328, ASP258, ARG193. | Suppressed NF-κB activation in TNF-α-induced hepatocytes | 44 |
| Gedunin | −7.8 | ASN295, THR365, ARG298, GLU299, GLY296, ASP259. | Hsp90 inhibitor, Under investigation as anti-cancer agent and antimalarial activity. | 45 |
| Limonin glucoside | −7.8 | ASN295, TRP328, SER368, ASP258, HIS194, ARG298, ARG193, ARG197. | Decreases liver enzymes expression associated with chronic inflammatory disease. | 46 |
| Epigallocatechin | −7.7 | HIS194, ASP191, HIS364, SER368, ASN295, MET189, ARG193, ASN190. | Potential nutraceutical agent. | 47 |
| Betulinic acid | −7.2 | ASN295, HIS194, TRP328, GLY366, ASP258, ARG298, GLU257, SER368. | Potential agent against cancer and HIV infection. | 48 |
| Epicatechin | −7.1 | HIS194, SER368, ASP191, HIS364, THR365, MET189, ASN190, ARG197. | Antioxidant. | 49 |
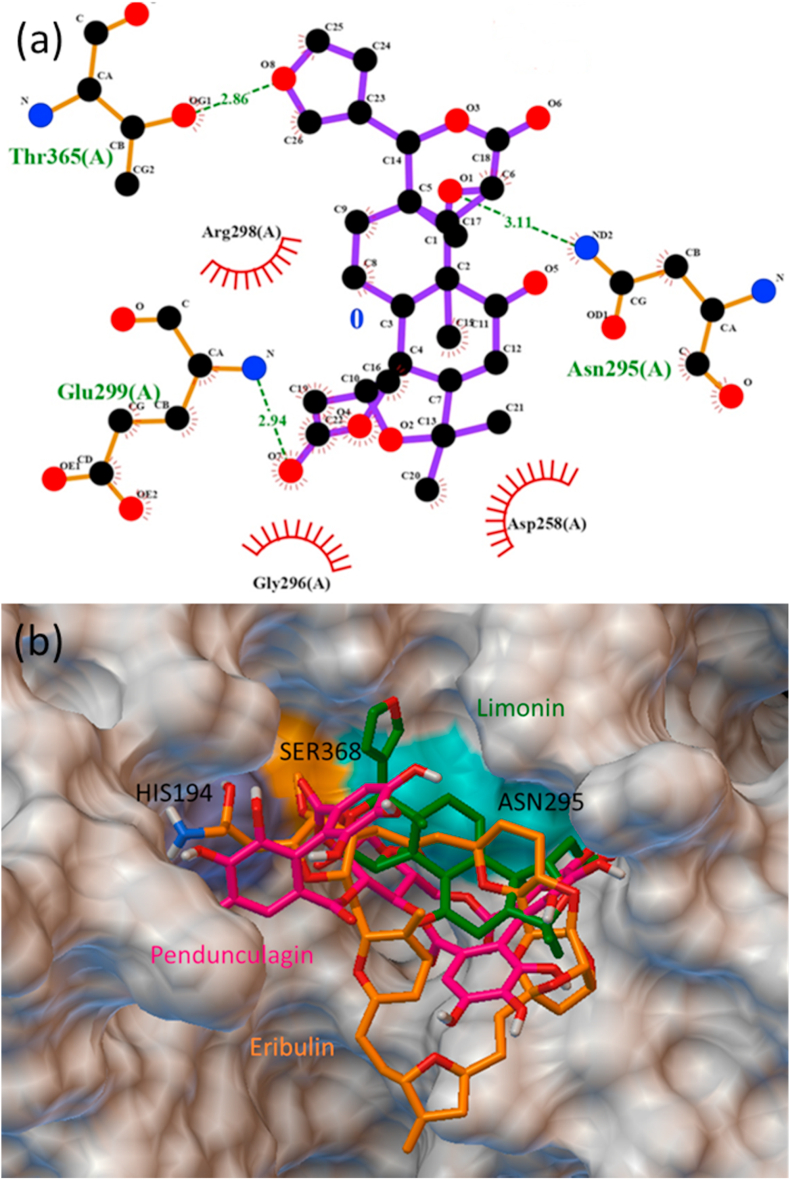
Limonin, the triterpenoid based phytochemical is the first isolated limonoids found mainly in the citrus fruit is known for inhibiting colon cancer cell proliferation through apoptosis and antiviral against HIV-1 and HTLV-1 replication.50 Limonin was proposed as potential phytochemicals that bind at the active site of the main protease Mpro and RdRp of SARS-CoV-2 with the dock score −8.7 and −8.2 kcal/mol, respectively.18 The present study also revealed that limonin is binding at the active site of furin with a higher dock score of −8.7 kcal/mol. Limonin is forming multiple interactions with the residues like ASN295, ARG298, GLU299, THR365, ASP259, ASP258, TRP328 and GLY296 (Fig. 5a). Similarly, eribulin, an anti-cancerous agent exhibiting strong affinity towards active site showing multiple interactions with the residues ASN295, ASP191, ASN190, ARG193, ASP259, HIS194, SER368, ARG197 with the binding energy of −8.6 kcal/mol. Pedunculagin also an anti-cancerous agent and carbonic anhydrase inhibitor found in the pericarp of pomegranate posing at the active site of furin and binding to ASN295, HIS194, ASP258, GLU299, GLY296, TRP328 residues with the conventional hydrogen bond and non-covalent interactions with the SER368, ARG193 residues.51 The binding of the best pose of the ligands limonin, pendunculagin and eribulin at the active site of furin is shown in Fig. 5b.
Fig. 5.
(a) Ligplot of limonin pose at the active site of furin, and (b) the surface image of furin active site docked with limonin, pendunculagin and eribulin.
The phenolic phytochemicals like ellagic acid, epigallocatechin, epicatechin also bound to the active site and inhibiting the furin activity through binding with HIS194, ARG193, ARG197, ASP191, ASN295, THR364, MET189 and THR365 residues posing π-sigma, π-anion and conventional hydrogen bonds. Ellagic acid is a well-known antioxidant that not only inhibiting furin52 and also inhibiting ACE2 active site with a better binding affinity.19 Epicatechin and epigallocatechin found in green tea leaves have a predominant role as antioxidant and antiviral properties. Several limonoids and triterpenoids based ligands like 