Abstract
Neutralizing antibodies elicited by HIV-1 coevolve with viral Envs in distinctive patterns, in some cases acquiring substantial breadth. Here we show that primary HIV-1 Envs, when expressed by simian-human immunodeficiency viruses in rhesus macaques, elicited patterns of Env-antibody coevolution strikingly similar to those in humans. This included conserved immunogenetic, structural and chemical solutions to epitope recognition and precise Env-amino acid substitutions, insertions and deletions leading to virus persistence. The structure of one rhesus antibody, capable of neutralizing 49% of a 208-strain panel, revealed a V2-apex mode of recognition like that of human bNAbs PGT145/PCT64-35S. Another rhesus antibody bound the CD4-binding site by CD4 mimicry mirroring human bNAbs 8ANC131/CH235/VRC01. Virus-antibody coevolution in macaques can thus recapitulate developmental features of human bNAbs, thereby guiding HIV-1 immunogen design.
One sentence summary:
Virus-antibody coevolution in rhesus macaques recapitulates developmental features of human antibodies.
Introduction
A major roadblock to rational HIV-1 vaccine design is the lack of a suitable primate model in which broadly neutralizing antibodies (bNAbs) can be commonly induced and the molecular, biological and immunological mechanisms responsible for such responses studied in a reproducible and iterative fashion. Since most HIV-1 bNAbs identified to date have come from humans chronically infected by HIV-1, we hypothesized that one means to elicit such antibodies in primates might be by infecting Indian rhesus macaques (RMs) with simian-human immunodeficiency virus (SHIV) strains that bear primary or transmitted/founder (T/F) HIV-1 Envs that induced bNAbs in humans (1-7). SHIV infected RMs could then be employed to assess the potential of particular HIV-1 Envs to elicit bNAbs and to characterize the coevolutionary pathways of bNAb lineages and the cognate Env intermediates that elicited them (2-8), thereby serving as a molecular guide for rational vaccine design. Recent innovations in SHIV design (9) now make this experimental strategy testable.
HIV-1 bNAbs target one of several conserved regions on the native Env trimer, including the CD4-binding site (CD4bs), V2 apex, V3 high mannose patch, gp120/gp41 interface, fusion peptide, glycosylated silent face, and membrane-proximal external region (MPER) (10, 11). Such antibodies generally share certain features such as exceptional HCDR3 length, extensive somatic hypermutation, autoreactivity or unusual mechanisms for binding glycans or glycopeptides (10-13). Long HCDR3s result from initial germline immunoglobulin gene rearrangements whereas the characteristic high frequencies of V(D)J mutations that lead to affinity maturation and neutralizing breadth result from persistent virus replication, epitope variation, and continued Env-Ab coevolution. In some subjects, cooperating antibody lineages that target the same epitope have been identified, and together they contribute to Env diversification and bNAb development (4, 6). A critical question in the HIV-1 vaccine field is if the relatively rare examples of high titer, broadly neutralizing antibodies that have been identified in a subset of chronically infected humans represent stochastic accidents of nature, not likely to be replicated by a vaccine, or if there are special properties of particular HIV-1 Envs that along with deterministic features of Env-Ab coevolution make HIV-1 vaccine development more plausible.
The current study is based on the premise that while Env diversity within HIV-1 group M is extraordinarily high, primary HIV-1 Envs [i.e., native Env trimers on infectious virions that allow for persistent replication and clinical transmission in humans (1)] are nonetheless constrained in their immediate evolutionary potential (14-16). This paradox – extreme Env diversity globally but constraints on immediate or near-term evolution in an individual – can be explained by competing evolutionary forces: positive selection imposed by immune evasion balanced by purifying (negative) selection acting to retain viral fitness. At least four interrelated Env properties, acting in concert, facilitate immune escape – occlusion of trimer-interface epitopes (17), glycan shielding (18), epitope variation (19), and conformational masking (20) – while conservation of structure and cell-entry properties and retention of protein stability constrain each primary Env’s immediate and short-term evolutionary trajectory. When introduced into a human as HIV-1 infection or into RMs as SHIV infections, each Env will have unique restrictions in its exposure of antigenic sites, thus favoring distinct molecular pathways of escape from neutralizing antibodies with each pathway exacting a different fitness cost from the virus. These considerations led us to hypothesize that SHIV infection of RMs will recapitulate HIV-1 infection of humans with respect to molecular patterns of Env-Ab coevolution, within bounds set by any particular Env sequence and the respective rhesus and human immunoglobulin gene repertoires that respond to it (21).
Results
Some of the best studied examples of HIV-1 Env-Ab co-evolution have come from the analysis of HIV-1 infected human subjects CH505, CH848 and CAP256, who developed bNAb responses targeting the CD4bs, V3 high mannose patch and V2 apex of HIV-1 Env, respectively (2-7). Subjects CH505 and CH848 were each productively infected by a single T/F virus, and previously we constructed SHIVs containing each of these T/F Envs (9). CAP256 was infected by a single virus (CAP256PI, primary infection) and then became superinfected by a second genetically-divergent virus (CAP256SU, superinfection) (22). The CAP256SU variant was thought to trigger the V2 bNAb lineage in this individual (5), so we constructed and analyzed a SHIV containing this Env (Fig. S1). SHIVs containing CH505, CH848 and CAP256SU Envs were modified at gp120 residue 375 to enhance binding and entry into rhesus CD4 T cells. The Env375 substitutions resulted in no discernible change in antigenicity or sensitivity to anti-HIV-1 NAbs compared with wild-type virus, but they were essential for replication in primary RM CD4+ T cells [Fig. S1 and (9)]. We inoculated 22 RMs (Table S1) with SHIV.CH505 (n=10), SHIV.CH848 (n=6) or SHIV.CAP256SU (n=6) by the intravenous route and followed them for an average period of two years (mean of 103 weeks, range 36-184) for viral replication kinetics, development of strain-specific (autologous) and broadly reactive (heterologous) NAbs, and patterns of Env sequence evolution. Eight of the animals were treated with anti-CD8 mAb at or near the time of SHIV inoculation in order to transiently deplete CD8+ cells and increase peak and set point plasma virus titers. One animal (RM5695), which was repurposed from an earlier study of HIV-1 gp120 protein immunization (23), was inoculated with plasma from three animals recently infected by SHIV.CH505 to increase early virus diversity.
SHIV Replication In Vivo and Elicitation of Neutralizing Antibodies
Figure S2 depicts the kinetics of virus replication and the development of autologous, strain-specific tier 2 NAbs as measured by plasma vRNA and inhibition of virus entry into TZM-bl cells (18). Acute virus replication kinetics were consistent for all SHIVs in all animals with peak viremia occurring about 14 days post-inoculation and reaching titers in the 105-108 vRNA/ml range [geometric mean (GM) = 4.2x106]. Peak vRNA titers were higher in anti-CD8 treated animals than in untreated animals [geometric mean titers (GMT) of 2.6x107 vs 1.5x106, respectively; p<0.0001]. Plasma virus load set points at 24 weeks post-infection varied widely between 100 and 988,110 vRNA/ml with a mean for anti-CD8 treated animals of 248,313 ± 136,660 vRNA/ml compared with 8,313 ± 3509 vRNA/ml for untreated animals (p=0.002). Set point vRNA levels in human subjects infected with the corresponding HIV-1 CH505, CH848 or CAP256 strains were 81,968, 132,481 and 750,000 vRNA/ml, respectively. Compared with two human cohort studies of acute and early HIV-1 infection (24, 25), peak and setpoint vRNA titers in SHIV infected animals were comparable, although several macaques developed viral loads near the lower limit of detection (100 vRNA/ml), which is rare in humans. Six macaques developed AIDS-defining events and were euthanized 36, 40, 60, 65, 89 or 136 weeks post-SHIV infection; four of these animals had been treated with anti-CD8 mAb (Table S1).
