Significance
The caudal hematopoietic tissue (CHT) is characterized as a hematopoietic organ for fetal hematopoietic stem and progenitor cell (HSPC) expansion in zebrafish. In this study, we used scRNA-seq combined with functional assays to decode the developing CHT. First, we resolved fetal HSPC heterogeneity, manifested as lineage priming and metabolic gene signatures. We further analyzed the cellular interactions among nonhematopoietic niche components and HSPCs and identified an endothelial cell-specific factor, Gpr182, followed by experimental validation of its role in promoting HSPC expansion. Finally, we uncovered the conservation and divergence of developmental hematopoiesis between human fetal liver and zebrafish CHT. Our study provides a valuable resource for fetal HSPC development and clues to establish a supportive niche for HSPC expansion in vitro.
Keywords: zebrafish, HSPC expansion, single-cell RNA-seq, caudal hematopoietic tissue, Gpr182
Abstract
During vertebrate embryogenesis, fetal hematopoietic stem and progenitor cells (HSPCs) exhibit expansion and differentiation properties in a supportive hematopoietic niche. To profile the developmental landscape of fetal HSPCs and their local niche, here, using single-cell RNA-sequencing, we deciphered a dynamic atlas covering 28,777 cells and 9 major cell types (23 clusters) of zebrafish caudal hematopoietic tissue (CHT). We characterized four heterogeneous HSPCs with distinct lineage priming and metabolic gene signatures. Furthermore, we investigated the regulatory mechanism of CHT niche components for HSPC development, with a focus on the transcription factors and ligand–receptor networks involved in HSPC expansion. Importantly, we identified an endothelial cell-specific G protein–coupled receptor 182, followed by in vivo and in vitro functional validation of its evolutionally conserved role in supporting HSPC expansion in zebrafish and mice. Finally, comparison between zebrafish CHT and human fetal liver highlighted the conservation and divergence across evolution. These findings enhance our understanding of the regulatory mechanism underlying hematopoietic niche for HSPC expansion in vivo and provide insights into improving protocols for HSPC expansion in vitro.
In vertebrates, the earliest definitive hematopoietic stem and progenitor cells (HSPCs) generated during embryogenesis can give rise to multiple blood lineages and exhibit a self-renewal property (1–3). For the establishment of the HSPC pool, nascent HSPCs will first migrate into a transitory hematopoietic organ, termed as fetal liver (FL in mammals) or caudal hematopoietic tissue (CHT in zebrafish), for rapid expansion (4, 5). Clinically, in vitro HSPC expansion is a feasible approach to obtain sufficient transplantable HSPCs (6) but remains technically challenging. Thus, decoding the complex regulatory mechanism of HSPC expansion within the hematopoietic organ is essential.
Zebrafish CHT is a highly vascularized niche, and the rapidly increased vascular endothelial cells (ECs) form a complex structure composing the dorsal artery and honeycomb-like venous network (4). Its formation also involves other types of cell components, such as fibroblastic reticular cells and hematopoietic cells (HC). Therefore, the maintenance of CHT function relies on an orderly and precise process of interaction among all cell types inside. Previously, it has been shown that the caudal vascular ECs are able to attract HSPCs and support HSPC expansion via secreting chemokines and cytokines, such as Cxcl12a, Ccl25b, Cxcl8, Kit ligand b, thrombopoietin, Csf1a, and erythropoietin (7–10). Recently, macrophages have been reported as “ushers” for HSPC homing to the CHT (11), and neutrophils can promote CHT-resident HSPC egression and colonization into the following niche by releasing matrix metalloproteinase 9 (12). Moreover, the stromal-cell maturation is crucial for CHT niche formation (13). However, it remains to be resolved how global niche components orchestrate HSPC development.
Recent advances in bulk RNA-sequencing (RNA-seq) and geographical position sequencing have helped identify the spatiotemporal characteristics of HSPC expansion in the CHT region (14), however, our understanding of molecular and cellular dynamics in CHT hematopoiesis at single-cell resolution remains elusive. Here, we used single-cell RNA-seq (scRNA-seq) to map the transcriptional profiles of 28,777 cells, which were sorted from the CHT spanning three successive developmental stages. With these profiles, we characterized HSPC transcriptional heterogeneity and identified an EC-specific regulator, G protein–coupled receptor 182 (Gpr182), functionally supporting HSPC expansion in cell type- and stage-specific manners. Moreover, cross-species analysis of zebrafish CHT and human FL revealed the conserved cellular components and transcriptional programs across evolution. Our study provides potential clues to establish a supportive niche for HSPC expansion in vitro.
Results
A Single-Cell Resolution Atlas of Developing Zebrafish CHT.
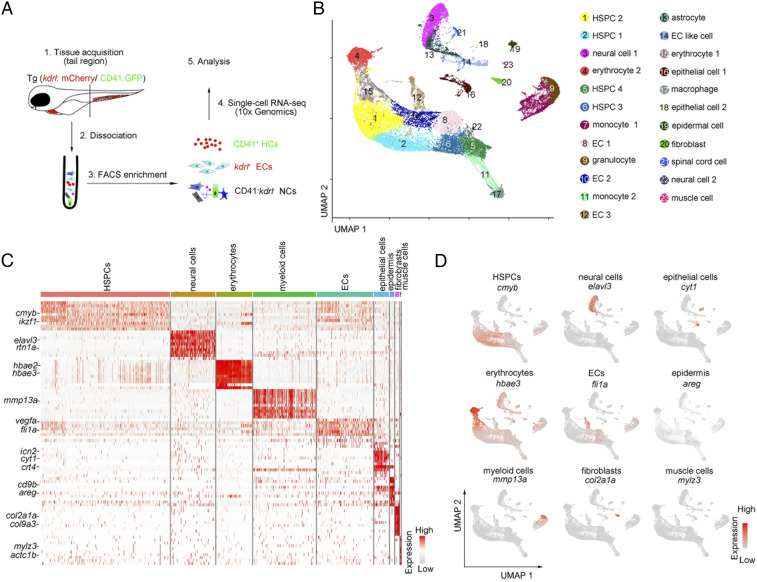
In zebrafish, HSPCs exhibit the spatiotemporal characteristics in the CHT niche. At 55 h postfertilization (hpf), HSPCs migrate into the caudal vein plexus (CVP) from the caudal artery; at 3 d postfertilization (dpf), HSPCs seed the dorsal wall of the caudal vein for expansion; and at 4 dpf, the expanding HSPCs differentiate into multiple blood lineages (9). To investigate the molecular and cellular dynamics in this process at single-cell resolution, we complemented a previous dataset at 55 hpf (14) by additionally generating two sets of scRNA-seq profiles of CHT at 3.5 dpf and 4.5 dpf (Fig. 1A). We employed the transgenic line (Tg) (kdrl:mCherry/CD41:GFP) (14–16) for sorting ECs (kdrl:mCherry+), HCs (CD41:GFP+), and double-negative cells (NCs) (CD41:GFP−kdrl:mCherry−). We used the Seurat package (17) for scRNA-seq data analysis, and the batch effect was removed by using the canonical-correlation analysis algorithm (18). The scRNA-seq datasets contained a total of 28,777 high-quality cells, including 4,587 ECs, 18,577 HC, and 5,613 NCs, with a median of ∼1,800 genes detected per cell (SI Appendix, Fig. S1 A and B and Table S1). Based on uniform manifold approximation and projection (UMAP) analysis, we clustered and annotated nine major cell types (HSPCs, erythrocytes, myeloid cells, ECs, neural cells, epithelial cells, epidermis, fibroblasts, and muscle cells), including 23 clusters (Fig. 1 B and C and SI Appendix, Fig. S1C). HSPC clusters were characterized by enriched expression of cmyb and hdr. Myeloid genes mmp13a, mfap4, mpx, and pu.1 were highly expressed in myeloid clusters. Erythrocyte clusters were identified by specific expression of hbae3 and hemgn. Moreover, nonhematopoietic niche components were also identified by their corresponding marker genes. For example, EC clusters were featured by high fli1a and vegfc expression, and neural cell clusters were characterized by elavl3, tuba1c, and snap25a (Fig. 1 C and D and SI Appendix, Fig. S1C and Table S2). Taken together, the scRNA-seq datasets provided a reference atlas to examine the cell population and gene-expression dynamics within the developing zebrafish CHT niche.
