Abstract
The emerging pathogen SARS-CoV2 causing coronavirus disease 2019 (COVID-19) is a global public health challenge. To the present day, COVID-19 had affected more than 40 million people worldwide. The exploration and the development of new bioactive compounds with cost-effective and specific anti-COVID 19 therapeutic power is the prime focus of the current medical research. Thus, the exploitation of the molecular docking technique has become essential in the discovery and development of new drugs, to better understand drug-target interactions in their original environment. This work consists of studying the binding affinity and the type of interactions, through molecular docking, between 54 compounds from Moroccan medicinal plants, dextran sulfate and heparin (compounds not derived from medicinal plants), and 3CLpro-SARS-CoV-2, ACE2, and the post fusion core of 2019-nCoV S2 subunit. The PDB files of the target proteins and prepared herbal compounds (ligands) were subjected for docking to AutoDock Vina using UCSF Chimera, which provides a list of potential complexes based on the criteria of form complementarity of the natural compound with their binding affinities. The results of molecular docking revealed that Taxol, Rutin, Genkwanine, and Luteolin-glucoside have a high affinity with ACE2 and 3CLpro. Therefore, these natural compounds can have 2 effects at once, inhibiting 3CLpro and preventing recognition between the virus and ACE2. These compounds may have a potential therapeutic effect against SARS-CoV2, and therefore natural anti-COVID-19 compounds.
Keywords: Molecular docking, SARS-CoV2, anti-COVID 19, Moroccan plants
Introduction
Emerging and re-emerging pathogens are global public health challenges.1 Coronaviruses are non-segmented, enveloped, positive RNA viruses belonging to the family Coronaviridae and the order Nidovirales and are widely distributed in humans and other mammals.2 The coronavirus of severe acute respiratory syndrome 2 (SARS-CoV-2), the most recently discovered, which causes COVID-19, has spread rapidly in China and other countries.3-11 Thus, by October 20, 2020, COVID-19 had affected more than 40 million people worldwide.12
The development of novel, cost-effective, and specific anti-COVID 19 drugs is the prime focus of the current medical research.13 The exploration of new bioactive compounds with anti-COVID 19 therapeutic power must target nature directly. Multiple scientific research works have revealed the enormous therapeutic antiviral potential of several medicinal plants and algae.14,15 Indeed, infection by SARS-CoV 2 may be limited by different stratagems. One can consider blocking the entry of the virus into the host cell, by targeting either the spike protein S2 or the ACE2 protein of the plasma membrane to which the virus binds. However, another strategy would be to prevent the formation of viral RNA. Blocking the virus protease could prevent it from cutting the viral poly-protein synthesized by the infected cell. The viral particles could not assemble in the cell, which will stop the infection.16-19
In recent years, molecular modelling more precisely molecular docking has very quickly integrated the fields of biology, pharmacy, and medicine, seen its impact on improving efficiency and reducing the cost of research.20,21 The increasing number of protein crystal structures, especially in the most popular public protein structure database: the database Protein Data Bank (PDB) shows the interest and development of research on molecular docking.22 Also, the public databases such as DrugBank,23 ChEMBL,24 ZINC,25 and PubChem26 give the public the opportunity of accessing the structures and biological activities of millions of chemical compounds. Molecular docking has become an indispensable technique and has begun to have an impact on drug discovery and development, by studying drug-target interactions in their native environments.27
The objective of our work is the use of bioinformatics tool ‘Molecular Docking’, which anchors small molecules in the target structures of macromolecules and note potential complementarity with the binding sites to identify the drugs hits and optimize therapeutic leads. Thus, the molecular docking of natural molecules derived medicinal plants could be a therapeutic alternative to fight the SARS-CoV2.
Materials and Methods
To meet our goal, we have adopted the molecular docking approach. This approach consists of studying the binding affinity and the type of interactions, between 54 compounds from Moroccan medicinal plants (ligands) and the 3 key proteins involved in SARS-CoV2 infection: 3CL protease (3CLpro)-SARS-CoV-2, human angiotensin-converting enzyme (ACE2), and the post-fusion core of 2019-nCoV S2 subunit (receptors).
Data collection
We selected several plants from the Moroccan flora used in traditional medicine. These plants are known for their antimicrobial and anti-cancer power. The list is obtained by consulting several scientific publications.28-33 Regarding heparin and dextran sulfate (compounds not derived from medicinal plants), their antiviral activity has already been proven in the literature, and can thus be proposed as an anti-COVID 19 molecule.34,35 In total, we have listed 54 compounds from different medicinal plants and heparin and dextran sulfate (Table 1). The PubChem database26 was consulted to assign each compound its ID and retrieve their chemical structure.