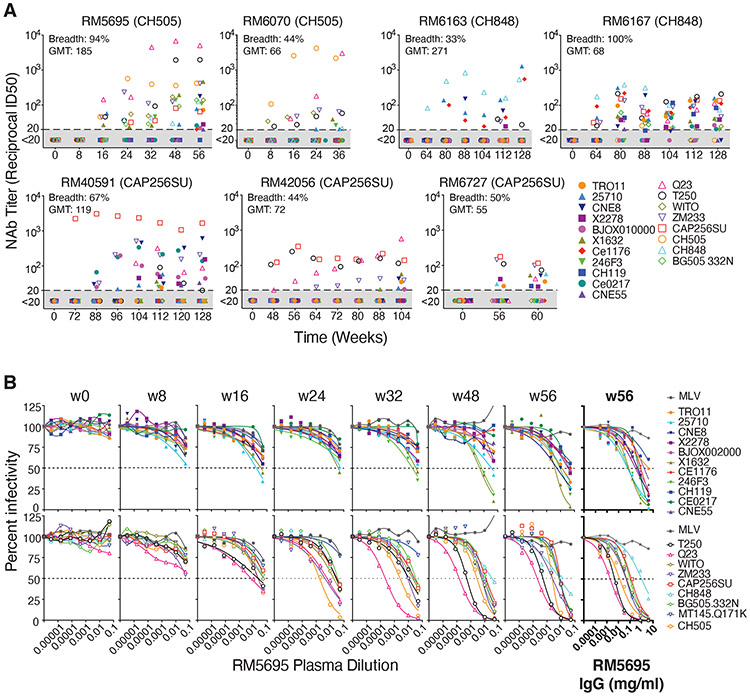
Autologous tier 2 NAb responses to the three SHIVs were detected as early as 8 weeks post-infection and peaked between 24 and 80 weeks with 50% inhibitory dilutions (ID50) between 0.05 (1:20 dilution) and 0.000125 (1:8,000 dilution) (Fig. S2). The kinetics of appearance and titers of autologous tier 2 Nabs that developed in response to SHIV infections were generally comparable to those observed in humans infected by viruses bearing the homologous Envs (2, 7, 22). Among all animals, there was a significant association between higher setpoint virus titers and higher autologous tier 2 NAb titers (Spearman correlation rank correlation coefficient rs=0.74, p<0.0001). Heterologous plasma neutralizing activity was assessed against a diverse 22-member global panel of tier 1a (n=3) or tier 2 (n=19) HIV-1 strains (26-28) (29) (Table S2). All animals developed potent neutralizing responses to the three tier 1a viruses (GMT ID50 = 0.0004). Tier 1a viruses have “open” Env trimers that spontaneously expose linear V3 and conformational CD4 induced (CD4i) bridging sheet epitopes, thus explaining their extreme sensitivity to what are otherwise non-neutralizing antibodies. Such “non-neutralizing” antibodies that target linear V3 and CD4i epitopes are elicited in virtually all HIV-1 infected humans, thereby selecting for Envs with an open-closed equilibrium that strongly favors the closed configuration (20, 30-32). Tier 2 viruses typify primary or T/F viruses that are generally resistant to neutralization by heterologous plasma antibodies except for those that target one of the canonical bNAb epitope clusters (1, 10, 11, 26). In this study, we used a reciprocal neutralization titer cutoff of ID50 ≤ 0.05 against ≥33% of heterologous tier 2 viruses as an indication of neutralization breadth. This is a conservative threshold consistent with other studies that have characterized bNAb prevalence in human cohorts (28, 33, 34). Seven of 22 RMs developed antibody responses that neutralized between 6 and 18 of 18 heterologous HIV-1 tier 2 viruses in our test panel (Fig 1A; Table S2). Heterologous neutralization was first detected as early as 8-16 weeks post-SHIV infection in two animals and as late as 88 weeks post-SHIV infection in others. Heterologous NAbs reached ID50 titers as high as 0.0001, with GMTs in the seven animals ranging from 0.018 (1:55) to 0.004 (1:271) (Fig 1A). Peak and setpoint plasma virus titers were higher in the seven animals that developed bNAbs (GMT=18,150,586 and 47,471 vRNA/ml, respectively) than in animals that did not (GMT=2,101,814 and 1,945 vRNA/ml, respectively; p<0.015 for both). Two animals (RM5695 and RM6070) with bNAbs were infected by SHIV.CH505, two (RM6163 and RM6167) by SHIV.CH848, and three (RM40591, RM42056 and RM6727) by SHIV.CAP256SU. The remaining 15 animals in the study showed either no or very limited, low titer neutralization of heterologous tier 2 viruses (Table S2). Altogether, the findings show that SHIVs bearing primary T/F Envs reproduce in RMs key features of virus replication dynamics and NAb elicitation that are characteristic of HIV-1 infection in humans, including the potential to elicit bNAbs.
Fig. 1. Broadly neutralizing antibodies in seven rhesus macaques.
(A) RMs 5695 and 6070 were infected by SHIV.CH505, RMs 6163 and 6167 by SHIV.CH848, and RMs 40591, 42056 and 6727 by SHIV.CAP256SU. Neutralizing antibody titers (ID50, 50% inhibitory dilution) from longitudinal plasma specimens against 19 global tier 2 HIV-1 strains (26, 27, 86) are depicted. Full designations of target viruses are provided in Fig. S22 and Table S3. Maximum neutralization breadth across all time points and maximum geometric mean titer (GMT) of neutralization at any one time point are indicated for each animal. (B) Neutralization curves for longitudinal plasma specimens and purified plasma IgG (highlighted bold) from RM5695 show the development of broad and potent neutralization. MT145K.Q171K is a chimpanzee-derived SIVcpz strain that shares antigenic cross reactivity with HIV-1 in the V2 apex (35, 134). Dashed lines indicate 50% reduction in virus infectivity. MLV, murine leukemia virus.
Figure 1B highlights the kinetics, potency and breadth of neutralization by plasma from the SHIV.CH505 infected animal RM5695 and identifies immunoglobulin G (IgG) as the active component. By 16 weeks post-infection, an autologous NAb response to SHIV.CH505 was detectable at an ID50 titer of 0.02 along with heterologous responses to viruses bearing HIV-1 25710, X1632, Q23, ZM233, T250 and WITO Envs at titers of 0.05 – 0.01. NAbs increased rapidly in breadth and titer thereafter. By week 48, bNAbs targeting Q23 and T250 jumped in titer to 0.0002 – 0.0005 and against X1632, 246F3, ZM233 and WITO to titers of 0.005. By week 56, bNAbs in the plasma of RM5695 neutralized 17 of 18 viruses in the heterologous HIV-1 test panel at titers ranging from 0.05 – 0.0002, along with a divergent tier 2 simian immunodeficiency virus strain (SIVcpz.MT145.Q171K) that shares selective antigenic cross-reactivity with HIV-1 in the V2 apex bNAb epitope cluster (35) at a titer of 0.006. IgG was purified from RM5695 week 56 plasma and assayed for neutralizing activity against the same 19 heterologous viruses: IgG concentrations between 0.002 mg/ml (corresponding to a ~1:10,000 dilution of rhesus plasma) and 4 mg/ml neutralized 18 of the 19 viruses in a rank order similar to the polyclonal plasma (Figure 1B, far right panels). Neither plasma nor purified IgG neutralized control viruses pseudotyped with the murine leukemia virus (MLV) Env. Thus, anti-HIV-1 specific IgG accounted for all of the autologous and heterologous neutralizing activity in the RM5695 plasma. Of note, RM5695 plasma and plasma IgG from week 56 reached ID90 or ID95 thresholds against most viruses and exhibited steep inflections at the ID50 midpoint, indicating potent neutralization. Neutralization breadth detected as early as 16 weeks post-infection is unusual in HIV-1 infection but not unprecedented (36) and occurs most often with V2 apex bNAbs, likely because their activity depends more on long HCDR3s than on extensive somatic hypermutation. We show below that the bNAb activity in RM5695 plasma and its isolated IgG fraction as well as monoclonal bNAbs derived from RM5695, all targeted a bNAb epitope in the V2 apex that included the conserved lysine rich C-strand and N160 glycan. The kinetics of appearance, breadth, titers and potency of bNAbs elicited in the six other SHIV-infected RMs, including three animals (RM6070, RM40591 and RM42056) whose bNAbs also targeted the V2 apex C-strand, are summarized in Figure 1A and Table S2.
Env Evolution in SHIV Infected Macaques and HIV-1 Infected Humans
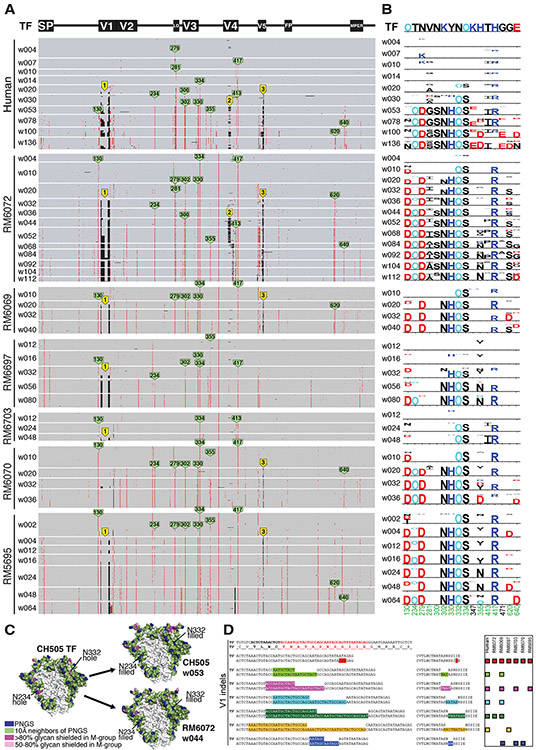
To explore if SHIV Env evolution in RMs recapitulates that of HIV-1 in humans in a strain-specific manner, we analyzed Env sequences in the 22 SHIV infected animals over one to three years of follow-up and compared them with the evolution of the homologous Envs in humans infected by HIV-1. We used single genome sequencing (SGS) for this analysis, since it allows for retention of genetic linkage across intact viral genes and enables precise molecular tracking of sequence diversification from unambiguous T/F genomes (1, 37-39). We analyzed cDNA sequences derived from plasma virion RNA, since the half-life of circulating plasma virus is <1 hour (40) and that of cells producing most of this virus is <1 day (40). This short composite half-life (<1 day) reflects the lifespan of >99.9% of circulating plasma virus in individuals not receiving antiretroviral therapy (41, 42), making the genetic composition of the plasma virus quasispecies an exquisitely sensitive real time indicator of in vivo selection pressures acting on virus and virus-producing cells, including that exerted by NAbs, cytotoxic T cells (CTLs) or antiretroviral drugs (18, 37, 39, 42, 43). Figure 2A illustrates Env amino acid substitutions in virus from sequential plasma specimens from human subject CH505 and from six SHIV.CH505 infected RMs. By inspection and the algorithm-based statistical methods previously described [(37, 38) and Supplementary Materials], amino acid substitutions in Env were found to be nonrandomly distributed in patterns that evolved over time beginning as early as 4 weeks post-infection. Using LASSIE [Longitudinal Antigenic Sequences and Sites from Intra-Host Evolution (44)], a computational tool that systematically identifies amino acid differences in a set of evolved sequences that reach a predetermined frequency threshold compared to a founder sequence, we identified in human subject CH505 fifteen Env residues where mutations altered the encoded amino acid in at least 75% of sequences at one or more subsequent time points. Thirteen of these 15 sites varied by >75% in one or more monkeys and 10 sites varied in at least half of the animals (Fig. 2B). A remarkably parallel Env evolutionary trajectory was observed between the human host and monkey RM6072. Surprisingly, the substituted amino acids at many of the variable sites were identical in human and rhesus sequences, including mutations that created potential N-linked glycans (PNGs) that filled “glycan holes” (Figs. 2C, S3) (45). The conserved patterns of amino acid substitutions in CH505 Envs were very different from mutational patterns of Env sequences observed in the human subject CH848 and in RMs infected by SHIVs bearing the CH848 Env (Fig. S4). Twenty-nine variable sites were identified in Envs from the human subject CH848 and 16 of these were shared in SHIV.CH848 infected RMs, often with identical amino acid substitutions including some that altered the distribution of PNGs and glycan holes (Fig. S5). As a control, we compared patterns of Env evolution in CH848 infected RMs against selected sites in the CH505 infected human, and vice versa. Many fewer sites of shared evolution were observed in either discordant Env pairing (p < 0.01 for both comparisons) (Fig. S6). The Env strain-specific mutational patterns observed in humans and rhesus animals infected by CH505 and CH848 viruses were different still from the CAP256SU infected RMs in which 45 selected sites were identified, including 25 that were shared in more than one animal (Figs. S7A,B and S8). A comparable Env-wide analysis in the human subject CAP256 could not be performed because this individual was infected and then superinfected by two widely divergent viruses whose progeny underwent extensive recombination (22) (Fig. S7C). Still, we could identify common sites of positive selection in human and rhesus sequences, including a key mutation at residue 169 that led to virus escape from V2 apex C-strand targeted bNAbs (Fig. S7D). Overall, we identified 89 selected Env residues in CH505, CH848 or CAP256SU T/F viruses and 50 of these sites were shared among humans and RMs that were infected by viruses bearing the same Env strain. This result was markedly different for individuals infected by different virus strains where no more than 6 of these 89 variable sites were shared by any CH505, CH848 or CAP256SU pairing (p<0.0001) and only 14 sites were shared overall (p<0.0001). These findings thus show striking examples of conserved strain-specific evolution of HIV-1 Env sequences in humans and RMs.