17-hydroxyazadiradione, gedunin, azadiradione, glycyrrhitinic acid and limoninglucoside showed binding affinity towards HIS194, SER368, ASN295 residues and also interacting with TRP328, HIS300, ALA332, ALA326, ALA517 with covalent interactions and ASN407, ALA408, ASN409, ASN325, PRO327, SER330, GLY297, GLU331, SER302, ASP301 posing non-covalent interactions of the substrate binding site. Glycyrrhitinic acid commonly known as enoxolone reported for potential activity against peptic ulcers.53 The enoxolone structure poses pharmacological properties that may exhibit antiviral, antibacterial and anti-cancerous properties.54 Importantly, enoxole is the hydrolysis product of glycyrrhizic acid found in the herb licorice, and the glycyrrhizic acid is claimed to be an important phytochemical against SARS-CoV-2.18 Limonin glucoside inhibiting the colon adenocarcinoma cells and 17-hydroxyazadiradione known for antifungal activity. In addition, the betunilic acid-like compound bevirimat well known for anti-HIV activity binds effectively at the active site of furin.
3.3. Docking with TMPRSS2
TMPRSS2 is a serine protease that activates the SARS-CoV and SARS-CoV-2 S protein through N-terminal and C-terminal cleavage sites. TMPRSS2 cleaves the S protein in the lungs of SARS-CoV-2 infected person and promotes pathogenicity. SARS-CoV-2 S protein S1/S2 cleaved by furin, and subsequently, TMPRSS2 mediates the cleavage and activation of the S protein. TMPRSS2 cleaved the S protein at two potential sites, i.e., N-terminal (ARG685, SER686) and C terminal (ARG815, SER816). So, the TMPRSS2 serine protease opted as a potential therapeutic target. The TMPRSS2 possesses catalytically active residues HIS296, ILE346 and SER441, and the substrate binding residues ASP345, ASP435, SER460, SER465 and LYS467. For virtual screening of potential compounds, the structurally refined TMPRSS2 protein was used for molecular docking with 163 ligands by adjusting the grid dimensions x: 1.6407, y: 2.5071, z: 40.3205 that covered the entire substrate binding and catalytic sites. The protein-ligand interaction study revealed that total 90 ligands bind with the catalytic/substrate binding residues of TMPRSS2 at the active site and their dock score observed between −8.7 and −5.0 kcal/mol (Table S1). It is important to mention here that the approved drug nafamostat (−8.1 kcal/mol) binds at the active site of TMPRRSS2 with a higher binding affinity than the drug camostat (−6.6 kcal/mol), and their best pose at the active site of TMPRSS2 is shown in Fig. 6. Based on the computational results, the use of nafamostat is expected to be more beneficial than the camostat.20 Also, the nafamostat binds more efficiently at the active site of TMPRSS2 compared to the protease inhibitors aloxistatin (−5.2 kcal/mol) and nelfnavir (−7.0 kcal/mol).21,23 Therefore, considering nafamostat as control, fourteen best dock score ligands were selected (Fig. S3) and their interactions at the active site of TMPRSS2 along with medicinal properties are summarized in Table 2.
