Abstract
Glycosylation of nuclear and cytoplasmic proteins is an essential post-translational modification in mammals. O-GlcNAc transferase (OGT), the sole enzyme responsible for this modification, glycosylates over a thousand unique nuclear and cytoplasmic substrates. How OGT selects its substrates is a fundamental question that must be answered to understand OGT’s unusual biology. OGT contains a long tetratricopeptide repeat (TPR) domain that has been implicated in substrate selection, but there is almost no information about how changes to this domain affect glycosylation of individual substrates. By profiling O-GlcNAc in cell extracts and probing glycosylation of purified substrates, we show here that ladders of asparagines and aspartates that extend the full length of OGT’s TPR lumen control substrate glycosylation. Different substrates are sensitive to changes in different regions of OGT’s TPR lumen. We also found that substrates with glycosylation sites close to the C-terminus bypass lumenal binding. Our findings demonstrate that substrates can engage OGT in a variety of different ways for glycosylation.
Graphical Abstract
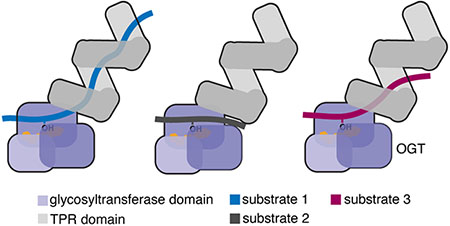
O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) is an essential nutrient- and stress-responsive enzyme that transfers GlcNAc to serine or threonine side chains of thousands of nuclear and cytoplasmic proteins.1–6 OGT’s substrates are structurally and functionally diverse and are involved in most cellular processes.7–15 A longstanding question in the field is how OGT chooses its substrates. OGT contains a C-terminal glycosyltransferase domain that performs chemistry and an N-terminal tetratricopeptide repeat (TPR) domain that has been implicated in substrate selection (Figure 1).3, 16–22 OGT’s TPR domain, which contains 13.5 repeats, forms a superhelix with conserved asparagine and aspartate ladders that extend the length of the lumenal surface (Figure 1 & Tables S1–2).18, 23 A structure of OGT complexed with a peptide that binds in the active site-proximal TPR lumen revealed a network of bidentate contacts from five sequential asparagine amides to the peptide backbone; side chains from three aspartates contacted polar side chains of the peptide (Figure 1C).24 Proteome-wide studies showed that the active site-proximal asparagines and aspartates are important in substrate glycosylation, with the former contributing to sequence-independent binding and the latter controlling selectivity.23, 25 Although these studies identified functions of residues in the proximal TPR lumen in substrate glycosylation, the TPR lumen is more than 100 Å long. The functions of conserved asparagine and aspartate residues in the medial and distal lumen are not well understood. Elucidating the mechanisms of substrate selection is critical because elevated O-GlcNAc levels have been implicated in cancers and metabolic diseases.26–31 To evaluate opportunities for therapeutic intervention, we need information on how different regions of the OGT’s TPR domain are involved in glycosylation of different substrates.
Figure 1.
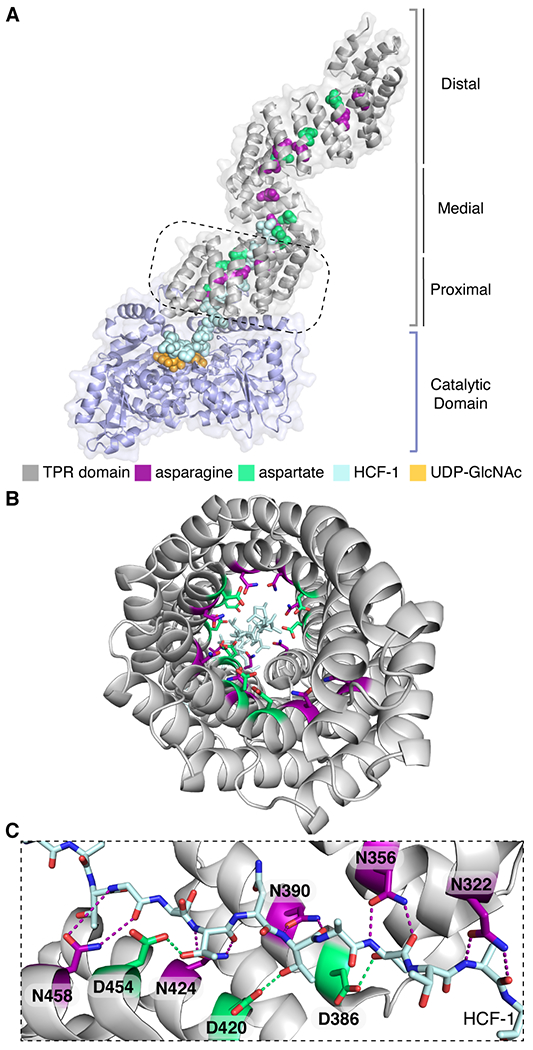
Conserved asparagine and aspartate ladders line the entire TPR lumen of OGT. (A) Composite structure (PDB 1W3B and 4N3B) of full-length OGT complexed with a peptide (HCF-1pro; light blue) that extends from the active site into the proximal TPR lumen (boxed region).18,24 The TPR lumen is lined by conserved asparagine (magenta) and aspartate (green) ladder motifs. (B) View through OGT’s TPR domain from the N-terminus. The peptide binds towards the C-terminal TPRs. (C) TPR asparagine side chains anchor the HCF-1pro peptide backbone; TPR aspartates contact polar groups on side chains of the bound peptide (the orientation shown is 90° from that in the dashed boxed in A).