Table 1.
List of 54 compounds from different medicinal plants and 2 synthetic compounds.
| PubChem CID | Name | Plant |
|---|---|---|
| 3084656 | Dextran sulfatea | – |
| 772 | Heparina | – |
| 5280450 | Linoleic acid | Marrubium vulgare |
| 11601669 | CBDV | Cannabis sativa L. |
| 16078 | THC | Cannabis sativa L. |
| 644019 | CBD | Cannabis sativa L. |
| 14986 | Tocopherols | Argania spinosa L. |
| 11027 | 3-Thujanone | Artemisia herba-alba |
| 2537 | Camphor | Artemisia herba-alba |
| 88556 | Davanone D | Artemisia herba-alba |
| 442463 | Chrysanthenone | Artemisia herba-alba |
| 520909 | α-Himachalene | Cedrus atlantica Mannerti |
| 577062 | γ-Himachalene | Cedrus atlantica Mannerti |
| 11586487 | β-Himachalene | Cedrus atlantica Mannerti |
| 91698329 | γ-Atlantone | Cedrus atlantica Mannerti |
| 2758 | 1,8-Cineole | Laurus nobilis & Laurus azorica |
| 6549 | Linalool | Laurus nobilis & Laurus azorica |
| 111037 | α-Terpinyl acetate | Laurus nobilis & Laurus azorica |
| 5280445 | Luteolin | Rosmarinus officinalis L. |
| 5280443 | Apigenin | Rosmarinus officinalis L. |
| 5281612 | Diosmetin | Rosmarinus officinalis L. |
| 5281628 | Hispidulin | Rosmarinus officinalis L. |
| 161271 | Salvigenin | Rosmarinus officinalis L. |
| 442018 | Genkwanine | Rosmarinus officinalis L. |
| 339816 | Diterpene II (lactone) | Rosmarinus officinalis L. |
| 5281792 | Rosmarinic acid | Rosmarinus officinalis L. |
| 1794427 | Chlorogenic acid | Rosmarinus officinalis L. |
| 13966122 | Rosmanol | Rosmarinus officinalis L. |
| 15801061 | Rosmadial | Rosmarinus officinalis L. |
| 442009 | Carnosol | Rosmarinus officinalis L. |
| 73170 | Alpha-Amyrin | Rosmarinus officinalis L. |
| 64945 | Ursolic acid | Rosmarinus officinalis L. |
| 10494 | Oleanolic acid | Rosmarinus officinalis L. |
| 222284 | β-Sitosterol | Taxus baccata L. |
| 44351600 | 11-Taxadiene | Taxus baccata L. |
| 167825 | Taxusin | Taxus baccata L. |
| 15378021 | Baccatin VI | Taxus baccata L. |
| 65366 | Baccatin III | Taxus baccata L. |
| 5318150 | Hydroxybaccatin I | Taxus baccata L. |
| 36314 | Taxol | Taxus baccata L. |
| 442495 | Pulegone | Satureja calamintha spp. nepeta |
| 6986 | Isomenthone | Satureja calamintha spp. nepeta |
| 22311 | Limonene | Satureja calamintha spp. nepeta |
| 9064 | Catechin | Satureja |
| 21550 | Caffeine | Satureja |
| 5280805 | Rutin | Satureja |
| 72378 | Lycorine | Satureja |
| 10364 | Carvacrol | Satureja |
| 7461 | γ-Terpinene | Satureja |
| 6989 | Thymol | Satureja |
| 7463 | p-Cymene | Satureja |
| 445858 | Ferulic acid | Satureja |
| 338 | Salicylic acid | Satureja |
| 370 | Gallic acid | Satureja |
| 72276 | Epicatechin | Satureja |
| 5280637 | Luteolin-glucoside | Satureja |
Dextran sulfate and heparin are compounds not derived from medicinal plants.
