Vijay et al. identify that phosphatidylserine exposed on dead or dying red blood cell membranes signals via Axl on B cells to promote polyclonal B cell activation and plasmablast differentiation during infections that are associated with hemolysis.
Abstract
Antimalarial antibody responses are essential for mediating the clearance of Plasmodium parasite–infected RBCs from infected hosts. However, the rapid appearance of large numbers of plasmablasts in Plasmodium-infected hosts can suppress the development and function of durable humoral immunity. Here, we identify that the formation of plasmablast populations in Plasmodium-infected mice is mechanistically linked to both hemolysis-induced exposure of phosphatidylserine on damaged RBCs and inflammatory cues. We also show that virus and Trypanosoma infections known to trigger hemolytic anemia and high-grade inflammation also induce exuberant plasmablast responses. The induction of hemolysis or administration of RBC membrane ghosts increases plasmablast differentiation. The phosphatidylserine receptor Axl is critical for optimal plasmablast formation, and blocking phosphatidylserine limits plasmablast expansions and reduces Plasmodium parasite burden in vivo. Our findings support that strategies aimed at modulating polyclonal B cell activation and phosphatidylserine exposure may improve immune responses against Plasmodium parasites and potentially other infectious diseases that are associated with anemia.
Introduction
Malaria caused an estimated 409,000 deaths in 36 sub-Saharan African countries in 2019 (World Health Organization, 2020). Although severe anemia and cerebral malaria are significant causes of morbidity and mortality among infected children (Moxon et al., 2020; White, 2018), malaria-associated disease severity and Plasmodium pathogenesis can be limited by an effective humoral immune response. However, protective antibody responses are not efficiently induced by Plasmodium infections, and such humoral responses take years to develop among those living in malaria endemic areas (Tran et al., 2013), which leaves individuals susceptible to chronic and repeated bouts of infection.
Potent humoral immune responses elicited by vaccination and infection consist of temporally and spatially distinct B cell activation events that culminate in the formation of germinal center (GC)–derived memory B cells and long-lived plasma cells that are capable of producing class-switched high-affinity antibodies. The reasons for delayed and inefficient acquisition of antimalarial humoral immune responses are multifactorial (reviewed in Ly and Hansen, 2019), but recent data support that GC responses induced by blood-stage Plasmodium infection are constrained by the rapid emergence of metabolically hyperactive populations of plasmablasts that numerically dominate the initial B cell response and suppress humoral immunity (Vijay et al., 2020). Using specific immunoassays, these studies further showed that only a small fraction (<1%) of the plasmablast population appeared parasite specific, consistent with blood-stage Plasmodium infection–associated polyclonal B cell activation.
Polyclonal B cell activation has been described as an immune evasion mechanism that multiple pathogens, including Plasmodium, have evolved to exploit in order to establish persistent infection, thereby increasing the likelihood for transmission. Initial work showed that the CIDR1α subdomain of Plasmodium falciparum erythrocyte membrane protein-1 (PfEMP-1) could trigger B cell proliferation independent of B cell receptor (BCR) specificities (Donati et al., 2006; Donati et al., 2004; Simone et al., 2011). Additionally, controlled human malaria infection studies showed that the B cell growth and survival factor BAFF is potently induced during P. falciparum infection of malaria-naive individuals (Scholzen et al., 2014), which may additionally contribute to polyclonal B cell proliferation. Relevant to these sets of observations, Plasmodium blood-stage infection has been linked to the activation of self- and lipid-specific B cells (Rivera-Correa et al., 2017). The secretion of phosphatidylserine (PtS)-specific antibodies exacerbates anemia in experimental malaria models and is associated with severe malarial anemia in P. falciparum–infected individuals (Fernandez-Arias et al., 2016). As a composite, these reports suggest that the numerical over-representation of immunosuppressive and potentially self-reactive plasmablast populations triggered by Plasmodium blood-stage infection may be linked to polyclonal B cell activation events. However, the pathways and mechanisms that lead to the formation of rapid, high-magnitude plasmablast responses and whether the appearance of plasmablasts relates to distinct pathophysiologic features of malaria are not understood. Moreover, whether other infections associated with polyclonal B cell activation events also trigger high-magnitude plasmablast expansions has not been investigated.
Herein, we used comparative studies of microbial infections and combinations of transcriptomic analyses and biochemical and genetic approaches to explore these knowledge gaps. Our results reveal previously unrecognized stimuli that promote the differentiation and expansion of immunosuppressive plasmablast responses that include polyclonal B cell activation via IFN-γ and pathogen recognition receptor (PRR) signaling and hemolysis-associated exposure of PtS on RBC membranes. We further show exuberant plasmablast responses were also induced by virus and Trypanosoma infections that are associated with RBC damage and hemolysis and that blocking PtS in vivo during experimental malaria markedly reduced the formation of plasmablasts and improved parasite control. B cell–intrinsic expression of the PtS receptor Axl was also essential for optimal plasmablast expansion. Collectively, our data identify additional pathways that regulate nonspecific, polyclonal B cell activation and pathogen evasion of humoral immunity and reveal potential new targets to improve immune-mediated resistance to malaria and potentially other infectious diseases.
Results and discussion
Polyclonal B cell activation and CD138hi plasmablast differentiation occur independently of antigen recognition by the BCR
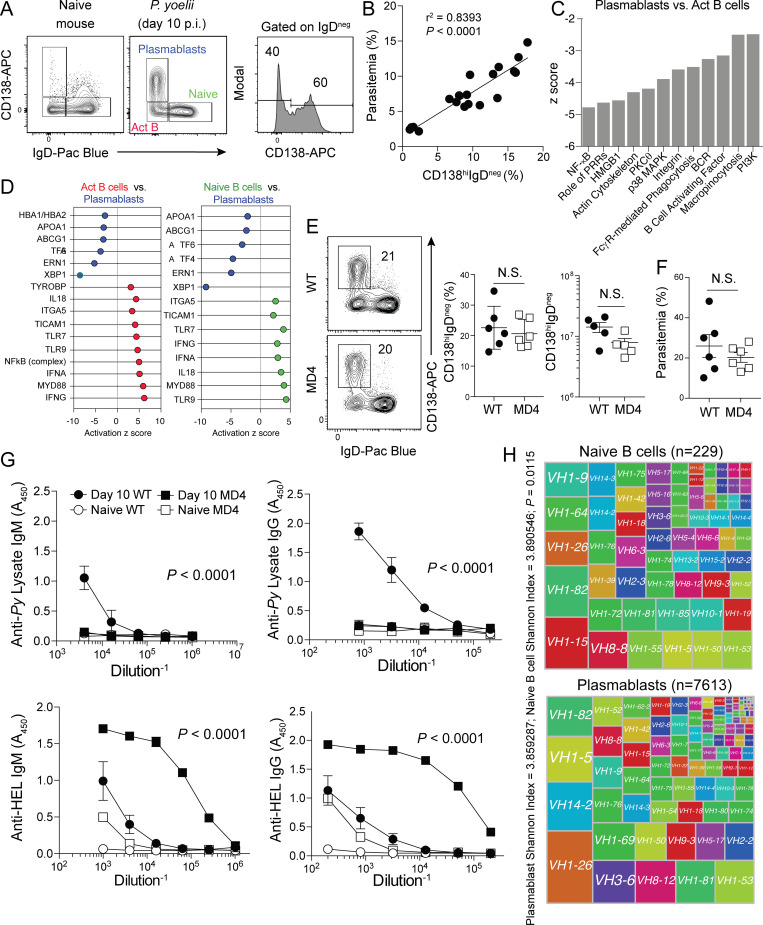
10 d after the induction of experimental malaria in WT C57BL/6 mice, 50–70% of the entire activated (IgDneg) splenic B cell pool is composed of CD138hi immunosuppressive plasmablasts (Fig. 1 A and Fig. S1 A; Vijay et al., 2020). To investigate factors that promote the enormous expansions of malaria-associated plasmablasts, we first examined whether their appearance was linked to parasite burden. Across multiple time points, we observed a strong positive correlation between the proportion of CD138hi plasmablasts among total B cells and parasite burden (Fig. 1 B), suggesting that pathogen-associated molecular patterns and/or antigen and BCR-associated activation events contribute to CD138hi plasmablast expansion. However, RNA sequencing analyses revealed transcriptional signatures that were consistent with blunted BCR-mediated activation in CD138hi plasmablasts compared with CD138loIgDneg activated B cells. Moreover, key pathways that include PI3K and BCR signaling showed lower activation scores (z score less than −1.5) in CD138hi plasmablasts (Fig. 1 C and Table S1). Blunted BCR signaling was also evident by reduced phosphorylation of the tyrosine kinase Syk in CD138hi plasmablasts compared with CD138loIgDneg activated B cells (Fig. S1 B). CD138hi plasmablasts also exhibited expected signatures of the unfolded protein response, yet substantially lower activation scores for genes associated with IFN-γ, TLR, and other PRR signaling pathways compared with activated B cells and naive B cells (Fig. 1 D, left and right, respectively; and Table S1).
Figure 1.