Fig. 1.
Single-cell transcriptome map of zebrafish CHT. (A) A schematic paradigm of tissue processing and fluorescence-activated cell sorting for scRNA-seq profiling of CHT. ECs, HCs, and NCs were sorted from Tg (kdrl:mCherry/CD41:GFP) embryos across three developmental stages (55 hpf, 3.5 dpf, and 4.5 dpf). (B) UMAP plot showing 23 CHT cell clusters across three developmental stages (28,777 cells). Cells are colored by their cell-type annotation and numbered according to the legend beside. (C) Heatmap showing blocks of DEGs (top 10 genes) in HSPCs, neural cells, erythrocytes, myeloid cells, ECs, epithelial cells, epidermis, fibroblasts, and muscle cells in the CHT. (D) UMAP visualization of the expression of curated feature genes for cell-type identification.
Cell Type and Gene-Expression Dynamics in Developing CHT.
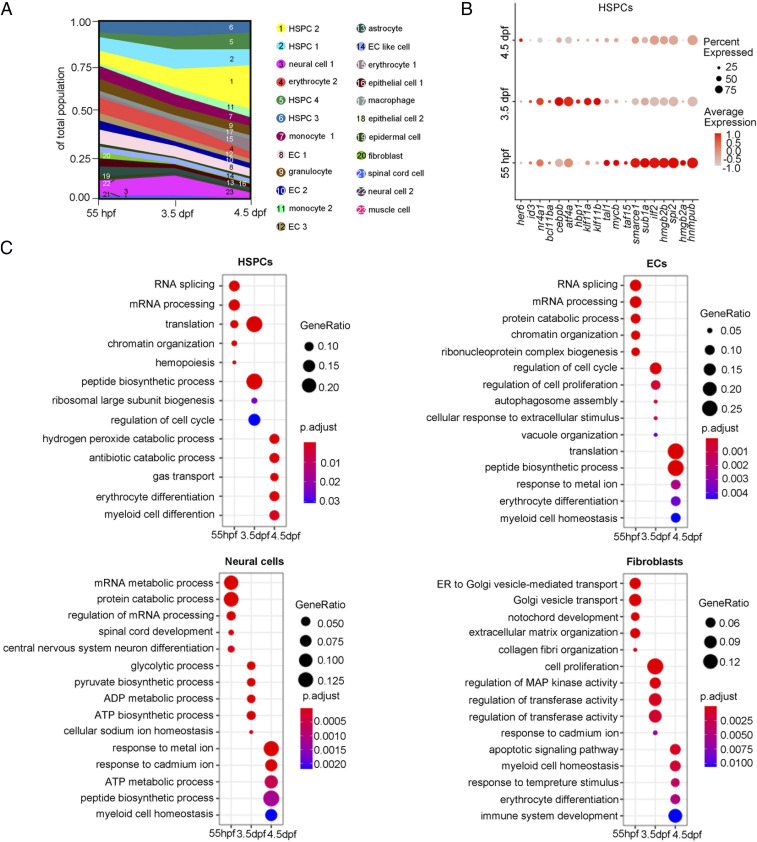
Next, we analyzed the transcriptome atlas to assess the fraction dynamics and gene-expression changes in developing CHT. Integrated analysis of cell atlases from three stages revealed that most of the annotated cell clusters were overlapped (SI Appendix, Fig. S1B), but the fraction of hematopoietic components changed dynamically (Fig. 2A). The fraction of HSPCs and HSPC-derived HC (monocyte 2, macrophage, and erythrocyte 1) increased but that of the erythro-myeloid progenitor-derived HC (early granulocyte, early monocyte 1, and erythrocyte 2) gradually decreased over time, indicating that definitive HSPCs are undergoing rapid expansion and differentiation, whereas early definitive hematopoiesis is being replaced by definitive hematopoiesis in developing CHT (Fig. 2A). Transcription factors (TFs) and gene-ontology (GO) analysis of differentially expressed genes (DEGs) across three developmental stages revealed the stage-specific dynamics of hematopoietic and niche cells. For HSPCs at 55 hpf, tal1 and smarce1 were highly expressed, and the DEGs were enriched in “messenger RNA (mRNA) processing” related terms, and at 3.5 dpf, bcl11ba and cebpb were highly expressed and DEGs were enriched in “translation” and “peptide biogenesis process” related terms, whereas at 4.5 dpf, her6 was highly expressed and DEGs were enriched in “erythrocyte differentiation” and “myeloid cell differentiation” related terms, suggesting that rapidly proliferative HSPCs (55 dpf and 3.5 dpf) initiate differentiation at 4.5 dpf (Fig. 2 B and C). Likewise, niche populations within the CHT exhibited notable dynamic profiles. From 55 hpf to 4.5 dpf, all of the niche cells were experiencing the processes of cell proliferation, tissue organization, and functional enhancement. Interestingly, at 4.5 dpf, in addition to strengthening their own functions, most of the niche populations showed a coordinated change of transcriptomes that were similar to HSPCs (except the population of muscle cells) (Fig. 2C and SI Appendix, Fig. S2A). For instance, the DEGs of ECs, neural cells, fibroblasts, epithelial cells, and epidermis were enriched in GO terms related to “hemopoiesis,” “myeloid cell homeostasis,” and “erythrocyte differentiation,” indicating that the coordinated development of HSPCs and niche components is likely coregulated by shared transcriptional programs (Fig. 2C and SI Appendix, Fig. S2A and Dataset S1) (14). Taken together, global transcriptome profiling of the developing CHT revealed the dynamic features of cellular composition and gene expression of niche components during HSPC development across three stages.
Fig. 2.
Cell composition and gene-expression dynamics during CHT development. (A) Fraction of cell clusters per developmental stage, displaying a dynamic change in cell-type complexity throughout our sampling. (B) Dot plot showing the differentially expressed TFs within the HSPCs at each development stage. (C) Dot plots showing the differentially enriched GO terms within HSPCs, ECs, neural cells, and fibroblasts at each developmental stage.
HSPC Heterogeneity in Lineage Priming and Metabolic Gene Signatures.
