Significance
Receptor-interacting protein kinases 3 (RIPK3), a hub player in necrotic cell death, forms amyloid-like polymers in response to upstream mediators. This work, by using cryo-EM and solid-state NMR, reports the structure of human RIPK3 amyloid fibril core. The fibril core is mainly composed of the consensus RHIM domain and exhibits distinctive properties: an exceptionally small fibril core and existence in both handedness. These traits may provide a favorable geometry for the arrangement of kinase domain on the fibril surface to boost RIPK3 phosphorylation and the following activation of downstream MLKL. This work sheds light on how functional fibrils utilize the amyloid state to spatially regulate their functional domains, which is usually not a concern of pathological fibrils.
Keywords: necroptosis, amyloid fibril, Cryo-EM, ssNMR
Abstract
Receptor-interacting protein kinases 3 (RIPK3), a central node in necroptosis, polymerizes in response to the upstream signals and then activates its downstream mediator to induce cell death. The active polymeric form of RIPK3 has been indicated as the form of amyloid fibrils assembled via its RIP homotypic interaction motif (RHIM). In this study, we combine cryogenic electron microscopy and solid-state NMR to determine the amyloid fibril structure of RIPK3 RHIM-containing C-terminal domain (CTD). The structure reveals a single protofilament composed of the RHIM domain. RHIM forms three β-strands (referred to as strands 1 through 3) folding into an S shape, a distinct fold from that in complex with RIPK1. The consensus tetrapeptide VQVG of RHIM forms strand 2, which zips up strands 1 and 3 via heterozipper-like interfaces. Notably, the RIPK3-CTD fibril, as a physiological fibril, exhibits distinctive assembly compared with pathological fibrils. It has an exceptionally small fibril core and twists in both handedness with the smallest pitch known so far. These traits may contribute to a favorable spatial arrangement of RIPK3 kinase domain for efficient phosphorylation.
Necroptosis is an important form of regulated necrotic cell death, dysregulation of which is closely associated with a variety of human diseases, including neurodegenerative diseases (1, 2), inflammatory disorders (3–5), and cancers (6, 7). RIPK3 (receptor-interacting protein kinase 3) serves as the central node to converge multiple upstream signals to induce necroptosis (8–11). RIPK3 is activated via interactions with proteins that contain the RIP homotypic interaction motif (RHIM) such as RIPK1 (receptor-interacting protein kinase 1), TRIF (TIR-domain-containing adapter-inducing interferon-β), and ZBP1/DAI (Z-DNA-binding protein 1/DNA-dependent activator of IFN-regulatory factors). RIPK1 mediates RIPK3 activation downstream of death receptors, such as TNFR1 (12). TRIF links RIPK3 to the TLR3 and TLR4 signaling pathway (8). ZBP1/DAI mediates RIPK3 activation in response to certain viruses, such as influenza A virus (9, 10). RIPK3 is composed of a well-defined N-terminal kinase domain and a RHIM-containing C-terminal domain (CTD) (13). Previous studies show that RHIM plays an important role in the interactions of RIPK3 with its upstream mediators and amyloid fibrillation of RIPK3 (9, 10, 14, 15). A previous solid-state NMR (ssNMR) study has revealed the structure of a heterofibril core formed by the CTDs of RIPK3 and RIPK1, where the RHIM domains of both proteins adopt a serpentine fold and stack alternatively along the fibril axis (15). The structure provides insights into how RIPK1 recruits and activates RIPK3 for signaling transduction. However, it remains unknown how RIPK3 assemblies into fibril in the absence of RIPK1.
In this work, by using cryo-EM and ssNMR, we determined the structures of two amyloid fibrils formed by RIPK3-CTD. Despite the different fibril preparation, the RIPK3-CTD fibrils present a nearly identical structure. The fibril core exhibits an exceptionally small S-shaped fold of RHIM, which is distinct from that in the heterofibril of RIPK1 and RIPK3 CTDs. The consensus tetrapeptide VQVG forms the central strand 2 of the S-shaped structure and forms heterosteric zipper interfaces with the adjacent strands 1 and 2 within the same subunit. Intriguingly, the RIPK3-CTD fibril presents in both left and right handedness and features a minimum fibril core among the 50 different cryo-EM fibril structures reported previously and also represents the smallest fibril pitch and largest twist angle. By analyzing the reported cryo-EM fibril structures, we observed a strong positive correlation between the size of fibril core and the fibril pitch. Furthermore, we discussed how the small RIPK3 fibril core leads to a highly twisted fibril, which may display the N-terminal kinase domains in a favorable geometry to increase the efficiency of RIPK3 phosphorylation.
Results
Structural Characterization of RIPK3-CTD Fibril by ssNMR.
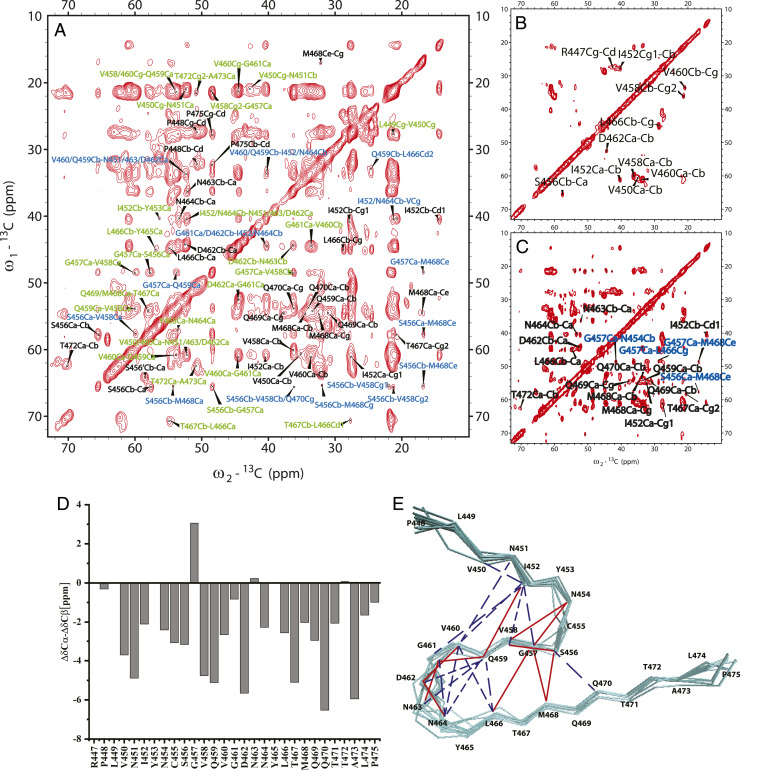
Magic angle spinning (MAS) ssNMR was applied to investigate the fibril structure of RIPK3-CTD containing residues 418 through 518 (referred to as RIPK3-CTDNMR). Lyophilized RIPK3-CTDNMR powder was first dissolved in 6 M Guanidine hydrochloride to ensure a complete dissolution. The protein solution was then dialyzed into Mili-Q water with pH adjusted to 7.5. This preparation provided fibrils with a good resolution in ssNMR spectra. An EM image of the fibrils is shown in SI Appendix, Fig. S1A. Atomic force microscopic (AFM) measurement showed that the RIPK3-CTDNMR fibrils contain both left- and right-handed fibrils with a predominant proportion of left-handed fibrils (71.9%) (SI Appendix, Fig. S1B). Extensive two-dimensional (2D) and three-dimensional (3D) ssNMR studies were carried out using fibrils grown from RIPK3 monomers uniformly labeled with 15N and 13C (Fig. 1A) or sparsely 13C-labeled using [1,3-13C] or [2-13C] glycerol (Fig. 1 B and C). Only residues from P448 to P475 gave ssNMR signals, indicating the location of the rigid fibril core (chemical shift assignment was shown in SI Appendix, Table S1). The secondary chemical shifts (Δδ13Cα-Δδ13Cβ) were mostly negative values (Fig. 1D), indicating β-strand conformation of RHIM domain with 458VQVG461 at the center. The backbone dihedral angles were predicted by TALOS-N (16) and listed in SI Appendix, Table S2. Although most of these residues presented single assignments, I452, C455, and S456 exhibited a minor second assignment. The residues at the N-terminal, including P448, L449, and V450, only had partial assignments probably caused by the dynamics at this region of the fibril. Both Y453 and Y465 have ambiguous assignments because of the low resolution of the aromatic residue peaks. The fibril structure was calculated using xplor-NIH (17) with NMR restraints (SI Appendix, Tables S1–S3) supplemented with the additional information from other experimental techniques. The X-ray diffraction of fibril showed the diffraction at about 4.8 Å (SI Appendix, Fig. S1C), giving the distance restraint between monomers. Dark-field beam tilted–transmission electron microscopy (BT–TEM) was used to obtain mass-per-length (MPL) value for the fibril (18), indicating a single monomer in the cross-β unit (SI Appendix, Fig. S1 D and E). The RIPK3-CTDNMR fibril structure model calculated from NMR restraints was shown in Fig. 1E and SI Appendix, Fig. S2, which will be further described in combination with the cryo-EM fibril structure below.