To determine whether the entire asparagine ladder plays a role in glycosylation, we tested the protein glycosylation activity of three OGT constructs in which groups of five asparagines were mutated to alanine (Figure 2A, referred to as N5A mutants). We used HeLa extracts for these studies because they are metabolically active and mimic a cellular environment but enable precise control of the concentrations of OGT added.23, 25, 32 Glycosylation was detected by immunoblotting with a pan O-GlcNAc antibody. While the antibody does not recognize all O-GlcNAcylated proteins equally, it is useful for reporting on relative changes in band intensities. As reported previously, N5Aprox showed reduced global glycosylation compared to wildtype OGT.25 The N5Amed and N5Adist mutants also showed lower glycosylation activity than wildtype, but glycosylation gradually increased as the asparagine mutations moved further from the active site (Figure 2B). Although the asparagines closest to the active site were more important, these results showed that the entire asparagine ladder contributes to global protein glycosylation.
Figure 2.
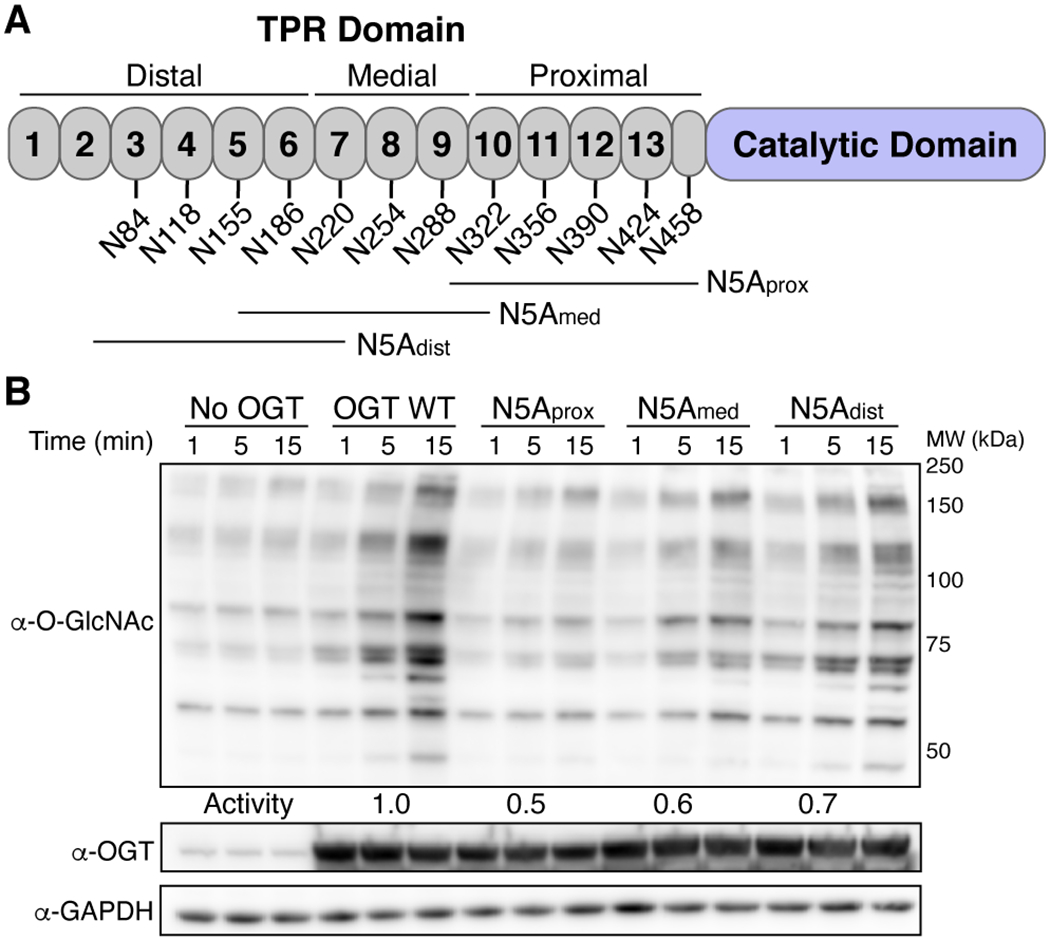
OGT requires the entire asparagine ladder for global glycosylation of HeLa extracts. (A) Cartoon schematic of asparagine ladder mutations. (B) HeLa extracts show loss of global glycosylation by OGT N5A mutants. Representative blot for two independent experiments. See methods for activity calculations.
