VanBlargan et al. describe a novel panel of neutralizing monoclonal antibodies against the emerging tick-borne Powassan virus, including domain III– and fusion loop–binding cross-reactive antibodies that confer broad protection in vivo against Powassan and other tick-borne flaviviruses.
Abstract
Although Powassan virus (POWV) is an emerging tick-transmitted flavivirus that causes severe or fatal neuroinvasive disease in humans, medical countermeasures have not yet been developed. Here, we developed a panel of neutralizing anti-POWV mAbs recognizing six distinct antigenic sites. The most potent of these mAbs bind sites within domain II or III of the envelope (E) protein and inhibit postattachment viral entry steps. A subset of these mAbs cross-react with other flaviviruses. Both POWV type–specific and cross-reactive neutralizing mAbs confer protection in mice against POWV infection when given as prophylaxis or postexposure therapy. Several cross-reactive mAbs mapping to either domain II or III also protect in vivo against heterologous tick-transmitted flaviviruses including Langat and tick-borne encephalitis virus. Our experiments define structural and functional correlates of antibody protection against POWV infection and identify epitopes targeted by broadly neutralizing antibodies with therapeutic potential against multiple tick-borne flaviviruses.
Introduction
Powassan virus (POWV) is an emerging tick-borne flavivirus (TBFV) that circulates in parts of North America and Russia. Although human disease associated with POWV is relatively rare, the last two decades have been characterized by a >600% rise in cases compared with the four prior decades (Fatmi et al., 2017). POWV can cause severe neurological disease in humans after a tick bite or blood transfusion from an infected donor (Taylor et al., 2021). Neuroinvasive POWV infection has a high case fatality rate (10–15%), with over 50% of survivors experiencing substantive long-term neurological sequelae (Ebel, 2010; Hermance and Thangamani, 2017).
The two serologically and clinically indistinguishable distinct lineages of POWV circulating in North America, lineages I and II (also called deer-tick virus [DTV]), share at least 96% amino acid–sequence identity in their envelope (E) glycoproteins. Despite this genetic similarity, the two lineages are transmitted by different tick vectors; POWV lineage I strains are maintained predominantly in Ixodes cookei ticks, whereas lineage II strains are carried primarily by Ixodes scapularis (deer ticks; Ebel et al., 2001). Since deer ticks are more aggressive at biting humans, lineage II viruses cause the majority of infections in North America (Hermance and Thangamani, 2017).
TBFVs are classified phylogenetically into three groups: the mammalian, seabird, and Kadam virus groups (Grard et al., 2007). The mammalian TBFVs include viruses that cause encephalitis (including tick-borne encephalitis virus [TBEV] and POWV) or hemorrhagic fever in humans, as well as viruses not linked to human disease, such as Gadgets Gully virus (GGYV). The E proteins of the mammalian TBFV group share ≥70% amino acid identity, whereas they share only ∼38–45% identity with E proteins of mosquito-borne flaviviruses (MBFVs).
There are no licensed vaccines or therapies for POWV infection. Although there are effective inactivated virus vaccines against TBEV, there are still ∼10,000 cases annually due to variable vaccination rates (Kubinski et al., 2020). Anti-TBEV serum Ig has been used as postexposure treatment for TBEV, although its current use is limited by safety concerns (Bröker and Kollaritsch, 2008; Charrel et al., 2004). Antibodies elicited by TBEV vaccination, while capable of cross-neutralizing more closely related TBFVs including Omsk hemorrhagic fever, Kyasanur forest disease, and Alkhurma hemorrhagic fever viruses, showed limited capacity to neutralize the more distantly related POWV (McAuley et al., 2017).
Previously, we demonstrated that an mRNA vaccine encoding the prM and E proteins of POWV lineage II strain Spooner (POWV-SPO) induced neutralizing antibodies in mice that inhibited infection in vitro of several strains of POWV and other TBFVs (VanBlargan et al., 2018). The humoral immune response elicited by the vaccine was sufficient to mediate protection, as passive transfer of immune serum to naive mice prevented lethality and viremia following POWV challenge. Here, we further evaluated the protective components of the humoral immune response to POWV infection by developing a panel of neutralizing mAbs isolated from mice immunized with the POWV mRNA vaccine or infected with POWV-SPO. The most protective antibodies in vivo block postattachment steps in the viral life cycle and recognize epitopes on the lateral ridge/C-C′ loop or A-strand of domain III (DIII) or the fusion loop of domain II (DII) in the E protein. Several of these antibodies cross-react with other TBFVs and protect against heterologous virus challenge. Finally, structural analysis of POWV DIII bound by POWV-80 reveals the molecular determinants of cross-reactivity and cross-protection.
Results
Development of anti-POWV mAbs
To gain insight into the protective humoral immune response against POWV, we generated a panel of mAbs from C57BL/6J mice that were infected with POWV strain SPO or had received a POWV mRNA vaccine (VanBlargan et al., 2018). 84 hybridomas producing anti-POWV antibodies were isolated using flow cytometry and ELISA-based screens with infected cells and recombinant POWV E protein. These mAbs were cloned by limiting dilution, and 21 mAbs were selected for further study (Table 1) based on neutralization of POWV by hybridoma supernatants.
Table 1. POWV mAb competition group and neutralization potency.
| Competition group | mAb | VH gene | Isotype | ELISA reactivitya | KD kinetic (nM ± SD)b | t1/2 (s) | EC50 (ng/ml)c | |||
|---|---|---|---|---|---|---|---|---|---|---|
| POWV RVP | Lineage II SPO | Lineage II MA51240 | Lineage I LB | |||||||
| None | POWV-16 | IGHV1-39*01 | IgG2c | None | N/A | N/A | 10 | >20,000 | >20,000 | >20,000 |
| A | POWV-4 | IGHV1-62-2*01 | IgG2c | E | 9.7 ± 1.7 | 285 | 43 | 3,658 | 368 | 10,856 |
| B | POWV-5 | IGHV1-50*01 | IgG2c | E | 2.8 ± 0.47 | 2,214 | 311 | 6,631 | 1,049 | 5,269 |
| POWV-18 | IGHV1-50*01 | IgG2b | E | 3.2 ± 0.81 | 2,390 | 803 | 8,507 | 1,578 | 10,504 | |
| POWV-54 | IGHV1-26*01 | IgG2c | E | 1.3 ± 0.2 | 3,536 | 9 | 1,757 | 231 | 2,260 | |
| POWV-63 | IGHV1-26*01 | IgG2c | E | 3.2 ± 0.4 | 721 | 6 | 5,018 | 223 | 2,999 | |
| POWV-56 | IGHV1-53*01 | IgG2c | E | 2.0 ± 0.41 | 1,724 | 6 | 2,612 | 310 | 726 | |
| C | POWV-14 | IGHV1-62-2*01 | IgG2c | E | 1.7 ± 0.98 | 2,740 | 122 | 4,085 | 340 | 12,248 |
| POWV-15 | IGHV1-15*01 | IgG2c | E | 23 ± 1.91 | 171 | 358 | 10,840 | 660 | >20,000 | |
| POWV-23 | IGHV1-67*01 | IgG2c | E | 4.0 ± 0.68 | 1,920 | 111 | 3,800 | 318 | >20,000 | |
| D | POWV-48 | IGHV1-34*01 | IgG2b | E | 3.6 ± 0.12 | 1,075 | 234 | 5,395 | 434 | 3,149 |
| POWV-62 | IGHV1-34*01 | IgG2c | E | 0.6 ± 0.43 | 1,998 | 83 | 2,412 | 226 | 843 | |
| POWV-70 | IGHV1-34*01 | IgG2c | E | 1.6 ± 0.39 | 1,853 | 87 | 1,358 | 202 | 1,054 | |
| E | POWV-33 | IGHV1-75*01 | IgG2c | E, DIII | 2.5 ± 0.05 | 1,066 | 95 | 5,307 | 162 | 7,577 |
| POWV-37 | IGHV1-75*01 | IgG2c | E, DIII | 1.8 ± 0.17 | 837 | 51 | 3,455 | 127 | 8,411 | |
| POWV-65 | IGHV1-22*01 | IgG2c | E, DIII | 1.5 ± 0.17 | 2,840 | 102 | 7,955 | 110 | 4,854 | |
| POWV-71 | IGHV1-54*01 | IgG2b | E, DIII | 0.6 ± 0.26 | 4,151 | 27 | 14,924 | 339 | 6,789 | |
| POWV-80 | IGHV5-16*02 | IgG2c | E, DIII | 34.7 ± 4.47 | 84 | 104 | 12,060 | 83 | 17,962 | |
| F | POWV-55 | IGHV14-3*01 | IgG2c | E, DIII | 1.2 ± 0.35 | 1,066 | 16 | 1,764 | 23 | 5,410 |
| POWV-60 | IGHV14-3*01 | IgG2c | E, DIII | 1.9 ± 0.58 | 857 | 19 | 2,621 | 21 | 6,913 | |
| POWV-61 | IGHV14-3*01 | IgG2c | E, DIII | 2.1 ± 0.19 | 1,122 | 16 | 1,839 | 21 | 4,463 | |
N/A, not available.
mAbs were assessed for binding to recombinant POWV E protein and DIII of the E protein by ELISA.
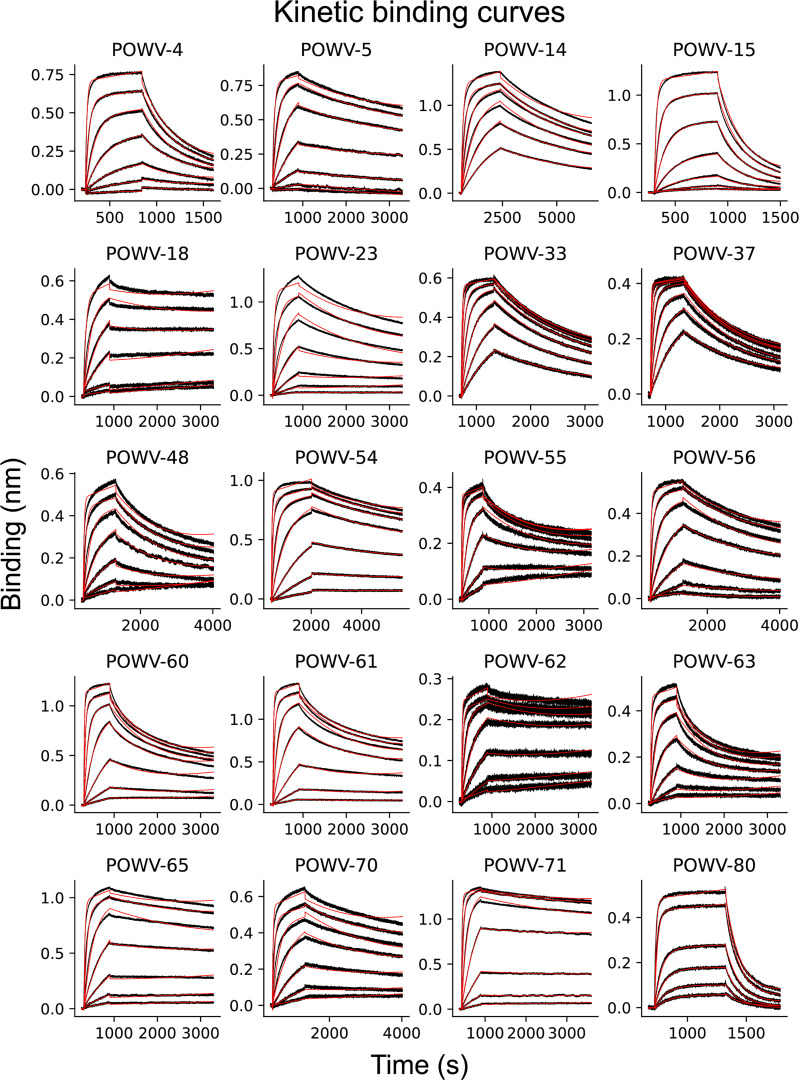
Example kinetic binding curves with fits shown in Fig. S4.
Neutralization of POWV strains was assessed by FRNT or an RVP-based assay where noted. EC50 values were determined by nonlinear regression. Geometric mean from three or four independent experiments.