Fig. 1.
ssNMR studies of human RIPK3-CTDNMR fibril. (A) 2D 13C-13C spectrum of uniformly [13C/15N]-labeled RIPK3-CTDNMR fibril using 500 ms DARR mixing. The spectrum exhibits many cross-peaks with intraresidue peaks in black, peaks originated from correlations of sequential residues in green, and peaks from residues equal to or more than two residues apart in blue. (B) 2D 13C-13C spectrum of sparsely 13C-labeled RIPK3-CTDNMR fibril using [2-13C] glycerol with 50 ms DARR mixing. The spectrum displays very few intraresidue cross-peaks. (C) 2D 13C-13C spectrum of sparsely 13C-labeled RIPK3-CTDNMR fibrils using [2-13C] glycerol with 500 ms DARR mixing. The spectrum shows more cross-peaks. Those supposedly intraresidue cross-peaks (Q459, D462, N463, N464, L466, M468, Q469, Q470, shown in black) are from correlations of neighboring monomers, indicating a parallel in-register conformation of the fibril. Some correlations from distant residues (G457Cα-N454Cβ, G457Cα-L466Cγ, G457Cα-M468Cε, S456Cα-M468Cε, in blue) are also indicated. The experiments were carried on a Bruker 700 MHz MAS NMR spectrometer with ωr = 15 kHz, T = 303 K, and 83.33 kHz 1H decoupling field applied during acquisition. (D) Secondary chemical shifts calculated from assigned resonance shifts and random coil values predicating mostly β-strand conformation with breaks at near S456 and N463. (E) The structure model (Protein Data Bank: 7DAC, an ensemble of 12 structures) calculated using xplor-NIH with NMR restraints indicated. The solid red lines show the unambiguous assignments, and the dotted blue lines show the ambiguous assignments.
Cryo-EM Structure Determination of RIPK3-CTD Fibril.
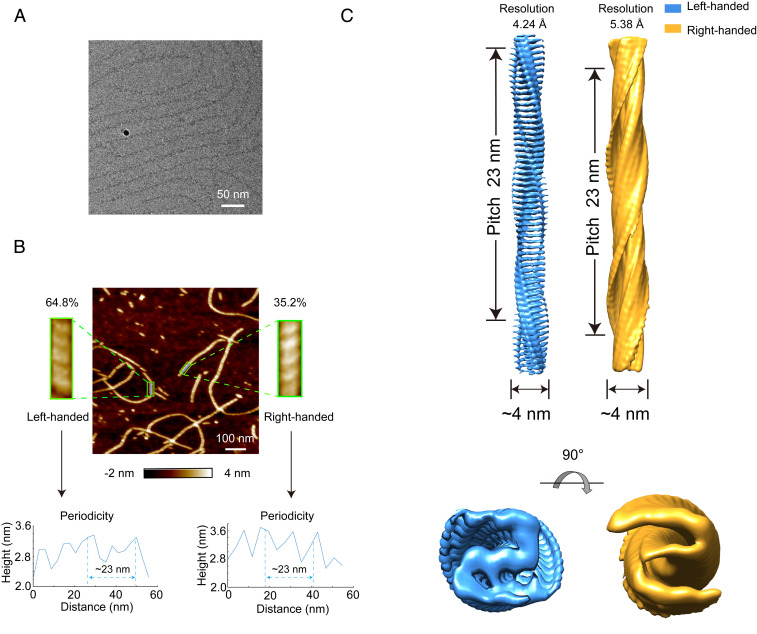
We further explored the structural details of the overall architecture of RIPK3-CTD fibril by cryo-EM. For the cryo-EM study, well-dispersed and morphologically homogeneous fibrils are essential for high-resolution 3D helical reconstruction, but the RIPK3 fibrils used for ssNMR study are prone to bundle together. To obtain the fibril sample suitable for cryo-EM structure determination, we examined a few RIPK3-CTD constructs, and the construct containing residues 388 through 518 (referred to as RIPK3-CTDEM) provided a polydispersed and highly homogenous fibril sample (Fig. 2A). The fibril was grown in 50 mM acetate buffer pH 5.0 instead of water used in ssNMR fibril preparation. AFM measurement showed that the RIPK3-CTDEM fibril twists in both handedness with ∼64.8% left-handed twist and ∼35.2% right-handed twist (Fig. 2B and SI Appendix, Fig. S3A). Both fibrils have a twist pitch of ∼23 nm (Fig. 2B and SI Appendix, Fig. S3B). Next, we collected totally 3,460 micrographs from one cryo-EM grid at ×22,500 magnification on a 300 keV Titan Krios microscope equipped with a K3 camera (SI Appendix, Table S4); 15,505 fibril segments were picked up from the micrographs (SI Appendix, Table S4). 2D classification revealed a major fibril polymorph with a population of over 88% of the total fibril segments (SI Appendix, Fig. S4A). As we cannot distinguish fibrils with different handedness from the cryo-EM images or 2D classification, we tested both handedness for 3D classification and refinement with helical symmetry by RELION. For both reconstructions, all particles after 2D classification were used. An initial twist angle of −7.68° and initial helical rise of 4.8 Å was used for left-handed reconstruction, and right-handed reconstruction was performed in the same way with an initial twist angle of 7.68° and initial helical rise of 4.8 Å. However, limited by the classification method, the particles we selected after 3D classification should represent a mixture of both left- and right-handed fibrils, which restricted us from high-quality reconstruction. Consistent with the dominant proportion of left-handed fibrils measured by AFM (Fig. 2B), the final resolution of left-handed reconstruction is better than the other one. We generated 3D cryo-EM density maps for a left-handed RIPK3-CTDEM fibril with an overall resolution of 4.24 Å and a right-handed fibril with an overall resolution of 5.38 Å (Fig. 2C and SI Appendix, Fig. S4B). Both fibrils exhibited a width of 4 nm and a pitch of 23 nm (Fig. 2C). For the left-handed fibril, the twist between neighboring subunits is −7.53° and the rise is 4.80 Å (SI Appendix, Fig. S3C and Table S4).