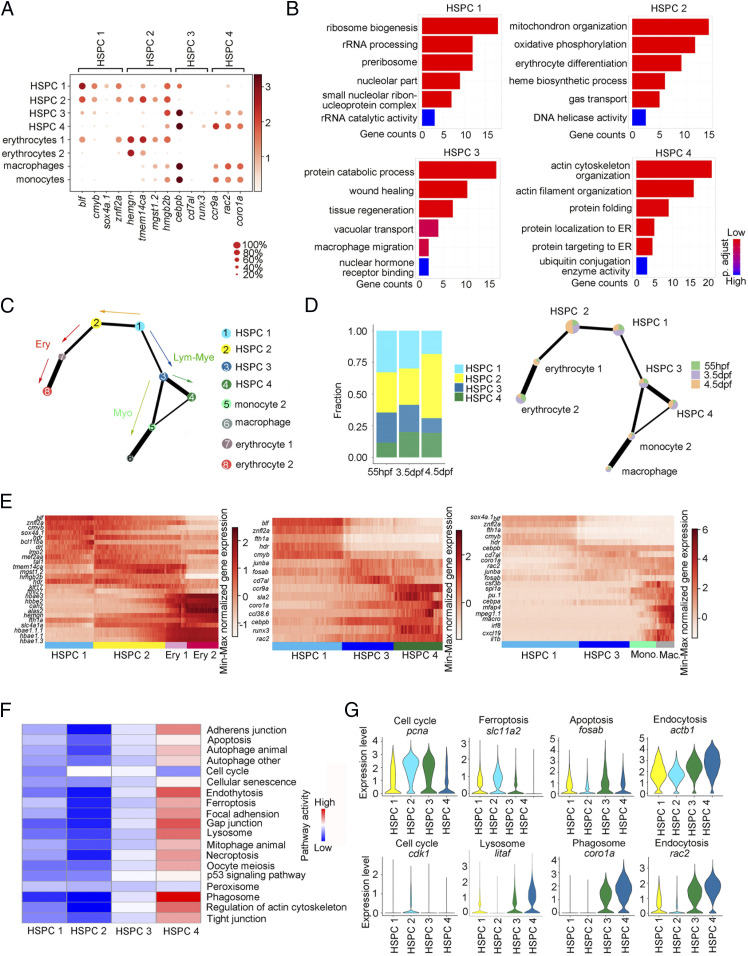
It has been well-known that HSPCs are the heterogeneous population in mammalian hematopoietic tissues (19–22). To further characterize the HSPC heterogeneity in developing CHT, we first compared their transcriptomes. GO and DEG analysis showed that HSPC 1 exhibited a transcriptional activation and highly expressed stemness gene cmyb (23); HSPC 2 was related to erythrocyte differentiation and highly expressed erythroid genes hemgn and tmem14ca (24, 25); HSPC 3 was associated with wound healing, tissue regeneration, and macrophage migration and highly expressed myeloid genes cebpb and runx3 (26, 27); and HSPC 4 was involved in actin cytoskeleton organization and protein folding and highly expressed lymphoid-myeloid genes coro1a, ccr9a, and rac2 (Fig. 3 A and B) (28, 29). Next, we sought to organize these populations using a trajectory model to delineate their lineage relationship. The partition-based graph abstraction (PAGA) and diffusion map analysis identified the organized and branched differentiation routes from the root state (HSPC 1) to the mature HC passing through HSPC 2, HSPC 3, and HSPC 4, respectively (Fig. 3C and SI Appendix, Fig. S3 A and B). The fraction of HSPC 1 is more than that of lineage-biased HSPC 2, HSPC 3, and HSPC 4 at 55 hpf, which means that the expansion of the HSPC 1 pool is to meet the CHT demand for hematopoietic differentiation. At 3.5 dpf and 4.5 dpf, the fraction of HSPC 1 is reduced, while the fraction of lineage-biased HSPC 2, HSPC 3, and HSPC 4 is increased during the progression of differentiation, indicating that the level of heterogeneity of HSPCs was correlated with the developmental timepoints (Fig. 3D). Additionally, heatmap analysis also uncovered the expression dynamics of erythrocyte, lymphoid-myeloid progenitor, and monocyte genes alongside the differentiation trajectories (Fig. 3E). Collectively, we concluded that HSPC 1 is an uncommitted and proliferative population, but HSPC 2 and HSPC 3/4 have primed to differentiation toward their progenies to exert specialized functions.
Fig. 3.
HSPC heterogeneity in lineage priming and metabolic gene signatures. (A) Dot plot of 14 selected DEGs for HSPC 1 through 4 and blood lineages (erythrocytes 1/2, macrophage, and monocyte 2). The size of the dot corresponds to the percentage of cells expressing the gene in each cluster. The color represents the average expression level. (B) The major GO terms enriched in HSPC 1 through 4. (C) PAGA visualization layout of HCs derived from HSPCs (15,084 cells). PAGA showing the putative developmental process from the root state (HSPC 1) to the branches (erythrocyte, myelocyte, and HSPC 4) passing through HSPC 2 and 3. (D) The fraction of four HSPCs at each development stage. (E) Heatmap showing the dynamic gene expressions based on PAGA-inferred trajectories from HSPC 1 to erythrocytes, HSPC 4, and macrophages, respectively. (F) The major metabolic pathway–activity score in HSPC 1 through 4. (G) Violin plots showing the expression of representative metabolic marker genes for cell cycle, ferroptosis, apoptosis, endocytosis, lysosome, and phagosome in HSPC 1, HSPC 2, HSPC 3, and HSPC 4.
Metabolic cues have been reported as a key regulator of hematopoietic stem cell (HSC) fate decisions (30, 31). To study the metabolic states of the four heterogeneous HSPCs, we used the single-cell computational pipeline (32). Interestingly, we found that the four HSPC subpopulations could be clearly distinguished by metabolic genes (SI Appendix, Fig. S3C), suggesting that the metabolic transcriptome signature contributed to HSPC heterogeneity. By analyzing the metabolic genes from the Kyoto Encyclopedia of Genes and Genomes database to obtain the pathway activity scores of four HSPCs, we observed that heterogeneous HSPCs up-regulated the expression of distinct sets of metabolic genes. HSPC 2 exhibited active cell-cycle progression and mitochondrial activity with up-regulated cell-cycle–related genes (pcna and cdk1) and ferroptosis gene (slc11a2), explaining the increased numbers of HSPC-derived erythrocytes during development. HSPC 3 highly expressed phagosome and apoptosis marker gene fosab, supporting its myeloid differentiation potential. Lymphoid-myeloid–biased HSPC 4 showed active endoplasmic reticulum and Golgi activity with up-regulated endocytosis marker genes actb1 and rac2, and lysosome and phagosome marker genes litaf and coro1a, consistent with its function of adaptive immunity (Fig. 3 F and G and SI Appendix, Fig. S3D). In conclusion, HSPCs showed heterogeneity, manifested as different lineage priming and metabolic gene signatures in developing CHT.
Identification of Crucial Niche Regulators in Facilitating HSPC Expansion.
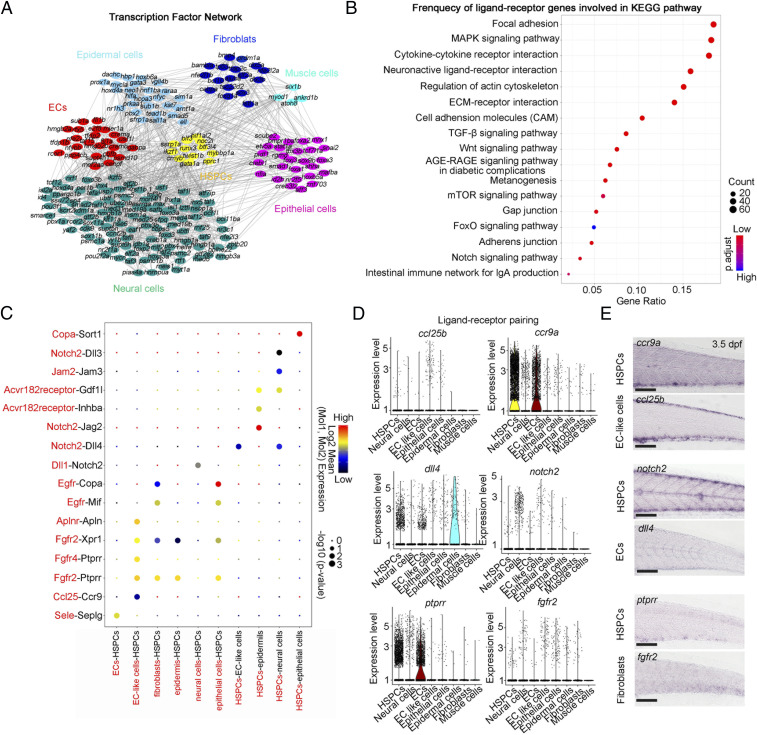
HSPC expansion has been demonstrated to be a crucial feature occurring in the CHT and regulated by the extrinsic complex niche components, including vascular ECs, stromal cells, and macrophages (7, 9–11). To further decode the regulatory mechanism of nonhematopoietic niche components (ECs, neural cells, epithelial cells, epidermal cells, muscle cells and fibroblasts) for HSPC expansion, we first constructed a genetic network that underlies each nonhematopoietic niche-component development. We identified 196 differentially expressed TFs, and the analytic results showed that the enriched expression of cell type-specific TFs in corresponding niche components may support HSPC expansion via regulating CHT niche formation (Fig. 4A). For example, ETS family factor fli1b specifically expressed in ECs has been demonstrated to be required for angiogenesis (33, 34); hoxd4a specifically expressed in neural cells has been reported to be involved in vasculogenesis and angiogenesis in zebrafish CVP (Fig. 4A) (35).
