Significance
At the mid-blastula transition, rapid cell divisions are slowed down and global activation of zygotic genes occur, preparing the early embryo for differentiation and further development. Using live imaging of embryos with reduced ploidy and cell cycle lengths, we examined the effects of the nuclear content to cytoplasm (N/C) ratio on the timing of zygotic genome activation. While some genes are predominantly affected by the N/C ratio–dependent changes in cell cycle durations, other genes demonstrate direct regulation by the N/C ratio through changes in the transcriptional kinetics and in the probability of gene activation.
Keywords: transcription, ZGA, cell cycle, N/C ratio, mid-blastula transition
Abstract
Early embryos must rapidly generate large numbers of cells to form an organism. Many species accomplish this through a series of rapid, reductive, and transcriptionally silent cleavage divisions. Previous work has demonstrated that the number of divisions before both cell cycle elongation and zygotic genome activation (ZGA) is regulated by the ratio of nuclear content to cytoplasm (N/C). To understand how the N/C ratio affects the timing of ZGA, we directly assayed the behavior of several previously identified N/C ratio–dependent genes using the MS2-MCP reporter system in living Drosophila embryos with altered ploidy and cell cycle durations. For every gene that we examined, we found that nascent RNA output per cycle is delayed in haploid embryos. Moreover, we found that the N/C ratio influences transcription through three overlapping modes of action. For some genes (knirps, fushi tarazu, and snail), the effect of ploidy can be primarily attributed to changes in cell cycle duration. However, additional N/C ratio–mediated mechanisms contribute significantly to transcription delays for other genes. For giant and bottleneck, the kinetics of transcription activation are significantly disrupted in haploids, while for frühstart and Krüppel, the N/C ratio controls the probability of transcription initiation. Our data demonstrate that the regulatory elements of N/C ratio–dependent genes respond directly to the N/C ratio through multiple modes of regulation.
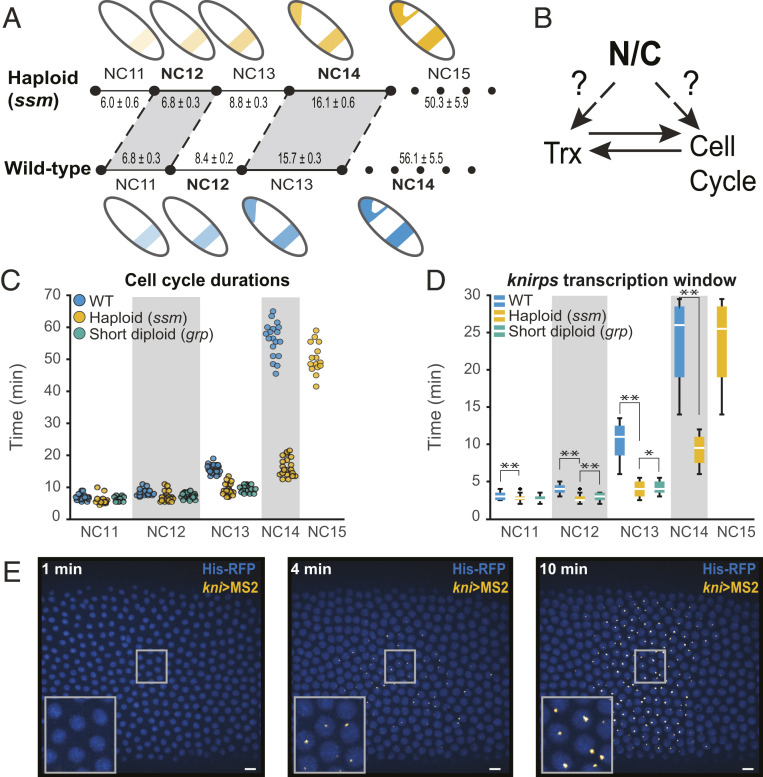
The early embryo of many fast, externally developing species is largely transcriptionally silent during the rapid cleavage stage preceding a developmental transition known as the mid-blastula transition (MBT) (1, 2). At the MBT, the cell cycle slows, and large-scale zygotic genome activation (ZGA) occurs (3–10). The timing of both cell cycle slowing and ZGA are controlled by the ratio of nuclear material, likely DNA, to cytoplasm (N/C ratio), as seen from previous studies that altered this ratio through manipulations in ploidy, injection of exogenous DNA, removal of cytoplasm, or changes in cell size (Fig. 1A) (1, 11–18).
Fig. 1.
The N/C ratio regulates cell cycle and transcription duration. (A) Cell cycle elongation and transcription activation (as illustrated by a cartoon of the kni expression pattern) are both delayed by a reduction of the N/C ratio. Haploid (ssm) embryos undergo one additional fast cell cycle to restore the correct N/C ratio before slowing, and all previous NCs are correspondingly shortened. Transcription is similarly delayed. The mean cell cycle durations ± the SEM are given (in minutes) for both genotypes. (B) Since the N/C ratio affects both cell cycle and transcription, it is difficult to disentangle which event is upstream or if both sense the N/C ratio independently. (C) Scatterplot of cell cycle duration from WT (blue), haploid (yellow), and short-cycle diploid (green) embryos illustrate how ploidy affects the length of interphase and therefore the maximum potential transcriptional window. The number of embryos analyzed in WT, haploid (ssm), and short-cycle diploid (grp) in each NC is as follows: NC11 [26, 20, 18], NC12 [30, 25, 18], NC13 [30, 25, 18], NC14 [24, 26, N/A], and NC15 [N/A, 16, N/A]. (D) Boxplots showing that kni(5′+int)>MS2 transcription duration is longer in WT than haploid and short-cycle diploid embryos. Boxplots show minimum (10%), lower (25%), median, upper (75%), and maximum (90%) quantiles. Outliers are shown as “+.” *P < 0.05 and **P < 0.005 from Student’s t test. 47 NC11, 184 NC12, 658 NC13, and 1051 NC14 nuclei from six replicate kni(5′+int)>MS2 WT embryos; 42 NC11, 121 NC12, 393 NC13, 755 NC14, and 1,218 NC15 nuclei from four replicate kni(5′+int)>MS2 haploid embryos; and 25 NC11, 68 NC12, and 267 NC13 nuclei from four replicate kni(5′+int)>MS2 short-cycle diploid embryos were analyzed. (E) Representative images of MS2-foci in an embryo expressing kni(5′+int)>MS2 reporter (yellow). Nuclei are marked with His2Av-mRFP (shown in blue). Images show kni(5′+int)>MS2 expression at 0, 4, and 10 min after the onset of NC14. The insets are magnifications of the embryo within the rectangle. Pixel histogram was adjusted for visualization purposes only, and images were rotated to orient the embryo (left-anterior, right-posterior).