Fig. 2.
Structural characterization of RIPK3-CTDEM fibril by AFM and cryo-EM. (A) A representative cryo-EM image of RIPK3-CTDEM fibril. (B) AFM measurement of RIPK3-CTDEM fibril. An AFM 2D image and zoom-in views of fibrils with different handedness are shown (Top). The twist periodicity of each fibril was measured (Bottom). (C) Cryo-EM reconstruction density maps of left- and right-handed RIPK3-CTDEM fibrils. Fibril width and pitch are indicated. Graphing was performed with UCSF Chimera version 1.13.
Although the RIPK3-CTDEM and RIPK3-CTDssNMR fibrils were formed by different constructs under different conditions (Fig. 3A), their structures look very similar. The cryo-EM structure is composed of one single protofilament, which is the same as the ssNMR structure (Figs. 1E and 3B). The density map of each RIPK3-CTDEM subunit features an S-shaped fold, which may well accommodate the fibril model derived from ssNMR restraints (SI Appendix, Fig. S5) In order to confirm the structural similarity between the two structures, we carried out another ssNMR study using the RIPK3-CTDEM fibrils. The results showed an almost identical 13C-13C 2D spectra of the two fibrils, which confirms that the RIPK3-CTDEM fibril has the same fibril core as the RIPK3-CTDNMR fibril (SI Appendix, Fig. S6). So we built a structure model based on the density map of the left-handed fibril with the ssNMR structure as the initial model. After refinement, the alignment of Cα atoms of the final two structures showed an rmsd of 2.035 Å (Fig. 3C).
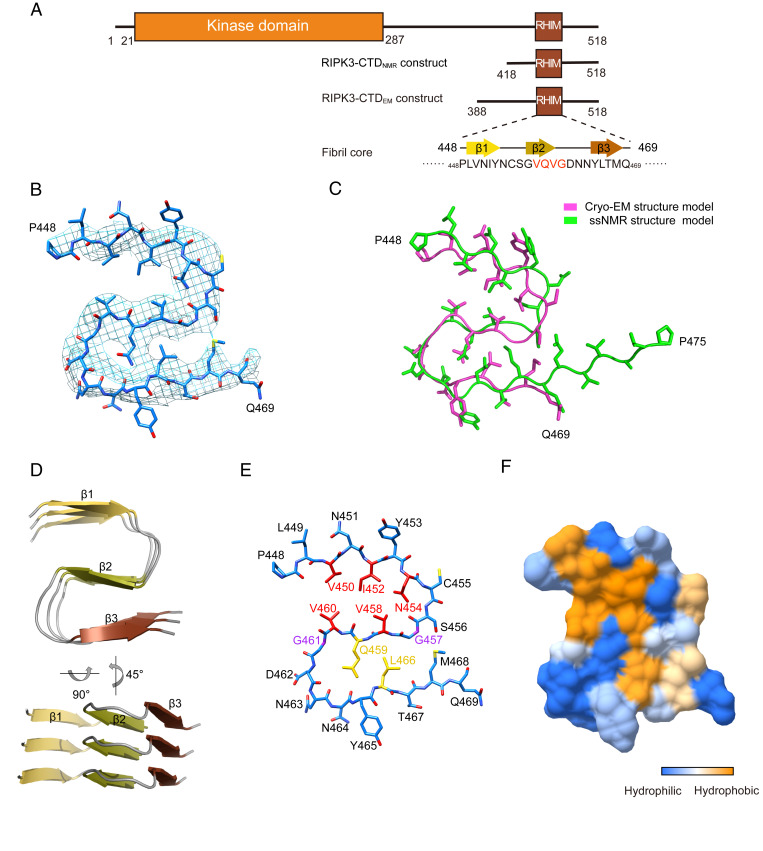
Fig. 3.
Cryo-EM structure of the RIPK3-CTDEM fibril. (A) Domain organization of RIPK3. RIPK3-CTD constructs used for cryo-EM and ssNMR are shown. The primary sequence and secondary structure of the fibril core are shown. (B) Overlay of the RIPK3-CTD fibril structure models determined by cryo-EM (light magenta) and ssNMR (green) with an rmsd of 2.035 Å. A total of 19 Cα atoms in the fibril core (451NIYNCSGVQVGDNNYLTMQ469) are aligned for rmsd calculation. (C) Density map with a structure model of RIPK3-CTDEM fibril. (D) Views of three layers of RIPK3-CTDEM fibril are shown in cartoon. The β-strands are numbered and labeled in different colors. (E) Structure model of RIPK3-CTDEM fibril (one layer). Individual residues are labeled. Key residues for the formation of this fold are colored in red, yellow, and purple, respectively. (F) The surface of one subunit of RIPK3-CTDEM is shown according to the hydrophobicity of each residue.
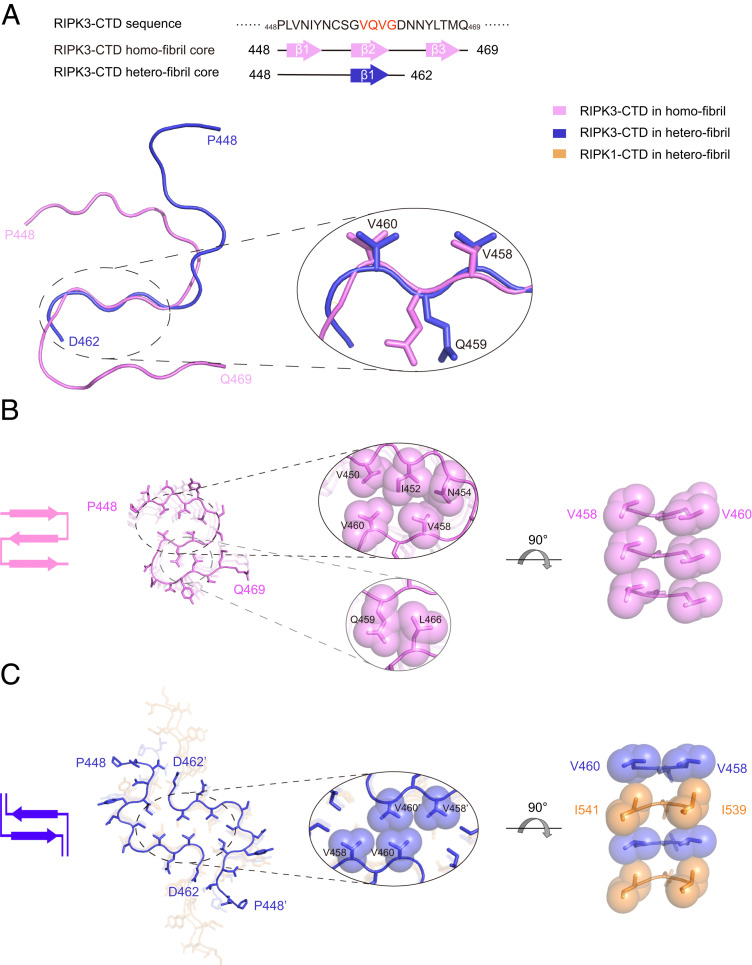
The ssNMR and cryo-EM structures show that the core of the RIPK3-CTD fibril is very small, consisting of only ∼20 residues out of a total of over 100 residues of RIPK3-CTD (Fig. 3A). The rest of the residues in the CTD remain flexible and are invisible by either cryo-EM or ssNMR. The RIPK3-CTD fibril core is highly ordered and compact featuring an S-shaped architecture, consisting of three β-strands (strands 1 through 3) stacking along the fibril axis in a parallel and in-register fashion (Fig. 3D). Notably, the consensus tetrapeptide 458VQVG461 of the RHIM domain comprises strand 2, which plays a central role in the fibril core structure (Fig. 3E). Specifically, V458 and V460 form a hydrophobic steric-zipper-like interface with V450, I452, and N454 from strand 1 (Fig. 3 E and F). Q459 interacts with L466 from strand 3 and forms a complimentary interface to hold strands 2 and 3 together (Fig. 3E). G457 and G461 flanking strand 2 facilitates turns of the peptide chain so that strands 1 and 3 can contact with strand 2 (Fig. 3E).