Fig. 4.
Cell–cell interaction network between HSPCs and nonhematopoietic niche components. (A) The interaction network of TFs of each component. All differentially expressed TFs in each cell type are used to construct network; nodes (TFs) with more than one edge are shown. (B) Dot plot showing the frequency of ligand–receptor genes involved in Kyoto Encyclopedia of Genes and Genomes pathways. (C) Dot plot showing the ligand (red)–receptor (black) pairs between HSPCs and ECs, EC-like cells, fibroblasts, neural cells, epidermal cells, and epithelial cells. (D) Violin plot showing the normalized expression of the ligand and its receptor genes in HSPCs and ECs, fibroblasts, neural cells, spinal cord cells, epidermis, and epithelial cells for each indicated pairing. (E) WISH indicating the expression patterns of ccr9a, notch2, and ptprr in HSPCs, ccl25b and dll4 in ECs, and fgfr2 in fibroblasts at 3.5 dpf. (Scale bar, 100 μm.)
Previous studies showed that vascular ECs, muscle cells, and neural cells are involved in the short-range and/or long-range regulation of CHT-HSPC development (14). To further explore the dynamic cell communication network governing HSPC development, we performed unbiased ligand–receptor analysis of CHT HSPCs and niche cells, including vascular ECs, fibroblasts, neural cells, and epidermal cells using CellPhoneDB (SI Appendix, Fig. S4 A and B) (36). We identified distinct ligand–receptor pairs, including the autocrine and bidirectional effects of HSPCs on the niche cells (SI Appendix, Fig. S4 A and B). There were strong interactions related to “cytokine–cytokine receptor interaction” and “neuron-active ligand–receptor interaction” between HSPCs and their niches, suggesting their important roles during niche development and HSPC expansion (Fig. 4B). Among them, we focused on the interaction between receptors expressed by HSPCs and ligands expressed by other niche cell types. For example, the interaction between EC-like cells and HSPCs via Ccl25b-Ccr9a has been demonstrated to be required for HSPC expansion in CHT (Fig. 4C) (9). We also predicted two unrecognized ligand–receptor pairs, Notch-mediated cell–cell contact regulation between ECs and HSPCs (Dll4-Notch2) and fibroblast related short-range regulation between fibroblasts and HSPCs (Fgfr2 and Ptprr) (Fig. 4 C and D). Whole-mount in situ hybridization (WISH) confirmed their cell-type–specific expression patterns, implying the potential regulatory role of chemokine signaling, Notch signaling, and fibroblast growth factor receptor (FGFR) signaling in HSPC expansion (Fig. 4E).
Gpr182 Enhances Functional EC Niche to Support HSPC Expansion.
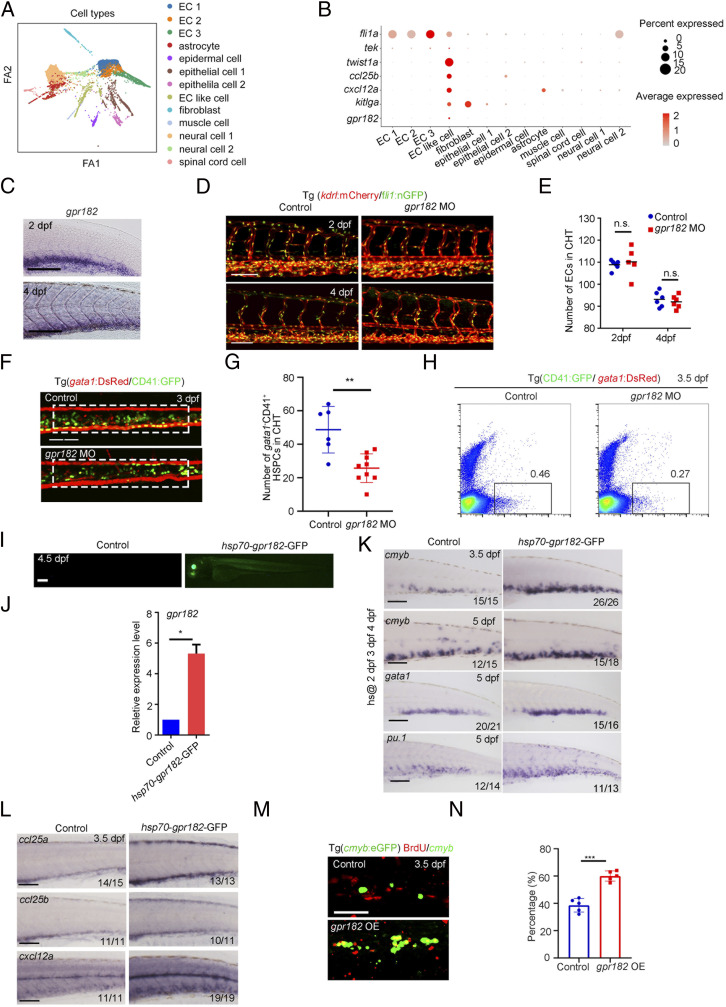
To explore the functional plasticity of CHT based on our aforementioned bioinformatic analysis, we focused on a type of previously unrecognized cells coexpressing EC genes, fli1a and tek, and stromal cell gene, twist1a, which were termed as EC-like cells (Fig. 5 A and B) (37). This cell type also expressed cytokine- and chemokine-related genes, kitlga, cxcl12a, and ccl25b, as well as a specific and uncharacterized G protein–receptor encoding gene, gpr182 (Fig. 5B). Gpr182, formerly known as adrenomedullin receptor, has been recently reported to regulate HSPC generation and myeloid cell differentiation (38, 39). However, it was unknown whether and how gpr182 was spatiotemporally involved in HSPC expansion in the CHT region. From the WISH result, we found that gpr182 is not maternally expressed and is first expressed at 1k-cell stage at which the zygotic genome becomes transcriptionally active. At 28 and 36 hpf, gpr182 is expressed in the ventral wall of dorsal aorta (VDA) and posterior caudal vein. At 2 and 4 dpf, the expression level of gpr182 was decreased in the VDA and specifically maintained in the CHT (Fig. 5C and SI Appendix, Fig. S5A). This result indicated that gpr182 showed a spatiotemporally specific expression pattern in the VDA and CVP. To further characterize the cellular identity of gpr182-expressing cells, we examined its expression in cloche and etsrp mutants, which lack hemato-vascular system and ECs, respectively. The results showed that gpr182 expression was barely detected in both cloche and etsrp mutants, indicating their EC identity (SI Appendix, Fig. S5B).
Fig. 5.