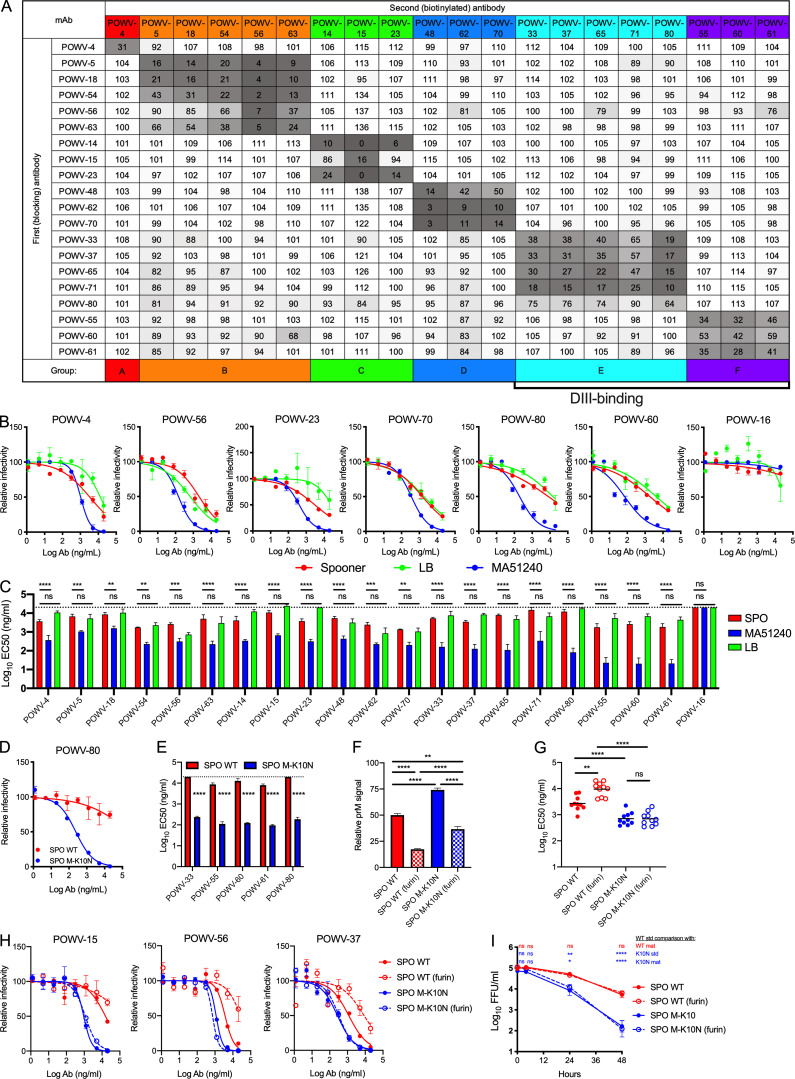
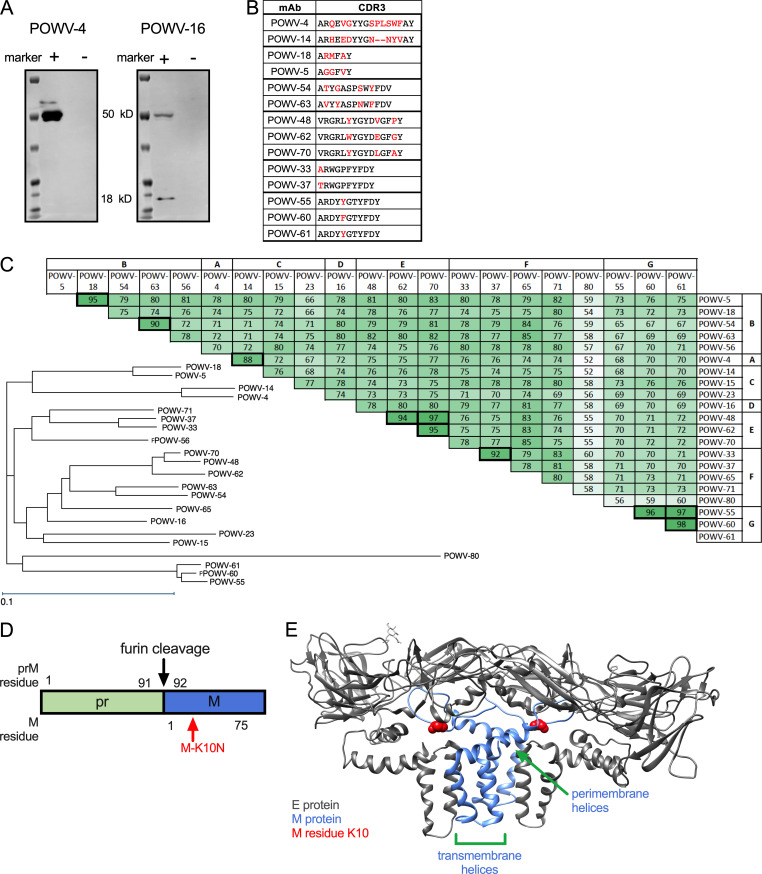
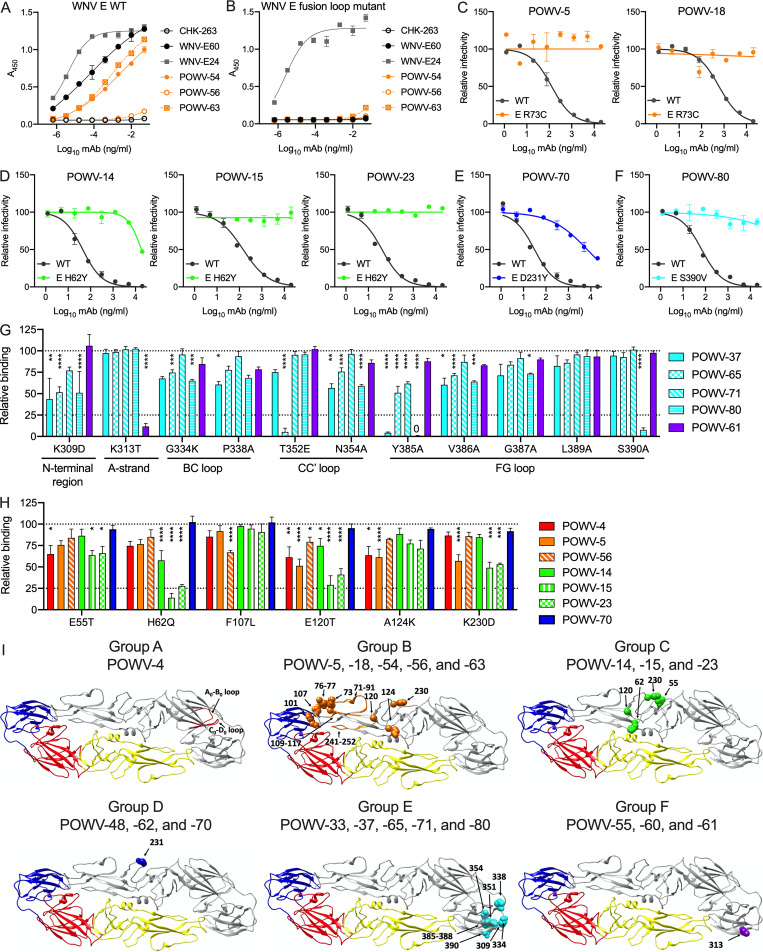
The 21 selected mAbs were purified and isotyped. Most mAbs were IgG2c, although three (POWV-18, POWV-48, and POWV-71) were of the IgG2b subclass (Table 1). These 21 mAbs were characterized for competitive binding to E protein by ELISA. The mAbs separated into six competition groups (A–F) based on their ability to block binding of other mAbs to E protein (Fig. 1 A and Table 1). POWV-16 did not bind E protein by ELISA, and thus it was not placed in a competition group. Western blotting of POWV-infected cell lysates with POWV-16 revealed two bands at ∼55 and 18 kD, suggesting it likely engages an epitope spanning the prM and E proteins (Fig. S1 A). This panel of mAbs also was tested for their ability to bind recombinant DIII of the E protein; all mAbs in groups E and F bound DIII, indicating that two distinct epitopes on DIII were targeted.
Figure 1.
Anti-POWV neutralizing mAbs cluster into six competition groups. (A) Competition ELISA for mAb binding to POWV E protein. Binding of each biotinylated antibody in the presence of each blocking antibody is expressed relative to binding to POWV E in the absence of a blocking antibody. Mean of three experiments. (B and C) POWV mAbs were assayed for neutralization by FRNT against POWV strains SPO, MA51240, and LB. (B) Representative dose-response curves. Error bars represent the range from two technical replicates. (C) Mean EC50 values. Error bars represent SEM from three to four experiments. (D and E) POWV mAbs were assayed for neutralization by FRNT against POWV-SPO encoding WT sequence or an M protein K10N mutation. (D) Representative dose-response curves. Error bars represent the range from two technical replicates. (E) Mean EC50 values for each mAb. Error bars represent SEM from three experiments. (F) The amount of prM on virions was measured for WT and K10N infectious clone–derived virus produced in Vero cells or Vero-furin cells (furin) by ELISA. Error bars represent SEM from three experiments. (G and H) POWV mAbs were assayed for neutralization against standard and Vero-furin stocks of WT and M-K10N infectious clone–derived POWV-SPO. (G) Mean EC50 values of all tested mAbs from three experiments. (H) Representative dose-response curves. Error bars represent the range from two technical replicates. (I) The loss of virus infectivity was determined by incubation at 37°C before determination of viral titer by FFA. Error bars represent SEM from three experiments. (C and E) Dotted lines represent the limit of detection of the assay. Statistical analysis: ANOVA with Tukey’s post-test (C, F, and G); ANOVA with Sidak’s post-test (E); ANOVA with Dunnett’s post-test (I). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Ab, antibody; mat, mature; std, standard.
Figure S1.
Characteristics of POWV mAbs and POWV E-M heterodimer. (A) POWV-infected cell lysates were tested for reactivity with POWV-4 and POWV-16 by Western blotting. The E protein is ∼50 kD and prM is ∼18 kD. Blots shown are representative data from three experiments. (B) Alignment of CDR3 sequences of related mAbs; differences are highlighted in red. (C) Sequence conservation and phylogram of antibody VH sequences. Numbers indicate the percent amino acid identity shared between VH sequences of respective mAbs. (D) Schematic of pr(M), showing the furin cleavage site and the relative position of the M-K10 residue. (E) The residue corresponding to POWV M residue K10 is highlighted on mature structure of the TBEV E-M heterodimer (PDB accession no. 5O6A).
The variable region of each mAb gene was sequenced. Several mAbs shared VH gene usage and had high sequence similarity (90% or greater; Fig. S1, B and C; and Table 1). Generally, mAbs that shared high sequence similarity were in the same competition groups. The exception was POWV-4 in group A and POWV-14 in group B, which shared VH gene usage but fell into separate competition groups. Notably, these two mAbs shared lower sequence homology than other related antibodies (∼88%) and had more divergent CDR3 sequences (Fig. S1, B and C), which likely explains their distinct binding specificities.
Neutralizing activity of anti-POWV mAbs
We tested the panel of 21 mAbs for neutralization potency using a reporter virus particle (RVP)–based neutralization assay. RVPs were produced with the C-prM-E proteins of the POWV lineage II strain P0375 and were used for infection of Raji-DCSIGN-R cells (VanBlargan et al., 2018). All 21 mAbs neutralized POWV RVP infection, with half-maximal effective concentration (EC50) values ranging from 6 to 803 ng/ml (Table 1). Group B mAbs POWV-54, -56, and -63 were the most potently inhibitory (EC50 < 10 ng/ml), followed by POWV-16 (10 ng/ml) and group F mAbs POWV-55, -60, and -61 (16–19 ng/ml).
We next evaluated neutralization potency using focus-reduction neutralization tests (FRNTs) against three authentic viral isolates: POWV lineage I strain LB (1958; Ontario, Canada), lineage II strain SPO (1997; Spooner, WI), and lineage II strain MA51240 (2008; Spooner, WI). As seen with other flaviviruses (Dowd et al., 2016a), the neutralization potency of mAbs was less when determined by FRNT than by RVP-based assay (Table 1). Nonetheless, all mAbs inhibited infection of all three POWV strains by FRNT with the exception of POWV-16 (which did not neutralize any strain) and POWV-15 and -23 (which did not neutralize POWV-LB; Fig. 1, B and C; and Table 1). Overall, the mAbs showed weaker neutralization potency against POWV-SPO and POWV-LB by FRNT (EC50 > 1 µg/ml) than against POWV-MA51240, even though they were raised against viral proteins from POWV-SPO.
To understand the basis for the differential potency of mAb neutralization of POWV-MA51240 and POWV-SPO, we sequenced both viruses and identified three coding differences in the viral polyprotein: K213N, T2369S, and R2903K. Only one of the changes, K213N, is in a structural protein, whereas the other substitutions are in NS4B and NS5, respectively. Polyprotein residue K213 corresponds to prM residue K101 and is retained on the virion as M residue K10 following pr cleavage and dissociation during viral maturation (Fig. S1 D). To test whether the K10N substitution in M modulates neutralizing activity of anti-E mAbs against POWV-MA51240, we introduced the change into an infectious clone of POWV-SPO (Kenney et al., 2018) and assessed activity of several DIII mAbs. Remarkably, all five mAbs tested (POWV-33, -55, -60, -61, and -80) showed greater neutralizing activity against the M-K10N mutant than WT POWV-SPO (80- to 100-fold; Fig. 1, D and E). Thus, variation at residue 10 of the M protein affects the neutralizing potency of anti–E protein mAbs against POWV.
Although a structure of POWV M is not available, the E-M dimer structure of the related TBEV shows that the M protein consists of an N-terminal loop, one perimembrane helix, and two transmembrane helices (Fig. S1 E; Füzik et al., 2018). The M protein is not exposed on the surface of the virion. Most of the M protein is adjacent to or embedded within the viral membrane, with the N-terminal loop located between the viral membrane and the E protein. This N-terminal loop, where M residue 10 is located, interacts with DII of the E protein (Füzik et al., 2018).
Due to the proximity of K10N to the site of prM cleavage, we hypothesized that the substitution might impact POWV maturation, which globally can modulate neutralization sensitivity of anti–E protein mAbs (Nelson et al., 2008). We assessed the impact of virion maturation on the neutralization sensitivity of the two POWV strains by producing viruses in Vero cells overexpressing furin (Mukherjee et al., 2016); viruses propagated in these cells have greater prM cleavage and more mature virus populations than stocks produced in conventional Vero cells. The maturation state of the viral stocks was assessed by ELISA using the anti–prM-E antibody POWV-16. Both WT and M-K10N POWV-SPO produced in Vero-furin cells had lower prM content than either standard preparation (Fig. 1 F). A standard preparation of POWV-SPO M-K10N retained more uncleaved prM than WT POWV-SPO; similarly, Vero-furin cell–derived M-K10N virus had more uncleaved prM than Vero-furin cell–derived WT POWV-SPO (Fig. 1 F). These data indicate that the M-K10N substitution results in higher levels of uncleaved prM in virions, even in the presence of furin overexpression. To probe the functional effect of pr cleavage on neutralization sensitivity, we assessed neutralization of standard and Vero-furin cell preparations of WT and M-K10N POWV-SPO with 10 mAbs (POWV-4, -5, -14, -15, -23, -37, -56, -60, -65, and -70) with at least one from each competition group. Vero-furin cell–derived WT POWV-SPO was neutralized less efficiently than a standard preparation of WT POWV-SPO. However, standard and Vero-furin cell–derived M-K10N POWV-SPO was neutralized equivalently, and both M-K10N–containing viral stocks were inhibited more efficiently than WT POWV-SPO (Fig. 1, G and H). Together, these data suggest that differences in virus maturation between WT and M-K10N may contribute to differences in sensitivity to antibody neutralization. However, this finding does not explain the entire effect, as Vero-furin cell–derived M-K10N still is neutralized more efficiently than standard preparation of WT POWV-SPO even though it has less uncleaved prM.
Due to the role of the N-terminal loop in regulating stability of the mature virus particle (Füzik et al., 2018; Zhang et al., 2013), we tested the effect of the M-K10N substitution on the decay rate of virus infectivity. WT and K10N POWVs produced in either Vero-furin or standard Vero cells were incubated at 37°C for up to 48 h and then tested for infectivity using a focus-forming assay (FFA). M-K10N POWV lost infectivity more rapidly over time than WT POWV, and the maturation state of either virus did not affect decay rates (Fig. 1 I). Thus, the M-K10N substitution alters virus stability independent of virus maturation state. Loss of virus infectivity in solution over time, or intrinsic decay, has been hypothesized to be a consequence of viral “breathing” (the conformational flexibility of virion surface proteins), which results in virion structural proteins sampling ensembles of conformations, a subset of which leads to an irreversible loss of infectivity (Carson, 2014; Goo et al., 2017; Organtini et al., 2014). Changes in viral stability resulting from the M-K10N substitution may reflect inherent differences in viral breathing, which could in turn affect epitope exposure, explaining the altered neutralization sensitivity patterns.
Mechanism of neutralization by anti-POWV mAbs
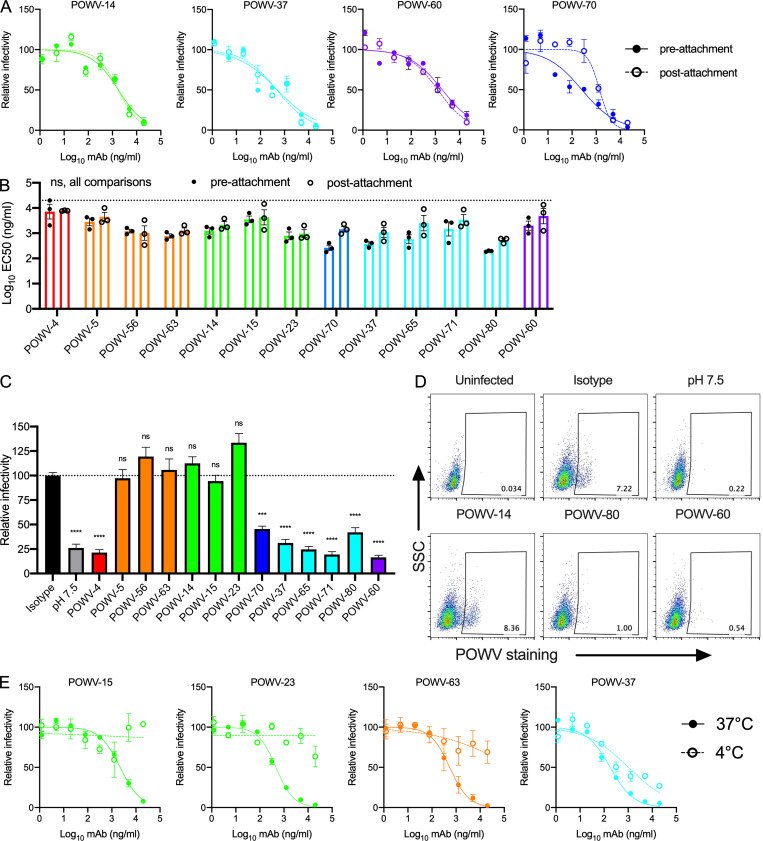
To probe the mechanisms of mAb neutralization, we evaluated their capacity to inhibit POWV-MA51240 infection when added after viral attachment to cells. A subset of mAbs was tested; for mAbs with high sequence similarity, only one representative mAb was selected for evaluation. POWV-70 had a fivefold reduction in neutralization potency when added at a postattachment step, indicating it may inhibit more efficiently at a preattachment step (Fig. 2, A and B). As all other mAbs tested efficiently neutralized POWV after virus attachment to cells, they likely block an entry step after cellular engagement.