Since transcription and the cell cycle progression are tightly coupled, disentangling which is upstream during the MBT has remained difficult (6, 19). On the one hand, transcript accumulation is necessarily dependent on the length of the transcriptional window (i.e., interphase duration) (5, 20, 21). Indeed, artificial manipulation of the cell cycle results in corresponding changes to the timing of ZGA in Xenopus, zebrafish, and Drosophila (22–24), while pharmacological inhibition of transcription does not affect cell cycle behavior in Xenopus or zebrafish (25–28). In Drosophila and zebrafish, it has been observed that the early interphases simply are not long enough to sustain robust transcription of most genes (20, 21, 29–32). One recent study found that if the cell cycle is artificially paused, the vast majority of ZGA genes can be induced one or two cell cycles prematurely in Drosophila (33). On the other hand, ZGA has been implicated as upstream of cell cycle slowing. Premature activation of transcription does lead to earlier cell cycle slowing in Drosophila, while transcription inhibition leads to delayed cycle lengthening (in contrast to Xenopus and zebrafish) (34–36).
Given this complex interdependence between the cell cycle and transcription, the question of whether the N/C ratio directly or indirectly affects transcription has remained unanswered (Fig. 1B). In vitro, at least one transcript is directly sensitive to the N/C ratio in cell cycle–arrested Xenopus egg extracts, but it is unclear if this direct relationship is maintained in vivo for any or all genes (37). In vivo, manipulations in ploidy coupled with RNA-sequencing (RNA-seq), microarrays, or qPCR have found that haploid Drosophila, Xenopus, and zebrafish embryos have reduced gene expression when compared to their wild type (WT) counterparts with a spectrum of N/C dependence across transcribed genes (15, 18, 38). Yet, such sampling-based experiments are ill suited to determine if the observed changes in transcription are a direct response to the altered N/C ratio or an indirect response to changes in cell cycle duration since they lack the temporal resolution required to properly account for the cumulative changes in interphase length. Moreover, examination of endogenous genes in ploidy manipulated embryos are inherently confounded by the inevitable effect on template availability. Carefully designed experiments have attempted to account for this effect by making the assumption that halving the template should result in half the output (18). However, this assumes that the template is the sole limiting factor for transcription, which may not be the case for all genes (39–42). In addition, whole embryo sequencing–based approaches also destroy the spatial information within an embryo, which is important since many of the early genes are spatially patterned. Fixed tissue imaging–based approaches, such as in situ hybridization or labeled ribonucleotide incorporation, can circumvent the latter issue but have limited temporal resolution to fully address if and how transcriptional dynamics respond to the N/C ratio.
In this study, we have employed the MS2 stemloop-MS2 coat protein (MS2-MCP) system to directly and quantitatively investigate the effects of the N/C ratio on real-time transcriptional dynamics in the early Drosophila embryos in single cells. This system allows us to follow the transcription of candidate genes over the course of several cell cycles with <30 s temporal resolution. We find that for all of the genes studied, N/C ratio–dependent changes in interphase duration result in changes in total transcription output within a given cycle. For some genes, such as knirps (kni), fushi tarazu (ftz), and snail (sna), the change in cell cycle duration is the dominant mechanism by which the N/C ratio impinges on transcriptional output. However, other genes directly sense the N/C ratio through two distinct mechanisms independent of the interphase length. Some genes, such as giant (gt) and bottleneck (bnk), significantly alter the kinetics of transcription activation such that transcription is slower at lower N/C ratios. On the other hand, frühstart (frs) and Krüppel (Kr) dramatically reduce their probability of transcription initiation such that fewer nuclei are activated in haploid embryos of the same cycle. We conclude that while all genes are affected by cell cycle duration, some genes, including gt, bnk, frs, and Kr, contain regulatory elements that respond directly to the N/C ratio. Moreover, different N/C ratio–responsive genes employ a combination of mechanisms to regulate transcription in response to ploidy.
Results
Cell Cycle Duration Controls the Transcriptional Window, which Determines Total Messenger RNA Output.
To investigate how the N/C ratio regulates transcription, we visualized nascent messenger RNA (mRNA) production in developing embryos through the MS2-MCP–based live imaging system (43–47). When the inserted 24 MS2 repeats are transcribed, the nascent RNA is bound by maternally provided MCP:green fluorescent protein (GFP), allowing us to detect nascent transcripts (Fig. 1E), and the integral of fluorescent intensity over time is used as a proxy for cumulative mRNA production (47, 48). MS2 repeats were integrated either into the yellow transgene driven by well-defined enhancers of early zygotic genes or within the endogenous locus of the gene of interest (49, 50). To remove the confounding effects of copy number on transcriptional output, we compared WT embryos that were heterozygous for a given MS2 construct to haploid embryos, which also contained only a single copy of the construct (see SI Appendix for details).
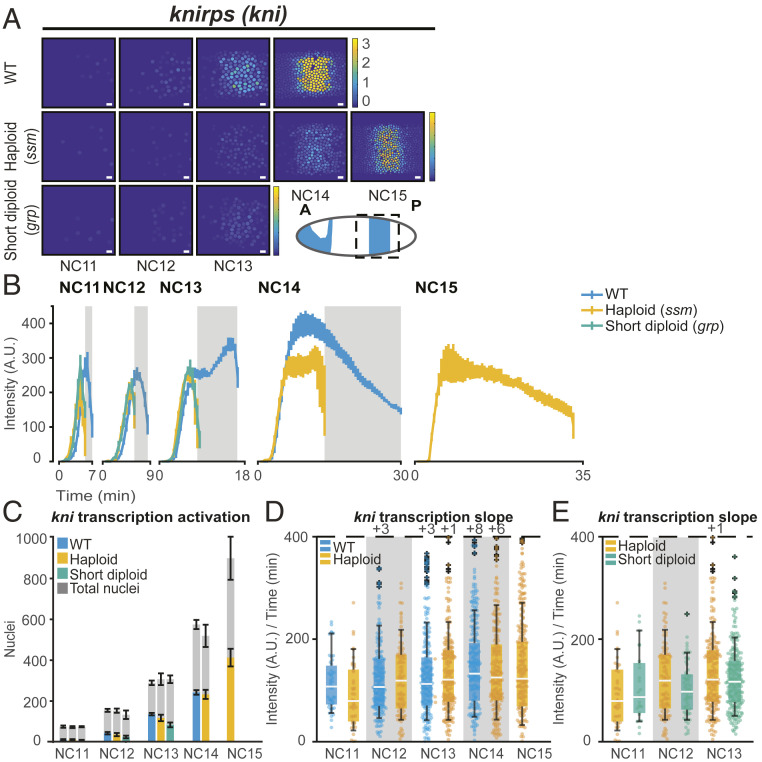
Since Drosophila forgo cytokinesis until the MBT when the resulting syncytium is cellularized, the first 13 divisions are referred to as nuclear cycles (NCs, not to be confused with the N/C ratio). We used the sesame/Hira185b mutation to generate haploid embryos (referred to hereafter as haploids) (51–53). The cell cycle lengths of haploids are shifted by one nuclear cycle (e.g., duration of diploid NC13 ≈ haploid NC14) and undergo one additional nuclear division in order to reach the same N/C ratio (where N is proportional to the total amount of DNA, not the number of nuclei) as their diploid counterparts before the MBT (Fig. 1 A and C) (11, 15, 54–57). We found that the duration of active transcription scales directly with cell cycle length for all genes studied (kni, ftz, sna, gt, bnk, frs, and Kr) (Fig. 1D and SI Appendix, Fig. S1). Regardless of each gene’s previous N/C ratio categorization (15), the change in the duration of the cell cycle, and hence, the transcriptional window, had a profound effect on the total transcriptional output (SI Appendix, Fig. S2). For example, the cumulative kni(5′+int)>MS2 mRNA output of each haploid cycle is delayed by precisely one cycle as compared to WT embryos (Fig. 2A and SI Appendix, Fig. S2A).