Identification of crucial niche regulator for HSPC expansion. (A) ForceAtlas2 plot of nonhematopoietic niche cells (10,200 cells). Cells are colored by their cell-type annotation. (B) Dot plot showing the expression of selected known EC, muscle cell, and epithelial cell genes (fli1a, tek, twist1a, ccl25b, cxcl12a, kitlga, and gpr182) for niche cell clusters. (C) WISH showing the expression of gpr182 in CHT at 2 dpf and 4 dpf. (D) Confocal imaging of the Tg (kdrl:mCherry/fli1:nGFP) showing the structure of ECs in the CHT region at 2 dpf and 4 dpf in control and gpr182 morphants. (Scale bar, 50 mm.) (E) The statistical data of the CHT-EC number in control and morphants at 2 dpf and 4 dpf. The dashed boxes indicate the region of EC counting. (F) Confocal imaging of the Tg (CD41:GFP/gata1:DsRed) showing the number of CD41 +gata1− HSPCs in the CHT region at 3 dpf in control and gpr182 morphants. (Scale bar, 50 mm.) (G) The statistical data of the CHT CD41 +gata1− HSPC number in F. The dashed boxes indicate the region of CD41+gata1− HSPC counting. (H) Fluorescence-activated cell sorting analysis showing the number of CD41+ gata1− HSPCs in control and gpr182 morphants at 3.5 dpf. (I) The imaging of GFP fluorescence in Tg (hsp70-gpr182-GFP) embryos at 4.5 dpf. Heat shock was performed at 2 dpf, 3 dpf, and 4 dpf. (J) qPCR result of gpr182 expression in control and hsp70-gpr182-GFP-positive embryos. (K) WISH showing the expression of cmyb, gata1, and pu.1 in the CHT of the control and hsp70-gpr182-GFP-injected embryos at 3.5 dpf and 5 dpf. (L) WISH showing the expression of ccl25a, ccl25b, and cxcl12a in control and gpr182-overexpressed embryos at 3.5 dpf. (M) The double staining imaging of anti-BrdU and anti-GFP antibodies in control and gpr182-overexpressed embryos at 3.5 dpf. OE, overexpression. (N) The statistical data of the percentage of cmyb:GFP+BrdU+/cmyb:GFP+ cells in M. (Scale bar (C, I, L, and M), 100 μm.) The results are represented as means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Student’s t test.
To determine whether gpr182 is involved in CHT hematopoiesis, we used an ATG morpholino (MO) to knock down gpr182 expression in zebrafish. After validation of the efficiency of MO knockdown by Western blotting (SI Appendix, Fig. S5C), we determined the effect of gpr182 knockdown on CHT hematopoiesis. We found that although the structure of CVP and the number or ECs were normal in the gpr182 morphants (Fig. 5 D and E), the number of CD41+gata1− HSPCs were reduced within the CVP of gpr182 morphants compared with control embryos at 3 dpf (Fig. 5 F and G). Fluorescence-activated cell sorting analysis also confirmed that the number of CD41+gata1− HSPCs decreased significantly in the morphants of Tg (CD41:GFP/gata1:DsRed) background at 3.5 dpf (Fig. 5H), indicating an indispensable function of Gpr182 during HSPC development in the CHT.
To further determine whether gpr182 plays a stage-specific role in CHT-HSPC development, we performed heat-shock induction of gpr182 after 2 dpf, when VDA-derived HSPCs initiate the colonization in the CHT (Fig. 5 I and J). WISH results showed that overexpression of gpr182 at 2 dpf, 3 dpf, and 4 dpf enhanced the expression of HSPC gene cmyb at 3.5 dpf and 5 dpf, erythrocyte gene gata1, and myeloid cell gene pu.1 at 5 dpf (Fig. 5K), suggesting the positive regulation of gpr182 in CHT hematopoiesis. Given that hemogenic endothelium (HE)/HSPC formation in the zebrafish VDA has been reported to be negatively regulated by gpr182 (38), we performed gpr182 overexpression with heat-shock induction at 28 hpf, when HE/HSPC formation occurs, to validate its negative regulatory role. WISH results showed that overexpression of gpr182 decreased cmyb expression in the VDA at 36 hpf (SI Appendix, Fig. S5D), which is similar to the phenotype reported in the previous study (38). Previous studies in mice have reported that chemokines secreted by ECs play roles in guiding HSPC migration into the CVP and promoting HSPC proliferation and differentiation (40). To dissect the underlying mechanism of Gpr182+ ECs in facilitating HSPC development, we examined the expression of genes related to chemokines, such as cxcl12a, ccl25a, and ccl25b (9, 10). WISH results showed that their expression was significantly increased upon gpr182 overexpression (Fig. 5L). Meanwhile, qPCR result showed that the genes associated with cell division, ccna2, cdk1, and stil, were significantly increased upon gpr182 overexpression (SI Appendix, Fig. S5E). We further examined the proliferation state of HSPCs in the control and gpr182-overexpressed embryos by 5-bromodeoxyuridine (BrdU) staining experiments. The percentage of cmyb+BrdU+ double-positive cells in the CHT was increased significantly in the gpr182-overexpressed embryos (Fig. 5 M and N). Collectively, these studies suggested that an EC-specific factor, Gpr182, plays a positive role to promote HSPC expansion in the CHT niche, via increasing the expression of chemokine genes.
A recent study showed that the deletion of gpr182 in zebrafish embryos leads to an increased number of HE/HSPCs in the VDA (38). We next generated the gpr182-F0 mutant using a highly efficient two-RNA component (CRISPR-derived RNA:transactivating RNA) version of the CRISPR/Cas9-mediated mutagenesis system (41). Duplex guide ribonucleoproteins were designed to target sequences in the second exon of gpr182 for generating the CRISPR/Cas9 mutants (SI Appendix, Fig. S5F). Western blotting showed that the level of Gpr182 protein was decreased markedly in the F0 mutants (SI Appendix, Fig. S5G). WISH results showed that the expression of cmyb was increased in the VDA at 36 hpf, and at 4.5 dpf, the expression of cmyb and pu.1 was increased obviously in the CHT region of gpr182-F0 mutants (SI Appendix, Fig. S5H). Moreover, the gpr182-F1 mutants were generated using conventional the CRISPR-Cas9 approach (SI Appendix, Fig. S5I). The increased expression of cmyb in the 36 hpf VDA and 4 dpf CHT was also observed in the gpr182-F1 mutants (SI Appendix, Fig. S5J). Overall, the hematopoietic phenotype of our newly generated gpr182 mutants is similar to that of gpr182 mutants reported in the previous study (38). Given that the HSPC phenotype in the gpr182 mutants is different from that observed in the gpr182 morphants, a possible explanation for this discrepancy is due to the genetic compensation response (GCR) triggered by nonsense mRNA decay in the mutants but not morphants (42–44). Indeed, the decrease of gpr182 transcript levels in the gpr182 mutants compared with control embryos was observed by qPCR (SI Appendix, Fig. S5K). Notably, the transcripts of G protein–receptor family members, including gpr183a, gpr37b, and gpr132b, were increased in the gpr182 mutants but not in the gpr182 morphants (SI Appendix, Fig. S5L), implying the possible GCR in the gpr182 mutants.
EC-Specific Gpr182 Signaling Is Required for HSPC Expansion in Mouse FL.