Figure 2.
Mechanism of neutralization by anti-POWV mAbs. (A and B) POWV mAbs were assayed for pre- or postattachment inhibition using a modified FRNT against POWV-MA51240. (A) Representative dose-response curves. Error bars represent the range from two technical replicates. (B) Mean EC50 values for each mAb. Error bars represent SEM from three experiments (ANOVA with Sidak’s post-test: ns, all comparisons). Dotted line represents the limit of detection of the assay. (C and D) FFWO neutralization assays. (C) Infection levels are expressed relative to cells incubated with an isotype control mAb. Error bars represent SEM from five independent experiments (ANOVA with Dunnett’s post-test: ***, P < 0.001; ****, P < 0.0001). Dotted line represents the isotype control value. (D) Representative flow cytometry plots are shown. (E) Antibodies were assayed for neutralization of POWV-MA51240 by FRNT at 37°C and 4°C. Representative dose-response curves are shown from two independent experiments. Error bars represent the range from two technical replicates. Ab, antibody; SSC, side scatter.
The mAbs were next evaluated for their ability to inhibit viral fusion using a fusion-from-without (FFWO) assay (Fernandez et al., 2018). MAbs from competition group A (POWV-4), group D (POWV-70), group E (POWV-37, -65, -71, and -80), and group F (POWV-60 and -61) all inhibited viral fusion (Fig. 2, C and D). Although mAbs from competition group B (POWV-5, -56, and -63) and group C (POWV-14, -15, and -23) did not inhibit viral fusion, this outcome might be confounded by their temperature dependence of binding; these mAbs lacked the ability to neutralize POWV in solution when they were added at 4°C but were removed before the shift in temperature and incubation at 37°C (Fig. 2 E). Thus, these mAbs may fail to block fusion in the FFWO assay because they do not bind efficiently to virus at 4°C.
Cross-reactivity of POWV mAbs against other flaviviruses
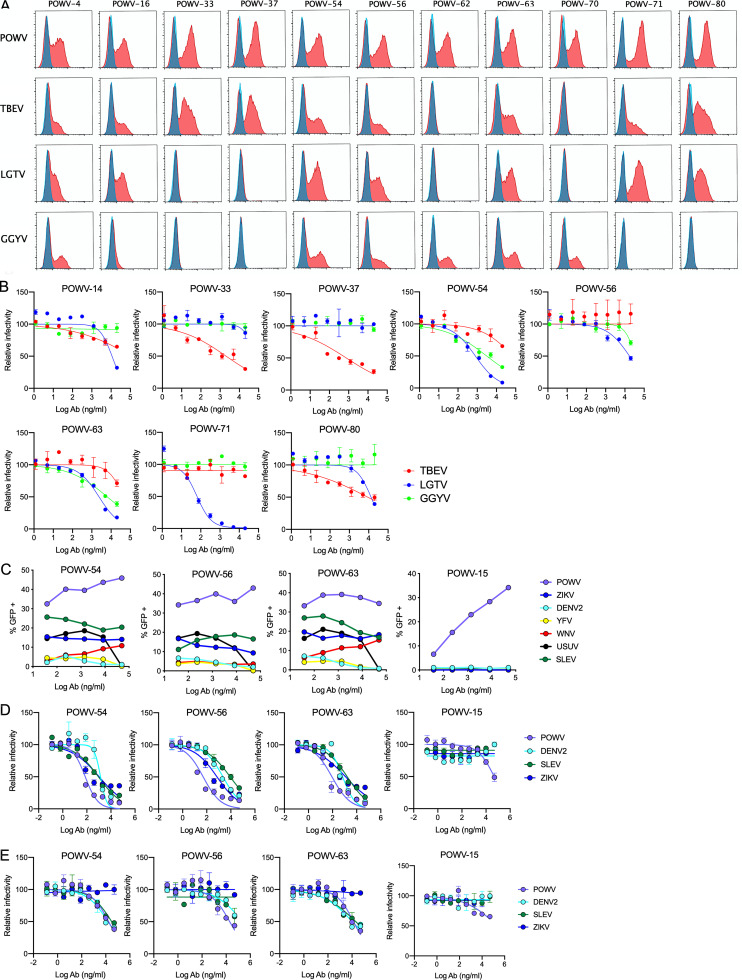
To assess cross-reactivity of the anti-POWV mAbs against other TBFVs, we tested three other viruses in the TBFV clade: TBEV, Langat virus (LGTV), and GGYV. POWV shares ∼77–78% amino acid identity in the E protein with LGTV and TBEV and 72% identity with GGYV. The POWV mAbs first were tested for their ability to bind to cells transfected with the structural genes of TBEV, LGTV, or GGYV. Several group B mAbs (POWV-54, -56, and -63) as well as group A mAb POWV-4 cross-reacted with all three TBFVs tested (Fig. 3 A and Table 2). Some mAbs exhibited cross-reactivity to a subset of the TBFVs, including mAbs in groups D and E, and the prM-E mAb POWV-16. In comparison, mAbs in groups C and F and the remaining mAbs in group B showed type-specific binding patterns. Thus, subsets of DIII- and non–DIII-binding anti-POWV mAbs recognize conserved epitopes in several TBFVs.
Figure 3.
Cross-reactivity of POWV mAbs against other flaviviruses. (A) Flow cytometric analysis of HEK-293T cells transfected with the C-prM-E structural genes of POWV, TBEV, LGTV, or GGYV; representative histograms from one of three experiments is shown. Untransfected cells (blue); transfected cells (red). (B) Neutralization by mAbs was assessed using an RVP-based assay (TBEV and GGYV) or FRNT (LGTV) on Vero cells. Representative dose-response curves from two to three experiments. Error bars represent the range from two technical replicates. (C) ADE of the indicated MBFV by POWV mAbs was assessed by an RVP-based assay using K562 cells. Dose-response curves from two to three experiments. (D and E) Standard (D) and mature (E) RVP preparations were assessed for sensitivity to mAb neutralization on Raji-DCSIGNR cells. Dose-response curves are shown from three to four independent repeats. Error bars represent the range from two technical replicates. Ab, antibody.
Table 2. Cross-reactivity of anti-POWV mAbs against TBFVs.
| Competition group | mAb | Binding (flow cytometry)a | EC50 (ng/ml)b | ||||
|---|---|---|---|---|---|---|---|
| TBEV | LGTV | GGYV | TBEV | LGTV | GGYV | ||
| None | POWV-16 | ++ | ++ | ||||
| A | POWV-4 | ++ | ++ | ++ | |||
| B | POWV-5 | ||||||
| POWV-18 | |||||||
| POWV-54 | ++ | ++ | ++ | 1,480 | 7,818 | ||
| POWV-56 | ++ | ++ | ++ | 11,477 | |||
| POWV-63 | ++ | ++ | ++ | 2,520 | 11,183 | ||
| C | POWV-14 | 10,289 | |||||
| POWV-15 | |||||||
| POWV-23 | |||||||
| D | POWV-48 | ||||||
| POWV-62 | ++ | ||||||
| POWV-70 | ++ | ||||||
| E | POWV-33 | ++ | 1,615 | ||||
| POWV-37 | ++ | 587 | |||||
| POWV-65 | |||||||
| POWV-71 | ++ | ++ | 44 | ||||
| POWV-80 | ++ | ++ | 8,475 | 10,354 | |||
| F | POWV-55 | ||||||
| POWV-60 | |||||||
| POWV-61 | |||||||
Cross-reactivity to other TBFVs was determined by mAb binding of C-prM-E–transfected cells by flow cytometry.
Cross-neutralization was assessed by FRNT (LGTV) or an RVP-based assay (TBEV and GGYV) in Vero cells. Geometric mean EC50 values are from two to three independent experiments.
We next tested whether the cross-reactive mAbs could inhibit infection of the other TBFVs (Fig. 3 B and Table 2). POWV-54, -56, and -63 in group B all neutralized LGTV, albeit weakly (EC50 of 1.5, 11.5, and 2.5 µg/ml, respectively), with POWV-54 and -63 also exhibiting some cross-neutralization of GGYV RVPs (7.8 µg/ml and 11.2 µg/ml, respectively). The cross-reactive DIII-binding mAbs in group E also cross-neutralized other TBFVs. POWV-33, -37, and -80 inhibited TBEV RVPs (EC50 of 1.6, 0.6, and 8.5 µg/ml, respectively), and POWV-71 and POWV-80 neutralized LGTV infection (EC50 of 0.04 and 10.3 µg/ml, respectively).
We evaluated whether the cross-reactivity extended to the MBFV clade using a panel of RVPs displaying the structural proteins of dengue virus serotype 2 (DENV-2), St. Louis encephalitis virus (SLEV), Zika virus (ZIKV), yellow fever virus (YFV), West Nile virus (WNV), and Usutu virus (USUV). We initially assessed cross-reactivity using an antibody-dependent enhancement (ADE) assay because the required stoichiometry of binding for ADE is less than that for neutralization (Pierson et al., 2007). The group B cross-reactive mAbs POWV-54, POWV-56, and POWV-63 all enhanced MBFV infection of K562 cells, whereas a type-specific antibody POWV-15 did not (Fig. 3 C). Minimal to no enhancement of the MBFVs was observed for any of the other TBFV–cross-reactive or POWV-type–specific mAbs. The MBFV–cross-reactive mAbs subsequently were tested for neutralization activity against DENV-2, SLEV, and ZIKV RVPs. POWV-54, POWV-56, and POWV-63 all cross-neutralized these RVPs (Fig. 3 D), although the potency of inhibition was decreased against mature particles produced in Vero-furin cells (Fig. 3 E). Thus, these pan–flavivirus-neutralizing mAbs are maturation state sensitive.
Epitopes recognized by anti-POWV mAbs
Because of their pan-flavivirus cross-reactivity and maturation state dependence of neutralization, we hypothesized that POWV-54, POWV-56, and POWV-63 might recognize an epitope in the conserved fusion loop in DII of the E protein. To test this idea, we performed ELISAs using recombinant WNV E protein, with a WT protein or a variant with four mutations in the fusion loop epitope (FLE): T76A, M77G, W101R, and L107R (Oliphant et al., 2007; Vogt et al., 2011). Fusion loop mAb WNV-E60, DIII mAb WNV-E24, and POWV-54 and POWV-63 all bound avidly to WT WNV E protein (Fig. 4 A). POWV-56 displayed minimal binding to WNV E, which is consistent with its inability to promote ADE of WNV RVPs (Fig. 3 C). Whereas the DIII-specific mAb WNV-E24 bound to the WNV E fusion loop mutant, WNV-E60 and the POWV mAbs did not (Fig. 4 B). Thus, POWV-54 and POWV-63 recognize an epitope that requires an intact DII fusion loop.
Figure 4.
Mapping of anti-POWV mAb epitopes. (A and B) The indicated mAbs were tested for binding by ELISA to WT WNV E protein (A) or a WNV E fusion loop variant (B) with T76A, M77G, W101R, and L107R mutations. Error bars represent the range from two technical replicates. Data are representative of three experiments. (C–F) POWV RVPs encoding escape mutations were tested for neutralization by the indicated mAbs on Raji-DCSIGNR cells. WT RVPs were compared with R73C (C), H62Y (D), D231Y (E), and S390V (F). Dose-response curves from two to four experiments. Error bars represent the range from two technical replicates. (G and H) POWV mAb binding to a panel of E protein mutants. Only selected mutations in DIII (G) or DI and DII (H) of the E protein are shown; see Fig. S2 for all tested mutations. Mean binding relative to WT is shown. Error bars represent SEM from three experiments (ANOVA with Dunnett’s post-test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Dotted lines represent the isotype control value. (I) Residues determined in Fig 4, A–H and Fig. S3 as important for binding by the indicated mAbs are highlighted on the E protein dimer of TBEV (PDB accession no. 5O6A). Residues are highlighted red, orange, green, blue, cyan, or purple in the top E protein (in gray). The bottom E protein is colored by domain: DI is red, DII is yellow, and DIII is blue.
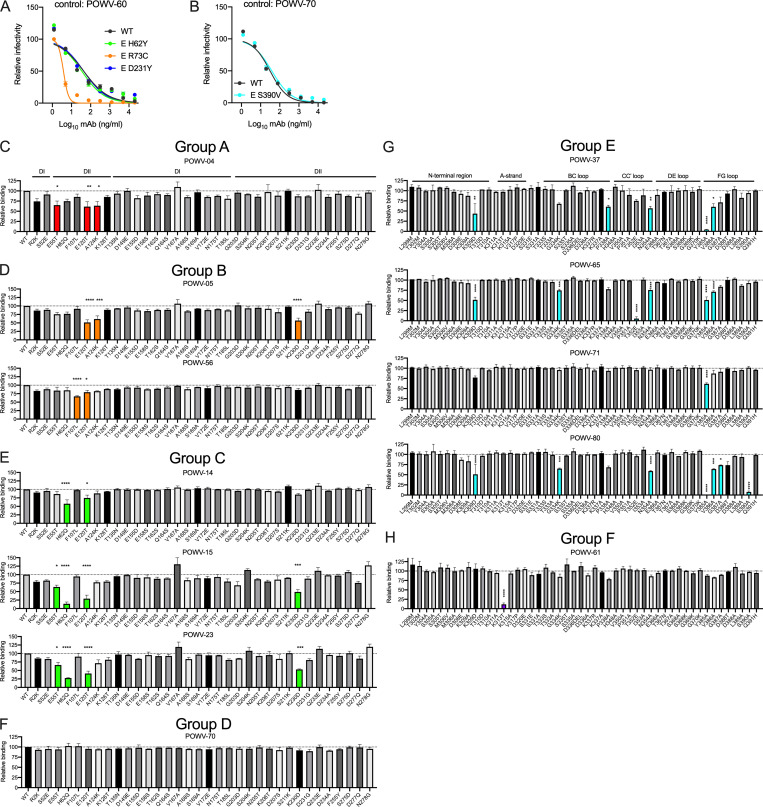
To identify additional residues that are important for recognition by the anti-POWV mAbs, we selected for neutralization escape mutations by serially passaging POWV-MA51240 in the presence of antibody. The resulting viruses were sequenced, and mutations were introduced into the POWV C-prM-E expression plasmid. POWV RVPs bearing these mutations were produced, and the variants were tested for neutralization by the selecting antibody. Both type-specific mAbs in group B, POWV-5 and POWV-18, selected for mutation at R73C in the DII b-c loop near the fusion peptide, which conferred resistance to neutralization by either antibody (Fig. 4, C and I). All three mAbs in group C (POWV-14, POWV-15, and POWV-23) selected for mutation at H62Y in DII, close to the E dimer interface, which conferred resistance to neutralization by all three mAbs (Fig. 4, D and I). Group D mAb POWV-70 selected for and did not neutralize virus with mutation D231Y in DII, which is in the h-i loop on the face opposite to the inter-dimer interface (Fig. 4, E and I). These data indicate that epitopes recognized by mAbs in groups B, C, and D map, at least in part, to residues in DII. POWV-80 selected a variant S390V, which is located in the F-G loop of the lateral ridge of DIII, and conferred resistance to neutralization of this mAb (Fig. 4, F and I). As a control, all escape mutants also were tested for neutralization by an antibody in a different competition group than the selecting antibody; all escape mutants were inhibited by these other mAbs (Fig. S2, A and B)
Figure S2.