Fig. 2.
N/C ratio regulates kni transcription by shortening the cell cycle. (A) Heat map showing that total RNA output is greater in representative WT embryos expressing kni(5′+int)>MS2 in NC11-NC14 than haploid or short-cycle diploid. Haploids catch up as cell cycle slows in NC15. The color bar represents the total cumulative output per nucleus per NC (A.U.). The cartoon shows endogenous kni pattern at NC14 and the dashed box indicates the area under analysis. (Scale bar, 10 µm.) (B) Average kni(5′+int)>MS2 transcriptional trajectory over time for all transcribing nuclei per NC. Data are represented as mean ± SEM of replicate embryos. Gray boxes represent haploid mitoses. (C) Bar chart showing that the number of nuclei transcribing kni(5′+int)>MS2 in a given nuclear cycle is similar between WT (blue), haploid (yellow), and short-cycle diploid (green) embryos. Gray bars represent the total number of nuclei analyzed in each cycle, and colored bars represent the number of active nuclei. Values are similar across all genotypes. 55 NC11, 257 NC12, 816 NC13, and 1454 NC14 nuclei from six replicate kni(5′+int)>MS2 WT embryos; 47 NC11, 144 NC12, 470 NC13, 931 NC14, and 1,652 NC15 nuclei from four replicate kni(5′+int)>MS2 haploid embryos; and 31 NC11, 97 NC12, and 334 NC13 nuclei from four replicate kni(5′+int)>MS2 short-cycle diploid embryos were analyzed. Data in B and C are represented as mean ± SEM of six WT, four haploid (ssm), and four short-cycle diploid (grp), biologically replicate kni(5′+int)>MS2 embryos. (D and E) Boxplots showing no difference in the rate of transcriptional activation of kni(5′+int)>MS2 for all actively transcribing nuclei for (D) WT versus haploid (ssm) and (E) haploid versus short-cycle diploid (grp) embryos. Individual data points (up to 200 points) in each nuclear cycle are overlaid on the respective boxplots. The rate was quantified by measuring the slope of transcription initiation. The dashed line represents the cutoff for outlier values. The numbers of outlier values above the cutoff are given after “+.” The number of nuclei analyzed in each NC and for each genotype in B, D, and E is the same as shown in Fig. 1D.
The shorter cell cycle in haploids makes it difficult to disentangle the effect of cell cycle duration versus N/C ratio on transcription. To quantify changes in transcriptional activity independent of cell cycle length, we compared haploids to diploid embryos with similarly shortened cell cycles produced by mutation of checkpoint kinase 1 (grp/chk1, referred to hereafter as short-cycle diploids) (58–60). These embryos undergo the early nuclear cycles normally but fail to slow the cell cycles leading up to the MBT, resulting in a catastrophic 13th mitosis, and therefore cannot be analyzed after NC13 (60). We found that interphase length was more similar between haploids and short-cycle diploids than either was to the WT, allowing us to more directly compare the effect of N/C ratio on transcription (Fig. 1 C and D). The total mRNA output for kni(5′+int)>MS2 was comparable between haploids and short-cycle diploids, suggesting that the transcription of kni(5′+int)>MS2 is primarily affected by cell cycle length (Fig. 2A and SI Appendix, Fig. S2A). However, not all genes responded to haploids and short-cycle diploids in an N/C ratio–independent manner.
We next asked if the observed N/C ratio effects on transcription output could be attributed solely to the change in cell cycle length or if any of these effects were direct results of N/C ratio sensing. If the regulatory elements of a given gene directly sense the N/C ratio, they could change the rate of polymerase loading, the rate of elongation, and the median transcriptional signal, irrespective of cell cycle length. Or, the timing and/or probability of gene activation could be affected based on the respective N/C ratio. To this end, we measured the following three parameters of transcriptional activity that are independent of the cell cycle duration: the number of actively transcribing nuclei, the rate of transcriptional activation (slope), and the median amplitude of transcription in each cell cycle. Again, kni(5′+int)>MS2 behaved in a primarily time-dependent manner with no evidence of direct N/C ratio effects on transcription. The number of active nuclei and the rate of transcription activation within a given cell cycle were indistinguishable between WT, haploid, and short-cycle diploid (Fig. 2 C–E).
Endogenous ftz-MS2 and sna-distalE>MS2 transgene displayed a similar N/C ratio–independent transcriptional pattern in which the number of nuclei was unaffected by ploidy. Although the rate of transcription activation was mildly increased in haploid ftz-MS2 and slightly reduced in sna-distalE>MS2 haploid embryos compared to WT, the relative contribution of each parameter indicates that the dominant effect of N/C ratio on ftz-MS2 and sna-distalE>MS2 is cell cycle duration (see Fig. 5 C–F and SI Appendix, Figs. S3 and S4). We note that the number of active nuclei decreased in NC13 of sna-distalE>MS2 short diploids; however, this may be a result of experimental error since the observation of the sna expression domain is highly dependent on embryo orientation. There was also a small decrease in the median amplitude of kni(5′+int)>MS2 and sna-distalE>MS2 transcription in haploid cell cycles (SI Appendix, Fig. S7 A–D). However, since the signal in the early cycles never reaches steady state, we reasoned that the shortened cell cycle in haploids may prematurely truncate transcription during the rising phase. Indeed, the average transcriptional trajectory of all transcribing nuclei over time confirmed a premature termination of transcription in both haploids and short-cycle diploids (Fig. 2B). When fluorescent signal was aligned by equivalent N/C ratio, and therefore equivalent cell cycle length between WT and haploids (e.g., WT NC13 and haploid NC14), the transcriptional trajectories were better aligned, suggesting that cell cycle duration plays a major role in transcriptional regulation of kni, ftz, and sna (Fig. 2B and SI Appendix, Figs. S3B, S4B, and S6 A–C).
Fig. 5.