Fig. 6.
The surface image of TMPRSS2 active site docked with the best pose of nafamostat and camostat.
Table 2.
Compounds pose at the TMPRSS2 active site with their binding energy (B.E., kcal/mol) and medicinal properties.
| Compounds | B.E. | Residues interacting at active site | Medicinal and biological properties | Ref. |
|---|---|---|---|---|
| Gedunin | −8.7 | HIS296, CYS465, GLY462, SER460, SER441, GLY464, CYS437, CYS297. | Hsp90 inhibitor, Under investigation as anti-cancer agent and antimalarial activity. | 45 |
| Deoxyobacunone | −8.4 | HIS296, CYS465, GLY464, GLY462, CYS297, SER441, SER460. | Stimulatory activity in strigahermonthica seeds | 55 |
| 2-hydroxyseneganolide | −8.4 | HIS296, TRP461, SER460, SER463, GLY462, LYS342, THR341, LY464. | Anti-fungal agent. | 56 |
| Pedunculagin | −8.4 | SER441, HIS296, GLN438, GLY464, GLY462, CYS297, SER436. | Carbonic anhydrase inhibitor, anti-human myelogenous leukemia. | 30,31 |
| 7-deacetyl-7-benzoylgedunin | −8.3 | HIS296, CYS465, SER436, SER441, SER460, TRP461, GLY462, SER436, | Cytotoxic activity against HL60 leukemia cells and antineoplastic agent. | 57 |
| 7-deacetylgedunin | −8.3 | HIS296, CYS465, CYS437, GLY462, SER441, SER460, TRP461, GLY462 | Activates Keap1/Nrf2/HO-1 signaling and suppress inflammatory response. | 58 |
| Limonin | −8.3 | HIS296, SER441, LYS342, TRP461, CYS437, CYS465, GLY462, GLY464 | Anti-viral properties, Inhibiting HIV-I, HTLV-I replication, and inhibiting proliferation of colon cancer cell. | 27,28 |
| Solamargine | −8.3 | HIS296, SER441, GLN438, GLU299, LEU419, TRP461, GLY439 | Induces non-selective cytotoxicity and inhibits P-glycoprotein. | 59 |
| Betulinic acid | −8.3 | SER460, SER441, HIS296, LYS342, VAL280, LEU302, GLN438 | Antimalarial, antiretroviral and anti-inflammatory agent. | 39 |
| Eribulin | −8.2 | SER441, LYS340, GLN438, LYS342, HIS296, SER460, LYS342 | Anticancer agent, treats breast cancer and liposarcoma. | 29 |
| Baicalein | −8.2 | HIS296, LN438, GLY464, LYS342, TRP461, LEU419, SER441, SER460. | Anti-inflammatory and anti- proliferative agent. | 60 |
| Limonin glucoside | −8.1 | GLY464, GLY462, HIS296, LYS342, TRP461, GLY439, SER441, THR341. | Decreases liver enzymes expression associated with chronic inflammatory disease. | 37 |