Plasmablasts can expand independently of antigen–BCR interactions. (A–D) WT mice were infected with 106 Py pRBCs. On days 4, 7, and 10 p.i., parasitemia, plasmablasts (CD138hiIgDneg), activated B cells (CD138loIgDneg), and naive splenic B cell populations (CD138loIgDhi) were quantified (n = 19). Using other WT mice (n = 4), the three B cell populations were sort-purified on day 10 p.i. and RNA sequenced for transcriptomic analysis. (A) Representative gating strategy showing plasmablasts, activated B (Act B) cells, and naive splenic B cells in either naive (left panel) or Py-infected mice (middle panel) on day 10 p.i. (B) Correlation graph showing frequency of plasmablasts (x axis) compared with frequency of infected RBCs (% parasitemia, y axis). (C and D) Canonical pathways and upstream regulators of each phenotype were identified using Ingenuity Pathway Analysis software. (C) Pathways down-regulated (negative activation z score) in CD138hi plasmablasts compared with activated B cells. (D) Left panel: Upstream regulators defining activated B cells (red) in comparison to CD138hi plasmablasts (blue). Right panel: Upstream regulators defining naive B cells (green) in comparison to CD138hi plasmablasts (blue). (E) WT and MD4 mice were infected with 106 Py pRBCs. Representative plots, proportions, and total numbers of CD138hiIgDneg plasmablasts on day 10 p.i. n = 5 or 6 mice/group, mean ± SEM, representative of two biologically independent experiments, analyzed by Mann-Whitney. (F) Parasitemia on day 10 p.i. (G) Anti–parasite lysate–specific IgM and IgG (top row) and anti-HEL–specific IgM and IgG (bottom row) antibody responses in sera from day 10 p.i. Data are mean ± SEM, representative of six independent experiments, analyzed using two-way ANOVA. n = 6 mice/group, mean ± SEM, representative of two biologically independent experiments, analyzed by Mann-Whitney. (H) Treemap diagrams of VH gene usage by naive B cells and plasmablasts recovered from infected mice on day 10 p.i. Shannon index indicates the diversity of the B cells within each population, and the number of cells within each population is listed above the diagrams. P value represents the median P value of 1,000 runs of Jack knife resampling of χ2 tests.
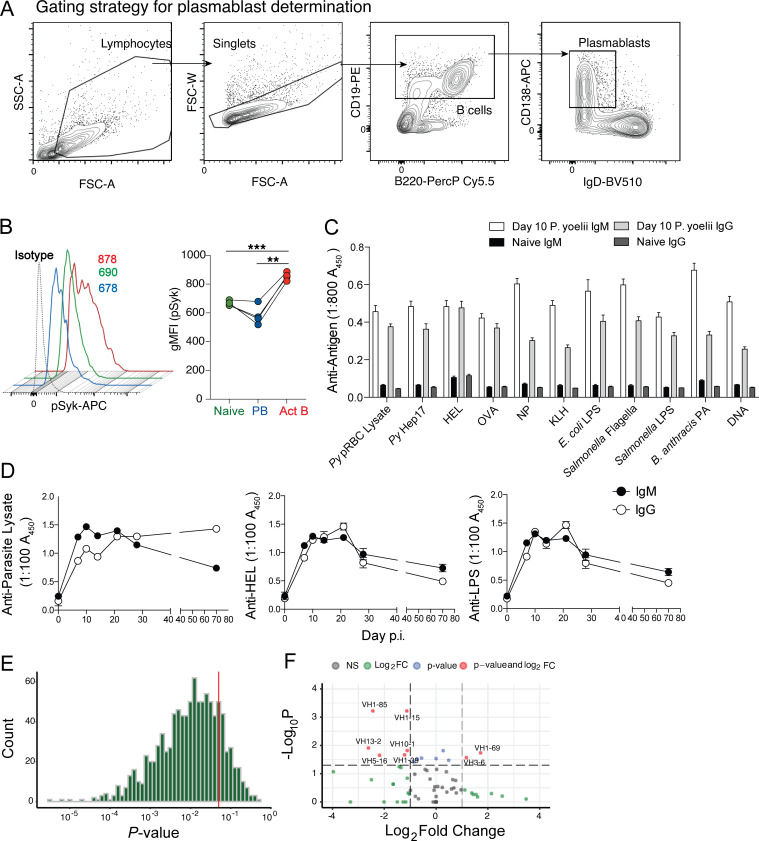
Figure S1.
Cytometry gating strategy, phosphoflow, serum ELISA data, and statistical analyses of single-cell RNA sequencing data. (A) Gating strategy for identifying plasmablasts is shown. (B) Py-infected WT mice were harvested on day 6 p.i., and the B cell populations were evaluated for phosphorylation of Syk. Representative histograms and summary graphs showing geometric mean fluorescence intensity (gMFI) of pSyk on B cell subsets are shown. (C) Sera were collected from Py-infected WT mice on day 10 p.i., diluted 1:800, and reacted against the indicated parasite-, model-, microbial-, and self-specific antigens. n = 20 mice. Data are mean ± SEM, pooled from five biologically independent experiments. (D) Longitudinal analyses of parasite lysate- (top), HEL- (bottom left), and LPS- (bottom right) specific serum IgM and IgG titers in Py-infected mice. n = 6 mice/time point; mean ± SEM pooled from two biologically independent experiments, analyzed by Mann-Whitney. (E) Histogram of P value results of 1,000 Jack knife resampling χ2 tests. (F) Volcano plot of VH loci expressed by plasmablast population relative to naive B cell repertoire. Act B, activated B; FC, fold change; FSC-A, forward scatter area; FSC-W, forward scatter width; NP, (4-hydroxy-3-nitrophenyl)acetyl antigen; PA, protective antigen; PB, plasmablast; PerCP, peridinin-chlorophyll-protein; scRNA, single-cell RNA; SSC-A, side scatter area.
These data raised the possibility that the differentiation of a proportion of Plasmodium infection–induced plasmablasts is either independent of BCR-antigen recognition and IFN-γ and PRR signaling or that plasmablasts are terminally differentiated and no longer express or engage these pathways, as do activated and naive B cells. To explore this hypothesis, we infected MD4 transgenic mice, in which the B cell repertoire is enriched for cells expressing a single BCR specific for hen egg lysozyme (HEL). Strikingly, numerically equivalent plasmablast expansions were observed (Fig. 1 E), and parasite control was equivalent through day 10 post-infection (p.i.; Fig. 1 F) in both Plasmodium-infected WT and MD4 mice. Expectedly, the sera of WT, but not MD4 mice, contained parasite-specific IgM and IgG antibody (Fig. 1 G, top). However, both genotypes harbored elevated HEL-specific antibody (Fig. 1 G, bottom), which is additionally consistent with polyclonal B cell activation following blood-stage Plasmodium infection. We also reacted sera from Plasmodium-infected WT mice against a panel of model antigens and bacterial components and identified robust IgM and IgG responses toward all antigens compared with naive mice (Fig. S1 C). Although parasite lysate–specific IgG titers remained elevated for 10 wk in Plasmodium-infected WT mice, anti-HEL and anti-LPS IgM and IgG responses markedly waned (Fig. S1 D), supporting that these off-target secreted antibody responses likely derive from short-lived, polyclonally activated plasmablasts. Consistent with nonspecific, polyclonal CD138hi plasmablast activation and differentiation in Plasmodium-infected mice, single-cell transcriptomic analyses of the CD138hi plasmablast BCR repertoire showed that it was as diverse as the naive B cell repertoire (Fig. 1 H and Fig. S1 E). Moreover, only two unique VH genes were selected within the plasmablast population (Fig. S1 F), suggesting that a very limited proportion of plasmablasts likely underwent BCR/antigen–dependent expansion. Together, these transcriptomic, genetic, and functional data support that a substantial proportion of the immunosuppressive CD138hi plasmablast population expanded polyclonally during experimental malaria independently of canonical antigen recognition by BCR.
Optimal CD138hi plasmablast expansion requires inflammatory cytokine and PRR signaling and is associated with hemolysis and RBC damage
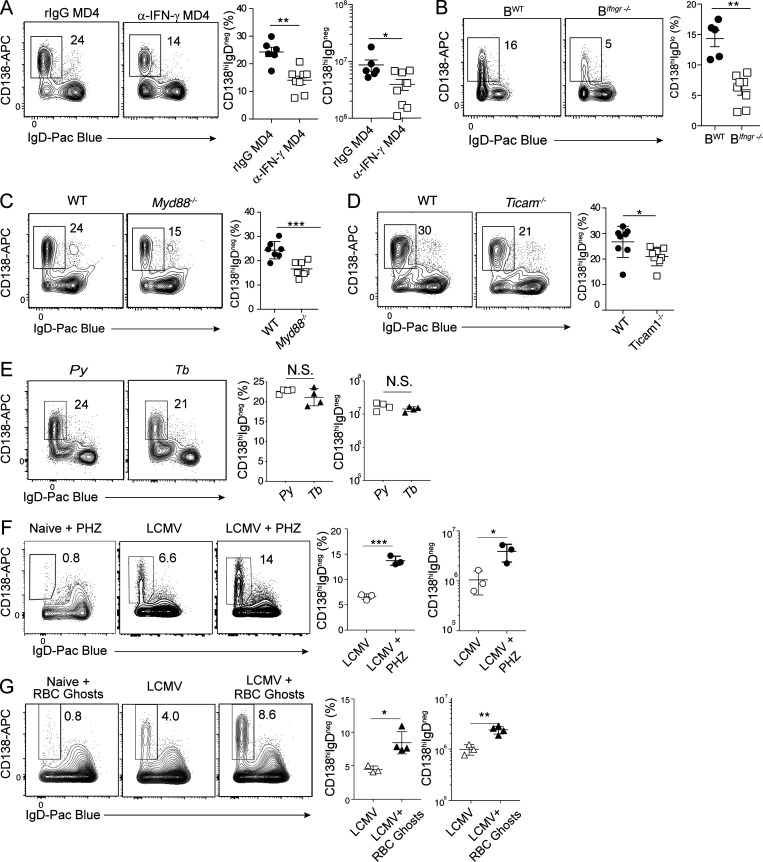
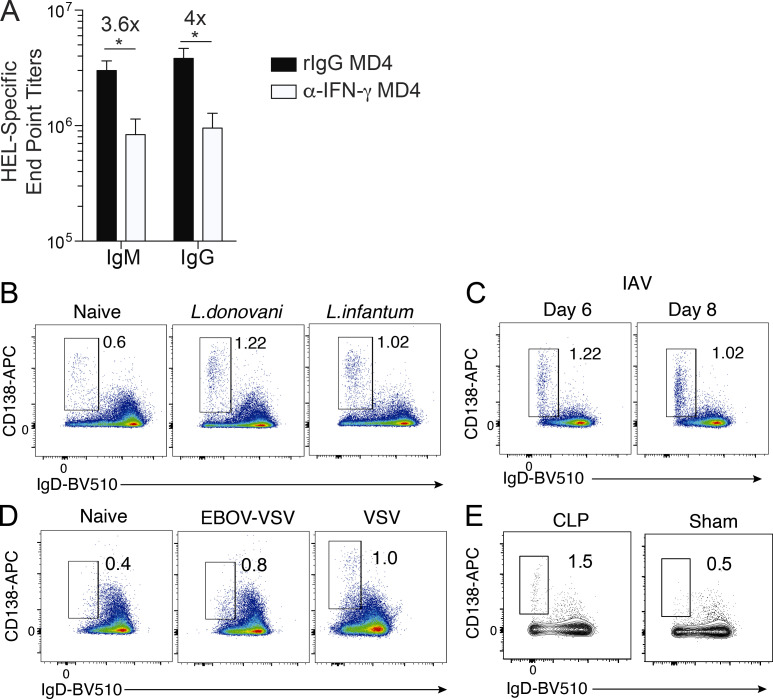
Given that CD138hi plasmablast expansion appeared to occur, in part, independently of antigen recognition by the BCR, we next tested whether other triggers of polyclonal B cell activation, including cytokine and PRR signaling, contribute to their expansion. Robust IFN-γ responses are key features of both clinical and experimental malaria (De Souza et al., 1997; King and Lamb, 2015). Although terminally differentiated plasmablasts exhibit reduced signatures of IFN-γ signaling (Fig. 1 D), the neutralization of IFN-γ in Plasmodium-infected MD4 mice reduced the number of CD138hi plasmablasts by more than twofold (Fig. 2 A) and blunted anti-HEL–secreted IgM and IgG antibody titers by threefold to fourfold (Fig. S2 A). Chimeric mice in which Ifnγr1 deficiency was restricted to the B cell compartment (BIfngr−/−)also exhibited threefold fewer CD138hi plasmablasts compared with BWT chimeric mice (Fig. 2 B). Moreover, the absence of TLR- and PRR-associated signaling adaptors MyD88 and TRIF reduced the differentiation and accumulation of CD138hi plasmablasts by 30–40% (Fig. 2, C and D, respectively). The splenic B cell compartments in MyD88−/− and Ticam1−/− mice were 40–60% larger than those of WT mice, so the absolute number of plasmablasts was not different among these three groups (not shown). Together, these data support that the activity of polyclonal B cell activators, including IFN-γ and PRR signaling, and potentially other pathophysiological aspects of blood-stage malaria may act on precursor B cell populations to promote the induction and accumulation of terminally differentiated CD138hi plasmablasts.
