Figure 4.
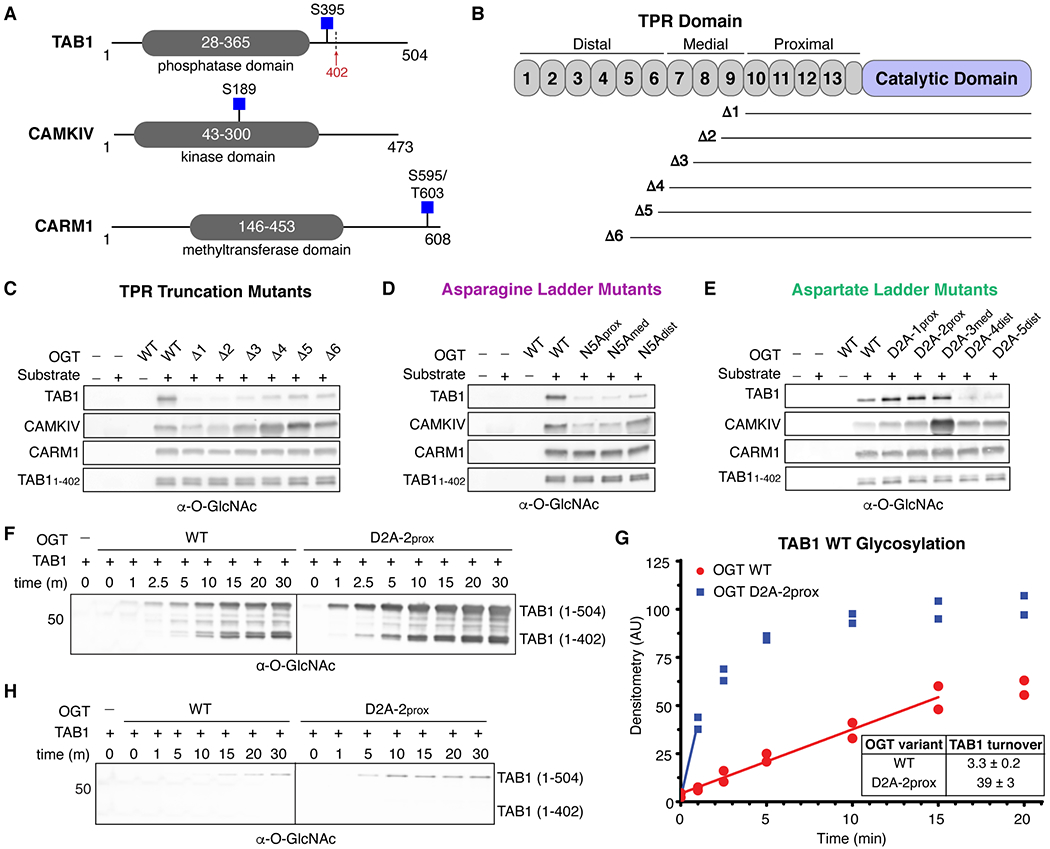
OGT protein substrates have different dependencies on the TPR domain for glycosylation. (A) Cartoon schematics of TAB1, CAMKIV, and CARM1 protein architecture. Glycosites are denoted by blue squares. TAB1 is proteolytically cleaved at residue K402 (red arrow).19, 43 (B) Cartoon schematic of OGT N-terminal TPR truncation mutants. (C) In vitro glycosylation of substrates by TPR truncation mutants. (D) In vitro glycosylation of substrates by asparagine ladder mutants. (E) In vitro glycosylation of substrates by aspartate ladder mutants. (F) Time-dependent in vitro glycosylation of TAB1 by wildtype OGT and D2A-2prox. Subpanels are from the same blot and image. (G) Densitometry vs. time plot of in vitro TAB1 glycosylation. Best-fit lines are shown in the linear range for each activity curve. (H) Time-dependent in vitro glycosylation of TAB1 S395A by wildtype OGT and D2A-2prox. Subpanels are from the same blot and image. Images in panels F and H were acquired under identical conditions. Representative blots are shown for two independent experiments for each data set.