| Nafamostat | −8.1 | HIS296, SER463, ASP435, CYS437, SER441, CYS465, GLN438, TRP461 | Serine protease inhibitor, antiviral and anti-cancerous agent. | 61 |
| Hesperidin | −8.0 | SER441, HIS296, TRP461, LYS342, GLN438, SER436, CYS437. | Antioxidant and anti-allergen. | 62 |
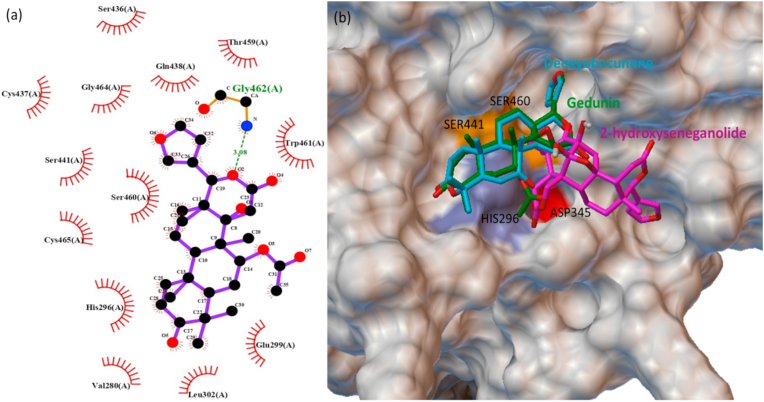
The phytochemicals such as gedunin, deoxyobacunone, 2-hydroxyseneganolide, 7-deacetyl-7-benzoylgedunin and 7-deacetylgeduin proven to be antiviral agents were actively binding at the catalytic site of TMPRSS2. Gedunin is extracted mainly from the neem (azadirachtaindica) is computationally proposed as a potential phytochemical, and binds firmly at the active site of receptor-binding domain of S protein and ACE2.18 Gedunin is also binding firmly at the active site of TMPRSS2 by forming non-covalent interactions with the residues HIS296, CYS465, GLY462, SER460, SER441, GLY464, CYS437 and CYS297 (Fig. 7a). The binding of the three best phytochemicals gedunin, deoxyabacunone and 2-hydroxyseneganolide at the active site of TMPRSS2 is shown in Fig. 7b.
Fig. 7.
(a) Ligplot of gedunin posing at the active site of TMPRSS2, and (b) the surface image of TMPRSS2 active site docked with gedunin, deoxyabacunone and 2-hydroxyseneganolide.
Limonin and limonin glucoside derivative known for antiviral and anti-cancerous properties can also potentially inhibit the TMPRSS2 activity by interacting with the residues LYS342, GLN438 by forming a conventional hydrogen bond, and π-alkyl and π-π interactions with CYS465, HIS296 and CYS437, TRP461. The catalytic residue SER441 posing non-covalent interaction with limonin. Betulinic acid and bevirimat are also showed potential to inhibit serine protease-2 activity, and these compounds may be potential to use against SARS-CoV-2 because of their anti-HIV properties. Baicalein is known to suppress the androgen receptor (AR) target genes like TMPRSS2 in prostate cancer (PCa) progression,63 and its effective binding with at the active site can be a potential target to use against COVID-19. Hesperidin is a flavone glycoside that showed a similar binding characteristic to nafamostat, a TMPRSS2 inhibitor. It is binding to the catalytic site and forming hydrogen bonds to GLN438, SER441, SER436, CYS437, SER460, π-alkyl, π-π and π-cation interactions with LYS342, TRP461 and HIS296 residues.
