Significance
RNA triple helices were characterized in vitro over 60 y ago, but only recently implicated in diverse biological activities. The element for nuclear expression (ENE) protects RNA from degradation via triplex formation with the poly(A) tail on the 3ʹ ends of cellular RNAs. ENEs appear in diverse genomes, including a category containing two ENEs separated by a double-stranded region (dENEs). Using structure probing, SAXS, and cryo-EM, we show that poly(A) binding to a rice dENE induces local conformational changes, like formation of a motif that protects the extreme 3ʹ end of the poly(A). These analyses support the relevance of the recent crystal structure of the dENE+poly(A) complex and demonstrate the applicability of other techniques for understanding relatively small RNA structures.
Keywords: RNA triple helix, RNA stability, poly(A), SAXS, cryo-EM
Abstract
Cis-acting RNA elements are crucial for the regulation of polyadenylated RNA stability. The element for nuclear expression (ENE) contains a U-rich internal loop flanked by short helices. An ENE stabilizes RNA by sequestering the poly(A) tail via formation of a triplex structure that inhibits a rapid deadenylation-dependent decay pathway. Structure-based bioinformatic studies identified numerous ENE-like elements in evolutionarily diverse genomes, including a subclass containing two ENE motifs separated by a short double-helical region (double ENEs [dENEs]). Here, the structure of a dENE derived from a rice transposable element (TWIFB1) before and after poly(A) binding (∼24 kDa and ∼33 kDa, respectively) is investigated. We combine biochemical structure probing, small angle X-ray scattering (SAXS), and cryo‐electron microscopy (cryo-EM) to investigate the dENE structure and its local and global structural changes upon poly(A) binding. Our data reveal 1) the directionality of poly(A) binding to the dENE, and 2) that the dENE-poly(A) interaction involves a motif that protects the 3ʹ-most seven adenylates of the poly(A). Furthermore, we demonstrate that the dENE does not undergo a dramatic global conformational change upon poly(A) binding. These findings are consistent with the recently solved crystal structure of a dENE+poly(A) complex [S.-F. Torabi et al., Science 371, eabe6523 (2021)]. Identification of additional modes of poly(A)–RNA interaction opens new venues for better understanding of poly(A) tail biology.
RNA stabilization elements counteract RNA decay, a robust and conserved process that controls the maturation and abundance of cellular transcripts. The rate-limiting step in poly-adenylated RNA turnover is often the removal of the poly(A) tail by deadenylases (1, 2). A growing body of evidence suggests that structured RNA sequences near the 3′ ends of polyadenylated transcripts can enhance their stability (3, 4). One stabilization mechanism involves triplex-forming RNA motifs, called elements for nuclear expression (ENEs). Polyadenylated nuclear (PAN) RNA, a long noncoding RNA (lncRNA) produced by Kaposi’s sarcoma-associated herpesvirus (KSHV), was the first RNA discovered to employ an ENE to control nuclear RNA decay (5). The ENE, which folds into a U-rich internal loop (URIL) flanked by short double helices, is a cis-acting element that sequesters the downstream 3′-poly(A) tail via formation of a triple helix composed of U-A•U major-groove base triples (6). This triplex structure inhibits a rapid nuclear deadenylation process (7), the first step in RNA decay (1), and stabilizes a reporter RNA when inserted into its 3′-untranslated region (UTR) (8, 9).
Determination of the PAN ENE triple helix structure, revealing its functionally important sequence and structural features, facilitated identification of multiple additional ENE-like elements through bioinformatic strategies (10, 11). These stabilization elements are located near the 3′ ends of messenger RNA (mRNAs) and lncRNAs expressed by the genomes of various viruses, fungi, and plants. In addition, ENEs were identified in two abundant vertebrate lncRNAs, human metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and multiple endocrine neoplasia β (MENβ) (12, 13). Another class of ENEs containing two distinct ENE motifs separated by a double-helical region, and hence called double ENEs (dENEs), were identified in transposable element mRNAs, mostly in fungi and plants. All dENEs contain three highly conserved adenosines (the A triad) in the lower dENE domain and exhibit a pyrimidine/purine (Y/R) bias in the composition of the stems that had not been predicted to interact with poly(A) (11, 14), based on the crystal structures of the PAN and MALAT1 triple helices (6, 15). Thus, the striking conservation of Y/R bias in those stems suggested that they might be involved in dENE–poly(A) interaction.
To gain insight into the interaction between dENEs and the poly(A) tail, we used in-line probing assays in combination with small-angle X-ray scattering (SAXS) and cryo‐electron microscopy (cryo-EM) to study the structure of a dENE from rice TWIFB1 (Fig. 1), an hAT DNA transposon, before and after complex formation with poly(A) in trans. Our data suggest that, despite local conformational changes, the dENE does not undergo extensive structural rearrangement upon poly(A) binding. Using SAXS-derived reconstructions, we determined the orientation of poly(A) binding to a relatively small dENE molecule (∼24 kDa), which is important for establishing poly(A) protection from deadenylases. While this study was underway, we determined the 2.89-Å crystal structure of the dENE core (Xtal in Fig. 1) complexed with a 28-mer poly(A) (14). The results presented here complement that work (14) by providing direct in-solution structural evidence from SAXS experiments 1) on the directionality of the poly(A)-bound dENE interaction, and 2) in combination with cryo-EM data and structure-probing assays, to confirm that the atomic model obtained from X-ray crystallography is consistent with structures formed in dilute aqueous solutions.
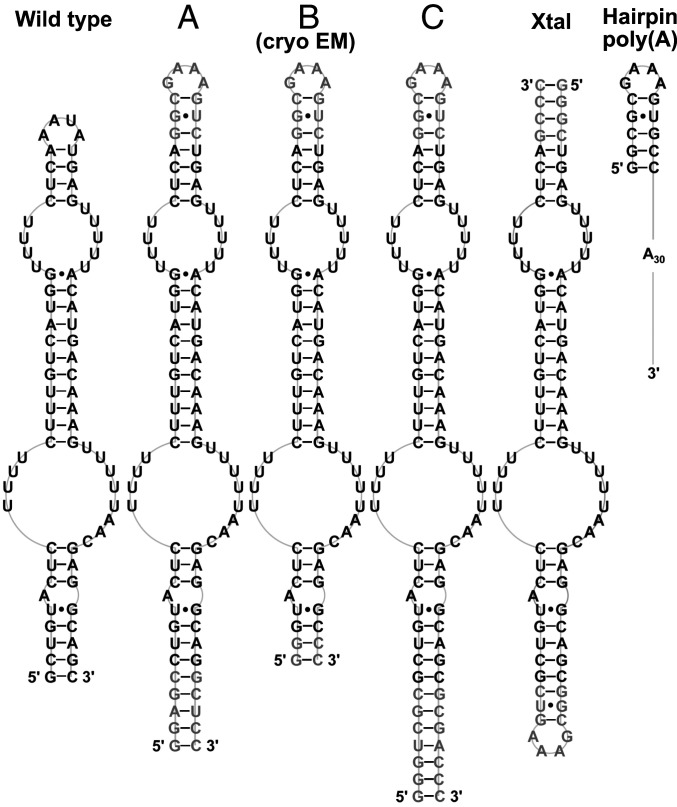
Fig. 1.
Schematic diagrams of the secondary structures of the TWIFB1 dENE constructs (including wild type, A, B, C, and Xtal) and the hairpin poly(A) used in SAXS experiments. The B dENE is also used in our cryo-EM studies. Shown in black are wild-type sequences while nonnative residues in other dENE constructs are shown in gray.
Results
Biochemical and Cellular Evidence for the Direction of poly(A) Interacting with the dENE.
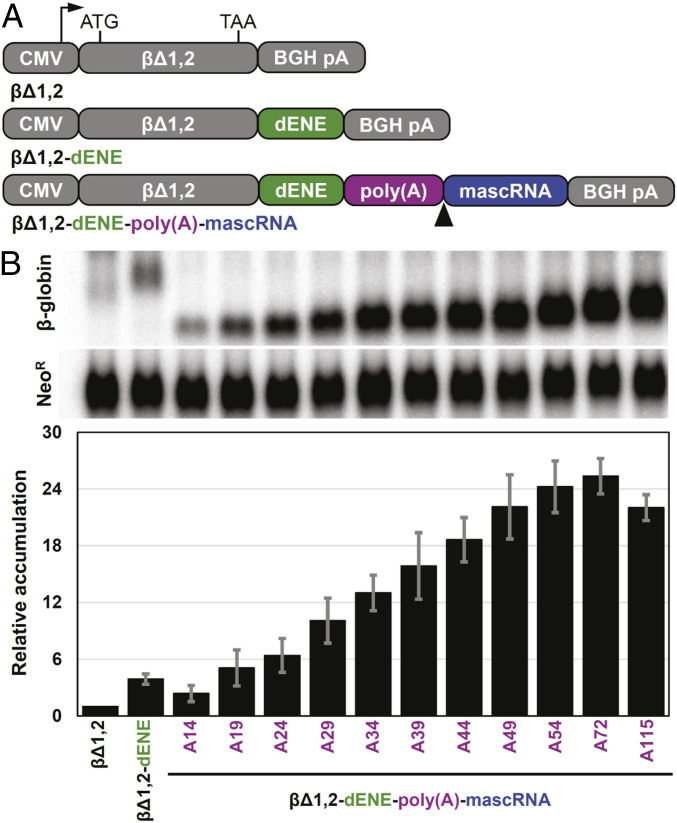
A cell-based intronless β-globin (βΔ1,2) reporter assay has been developed to assess the RNA stabilization activity of ENEs in vivo (5). We evaluated cellular accumulation of the βΔ1,2 reporter transcript containing the wild-type TWIFB1 dENE inserted upstream of 3ʹ poly(A) tails of defined lengths (ranging from 14 to 115 nucleotides [nt]) in the reporter plasmid (Fig. 2A). As shown in Fig. 2B, maximum stabilization requires the poly(A) to be ∼70 nt long. Yet, we previously determined that, when poly(A) forms a complex in trans, ∼30 to 35 nt are sufficient to achieve high binding affinity (14). Together, these results suggest that an ∼35-nt linker between the dENE-bound region of poly(A) and the 3ʹ end of the lower dENE stem is needed to obtain maximum stabilization activity in vivo where the complex forms in cis. Therefore, we hypothesized that, in vivo, an ∼35-mer tract immediately downstream of the dENE, serving as a linker, is required for poly(A) to reach and interact with the upper dENE domain.
