Significance
Several neurodegenerative and neuropsychiatric disorders with cognitive symptoms show an abnormal expression of microRNA-218 (miR-218). In this study, we demonstrated that miR-218 regulated presynaptic functions and learning related behaviors through targeting the 3′-untranslated region (UTR) of the mRNA of complement component 3 (C3), a central component of the immune system. This finding may represent a common mechanism for the defective cognition of multiple neuronal disorders with miR-218 deficits and provide a strategy for the development of new therapy.
Keywords: miR-218, synapse, hippocampus, LTP, C3
Abstract
microRNA-218 (miR-218) has been linked to several cognition related neurodegenerative and neuropsychiatric disorders. However, whether miR-218 plays a direct role in cognitive functions remains unknown. Here, using the miR-218 knockout (KO) mouse model and the sponge/overexpression approaches, we showed that miR-218-2 but not miR-218-1 could bidirectionally regulate the contextual and spatial memory in the mice. Furthermore, miR-218-2 deficiency induced deficits in the morphology and presynaptic neurotransmitter release in the hippocampus to impair the long term potentiation. Combining the RNA sequencing analysis and luciferase reporter assay, we identified complement component 3 (C3) as a main target gene of miR-218 in the hippocampus to regulate the presynaptic functions. Finally, we showed that restoring the C3 activity in the miR-218-2 KO mice could rescue the synaptic and learning deficits. Therefore, miR-218-2 played an important role in the cognitive functions of mice through C3, which can be a mechanism for the defective cognition of miR-218 related neuronal disorders.
Patients of neurodegenerative and neuropsychiatric disorders, such as Alzheimer’s disease and schizophrenia, often show severe cognitive symptoms. However, it remains poorly understood whether these diseases share common mechanisms directly underlying the defective cognition in the patients. In recent years, accumulating evidence has shown that the expression and activity of some microRNAs (miRNAs), such as miR-128b, miR-34c, and miR-132, are directly involved in the cognitive functions of invertebrate and vertebrate animals (1–7). miRNAs are short noncoding RNAs (∼19 to 22 nucleotides long) that function through binding to the 3′-untranslated region (UTR) of target messenger RNAs (mRNAs) to regulate gene expression at the posttranscriptional level (8, 9). Many miRNAs have been suggested to play important roles in the central nervous system (CNS), participating in a variety of neuronal functions, such as synaptic transmission, synaptic plasticity, neuronal morphology, and neural stem-cell differentiation (6–17). Particularly, it has been suggested that miRNAs may play a role in the cognitive functions of neurological disorders. For instance, miR-34c can negatively regulate the learning and memory in a rat model of epilepsy through modulating the N-methyl-d-aspartate receptors (NMDARs) (18).
Recently, miR-218 has been suggested to show abnormal expression in a variety of neurodegenerative and neuropsychiatric disorders, including Alzheimer’s disease, Parkinson’s disease, schizophrenia, depression, and anxiety (19–24). miR-218 includes miR-218-1 and miR-218-2, which are located in the introns of the tumor suppressor genes SLIT2 and SLIT3, respectively (25). The SLIT-family proteins mainly contribute to axon guidance and neuronal and leukocyte migration (26, 27). The miR-218 itself likely plays antitumor roles in diverse types of cancers, such as cervical cancer, colon cancer, and lung cancer (28–30). In the brain, miR-218 is highly expressed in astrocytes and thus impacts gliomagenesis (31, 32). Also, miR-218 has been found to have important impacts on the development of motoneurons in the spinal cord and dopaminergic neurons in the midbrain (33–35). For instance, inhibition of miR-218 in the developing spinal cord could repress motoneuron generation through the downstream Isl1-Lhx3 pathway (34), and mice with full loss of both miR-218-1 and miR-218-2 died neonatally and showed a significant loss of motoneurons (36). However, although miR-218 is linked to many cognition-related neurological disorders, it remains unclear whether miR-218 has a common function in these diseases.
In the present study, we found that miR-218 played an important role in the cognitive functions of mice. Using the miR-218-2 knockout (KO) mouse model and the miRNA sponge and gene overexpression approaches, we modulated the expression of miR-218 in the hippocampus of mice and found that miR-218-2 but not miR-218-1 could bidirectionally regulate the learning and memory of the animals. Using slice patch-clamp recording and Golgi staining techniques, we demonstrated that depletion of miR-218-2 in the hippocampus induced abnormalities in the morphology and presynaptic synaptic-vesicle (SV) release of the neurons, which consequently impaired the long term potentiation (LTP). Furthermore, combining the RNA sequencing (RNA-Seq) and luciferase reporter analyses, we identified that complement component 3 (C3), a central component of the immune system, was a main target gene of miR-218 in the hippocampal neurons to regulate the presynaptic functions. Finally, we demonstrated that restoring the C3 activity in the miR-218-2 KO mice could rescue the deficits in presynaptic functions and learning behaviors. We therefore concluded that miR-218-2 played an important role in the learning and memory of the mice via the C3-dependent pathway that regulates the presynaptic functions of hippocampal neurons. Our study helps understand the mechanism of cognitive defects involved in the neuronal disorders with miR-218 defects.
Results
miR-218-2 Deficiency Induces Impairments in Learning and Memory in Mice.
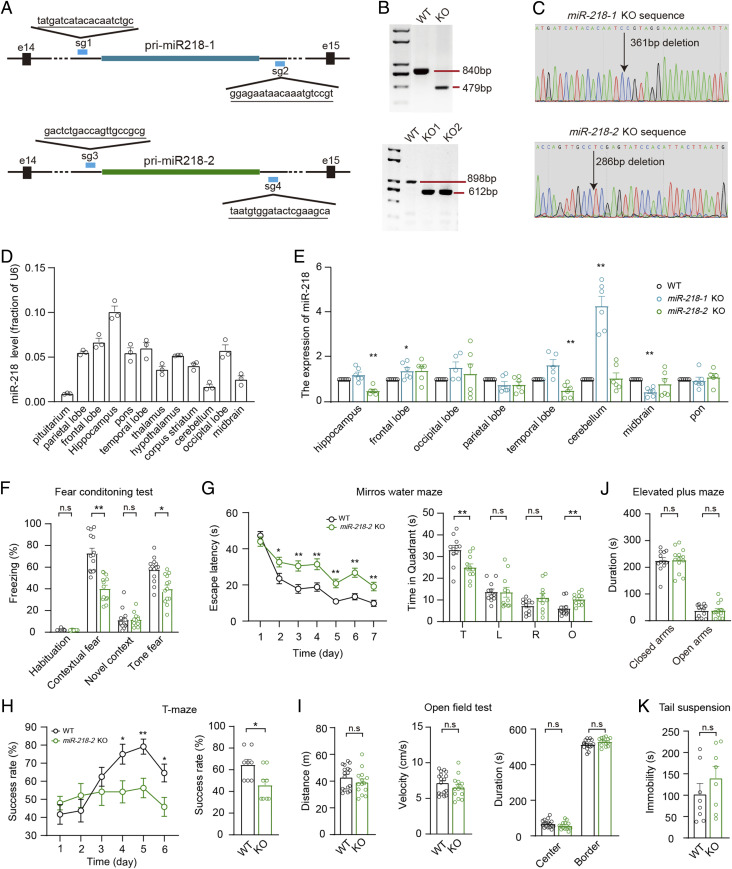
Previously, it has been suggested that the double KO of miR-218-1 and miR-218-2 resulted in severe motoneuron deficits (36), which makes it difficult to investigate the behavioral abnormalities in the mice. To explore the functions of miR-218 in the brain, we used CRISPR-Cas9 technology to generate the miR-218-1 and miR-218-2 knockout mouse strains separately (Fig. 1A and SI Appendix, Fig. S1 A and B). The deletion of miR-218-1 or miR-218-2 in the mice was verified with PCR genotyping (Fig. 1 B and C). In the wild-type (WT) mice, miR-218 was widely distributed in a variety of brain regions, with the highest expression level in the hippocampus and relatively low expression in the pituitarium, cerebellum, and midbrain (Fig. 1D). Hence, miR-218 might be involved in hippocampal functions. We observed that in the miR-218-2 KO mice, the expression of miR-218 was decreased by >50% in the hippocampus and temporal lobe, whereas the miR-218-1 KO mice showed a reduction in the midbrain and fivefold enhancement in the cerebellum (Fig. 1E). This result indicated that the two miR-218 isoforms probably showed a redundant expression pattern.
