Significance
Rift Valley fever virus (RVFV)–specific monoclonal antibodies from survivors of natural infection and vaccination were isolated to understand how RVFV is targeted for neutralization by the human immune system. These antibodies bind to specific regions of the viral surface, some of which are complex quaternary epitopes, and they block RVFV infection at extremely low concentrations. A new mechanism by which these mAbs can neutralize RVFV is described whereby the antibody may prevent necessary structural rearrangements in the viral proteins for infection. The antibodies isolated here have potential use in pre-exposure prophylaxis or post-exposure therapy against RVFV infection and should be studied further in that context.
Keywords: Rift Valley fever virus, antibodies, monoclonal, adaptive immunity, virus internalization, Phlebovirus
Abstract
Rift Valley fever virus (RVFV), an emerging arboviral and zoonotic bunyavirus, causes severe disease in livestock and humans. Here, we report the isolation of a panel of monoclonal antibodies (mAbs) from the B cells of immune individuals following natural infection in Kenya or immunization with MP-12 vaccine. The B cell responses of individuals who were vaccinated or naturally infected recognized similar epitopes on both Gc and Gn proteins. The Gn-specific mAbs and two mAbs that do not recognize either monomeric Gc or Gn alone but recognized the hetero-oligomer glycoprotein complex (Gc+Gn) when Gc and Gn were coexpressed exhibited potent neutralizing activities in vitro, while Gc-specific mAbs exhibited relatively lower neutralizing capacity. The two Gc+Gn–specific mAbs and the Gn domain A-specific mAbs inhibited RVFV fusion to cells, suggesting that mAbs can inhibit the exposure of the fusion loop in Gc, a class II fusion protein, and thus prevent fusion by an indirect mechanism without direct fusion loop contact. Competition-binding analysis with coexpressed Gc/Gn and mutagenesis library screening indicated that these mAbs recognize four major antigenic sites, with two sites of vulnerability for neutralization on Gn. In experimental models of infection in mice, representative mAbs recognizing three of the antigenic sites reduced morbidity and mortality when used at a low dose in both prophylactic and therapeutic settings. This study identifies multiple candidate mAbs that may be suitable for use in humans against RVFV infection and highlights fusion inhibition against bunyaviruses as a potential contributor to potent antibody-mediated neutralization.
Rift Valley fever virus (RVFV) is an emerging arbovirus and zoonotic threat to human and animal health with pandemic potential because of the global presence of its vectors and hosts (1). First identified in 1931 (2), RVFV causes ongoing infections in Africa, with occasional rises in incidence related to the propagation of mosquitos due to alternating weather patterns (3). These outbreak episodes are characterized by mass livestock die-off events, particularly in young animals and abortions in pregnant females. Spillover events to humans occur by infected mosquito exposures or contact with the blood and/or organs of infected animals (4). Human-to-human transmission of RVFV has not been documented, but concern is growing that RVFV can be transmitted from mother to fetus in utero (5, 6). In humans, disease presentation ranges from a mild influenza-like illness to a potentially lethal hemorrhagic fever syndrome. The World Health Organization (W.H.O.) has reported more than 4,600 cases and 957 deaths from 2000 to 2016 due to RVFV infections, with a case fatality rate of more than 20% (7). RVFV outbreaks typically occur in sub-Saharan and North Africa, but in 2000, the first reported cases outside the African continent occurred in the Arabian Peninsula (8). Given the increasing observed mortality rate, spread to new regions, and potential use as a bioterrorist agent (9), the W.H.O. and the US National Institute of Allergy and Infectious Diseases have designated RVFV as a priority pathogen for urgent research and therapeutic development (10, 11).
RVFV, a member of the Phlebovirus genus in the Phenuiviridae family of the Bunyavirales order (12) has a tripartite RNA genome containing large (L), medium (M), and small (S) gene segments. The sequences in the M segment encode for the viral envelope glycoproteins (Gc and Gn). These two glycoproteins allow for viral attachment, entry, fusion, and assembly when oriented as a pentamer of heterodimers (13–16). Data from animal and human vaccine trials indicate that high-serum neutralizing titers and protection (in animals) are associated with responses to Gn, Gc, or entire virion particles (17–23). Protection against experimental RVFV challenge is observed in vaccinated animals with high-serum titers of neutralizing antibodies (22, 23).
On the viral surface, Gn and Gc organize into a pentamer or hexamers of Gc-Gn heterodimers with icosahedral symmetry (13, 14). Receptor engagement of DC-SIGN or possibly other receptors like heparan sulfate (24) by Gn allows for cellular attachment and initiates caveolae-mediated endocytosis (25, 26). Premature fusion and extension of the fusion loop is prevented by Gn, which provides a shield for the fusion loop in Gc (27). Upon exposure to acidic conditions in the late endosome, Gn repositions, and the class II fusion protein, Gc extends so that its fusion loop interacts with the host membrane (27–29). The extended Gc conformation subsequently rearranges so the membranes organize in close proximity to fuse (29). Given the broad host, tissue, and cell tropism of RVFV (24), inhibiting RVFV by blocking DC-SIGN engagement may not fully explain the neutralizing and protective capacity of potent mAbs. The fusion process, a common mechanistic feature of RVFV entry facilitating broad tropism, may be a desirable target to enable neutralization. These observations may explain why neutralization is observed in cells that do not express DC-SIGN.
Here, we report the isolation of 20 human monoclonal antibodies (mAbs) from the circulating B cells of RVFV MP-12 vaccines or survivors of natural RVFV infection using neutralization as the primary screening method. This panel of antibodies targets diverse antigenic sites on Gc, Gn, and an undefined epitope that may present only in the glycoprotein complex (Gc+Gn) of properly hetero-oligomerized Gc-Gn. Here, we refer to mAbs that bind to coexpressed full-length Gc-Gn proteins but not to monomeric Gc or Gn as “Gc+Gn–specific mAbs.” Four competition groups for binding to viral antigen on the RVFV surface were recognized by neutralizing antibodies, and all groups contained antibodies that cross-neutralized diverse strains of RVFV. Antibodies from both vaccinated and naturally infected individuals primarily neutralized virus by targeting domain A on the Gn protein. Previously, mAbs derived from a single patient in China inhibited receptor engagement by binding to Gn domain A (30). We sought to explore the role of fusion inhibition as a second major mechanism of neutralization, by which mAbs could contribute to the overall potent neutralizing activity observed and prevent infection even in cells and tissues that do not readily express DC-SIGN. Fusion-inhibiting activity of anti-RVFV–specific mAbs has not been reported to date but has been observed for murine mAbs (31). We observed two classes of potently neutralizing antibodies, those that target domain A and cause partial inhibition of fusion and Gc+Gn–specific mAbs that cause complete inhibition of fusion. Studies of a representative mAb from each group indicate the Gn domain A-specific mAb loses fusion inhibition activity when used as Fab fragments, whereas the Gc+Gn–specific mAb retains fusion inhibition activity as a Fab, suggesting different mechanisms of action for these two classes of mAbs. Antibodies that recognize domain A, domain B, and Gc+Gn–specific epitopes neutralized RVFV with varying potencies, and all provided protection against lethal ZH501 wild-type (wt) virus challenge in prophylactic or therapeutic mouse models of experimental infection when used as a monotherapy. These results suggest these mAbs can be investigated as therapies against RVFV infection and that multiple antigenic sites of vulnerability for neutralization by inhibition of viral fusion exist on the virion surface.
Results
Isolation of Neutralizing Antibodies for RVFV.
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood samples of two sets of immune individuals. Samples were obtained from individuals who had received MP-12 vaccine approximately 5 y before blood sample collection as part of an occupational health program at the United States Army Medical Research Institute of Infectious Diseases (USAMRIID). PBMCs were also obtained from individuals with prior history of laboratory-confirmed RVFV infection in Kenya due to natural exposure but who were healthy at the time of sample collection as a part of seroprevalence study in Kenya (32). The collected PBMCs were isolated, cryopreserved, and transferred to the Vanderbilt research site where they were thawed and transformed in vitro with Epstein–Barr virus (EBV) to generate B cell–derived lymphoblastoid cell lines (LCLs). The supernatants from the LCLs, containing soluble antibodies secreted by the LCLs, were screened using a focus forming assay (FFA) for the presence of RVFV neutralizing activity. Cultures with transformed B cells secreting antibodies that fully reduced infection in the FFA were selected and electrofused with a nonsecreting myeloma cell line. The resulting hybridoma cells were cloned by single-cell flow cytometric sorting to generate clonal hybridoma cells secreting fully human mAbs.