Fig. 2. Env evolution in SHIV.CH505 infected rhesus macaques recapitulates HIV-1 Env evolution in the human subject CH505.
(A) A PIXEL plot (https://www.hiv.lanl.gov/content/sequence/pixel/pixel.html) (https://www.hiv.lanl.gov/content/sequence/HIV/HIVTools.html) of amino acid alignments of longitudinal HIV-1 Env sequences obtained by single genome sequencing of plasma virion RNA. Between 15-60 individual sequences are grouped by time point and amino acids are colored red to highlight mutations relative to the infecting CH505 transmitted/founder strain illustrated schematically at the top. The image is highly compressed. Each row within a time block represents a single sequence, and each column represents an amino acid position in the alignment; thus each pixel represents a single amino acid that is colored grey if it matches the T/F sequence, red if it is a mutated residue, and black if it is an insertion or deletion (Indel) relative to the T/F virus. All SHIV sequences differ from the T/F at position 375, reflecting the SHIV design strategy that enables replication in rhesus macaques (9). Green tags indicate amino positions (HXB2 numbering) that are mutated in both human and rhesus. Yellow tags indicate three sites of identical Indels observed in both human and rhesus. (B) LASSIE analysis (44) of the same longitudinal Env sequences was used to characterize mutations under positive selection. If a T/F amino acid was retained in <25% of the sequences in the human CH505 infection, the site was considered to be under selective pressure and tracked in all hosts. The height of each amino acid mutation is proportional to its frequency at the respective time point. Red, dark blue and black indicate acidic, basic and neutral residues. “O” indicates asparagine (N) embedded in an N-linked glycosylation motif. Numbers at the bottom indicate residue positions (HXB2 numbering). Green numbers indicate mutations that reached 75% frequency in the human and in at least one animal. (C) Side projection of the CH505 Env trimer with potential N-linked glycans (PNGS) indicated in blue, 10Å neighbors of PNGs shown in green, and “glycan holes” that are typically covered by glycans in >80% and 50-80% of group M HIV-1 strains in magenta and pink, respectively. The light grey area in the central region of the trimer is the inter-protomer surface that forms a cleft with low glycan coverage (45). Two glycan holes in the CH505 T/F at positions 234 and 332 were filled by the addition of NXS/T motifs over time in the human subject CH505 and in RM6072 as well as in all other monkeys (see Fig. S3). (D) The V1 sequence of CH505 env is shown at the top of the panel with the hypervariable region indicated in red. Indels in V1 that arose in the first year of infection in the human host are illustrated in the sequences below the reference TF sequence. Indels occurred in nucleotide lengths divisible by three so as to maintain a viable Env open reading frame, and in the case of insertions, consisted of direct repeats. Indels that were replicated identically in one or more rhesus macaques are indicated by color-matched boxes to the right. Identical Indels in human and rhesus CH505 sequences were also found in V4 and V5 (Fig. S9).
Another type of Env variation common in HIV-1 infection that can affect immune recognition results from insertions and deletions (Indels). Indels occur as a consequence of polymerase slippage and template switching during reverse transcription of the retroviral genome (46, 47). Indels are most commonly observed in the surface-exposed variable loops 1, 2, 4 and 5, which can better accommodate changes in sequence length than structural or enzymatic elements of the viral proteome; this was the case for the human subject CH505 and macaques infected by SHIV.CH505 (Fig. 2A). Remarkably, we found multiple examples of identical Indels between human and rhesus CH505 sequences and among CH505 sequences from different rhesus animals (Figs. 2D and S9). A total of six distinct V1 Indels, three V4 Indels and one V5 Indel were identified in sequences from human subject CH505 that were replicated exactly in sequences from one or more monkey. Two of these Indels were found in the human and in all six monkeys. Additional identically replicated Indels in V1 and V5 were identified only in monkeys. Indels were also observed in sequences from the human subject CH848 (Figs. S4,S10). An exact replica of all but one of these Indels was present in one or more macaque. Additional Indels were replicated in multiple animals (Fig. S10). None of the Indels in CH505 sequences were found in CH848, and neither set was found in CAP256SU sequences. In both human and rhesus, Indels generally lengthened or shortened variable loops in a direction that approached the median lengths of globally circulating viruses (Figs. S9,S10). We performed statistical analyses to determine the likelihood that the conserved sets of Env specific Indels that we observed in different individuals could occur by chance and that likelihood was estimated to be vanishingly small (see Extended Discussion in Supplement).
NAbs select for mutations in Env
CH505.
To examine if NAb-mediated selection was a primary driving force in the Env-specific evolutionary patterns that we observed, early strain-specific NAb responses and later bNAb responses were mapped using site-directed mutagenesis to introduce observed mutations into neutralization sensitive autologous and heterologous Envs. We then tested these mutant Envs, compared with their wild-type counterparts, for sensitivity to polyclonal plasma and mAbs isolated from the infected RMs or human subjects. Variable sites in the CH505 Env (Figs. 2A,B) were interrogated alone or in combination for their effects on Env sensitivity to autologous antibodies. Aside from variable residues in the leader sequence of gp160 that generally represent CTL escape mutations (37, 43), CTL epitope reversions to global consensus at or near residue 417 (4), and mutations at residues 300, 620 and 640 that occurred inconsistently and late in the course of infection, most of the variable residues were found to represent escape mutations from autologous NAbs (Fig. S11A). These included residues 234 and 334 where mutations restored PNGs that filled T/F glycan holes (Fig. S3), loop D residues 279 and 281 involved in CD4 binding, CD4 contact residues in V5 (460/ΔV5), and residue 130. The temporal appearance of these NAbs coincided with the appearance of phenotypically demonstrable NAb escape mutations in Env (Fig. 2A,B). These results were corroborated by neutralization patterns of V3 targeted mAbs DH647 and DH648 and CD4bs targeted mAbs DH650.UCA and DH650 (all isolated from the SHIV.CH505 infected RM6072) and the human CD4bs targeted mAbs CH235.UCA, CH235.IA3 and CH235.9 isolated from human subject CH505 (6) (Figs. S11B,C). Human subject CH505 developed two cooperative lineages of CD4bs antibodies that targeted epitopes that included loop D residues 279-281 and residues in V5, eventually leading to neutralization breadth by both antibody lineages (4, 6). In RM6072, a mAb lineage, termed DH650, was isolated that targeted these same epitopes but it never developed neutralization breadth, a finding for which we found a structural explanation (see below).
