Abstract
Circular RNAs (circRNAs) belong to a highly conserved subtype of non-coding RNAs, produced by the back-splicing of specific regions of pre-mRNA. CircRNAs have wide-ranging effects on eukaryotic physiology and pathology by acting as transcription regulators, miRNA sponges, protein sponges, and templates for translation. Skeletal and chondral disorders are the leading causes of pain and disability, especially for elders, affecting hundreds of millions of people worldwide. Plenty of evidence have shown that circRNAs are dysregulated and play vital roles in the occurrence and progression of skeletal and chondral disorders. Herein, we systematically summarize the emerging roles and underlying molecular mechanisms of hub circRNAs in the pathogenesis of several representative skeletal and chondral disorders. Our findings may provide further insight into the mechanistic details of the role of circRNA in bone or cartilage metabolism, and highlight the promising application of circRNAs in serving as potential diagnostic or therapeutic targets for the prevention and treatment of skeletal and chondral disorders.
Keywords: Circular RNAs, skeletal and chondral disorders, regulatory mechanism, biomarker, therapeutic target.
Introduction
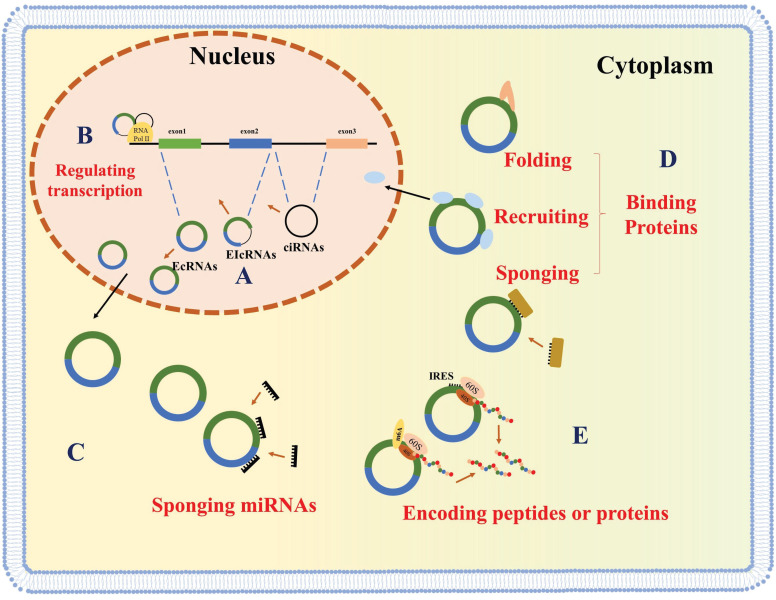
Circular RNAs (circRNAs) are recently discovered non-coding RNAs (ncRNAs) that consist of a specially covalently closed ring structure and can be stably expressed in multiple cell lines 1, 2. CircRNAs derive from canonical splice sites, upon suppression or slowing down of the splicing of pre-mRNA 3, 4. CircRNAs are classified based on their sequence. There three types of circRNAs are exonic circRNAs (EcRNAs), intronic circRNAs (ciRNAs) and exon-intron circRNAs (EIcRNAs) (Figure 1A) 5, 6. EcRNAs are mainly found in the cytoplasm, while ciRNAs and EIcRNAs are predominantly found in the nucleus. EcRNAs are the most abundant circRNAs and account for over 80% of the known circRNAs 1, 7, 8.
Figure 1.
Biogenesis and molecular functions of circRNAs. CircRNAs are generated by back-splicing (A) and regulate gene expression at the level of transcription (B), sponging miRNAs (C), interacting with proteins (D), or encoding peptides (E).
CircRNAs were first discovered in pathogens in the 1970s 9 and were initially thought to be transcriptional splicing intermediates or byproducts of pre-mRNA splicing errors 10, 11. Since then, due to the advancements in RNA sequencing technologies and bioinformatic analyses, thousands of circRNAs have been identified 12. Multiple lines of evidence have highlighted the significant impact of circRNAs on various eukaryotic physiological and pathological processes 13. CircRNAs in the nucleus (ciRNAs and EIcRNAs) play roles in host gene transcription regulation (Figure 1B) 5, 14, 15, while cytoplasmic circRNAs have been shown to function as miRNA sponges, thereby promoting the expression of downstream mRNAs (Figure 1C) 7, 16, 17. Recently, several circRNAs have been implicated in the function and localization of proteins by serving as a decoy, and facilitating protein folding and recruitment (Figure 1D) 7, 16-20. Some studies have also shown that coding circRNAs can play critical roles in human diseases (Figure 1E) 21-24.
CircRNAs function as pervasive regulators of cellular and physiological processes and have been proven to be vital in many diseases, such as skeletal and chondral disorders 25-27. Skeletal and chondral disorders are among the leading debilitating factors and represent an increasing societal and economic burden in the context of aging population and increasing life expectancy. The most common of these diseases are osteoarthritis, osteonecrosis of the femoral head (ONFH), osteoporosis, and rheumatoid arthritis (RA), affecting hundreds of millions of people worldwide 28-31. Several factors have been associated with the occurrence and progression of skeletal and chondral disorders, including inflammation, apoptosis, degradation of extracellular matrix (ECM), and an imbalance between osteogenesis and adipogenesis 30, 32-36. However, the underlying molecular mechanisms of the pathology of these diseases still remains elusive, and an understanding of these mechanisms is essential for both the prevention and treatment of bone and cartilage disorders. Studies have begun to highlight the differential expression of circRNAs in bone and cartilage diseases as well as their regulatory roles 37. Further characterizing the roles of circRNAs in the pathological processes of skeletal and chondral disorders could provide new avenues for both diagnosis and treatment.
In this review, we summarize recent studies of circRNAs involved in common skeletal and chondral disorders, such as osteoarthritis, ONFH, osteoporosis, and RA, and highlight the potential applications of these circRNAs in the prevention and treatment of these disorders.
CircRNA and osteoarthritis
Osteoarthritis is recognized as the most common disease of the musculoskeletal system, with up to 250 million cases globally 38. Cartilage damage associated with osteoarthritis can be caused by mechanical, inflammatory, or metabolic stresses that alter cartilage properties 39-41. As a result, hypertrophic chondrocytes attempt to repair eroded cartilage, but in turn release more matrix degradation products and proinflammatory factors, which then leads to structural alterations in articular cartilage, subchondral bone, synovium, capsule, ligaments, and periarticular muscles 42-44. Here, we discuss osteoarthritis-related circRNAs and their regulatory effects on inflammatory and matrix metabolic factors in osteoarthritis-related cells. Understanding the role of these circRNAs can reveal new strategies for intervening the progression of osteoarthritis.
Diagnostic circRNAs of osteoarthritis
CircRNAs have been widely recognized as stable biomarkers due to their resistance from degradation by RNases 45. The use of diagnostic circRNAs in peripheral blood and synovial fluid could have a significant impact on osteoarthritis diagnosis. Studies have indicated that hsa_circ_0104873, hsa_circ_0104595, and hsa_circ_0101251 are steadily expressed in synovial fluid, while hsa_circ_0032131_CBC1 is significantly over-expressed in the blood samples of osteoarthritis patients (Table 1) 46, 47. Further investigation of these circRNAs could set the groundwork for the advancement of minimally invasive, highly specific, easily accessible, and rapid diagnostic biomarkers.
Table 1.