Total RNA was obtained from hybridoma cells to perform sequence analysis of antibody variable regions using a 5′ rapid amplification of cDNA ends (5′RACE) methodology (33) (SI Appendix, Table S1). Denoted mAbs (regardless of original isotype of IgG, IgA, or IgM) was converted to a recombinant IgG1 isotype molecule for the remainder of the studies, while others were derived from hybridoma cells. In vitro neutralization activity of each mAb was tested to determine the half maximal inhibitory concentration (IC50) of 80 foci forming units for RVFV vaccine strain MP-12. Eight mAbs exhibited ultra-potent (IC50 <10 ng/mL) neutralizing activity for MP-12, with mAb RVFV-268 exhibiting the strongest neutralization activity (a remarkable IC50 value of ∼0.1 ng/mL) (Table 1 and SI Appendix, Fig. S1). It was difficult to calculate an accurate IC50 value for the RVFV-268 antibody since the activity is so high, and the 95% confidence intervals for results were wide, but it is clear that it is an ultra-potent antibody with superior potency. MAb CCHFV-4 (a similarly prepared human mAb directed to Crimean–Congo hemorrhagic fever orthonairovirus that we isolated previously) was used as a negative control. These mAbs had similar IC50 values for neutralization of wt strains ZH501 (genetic lineage A) or SA51 (genetic lineage G), except mAb RVFV-144 which lost activity to strains SA51 and ZH501 (Table 1). This finding may be explained in part by amino acid differences between the strains. A subset of mAbs (RVFV-326 and RVFV-121) saw a minor reduction in neutralization potency for wt strains compared to the MP-12 vaccine strain. A wide range of IC50 values in the mAbs obtained from both vaccinated and naturally infected individual is present in the panel, with some mAbs from both donors achieving ultra-potent (IC50 values <10 ng/mL) or potent (IC50 values <120 ng/mL) activity.
Table 1.
Neutralization of RVFV wild-type (wt) or vaccine strains by human mAbs
| MAb (RVFV-) | Antigenic target | IC50 value for neutralization of indicated strain (ng/mL)*** | Source of human B cells | |||
| ZH501 wt strain | SA51 wt strain | MP-12 vaccine | Individual ID | Infection type | ||
| 268 | Gn | ∼0.2 | ∼0.5 | ∼0.1 | 0006 | Vaccine |
| 142 | Gn | 1.5 | 1.5 | 5.9 | 0002 | Vaccine |
| 436 | Gn | 4.6 | 4.6 | 2.5 | 445 102 | Wild-type |
| 429 | Gn | 4.6 | 4.6 | 0.9 | 311 802 | Wild-type |
| 379 | Gn | 4.6 | 13 | 1.3 | 442 | Wild-type |
| 140 | * | 13 | 4.6 | 1.1 | 0002 | Vaccine |
| 426 | Gn | 41 | 120 | 9.1 | 311 802 | Wild-type |
| 296 | Gn | 120 | 120 | 638 | 0006 | Vaccine |
| 220 | * | 120 | 120 | 157 | 0002 | Vaccine |
| 250 | Gc | 120 | 120 | 51 | 0006 | Vaccine |
| 401 | Gn | 370 | 1,110 | 2,120 | 325 001 | Wild-type |
| 128 | Gc | 370 | 370 | 38 | 0002 | Vaccine |
| 121 | Gc | 1,110 | 1,110 | 35 | 0002 | Vaccine |
| 326 | Gc | 1,110 | 370 | 70 | 0006 | Vaccine |
| 144 | * | 3,330 | 3,330 | 1.9 | 0002 | Vaccine |
| 381 | Gn | 10,000 | 10,000 | 4,435 | 442 | Wild-type |
| 226 | Gn | 10,000 | 3,330 | 7,692 | 0002 | Vaccine |
| 229 | NT** | 10,000 | 10,000 | 3,040 | 0002 | Vaccine |
| 405 | Gn | > | > | > | 325 001 | Wild-type |
| 249 | Gc | > | > | 6,687 | 0006 | Vaccine |
* indicates antigenic target could not be mapped. ** NT indicates not tested. *** MP-12 neutralization assay was performed three times with three technical replicates in each assay. SA51 and ZH501 neutralization assays were performed three times with two duplicates. Results were similar between biological replicates; data shown are the mean values of technical replicates from one assay. > indicates neutralization was not detected even at the highest concentration tested of 10 µg/mL.
Antigen Specificity and Epitope Determination of RVFV-Reactive mAbs.
To determine antigenic specificity of these mAbs, we performed enzyme-linked immunosorbent assay (ELISA) with recombinant Gc or Gn proteins (34, 35) and flow cytometry with M gene–transfected cells. These results reveal three binding patterns: mAbs that bound 1) in a Gc-specific manner, 2) in a Gn-specific manner, or 3) only to cells expressing both Gc and Gn simultaneously (SI Appendix, Figs. S2 and S3). MAbs that recognized Gc-Gn–coexpressing cells but not monomeric Gn or Gc independently expressed were designated Gc+Gn–specific mAbs.
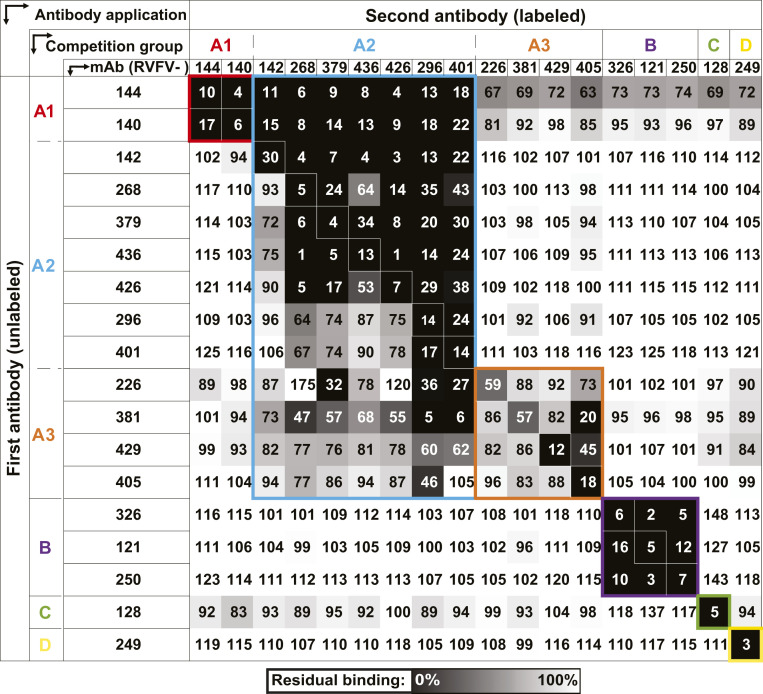
To identify the number of major antigenic sites on the viral surface that can be bound by human mAbs, we performed competition-binding analysis. Identification of antibody groups with similar epitopes was achieved by competing antibodies for binding to 293F cells that were transfected transiently with full-length M gene segment to recapitulate the quaternary structures formed by Gc-Gn. For competition-binding experiments, M-segment complementary DNA (cDNA)–transfected 293F cells were stained with a high concentration of the first mAb (unlabeled). Following this step, a second mAb conjugated to Alexa Fluor 647 was added in a lower concentration. The ability of the second mAb to bind in the presence of the first mAb was determined. We calculated the results as a percentage of signal for the second mAb in the presence of the first mAb compared to maximal binding of the second mAb alone. This analysis identified four major competition-binding groups, with group A also containing three subgroups (designated group A1, A2, and A3; Fig. 1). Group A1 contained the Gc+Gn–specific mAbs RVFV-140 and RVFV-144, indicating these mAbs recognized similar sites. Interestingly, group A1-specific mAbs competed with mAbs from group A2 when group A1 antibodies were added first. Group A2 and A3 mAbs partially competed with each other, and those mAbs bound to Gn, while mAbs in group A3 competed very weakly with each other. MAbs from group B, C, and D all recognize different regions on Gc.
Fig. 1.
Competition binding of RVFV-specific mAbs on Gc-Gn coexpressing cells. We tested 18 mAbs in competition-binding assays. The mAbs are displayed in four groups (designated A [with A1, A2, or A3 subgroups], B, C, or D) based on their ability to compete for binding with each other. The values shown are the percentage of binding that occurred during competition compared to noncompeted binding of the mAb and derived from the mean fluorescence intensity (MFI). MFI values were normalized against a mock transfected control. The values are also indicated by the box fill color; darker colors toward black indicate higher competition, and lighter colors toward white indicate less competition, on a gradient scale. Values shown are the average of three technical replicates from three independent experiments.