Molecular docking
The crystal structure of the human ACE2 (ID: 1R4L), 3CLpro-SARS-CoV-2 (ID: 6M2N), and the post-fusion core of 2019-nCoV S2 subunit (ID: 6LXT) was recovered by the PDB RCSB database.22 Ligands and ACE2, 3CLpro-SARS-CoV-2, and spike protein S2 were prepared for docking using UCSF Chimera.36 The steps for preparing ligands and proteins for docking protocol were done employing default settings. Afterwards, the PDB files of the target proteins and prepared compounds (ligands) were subjected to AutoDock Vina37 to predict the structure of the protein-ligand complexes and to evaluate the binding energy. To predict the ideal mode of binding between the target proteins (IDs: 1R4L, 6M2N, and 6LXT) and each compound shown in Table 1, the results of molecular docking are analysed using Discovery Studio 2020.38
Results and Discussion
In this study, the molecular docking analysis was used to identify the anti-COVID-19 potential of natural compounds, derived from medicinal plants, and other synthetics molecules such as dextran sulfate and heparin.
In the docking analysis, chemical compounds and the structure of ACE2, 3CLpro, and spike protein S2 were submitted to AutoDock Vina, which provides a list of potential complexes based on the criteria of form complementarity of the chemical compound with their binding affinities. The binding affinity values of docked natural compound-protein complex were calculated according to the binding affinity energies.
ACE2-ligands docking analysis
The human ACE2 structure of SARS-CoV-2 recovered by the RCSB PDB server with the pdb identifier 1R4L revealed that the ACE2 has a resolution of 3.00 Å, a total structural weight 76.98 kDa and a residue number 655.
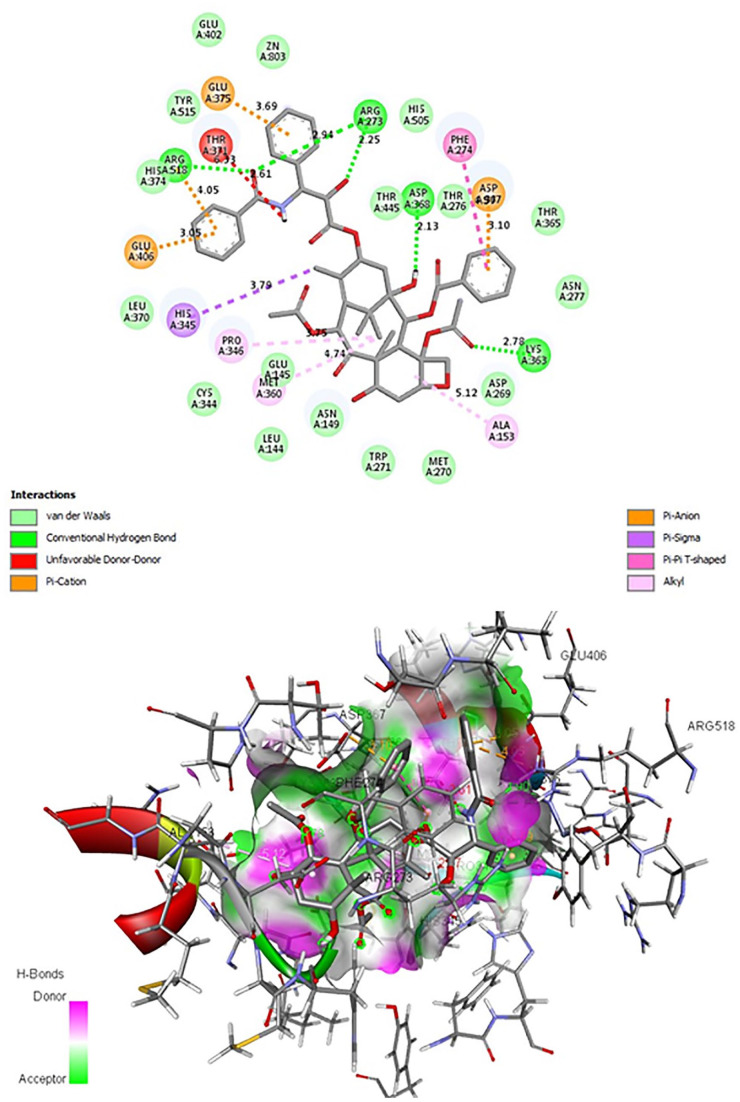
The ACE2-ligands docking analysis revealed that among the 56 chemical compounds tested (Table 1), 14 have a low binding energy and therefore a good interaction with ACE2 (Table 2). The compounds Taxol, Rutin, Baccatin III, Genkwanine, Ursolic acid, Luteolin-glucoside, and Alpha-Amyrin have the lowest binding energies (Table 2). Taxol has the lowest binding energy (equal to −12.2 Kcal/mol) (Figure 1, Table 2). The Rutin has a value of −11.4 Kcal/mol with 3 hydrogen bonds ALA 348, GLU 375, and GLU 402 (Table 2). Baccatin III has a value of −11.3 Kcal/mol and a hydrogen bond with ASP 368 (Table 2).