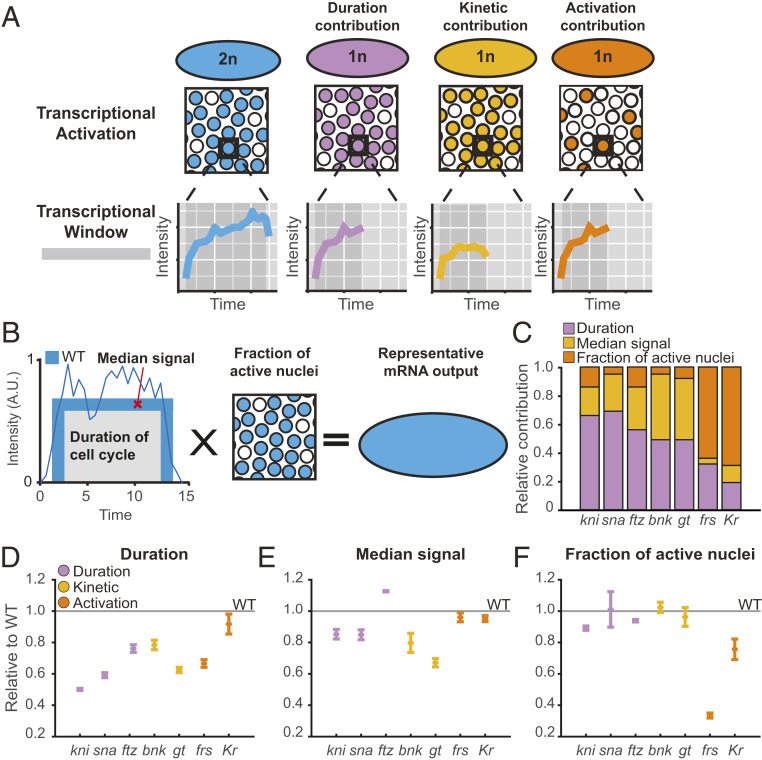
The N/C ratio affects transcription through multiple mechanisms. (A) The model depicting the different modes by which N/C ratio modulates transcription. The WT (2n) for all the genes has a number of actively transcribing nuclei each with a trace over a period of time indicated by the generic curve. The duration contribution only shortens the cell cycle duration, while the fraction of active nuclei and the rate of transcription remain identical to WT. In kinetic-contribution genes, the rate at which transcription occurs in haploids is slower than WT. In the N/C activation category, the number of active nuclei is significantly lower than WT, while the rate of transcription remains comparable to WT. (B) Model illustrating an estimate of RNA output of a WT embryo to determine contributions of each of the following three parameters: duration, median signal, and fraction of active nuclei. A single nuclear MS2 trajectory from a WT embryo is shown overlaid on the schematic. (C) Bar chart showing relative contributions of duration (purple), median signal (yellow), and fraction of active nuclei (orange) for each gene. Each bar is normalized by the sum of percent differences of the three parameters. While all the genes show some dependency on each parameter, kni, sna, and ftz exhibit significant duration contribution. gt and bnk have higher median signal contribution than other genes. The fraction of active nuclei has the highest contribution in frs and Kr. (D–F) The fraction of contribution of each parameter compared to WT for (D) duration, (E) median signal, and (F) fraction of active nuclei. All of the genes are significantly affected by the cell cycle duration. In addition to the duration contribution, gt and bnk show significant reduction in median signal, while frs and Kr are largely affected by the fraction of active nuclei compared to all other genes. Data in D–F are represented as mean ± SEM of four kni(5′+int)>MS2, three sna-distalE>MS2, three ftz-MS2, four bnk>MS2, five gt>MS2, four frs-MS2, and two KrCD2>MS2 replicate embryos.
We also noted that all genotypes displayed a small increase in the median signal with age, consistent with transcriptional memory or increased translation of transcriptional activators (42, 61, 62). This indicates that the small change in the transcriptional amplitude observed between WT and haploids can be attributed to changes in cell cycle duration or cell cycle state, not changes in the underlying competency of the transcription machinery in response to the N/C ratio. Therefore, for one category of genes, represented by kni, ftz, and sna, the dominant effect of the N/C ratio on transcriptional output is due to changes in cell cycle length.
The N/C Ratio Affects Transcriptional Kinetics for a Subset of Genes.
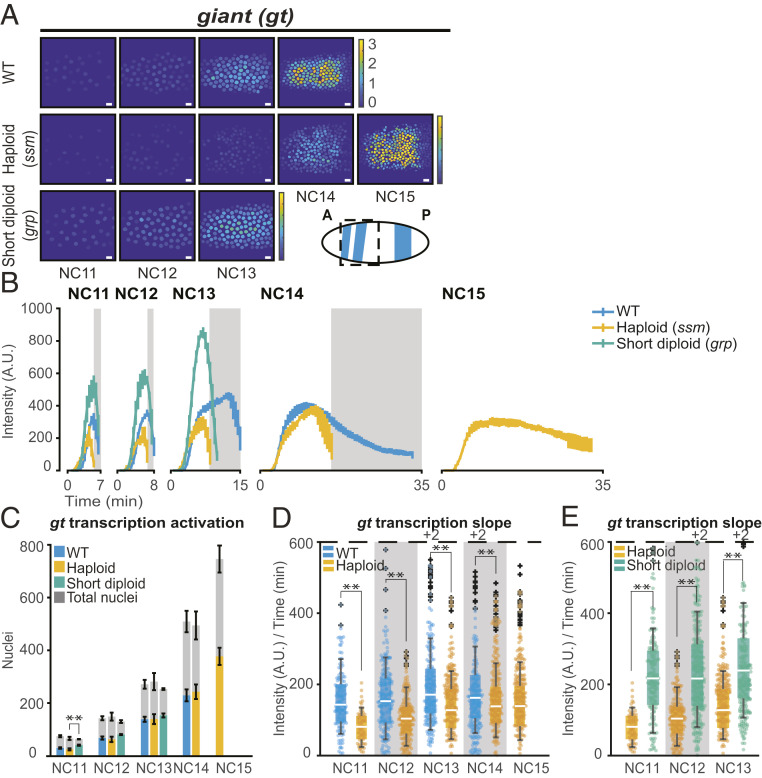
In contrast to kni, ftz, and sna, the effects of the N/C ratio on other genes cannot be explained purely by changes in cell cycle durations. We found two categories of direct N/C-dependent gene behavior. In the first, the N/C ratio regulates the kinetics of transcription activation. The gap gene gt and the cellularization gene bnk are examples of such regulation. Similar to kni, ftz, and sna, transcriptional activity of the MS2-yellow transgene driven by the gt and bnk enhancers displayed a delayed accumulation of nascent RNA with each nuclear cycle (Fig. 3A and SI Appendix, Fig. S2 D and E). Additionally, there was no delay in global activation of these genes, as WT and haploids both showed comparable numbers of actively transcribing nuclei in each cell cycle (Fig. 3C and SI Appendix, Fig. S5C).