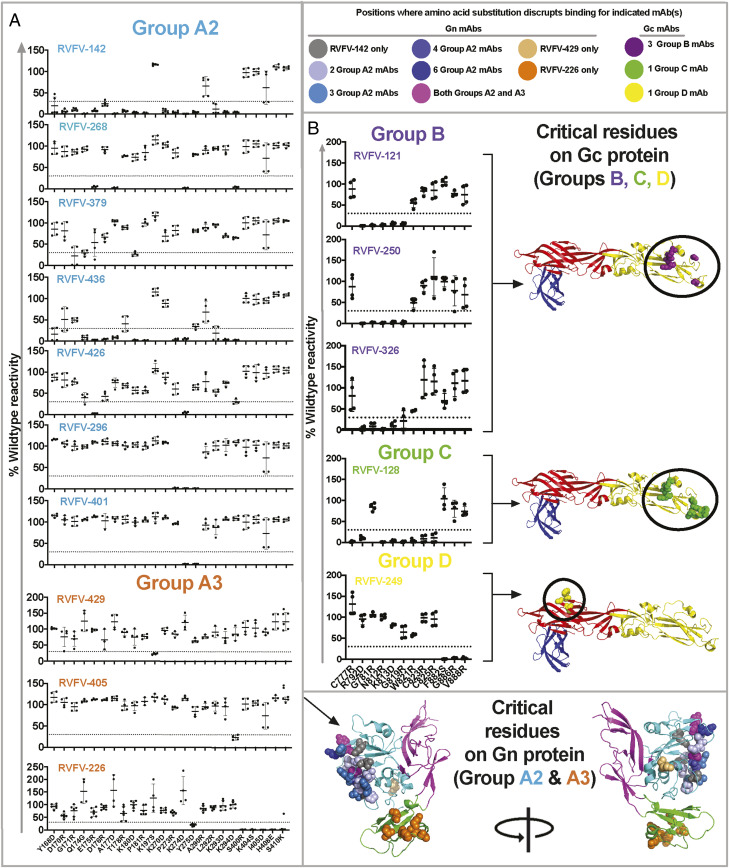
To map the epitopes of mAbs that bound to monomeric Gc or Gn, a mutagenesis screening approach was used with Gc and Gn independently expressed in 293T cells. Gn-specific mAbs were screened for the loss of binding to variants from a library in which individual surface-exposed residues in Gn had been systematically mutated (Fig. 2A and SI Appendix, Fig. S4). The highest number of critical residues for a single mAb was noted for mAb RVFV-142, a vaccine-induced antibody, which was affected by multiple residues in the amino acid spans of 164 to 186 and 270 to 294. Amino acids E175, A177, K274, and K294 were critical for binding of the most potently neutralizing mAb, RVFV-268. MAb RVFV-226 was affected by changes in domain B amino acids and Y275. Comparison of the antigenic sites identified by the loss-of-binding and competition-binding analysis demonstrate that competition group A2 relies on amino acids in the surface-exposed residues of domain A, whereas group A3 may recognize both domain A and B but may bind in a different orientation (RVFV-429) or primarily to domain B (RVFV-226). The weak competition observed in group A3 is explained by the diverse amino acid recognition throughout the Gn protein, suggesting these antibodies may not directly compete for binding to the same epitope. Antibodies binding to Gc were mapped using a similar strategy (Fig. 2B and SI Appendix, Fig. S5). Changes in amino acids R792, G781, N812, K813, and G819 (domain II region adjacent to the fusion loop) caused the loss of binding of RVFV-121, RVFV-250, and RVFV-326, all in group B. RVFV-128, in group C, bound a unique epitope recognizing amino acids (C777, R792, N812, K813, G819, W821, C823, and C825), thus mapping to the fusion loop and adjacent region in domain II. The epitope for RVFV-249 (group D) mapped to domain I and decreases binding significantly when amino acids F882, G886, or V888 were mutated. Comparison of amino acid mutational mapping with competition-binding group analysis indicates that group B and C primarily recognize amino acids near the fusion loop of domain II, while group D mAb (RVFV-249) bind domain I of Gc.
Fig. 2.
Mutation library screening for loss-of-binding to identify critical residues. Expression clones containing key mutations in (A) Gn (for groups A2 and A3) and (B) Gc (for groups B, C, and D) from loss-of-binding screening are shown. The color of the mAb clone names corresponds to the competition group assigned in Fig. 1. The reactivity of each mAb tested for mutants is shown as a percentage of the reactivity of that mAb to wt Gn or Gc. A larger library of mutants was tested, but here, results are shown only for residues that reduced binding of at least one mAb. Amino acids were considered critical if the level of binding was <30% of wt binding. Data shown are the average of technical duplicates from two independent experiments. The location of residues at which amino acid substitution disrupts mAb binding are mapped onto the crystal structure of Gn (A) or Gc (B) and are designated by color-coded spheres. Color coding is as follows: gray (RVFV-142), shades of blue (residues that influence the binding of multiple competition group A2 mAbs), magenta (residues that influence the binding of mAbs in both A2 and A3 competition groups), light orange (RVFV-429), dark orange (RVFV-226), purple (group B mAbs), light green (group C mAb, RVFV-128), and yellow (group D mAb, RVFV-249). A side-view of Gn (A) is shown and is also shown rotated 180°. Gn (domain A, B, or C in cyan, green, or magenta) or Gc (domain I in red, domain II in yellow, or domain III in blue) are shown as cartoon. Gc models has been adapted from PDB (Protein Data Bank):4HJ1, and the Gn model has been adapted from PDB:5Y0W as their respective templates.
Fusion Inhibition of RVFV by Human mAbs.
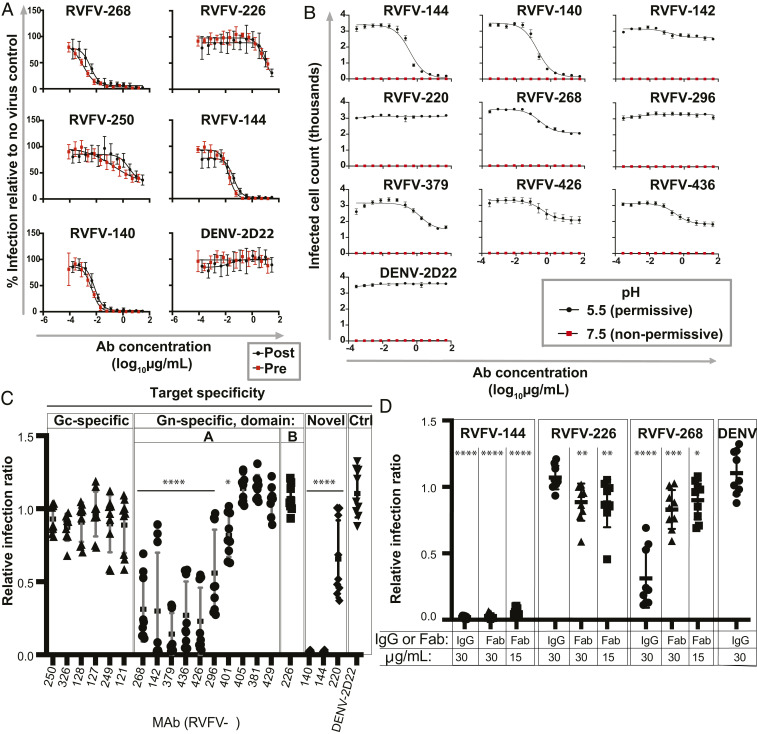
Previously, investigators reported blocking activity for binding of virus to the receptor DC-SIGN by mAbs that bound the Gn domain A of RVFV (30). Given evidence that RVFV uses diverse receptors and entry factors, we sought antibodies that could inhibit virus replication at a stage after attachment of the virus to permissive cells. We performed pre- versus postattachment neutralization assays in Vero cell monolayer cultures (Fig. 3A). All antibodies tested neutralized MP-12 in Vero cells with similar IC50 value when the pre- and postattachment assays were compared. These data suggest that a significant level of neutralization is contributed at the postattachment step and may be independent of receptor blocking activity.
Fig. 3.
Fusion inhibition is mediated by RVFV mAbs against Gn domain A or Gc+Gn epitopes but not Gn domain B or Gc mAbs. (A) Representative mAbs from each domain of Gn, one Gc binding mAb, and Gc+Gn–specific mAbs were titrated in a pre- versus postattachment assay to determine relative contribution of neutralization before or after attachment in Vero cells. (B–D) A FFWO assay was used to assess antibody inhibition of viral fusion with Vero cell membranes under low-pH conditions at the cell surface to mimic acidified endosomes. Virus was adsorbed on a Vero cell monolayer at 4 °C for 45 mins. Virus then was removed, and RVFV-specific mAbs were added for 30 min. Cells were exposed to a prewarmed pH 5.5 medium or a control neutral pH medium for 2 min at 37 °C. The medium was removed, and the cells were incubated in supplemented DMEM for 16 h before fixing on plate, permeabilizing, and staining for viral antigen. Quantification of intracellular viral antigens was done by counting infected cells relative to a virus-only control on a BioSpot CTL plate reader. Three independent experiments were performed in triplicate for each antibody for all assays. (A) Pre- versus postattachment of representative mAbs. (B) Full dilution curves for 10 IgG mAbs in the FFWO assay. (C) Relative infection ratio at 30 µg/mL for 20 IgG mAbs in the FFWO assay. (D) Relative infection ratio at 15 or 30 µg/mL for 3 mAbs tested as Fab fragments in the FFWO assay.