Fig. 1.
miR-218-2 KO mice show impaired learning and memory. (A) Illustration of sgRNA designed for miR-218-1 and miR-218-2 KO mice. The sgRNA-targeting sequences are underlined. (B, C) Genotyping (B) and Sanger-sequencing (C) analyses of miR-218-1 (Upper) and miR-218-2 (Lower) KO mice. For miR-218-1, expected fragment size is as follows: WT, 840 bp; KO, 479 bp. For miR-218-2, WT, 898 bp; KO, 612 bp. (D) qRT-PCR analysis of miR-218 expression in brain regions of WT mice. n = 3. (E) qRT-PCR analysis of miR-218 expression in brain regions of miR-218-1 KO and miR-218-2 KO mice. n = 6. (F) Percentage of freezing levels in the habituation, training context, novel context, and cued-tone FC test. WT, n = 14; KO, n = 13. (G) Analysis of MWM test. Average time spent to locate the submerged escape platform during the 7 d training session (Left). Average time spent in the four quadrant areas when the platform was absent (Right). T, target; L, left; R, right; O, opposite. n = 11. (H) Success rate of mice entering the right arm in the TM test. n = 8. (I) Total distance (Left), velocity (Middle), and time in the center/border zone (Right) in the OFT. WT, n = 17; KO, n = 14. (J) Total time spent in the closed arms and open arms of the EPM test. WT, n = 12; KO, n = 13. (K) Total immobile time in the TS test. n = 8. Student’s t test. *P < 0.05; **P < 0.001; error bars, SEM.
As the miR-218 expression in the hippocampus was largely reduced in the miR-218-2 KO mice, we investigated the effects of miR-218 on learning and memory using the miR-218-2 KO mice as a model. To evaluate the associative learning ability, we performed the fear conditioning (FC) test, which has been suggested to involve hippocampal functions (37, 38), to analyze the freezing time of the miR-218-2 KO mice. We observed that compared to the WT controls, the KO mice showed a significant attenuation in the freezing ratio in response to the conditioned and cued stimulus (Fig. 1F). We then measured the spatial memory of the mice using the Morris water maze (MWM) paradigm. We found that compared to the WT group, the miR-218-2 KO mice spent a much longer time seeking for the hidden platform during the training phase; moreover, the time spent in the target quadrant in the final test was significantly decreased (Fig. 1G). We also tested reward long-delay learning using the T-maze (TM) test. The result indicated that compared to the WT animals, the miR-218-2 KO mice showed a lower rate of correct alternation of entry into the unbaited arm starting from day four, leading to a significantly lower rewarded alternation rate (Fig. 1H). These deficits in learning and memory were not caused by defective locomotor activity, because the WT and miR-218-2 KO mice showed similar moving velocity and distance during an open field test (OFT) (Fig. 1I).
It has been suggested that insufficient miR-218 expression in the medial prefrontal cortex (mPFC) of mice would promote the susceptibility to depression-like behaviors (21, 22). Although the miR-218 expression was unchanged in the mPFC of the miR-218-2 KO mice (Fig. 1E), we still investigated the neuropsychiatry-related behaviors in the miR-218-2 KO mice using the OFT, elevated plus maze (EPM), and tail suspension (TS) paradigms. The OFT and EPM analyses revealed that the WT and miR-218-2 KO mice spent similar time in the center of the OF and in the open arm of the maze, indicating that the KO animals failed to show an “anxiety-like” phenotype (Fig. 1 I and J). In the TS test, the miR-218-2 KO mice also showed a similar immobility time compared to the WT group, indicating that these animals had normal responses to acute stress (Fig. 1K).
In addition, we investigated the behaviors of miR-218-1 KO mice and found that these mice did not show any significant changes in the learning- or neuropsychiatry-related behaviors compared to the WT controls (SI Appendix, Fig. S1 C–H).
Together, our data indicated that miR-218 was enriched in the hippocampus of the mice, and the deletion of miR-218-2 induced impairments in the learning and memory of the mice while the locomotion and emotion of the animals were unaffected.
miR-218-2 KO Mice Show Impaired Excitatory Synaptic Transmission and Synaptic Plasticity in the Hippocampus.
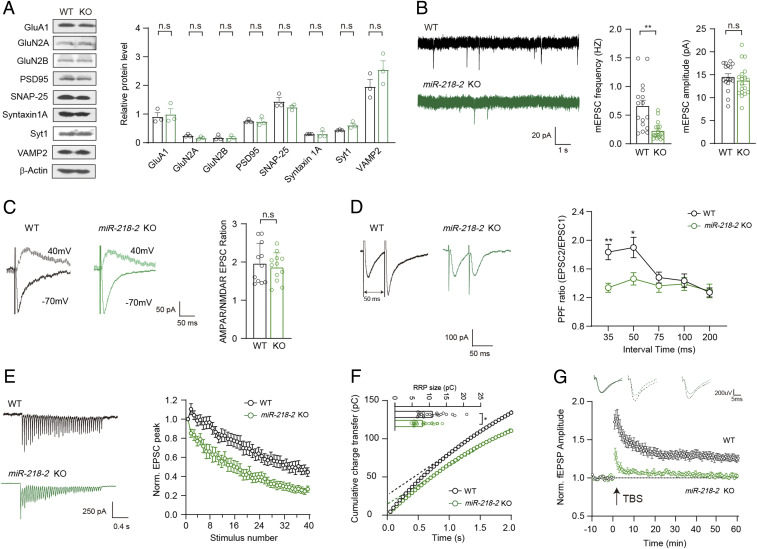
To make clear the molecular mechanisms underlying the defective learning and memory in the miR-218-2 KO mice, we investigated the synaptic functions of the hippocampus in these animals. Immunoblot analysis revealed that the expression of major pre- and postsynaptic proteins directly involved in synaptic transmission, such as Syt1, SNAREs, and subunits of glutamate receptors, were unchanged in the hippocampus of the miR-218-2 KO mice compared to the WT animals (Fig. 2A). We then performed whole-cell patch-clamp recordings in the acutely dissected hippocampal slices of the miR-218-2 KO mice. Analysis of spontaneous SV release in the CA1 region, which was evaluated by miniature excitatory postsynaptic currents (mEPSCs), revealed that compared to the WT slices, the neurons of the miR-218-2 KO mice showed a largely decreased mEPSCs frequency, indicating a significant reduction in the presynaptic SV release rate (Fig. 2B). However, the mEPSCs amplitude was unchanged, indicating that the density of the postsynaptic AMPA receptors (AMPARs) was unaffected. Next, we recorded the action potential (AP)-evoked EPSCs in the Schaffer collateral-CA1 (SC-CA1) synapses of the hippocampus. Analysis of single AP-evoked EPSCs revealed that compared to the WT control, the miR-218-2 KO mice showed a similar ratio of EPSCAMPAR/EPSCNMDAR (Fig. 2C). Combined with the unchanged mEPSCs amplitude, these results indicated that the total synapse number per neuron was probably similar between the WT and KO neurons. However, we observed that the paired-pulse facilitation (PPF) ratio was significantly reduced in the KO neurons when the time interval was no more than 50 ms (Fig. 2D). Thus, we further analyzed the EPSCs evoked by a 2 s 20 Hz train stimulation in the SC-CA1 synapses. The results indicated that consistent with the PPF experiment, the miR-218-2 KO neurons showed a much faster depression in the EPSC amplitude compared to the WT group (Fig. 2E). Based on this data, we estimated the size of the readily releasable SV pool (RRP) of the SC-CA1 synapses and found that the miR-218-2 KO neurons showed a significantly smaller RRP size compared to the WT control (Fig. 2F). To test whether the reduced presynaptic glutamate release contributed to the impairments in the learning and memory in the miR-218-2 KO mice, we assayed the theta burst stimulation (TBS)-induced LTP in the SC-CA1 synapses through recording the field excitatory postsynaptic potentials (fEPSPs). In the slices of WT mice, the fEPSPs were augmented in response to the TBS and such enhancement was maintained for more than 1 h. However, in the miR-218-2 KO mice, the LTP could not be efficiently induced or maintained (Fig. 2G). Thus, the LTP was impaired in the hippocampus of miR-218-2 KO mice.