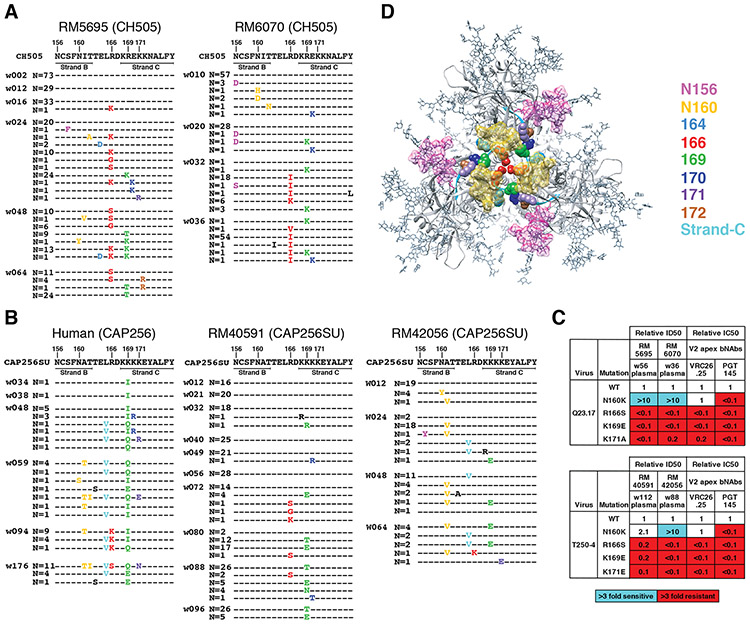
In addition to the strain-specific NAb responses that we identified in SHIV.CH505 infected RMs, we found in two CH505 animals (RM5695 and RM6070) neutralizing antibodies that targeted heterologous tier 2 viruses (Fig. 1, Table S2). These bNAbs were first detected at weeks 8 (RM6070) and 16 (RM5695) post-infection and their development was temporally associated with the appearance of mutations in the Env V2 apex, including residues 166 or 169 (Figs. 3A). These are common contact residues of human V2 apex bNAbs (5, 8, 48). A minor fraction of sequences lost PNGs at 156 and 160. By 24 weeks post-infection in RM5695 and 32 weeks post-infection in RM6070, most circulating virus contained mutations at residues 166 or 169; by 36-48 weeks post-infection, all sequences were mutant at one or the other of these positions. We corroborated this single genome sequence evidence of strong virus selection in the central cavity of the V2 apex of RM5695 by next generation sequencing, which revealed that 99.3% of 10,000 sequences sampled between weeks 48 and 64 post-infection contained mutations at residues 166 or 169. When these residues were mutated in the wild type versions of heterologous primary virus strains T250, Q23, MT145K, 246F or BG505 that were otherwise neutralized by RM5695 and RM6070 plasma, neutralization was abrogated (Figs. 3C,S12). Neutralization of these heterologous strains was variably dependent on the glycan at N160 (Fig. S13), similar to what has been reported for V2 apex bNAbs in subject CAP256SU (49). These findings indicated the presence of potent V2 apex C-strand targeted bNAbs in RM5695 and RM6070. In summary, the mapping of autologous and heterologous Nab responses in RMs infected by SHIV.CH505 indicated that most mutations in Env that could not be ascribed to CTL selection were the result of NAb selection.
Fig. 3. bNAb responses in four rhesus macaques map to the V2 apex.
(A) Single genome sequencing (N = number of sequences; w = weeks post-SHIV infection) in SHIV.CH505 infected animals RM5695 and RM6070 reveals selection and fixation of mutations in and immediately proximal to strand C and additional mutations that eliminate PNG sites at 156 and 160. (B) Sequential Env sequences from human subject CAP256 and SHIV.CAP256SU infected RMs 40591 and 42056 showed selection and fixation of mutations in and proximal to strand C, with fewer mutations eliminating the PNG site at 160. (C) Heterologous neutralization of Q23.17 and T250-4 Env-pseudotyped viruses by rhesus plasma is drastically reduced (expressed as fold-change from wildtype) by mutations at residues 166, 169 and 171 similar to human V2 apex bNAbs. Enhanced neutralization against the N160K mutants is illustrated in Fig. S13 and further described in the Supplement. (D) Escape mutations from rhesus V2 apex bNAbs are displayed on the 5FYL structure of BG505.N332 SOSIP (top view).
CH848.
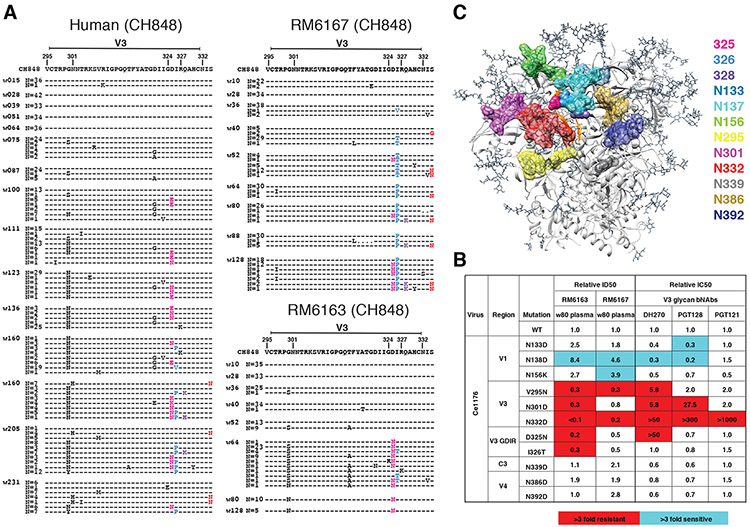
We also examined Env mutations shared between SHIV.CH848 infected RMs and the HIV-1 infected CH848 human subject as potential sites targeted by NAbs. The earliest Env substitution in the human subject that was also mutated in sequences from all six RMs was at position R336, which is surface exposed on the HIV-1 trimer. The R366G substitution in RM6167 occurred at week 10 post-SHIV infection when autologous NAb titers to the T/F SHIV were 1:220. NAb titers to a site-directed mutant of the T/F virus that contained the R366G substitution fell three-fold to 1:72 (Fig. S14A), indicative of an early immunodominant epitope-focused NAb response. Autologous Nab titers in RM6167 increased to 1:435 by week 36 and this was accompanied by strong selection on the evolving virus quasispecies resulting in deletions in V1 and V5 (Figs. S4, S10) and amino acid substitutions at positions indicated in Fig. S4. When these consensus mutations were introduced into the CH848 T/F Env and tested for neutralization sensitivity to week 10 and 36 plasma, the mutant virus showed complete escape (Fig. S14A). In the human subject CH848, an early strain-specific NAb response targeted many of these same sites including V1 (7). The virus quasispecies in subject CH848 escaped by deleting ~10 amino acids in V1, which was followed by the development of the V3 glycan focused bNAb lineage DH270 (7, 50). Remarkably, the same sequence of events occurred in SHIV.CH848 infected animals RM6163 and RM6167 (Fig. S10). V1 was first targeted by an early autologous NAb response (Fig. S14), followed by deletions in V1, which was in turn followed by the development of V3 glycan targeted bNAbs (Fig. 1A, Table S2). We mapped the epitopes of these bNAb responses to the glycan at residue N332 and the Gly-Asp-Ile-Arg (GDIR) motif at positions 324-327 (Fig. 4B). Consistent with this epitope mapping, we identified in the evolving viral quasispecies of both RMs, mutations at residues 332-334 and GDIR residues 324-327 (Fig. 4A). Like the prototypic human V3 glycan bNAbs DH270, PGT121 and PGT128, the bNAbs that we identified in RMs 6163 and 6167 were strictly N332 dependent, and mutations in spatially-associated surface residues V295, N301, N138 and N133 had similar effects on neutralization potency of both rhesus and human bNAbs (Figs. 4B,C).
Fig. 4. bNAb responses in two rhesus macaques map to the V3 glycan high mannose patch.
(A) Sequential Env sequences from human subject CH848 and SHIV.CH848 infected RMs 6167 and 6163 showed selection and fixation of mutations in the GDIR motif. Ser to Asn substitutions at position 334 shift the PNG from residue 332 to 334 in the human subject and in RM6167. (B) Heterologous neutralization of Ce1176 Env-pseudotyped virus by RMs 6163 and 6167 plasma is reduced (expressed as fold-change from wildtype) by mutations at N332D and V295N, consistent with human V3 glycan bNAbs. Neutralization of GDIR mutants is reduced by 2-5-fold. Elimination of a glycan at position N138 enhances neutralization in RMs 6163 and 6167 and for some human V3 glycan bNAbs. (C) V3 glycan bNAb escape mutations are displayed on the 5FYL structure of BG505.N332 SOSIP (side view) highlighting their close spatial proximity.
CAP256.
In the human subject CAP256, recombination between PI and SU lineage sequences (Fig. S7C) precluded a gp160-wide Env analysis. We focused instead on mutations in and near the V2 C strand, since the human subject and two of six RMs (40591 and 42056) developed V2 apex targeted bNAbs (Fig. 1A and Table S2). In both human and RMs, very similar patterns of Env V2 sequence evolution occurred (Fig. 3B). These mutations included substitutions at the canonical residues 160, 166, 169 and 171 shown to be contact residues for other prototypical human V2 apex bNAbs (5, 8, 51-53). When introduced into heterologous neutralization-sensitive Envs, mutations at 166, 169 and 171 abrogated neutralization by plasma from RMs 40591 and 42056 as it did for control human bNAbs PGT145 and VRC26.25, the latter having been isolated from subject CAP256 (49, 53) (Fig 3C). In RMs 40591 and 42056, we observed that V2 apex targeted antibodies were variably dependent on binding to the glycan at Env residue 160 for neutralizing activity, a pattern that was similar to antibodies from animals RM5695 and RM6070 and the human subject CAP256 (49). This variable dependence on N160 for bNAb activity is further shown in Fig. S13 and is discussed in Supplementary Materials. Overall, the molecular and temporal patterns of Env sequence evolution reported here for SHIV-infected macaques, and previously in humans (18, 37-39, 43), highlight the utility of dynamic measurements of localized sequence variation as a highly sensitive indicator of epitope-specific adaptive immune responses.