We reasoned that since these mAbs function at a postattachment step and conformational rearrangements in the Gc-Gn interface occur during fusion (26–28), these mAbs may inhibit the pH-dependent fusion events of RVFV infection. To measure this activity, we used a fusion-from-without (FFWO) experimental design where fusion occurs on the plasma membrane to mimic endosomal fusion. Normally, viral fusion to cells occurs in the acidic environment of endosomes (27), but fusion of RVFV can be induced by exposing the virus to low pH on the surface of the plasma membrane, although the efficiency of fusion in this environment is thought to be relatively low compared to that during the natural modes of infection, as has been seen for flaviviruses (36). RVFV-specific mAbs were tested for their ability to block fusion on the plasma membrane in low pH conditions. RVFV was allowed to attach to the surface of Vero cell culture monolayer at 4 °C and then incubated at 4 °C with an RVFV mAb IgG protein, no mAb, or a control mAb DENV-2D22 directed to an unrelated virus (dengue, DENV); a Fab form of RVFV mAbs was also used in some experiments. Cells then were warmed to 37 °C in pH 5.5 or pH 7.5 medium. At pH 5.5, viral–plasma membrane fusion is induced, and the cell monolayer was assessed for viral infection 18 h later by staining for viral antigen. Importantly, in all conditions, 20 mM NH4Cl was added to ensure the decoupling of the proton pumps of the endosomes (37) to prevent de novo infection via the canonical receptor-mediated endocytic route. We defined the pertinent result as a relative infection ratio (normalized permissive pH cell count/normalized nonpermissive pH cell count). For all antibodies tested, we observed negligible counts of infected cells at the nonpermissive pH of 7.5 (<3% of total positive cells compared to permissive pH in the absence of mAb). At the permissive pH of 5.5, the presence of the heterologous mAb DENV-2D22 did not inhibit fusion detectably. RVFV-140 and RVFV-144, the Gc+Gn–specific neutralizing mAbs, fully reduced infection in the FFWO assay. A panel of Gn domain A-specific antibodies showed limited ability to reduce infection (Fig. 3 B and C). Similarly, when antibodies were tested at a concentration of 30 µg/mL, we observed complete reduction of viral infection in the FFWO assay for RVFV-140 and 144, with limited reduction for a subset of Gn domain-A antibodies. Surprisingly, neither Gn-specific domain B (RVFV-226) nor Gc-specific antibodies prevented infection in the FFWO assay (Fig. 3 B and C).
We then tested whether the neutralizing mAbs require bivalent attachment to glycoprotein pentamers to inhibit fusion. We reasoned that if a mAb was inhibiting fusion by attaching to two Gn protomers in the pentameric ring, this configuration could lock the pentamer and prevent subsequent conformational rearrangements necessary to allow fusion. To test this activity, we repeated the FFWO assay with Fab fragments of representative antibodies (RVFV-140, RVFV-268, and RVFV-226) from three competition-binding groups. A marked reduction in infection was detected for RVFV-140 Fab at a 15 or 30 µg/mL concentration, which is consistent with the activity of the corresponding full-length IgG molecule. Furthermore, RVFV-268 Fab showed a reduction of fusion inhibiting activity as a Fab (Fig. 3D).
Prophylactic or Therapeutic Effects of RVFV-Specific Neutralizing mAbs in Murine Models of Infection.
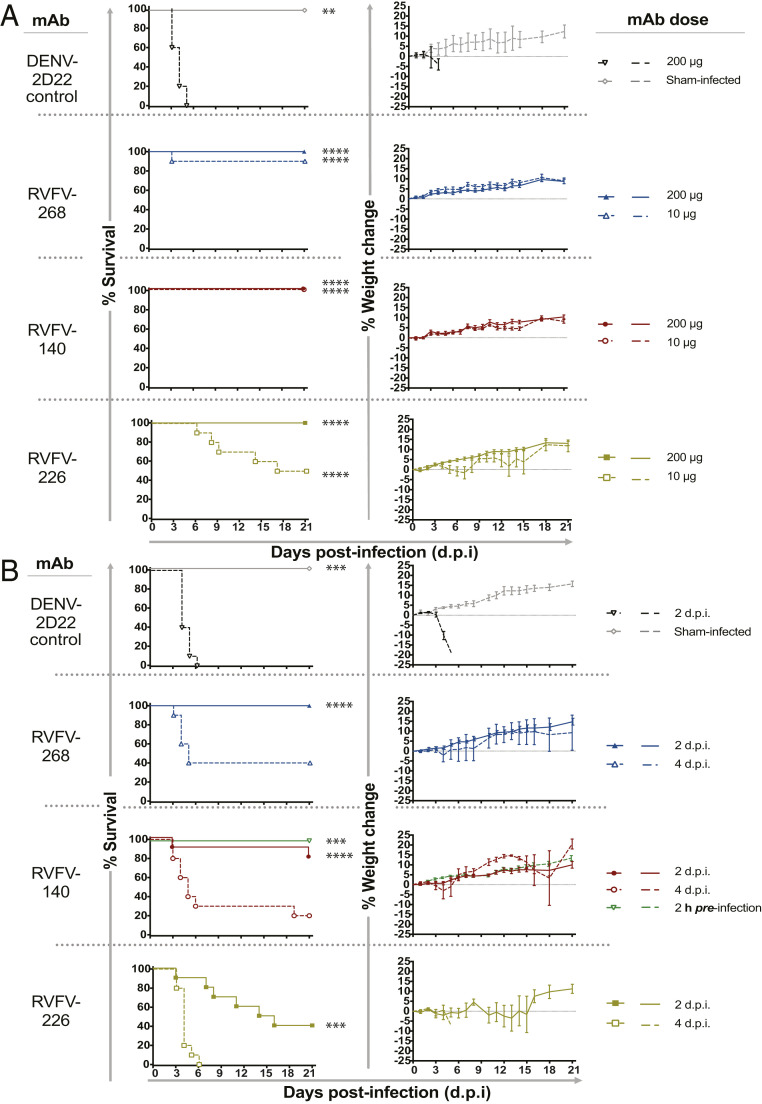
We next sought to determine whether representative mAbs could prevent or treat RVFV infection or disease in murine models of infection. We used two mixed-sex models of infection that were established previously (38, 39), measuring survival and viral titers in liver, spleen, and blood. We tested the ultra-potent domain A mAb RVFV-268, the weakly neutralizing domain B mAb RVFV-226, and the Gc+Gn–specific mAb RVFV-140. First, we tested whether antibodies could reduce viremia and increase survival in a prophylactic model of mAb administration before RVFV infection of inbred C57BL/6 mice. Here, each mAb was administered by the intraperitoneal (IP) route at 10 or 200 µg per mouse 2 h before subcutaneous (SC) challenge with 300 plaque-forming units (PFU) of RVFV wt strain ZH501. Subsets of animals were euthanized 3 d postinfection (d.p.i.) for viral titer analysis of serum and liver or spleen homogenates. The remaining mice were observed for morbidity and mortality for 21 d after inoculation. Each of the mAbs RVFV-268, RVFV-140, and RVFV-226 administered prophylactically 2 h before viral inoculation provided sterilizing immunity at a dose of 200 µg, and similar effects were observed at the dose of 10 µg, except for the detection of some virus in spleen and liver of animals treated with RVFV-226 (SI Appendix, Fig. S6). Each antibody and dose tested provided significant improvements in survival and weight maintenance in the prophylactic setting, with RVFV-226 at 10 µg showing the weakest protective capabilities (Fig. 4A).
Fig. 4.
RVFV-140, RVFV-268, and RVFV-226 increase survival when used as prophylaxis or therapy against lethal RVFV infection in mice. (A) Prophylaxis study in a C57BL/6 mouse model of RVFV infection. MAb (200 or 10 µg) was administered once by the IP route to mice (n = 14) 2 h prior to SC inoculation of 300 PFU of RVFV (ZH501 strain). (B) Therapeutic study in a BALB/c mouse model of RVFV infection. MAb (200 µg) was administered once by the IP route to mice (n = 14) either 2 or 4 d after SC inoculation of 100 PFU of RVFV (ZH501 strain). Kaplan–Meier survival curves were assessed by a Mantel–Cox logrank test. Weight data are represented as the group mean and SE (SEM) of the percent change in weight of surviving animals relative to starting weight. Error bars represent the SEM. Viral titer data were assayed using an infectious cell culture assay in technical triplicate. In both studies, human mAb DENV 2D22 (specific to an unrelated target, dengue virus) was used as the negative control, and mice were monitored daily from day 0 to 21 d.p.i. for survival and body weight. **P = 0.0021; ***P = 0.0002; ****P < 0.0001.