4. Conclusions
In summary, the binding and conformational details of the proteases furin and TMPRSS2 with the S protein of SARS-CoV-2 was examined in silico by performing protein-protein docking, and the residues involved in catalytic or substrate binding activity were discussed. Furin showed lower Cluspro energy than TMPRSS2 in binding with S protein, and the role of furin may be investigated in details to search potential vaccines or drugs against the COVID-19. Virtual screening of 163 ligands by performing protein-ligand docking by targeting the active site of furin and TMPRSS2 resulted several key compounds that can suppress the proteolytic activity. Limonin and gedunin found mainly in the citrus fruits and neem showed the higher binding energy at the active site of furin and TMPRSS2, respectively among all the examined compounds. The compounds like limonin, eribulin, pedunculagin, gedunin, limonin glycoside and betunilic acid bind at the active sites of both furin and TMPRSS2. The polyphenols found in green tea can also be useful in suppressing the furin activity. For TMPRSS2, the approved drug nafamostat may be more beneficial than the camostat. The effective binding of glycyrrhetinic acid and baicalein with the proteases, along with their effective binding to the proteins target associated with SARS-CoV-2 makes them the potent phytochemicals similar to limonin and geduninin for formulating traditional medicines. Overall, the current in silico outcomes will be useful in selecting the key targets for further experimental studies that can be beneficial against COVID-19 by inhibiting the proteolytic activity of the proteases furin and TMPRSS2.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors are thankful to Director, SVNIT for providing necessary research facilities and infrastructure.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtcme.2021.04.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 2020;9(7):529. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30(7):1346–1351. doi: 10.1016/j.cub.2020.03.022. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naqvi A.A.T., Fatima K., Mohammad T., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta (BBA) - Mol Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki A., Al-Karmalawy A., El-Amier Y., Ashour A. Molecular docking reveals the potential of Cleome amblyocarpa isolated compounds to inhibit COVID-19 virus main protease. New J Chem. 2020;44(39):16752–16758. doi: 10.1039/d0nj03611k. [DOI] [Google Scholar]
- 5.Alnajjar R., Mostafa A., Kandeil A., Al-Karmalawy A.A. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon. 2020;6(12) doi: 10.1016/j.heliyon.2020.e05641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmaaty A.A., Alnajjar R., Hamed M.I.A., Khattab M., Khalifa M.M., Al-Karmalawy A.A. Revisiting activity of some glucocorticoids as a potential inhibitor of SARS-CoV-2 main protease: theoretical study. RSC Adv. 2021;11:10027–10042. doi: 10.1039/D0RA10674G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ysrafil Astuti I. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiefer M., Tucker J. Identification of a second human subtilisin-like protease gene in the fes/fps region of chromosome 15. DNA Cell Biol. 1991;10(10):757–769. doi: 10.1089/dna.1991.10.757. [DOI] [PubMed] [Google Scholar]
- 9.Checkley M., Luttge B., Freed E. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410(4):582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutard B., Valle C., de Lamballerie C., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia S., Lan Q., Su S., et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther. 2020;5(1):92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia S., Liu M., Wang C., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Böttcher-Friebertshäuser E., Freuer C., Sielaff F., et al. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J Virol. 2010;84(11):5605–5614. doi: 10.1128/jvi.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing Y., Li X., Gao X., Dong Q. Natural polymorphisms are present in the furin cleavage site of the SARS-CoV-2 spike glycoprotein. Front Genet. 2020;11:783. doi: 10.3389/fgene.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng X.Y., Zhang H.X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vardhan S., Sahoo S. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput Biol Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vardhan S., Dholakiya B.Z., Sahoo S. 2020. Protein-ligand Interaction Study to Identify Potential Dietary Compounds Binding at the Active Site of Therapeutic Target Proteins of SARS-CoV-2. arXiv:2005.11767vol. 1. [Google Scholar]