Figure 2.
Hemolysis augments plasmablast differentiation. (A) MD4 mice were infected with 1 × 106 Py pRBCs and administered 500 µg of α-IFN-γ (clone XMG1.2) or rIgG on days 0, 3, 6, and 9 p.i. Representative plots, proportions, and total numbers of CD138hiIgDneg plasmablasts on day 10 p.i. n = 6 (rIgG) and n = 8 (α-IFN-γ) mice/group; mean ± SEM representative of six biologically independent experiments. (B) Representative flow plots and summary of WT or Ifnγr1−/− B cells that differentiate into CD138hiIgDneg plasmablasts in individual BWT and BIfnγr1−/− competitive mixed bone marrow chimeric on day 10 p.i. n = 9 mice, data derived from two pooled experiments. n = 9 mice/group, mean ± SEM, data derived from two pooled experiments. (C) Representative plots and proportions of CD138hiIgDneg plasmablasts on day 10 p.i. in n = 7 (WT) and n = 7 (Myd88−/−) mice/group; mean ± SEM, representative of two biologically independent experiments. (D) Representative plots and proportions of CD138hiIgDneg plasmablasts on day 10 p.i. in n = 8 (WT) and n = 8 (Ticam1−/−) mice/group; mean ± SEM, representative of two biologically independent experiments. (E) Representative plots and summary graphs showing splenic plasmablasts (CD138hiIgDneg) in Py and Tb infection on day 10 p.i. n = 4 mice/group; mean ± SD, representative of two biologically independent experiments. (F) Representative plots and summary graphs showing splenic plasmablasts (CD138hiIgDneg) on day 10 p.i. following LCMV infection with or without PHZ treatment. n = 3 mice/group; mean ± SD, representative of two biologically independent experiments. (G) Representative plots and summary graphs showing splenic plasmablasts (CD138hiIgDneg) on day 10 p.i. following LCMV infection with or without RBC ghost administration. n = 4 mice/group; mean ± SD, representative of two biologically independent experiments. Significance was calculated using Mann-Whitney (A, C–E, and G), paired t test (B), and Student’s t test (F). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S2.
Serum ELISA data from infected MD4 mice and plasmablast responses in Leishmania, influenza, and VSV-infected mice, and mice subjected to cecal ligation and puncture. (A) MD4 mice were infected with 106 Py pRBCs and administered 500 µg of α-IFN-γ (n = 8; clone XMG1.2) or rIgG (n = 6) on days 0, 3, 6, and 9 p.i. HEL-specific IgM and IgG serum end point titers as measured by ELISA on day 10 p.i. Data are mean ± SEM, representative of four independent experiments, analyzed using unpaired Mann-Whitney tests. (B–E) CD138hiIgDneg splenic plasmablast responses were evaluated on day 10 p.i. in WT mice infected i.v. with 2.5 × 107 of either L. donovani or L. infantum parasites (n = 10 mice/group; B) or intranasally with 100 IU of influenza A virus (IAV; n = 5; C); in IFNAR-deficient mice (n = 6) infected i.p. with 100 PFUs of recombinant VSV (EBOV-VSV; D, middle panel) or WT mice infected with high-dose (1 × 106 PFUs) VSV (D, right panel); or WT mice subjected to cecal ligation and puncture (CLP; n = 4) or sham surgery (n = 4; E). *, P < 0.05.
Because IFN-γ and two key PRR signaling pathways were required for optimal expansion of CD138hi plasmablasts during experimental malaria, we hypothesized that high-magnitude CD138hi plasmablast expansions would be observed in other systemic, pathological, and inflammatory immune insults. However, we observed minimal induction of splenic CD138hi plasmablast responses in WT mice infected with either Leishmania donovani or Leishmania infantum (Fig. S2 B) or influenza A virus (Fig. S2 C) in type I IFN receptor–deficient mice infected with a recombinant vesicular stomatitis virus (VSV) or in WT mice infected with high-dose VSV (Fig. S2 D) or in septicemic WT mice subjected to cecal ligation and puncture (Fig. S2 E). By contrast, we observed marked CD138hi plasmablast expansions in mice infected with either Trypanosoma brucei (Tb) or lymphocytic choriomeningitis virus strain Armstrong (LCMV-Arm; Fig. 2, E and F). These data led us to further hypothesize that in addition to IFN-γ and PRR signaling, other key pathological features of malaria, trypanosomiasis, and acute LCMV-Arm infection may also regulate the induction and expansion of CD138hi plasmablasts. Indeed, polyclonal B cell activation, hemolysis, and anemia are features of both Plasmodium and Tb blood infections in mice and humans (Ikede et al., 1977; Lamikanra et al., 2007; Stijlemans et al., 2018) and LCMV-Arm infection in mice (Stellrecht and Vella, 1992; von Herrath et al., 1999).
To test whether exacerbating hemolysis could augment the appearance and accumulation of CD138hi plasmablasts, we induced acute hemolysis by administering phenylhydrazine hydrochloride (PHZ) to LCMV-infected mice. Administration of PHZ resulted in twofold to threefold increases in the frequency and number of CD138hi plasmablasts in LCMV-infected mice but not naive mice (Fig. 2 F). PHZ-mediated hemolysis may stimulate CD138hi plasmablast expansions via the release and recognition of either RBC cytoplasmic contents or damaged RBC membranes. To address whether membrane components were promoting plasmablast responses, we administered RBC membrane ghosts to LCMV-infected mice and found that RBC ghosts were sufficient to expand the number and proportion of CD138hi plasmablasts by twofold (Fig. 2 G), which effectively phenocopied PHZ treatment. Together, these data suggest that the accumulation of infection- and inflammation-induced polyclonal CD138hi plasmablast responses may be mechanistically linked to RBC damage and sensing of RBC membrane-associated factors.