Homologous B Cell Responses in Humans and Macaques
The observation that homologous Envs evolved in similar molecular patterns in humans and macaques could be explained by limited numbers of antigenic sites accessible for antibody binding and restricted pathways of virus escape. In addition, homologous human and rhesus germline B cell receptors could favor binding to common HIV-1 Env epitopes and follow similar patterns of Ab-Env coevolution. We explored the latter possibility by isolating and characterizing neutralizing mAbs from two RMs that were infected by SHIV.CH505. We selected these animals for study because both exhibited favorable virus replication kinetics, comparable early NAb responses, and an overall pattern of Env evolution similar to that in the human subject CH505; nonetheless, one animal (RM5695) developed bNAbs and the other (RM6072) did not. We asked what might be the genetic and structural similarities and dissimilarities between the NAbs elicited in these animals.
CD4bs-directed antibodies.
Two broadly neutralizing mAb lineages (CH235 and CH103) targeting the HIV-1 CD4bs were previously isolated from the human subject CH505 and their evolutionary pathways from germline receptors to mature antibodies determined (2, 4, 6). In animal RM6072, serological analysis of early plasma samples suggested the presence of CD4bs targeted Abs based on selective Ab binding to CH505 T/F gp120 and resurfaced Env core but not to their isogenic Δ371I mutants, which do not bind CD4 (Fig. S15A). Additional evidence of CD4bs targeted Abs in the plasma of this animal included competitive blocking of soluble CD4 (sCD4) and CH235 and CH106 mAbs (Fig. S15B). We sorted memory B cells from RM6072 from longitudinal timepoints 20, 24 and 32 weeks post-SHIV infection and selected cells that reacted with CH505 T/F gp120 but not with the Δ371I mutant. We isolated a 15-member B cell clonal lineage designated DH650 (Figs. S15C,D) that selectively bound the autologous CH505 T/F Env gp120 but not the Δ371I mutant (Figs. S15E). Some of these mAbs competed with sCD4 and CH235 and CH106 mAbs for binding CH505 T/F gp120 (Figs. S15F). Mature DH650 lineage mAbs, but not the inferred germline UCA, bound CH505 T/F gp140 SOSIPv4.1 trimer (Fig. S16A). Most members of the DH650 lineage neutralized a glycan-deficient mutant of CH505 Env (gly4) and two-thirds of them neutralized the wildtype CH505 T/F strain (Fig. S16B). The DH650 UCA neutralized neither. None of the DH650 lineage mAbs neutralized heterologous viruses (Fig. S16C and Table S3).
The immunogenetic features of the DH650 lineage mAbs suggest how they recognize HIV-1 Env. The lineage comes from V(D)J recombination of the macaque VH1-h gene (Fig. S15D), which is 91% similar to an orthologous human gene VH1-46 used by the CD4bs bNAbs CH235 and 8ANC131 (6, 54). DH650 antibodies share key VH residues with CH235 (Fig. S17A), which were shown previously to be contact sites with gp120 Env (6, 55). These included residue 57, which in both the CH235 UCA and DH650 UCA underwent affinity maturation to R57, which is important for CH235 bNAb activity and is shared among CD4bs bnAbs using VH1-46 (6, 55). We found this N57R substitution in DH650 to be essential for binding to CH505 T/F Env (Fig. S17B). Other DH650 VH1-h gene residues that we found to be important for Env binding included N35, Q62 and R72 (Figs. 5, S17C,D). A distinguishing feature of DH650 lineage antibodies was the IGVk2 light chain, which has an exceptionally long LCDR1 of 17 amino acids (Fig. S17E) that we explored by structural studies.
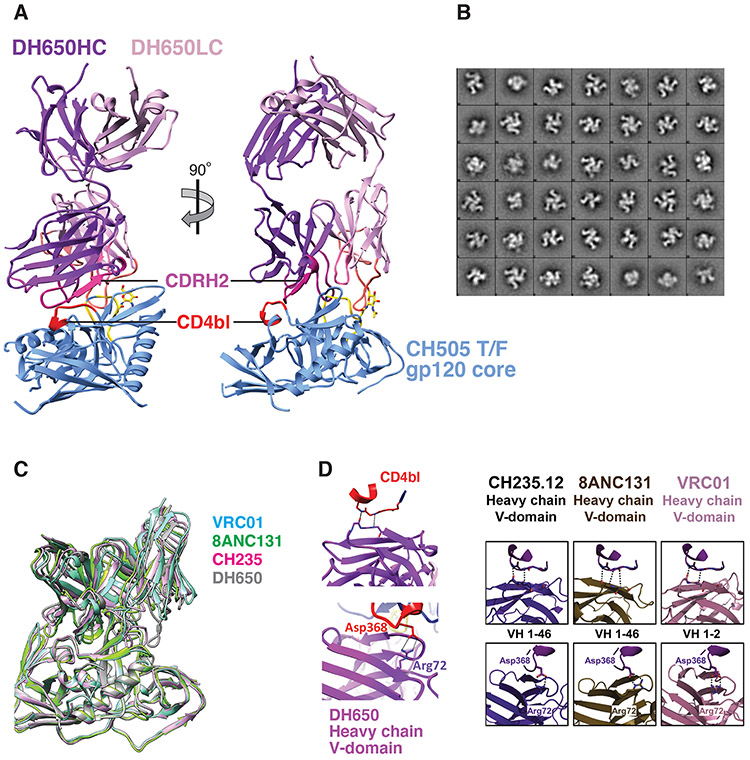
Fig. 5. Structure of Fab DH650 bound to CH505 gp120 core mimics human CD4bs antibodies.
(A) Ribbon representations of Fab DH650 heavy chain (purple) and light chain (pink) and CH505 gp120 core (blue). The heavy-chain CDRH2 is in magenta, the light-chain CDRL1 is in orange, and the CD4 binding loop (CD4bl) is in red. Loop D of gp120, which shifts from its position in other complexes to accommodate the long CDRL1 of DH650, is in yellow; the first N-acetyl glucosamine of glycan 276, which also shifts to accommodate CDRL1, is in stick representation (carbon, yellow; nitrogen, blue; oxygen, red). (B) 2D class averages after negative stain EM analysis of DH650-CH505 DS-SOSIP trimer showing three Fab bound per trimer. (C) Superposition of Fab DH650-CH505 gp120 complex with other CD4 binding site Fab-gp120 complexes, with the essentially identical structures of gp120 as the common reference. The view corresponds to the orientation in the right-hand part of panel A. Only the Fv modules of the Fabs are shown. Complex of DH650 (this study) is shown in grey, VRC01 (PDB-3NGB) in cyan, CH235.12 (PDB-5F96) in magenta, and 8ANC131 (PDB-4RWY) in green. DH650-bound gp120 core Cαs have rmsds of 0.78Å, 0.82 Å and 0.71Å from those of the VRC01-, CH235.12- and 8AC131-bound gp120 structures, respectively. (D) Left panel: Recognition of CD4 binding loop of CH505 T/F gp120 by variable heavy domain of DH650 (top). Coordination of Asp368 by Arg72 of DH650 (bottom). Right panel: Comparison with other CD4 binding site antibodies (6).
The crystal structure of DH650 bound to the gp120 Env core of the CH505 T/F virus showed that its interactions with the gp120 CD4bs closely resembled those of the human CD4bs mAbs CH235, 8ANC131 and VRC01 (Fig. 5A-D, Table S4). This similarity included conserved HCDR2-mediated CD4-mimicry and coordination of Env Asp368 by Arg72. An important difference between the rhesus and human antibody lineages was in the light chains (DH650, macaque IGVk2; CH235, human IGVλ3), in which the LCDR1 of the DH650 light chain was six residues longer than its CH235 counterpart (Fig. S17E). The structure showed that in the Env-Ab complex, the CH505 gp120 loop D had undergone a conformational change to accommodate the longer DH650 LCDR1. As we show elsewhere (Chug et al., to be submitted), this shift could occur only because of the absence of a commonly found glycan at gp120 position 234 in the CH505 TF virus. Moreover, addition of that glycan, which occurred in both the human donor and RM6072 by 30-36 weeks post infection, conferred resistance to DH650 (Fig. S11C) and likely eliminated selective pressure in the monkey to enforce deletions in LCDR1. Thus, infection of RM6072 with SHIV.CH505 expanded a B-cell clone bearing an antigen receptor encoded by the RM VH1-h gene segment that is orthologous to the human VH1-46 gene. This B-cell lineage underwent affinity maturation, including selection for a critical R57 VH mutation that is also found in the human CH235 bNAb lineage. Zhou et al. have shown that maturation of VRC01-class CD4bs bnAbs generally includes deletions in LCDR1 or mutations to glycine that confer flexibility (56). Evolution of the DH605 lineage in RM6072 failed to include deletions or flexibility in LCDR1 and hence neutralization breadth did not develop.
V2 apex-directed antibodies.