Next, we assessed the effects of these mAbs when used as a postexposure therapy. To assess efficacy, we used a BALB/c mouse model of infection, since the time to death is longer in this strain than in C57BL/6 mice, which allowed us to test the mAbs during a brief therapeutic window after viral inoculation. SC administration of RVFV was followed with the administration of a mAb on 2 or 4 d.p.i. by the IP route with 200 µg of RVFV-140, RVFV-268, or RVFV-226. Subsets of animals were euthanized on 5 d.p.i. to assess viral titers. Each of the antibodies mediated reduction in viral titers in the serum, liver, and spleen with significant therapeutic benefit when administered 2 d.p.i. When the antibody was administered 4 d.p.i., a significant reduction in survival was observed, and for those animals that survived, weight gain was normal except in the RVFV-226 treatment group (Fig. 4B). We were unable to obtain sufficient data for viral titers in animals treated with RVFV-226 on 4 d.p.i. due to the rapid death that occurred at this time point, but a small therapeutic benefit was observed for animals treated with RVFV-268 or RVFV-140 on 4 d.p.i. (SI Appendix, Fig. S7)
Discussion
These studies reveal common features of antigenic recognition of RVFV among human B cells secreting neutralizing antibodies in vaccinated or naturally infected donors. The neutralizing antibodies from individuals in both groups recognized similar sites of vulnerability and exhibited an interesting mechanism of neutralization of RVFV through an indirect mode of fusion inhibition that prevents exposure of the viral fusion loop. The structure of bunyavirus surface glycoproteins in Gc-Gn hetero-oligomers is complex and incompletely understood, which posed a challenge for designing the antigens for an effective mAb discovery campaign. To identify neutralizing antibodies, we did not screen with binding assays using monomeric Gc or Gn proteins (which do not recapitulate the complex oligomeric structure of the Gc-Gn complex), rather, we chose to use an unbiased screening approach based on neutralization of the MP-12 vaccine strain. This approach was successful and identified potently neutralizing antibodies that bind to both known Gn-specific, Gc-specific, and Gc+Gn–specific epitopes. Gc-reactive mAbs typically did not fully reduce viral infection in the neutralization assay but instead allowed some residual viral focus formation even at the highest mAb concentrations tested. These findings highlight the importance of Gn-specific neutralizing mAbs in prevention of viral infection. RVFV Gn-specific domain A-specific mAbs exhibited the highest neutralizing capacity, and Gc+Gn mAbs also possessed potent neutralizing abilities. This previously unknown pattern of binding to an unmapped Gc+Gn antigenic site that is likely quaternary in nature and is only present in the full Gc-Gn heterodimer glycoprotein complex is of interest and merits structural definition in future studies.
This panel of human mAbs appears to identify a substantial portion of the diversity of antigenic recognition patterns for the surface proteins, as competition-binding analysis suggested three major sites of recognition in the Gc glycoprotein, two major overlapping sites on the Gn glycoprotein, and an unmapped Gc+Gn–specific site recognized by mAbs of group A1. Both Gc and Gn antibodies can bind to the viral surface simultaneously, since they did not compete with each other for binding to proteins expressed from the full-length M-segment gene transfection system we used to recapitulate the viral pentameric heterodimer structure on the surface of cells. The competition between group A1 and group A2 mAbs was asymmetric, possibly due to differences in affinity or binding pose. The competition-binding study used full-length IgG molecules, so the unilateral competition between groups A1 and A2 also might be explained by indirect effects in which one antibody may prevent access to the antigenic site recognized by the other group of mAbs by Fc-related steric hindrance. Mutagenesis of Gn or Gc proteins to determine critical residues for binding supported the groupings assigned by the interpretation of competition-binding experiments. Antibodies that recognize Gn domain A (group A2) rely heavily on amino acids from two pockets on Gn (amino acids: 164 to 186 and 270 to 294) with K274 contributing to an especially critical antigenic patch. Antibodies in group A3 appear to bind in different orientations in domain A or to domain B but regardless appear to compete weakly with mAbs from group A2 with some overlap in amino acid usage. MAbs within the competition group A3 weakly impede each other from accessing their epitopes, and their diverse amino acid recognition explains this weak competitive observation. Antibodies in competition groups B and C on the Gc glycoprotein mapped to residues on or adjacent to the fusion loop, and mAb RVFV-249 (group D) mapped to domain I of Gc. In the cases of Gc and Gn domain A, a high level of sequence conservation is observed for different isolates of RVFV (40). MAbs mapping to these sites cross-neutralized strains from different clades of wt RVFV that cause differing levels of disease severity in mice (41), suggesting the feasibility of developing a pan-RVFV mAb therapy. Further analysis against contemporary strains of RVFV could be of interest in the future.
The neutralization mediated by antibodies recognizing Gn domain A that block receptor engagement on cells expressing DC-SIGN have been reported (30), but infection of cells and tissues lacking DC-SIGN also have been observed (24). Because of the wide tropism of RVFV infection for diverse species and tissues, it is desirable to identify antibodies that neutralize virus independent of the specific attachment factor(s) used for entry to cells. The fusion process is a step in the virus life cycle that could be more generally vulnerable to antibodies in diverse tissues. Since many of the mAbs isolated here potently prevent infection at a postattachment step on cells lacking DC-SIGN, we explored fusion inhibition as a potential mechanism of neutralization. We observed potent fusion inhibition by Gc+Gn mAbs targeting a complex quaternary site, while the fusion inhibition mediated by mAbs binding to Gn domain A was less potent. We suggest that receptor blockade activity together with fusion inhibition activity likely explains the ultra-potent (<10 ng/mL IC50) neutralizing capabilities observed in these Gc+Gn mAbs.
It may not seem logical that antibodies against Gn prevent fusion, while Gc contains the critical fusion loop. However, it has been established that the Gn protein of bunyaviruses shields the fusion loop of Gc protein from premature exposure and aberrant fusogenic rearrangements. Upon exposure to acidic pH, Gn shifts position and allows Gc to extend its fusion loop and engage with host membranes (27). Given the role Gn plays as a shield and the requirement to conformational rearrangements in the Gc-Gn hetero-oligomer for fusion to occur, we reasoned that antibodies against RVFV likely inhibit fusion by providing an impediment for this rearrangement process to occur. Given the reduction in fusogenic activity of RVFV in the surrogate FFWO assay that we observed for group A1, A2, and A3 mAbs, it is possible that they function to prevent the Gc-Gn heterodimer fusion-related rearrangements in the cellular infection process and thus prevent the exposure of the Gc fusion loop following acidification in endosomes. Furthermore, Gn domain A mAb RVFV-268 functioned only as a full-length IgG, indicating that bivalent recognition is necessary to prevent fusion, which might suggest a “stapling” mechanism that prevents Gn movement or dissociation. Conversely, competition group A1 mAb RVFV-140 prevented fusion as a Fab, indicating a different mechanism of inhibition. Obtaining structural information is of interest for both antibody classes to further elucidate these mechanisms. Furthermore, the Gc+Gn mAbs RVFV-140 and RVFV-144 potently inhibit fusion in the FFWO assay. Further investigation into the specificity and epitopes for these Gc+Gn–specific antibodies will be of interest for future studies.
The development of vaccines and antivirals against RVFV is a high priority (10, 11), and testing of the antibodies in larger animal models that are also natural hosts of RVFV, such as sheep, are warranted as a future direction. Demonstration of in vivo protective efficacy of three mAbs targeting different sites on the RVFV surface provides evidence that mAbs to diverse epitopes on the surface of RVFV contribute to protection. These studies support the investigation of the MP-12 vaccine or other vaccines that display multiple protective epitopes as a method for vaccination over single-epitope display strategies. Furthermore, antibodies, such as RVFV-140 and RVFV-268 that have fusion inhibition properties, likely would potently reduce viral replication in vivo because of this desirable mechanism of action. Overall, the studies presented here show that natural infection or MP-12 vaccination of humans induces B cells encoding diverse potent neutralizing antibodies that can provide preventative and therapeutic benefit. This work lays the foundation for future therapeutic antibody development as an antiviral agent against RVFV.
Materials and Methods
Human Samples.
Human PBMCs were obtained from two sets of immune individuals. First, PBMCs were collected from survivors of RVFV in Kenya well after the illness had resolved, following informed written consent through a program maintained by the Technical University of Mombasa Institutional Review Board (IRB-P183/04/2107). Second, human PBMCs of vaccinated donors were obtained from individuals who had received MP-12 vaccine approximately 5 y before collection as part of an occupational health program at the USAMRIID. The studies were approved by the Institutional Review Boards of Vanderbilt University Medical Center, USAMRIID, and the Kenyatta National Hospital/University of Nairobi Ethics and Research Committee.
Cell Culture.
Vero (monkey, female) cell lines were obtained from the American Type Culture Collection (ATCC CCL-81), and HEK-293T (human, female) were maintained at 37 °C in 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific) containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) (HyClone), 10 mM Hepes pH 7.3, 1 mM sodium pyruvate, 1× nonessential amino acids, and 100 U mL of penicillin–streptomycin. Expi293F cells (Thermo Fisher Scientific, A1452) were maintained at 37 °C in 8% CO2 in Expi293F Expression Medium (Thermo Fisher Scientific, A1435102). The HMMA 2.5 nonsecreting mouse–human heteromyeloma cell line (female mouse and female human) was provided by L. Cavacini and M. Posner and was cultured as previously described (42). Mycoplasma testing of Expi293F and Vero E6 cultures was performed on a monthly and bimonthly basis, respectively, using a PCR-based mycoplasma detection kit (ATCC, 30 to 1012 K), and all tests were negative during the time of study.
Viruses.
The RVFV vaccine strain MP-12 was used at the Vanderbilt University site. The RVFV strains ZH501 and SA51 were obtained from Dr. Tetsuro Ikegami at the University of Texas Medical Branch. The virus was passaged and titrated by plaque assay in Vero E6 cells. Infectious work was approved by the University of Texas Medical Branch Institutional Biosafety Committees and conducted in approved biosafety level 4 (BSL-4) facilities. For the animal efficacy studies, we used the molecular clone of RVFV ZH501, kindly provided by Dr. Stuart Nichol (Centers for Disease Control and Prevention, Atlanta, GA).
Mouse Models of Infection.