Fig. 2.
Comparative accumulation of β-globin reporter transcripts containing different poly(A) lengths stabilized by the dENE. (A) Schematic of the β-globin reporter constructs containing a cytomegalovirus (CMV) promoter, a human intronless β-globin gene (βΔ1,2), and the dENE, followed by either a bovine growth hormone poly(A) site (BGH pA) (Middle) or a poly(A) sequence of different lengths as indicated on the x-axis of the graph in B and mascRNA (Bottom). The arrowhead indicates the RNase P cleavage site upstream of mascRNA. (B) Northern blot analyses of transcripts expressed in HEK293T cells upon transfection of different β-globin reporter constructs. Blots (Top) were probed for β-globin and neomycin resistance (NeoR) sequences. NeoR serves as a transfection and loading control. Quantification of the Northern blots (Bottom) with the β-globin signals normalized to those of NeoR. Accumulation of βΔ1,2 was set at 1. Accumulations relative to no ENE control are the average of at least three independent experiments ± SD.
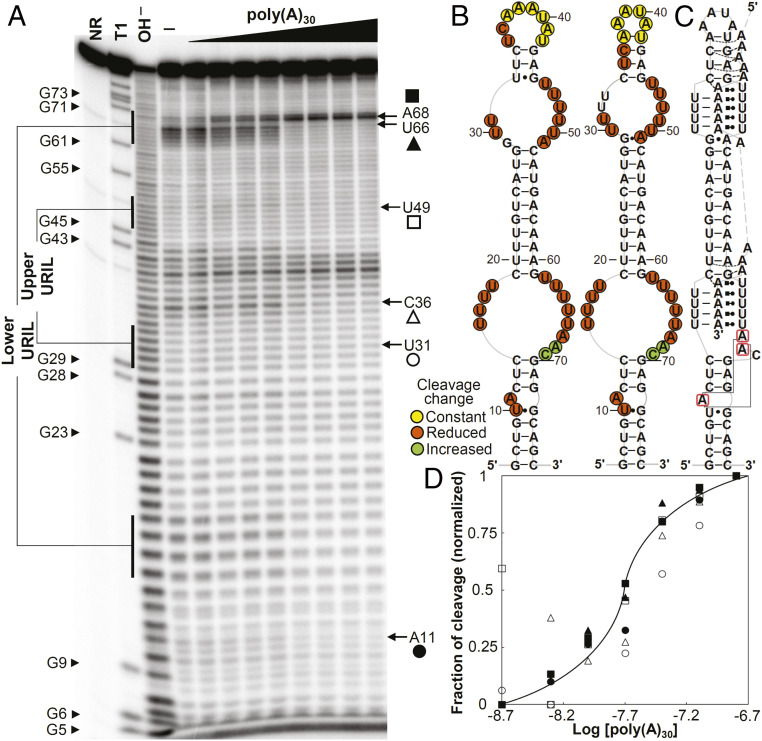
We used in-line probing analyses to further investigate the directionality of the poly(A) interaction with the dENE in vitro when it binds poly(A) in trans. [32P] 5ʹ end-labeled poly(A) complexed with the dENE was incubated under in-line probing conditions (16), and the cleavage profile of the bound poly(A) was compared to that of free poly(A). In the presence of the dENE, the least relative cleavage was observed near the 3ʹ end of the poly(A) (Fig. 3 and SI Appendix, Fig. S1). This difference is independent of poly(A) length (SI Appendix, Fig. S2), suggesting formation of a structure that engages the extreme 3ʹ end of poly(A). In contrast, increased cleavage in the middle of poly(A) argues against engagement of the middle region in a rigid structure.
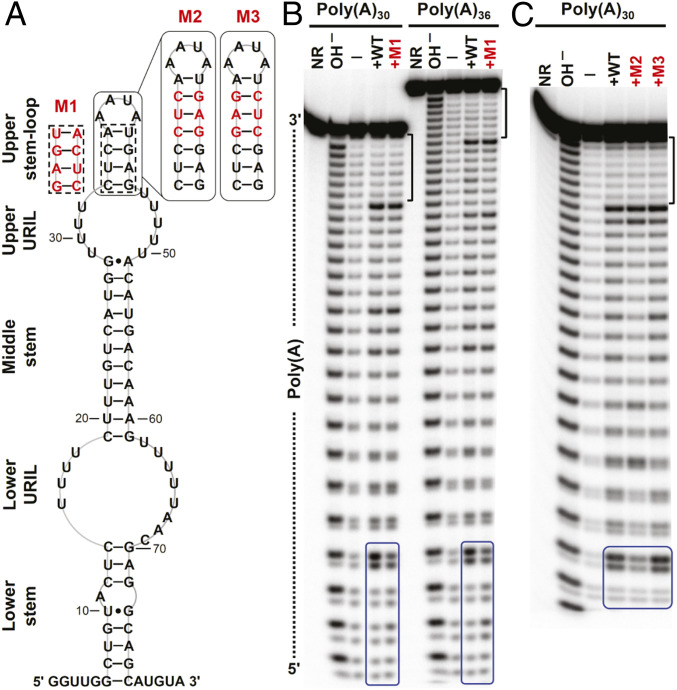
Fig. 3.
In-line probing analyses of poly(A) RNAs complexed with dENE variants. (A) Sequence and secondary structure model for the wild-type and mutant (M1 to M3) dENEs used in the experiment. (B) In-line probing analyses of 32P 5ʹ end-labeled poly(A)30 or poly(A)36 in free form (—) or complexed with the wild-type (WT) or M1 dENE. The dotted line indicates the directionality of poly(A) hydrolysis products. (C) In-line probing analyses of poly(A)30 complexed with the WT, M2, and M3 dENEs. NR and OH – designate no reaction and partial digestion with alkali, respectively. The 7-nt 3ʹ-end protected region of the dENE-bound poly(A) is marked by brackets. The 5ʹ-end region of poly(A), whose cleavage pattern responds to the upper stem mutations, is boxed in blue.
This selective cleavage pattern for the dENE-bound poly(A) provided a means to explore the direction of poly(A) binding to the dENE. First, we studied the effect of a strand-swap mutation in the upper stem, M1 dENE, on the poly(A) cleavage pattern, rationalizing that changes might occur if this part of the dENE molecule interacts with the poly(A) strand. While the cleavage pattern near the 5ʹ end of poly(A) changed (boxed in Fig. 3B), we observed no change in the protection of the 3ʹ-end region (Fig. 3B and SI Appendix, Fig. S1B). Moreover, we tested two other dENE mutants containing an extended upper stem (M2 and M3 with continuous or disrupted Y/R bias in the upper stem, respectively). Similarly, no change was observed in the cleavage pattern of poly(A) near its 3ʹ end while significant changes were detected close to the 5ʹ end (Fig. 3C and SI Appendix, Fig. S1C). Therefore, our structure-probing results argue that poly(A) interacts with the upper dENE domain and then the lower dENE domain in a 5ʹ-to-3ʹ direction. In addition, structure-probing results suggested that the poly(A) 5ʹ end interacts with the upper stem of the dENE, foreshadowing our SAXS results (see Structural Insight into the Directionality of the dENE-Bound poly(A)).
Analysis of dENE+poly(A) Complexes Using Steady-State SAXS.