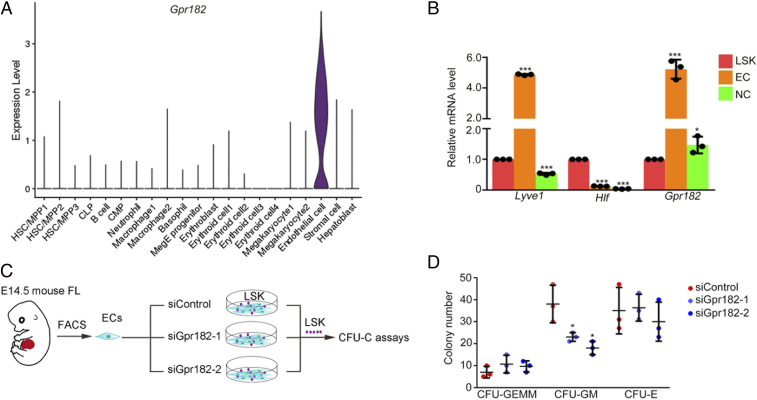
To investigate whether Gpr182 plays an evolutionarily conserved role in mammals, we first examined the expression pattern of Gpr182 in mouse FL cells by analyzing the scRNA-seq data in mouse FL. A violin plot showed that the expression of Gpr182 was highly enriched in mouse FL ECs (Fig. 6A). We also determined the expression of Gpr182 in sorted ECs and Lineage −Sca-1+c-Kit+ (LSK) cells from embryonic day 14.5 (E14.5) FL by qPCR and found that Gpr182 expression was enriched in ECs (Fig. 6B and SI Appendix, Fig. S6A). We next performed in vitro functional experiments with HSPCs sorted from the mouse FL (Fig. 6C). A previous study showed that GPR182 is a marker for human liver sinusoidal ECs (39). To examine whether EC-specific Gpr182 is required for HSPC expansion, we performed the small interfering RNA (siRNA) knockdown assay specifically in ECs and then cocultured them with HSPCs (LSK cells) (Fig. 6 C and D). The efficiency of siRNA knockdown was validated by qPCR and Western blotting (SI Appendix, Fig. S6 B and C). The results showed that knockdown of Gpr182 in ECs could significantly reduce the formation ability of granulocyte-macrophage colonies of HSPCs (Fig. 6D). These results, together with the data obtained in zebrafish, demonstrated the conserved Gpr182 signaling cascade as an essential regulatory mechanism for HSPC expansion in vertebrates.
Fig. 6.
EC-specific Gpr182 is required for HSPC expansion in mouse FL. (A) The expression pattern of Gpr182 in mouse FL scRNA-seq data. (B) qPCR analysis of Gpr182 in ECs, LSK cells, and NC cells. Lyve1 and Hlf as positive markers for ECs and HSPCs, respectively. (C) The flowchart of ex-vivo siGpr182 knockdown and HSPC culture. (D) CFU-cell assays to detect the colony-forming ability of HSPCs following siRNA treatment.
Cross-Species Analysis of Human FL and Zebrafish CHT.
To better understand the evolutionary conservation and divergence between the mammalian FL and zebrafish CHT, we compared our data with the recently published scRNA-seq data of human FL (19). After quality control and batch correction, a total of 8,432 single cells from human FL at 13 postconception weeks and 10,010 single cells from zebrafish CHT at 3.5 dpf were integrated and subjected to UMAP analysis (Fig. 7A). First, we found that HSPCs, HCs (macrophages, neutrophils, erythrocytes, and thrombocytes), and nonhematopoietic niche cells (ECs and fibroblasts) were mostly conserved in two species. Then, we also found that several clusters were divergent between human FL and zebrafish CHT. For instance, neural cells (cluster 6), epidermis (cluster 11), and epithelial cells (cluster 12) were identified in the zebrafish CHT, while hepatocytes (cluster 9) and lymphoid cells (cluster 13) were found in the human FL (Fig. 7A and SI Appendix, Fig. S7 A and B). Overall, the conservation of cellular components indicated that HSPC development in the mammalian FL and zebrafish CHT was mainly associated with EC-forming vascular niche, whereas the divergence of cellular components also demonstrated the existence of species–specific cellular regulatory mechanisms for HSPC development (14, 45).
Fig. 7.
Cross-species analysis between human FL and zebrafish CHT reveals conserved and divergent gene-expression profiles. (A, Left) Unsupervised graph-based clustering of single cells from human FL (n = 8,432) and zebrafish CHT (n = 10,010) based on homologous gene-expression profiles; color labels for different species. (A, Right) Color labels for identified cell clusters. (B) Similarity matrix showing the Pearson correlations between each pair of corresponding cell clusters from human FL and zebrafish CHT based on homologous gene-expression profiles. (C) Dot plot showing the conserved signature genes of each cluster in human and zebrafish. (D) Identification of GPR182, DLL4, and NOTCH2 expression in human FL cell clusters. (E) The major GO terms of divergent HSPC genes in human FL and zebrafish CHT.
Furthermore, we examined the conservation and divergence of transcriptome features between human FL and zebrafish CHT. First, we performed correlation analysis of homologous genes among the two species and found that the gene expression of corresponding cell clusters was conserved between human and zebrafish (Fig. 7B). We also examined the conserved expression of several marker genes. Specifically, MYB serving as an HSPC development regulator showed a conserved gene expression in HSPCs of human and zebrafish (46, 47); ANGPT4 has been demonstrated to play an essential role in HSPC development and displayed a conserved EC-specific expression pattern in humans (Fig. 7C and SI Appendix, Fig. S7 A–C) (48). In addition, we found that GPR182 was specifically expressed in human FL ECs, and ligand–receptor pair Dll4-Notch2 was also expressed in human FL ECs and HSPCs, implying their conserved role in HSPC expansion across species (Fig. 7D). Together, our bioinformatics and functional experiments support that the regulation of HSPC expansion by EC-specific factor Gpr182 is conserved between zebrafish and mammals.
Furthermore, we observed the transcriptional divergence in hematopoietic components. For example, zebrafish HSPCs showed the characteristics of responses of hypoxia and temperature homeostasis, while human HSPCs were associated with leukocyte differentiation and inflammatory response (Fig. 7E). Taken together, our bioinformatic analysis revealed the conservation and divergence of gene expression between human FL and zebrafish CHT. More importantly, the divergent gene expression of HSPCs is potentially associated with their evolutionarily adaptive response to microenvironment.
Discussion
Zebrafish CHT is characterized as a unique hematopoietic organ with multiple biological functions (9, 14, 49–51). In this study, we used scRNA-seq, combined with functional assays, to reveal the global transcriptome landscape and cellular organization of developing CHT in zebrafish. First, we revealed the dynamics of cellular components and transcriptional features and resolved HSPC heterogeneity in developing CHT. Then, we dissected the complexed nonhematopoietic niche components and identified an EC-specific factor, Gpr182, and validated its role in strengthening the niche function for HSPC expansion in zebrafish and mice. Finally, we comprehensively uncovered the conservation and divergence of developmental hematopoiesis, in terms of cellular components and gene expression, between human FL and zebrafish CHT.
HSPC heterogeneity is being unveiled with the advance of single-cell technology; however, the causes of HSPC heterogeneity remain unclear (31, 52). A combination of various factors, including the cell extrinsic factors derived from HSPC-resident niches, and cell intrinsic factors, such as cell cycle and transcriptomic features, likely shape the heterogeneity within the stem and progenitor compartment (31). In our study, we identified the heterogeneous HSPCs by different lineage-specific genes in zebrafish CHT. Of note, the four HSPC subpopulations could also be clearly distinguished by metabolic genes, suggesting that the metabolic signature might contribute to HSPC heterogeneity. Recent studies have reported that metabolism is the regulator of HSC heterogeneity and that metabolic defects can shift HSC symmetric self-renewal toward commitment, which leads to HSC exhaustion (31, 53). For example, compared to the multipotent progenitor (MPP) and other progenitors, HSCs remain relatively quiescent to maintain their undifferentiated and multipotent state (54). Furthermore, a mitochondrial fusion regulator, mitofusion-2(mfn2), is required for HSC maintenance and is especially critical to cell differentiation to the lymphoid lineages (55). However, it remains to be investigated whether metabolic state can act as a key regulator of HSPC fate decision during embryogenesis, which awaits further investigation.