PtS potentiates plasmablast differentiation and accumulation
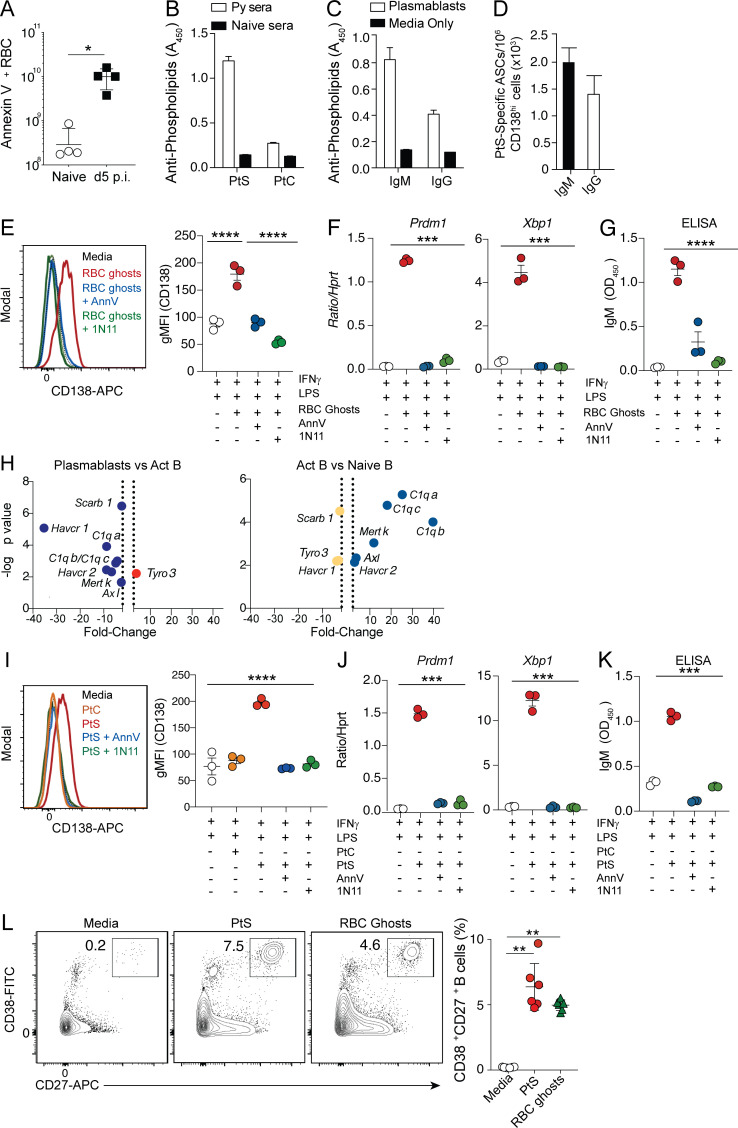
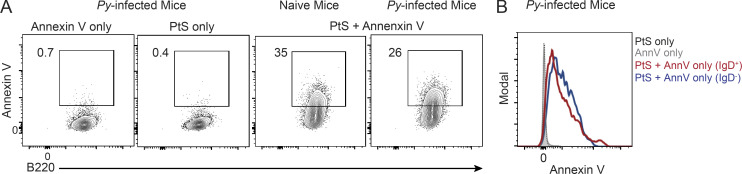
Malaria induces RBC stress and associated translocation of PtS to the surface of both infected and noninfected RBCs (Fernandez-Arias et al., 2016), and PtS is a known antigenic target of self-phospholipid–specific, autoimmune antibody responses (Fernandez-Arias et al., 2016; Rivera-Correa et al., 2017; Rivera-Correa et al., 2019). Consistent with these reports, we found that substantial numbers of RBCs exposed PtS in our experimental Plasmodium-infected mice, as evidenced by Annexin V staining (Fig. 3 A), and appreciable levels of PtS-specific serum IgM and IgG antibodies were present on day 10 p.i. (Fig. 3 B), which coincides with the numerical peak of the immunosuppressive CD138hi plasmablast response (Vijay et al., 2020). Sort-purified CD138hi plasmablasts secreted detectable PtS-specific antibodies in both ELISA (Fig. 3 C) and ELISpot assays (Fig. 3 D), suggesting that a minor proportion of the polyclonal plasmablast response (∼0.4%) includes cells expressing a BCR that targets self-lipid antigens.
Figure 3.
PtS promotes plasmablast formation. (A) Summary graph showing the number of Annexin V–positive RBCs/ml of blood in naive and Py-infected mice as a measure of PtS exposure. n = 4 mice/group; mean ± SD, representative of two biologically independent experiments. (B) Detection of PtS- and PtC-specific IgM antibodies in sera (1:100 dilution) of day 10 Py–infected mice or naive WT mice. n = 10 mice; mean ± SEM, pooled from four independent experiments. (C and D) Sort-purified CD138hiIgDneg plasmablasts from day 10 Py-infected mice were cultured for 20 h, and parasite lysate–specific IgM- and IgG-secreting antibody-secreting cells were detected by ELISA (C) or ELISPOT (D). n = 6 mice; mean ± SEM, pooled from two independent experiments. (E–G) B cells from naive mice were cultured ex vivo for 3 d in media containing IFN-γ and LPS in the presence or absence of RBC ghosts or RBC ghosts plus Annexin V (AnnV) or RBC ghosts plus mch1N11. Representative histograms and summary data showing geometric mean fluorescence intensity (gMFI) of CD138 expression (E), summary data comparing mRNA expression of Prdm1 and Xbp1 relative to Hprt (F), and IgM antibody titers in the supernatant as detected by ELISA (G). n = 3 mice/group, mean ± SEM. Each data point reflects the mean of two technical replicates; data are representative of four biologically independent experiments. (H) Volcano plot showing relative expression of receptors capable of binding to PtS when comparing either plasmablasts and activated B (Act B) cells (left) or activated B cells and naive B cells (right). Dotted vertical lines demarcate positive versus negative log2(fold change). (I–K) B cells from naive mice were cultured ex vivo for 3 d in media containing IFN-γ and LPS in the presence or absence of PtC or PtS liposomes or PtS liposomes plus AnnV or PtS liposomes plus mch1N11. Representative histograms and summary data showing gMFI of CD138 expression (I), summary data comparing mRNA expression of Prdm1 and Xbp1 relative to Hprt (J), and IgM antibody titers in the supernatant as detected by ELISA (K). n = 3 mice/group, mean ± SEM. Each data point reflects the mean of two technical replicates; data are representative of four biologically independent experiments. (L) Representative plots showing the differentiation of CD38+CD27+ human plasma cells from naive peripheral B cells cultured ex vivo in the presence of PtS liposomes or autologous RBC ghosts. n = 2 human leukocyte reduction system (LRS) blood cones with three wells/treatment group; mean ± SEM, data are representative of two biologically independent experiments. Significance was calculated using one-way ANOVA (E–G and I–K) and Mann-Whitney test (A and L). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Consistent with our in vivo data showing that IFN-γ and PRR signaling and hemolysis individually contribute to promoting plasmablast responses, we identified that naive B cells stimulated ex vivo with LPS and IFN-γ and in the presence of RBC ghosts up-regulated surface CD138 expression (Fig. 3 E) and Prdm1 and Xbp1 mRNA levels (Fig. 3 F), a gene signature that is characteristic of plasmablast differentiation (Iwakoshi et al., 2003a; Iwakoshi et al., 2003b; Shaffer et al., 2004). ELISA assays also confirmed that naive B cells stimulated with LPS, IFN-γ, and RBC ghosts secreted IgM (Fig. 3 G), supporting the notion that RBC membrane–associated factor(s) promote the induction and differentiation of CD138hi plasmablasts.
Because both malaria (Fernandez-Arias et al., 2016) and PHZ treatment (Adams et al., 2002; White et al., 2013; Wu et al., 2014; Zhu et al., 2015) trigger PtS exposure in the outer leaflet of RBC membranes, we hypothesized that direct sensing of PtS exposed on damaged RBC membranes may promote the differentiation of CD138hi plasmablasts, independently of BCR-PtS recognition. Indeed, we observed that a number of receptors documented to bind to PtS either directly (Havcr1 encoding Tim1) or indirectly (Mertk, Axl, C1qa, C1qb, C1qc, Scarb1, and Havcr2 encoding Tim2) were more highly expressed in naive or activated B cells compared with CD138hi plasmablasts (Fig. 3 H). We also confirmed that splenic B cells recovered from both naive and Plasmodium-infected mice bound to PtS liposomes ex vivo, as detected by Annexin V labeling (Fig. S3 A), and that both resting (IgDhi) and activated B cells (IgDneg) from infected mice were equivalently labeled (Fig. S3 B). In further support of our hypothesis, ex vivo polyclonal activation of naive murine B cells in the presence of PtS, but not phosphatidylcholine (PtC), liposomes induced surface expression of CD138 (Fig. 3 I), up-regulation of Prdm1 and Xbp1 mRNA (Fig. 3 J), and IgM secretion (Fig. 3 K). Importantly, blocking either RBC ghosts or PtS liposomes with either Annexin V or the mouse chimeric monoclonal antibody (mch1N11) fully abrogated surface expression of CD138, induction of Prdm1 and Xbp1 mRNA, and IgM secretion (Fig. 3, E–G and I–K). We also found that substantial fractions of human peripheral B cells also acquired a plasmablast-like phenotype (CD27+CD38+) upon polyclonal stimulation and co-culture with either PtS liposomes or autologous RBC ghosts (Fig. 3 L). Collectively, these data support a model in which polyclonal B cell activation via IFN-γ and PRR signaling, coupled with either hemolysis- or RBC damage–associated PtS exposure and sensing, potentiates the differentiation and accumulation of plasmablasts.
Figure S3.
Annexin V staining of B cells recovered from naive and Py-infected mice. (A and B) Representative plots (A) and histograms showing geometric mean fluorescence intensity (B) of Annexin V (AnnV) staining, probing for PtS binding to B cells recovered from naive mouse (A, third panel) or on day 4 after Py infection (A,fourth panel, and B). n = 3 mice/group, representative of two biologically independent experiments.
Therapeutic blockade of PtS limits plasmablast responses and improves humoral immunity
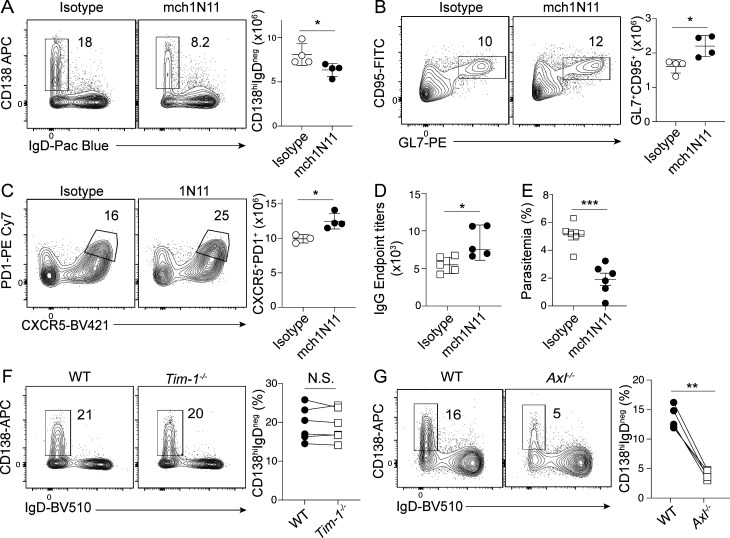
PtS broadly functions as an immunosuppressive signal in the tumor microenvironment, and biologics that block PtS can boost immune activity and tumor control (Dayoub and Brekken, 2020). Our studies suggest that PtS may additionally function as an immune checkpoint via its capacity to promote the induction of immunosuppressive CD138hi plasmablast responses. To directly test the relevance of PtS exposure in promoting CD138hi plasmablast responses during experimental malaria, we employed both biological and genetic approaches to manipulate PtS exposure and PtS receptor signaling. Consistent with our ex vivo B cell differentiation data, administration of mch1N11 to Plasmodium-infected mice reduced the absolute number of CD138hi plasmablasts (Fig. 4 A) and increased the total numbers of GC B cells (Fig. 4 B) and GC–T follicular helper (Tfh) cells (Fig. 4 C) by 20–40%. Notably, mch1N11-mediated abrogation of CD138hi plasmablast accumulation and augmented GC reactions were further associated with elevated class-switched, parasite-specific serum IgG antibody responses (Fig. 4 D) and enhanced parasite control (Fig. 4 E), which is in line with our recent report establishing the immunosuppressive function of CD138hi plasmablasts during Plasmodium infection (Vijay et al., 2020).