We took a similar approach to assessing the roles of the aspartate ladder residues in global substrate selection. For these studies, we mutated conserved aspartates to alanines in groups of two (Figure 3A, referred to as D2A mutants). As previously reported, global glycosylation increased when aspartates in the proximal region of the TPR domain were mutated to alanines (D2A-2prox, Figures 3A & S1).23 Global glycosylation also increased for other D2A mutants along the TPR lumen (Figure S1), with the greatest increase observed for mutations in the medial lumen (D2A-3med; Figure 3B). We observed both new bands and intensity changes in bands that were also seen with wildtype OGT. Notably, there were pronounced differences in the banding pattern for different D2A constructs (compare D2A-2prox to D2A-3med), suggesting that aspartates in different regions of the TPR lumen differentially regulate substrate glycosylation. When pairs of D2A mutants in the proximal and medial lumen were combined (D4A), bands observed for the individual pairs remained in the quadruple mutant (Figure 3B, see asterisks), suggesting that the contributions of different aspartate pairs in controlling substrate preferences are additive. Overall, the results for the aspartate ladder mutants implicate the entire TPR lumen in global glycosylation of cell extracts, but show that the effects are largest for aspartates in the five TPRs closest to the active site (i.e., TPRs 9 to 13).
Figure 3.
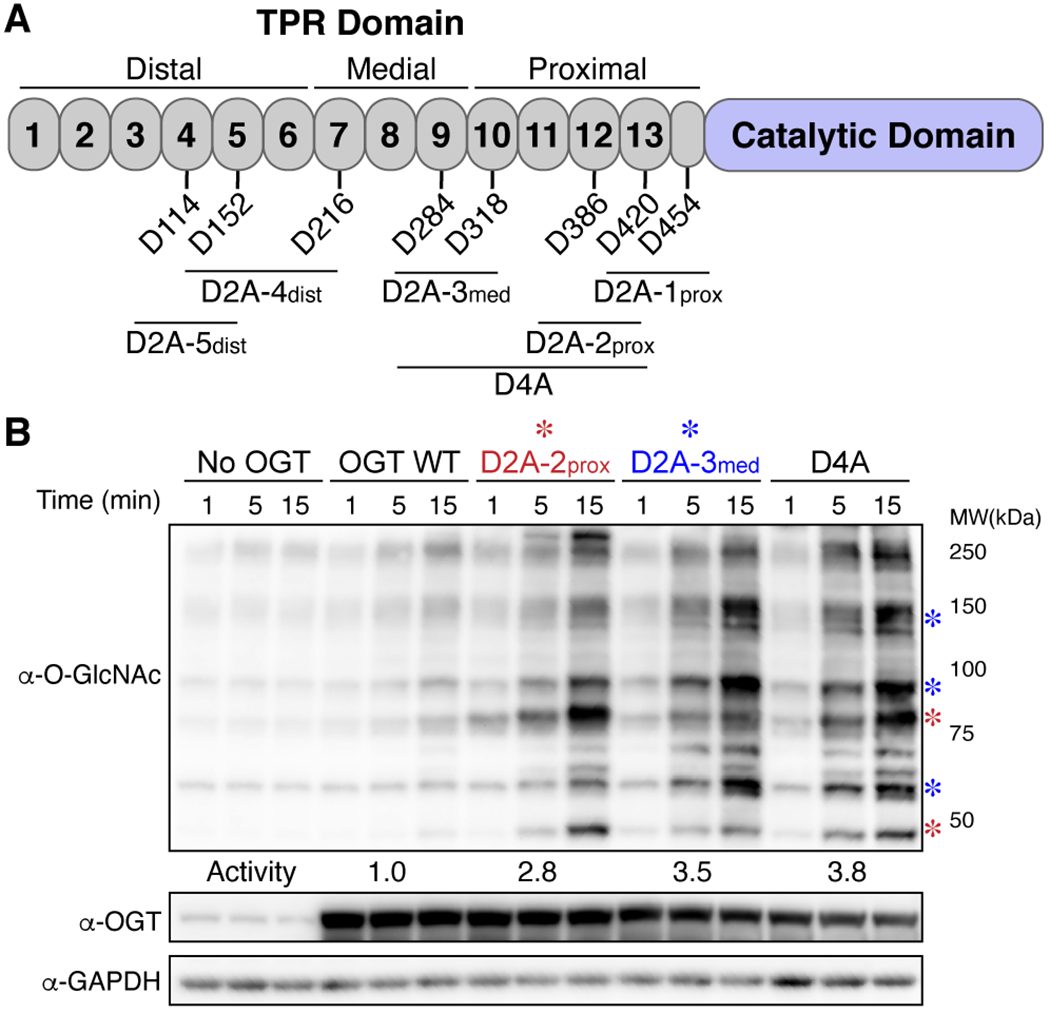
Aspartate ladder regulation of substrate selection is additive. (A) Cartoon schematic of aspartate ladder mutations. (B) Glycosylation of HeLa extracts by D2A-2prox and D2A-3med mutants shows large global increase in glycosylation and preferential increases in different O-GlcNAc bands (see asterisks). These preferences are additive in the D4A mutant. Representative blot for two independent experiments. See methods for activity calculations.