Table 2.
List of natural compounds with the lowest binding energy and their hydrogen bond interactions with ACE2, the post fusion core of 2019-nCoV S2 subunit, and 3CLpro.
| Name | H bonds | Binding energy (Kcal/mol) | |
|---|---|---|---|
| ACE2-ligands | Taxol | 0 | –12.2 |
| Rutin | ALA 348, GLU 375, GLU 402 | –11.4 | |
| Baccatin III | ASP 368 | –11.3 | |
| Genkwanine | 0 | –10.5 | |
| Ursolic acid | 0 | –10.5 | |
| Luteolin-glucoside | ASP 368 | –10.2 | |
| Alpha-amyrin | 0 | –10.2 | |
| Rosmanol | 0 | −9.9 | |
| β-Sitosterol | 0 | −9.8 | |
| Oleanolic acid | THR 371 | −9.2 | |
| Carnosol | 0 | −9.2 | |
| THC | 0 | −9.2 | |
| Hispidulin | TYR 515 | −9.0 | |
| Diterpene II (lactone) | 0 | −9.0 | |
| Spike protein S2-ligands | Genkwanine | 2× ARG 1185A | −5.0 |
| Rosmanol | LYS 1181A | −4.8 | |
| Luteolin-glucoside | GLU 1188A | −4.5 | |
| Rosmarinic acid | 0 | −4.5 | |
| Oleanolic acid | GLU 1188A | −4.4 | |
| Rutin | ASP 1184A | −4.4 | |
| Diterpene II (lactone) | 0 | −4.4 | |
| Alpha-amyrin | 0 | −4.4 | |
| Carnosol | 0 | −4.3 | |
| Ursolic acid | 0 | −4.3 | |
| THC | 0 | −4.2 | |
| Chlorogenic acid | ARG 1185A | −4.0 | |
| 3CLpro-ligands | Genkwanine | GLN 192A | –8.5 |
| Luteolin-glucoside | 3 H Bonds ASN 142A | –8.4 | |
| Rutin | ASN 142A, HIS 164A, 2× GLU 166A | –8.0 | |
| Luteolin | 0 | −7.8 | |
| Diterpene II (lactone) | CYS 145A | −7.8 | |
| Rosmarinic acid | LEU 141A, GLU 166A | −7.7 | |
| Catechin | LEU 141A, CYS 145A | −7.5 | |
| Taxol | ASN 142A, GLU 166A | −7.5 | |
| Diosmetin | LEU 141A, CYS 145A | −7.4 | |
| Hispidulin | 0 | −7.3 | |
| Dextran sulfate | THR 26A, HIS 41A, GLY 143A, ASP 187A | −7.2 | |
| Apigenin | 0 | −7.2 | |
| Salvigenin | ASN 142A, GLN 192A | −7.2 | |
| Chlorogenic acid | GLU 166A, CYS 145A, 2× THR 190A | −7.2 |
The values in bold correspond to molecules which have a high affinity with the receptor in question.
Figure 1.
Interaction of the ligand Taxol with the active site of ACE2.
These compounds can bind directly to the ACE2 receptor with high affinity which may suggest for competition with SARS-CoV-2. Thus, this strategy turns out to be difficult to implement because the receptor in question is necessary for other vital cellular functions.39,40
Spike protein S2-ligands docking analysis
The post-fusion core of 2019-nCoV S2 subunit structure of SARS-CoV-2 recovered by the RCSB PDB server with the pdb identifier 6LXT revealed that the spike protein S2 has a resolution of 2.90 Å, a total structural weight 84.66 kDa, and a residue number 686.
The spike protein S2-ligands molecular docking analysis revealed that the chemicals compounds tested (Table 1) have high binding energy scores. Genkwanine has binding energy equal to −5.0 Kcal/mol, and two hydrogen bonds with ARG 1185. Rosmanol (equal to −4.8 Kcal/mol) has a hydrogen bond with LYS 1181 (Table 2). These values of binding energies (Table 2) reflect the instability of the interactions between the site of inhibition of spike protein S2 and the ligands tested (Table 1). These results suggest that the natural compounds tested (Table 1) do not have a strong affinity with the spike protein and are therefore unable to hinder the entry of SARS-CoV2 to the host cell.