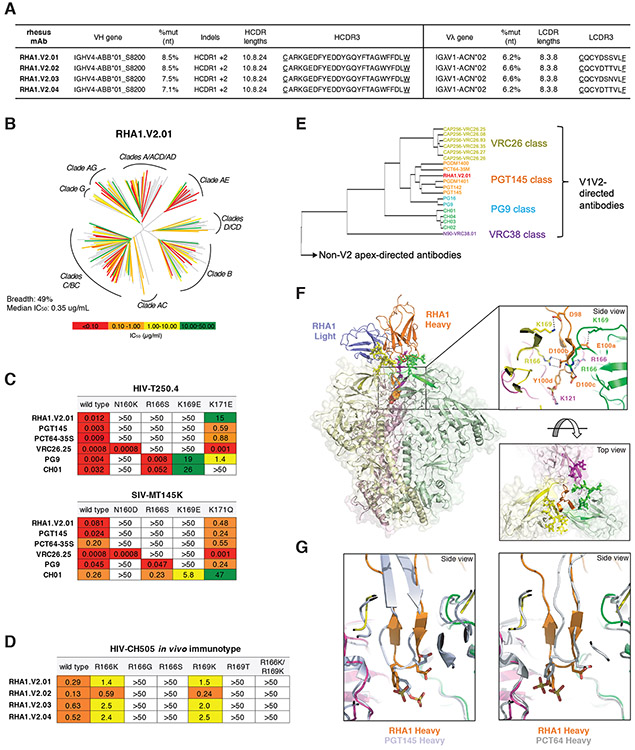
RM5695, infected with an early SHIV.CH505 plasma virus quasispecies (see Supplementary Materials), developed broadly neutralizing antibodies which, based on Env escape patterns in vivo and neutralization phenotypes of site-directed Env mutants in vitro, targeted the V2 apex (Fig. 3). We used an unbiased FACS strategy to isolate 20,000 individual memory B cells from peripheral blood mononuclear cells (PBMCs) 65 weeks post-SHIV infection and expanded these cells in 384-well plates. Culture supernatants were screened for neutralizing activity against two heterologous virus strains (T250-4 and Q23.17). Five wells scored positive and paired heavy and light chain immunoglobulin genes were successfully amplified from cellular mRNA from four of them (Figs. 6A,S18). All four rhesus mAbs belonged to a single lineage that we designated RHA1.V2 (Rhesus HIV Antibody 1 targeting a V2 epitope, with lineage members RHA1.V2.01-04). The IGVH and IGVL genes of the four RHA1.V2.01-04 mAbs were closely related with maximum nucleotide diversities of 5.3% and 3.9%, respectively. We employed NextGen sequencing to characterize immunoglobulin transcripts expressed by naïve IgD+IgM+IgG- B cells from RM5695 and used IgDiscover (57) and a recent database of RM Ig alleles (21) to identify a personalized immunoglobulin gene repertoire (Table S5). From this analysis, we determined the germline origins of the mature RHA1 mAbs to include a novel IGVH4 allele, IGHV4-ABB-S*01_S8200 (Fig. S18A), and IGλV1-ACN*02 (Fig. S18B). Somatic hypermutation within the lineage was modest, with nucleotide divergence from germline of 7.1 - 8.5% for VH and 6.2 - 6.6% for VL (Fig. 6A). These values are comparable to some human V2 apex bNAbs, which typically have lower frequencies of VH mutations than members of other human bNAb classes. The mature rhesus bNAb heavy chains contained a two amino acid insertion within HCDR1 (Figs. S18A,19A) and a 24 residue long HCDR3 (IMGT numbering) that was derived from rearrangement of the VH4-ABB-S*01_S8200, DH3-9 and JH2-P genes plus six non-templated amino acids (Fig. S18A,19B). This HCDR3 was rich in aromatic and negatively charged residues, like HCDR3s of human V2 apex bNAbs (Fig. S19C). Despite this long HCDR3, the mature rhesus bNAb RHA1.V2.01 was not auto- or polyreactive (Fig. S20). The HCDR3 of the RHA1 lineage antibodies was similar in length to the human V2 apex bNAb PCT64-35S (24 vs 25 amino acids, respectively), and the two broadly neutralizing mAbs contained conserved motifs within their respective HCDR3s including a negatively charged “DDY” segment (Fig. S19C), which in PCT64-35S was shown to be tyrosine-sulfated and to interact with positively charged V2 apex-hole residues of Env (8, 58). All four rhesus mAbs were tested for neutralization against the 19-member global panel of tier 2 viruses and showed similar patterns of reactivity, neutralizing 15 - 17 strains (Fig. S21). One antibody (RHA1.V2.01) was tested for neutralization against a 208 member global virus panel and was found to neutralize 102 heterologous virus strains, or 49%, at a maximum concentration threshold of 50 μg/ml (Figs. 6B,S22). Neutralization of heterologous virus strains depended on Env residues N160, R/K166 and R/K169, with partial dependence on K171 (Figs. 6C,S23). This precise pattern of neutralization sensitivity to N160, R/K166 and C-strand residue R/K169 and K171 mutations was shared by the human V2 apex bNAbs PCT64-35S and PGT145 but was different from that of PG9, VRC26.25 and CH01. CH505 Envs that evolved in RM5695 in vivo coincident with the development and maturation of RHA1 lineage antibodies showed evidence of strong selection at residues 166 and 169 (Fig. 3A). Introduction of these mutated residues into the CH505 T/F Env resulted in loss of neutralization sensitivity to RHA1 mAbs (Fig 6D).
Fig. 6. Rhesus bNAb lineage RHA1 targets the V2 apex, is broadly reactive, and contains a sulfated tyrosine in HCDR3 that shows precise chemical mimicry to human bNAbs PCT64-35S and PGT145.
(A) Immunogenetic characteristics of four RHA1 lineage broadly neutralizing monoclonal antibodies. A key feature is the 24 amino acid long CDRH3 that contains an acidic EDDY core motif. (B) Neutralization breadth and potency of RHA1.V2.01 against a 208 strain global virus panel. The dendrogram depicts phylogenetic relatedness of the HIV-1 Envs tested. (C) Neutralization expressed as IC50 (μg/ml) of wildtype heterologous viruses and their V2 apex mutants by RHA1.V2.01 and by prototypic human V2 apex bNAbs. Like most human V2 apex bNAbs, RHA1.V2.01 is strictly dependent on N160 and positively charged residues at 166 and 169. (D) Neutralization of CH505 T/F (wildtype) virus and C-strand variants, or “immunotypes,” that evolved in vivo in RM5695. Predominant mutations at 24 wks post-SHIV infection in RM5695 were R166K or R169K; at 48 wks R166S, R166G, R169T, and R166K plus R169K were prevalent; at 64 wks R166S or R169T became fixed (see Fig. 3A). Panel D shows progressive loss in neutralization sensitivity to RHA1 bNAbs by the evolving CH505 Envs, beginning with CH505 T/F wild type (most sensitive), CH505.R166K or R169K (intermediately sensitive), and ending with CH505.R166G, R166S, R169T or R166K+R169K (all resistant). Results are expressed as IC50 (μg/ml). (E) Neutralization fingerprint for RHA1.V2.01 shows it to cluster within the PGT145 class. (F) Cryo-EM structure (side view) of RHA1.V2.01 in complex with BG505 DS-SOSIP at 4-Å resolution. Inset (top) highlights electrostatic contacts of the HCDR3 with Env protomers, including interactions of the tyrosine-sulfated 100d residue with Env K121. Inset (bottom) shows the trimer apex cavity highlighting glycans at N160 and the C-strands. (G) Alignment of gp120 from the complex trimer structure with RHA1.V2.01 Fab to trimer complexes with human Fabs PGT145 (PDB-5V8L) and PCT64-35S (modeled with PDB-6CA6 fit to EMD-7865) reveals alignment of tyrosine sulfated residues within the respective HCDR3 tips, which insert into the hole at the V2 apex of the Env trimer.
Bioinformatical comparisons of human and RM V2-apex bnAbs.
Using a neutralization “fingerprint” analysis (59), which compares the potency of individual bNAbs against a large set of HIV-1 strains (Fig. S22), we observed clustering of RHA1.V2.01 within the PGT145 class of V2 apex bNAbs that includes PCT64-35M and PGDM1400 (Figs. 6E,S19D,E). In a hierarchical clustering analysis of the neutralization profiles of RHA1.V2.01 and other prototypic human V2 apex bNAbs measured against the 208 virus panel, RHA1.V2.01 grouped most closely with PCT64-35M (Fig. S24A). This finding was strongly supported statistically by the overlap of viruses that were sensitive or resistant to those antibodies [Fisher’s exact test p=2x10−16; odd’s ratio = 13.14; accuracy ratio (all concordant = 1, none = 0) = 0.78] (Fig. S24B) and by correlation of IC50 titers of RHA1.V2.01 and PCT64-35M (R2=0.4467; p=1.4x10−10) compared with other V2 apex bNAbs (Fig. S24C). We next examined neutralization profiles across different HIV-1 group M subtypes (Fig. S25). We found that RHA1.V2.01 was more subtype-independent than any of the human V2 apex bNAbs but was otherwise most similar to PCT64-35M and CAP256.VRC26. Finally, we performed a neutralization signature analysis using GenSig (https://www.hiv.lanl.gov/content/sequence/GENETICSIGNATURES/gs.html) (Fig. S26). Neutralization “signatures” identify individual Env residues that contribute directly or indirectly to antibody binding, including those potentially involved in selecting for affinity maturation (60). The signature analysis was performed using Fisher’s test with a binary IC50 threshold greater or less than 50 μg/ml and a Wilcoxon test that measures the difference in IC50 distributions with and without a given amino acid residue. High-confidence signature sites were defined as those meeting at least two of three criteria: i) contact sites; ii) at least one phylogenetically corrected signature at the site; or iii) at least one signature at the site that had a false discovery rate q < 0.1. For the rhesus mAb RHA1.V2.01, statistically robust signatures were identified at residues 130, 160, 166, 167 and 169. Among all of the V2 apex bNAbs analyzed, only PCT64-35M shared all five of these signature sites.