Figure 4.
PtS blockade inhibits plasmablast responses and improves humoral immunity. (A–E) C57BL/6 mice were infected with Py and treated with mch1N11 or isotype control from days 5 to 7 p.i. Representative plots and summary graphs showing splenic plasmablasts (CD138hiIgDneg) on day 10 p.i. (A), CD95+GL7+ B (GC) cells (B), and CXCR5+PD1+ CD4 (GC-Tfh) cells on day 21 p.i. (C). n = 4 mice/group; mean ± SD, representative of two biologically independent experiments. (D) Summary graphs showing parasite-specific serum IgG antibody end point titers on day 21 p.i. n = 5 mice/group; mean ± SD, representative of two biologically independent experiments. (E) Summary graph showing frequency of infected RBCs on day 10 p.i. n = 6 mice/group, mean ± SD, pooled from two biologically independent experiments. (F and G) 50:50 bone marrow chimeras using Tim1−/− and WT cells (F) or Axl−/− and WT cells (G) were generated and infected with 1 × 106 Py pRBCs. Representative plots and summary graphs showing frequency of splenic plasmablasts (CD138hiIgDneg) derived from each genotype on day 10 p.i. are shown. Connecting lines match gene knockout and WT B cells within each individual chimeric mouse. Significance was calculated using Mann-Whitney test (A–E) and paired t test (F and G). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
PtS can either directly or indirectly engage a number of receptors to mediate its functions, including Tim1 and Axl. To distinguish between direct and indirect PtS signaling and begin to interrogate the biological relevance of specific PtS receptors, we generated mixed bone marrow chimeric mice containing either WT and Tim1−/− or WT and Axl−/− compartments at 1:1 ratios, which allowed us to dissect the role of B cell–intrinsic PtS signaling in the same infected host. While plasmablast differentiation was equivalent among both Tim−/− and WT compartments (Fig. 4 F), we observed greater than fivefold reductions in plasmablast differentiation among Axl−/− B cells compared with WT B cells in the same Plasmodium-infected host (Fig. 4 G). Together, the composite of our ex vivo and in vivo biochemical, transcriptomic, and genetic studies supports the hypothesis that either RBC damage or destruction may expose PtS that can signal via Axl and function together with polyclonal B activators, inflammatory cytokines, and PRR signaling cascades to program the appearance and accumulation of immunosuppressive CD138hi plasmablast populations.
Plasmablasts numerically dominate the initial B cell response during both Trypanosoma and Plasmodium blood-stage infections, and prominent features of both human and experimental malaria and trypanosomiasis include polyclonal B cell activation and the induction of nonparasite-specific antibody responses and hyperglobulinemia (Freeman and Parish, 1978; Rosenberg, 1978; Wenisch et al., 1996). Our recent studies identified the immunosuppressive mechanisms and consequences of these massive plasmablast expansions (Vijay et al., 2020), and our current experiments support the model that IFN-γ and PRR signaling in combination with hemolysis-associated PtS exposure promote the differentiation of these immunosuppressive B cells. Our data further support that during pathological insults characterized by inflammation and hemolysis, PtS blockade has the potential to boost the immune function and protective immunity.
Our findings are particularly timely given multiple reports describing poor clinical outcomes associated with high-magnitude plasmablast responses (Thevarajan et al., 2020; Woodruff et al., 2020 Preprint) and reduced GC responses (Kaneko et al., 2020; Mathew et al., 2020) in COVID-19 patients. The appearance of SARS-CoV-2–induced plasmablast responses was correlated not only to systemic inflammation, suggesting that other B cell activation stimuli, possibly PtS, may contribute to their differentiation and expansion. Indeed, hemolytic anemia has also been well documented during various stages of COVID-19 disease (Hindilerden et al., 2020a; Hindilerden et al., 2020b; Lazarian et al., 2020). It will be of interest to determine whether the mechanisms governing plasmablast formation following SARS-CoV-2 infection is driven by hemolysis as well as determine whether plasmablasts exert either protective functions or immunosuppressive functions during COVID-19.
Our finding that polyclonal B cell activation signals, such as inflammatory cytokine signaling and/or TLR and PRR activation cascades, act in concert with PtS receptor signaling to promote optimal CD138hi plasmablast differentiation is also consistent with reports showing that PtS-mediated modulation of leukocyte biology requires multiple inputs (Bosurgi et al., 2017; Chen et al., 2012). Our transcriptomic analyses show that compared with CD138hi plasmablasts, both resting and activated B cells expressed higher levels of several PtS receptors and that these expression patterns are consistent with the model that CD138hi plasmablasts are terminally differentiated cells that likely no longer respond to PtS-associated signaling; the analyses also show that PtS acts on precursor populations, including other activated B cell subsets and potentially naive B cells, to potentiate their differentiation into plasmablasts.
To identify the specific PtS receptors that contribute to plasmablast differentiation, we focused on Tim1, which is reported to shape B cell differentiation (Ma et al., 2011), and Axl, which may be induced under conditions prevalent during Plasmodium infection. Indeed, hypoxia, which is a key feature of blood-stage Plasmodium infection (unpublished data) has been shown to stabilize and enhance Axl signaling (Mishra et al., 2012). Moreover, Axl signaling via PtS requires GAS6, which binds PtS via its “Gla” domain to activate the PI3K/AKT pathway and transduce intracellular signals that promote cell proliferation (Ruan and Kazlauskas, 2012), and PI3K signaling is essential for the differentiation of plasmablasts (Omori et al., 2006). While these reports are consistent with our finding that B cell–intrinsic Axl expression is critical for optimal plasmablast differentiation, our data do not rule out the potential contributions of other PtS receptors that share intracellular signaling cascades, such as Tyro3 (Stitt et al., 1995). Notably, multiple drugs targeting Axl have entered clinical trial, which raises the possibility that the PtS–GAS6–Axl pathway might serve as a therapeutic target to improve patient outcomes during malaria or other inflammatory pathologies associated with anemia and hemolysis and the exuberant expansion of plasmablasts.
In summary, we find that the preferential induction and differentiation of plasmablasts was associated with polyclonal B cell activation stimuli that include IFN-γ and PRR signaling and PtS exposure. Targeting these pathways significantly reduced plasmablast expansions, and blocking PtS exposure in vivo improved host resistance to Plasmodium infection. Collectively, our data support that immunotherapeutic strategies that include combinatorial targeting of multiple immune checkpoint pathways, including PtS, may prove effective for promoting resistance to a variety of infections.
Materials and methods
Animals, infections, and parasitemia quantification
The University of Iowa institutional animal care and use committee approved all experiments. C57BL/6 WT (JAX stock 002288), Ifngr1−/− (JAX stock 025545), and MD4 (JAX stock 002595) mice were purchased from The Jackson Laboratory. C57BL/6 Myd88−/− and Ticam1−/− were generous gifts from Michelle Callegan (University of Oklahoma Health Sciences Center, Oklahoma City, OK); Tim1−/− mice were a gift from Paul Rothman (Johns Hopkins, Baltimore, MD); and Axl−/− bone marrow was a generous gift from Carla Rothlin (Yale University, New Haven, CT). Infections in experimental mice were initiated by either serial transfer (i.v.) of 106 Plasmodium yoelii (Py; American Type Culture Collection) parasitized RBCs (pRBCs) derived from a single donor C57BL/6 mouse, or i.p. injections of either 2.5 × 106 PFUs of LCMV-Arm or 5,000 Tb Antat1.1 parasites (Tb; a generous gift from Kent Hill, University of California, Los Angeles, Los Angeles, CA). In some experiments, Py-infected mice were treated with 500 µg of either α-IFN-γ (clone XMG1.2; BioXcell) or rat IgG1 isotype control or with 400 µg of either mouse chimeric PtS-blocking antibody clone mch1N11 or mouse IgG2a (clone 1.18.4; BioXcell) isotype control, as indicated in the figure legends. Parasitemia was measured using flow cytometry as described (Malleret et al., 2011; Villarino et al., 2016).