RIPK3-CTD Forms a Highly Twisted Fibril with Distinctively Short Fibril Pitch.
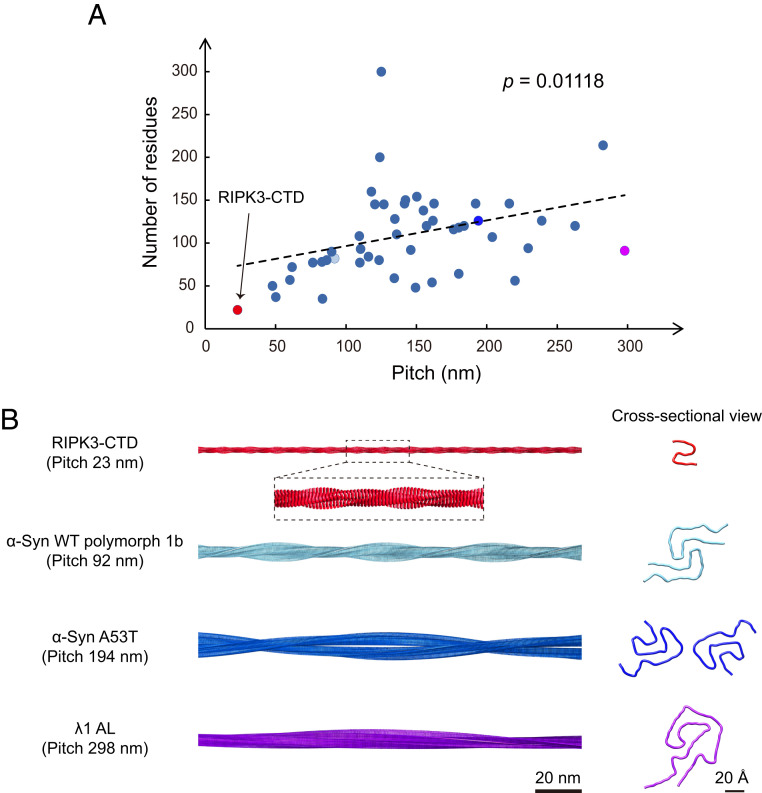
The cryo-EM structure reveals that the RIPK3-CTD fibril features a small fibril pitch of ∼23 nm and a large twist angle of −7.53° (Fig. 2C and SI Appendix, Fig. S3C). Given that the fibril contains only one protofilament and only 22 residues per layer spaced by 4.8 Å along the fibril axis, the fibril appears exceptionally narrow with a width of ∼4 nm. Notably, as we surveyed the cryo-EM structures of totally 50 different fibrils reported to date, the RIPK3-CTD fibril presents the shortest fibril pitch among them (Fig. 4 and SI Appendix, Fig. S7). The fibril pitch of different amyloid fibrils ranges from 23 nm to 297.9 nm, and the majority is within 100 to 200 nm (Fig. 4A). Intriguingly, as we plotted the pitch size and the number of residues per layer in the fibril cores, we found a positive correlation between these two parameters with a P value of 0.01118 (Fig. 4A), suggesting that a smaller fibril is favorable for the formation of a more twisted fibril with a shorter fibril pitch.
Fig. 4.
Statistical analysis of cryo-EM fibril structures formed by different proteins. (A) Statistics of the fibril pitch and number of residues within fibril core per 4.8 Å layer for 50 different cryo-EM fibril structures. The regression model is statistically significant. Equation for linear regression: Number of residues = 0.2995 × Pitch + 66.481. The exact P value of the linear regression is shown. (B) Four representative fibril structures with distinct pitches are shown. Protein Data Bank IDs: RIPK3-CTD (7DA4), α-syn WT (wild-type) polymorph 1b (6CU8), α-syn A53T (6LRQ), and λ1 AL (6IC3).
RIPK3-CTD Forms Distinct Structures in the Homofibril and Heterofibril Formed with RIPK1-CTD.
The cryo-EM and ssNMR structures demonstrate that RIPK3-CTD forms a conserved S-shaped fibril structure under different conditions, suggesting this fibril polymorph is preferentially adopted by RIPK3-CTD. Whereas in the heterofibril formed by RIPK3-CTD and RIPK1-CTD, RIPK3-CTD displays a totally different serpentine fold (Fig. 5A). Instead of forming three interconnected β-strands observed in the RIPK3-CTD homofibril, RIPK3-CTD utilizes only the VQVG motif to form a single β-strand in the heterofibril (SI Appendix, Fig. S8). Notably, the conformations of VQVG in both the homo- and heterofibrils are highly conserved, demonstrating the essential role of VQVG in mediating both fibrils (Fig. 5A). However, rather than forming intramolecular interactions with the adjacent β-strands in the same subunit in the homofibril (Fig. 5B), VQVG forms intermolecular interactions with the same segment in the mating subunit in the heterofibril, which provides an essential force to zip up two protofilaments together (Fig. 5C). Unfortunately, limited by the ssNMR technology, the fibril twist parameters are absent in the structure of the heterofibril (15), which precludes further comparison of the fibril structures along the fibril axis. Nevertheless, these structures demonstrate that RIPK3-CTD forms distinct structures in the homofibril and the heterofibril formed with RIPK1-CTD, while in both fibrils, RHIM, in particularly segment VQVG, is vital in the fibril core formation.
Fig. 5.
Structural comparison between RIPK3-CTD homofibril and heterofibril in complex with RIPK1-CTD. (A) The fibril core sequences of RIPK3-CTD homofibril and heterofibril are shown (Top). Overlay of one RIPK3-CTD subunit from RIPK3-CTD homofibril and heterofibril structures is shown (Bottom). The structure of the best-aligned VQVG is zoomed in. The rmsd of the four Cα atoms is 0.646 Å. (B) Top view of the RIPK3-CTD homofibril. The fibril contains one protofilament. The intramolecular interfaces within one RIPK3-CTD subunit are zoomed in. (C) Top view of the RIPK3-CTD heterofibril. The fibril contains two protofilaments. The intermolecular interface between two RIPK3-CTD subunits is zoomed in. The structures are shown in sticks. The sidechains in zoom-in views are shown in spheres. Topology of the fold in one layer is shown on the left.
