Hepatitis B virus (HBV) is a widespread human pathogen, especially in China, and is known to cause liver inflammation, cirrhosis, and hepatocellular carcinoma. A total of ten different genotypes of HBV (A–J) have been reported worldwide. HBV has partially double-stranded DNA, and the 3.2-kb viral genome is located in the nucleus of infected hepatocytes. All of the viral RNAs are transcribed from the covalently closed circular DNA (cccDNA), which acts as the transcription template. The seven main proteins of HBV include Core, pre-Core, Small S, Middle S, Large S, Polymerase, and the critical HBV-encoded regulatory protein hepatitis B virus X protein (HBx). HBx is a 154-amino acid (aa) protein that facilitates the efficient replication of HBV by stimulating HBV gene expression from the cccDNA template. However, the mechanism by which HBx interacts with host proteins and facilitates HBV replication is not precisely known.1,2
In our study, sought to identify several host factors, which are interferon-stimulated genes (ISGs) that regulate HBV replication and infection by targeting HBx.3–5 Using a bimolecular fluorescence complementation assay, we screened a group of ISGs that interacted with the HBx protein to inhibit HBV replication and found that CBFβ was the top-ranking ISG protein. CBFβ is a small molecule found to facilitate HIV replication by increasing the steady-state level of the viral infectivity factor protein.6 In our further study to clarify the mechanism involved in CBFβ induction by IFN, we found that CBFβ could not be upregulated by type I IFN in HepG2 cells. However, it has been previously reported that type III IFN production is induced after HBV infection;7 thus, we speculated that type III interferons might induce CBFβ. As we expected, CBFβ induction was confirmed in both the qPCR and immunoblot analyses, and we thus focused our research on the IFN-III-CBFβ-HBx-HBV axis.
Interestingly, we found that type III interferon-induced IL-10 plays a critical role in CBFβ upregulation. The binding of CBFβ to HBx blocked the formation of the HBx-CUL4-DDB1-SMC complex and rescued the expression of SMC5/6 needed for the structural maintenance of chromosomes (SMC).3 SMC5/6 is a host restriction factor that suppresses HBV transcription from the cccDNA template.8
The CBFβ-HBx dimer not only blocks HBx-induced SMC5/6 degradation but also stabilizes the CBFβ protein by deubiquitination.3 This process is promising in revealing a new pathway by which HBx may act as a deubiquitinase (DUB). However, HBx does not have DUB activity, so it might act as an adapter and hijack DUBs. We are trying to characterize the interaction of the HBx-CBFβ dimer, which might reveal new directions regarding the interaction between HBx and the host.
From our list, we also discovered that TRIM and IFIT family members were consistently found to act as partners of HBx. We were encouraged to find that K90 and K95 are sites for HBx ubiquitination and degradation induced by two host factors, TRIM31 (K95)4 and TRIM25 (K90) (unpublished data). Interestingly, TRIM5γ plays an indispensable role in TRIM31-mediated HBx degradation by acting as an adaptor.4 IFIT family members were also found to interact with HBx, and HBx-activated NF-kB-induced IFIT3 expression, which promoted HBV replication. Furthermore, we reported another protein, TRIM14, which played a similar role as CBFβ and suppressed Smc5/6 expression by targeting HBx.5 These protein–protein interactions are a good source of targeting sites for further drug development in the future.
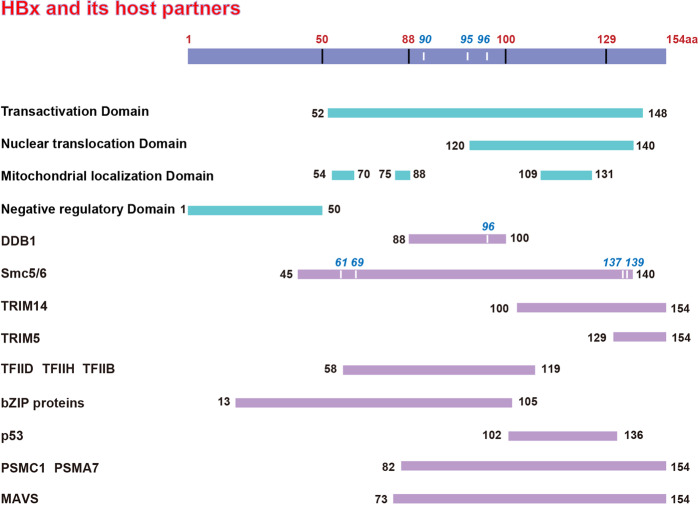
Structurally, HBx has an N-terminal negative regulatory domain, whereas transactivation or coactivation of the C-terminal domain interferes with host cell signaling transduction pathways to benefit HBV replication. The best-characterized HBx binding partner is cellular damage-specific DNA binding protein 1 (DDB1). The HBx–DDB1 interaction is essential for HBV replication9 via targeting of the SMC5/6 complex.8 aa 45–140 and C61, C69, C137, and H139 in this region are the critical residues of HBx required for SMC5/6 degradation,10 and aa 88–100 (HBx88–100) have been identified as the minimal DDB1 binding domain in HBx. It was reported earlier that the HBx R96E point mutant no longer bound to DDB1 (HBx R96E) and failed to restore HBx-deficient HBV replication.9 Interestingly, HBx always regulates gene expression via an indirect mechanism by interacting with cellular proteins. Additionally, ERCC3 and ERCC2, the DNA helicase subunits of the holoenzymes TFIIH and TFIIB/D, are targeted by HBx and activated by transcription. Furthermore, HBx interacts with TATA-binding protein and basic