Epitopes recognized by anti-POWV antibodies. (A and B) Escape mutants from Fig 4, C–F were tested for neutralization by mAbs that bind an epitope distinct from the antibody that selected for their growth. (A) Mutations in DII were tested for their effect on POWV RVP neutralization by DIII mAb POWV-60. (B) DIII mutant S390V was tested for neutralization by non-DIII mAb POWV-70. (C–H) Anti-POWV mAbs were tested for binding to the indicated E protein mutants encoding single amino acid changes in DI and DII (C–F) or DIII (G and H); binding relative to WT is shown. Error bars represent SEM from three experiments (ANOVA with Dunnett’s post-test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Colored bars represent statistically significant decreases in binding, P < 0.05. Dotted lines represent the isotype control value.
To complement our escape mutant studies, we identified residues important for anti-POWV mAb binding using site-directed mutagenesis of the E protein followed by screening for loss of mAb binding by flow cytometry. Group E mAbs POWV-37, POWV-65, and POWV-80 showed reduced binding to E proteins harboring mutations in the N-terminal region, B-C loop, C-C′ loop, and F-G loop of DIII (Fig. 4, G and I; and Fig. S2 G). Group E mAb POWV-71 exhibited reduced binding to the Y385A mutant alone, which maps to the F-G loop in DIII (Fig. 4 G and Fig. S2 G). Group F mAb POWV-61 lost binding only to a mutation at K313 in the A-strand of DIII (Fig. 4, G and I; and Fig. S2 H).
Group A mAb POWV-4 had decreased binding to residues 55, 120, and 124 in DII, although none of these were by >50% (Fig. 4, H and I; and Fig. S2 C). Group B mAb POWV-5 exhibited diminished binding when mutations were introduced at residues 120, 124, and 230 of DII (Fig. 4, H and I; and Fig. S2 D). Group B mAb POWV-56 showed reduced binding to DII mutations in the fusion loop at residue F107 and at residue E120, establishing fusion loop binding by this mAb. Group C mAbs POWV-14, POWV-15, and POWV-23 all showed decreased binding to the H62Q mutant, as well as variable reduction with mutations at residues 55, 120, and 230 in DII (Fig. 4 H and Fig. S2 E). Group D mAb POWV-70 did not exhibit reduced binding against any of the domain I (DI)/DII mutants (Fig. S2 F).
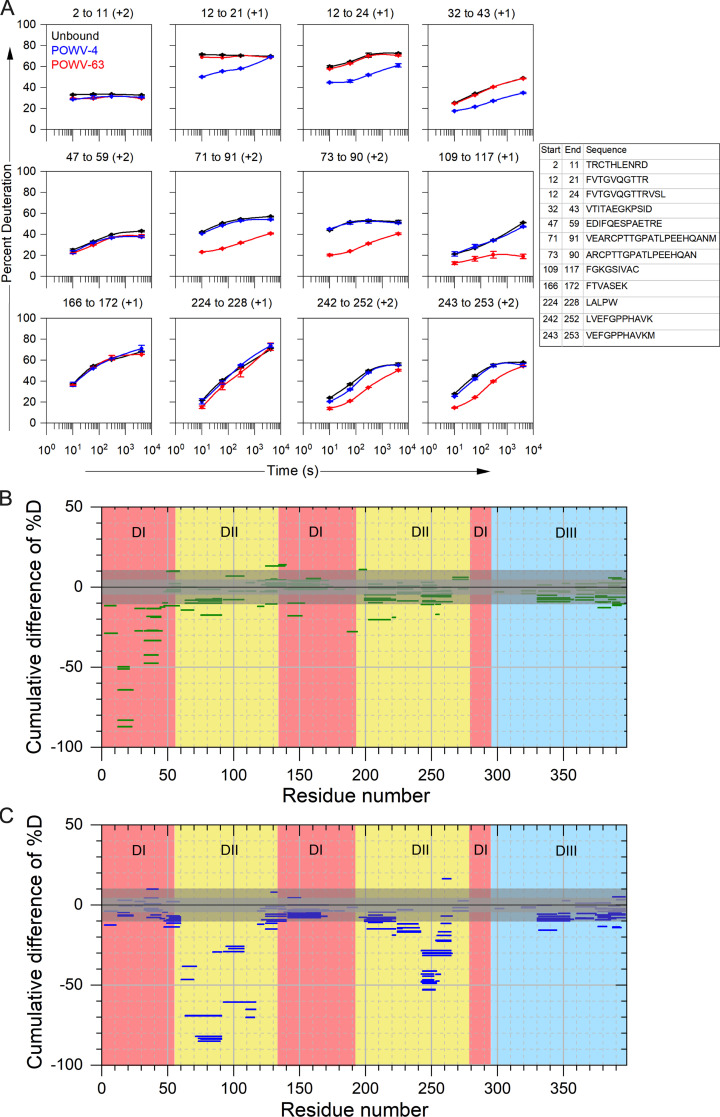
Because critical residues for POWV-4 binding were not selected in the site-directed mutagenesis screen, we performed hydrogen-deuterium exchange (HDX) mass spectrometry (MS) with recombinant POWV E protein and POWV-4, as well as with POWV-63 to confirm DII FLE binding. MS analysis revealed that POWV-4 binding protected peptide epitopes in DI of the E protein, specifically peptides containing residues 12–24 near the A0-B0 loop and residues 32–43 near the C0-D0 loop (Fig. S3, A and B; and Fig. 4 I). In contrast, POWV-63 binding protected peptides near the DII FLE, at residues 71–91 and 109–117 as well as at residues 243–253, which are structurally adjacent to the FLE (Fig. S3 A and C; and Fig. 4 I).
Figure S3.
HDX of E protein bound to POWV-4 and POWV-63. (A) Representative kinetic plots for 12 different peptides showing effects of antibody binding on HDX. Black, blue, and red lines are for POWV E in the absence of antibody, in the presence of POWV-4 mAb, and in the presence of POWV-63 mAb, respectively. At the top of each panel are the residue numbers and charge states of the peptide, and sequences of each peptide are given as a table. Error bars represent SEM from duplicate measurements. (B and C) Woods plots showing accumulated difference in %D (bound state–unbound state) across all time points for each analyzed peptide for POWV-4 (B) and POWV-63 (C). Central and outer gray shaded areas indicate 95% and 99% confidence intervals, respectively, for the global SEM calculated for each differential HDX experiment.
Taken together, these data demonstrate that the non–DIII-binding mAbs in groups B, C, and D recognize epitopes within DII of the E protein. Group B mAbs recognize epitopes containing or proximal to the DII fusion loop, whereas group C and D mAbs bind residues in the middle section of DII. The sole group A member, POWV-4, binds to residues in DI of the E protein. MAbs in group E bind an epitope composed of residues in both the C-C′ loop and the lateral ridge of DIII, whereas mAbs in group F bind a DIII epitope that includes the A-strand.
The cross-reactive mAb POWV-80 targets the C-C′ loop of DIII
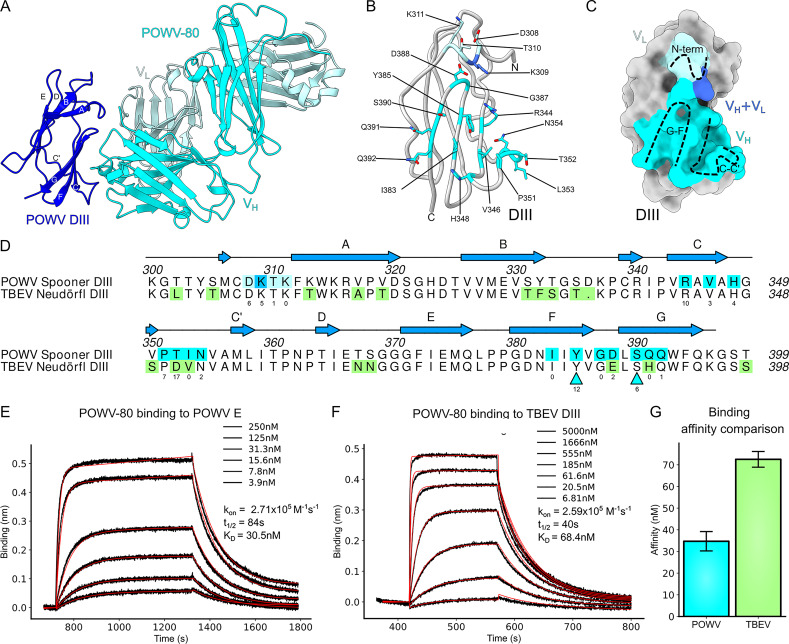
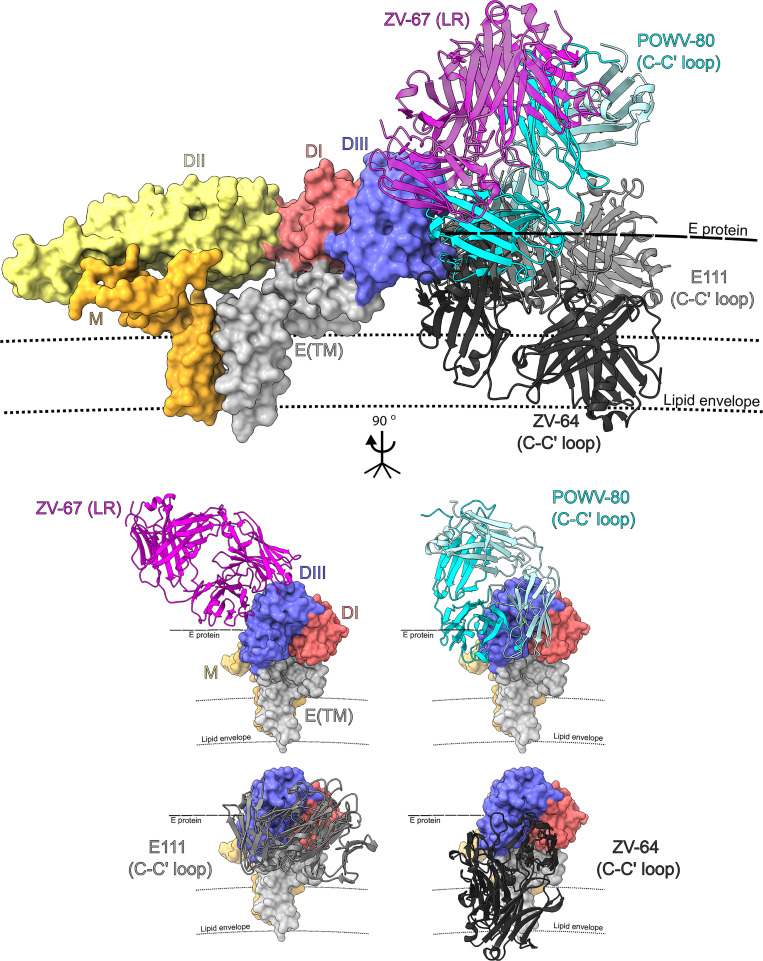
To elucidate how POWV-80 can cross-react to other TBFVs, we solved the crystal structure of a complex of POWV DIII bound by the antigen-binding fragment (Fab) of POWV-80 at 2.0-Å resolution (Fig. 5 A and Table S1 A). POWV-80 targets the conserved, cryptic C-C′ loop epitope, which previously was described for DENV and ZIKV mAbs (Austin et al., 2012; Zhao et al., 2016). The majority of the contact residues in the epitope lie in a β sheet formed by the C-C′ and F-G strands and loops, with some involvement from N-terminal residues (Fig. 5 B). The POWV-80 paratope is formed mostly by the heavy chain, with minor light chain involvement (buried surface area of 631 Å2 versus 55 Å2, respectively; Fig. 5 C). Despite overall similarity to other C-C′ loop mAbs, POWV-80 has more contacts in the F-G loop than anti–DENV-2 mAb E111 and anti-ZIKV mAb ZV-64. Furthermore, POWV-80 adopts a unique binding orientation to DIII that aligns closely with potently neutralizing lateral ridge antibodies (Nybakken et al., 2005; Zhao et al., 2016) rather than other C-C′ loop mAbs (Fig. 6).
Figure 5.
The cross-reactive mAb POWV-80 binds to the C-C′ loop of DIII. (A) 2.0-Å crystal structure of POWV DIII (SPO strain) bound by POWV-80 Fab, depicted as a ribbon diagram. POWV-80 heavy and light chains are colored in cyan and turquoise, respectively. DIII is depicted in blue. (B) Ribbon diagram of POWV DIII with epitope contact residue side chains shown as sticks colored cyan for heavy chain contacts, pale turquoise for light chain contacts, or blue for dual heavy/light chain contacts. (C) Surface depiction of POWV DIII showing the buried surface area covered by POWV-80, colored as in B. (D) Sequence alignment of POWV DIII and TBEV DIII. Contact residues in the POWV-80 epitope are colored as in B. Residues that cause loss of binding are indicated by cyan triangles. Residues highlighted in green in the TBEV DIII sequence represent substitutions relative to POWV DIII. Numbers below contact residues indicate the number of close contacts (<3.9-Å distance between atoms). (E) Representative BLI plot showing association-dissociation curves of POWV-80 binding to POWV E protein. Raw traces are shown in black, with fitted curves used to generate binding constants shown as red lines. (F) Representative BLI plot showing association-dissociation curves of POWV-80 binding to TBEV DIII. (G) Bar graph summarizing the dissociation constants calculated in E and G. Error bars represent SD from three independent experiments.
Figure 6.
Binding orientation of POWV-80 compared with other C-C′ loop antibodies. Structures of POWV-80 (cyan and pale turquoise, C-C′ loop antibody), ZV-67 (magenta, lateral ridge antibody), E111 (light gray, C-C′ loop antibody), and ZV-64 (dark gray, C-C′ loop antibody) docked onto the structure of TBEV E (PDB accession no. 5O6A). The bottom set of dashed lines (smaller dashes) represents the lipid bilayer that comprises the viral envelope. The thicker dashed line above represents the E protein shell of the virion. E(TM), E protein transmembrane domain.
Several residues in the epitope targeted by POWV-80 are involved in hydrogen bonding and salt-bridge formation with the paratope (Table S1 B). Most notably, Y385 forms hydrogen bonds with heavy chain residues H33 and Y55 on the CDR-1 and -2 of POWV-80. Additionally, K309 and R344 form multiple salt bridges with basic residues E102 and D114 in the POWV-80 CDR3. Whereas Y385, K309, and R344 are identical between POWV and TBEV DIII, several other epitope residues at the binding interface are substituted (Fig. 5 D). To understand the impact of these substitutions on POWV-80 cross-reactivity, we measured the binding affinity of POWV-80 to recombinant TBEV DIII using biolayer interferometry (BLI) and compared it to POWV E. POWV-80 binds POWV E with an affinity (KD) of ∼30 nM (Fig. 5 E and Fig. S4). POWV-80 also binds TBEV DIII strongly, with a KD of ∼75 nM, only a small decrease relative to POWV (Fig. 5, F and G). These results show that POWV-80 efficiently binds TBEV DIII despite multiple substitutions in its epitope, explaining why POWV-80 neutralizes TBEV RVPs (Fig. 3B). Almost all of the loss in affinity is due to an increased dissociation rate (decreased half-life) for POWV-80 binding to TBEV DIII, as the association rates (kon) for both complexes are nearly identical (Fig. 5, E and F).
Figure S4.
Association-dissociation curves for POWV mAbs binding to POWV E protein. (A) Representative association-dissociation curves for all POWV mAbs that bound to recombinant E protein. Black lines indicate raw data; red lines show fits used to derive binding affinity constants. Each plot represents one of at least three independent experiments used to generate summary dissociation constants and binding half-lives in Table 1.