Fig. 3.
giant responds to the N/C ratio in a kinetic-dependent manner. (A) Heat map of total RNA output for a representative embryo expressing gt>MS2. RNA output is comparable between WT and short-cycle diploid and is lower in haploids. The color bar represents the total cumulative output per nucleus per NC (A.U.). The cartoon shows endogenous gt pattern at NC14, and the dashed box indicates the area under analysis. (Scale bar, 10 µm.) (B) Average gt>MS2 transcriptional trajectory over time for all transcribing nuclei per NC. Data are represented as mean ± SEM of replicate embryos. Gray boxes represent haploid mitoses. (C) Bar chart showing that the number of nuclei transcribing gt>MS2 in a given nuclear cycle is similar between WT (blue), haploid (yellow), and short-cycle diploid (green) embryos. Gray bars represent the total number of nuclei analyzed in each cycle, and colored bars represent the number of active nuclei. Data in B and C are represented as mean ± SEM of six WT, five haploid (ssm), and four short-cycle diploid (grp), biologically replicate gt>MS2 embryos. (D and E) Boxplots showing the rate of transcriptional activation (initial slope) of gt>MS2 for all actively transcribing nuclei for (D) WT versus haploid (ssm) and (E) haploid versus short-cycle diploid (grp) embryos. The initial slope is lower in haploids compared to WT and short-cycle diploids. The dashed line represents the cutoff for outlier values. The numbers of outlier values above the cutoff are given after “+.” Individual data points (up to 200 points) in each nuclear cycle are overlaid on the respective boxplots. **P < 0.005 from Student’s t test. The numbers of nuclei analyzed in C and B, D, and E in each NC are as follows: NC11 [176, 164], NC12 [403, 362], NC13 [830, 730], and NC14 [1,332, 1,077] nuclei from six replicate gt>MS2 WT embryos; NC11 [97, 80], NC12 [311, 270], NC13 [695, 587], NC14 [1,214, 1,013], and NC15 [1,885, 1,513] nuclei from five replicate gt>MS2 haploid embryos; and NC11 [160, 158], NC12 [321, 308], and NC13 [611, 581] nuclei from four replicate gt>MS2 short-cycle diploid embryos.
However, unlike kni, ftz, and sna, where cell cycle duration was the main factor of N/C ratio–dependent changes in transcription, the MS2-yellow transgene expression driven by the gt and bnk enhancers in haploids demonstrated an additional form of N/C ratio–dependent regulation. We observed a significant decrease in the rate of transcriptional activation compared to both WT and short-cycle diploids in the early cell cycles for both genes (Fig. 3 D and E and SI Appendix, Fig. S5 D and E). This N/C ratio–dependent initial trajectory suggests that the kinetics of gt and bnk transcription initiation are directly responsive to embryo ploidy. We further confirmed that the initial rate of transcription and average transcriptional amplitude were lower in haploids in the early cell cycles (SI Appendix, Fig. S7 E–H). The reduction in median transcriptional amplitude was also observed in the average nuclear trajectories, although the nuclei-to-nuclei variability in the timing of transcription activation obscured this distinct difference (Fig. 3B and SI Appendix, Figs. S5 A and B and S6 D and E). This result suggests that the kinetics of polymerase loading are directly sensitive to the N/C ratio for a class of genes. Surprisingly, we observed a much higher gt>MS2 signal in the short-cycle diploids compared to haploids and WT (Fig. 3 B and E). A less dramatic increased slope is also observed in sna and bnk and short-cycle diploids (SI Appendix, Figs. S4E and S5E). This may be due to the faster replication and therefore greater early template availability in the short cell cycle conditions or some other, more direct effect of chk1 activity, though this effect is not as striking or not observed in other genes we analyzed. Nonetheless, the fact that the short-cycle diploid is more, not less, active than WT strongly indicates that the reduced rate of transcriptional activation observed in haploids is not a result of cell cycle duration alone.
The N/C Ratio Regulates Binary Activation Probability for a Subset of Genes.
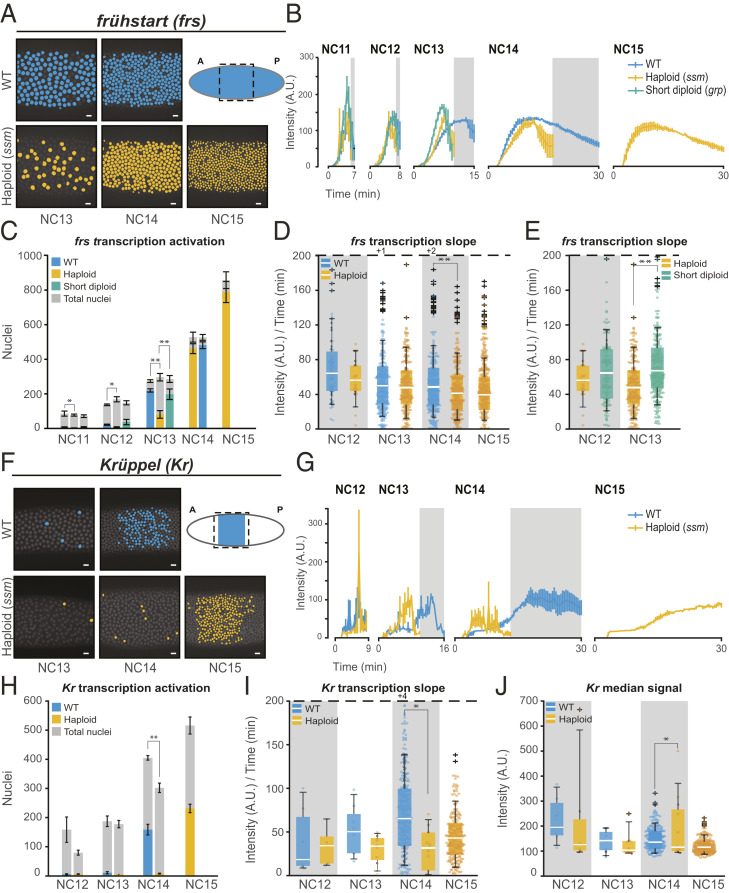
The second category of direct N/C ratio–responsive transcription behavior was more striking than the change in the rate of transcription observed in gt and bnk. In this class, which consists of the cell cycle regulator frs and the gap gene Kr, the global probability of gene activation was directly N/C ratio sensitive. While kni, ftz, sna, gt, and bnk enhancer-mediated transcription showed gradual activation from NC11 to NC14, endogenous frs-MS2 and KrCD2>MS2 transgene showed a dramatic switch in the number of active nuclei between cell cycles in WT, going from 16% in WT NC12 to 80% in WT NC13 for frs and 6% in WT NC13 to 39% in WT NC14 for Kr (Fig. 4 A, C, F, and H). This is consistent with the switch-like RNA accumulation observed for frs and Kr by time-course RNA-seq (63, 64). We found the switch from majority inactive to majority active nuclei was delayed by one cell cycle in haploids, occurring between NC13 and NC14 rather than between NC12 and NC13 for frs and between NC14 and NC15 for KrCD2 (Fig. 4 C and H). Notably, the number of active nuclei was comparable between WT and short-cycle diploids in NC13 for frs-MS2 (Fig. 4C), demonstrating that the switch-like activation of frs is a direct effect of ploidy as opposed to cell cycle duration. Since KrCD2 does not become activated until NC14 and short-cycle diploid embryos do not progress past mitosis of NC13, we were not able to compare WT and short-cycle diploids for KrCD2>MS2. During haploid NC14, frs was fully activated and the fraction of actively transcribing nuclei stayed constant in haploid NC15, as we observed in the other genes (Fig. 4C). Similarly, localized KrCD2>MS2 expression was delayed until haploid NC15 (Fig. 4 F and H). It is important to note that ploidy does not change the total number of nuclei in a given cycle and only alters the amount of DNA in each nucleus (Figs. 3C and 4C).
Fig. 4.