region/leucine zipper proteins to modulate the transcriptional pathway. In addition, HBx has been claimed to inhibit p53-specific DNA binding and transcriptional activity.11–13 Interestingly, HBx was reported to inhibit IFN signaling by targeting and downregulating mitochondrial antiviral-signaling protein.14 By targeting PSMA7 and PSMC1, HBx was proven to disrupt the proteasome pathway15 (Fig. 1). While the role of HBx is expansive, many critical HBx-related findings are not reviewed here due to space limitations. We believe that new targets of HBx will continually emerge in the near future. Previous studies have demonstrated that the interaction between HBx and its host partners plays an indispensable role in understanding the host–virus interaction mechanism. This is because HBx potentially captures many host factors that will eventually play a series of irreplaceable roles during the host–virus interaction.
Fig. 1.
Functional domains of HBx and the regions interacting with host proteins. The 154-amino-acid (aa) HBx protein is shown as indicated. Functional domains are shown as blue lines, the target regions for the host proteins are shown as pink lines, and the numbers indicate the amino acid position
In addition, SARS-CoV-2 has been rampant worldwide for more than a year, and more than 100 million people have developed COVID-19. However, the infection and spread of this virus have not been effectively controlled. Therefore, several questions arise: What will happen to SARS-CoV-2-infected HBV patients? Is there any interaction between HBx and proteins encoded by the SARS-CoV-2 genome? Do the interactions benefit HBV or SARS-CoV-2? Thus, HBx is expected to have new partners and interactions, opening a new research area.
Acknowledgements
This work was supported by the National Natural Science Foundation, China (Grant Nos. 81901592 to H.S., 81801563 to Q.X., and 81801565 to F.X.), and the 68th batch of first-class funding from China Postdoctoral Science Foundation (Grant No. 2020M680044 to G.T.).
Competing interests
The authors declare no competing interests.
References
- 1.Keasler VV, et al. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J. Virol. 2007;81:2656–2662. doi: 10.1128/JVI.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan G, et al. When hepatitis B virus meets interferons. Front. Microbiol. 2018;9:1611. doi: 10.3389/fmicb.2018.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu F, et al. Type III interferon-induced CBFbeta inhibits HBV replication by hijacking HBx. Cell Mol. Immunol. 2019;16:357–366. doi: 10.1038/s41423-018-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan G, et al. Type-I-IFN-stimulated gene TRIM5gamma Inhibits HBV replication by promoting HBx degradation. Cell Rep. 2019;29:3551–3563. doi: 10.1016/j.celrep.2019.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan G, et al. Identification of TRIM14 as a type I IFN-stimulated gene controlling hepatitis B virus replication by targeting HBx. Front. Immunol. 2018;9:1872. doi: 10.3389/fimmu.2018.01872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jager S, et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2011;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato S, et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity. 2015;42:123–132. doi: 10.1016/j.immuni.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Decorsiere A, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 9.Li T, et al. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat. Struct. Mol. Biol. 2010;17:105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramakrishnan D, et al. Hepatitis B Virus X Protein Function Requires Zinc Binding. J Virol. 2019;93:e00250–19. doi: 10.1128/JVI.00250-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang XW, et al. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl Acad. Sci. USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haviv I, et al. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol. Cell Biol. 1998;18:1562–1569. doi: 10.1128/MCB.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perini G, Oetjen E, Green MR. The hepatitis B pX protein promotes dimerization and DNA binding of cellular basic region/leucine zipper proteins by targeting the conserved basic region. J. Biol. Chem. 1999;274:13970–13977. doi: 10.1074/jbc.274.20.13970. [DOI] [PubMed] [Google Scholar]
- 14.Wei C, et al. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J. Immunol. 2010;185:1158–1168. doi: 10.4049/jimmunol.0903874. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, et al. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 2000;275:15157–15165. doi: 10.1074/jbc.M910378199. [DOI] [PubMed] [Google Scholar]