Fig. 2.
miR-218-2 KO mice show impaired excitatory synaptic transmission and LTP in the hippocampus. (A) Immunoblots (Left) and quantification (Right) of synaptic proteins in WT and miR-218-2 KO hippocampus neurons. n = 3. (B) mEPSCs in acutely dissected hippocampal slices from 4 wk old WT and miR-218-2 KO mice. Sample traces (Left); frequency (Middle); amplitude (Right). WT, n = 16; KO, n = 18. (C) Sample traces (Left) and ratio (Right) of EPSCAMPAR/EPSCNMDAR recorded in hippocampal slices. WT, n = 11; KO, n = 12. (D) Sample traces (Left) and quantitative analysis (Right) of PPF recordings at different intervals. WT, n = 15; KO, n = 12. (E) Sample traces (Left) and amplitude depression analysis (Right) of train-stimulation–evoked EPSCs. WT, n = 26; KO, n = 15. (F) RRP size of WT and KO hippocampal neurons. Cumulative charge transfer curve (Left). Dashed lines represent extrapolation of cumulative charge to the y-axis. RRP size represented by the intercept of the dashed line at the y-axis (Right), n = 21. (G) Representative traces of fEPSPs (Upper) and time course of the fEPSPs amplitude (Lower) before and after LTP induction by a TBS protocol. WT, n = 12; KO, n = 19. Student’s t test. *P < 0.05; **P < 0.001; error bars, SEM.
In summary, our results indicated that the miR-218-2 KO mice showed deficits in the presynaptic glutamate release in the hippocampal neurons, which consequently induced defective LTP that probably contributed to the impaired learning and memory in the miR-218-2 KO mice.
Overexpressing miR-218 in the Hippocampus Can Promote Learning and Memory.
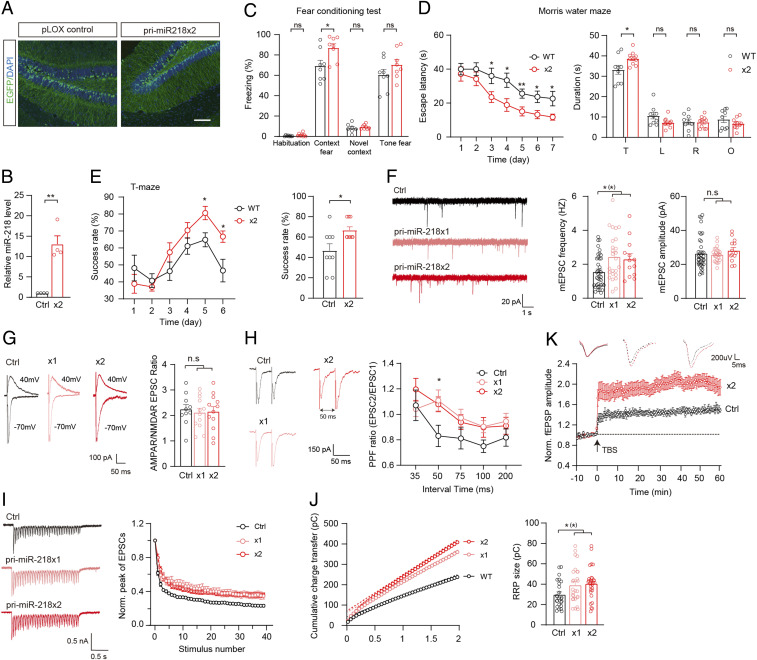
To further verify the effects of miR-218 on the functions of hippocampal neurons, we next up-regulated the expression of miR-218 in the hippocampus and investigated its impact on learning and memory. We overexpressed two copies of the miR-218 precursor (premiR-218 ×2) in the hippocampal dentate gyrus (DG) of juvenile WT mice through bilateral infusion of lentivirus. Enhanced green fluorescence protein (EGFP) was employed as an in-vector marker to indicate the expression of premiR-218 ×2 or a control vector (Fig. 3A and SI Appendix, Fig. S2 A–C). The overexpression of miR-218 was verified by qRT-PCR analysis (Fig. 3B). After the mice became mature, we investigated the behavioral phenotypes of the premiR-218 ×2-expressing animals compared to the control group. In the FC test, the premiR-218 ×2-expressing mice showed an increased freezing ratio in response to the conditioned stimulus (Fig. 3C). In the MWM task, the premiR-218 ×2-expressing mice spent less time overall in finding the escape platform during the training period and spent more time in the target quadrant in the final test (Fig. 3D). In the TM test, the premiR-218 ×2-expressing mice showed a significantly higher rate of correct alternation of entry into the unbaited arm (Fig. 3E). These results indicated that overexpression of miR-218 in the hippocampus could enhance the learning and memory of the animals. In addition, we also tested the locomotion, depression-, and anxiety-like behaviors of the premiR-218 ×2-expressing mice. Similar to the miR-218-2 KO mice, the mice overexpressing miR-218 did not show any significant changes compared to the controls (SI Appendix, Fig. S2 D–F).
Fig. 3.
Overexpression of miR-218 in the hippocampus promotes learning and memory. (A and B) Investigation of miR-218 overexpression in the hippocampus. (A) Sample immunofluorescence images showing the expression of lentivirus-introduced EGFP. (Scale bar, 100 μm.) (B) qRT-PCR analysis of premiR-218 ×2 or control pLOX Syn-DsRed-Syn-GFP expression in the DG. n = 4. (C) Percentage of freezing levels in the habituation, training context, novel context, and cued-tone FC test. n = 8. (D) MWM test. Average time spent to locate the submerged escape platform during the 7 d training session (Left). Average time spent in the quadrant areas when the platform was absent (Right). T, target; L, left; R, right; O, opposite. Control, n = 9; premiR-218 ×2, n = 10. (E) Success rate of mice entering the right arm in the TM test. n = 9. (F) Sample traces (Left), frequency (Middle), and amplitude (Right) of mEPSCs recorded in cultured neurons. Control, n = 39, premiR-218 ×1, n = 25, premiR-218 ×2, n = 13. (G) Sample traces (Left) and ratio (Right) of EPSCAMPAR/EPSCNMDAR. Control, n = 10; ×1, n = 12; ×2, n = 12. (H) Sample traces (Left) and quantitative analysis (Right) of PPF recordings at different time intervals. Control, n = 9; ×1, n = 11; ×2, n = 13. (I) Train-evoked EPSCs. Sample traces (Left) and depression of EPSC amplitude (Right). Control, n = 34; ×1, n = 15; ×2, n = 30. (J) Analysis of RRP size. Control, n = 25; ×1, n = 27; ×2, n = 30. (K) Representative traces of fEPSPs (Upper) and time course of the fEPSPs amplitude (Lower) before and after LTP induction. Control, n = 20; ×2, n = 19. Student’s t test. *P < 0.05; **P < 0.001; error bars, SEM.