The extract experiments provided a convenient way to assess global glycosylation, but mechanistic interpretation is challenging. Adaptors, or proteins that recruit substrates to OGT’s active site, are proposed to play a substantial role in substrate selection.33–38 Thus, changes in global glycosylation with the TPR mutants could reflect changes in adaptor recognition rather than intrinsic differences in substrate recognition. To assess how individual substrates are affected by mutations in different regions of the TPR domain, we tested three previously studied OGT substrates, TAB1, CAMKIV, and CARM1.33, 39–42 These proteins are among the few substrates that have been characterized kinetically,43 and each is approximately 55 kDa with an enzymatic domain and a long, C-terminal region that is predicted to be disordered. Each also has one major glycosite that is located in a different region of the protein, either in a long loop in the folded domain (CAMKIV), in the unstructured region but immediately adjacent to the folded domain (TAB1), or in the unstructured region close to the C-terminus of the protein (CARM1) (Figure 4A).39–42 To identify large differences in how changing OGT’s TPR domain affects these substrates, we ran glycosylation assays under forcing conditions that would obscure small effects.
Figure 4.
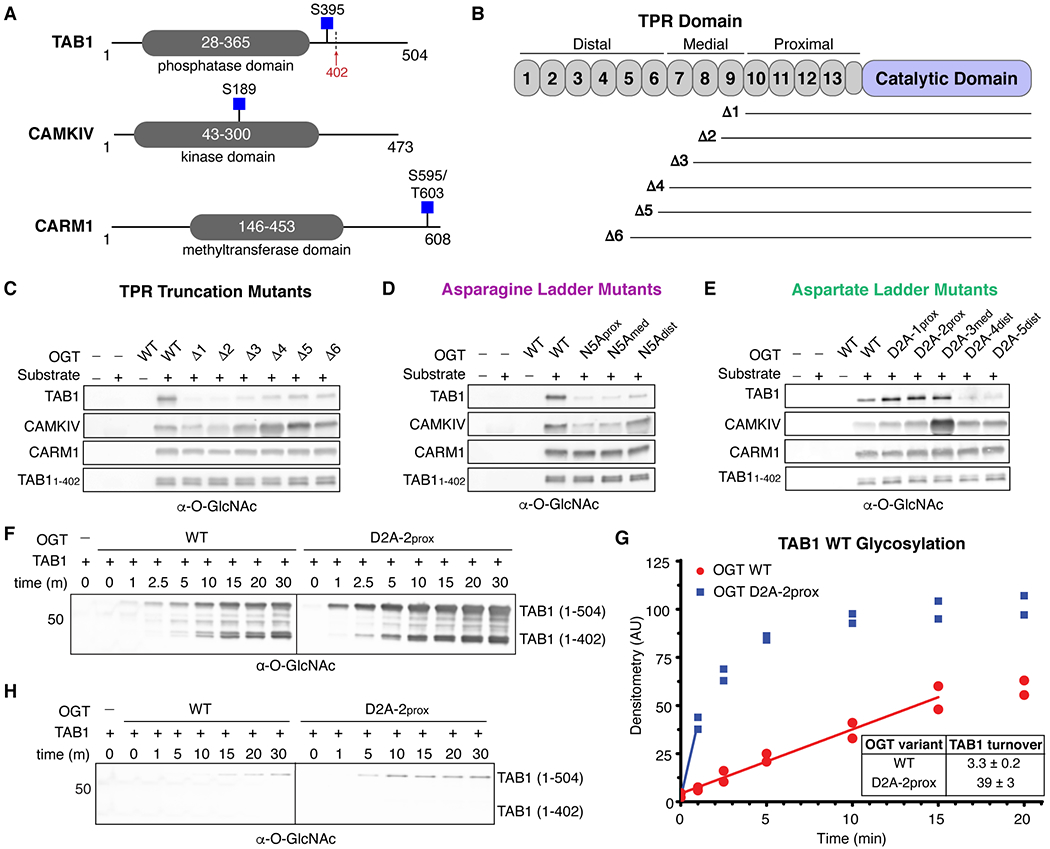
OGT protein substrates have different dependencies on the TPR domain for glycosylation. (A) Cartoon schematics of TAB1, CAMKIV, and CARM1 protein architecture. Glycosites are denoted by blue squares. TAB1 is proteolytically cleaved at residue K402 (red arrow).19, 43 (B) Cartoon schematic of OGT N-terminal TPR truncation mutants. (C) In vitro glycosylation of substrates by TPR truncation mutants. (D) In vitro glycosylation of substrates by asparagine ladder mutants. (E) In vitro glycosylation of substrates by aspartate ladder mutants. (F) Time-dependent in vitro glycosylation of TAB1 by wildtype OGT and D2A-2prox. Subpanels are from the same blot and image. (G) Densitometry vs. time plot of in vitro TAB1 glycosylation. Best-fit lines are shown in the linear range for each activity curve. (H) Time-dependent in vitro glycosylation of TAB1 S395A by wildtype OGT and D2A-2prox. Subpanels are from the same blot and image. Images in panels F and H were acquired under identical conditions. Representative blots are shown for two independent experiments for each data set.