Anti-POWV mAbs protect against POWV challenge in vivo
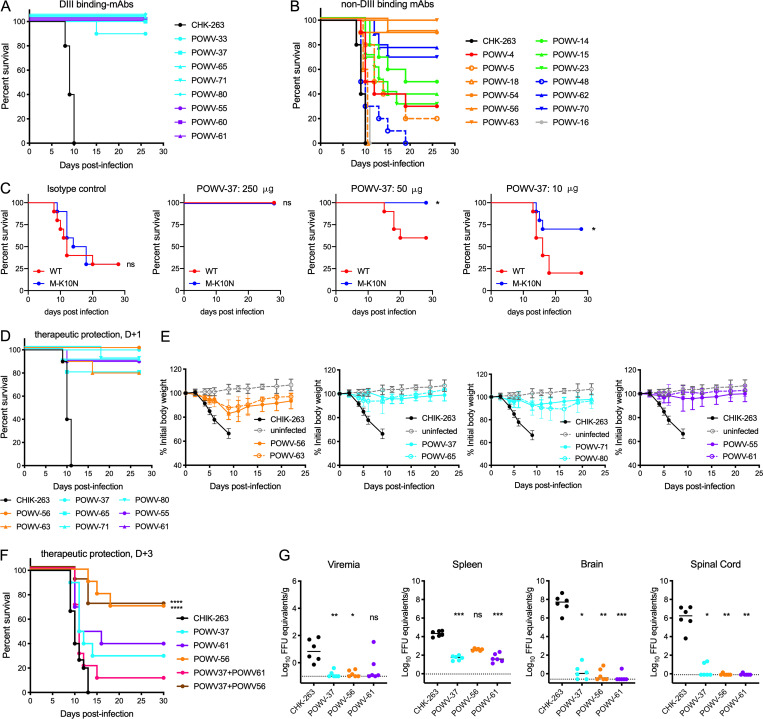
We tested the anti-POWV mAbs for protection against lethal virus challenge in vivo. 7-wk-old C57BL/6J mice were administered a 250-µg dose (12.5 mg/kg) of anti-POWV mAb as prophylaxis, as well as 500 µg of an anti-Ifnar1 mAb to enhance replication and lethality (VanBlargan et al., 2018). 1 d later, mice were inoculated with POWV-SPO. Isotype control mAb (CHK-263)–treated mice succumbed to infection 8–10 d after infection, whereas mice treated with any of the DIII-binding anti-POWV mAbs were protected (Fig. 7 A). Mice treated with non–DIII-binding POWV mAbs showed variable outcomes (Fig. 7 B). Treatment with group B mAbs POWV-63, -56, and -54 resulted in 90–100% survival of infected mice, whereas other group B mAbs (POWV-5 and -18) were not protective (0–20% survival). Two group D mAbs, POWV-70 and -62, partially protected against lethal POWV infection (70–80% survival), but another group D mAb (POWV-48) did not protect (0% survival), even though POWV-48, -62, and -70 share sequence homology and bind a similar epitope (Table 1). Group A mAb POWV-4 and all group C mAbs (POWV-14, -15, and -23) were partially protective against POWV challenge (30–50% survival). Lastly, POWV-16, the sole prM-E-binding mAb, did not confer any protection against mortality.
Figure 7.
Anti-POWV mAbs protect against POWV challenge in vivo. (A and B) 7-wk-old C57BL/6J mice were administered 250 µg of the indicated mAb and 500 µg of anti-Ifnar1 mAb by intraperitoneal injection 24 h before subcutaneous inoculation with 102 FFU of POWV-SPO. Survival of mice receiving DIII-binding mAb treatment (A) and non–DIII-binding mAb treatment (B) are shown relative to mice receiving isotype control mAb (CHK-263; n = 10 per group). (C) 4–5-week-old mice were administered indicated doses of POWV-37 (or 250 µg isotype control mAb) by intraperitoneal injection 24 h before subcutaneous inoculation of 102 FFU of POWV WT or M K10N virus generated from the POWV-SPO infectious clone. n = 9 or 10 per group (log-rank test: *, P < 0.05). (D and E) 7-wk-old C57BL/6J mice were administered 500 µg of anti-Ifnar1 mAb by intraperitoneal injection 24 h before subcutaneous inoculation of 102 FFU POWV-SPO, followed by administration of 250 µg of the indicated mAb 24 h after inoculation. Mice were monitored for mortality (D) and weight loss (E) for 4 wk after viral challenge; n = 10 per group. Mean weight change is shown; error bars represent ± SD. (F) 5-wk-old mice were inoculated with POWV-SPO and 72 h later were administered 250 µg of each indicated mAb or 125 µg of each mAb when administered as a combination; n = 10 per group (log-rank test with Bonferroni correction: ns when not listed; ****, P < 0.0001). (G) 5-wk-old C57BL/6J mice were administered 250 µg of the indicated mAb before subcutaneous inoculation with POWV-SPO. At 8 d after infection, the indicated tissues were harvested, and viral load was determined by qRT-PCR. Median viral titers are shown (Kruskal-Wallis test with Dunn’s post-test: ****, P < 0.0001). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Dotted lines represent the limit of detection of the assay. (A–G) Data are from two experiments.
To probe whether the increased neutralization by mAbs against the M-K10N virus was associated with enhanced protection in vivo, varying doses of POWV-37 were administered to mice 1 d before inoculation with infectious clone-derived WT or M-K10N POWV-SPO. Following treatment with 250 µg of an isotype control mAb, infection with either WT or M-K10N POWV resulted in 70% lethality (Fig. 7 C). A 250-µg dose of POWV-37 was fully protective against both WT and M-K10N POWV challenge. However, although a 50-µg dose fully protected against POWV M-K10N challenge, this dosage conferred less protection against WT POWV. Similarly, a 10-µg dose of POWV-37 conferred greater protection against challenge with POWV M-K10N than WT POWV. Thus, the increased neutralization sensitivity of POWV M-K10N translates to increased mAb protection in vivo. Despite the differences in virus stability in vitro noted previously, WT and M-K10N POWV infection in vivo resulted in a similar level of lethality with equivalent kinetics (Fig. 7 C, left panel).
We next tested the efficacy of mAbs that performed well in the prophylaxis model in a postexposure therapeutic setting using POWV-SPO. Mice were administered 500 µg anti-Ifnar1 mAb 1 d before virus inoculation and 250 µg anti-POWV mAb 1 d after infection with POWV-SPO. All the mAbs tested prevented lethal POWV infection, with POWV-37, POWV-56, and POWV-61 conferring virtually complete protection (Fig. 7, D and E).
We extended these results by assessing whether POWV-37, -56, and -61 could protect when administered 3 d after infection. Treatment with 250 µg of the DIII-binding mAb POWV-37 or POWV-61 offered moderate protection against POWV challenge (30–40% survival; Fig. 7 F). Because POWV-37 and POWV-61 are in different competition groups, we tried combination therapy to potentially limit the generation of escape mutants, as was seen with monotherapy with DIII-binding mAbs against WNV (Zhang et al., 2009). A combination of POWV-37 and POWV-61 (125 µg of each) afforded less protection against infection than monotherapy (10% survival), possibly reflecting the lower dose of each individual mAb. In comparison, therapeutic treatment at day +3 with the fusion loop mAb POWV-56 substantially protected mice against POWV infection (70% survival), and a combination therapy of POWV-56 and POWV-37 (125 µg of each) showed a similar level of protection as monotherapy with POWV-56. Thus, POWV-56, a fusion loop mAb, offered more protection than DIII-binding mAbs when administered 3 d after infection.
We next evaluated whether POWV-37, -61, and -56 limit viral burden using the prophylaxis model. Serum, spleen, brain, and spinal cord tissues were collected 8 d after virus inoculation and assessed by quantitative RT-PCR (qRT-PCR). Prophylaxis with POWV-37, -56, and -61 all resulted in a ∼50-fold reduction in viremia relative to control mAb-treated mice (Fig. 6 G). Treatment with POWV-37 and -61 decreased the viral load in the spleen by ∼400–600-fold, whereas POWV-56 treatment reduced infection ∼50-fold. Treatment with any of the POWV mAbs greatly reduced viral RNA levels in the central nervous system with a 6-log10 reduction in the brain and spinal cord relative to control mAb-treated mice. Thus, POWV mAbs likely protect against mortality because they prevent infection in the central nervous system.
Cross-protection by POWV mAbs against TBFV challenge in vivo
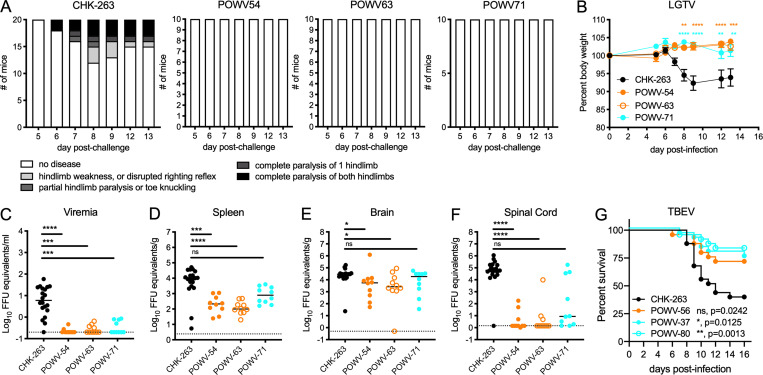
We assessed whether some of the cross-reactive mAbs could protect against challenge with heterologous TBFVs in vivo. POWV-54, POWV-63, and POWV-71 were administered to C57BL/6J mice treated with anti-Ifnar1 mAb before challenge with LGTV. Mice treated with POWV-54, POWV-63, or POWV-71 were protected from clinical disease, whereas 40% of control mAb-treated mice developed paralysis (Fig. 8 A). Additionally, 50% of isotype control mAb-treated mice lost over 5% body weight, with 30% of mice losing 15–30% body weight. In comparison, POWV-54– and POWV-63–treated mice maintained 95% body weight or more, and for POWV-71–treated mice, only 20% displayed minor (5–15%) weight loss (Fig. 8 B). POWV-54–, POWV-63–, or POWV-71–treated mice all had lower viremia than isotype control mAb-treated mice with 20- to 30-fold reductions (Fig. 8 C). Decreased viral titers also were observed in the spleen following POWV-54 (30-fold) and POWV-63 (50-fold) treatment, whereas POWV-71 was less protective (Fig. 8 D). Unexpectedly, viral titers in the brain were not greatly reduced following treatment with POWV-54 (sixfold), POWV-63 (ninefold), or POWV-71 (fourfold; Fig. 8 E), although large reductions in the spinal cord were observed in mice treated with POWV-54 (13,000-fold) and POWV-63 (10,000-fold; Fig. 8 F). POWV-71 treatment resulted in a smaller reduction in viral titer in the spinal cord (500-fold). Thus, the cross-reactive mAbs POWV-54, POWV-63, and POWV-71 protect mice against LGTV infection and disease, although POWV-71 had less activity than POWV-54 and POWV-63 despite showing higher neutralizing titers (Table 2).
Figure 8.
Cross-protection by POWV mAbs against LGTV challenge in vivo. (A and B) 7-wk-old C57BL/6J mice were administered 250 µg of the indicated mAb and 500 µg of anti-Ifnar1 mAb by intraperitoneal injection 24 h before subcutaneous inoculation with 102 FFU of LGTV. (A) Mice were scored for paralysis following viral infection. (B) Mice were monitored for weight change for 13 d following viral infection. Mean weight change is shown. Error bars represent SEM (ANOVA with Sidak’s post-test: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (C–F) At day 14 after viral challenge, serum (C), spleen (D), brain (E), and spinal cord (F) were harvested, and viral load was determined by qRT-PCR. Median viral titers (Kruskal-Wallis ANOVA with Dunn’s post-test: *, P < 0.05; ***, P < 0.001; ****, P < 0.0001). Data are from two experiments. Dotted lines represent the limit of detection of the assay. (G) C57BL/6J mice were administered 250 µg of the indicated mAb by intraperitoneal injection 24 h before subcutaneous inoculation with 104 FFU of TBEV; n = 25 per group (log-rank test with Bonferroni correction: ns, P > 0.0167; *, P < 0.0167; **, P < 0.01). Data are from three experiments.
TBEV–cross-reactive mAbs POWV-56, POWV-37, and POWV-80 were tested for their ability to protect against lethal challenge with TBEV in WT C57BL/6J mice when administered 24 h before infection. While only 40% of mice survived TBEV infection following treatment with the isotype control mAb CHK-263, treatment with POWV-56, POWV-37, and POWV-80 resulted in 70–85% survival (Fig. 8 G).
Discussion
Neutralizing mAbs with therapeutic potential against flaviviruses have been previously characterized, although no mAbs raised specifically against POWV have been described. We developed a panel of mAbs that recognize multiple epitopes on the POWV E glycoprotein, as well as one that engages an epitope across prM and E. Anti-E mAbs against POWV separated into six competition groups, with one group binding DI, three groups recognizing epitopes on DII, and two groups binding epitopes on DIII. Regions targeted by the DIII-binding mAbs included the lateral ridge, C-C′ loop, and A-strand. Non–DIII-binding mAbs principally targeted epitopes on E-DII, including the FLE, the DII dimer interface, and the DII central interface. MAbs from all six groups were neutralizing against POWV lineage I and lineage II viruses. All DIII-binding mAbs protected against lethal POWV-SPO challenge when administered as prophylaxis, whereas non–DIII-binding mAbs showed a broader range of activity, with only a subset (DII FLE mAbs) demonstrating complete protection. DIII mAbs and DII FLE mAbs exhibited protective activity when administered as therapy after POWV-SPO infection. MAbs from DI, DII, and DIII epitope groups cross-reacted with other TBFVs and, in the case of the DII FLE mAbs, against MBFV as well. Both DIII and DII FLE cross-reactive mAbs conferred protection against LGTV or TBEV infection in vivo.
Most mAbs neutralized lineage I (LB) and lineage II (SPO) POWV strains equivalently. However, when tested against another lineage II strain, MA51240, a more recent isolate from the same geographic region as POWV-SPO, the anti-E mAbs were more inhibitory despite being raised against POWV-SPO. Because mAbs from six unique competition groups were more inhibitory against the POWV-MA51240 strain, we hypothesized that a global difference in E protein epitope exposure might broadly impact antibody neutralization, as was described for WNV (Goo et al., 2017). Indeed, the POWV-SPO and POWV-MA51240 strains have identical E protein sequences but vary at M protein residue 10. Using a reverse genetics approach, we confirmed that the K10N substitution of the M protein mediated the difference in neutralization sensitivity between the two lineage II strains.
Multiple factors can impact flavivirus neutralization sensitivity to antibodies independent of sequence differences within epitope–paratope interactions. The degree of virus maturation can affect accessibility of epitopes for antibody binding, including those targeting the highly conserved fusion loop, which often is cryptic on fully mature virions (Nelson et al., 2008; Stiasny et al., 2006). Although the M-K10N mutation affected the degree of prM cleavage, this result did not explain the discrepancy in neutralization sensitivity between WT and M-K10N viruses. Changes in viral breathing, the ensemble of structural conformations sampled by the virus particle, also can impact epitope accessibility and neutralization sensitivity. A consequence of extensive viral breathing is an enhanced decay of virus infectivity over time (Kuhn et al., 2015). We demonstrated that POWV-SPO M-K10N has a faster rate of intrinsic decay than WT POWV. While most of the M protein closely associates with the viral membrane, the M-K10 residue is located in the N-terminal loop and is thought to stabilize the E protein dimer by associating with DII (Füzik et al., 2018; Zhang et al., 2013). We hypothesize that the M-K10N substitution alters the conformations sampled by the E protein, which increases the exposure of both DII and DIII epitopes for antibody binding and decreases the inherent stability of the virus. Remarkably, the M-K10N variation altered neutralization sensitivity to all mAbs tested, including DII- and DIII-specific mAbs; thus, substitutions in the flavivirus M protein globally can affect neutralization of anti–E protein antibodies. Although the M-K10N substitution was detected in a virus isolated from ticks in a POWV-endemic region (Pesko et al., 2010), its prevalence appears low; it is the only strain of 27 available full-length POWV sequences with this change in the Virus Pathogen Database and Analysis Resource.