To investigate the impact of miR-218 overexpression on the functions of the hippocampal synapse, we overexpressed one or two copies of premiR-218 (premiR-218 ×1, ×2) in cultured hippocampal neurons (SI Appendix, Fig. S2 B and C) and carried out patch-clamp recordings to evaluate neurotransmitter release and synaptic plasticity. Analysis of mEPSCs revealed that compared to the control group, the frequency of mEPSCs was increased in the premiR-218 ×1- and ×2-expressing neurons while the mEPSC amplitude was unchanged (Fig. 3F). Furthermore, in the two miR-218 overexpression groups, analysis of the single AP-evoked EPSCs did not show any significant changes in the ratio of EPSCAMPAR/EPSCNMDAR (Fig. 3G). We also investigated the EPSCs evoked by multiple APs. Analysis of PPF revealed that compared to the control group, the premiR-218 ×1/×2-expressing neurons showed significantly enhanced PPF ratios when the time interval was 50 ms (Fig. 3H). Similarly, analysis of train-stimulation (2 s 20 Hz)–evoked EPSCs revealed that compared to the control group, the diminishing of the EPSC amplitude was obviously decelerated in the premiR-218 ×1/×2-expressing neurons (Fig. 3I). Moreover, the RRP size was increased in these overexpressing neurons (Fig. 3J). These results indicated that miR-218 overexpression could enhance the SV release probability, which is in agreement with our findings in the miR-218-2 KO neurons. Finally, we carried out fEPSP recordings in the SC-CA1 synapses of the mice with the premiR-218 ×2 lentivirus injected into the CA3 region of the hippocampus. We found that compared to the control group, the premiR-218 ×2-expressing mice showed a greater LTP induction following the TBS compared to the control mice. Moreover, this fEPSP enhancement could be maintained for more than 1 h as we observed in the control animals (Fig. 3K). These results indicated that overexpressing miR-218 could generate an enhanced LTP compared to the WT mice and thus might underlie the enhanced learning and memory in the mice.
In summary, combining the results of the miR-218 depletion and overexpression experiments, our data showed that miR-218 could bidirectionally regulate learning and memory in the mice.
Morphological Abnormalities Induced by miR-218-2 KO.
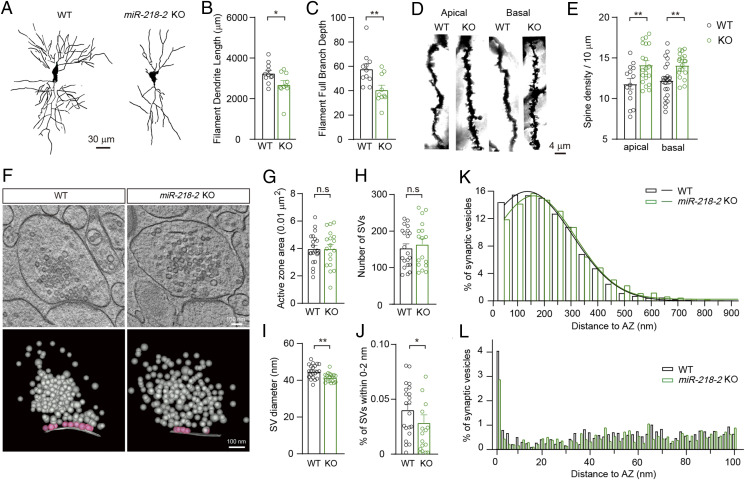
As miR-218 can regulate the development of dopaminergic and motoneurons (33, 34), we were interested to know whether the development of hippocampal neurons and synapses could be regulated by miR-218. To this end, we investigated the in vivo morphology of hippocampal CA1 neurons of the miR-218-2 KO mice using the Golgi staining technique (Fig. 4A). We observed that compared to the WT control, the CA1 neurons of the miR-218-2 KO mice showed a significantly decreased dendrite length and branch depth (Fig. 4 B and C). As there was still ∼50% expression of miR-218 in the KO neurons (Fig. 1E), we injected an miR-218 sponge lentivirus into the hippocampus of the WT mice to achieve a nearly complete depletion of miR-218 (SI Appendix, Fig. S3A). Compared to the WT control mice expressing a control vector, the sponge-expressing mice showed remarkable reductions in dendrite length and branch depth (SI Appendix, Fig. S3 B–D). To monitor the effects of miR-218 at different developmental stages, we labeled the cultured neurons with an EGFP-encoding lentivirus at 0 day in vitro (DIV) and traced the neuronal morphology in a 14 d time period (SI Appendix, Fig. S3E). Compared to the WT controls, the dendrite length and branch depth of the cultured miR-218-2 KO neurons and WT neurons expressing miR-218 sponge became significantly shorter starting from 7 DIV, and the morphological deficits became greater along with the development (SI Appendix, Fig. S3 F and G). Moreover, neurons expressing miR-218 sponge showed significantly aggravated morphological abnormalities compared to the miR-218-2 KO neurons. These results indicated that miR-218 had significant impacts on the development of the hippocampal neurons.
Fig. 4.
Morphological changes of neurons and synapses induced by miR-218-2 KO. (A) Representative Golgi-stained CA1 pyramidal neurons showing decreased total dendrite length and branch depth in miR-218-2 KO mice. (Scale bar, 30 μm.) (B, C) Average dendrite length (B) and dendritic complexity (C) per neuron. WT, n = 11; miR-218-2 KO, n = 10. (D) Representative images of dendritic branches from Golgi-stained CA1 pyramidal neurons. (Scale bar, 4 μm.) (E) Spine density of the dendrites of the pyramidal neurons in the hippocampal CA1 region. Apical: WT, n = 14; KO, n = 20. Basal: WT, n = 24; KO, n = 19. (F) Sample electron tomography images of presynaptic boutons. ET subvolumes of WT and miR-218-2 KO synapses (Upper). 3D models of synaptic profiles including orthogonal views of SV distribution (Lower). (Scale bar, 100 nm.) (G–J) Quantification of AZ area (G), SV number (H), SV diameter (I), and the number of SVs within 0 to 2 nm to the AZ (J). (K, L) Spatial distribution of SVs with 900 nm (K) and 100 nm (L) to the AZ. (G–L) WT, n = 21; KO, n = 16. Student’s t test. *P < 0.05; **P < 0.001; error bars, SEM.
We then investigated the density of dendritic spines in the Golgi-stained miR-218-2 KO CA1 neurons (Fig. 4D). Compared to the WT group, the densities of the apical and basal dendritic spines were both significantly increased in the miR-218-2 KO CA1 neurons (Fig. 4E). Combining this data with the result of dendrite-length analysis, the WT and miR-218-2 KO neurons showed similar total numbers of dendritic spines, which is in agreement with our electrophysiological observations that the amplitude of evoked EPSCs was unaffected by miR-218.
We also carried out electron microscopy and three-dimensional (3D) tomography analyses to investigate the morphology of single presynaptic boutons of the miR-218-2 KO neurons (Fig. 4F). The results indicated that compared to the WT control, the miR-218-2 KO neurons showed a similar active zone (AZ) area and total SV number, while the SV size was reduced (Fig. 4 G–I). We then analyzed the distribution of SVs. We found that compared to the WT control, the number of tightly docked or primed SVs (∼0 to 2 nm to the AZ membrane) was significantly reduced in the miR-218-2 KO synapses (Fig. 4J). Moreover, the overdistribution of SVs was obviously shifted away from the AZ (Fig. 4 K and L). Hence, the SV transportation could be decelerated in the miR-218-2 KO neurons, which is in agreement with our electrophysiological finding that the SV-release probability was reduced in these neurons.