However, despite the weak interactions of heparin with the spike protein S2 inhibition site (−3.4 Kcal/mol), heparin showed a high affinity (−8.0 Kcal/mol) with the protein outside the site of inhibition (data not shown). Therefore, this molecule can inhibit SARS-CoV-2 either by changing the three-dimensional structure of spike protein S2 or else hiding the active site of the latter. These results are consistent with the study of Kwon et al,41 which showed that heparin effectively inhibits SARS-CoV-2 in vitro.
3CLpro-ligands docking analysis
The 3CLpro structure of SARS-CoV-2 recovered by the RCSB PDB server with the pdb identifier 6M2N revealed that the 3CLpro-SARS-CoV-2 has a resolution of 2.20 Å, a total structural weight 136.38 kDa, and a residue number 1223.
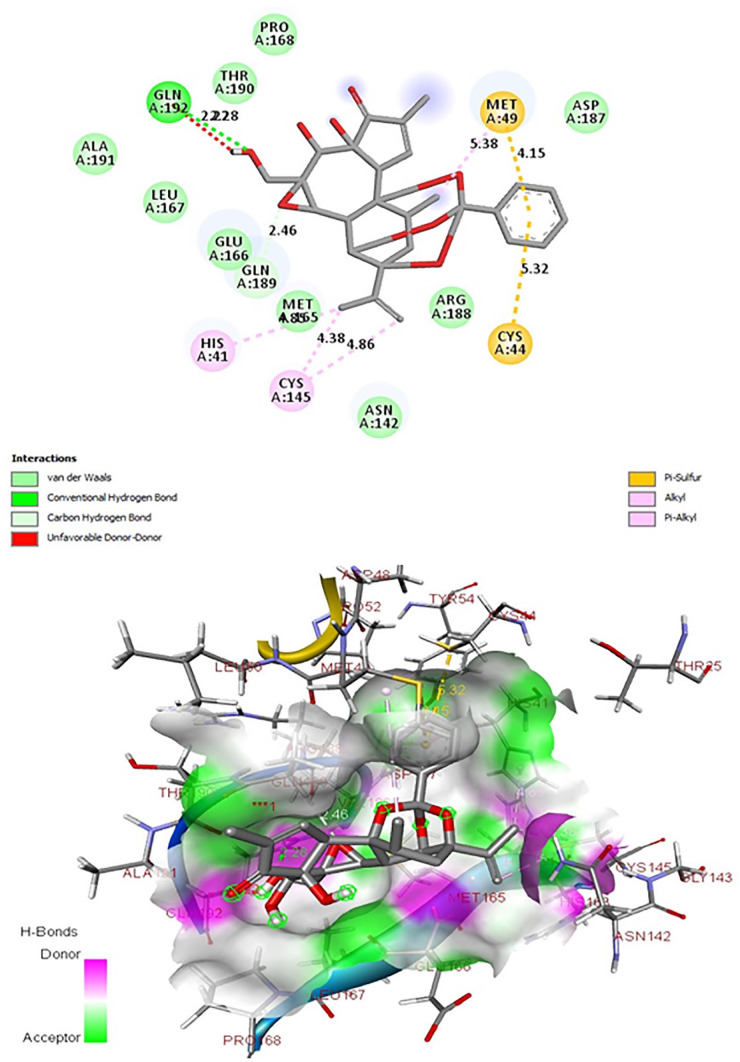
The molecular docking analysis revealed that among the 56 chemical compounds tested, 17 have a low binding energy and therefore a good interaction with 3CLpro (Table 2). The compounds Genkwanin, Luteolin-glucoside, and Rutin have the lowest binding energies (Table 2). Genkwanin has the lowest binding energy (equal to −8.5 Kcal/mol) and a hydrogen bond with GLN 129 (Table 2), and interacts with the active site (His-41, Cys-145 catalytic dyad), essential residues for the activity of SARS-CoV-2 3CLpro (Figure 2).42,43 Luteolin-glucoside has a value of −8.4 Kcal/mol and 3 hydrogen bonds with ASN 142 (Table 2). The Rutin has a value of −8.4 Kcal/mol and 4 hydrogen bonds ASN 142, HIS 164, and 2× GLU 166 (Table 2). Moreover, Luteolin-glucoside and Rutin interact only with His-41 of the catalytic dyad (Figures 3 and 4).
Figure 2.