The N/C ratio regulates frs and Kr in an activation-mediated manner. (A) frs nuclei that ever showed an active transcription are false colored in blue (WT) from NC13-NC14 and yellow (haploids) from NC13-NC15. Note the decreased number of active nuclei in haploid NC13. The cartoon shows endogenous ubiquitous frs pattern at NC14, and the dashed box indicates the area under analysis. (Scale bar, 10 µm.) (B) Average frs-MS2 transcriptional trajectory over time for all transcribing nuclei per nuclear cycle. Data are represented as mean ± SEM of replicate embryos. Gray boxes represent haploid mitoses. (C) Bar chart showing a delay in the switch from mostly inactive to mostly actively transcribing frs-MS2 in haploid (yellow) compared to WT (blue) embryos. Short-cycle diploids (green) activate similarly to WT in NC13. Gray bars represent the total number of nuclei analyzed in each cycle, and colored bars represent the number of active nuclei. Data in B and C are represented as mean ± SEM of five WT, four haploid (ssm), and four short-cycle diploid (grp) biologically replicate frs-MS2 embryos. (D and E) Boxplots showing that the rate of transcriptional activation (initial slope) of frs-MS2 is similar for all actively transcribing nuclei for (D) WT versus haploid (ssm) and (E) haploid versus short-cycle diploid (grp) embryos. The number of nuclei analyzed in A and C and B, D, and E in each NC is as follows: NC 11 [38, 23], NC12 [108, 55], NC13 [1,100, 791], and NC14 [2,406, 1,921] nuclei from five replicate frs-MS2 WT embryos; NC 11 [11,10] NC12 [33, 17], NC13 [324, 177], NC14 [1,846,1,355], and NC15 [2,155, 1,510] nuclei from four replicate frs-MS2 haploid embryos; and NC11 [28, 24] NC12 [148, 97], and NC13 [791, 546] nuclei from four replicate frs-MS2 short-cycle diploid embryos. (F) Kr nuclei that ever showed an active transcription are false colored in blue (WT) from NC13-NC14 and yellow (haploids) from NC13-NC15. Note the decreased number of active nuclei in haploid NC14. The cartoon shows endogenous Kr pattern at NC14, and the dashed box indicates the area under analysis. (Scale bar, 10 µm.) (G) Average KrCD2>MS2 transcriptional trajectory over time for all transcribing nuclei per nuclear cycle. Data are represented as mean ± SEM of replicate embryos. Gray boxes represent haploid mitoses. (H) Bar chart showing a delay in the switch from mostly inactive to mostly actively transcribing KrCD2>MS2 in haploid (yellow) compared to WT (blue) embryos. Gray bars represent the total number of nuclei analyzed in each cycle, and colored bars represent the number of active nuclei. Data in G and H are represented as mean ± SEM of three WT and two (NC12 and NC13) or three (NC14 and NC15) haploid (ssm) biologically replicate KrCD2>MS2 embryos. (I) Boxplots showing that the rate of transcriptional activation (initial slope) of KrCD2>MS2 is similar for all actively transcribing nuclei for WT and haploid (ssm) embryos. (J) Boxplots showing median transcriptional activity of KrCD2>MS2 from all transcribing nuclei. The average transcription amplitude is comparable between WT and haploid (ssm) embryos. The number of nuclei analyzed in F and H and G, I, and J in each NC is as follows: NC12 [11, 7], NC13 [32, 9], and NC14 [476, 311] nuclei from three replicate KrCD2>MS2 WT embryos and NC12 [12 and 7], NC13 [10 and 8], NC14 [23 and 12], and NC15 [694 and 504] nuclei from two to three replicate KrCD2>MS2 haploid (ssm) embryos. For all boxplots, the dashed line represents the cutoff for outlier values. The number of outlier values above the cutoff are given after “+.” Individual data points (up to 200 points) in each nuclear cycle are overlaid on the respective boxplots. *P < 0.05 and **P < 0.005 from Student’s t test.
For those nuclei that are active in NC13 haploids and short-cycle diploids (frs) and in NC14 haploids (KrCD2), the total per nucleus output of frs-MS2 and KrCD2>MS2 is highly reflective of cell cycle duration, similar to other genes in this study (SI Appendix, Fig. S2 F and G). This suggests that the N/C ratio may primarily regulate the probability of activation rather than affecting the entire transcriptional kinetics. Although the few actively transcribing nuclei in the earlier cell cycles cause high variability, the average transcriptional trajectories seem to indicate comparable rates of transcription between the genotypes (Fig. 4 B and G and SI Appendix, Fig. S6 F and G). Indeed, the rate of transcriptional activation and the median amplitude of transcriptional activity in those nuclei that do initiate transcription in the earlier cycles are only mildly affected by ploidy (Fig. 4 D, E, I, and J and SI Appendix, Fig. S7 I and J). These results indicate that the regulatory elements of frs and Kr are directly responsive to the N/C ratio in a binary on/off manner and that the N/C ratio has limited additional detectable effects on transcription kinetics.
A Single Gene Can Be Regulated by Multiple Mechanisms.
To better characterize the relative contribution of the cell cycle duration and the N/C ratio on zygotic transcription, we estimated a representative RNA output of a given gene per embryo. First, we calculated the RNA output in WT embryos by multiplying the average duration of each cell cycle, median amplitude, and the fraction of active nuclei within the gene expression domain (Fig. 5B). To assess the contribution of each parameter (duration, median signal, or fraction of active nuclei), we replaced the WT parameter with the haploid parameter value and measured the change in RNA output compared to the WT (Fig. 5 D–F). Using this method, we quantified the relative contribution of each parameter for each gene (Fig. 5C). Albeit a semiquantitative estimate that does not include gene length, this analysis succinctly summarizes our findings and demonstrates relative contribution of cell cycle duration, N/C ratio–responsive median amplitude, and N/C ratio–mediated activation on gene regulation (Fig. 5C). Indeed, cell cycle duration plays a major role for kni, ftz, and sna. However, genes whose kinetics were significantly altered by the N/C ratio (gt and bnk) exhibit a strong additional contribution from the changes in the median amplitude. Moreover, genes with significant N/C ratio–dependent changes in the activation probability (frs and Kr) were dominated by the contribution of the fraction of active nuclei in their total transcriptional output when comparing haploids to WT.
Discussion
Here, we have shown that the total transcriptional output during the MBT is a function of the N/C ratio for all measured genes in Drosophila and classified three nonmutually exclusive categories of N/C ratio control. For some genes (kni, ftz, and sna), the primary effect of the N/C ratio is due to changes in the length of the cell cycle and hence the length of the transcriptional window. This results in a reduction in total mRNA production within a given cycle due to the abortion of transcription at mitosis (20, 21, 32, 65). However, the activation of other genes (gt, bnk, frs, and Kr) is directly sensitive to the N/C ratio despite the effect of the cell cycle duration. We show that this direct sensing of the N/C ratio is manifested as a contribution from the following two different gene expression regulatory paradigms: 1) altering the kinetics of transcriptional activation of actively transcribing nuclei and 2) regulating the probability of gene activation in a given cell cycle (Fig. 5A).