- 20.Weissenstein A. TMPRSS2-Inhibitors play a role in cell entry mechanism of COVID-19: an insight into camostat and nafamostat. J Regen Biol Med. 2020;2(2):1–3. doi: 10.37191/mapsci-2582-385x-2(2)-022. [DOI] [Google Scholar]
- 21.Musarrat F., Chouljenko V., Dahal A., et al. The anti-HIV drug nelfinavir mesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARSCoV-2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID-19 infections. J Med Virol. 2020;92(10):2087–2095. doi: 10.1002/jmv.25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Y.W., Chao T.L., Li C.L., et al. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2) doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J., Gao Q., He C., Huang A., Tang N., Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020;7(4):551–557. doi: 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuzimoto A.D., Isidoro C. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds - additional weapons in the fight against the COVID-19 pandemic? J Tradit Complement Med. 2020;10(4):405–419. doi: 10.1016/j.jtcme.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Řezáč J., Fanfrlík J., Salahub D., Hobza P. Semiempirical quantum chemical PM6 method augmented by dispersion and H-bonding correction terms reliably describes various types of noncovalent complexes. J Chem Theor Comput. 2009;5(7):1749–1760. doi: 10.1021/ct9000922. [DOI] [PubMed] [Google Scholar]
- 26.Kozakov D., Hall D., Xia B., et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozakov D., Beglov D., Bohnuud T., et al. How good is automated protein docking? Proteins. 2013;81(12):2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dassault Systmes Biovia . 2017. Discovery Studio Modeling Environment.http://accelrys.com/products/collaborative-science/biovia-discovery-studio/ Release. (2016) [Google Scholar]
- 29.Trott O., Olson A. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009 doi: 10.1002/jcc.21334. NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski R., Swindells M. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 32.Basu A., Sarkar A., Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-74715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jena A.B., Kanungo N., Nayak V., Gbn Chainy, Dandapat J. Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: insights from computational studies. Sci Rep. 2021;11(1):2043. doi: 10.1038/s41598-021-81462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balestrieri E., Pizzimenti F., FerlazzoA, et al. Antiviral activity of seed extract from Citrus bergamia towards human retroviruses. Bioorg Med Chem. 2011;19(6):2084–2089. doi: 10.1016/j.bmc.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 37.Chidambara Murthy K., Jayaprakasha G., Kumar V., Rathore K., Patil B. Citrus limonin and its glucoside inhibit colon adenocarcinoma cell proliferation through apoptosis. J Agric Food Chem. 2011;59(6):2314–2323. doi: 10.1021/jf104498p. [DOI] [PubMed] [Google Scholar]
- 38.Swami U., Shah U., Goel S. Eribulin in cancer treatment. Mar Drugs. 2015;13(8):5016–5058. doi: 10.3390/md13085016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang J., Cho J., Kim H., et al. Antitumor activity of pedunculagin, one of the ellagitannin. Arch Pharm Res (Seoul) 1995;18(6):396–401. doi: 10.1007/bf02976342. [DOI] [Google Scholar]
- 40.Satomi H., Umemura K., Ueno A., Hatano T., Okuda T., Noro T. Carbonic anhydrase inhibitors from the pericarps of punicagranatum L. Biol Pharm Bull. 1993;16(8):787–790. doi: 10.1248/bpb.16.787. [DOI] [PubMed] [Google Scholar]
- 41.Yu T., Bai L., Clade D., et al. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnemapretiosum. Proc Natl Acad Sci Unit States Am. 2002;99(12):7968–7973. doi: 10.1073/pnas.092697199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith P., Ogundele A., Forrest A., et al. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-O-(3′,3′-Dimethylsuccinyl)Betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob Agents Chemother. 2007;51(10):3574–3581. doi: 10.1128/aac.00152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Priyadarsini K., Khopde S., Kumar S., Mohan H. Free radical studies of ellagic acid, a natural phenolic antioxidant. J Agric Food Chem. 2002;50(7):2200–2206. doi: 10.1021/jf011275g. [DOI] [PubMed] [Google Scholar]
- 44.Chen H., Kang S., Lee I., Lin Y. Glycyrrhetinic acid suppressed NF-κB activation in TNF-α-induced hepatocytes. J Agric Food Chem. 2014;62(3):618–625. doi: 10.1021/jf405352g. [DOI] [PubMed] [Google Scholar]
- 45.Brandt G., Schmidt M., Prisinzano T., Blagg B. Gedunin, a novel Hsp90 inhibitor: semisynthesis of derivatives and preliminary Structure−Activity relationships. J Med Chem. 2008;51(20):6495–6502. doi: 10.1021/jm8007486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley D., Adkins Y., Zunino S., et al. Citrus limoninglucoside supplementation decreased biomarkers of liver disease and inflammation in overweight human adults. J Funct Foods. 2015;12:271–281. doi: 10.1016/j.jff.2014.11.026. [DOI] [Google Scholar]