CircRNAs as potential biomarkers of skeletal and chondral disorders.
| Diseases | CircRNAs | Samples | Expression | Methods | Ref. |
|---|---|---|---|---|---|
| OA | Hsa_circ_0104873 | synovial fluid | up | CircRNA-array | 46 |
| OA | Hsa_circ_ 0104595 | synovial fluid | up | CircRNA-array | 46 |
| OA | Hsa_circ_0101251 | synovial fluid | up | CircRNA-array | 46 |
| OA | Hsa_circ_0032131 | peripheral blood | up | CircRNA-array | 47 |
| OP | Hsa_circ_0002060 | serum and plasma | up | CircRNA-array | 86 |
| OP | Hsa_circ_0001445 | plasma | down | qRT-PCR | 87 |
| RA | Hsa_circ_0044235 | peripheral blood | down | qRT-PCR | 106 |
| RA | Hsa_circ_102594 | PBMSs | down | CircRNA-array | 107 |
| RA | Hsa_circ_104194 | PBMSs | up | CircRNA-array | 107 |
| RA | Hsa_circ_104593 | PBMSs | up | CircRNA-array | 107 |
| RA | Hsa_circ_103334 | PBMSs | up | CircRNA-array | 107 |
| RA | Hsa_circ_101407 | PBMSs | up | CircRNA-array | 107 |
| RA | Has_circ_0008360 | PBMSs | down | RNA-seq | 108 |
| RA | Has_circ_0001200, | PBMSs | up | RNA-seq | 108 |
| RA | Has_circ_0001566 | PBMSs | up | RNA-seq | 108 |
| RA | Has_circ_0003972 | PBMSs | up | RNA-seq | 108 |
| RA | Hsa_circ_0002715 | peripheral blood | up | qRT-PCR | 109 |
| RA | Hsa_circ_0035197 | peripheral blood | up | qRT-PCR | 109 |
| RA | Hsa_circ_0000175 | PBMSs | down | qRT-PCR | 110 |
| RA | Hsa_circ_0008410 | PBMSs | up | qRT-PCR | 110 |
| RA | circRNA_104871 | PBMSs | up | CircRNA-array | 111 |
| RA | circRNA_003524 | PBMSs | up | CircRNA-array | 111 |
| RA | circRNA_101873 | PBMSs | up | CircRNA-array | 111 |
| RA | circRNA_103047 | PBMSs | up | CircRNA-array | 111 |
OA: osteoarthritis; OP: osteoporosis; RA: rheumatoid arthritis.
Regulatory circRNAs of osteoarthritis
Articular cartilage degeneration is the vital process in the pathogenesis of osteoarthritis 48. The imbalance of cell proliferation and apoptosis of chondrocytes finally results in the increased catabolism and decreased anabolism of ECM, thereby leading to the inflammation and degeneration of cartilage 48.
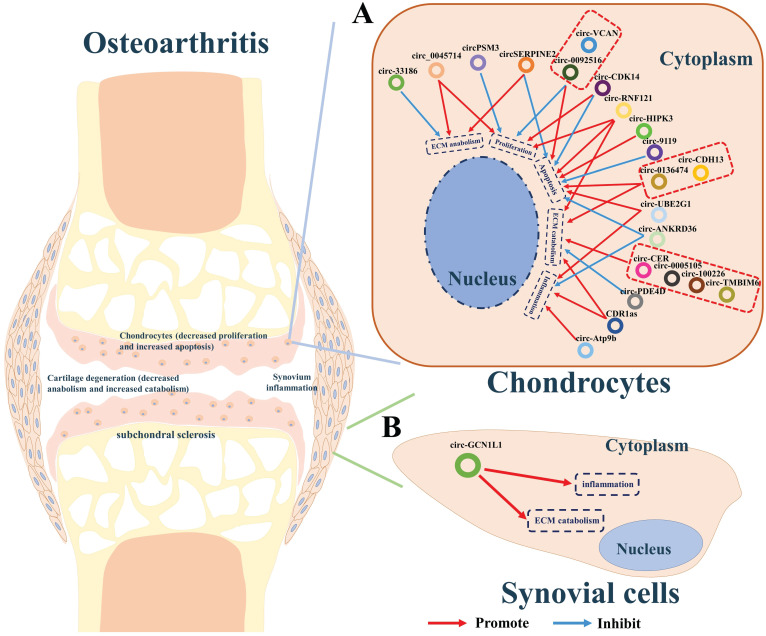
Increasing evidence indicates that some circRNAs plays pivotal roles in the development of osteoarthritis. We summarized the circRNAs dysregulated in articular cartilage and synovium (Table 2). Furthermore, the downstream pathways caused by these dysregulated circRNAs finally result in inflammation, the imbalance between anabolism and catabolism of ECM, inhibition of cells proliferation, or apoptosis. Studies have shown that CDR1as 49, circ-0005105 50, circ-33186 51, circ-0136474 52, circ-100226 32, circ-CER 53, circ-PSM3 54, circ-Atp9b 55, circ-UBE2G1 56, circ-0092516 57, circ-CDH13 58, circ-TMBIM6 59, circ-RNF121 60, circ-VCAN 61, and circ-HIPK3 62, which were found over-expressed in cartilage of osteoarthritis, could significantly promote the expression of osteoarthritis-related genes including MMP13, PTEN, FGF2, NAMPT, TNFα, TLR4, HIFα, MYD88, and SOX8, thereby accelerating the progress osteoarthritis. While circ-SERPINE2 63, 64, circ-CDK14 65, circ-ANKRD36 66, circ-PDE4D 67 circ-0045714 68, and circ-9119 69 were found down-expressed and have positive effects on alleviating the progress of osteogenesis (Figure 2A).
Table 2.
Regulatory circRNAs and their roles in skeletal and chondral disorders.
| Disease | CircRNA | Tissue | Expression | miRNA/gene | Roles | Ref. |
|---|---|---|---|---|---|---|
| OA | circ-100226 | cartilage | up | miR-875/TNFα | ECM catabolism ↑ | 32 |
| OA | CDR1as | cartilage | up | miR-641/FGF2 | inflammation ↑, ECM catabolism ↑ | 49 |
| OA | circ-0005105 | cartilage | up | miR-26a/NAMPT | ECM catabolism ↑ | 50 |
| OA | circ-33186 | cartilage | up | miR-217-5p/MMP13 | cell proliferation ↓ | 51 |
| OA | circ-0136474 | cartilage | up | miR-217-5p/MMP13 | ECM catabolism ↑, apoptosis ↑ | 52 |
| OA | circ-CER | cartilage | up | miR-136/MMP13 | ECM catabolism ↑ | 53 |
| OA | circ-PSM3 | cartilage | up | miR-296-5p | cell proliferation ↓ | 54 |
| OA | circ-Atp9b | cartilage | up | miR-138-5p/TLR4 | inflammation ↑ | 55 |
| OA | circ-UBE2G1 | cartilage | up | miR-373/HIFα | inflammation ↑, apoptosis ↑ | 56 |
| OA | circ-0092516 | cartilage | up | miR-337-3p/PTEN | apoptosis ↑, cell proliferation ↓ | 57 |
| OA | circ-CDH13 | cartilage | up | miR-296-3p/PTEN | ECM catabolism ↑, apoptosis ↑ | 58 |
| OA | circ-TMBIM6 | cartilage | up | miR-27a/MMP13 | ECM catabolism ↑ | 59 |
| OA | circ-RNF121 | cartilage | up | miR-665/MYD88 | ECM catabolism ↑, apoptosis ↑, cell proliferation ↓ | 60 |
| OA | circ-VCAN | cartilage | up | NF-κB | apoptosis ↑, cell proliferation ↓ | 61 |
| OA | circ-HIPK3 | cartilage | up | miR-124/SOX8 | apoptosis ↑ | 62 |
| OA | circ-SERPINE2 | cartilage | down | miR-1271-5p/ERG | ECM anabolism ↑ | 63 |
| cartilage | down | miR-495/ TGFBR2 | apoptosis ↓ | 64 | ||
| OA | circ-CDK14 | cartilage | down | miR-125a-5p/SMAD2 | apoptosis ↓, cell proliferation ↑ | 65 |
| OA | circ-ANKRD36 | cartilage | down | miR-599/CAS21 | inflammation ↓, apoptosis ↓ | 66 |
| OA | circ-PDE4D | cartilage | down | miR-103a-3p/FGF18 | ECM catabolism ↓ | 67 |
| OA | circ-0045714 | cartilage | down | miR-1936/IGF1R | cell proliferation ↑, ECM anabolism ↑ | 68 |
| OA | circ-9119 | cartilage | down | miR-127-5p/PTEN | apoptosis ↓ | 69 |
| OA | circ-GCN1L1 | synovium | up | miR-330-3p/TNFα | inflammation ↑, ECM catabolism ↑ | 71 |
| ONFH | CDR1as | BMSCs | up | miR-7-5p/WNT5B | osteogenesis ↓, adipogenesis ↑ | 36 |
| ONFH | circ-USP45 | BMSCs | up | miR-127-5p/PTEN | osteogenesis ↓ | 80 |
| OP | circ-RUNX2 | bone | down | miR-203/RUNX2 | osteogenesis ↑ | 88 |
| OP | circ-VCNGL1 | serum | down | miR-217-5p/RUNX2 | osteogenesis ↑ | 89 |
| OP | circ-0011269 | serum | down | miR-122/RUNX2 | osteogenesis ↑ | 90 |
| OP | circ-0076906 | serum/bone | down | miR-1305/OGN | osteogenesis ↑ | 91 |
| OP | circ-0016624 | serum | down | miR-98/BMP2 | osteogenesis ↑ | 92 |
| OP | circ-0006393 | BMSCs | down | miR-145-5p/FOXO1 | osteogenesis ↑ | 93 |
| OP | circ-0048211 | BMSCs | down | miR-93-5p/BMP2 | osteogenesis ↑ | 94 |
| OP | circ-SLC8A1 | Bone | down | miR-516b-5p/APAK2 | osteogenesis ↑ | 95 |
| OP | circ-YAP1 | BMSCs | down | miR-376b-3p/YAP1 | osteogenesis ↑ | 96 |
| OP | circ-0076690 | serum | down | miR-152/RUNX2 | osteogenesis ↑ | 97 |
| OP | circ-RTN4 | exosome | down | miR-1446a/TNFα | osteogenesis ↑ | 101 |
| OP | circ-0026827 | DPSCs | down | miR-188-3p/RUNX1 | osteogenesis ↑ | 102 |
| OP | circ-FOXP1 | ADMSCs | down | miR-330-5P/FOXP1 | osteogenesis ↑ | 103 |
| OP | circ-28313 | BMM | up | miR-195a/ CSF1 | osteoclastogenesis ↑ | 104 |
| RA | CDR1as | PBMCs | up | miR-7-5p/ mTOR | inflammation ↑ | 116 |
| RA | circ-FADS2 | cartilage | down | miR-498/mTOR | apoptosis↓, inflammation ↓ | 117 |
| RA | circ-09505 | PBMCs | up | miR-6089/AKT1 | inflammation ↑ | 118 |
| RA | circ-0001859 | synovium | up | miR-204/211/ATF2 | inflammation ↑ | 119 |
| RA | circ-0088036 | synovium | up | miR-140-3p/SIRT1 | inflammation ↑ | 120 |
OA: osteoarthritis; ONFH: osteonecrosis of the femoral head; OP: osteoporosis; RA: rheumatoid arthritis.