Interactions of the ligand Genkwanine with the active site of 3CLpro.
Figure 3.
Interactions of the ligand Luteolin-glucoside with the active site of 3CLpro.
Figure 4.
Interactions of the ligand Rutin with the active site of 3CLpro.
The lowest binding energies and the presence of hydrogen bonding interactions in our ligand 3CLpro complex show the existence of strong interactions between these 3 natural compounds and our target protein. However, inhibition of 3CLpro’s activity is among the most important therapeutic targets against coronaviruses, since this enzyme is crucial during viral replication.44,45 The enzyme’s active site contains a catalytic dyad (His-41, Cys-145) where a cysteine residue acts as a nucleophile in the proteolytic process and a histidine residue acts as the general acid base.42,46-48 The molecular docking analysis revealed that Luteolin-glucoside and Rutin interact only with His-41 of the catalytic dyad (Figures 3 and 4). However, the Genkwanin can target 3CLpro through interactions regulating the catalytic dyad (Cys-145, His-41) of SARS-CoV-2 3CLpro (Figure 2), and in this sense, it is believed to hinder protease activity, and therefore natural anti-COVID-19 compound.
Taken together, the analysis of the molecular docking of the 2 membrane receptors (spike protein S2 and ACE2) as well as the viral protease 3CLpro revealed that the natural compounds Taxol, Rutin, Baccatin III, Genkwanine, Ursolic acid, Luteolin-glucoside, and Alpha-Amyrin have strong affinities with ACE2, and therefore can be suggested for competition with SARS-CoV-2. The compounds Genkwanine, Luteolin-glucoside, and Rutin can be potential inhibitors of 3CLpro, and therefore natural anti-COVID-19 compounds. During this study, the natural compounds listed in Table 1 showed high affinity to the therapeutic targets 3CLpro SARS-CoV2 and human ACE2, compared with dextran sulfate and heparin. However, the results of our study revealed that heparin has a potential to inhibit the function of the spike membrane receptor, compared with natural compounds (Table 1). These have an unstable interaction with the spike protein, and therefore cannot hinder the entry of SARS-CoV2 into the host cell.
In our study, Taxol, Rutin, Genkwanine, and Luteolin-glucoside have high affinities with ACE2 and 3CLpro. These results agree with other experimental studies, showing the important antiviral power of these molecules of natural origin. Taxol (Taxus baccata L.) has been reported to have antiviral activity.49,50 Thus, Taxol has the capacity to inhibit the entry of HIV-1 virus into cells. Moreover, this molecule not only acts on the process before the virus invades, but also has an inhibitory effect on HIV-1 protease and integrase activity.51 Rutin (Satureja) has a strong interaction with 3CLpro and ACE2 and has significant antibacterial and antiviral power.52,53 This molecule has demonstrated an antiviral effect against avian influenza strain H5N1.54 An in vitro study has shown that Genkwanine (Rosmarinus officinalis L) has an antiviral activity. It was identified as a potent anti-ASFV compound that targets viral entry and replication.55 Thus, the present study revealed that Genkwanine is able to bind with the catalytic dyad, and is thereby anticipated to interrupt the 3CLpro activity. Luteolin-glucoside (Satureja) activity was demonstrated as a neuraminidase inhibitor against influenza A virus infection in cell culture and mice.56 Ultimately, based on the available evidence, these natural compounds can have 2 effects at once, inhibiting 3CLpro and preventing recognition between the virus and ACE2.
Conclusion
Molecular docking analysis revealed that Taxol, Rutin, Genkwanine, and Luteolin-glucoside exhibit strong interactions, in terms of binding energy, with the targets of SARS-CoV2 and human ACE2. The results of our study are consistent with other experimental studies that have demonstrated antiviral potency. Therefore, we suggest that these naturally occurring molecules could be effective against COVID-19.
Acknowledgments
Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: All authors contributed to the conception and design of the study, and drafting of the manuscript.
ORCID iD: Badreddine Nouadi  https://orcid.org/0000-0001-5175-4601
https://orcid.org/0000-0001-5175-4601
References
- 1. Gao GF. From ‘A’IV to ‘Z’IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157-1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peiris JSM. Coronaviruses. In: Richman DD, Whitley RJ, Hayden FG, eds. Clinical Virology. New York, NY: John Wiley & Sons, Ltd; 2016:1243-1265. doi: 10.1128/9781555819439.ch52. [DOI] [Google Scholar]
- 3. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929-936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970-971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Young BE, Ong SWX, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488-1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guan W, Ni Z, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;328:1708-1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382:1199-1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061-1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int. Accessed October 20, 2020.