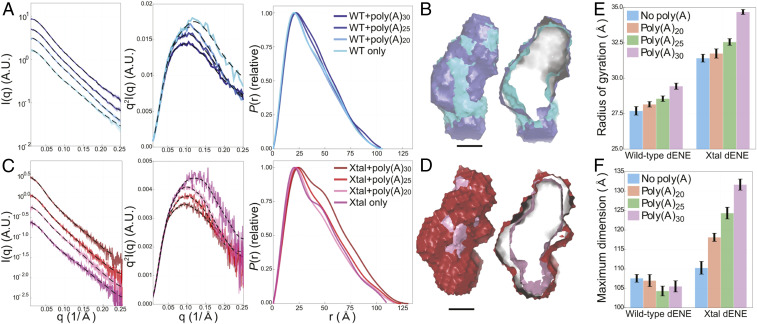
To evaluate the global structural and conformational properties of the dENE and the dENE+poly(A) complex, we performed steady-state SAXS experiments under conditions that support formation of a conformationally homogenous complex, as judged by native gel mobility-shift assays (SI Appendix, Fig. S3). Different dENE constructs, including the wild-type, crystallization (Xtal), and other dENE constructs, were examined (Fig. 1). Raw SAXS profiles, Kratky plots, and pair-distance distributions [P(r)s] for the wild-type and Xtal dENE constructs, with and without added poly(A), are shown in Fig. 4 A and C. These data show that poly(A) binding induces limited conformational changes in each dENE construct. To interpret these changes, we focused on the P(r), which is generated through calculations that involve an indirect Fourier transform of scattering plots and reports the relative probability associated with sets of distances between all pairs of atoms (17). It is standard procedure when using the GNOM program (18) to back calculate the data from the P(r) as a quality check. The black dashed lines (Fig. 4 A and C) that are superimposed on the data are the recomputed profiles from the corresponding P(r) functions. For each condition, the two curves are in excellent agreement. With confidence gained by the quality of P(r), the information can be interpreted to provide a real-space representation of the RNA structural changes. Three key features of these P(r) curves will be discussed here: 1) the position of the first peak, which reflects the most probable distance between parts of the structure; 2) the appearance of a “shoulder” in P(r) near 40 Å, indicative of a bend in the structure; and 3) the curve’s tail, terminating in the maximum linear dimension of the molecule, Dmax.
Fig. 4.
SAXS experimental data, Kratky plots, and P(r)s for two dENE constructs and their complexes: wild-type dENE (A) and Xtal dENE (C) constructs. A.U., arbitrary units. The reconstructions of the wild-type, Xtal dENE constructs and their poly(A)30 complexes are shown in B and D (cross-sections on the Right). (Scale bars: 20 Å.) The color coding presented in A and C apply to B and D as well. The histograms compare the obtained values for Rg (E) and Dmax (F) for the wild-type and Xtal dENE constructs. The error bars in E were obtained directly from the Guinier analysis. The error bars in F were computed by sampling 25 values close to Dmax, with individual weights based on the metric α (18).
For duplex-like extended RNA constructs, the most probable pair distance (peak position) represents the averaged diameters of the base-paired stems (19). For each dENE construct, the position of this peak shifts to larger values following the addition of poly(A), indicating that poly(A) binding increases this dimension of the molecules. Such behavior is consistent with de novo formation of major-groove triple helices and A-minor interactions upon poly(A) binding (14). For example, the triple-stranded helix composed of stacked U-A•U base triples appears larger in diameter than a double helix. Interestingly, the peak positions of the dENE complexes increase further as longer strands of poly(A) are bound. One possible interpretation is that triplex formation does not extend into the entire region between the two ENE domains. Rather, the poly(A) loops out between the two triple helices, with longer poly(A) RNAs forming bigger loops, slightly expanding the average molecular diameter.
The shoulder at ∼40 Å reflects a kink or bend in the dENE molecule, a feature associated with such noncoaxially aligned helices that has been observed in other RNAs (19). The P(r) curves for the free wild-type and Xtal dENE constructs display the shoulder (Fig. 4 A and C), suggesting the presence of intrinsic kinks in the dENE RNA in the absence of poly(A). This feature in the P(r) depends on the length of the helical units: dENE constructs with longer flanking stems and hairpins (e.g., A, C, and Xtal) exhibit more pronounced shoulders. Moreover, Kratky plots of free dENE have q > 0.25 Å−1 features characteristic of local disorder (20). Upon poly(A) binding, the shoulder in the P(r) curves smears out, consistent with the creation of a more conformationally rigid structure. Changes in the Kratky plots also reflect transitions to more ordered structures, demonstrating local structural rearrangements upon poly(A) binding.
Changes in the Dmax and radius of gyration (Rg) provide information about the global shape of a molecule although their values do not necessarily positively correlate with each other (21). The Rg values increase upon addition of poly(A) to the wild-type and Xtal dENEs (Fig. 4E) while an increase in Dmax is observed only for the Xtal dENE construct (Fig. 4F). Since the wild-type dENE contains the shortest apical hairpin among all the examined dENE constructs (Fig. 1), the number of adenylates that can interact with the upper stem is expected to be fewer than those of other dENE constructs. We speculate that, for the wild-type dENE, additional residues in the longer poly(A) RNAs, compared to the shorter ones, loop out between the two major-groove triple helices and expand the width of the complex (SI Appendix, Fig. S4). In contrast, the Xtal dENE construct possesses a longer upper stem (Fig. 1) and could associate with a longer tract of poly(A). Therefore, the observed increase in the Dmax might be explained by the formation of an extended RNA structure. Finally, low-resolution ab initio reconstructions of the wild-type and Xtal dENE constructs with and without poly(A)30 are shown in Fig. 4 B and D using surface representations. These models represent the overall molecular shapes directly generated from P(r) functions using DAMMIF (22). The reconstructions have normalized spatial discrepancy (NSD) values of <0.9 (SI Appendix, Table S1), indicating consistency. As illustrated by the surface models, complex formation upon poly(A) addition does not significantly affect the molecular shape.
To address the effect of the positioning the GAAA tetraloop at one or the other end of the construct on the SAXS data, we tested the A dENE construct (Fig. 1). As in the Xtal construct, A dENE is 86 nt long, but the GAAA tetraloop has been relocated to the opposite end of the structure. The raw SAXS profiles, Kratky plots, and P(r) distribution for the A dENE construct are shown in SI Appendix, Fig. S5. The SAXS-derived parameters of the A dENE construct with bound poly(A) of different lengths are the same as those of the Xtal dENE construct (SI Appendix, Fig. S6), indicating that the position of the GAAA tetraloop has no significant impact on the overall structure of the RNA. More comprehensive comparisons of SAXS reconstructions of the wild-type, A dENE, and Xtal dENE constructs are shown in SI Appendix, Fig. S4. These reconstructions indicate that all three dENE constructs are globally compact and the conformational changes observed upon poly(A) binding are modest and limited to small regions.
Structural Insight into the Directionality of the dENE-Bound poly(A).
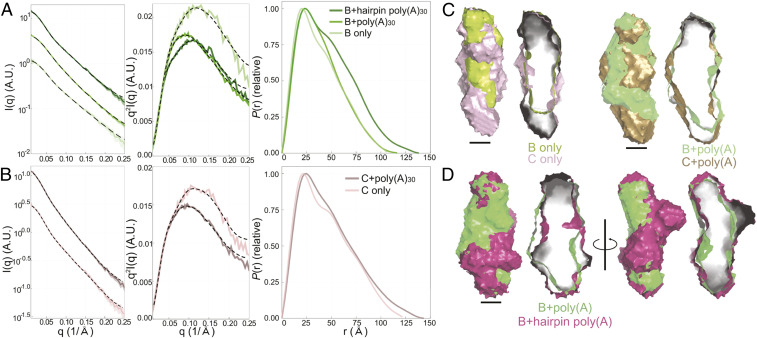
Encouraged by the quality of the dENE+poly(A) SAXS data, we designed two other dENE constructs (B and C in Fig. 1) to distinguish the location of the head (hairpin) and tail (lower stem) of dENEs in SAXS reconstructions. The two constructs have an identical hairpin, but the lower stem of construct C is extended by 8 base pairs (bp) (Fig. 1). By comparing the SAXS reconstructions of B and C dENEs and of their poly(A)-bound complexes, we can distinguish the position of the lower stem from that of the upper hairpin. In Fig. 5 A and B, the black dashed lines show recalculation of SAXS profiles from the real space interpretations, which nicely match the experimental data. The P(r) of construct B dENE (Fig. 5A) resembles that of the wild type, consistent with the presence of a shorter lower stem. Similar to the A dENE and Xtal dENE constructs, the C dENE shows a significant ∼40-Å shoulder, suggesting the presence of a kink in the molecule in the absence of poly(A). This feature disappears when poly(A)30 is added (Fig. 5B). Shifts in the peak position of the P(r) curves indicate an expansion of the overall diameter of both RNAs when complexed with poly(A)30, as observed for the other dENEs. In Fig. 5C, we first aligned constructs B (lime green) and C (light purple) in the absence of poly(A). We further aligned the B+poly(A) (green) with C+poly(A) (gold) complexes to determine the orientation of the dENE in SAXS reconstructions by identifying the 8-bp extension of the lower stem in the C dENE construct. As shown in Fig. 5C, the extended lower stem is located at the bottom; hence, the hairpin is at the top of the structure.
Fig. 5.
SAXS experimental data, Kratky plots, and P(r)s of two designed dENE constructs and their complexes: construct B (A) and construct C (B) (see Fig. 1). (C) The aligned comparisons of the SAXS reconstructions of B (lime green) and C (light purple) in the absence of poly(A) and the aligned reconstructions of their corresponding complexes: B+poly(A) (green) and C+poly(A) (gold). Using these reconstructions, we identified the longer stem in construct C and its poly(A)-bound complex and, therefore, established the orientation of both constructs. (Scale bars: 20 Å.) The hairpin of the hairpin poly(A) can be located using the known orientation of B+poly(A) and aligning it with B+hairpin poly(A) (magenta). The first two alignments in D demonstrate that these two complexes have similar lengths while the other alignments, rotated 90 degrees, reveal that the added hairpin extrudes from the complex near the top. These models indicate that the 5ʹ-to-3ʹ directionality of poly(A) is from top to bottom of construct B and that the interaction of the 3ʹ poly(A) terminus is with the lower ENE domain. Cross-sections of each surface model are on the Right.