Figure 2.
A summary diagram of circRNAs in regulating osteoarthritis. (A) CircRNAs CDR1as, circ-0005105, circ-33186, circ-0136474, circ-100226, circ-CER, circ-PSM3, circ-Atp9b, circ-UBE2G1, circ-0092516, circ-CDH13, circ-TMBIM6, circ-RNF121, circ-VCAN, and circ-HIPK3 could significantly promote the progress of osteoarthritis by stimulating ECM catabolism, apoptosis, inflammation, but suppressing cell proliferation and ECM anabolism of chondrocytes thereby aggravating the progress of osteoarthritis. While circ-SERPINE2, circ-CDK14, circ-ANKRD36, circ-PDE4D, circ-0045714, and circ-9119 could inhibit the progress of osteogenesis by stimulating cell proliferation, ECM anabolism, but suppressing ECM catabolism, apoptosis, inflammation of chondrocytes thereby hindering the progress of osteoarthritis. (B) CircGCN1L1 could promote synoviocyte proliferation and chondrocyte apoptosis in osteoarthritis thereby aggravating the progress of osteoarthritis.
Additionally, synovitis is the main cause of joint pain in osteoarthritis 70. CircGCN1L1 was found upregulated in the synovium and play its roles by promoting synoviocyte proliferation and chondrocyte apoptosis in osteoarthritis. Silencing of circGCN1L1 attenuates the loss of condylar cartilage and subchondral bone via the circGCN1L1-miR-330-3p-TNF axis (Figure 2B) 71.
These osteoarthritis-related circRNAs may function as novel therapeutic targets for the treatment and prevention of osteoarthritis. Future work exploring the upstream regulation of aberrantly expressed circRNAs in osteoarthritis, as well as the molecular details of circRNA-protein interactions in osteoarthritis, will be required to further understand the therapeutic potential of osteoarthritis -related circRNAs.
CircRNAs and ONFH
Osteonecrosis is a class of orthopedic diseases that is caused by the interruption of blood flow, affecting over 20 million people worldwide 72-74. Due to its anatomical structure, the femoral head is particularly likely to undergo osteonecrosis 75. Common risk factors of ONFH include excessive use of steroids or alcohol, trauma, or sickle cell anemia 76, all of which reduce blood supply to the femoral head, thus resulting in bone necrosis and alteration 77. Important to the prevention of bone necrosis is bone regeneration via the differentiation and proliferation of bone marrow stromal cells (BMSCs) 78. However, an imbalance of osteogenesis and adipogenesis of BMSCs has been observed to disrupt bone remodeling. Studies have revealed that circRNAs are dysregulated during osteogenesis. Here, we review circRNAs that have been shown to play critical roles in regulating osteogenesis and adipogenesis, to shed light on a regulatory mechanism for steroid-induced osteonecrosis of the femoral head (SONFH).
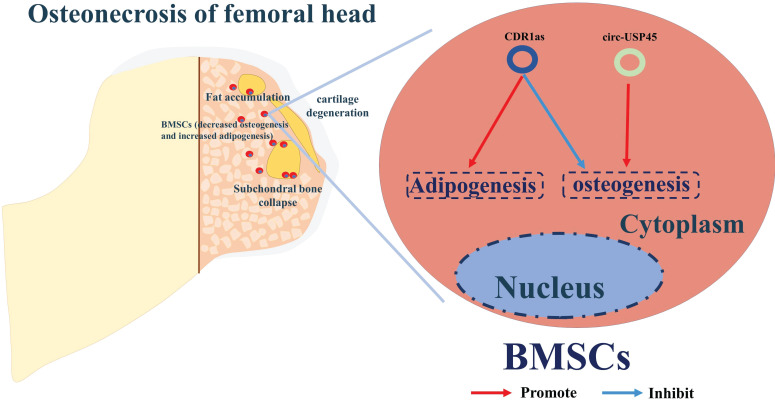
The correlation between circRNAs and ONFH is still in its infancy. To date, studies of the ONFH-related circRNAs have mainly highlighted their roles in osteogenesis or adipogenesis of BMSCs, the imbalance of which has a critical role in the progression of ONFH (Table 2) 36, 79, 80. By screening the circRNA expression profiles in BMSCs from patients with SONFH, and using bioinformatics and functional characterization assays, CDR1as was found up-regulated in SONFH-BMSCs, and thus could play a critical role in osteogenic/adipogenic differentiation disorders via the miR-7-5p/WNT5B axis of regulation (Figure 3) 36. Kuang et al. identified circUSP45 as an upregulated circRNA in BMSCs isolated from SONFH patients 80. RNA pull-down and dual luciferase reporter assays were performed to confirm that circUSP45 mainly localizes in the cytoplasm and directly interacts with miR-127-5p. Further experiments verified that circUSP45 upregulates the expression of PTEN and inhibits AKT pathway by sponging miR-127-5p, thereby suppressing the expression of osteogenic genes, such as bone morphogenetic protein-2 (BMP2) and runt-related transcription factor 2 (RUNX2). Additionally, the anti-bone metabolism function of circUSP45 was verified in vivo by a SONFH rat model.
Figure 3.
A summary diagram of circRNAs in regulating ONFH. CircUSP45 and CDR1as could promote the progress of ONFH by inhibiting osteogenesis but enhancing adipogenesis in BMSCs, thereby aggravating the progress of ONFH.
Therefore, silencing CDR1as and circUSP45 may promote bone metabolism and improve bone mass in SONFH progression. However, future work is required to understand more mechanistic details of regulatory circRNAs and their roles in the pathogenesis of ONFH (Figure 3).
CircRNAs and osteoporosis
Osteoporosis is a systemic and metabolic skeletal disorder affecting with more than 200 million individuals affected worldwide 81, 82. Osteoblastic bone formation and osteoclastic bone resorption dynamically maintain bone homeostasis 83. Osteoporosis is typically characterized by decreased osteoblastic activity and increased osteoclastic activity 84. During osteoporosis, BMSCs, which are precursors to osteoblasts, have been shown to have a lower osteogenic differentiation potential 85. Regulatory mechanisms underlying osteoporosis are complex and involve a number of pathways, including circRNA mediation pathway.
Diagnostic circRNAs of osteoporosis
Here, we summarized circRNAs that are abnormally expressed in human bodily fluids from osteoporosis patients. These circRNAs may be promising candidates for clinical diagnosis of osteoporosis. In particular, circ_0002060 was shown by Huang et al. via microarray and bioinformatic analyses, to be upregulated in clinical samples, suggesting its use as a potential biomarker (Table 1) 86, 87.