- 13. Wu R, Wang L, Kuo H-CD, et al. An update on current therapeutic drugs treating COVID-19 [published online ahead of print May 11, 2020]. Curr Pharmacol Rep. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahmadi A, Zorofchian Moghadamtousi S, Abubakar S, Zandi K. Antiviral potential of algae polysaccharides isolated from marine sources: a review. Biomed Res Int. 2015;2015:825203. doi: 10.1155/2015/825203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogbole OO, Akinleye TE, Segun PA, Faleye TC, Adeniji AJ. In vitro antiviral activity of twenty-seven medicinal plant extracts from Southwest Nigeria against three serotypes of echoviruses. Virol J. 2018;15:110. doi: 10.1186/s12985-018-1022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahmoud Jarrar YB, Alshaer W, Ismail S. SARS-CoV-2 entry in host cells-multiple targets for treatment and prevention. Biochimie. 2020;175:93-98. doi: 10.1016/j.biochi.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gelman R, Bayatra A, Kessler A, Schwartz A, Ilan Y. Targeting SARS-CoV-2 receptors as a means for reducing infectivity and improving antiviral and immune response: an algorithm-based method for overcoming resistance to antiviral agents. Emerg Microbes Infect. 2020;9:1397-1406. doi: 10.1080/22221751.2020.1776161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jamshaid H, Zahid F, ud Din I, et al. Diagnostic and treatment strategies for COVID-19. AAPS Pharmscitech. 2020;21:222. doi: 10.1208/s12249-020-01756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rehman M, Tauseef I, Aalia B, Shah SH, Junaid M, Haleem KS. Therapeutic and vaccine strategies against SARS-CoV-2: past, present and future. Future Virol. 2020;15:471-482. doi: 10.2217/fvl-2020-0137. [DOI] [Google Scholar]
- 20. Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935-949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 21. Fan J, Fu A, Zhang L. Progress in molecular docking. Quant Biol. 2019;7:83-89. doi: 10.1007/s40484-019-0172-y. [DOI] [Google Scholar]
- 22. Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668-D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bento AP, Gaulton A, Hersey A, et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083-D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Irwin JJ, Shoichet BK. ZINC – a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177-182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S, Thiessen PA, Bolton EE, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202-D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fang Y. Combining label-free cell phenotypic profiling with computational approaches for novel drug discovery. Expert Opin Drug Discov. 2015;10:331-343. doi: 10.1517/17460441.2015.1020788. [DOI] [PubMed] [Google Scholar]
- 28. Adiguzel A, Ozer H, Kilic H, Cetin B. Screening of antimicrobial activity of essential oil and methanol extract of Satureja hortensis on foodborne bacteria and fungi. Czech J Food Sci. 2007;25:81-89. https://agris.fao.org/agris-search/search.do?recordID=CZ2007000420. Accessed October 28, 2020. [Google Scholar]
- 29. Četojević-Simin DD, Velićanski AS, Cvetković DD, et al. Bioactivity of lemon balm Kombucha. Food Bioprocess Technol. 2012;5:1756-1765. doi: 10.1007/s11947-010-0458-6. [DOI] [Google Scholar]
- 30. Nebia B, Khalladi Mederbal DB. Chemical composition of the essential oil of Satureja calamintha subsp. Nepeta of west Algerian. Moroc J Chem. 2018;6:213-217. [Google Scholar]
- 31. Andre CM, Hausman J-F, Guerriero G. Cannabis sativa: the plant of the thousand and one molecules. Front Plant Sci. 2016;7:19. doi: 10.3389/fpls.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Talbaoui A, Hamdaoui L, Bouyahya A, El Moussaouiti M, Bakri Y. Chemical composition, in vitro cytotoxic, and antibacterial activities of Moroccan medicinal plants Euphorbia resinifera and Marrubium vulgare. Biointerface Res Appl Chem. 2020;10:7343-7355. doi: 10.33263/BRIAC106.73437355. [DOI] [Google Scholar]
- 33. Zrira S. Some important aromatic and medicinal plants of Morocco. In: Neffati M, Najjaa H, Máthé Á, eds. Medicinal and aromatic plants of the world – Africa Volume 3. Medicinal and aromatic plants of the world. Dordrecht, The Netherlands: Springer; 2017:91-125. doi: 10.1007/978-94-024-1120-1_5. [DOI] [Google Scholar]