With the molecular orientation of the dENE established, we embarked on identifying the 5ʹ-to-3ʹ orientation of the dENE-bound poly(A). To do so, we designed and in vitro transcribed a poly(A) RNA containing a stable RNA hairpin at its 5ʹ end [hairpin poly(A) in Fig. 1]. First, we confirmed that the hairpin poly(A) functions similarly to poly(A) in dENE binding by using native gel-shift assays and in-line probing experiments (SI Appendix, Fig. S7). Having established the position of the hairpin and lower stem in the dENE B+poly(A) reconstruction (green in Fig. 5D), we aligned the model with the dENE B+hairpin poly(A) reconstruction (magenta in Fig. 5D). The overall shapes are similar, with the exception of the additional density that extends from the side of the molecule near the top in the dENE B+hairpin poly(A) reconstruction. This density corresponds to the stem loop on the 5ʹ end of the hairpin poly(A). The hairpin also appears in the P(r) curve (Fig. 5A) as an additional shoulder near 50 Å compared to the P(r) of dENE B+poly(A). Therefore, our SAXS measurements reveal that the 5ʹ end of poly(A) is located at the top of the complex near the upper stem. Namely, the 5ʹ-to-3ʹ direction of poly(A) binding to the dENE is from head to tail, in agreement with the cell-based stability assay and structure-probing results.
Exploring the Fold of the dENE before and after poly(A) Binding.
The predicted secondary structure of the wild-type TWIFB1 dENE (Fig. 3A) comprises two URILs flanked by double helices, an apical AAUA tetraloop, and a single bulged adenylate located in the lower stem. To evaluate dENE structural changes in response to poly(A) binding, we used in-line probing. Overall, the pattern of RNA cleavage products separated by denaturing gel electrophoresis is in agreement with our predicted secondary structure model (Fig. 6 A and B). In the absence of poly(A), it appears that the dENE folds into a slightly different conformation, with the upper stem loop rearranged (Fig. 6B). Upon poly(A) binding, in addition to the uracil nucleotides in URILs, the bulged adenylate and its upstream uridylate exhibit reduced cleavage (SI Appendix, Fig. S8). Reduction in cleavage upon poly(A) binding is more pronounced in the lower URIL than in the upper URIL. In addition, two nucleotides, A68 and C69, exhibit increased cleavage in the presence of poly(A) (SI Appendix, Fig. S8). Dissociation constants (KDs) of ∼15 to 30 nM for the dENE-poly(A) complex were calculated by plotting the fraction of cleavage in multiple regions, including both the lower and upper dENE domains, against the logarithm of the poly(A) concentration (Fig. 6D). Based on the KD values, the binding affinity of the lower dENE domain for poly(A) is approximately two times greater than that of the upper dENE domain.
Fig. 6.
In-line probing analyses of the wild-type dENE. (A) In-line probing analyses of 32P 5ʹ end-labeled dENE in the absence (—) and presence of varying concentrations of poly(A) (2 to 320 nM) at room temperature (22 °C). Lane NR shows no reaction, lane T1 indicates RNase T1-digested RNA (G-specific cleavage), and lane OH− shows alkaline-mediated partial digestion. (B) Sequence and secondary structure models for the wild-type dENE (derived from in-line probing data in the absence of poly(A) on the Left and the predicted from comparative sequence analysis (11) on the Right). Nucleotide linkages that undergo constant, reduced, or increased cleavage upon poly(A) binding as determined by their relative band intensities in A are circled in yellow, red, or green, respectively. (C) Schematic of the poly(A)-bound dENE structure derived from the crystal structure (14). Long-range interactions between the A-triad nucleotides (boxed in red) are indicated by solid gray lines. (D) Dependence of the fraction of cleavage at selected sites (marked in A) in the lower dENE domain (closed symbols) and the upper dENE domain (open symbols) on poly(A) concentration. Calculation of fraction modulated is described in Materials and Methods. Data from selected cleavage sites were plotted against the logarithm of poly(A) concentration showing an apparent KD of ∼15 to 30 nM. The solid line indicates a theoretical binding curve for a 1:1 dENE-poly(A) interaction with a KD of 20 nM.
To determine whether modest conformational changes in the apical hairpin of the TWIFB1 dENE upon poly(A) binding are a common feature of dENEs, we studied the in-line probing pattern of two other dENE complexes: the hAT-15 dENE from a maize hAT DNA transposon and the ATGP5 dENE from an Arabidopsis long terminal repeat (LTR) retrotransposon. Although both dENEs are homologous to the TWIFB1 dENE based on sequence similarity, the latter deviates more substantially (SI Appendix, Fig. S9). Overall, their cleavage patterns by in-line probing are similar to that of the TWIFB1 dENE; the URILs of the dENE alone exhibit high cleavage that decreases significantly upon complex formation with poly(A) while the loop regions of hairpins remain unstructured (SI Appendix, Fig. S10). These observations are consistent with predicted secondary structures, confirming that the examined dENEs exhibit structural as well as sequence homology (SI Appendix, Fig. S10).
Next, we evaluated changes caused by poly(A) binding to Xtal dENE, the construct used to determine the dENE+poly(A) complex structure using X-ray crystallography (14). In the Xtal construct, the upper stem loop of the wild-type dENE is replaced with a 4-bp stem, producing a blunt-ended helix, and the lower stem is closed by a hairpin structure with a GAAA tetraloop. As shown in SI Appendix, Fig. S11, the in-line probing pattern of the Xtal construct resembles that of the wild-type dENE. Robust cleavage in the hairpin loop regions, as observed for other dENEs, is absent in the Xtal dENE, probably due to the strong folding of the GAAA tetraloop (23). Finally, a similar in-line probing profile was observed for an ENE variant that contains only the lower dENE domain, LD (SI Appendix, Fig. S12). These results further indicate that folding of the dENE core complexed with poly(A) is independent of the flanking stem and hairpin.
Global Structure of the dENE and the dENE+poly(A) Complex Resolved by Cryo-EM.
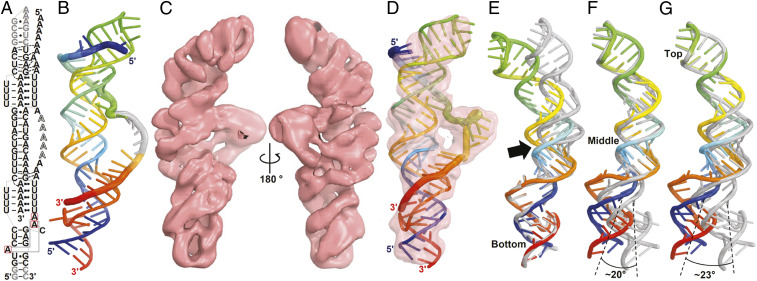
We also used single-particle cryo-EM to explore the noncrystalline structure of the poly(A)-bound dENE (Fig. 7 and SI Appendix, Fig. S13). First, we computationally modeled the structure of the cryo-EM construct (B dENE) based on the high-resolution crystal structure of the Xtal+poly(A)28 complex (Fig. 7 A and B and Movie S1) (14). Building the structure model involved relocating the stem loop from the bottom to the top of the Xtal dENE construct (Fig. 1, gray hairpin), as well as adding the adenylates that must bridge from the upper to the lower dENE domains, where poly(A) electron density is missing from the crystal structure (14). Second, we used cryo-EM to determine the structure of the B dENE+poly(A)28 complex at 5.6-Å resolution (Fig. 7C). Then the crystallographic model-derived structure (we call it the crystallographic structure) was fit into the cryo-EM density map (Fig. 7D and Movie S2). Similarity between the two models was evaluated based on their superposition (Fig. 7 E–G, SI Appendix, Fig. S14, and Movie S3). In addition, the cryo-EM map was compared with the calculated electrostatic potential (ESP) map of the atomic model, generated based on the crystal structure and blurred to 5.6-Å resolution (SI Appendix, Fig. S15 and Movie S4). Comparison shows modest differences between the two models and the two maps in the region including the middle stem. The major difference is that the presumably disordered region of the poly(A), which is invisible in the crystal structure, is clearly detected in the cryo-EM map (Fig. 7C). Superposition of the cryo-EM model with the crystallographic model reveals high similarity between comparable dENE domains (bottom, middle, and top) in the two models. However, a slight bend in the middle stem near the lower triple helical structure is observed in the cryo-EM model, ∼23° relative to the crystal structure (Fig. 7G). Y/R bias in the composition of the bent stem perhaps contributes to stem curvature as sequence-dependent double-stranded RNA (dsRNA) bending has been previously observed (24). Together, structural features observed in the upper and lower dENE domains in the cryo-EM structure, which was obtained in the absence of crystal packing constraints, resemble those of the crystal structure.
Fig. 7.
Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A) at 5.6-Å resolution. (A) Schematic of the B dENE+poly(A)28 structure derived from the crystal structure (14). Five disordered adenylates in the crystal structure are indicated by outlined letters. Non-native sequences are in gray. Dashed lines indicate regions of poly(A) interaction with the minor grooves of the stems. Long-range interactions between the A-triad nucleotides (boxed in red) are indicated by solid gray lines. (B) Computational atomic model derived from the crystal structure of the Xtal dENE+poly(A)28 complex (14) in rainbow colors (5ʹ, blue; 3ʹ, red). The disordered adenylated are added arbitrarily to the structure without any modeling (gray). (C) Two views of the cryo-EM density contoured at 3σ. (D) Real-space refined fitting of the crystallographic model to the cryo-EM density map. The cryo-EM (rainbow) and crystallographic (gray) structures were aligned using the bottom one-third (E), the middle one-third (F), or the top one-third of the structure (G) of the structure, which shows displacement of the opposite ends by ∼23° rotation, and a bend between the lower dENE domain and the middle stem (black arrow). Alignment of the poly(A)-bound structures is shown in SI Appendix, Fig. S14.
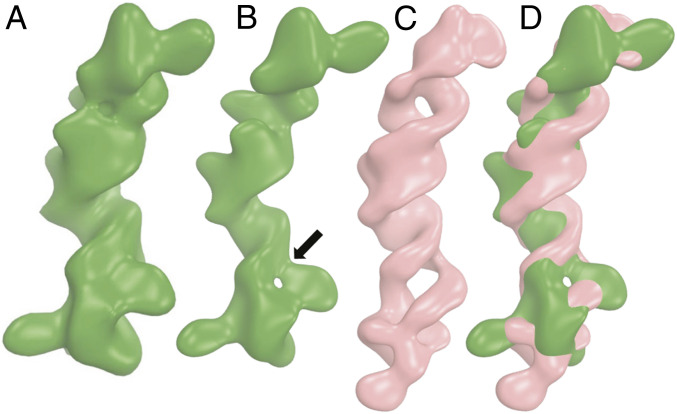
The cryo-EM structure of the free B dENE construct was determined at 8.7-Å resolution (Fig. 8 and SI Appendix, Fig. S16). The handedness of the middle stem is the most recognizable feature observed in the map. The resolution of the free dENE structure is considerably poorer than that of the dENE+poly(A) complex, possibly due to its smaller size and more dynamic nature in the absence of poly(A). At a higher contour level (3.5σ), a hole appears in the density map near the lower end of the dENE (Fig. 8B). Map-to-map alignment of the free dENE with the dENE+poly(A) complex data reveals that aligning the hole with the lower dENE domain results in a 10% higher correlation coefficient than with the upper dENE domain. The maximum dimension of the free dENE structure is ∼8 Å less than that of the dENE+poly(A) complex (Fig. 8 C and D and Movie S5), consistent with our SAXS data for the same construct (SI Appendix, Table S1). In addition, the alignment reveals differences in the lower part of the molecules (Fig. 8D). Based on the SAXS data, a kink exists in the free dENE structure, which is reflected by a significant shoulder in the P(r) curve when the lower stem is longer than 8 bp: It disappears upon poly(A) binding. The presence of the hole in the free dENE cryo-EM map and local structural rearrangements in the lower dENE domain upon poly(A) binding (Fig. 8) argue that a kink forms in free dENE between the lower and the middle stems in the same position as the hole forms in the lower URIL in the free dENE density map. Addition of poly(A) resolves these features and straightens the lower dENE domain, consistent with the SAXS and structure-probing data.
Fig. 8.
Cryo-EM structure of the B dENE construct without poly(A) at 8.7-Å resolution. (A and B) Density maps displayed at two contour levels (2.5σ, A; 3.5σ, B). The arrow indicates a hole in the lower dENE domain. (C) The dENE+poly(A) complex cryo-EM map with 8.7-Å resolution low-pass filtering, with poly(A) density subtracted (contoured at 3σ, salmon) and (D) its alignment with the free dENE map (contoured at 3.5σ, green). The two holes in C are in the upper and the lower URILs that form major-groove triple helices with poly(A). Upon poly(A) binding, local conformational changes occur in the lower dENE domain.
Poly(A) 3ʹ-End Protection by the Lower dENE Domain.
Our recent dENE+poly(A)28 crystal structure revealed formation of a 3ʹ-end RNA binding pocket that clamps and protects the extreme 3ʹ end of the poly(A) RNA (14). To gain insight into the role of the dENE pocket motif in determining the structure of the dENE-bound poly(A), we studied the in-line cleavage profile of a 28-mer poly(A) bound to different ENE variants. First, the effect of deleting the pocket motif in the TWIFB1 dENE or LD variant (SI Appendix, Fig. S17A) on the poly(A) 3ʹ-end cleavage pattern was assessed by in-line probing assays. As shown in SI Appendix, Fig. S17B, the cleavage pattern near the poly(A) 3ʹ end is identical for the wild-type dENE and LD ENE, but it changes when poly(A) is bound to either the pocket-deleted mutant (Δp) dENE or Δp LD ENE. Second, we tested different LD ENE variants with differing numbers of uridylates in the URIL. These constructs are predicted to form 3, 4, 5, or 6 U-A•U triples. As expected, the length of the protected region at the poly(A) 3ʹ end changes proportional to the number of predicted U-A•U triples (SI Appendix, Fig. S17C). Finally, we examined the effect of creating steric clashes, predicted—based on the crystal structure—to occur when the poly(A) 3ʹ end is extended beyond the major-groove triplex (SI Appendix, Fig. S18A). In-line probing assays were performed using the [32P] 5ʹ end-labeled Xtal dENE construct complexed with poly(A)28, poly(A)28G, or poly(A)26G(A)2 (SI Appendix, Fig. S18B). The single G nucleotide in the two latter poly(A) RNAs cannot engage in base triple formation and is expected to create a 3ʹ overhang leading to a steric clash with the dENE pocket motif (14). As shown in SI Appendix, Fig. S18B, the extent of cleavage is indeed increased within the pocket motif for complexes formed with G-containing poly(A) RNAs, indicating partial disruption of the structure.
Discussion
ENEs are RNA stabilization elements located upstream of the poly(A) tails of mRNAs and lncRNAs in a wide range of species, including viruses, fungi, plants, and vertebrates (5, 10–12). They retard RNA decay initiated by poly(A) tail shortening through sequestering a portion of the poly(A) sequence within a triple helical structure, as previously described for PAN or MALAT1 lncRNAs (7, 8, 15). Similar to PAN and MALAT1 ENEs, the TWIFB1 dENE stabilizes RNA by clamping the poly(A) tail (14). However, the dENE engages poly(A) in a 5ʹ-to-3ʹ direction (SI Appendix, Fig. S19) opposite to that established for PAN or MALAT1 lncRNAs (6, 15, 25). More interestingly, the dENE contains a pocket motif, whose formation relies on long-range interactions between A-triad nucleotides (bulged A11 in the lower stem, A67 and A68 in the lower URIL) (Fig. 6C). The pocket protects the extreme 3ʹ end of a poly(A) RNA by formation of a blunt-ended triple helix through a steric mechanism (14). This mechanism of poly(A) 3ʹ-end protection is distinct from that of the MALAT1 triple helix, which forms a 3ʹ blunt-ended structure due to fixing the register of its A-rich tract (15). This difference suggests independent evolutionary origins for these related RNA stabilization elements.
We obtained biochemical data validating our secondary structure models for different dENEs in free and poly(A)-bound forms using in-line probing. The fact that the dENE binds poly(A) in a manner that fixes its extreme 3ʹ end provides a unique opportunity to study structural changes induced when poly(A) RNA binds a pocket-containing ENE. Typically, during in-line probing assays, phosphodiester linkages in structured regions are held tightly in a conformation that prevents formation of the in-line geometry required for strand cleavage and yields relatively low cleavage levels (16, 26). The in-line probing patterns of dENE and poly(A) and their changes upon dENE+poly(A) complex formation can inform on the secondary structure and conformation of the dENE and poly(A) molecules, both before and after complex formation. Significant changes in the in-line probing pattern of poly(A) upon complex formation indicate protection of the poly(A) 3ʹ end by the lower dENE domain (Fig. 3 and SI Appendix, Figs. S1, S2, and S17). Poly(A) 3ʹ-end protection is consistent with our high-resolution crystal structure of a complex formed between the dENE core and poly(A)28, which reveals formation of a poly(A) 3ʹ-end binding pocket (14) and is supported by the cryo-EM structure presented in this study. Based on the crystal structure, the pocket motif recognizes the 3ʹ-most adenosine of the poly(A) via hydrogen bonding between the 3ʹ-OH group of poly(A) and a backbone phosphate of the dENE. It thereby prevents emergence of a 3ʹ poly(A) overhang through a steric mechanism (14) and is critical for protection of the poly(A) 3ʹ end through formation of a blunt-ended RNA triplex. Abolition of the observed poly(A) 3ʹ-end protection in the in-line probing assays of Δp ENE variants (SI Appendix, Fig. S17 A and B) confirms the importance of the residues that are critical for the formation of the pocket. Furthermore, our observation that the length of poly(A) that is protected at the 3ʹ end correlates with the number of U-A•U triples formed by the lower dENE domain (SI Appendix, Fig. S17C) supports the atomic model showing that the extreme 3ʹ end of poly(A) is rigidly held within the triplex structure in a pocket-dependent manner.