Discussion
As a central regulator of necroptosis, RIPK3 receives death signals from various upstream mediators and polymerizes into amyloid-like aggregates to trigger mixed lineage kinase domain-like protein (MLKL) activation, membrane permeabilization, and cell death (8–12). Structural characterization on the amyloid assembly of RIPK3 may provide mechanistic understanding for its function in mediating necroptosis. In this work, we determined the structures of two RIPK3-CTD fibrils formed under different conditions by using ssNMR and cryo-EM, respectively. Both fibrils exhibit an ordered fibril core featuring a conserved S-shaped fold formed by nearly the same sequence spanning residues 448 through 469 (cryo-EM) and 448 through 475 (ssNMR). Note that although the two structures are similar, an rmsd of 2.035 Å calculated from the alignment of 19 Cα atoms of residues 451 through 469 indicates slight differences between the two structures, which may account for the influence of different noncore residues and buffer conditions. To this point, we speculate that as the full-length RIPK3 forms fibrils, the N-terminal kinase domain may have some influence on the geometric packing of the fibril but most likely would not change much of the sequence and fold of the fibril core. Rather, the interaction with different mediators would contribute to a significant change of the fibril core. The structure of RIPK3-CTD in the homofibril is distinct from that in the complex fibril with RIPK1-CTD (Fig. 5). RIPK3-CTD seems more ordered and compact in the homofibril with the formation of three β-strands and intramolecular hydrophobic interactions between the side chains of the strands. In contrast, RIPK3-CTD in the heterofibril is more extended with only one β-strand formed by the VQVG motif. Interestingly, Kajava et al. predicted that RIPK3 can also adopt an S-shaped fold in the heterofibril based on the evolutionary relationship between mammalian proteins and the Heterokaryon-s protein (HET-s) prion of Podospora anserine (19). The predicted S-shaped fold is similar to that we determined in the homofibril of RIPK3-CTD. This indicates that the S-shaped fold might represent a conserved fold of RIPK3 in different fibrils. On the other hand, different folds of RIPK3 in response to different mediators indicate structural diversity of RIPK3 fibrils, which may thus regulate the activity of RIPK3 and the fate of cells. Notably, the VQVG motif in the RHIM domain adopts a highly conserved β-strand conformation and plays an essential role in the fibril core formation in both RIPK3-CTD homofibril and RIPK1-RIPK3-CTD heterofibril (SI Appendix, Fig. S8). Thus, the VQVG motif may serve as a common key element in triggering RIPK3 fibrillation in different signaling pathways. In addition to RIPK1, several other mediators have been identified to directly interact with RIPK3 to pass on or block the death signals in different signaling pathways. For instance, DAI binds with RIPK3 under RIPK1-lacking conditions to drive perinatal death as well as skin inflammation (9, 10). TRIF interacts with RIPK3 for TLR3/TLR4-induced necroptosis in response to viral and bacterial infections (8). Infected cell polypeptides 6 (ICP6) from herpes simplex virus type 1 inhibits virus-induced RIPK3 necroptosis, and the ICP6 homolog M45 from murine cytomegalovirus inhibits the DAI-RIPK3 necroptosis pathway (11, 20). Thus, RIPK3 may form new fibril structures in complex with other mediators, which may link to the divergent signal transduction and activation of different signal pathways for RIPK3-mediated necroptosis. Further studies to this point would be of high interest.
Recent advances in cryo-EM and ssNMR technologies enable structural determination of a variety of amyloid fibrils formed by different proteins (e.g., α-syn, Tau, and prion) (14, 15, 21–30), which greatly enriches our knowledge on the structural basis underlying protein amyloid fibrillation and its pathology in diseases. By analyzing 50 cryo-EM structures of amyloid fibrils, we observed a positive correlation between the fibril core size and fibril pitch (Fig. 4). Eran’s group studied the mechanical process of seed pods opening in Bauhinia variegate and found a chirality-creating mechanism, which turns an initially flat pod valve into a helix (31). Using the mathematical framework of “incompatible elasticity,” they modeled the pod as a thin strip with a flat intrinsic metric and a saddle-like intrinsic curvature. The theoretical analysis quantitatively predicts all observed configurations, thus linking the pod’s microscopic structure and macroscopic conformation. In the case of our research system, the configurations of self-assembled ribbons in the narrow regime are twisted helices. They result from a compromise between stretching and bending energies, thus the pitch increases with the width. We suggest that the twisted ribbons self-assembled by amyloid proteins belongs to this type of incompatible strip. Unlike the pathological fibrils, the RIPK3-CTD fibril exhibits a distinctively small fibril core, accompanied by which the fibril pitch is the smallest and the fibril twist angle is the largest (Fig. 4). This trait may play an important role in accommodating the N-terminal kinase domain in a favorable geometry on the surface of the RIPK3 fibril to not only avoid steric clash but also facilitate effective phosphorylation (SI Appendix, Fig. S9). The latter is usually not a consideration of pathological fibrils. In addition, most reported pathological fibrils exhibit a left-handed twist except for a brain-derived Aβ fibril (32) and a fibril formed by E46K mutant α-syn (33). Intriguingly, the RIPK3-CTD fibril is a mixture of left- and right-handed twist fibrils (SI Appendix, Fig. S3). Although the role of fibril twist handedness is unclear, the special handedness of the RIPK3-CTD fibril is likely to relate to its physiological function, which needs further investigation. Taken together, the fibril twist parameters are important to determine both the fibril morphology and the spatial arrangement of the flanking functional domains. In other words, amyloid fibril assembly can add an additional dimension on the regulation of kinase activities in signal transduction. It will be interesting to further investigate in more details on how the different fibril assembly regulates the activity of a given protein like we see in the case of RIPK3 homo- and heterofibrils.
Methods
Human RIPK3-CTD Protein Expression and Purification for NMR.
The NMR constructs (human RIPK3-CTD residue 418 through 518) were subcloned into pSMT3 vector with a N-terminal 6His tag. All proteins were expressed in Escherichia coli Rosetta (DE3) cells and were purified by Ni-affinity and high-performance liquid chromatography. Unlabeled protein were obtained by growing the cells in 1 L Luria broth medium (Sangon Biotech) supplemented with 100 mg/mL ampicillin (Sangon Biotech) and the induction of protein expression at OD600 (optical density at 600 nm) of 0.8 to 1.0 (determined with a BIOMATE 3S UV-Visible Spectrophotometer) by 0.8 mM isopropyl b-D-thiogalactoside (Sangon Biotech). After 5 h of induction at 37 °C, cells were harvested by centrifugation at 9,000 rpm for 10 min and lysed by high pressure nano homogenizer (FB-110×, Shanghai Litu) of about 850 bar in a buffer containing 50 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 2 mM phenylmethylsulfonyl fluoride (BBI Life Science), and 1 mM -mercaptoethanol (Sigma). After being centrifuged at 12,000 rpm for 30 min, the pellet was dissolved in 20 mL dissolving buffer containing 6 M Guanidine hydrochloride (GuHCl; General-Reagent, Shanghai Titan Scientific), 50 mM Tris⋅HCl (pH 8.0), and 300 mM NaCl. After enough dissolution and being centrifuged at 12,000 rpm for 30 min, the supernatant was incubated with Ni-NTA beads 6 FF (Smart-Life Science) at 4 °C for 30 min. The protein was eluted with 15 to 20 mL dissolving buffer containing 250 mM imidazole (Sangon Biotech). Then the protein solution was dialyzed using 1 L pure water at 4 °C with Spectra/por 6 Dialysis Membranes (Molecular weight cut off 3.5 K, width 45 mm, diameter 29 mm; BBI Life Science). The water was replaced twice after every 6 h. Later, the protein precipitation was harvested at 12,000 rpm for 10 min. The pellet was dissolved with 25% (vol/vol) acetic acid solution. The protein was further purified by reverse phase high-performance liquid chromatography run on a Waters HPLC (2545) at room temperature with a linear gradient of 30 to 70% aqueous-organic solvent over 10 min at 10 mL/min. using the XBridge Peptide BEH C18 column (130 Åpore, 5.0 μm beads, 19 100 mm column, Waters). Aqueous phase was Milli-Q H2O with 0.05% trifluoroacrtic acid (TFA) and the organic solvent was acetonitrile (ACN) with 0.05% TFA. The purified protein was flash frozen in liquid nitrogen and dried at −80 °C. Protein concentrations were determined by absorbance at wavelength of 280 nm with Nanodrop Spectrometer.