The most protective anti-POWV mAbs targeted the DIII lateral ridge/C-C′ loop, the DIII A-strand, and the DII FLE. Many potently neutralizing and protective anti-flavivirus mAbs recognize DIII epitopes (Oliphant et al., 2007; VanBlargan et al., 2016; Vratskikh et al., 2013; Wahala et al., 2009). In comparison, mAbs targeting the highly conserved DII FLE often are cross-reactive yet weakly neutralize owing to the cryptic nature of their epitope (Nelson et al., 2008; Stiasny et al., 2006). Though DII FLE mAbs can confer protection in vivo, they often require Fc-dependent effector functions (Vogt et al., 2011). One exception to this is the highly protective and neutralizing E-dimer epitope mAbs that bind a complex quaternary epitope that includes the DII FLE (Rouvinski et al., 2015); however, antibodies against this epitope have not yet been described for TBFVs. Notably, in our study, the DII FLE mAb POWV-56 was more protective than DIII-specific mAbs POWV-37 and POWV-61 when administered after infection. The cross-reactive DII FLE mAbs POWV-54, POWV-56, and POWV-63 also protected against heterologous viral challenge with either LGTV or TBEV, despite lower neutralization potency against those viruses. Whether the effector functions of DII FLE mAbs are required for their therapeutic efficacy requires further study.
In addition to the DII FLE mAbs, several of the DIII-specific mAbs that recognize a lateral ridge/C-C′ loop epitope were cross-reactive, cross-neutralizing, and protective against LGTV and TBEV. Protective anti-ZIKV mAbs similarly were identified that bind epitopes on the DIII-LR and C-C′ loop on ZIKV and cross-react with DENV1 (Robbiani et al., 2017; Sapparapu et al., 2016; Zhao et al., 2020). Thus, the DIII–LR/C-C′ loop epitope represents an antigenic site that can be targeted by multiple MBFV and TBFV members.
In our panel of anti-POWV mAbs, DI-, DII-, and DIII-specific mAbs neutralized POWV infection at a postattachment step, likely through inhibition of fusion to the host cell membrane. Many anti-flavivirus antibodies, including both DIII and DII-FLE mAbs, previously were shown to block infection at postattachment steps (Füzik et al., 2018; Gollins and Porterfield, 1986; Lai et al., 2007; Nybakken et al., 2005; Thompson et al., 2009). Thus, this common mechanism of inhibition for highly neutralizing anti-flavivirus antibodies also extends to anti-POWV mAbs.
Structural analysis of POWV-80, an antibody that neutralizes in vitro and protects in vivo, revealed that it targets the C-C′ loop epitope on DIII, with some engagement of the F-G loop. Alignment of the structure of POWV-80 bound to POWV DIII with those of several other antibodies to heterologous flaviviruses showed that POWV-80 adopts a binding orientation more similar to the potently neutralizing lateral ridge antibodies than other C-C′ loop mAbs. This orientation may be sterically more favorable for binding of POWV-80 than other C-C′ loop antibodies, as the bulk of the constant region is oriented perpendicular to the surface of the virus, whereas other C-C′ loop mAbs bind in the plane of the viral envelope. This may explain why POWV-80 is more potently neutralizing than other C-C′ loop antibodies against DENV and ZIKV (Shrestha et al., 2010; Zhao et al., 2016).
Despite several substitutions in the epitope footprint of POWV-80 between POWV and TBEV, POWV-80 still binds with relatively high affinity to TBEV DIII and can neutralize TBEV in vitro and protect from TBEV infection in vivo. Although 4 of the 18 residues in the epitope are substituted, only one of these is conservatively changed (D388E). This shows that POWV-80 maintains high-affinity binding due to the conserved nature of the major contact residues. LGTV contains only one substitution in these residues (S390N), and POWV-80 displays a similar neutralization potency against LGTV as TBEV, suggesting that multiple substitutions may be necessary to abrogate the cross-reactivity of POWV-80 binding.
In summary, we have described a unique panel of neutralizing mAbs with therapeutic potential against an emerging tick-borne viral infection. Antibodies against both the DII FLE and the DIII lateral ridge/C-C′ loop are cross-reactive and broadly neutralizing and confer protection in vivo against multiple TBFVs. These results provide insight into the protective components of the humoral immune response to POWV, which may inform the design of vaccines and antibody therapeutics against POWV and other clinically relevant TBFVs.
Materials and methods
Viruses
POWV lineage I strain LB (Mandl et al., 1993) was isolated from a human brain sample. POWV lineage II (DTV) strain SPO was isolated from an adult deer tick (Ebel et al., 1999). LGTV strain E5 was derived from strain TP21 (Campbell and Pletnev, 2000). POWV-LB, POWV-SPO, and LGTV were obtained from World Reference Center for Emerging Viruses and Arboviruses (K. Plante and S. Weaver, University of Texas Medical Branch, Galveston, TX). POWV strain MA51240 was isolated from ticks collected in Wisconsin (Brackney et al., 2008). Virus stocks were propagated on Vero cells and used at passages 3 and 4. Infectious clone–derived viruses for WT and M-K10N POWV-SPO were produced in BHK21 cells as previously described (Kenney et al., 2018), followed by propagation for one passage on Vero cells. Mature virus stocks were propagated in Vero-furin cells (Mukherjee et al., 2016). Viral titer was determined by FFA on Vero cells as described (VanBlargan et al., 2018). TBEV strain Sofjin was provided by Dr. Michael Holbrook (National Institute of Allergy and Infectious Disease/National Institutes of Health [NIH], Bethesda, MD) and propagated in Vero cells. Viral titer was determined by limiting dilution plaque assay. All work with infectious TBEV was performed in the BSL4 facility at NIH Rocky Mountain Laboratories in accordance with approved biosafety protocols.
Cells
Cell lines were maintained at 37°C in the presence of 5% CO2. HEK-293T and BHK-21 cells were passaged in DMEM (Invitrogen) supplemented with 10% FBS (Omega Scientific) and 100 U/ml penicillin-streptomycin (P/S; Invitrogen). Vero cells were passaged in DMEM supplemented with 5% FBS and 100 U/ml P/S. Vero cells that overexpress furin (Vero-furin; Mukherjee et al., 2016) were maintained as WT Vero cells with the addition of 5 µg/ml blasticidin. Raji B lymphoblast cells stably expressing the C-type lectin DC-SIGNR (Raji-DCSIGNR; Davis et al., 2006) were cultured in RPMI 1640 medium (Invitrogen) supplemented with 7% FBS and 100 U/ml P/S.
Mice
Animal studies were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (Assurance number A3381-01). Protocols with TBEV in mice were approved by the NIH Rocky Mountain Laboratory Institutional Animal Care and Use Committee. Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
C57BL/6J mice were purchased from The Jackson Laboratory and housed in a pathogen-free animal facility at Washington University in St. Louis or in the ABSL4 facility for TBEV studies. For passive transfer studies, mAbs were diluted in PBS and administered to mice via intraperitoneal injection in a 100-µl total volume. Viral infections with POWV-SPO or LGTV were performed via subcutaneous inoculation in the footpad with 102 focus-forming units (FFU) of virus. For LGTV and POWV-SPO (where indicated) infection, 0.5 mg anti-Ifnar1–blocking mAb MAR1-5A3 (Sheehan et al., 2006) was administered 1 d before inoculation via intraperitoneal injection. Animals were monitored for mortality (POWV) or weight loss and clinical score (LGTV). To evaluate clinical disease following LGTV challenge, cages were blinded, and mice were scored each day as having (a) no disease, (b) hind limb weakness or disrupted righting reflex, (c) partial hind limb paralysis, (d) complete paralysis of one hind limb, or (e) complete paralysis of both hind limbs. For TBEV, 104 PFUs of virus was inoculated into the ear pinnae in 20 µl. Animals were weighed daily and euthanized when an animal showed signs of limb paresis or lost 20% or more of its starting weight.
Plasmids
Plasmids DTVp1 and DTVp2 for the construction of the POWV-SPO infectious clone were described previously (Kenney et al., 2018). For the production of RVPs, the plasmid encoding a subgenomic replicon of the WNV lineage II strain 956 that expresses GFP (WNVrepG/Z) and plasmids expressing the C-prM-E structural genes of POWV strain P0375 (GenBank accession no. KU886216), TBEV strain Neudoerfl (GenBank accession no. U27495), LGTV strain TP21 (GenBank accession no. NC_003690), GGYV strain Macquarie Island (GenBank accession no. DQ235145), DENV-2 strain 16681 (GenBank accession no. U87411), ZIKV strain H/PF/2013 (GenBank accession no. KJ776791), and WNV strain NY99 (GenBank accession no. KC407666) have been reported (Ansarah-Sobrinho et al., 2008; Dowd et al., 2016b; Pierson et al., 2006; VanBlargan et al., 2018). Plasmids that express the C-prM-E structural genes of SLEV strain Kern 217 (GenBank accession no. DQ525916), YFV strain 17D-204 (GenBank accession no. KF769015), and USUV strain Vienna 2001 (GenBank accession no. AY453411) were cloned into the expression vector pcDNA3.1 (Invitrogen). All plasmid propagation and cloning procedures were performed using Stbl2 bacteria grown at 30°C (Invitrogen).
mAb generation
One group of C57BL/6J mice was inoculated with 102 FFU of POWV-SPO, boosted 6 wk later with 2 × 102 FFU POWV-SPO, and given a final intravenous boost with 5 × 106 FFU of POWV-SPO 3 d before splenocyte collection. A second group of C57BL/6J mice was immunized twice (4 wk apart) with a lipid nanoparticle–encapsulated modified mRNA vaccine encoding the POWV prM and E genes (VanBlargan et al., 2018) and given a final intravenous boost of recombinant E (ectodomain) or DIII proteins 3 d before splenocyte collection. Subsequently, splenocytes were fused with P3X63.Ag.6.5.3 myeloma cells. Hybridomas producing antibodies that were bound to POWV-infected Raji-DCSIGNR cells by flow cytometry and/or POWV E by direct ELISA were cloned by limiting dilution. All hybridomas were screened initially with a single-endpoint RVP-based neutralization assay using neat hybridoma supernatant incubated with POWV RVPs for 1 h at 37°C. Raji-DCSIGNR cells were added to mAb-virus, incubated for 2 d, and assayed for GFP expression by flow cytometry. Hybridoma supernatants with >60% neutralization were purified commercially (Bio-X Cell) after adaptation for growth under serum-free conditions.
POWV E protein preparation
POWV-SPO soluble E ectodomain (E residues 1–397) and DIII (E residues 297–397 of E) were cloned into pET21a (Invitrogen) by Gibson assembly. Plasmids were transformed into BL21(DE3) cells (Invitrogen), and protein expression was induced by adding a 1:5 mixture of glucose and lactose to the culture and growing on a shaker overnight at 37°C. Proteins were expressed as inclusion bodies, which were purified, denatured, and refolded by slow dilution into a large volume of oxidative refolding buffer (400 mM arginine, 100 mM Tris-HCl, 1 mm EDTA, pH 8.3, 2.5% wt/vol glycerol, 10 mM reduced glutathione, 1 mM oxidized glutathione, and 2.5 mM phenylmethylsulfonyl fluoride). The refolded E protein was concentrated and dialyzed against 10 mM Tris, pH 8.0, overnight before being purified across a HiTrap Q HP column (GE Life Sciences) followed by size-exclusion chromatography on an S75 increase column (GE Life Sciences). DIII was purified by affinity capture using Ni-NTA resin (GoldBio) followed by size exclusion chromatography on an S75 column (GE Life Sciences).
Competition ELISA
Nunc MaxiSorp plates (Thermo Fisher Scientific) were coated with 4 µg/ml POWV-SPO E protein in 100 µl sodium bicarbonate coating buffer (0.1 M Na2CO3, 0.1 M NaHCO3, and 0.02% NaN3, pH 9.6) and incubated overnight at 4°C. Plates were washed three times with ELISA wash buffer (PBS containing 0.05% Tween-20) and then incubated with 200 µl blocking buffer (PBS, 2% BSA, 0.05% Tween-20, and 0.025% NaN3) for 2 h at room temperature. Blocking buffer was removed, and plates were incubated with 25 µl of each anti-POWV mAb (20 µg/ml) diluted in blocking buffer. After an incubation for 1 h at room temperature, 25 µl of a biotinylated version of each anti-POWV mAb (2 µg/ml) was added directly to the plates and incubated for 1 h at room temperature. Biotinylation was performed using an EZ-Link NHS-PEG4-Biotin kit (Thermo Fisher Scientific) according to manufacturer instructions. Plates were washed three times with ELISA wash buffer and incubated with 50 µl streptavidin-HRP (Invitrogen; diluted 1:40,000 in ELISA wash buffer) for 1 h at room temperature. Plates were washed three times and then incubated with 100 µl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific) for 3 min at room temperature before quenching by addition of 50 µl of 2 N H2SO4 and measuring with a microplate reader at OD 450 nm.
Virion prM content
Nunc MaxiSorp plates (Thermo Fisher Scientific) were coated with 5 µg/ml POWV-55 in 100 µl NaHCO3 (pH 9.6) coating buffer and incubated overnight at 4°C. Plates were washed three times with ELISA wash buffer (PBS containing 0.05% Tween-20) and then incubated with 200 µl blocking buffer (PBS, 2% BSA, and 0.05% Tween-20) for 2 h at room temperature. Plates were incubated with 3 × 105 FFU of virus diluted in DMEM supplemented with 2% FBS for 1 h at 37°C. Plates were fixed with 1% paraformaldehyde (PFA) for 10 min at room temperature, followed by extensive washing with PBS. Plates then were incubated with 100 µl biotinylated POWV-16 (anti-prM) or POWV-56 (anti-E) mAbs diluted in blocking buffer for 1 h at 37°C. Plates were washed three times with ELISA wash buffer and incubated with 50 µl streptavidin-HRP diluted (1:625) in PBS for 20 min at room temperature. Plates were washed again and then incubated with 100 µl TMB substrate (Thermo Fisher Scientific) for 3 min at room temperature before quenching with the addition of 50 µl of 2 N H2SO4 and measuring at OD 450 nm. The relative prM signal was calculated by dividing the OD450 measurement of POWV-16 signal by the OD450 measurement for POWV-56, multiplied by 100.
RVPs
RVPs were produced by the genetic complementation of a DNA-launched, WNV subgenomic replicon with a C-prM-E expression plasmid, as described previously (Ansarah-Sobrinho et al., 2008). Briefly, preplated HEK-293T cells were cotransfected with WNVrepG/Z, a WNV replicon expressing GFP, and the C-prM-E expression plasmids described above using FugeneHD (Promega) in accordance with the manufacturer’s instructions. Transfected cells were incubated at 37°C, and supernatant was harvested at 48 h after transfection, filtered using a 0.22-µm syringe filter, and stored at −80°C. RVPs were produced using the genetic complementation of plasmids as described above; however, HEK-293T cells were transfected using Lipofectamine 3000 transfection reagent (Invitrogen). These transfected cells were incubated at 30°C, and RVP-containing supernatants were harvested daily, pooled, and then filtered through a 0.22-mm filter (Millipore) and stored at −80°C. The infectious titer of RVPs was assayed using Raji-DCSIGNR cells or Vero cells, as described previously (Mukherjee et al., 2014).