Similar to other RNA structures that do not undergo global structural rearrangements upon association with their cognate binding partners, including small molecules and proteins (27–30), dENEs show only local conformational changes upon poly(A) binding. In-line probing results suggest a minor rearrangement in the upper stem loop of the TWIFB1 dENE upon association with poly(A) (Fig. 6B). This local conformational change is absent from the two other dENEs tested in this study, those residing in the maize hAT-15 and Arabidopsis ATGP5 transposons (SI Appendix, Fig. S10), perhaps due to the presence of stable apical stem-loop structures (31) that prevent formation of alternative conformations. In the ATGP5 dENE, the upper domain—based on its structural homology to the PAN ENE (6)—is predicted to have its GU nucleotides on the 3ʹ side of the URIL (G37 and U38 in SI Appendix, Fig. S10C) flipped out. Based on the recent TWIFB1 dENE+poly(A)28 crystal structure (14), it is expected that U75 and U70 in the lower URILs of hAT-15 and ATGP5 dENE+poly(A) complexes, respectively, are flipped out (SI Appendix, Fig. S10). Accordingly, upon poly(A) binding, all these flipped-out nucleotides show increased signal in in-line probing assays (SI Appendix, Fig. S10). Generally, the changes observed in the in-line probing pattern of different ENEs tested in this work upon poly(A) binding are consistent with the atomic model obtained from X-ray crystallography (14) (Fig. 6C) and the global fold revealed by cryo-EM (Fig. 7): These are 1) an overall reduction in the cleavage rate within both URILs, upon formation of the triplex structure with poly(A); 2) a decrease in the cleavage of the bulged adenylate (A11 in the TWIFB1 dENE) in the lower stem as it participates in the formation of the pocket motif in the complex (Fig. 6C); and 3) a significant increase in the cleavages of the flipped-out pyrimidine nucleotide and its neighboring adenosine in URIL of the lower dENE domain. Overall, in-line probing data support formation of triple helical structures between URILs and poly(A), and of the poly(A) 3ʹ-end binding pocket through local structural rearrangements (e.g., the A triad interaction).
SAXS studies of the structure of the poly(A)-bound forms of different TWIFB1 dENE variants (wild type, A, B, C, and Xtal) containing the same dENE core demonstrate that they adopt relatively straight extended conformations. In addition, reconstructions reveal that the dENE does not undergo a dramatic global conformational change upon poly(A) binding (Fig. 4), consistent with our cryo-EM data (Fig. 8). Yet, SAXS data reveal local conformational changes induced by poly(A) binding, such as straightening of the bend between the lower and the middle stems, consistent with pocket formation and supported by the structure-probing data. Therefore, we speculate that dENEs have evolved a structure featuring accessible URILs with flanking double helices to bring the A-triad nucleotides into close proximity for recognition of the poly(A) tail, with little if any need for a prior slow conformational rearrangement and search. Moreover, indications of local disorder in the SAXS data for free dENE samples are in agreement with structure-probing results demonstrating that the URILs are not rigidly structured in the absence of poly(A).
To overcome uncertainties in identification of the location of domains and orientation of the dENE molecule due to the low resolution of SAXS, we used the strategy of expanding a portion of the molecule residing outside the dENE core. By addition of an 8-bp duplex to the lower stem, we established the orientation of the molecule with high confidence. Using a similar approach, we determined the directionality of poly(A) interaction with the dENE (Fig. 5). The conclusions are consistent with our structural probing data and crystal structure model. Based on the crystal structure of the dENE+poly(A) complex (Xtal dENE), we computationally built atomic models that match the solution constructs by adjusting the length of the upper and lower stems and relocating the GAAA tetraloop or replacing it with the wild-type TWIFB1 tetraloop. Interestingly, the theoretical scattering profiles calculated from computationally built atomic models that match the solution constructs are in good agreement with the experimental SAXS curves (SI Appendix, Fig. S20A and Table S2). For example, the NSD, which shows the agreement between the atomic model and SAXS reconstruction, is ∼1 for the wild-type dENE. In addition, the shape complementarity of SAXS reconstructions to our dENE+poly(A) complex models (SI Appendix, Fig. S20B) demonstrates the consistency of the SAXS-derived solution structure of the RNA under physiological conditions with the crystal structure.
In recent years, cryo-EM has become a powerful technique to determine high-resolution structures of macromolecular complexes (32, 33) and of relatively small RNA molecules (34–36). Our cryo-EM data provide structural insights into the molecular architecture of the dENE and dENE+poly(A) complex, which are among the smallest macromolecules (76 nt ∼24 kDa and 104 nt ∼33 kDa, respectively) so far determined by single-particle cryo-EM at subnanometer resolution levels (Figs. 7 and 8). Three-dimensional (3D) structural data obtained by cryo-EM in this study not only show high levels of consistency with our SAXS and crystal structure data but also complement them. Local resolution maps of free dENE and its complex with poly(A) (SI Appendix, Fig. S21) support SAXS and structure-probing data in demonstrating the presence of local flexibility in the lower URIL of the free dENE and in the region of the dENE-bound poly(A) bridging the two triple-helical domains. While electron density for five adenylates that bridge between the two triple helical features is missing in our crystal structure (14), this portion of the poly(A) strand is detected in the cryo-EM structure. Based on the crystal structure, seven adenylates that do not interact with the middle stem would be expected to bridge the two triple helices (Fig. 7A) whereas our cryo-EM structure suggests that at least eight adenylates could fit into the cryo-EM density for the corresponding region. In addition, poly(A) association with the minor groove of the upper stem extends into the nonnative GC-rich residues of the stem in the Xtal dENE construct (gray in Fig. 1) (14). However, such an extended interaction may not be observed in the cryo-EM structure, due to the absence of the GC-rich stem in the cryo-EM dENE construct. Furthermore, in agreement with our structure-probing results, cryo-EM data suggest that poly(A) does not interact with the middle stem in the region between the two triple-helices of the dENE (Fig. 7 C and D). While the 3ʹ end of the poly(A) is securely held in the lower dENE domain, the 5ʹ region of the poly(A) can slide relative to the upper dENE domain in either direction. Our SAXS and structure-probing data support the ability of the poly(A) 5ʹ region to alter the position of its sequestered tract relative to the upper dENE domain and to vary the number of adenylates that comprise the bridging region. We speculate that dENEs may associate with poly(A) binding proteins (PABPs) in nonadjacent regions of the poly(A) tail (the linker and bridging regions) (SI Appendix, Fig. S19), thereby preventing formation of the PABP–PABP interface, which is required for poly(A) tail recognition by Pan2-Pan3 deadenylases (37). Therefore, dENEs might impede RNA decay through several different mechanisms.
Molecular reconstructions using SAXS data provide valuable insights into the global shape and conformation of molecules under biologically relevant conditions in dilute aqueous solutions (17, 38). Even though the resolution of SAXS reconstructions is not comparable to that of structures obtained by crystallography or cryo-EM, it is a powerful and readily employed technique that provides valuable insights into the solution structures of RNA molecules to support biochemical and structure-probing data (17, 39). After several years of effort examining a large number of RNA constructs by X-ray crystallography, we eventually determined the high-resolution crystal structure of the dENE+poly(A) complex (14). In addition, we determined a subnanometer-resolution cryo-EM structure for the same complex; determining such small structures using cryo-EM is challenging despite recent breakthroughs in the field (40, 41). In this study, we demonstrated that proper design of RNA constructs (e.g., extension of the molecule outside the RNA core or incorporation of bulky groups) can directly address challenging structural questions, such as orientation of the molecule and directionality of the binding of another RNA to a small RNA element. Consistency of the SAXS data with the high-resolution crystal structure, subnanometer-resolution cryo-EM structures, and structure-probing data not only validate our findings but also demonstrate the reliability of low-resolution SAXS reconstructions to resolve challenges associated with understanding the structure and biology of small RNA molecules.
Materials and Methods
RNA Preparation.
ENE RNAs used in the study were in vitro transcribed using homemade T7 RNA polymerase. DNA templates for different TWIFB1 dENE constructs used in SAXS and cryo-EM experiments were cloned in a plasmid with a downstream hepatitis delta virus (HDV) ribozyme. The template for the Xtal dENE construct included an upstream hammerhead (HH) ribozyme and a downstream HDV ribozyme (14). Flanking ribozyme sequences were added to produce RNA with defined 5ʹ and 3ʹ ends (42). Similar to the TWIFB1 dENE, the template for hairpin poly(A) RNA included a downstream HDV ribozyme to ensure homogeneous poly(A) tract production. All DNA plasmids were linearized with EcoRI before in vitro transcription with T7 RNA polymerase. Double-stranded DNA templates for wild-type dENE RNAs and their mutants, which were used in native gel and in-line probing assays, were prepared via PCR using primers that contain the T7 RNA polymerase promoter. RNAs used for in vitro experiments were in vitro transcribed to start with G at their 5ʹ end to overcome low transcription yields (i.e., the extreme 5ʹ nucleotides were substituted by G in different dENEs). In constructs used in SAXS and cryo-EM experiments, the lower stem-flanking nucleotides (5ʹ-GGUUG and AUGUA-3ʹ) were deleted. All RNA samples were polyacrylamide gel electrophoresis (PAGE) purified after in vitro transcription reactions. Poly(A) RNAs were purchased from Dharmacon and PAGE purified using a 12% to 16% denaturing gel. RNA samples prepared for SAXS and cryo-EM experiments were exchanged into a buffer containing 10 mM 3-(N-morpholino) propanesulfonic acid (MOPS), pH 7.0, at 22 °C, 50 mM KCl, 1 mM MgCl2, and 0.025 mM (ethylenedinitrilo)tetraacetic acid (EDTA). Other RNA samples were dissolved in ddH2O after PAGE purification.