The uniformly labeled protein, [U-99% 13C; U-99% 15N]-labeled RIPK3-CTD, was expressed in fresh prepared M9 medium containing 1.5 g/L 15N-ammonium chloride and 2 g/L 13C-glucose (purchased from Cambridge Isotope Laboratories), 1 mL/L BME vitamins (Sigma-Aldrich B6891), 0.2 M CaCl2, 2 M MgCl2, and 50 mg/L Thiamine. The culture first grew in 500 mL Luria-Bertani medium with shaking (220 rpm) at 37 °C to a cell density of about OD600 = 1.0. Cells were then collected by centrifugation at 25 °C and resuspended in the fresh M9 medium. After a brief incubation, the protein expression was induced with 0.8 mM isopropyl -d-1-thiogalactopyranoside for 5 h at 37 °C.
For [natural abundance 13C, U-50% 15N]-labeled protein expression, add 1.5 g/L 15NHCl and 2 g/L [12C]-glucose as nitrogen and carbon source. For [U-50% 13C; 14N], add 1.5 g/L 14NHCl and [13C]-glucose as nitrogen and carbon source. And for sparsely 13C-labeled protein, the [2-13C]-glycerol and [1,3-13C]-glycerol was used as a carbon source instead.
Human RIPK3-CTD Protein Expression and Purification for Cryo-EM.
Human RIPK3-CTD (residues 388 through 518) with a C-terminal 6His tag was inserted into pET28a vector and transformed into E. coli expression cell line BL21(DE3) (TransGen Biotech, Cat. # CD601-02) for protein purification. A total of 1 mM isopropyl-1-thio-D-galactopyranoside (IPTG) was used to induce protein expression at 37 °C overnight. Cells were then harvested by centrifugation at 5,000 rpm for 20 min at 4 °C and resuspended with buffer A (6 M Guanidine hydrochloride, 50 mM Tris⋅HCl (pH8.0), 50 mM sodium chloride, and 2 mM dithiothreitol). The mixture was sonicated for 30 min (1 s per time, 0.5 s interval) on ice by JY92-IIN sonicator at a power of 45%. After centrifugation at 15,000 rpm for 40 min at 4 °C, the supernatant was filtered with a 0.22 μm filter and then loaded to Ni-column with buffer A. Proteins were eluted with a gradient (10%, 20%, and 50%) of buffer B (6 M Guanidine hydrochloride, 50 mM Tris⋅HCl [pH 8.0], 50 mM sodium chloride, 500 mM imidazole, and 2 mM dithiothreitol). The eluted RIPK3-CTD protein was flash frozen in liquid nitrogen and stored at −80 °C for subsequent fibril formation.
Fibrils Sample Preparation.
For NMR sample preparation, lyophilized protein powder was dissolved in 6 M GuHCl buffer at a concentration of 5 mg/mL (determined by absorbance at wavelength 280 nm with Nanodrop Spectrometer). The solution was placed at room temperature for 1 h to ensure a complete dissolution and was dialyzed overnight in Milli-Q water with Spectra/por 6 Dialysis Membranes (MWCO 3.5 K, W.45 mm, Diam 29 mm; BBI Life Science). The water (adjusted pH to 7.5) was changed three times after every 6 h. After dialysis for 3 d, Fibril pellets was collected by ultracentrifugation at 55,000 rpm, 25 °C for 1 h (Optima Max-TL, BECKMAN COULTER).
For cryo-EM fibril sample preparation, the eluted RIPK3-CTD protein in 10% buffer B was dialyzed into 1 L acetic acid-sodium acetate buffer solution (pH 5.0) at 4 °C with Slide-A-Lyzer Dialysis Cassette G2 (MWCO 3.5 K, 3 mL Capacity, Thermo Scientific). After 16 h of dialysis, the dialyzate was then collected and incubated at 4 °C for 2 to 3 d to obtain mature fibrils for cryo-EM sample preparation. A uniformly [13C/15N]-labeled RIPK3-CTDEM fibrils using the cryo-EM sample preparation protocol was also carried out for additional ssNMR studies.
X-ray Diffraction Measurement.
Fibrils for NMR measurement were also tested using X-ray diffraction. The fibrils were mounted in a loop and exposed to Cu radiation from a Bruker D8 VENTURE X-ray generator at 0.154184 nm wavelength, distance 50 mm. Data were collected at room temperature for 1 min on a Bruker D8 VENTURE imaging plate detector.
AFM.
RIPK3-CTD Fibril samples were diluted to a final concentration of 5 μM (ssNMR sample) and 20 μM (cryo-EM fibril sample) for AFM measurement. A total of 5 μL fibril sample was loaded onto a clean mica surface and remained for 3 min at room temperature before being rinsed with Milli-Q water. Then the samples were dried in air and recorded with ScanAsyst air mode using Nanoscope V Multimode 8 (Bruker). SCANASYST-Air probe was taken with a constant of 0.4 N/m for scanning. Measurements were recorded at 512 × 512 pixels at a line rate of 0.98 Hz. Data were processed and analyzed by NanoScope Analysis software (version 1.5).
MPL Measurement of RIPK3-CTDNMR Fibril Using BT-TEM.
Samples (fibrils using the preparation for NMR measurement) were adsorbed onto 3 to 5 nm thick carbon films supported on 200 mesh copper grids (Beijing Zhongjingkeyi Technology). Images were acquired by a Talos L120C TEM at an acceleration voltage of 120 keV. BT-TEM (18) images were taken at 36,000 magnification, using a beam tilt of 1.2°, a 70 μm diameter objective aperture, a 150 μm diameter condenser aperture, and a spot 2 with a filament current of about 5 μA. The dose rate was 15 to 20 e/nm2-s. The images were stored as 8-bit tiff files and analyzed using ImageJ (NIH) (34).
ssNMR Experiments.
All MAS ssNMR experiments were carried out at 16.45 T (700 MHz 1H frequency) on a Bruker 700 MHz spectrometers with 3.2-mm ssNMR rotor. All experiments were conducted at 303 K. For the 2D DARR (35) experiments, the Hartman–Hahn cross polarizations (CPs) were carried out with a 13C field strength of 51.2 kHz, adjusting the 1H field strength to near the n = 1 Hartman–Hahn condition. The CP contact time was 1.5 ms. The 1H and 13C π/2 pulses were 3 μs and 3.3 μs, respectively. The DARR mixing time was 50 ms, 200 ms, and 500 ms. TPPM (36) decoupling was employed during t1 and t2 increment with an radio frequency-field strength of 83.3 kHz (14).
For 2D and 3D NCACX (N-Cα-Ci,sidechain) and NCOCX (N-C’-Ci-1,sidechain), 1H and 15N π/2 pulses were 3 μs and 4.8 μs, respectively. For the NCOCX-specific CP, a contact time of 5.5 ms was applied with the rf-field strengths of 26.6 kHz and 45.6 kHz on 15N and 13C, respectively. For the NCACX-specific CP, the rf-field strengths of 39.1 kHz and 25.9 kHz on 15N and 13C were used, respectively. The DARR mixing was 50 ms for both experiments. During acquisition, a two-pulse phase-modulated (TPPM) 1H decoupling field of 87.72 kHz was applied (14).