The existence of the first category of direct N/C ratio sensing, which affects the rate of transcription without changing the number of actively transcribing nuclei per cycle, suggests that the cis regulatory elements of these genes directly respond to the N/C ratio. In particular, the rate of polymerase elongation during transcription or the rate of polymerase recruitment to the promoter is likely affected in haploids, independent of cell cycle length. We found that such N/C kinetic–mediated genes like gt and bnk showed faster rates of transcription in WT compared to other genes, implying that the rapidly transcribing genes may be more sensitive to the kinetic-mediated transcription regulated by the N/C ratio (SI Appendix, Fig. S8). Interestingly, both gt and bnk also showed increased slopes in the short-cycle diploids, suggesting that there may be a compensatory mechanism for fast-activating genes when the cell cycle is shorted in diploids or the activity of Chk1 itself may contribute to their activation. In contrast, the activation probability N/C ratio–sensing genes, frs and Kr, are much more switch-like in our study. We speculate that this switch-like behavior may be a consequence of its responsiveness to the exponentially increasing N/C ratio (Fig. 4 A, C, F, and H). This process may set a threshold for the recruitment of polymerase to the promoter or, more likely, its release from pausing at the promoter. The promoter of frs and Kr, as well as other genes we assayed, are already nucleosome free by NC11, indicating chromatin opening alone cannot explain their differences in timing (56).
In our study, we used a combination of transgenes and endogenous genes to assay the role of regulatory regions in sensing and responding to the N/C ratio. Both the N/C-independent and the N/C-dependent categories contain a mixture of transgenes and endogenous genes, indicating that our findings are not the result of how the reporters were constructed. The endogenously tagged genes (ftz and frs) presumably have access to all endogenous enhancers. For some reporter genes like kni and gt, we used a transgene with multiple enhancers, while for other transgenes like sna and Kr, a single enhancer (the distal enhancer for sna and the CD2 enhancer for Kr) was used to drive transcription. In the future, it will be interesting to elucidate the role of individual enhancers of a single gene in directly sensing the N/C ratio.
We also note that most manipulations of the N/C ratio are, in fact, manipulations in ploidy. In these cases, since the amount of template for a given transcript is altered, we speculate that the transcript accumulation is much more dramatically affected than a simple response to the change in N/C ratio–dependent cell cycle duration. Template reduction could also directly affect the process of cell cycle slowing. For example, in the model where RNA-polymerase on the DNA acts as a source of replication stress, simply halving the amount of DNA without changing other aspects of transcription would halve the number of such conflicts embryo wide (60). This may permit an additional cell cycle in haploids to allow for the critical number of global conflicts to induce the checkpoint response to slow the cell cycle.
Recent RNA-seq has shown that the vast majority of genes, including the genes used in this study, are capable of initiating detectable transcription if the cell cycle is stopped one or two cycles prematurely (33). This demonstrates that pre-ZGA N/C ratios are not strictly incompatible with transcription. However, it is unclear if the dynamics of transcription initiation and the molecular state of the genome are the same in N/C ratio–stalled embryos as those with a normal N/C ratio at ZGA. Indeed, all of the ZGA genes assayed by live imaging in this study show at least some transcription before NC14; therefore, mRNAs would be expected to accumulate if the rate of transcription were unchanged and cell cycle was simply lengthened. For a hypothetical pure N/C ratio–independent gene, cell cycle duration would be the predominant parameter for transcriptional activity. For a pure kinetic-mediated gene, the effect of the N/C ratio–dependent initial rate of transcription and amplitude of transcription would be reduced as the cell cycle gets longer. However, the effect of activation-mediated N/C ratio response would be expected to remain proportional regardless of cell cycle length. Nonetheless, given that some transcription is detected for both activation-mediated genes in this study (frs and Kr), we would expect transcripts to accumulate if the cell cycle were arrested in interphase. Further studies will be required to decipher the complex relationship between N/C ratio, cell cycle, and transcription. Our findings serve to constrain the available models by demonstrating multiple effects of the N/C ratio on transcription activation and kinetics independent of cell cycle duration.
Materials and Methods
All fly stocks were maintained by standard method at 25 °C and were grown on standard cornmeal media. Live imaging with a confocal microscope was performed with the same exposure and laser settings for each comparison group (e.g., frs-MS2 WT versus haploid versus short diploid). All the image processing analyses were implemented in MATLAB (R2018b, MathWorks), and Student’s t test was used to calculate P values in each plot. Details on WT, haploid, and short-cycle diploid embryo generation, plasmid construction, and image analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
We are grateful to Gary Laevsky and the Molecular Biology Confocal Imaging Facility and Gordon Gray and the Drosophila Media Core Facility at Princeton University for technical support. We thank Michal Levo for reagents. We thank Eric Wieschaus, Stas Shvartsman, and Mike Levine for discussion. Stocks obtained from the Bloomington Drosophila Stock Center (NIH Award P40OD018537) were used in this study (66). We also thank the FlyBase for providing useful information (67). S.S. and B.L. are supported by NIH Award R35GM133425.