HSPC expansion is tightly controlled by various cell-intrinsic and cell-extrinsic factors, which have not been fully understood yet. To achieve efficient HSPC expansion in vitro or in vivo, a better understanding of HSPC expansion in their native niches is required. A recent in vitro study reported that a 899-fold increase of mouse functional HSCs via optimizing culture conditions could be accomplished (56), which further emphasizes the significance of microenvironmental effects on HSC expansion. The complexed HSPC niche components provide physical support as well as biological signals to sustain HSPC lodgment and expansion. For example, in the bone marrow, Leptin receptor-positive perivascular stromal cells are the major source of Scf and Cxcl12 (40, 57, 58) that can promote HSC localization and maintenance within the perivascular niche. In zebrafish, CHT is a highly vascularized niche, and the rapidly increased vascular ECs form a complex structure composing the caudal artery, caudal vein, and CVP. Moreover, HSPC development in the CHT is also regulated by other types of cell components, such as fibroblasts, neural cells, and muscle cells (4). In this study, we first presented a coordinated regulation of diverse niche cells for HSPC development at cellular level. Next, we analyzed the transcriptional network of different niche components and further identified cell–cell interactions governing HSPC development at molecular level. Moreover, we characterized an EC-specific factor Gpr182, which plays a positive role in remodeling of the EC niche that favors CHT-HSPC expansion. Given that Gpr182 has been reported to be a negative regulator for HSPC formation and myeloid differentiation by a recent paper (38), we reason that Gpr182 serves as a stage-specific regulator for HSPC development at different developmental windows. Taken together, these results implied the functional plasticity of the CHT microenvironment via genetic manipulation.
In summary, we build up a single-cell transcriptomic atlas of HSPC expansion in the zebrafish CHT, which provides an essential resource for understanding regulatory mechanisms of HSPC expansion and differentiation in the niche. Importantly, revealing the conservation and divergence between human FL and zebrafish CHT will uncover the underlying key factors in HSPC expansion, which will help improve current protocols in functional HSPC expansion in vitro.
Materials and Methods
Zebrafish and Mice.
Zebrafish strains including AB, Tubingen, etsrpy11 mutant (59), clochem378 mutant (60), and Tg (kdrl:mCherry) (61), Tg (CD41:GFP) (62), Tg (gata1:DsRed) (63), Tg (cmyb:GFP) (64), and Tg (fli1a-nuclear-GFP) (65) were raised in 28.5 °C system water and staged as previously described (66). Wild-type C57BL/6 mice were housed under specific pathogen-free conditions and handled in accordance with institutional guidelines. The embryos from crosses between adult males and females of zebrafish or mice were used for all experiments. This study was approved by the Ethical Review Committee in the Institute of Zoology, Chinese Academy of Sciences, China.
MOs, Plasmid Construction, and Microinjection.
The antisense MO of gpr182 was purchased from GeneTools and prepared as 1 mM stock solutions using RNase-free H2O. The sequence of gpr182-ATG MO is 5′-TGAGTTGTGAATATCATGCGTCATG-3′ (4 ng for gpr182 MO). gpr182-ATG MO was injected at one-cell-stage zebrafish embryos at the yolk/blastomere boundary. For the overexpression experiment of gpr182 in the whole embryos, the full-length complementary DNA (cDNA) was cloned and assembled into the pDestTol2pA2 vector with an hsp70 promoter and an EGFP reporter by Gateway systems (67). Overexpression plasmids (50 pg) and Tol2 mRNA (25 pg) were injected alone or in combination, into one-cell-stage zebrafish embryos at the yolk/blastomere boundary.
WISH and Fluorescence In Situ Hybridization.
WISH for zebrafish embryos was performed with probes, including gpr182, runx1, cmyb, gata1, pu.1, cxcl12a, ccl25a, ccl25b, ccr9a, fgfr2, ptprr, notch2, and dll4, as previously described (14). Fluorescence in situ hybridization was used to detect the expression of cmyb. The protocol was performed similarly to the WISH before the antibody incubation. After removing antibody and washing embryos with phosphate buffered saline with 0.1% Tween-20 (PBST), embryos were stained with Tyramide signal amplification (TSA)-FITC amplification reagent (1:100). Before immunofluorescence staining, the first color reaction was stopped by gradient methanol. After removing the reaction buffer, embryos were washed sequentially by 25, 50, and 75% methanol/PBST (10 min/each buffer), 1% H2O2/methanol (30 min), 75, 50, and 25% methanol/PBST (10 min/each buffer), and PBST (2 × 35 min).
BrdU Staining.
BrdU labeling was performed as described previously (9). In brief, embryos were injected with BrdU (10 mM) at 3.5 dpf and were fixed using 4% paraformaldehyde at 2 h after injection. After washing, embryos were digested with the Proteinase K (10 mg/mL), then treated with HCl (2 mol/L) for 1 h. After blocking with 1% bovine serum albumin (BSA) for 1 h, the embryos were incubated with anti-BrdU antibody (1:800) (5-Bromo-20-deoxy-uridine Labeling and Detection Kit, Roche) at 4 °C overnight. After washing with PBST, the embryos were incubated with Alexa Fluor 555 Goat Anti-Mouse IgG (H+L) Antibody (1:500).
qPCR.
For zebrafish embryos, total RNAs of the tail region (20 to 40 embryos pooled for each sample) at 4 dpf were extracted by TRIzol and reverse transcribed using M-MLV Reverse Transcriptase. The cDNA was diluted fivefold to be used as the template for qPCR amplification (9). The qPCR primers are listed in SI Appendix, Table S3.
Confocal Microscopy.
For the confocal microscopy, zebrafish embryos were mounted in 1% low melting agarose and transgenic zebrafish embryos were imaged by Nikon confocal A1 laser microscope with 20× objective. The imaging was edited by ImageJ and Photoshop CS6.
Western Blotting.
The samples were manually homogenized with a 1 mL syringe and needle in lysis buffer (10 mM Tris HCl, pH 8.0, 10 mM NaCl, and 0.5% Nonidet P-40) containing 1× protease inhibitor. Lysate was centrifuged at 12,000 g for 2 min at 4 °C, and the supernatant was loaded as protein sample. The following antibodies were used: anti-GPR182 antibody (Abcam, ab182555) and anti–β-Actin antibody (Cell Signaling Technology, 4967). Quantification of each band was carried out by using Quantity One software (Bio-Rad).
siRNA Interference.
Control and specific siRNAs were designed and synthesized by Genepharma Corporation. Flow cytometry sorted E14.5 FL ECs (Ter119− CD45− CD31+) were first cultured in 24-well plate, and siRNAs were transfected with RNAiMAX Transfection Reagent (Invitrogen, 13778030) according to the manufacturer’s instructions. Then, E14.5 FL LSKs were pooled together with siRNA-treated ECs at a 1:10 ratio for coculture. The sequences of siRNA used in the present study are listed in SI Appendix, Table S4.
scRNA-Seq Analysis.
The details on scRNA-seq analysis are provided in SI Appendix, Materials and Methods.
Colony-Forming Unit Cell Assay.
The cells treated with control and Gpr182 siRNAs were harvested and added into MethoCult GF M3434 medium (Stem Cell Technologies, 03434) in ultra-low attachment 24-well plates (Corning, 3473). After being cultured at 37 °C in 5% CO2 for 7 to 10 d, the number for each type of colonies including colony-forming unit (CFU)-erythroid, CFU-granulocyte, macrophage, and CFU-granulocyte, erythrocyte, macrophage, megakaryocyte was counted.
Statistical Analysis.
All statistical analyses of qPCR and confocal imaging were performed on at least three independent biological or experimental replicates. Student’s two-tailed unpaired t test was used for statistical comparisons, and data were shown as mean ± SD; P values were used to indicate the significance.
Supplementary Material
Acknowledgments
We thank Prof. Cheng Li of Peking University for helpful discussions. This work was supported by grants from the National Key Research and Development Program of China (2018YFA0800200, 2018YFA0801000, and 2016YFA0100500), the Strategic Priority Research Program of the Chinese Academy of Sciences, China (XDA16010207), the National Natural Science Foundation of China (31830061, 81530004, and 31425016), and Youth Innovation Promotion Association Chinese Academy of Sciences (CAS) (2016083).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015748118/-/DCSupplemental.