We first assessed how truncating the TPR domain affected glycosylation of these three substrates (Figure 4B). Previous studies of two purified proteins showed that glycosylation was largely abolished when three N-terminal TPRs were removed, while glycosylation of a short peptide was unaffected.44, 45 These studies were limited to two substrates, but it has largely been assumed that all protein substrates must interact with a large part of the TPR domain to be glycosylated. We found that TAB1 behaved like the proteins in earlier studies in that removal of four or more TPRs greatly attenuated glycosylation (Figure 4C). In contrast, removing four to six N-terminal TPRs increased glycosylation of CAMKIV (Figures 4B,C: compare Δ6, which has 9 TPRs, to Δ4 which has 7 TPRs), although further TPR trimming led to reduced glycosylation levels compared to wildtype OGT. For CARM1, removal of as many as nine TPRs had no impact on glycosylation. Similar to CARM1, a TAB1 proteolysis product that co-purified with full-length TAB119, 43 (TAB11-402; Figure 4A) was glycosylated to the same extent by all TPR truncation mutants. The results for CARM1 and TAB11-402 suggest that proteins with glycosites very close to the C-terminus behave like peptide substrates in that they do not require contacts to the TPR domain to access the OGT active site for glycosylation.17, 44–46 Overall, the data for the three full-length protein substrates showed that each relies on contacts to different regions of OGT.
Although the truncation studies revealed differences in how proteins interact with the TPR domain, they did not provide information on whether contacts to the TPR lumen were important. We used the asparagine ladder mutants to assess whether substrates bind in the TPR lumen (Figure 2A, 4D). As expected from the truncation studies, CARM1 and TAB11-402 glycosylation was unaffected by the lumenal mutations. However, glycosylation of full-length TAB1 was largely abolished for all three of the N5A mutants. CAMKIV glycosylation was reduced for the N5Aprox and N5Amed mutants, but not for N5Adist. Therefore, both TAB1 and CAMKIV engage the TPR lumen, but TAB1 requires asparagine ladder residues in the distal region of the TPR lumen for glycosylation, whereas CAMKIV glycosylation depends only on contacts to the proximal and medial regions of the TPR lumen.
TAB1 and CAMKIV, the two substrates that interacted with the TPR lumen, also showed notable differences in glycosylation with the aspartate mutants (Figure 3A,4E). Whereas glycosylation of full-length TAB1 decreased when aspartates in the distal TPR lumen were mutated to alanine (D2A-4dist – D2A-5dist), it increased for D2A mutants elsewhere (D2A-1prox – D2A-3med). CAMKIV showed a large increase in glycosylation when two aspartates in the medial lumen were changed to alanines, but the other aspartate mutants showed only modest increases compared to wildtype OGT. Taken together, these findings are generally consistent with the cell extract studies in showing that replacing aspartates in the proximal and medial TPR lumen with alanine increases glycosylation. Increased glycosylation could be due to either new glycosites or to increased glycosylation at existing glycosites. Using glycosite mapping, we have previously reported that D2A-2prox can modify new sites compared with wildtype OGT.23 The new product bands that appeared in the extract studies imply that the D2A mutants are able to glycosylate some new protein substrates. To test whether D2A mutants can increase glycosylation at existing sites, we quantified turnover for full-length TAB1 using wildtype OGT and the D2A-2prox, and we observed that the O-GlcNAc signal for TAB1 appeared earlier with D2A-2prox and increased more over time than with wildtype OGT (Figure 4F–G). To confirm that increased glycosylation was due to modification at S395, we performed similar experiments for a TAB1 S395A mutant (Figure 4H). The O-GlcNAc signal for the 1–402 cleavage product disappeared completely. Upon treatment with either wildtype OGT or D2A-2prox, we observed a weak signal growing in over time for full-length TAB1. We conclude that S395 is the primary site of glycosylation for both wildtype OGT and the D2A mutant, although there is a weak secondary site that appears to be in the disordered region C-terminal to S395. We quantified the rate of glycosylation for full-length TAB1 and found a >10-fold increase in rate for D2A-2prox compared with wildtype OGT (Figure 4G). Similar time-dependent experiments were run for CAMKIV WT and a variant with the major glycosite, S189, changed to alanine. As with TAB1, the S189A mutant was poorly glycosylated by wildtype OGT, although we observed a weak signal at longer time points (Figure S6A). We also tested the D2A mutant that was found to have the largest effect on CAMKIV WT glycosylation (D2A-3med) against CAMKIV S189A. We observed substantial glycosylation of CAMKIV S189A with this mutant, although CAMKIV WT was still the better substrate (Figure S6B). These studies confirm S189 as the major glycosite in CAMKIV for wildtype OGT, but show that there is at least one other glycosite. Furthermore, D2A-3med glycosylates both the major and secondary sites more effectively than wildtype OGT. These findings support the conclusion that the conserved aspartates in the TPR lumen act as “gatekeeper” residues, but we now know that they can not only control what sites are glycosylated,23 but how rapidly they are modified.