Structure of V2-apex bnAb RHA1.V2.01.
The structure of mAb RHA1.V2.01 in complex with the BG505 DS-SOSIP Env trimer, determined by cryo-EM at 3.9Å resolution, showed striking similarity to PGT145 and PCT64-35S (Figs. 6F,G and Table S6). These antibodies bind Env with a 1:1 stoichiometry near the trimer 3-fold axis and are surrounded by the three N160 glycans. Their respective CDRH3s adopt a needle-like antiparallel β-hairpin conformation that extends from the combining surface of the Fab and inserts into a cationic hole at the trimer apex. The N-terminal ends of each of the three C-strands abut the apex hole and are oriented perpendicular to the inserting HCDR3. Like PCT64-35S and PGT145, the acidic EDDY motif of RHA1.V2.01 was tyrosine-sulfated and made key contacts with Env residues 121, 166 and 169. (Fig. 6F boxed insert). When the Env-bound structures of RHA1.V2.01, PCT64-35S and PGT145 were overlaid, the respective EDDY motifs aligned at the tips of their respective HCDR3 loops around the β-hairpin turn (Fig. 6G). Otherwise, the overall Fab orientations differed, indicating the HCDR3 tip structural mimicry to be the main source of the neutralization similarity among these antibodies. The HCDR1 of RHA1.V2.01, which contained a non-templated two amino acid insertion in addition to other strongly selected mutations, was sandwiched between the Env N160 glycans of two protomers and proximal to the C-strand of one, with buried surface area of 52 Å2 and 49 Å2 for the two glycans and a key electrostatic interaction between D29 and K171 (Figs. 6F,S19A,S27F). Thus, the V2 apex bNAb lineage in RM5695 exhibits genetic, chemical and structural solutions to epitope recognition that are shared with human V2 apex targeted bNAbs, especially PCT64-35S and PGT145.
DISCUSSION
A principal finding of this study is that SHIVs bearing primary T/F HIV-1 Envs elicit strain-specific and heterologous NAbs in RMs that can replicate to a striking degree responses to HIV-1 in humans. This mimicry includes the frequency, kinetics, titers, immunogenetics, structures and target epitopes of elicited antibodies; structural and chemical features of epitope recognition; and coevolutionary pathways of antibody maturation and Env escape. All are key features to be considered in vaccine design. Our findings add substantially to earlier reports of sporadic neutralization of heterologous tier 2 viruses elicited in RMs by SHIVs bearing lab-adapted or animal passaged HIV-1 Envs (61-64) or Env immunogens (65-72). The current results, together with a recent report by Wang and colleagues (73), show how closely neutralizing antibody responses in RMs can mirror responses in humans and indicate the extent to which protective responses elicited by reverse engineered or lineage-based vaccines in RMs might be expected to predict human responses to candidate vaccines (10, 11, 74, 75).
While bNAbs from animal RM5695 were exceptional, the breadth and potency of bNAbs from the other six animals were more in keeping with a large cross-section of chronically-infected humans who were studied for neutralization breadth generally after many years of infection (28, 33, 34). In those studies, “elite neutralizers” with the most broadly reactive antibodies and from whom many of the best bNAb mAbs have been isolated, actually represented a very small fraction (<5%) of infected individuals. What is most notable about our study is not the overall breadth and potency of rhesus bNAb responses, but the fact that we observed bNAbs arising in 7 of 22 prospectively studied animals after less than one or two years of SHIV infection and that the epitope specificities of these antibodies could be mapped to canonical HIV-1 bNAb sites in six. These findings suggest that the rhesus macaque is a favorable model for HIV-1 vaccine studies where the key variables of priming, boosting, affinity maturation and antibody durability can be explored rapidly and iteratively.
A surprising observation was the extent to which Env evolution in macaques recapitulated evolution of homologous Envs in human infections. Similarities included site-specific and amino acid-specific mutations and identities or near identities of insertions and deletions. These similarities likely resulted from: (i) the highly evolved and functionally constrained nature of primary T/F Env trimers; (ii) limited sites of antibody accessibility and variable fitness costs of escape mutations; and (iii) homologous germline B cell responses in different animals and humans to conserved Env epitopes. Equally surprising were the genetic and structural similarities between rhesus and human antibodies that targeted CD4bs or V2 apex epitopes and their conserved mechanisms of epitope recognition. This included the HCDR2-mediated CD4 mimicry of the rhesus antibody DH650 and the tyrosine-sulfated, 24 residue long HCDR3 of the rhesus antibody RHA1.V2.01, which bound N160 glycans and positively charged Env residues at positions 121, 166, 169 and 171. Together, the conserved patterns of Env-specific sequence variation and the homologous and orthologous B cell responses in humans and rhesus represent remarkable examples of convergent evolution (14) that may aid in the design and testing of novel HIV-1 vaccines.
Our findings suggest that HIV-1 Envs are not equal in their propensity for eliciting epitope specific bNAb responses. For example, we found that CAP256SU Env, which elicited V2 apex bNAbs in the human subject CAP256 (3, 5, 22), induced bNAbs of the same specificity in 2 of 6 SHIV infected RMs. CH848 Env, which elicited V3 glycan targeted bNAbs in a human subject (7), did the same in 2 of 6 SHIV infected RMs. And CH505 Env, which elicited CD4bs targeted bNAbs in a human subject (2, 4, 6), induced homologous strain-specific CD4bs targeted NAbs in RM6072. CH505 Env also elicited V2 apex targeted NAbs with variable breadth in vaccinated CH03 heavy chain Ig knock-in mice and an immunized RM (69). In the present study, it did so as well in 2 of 10 SHIV infected RMs. In other work (G.M.S., unpublished), we have identified SHIV infected RMs that have developed fusion peptide targeted bNAbs and broadly reactive and potent CD4bs bNAbs. These findings highlight the potential for RMs to develop antibody responses targeting an array of different canonical bNAb epitope clusters and suggest a tendency for certain Envs to preferentially elicit bNAb responses targeting particular epitopes.
SHIV replication in RMs is the only model system other than naturally infected humans where the immunogen (Env) coevolves with antibodies. The high mutability and dynamic replication of HIV-1 and SHIV result in a constantly evolving virus quasispecies (9, 18, 38, 39, 42, 43), which means that Envs with binding affinities sufficient to drive bNAb lineage affinity maturation are constantly being generated. The SHIV infected macaque can therefore be particularly informative for vaccine design by enabling the identification, and then rapid testing, of Env intermediates that guide the evolution of germline bNAb precursor B cells through stages of affinity maturation to acquire breadth and potency. In the CAP256 (5) and PCT64 (8) infected human subjects, viral sequences showed very similar patterns of Env evolution at residues 166 and 169, which in turn were similar to the pattern in the SHIV.CH505 infected RM5695. Deep sequencing of RM5695 plasma vRNA covering the V1V2 region revealed selection focused primarily on residues 166 and 169 with mutations at these two sites rapidly and completely replacing the infecting virus strain. The infecting SHIV.CH505 virus had an Arg at these two positions, which evolved progressively to R166K or R169K and then to R166G, R166S or R166T (Fig. 3). The earliest mutants, R166K or R169K, were approximately 5-fold more resistant to the mature rhesus bNAb mAbs than the infecting virus, whereas the subsequent R166G, R166S or R166T mutants were >100-fold more resistant (Fig. 6D). Thus, sequential Envs that varied at residues 166 and 169 in animal RM5695 showed progressive phenotypic escape from V2 apex bNAb antibodies, closely resembling the viral Env-bNAb coevolution observed in humans CAP256 and PCT64. Cryo-EM analysis of the RHA1.V2.01 mAb provided a structural explanation for this loss of antibody recognition by showing that Env residues 166 and 169 were primary electrostatic contacts with the antibody. Mutations in these two residues in the V2 apex appear to be largely or solely responsible for driving affinity maturation of diverse antibody lineages to breadth in multiple rhesus animals and humans within a relatively short time (<1 yr), suggesting that Env intermediates or “immunotypes” (5) required for V2 apex bNAb elicitation may be few and simple. This hypothesis has important implications for V2 apex targeted HIV-1 vaccine design, which can be tested rapidly in Ig knock-in mice and outbred macaques using immunogens designed from V2 apex variants of CH505 and other primary Envs. The goal of such research would be to learn the “rules” governing consistent bNAb induction in RMs and then to translate these findings to human studies using SOSIP, mRNA or other non-SHIV based vaccine platforms.
V3 high mannose patch glycopeptides are also commonly targeted by bNAbs in HIV-1 infected humans (33) and are of high interest for HIV-1 vaccine development (50, 76, 77). Site-directed mutagenesis coupled with antibody neutralization showed that polyclonal bNAb responses in SHIV.CH848 infected RMs 6167 and 6163 targeted canonical N332 and 324GDIR327 motifs, similar to human V3 glycan bNAbs (78, 79). Deletions in V1 of SHIV.CH848 sequences preceded the development of V3 glycan bNAbs in both monkeys and in the human subject CH848. Long, glycosylated (N133, N138) V1 segments obstruct access of V3 glycan bNAbs (7, 80), and germline-targeted Env immunogens with shortened V1 segments depleted of glycans enhance Ab access to V3 high mannose patch epitopes (50, 76). Because Env-Ab coevolution leading to V3 glycan bNAbs generally requires more extensive somatic hypermutation compared with V2 apex bNAbs (10, 11), a rhesus model in which vaccinations with germline-targeted Envs (50, 76, 77) is followed by infection with SHIVs whose Envs are similarly targeted, offers a novel strategy for identifying the “finishing” or “polishing” immunogens necessary for bNAb affinity maturation and an outbred primate model system to test them.