Together, our results indicated that the miR-218-2 KO mice showed obvious deficits in neuronal and synaptic morphology, which support our electrophysiological results.
Identification of C3 as a Main Target Gene of miR-218 in Hippocampal Neurons.
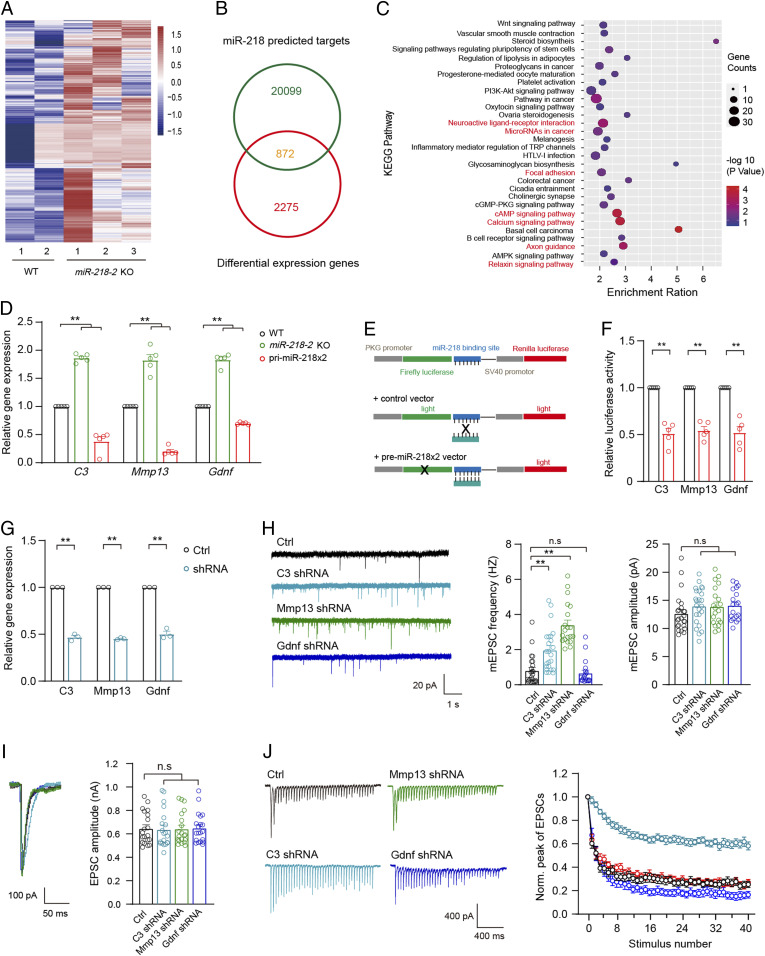
In the next step, we aimed to make clear the downstream mechanism by which miR-218 regulated the learning and memory in the mice. MiRNAs usually have effects through downregulating the mRNA translation of their target genes. To identify the target genes of miR-218 in hippocampal neurons, we carried out RNA-seq analysis in the hippocampus of miR-218-2 KO mice and found 2,275 up-regulated genes compared to the WT control (Fig. 5A). By searching several target-gene-predicting databases (see Materials and Methods), we found that of the 2,275 genes, 872 could potentially be the targets of miR-218 (Fig. 5B). To elucidate the functional roles of these differentially expressed genes, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (SI Appendix, Table S1) using the DAVID online analysis database and selected seven pathways that were closely related to the functions of neurons, miRNAs, and synapses for further analysis (Fig. 5C). We verified the changes in the expression of genes involved in these pathways using qRT-PCR analysis and determined that 23 genes were significantly up-regulated at the mRNA level. We then carried out the luciferase reporter assay in the Hek293 cells to test the direct suppression of miR-218 on the 23 genes and eventually identified 3 highly potential genes, including C3, Mmp13 (matrix metallopeptidase 13), and Gdnf (glial cell line-derived neurotrophic factor) (Fig. 5 D–F). Previously, these genes have been suggested to be involved in Alzheimer’s disease and spatial memory (39–41). We investigated the effects of these genes on the synaptic transmission and synaptic plasticity of cultured hippocampal neurons using the lentivirus-based short hairpin RNA (shRNA) knock down (KD) approach (Fig. 5G). The results indicated that compared to the WT group, the mEPSCs frequency was increase in the C3 and Mmp13 KD neurons but remained unchanged in the Gdnf KD neurons. Moreover, the mEPSC amplitude was unchanged in all groups (Fig. 5H). The analysis of single AP-evoked EPSCs revealed that compared to the control group, none of the three KD neuron groups showed significant changes in the amplitude of EPSC (Fig. 5I). However, during the 2 s 20 Hz train stimulation, the C3 KD neurons showed a significantly decelerated EPSC depression compared to the control group, whereas the KD of Gdnf or MMP13 did not show any obvious changes (Fig. 5J). These results indicated that of the three potential target genes, the KD of C3 could induce electrophysiological phenotypes similar to the miR-218 overexpressing neurons. Moreover, previously it has been reported that enhanced C3 activity could induce defective dendritic morphology in cortical neurons (39), which is consistent with our observations in the miR-218–deficient hippocampal neurons. We therefore concluded that C3 was likely a main target gene of miR-218 to regulate the presynaptic SV release in hippocampal neurons.
Fig. 5.
Identification of C3 as a target gene of miR-218 in hippocampal neurons. (A–C) RNA-seq analysis of gene expression profiles in miR-218-2 KO hippocampal neurons. (A) Heat map of differentially expressed genes (false discovery rate no. 0.05, n = 2,275). (B) 872 genes are overlapped in the differentially expressed genes and the predicted miR-218 target genes. (C) KEGG analysis of the 872 genes. Red colored pathways were selected for further analysis. (D) qRT-PCR analysis of the expression of candidate genes in miR-218 overexpression and KO hippocampal neurons. n = 5. (E) Illustration of the reporter luciferase assay for the candidate target genes. (F) Relative luciferase activity by the candidate target genes. n = 5. (G) qRT-PCR analysis of the efficiency of C3, Mmp13, and Gdnf shRNAs. n = 3. (H) Sample traces (Left), frequency (Middle), and amplitude (Right) of mEPSCs recorded in cultured hippocampal neurons infected with lentivirus encoding the shRNAs for C3, Mmp13, or Gdnf. Control, n = 24; C3 shRNA, n = 24; Mmp13 shRNA, n = 20; Gdnf shRNA, n = 19. (I) Sample traces (Left) and amplitude of AP-evoked EPSCs. n = 20. (J) Sample traces (Left) and amplitude depression analysis (Right) of train-stimulation–evoked EPSCs. Control, n = 21; C3 shRNA, n = 19; Mmp13 shRNA, n = 17; Gdnf shRNA, n = 14. Student’s t test. *P < 0.05; **P < 0.001; error bars, SEM.
Modulating the C3 Activity Can Reverse the Phenotypes of miR-218-2 KO Neurons.