Regulatory circRNAs of osteoporosis
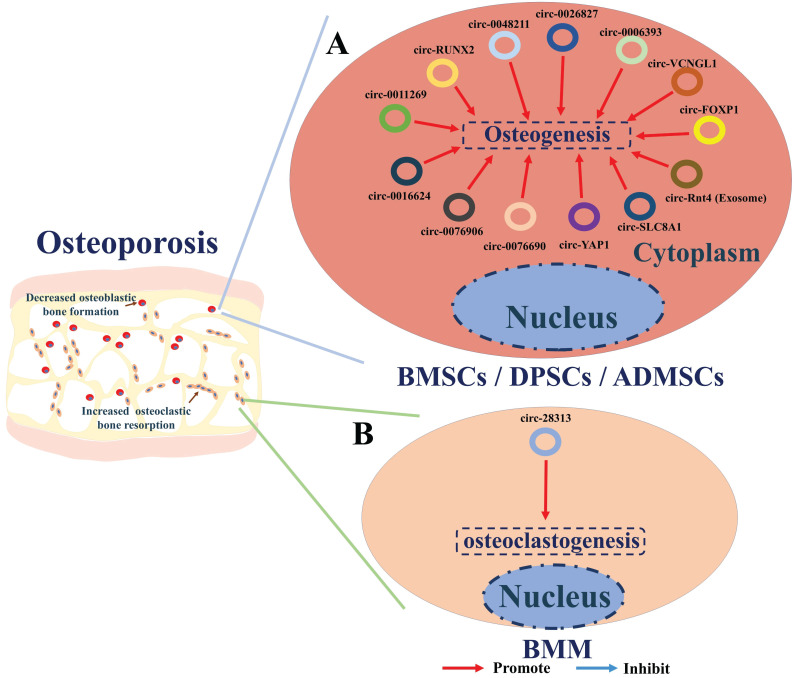
CircRNAs participate in the regulation of osteoporosis via several ways (Table 2). Recent researches have screened the circRNAs expression profile in bone and serum of patients with osteoporosis. The majority of osteoporosis -related circRNAs are involved in regulating osteogenesis by sponging miRNAs and consequently regulating the expression or activity of downstream osteogenesis genes. Circ-RUNX2 88, circ-VANGL1 89, circ-0011269 90, circ-0076906 91, circRNA-0016624 92, circ-0006393 93, circRNA-0048211 94, circ-SLC8A1 95, circ-YAP1 96 and circ-0076690 97 are found down-regulated in osteoporosis and could enhance osteogenesis of BMSCs by sponging miRNAs and subsequently upregulating the expression and activities of osteogenetic genes (such as RUNX2, BMP2, OPN, OCN, OGN, FOXO1, APAK2 or ALP) (Figure 4A). Exosomes of BMSCs (BMSCs-Exos) have also been shown to function in bone regeneration 98-100, while exosomes-derived from circ-Rtn4 have been shown to promote osteogenesis by targeting miR-146a 101. Studies performed by Cao et al. revealed that the expression of miR-146a is positively correlated with TNF-α, a cytokine that serves as a key regulator of osteoporosis pathology. Additionally, they showed that circ-Rtn4 attenuates TNF-α-induced cytotoxicity and apoptosis in MC3T3-E1 cells by acting as a sponge for miR-146a, implicating Rtn4-Exos as a promising therapeutic candidate for osteoporosis (Figure 4A) 101.
Figure 4.
A summary diagram of circRNAs in regulating osteoporosis. (A) CircRNAs circ-RUNX2, circ-VANGL1, circ-0011269, circ-0076906, circRNA-0016624, circ-0006393, circRNA-0048211, circ-SLC8A1, circ-YAP1, circ-0076690, and circ-Rtn4 (exosomes) could promote the osteogenesis of BMSCs thereby hindering the progress of osteoporosis. Circ_0026827 could promote the osteogenesis of human dental pulp stem cells (DPSCs) thereby hindering the progress of osteoporosis. CircFOXP1 could promote the osteogenesis of adipose-derived mesenchymal stem cells (ADMSCs) thereby hindering the progress of osteoporosis. (B) While circ-28313 could promote the osteoclastogenesis of bone marrow monocyte /macrophage (BMM) thereby aggravating the progress of osteoporosis.
Some studies have also shown the regulatory mechanism of circRNAs in other types of stem cells. The effect of circ_0026827 on human dental pulp stem cells (DPSCs) during osteogenesis has been examined by Ji et al. 102 to seek novel therapeutics for osteoporosis. These studies showed that circ_0026827 function as a sponge of miR-188-3p to promote osteoblastic differentiation of DPSCs via the upregulation of Beclin-1-mediated autophagy and RUNX1 signaling pathways. CircFOXP1 were found to play critical roles in promoting osteogenic differentiation of adipose-derived mesenchymal stem cells (ADMSCs) in osteoporosis by targeting miR-33a-5p. In vivo and in vitro evidence indicates circFOXP1 could enhance the expression of FOXP1, thereby promote the osteogenic differentiation of ADMSCs and bone formation in osteoporosis (Figure 4A) 103.
To date, circRNA_28313 is the only circRNA that has been shown to be a regulator of osteoclast resorption. Recent studies by Chen et al. 104 have examined mechanisms, involving circRNA regulation, of excessive bone resorption caused by osteoclasts during their differentiation. The authors used RANKL+CSF1-treated bone marrow monocyte/macrophage (BMM) cells to mimic osteoclasts during the progression of osteoporosis. Their results showed that circRNA_28313 is dramatically overexpressed upon RANKL+CSF1 stimulation, while miR-195a is down-expressed. Further bioinformatics and experimental validation both in vitro and in vivo showed that circRNA_28313 interacts with miR-195a and consequently regulates the colony-stimulating factor (CSF1) gene. High expression level of CSF1 in turn, promotes the expression of downstream proteins such as PU.1 (a factor related to osteoclast differentiation), nuclear factor of activated T cells c1 (NF-ATc1), tartrate-resistant acid phosphatase (TRAP), and Cathepsin K (CTSK), thereby enhancing osteoclast-induced bone absorption (Figure 4B).
Thus, circRNAs can serve as powerful therapeutic candidates to promote bone regeneration and thus reverse the progression of osteoporosis. Moreover, a deeper understanding of these circRNAs can shed light on the mechanisms underlying regulation of circRNAs in bone metabolism.
CircRNAs and rheumatoid arthritis
Rheumatoid arthritis (RA) is a common chronic autoimmune disease characterized by inflammatory destruction that can cause serious cartilage and bone damage, affecting approximately 1% of the population worldwide 30. Several non-coding RNAs have been identified as regulators of RA via different pathways, but the exact mechanisms underlying the role of circRNAs in RA remain to be understood.
Diagnostic circRNAs of rheumatoid arthritis
Early diagnosis is critical to optimal therapeutic success for rheumatoid arthritis (RA). Existing studies have identified a several circRNAs that could serve as diagnostic biomarkers of RA, partially due to their increased stability in plasma, serum, or other biofluids 105. Here, we summarizes potential biomarkers identified by various studies (Table 1) 106-111.
Regulatory circRNAs of rheumatoid arthritis
The main pathological features of RA are autoimmune response and inflammation 112. Recently, a number of circRNAs have been implicated in RA pathogenesis (Table 2), but their function and hidden molecular mechanism in immune and inflammation regulation still remains little known 30.
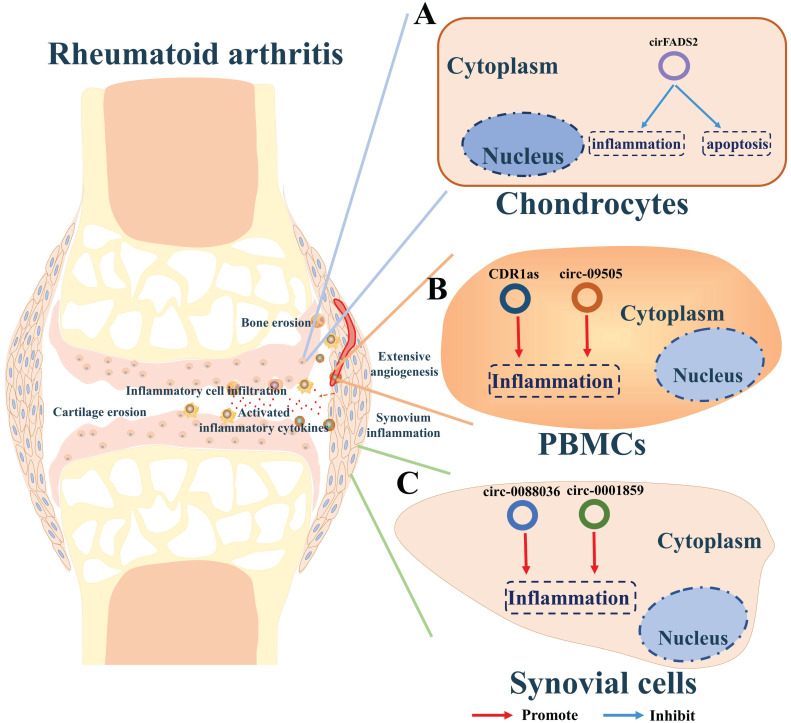
Given that the phosphatidylinositol-3-kinase/AKT/mTOR (PI3K/AKT/mTOR) signaling pathway plays a crucial role in cellular proliferation and inflammatory responses 113, 114, circFADS2 and ciRS-7 have been implicated in the initiation and progression of RA by regulating these pathways. CiRS-7 promotes the inflammation of PBMCs by sponging miR-7 to upregulate mTOR while circFADS2 protects LPS-treated chondrocytes (RA model cells) from apoptosis by mediating mTOR expression via sponging miR-498 (Figure 5A and B) 115-117. CircRNA_09505 is an up-regulated circRNA which can promote AKT1 expression via miR-6089/IκBα/NFκB signaling pathway in macrophages, thereby aggravating inflammation and joint damage in RA (Figure 5B) 118.