Seven- to eight-week-old male and female C57B/6 and BALB/c mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and Charles River Laboratories (Wilmington, MA), respectively. Mice were housed in microisolator cages and provided water and food ad libitum. The mouse RVFV challenge efficacy studies were approved by the Utah State University Institutional Biosafety Committees and conducted in Select Agent–approved animal BSL-3 facilities.
Virus Neutralization Assay.
Virus neutralization assays were performed in the FFA format using the MP-12 vaccine strain of RVFV. The virus was incubated with increasing concentrations of mAb in triplicate for 1 h at 37 °C, and then each suspension was added to a monolayer of Vero cells of a 96-well plate for 1 h at 37 °C. Following incubation, a 1:1 mixture of fully supplemented (5% FBS) DMEM and 2.4% methylcellulose mixture was added onto the cells. After a 3-d incubation in 5% CO2 at 37 °C, cells were fixed for 1 h with 1% paraformaldehyde and stained with a 1:3,000 dilution of mAb-1D8 (BEI Resources) for 1 h in 2% milk in phosphate-buffered saline (PBS) containing Tween (PBST). Following primary incubation and washing, a 1:3,000 dilution of anti-human HRP conjugated secondary antibodies was added for one h in the same buffer. Cells were washed and stained with TrueBlue peroxidase substrate (SeraCare5510-0030) for 30 min. Cells were washed in dH2O, imaged on a BioSpot CTL plate reader, and foci were counted. The percent relative infection was determined based on the virus-only control. Assays for ZH501 and SA51 were performed in the plaque reduction neutralization format. After the virus/mAb incubation step, the suspension was added to a monolayer of Vero cells in a 24-well plate for 1 h at 37 °C. Following incubation, cells were overlaid with MEM containing 2% FBS, 1% penicillin–streptomycin, and 0.5% methylcellulose. After a 5-d incubation in 5% CO2 at 37 °C, cells were fixed in 10% buffered formalin for 30 min and stained with crystal violet. Plaques were enumerated by visual examination. Tests were performed for each antibody using MP-12 (triplicate), ZH501 (duplicate), or SA51 (duplicate) strains, respectively. IC50 values were determined using a sigmoidal, four parameter logistic nonlinear fit analysis in Prism software version 8 (GraphPad).
Hybridoma Generation.
Vaccinated individuals, who had received MP-12 vaccine as part of an occupational health program at USAMRIID, were identified. Naturally infected individuals in Kenya were identified with prior history of laboratory-confirmed RVFV infection but who were healthy at the time of sample collection. After written informed consent was obtained, peripheral blood was collected and stored at room temperature until PBMCs could be purified using SepMate tubes (STEMCELL Technologies) per the manufacturer’s protocol and then cryopreserved in 10% (vol/vol) dimethyl sulfoxide in FBS and stored in the vapor phase of liquid nitrogen. Samples derived in Kenya were transferred to the Vanderbilt site. Approximately 107 cryopreserved PBMCs were thawed, and LCLs were generated as previously described (42) from memory B cells within the PBMCs by transformation with EBV (obtained from B95.8 cells) and supplemented with cell cycle checkpoint kinase 2 inhibitor (Sigma-Aldrich), CpG (Sigma-Aldrich), and cyclosporin A (Sigma-Aldrich) in Medium A (STEMCELL Technologies). One week later, LCLs were counted and then expanded on a feeder layer of gamma-irradiated, human PBMCs from discarded leukofiltration devices. After 7 d, LCL supernatants were screened for the presence of RVFV neutralizing mAbs by an adaptation of the neutralization assay with MP-12 previous described without overlay. LCLs from wells containing virus-neutralizing antibodies were fused to HMMA 2.5 myeloma cells by an established electrofusion technique (43). After fusion, hybridoma lines were cultured in a selection medium with hypoxanthine, aminopterin, and thymidine medium supplements (Sigma-Aldrich) and ouabain (Sigma-Aldrich) in 384-well cell culture plates before screening for RVFV-specific antibody production in supernatants. Two weeks later, supernatants from the hybridoma cell lines were screened by neutralization and then cloned by single-cell flow cytometric sorting on a BD FACSAria III sorting cytometer with aerosol containment in 384-well plates. These cloned cells were expanded in Medium E in 12-well tissue culture-treated plates (Corning) upon reaching 50% confluence, and their supernatants were screened for neutralizing activity. MAb-producing hybridoma cell lines were selected from wells displaying neutralizing activity.
Antibody Production and Purification.
For hybridoma-derived mAb, clonal cells were grown in 75 cm2 flasks to 70% confluency in hybridoma growth medium (ClonaCell-HY medium E from STEMCELL Technologies, 03805). The hybridoma cells were grown to exhaustion in Hybridoma-SFM (1×) serum-free medium (Gibco Hybridoma-SFM, Invitrogen, 12045084) in four 225 cm2 flasks. Exhausted hybridoma supernatant was harvested after one month. For the recombinant mAb production, the genes of heavy and light chains were synthesized into cDNA. The fragments were cloned into a full-length IgG1 DNA plasmid expression vector (44). The heavy and light chains were transformed into Escherichia coli cells to produce large amounts of DNA. Following the manufacturer’s protocol, plasmids encoding heavy and light antibody chains were transiently transfected into Expi293F cells to produce mAb proteins. Secreted IgGs from recombinant and hybridomas were purified from filtered supernatants by affinity chromatography using Protein G columns (Cytiva, HiTrap Protein G HP columns) on an ÄKTA pure instrument. Purified mAbs were processed by buffer-exchanging into PBS using buffer-exchanged into PBS using Zeba Spin Desalting Columns (Thermo Fisher Scientific), filtered using sterile 0.45 μm Millipore filter devices, concentrated using Amicon Ultra-4 50 kDa Centrifugal Filter Units (MilliporeSigma), and stored at −80 °C. Recombinant mAbs were used for all in vivo experiments, and designated mAbs were either hybridoma or recombinant derived in vitro experiments.
Fab Fragment Production.
DNAs encoding heavy and light chains of Fabs were inserted into Fab DNA plasmid expression vectors, processed, and transiently expressed similarly as full-length IgG expression. Fab fragments were purified using CaptureSelect Anti-CH1 columns (Thermo Fisher) on an ÄKTA pure instrument. Final Fab fragments were buffer-exchanged into PBS, concentrated, and stored at 4 °C until use.
MAb Isotype and Gene Sequencing Analysis.
The antibody heavy- and light-chain variable region genes were obtained from hybridoma cell lines that had been cloned by flow cytometric single-cell sorting. Total RNA extraction was performed using RNeasy Mini Kit (QIAGEN) per the manufacturer’s protocol. Amplification of cDNA ends was done using a modified 5′ rapid amplification approach (33). Briefly, a mixture of 5 μL of total RNA and cDNA synthesis primer mix (10 μM each) was incubated for at 70 °C (2 min), followed by a 42 °C incubation step (1 to 3 min) for synthesis primer annealing. Post incubation, added to the total RNA reaction was a mixture of 5× first-strand buffer (Clontech), dithiothreitol (20 mM), 5′ template switch oligo (10 μM), deoxynucleotide triphosphate (dNTP) solution (10 mM each), and 10× SMARTScribe Reverse Transcriptase (Clontech) (60 min incubation) at 42 °C. The first-strand synthesis reaction was purified using Ampure Size Select Magnetic Bead Kit (ratio of 1.8× [Beckman Coulter]). After purification, a single PCR amplification reaction with 5 μL of first-strand cDNA, 2× Q5 High-Fidelity Master Mix (New England Biolabs), dNTP (10 mM each), forward universal primer (10 μM), and reverse primer mix (0.2 μM each in heavy-chain mix and 0.2 μM each in light-chain mix) was subjected to the following: initial denaturation for 90 s followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 60 °C for 20 s, and extension at 72 °C for 40 s, followed by a final extension step at 72 °C for 4 min. Primers used here were previously detailed (33). Using the AMPure Size Select Magnetic Bead Kit (ratio of 0.6× [Beckman Coulter]), the first PCR was purified. Amplicon libraries were prepared per the Multiplex SMRT Sequencing protocol (Pacific Biosciences) and sequenced on a Sequel instrument (Pacific Biosciences). Raw sequences were demultiplexed, and circular consensus sequences were determined using SMRT Analysis tool suite (Pacific Biosciences). The identities of gene segments, complementarity-determining regions (CDRs), and mutations were determined using the ImMunoGeneTics database (45).
Gc and Gn Recombinant Protein Production and ELISA.
A gene encoding the ectodomain of Gc and Gn was synthesized, cloned, and expressed in SF9 cells described previously (34, 35). These recombinant proteins were isolated by metal affinity chromatography on HisTrap Excel columns (GE Healthcare). For ELISA binding assessment of mAbs, 384-well ELISA plates were directly coated with either Gc or Gn at 1 μg/mL (diluted in PBS) and incubated overnight at 4 °C. Plates were washed three times with PBST using an EL406 combination washer dispenser instrument (BioTek) and blocked for 1 h at room temperature with 5% milk powder and 2% goat serum (diluted in PBS). After washing three times with PBST, 30 µL antibody diluted in DMEM was added to plates and incubated for 1 h at room temperature. Plates were then washed three times, and 30 μL of goat anti-human HRP-conjugated secondary antibodies (Meridian Life Science) diluted 1:3,000 in PBS was added to plates and incubated for 1 h at room temperature. Plates were washed three times, and 25 μL of 1-Step Ultra TMB-ELISA (Thermo Fisher Scientific-34029) was added for 7 mins, and the reaction was quenched with 25 μL of 1 M HCl. Plates were read for optical density at 405 nm immediately using a BioTek plate reader.