For z-filtered transferred echo double resonance (z-filtered TEDOR) experiment, the 13C and 15N hard pulse were 75.7 kHz and 50 kHz, respectively. During the magnetization transfer, 15N π/2 pulse length was phase cycled according to the xy-4 scheme. The z-filtered time was 200 μs. 13C -15N TEDOR mixing time was set to 6.4 ms and 8.5 ms, respectively (37).
13C chemical shifts were referenced using the published shifts of adamantine relative to 4, 4-dimethyl-4-silapentane-1-sulfonic acid (DSS), and 15N chemical shifts were referenced using the International Union of Pure and Applied Chemistry (IUPAC) relative frequency ratios between DSS (13C) and liquid ammonia (15N). All spectra were processed using topspin and analyzed using the program Sparky (38).
Calculation of Structural Models for RIPK3-CTDNMR Fibrils.
Structure calculations were performed using simulated annealing with the Xplor-NIH package (17). Two rounds of calculations were carried out. RIPK3 molecule residues R447 through P475 were used in the calculation since those residues had chemical shifts assignments. In these calculations, each RIPK3 subunit is assumed to have the exactly same conformation. To enforce this condition, the strict symmetry module (symSimulation) in the Xplor-NIH package (2.48) was utilized to reduce the computational cost, where only a single copy of protomer coordinates was maintained (39). In total, five copies of the monomer subunit were used in the calculation to represent a short fibril segment, where four subunit copies were generated from a protomer using rigid body translations; approximate twist angle of 7.4° was also used considering both AFM and EM results.
In the first round of calculation, 240 independent structures were calculated from starting coordinates having different, random torsion angles and packing. The protocol contains first torsion-angle dynamics for a duration of 10 ps or 5,000 timesteps at 4,000 K, followed by annealing to 25 K in decrements of 12.5 K for 20 ps or 2,000 timesteps of torsion-angle dynamics at each temperature and finally 500 steps of energy minimizations in torsion angle and Cartesian coordinates. The calculation was done on the high-performance calculation platform of ShanghaiTech University. Based on MPL data, there is only one protofibril in a mature fibril structure. Backbone torsion angles (using the CDIH potential) were restrained using predictions from both TALOS-N (16) and TALOS+ (40). Although the TALOS-N predicted values were used for the structure calculations, only those predictions whose values agreed within 20° by both methods were used and the uncertainties were expanded to accommodate the differences between the two methods. Intermolecular distance restraints (using the nuclear overhauser effect potentials, NOEPot) were applied between neighboring subunits for the I452Cα, G457Cα, and L466CO atoms, using a carbon–carbon distance of 4.75 ± 0.1 Å, and explicitly representing intermolecular hydrogen bonds between I452NH and N451CO, Q459NH and V458CO, V460NH and Q459CO, T467NH and L466CO, Q469NH and M468CO, T471NH and Q470CO using hydrogen–oxygen distances of 2.3 ± 0.1 Å and nitrogen–oxygen distances of 3.3 ± 0.1 Å. These intermolecular bonds were employed so that the resulting fibrils are consistent with the 4.8 Å peak seen in X-ray powder diffraction. Intramolecular long-range distance restraints were obtained from 2D 13C-13C correlation using 200 ms or 500 ms dipolar assisted rotational resonance (DARR) mixing (distance restraint values: 5.5 ± 1.5 Å) and z-filtered TEDOR with 13C -15N recoupling time 6.4 ms (distance restraint values: 4.5 ± 2.5 Å) and comprised 22 unambiguous nonsequential restraints. Long-range distance restraints with low ambiguity from DARR and TEDOR were also used in the calculation, usually with one site having a unique assignment and the other site having two possible assignments. Aside from the experiment-based dihedral and distance restraint terms, the knowledge-based TorsionDB (41), low-resolution residueAff (42) contact terms, along with the standard purely repulsive nonbonded RepelPot (39) and covalent bond, bond-angle, and improper dihedral terms were used in this initial docking calculation. The best 12 structural models with the lowest energy were retained for the second round of structure calculation.
The second refinement round of calculation was similar to the initial folding calculation except that the RepelPot term was replaced by the EEFx (43) implicit solvent force field. Thirteen side-chain χ values from TALOS-N predictions were also added into the CDIH potential term to improve the side-chain conformation. A total of 96 structures were calculated, and the best 12 structures with the lowest energy were obtained. An all-atom energy minimization of 3 ps was then carried out in which the positions of the backbone atoms of the two RIPK3 subunits were restrained to remain within 1 Å of their initial positions using PosDiffPot. In the energy minimization, the XplorPot, TorsionDB, implicit solvent (EEFxPot), and covalent energy terms were also included.
Cryo-EM Imaging and Data Processing.
An aliquot of 4 μL RIPK3 fibril solution (2 μM) was applied to a glow-discharged holey carbon grid (Quantifoil R2/1, 300 mesh) and incubated for 10 s before blotted for 1 s and plunge-frozen in liquid ethane using FEI Vitrobot Mark IV with 95% humidity at 16 °C. Cryo-EM images were collected on an FEI Titan Krios microscope operated at 300 kV (Thermo Fisher) with magnification of 22,500, and a Gatan K3 camera was used to record the micrographs with a pixel size of 0.53 Å in superresolution mode. All the micrographs were dose-fractionated to 32 frames. Defocus values were set from −1.3 to −2.3 μm, and the electron dose rate was set at ∼20 e−/Å2/s with an exposure time of 3.11 s to make sure the total dose was ∼55 e−/Å2 per micrograph. Data were collected by the Serial EM software.
As for image preprocessing, MotionCorr2 was used to correct beam induced sample motion and implement dose weighting for all 32 frames (44). The pixel size was further binned to 1.06 Å. CTFFIND4.1.8 was used to estimate the defocus values of dose-weighted micrographs (45). RELION3.0 was used for manual picking, particle extraction, 2D classification, 3D classifications, 3D refinement, and postprocessing (46). A total of 15,505 filaments were picked manually from 3,460 micrographs. Particles were extracted into 686- and 288-pixel boxes using the 90% overlap scheme.
Helical Reconstruction.
Several iterations of 2D classification were performed to select particles which belong to the same polymorph. 3D initial model was built by selected 2D classes. AFM data, cryo-EM micrographs, and 2D classification were used together to calculate an initial twist angle. A value of 4.8 Å was used as an initial value of helical rise. Local searches of symmetry in 3D classification were used to find the final twist angle and rise value. For left-handed reconstruction, all 294,736 particles after 2D classification were used for 3D classification. Initial twist angle of −7.68° and initial helical rise of 4.8 Å was used for the first round of 3D classification. After several round of 3D classification with K = 3, the best 3D class with 45,526 particles was selected and used for 3D refinement. Right-handed reconstruction was performed in the same way with an initial twist angle of 7.68° and initial helical rise of 4.8 Å. A total of 37,016 particles were selected from all 294,736 particles for 3D refinement. 3D refinement of the selected 3D classes with appropriate reference was performed to obtain final reconstruction.
The final map of RIPK3-CTDEM fibril was convergence with the rise of 4.80 Å and the twist angle of −7.53°. Postprocessing was preformed to sharpen the map with a B-factor of −271.84 Å2. Based on the gold-standard Fourier shell correlation = 0.143 criteria, the overall resolution was reported as 4.24 Å.
Atomic Model Building and Refinement.
Atomic model was built into the refined map with COOT (47). The model of RIPK3-CTDEM fibril was built based on the ssNMR model. A three-layer model was generated for structure refinement. Structure models were refined by using the real-space refinement program in PHENIX (48).