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010210118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Newport J., Kirschner M., A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686 (1982). [DOI] [PubMed] [Google Scholar]
- 2.Newport J., Kirschner M., A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30, 687–696 (1982). [DOI] [PubMed] [Google Scholar]
- 3.Harrison M. M., Eisen M. B., “Transcriptional activation of the zygotic genome in Drosophila” in Current Topics in Developmental Biology, Lipshitz H. D. Ed. (Elsevier, 2015), 113, pp. 85–112. [DOI] [PubMed] [Google Scholar]
- 4.Blythe S. A., Wieschaus E. F., “Coordinating cell cycle remodeling with transcriptional activation at the Drosophila MBT” in Current Topics in Developmental Biology, Lipshitz H. D. Ed. (Elsevier, 2015), 113, pp. 113–148. [DOI] [PubMed] [Google Scholar]
- 5.Yuan K., Seller C. A., Shermoen A. W., O’Farrell P. H., Timing the Drosophila mid-blastula transition: A cell cycle-centered view. Trends Genet. 32, 496–507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jukam D., Shariati S. A. M., Skotheim J. M., Zygotic genome activation in vertebrates. Dev. Cell 42, 316–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B., Grosshans J., Link of zygotic genome activation and cell cycle control. Methods Mol. Biol. 1605, 11–30 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Lefebvre F. A., Lécuyer É., Flying the RNA nest: Drosophila reveals novel insights into the transcriptome dynamics of early development. J. Dev. Biol. 6, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vastenhouw N. L., Cao W. X., Lipshitz H. D., The maternal-to-zygotic transition revisited. Development 146, dev161471 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Schulz K. N., Harrison M. M., Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 20, 221–234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar B. A., Kiehle C. P., Schubiger G., Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44, 365–372 (1986). [DOI] [PubMed] [Google Scholar]
- 12.Almouzni G., Wolffe A. P., Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J. 14, 1752–1765 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee D. R., Lee J. E., Yoon H. S., Roh S. I., Kim M. K., Compaction in preimplantation mouse embryos is regulated by a cytoplasmic regulatory factor that alters between 1- and 2-cell stages in a concentration-dependent manner. J. Exp. Zool. 290, 61–71 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Dekens M. P. S., Pelegri F. J., Maischein H. M., Nüsslein-Volhard C., The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development 130, 3907–3916 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Lu X., Li J. M., Elemento O., Tavazoie S., Wieschaus E. F., Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development 136, 2101–2110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jevtić P., Levy D. L., Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr. Biol. 25, 45–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H., Einstein L. C., Little S. C., Good M. C., Spatiotemporal patterning of zygotic genome activation in a model vertebrate embryo. Dev. Cell 49, 852–866.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan S. H., et al., Brd4 and P300 confer transcriptional competency during zygotic genome activation. Dev. Cell 49, 867–881.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferree P. L., Deneke V. E., Di Talia S., Measuring time during early embryonic development. Semin. Cell Dev. Biol. 55, 80–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shermoen A. W., O’Farrell P. H., Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67, 303–310 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothe M., Pehl M., Taubert H., Jäckle H., Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature 359, 156–159 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Edgar B. A., Schubiger G., Parameters controlling transcriptional activation during early Drosophila development. Cell 44, 871–877 (1986). [DOI] [PubMed] [Google Scholar]
- 23.Kimelman D., Kirschner M., Scherson T., The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell 48, 399–407 (1987). [DOI] [PubMed] [Google Scholar]
- 24.Collart C., Allen G. E., Bradshaw C. R., Smith J. C., Zegerman P., Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 341, 893–896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newport J., Dasso M., On the coupling between DNA replication and mitosis. J. Cell Sci. 1989, 149–160 (1989). [DOI] [PubMed] [Google Scholar]
- 26.Clute P., Masui Y., Regulation of the appearance of division asynchrony and microtubule-dependent chromosome cycles in Xenopus laevis embryos. Dev. Biol. 171, 273–285 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Müller F., Lakatos L., Dantonel J., Strähle U., Tora L., TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr. Biol. 11, 282–287 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Hadzhiev Y., et al., A cell cycle-coordinated Polymerase II transcription compartment encompasses gene expression before global genome activation. Nat. Commun. 10, 691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKnight S. L., Miller O. L. Jr, Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell 8, 305–319 (1976). [DOI] [PubMed] [Google Scholar]
- 30.De Renzis S., Elemento O., Tavazoie S., Wieschaus E. F., Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5, e117 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heyn P., et al., The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep. 6, 285–292 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Kwasnieski J. C., Orr-Weaver T. L., Bartel D. P., Early genome activation in Drosophila is extensive with an initial tendency for aborted transcripts and retained introns. Genome Res. 29, 1188–1197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strong I. J. T., Lei X., Chen F., Yuan K., O’Farrell P. H., Interphase-arrested Drosophila embryos activate zygotic gene expression and initiate mid-blastula transition events at a low nuclear-cytoplasmic ratio. PLoS Biol. 18, e3000891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar B. A., Datar S. A., Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes Dev. 10, 1966–1977 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Shermoen A. W., McCleland M. L., O’Farrell P. H., Developmental control of late replication and S phase length. Curr. Biol. 20, 2067–2077 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung H. W., Spangenberg S., Vogt N., Großhans J., Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Curr. Biol. 23, 133–138 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Amodeo A. A., Jukam D., Straight A. F., Skotheim J. M., Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc. Natl. Acad. Sci. U.S.A. 112, E1086–E1095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jevtić P., Levy D. L., Both nuclear size and DNA amount contribute to midblastula transition timing in Xenopus laevis. Sci. Rep. 7, 7908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pritchard D. K., Schubiger G., Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 10, 1131–1142 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Veenstra G. J. C., Destrée O. H. J., Wolffe A. P., Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol. Cell. Biol. 19, 7972–7982 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guven-Ozkan T., Nishi Y., Robertson S. M., Lin R., Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 135, 149–160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada S., et al., The Drosophila pioneer factor zelda modulates the nuclear microenvironment of a dorsal target enhancer to potentiate transcriptional output. Curr. Biol. 29, 1387–1393.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertrand E., et al., Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Alexander J. M., et al., Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 8, 1159–1168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golding I., Paulsson J., Zawilski S. M., Cox E. C., Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Larson D. R., Zenklusen D., Wu B., Chao J. A., Singer R. H., Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332, 475–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia H. G., Tikhonov M., Lin A., Gregor T., Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr. Biol. 23, 2140–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukaya T., Lim B., Levine M., Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El-Sherif E., Levine M., Shadow enhancers mediate dynamic shifts of gap gene expression in the Drosophila embryo. Curr. Biol. 26, 1164–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim B., Fukaya T., Heist T., Levine M., Temporal dynamics of pair-rule stripes in living Drosophila embryos. Proc. Natl. Acad. Sci. U.S.A. 115, 8376–8381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loppin B., Docquier M., Bonneton F., Couble P., The maternal effect mutation sésame affects the formation of the male pronucleus in Drosophila melanogaster. Dev. Biol. 222, 392–404 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Loppin B., Berger F., Couble P., The Drosophila maternal gene sésame is required for sperm chromatin remodeling at fertilization. Chromosoma 110, 430–440 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Loppin B., et al., The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437, 1386–1390 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Di Talia S., et al., Posttranslational control of Cdc25 degradation terminates Drosophila’s early cell-cycle program. Curr. Biol. 23, 127–132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farrell J. A., O’Farrell P. H., Mechanism and regulation of Cdc25/Twine protein destruction in embryonic cell-cycle remodeling. Curr. Biol. 23, 118–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blythe S. A., Wieschaus E. F., Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. eLife 5, e20148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shindo Y., Amodeo A. A., Dynamics of free and chromatin-bound histone H3 during early embryogenesis. Curr. Biol. 29, 359–366.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Fogarty P., Kalpin R. F., Sullivan W., The Drosophila maternal-effect mutation grapes causes a metaphase arrest at nuclear cycle 13. Development 120, 2131–2142 (1994). [DOI] [PubMed] [Google Scholar]
- 59.Sibon O. C. M., Stevenson V. A., Theurkauf W. E., DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388, 93–97 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Blythe S. A., Wieschaus E. F., Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160, 1169–1181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foo S. M., et al., Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr. Biol. 24, 1341–1346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferraro T., et al., Transcriptional memory in the Drosophila embryo. Curr. Biol. 26, 212–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lott S. E., et al., Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 9, e1000590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chari S., Wilky H., Govindan J., Amodeo A. A., Histone concentration regulates the cell cycle and transcription in early development. Development 146, dev177402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Djabrayan N. J. V., et al., Metabolic regulation of developmental cell cycles and zygotic transcription. Curr. Biol. 29, 1193–1198.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitworth C., “The Bloomington Drosophila Stock Center” in The Biological Resources of Model Organisms, Jarret R. L., McCluskey K., Eds. (CRC Press, 2019), pp. 145–162. [Google Scholar]
- 67.Gramates L. S.et al.; the FlyBase Consortium , FlyBase at 25: Looking to the future. Nucleic Acids Res. 45, D663–D671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.