MAb Competition-Binding Assay Using a Gc-Gn Cell-Surface Display System.
For antibody preparation, mAbs were directly fluorescently labeled. Briefly, mAbs were labeled with Alexa Fluor 647 NHS ester (Thermo Fisher) by following the manufacturer’s protocol. Labeled mAbs were purified and buffer exchanged into PBS using desalting Zeba columns (Thermo Fisher) and stored at 4 °C until use. For cell display of Gc-Gn, plasmid encoding full-length M segment of RVFV ZH501 [a gift from the laboratory of Dr. Friedemann Weber (46)] was transiently transfected into Expi293F cells per the manufacturer’s protocol. Cells were cultured to produce antigen for 3 d, and cells were either processed for staining or frozen and stored in the vapor phase of liquid nitrogen until use. Cells (1 × 107) were fixed and permeabilized using BD Cytofix Cytoperm according to the manufacturer’s protocol. Cells (50,000/well) then were plated into 96-well V-bottom plates in permeabilization buffer. Cells were incubated with a saturating concentration (typically 20 µg/mL) of the first unlabeled mAb at room temperature for 30 min. Second, fluorescently labeled mAb (5 µg/mL) was added for 30 min without prewashing to minimize the dissociation of the first mAb. Cells were washed, fixed in 1% paraformaldehyde, and resuspended in 30 µL of fluorescence activated cell sorting (FACS) buffer (Dulbecco’s phosphate buffered saline [DPBS], 2 mM EDTA, and 2% FBS). Staining was analyzed using an Intellicyt IQue flow cytometer, and mean fluorescence intensity values were used for analysis. Background values were determined from binding of the second (labeled) mAb to mock-transfected expi293F cells. Results were expressed as the percent of binding in the presence of competitor mAb minus the background signal over the second mAb-only (maximal binding) value minus the background signal. Antibodies were considered competing if the presence of the first antibody reduced the signal of the second antibody to less than 30% of its maximal binding or noncompeting if the signal was greater than 70%.
MAb Binding Assay Using a Gc-Gn Cell-Surface Display System.
RVFV-144 and RVFV-140 were prepared by using Alexa Fluor 647 NHS ester (Thermo Fisher) per the manufacturer’s protocol and used to stain RVFV M-segment transfected 293F cells. Briefly, cells were stained with 30 µg/mL of primary antibody for 20 min at room temperature and then assessed using the IQue flow cytometer. Untransfected cells were used as the negative control.
Mutagenesis Epitope Mapping.
Independently expressed Gn and Gc expression constructs (based on RVFV ZH501) were synthesized with designated mutations to generate a mutation library focusing on predicted surface-exposed residues. In total, 76 or 98 mutants were generated for Gn or Gc, respectively. Each RVFV Gn or Gc mutant was transfected into 293T cells with FuGENE 6 transfection reagent and allowed to express for 40 h. Mock-transfected cells were included as control. Cells were fixed in 4% (vol/vol) PFA, washed with PBS, and permeabilized with staining buffer (PBS supplemented with 0.1% [wt/vol] saponin [Sigma-Aldrich] and 1% bovine serum albumin [BSA]). Cells then were incubated with purified mAbs at concentration of 1 or 2 µg/mL for 1 h at 4 °C, washed twice, and stained with FITC-conjugated goat anti-Human IgG, IgM, IgA (H+L) secondary antibody (Invitrogen, 1:500). Mouse anti-HA antibody (Covance, 1:500) was included to monitor Gn or Gc protein expression. Flow cytometry was performed on a MACSQuant Analyzer (Miltenyi Biotec), and the data were analyzed using FlowJo software. Binding to mutant gene-transfected cells was calculated relative to wt Gn- or Gc-transfected cells. Mutations were categorized as critical if they supported binding of other RVFV-specific mAbs but showed <15% (RVFV-142) or <30% (most of the mAbs) reactivity relative to wtprotein. The incorporation of this strategy assists in the exclusion of mutants that are misfolded or cannot properly express.
Pre- or Postattachment Inhibition Assays.
96-well tissue culture plates were treated with poly-D lysine at 50 µg/mL overnight at 4 °C and dried before seeding 20,000 Vero cells/well the day before use in the assay. In the preattachment assay, 600 PFUs of virus was added with antibody for 1 h. Cells were washed and incubated at 4 °C for 15 min. Virus and mAb mixture was added to cells for 1 h at 4 °C and then washed and incubated at 37 °C. A methylcellulose layer was added, and cells were allowed to incubate for three days. In the postattachment assay, Vero cells were plated as before and washed twice and allowed to cool in 4 °C for 15 min. In total, 300 PFUs of RVFV was added to each well and allowed to incubate for 1 h at 4 °C. Diluted mAb was added to cells after free virus was washed and allowed to incubate for 1 h at 4 °C. Antibody was washed off, and cells were allowed to incubate for 15 min at 37 °C before adding methylcellulose and incubating for 3 d. Both pre- and postattachment plates were then fixed and processed as described before.
FFWO Assay.
Cells were plated as described in the pre- versus postattachment section. The supernatant was decanted and washed with binding medium (Roswell Park Memorial Institute Medium [RPMI] 1640, 0.2% BSA, 10 mM Hepes pH 7.4, and 20 mM NH4Cl) and incubated 4 °C for 15 min. RVFV MP-12 was prepared to an multiplicity of infection (MOI) of 1 in binding medium and added to cells for 45 min at 4 °C, and then free virus was removed. Following incubation, dilution medium (DMEM with 2% FBS) with or without RVFV-specific or DENV-2D22 control mAbs was added to cells for 30 mins at 4 °C. FFWO was induced by adding prewarmed fusion media (RPMI 1640, 0.2% BSA, 10 mM Hepes, and 30 mM succinic acid at pH 5.5) for 2 mins at 37 °C. In identical wells, control media (RPMI 1640, 0.2% BSA, and 10 mM Hepes at pH 7.4) was added for 2 min at 37 °C to ensure infection occurs only via the pH-dependent plasma membrane fusion. The medium was removed, and cells were incubated at 37 °C for 16 h in DMEM with 5% FBS, 10 mM Hepes, and 20 mM NH4Cl (pH 7.4). Following incubation, cells were fixed in 1% PFA for 2 h at room temperature, washed three times, stained with a mixture of murine mAbs (3C10, 1D8, and 1F6 [BEI Resources]) with a 1:3,000 dilution in permeabilization-wash buffer (1× DPBS, 0.1% BSA, and 0.1% saponin). Plates were washed three times, and 50 µL of 1:3,000 dilution of anti-mouse HRP-conjugated antibodies in permeabilization wash buffer was added, and plates were incubated for 1 h at room temperature. Following incubation, plates were washed three times, and 40 µL of TrueBlue peroxidase substrate (SeraCare) was added. Cells were allowed to develop, washed with dH2O, and imaged as described previously for neutralization assays.
RVFV Challenge in Mice and Viral Titer.
Experimental groups (n = 14 for the treatment group with four animals per group for the 3 d.p.i sacrifice for viral titers) in the prophylactic study consisted of 7- to 8-wk-old male and female C57BL/6 mice (n = 5 for sham-infected controls, with two euthanized for viral titer controls). Animals were treated once by IP injection with either 200 µg (∼10 mg/kg) or 10 µg (∼0.5 mg/kg) of individual mAb at a time point 2 h before SC inoculation of 300 PFU of RVFV strain ZH501. Experimental groups (n = 14 for the treatment group with four animals per group for 5 d.p.i. sacrifice for viral titers) in the therapeutic study consisted of 7- to 8-wk-old male and female BALB/c mice (n = 5 for sham-infected controls, with two euthanized for viral titer controls). Animals were inoculated with 100 PFU of RVFV (ZH501 strain) by the SC route. Animals were treated with 200 µg antibody once IP on 2 or 4 d.p.i. In both prophylactic and treatment studies, human mAb DENV 2D22 (specific to an unrelated target, dengue virus) was used as the negative control. Mice were monitored daily from 0 to 21 d.p.i. for survival and body weight, and survivors were euthanized on 21 d.p.i.
Viral titers were assayed using an infectious cell culture assay, previously described (47). Here, a volume of tissue homogenate or serum was diluted and added to triplicate wells of Vero cell monolayer cultures. Viral cytopathic effect was determined 7 d after plating to calculate 50% endpoints. Lower limits of detection were 1.49 log10 50% cell culture infectious dose (CCID50)/mL in serum and 2.1 log10 CCID50/g in tissue.
Quantification and Statistical Analysis.