Neutralization assays
(a) RVP neutralization assays were performed as described (Mukherjee et al., 2014). Briefly, RVP stocks were diluted so that ∼3–5% of cells become infected and incubated with serial dilutions of serum for 1 h at 37°C before the addition of Raji-DCSIGNR cells or Vero cells. Infections were performed at 37°C for 48 h, and infectivity was scored as the percentage of GFP-expressing cells determined by flow cytometry. (b) FRNTs were performed as described for other flaviviruses (Brien et al., 2013). Briefly, serial dilutions of serum were incubated with 2 × 102 FFU of POWV for 1 h at 37°C. Immune complexes were added to Vero cell monolayers and incubated for 1 h at 37°C before the addition of 1% (wt/vol) methylcellulose in MEM. Following incubation for 3 d at 37°C, cells were fixed with 1% PFA, permeabilized with 0.1% saponin, and stained for infection foci with POWV-16 (1 µg/ml). For both RVP neutralization assays and FRNT, antibody-dose response curves were analyzed using nonlinear regression analysis (with a variable slope and the top of the curve constrained to 100; GraphPad Software). Data are expressed as the serum dilution required to reduce infection by half (EC50).
Intrinsic virus decay
Loss of viral infectivity was measured by incubating 105 FFU of virus (diluted in DMEM supplemented with 2% FBS) at 37°C for various lengths of time before collection of media and storage at −80°C. Viral titer was determined by FFA.
Pre- and postattachment neutralization assays
For preattachment assays, serial dilutions of mAbs were prepared at 4°C in DMEM with 2% FBS and preincubated with 102 FFU of POWV-MA51240 for 1 h at 4°C. mAb–virus complexes were added to a monolayer of Vero cells for 1 h at 4°C. Virus was allowed to internalize during a 37°C incubation for 1 h. Cells were overlaid with 1% (wt/vol) methylcellulose in MEM supplemented with 4% FBS. For postattachment assays, 5 × 102 FFU of POWV-MA51240 was adsorbed onto a monolayer of Vero cells for 1 h at 4°C. After removal of unbound virus, serial dilutions of mAbs were added to virus-adsorbed cells for 1 h at 4°C. Virus then was allowed to internalize for 1 h at 37°C, and subsequently cells were overlaid with methylcellulose as described above. 3 d later, the plates were fixed with 1% PFA and analyzed for antigen-specific foci as described above.
FFWO neutralization assay
FFWO neutralization assays were performed by incubating prechilled Vero cells with POWV MA51240 (multiplicity of infection of 200) for 2 h at 4°C. Unbound virus was removed, and 50 µg of the indicated anti-POWV mAbs was incubated with the cells for 1 h at 4°C. Cells were washed and incubated with RPMI supplemented with 0.2% BSA, 10 mM Hepes, and 30 mM succinic acid and adjusted to pH 5.5. RPMI without succinic acid at pH 7.4 was used as a control to determine the background level of infection. Cells were incubated with the acidic media for 7 min at 37°C. After the cells were rinsed with DMEM supplemented with 2% FBS and 10 mM Hepes, cells were incubated with DMEM supplemented with NH4Cl for 24 h at 37°C. Cells were then trypsinized, fixed with 1% PFA, permeabilized with HBSS with 5% FBS and 1% (wt/vol) saponin, stained with POWV-16 followed by Alexa Fluor 647–conjugated secondary antibody (Invitrogen), and analyzed for infection by flow cytometry.
Generation of escape mutants
Escape mutants were generated by blind passaging of POWV-MA51240 in the presence of increasing concentrations of mAb for five passages on Vero cells. Virus from passage 5 was tested for neutralization sensitivity by FRNT, and subsequently viral RNA was isolated and used to make cDNA, which was sequenced. Amino acid mutations detected in the sequencing reactions were introduced in the POWV C-prM-E plasmid and used to produce RVPs, which were tested for neutralization as described above.
Site-directed mutagenesis epitope mapping
To identify amino acids on the E protein important for mAb binding, we narrowed down target residues for mutagenesis by selecting residues that differed between POWV and LGTV or GGYV, as many of the antibodies we mapped did not cross-react against subsets of these viruses. In the case of mAbs that did cross-react against both (POWV-56 and -63), mAb binding was decreased against GGYV. We focused on residues predicted to be exposed on the surface of the virion. Surface accessibility was estimated using solvent-accessible surface areas of the residues determined from the structure of the TBEV E protein dimer (Protein Data Bank [PDB] accession no. 5O6A), with a cutoff value of 30 Å2 (University of California, San Francisco Chimera package). We expanded our mutagenesis of DIII to include additional residues on the lateral ridge. Residues were mutated to the corresponding residue in GGYV or LGTV or, in the case of the additional DIII residues, to alanine or lysine (when the residue was a glycine). Mutations were introduced into the POWV C-prM-E plasmid described above. In total, 98 mutants were generated, although some were excluded due to low transfection efficiency. Each mutant was transfected into HEK-293T cells; 48 h later, cells were fixed with 1% PFA, permeabilized with HBSS with 5% FBS and 1% (wt/vol) saponin (referred to as permeabilization buffer), and incubated with mAbs at concentrations optimized for staining (range, 14–4,000 ng/ml) and Alexa Fluor 647–conjugated secondary antibody (Invitrogen) in permeabilization buffer. Fluorescence signal was detected by flow cytometry (MacsQuant) and analyzed using FlowJo software. The percentage of positively stained cells was calculated relative to mock-transfected cells. Transfection efficiency for each mutant was normalized using the percentage of cells stained with anti-POWV polyclonal mouse serum. Antibody reactivity against each mutant was compared with that of the WT C-prM-E after subtracting the signal from mock-transfected controls.
Cross-reactivity of mAbs
Cross-reactivity against other TBFVs was assessed by transfecting HEK-293T cells with the POWV, TBEV, LGTV, or GGYV C-prM-E plasmids described above. 48 h later, cells were fixed and permeabilized as described above and then incubated with mAbs at 10 µg/ml and Alexa Fluor 647–conjugated secondary antibody (Invitrogen) in permeabilization buffer. Fluorescence signal was detected by flow cytometry. Cross-reactivity against MBFVs was assessed by ADE assays as described previously (Mukherjee et al., 2014). Briefly, serial dilutions of Mab were incubated with RVPs of the indicated MBFVs for 1 h at 37°C and subsequently added to K562 cell monolayers. Cells were incubated at 37°C for 2 d, and infectivity was scored as the percentage of GFP-expressing cells determined by flow cytometry.
BLI
Biotinylated POWV mAbs were diluted to 5 µg/ml in BLI buffer (HBS-EP+, 3% wt/vol BSA) and loaded onto streptavidin pins in an Octet Red 96 system (ForteBio). Subsequently, the pins were dipped into various dilutions of recombinant POWV E protein or TBEV DIII in BLI buffer to measure association kinetics followed by blank BLI buffer to measure dissociation kinetics. Binding affinities were calculated as the ratio of the off-rate to the on-rate determined from curves fitted to the association-dissociation data.
Crystallization of POWV-80 with DIII
Concentrated POWV Fab fragments were mixed with a stoichiometric excess of recombinant untagged POWV DIII at 4°C for >2 h. The mixture was then purified over an S75 size-exclusion column to isolate Fab:DIII complexes in 20 mM Hepes, pH 7.4, and 150 mM NaCl. The complex was concentrated to 13.5 mg/ml. POWV-80–DIII complexes were crystallized by hanging drop vapor diffusion by adding 0.5 µl of the complex to 0.5 µl of 0.16 M NH4SO4, 0.08 M NaCH3COOH, 20% wt/vol glycerol, and 16% wt/vol polyethylene glycol 4000, pH 4.6.
Structure determination and refinement
Diffraction data were collected at Advanced Light Source beamline 4.2.2 (Molecular Biology Consortium) at 100 K using a wavelength of 1.0 Å, with a CMOS detector in shutter-less mode. The diffraction data were processed with XDS, then scaled and merged with Aimless. Molecular replacement was done in PHENIX using the PHASER GUI (Adams et al., 2010; Evans and Murshudov, 2013; Kabsch, 2010; McCoy et al., 2007). Molecular replacement of POWV-80 and DIII was accomplished by truncating the structure of the TBEV envelope (PDB accession no. 1SVB) to DIII and mutating residues to match the sequence of POWV. For the Fab, the sequence of POWV-80 heavy chain was used as a search term for BLAST to find a structure of a sequentially homologous Fab (PDB accession no. 3UO1), which was subsequently split into N-terminal and C-terminal variable and constant-domain search models. A model fitting the electron density map was built manually using Coot (Emsley et al., 2010) and refined with PHENIX. A summary of data collection and refinement statistics is provided in Table S1. Structures were visualized using University of California, San Francisco ChimeraX (Goddard et al., 2018). Buried surface area was calculated using ChimeraX. Interfacial residues were identified using QtPISA. Structural data have been deposited in the PDB under accession no. 7KYL.
HDX-MS
Peptide mapping
To prepare for acquisition and analysis of HDX data, a peptide map (in triplicate) of POWV E protein was generated by liquid chromatography/MS/MS using a Thermo LTQ Orbitrap XL mass spectrometer. POWV E (100 pmol) was admitted to the mass spectrometer operating in a data-dependent fragmentation mode and monitoring the six most abundant peptides. Data were analyzed by Byonic (Protein Metrics) for sequencing and accurate precursor mass (±5 ppm), and the peptides were manually curated.
Given three disulfide bonds of POWV E, digestion had to be optimized for sequence coverage by adjusting the reducing agent concentration (Tris (2-carboxyethyl) phosphine hydrochloride [TCEP]), denaturant (guanidine hydrochloride [GdnHCl]), and incubation time. The final optimized conditions were 1:1 dilution of HDX reaction volume (100 µl) with quench buffer containing 500 mM TCEP and 4 M GdnHCl, pH 2.4 (250 mM TCEP, 2 M GdnHCl and pH 2.6 final), 3-min incubation, 25°C.
HDX
POWV E was equilibrated with or without antibody in PBS (pH 7.0) at 10°C for 30 min and then exchanged in with D2O in the presence and absence of antibody by diluting the protein (100 pmol, 10 µl) by 10-fold with PBS in D2O at 10°C (90 µl, pH 7.0). HDX was measured at 0 (undeuterated control), 10, 60, 300, and 4,100 s at 10°C. For the undeuterated control, the conditions were the same except the added buffer solution was H2O instead of D2O. The HDX was quenched by adding an equal volume of quench buffer equilibrated at 45°C followed by mixing and incubation at room temperature to give a final temperature after mixing of ∼25°C.
The quenched sample was digested by passing through a custom-packed column (2 mm × 20 mm) of immobilized pepsin beads followed by a column of immobilized Fungal XIII beads at 200 µl/min flow rate. The resulting peptides were captured and desalted on a Zorbax Eclipse XDB-C8 trap (Agilent) column by using 0.1% formic acid in water for 4.7 min. Desalted peptides were loaded on a C18 analytical column (2.1 × 50 mm in size, 2.5 µm Xselect-CSH from Waters) where peptides were separated using a gradient of acetonitrile (ACN) in 0.1% formic acid (most peptides eluted during the linear part of gradient from 5 min [4% ACN] to 15 min [40% ACN]). To minimize back exchange, the trap and analytical columns were kept in an ice slush. The isotope distributions of the exchanged peptides were measured with a Thermo Fisher Scientific LTQ Orbitrap XL MS (MS only mode) for duplicate samples.
HDX data analysis
Liquid chromatography–MS HDX data acquisition (retention time, isotopic distribution, and observed m/z) was directed by the peptide map, and data were analyzed by HDExaminer (Sierra Analytics). The maximum deuterium level was normalized to 90%, and the data were displayed as kinetic plots for each peptide for HDX. To elucidate those regions where HDX changed upon antibody binding, the mean cumulative differences (bound–unbound) across all the time points for each peptide were calculated and plotted as a Woods plot. To identify significant difference upon binding, global SEM was calculated according to published methods (Hageman and Weis, 2019). Protected regions upon binding showed negative cumulative percent deuteration (%D), whereas regions exposed upon binding gave positive cumulative %D.
Measurement of viral burden
Mice were sacrificed on the indicated days after infection and perfused extensively with PBS, and the indicated organs were collected. Organs were weighed and homogenized using a MagNA Lyser (Roche). Viral RNA from homogenized organs or serum was isolated using the MagMAX Viral RNA Isolation Kit (Thermo Fisher Scientific) and measured by TaqMan one-step qRT-PCR on an ABI 7500 Fast Instrument. Viral burden is expressed on a log10 scale as viral RNA equivalents per gram or milliliters after comparison with a standard curve produced using serial 10-fold dilutions of viral RNA from known quantities of infectious virus. POWV-SPO primers (Platt et al., 2018) were 5′-GCAGCACCATAGGTAGAATGT-3′, 5′-CCACCCACTGAACCAAAGT-3′, and probe 5′-/56-FAM/TCTCAGTGG/Zen/TTGGAGAACACGCAT/3IABkFQ-3′. LGTV primers (VanBlargan et al., 2018) were 5′-GGAACTAGGCCTTGCAGAAT-3′, 5′-TGTTCTCCATTGTCGGGTTAG-3′, and probe 5′-/56-FAM/TGAGGTTAA/Zen/CGTGGCCATGCTCAT/3IABkF-3′.
Statistical analysis
Data analyses were performed using Prism software Version 7.0 (GraphPad Software). Log EC50 values were compared by one-way ANOVA followed by Sidak’s multiple comparisons test (when comparing a subset of values) or Tukey’s multiple comparisons test (when comparing all values). For survival analysis, Kaplan-Meier curves were plotted and analyzed by the log-rank test with a Bonferroni correction for multiple comparisons. Weight loss experiments were compared by an ANOVA with Sidak’s multiple comparisons test. Viremia and viral burden in tissues were compared by Kruskal-Wallis test followed by Dunn’s post-test.
Online supplemental material
Fig. S1 shows Western blotting of POWV-infected cell lysates with POWV-16, mAb variable region sequence homology, and the location of residue M-K10N within (pr)M. Fig. S2 shows complete site-directed mutagenesis mapping results and neutralization of escape mutants with a control antibody. Fig. S3 shows HDX-MS results. Fig. S4 shows kinetic binding curves of mAbs to POWV E protein. Table S1 shows x-ray crystallography data and contact residues between POWV-80 and POWV DIII.
Supplementary Material
shows x-ray crystallography dataset processing, model refinement, and validation statistics and summarizes interfacial contacts between POWV-80 and POWV DIII.
Acknowledgments
This study was supported by National Institutes of Health grants and contracts (R01 AI073755, P41GM103422 and R24GM136766, T32AI007172, 75N93019C00062, HHSN272201400058C, HHSN272201700060C, and HHSN272201400018C).
Author contributions: Conceptualization, L.A. VanBlargan, M.S. Diamond, J.M. Errico, and D.H. Fremont; Methodology, L.A. VanBlargan, M.S. Diamond, J.M. Errico, D.H. Fremont, and C.A. Nelson; Investigation, L.A. VanBlargan, J.M. Errico, N.M. Kafai, K.E. Burgomaster, P.N. Jethva, R.M. Broeckel, and K. Meade-White; Formal Analysis, J.M. Errico, C.A. Nelson, D. Wang, and S.A. Handley; Resources, S. Himansu; Supervision, M.S. Diamond, D.H. Fremont, T.C. Pierson, S.M. Best, and M.L. Gross; Writing—Original Draft, L.A. VanBlargan, J.M. Errico, and M.S. Diamond; Writing—Review and Editing, all authors.