To generate 32P 5ʹ end-labeled ENE RNAs, in vitro transcribed samples were first treated with calf intestinal phosphatase (CIP), extracted by phenol-chloroform, precipitated by ethanol, and then labeled with [γ-32P]ATP and T4 polynucleotide kinase. 32P 5ʹ-end labeling reactions for poly(A) RNAs were carried out directly on chemically synthesized RNAs. All freshly radio-labeled RNA samples were PAGE purified before experiments.
Cell-Based Intronless β-Globin Reporter Assays.
The βΔ1,2 plasmid used in the reporter assay was previously described (5). The wild-type TWIFB1 dENE-containing plasmid was generated by inserting the ENE into the ApaI site of the βΔ1,2 reporter construct as described (11). To study the effect of poly(A) length on the cellular accumulation of the βΔ1,2 reporter transcript, a defined length of poly(A) (ranging from 14 to 115 nucleotides) was added downstream of the TWIFB1 dENE followed by a transfer RNA (tRNA)-like structure known as MALAT1-associated small cytoplasmic RNA (mascRNA). The 3′ end of the reporter transcript with a defined 3′-terminal poly(A) tract was generated upon RNase P cleavage, which occurs at the 5′ end of the mascRNA (43). The TWIFB1 dENE-containing βΔ1,2 reporter constructs were transfected into HEK293T cells, and accumulation of the transcript was evaluated using Northern blot analyses as described (14). Blots were exposed to a phosphorimager screen, scanned using a Storm 860 (GE Healthcare), and quantitated using ImageQuant (Molecular Dynamics) software. Fold accumulation of β-globin transcripts was calculated by first dividing the β-globin signal in each lane by that of NeoR, which was expressed from the same plasmid and provided a loading and transfection control. The signal from no ENE-containing βΔ1,2 plasmid was set at an arbitrary value of 1.0 to calculate the normalized fold accumulation of β-globin transcript for each ENE-containing construct (14).
Native Gel-Shift Assays.
To evaluate poly(A)-binding affinity of the TWIFB1 dENE and the conformational homogeneity of the complexes studied in SAXS and cryo-EM experiments, native gel mobility-shift assays were performed using 8% native polyacrylamide gels prepared with Tris-Hepes buffer (pH ∼7.5) including 1 mM MgCl2 in the gel and running buffer. Experiments were carried out with 5ʹ-radiolabeled poly(A) RNAs containing trace amounts of radio-labeled poly(A) with final concentration ∼1 nM in a binding buffer containing 10 mM MOPS, pH 7.0, 50 mM KCl, 1 mM MgCl2, 0.025 mM EDTA, and 0.5 mg/mL Escherichia coli carrier RNA, and 5% vol/vol glycerol. Before resolving the complexes from unbound poly(A) on a native gel, samples were heated at 95 °C for 2.5 min, snap-cooled on ice for 10 min, and incubated at room temperature for ∼1 h (14). Gels were dried, exposed to a phosphorimager screen, scanned using Storm 860, and quantitated using ImageQuant software. Apparent KDs were determined by plotting the fraction of ENE-bound poly(A) complex versus the initial ENE concentration and fitting the data to the following equation (14): Fraction of bound poly(A) = 0.5(KD + E + A) − 0.5[(KD + E + A)2 − 4EA]1/2. In this equation, E and A are the initial concentrations of ENE and poly(A), respectively.
In-Line Probing Assays.
In-line probing reactions were conducted based on a previously published protocol (26) with slight modifications. For each reaction, ∼1 nM 32P 5ʹ end-labeled RNA was annealed as described in the previous section. In reactions that 32P 5ʹ end-labeled poly(A) was used in in-line probing assays, poly(A):ENE ratio of <<1 was used unless stated otherwise. RNA samples were incubated for ∼70 h at 22 °C in a mixture of 50 mM Tris⋅HCl (pH 8.3 at 22 °C), 20 mM MgCl2, and 100 mM KCl. To prepare alkali ladders, 5ʹ end-labeled RNAs were partially alkali digested in 50 mM Na2CO3 (pH 9.0 at 22 °C) and 10 mM EDTA at 95 °C for 5 min. RNase T1 partial digestion to generate a G-specific RNA ladder was carried out by incubating 5ʹ end-labeled RNAs in a solution containing 25 mM sodium citrate (pH 5.0 at 22 °C), 3.5 M urea, and 0.1 unit/μL of RNase T1 at 60 °C for ∼10 min. RNA cleavage products were separated by 10% or 18% denaturing PAGE, dried, exposed to a phosphorimager screen, and analyzed using ImageQuant software. RNA ladders were prepared immediately before running denaturing gels. Intensities of cleavage bands were quantified after subtracting background. To obtain the fraction cleaved for each cleavage product, the band’s intensity was divided by the total intensity of the lane. Cleavage bands showing robust cleavage, i.e., a cleavage fraction above 0.0045, were used to track changes occurring upon ENE+poly(A) complex formation. Fraction of RNA cleavage altered (decrease or increase) at a particular site was calculated by quantification of fraction cleaved normalized to the maximum change detected upon poly(A) binding. Maximum alteration observed in response to poly(A) binding was set at 1.
SAXS Experiments.
To prepare dENE+poly(A) complexes, dENE constructs were mixed with poly(A) RNAs in a 1:1.05 ratio in a buffer containing 50 mM KCl, 1.0 mM MgCl2, 10 mM MOPS (pH 7.0 at 22 °C), and 0.025 mM EDTA. Samples were annealed at a final concentration of 25 µM using a programmable PCR machine and subsequently equilibrated at room temperature for an hour at the beamline prior to SAXS measurements. Except for the Xtal dENE samples, SAXS data were acquired at the 16-ID (Life Science X-ray Scattering [LiX]) beamline of National Synchrotron Light Source II (NSLS-II) at Brookhaven National Laboratory (44). Sixty microliters of each sample or buffer was manually loaded and continuously flowed through the beam while five 1-s exposures were acquired using vacuum detectors: an Eiger X 1M and two Pilatus 300K detectors (Dectris). The magnitude of X-ray photon momentum transfer , where is the wavelength of incident X-ray photons in Å and 2θ is the scattering angle. The experimental setup achieved a range from 0.005 to above 2.000 Å−1. To control for systematic errors, consistency in scattering profiles of buffer frames that bracket the sample-containing exposure was monitored using the py4xs Python package on site. SAXS data on the Xtal dENE samples were acquired at sector 7 of the Cornell High Energy Synchrotron Source (CHESS) over a range from 0.01 to 0.28 Å−1 using an Eiger X 1M detector. The data were monitored in real time and later fully analyzed using the BioXTAS RAW version 1.6.0 (45). The buffer-subtracted SAXS profile of each sample was reproducible over multiple beamtimes and across the different sources. The final SAXS profiles represent the average of two independent sets of measurements.
We analyzed the averaged SAXS profiles to obtain the Rgs and P(r) functions for all dENE constructs and their complexes using Guinier analysis (SI Appendix, Fig. S22) and the GNOM package, respectively (18). Kratky plots of Iq2 vs. q were also computed using the Julia language 1.6.0 and the Plots package version 1.10.0. They emphasize the higher angle scattering, which contains useful information about molecular conformations on length scales as short as ∼20 Å. Features of these curves can be used to assess molecular compactness and provide details of internal structures. The Dmax is calibrated for goodness-of-fit by enforcing a smooth zeroing of P(Dmax). We explored 25 close values of the Dmax to build statistics. We ran 20 ab initio reconstructions in parallel using DAMMIF (22). Reconstructions were averaged and superimposed using DAMAVER (46) and SUPCOMB (47) packages, respectively. Programs used for analysis of the SAXS data are available in the ATSAS 2.7.2 suite. Reconstructions were visualized with PyMOL 2.0.6 using the surface mode.
Cryo-EM Data Acquisition, Image Processing, and 3D Reconstruction.