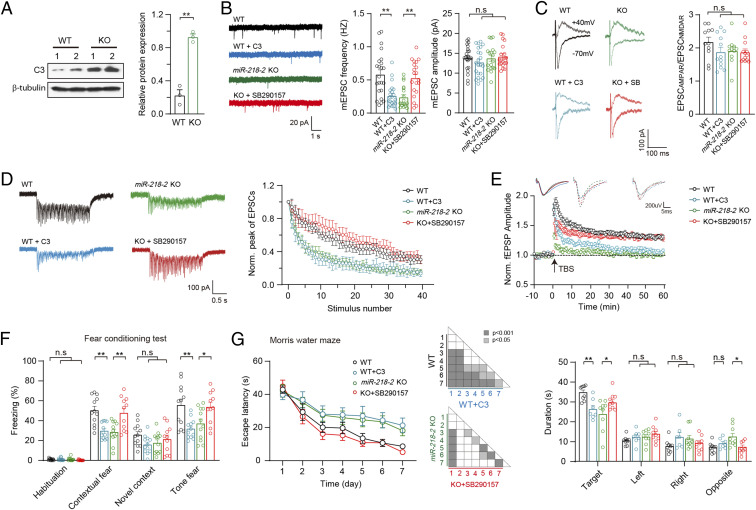
We further investigated the role of C3 in miR-218–dependent synaptic functions and learning-related behaviors. Immunoblot analysis confirmed that the protein expression of C3 was significantly increased in the miR-218-2 KO neurons (Fig. 6A). We infused the purified C3 protein into the hippocampal CA1 of the WT mice or SB290157, a C3a receptor (C3aR)-specific inhibitor, into the miR-218-2 KO mice and investigated the in-vivo morphology of hippocampal CA1 neurons 30 min after the drug treatment using the Golgi staining technique. We observed that compared to the untreated WT and KO neurons, the treatments of C3 and SB290157 failed to induce any significant changes in either the dendrite morphology or the dendritic-spine density (SI Appendix, Fig. S4).
Fig. 6.
C3 is a target gene of miR-218 to regulate presynaptic functions and learning behaviors in mice. (A) Immunoblots (Left) and quantification (Right) of C3 in WT and miR-218-2 KO neurons. n = 3. (B) Sample traces (Left), frequency (Middle), and amplitude (Right) of mEPSCs recorded in hippocampal slices of WT mice treated with C3 peptide, or miR-218-2 KO slices treated with SB290157. WT, n = 23; WT + C3, n = 22; miR-218-2 KO, n = 19; KO + SB290157, n = 20. (C) Sample traces (Left) and ratio (Right) of EPSCAMPAR/EPSCNMDAR recorded in acute slices. WT, n = 11; WT + C3, n = 12; miR-218-2 KO, n = 11; KO + SB290157, n = 13. (D) Train-stimulation–evoked EPSCs. WT, n = 12; WT + C3, n = 15; miR-218-2 KO, n = 13; KO + SB290157, n = 14. (E) Sample traces of fEPSPs (Upper) and time course of the fEPSPs amplitude (Lower) before and after LTP induction. WT, n = 14; WT + C3, n = 24; miR-218-2 KO, n = 19; KO + SB290157, n = 31. (F) Percentage of freezing ratio in the FC test. WT, n = 12; C3, n = 12; miR-218-2 KO, n = 12; KO + SB290157, n = 11. (G) Analysis of MWM test. Average time spent to locate the submerged escape platform during the 7 d training session (Left); ANOVA. Average time spent in the four quadrant areas when the platform was absent (Right). WT, n = 9; C3, n = 7; miR-218-2 KO, n = 9; KO + SB290157, n = 9. Student’s t test. *P < 0.05; **P < 0.001; error bars, SEM.
We then carried out brain-slice patch-clamp recordings in WT mice treated with C3 and miR-218-2 KO mice treated with SB290157. Analysis of mEPSCs revealed that compared to the untreated WT controls, the WT neurons treated with C3 showed a significantly reduced mEPSC frequency, whereas in the miR-218-2 KO neurons, the treatment of SB290157 induced an obvious enhancement (Fig. 6B). Moreover, in all groups of neurons, the mEPSC amplitude was similar. Further analysis of single AP-evoked EPSCs revealed that compared to their own control, the treatments of C3 and SB290157 did not induce any significant changes in the ratio of EPSCAMPAR/EPSCNMDAR (Fig. 6C). We also investigated the EPSCs evoked by the 2 s 20 Hz train stimulation (Fig. 6D). Compared to the untreated WT neurons, the WT neurons treated with C3 showed an accelerated depression of EPSC amplitude, whereas compared to the untreated miR-218-2 KO neurons, the treatment of SB290157 induced a slower EPSC depression. To investigate whether C3 could affect the TBS-induced LTP, we recorded fEPSPs in acutely dissected hippocampal slices perfused with C3 or its inhibitor. We observed that compared to the untreated WT group, the WT slices treated with C3 showed a significantly attenuated LTP, whereas compared to the untreated miR-218-2 KO group, the KO slices perfused with SB290157 showed an obvious enhancement (Fig. 6E).
Next, we carried out behavioral tasks to investigate the impact of C3 on the learning-related behaviors. We bilaterally infused C3 into the hippocampal DG of the WT mice or SB290157 into the miR-218-2 KO mice during the training period of the FC and MWM tests. In the FC test, compared to the untreated WT group, the C3-treated WT mice showed a decreased freezing ratio in response to the conditioned and cued stimulus, whereas compared to the untreated miR-218-2 KO control, the SB290157-treated KO mice showed an increased freezing ratio (Fig. 6F). Furthermore, in the MWM task, compared to the untreated WT control, the C3-treated WT mice showed a longer time to find the escape platform, a shorter time of staying on the platform, and the target quadrant. Compared to the untreated miR-218-2 KO control mice, the SB290157-treated KO mice showed a shorter time to find the escape platform and a longer staying time on the platform and in the target quadrant (Fig. 6G).
Together, our results indicated that C3 mediated the miR-218–dependent presynaptic functions and learning behaviors in the mice and thus was a main target gene of miR-218 in mouse hippocampal neurons. We note that although the short-term treatment of C3 and SB290157 did not change the morphology of the neurons, it is possible that C3 could have long-term effects on the morphology of the neurons, which makes further contributes to the learning and memory of the mice.
Discussion
miR-218 has been found to show abnormal expression in a variety of cognition-related neurodegenerative and neuropsychiatric disorders. However, it remains unclear whether miR-218 plays a direct role in the cognitive defects involved in these diseases. In the present study, using the mouse hippocampus as a model, we demonstrated that modulating the expression of miR-218 in the hippocampus could bidirectionally regulate the learning and memory of the mice. We investigated the morphology and synaptic transmission of the hippocampal neurons of miR-218-2 KO mice and found that miR-218-2 deficiency resulted in abnormalities in dendritic morphology, presynaptic SV release, and synaptic plasticity, which probably underlie the impairments in the learning and memory of miR-218-2 KO mice.
miR-218 was initially regarded as a tumor-related miRNA that could be used as a prognostic biomarker of multiple cancers (42–44). Later, several studies suggested that miR-218 probably played important roles in the CNS. For instance, a low expression of miR-218 in the mPFC of mice could confer susceptibility to depression-like behaviors (21, 22). In the present study, the miR-218-1 KO and miR-218-2 KO mice did not show any significant abnormalities in depression-like behaviors, probably because the miR-218 expression level was not decreased in the mPFC of the miR-218-1 or miR-218-2 KO mice (Fig. 1E). In addition, it has been reported that the miR-218 expression was down-regulated in the hippocampus in the rat model of temporal-lobe epilepsy and the patients suffering from hippocampal sclerosis (45, 46). In support of these reports, our study indicated that miR-218 deficits might exacerbate the cognitive deficits involved in the diseases.