Figure 5.
A summary diagram of circRNAs in regulating RA. (A) CircRNA circFADS2 could protect the chondrocytes from apoptosis and inflammation thereby hindering the progress of rheumatoid arthritis. (B) CDR1as and circ-09505 could enhance the inflammation of PBMCs thereby promoting the progress of rheumatoid arthritis. (C) Circ-0001859 and circ-0088036 could promote the inflammation of synovium in rheumatoid arthritis.
Joint swelling and pain reflect synovial membrane inflammation in RA, resulting from the immune activation and infiltration of leucocyte. Li and colleagues examined the expression of circRNAs in synovial tissues and screened out circ-0001859 as a critical RA-related circRNA 119. Their further studies indicated that circ-0001859 could up-regulate ATF2 via sponging miR-204/211 in synovial sarcoma cells (SW982 cells). Silencing circ_0001859 reduced the hyper inflammatory activity in the synovial tissue and alleviates the pathogenesis of RA (Figure 5C). Circ-0088036 was found to be aberrantly upregulated in fibroblast-like synoviocytes (FLSs) in RA 120. Circ-0088036 could promote the proliferation and migration of RA-FLSs via miR-140-3p/SIRT1 axis, subsequently promoting RA progression (Figure 5C).
The studies described above highlight the effects of circRNAs on RA, but a great deal remains to be understood. Future studies are required to further understand the underlying mechanisms of regulation of RA progression by circRNAs.
Conclusions and future perspective
CircRNA is a class of noncoding RNA molecules with a closed loop structure formed by covalent bonds, which can protect them from degradation by most RNases 121. Emerging studies have exploited highly-accurate circRNA biomarkers in human body fluids for diagnosis and prognosis of some diseases, such as cancer and cardiovascular diseases 122, 123. In recent years, studies have also revealed the potential value of circRNAs in clinical treatment of various diseases, including skeletal and chondral disorders. In this review, we have reviewed the regulatory role of circRNA in skeletal and chondral disorders and summarized findings of several circRNAs that play critical roles in the process of osteoarthritis, ONFH, osteoporosis, or RA. These findings may provide further insight into the mechanistic details of the role of circRNAs in bone or cartilage metabolism, and highlight potential targets for the clinical treatment of orthopedic diseases. Nevertheless, the molecular functions of circRNAs in skeletal and chondral disorders still remain largely enigmatic. Their regulatory mechanism in skeletal and chondral disorders by regulating transcription, binding protein, and encoding protein or peptides still remains unknown, which is worth to be further explored in later research on skeletal and chondral disorders.
Despite recent discoveries summarized above, many challenges remain to be overcome. The functional verification of circRNAs implicated in skeletal and chondral disorders, has been mostly in vitro. In vivo studies are a greater challenge due to the lack of stable and specific delivery vehicles. Recent studies have discovered some biological vehicle that can be loaded with circRNA (or si-circRNA) for in vivo therapy, including extracellular vesicles (EVs) and adeno-associated viral (AAV) vectors. EVs are a heterogeneous group of lipid bilayer-enclosed nanosized vesicles releasing from various types of cells 124. Many studies have highlighted the application of EVs in transporting circRNA to specific tissues by modification 125, 126. AAV is a non-pathogenic member of the Parvovirus family which can deliver circRNA producing transgenes 127. Studies have also revealed the potential value of recombinant AAV in targeting therapy via genetic modification 128. These advances may shed new lights on the application of circRNA in the treatment of skeletal and chondral disorders. We believe that an in-depth understanding and correct application of circRNAs in clinical practice will make a giant progress in the treatment of skeletal and chondral disorders in the near future.
Acknowledgments
This work was supported by Shenzhen Municipal Science and Technology Innovation Committee Project (JCYJ20180305164659637, JCYJ20190806160407178, JCYJ20180305164544288, JSGG20180504170427135, SGLH20180625141602256, JCYJ2017041362104773).
Author contributions
Conception and first draft written by Gaoyang Chen and Wanze Tang with contributions from Shang Wang, Canling Long, Dazhi Yang and Songlin Peng. Gaoyang Chen, Wanze Tang, Shang Wang, Canling Long, Dazhi Yang and Songlin Peng revised the manuscript and approved the final version of the manuscript.
Abbreviations
- circRNAs
circular RNAs
- ncRNAs
non-coding RNAs
- EcRNAs
exonic circRNAs
- ciRNAs
intronic circRNAs
- EIcRNAs
exon-intron circRNAs
- OA
osteoarthritis
- ONFH
osteonecrosis of the femoral head
- OP
osteoporosis
- RA
rheumatoid arthritis
- ECM
extracellular matrix
- ERG
E26 transformation-specific-related gene
- IL-1β
interleukin-1 beta
- NAMPT
nicotinamide phosphoribosyltransferase
- Col II
type II collagen
- TNF-α
tumor necrosis factor
- IGF1R
insulin-like growth factor 1 receptor
- LPS
lipopolysaccharide
- BMSCs
bone marrow stromal cells
- PTEN
phosphate and tension homology deleted on chromsome ten
- BMP2
bone morphogenetic protein-2
- RUNX2
runt-related transcription factor 2
- BMM
bone marrow monocyte/macrophage
- RANKL
receptor activator of nuclear factor-κB ligand
- CSF1
colony-stimulating factor
- NF-ATc1
nuclear factor of activated T cells c1
- TRAP
tartrate-resistant acid phosphatase
- CTSK
cathepsin K
- OCN
osteocalcin
- OPN
osteopontin
- BSP
bone sialoprotein
- ALP
alkaline phosphatase
- OGN
osteoglycin
- DPSCs
human dental pulp stem cells
- FOXO1
forkhead box o1
- BMSCs-Exos
exosomes of BMSCs
- PBMCs
peripheral blood mononuclear cells
- ATF2
activating transcription factor 2
References
- 1.Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei L, Sun J, Zhang N. et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19(1):62. doi: 10.1186/s12943-020-01185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashwal-Fluss R, Meyer M, Pamudurti NR. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Liang D, Tatomer DC, Luo Z. et al. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol Cell. 2017;68(5):940–954.e3. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 6.Zaiou M. Circular RNAs as Potential Biomarkers and Therapeutic Targets for Metabolic Diseases. Adv Exp Med Biol. 2019;1134:177–191. doi: 10.1007/978-3-030-12668-1_10. [DOI] [PubMed] [Google Scholar]
- 7.Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9-10):639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–40. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 10.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–60. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 11.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–6. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 14.Conn VM, Hugouvieux V, Nayak A. et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Su H, Zhang J, Liu Y, Feng C, Han F. Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol. 2020;18(1):e3000582. doi: 10.1371/journal.pbio.3000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- 17.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146(3):353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holdt LM, Stahringer A, Sass K. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun YM, Wang WT, Zeng ZC. et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. 2019;134(18):1533–1546. doi: 10.1182/blood.2019000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legnini I, Di Timoteo G, Rossi F. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pamudurti NR, Bartok O, Jens M. et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9(1):2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Fan X, Mao M. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 26.Huang X, Cen X, Zhang B. et al. Prospect of circular RNA in osteogenesis: A novel orchestrator of signaling pathways. J Cell Physiol. 2019;234(12):21450–21459. doi: 10.1002/jcp.28866. [DOI] [PubMed] [Google Scholar]