ELISA and ELISpot
ELISA plates (Nunc) were coated overnight with either Py parasite lysate (18 µg/ml; in-house), Py Hep17 (0.5 µg/ml; Dr. Scott Lindner, Penn State University, State College, PA), HEL (0.5 µg/ml; Sigma-Aldrich), ovalbumin (0.5 µg/ml; Sigma-Aldrich), 4–Hydroxy-3-nitrophenylacetyl (26) conjugated to BSA (0.5 µg/ml; Biosearch Technologies), LPS (Escherichia coli or Salmonella enterica serovar Minnesota; 1 µg/ml; Sigma-Aldrich), flagellin from S. enterica serovar Typhimurium (1 µg/ml; Dr. Matam Vijay-Kumar, Penn State University, State College, PA), anthrax protective antigen (recombinant from Bacillus anthracis; 0.5 µg/ml; List Biological Laboratories, Inc.), and DNA, MB grade, from fish sperm (1 µg/ml; Roche). For lipid ELISA, plates were coated with PtC or PtS (Avanti Polar Lipids; 40 µg/ml each in chloroform). For IgM ELISA from naive B cell cultures, Nunc plates were coated with goat anti-mouse IgM antibody (1 µg/ml) overnight. Plates were blocked with 2.5% BSA + 5% normal goat serum, and serum and culture supernatant samples were serially diluted and reacted for 24 h, and antigen- or Py-specific or IgM antibodies were detected with (1:500) HRP-conjugated goat anti-mouse IgM or IgG. The SureBlue reserve TMB Kit (KBL) was used as a substrate, and absorbance was assessed with a Spectra Max 340 (Molecular Devices). End point titers were extrapolated from sigmoidal 4PL (where X is log concentration) standard curve for each sample. The threshold for end point titers is the mean plus 2× SD of signal detected using sera from naive mice. For ELISpot, clear polypropylene plates (Nunc) were coated with PtS (50 µg/ml) in chloroform (Avanti Polar Lipids). Plates were washed with PBS and blocked for at least 2 h with PBS/2.5% BSA/5% goat serum. CD19+CD138hiIgDlo B cells were sort-purified from day 10 Py-infected mice. Serial dilutions of cells in supplemented IMDM were added to each well for 20 h at 37°C with 5% CO2. Following extensive washes with PBS/0.05% Tween 20, HRP-conjugated goat anti-mouse IgM and IgG was added overnight at 4°C. Spots were developed with 3-amino-9-ethylcarbazole.
Bone marrow chimeras
For BWT and BIfnγr1−/− chimeras, WT recipient (CD45.1) mice were irradiated with 475 rads twice separated by 4 h and bone marrow from either WT (CD45.2) or IFNγR−/− (CD45.2); μMT (immunoglobulin heavy chain mu-deficient; Ighm−/−) donors were mixed in 1:9 ratios, and 107 cells were injected i.v. into the recipients. For WT:Axl−/− (50:50) chimeras, CD45.1 recipient mice were irradiated with 475 rads twice separated by 4 h, bone marrow from WT (CD45.1) and Axl−/− (CD45.2) was mixed 1:1, and 107 cells were injected i.v. into recipients. For WT:Tim1−/− (50:50) chimeras, Rag1−/− recipient mice were irradiated once with 475 rads, bone marrow from WT (CD45.1) and Tim1−/− (CD45.2) was mixed 1:1, and 107 cells were injected i.v. into recipients. All mice were maintained on Uniprim diet (Envigo) for 2–3 wk. Chimerism was assessed at 6 wk, and mice were infected with Py at 8 wk.
Bulk RNA sequencing and GSEA analyses
The three different B cell populations (resting B cells, activated B cells, and plasmablasts) were flow-sorted from four Py-infected mice on day 10 p.i., and RNA was extracted using the NucleoSpin RNA kit (Takara Bio USA Inc.) following the manufacturer’s protocol. RNA integrity and quantity were determined using Tape Station Bioanalyzer (Agilent), with all samples showing RNA integrity numbers >8. Illumina library creation protocol was used to create a strand-specific RNA sequencing library. After pooling, the indexed libraries were sequenced using an Illumina NextSeq 550 sequencer using paired-end chemistry with 50-bp read length. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess the quality of sequence reads, which were aligned using STAR aligner (Dobin et al., 2013) to the mouse genome version mm10. Gene expression profiles were computed using featureCounts (Liao et al., 2014). Differentially expressed genes were identified using Partek GS software, and genes with significant changes (adjusted P value <0.05 and fold change >2 or less than −2) were filtered. Differentially expressed gene lists were generated using Partek GS software, and Ingenuity Pathway Analysis software (Qiagen Bioinformatics) was used to identify the underlying molecular pathways involving the differentially expressed genes. Sequences are deposited in the Gene Expression Omnibus under accession no. GSE134548.
Single-cell RNA sequencing and repertoire analyses
Naive B cells (CD19+CD138loIgD+) and plasmablasts (CD19+CD138+IgDneg) from day 10 Py–infected mice (n = 3) were sort-purified (BD FACSAria II). Cells were partitioned into nanoliter-scale Gel Bead-In-EMulsions to achieve single-cell resolution for a maximum of 10,000 individual cells per sample. Using the Chromium Single Cell 5′ Library and Gel Bead Kit (1000006; 10x Genomics), poly-adenylated mRNA from an individual cell was tagged with a unique 16-bp 10× barcode and 10-bp Unique Molecular Identifier. Full-length cDNA was generated, and V(D)J target enrichment was performed using the Chromium Single Cell V(D)J Enrichment Kit, Mouse B Cell (PN-1000072; 10x Genomics). The cDNA amplicon size for the library (∼700 bp) was optimized using enzymatic fragmentation and size selection, and the concentration of the 10x single-cell library was determined through the Agilent High Sensitivity DNA Kit (Agilent). The single-cell V(D)J target enrichment libraries were sequenced on the Illumina HiSeq-4000 sequencer (The NUSeq Core facility; Northwestern University) at 150 × 150 bp paired end to target >5,000 reads per cell. We adopted Cell Ranger 3.0.2 (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger) for raw sequencing process. All raw short reads were assembled by Cell Ranger, and then the assembled reads were mapped to the mm10 mouse genome using cellranger vdj. V(D)J genes were exported into FASTA files. VH gene usage by naive B cells and plasmablasts was visualized using the “ggplot2” and “treemapify” packages, and the Shannon Index for each dataset was determined using the “vegan” package in R. For comparison of v gene locus usage of naive B cell and plasmablast populations, we calculated log2 fold changes and statistical significance, via χ2 tests, based on percentage of each v locus used by each population. In the volcano plot, the P value cutoff was equal to or less than 0.05, and the cutoff of log2(fold change) was 1. All up-regulated and down-regulated v gene loci are labeled on the plot. Sequences are deposited in the Gene Expression Omnibus under accession no. GSE154910.
Quantitative real-time PCR
The three different B cell populations (resting B cells, activated B cells, and plasmablasts) were flow-sorted from four Py-infected mice on day 10 p.i., and RNA was extracted as described above. 2 μg of RNA was used for cDNA synthesis. 1 μl cDNA was added to 19 µl of PCR mixture containing 2X PowerUp SYBR Green Master Mix and 0.2 µM of forward or reverse primers. Amplification was performed in a QuantStudio3 thermocycler (Applied Biosystems). Primer sequences are as follows: housekeeping gene hypoxanthine phosphoribosyltranferase (Hprt), (forward) 5′-ACCCAGTGCTGGAGAAATTG-3′ and (reverse) 5′-CTCGAGCAAGTCTTTCAGTCC-3′; Prdm-1, (forward) 5′-TAGACTTCACCGATGAGGGG-3′ and (reverse) 5′-GTATGCTGCCAACAACAGCA-3′; and Xbp-1, (forward) 5′-CCTGAGCCCGGAGGAGAA-3′ and (reverse) 5′-CTCGAGCAGTCTGCGCTG-3′. Cycle threshold (Ct) values were normalized to those of Hprt by the following equation: ΔCt = Ct(gene of interest) − Ct(HPRT). All results are shown as a ratio of HPRT calculated as 2−(ΔCt).
PHZ preparation and treatment
PHZ was purchased from Sigma-Aldrich (114715). 1 mg of PHZ (as supplied) was suspended in 1 ml of sterile PBS to achieve a concentration of 0.67 mg/ml. LCMV-Arm–infected mice were treated with 186 µl of the solution i.p. on day 3 p.i.
PtS and PtC liposome preparation
1, 2-Diacyl-sn-glycero-3-phospho-L-serine (PtS; P7769) and L-α-PtC (P7443) were purchased from Sigma-Aldrich. 5 mg of lyophilized PtS or PtC (as supplied) was resuspended in 3.2 ml of sterile PBS to achieve 2-mM concentration and sonicated on ice for 15 min until the micelles were formed. The solution was stored at −20°C until used.
RBC ghosts preparation
WT C57BL/6 mice were exsanguinated using heparinized Natelson blood collection tubes under anesthesia. RBC ghosts were prepared as previously described (Gupta et al., 2014). Briefly, blood was rinsed with twice the volume of osmotically balanced 0.15M salt solution. The tube was centrifuged at highest speed for 1 min at 4°C to pellet the RBCs. The top layer containing plasma was removed, and the remaining pellet was resuspended in osmotically balanced 0.15M salt solution and centrifuged at highest speed for 1 min at 4°C. After removing the supernatant, the blood pellet was resuspended in sterile deionized water by vigorous agitation to lyse the RBCs. The suspension was centrifuged at maximum speed for 5 min at 4°C. The top layer containing hemoglobin was removed, and the pellet was resuspended in sterile PBS at 30 × 106 RBCs/ml.
Ex vivo murine B cell cultures
Naive splenic B cells were enriched from single-cell suspension using a mouse B cell Isolation kit (Cat #130–090-862; Miltenyi Biotech) according to the manufacturer’s protocol. 105 cells were plated per well in a 96-well plate in triplicates/group and cultured in the presence of IFN-γ (50 ng/ml; Peprotech) and LPS (1 μg/ml; Sigma-Aldrich) in IMDM10 (IMDM + 10% FBS) in the presence or absence of either PtS liposomes (10 μM; Sigma-Aldrich) or 50 µl RBC ghosts for 3 d at 37°C (5% CO2 humidified incubator). Plates were spun down and stained with appropriate fluorochrome-conjugated antibodies (as described elsewhere), and data were acquired on a FACSVerse flow cytometer and analyzed as described elsewhere.