Three microliters of the B dENE (25 μM) and B dENE+poly(A)28 complex (25 μM) samples were applied to glow-discharged 200-mesh R2/1 Quantifoil copper grids. The grids were blotted for 4 s and rapidly cryocooled in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) at 4 °C and 100% humidity. The final datasets in this study were imaged in a Titan Krios cryo-electron microscope (Thermo Fisher Scientific) operated at 300 kV with GIF energy filter (Gatan) at a magnification of 130,000× (corresponding to a calibrated sampling of 1.06 Å per pixel) for dENE, and at a magnification of 165,000× (corresponding to a calibrated sampling of 0.82 Å per pixel) for the complex. Micrographs were recorded by EPU software (Thermo Fisher Scientific) with a Gatan K2 Summit direct electron detector, where each image was composed of 30 individual frames with an exposure time of 6 s and an exposure rate of 7.6 electrons per second per Å2 for dENE and 8.3 electrons per second per Å2 for the complex. A total of 2,562 movie stacks for dENE and 6,125 movie stacks for the complex were collected. All micrographs were first imported into Relion for image processing. The motion correction was performed using MotionCor2 (48), and the contrast transfer function (CTF) was determined using CTFFIND4 (49). All particles were autopicked using the NeuralNet option in EMAN2.3 (50). Then, particle coordinates were imported into Relion 3.0.2 (51) where the poor two-dimensional (2D) class averages were removed by several rounds of 2D classification. Then, 236,892 particles were picked, and 183,155 were selected after 2D classification for the dENE and 1,003,724 particles were picked and 593,875 were selected after 2D classification for the complex, respectively. Next, the particles were imported into cryoSPARC V2.15.0 (52) to perform 3D classification using the ab initio reconstruction option. Only classes that clearly exhibit RNA-like features (e.g., major groove) were further processed to produce interpretable and trustworthy 3D RNA structures (34). Then, the best classes were selected for final refinement, and an 8.7-Å map of the dENE and a 5.6-Å map of the complex were obtained. The resolution for the final maps was estimated with the 0.143 criteria of the Fourier shell correlation curve. For more information, see SI Appendix, Figs. S13 and S16 and Table S3. The charge density maps were calculated as described (53). The initial structure of the cryo-EM construct (B dENE) was generated based on the high-resolution crystal structure of the Xtal+poly(A)28 complex (14) using Coot (54). To relocate the stem loop (see gray stem loop in Fig. 1) from one end to the opposite end, we included two Watson–Crick base pairs flanking the tetraloop and used these two base pairs as a reference by least-square superimposing them on the corresponding base pairs at the opposite end. Then, we fitted the initial model to the cryo-EM map using the reciprocal space model refinement using structure factors derived from two-halve maps prior to postprocessing, which was carried out in Refmac5 from the CCP4 package (55). The ESP map of the crystallographic structure was calculated using electron form factors of ionized atoms from the atomic model (56).
Supplementary Material
Acknowledgments
We thank K.T. Tycowski for critical reading of the manuscript; M.-D. Shu for technical assistance; A. Miccinello for editorial work; S. Chodankar and L. Yang for SAXS experimental assistance; and all J.A.S., L.P., and W.C. laboratory members for helpful discussion. Support for work performed at the Center for BioMolecular Structure beamline LiX (16-ID) at NSLS-II was provided by NIH Grants P30-GM133893, S10 OD012331, and BER- BO070. NSLS-II is supported by Department of Energy Grant BES-FWP-PS001. This work was also conducted at the Center for High Energy X-ray Sciences (CHEXS). CHEXS is supported by NSF Award DMR-1829070, and the Macromolecular X-ray science at the CHESS resource is supported by National Institute of General Medical Sciences Award 1-P30-GM124166-01A1 and the Empire State Development's Division of Science, Technology and Innovation. This work was funded by NIH Grants P01CA16038 (to J.A.S.), R35-GM122514 (to L.P.), and P41GM103832, P01AI120943, and S10OD021600 (to W.C.). S.-F. T. was supported by Damon Runyon Cancer Research Foundation Postdoctoral Fellowship DRG225716. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026656118/-/DCSupplemental.
Data Availability
Cryo-EM maps of the free and poly(A)-bound dENEs were deposited in the Electron Microscopy Data Bank under accession codes EMD-23401 and EMD-23402. The atomic model of the poly(A)-bound dENE complex can be accessed in the Protein Data Bank (PDB ID code 7LJY). All other study data are included in the article and/or supporting information.
References
- 1.Chen C. Y., Shyu A. B., Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2, 167–183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer S., Temme C., Wahle E., Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 39, 197–216 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Geisberg J. V., Moqtaderi Z., Fan X., Ozsolak F., Struhl K., Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156, 812–824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X., Bartel D. P., Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell 169, 905–917.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad N. K., Steitz J. A., A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 24, 1831–1841 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitton-Fry R. M., DeGregorio S. J., Wang J., Steitz T. A., Steitz J. A., Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science 330, 1244–1247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad N. K., Mili S., Marshall E. L., Shu M. D., Steitz J. A., Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell 24, 943–953 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Conrad N. K., Shu M. D., Uyhazi K. E., Steitz J. A., Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc. Natl. Acad. Sci. U.S.A. 104, 10412–10417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pawlicki J. M., Steitz J. A., Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 182, 61–76 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tycowski K. T., Shu M. D., Borah S., Shi M., Steitz J. A., Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2, 26–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tycowski K. T., Shu M. D., Steitz J. A., Myriad triple-helix-forming structures in the transposable element RNAs of plants and fungi. Cell Rep. 15, 1266–1276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown J. A., Valenstein M. L., Yario T. A., Tycowski K. T., Steitz J. A., Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 109, 19202–19207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilusz J. E., et al., A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 26, 2392–2407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torabi S. F., et al., RNA stabilization by a poly(A) tail 3′-end binding pocket and other modes of poly(A)-RNA interaction. Science 371, eabe6523 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown J. A., et al., Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 21, 633–640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soukup G. A., Breaker R. R., Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantara W. A., Olson E. D., Musier-Forsyth K., Analysis of RNA structure using small-angle X-ray scattering. Methods 113, 46–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svergun D. I., Determination of the regularization parameter in indirect‐transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 (1992). [Google Scholar]
- 19.Chen Y. L., Lee T., Elber R., Pollack L., Conformations of an RNA helix-junction-helix construct revealed by SAXS refinement of MD simulations. Biophys. J. 116, 19–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulshina N., Baird N. J., Ferré-D’Amaré A. R., Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 16, 1212–1217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant T. D., et al., The accurate assessment of small-angle X-ray scattering data. Acta Crystallogr. D Biol. Crystallogr. 71, 45–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke D., Svergun D. I., DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferré-D’Amaré A. R., Doudna J. A., RNA folds: Insights from recent crystal structures. Annu. Rev. Biophys. Biomol. Struct. 28, 57–73 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Marin-Gonzalez A., et al., Double-stranded RNA bending by AU-tract sequences. Nucleic Acids Res. 48, 12917–12928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown J. A., Unraveling the structure and biological functions of RNA triple helices. Wiley Interdiscip. Rev. RNA 11, e1598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regulski E. E., Breaker R. R., In-line probing analysis of riboswitches. Methods Mol. Biol. 419, 53–67 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Launer-Felty K., Wong C. J., Cole J. L., Structural analysis of adenovirus VAI RNA defines the mechanism of inhibition of PKR. Biophys. J. 108, 748–757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serganov A., Huang L., Patel D. J., Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455, 1263–1267 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montange R. K., Batey R. T., Riboswitches: Emerging themes in RNA structure and function. Annu. Rev. Biophys. 37, 117–133 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Shelke S. A., et al., Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif. Nat. Commun. 9, 4542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu Z., Bevilacqua P. C., Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochemistry 38, 15369–15379 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Wang H. W., Fan X., Challenges and opportunities in cryo-EM with phase plate. Curr. Opin. Struct. Biol. 58, 175–182 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wang H. W., Wang J. W., How cryo-electron microscopy and X-ray crystallography complement each other. Protein Sci. 26, 32–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kappel K., et al., Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat. Methods 17, 699–707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K., et al., Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 Å resolution. Nat. Commun. 10, 5511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang K., et al., Cryo-electron microscopy and exploratory antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. bioRxiv [Preprint] (2020). 10.1101/2020.07.18.209270 (Accessed 20 July 2020). [DOI]
- 37.Schafer I. B., et al., Molecular basis for poly(A) RNP architecture and recognition by the Pan2-Pan3 deadenylase. Cell 177, 1619–1631.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baird N. J., Ferré-D’Amaré A. R., Analysis of riboswitch structure and ligand binding using small-angle X-ray scattering (SAXS). Methods Mol. Biol. 1103, 211–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., Pollack L., SAXS studies of RNA: Structures, dynamics, and interactions with partners. Wiley Interdiscip. Rev. RNA 7, 512–526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu M., Lander G. C., How low can we go? Structure determination of small biological complexes using single-particle cryo-EM. Curr. Opin. Struct. Biol. 64, 9–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K., et al., Structure of the 30 kDa HIV-1 RNA dimerization signal by a hybrid cryo-EM, NMR, and molecular dynamics approach. Structure 26, 490–498.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avis J. M., Conn G. L., Walker S. C., Cis-acting ribozymes for the production of RNA in vitro transcripts with defined 5′ and 3′ ends. Methods Mol. Biol. 941, 83–98 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Wilusz J. E., Freier S. M., Spector D. L., 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135, 919–932 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L., et al., Solution scattering at the life science X-ray scattering (LiX) beamline. J. Synchrotron Radiat. 27, 804–812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hopkins J. B., Gillilan R. E., Skou S., BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Cryst. 50, 1545–1553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkov V. V., Svergun D. I., Uniqueness of ab initio shape determination in small‐angle scattering. J. Appl. Cryst. 36, 860–864 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozin M. B., Svergun D. I., Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 34, 33–41 (2001). [Google Scholar]
- 48.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang G., et al., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Wang J., Experimental charge density from electron microscopic maps. Protein Sci. 26, 1619–1626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winn M. D., et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J., Liu Z., Frank J., Moore P. B., Identification of ions in experimental electrostatic potential maps. IUCrJ 5, 375–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps of the free and poly(A)-bound dENEs were deposited in the Electron Microscopy Data Bank under accession codes EMD-23401 and EMD-23402. The atomic model of the poly(A)-bound dENE complex can be accessed in the Protein Data Bank (PDB ID code 7LJY). All other study data are included in the article and/or supporting information.