- 27.Li HZ, Lin Z, Xu XH, Lin N, Lu HD. The potential roles of circRNAs in osteoarthritis: a coming journey to find a treasure. Biosci Rep. 2018. 38(5) [DOI] [PMC free article] [PubMed]
- 28.Appleton CT. Osteoarthritis year in review 2017: biology. Osteoarthritis Cartilage. 2018;26(3):296–303. doi: 10.1016/j.joca.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Yang B, Weng X, Tse G, Chan M, Wu W. Emerging roles of MicroRNAs in osteonecrosis of the femoral head. Cell Prolif. 2018. 51(1) [DOI] [PMC free article] [PubMed]
- 30.Wang J, Yan S, Yang J, Lu H, Xu D, Wang Z. Non-coding RNAs in Rheumatoid Arthritis: From Bench to Bedside. Front Immunol. 2019;10:3129. doi: 10.3389/fimmu.2019.03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song W, Xie J, Li J, Bao C, Xiao Y. The Emerging Roles of Long Noncoding RNAs in Bone Homeostasis and Their Potential Application in Bone-Related Diseases. DNA Cell Biol. 2020;39(6):926–937. doi: 10.1089/dna.2020.5391. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q, Zhang X, Hu X. et al. Emerging Roles of circRNA Related to the Mechanical Stress in Human Cartilage Degradation of Osteoarthritis. Mol Ther Nucleic Acids. 2017;7:223–230. doi: 10.1016/j.omtn.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin D, Wu X, Yu H. et al. Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am J Transl Res. 2018;10(5):1498–1510. [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Yang Q, Chen G. et al. LncRNA expression profiling of BMSCs in osteonecrosis of the femoral head associated with increased adipogenic and decreased osteogenic differentiation. Sci Rep. 2018;8(1):9127. doi: 10.1038/s41598-018-27501-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou ZB, Huang GX, Fu Q. et al. circRNA.33186 Contributes to the Pathogenesis of Osteoarthritis by Sponging miR-127-5p. Mol Ther. 2019;27(3):531–541. doi: 10.1016/j.ymthe.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G, Wang Q, Li Z. et al. Circular RNA CDR1as promotes adipogenic and suppresses osteogenic differentiation of BMSCs in steroid-induced osteonecrosis of the femoral head. Bone. 2020;133:115258. doi: 10.1016/j.bone.2020.115258. [DOI] [PubMed] [Google Scholar]
- 37.Ren S, Lin P, Wang J. et al. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as an miRNA Sponge. Mol Ther Methods Clin Dev. 2020;18:215–229. doi: 10.1016/j.omtm.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panoutsopoulou K, Zeggini E. Advances in osteoarthritis genetics. J Med Genet. 2013;50(11):715–24. doi: 10.1136/jmedgenet-2013-101754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang HS, Kim HA. Chondrocyte Apoptosis in the Pathogenesis of Osteoarthritis. Int J Mol Sci. 2015;16(11):26035–54. doi: 10.3390/ijms161125943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Adamo S, Cetrullo S, Minguzzi M, Silvestri Y, Borzì RM, Flamigni F. MicroRNAs and Autophagy: Fine Players in the Control of Chondrocyte Homeostatic Activities in Osteoarthritis. Oxid Med Cell Longev. 2017;2017:3720128. doi: 10.1155/2017/3720128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li C, Luo J, Xu X. et al. Single cell sequencing revealed the underlying pathogenesis of the development of osteoarthritis. Gene. 2020;757:144939. doi: 10.1016/j.gene.2020.144939. [DOI] [PubMed] [Google Scholar]
- 42.Lahner M, von Schulze Pellengahr C, Lichtinger TK. et al. The role of arthroscopy in patients with persistent hip pain after total hip arthroplasty. Technol Health Care. 2013;21(6):599–606. doi: 10.3233/THC-130761. [DOI] [PubMed] [Google Scholar]
- 43.Sokolove J, Lepus CM. Role of inflammation in the pathogenesis of osteoarthritis: latest findings and interpretations. Ther Adv Musculoskelet Dis. 2013;5(2):77–94. doi: 10.1177/1759720X12467868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komori T. Cell Death in Chondrocytes, Osteoblasts, and Osteocytes. Int J Mol Sci. 2016. 17(12) [DOI] [PMC free article] [PubMed]
- 45.Maass PG, Glažar P, Memczak S. et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 2017;95(11):1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu F, Xie C, Sun J, Feng H, Huang X. Circular RNA expression profiles in synovial fluid: a promising new class of diagnostic biomarkers for osteoarthritis. Int J Clin Exp Pathol. 2018;11(3):1338–1346. [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Wu C, Yang Y, Ren Z, Lammi MJ, Guo X. Preliminary Exploration of hsa_circ_0032131 Levels in Peripheral Blood as a Potential Diagnostic Biomarker of Osteoarthritis. Genet Test Mol Biomarkers. 2019;23(10):717–721. doi: 10.1089/gtmb.2019.0036. [DOI] [PubMed] [Google Scholar]
- 48.Glyn-Jones S, Palmer AJ, Agricola R. et al. Osteoarthritis. Lancet. 2015;386(9991):376–87. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Zhang C, Hu C, Luo C, Zhong B, Yu X. Circular RNA-CDR1as acts as the sponge of microRNA-641 to promote osteoarthritis progression. J Inflamm (Lond) 2020;17:8. doi: 10.1186/s12950-020-0234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Y, Zhang Y, Zhang Y, Wang JJ. CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol Int. 2017;41(12):1283–1289. doi: 10.1002/cbin.10761. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Z, Du D, Chen A, Zhu L. Circular RNA expression profile of articular chondrocytes in an IL-1β-induced mouse model of osteoarthritis. Gene. 2018;644:20–26. doi: 10.1016/j.gene.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 52.Li Z, Yuan B, Pei Z. et al. Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J Cell Mol Med. 2019;23(10):6554–6564. doi: 10.1111/jcmm.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Q, Zhang X, Hu X. et al. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 'Sponge' in Human Cartilage Degradation. Sci Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ni JL, Dang XQ, Shi ZB. CircPSM3 inhibits the proliferation and differentiation of OA chondrocytes by targeting miRNA-296-5p. Eur Rev Med Pharmacol Sci. 2020;24(7):3467–3475. doi: 10.26355/eurrev_202004_20805. [DOI] [PubMed] [Google Scholar]
- 55.Zhou ZB, Du D, Huang GX, Chen A, Zhu L. Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene. 2018;646:203–209. doi: 10.1016/j.gene.2017.12.064. [DOI] [PubMed] [Google Scholar]
- 56.Chen G, Liu T, Yu B, Wang B, Peng Q. CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis. Cell Cycle. 2020;19(13):1696–1705. doi: 10.1080/15384101.2020.1772545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Z, Ma W, Xiao J, Dai X, Ling W. CircRNA_0092516 regulates chondrocyte proliferation and apoptosis in osteoarthritis through the miR-337-3p/PTEN axis. J Biochem. 2020. [DOI] [PubMed]
- 58.Zhou Z, Ma J, Lu J, Chen A, Zhu L. Circular RNA CircCDH13 contributes to the pathogenesis of osteoarthritis via CircCDH13/miR-296-3p/PTEN axis. J Cell Physiol. 2020. [DOI] [PubMed]
- 59.Bai ZM, Kang MM, Zhou XF, Wang D. CircTMBIM6 promotes osteoarthritis-induced chondrocyte extracellular matrix degradation via miR-27a/MMP13 axis. Eur Rev Med Pharmacol Sci. 2020;24(15):7927–7936. doi: 10.26355/eurrev_202008_22475. [DOI] [PubMed] [Google Scholar]
- 60.Wang T, Hao Z, Liu C. et al. LEF1 mediates osteoarthritis progression through circRNF121/miR-665/MYD88 axis via NF-кB signaling pathway. Cell Death Dis. 2020;11(7):598. doi: 10.1038/s41419-020-02769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma HR, Mu WB, Zhang KY, Zhou HK, Jiang RD, Cao L. CircVCAN regulates the proliferation and apoptosis of osteoarthritis chondrocyte through NF-κB signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(12):6517–6525. doi: 10.26355/eurrev_202006_21635. [DOI] [PubMed] [Google Scholar]