Ex vivo human B cell cultures
Leukocyte reduction cones containing human peripheral blood drawn from unidentified infection-naive volunteers were obtained from DeGowin blood center at University of Iowa. Blood was diluted 1:1 with PBS (Ca− Mg−; HyClone) and carefully layered on top of Lymphocyte Separation Media (Corning) in 50-ml tubes, and lymphocytes were separated following the manufacturer’s protocol. Untouched B cells were separated from the lymphocytes using Mojosort Human pan B cell isolation kit (Biolegend) according to the manufacturer’s protocol. 105 naive human B cells in the presence of α-IgM (10 μg/ml; Jackson Immunoresearch) and α-CD40 (1 μg/ml; Biolegend) were cultured in IMDM10 (IMDM + 10% FBS) in the presence or absence of either PtS liposomes (10 μM; Sigma-Aldrich) or RBC ghosts for 3 d at 37°C (5% CO2 humidified incubator). Plates were spun down and stained with appropriate fluorochrome-conjugated antibodies, and data were acquired on a FACSVerse flow cytometer and analyzed as described below.
Cell staining and flow cytometry
For cellular analyses of splenic cells, mouse spleens were forced through a mesh to generate single-cell suspensions. The single-cell suspension of splenocytes was subjected to RBC lysis and was counted and suspended at 106 cells/100 µl. Fc receptors were blocked using Fc block/CD16/32 (clone 2.4G2) in FACS buffer (PBS + 0.09% sodium azide + 2% FCS) for 15 min at 4°C followed by cell-specific staining protocols as described below. For detecting GC-Tfh–like cells, cells were stained with purified rat α mouse CXCR5 (1:100; clone 2G8; BD Biosciences) in Tfh staining buffer (PBS + 0.09% sodium azide + 2% FCS + 0.5% BSA + 2% mouse serum) for 1 h at 4°C. Cell were washed in FACS buffer, spun at 300 ×g for 5 min at 4°C, and suspended in Biotin SP-conjugated Affinipure Goat–anti-rat IgG (H+L) F(ab) (1:1,000; Jackson Immunoresearch) in Tfh staining buffer and incubated for 30 min at 4°C. Cells were washed in FACS buffer, spun at 300 ×g for 5 min at 4°C, and suspended in an antibody cocktail containing CD4-PercP Cy5.5 (1:300; clone GK1.5; Biolegend), CD44-AF700 (1:1,000; clone IM7; BioLegend), CD11a-FITC (1:300; clone M17/4; BD Biosciences), PD-1-PE or PE-Cy7 (1:300; clone RMP1-14; BioLegend), and streptavidin-BV421 (1:500) and incubated 30 min at 4°C. The cells were washed with FACS buffer as before and suspended in FACS buffer before acquisition. For B cell staining, cells were stained with B220-PercP-Cy5.5 (1:300; clone RA3-6B2; BioLegend), CD19-AF700 or PE (1:300; clone 6D5; BioLegend), IgD–Pac Blue or BV510 (1:500; clone 11-26c.2a; BioLegend), CD138-APC or Pac Blue (1:250; clone 281–1; BioLegend; for plasmablast staining); GL7-PE (1:300; BioLegend), and CD95-FITC (1:300; clone Jo2; BD Biosciences; for GC B cell staining). For phospho-Syk (pSyk) staining, B cells were surface stained and permeabilized using the Transcription factor staining kit (Tonbo) and incubated with anti–mouse/human pSyk antibody (0.06 µg/106 cells; clone moch1ct; Invitrogen) or mouse IgG1 κ isotype (0.06 µg/106 cells; clone P3.6.2.8.1; Invitrogen) according to the manufacturer’s protocol. For Annexin V staining, Annexin V staining buffer kit (Cat #422201) was used according to the manufacturer’s protocol. For human B cell staining, 200 µl of blood was stained with surface antibodies to CD19 (FITC; clone HIB19), CD27 (AF700; clone M-T271), and CD38 (BV785; clone HIT2).
Statistical analysis
Statistical analyses, end point titers, and overall parasite burden, represented as area under the curve for rodent studies, were performed using GraphPad Prism 6 software (GraphPad). Specific tests of statistical significance are detailed in the figure legends. To calculate statistical differences in Fig. 1 G, we performed 1,000 independent runs of Jack knife resampling on plasmablast populations (randomly picked n = 225 samples from plasmablast population) and then performed χ2 test for each round between the naive B cell population and resampled the plasmablast population.
Data availability
All data are available from the corresponding author upon request.
Online supplemental material
Fig. S1 shows the cytometry gating strategy, pSyk phosphoflow, serum ELISA data, statistical analyses of single-cell RNA data, and a volcano plot of VH loci expression among plasmablasts and naive B cells. Fig. S2 shows serum ELISA data from infected MD4 mice and plasmablast responses in Leishmania, influenza, and VSV-infected mice, as well as mice subjected to cecal ligation and puncture. Fig. S3 shows Annexin V staining of B cells recovered from naive and Plasmodium-infected mice. Table S1 shows differentially enriched genes in plasmablasts and activated B cells.
Supplementary Material
shows differentially enriched genes in plasmablasts and activated B cells.
Acknowledgments
We thank members of the Butler laboratory for helpful discussions, Drs. Ali Jabbari (University of Iowa) and Martin Richer (McGill University) for critical review, and members of the University of Iowa Flow Cytometry Facility for cell sorting.
Research reported in this publication was supported by the National Cancer Institute (grant no. P30CA086862) and by the National Center for Research Resources of the National Institutes of Health (grant no. S10OD016199). This research was also supported by the National Institutes of Health/National Heart, Lung, and Blood Institute (T32HL007605 in support of J.J. Guthmiller) and the National Institutes of Health/National Institute of Allergy and Infectious Diseases (T32AI007511 in support of J.T. Johnson; R01AI134733 and R21AI139902 to W. Maury; P01AI097092 and HHSN272201400005C to P.C. Wilson; and R01AI125446 and R01AI127481 to N.S. Butler), and the US Department of Veterans Affairs (I01BX001983 and 2I01BX000536 to M.E. Wilson).
Author contributions: R. Vijay, J.J. Guthmiller, and A.J. Sturtz conceived, designed, and performed experiments and analyzed data. S. Crooks, J.T. Johnson, R.L. Pope, Y. Chen, K.J. Rogers, and N. Dutta performed experiments and provided technical assistance. R. Vijay, J.J. Guthmiller, L. Li, and L. Yu-Ling Lan contributed to bioinformatic analyses. J.E. Toombs, M.E. Wilson, P.C. Wilson, W. Maury, and R.A. Brekken provided critical reagents and technical assistance. R. Vijay and J.J. Guthmiller made the figures. R. Vijay wrote the manuscript. W. Maury, P.C. Wilson, R.A. Brekken, and N.S. Butler reviewed and edited the manuscript and figures. N.S. Butler supervised the studies.
References
- Adams, S., Brown H., and Turner G.. 2002. Breaking down the blood-brain barrier: signaling a path to cerebral malaria? Trends Parasitol. 18:360–366. 10.1016/S1471-4922(02)02353-X [DOI] [PubMed] [Google Scholar]
- Bosurgi, L., Cao Y.G., Cabeza-Cabrerizo M., Tucci A., Hughes L.D., Kong Y., Weinstein J.S., Licona-Limon P., Schmid E.T., Pelorosso F., et al. 2017. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science. 356:1072–1076. 10.1126/science.aai8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Liu Z., Wu W., Rozo C., Bowdridge S., Millman A., Van Rooijen N., Urban J.F. Jr., Wynn T.A., and Gause W.C.. 2012. An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat. Med. 18:260–266. 10.1038/nm.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayoub, A.S., and Brekken R.A.. 2020. TIMs, TAMs, and PS- antibody targeting: implications for cancer immunotherapy. Cell Commun. Signal. 18:29. 10.1186/s12964-020-0521-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza, J.B., Williamson K.H., Otani T., and Playfair J.H.. 1997. Early gamma interferon responses in lethal and nonlethal murine blood-stage malaria. Infect. Immun. 65:1593–1598. 10.1128/IAI.65.5.1593-1598.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati, D., Zhang L.P., Chêne A., Chen Q., Flick K., Nyström M., Wahlgren M., and Bejarano M.T.. 2004. Identification of a polyclonal B-cell activator in Plasmodium falciparum. Infect. Immun. 72:5412–5418. 10.1128/IAI.72.9.5412-5418.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati, D., Mok B., Chêne A., Xu H., Thangarajh M., Glas R., Chen Q., Wahlgren M., and Bejarano M.T.. 2006. Increased B cell survival and preferential activation of the memory compartment by a malaria polyclonal B cell activator. J. Immunol. 177:3035–3044. 10.4049/jimmunol.177.5.3035 [DOI] [PubMed] [Google Scholar]
- Fernandez-Arias, C., Rivera-Correa J., Gallego-Delgado J., Rudlaff R., Fernandez C., Roussel C., Götz A., Gonzalez S., Mohanty A., Mohanty S., et al. 2016. Anti-Self Phosphatidylserine Antibodies Recognize Uninfected Erythrocytes Promoting Malarial Anemia. Cell Host Microbe. 19:194–203. 10.1016/j.chom.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, R.R., and Parish C.R.. 1978. Polyclonal B-cell activation during rodent malarial infections. Clin. Exp. Immunol. 32:41–45. [PMC free article] [PubMed] [Google Scholar]
- Gupta, N., Patel B., and Ahsan F.. 2014. Nano-engineered erythrocyte ghosts as inhalational carriers for delivery of fasudil: preparation and characterization. Pharm. Res. 31:1553–1565. 10.1007/s11095-013-1261-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindilerden, F., Yonal-Hindilerden I., Akar E., and Kart-Yasar K.. 2020a. Covid-19 associated autoimmune thrombotic thrombocytopenic purpura: Report of a case. Thromb. Res. 195:136–138. 10.1016/j.thromres.2020.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindilerden, F., Yonal-Hindilerden I., Akar E., Yesilbag Z., and Kart-Yasar K.. 2020b. Severe Autoimmune Hemolytic Anemia in COVID-19 İnfection, Safely Treated with Steroids. Mediterr. J. Hematol. Infect. Dis. 12:e2020053. 10.4084/mjhid.2020.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikede, B.O., Lule M., and Terry R.J.. 1977. Anaemia in trypanosomiasis: mechanisms of erythrocyte destruction in mice infected with Trypanosoma congolense or T. brucei. Acta Trop. 34:53–60. [PubMed] [Google Scholar]