Kaplan–Meier survival curves were analyzed using the logrank test, and viraemia was compared using ANOVA with multiple comparisons test. Statistically significant differences were indicated by P < 0.05. Technical and biological replicates are indicated in the methods and figure legends. Error bars in figures represent SD unless otherwise indicated. Statistical analyses were performed using Prism version 8 (GraphPad).
Supplementary Material
Acknowledgments
We thank Philip Chebii, Priscilla Maina, Christine Njoroge, Omar Kallum, Saidi Malumbo, Charles Ng'ang'a, and Elton Mzungu and the communities of Milalani and Nganja, Kenya, for help with acquisition of critical samples and infrastructure support from Charles H. Ki and Uriel Kitron. The work was supported by National Institute of Allergy and Infectious Diseases (NIAID) US Department of Health & Human Services (HHS) Contracts 75N93019C00062 and HHSN2272201700041I and Fogarty International Center/NIH Grant R01 TW008067. The findings and conclusions in this report are those of the authors and do not represent the views of the US Department of Defense.
Footnotes
Competing interest statement: J.E.C. has served as a consultant for Luna Biologics and Eli Lilly; is on the Scientific Advisory Boards of CompuVax and Meissa Vaccines; is a recipient of previous or active unrelated research grants from Astra Zeneca, Takeda, and IDBiologics; and is a founder of IDBiologics. Vanderbilt University has applied for patents concerning RVFV antibodies that are related to this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025642118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
Change History
March 31, 2021: Fig. 3 has been updated.
References
- 1.Weaver S. C., Reisen W. K., Present and future arboviral threats. Antiviral Res. 85, 328–345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daubney R., Hudson J. R., Garnham P. C., Enzootic hepatitis or Rift Valley fever. An undescribed virus disease of sheep cattle and man from east africa. J. Pathol. 34, 545–579 (1931). [Google Scholar]
- 3.Anyamba A., et al., Prediction of a rift Valley fever outbreak. Proc. Natl. Acad. Sci. U.S.A. 106, 955–959 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO , Rift Valley fever. World Health Organization (2018). https://www.who.int/news-room/fact-sheets/detail/rift-valley-fever. Accessed 16 November 2020.
- 5.Baudin M., et al., Association of rift Valley fever virus infection with miscarriage in Sudanese women: A cross-sectional study. Lancet Glob. Health 4, e864–e871 (2016). [DOI] [PubMed] [Google Scholar]
- 6.McMillen C. M., et al., Rift Valley fever virus induces fetal demise in Sprague-Dawley rats through direct placental infection. Sci. Adv. 4, eaau9812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO , Emergencies preparedness, response; Rift Valley Fever. World Health Organization (2018). https://www.who.int/csr/don/archive/disease/rift_valley_fever/en/. Accessed 16 November 2020.
- 8.Balkhy H. H., Memish Z. A., Rift Valley fever: An uninvited zoonosis in the Arabian peninsula. Int. J. Antimicrob. Agents 21, 153–157 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Dar O., Hogarth S., McIntyre S., Tempering the risk: Rift Valley fever and bioterrorism. Trop. Med. Int. Health 18, 1036–1041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The WHO R&D Blueprint Team , Prioritizing diseases for research and development in emergency contexts. World Health Organization (2018). https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts. Accessed 16 November 2020.
- 11.NIAID Emerging Infectious Diseases/Pathogens , National Institute of Allergy and Infectious Diseases (2018). https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens. Accessed 16 November 2020.
- 12.Virus Taxonomy , ICTV (International Committee on Taxonomy of Viruses) (2019). https://talk.ictvonline.org/taxonomy/. Accessed 16 November 2020.
- 13.Freiberg A. N., Sherman M. B., Morais M. C., Holbrook M. R., Watowich S. J., Three-dimensional organization of Rift Valley fever virus revealed by cryoelectron tomography. J. Virol. 82, 10341–10348 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huiskonen J. T., Overby A. K., Weber F., Grünewald K., Electron cryo-microscopy and single-particle averaging of rift Valley fever virus: Evidence for GN-GC glycoprotein heterodimers. J. Virol. 83, 3762–3769 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phoenix I., et al., N-glycans on the Rift Valley fever virus envelope glycoproteins Gn and Gc redundantly support viral infection via DC-SIGN. Viruses 8, 149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiegel M., Plegge T., Pöhlmann S., The role of phlebovirus glycoproteins in viral entry, assembly and release. Viruses 8, 202 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niklasson B., Peters C. J., Bengtsson E., Norrby E., Rift Valley fever virus vaccine trial: Study of neutralizing antibody response in humans. Vaccine 3, 123–127 (1985). [DOI] [PubMed] [Google Scholar]
- 18.Pittman P. R., et al., Rift Valley fever MP-12 vaccine Phase 2 clinical trial: Safety, immunogenicity, and genetic characterization of virus isolates. Vaccine 34, 523–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spik K., et al., Immunogenicity of combination DNA vaccines for Rift Valley fever virus, tick-borne encephalitis virus, Hantaan virus, and Crimean Congo hemorrhagic fever virus. Vaccine 24, 4657–4666 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Morrill J. C., Peters C. J., Mucosal immunization of rhesus macaques with Rift Valley Fever MP-12 vaccine. J. Infect. Dis. 204, 617–625 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Faburay B., et al., A glycoprotein subunit vaccine elicits a strong Rift Valley fever virus neutralizing antibody response in sheep. Vector Borne Zoonotic Dis. 14, 746–756 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niklasson B. S., Meadors G. F., Peters C. J., Active and passive immunization against Rift Valley fever virus infection in Syrian hamsters. Acta Pathol. Microbiol. Immunol. Scand. [C] 92, 197–200 (1984). [DOI] [PubMed] [Google Scholar]
- 23.Papin J. F., et al., Recombinant Rift Valley fever vaccines induce protective levels of antibody in baboons and resistance to lethal challenge in mice. Proc. Natl. Acad. Sci. U.S.A. 108, 14926–14931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Boer S. M., et al., Heparan sulfate facilitates Rift Valley fever virus entry into the cell. J. Virol. 86, 13767–13771 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmon B., et al., Rift Valley fever virus strain MP-12 enters mammalian host cells via caveola-mediated endocytosis. J. Virol. 86, 12954–12970 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozach P. Y., et al., DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10, 75–88 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Halldorsson S., et al., Shielding and activation of a viral membrane fusion protein. Nat. Commun. 9, 349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Boer S. M., et al., Acid-activated structural reorganization of the Rift Valley fever virus Gc fusion protein. J. Virol. 86, 13642–13652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guardado-Calvo P., et al., A glycerophospholipid-specific pocket in the RVFV class II fusion protein drives target membrane insertion. Science 358, 663–667 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Wang Q., et al., Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus. Nat. Microbiol. 4, 1231–1241 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Besselaar T. G., Blackburn N. K., Study of the Surface Glycoprotein of Rift Valley Fever Virus Using Monoclonal Antibodies (University of Witwatersrand, Johannesburg, South Africa, 1992). [Google Scholar]
- 32.Grossi-Soyster E. N., et al., Rift Valley fever seroprevalence in coastal Kenya. Am. J. Trop. Med. Hyg. 97, 115–120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turchaninova M. A., et al., High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat. Protoc. 11, 1599–1616 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Dessau M., Modis Y., Crystal structure of glycoprotein C from Rift Valley fever virus. Proc. Natl. Acad. Sci. U.S.A. 110, 1696–1701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y., et al., Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope. Proc. Natl. Acad. Sci. U.S.A. 114, E7564–E7573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson B. S., et al., A therapeutic antibody against west nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 5, e1000453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohkuma S., Poole B., Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. U.S.A. 75, 3327–3331 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lathan R., et al., Innate immune basis for Rift Valley fever susceptibility in mouse models. Sci. Rep. 7, 7096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cartwright H. N., Barbeau D. J., McElroy A. K., Rift Valley fever virus is lethal in different inbred mouse strains independent of sex. Front. Microbiol. 11, 1962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen E. R., et al., A protective monoclonal antibody targets a site of vulnerability on the surface of Rift Valley fever virus. Cell Rep. 25, 3750–3758.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikegami T., et al., Distinct virulence of Rift Valley fever phlebovirus strains from different genetic lineages in a mouse model. PLoS One 12, e0189250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu X., McGraw P. A., House F. S., Crowe J. E. Jr, An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J. Immunol. Methods 336, 142–151 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith S. A., Crowe J. E. Jr, Use of human hybridoma technology to isolate human monoclonal antibodies. Microbiol. Spectr. 3, AID-0027–AID-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLean G. R., Nakouzi A., Casadevall A., Green N. S., Human and murine immunoglobulin expression vector cassettes. Mol. Immunol. 37, 837–845 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Giudicelli V., Lefranc M. P., IMGT/junctionanalysis: IMGT standardized analysis of the V-J and V-D-J junctions of the rearranged immunoglobulins (IG) and T cell receptors (TR). Cold Spring Harb. Protoc. 2011, 716–725 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Habjan M., et al., Efficient production of Rift Valley fever virus-like particles: The antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology 385, 400–408 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Gowen B. B., et al., In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrob. Agents Chemother. 51, 3168–3176 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.