Our findings show that the conserved asparagine and aspartate ladders that span the TPR lumen regulate glycosylation of a large fraction of OGT’s glycoproteome and reveal major differences in how different substrates engage the lumen. The three purified substrates tested here provide different working models for substrate engagement during glycosylation. In one model (CARM1-like), substrates behave like short peptide substrates in the sense that they access the active site without engaging the TPR lumen. In a second model (TAB1-like), substrates simultaneously engage the active site and the full TPR lumen to achieve glycosylation. The results for TAB1 are remarkable because they indicate an interaction with OGT that spans as much as 120 Å. Because removal of most of the C-terminal disordered domain removed TAB1’s dependence on lumenal contacts for glycosylation, we infer that the presence of this disordered domain somehow interferes with glycosylation. Therefore, one function of the TPR domain may be to chaperone unfolding of the disordered domain so that the glycosylation site is accessible. In other contexts, TPR domain-containing proteins are known to serve as co-chaperones to regulate protein fate.47–50 Given our finding that substrate proteolysis can alter requirements for glycosylation, it is also possible that proteolysis serves to regulate some O-GlcNAc modifications.
The CAMKIV results support a third model for TPR binding in which substrates rely on contacts to only part of the TPR lumen. Unlike the other model substrates tested, the glycosite of CAMKIV is located in a long, disordered loop within the kinase domain. The location of the glycosite in a long loop may place topological constraints on how the substrate engages the TPR lumen,42 and these constraints could explain the observation that removing distal TPRs increases glycosylation. The CAMKIV loop also contains a regulatory phosphorylation site, T200, that blocks its glycosylation.42, 43 This phosphorylation site is 11 residues C-terminal to the glycosite. Our earlier studies showed a strong bias against negatively-charged side chains several residues C-terminal to the site of glycosylation in OGT substrates. We interpreted this bias as reflecting unfavorable interactions between substrate side chains and lumenal aspartates.23 We speculate that phosphorylation may prevent engagement of CAMKIV with OGT’s TPR lumen by introducing a repulsive interaction, providing one possible mechanism to explain “crosstalk” between phosphorylation and glycosylation.51
Our work, along with a recent study on how glycosylation of O-GlcNAcase (OGA) is affected when lumenal residues in OGT’s TPR domain are altered,21, 52, 53 is beginning to clarify how different substrates use OGT’s TPR domain to access its active site, pointing the way for future work to develop a molecular understanding of substrate recognition. The work on OGA showed that it behaves like TAB1 in using lumenal interactions that involve most of the TPR domain for substrate recognition.21, 52, 53 Further progress in understanding OGT substrate recognition will require integration of glycosylation assays with quantitative assays to measure substrate binding affinities, which so far do not exist, and ideally with structural information. In the meantime, we note that modulating glycosylation of broad subsets of substrates by changing features of the TPR domain in cells or in animal models may help clarify the function of O-GlcNAcylation in health and disease.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. David Vocadlo for providing the pET28a-TAB1, pET28a-CARM1, and pET28a-CAMKIV plasmids.
Funding Sources
This work was supported by the NIH (GM0924263 to S.W. and F32GM129889 to C.M.J.). J.J. is a National Science and Engineering Research Council (NSERC) of Canada PGS-M and PGS-D3 fellowship recipient.
Footnotes
Supporting Information. The following files are available free of charge.
Experimental protocols used in this study, supplemental figures S1–S6, and tables S1–S7 (PDF)
The authors declare no competing financial interest.