- Iwakoshi, N.N., Lee A.H., and Glimcher L.H.. 2003a. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 194:29–38. 10.1034/j.1600-065X.2003.00057.x [DOI] [PubMed] [Google Scholar]
- Iwakoshi, N.N., Lee A.H., Vallabhajosyula P., Otipoby K.L., Rajewsky K., and Glimcher L.H.. 2003b. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321–329. 10.1038/ni907 [DOI] [PubMed] [Google Scholar]
- Kaneko, N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Massachusetts Consortium on Pathogen Readiness Specimen Working Group . 2020. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell. 183:143–157.e13: E13. 10.1016/j.cell.2020.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, T., and Lamb T.. 2015. Interferon-γ: The Jekyll and Hyde of Malaria. PLoS Pathog. 11:e1005118. 10.1371/journal.ppat.1005118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamikanra, A.A., Brown D., Potocnik A., Casals-Pascual C., Langhorne J., and Roberts D.J.. 2007. Malarial anemia: of mice and men. Blood. 110:18–28. 10.1182/blood-2006-09-018069 [DOI] [PubMed] [Google Scholar]
- Lazarian, G., Quinquenel A., Bellal M., Siavellis J., Jacquy C., Re D., Merabet F., Mekinian A., Braun T., Damaj G., et al. 2020. Autoimmune haemolytic anaemia associated with COVID-19 infection. Br. J. Haematol. 190:29–31. 10.1111/bjh.16794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Ly, A., and Hansen D.S.. 2019. Development of B Cell Memory in Malaria. Front. Immunol. 10:559. 10.3389/fimmu.2019.00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Usui Y., Takeda K., Harada N., Yagita H., Okumura K., and Akiba H.. 2011. TIM-1 signaling in B cells regulates antibody production. Biochem. Biophys. Res. Commun. 406:223–228. 10.1016/j.bbrc.2011.02.021 [DOI] [PubMed] [Google Scholar]
- Malleret, B., Claser C., Ong A.S., Suwanarusk R., Sriprawat K., Howland S.W., Russell B., Nosten F., and Rénia L.. 2011. A rapid and robust tri-color flow cytometry assay for monitoring malaria parasite development. Sci. Rep. 1:118. 10.1038/srep00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew, D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit . 2020. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 369:eabc8511. 10.1126/science.abc8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, A., Wang J., Shiozawa Y., McGee S., Kim J., Jung Y., Joseph J., Berry J.E., Havens A., Pienta K.J., and Taichman R.S.. 2012. Hypoxia stabilizes GAS6/Axl signaling in metastatic prostate cancer. Mol. Cancer Res. 10:703–712. 10.1158/1541-7786.MCR-11-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon, C.A., Gibbins M.P., McGuinness D., Milner D.A. Jr., and Marti M.. 2020. New Insights into Malaria Pathogenesis. Annu. Rev. Pathol. 15:315–343. 10.1146/annurev-pathmechdis-012419-032640 [DOI] [PubMed] [Google Scholar]
- Omori, S.A., Cato M.H., Anzelon-Mills A., Puri K.D., Shapiro-Shelef M., Calame K., and Rickert R.C.. 2006. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 25:545–557. 10.1016/j.immuni.2006.08.015 [DOI] [PubMed] [Google Scholar]
- Rivera-Correa, J., Guthmiller J.J., Vijay R., Fernandez-Arias C., Pardo-Ruge M.A., Gonzalez S., Butler N.S., and Rodriguez A.. 2017. Plasmodium DNA-mediated TLR9 activation of T-bet+ B cells contributes to autoimmune anaemia during malaria. Nat. Commun. 8:1282. 10.1038/s41467-017-01476-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Correa, J., Mackroth M.S., Jacobs T., Schulze Zur Wiesch J., Rolling T., and Rodriguez A.. 2019. Atypical memory B-cells are associated with Plasmodium falciparum anemia through anti-phosphatidylserine antibodies. eLife. 8:e48309. 10.7554/eLife.48309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, Y.J. 1978. Autoimmune and polyclonal B cell responses during murine malaria. Nature. 274:170–172. 10.1038/274170a0 [DOI] [PubMed] [Google Scholar]
- Ruan, G.X., and Kazlauskas A.. 2012. Axl is essential for VEGF-A-dependent activation of PI3K/Akt. EMBO J. 31:1692–1703. 10.1038/emboj.2012.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen, A., Teirlinck A.C., Bijker E.M., Roestenberg M., Hermsen C.C., Hoffman S.L., and Sauerwein R.W.. 2014. BAFF and BAFF receptor levels correlate with B cell subset activation and redistribution in controlled human malaria infection. J. Immunol. 192:3719–3729. 10.4049/jimmunol.1302960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. 10.1016/j.immuni.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Simone, O., Bejarano M.T., Pierce S.K., Antonaci S., Wahlgren M., Troye-Blomberg M., and Donati D.. 2011. TLRs innate immunereceptors and Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) CIDR1α-driven human polyclonal B-cell activation. Acta Trop. 119:144–150. 10.1016/j.actatropica.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellrecht, K.A., and Vella A.T.. 1992. Evidence for polyclonal B cell activation as the mechanism for LCMV-induced autoimmune hemolytic anemia. Immunol. Lett. 31:273–277. 10.1016/0165-2478(92)90126-9 [DOI] [PubMed] [Google Scholar]
- Stijlemans, B., De Baetselier P., Magez S., Van Ginderachter J.A., and De Trez C.. 2018. African Trypanosomiasis-Associated Anemia: The Contribution of the Interplay between Parasites and the Mononuclear Phagocyte System. Front. Immunol. 9:218. 10.3389/fimmu.2018.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt, T.N., Conn G., Gore M., Lai C., Bruno J., Radziejewski C., Mattsson K., Fisher J., Gies D.R., Jones P.F., et al. 1995. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell. 80:661–670. 10.1016/0092-8674(95)90520-0 [DOI] [PubMed] [Google Scholar]
- Thevarajan, I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B., et al. 2020. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 26:453–455. 10.1038/s41591-020-0819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, T.M., Li S., Doumbo S., Doumtabe D., Huang C.Y., Dia S., Bathily A., Sangala J., Kone Y., Traore A., et al. 2013. An intensive longitudinal cohort study of Malian children and adults reveals no evidence of acquired immunity to Plasmodium falciparum infection. Clin. Infect. Dis. 57:40–47. 10.1093/cid/cit174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay, R., Guthmiller J.J., Sturtz A.J., Surette F.A., Rogers K.J., Sompallae R.R., Li F., Pope R.L., Chan J.A., de Labastida Rivera F., et al. 2020. Infection-induced plasmablasts are a nutrient sink that impairs humoral immunity to malaria. Nat. Immunol. 21:790–801. 10.1038/s41590-020-0678-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino, N.F., LeCleir G.R., Denny J.E., Dearth S.P., Harding C.L., Sloan S.S., Gribble J.L., Campagna S.R., Wilhelm S.W., and Schmidt N.W.. 2016. Composition of the gut microbiota modulates the severity of malaria. Proc. Natl. Acad. Sci. USA. 113:2235–2240. 10.1073/pnas.1504887113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Herrath, M., Coon B., Homann D., Wolfe T., and Guidotti L.G.. 1999. Thymic tolerance to only one viral protein reduces lymphocytic choriomeningitis virus-induced immunopathology and increases survival in perforin-deficient mice. J. Virol. 73:5918–5925. 10.1128/JVI.73.7.5918-5925.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenisch, C., Wenisch H., Bankl H.C., Exner M., Graninger W., Looareesuwan S., and Rumpold H.. 1996. Detection of anti-neutrophil cytoplasmic antibodies after acute Plasmodium falciparum malaria. Clin. Diagn. Lab. Immunol. 3:132–134. 10.1128/CDLI.3.1.132-134.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, N.J. 2018. Anaemia and malaria. Malar. J. 17:371. 10.1186/s12936-018-2509-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, C., Yuan X., Schmidt P.J., Bresciani E., Samuel T.K., Campagna D., Hall C., Bishop K., Calicchio M.L., Lapierre A., et al. 2013. HRG1 is essential for heme transport from the phagolysosome of macrophages during erythrophagocytosis. Cell Metab. 17:261–270. 10.1016/j.cmet.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, M., Ramonell R., Cashman K., Nguyen D., Ley A., Kyu S., Saini A., Haddad N., Chen W., Howell J.C., et al. 2020. Critically ill SARS-CoV-2 patients display lupus-like hallmarks of extrafollicular B cell activation. medRxiv. https://doi.org/10.1101/2020.04.29.20083717 (Preprint posted May 3, 2020) 10.1101/2020.04.29.20083717 [DOI]
- World Health Organization . 2020. World Malaria Report 2020. https://www.who.int/publications/i/item/9789240015791 (accessed January 26, 2021).
- Wu, J., Tian L., Yu X., Pattaradilokrat S., Li J., Wang M., Yu W., Qi Y., Zeituni A.E., Nair S.C., et al. 2014. Strain-specific innate immune signaling pathways determine malaria parasitemia dynamics and host mortality. Proc. Natl. Acad. Sci. USA. 111:E511–E520. 10.1073/pnas.1316467111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Liu J., Feng Y., Pang W., Qi Z., Jiang Y., Shang H., and Cao Y.. 2015. Phenylhydrazine administration accelerates the development of experimental cerebral malaria. Exp. Parasitol. 156:1–11. 10.1016/j.exppara.2015.05.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows differentially enriched genes in plasmablasts and activated B cells.
Data Availability Statement
All data are available from the corresponding author upon request.