References
- Adams, P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansarah-Sobrinho, C., Nelson S., Jost C.A., Whitehead S.S., and Pierson T.C.. 2008. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology. 381:67–74. 10.1016/j.virol.2008.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, S.K., Dowd K.A., Shrestha B., Nelson C.A., Edeling M.A., Johnson S., Pierson T.C., Diamond M.S., and Fremont D.H.. 2012. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 8:e1002930. 10.1371/journal.ppat.1002930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackney, D.E., Nofchissey R.A., Fitzpatrick K.A., Brown I.K., and Ebel G.D.. 2008. Stable prevalence of Powassan virus in Ixodes scapularis in a northern Wisconsin focus. Am. J. Trop. Med. Hyg. 79:971–973. 10.4269/ajtmh.2008.79.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien, J.D., Lazear H.M., and Diamond M.S.. 2013. Propagation, quantification, detection, and storage of West Nile virus. Curr. Protoc. Microbiol. 31:15D.3.1–15D.3.18. 10.1002/9780471729259.mc15d03s31 [DOI] [PubMed] [Google Scholar]
- Bröker, M., and Kollaritsch H.. 2008. After a tick bite in a tick-borne encephalitis virus endemic area: current positions about post-exposure treatment. Vaccine. 26:863–868. 10.1016/j.vaccine.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Campbell, M.S., and Pletnev A.G.. 2000. Infectious cDNA clones of Langat tick-borne flavivirus that differ from their parent in peripheral neurovirulence. Virology. 269:225–237. 10.1006/viro.2000.0220 [DOI] [PubMed] [Google Scholar]
- Carson, S.D. 2014. Kinetic models for receptor-catalyzed conversion of coxsackievirus B3 to A-particles. J. Virol. 88:11568–11575. 10.1128/JVI.01790-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel, R.N., Attoui H., Butenko A.M., Clegg J.C., Deubel V., Frolova T.V., Gould E.A., Gritsun T.S., Heinz F.X., Labuda M., et al. 2004. Tick-borne virus diseases of human interest in Europe. Clin. Microbiol. Infect. 10:1040–1055. 10.1111/j.1469-0691.2004.01022.x [DOI] [PubMed] [Google Scholar]
- Davis, C.W., Mattei L.M., Nguyen H.Y., Doms R.W., and Pierson T.C.. 2006. The location of N-linked glycans on West Nile virions controls their interactions with CD209. J. Biol. Chem. 281:37183–37194. 10.1074/jbc.M605429200 [DOI] [PubMed] [Google Scholar]
- Dowd, K.A., DeMaso C.R., Pelc R.S., Speer S.D., Smith A.R.Y., Goo L., Platt D.J., Mascola J.R., Graham B.S., Mulligan M.J., et al. 2016a. Broadly Neutralizing Activity of Zika Virus-Immune Sera Identifies a Single Viral Serotype. Cell Rep. 16:1485–1491. 10.1016/j.celrep.2016.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd, K.A., Ko S.Y., Morabito K.M., Yang E.S., Pelc R.S., DeMaso C.R., Castilho L.R., Abbink P., Boyd M., Nityanandam R., et al. 2016b. Rapid development of a DNA vaccine for Zika virus. Science. 354:237–240. 10.1126/science.aai9137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel, G.D. 2010. Update on Powassan virus: emergence of a North American tick-borne flavivirus. Annu. Rev. Entomol. 55:95–110. 10.1146/annurev-ento-112408-085446 [DOI] [PubMed] [Google Scholar]
- Ebel, G.D., Foppa I., Spielman A., and Telford S.R. II. 1999. A focus of deer tick virus transmission in the northcentral United States. Emerg. Infect. Dis. 5:570–574. 10.3201/eid0504.990423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel, G.D., Spielman A., and Telford S.R.. 2001. Phylogeny of North American Powassan virus. J. Gen. Virol. 82:1657–1665. 10.1099/0022-1317-82-7-1657 [DOI] [PubMed] [Google Scholar]
- Emsley, P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, P.R., and Murshudov G.N.. 2013. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69:1204–1214. 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatmi, S.S., Zehra R., and Carpenter D.O.. 2017. Powassan Virus-A New Reemerging Tick-Borne Disease. Front. Public Health. 5:342. 10.3389/fpubh.2017.00342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, E., Kose N., Edeling M.A., Adhikari J., Sapparapu G., Lazarte S.M., Nelson C.A., Govero J., Gross M.L., Fremont D.H., et al. 2018. Mouse and Human Monoclonal Antibodies Protect against Infection by Multiple Genotypes of Japanese Encephalitis Virus. MBio. 9:e00008-18. 10.1128/mBio.00008-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füzik, T., Formanová P., Růžek D., Yoshii K., Niedrig M., and Plevka P.. 2018. Structure of tick-borne encephalitis virus and its neutralization by a monoclonal antibody. Nat. Commun. 9:436. 10.1038/s41467-018-02882-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., and Ferrin T.E.. 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27:14–25. 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollins, S.W., and Porterfield J.S.. 1986. The uncoating and infectivity of the flavivirus West Nile on interaction with cells: effects of pH and ammonium chloride. J. Gen. Virol. 67:1941–1950. 10.1099/0022-1317-67-9-1941 [DOI] [PubMed] [Google Scholar]
- Goo, L., VanBlargan L.A., Dowd K.A., Diamond M.S., and Pierson T.C.. 2017. A single mutation in the envelope protein modulates flavivirus antigenicity, stability, and pathogenesis. PLoS Pathog. 13:e1006178. 10.1371/journal.ppat.1006178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grard, G., Moureau G., Charrel R.N., Lemasson J.J., Gonzalez J.P., Gallian P., Gritsun T.S., Holmes E.C., Gould E.A., and de Lamballerie X.. 2007. Genetic characterization of tick-borne flaviviruses: new insights into evolution, pathogenetic determinants and taxonomy. Virology. 361:80–92. 10.1016/j.virol.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Hageman, T.S., and Weis D.D.. 2019. Reliable Identification of Significant Differences in Differential Hydrogen Exchange-Mass Spectrometry Measurements Using a Hybrid Significance Testing Approach. Anal. Chem. 91:8008–8016. 10.1021/acs.analchem.9b01325 [DOI] [PubMed] [Google Scholar]
- Hermance, M.E., and Thangamani S.. 2017. Powassan Virus: An Emerging Arbovirus of Public Health Concern in North America. Vector Borne Zoonotic Dis. 17:453–462. 10.1089/vbz.2017.2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney, J.L., Anishchenko M., Hermance M., Romo H., Chen C.I., Thangamani S., and Brault A.C.. 2018. Generation of a Lineage II Powassan Virus (Deer Tick Virus) cDNA Clone: Assessment of Flaviviral Genetic Determinants of Tick and Mosquito Vector Competence. Vector Borne Zoonotic Dis. 18:371–381. 10.1089/vbz.2017.2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinski, M., Beicht J., Gerlach T., Volz A., Sutter G., and Rimmelzwaan G.F.. 2020. Tick-Borne Encephalitis Virus: A Quest for Better Vaccines against a Virus on the Rise. Vaccines (Basel). 8:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, R.J., Dowd K.A., Beth Post C., and Pierson T.C.. 2015. Shake, rattle, and roll: Impact of the dynamics of flavivirus particles on their interactions with the host. Virology. 479-480:508–517. 10.1016/j.virol.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C.J., Goncalvez A.P., Men R., Wernly C., Donau O., Engle R.E., and Purcell R.H.. 2007. Epitope determinants of a chimpanzee dengue virus type 4 (DENV-4)-neutralizing antibody and protection against DENV-4 challenge in mice and rhesus monkeys by passively transferred humanized antibody. J. Virol. 81:12766–12774. 10.1128/JVI.01420-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl, C.W., Holzmann H., Kunz C., and Heinz F.X.. 1993. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology. 194:173–184. 10.1006/viro.1993.1247 [DOI] [PubMed] [Google Scholar]
- McAuley, A.J., Sawatsky B., Ksiazek T., Torres M., Korva M., Lotrič-Furlan S., Avšič-Županc T., von Messling V., Holbrook M.R., Freiberg A.N., et al. 2017. Cross-neutralisation of viruses of the tick-borne encephalitis complex following tick-borne encephalitis vaccination and/or infection. NPJ Vaccines. 2:5. 10.1038/s41541-017-0009-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., Dowd K.A., Manhart C.J., Ledgerwood J.E., Durbin A.P., Whitehead S.S., and Pierson T.C.. 2014. Mechanism and significance of cell type-dependent neutralization of flaviviruses. J. Virol. 88:7210–7220. 10.1128/JVI.03690-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., Sirohi D., Dowd K.A., Chen Z., Diamond M.S., Kuhn R.J., and Pierson T.C.. 2016. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology. 497:33–40. 10.1016/j.virol.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, S., Jost C.A., Xu Q., Ess J., Martin J.E., Oliphant T., Whitehead S.S., Durbin A.P., Graham B.S., Diamond M.S., and Pierson T.C.. 2008. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 4:e1000060. 10.1371/journal.ppat.1000060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken, G., Oliphant T., Johnson S., Burke S., Diamond M.S., and Fremont D.H.. 2005. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 437:764–769. 10.1038/nature03956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant, T., Nybakken G.E., Austin S.K., Xu Q., Bramson J., Loeb M., Throsby M., Fremont D.H., Pierson T.C., and Diamond M.S.. 2007. Induction of epitope-specific neutralizing antibodies against West Nile virus. J. Virol. 81:11828–11839. 10.1128/JVI.00643-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organtini, L.J., Makhov A.M., Conway J.F., Hafenstein S., and Carson S.D.. 2014. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J. Virol. 88:5755–5765. 10.1128/JVI.00299-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesko, K.N., Torres-Perez F., Hjelle B.L., and Ebel G.D.. 2010. Molecular epidemiology of Powassan virus in North America. J. Gen. Virol. 91:2698–2705. 10.1099/vir.0.024232-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson, T.C., Sánchez M.D., Puffer B.A., Ahmed A.A., Geiss B.J., Valentine L.E., Altamura L.A., Diamond M.S., and Doms R.W.. 2006. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology. 346:53–65. 10.1016/j.virol.2005.10.030 [DOI] [PubMed] [Google Scholar]
- Pierson, T.C., Xu Q., Nelson S., Oliphant T., Nybakken G.E., Fremont D.H., and Diamond M.S.. 2007. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe. 1:135–145. 10.1016/j.chom.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt, D.J., Smith A.M., Arora N., Diamond M.S., Coyne C.B., and Miner J.J.. 2018. Zika virus-related neurotropic flaviviruses infect human placental explants and cause fetal demise in mice. Sci. Transl. Med. 10:eaao7090. 10.1126/scitranslmed.aao7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani, D.F., Bozzacco L., Keeffe J.R., Khouri R., Olsen P.C., Gazumyan A., Schaefer-Babajew D., Avila-Rios S., Nogueira L., Patel R., et al. 2017. Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell. 169:597–609.e11. 10.1016/j.cell.2017.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinski, A., Guardado-Calvo P., Barba-Spaeth G., Duquerroy S., Vaney M.C., Kikuti C.M., Navarro Sanchez M.E., Dejnirattisai W., Wongwiwat W., Haouz A., et al. 2015. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 520:109–113. 10.1038/nature14130 [DOI] [PubMed] [Google Scholar]
- Sapparapu, G., Fernandez E., Kose N., Bin Cao J.M., Fox R.G., Bombardi H., Zhao C.A., Nelson A.L., Bryan T., Barnes, et al. 2016. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 540:443–447. 10.1038/nature20564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan, K.C., Lai K.S., Dunn G.P., Bruce A.T., Diamond M.S., Heutel J.D., Dungo-Arthur C., Carrero J.A., White J.M., Hertzog P.J., and Schreiber R.D.. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 26:804–819. 10.1089/jir.2006.26.804 [DOI] [PubMed] [Google Scholar]
- Shrestha, B., Brien J.D., Sukupolvi-Petty S., Austin S.K., Edeling M.A., Kim T., O’Brien K.M., Nelson C.A., Johnson S., Fremont D.H., and Diamond M.S.. 2010. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 6:e1000823. 10.1371/journal.ppat.1000823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny, K., Kiermayr S., Holzmann H., and Heinz F.X.. 2006. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J. Virol. 80:9557–9568. 10.1128/JVI.00080-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, L., Condon T., Destrampe E.M., Brown J.A., McGavic J., Gould C.V., Chambers T.V., Kosoy O.I., Burkhalter K.L., Annambhotla P., . 2021. Powassan virus infection likely acquired through blood transfusion presenting as encephalitis in a kidney transplant recipient. Clin. Infect. Dis. 72:1051–1054. 10.1093/cid/ciaa738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, B.S., Moesker B., Smit J.M., Wilschut J., Diamond M.S., and Fremont D.H.. 2009. A therapeutic antibody against west nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 5:e1000453. 10.1371/journal.ppat.1000453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan, L.A., Goo L., and Pierson T.C.. 2016. Deconstructing the Antiviral Neutralizing-Antibody Response: Implications for Vaccine Development and Immunity. Microbiol. Mol. Biol. Rev. 80:989–1010. 10.1128/MMBR.00024-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan, L.A., Himansu S., Foreman B.M., Ebel G.D., Pierson T.C., and Diamond M.S.. 2018. An mRNA Vaccine Protects Mice against Multiple Tick-Transmitted Flavivirus Infections. Cell Rep. 25:3382–3392.e3. 10.1016/j.celrep.2018.11.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, M.R., Dowd K.A., Engle M., Tesh R.B., Johnson S., Pierson T.C., and Diamond M.S.. 2011. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fcgamma receptor and complement-dependent effector mechanisms. J. Virol. 85:11567–11580. 10.1128/JVI.05859-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vratskikh, O., Stiasny K., Zlatkovic J., Tsouchnikas G., Jarmer J., Karrer U., Roggendorf M., Roggendorf H., Allwinn R., and Heinz F.X.. 2013. Dissection of antibody specificities induced by yellow fever vaccination. PLoS Pathog. 9:e1003458. 10.1371/journal.ppat.1003458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala, W.M., Kraus A.A., Haymore L.B., Accavitti-Loper M.A., and de Silva A.M.. 2009. Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology. 392:103–113. 10.1016/j.virol.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Vogt M.R., Oliphant T., Engle M., Bovshik E.I., Diamond M.S., and Beasley D.W.. 2009. Development of resistance to passive therapy with a potently neutralizing humanized monoclonal antibody against West Nile virus. J. Infect. Dis. 200:202–205. 10.1086/599794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Ge P., Yu X., Brannan J.M., Bi G., Zhang Q., Schein S., and Zhou Z.H.. 2013. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20:105–110. 10.1038/nsmb.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Fernandez E., Dowd K.A., Speer S.D., Platt D.J., Gorman M.J., Govero J., Nelson C.A., Pierson T.C., Diamond M.S., and Fremont D.H.. 2016. Structural Basis of Zika Virus-Specific Antibody Protection. Cell. 166:1016–1027. 10.1016/j.cell.2016.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Xu L., Bombardi R., Nargi R., Deng Z., Errico J.M., Nelson C.A., Dowd K.A., Pierson T.C., Crowe J.E., et al. 2020. Mechanism of differential Zika and dengue virus neutralization by a public antibody lineage targeting the DIII lateral ridge. J. Exp. Med. 217:e29101792. 10.1084/jem.20191792 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows x-ray crystallography dataset processing, model refinement, and validation statistics and summarizes interfacial contacts between POWV-80 and POWV DIII.