REFERENCES
- [1].Shafi R, Iyer SPN, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, and Marth JD (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny, Proc. Natl. Acad. U. S. A 97, 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].O’Donnell N, Zachara NE, Hart GW, and Marth JD (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability, Mol. Cell. Biol 24, 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Haltiwanger RS, Blomberg MA, and Hart GW (1992) Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide beta-N-acetylglucosaminyltransferase, J. Biol. Chem 267, 9005–9013. [PubMed] [Google Scholar]
- [4].Torres CR, and Hart GW (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc, J. Biol. Chem 259, 3308–3317. [PubMed] [Google Scholar]
- [5].Levine ZG, and Walker S (2016) The biochemistry of O-GlcNAc transferase: which functions make it essential in mammalian cells?, Annu. Rev. Biochem 85, 631–657. [DOI] [PubMed] [Google Scholar]
- [6].Levine ZG, Potter SC, Joiner CM, Fei GQ, Nabet B, Sonnett M, Zachara NE, Gray NS, Paulo JA, and Walker S (2021) Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase, Proc. Natl. Acad. U. S. A 118, e2016778118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tai H-C, Khidekel N, Ficarro SB, Peters EC, and Hsieh-Wilson LC (2004) Parallel identification of O-GlcNAc-modified proteins from cell lysates, J. Am. Chem. Soc 126, 10500–10501. [DOI] [PubMed] [Google Scholar]
- [8].Woo CM, Lund PJ, Huang AC, Davis MM, Bertozzi CR, and Pitteri S (2018) Mapping and quantification of over 2,000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (IsoTaG), Molecular & Cellular Proteomics 17, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, and Hart GW (2008) O-GlcNAc regulates FoxO activation in response to glucose, J. Biol. Chem 283, 16283–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Akhtar A, Sung Hwan K, Min Jun K, Mee Young C, Sang Soo K, Gyeong Jae C, Yoon Sook K, Jun-Young C, and and Wan Sung C (2017) O-GlcNAcylation of NF-κB promotes lung metastasis of cervical cancer cells via upregulation of CXCR4 expression, Mol. Cells 40, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lo P-W, Shie J-J, Chen C-H, Wu C-Y, Hsu T-L, and Wong C-H (2018) O-GlcNAcylation regulates the stability and enzymatic activity of the histone methyltransferase EZH2, Proc. Natl. Acad. U. S. A 115, 7302–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dias WB, Cheung WD, and Hart GW (2012) O-GlcNAcylation of kinases, Biochem. Biophys. Res. Commun 422, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kang ES, Han D, Park J, Kwak TK, Oh MA, Lee SA, Choi S, Park ZY, Kim Y, and Lee JW (2008) O-GlcNAc modulation at Akt1 Ser473 correlates with apoptosis of murine pancreatic beta cells, Exp. Cell Res 314, 2238–2248. [DOI] [PubMed] [Google Scholar]
- [14].Holt GD, Snow CM, Senior A, Haltiwanger RS, Gerace L, and Hart GW (1987) Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine, J. Cell. Biol 104, 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lewis BA, Burlingame AL, and Myers SA (2016) Human RNA Polymerase II promoter recruitment in vitro is regulated by O-linked N-acetylglucosaminyltransferase (OGT), J. Biol. Chem 291, 14056–14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kreppel LK, Blomberg MA, and Hart GW (1997) Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats, J. Biol. Chem 272, 9308–9315. [DOI] [PubMed] [Google Scholar]
- [17].Lazarus MB, Nam Y, Jiang J, Sliz P, and Walker S (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate, Nature 469, 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, and Conti E (2004) The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α, Nat. Struct. Mol. Biol 11, 1001–1007. [DOI] [PubMed] [Google Scholar]
- [19].Clarke AJ, Hurtado-Guerrero R, Pathak S, Schüttelkopf AW, Borodkin V, Shepherd SM, Ibrahim AFM, and van Aalten DMF (2008) Structural insights into mechanism and specificity of O-GlcNAc transferase, EMBO J. 27, 2780–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Joiner CM, Li H, Jiang J, and Walker S (2019) Structural characterization of the O-GlcNAc cycling enzymes: insights into substrate recognition and catalytic mechanisms, Curr. Opin. Struct. Biol 56, 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kositzke A, Fan D, Wang A, Li H, Worth M, and Jiang J (2021) Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe, International Journal of Biological Macromolecules 169, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hu C-W, Worth M, Fan D, Li B, Li H, Lu L, Zhong X, Lin Z, Wei L, Ge Y, Li L, and Jiang J (2017) Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase, Nat. Chem. Biol 13, 1267–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Joiner CM, Levine ZG, Aonbangkhen C, Woo CM, and Walker S (2019) Aspartate residues far from the active site drive O-GlcNAc transferase substrate selection, J. Am. Chem. Soc 141, 12974–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lazarus MB, Jiang J, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, Vocadlo DJ, Herr W, and Walker S (2013) HCF-1 is cleaved in the active site of O-GlcNAc transferase, Science 342, 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Levine ZG, Fan C, Melicher MS, Orman M, Benjamin T, and Walker S (2018) O-GlcNAc transferase recognizes protein substrates using an asparagine ladder in the tetratricopeptide repeat (TPR) superhelix, J. Am. Chem. Soc 140, 3510–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Slawson C, and Hart GW (2011) O-GlcNAc signalling: implications for cancer cell biology, Nat. Rev. Cancer 11, 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, and Reginato MJ (2012) Critical role of O-linked β-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis, J. Biol. Chem 287, 11070–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ma Z, and Vosseller K (2014) Cancer metabolism and elevated O-GlcNAc in oncogenic signaling, J. Biol. Chem 289, 34457–34465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hanover JA, Chen W, and Bond MR (2018) O-GlcNAc in cancer: An Oncometabolism-fueled vicious cycle, Journal of Bioenergetics and Biomembranes 50, 155–173. [DOI] [PubMed] [Google Scholar]