- 47.Zhu N., Huang T., Yu Y., LaVoie E., Yang C., Ho C. Identification of oxidation products of (−)-EpigallocatechinGallate and (−)-Epigallocatechin with H2O2. J Agric Food Chem. 2000;48(4):979–981. doi: 10.1021/jf991188c. [DOI] [PubMed] [Google Scholar]
- 48.Cichewicz R., Kouzi S. Chemistry, biological activity, and chemotherapeutic potential of betulinic acid for the prevention and treatment of cancer and HIV infection. Med Res Rev. 2003;24(1):90–114. doi: 10.1002/med.10053. [DOI] [PubMed] [Google Scholar]
- 49.Harada M., Kan Y., Naoki H., et al. Identification of the major antioxidative metabolites in biological fluids of the rat with ingested (+)-Catechin and (-)-Epicatechin. Biosci Biotechnol Biochem. 1999;63(6):973–977. doi: 10.1271/bbb.63.973. [DOI] [PubMed] [Google Scholar]
- 50.Chidambara Murthy K., Jayaprakasha G., Kumar V., Rathore K., Patil B. Citrus limonin and its glucoside inhibit colon adenocarcinoma cell proliferation through apoptosis. J Agric Food Chem. 2011;59(6):2314–2323. doi: 10.1021/jf104498p. [DOI] [PubMed] [Google Scholar]
- 51.Rahimi H.R., Arastoo M., Ostad S.N. A comprehensive review of punicagranatum (pomegranate) properties in toxicological, pharmacological, cellular and molecular biology researches. Iran J Pharm Res (IJPR) 2012;11(2):385–400. [PMC free article] [PubMed] [Google Scholar]
- 52.Brusselmans K., Schrijver E.D., Heyns W., Verhoeven G., Swinnen J.V. Epigallocatechin-3-gallate is a potent natural inhibitor of fatty acid synthase in intact cells and selectively induces apoptosis in prostate cancer cells. Int J Canc. 2003;106(6):856–862. doi: 10.1002/ijc.11317. [DOI] [PubMed] [Google Scholar]
- 53.Murray M. Glycyrrhizaglabra (licorice) Textbook of Natural Med. 2020:641–647. doi: 10.1016/b978-0-323-43044-9.00085-6. e3. [DOI] [Google Scholar]
- 54.Roohbakhsh A., Iranshahy M., Iranshahi M. Glycyrrhetinic acid and its derivatives: anti-cancer and cancer chemopreventive properties, mechanisms of action and structure- cytotoxic activity relationship. Curr Med Chem. 2016;23(5):498–517. doi: 10.2174/0929867323666160112122256. [DOI] [PubMed] [Google Scholar]
- 55.Rugutt J., Rugutt K., Berner D. Limonoids from NigerianHarrisoniaabyssinicaand their stimulatory activity againstStrigahermonthicaSeeds. J Nat Prod. 2001;64(11):1434–1438. doi: 10.1021/np0100183. [DOI] [PubMed] [Google Scholar]
- 56.Abdelgaleil S., Iwagawa T., Doe M., Nakatani M. Antifungal limonoids from the fruits of Khayasenegalensis. Fitoterapia. 2004;75(6):566–572. doi: 10.1016/j.fitote.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 57.Kikuchi T., Ishii K., Noto T., et al. Cytotoxic and apoptosis-inducing activities of limonoids from the seeds ofAzadirachtaindica(neem) J Nat Prod. 2011;74(4):866–870. doi: 10.1021/np100783k. [DOI] [PubMed] [Google Scholar]
- 58.Chen J., Zhu G., Su X., et al. 7-deacetylgedunin suppresses inflammatory responses through activation of Keap1/Nrf2/HO-1 signaling. Oncotarget. 2017;8(33):55051–55063. doi: 10.18632/oncotarget.19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burger T., Mokoka T., Fouché G., Steenkamp P., Steenkamp V., Cordier W. Solamargine, a bioactive steroidal alkaloid isolated from Solanumaculeastrum induces non-selective cytotoxicity and P-glycoprotein inhibition. BMC Compl Alternative Med. 2018;18(1) doi: 10.1186/s12906-018-2208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patwardhan R., Sharma D., et al. Sandur S. Baicalein exhibits anti-inflammatory effects via inhibition of NF-κB transactivation. Biochem Pharmacol. 2016;108:75–89. doi: 10.1016/j.bcp.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 61.Hoffmann M., Schroeder S., Kleine-Weber H., Müller M., Drosten C., Pöhlmann S. NafamostatMesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob Agents Chemother. 2020;64(6) doi: 10.1128/aac.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parhiz H., Roohbakhsh A., Soltani F., Rezaee R., Iranshahi M. Antioxidant and anti-inflammatory properties of the citrus flavonoids hesperidin and hesperetin: an updated review of their molecular mechanisms and experimental models. Phytother Res. 2014;29(3):323–331. doi: 10.1002/ptr.5256. [DOI] [PubMed] [Google Scholar]
- 63.Xu D., Chen Q., Liu Y., Wen X. Baicalein suppresses the androgen receptor (AR)-mediated prostate cancer progression via inhibiting the AR N-C dimerization and AR-coactivators interaction. Oncotarget. 2017;(8):105561–105573. doi: 10.18632/oncotarget.22319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.