- 62.Wu Q, Yuan ZH, Ma XB, Tang XH. Low expression of CircRNA HIPK3 promotes osteoarthritis chondrocyte apoptosis by serving as a sponge of miR-124 to regulate SOX8. Eur Rev Med Pharmacol Sci. 2020;24(15):7937–7945. doi: 10.26355/eurrev_202008_22476. [DOI] [PubMed] [Google Scholar]
- 63.Shen S, Wu Y, Chen J. et al. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann Rheum Dis. 2019;78(6):826–836. doi: 10.1136/annrheumdis-2018-214786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q, Qiao X, Xia W. CircSERPINE2 weakens IL-1β-caused apoptosis and extracellular matrix degradation of chondrocytes by regulating miR-495/TGFBR2 axis. Biosci Rep. 2020. 40(11) [DOI] [PMC free article] [PubMed]
- 65.Shen P, Yang Y, Liu G. et al. CircCDK14 protects against Osteoarthritis by sponging miR-125a-5p and promoting the expression of Smad2. Theranostics. 2020;10(20):9113–9131. doi: 10.7150/thno.45993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou JL, Deng S, Fang HS, Du XJ, Peng H, Hu QJ. Circular RNA circANKRD36 regulates Casz1 by targeting miR-599 to prevent osteoarthritis chondrocyte apoptosis and inflammation. J Cell Mol Med. 2020. [DOI] [PMC free article] [PubMed]
- 67.Wu Y, Hong Z, Xu W, Circular RNA circPDE4D Protects against Osteoarthritis by Binding to miR-103a-3p and Regulating FGF18. Mol Ther. 2020. [DOI] [PMC free article] [PubMed]
- 68.Li BF, Zhang Y, Xiao J. et al. Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Hum Cell. 2017;30(4):311–318. doi: 10.1007/s13577-017-0177-7. [DOI] [PubMed] [Google Scholar]
- 69.Chen C, Yin P, Hu S, Sun X, Li B. Circular RNA-9119 protects IL-1β-treated chondrocytes from apoptosis in an osteoarthritis cell model by intercepting the microRNA-26a/PTEN axis. Life Sci. 2020;256:117924. doi: 10.1016/j.lfs.2020.117924. [DOI] [PubMed] [Google Scholar]
- 70.Mathiessen A, Conaghan PG. Synovitis in osteoarthritis: current understanding with therapeutic implications. Arthritis Res Ther. 2017;19(1):18. doi: 10.1186/s13075-017-1229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu H, Hu Y, Wang C, Zhang X, He D. CircGCN1L1 promotes synoviocyte proliferation and chondrocyte apoptosis by targeting miR-330-3p and TNF-α in TMJ osteoarthritis. Cell Death Dis. 2020;11(4):284. doi: 10.1038/s41419-020-2447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang T, Azeddine B, Mah W, Harvey EJ, Rosenblatt D, Séguin C. Osteonecrosis of the femoral head: genetic basis. Int Orthop. 2019;43(3):519–530. doi: 10.1007/s00264-018-4172-8. [DOI] [PubMed] [Google Scholar]
- 73.Grecula MJ. CORR Insights®: Which Classification System Is Most Useful for Classifying Osteonecrosis of the Femoral Head. Clin Orthop Relat Res. 2018;476(6):1250–1252. doi: 10.1097/01.blo.0000533640.75452.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones LC, Solano M, Sterling RS, Oni JK, Khanuja HS. Moving Forward in Osteonecrosis: What Research Is Telling Us. Instr Course Lect. 2020;69:111–28. [PubMed] [Google Scholar]
- 75.Liu F, Wang W, Yang L. et al. An epidemiological study of etiology and clinical characteristics in patients with nontraumatic osteonecrosis of the femoral head. J Res Med Sci. 2017;22:15. doi: 10.4103/1735-1995.200273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lieberman JR, Engstrom SM, Meneghini RM, SooHoo NF. Which factors influence preservation of the osteonecrotic femoral head. Clin Orthop Relat Res. 2012;470(2):525–34. doi: 10.1007/s11999-011-2050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petek D, Hannouche D, Suva D. Osteonecrosis of the femoral head: pathophysiology and current concepts of treatment. EFORT Open Rev. 2019;4(3):85–97. doi: 10.1302/2058-5241.4.180036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhun W, Donghai L, Zhouyuan Y, Haiyan Z, Pengde K. Efficiency of Cell Therapy to GC-Induced ONFH: BMSCs with Dkk-1 Interference Is Not Superior to Unmodified BMSCs. Stem Cells Int. 2018;2018:1340252. doi: 10.1155/2018/1340252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Houdek MT, Wyles CC, Packard BD, Terzic A, Behfar A, Sierra RJ. Decreased Osteogenic Activity of Mesenchymal Stem Cells in Patients With Corticosteroid-Induced Osteonecrosis of the Femoral Head. J Arthroplasty. 2016;31(4):893–8. doi: 10.1016/j.arth.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 80.Kuang MJ, Xing F, Wang D, Sun L, Ma JX, Ma XL. CircUSP45 inhibited osteogenesis in glucocorticoid-induced osteonecrosis of femoral head by sponging miR-127-5p through PTEN/AKT signal pathway: Experimental studies. Biochem Biophys Res Commun. 2019;509(1):255–261. doi: 10.1016/j.bbrc.2018.12.116. [DOI] [PubMed] [Google Scholar]
- 81.Wang H, Zhou K, Xiao F. et al. Identification of circRNA-associated ceRNA network in BMSCs of OVX models for postmenopausal osteoporosis. Sci Rep. 2020;10(1):10896. doi: 10.1038/s41598-020-67750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beck-Cormier S, Lelliott CJ, Logan JG, Lafont DT, Merametdjian L, Leitch VD. et al. Slc20a2, Encoding the Phosphate Transporter PiT2, Is an Important Genetic Determinant of Bone Quality and Strength. J Bone Miner Res. 2019;34:1101–14. doi: 10.1002/jbmr.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alliston T, Derynck R. Medicine: interfering with bone remodelling. Nature. 2002;416(6882):686–7. doi: 10.1038/416686a. [DOI] [PubMed] [Google Scholar]
- 84.Tanaka S. RANKL-Independent Osteoclastogenesis: A Long-Standing Controversy. J Bone Miner Res. 2017;32(3):431–433. doi: 10.1002/jbmr.3092. [DOI] [PubMed] [Google Scholar]
- 85.Li CJ, Cheng P, Liang MK. et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125(4):1509–22. doi: 10.1172/JCI77716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Y, Xie J, Li E. Comprehensive circular RNA profiling reveals circ_0002060 as a potential diagnostic biomarkers for osteoporosis. J Cell Biochem. 2019;120(9):15688–15694. doi: 10.1002/jcb.28838. [DOI] [PubMed] [Google Scholar]
- 87.Xiang S, Wu Y, Shi H, Xue L, Luo K, Ding Y. Circular RNA hsa_circ_0001445 in plasma as a novel biomarker for osteoporosis in postmenopausal women. Biomark Med. 2020;14(16):1599–1607. doi: 10.2217/bmm-2020-0447. [DOI] [PubMed] [Google Scholar]
- 88.Yin Q, Wang J, Fu Q, Gu S, Rui Y. CircRUNX2 through has-miR-203 regulates RUNX2 to prevent osteoporosis. J Cell Mol Med. 2018;22(12):6112–6121. doi: 10.1111/jcmm.13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang L, Zeng Z, Kang N, Yang JC, Wei X, Hai Y. Circ-VANGL1 promotes the progression of osteoporosis by absorbing miRNA-217 to regulate RUNX2 expression. Eur Rev Med Pharmacol Sci. 2019;23(3):949–957. doi: 10.26355/eurrev_201902_16981. [DOI] [PubMed] [Google Scholar]
- 90.Xu X, Chen Y, Tan B, Wang D, Yuan Z, Wang F. Circular RNA circ_0011269 sponges miR-122 to regulate RUNX2 expression and promotes osteoporosis progression. J Cell Biochem. 2020. [DOI] [PubMed]
- 91.Wen J, Guan Z, Yu B, Guo J, Shi Y, Hu L. Circular RNA hsa_circ_0076906 competes with OGN for miR-1305 biding site to alleviate the progression of osteoporosis. Int J Biochem Cell Biol. 2020;122:105719. doi: 10.1016/j.biocel.2020.105719. [DOI] [PubMed] [Google Scholar]
- 92.Yu L, Liu Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem Biophys Res Commun. 2019;516(2):546–550. doi: 10.1016/j.bbrc.2019.06.087. [DOI] [PubMed] [Google Scholar]
- 93.Wang XB, Li PB, Guo SF. et al. circRNA_0006393 promotes osteogenesis in glucocorticoid-induced osteoporosis by sponging miR-145-5p and upregulating FOXO1. Mol Med Rep. 2019;20(3):2851–2858. doi: 10.3892/mmr.2019.10497. [DOI] [PubMed] [Google Scholar]
- 94.Qiao L, Li CG, Liu D. CircRNA_0048211 protects postmenopausal osteoporosis through targeting miRNA-93-5p to regulate BMP2. Eur Rev Med Pharmacol Sci. 2020;24(7):3459–3466. doi: 10.26355/eurrev_202004_20804. [DOI] [PubMed] [Google Scholar]