- 34. Nakashima H, Yoshida O, Baba M, De Clercq E, Yamamoto N. Anti-HIV activity of dextran sulphate as determined under different experimental conditions. Antiviral Res. 1989;11:233-246. doi: 10.1016/0166-3542(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 35. Conzelmann C, Müller JA, Perkhofer L, et al. “Inhaled and systemic heparin as a repurposed direct antiviral drug for prevention and treatment of COVID-19.” Clin Med. 2020;20(6):e218–e221. doi: 10.7861/clinmed.2020-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605-1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 2010;31:455-461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. BIOVIA. Dassault Systèmes. Discovery Studio Modeling Environment. San Diego, CA: Dassault Systèmes; 2020. [Google Scholar]
- 39. Newton J, Allen A, Westley B, May F. The human trefoil peptide, TFF1, is present in different molecular forms that are intimately associated with mucus in normal stomach. Gut. 2000;46:312-320. doi: 10.1136/gut.46.3.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vuille-dit-Bille RN, Camargo SM, Emmenegger L, et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids. 2015;47:693-705. doi: 10.1007/s00726-014-1889-6. [DOI] [PubMed] [Google Scholar]
- 41. Kwon PS, Oh H, Kwon S-J, et al. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020;6:50-54. doi: 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen L-R, Wang Y-C, Lin YW, et al. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg Med Chem Lett. 2005;15:3058-3062. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang C, Wei P, Fan K, Liu Y, Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568-4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 44. Bartlam M, Yang H, Rao Z. Structural insights into SARS coronavirus proteins. Curr Opin Struct Biol. 2005;15:664-672. doi: 10.1016/j.sbi.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chou K-C, Wei D-Q, Zhong W-Z. Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochem Biophys Res Commun. 2003;308:148-151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ghosh AK, Xi K, Grum-Tokars V, et al. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg Med Chem Lett. 2007;17:5876-5880. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289-293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 48. Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766-788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stebbing J, Wildfire A, Portsmouth S, et al. Paclitaxel for anthracycline-resistant AIDS-related Kaposi’s sarcoma: clinical and angiogenic correlations. Ann Oncol. 2003;14:1660-1666. doi: 10.1093/annonc/mdg461. [DOI] [PubMed] [Google Scholar]
- 50. Krawczyk E, Luczak M, Majewska A. [Antiviral and cytotoxic activities of new derivatives of natural sesquiterpenes and taxol]. Med Dosw Mikrobiol. 2005;57:93-99. [PubMed] [Google Scholar]
- 51. Ryang J, Yan Y, Song Y, Liu F, Ng TB. Anti-HIV, antitumor and immunomodulatory activities of paclitaxel from fermentation broth using molecular imprinting technique. AMB Express. 2019;9:194. doi: 10.1186/s13568-019-0915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carvalho OV, Botelho CV, Ferreira CGT, et al. In vitro inhibition of canine distemper virus by flavonoids and phenolic acids: implications of structural differences for antiviral design. Res Vet Sci. 2013;95:717-724. doi: 10.1016/j.rvsc.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 53. da Cruz RC, Agertt V, Boligon AA, et al. In vitro antimycobacterial activity and HPLC-DAD screening of phenolics from Ficus benjamina L. and Ficus luschnathiana (Miq.) Miq. leaves. Nat Prod Res. 2012;26:2251-2254. doi: 10.1080/14786419.2011.650637. [DOI] [PubMed] [Google Scholar]
- 54. Ibrahim AK, Youssef AI, Arafa AS, Ahmed SA. Anti-H5N1 virus flavonoids from Capparis sinaica Veill. Nat Prod Res. 2013;27:2149-2153. doi: 10.1080/14786419.2013.790027. [DOI] [PubMed] [Google Scholar]
- 55. Hakobyan A, Arabyan E, Kotsinyan A, et al. Inhibition of African swine fever virus infection by genkwanin. Antiviral Res. 2019;167:78-82. doi: 10.1016/j.antiviral.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 56. Ding Y, Cao Z, Cao L, Ding G, Wang Z, Xiao W. Antiviral activity of chlorogenic acid against influenza A (H1N1/H3N2) virus and its inhibition of neuraminidase. Sci Rep. 2017;7:45723. doi: 10.1038/srep45723. [DOI] [PMC free article] [PubMed] [Google Scholar]