- [30].Vasconcelos-dos-Santos A, de Queiroz RM, da Costa Rodrigues B, Todeschini AR, and Dias WB (2018) Hyperglycemia and aberrant O-GlcNAcylation: contributions to tumor progression, Journal of Bioenergetics and Biomembranes 50, 175–187. [DOI] [PubMed] [Google Scholar]
- [31].Bond MR, and Hanover JA (2013) O-GlcNAc cycling: a link between metabolism and chronic disease, Annu. Rev. Nutr 33, 205–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Merbl Y, and Kirschner MW (2009) Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays, Proc. Natl. Acad. U. S. A 106, 2543–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cheung WD, Sakabe K, Housley MP, Dias WB, and Hart GW (2008) O-linked β-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins, J. Biol. Chem 283, 33935–33941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yang X, Zhang F, and Kudlow JE (2002) Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression, Cell 110, 69–80. [DOI] [PubMed] [Google Scholar]
- [35].Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, and Fuks F (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS, Embo j 32, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hrit J, Goodrich L, Li C, Wang B-A, Nie J, Cui X, Martin EA, Simental E, Fernandez J, Liu MY, Nery JR, Castanon R, Kohli RM, Tretyakova N, He C, Ecker JR, Goll M, and Panning B (2018) OGT binds a conserved C-terminal domain of TET1 to regulate TET1 activity and function in development, eLife 7, e34870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, Zhao L, Bennett AM, Samuel VT, Wu J, Yates JR 3rd, and Yang X (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability, Cell Metab. 16, 226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Whisenhunt TR, Yang X, Bowe DB, Paterson AJ, Van Tine BA, and Kudlow JE (2006) Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development, Glycobiology 16, 551–563. [DOI] [PubMed] [Google Scholar]
- [39].Pathak S, Borodkin VS, Albarbarawi O, Campbell DG, Ibrahim A, and van Aalten DM (2012) O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release, EMBO J. 31, 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rafie K, Raimi O, Ferenbach AT, Borodkin VS, Kapuria V, and Aalten D. M. F. v. (2017) Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats, Open Biology 7, 170078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Charoensuksai P, Kuhn P, Wang L, Sherer N, and Xu W (2015) O-GlcNAcylation of co-activator-associated arginine methyltransferase 1 regulates its protein substrate specificity, Biochem. J 466, 587–599. [DOI] [PubMed] [Google Scholar]
- [42].Dias WB, Cheung WD, Wang Z, and Hart GW (2009) Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification, J. Biol. Chem 284, 21327–21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Shen DL, Gloster TM, Yuzwa SA, and Vocadlo DJ (2012) Insights into O-Linked N-Acetylglucosamine ([0–9]O-GlcNAc) Processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates, J. Biol. Chem 287, 15395–15408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Iyer SP, and Hart GW (2003) Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity, J. Biol. Chem 278, 24608–24616. [DOI] [PubMed] [Google Scholar]
- [45].Lubas WA, and Hanover JA (2000) Functional expression of O-linked GlcNAc transferase: domain structure and substrate specificity, J. Biol. Chem 275, 10983–10988. [DOI] [PubMed] [Google Scholar]
- [46].Pathak S, Alonso J, Schimpl M, Rafie K, Blair DE, Borodkin VS, Schüttelkopf AW, Albarbarawi O, and van Aalten DMF (2015) The active site of O-GlcNAc transferase imposes constraints on substrate sequence, Nature Structural & Molecular Biology 22, 744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Assimon VA, Southworth DR, and Gestwicki JE (2015) Specific binding of tetratricopeptide repeat proteins to heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) Is Regulated by Affinity and Phosphorylation, Biochemistry 54, 7120–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, and Moarefi I (2000) Structure of TPR domain;peptide complexes: critical elements in the assembly of the Hsp70;Hsp90 multichaperone machine, Cell 101, 199–210. [DOI] [PubMed] [Google Scholar]
- [49].Young JC, Hoogenraad NJ, and Hartl FU (2003) Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70, Cell 112, 41–50. [DOI] [PubMed] [Google Scholar]
- [50].Allan RK, and Ratajczak T (2011) Versatile TPR domains accommodate different modes of target protein recognition and function, Cell Stress Chaperones 16, 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hart GW, Slawson C, Ramirez-Correa G, and Lagerlof O (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease, Annu. Rev. Biochem 80, 825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li B, Li H, Hu C-W, and Jiang J (2017) Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase, Nature Communications 8, 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li B, Li H, Lu L, and Jiang J (2017) Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode, Nature Structural & Molecular Biology 24, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