- 95.Lin C, Zhong W, Yan W, Yang J, Zheng W, Wu Q. Circ-SLC8A1 regulates osteoporosis through blocking the inhibitory effect of miR-516b-5p on AKAP2 expression. J Gene Med. 2020;22(11):e3263. doi: 10.1002/jgm.3263. [DOI] [PubMed] [Google Scholar]
- 96.Huang Y, Xiao D, Huang S. et al. Circular RNA YAP1 attenuates osteoporosis through up-regulation of YAP1 and activation of Wnt/β-catenin pathway. Biomed Pharmacother. 2020;129:110365. doi: 10.1016/j.biopha.2020.110365. [DOI] [PubMed] [Google Scholar]
- 97.Han S, Kuang M, Sun C, Wang H, Wang D, Liu Q. Circular RNA hsa_circ_0076690 acts as a prognostic biomarker in osteoporosis and regulates osteogenic differentiation of hBMSCs via sponging miR-152. Aging (Albany NY) 2020;12(14):15011–15020. doi: 10.18632/aging.103560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao P, Xiao L, Peng J, Qian YQ, Huang CC. Exosomes derived from bone marrow mesenchymal stem cells improve osteoporosis through promoting osteoblast proliferation via MAPK pathway. Eur Rev Med Pharmacol Sci. 2018;22(12):3962–3970. doi: 10.26355/eurrev_201806_15280. [DOI] [PubMed] [Google Scholar]
- 99.Murphy C, Withrow J, Hunter M. et al. Emerging role of extracellular vesicles in musculoskeletal diseases. Mol Aspects Med. 2018;60:123–128. doi: 10.1016/j.mam.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie Y, Hu JH, Wu H, Huang ZZ, Yan HW, Shi ZY. Bone marrow stem cells derived exosomes improve osteoporosis by promoting osteoblast proliferation and inhibiting cell apoptosis. Eur Rev Med Pharmacol Sci. 2019;23(3):1214–1220. doi: 10.26355/eurrev_201902_17014. [DOI] [PubMed] [Google Scholar]
- 101.Zhang B, Yi J, Zhang CL. et al. MiR-146a inhibits proliferation and induces apoptosis in murine osteoblastic MC3T3-E1 by regulating Bcl2. Eur Rev Med Pharmacol Sci. 2017;21(17):3754–3762. [PubMed] [Google Scholar]
- 102.Ji F, Zhu L, Pan J. et al. hsa_circ_0026827 Promotes Osteoblast Differentiation of Human Dental Pulp Stem Cells Through the Beclin1 and RUNX1 Signaling Pathways by Sponging miR-188-3p. Front Cell Dev Biol. 2020;8:470. doi: 10.3389/fcell.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen W, Sun B, Zhou C, Ming W, Zhang S, Wu X. CircFOXP1/FOXP1 promotes osteogenic differentiation in adipose-derived mesenchymal stem cells and bone regeneration in osteoporosis via miR-33a-5p. J Cell Mol Med. 2020;24(21):12513–12524. doi: 10.1111/jcmm.15792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen X, Ouyang Z, Shen Y. et al. CircRNA_28313/miR-195a/CSF1 axis modulates osteoclast differentiation to affect OVX-induced bone absorption in mice. RNA Biol. 2019;16(9):1249–1262. doi: 10.1080/15476286.2019.1624470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Luo Q, Zhang L, Li X. et al. Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clin Exp Immunol. 2018;194(1):118–124. doi: 10.1111/cei.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zheng F, Yu X, Huang J, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep. 2017;16(6):8029–8036. doi: 10.3892/mmr.2017.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wen J, Liu J, Zhang P, RNA-seq reveals the circular RNA and miRNA expression profile of peripheral blood mononuclear cells in patients with rheumatoid arthritis. Biosci Rep. 2020. 40(4) [DOI] [PMC free article] [PubMed]
- 109.Luo Q, Liu J, Fu B. et al. Circular RNAs Hsa_circ_0002715 and Hsa_circ_0035197 in Peripheral Blood Are Novel Potential Biomarkers for New-Onset Rheumatoid Arthritis. Dis Markers. 2019;2019:2073139. doi: 10.1155/2019/2073139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo Q, Zeng L, Zeng L. et al. Expression and clinical significance of circular RNAs hsa_circ_0000175 and hsa_circ_0008410 in peripheral blood mononuclear cells from patients with rheumatoid arthritis. Int J Mol Med. 2020;45(4):1203–1212. doi: 10.3892/ijmm.2020.4498. [DOI] [PubMed] [Google Scholar]
- 111.Ouyang Q, Wu J, Jiang Z. et al. Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Rheumatoid Arthritis Patients. Cell Physiol Biochem. 2017;42(2):651–659. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 112.Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 113.Mitra A, Raychaudhuri SK, Raychaudhuri SP. IL-22 induced cell proliferation is regulated by PI3K/Akt/mTOR signaling cascade. Cytokine. 2012;60(1):38–42. doi: 10.1016/j.cyto.2012.06.316. [DOI] [PubMed] [Google Scholar]
- 114.Wu X, Long L, Liu J. et al. Gambogic acid suppresses inflammation in rheumatoid arthritis rats via PI3K/Akt/mTOR signaling pathway. Mol Med Rep. 2017;16(5):7112–7118. doi: 10.3892/mmr.2017.7459. [DOI] [PubMed] [Google Scholar]
- 115.Yu T, Li YJ, Bian AH. et al. The regulatory role of activating transcription factor 2 in inflammation. Mediators Inflamm. 2014;2014:950472. doi: 10.1155/2014/950472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tang X, Wang J, Xia X. et al. Elevated expression of ciRS-7 in peripheral blood mononuclear cells from rheumatoid arthritis patients. Diagn Pathol. 2019;14(1):11. doi: 10.1186/s13000-019-0783-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li G, Tan W, Fang Y. et al. circFADS2 protects LPS-treated chondrocytes from apoptosis acting as an interceptor of miR-498/mTOR cross-talking. Aging (Albany NY) 2019;11(10):3348–3361. doi: 10.18632/aging.101986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang J, Cheng M, Gu B, Wang J, Yan S, Xu D. CircRNA_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via miR-6089/AKT1/NF-κB axis. Cell Death Dis. 2020;11(10):833. doi: 10.1038/s41419-020-03038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li B, Li N, Zhang L. et al. Hsa_circ_0001859 Regulates ATF2 Expression by Functioning as an MiR-204/211 Sponge in Human Rheumatoid Arthritis. J Immunol Res. 2018;2018:9412387. doi: 10.1155/2018/9412387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhong S, Ouyang Q, Zhu D. et al. Hsa_circ_0088036 promotes the proliferation and migration of fibroblast-like synoviocytes by sponging miR-140-3p and upregulating SIRT 1 expression in rheumatoid arthritis. Mol Immunol. 2020;125:131–139. doi: 10.1016/j.molimm.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 121.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H. et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. 2021;20:13. doi: 10.1186/s12943-020-01298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu Z, Huang Q, Zhang Q, Wu H, Zhong Z. CircRNAs open a new era in the study of cardiovascular disease (Review) Int J Mol Med. 2021;47:49–64. doi: 10.3892/ijmm.2020.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–28. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 125.Li Z, Zhu X, Huang S. Extracellular vesicle long non-coding RNAs and circular RNAs: Biology, functions and applications in cancer. Cancer Lett. 2020;489:111–20. doi: 10.1016/j.canlet.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 126.Bai H, Lei K, Huang F, Jiang Z, Zhou X. Exo-circRNAs: a new paradigm for anticancer therapy. Mol Cancer. 2019;18:56. doi: 10.1186/s12943-019-0986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Meganck RM, Borchardt EK, Castellanos Rivera RM, Scalabrino ML, Wilusz JE, Marzluff WF. et al. Tissue-Dependent Expression and Translation of Circular RNAs with Recombinant AAV Vectors In Vivo. Mol Ther Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li C, Samulski RJ. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21:255–72. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 129.Luo ZW, Li FX, Liu YW. et al. Aptamer-functionalized exosomes from bone marrow stromal cells target bone to promote bone regeneration. Nanoscale. 2019;11(43):20884–20892. doi: 10.1039/c9nr02791b. [DOI] [PubMed] [Google Scholar]
- 130.Liang C, Guo B, Wu H. et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat Med. 2015;21(3):288–94. doi: 10.1038/nm.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]