As miRNA has ∼20 nucleotides, it can target a large number of genes that have similar binding sequences and consequently show divergent functions in the body. For instance, in the midbrain, miR-218 targets the Ebf3 gene to regulate the development of dopaminergic neurons (33), whereas in the periphery, miR-218 can target IKK-β to contribute to diabetic nephropathy (47). Previously, it has been reported that in hippocampal neurons, miR-218 can target the AMPAR subunit GluA2, which participates in the homeostatic plasticity (48). However, in the present study, we observed that the amplitude of mEPSCs was only slightly changed in the miR-218-2 KO and overexpressing neurons. In contrast, the frequency of mEPSCs was remarkably affected by the miR-218 expression. Combined with our observations of the unchanged PSD95 protein level and total synapse number, we concluded that miR-218 mainly functioned to regulate the probability of presynaptic SV release but not the density of postsynaptic AMPARs. The differential findings between the two studies might result from the approaches employed to regulate the miR-218 expression; the former chose to overexpress the miR-218-5P fragment, while we employed the full length miR-218 precursor (premiR-218) and miR-218 KO mice.
It is possible that at different developmental stages or under pathological conditions, miR-218 might function through different target genes. In the present study, we compared the outcome of searching target-predicting databases with the KEGG results of RNA-seq analysis and eventually determined that C3 was a major target gene of miR-218 for regulating the synaptic functions and learning related behaviors of mice. In recent years, more and more evidence has shown that the complement system showed a close relationship with the CNS (49–51). C3 is the central component of the immune system, of which all three complement cascade pathways required the activation of C3. C3 could be cleaved into C3a, C3b, iC3b, and C3d. Each product has its own receptors and thus functions differentially. The C3 KO mice have been demonstrated to show improved spatial and contextual memory (52) and exhibited a higher ratio of PPF and a slower decay of fEPSP magnitude compared to the WT mice (53). C3 has also been reported to inhibit neurite outgrowth (54). In the present study, our observations in the miR-218-2 KO mice are in agreement with these findings. In addition, C3 has been suggested to be a cerebrospinal fluid biomarker for amyotrophic lateral sclerosis (ALS) (55), in which C3 is up-regulated in motoneurons and neuromuscular junctions, whereas miR-218 is essential for motoneuron differentiation and lack of miR-218 is likely related with ALS. These studies also support that C3 is the intermediate of miR-218 signaling. However, the other two candidate target genes revealed by our experiments, Mmp13 and Gdnf, have been reported to be involved in cognitive deficits in Alzheimer’s disease (40, 41). Hence, it is possible that in Alzheimer’s disease, miR-218 can contribute to the learning and memory deficits through targeting Mmp13 and/or Gdnf via some unidentified downstream pathways. Furthermore, as miR-218 and C3 are also highly expressed in glial cells such as astrocytes and microglia (31, 32, 56), we note that in addition to functioning in neurons, miR-218-C3 could also act in glial cells and potentially other types of cells to contribute to the regulation of learning and memory.
Taken together, our study indicated that miR-218 deficiency in the hippocampus resulted in impairments in excitatory synaptic transmission and synaptic plasticity, which consequently induced defects in the learning and memory of the mice. We further elucidated that C3 was likely a major target gene of miR-218 in the hippocampus. Our results revealed a mechanism by which the miR-218–related neurodegenerative and neuropsychiatric disorders generate cognitive symptoms.
Materials and Methods
Animals.
miR-218-1 and miR-218-2 knockout mice were generated using CRISPR-Cas9 targeting, as previously described (36, 57). The single-guide RNAs (sgRNAs) specific for deletion of the precursor sequence of miR-218-1 and miR-218-2 are listed in SI Appendix, Table S2. sgRNAs were transcribed in vitro using the MEGAshortscript T7 kit (Life Technologies). Mouse oocytes were microinjected with Cas9 mRNA (invitrogen):gRNA:gRNA mixtures (100 ng/μL:50 ng/μL:50 ng/μL) and were reimplanted into C57BL/6 pseudopregnant females. Successful deletions were detected by PCR genotyping of mouse tails and confirmed by Sanger sequencing. miR-218-1 +/− and miR-218-2 +/− founders were maintained and used for breeding. PCR primers for miR-218-1 and miR-218-2 KO mouse genotyping are listed in SI Appendix, Table S3.
Stereotactic injections into the DG or the CA3 of adult mice were performed as described (58). C57BL/6 mice that were 4 wk old were bilaterally injected with 1 μL lentivirus. Before loading the virus into a glass micropipette (Sutter Instrument), the inside was coated with mineral oil. The stereotactic coordinates for dorsal DG injection were anterior/posterior: −1.8, medial/lateral: 1.8, and dorsal/ventral: −1.8. The stereotactic coordinates for CA3 injection were anterior/posterior: −1.8, medial/lateral: 2.4, and dorsal/ventral: −2.0. The total injection volume was 1 μL, injected at a rate of 0.1 μL/min.
Double cannula was transplanted into mice as described (59). All mice were implanted with dual cannula (RWD) guides for microinjections. The tips of the cannulae were at the DG region of 2-mo-old mice: 2.0 mm rostral to bregma, 2.0 mm ventral from the dura matter, and ±1.5 mm relative to the midline. Mice were allowed to recover for 2 wk after surgery before experimentation. The total microinjection volume was 0.5 μL, injected at a rate of 0.1 μL/min. The concentration for C3 was 25 μg/mL, and the concentration for SB290157 was 30 μM. In the FC and MWM experiments, the mice received the drug treatment each day, and functional experiments were performed 30 min after the microinjection.
Plasmids.
For overexpression of miR-218, premiR-218 was amplified by PCR and subcloned into a bicistronic lentiviral vector system, pLox Syn-DsRed-Syn-GFP. The DsRed coding sequence was replaced with premiR-218. The adjacent premiR-218 DNA was linked by a random sequence: GGAACCGGG-TACCGG-AACTGG. To generate the miR-218 sponge, oligonucleotides including miR-218 binding sites were annealed as double-stranded DNA and cloned to lentiviral vectors pLVX that contained EGFP. The sponge sequence is as follows: ACATGGTTACTGGAAGCACAAGACTACATGGTTAACGT. AAGCACAATCAGACA-TGGTTATTCGAAGCACAAGACTACATGGTTACCGTAAGCACAAACGTACATGGT-TATGCAAAGCACAAGTCAACATGGTTAAGAGAAGCACAA. The predicted regulation of C3, Mmp13, and Gdnf by miR-218 was investigated using the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega). Firefly luciferase was the primary reporter gene, and humanized Renilla luciferase-neomycin resistance cassette (hRluc -neo) was used as a control reporter for the normalization of gene expression and stable-cell-line selection. The 3′ UTR of the target genes was amplified by PCR and inserted at the sites 3′ of the firefly luciferase gene. For the knockdown of C3, Mmp13, and Gdnf, shRNA vectors including the shC3#3, shMmp13#4, and shGdnf#11 were ordered from the MISSION shRNA Library. pLKO.1-shScrambled was used as a control.
Mouse Primary Neuronal Culture.
Mouse hippocampal neurons were dissected from newborn WT and homozygous miR-218-2 KO mice and incubated in 0.25% trypsin-ethylenediaminetetraacetic acid (Life Technologies) for 15 min at 37 °C. After washing with Hank's Buffered Salt Solution plus 5 mM Hepes (Life Technologies), 20 mM d-glucose, and 2% fetal bovine serum (FBS) (Gibco), the neurons were mechanically dissociated in culture medium and plated on poly-d-lysine–coated glass coverslips at a density of 50,000 to 100,000 cells/cm2. Cells were grown in Neurobasal-A medium (Life Technologies) supplemented with 2% B-27 (Life Technologies) and 2 mM glutamax (Life Technologies). Cultures were maintained at 37 °C in a 5% CO2-humidified incubator.