Data Availability
The accession numbers for the scRNA-seq data reported in this paper are Gene Expression Omnibus (GEO): GSE120581 and GSE146404 and Genome Sequence Archive (GSA): CRA002374. All other study data are included in the article and/or supporting information.
References
- 1.Orkin S. H., Zon L. I., Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132, 631–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazo I. B., Massberg S., von Andrian U. H., Hematopoietic stem and progenitor cell trafficking. Trends Immunol. 32, 493–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wattrus S. J., Zon L. I., Stem cell safe harbor: The hematopoietic stem cell niche in zebrafish. Blood Adv. 2, 3063–3069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murayama E., et al., Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 25, 963–975 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Ema H., Nakauchi H., Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95, 2284–2288 (2000). [PubMed] [Google Scholar]
- 6.Bordignon C., Stem-cell therapies for blood diseases. Nature 441, 1100–1102 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Mahony C. B., Fish R. J., Pasche C., Bertrand J. Y., tfec controls the hematopoietic stem cell vascular niche during zebrafish embryogenesis. Blood 128, 1336–1345 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Tamplin O. J., et al., Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue Y., et al., The vascular niche regulates hematopoietic stem and progenitor cell lodgment and expansion via klf6a-ccl25b. Dev. Cell 42, 349–362.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Blaser B. W., et al., CXCR1 remodels the vascular niche to promote hematopoietic stem and progenitor cell engraftment. J. Exp. Med. 214, 1011–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D., et al., VCAM-1+ macrophages guide the homing of HSPCs to a vascular niche. Nature 564, 119–124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theodore L. N., et al., Distinct roles for matrix metalloproteinases 2 and 9 in embryonic hematopoietic stem cell emergence, migration, and niche colonization. Stem Cell Reports 8, 1226–1241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murayama E., et al., NACA deficiency reveals the crucial role of somite-derived stromal cells in haematopoietic niche formation. Nat. Commun. 6, 8375 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Xue Y., et al., A 3D atlas of hematopoietic stem and progenitor cell expansion by multi-dimensional RNA-seq analysis. Cell Rep. 27, 1567–1578.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Kissa K., et al., Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 111, 1147–1156 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Bertrand J. Y., Kim A. D., Teng S., Traver D., CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development 135, 1853–1862 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuart T., et al., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popescu D. M., et al., Decoding human fetal liver haematopoiesis. Nature 574, 365–371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Gao S., Xia J., Liu F., Hematopoietic hierarchy–An updated roadmap. Trends Cell Biol. 28, 976–986 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Velten L., et al., Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusi B. K., et al., Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Jin H., Li L., Qin F. X., Wen Z., cMyb regulates hematopoietic stem/progenitor cell mobilization during zebrafish hematopoiesis. Blood 118, 4093–4101 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Peters M. J., et al., Divergent Hemogen genes of teleosts and mammals share conserved roles in erythropoiesis: Analysis using transgenic and mutant zebrafish. Biol. Open 7, bio035576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson R., et al., Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 10, 119–130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyons S. E., et al., Molecular cloning, genetic mapping, and expression analysis of four zebrafish c/ebp genes. Gene 281, 43–51 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Kalev-Zylinska M. L., et al., Runx3 is required for hematopoietic development in zebrafish. Dev. Dyn. 228, 323–336 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Wang S., He Q., Ma D., Xue Y., Liu F., Irf4 regulates the choice between T lymphoid-primed progenitor and myeloid lineage fates during embryogenesis. Dev. Cell 34, 621–631 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Lu X., Zhang Y., Liu F., Wang L., Rac2 regulates the migration of T lymphoid progenitors to the thymus during zebrafish embryogenesis. J. Immunol. 204, 2447–2454 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Ito K., Ito K., Hematopoietic stem cell fate through metabolic control. Exp. Hematol. 64, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas S., Trumpp A., Milsom M. D., Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell 22, 627–638 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Xiao Z., Dai Z., Locasale J. W., Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 10, 3763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F., Patient R., Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ. Res. 103, 1147–1154 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Baltrunaite K., et al., ETS transcription factors Etv2 and Fli1b are required for tumor angiogenesis. Angiogenesis 20, 307–323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amali A. A., Sie L., Winkler C., Featherstone M., Zebrafish hoxd4a acts upstream of meis1.1 to direct vasculogenesis, angiogenesis and hematopoiesis. PLoS One 8, e58857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vento-Tormo R., et al., Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf A., et al., Zebrafish caudal haematopoietic embryonic stromal tissue (CHEST) cells support haematopoiesis. Sci. Rep. 7, 44644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon H. B., et al., The orphan G-protein coupled receptor 182 is a negative regulator of definitive hematopoiesis through leukotriene B4 signaling. ACS Pharmacol. Transl. Sci. 3, 676–689 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid C. D., et al., GPR182 is a novel marker for sinusoidal endothelial differentiation with distinct GPCR signaling activity in vitro. Biochem. Biophys. Res. Commun. 497, 32–38 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Ding L., Saunders T. L., Enikolopov G., Morrison S. J., Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoshijima K., et al., Highly efficient CRISPR-cas9-based methods for generating deletion mutations and F0 embryos that lack gene function in zebrafish. Dev. Cell 51, 645–657.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Brolosy M. A., et al., Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Brolosy M. A., Stainier D. Y. R., Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 13, e1006780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma Z., et al., PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Khan J. A., et al., Fetal liver hematopoietic stem cell niches associate with portal vessels. Science 351, 176–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin H., et al., Definitive hematopoietic stem/progenitor cells manifest distinct differentiation output in the zebrafish VDA and PBI. Development 136, 647–654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X., Angelis N., Thein S. L., MYB–A regulatory factor in hematopoiesis. Gene 665, 6–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng J., et al., Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature 485, 656–660 (2012).Correction in: Nature488, 684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perlin J. R., Robertson A. L., Zon L. I., Efforts to enhance blood stem cell engraftment: Recent insights from zebrafish hematopoiesis. J. Exp. Med. 214, 2817–2827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He S., et al., In vivo single-cell lineage tracing in zebrafish using high-resolution infrared laser-mediated gene induction microscopy. eLife 9, e52024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhan Y., et al., The caudal dorsal artery generates hematopoietic stem and progenitor cells via the endothelial-to-hematopoietic transition in zebrafish. J. Genet. Genomics 18, 30099–7 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Laurenti E., Göttgens B., From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito K., Bonora M., Ito K., Metabolism as master of hematopoietic stem cell fate. Int. J. Hematol. 109, 18–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weissman I. L., Anderson D. J., Gage F., Stem and progenitor cells: Origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell Dev. Biol. 17, 387–403 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Luchsinger L. L., de Almeida M. J., Corrigan D. J., Mumau M., Snoeck H. W., Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature 529, 528–531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkinson A. C., et al., Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature 571, 117–121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou B. O., Yue R., Murphy M. M., Peyer J. G., Morrison S. J., Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 15, 154–168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding L., Morrison S. J., Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pham V. N., et al., Combinatorial function of ETS transcription factors in the developing vasculature. Dev. Biol. 303, 772–783 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stainier D. Y., Weinstein B. M., Detrich H. W. III, Zon L. I., Fishman M. C., Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development 121, 3141–3150 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Bertrand J. Y., et al., Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin H. F., et al., Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood 106, 3803–3810 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hall C., Flores M. V., Storm T., Crosier K., Crosier P., The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7, 42 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.North T. E., et al., Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roman B. L., et al., Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development 129, 3009–3019 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 67.Kwan K. M., et al., The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the scRNA-seq data reported in this paper are Gene Expression Omnibus (GEO): GSE120581 and GSE146404 and Genome Sequence Archive (GSA): CRA002374. All other study data are included in the article and/or supporting information.