Supplementary Material
Acknowledgments
We thank the Biomolecular NMR Facility at the School of Life Science and Technology, ShanghaiTech University, for ssNMR studies and the Molecular and Cellular Biology Core Facility for ultracentrifugation instrumentation. TEM images were taken at the Bio-Electron Microscopy Facility of ShanghaiTech University. We also appreciate the support from the Analytical Instrumentation Center (Contract No. SPST-AIC 10112914), the School of Physical Science and Technology at ShanghaiTech University. We thank Na Yu for single-crystal crystallography and Charles D. Schwleters at the NIH for help in NMR structural calculation using Xplor-NIH. This work was supported by the Major State Basic Research Development Program (Grants 2016YFA0501902 to C.L. and 2017YFA0504804 to J. Lu), the National Natural Science Foundation (NSF) of China (Grants 91853113 and 31872716 to C.L. and 31770790 to J. Lu), the Science and Technology Commission of Shanghai Municipality (Grant 18JC1420500 to C.L.), the “Eastern Scholar” project supported by the Shanghai Municipal Education Commission, the Shanghai Municipal Science and Technology Major Project (Grant No. 2019SHZDZX02 to C.L.), the Shanghai Science and Technology Committee (Grant 20XD1425000 to C.L.), and the Yangfan program of the Shanghai municipal government (19YF1433500 to J.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022933118/-/DCSupplemental.
Data Availability
Cryo-EM density maps and the atomic model of RIPK3-CTDEM fibril are available through the Electron Microscopy Data Bank and Protein Data Bank with accession codes EMD-30622 and PDB 7DA4, respectively. The NMR, atomic coordinates, chemical shifts, and restraints of RIPK3-CTDNMR have been deposited in the Biological Magnetic Resonance Bank and the Protein Data Bank with accession codes 36392 and 7DAC.
References
- 1.Caccamo A., et al., Necroptosis activation in Alzheimer’s disease. Nat. Neurosci. 20, 1236–1246 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Ito Y., et al., RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science 353, 603–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasparakis M., Vandenabeele P., Necroptosis and its role in inflammation. Nature 517, 311–320 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Shan B., Pan H., Najafov A., Yuan J., Necroptosis in development and diseases. Genes Dev. 32, 327–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan J., Amin P., Ofengeim D., Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 20, 19–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Najafov A., Chen H., Yuan J., Necroptosis and cancer. Trends Cancer 3, 294–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seehawer M., et al., Necroptosis microenvironment directs lineage commitment in liver cancer. Nature 562, 69–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He S., Liang Y., Shao F., Wang X., Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 20054–20059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J., et al., RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature 540, 124–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton K., et al., RIPK1 inhibits ZBP1-driven necroptosis during development. Nature 540, 129–133 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Wang X., et al., Direct activation of RIP3/MLKL-dependent necrosis by herpes simplex virus 1 (HSV-1) protein ICP6 triggers host antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 111, 15438–15443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandenabeele P., Galluzzi L., Vanden Berghe T., Kroemer G., Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 11, 700–714 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Sun X., Yin J., Starovasnik M. A., Fairbrother W. J., Dixit V. M., Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J. Biol. Chem. 277, 9505–9511 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Li J., et al., The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150, 339–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mompeán M., et al., The structure of the necrosome RIPK1-RIPK3 core, a human hetero-amyloid signaling complex. Cell 173, 1244–1253.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y., Bax A., Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M., The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Chen B., Thurber K. R., Shewmaker F., Wickner R. B., Tycko R., Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 14339–14344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajava A. V., Klopffleisch K., Chen S., Hofmann K., Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci. Rep. 4, 7436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu H., et al., RIP3-mediated necroptosis is regulated by inter-filament assembly of RIP homotypic interaction motif. Cell Death Differ. 28, 251–266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schweighauser M., et al., Structures of α-synuclein filaments from multiple system atrophy. Nature 585, 464–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., et al., Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res. 28, 897–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao K., et al., Parkinson’s disease-related phosphorylation at Tyr39 rearranges α-synuclein amyloid fibril structure revealed by cryo-EM. Proc. Natl. Acad. Sci. U.S.A. 117, 20305–20315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzpatrick A. W. P., et al., Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L. Q., et al., Cryo-EM structure of an amyloid fibril formed by full-length human prion protein. Nat. Struct. Mol. Biol. 27, 598–602 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Tuttle M. D., et al., Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 23, 409–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dregni A. J., et al., In vitro 0N4R tau fibrils contain a monomorphic β-sheet core enclosed by dynamically heterogeneous fuzzy coat segments. Proc. Natl. Acad. Sci. U.S.A. 116, 16357–16366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelenter M. D., et al., The peptide hormone glucagon forms amyloid fibrils with two coexisting β-strand conformations. Nat. Struct. Mol. Biol. 26, 592–598 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seuring C., et al., The three-dimensional structure of human β-endorphin amyloid fibrils. Nat. Struct. Mol. Biol. 27, 1178–1184 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Wälti M. A., et al., Atomic-resolution structure of a disease-relevant Aβ(1-42) amyloid fibril. Proc. Natl. Acad. Sci. U.S.A. 113, E4976–E4984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armon S., Efrati E., Kupferman R., Sharon E., Geometry and mechanics in the opening of chiral seed pods. Science 333, 1726–1730 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Kollmer M., et al., Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 10, 4760 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao K., et al., Parkinson’s disease associated mutation E46K of α-synuclein triggers the formation of a distinct fibril structure. Nat. Commun. 11, 2643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray D. T., et al., Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takegoshi K., Nakamura S., Terao T., 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637 (2001). [Google Scholar]
- 36.Ernst M., Heteronuclear spin decoupling in solid-state NMR under magic-angle sample spinning. J. Magn. Reson. 162, 1–34 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Jaroniec C. P., Filip C., Griffin R. G., 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly (13)C,(15)N-labeled solids. J. Am. Chem. Soc. 124, 10728–10742 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Goddard T. D., Kneller D. G., SPARKY 3 (University of California, San Francisco, CA, 2008). https://www.cgl.ucsf.edu/home/sparky/. Accessed 28 March 2021.
- 39.Schwieters C. D., Bermejo G. A., Clore G. M., Xplor-NIH for molecular structure determination from NMR and other data sources. Protein Sci. 27, 26–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bermejo G. A., Clore G. M., Schwieters C. D., Smooth statistical torsion angle potential derived from a large conformational database via adaptive kernel density estimation improves the quality of NMR protein structures. Protein Sci. 21, 1824–1836 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryabov Y., Suh J. Y., Grishaev A., Clore G. M., Schwieters C. D., Using the experimentally determined components of the overall rotational diffusion tensor to restrain molecular shape and size in NMR structure determination of globular proteins and protein-protein complexes. J. Am. Chem. Soc. 131, 9522–9531 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian Y., Schwieters C. D., Opella S. J., Marassi F. M., High quality NMR structures: A new force field with implicit water and membrane solvation for xplor-NIH. J. Biomol. NMR 67, 35–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zivanov J., et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams P. D., et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density maps and the atomic model of RIPK3-CTDEM fibril are available through the Electron Microscopy Data Bank and Protein Data Bank with accession codes EMD-30622 and PDB 7DA4, respectively. The NMR, atomic coordinates, chemical shifts, and restraints of RIPK3-CTDNMR have been deposited in the Biological Magnetic Resonance Bank and the Protein Data Bank with accession codes 36392 and 7DAC.