All other materials and methods used in this study are described in detail in SI Appendix, Extended Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Songhai Shi, Yi Zhong, and Zhongfu Shen (Tsinghua University) for technical help. We thank all members in the laboratory for helpful discussion and technical assistance. This work was supported by National Natural Science Foundation of China (Grant No. 31830038 and 31771482) and National Key R&D Program of China (Grant No. 2016YFA0101900).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021770118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Griggs E. M., Young E. J., Rumbaugh G., Miller C. A., MicroRNA-182 regulates amygdala-dependent memory formation. J. Neurosci. 33, 1734–1740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin Q., et al., The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat. Neurosci. 14, 1115–1117 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Vetere G., et al., Selective inhibition of miR-92 in hippocampal neurons alters contextual fear memory. Hippocampus 24, 1458–1465 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Zovoilis A., et al., microRNA-34c is a novel target to treat dementias. EMBO J. 30, 4299–4308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen K. F., Sakamoto K., Wayman G. A., Impey S., Obrietan K., Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One 5, e15497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott H. L., et al., MicroRNA-132 regulates recognition memory and synaptic plasticity in the perirhinal cortex. Eur. J. Neurosci. 36, 2941–2948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J., et al., A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel D. P., MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 9.He L., Hannon G. J., MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Konopka W., Schütz G., Kaczmarek L., The microRNA contribution to learning and memory. Neuroscientist 17, 468–474 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Marcuzzo S., et al., Up-regulation of neural and cell cycle-related microRNAs in brain of amyotrophic lateral sclerosis mice at late disease stage. Mol. Brain 8, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park C. S., Tang S. J., Regulation of microRNA expression by induction of bidirectional synaptic plasticity. J. Mol. Neurosci. 38, 50–56 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Ryan M. M., et al., Temporal profiling of gene networks associated with the late phase of long-term potentiation in vivo. PLoS One 7, e40538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wibrand K., et al., Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur. J. Neurosci. 31, 636–645 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Q. H., et al., miR-26a and miR-384-5p are required for LTP maintenance and spine enlargement. Nat. Commun. 6, 6789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konopka W., et al., MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30, 14835–14842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remenyi J., et al., miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS One 8, e62509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y., et al., Role of miR-34c in the cognitive function of epileptic rats induced by pentylenetetrazol. Mol. Med. Rep. 17, 4173–4180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong Y. S., et al., Opposite effects of two estrogen receptors on tau phosphorylation through disparate effects on the miR-218/PTPA pathway. Aging Cell 14, 867–877 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You X., et al., Investigating aberrantly expressed microRNAs in peripheral blood mononuclear cells from patients with treatment-resistant schizophrenia using miRNA sequencing and integrated bioinformatics. Mol. Med. Rep. 22, 4340–4350 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres-Berrío A., et al., MiR-218: A molecular switch and potential biomarker of susceptibility to stress. Mol. Psychiatry 25, 951–964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres-Berrío A., et al., DCC confers susceptibility to depression-like behaviors in humans and mice and is regulated by miR-218. Biol. Psychiatry 81, 306–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurkiewicz M., et al., Integration of postmortem amygdala expression profiling, GWAS, and functional cell culture assays: Neuroticism-associated synaptic vesicle glycoprotein 2A (SV2A) gene is regulated by miR-133a and miR-218. Transl. Psychiatry 10, 297 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Sun Y., Chen J., Identification of critical genes and miRNAs associated with the development of Parkinson’s disease. J. Mol. Neurosci. 65, 527–535 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Tie J., et al., MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 6, e1000879 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickson B. J., Molecular mechanisms of axon guidance. Science 298, 1959–1964 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Brose K., et al., Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795–806 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Sugishima M., et al., Crystal structure of rat heme oxygenase-1 in complex with heme. FEBS Lett. 471, 61–66 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Yachie A., et al., Oxidative stress causes enhanced endothelial cell injury in human heme oxygenase-1 deficiency. J. Clin. Invest. 103, 129–135 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Z. M., et al., Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene 36, 2577–2588 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song L., et al., miR-218 inhibits the invasive ability of glioma cells by direct downregulation of IKK-β. Biochem. Biophys. Res. Commun. 402, 135–140 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Cheng M. W., Wang L. L., Hu G. Y., Expression of microRNA-218 and its clinicopathological and prognostic significance in human glioma cases. Asian Pac. J. Cancer Prev. 16, 1839–1843 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Baek S., Choi H., Kim J., Ebf3-miR218 regulation is involved in the development of dopaminergic neurons. Brain Res. 1587, 23–32 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Thiebes K. P., et al., miR-218 is essential to establish motoneuron fate as a downstream effector of Isl1-Lhx3. Nat. Commun. 6, 7718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoye M. L., et al., Motor neuron-derived microRNAs cause astrocyte dysfunction in amyotrophic lateral sclerosis. Brain 141, 2561–2575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amin N. D., et al., Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 350, 1525–1529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pignataro A., Middei S., Borreca A., Ammassari-Teule M., Indistinguishable pattern of amygdala and hippocampus rewiring following tone or contextual fear conditioning in C57BL/6 mice. Front. Behav. Neurosci. 7, 156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calandreau L., et al., Extracellular hippocampal acetylcholine level controls amygdala function and promotes adaptive conditioned emotional response. J. Neurosci. 26, 13556–13566 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lian H., et al., NFκB-activated astroglial release of complement C3 compromises neuronal morphology and function associated with Alzheimer’s disease. Neuron 85, 101–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu B. L., et al., MMP13 inhibition rescues cognitive decline in Alzheimer transgenic mice via BACE1 regulation. Brain 142, 176–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petukhova E. O., et al., Effects of transplanted umbilical cord blood mononuclear cells overexpressing GDNF on spatial memory and hippocampal synaptic proteins in a mouse model of Alzheimer’s disease. J. Alzheimers Dis. 69, 443–453 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Jin J., Cai L., Liu Z. M., Zhou X. S., miRNA-218 inhibits osteosarcoma cell migration and invasion by down-regulating of TIAM1, MMP2 and MMP9. Asian Pac. J. Cancer Prev. 14, 3681–3684 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Yang Y., et al., MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol. Cancer 16, 141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng M., et al., miR-218 suppresses gastric cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis in a feedback loop. Cancer Lett. 403, 175–185 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Gorter J. A., et al., Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol. Dis. 62, 508–520 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Kaalund S. S., et al., Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia 55, 2017–2027 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Li M., et al., miR-218 regulates diabetic nephropathy via targeting IKK-β and modulating NK-κB-mediated inflammation. J. Cell. Physiol. 235, 3362–3371 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Rocchi A., et al., Neurite-enriched MicroRNA-218 stimulates translation of the GluA2 subunit and increases excitatory synaptic strength. Mol. Neurobiol. 56, 5701–5714 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Boulanger L. M., Immune proteins in brain development and synaptic plasticity. Neuron 64, 93–109 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Lui H., et al., Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell 165, 921–935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coulthard L. G., Hawksworth O. A., Woodruff T. M., Complement: The emerging architect of the developing brain. Trends Neurosci. 41, 373–384 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Shi Q., et al., Complement C3-deficient mice fail to display age-related hippocampal decline. J. Neurosci. 35, 13029–13042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Alcazar M., et al., Altered cognitive performance and synaptic function in the hippocampus of mice lacking C3. Exp. Neurol. 253, 154–164 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Peterson S. L., Nguyen H. X., Mendez O. A., Anderson A. J., Complement protein C3 suppresses axon growth and promotes neuron loss. Sci. Rep. 7, 12904 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ganesalingam J., et al., Combination of neurofilament heavy chain and complement C3 as CSF biomarkers for ALS. J. Neurochem. 117, 528–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei Y., et al., The complement C3-C3aR pathway mediates microglia-astrocyte interaction following status epilepticus. Glia 69, 1155–1169 10.1002/glia.23955 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H., et al., One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siegert S., et al., The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 18, 1008–1016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amat J., et al., Medial prefrontal cortex determines how stressor controllability affects behavior and dorsal raphe nucleus. Nat. Neurosci. 8, 365–371 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.