It is generally believed that the development of an effective neutralizing antibody-based HIV-1 vaccine will require consistent activation of multiple germline precursor B cells that express immunoglobulin receptors specific for one or more of the canonical bNAb epitope clusters, followed by efficient antigen-driven selection for antibody affinity maturation (10, 11, 50, 74-77, 81). The present study demonstrates that the SHIV-infected rhesus model can inform both of these critical steps in bNAb elicitation. The fact that only a minority of SHIV infected animals in the current study developed bNAbs is a faithful reflection of the natural prevalence of bNAb responses in HIV-1 infected humans (28, 33, 34, 82) and further argues for the relevance of the rhesus model. A limitation of our study is that SHIV infection, of course, is not a viable vaccine strategy for humans, nor is CD8 depletion, which was employed to increase peak and setpoint virus loads. Nonetheless, it should be possible to combine established immunization platforms such as Env trimers, outer domain scaffolds, virus-like particles, or DNA/RNA expression followed by SHIV infection to identify optimized priming and boosting immunogens that elicit broad neutralization in macaques as a molecular guide for HIV-1 vaccine design in humans.
Materials and Methods
Nonhuman Primates and SHIV Inocula.
Indian RMs were housed at Bioqual, Inc., Rockville, MD, according to guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care standards. Experiments were approved by the University of Pennsylvania and Bioqual Institutional Animal Care and Use Committees. RMs were sedated for blood draws or excisional biopsies of lymph nodes, anti-CD8 mAb infusions, and SHIV inoculations. A subset of animals received an intravenous infusion of either 25-50 mg/kg of anti-CD8alpha mAb (MT807R1) or anti-CD8beta mAb (CD8beta255R1) one week prior to or at the time of SHIV inoculation. The intent of administering anti-CD8 mAbs was to transiently reduce CD8+ cells, thereby allowing for higher peak and setpoint viral loads. This was done in only a subset of animals because it was not known a priori if this might accelerate disease progression to an extent that would preclude long term follow-up of animals for bNAb induction. In humans, higher plasma virus loads and lower CD4+ T cell counts are correlated with the development of bNAbs (33, 34). SHIV infections were done intravenously. All animals in this study, with the exception of RM5695, were inoculated with molecularly cloned virus (50 or 500 ng p27Ag in DMEM or RPMI 1640 with 10% heat-inactivated fetal bovine serum) containing the preferred Env375 variant or a mixture of Env375 Env variants. Animals RM6069, 6070, 6072, 6163 and 6167 were repurposed from a previous study of SHIV replication dynamics and immunopathogenesis (9). Animal RM5695 was repurposed from a previous study of HIV-1 CH505 Env gp120 protein immunization (23). This animal was infected with 0.2 ml of SHIV-positive plasma from monkeys RM6069 (wks 10 and 20), RM6070 (wks 10 and 20) and RM6072 (wks 04, 10 and 20), for a total plasma inoculum of 1.4 ml. The rationale for this experiment was to increase early viral diversity and potentially include antigen-antibody immune complexes in the inoculum. The genetic composition of this plasma virus inoculum, and the particular viral genomes that were successfully transmitted to RM5695, can be seen in the Highlighter and LASSIE panels in Figs. 2A and 2B and the sequences can be accessed from GenBank (accession # MT484881-MT484977).
Processing and storage of clinical specimens.
All blood samples were collected in sterile vacutainers containing ACD-A anticoagulant. Forty ml of ACD-A anticoagulated blood was combined in a sterile 50 mL polypropylene conical tube, centrifuged at 2100 rpm (1000xg) for 10 min at 20°C, and the plasma collected in a fresh 50 mL conical tube without disturbing the buffy coat WBC layer and large red cell pellet. The plasma was centrifuged again at 2500 rpm (~1500g) for 15 minutes at 20°C in order to remove all platelets and cells. Plasma was collected and aliquoted into 1 ml cryovials and stored at −80°C. The RBC/WBC pellet was resuspended in an equal volume of Hanks balanced salt solution (HBSS) without Ca++ or Mg++ and containing 2mM EDTA and then divided into four 50 ml conical tubes. Additional HBSS-EDTA (2mM) buffer was added to bring the volume of the RBC/WBC mixture to 30 ml in each tube. The cell suspension was then carefully underlayered with 14 ml 96% Ficoll-Paque and centrifuged at 1800 rpm (725xg) for 20 min at 20°C in a swinging bucket tabletop centrifuge with slow acceleration and braking so as not to disrupt the ficoll-cell interface. Mononuclear cells at the ficoll interface were collected and transferred to a new 50ml centrifuge tube containing HBSS-EDTA (w/o Ca++ or Mg++) and centrifuged at 1000 rpm (~200 g) for 15 min at 20°C. This pellets PBMCs and leaves most of the platelets in the supernatant. The supernatant was removed and the cell pellet was resuspended in 40 ml HBSS (with Mg++/Ca++ and without EDTA) + 1% FBS. To remove additional contaminating platelets, the cell suspension was centrifuged again at 1000 rpm (~200 g) for 15 minutes at 20°C and the supernatant discarded. The cell pellet was tap-resuspended in the residual 0.1-0.3 ml of media and then brought to a volume of 10 ml HBSS (with Mg++/Ca++) +1%FBS. Cells were counted and viability assessed by trypan blue exclusion. Cells were centrifuged again at 1200rpm (300xg) for 10 min at 20°C, the supernatant discarded, and the cells resuspended at a concentration of 5-10x106 cells/ml in CryoStor cell cryopreservation media (Sigma Cat. C2999) and aliquoted into 1ml cryovials (CryoClear cryovials; Globe Scientific Inc., Cat. 3010). Cells were stored in a Mr. Frosty at −80°C overnight and then transferred to vapor phase liquid N2 for long-term storage. Mononuclear cells collected from lymph nodes (LN) and spleen were processed similar to blood mononuclear cells. LN nodes and spleen were excised and placed immediately into RPMI1640 medium on wet ice. LNs were diced with a sterile scalpel and spleen was homogenized and the material passed through a sterile mesh grid. Cells were collected from the pass-through and subjected to Ficoll density gradient purification as described above.
SHIV construction and characterization.
The experimental design for constructing SHIVs bearing primary or transmitted/founder Envs with allelic variation at gp120 residue 375 was previously described, including SHIV.CH505 and SHIV.CH848 (9). For the construction of SHIV.CAP256SU, we synthesized (GenScript) sequence KF996583.1 from GenBank (GenBank: KF996583.1) and cloned it into the pCRXL-TOPO-SIVmac766 backbone (9) by recombinant PCR. The QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) was used to create allelic variants (M, Y, F, W, or H) of the wild type Env375S codon. Wild type and mutant plasmids were transformed into MAX Efficiency Stbl2 Competent Cells (Invitrogen) for maxi-DNA preparations. Each 10-kb viral genome was sequenced in its entirety to authenticate its identity. Infectious SHIV stocks were generated in 293T cells as previously described (9). On day 0, five million 293T cells were plated in 100-mm tissue culture dishes in 10 mL of complete MEM growth media with 10% FBS. On day 1, 6 ug of SHIV plasmid DNA was combined with 18 uL of FuGENE 6 (Promega) in 500 μL of DMEM was added dropwise to tissue culture dishes. Media containing virus was harvested on day 3 and aliquoted for storage at −80C. Virus concentration was estimated by p27 antigen (p27Ag) ELISA (Zeptometrix) and infectious particle concentration was determined by entry into TZM-bl cells in the presence of DEAE–Dextran, as previously described (18). Typically, 293T-derived SHIV stocks contained >1,000 ng/ml p27Ag and >1,000,000 IU/ml on TZM-bl cells. The replication kinetics of each of the SHIV.CAP256SU Env375 variants in primary, activated human and rhesus CD4 T cells were determined as previously described (9). 293T supernatants containing 300 ng p27Ag of each variant, were added to 2 x 106 purified human or rhesus CD4 T cells in complete RPMI growth medium (RPMI1640 with 15% FBS (Hyclone), 100 U/mL penicillin–streptomycin (Gibco), 30 U/mL IL-2 (aldesleukin, Prometheus Laboratories) and 30 μg/ml DEAE-Dextran. 300 ng p27Ag is equal to ~3 x 109 virions, ~3 x 105 IU on TZM cells, or ~3 x 104 IU on primary CD4 T-cells, so the estimated MOI of this titration was estimated to be ≤ 0.1. The cell and virus mixtures were incubated for 2 hours under constant rotation at 37C to facilitate infection, washed three times with RPMI1640, and resuspended in complete RPMI1640 medium lacking DEAE-Dextran. Cells were plated into 24-well plates at 2 million cells in 1 ml and cultured for 11 days, with sampling of 0.2ml supernatant and media replacement every 2-3 days for 11 days. Supernatants were assayed for p27Ag concentration by ELISA (Zeptometrix).
Plasma vRNA quantification.
Plasma viral load measurements were performed by the NIH/NIAID-sponsored Nonhuman Primate Virology Core Laboratory at the Duke Human Vaccine Institute. This core facility is CLIA certified and operates a highly standardized, quality-controlled Applied Biosystems Real-Time SIV and HIV vRNA PCR assays. QIAsymphony SP and QIAgility automated platforms (QIAGEN) are used for high throughput sample processing and PCR setup. Viral RNA is extracted and purified from plasma, annealed to a target specific primer and reverse transcribed into cDNA. The cDNA is treated with RNase and added to a custom real-time PCR master mix containing target specific primers and a fluorescently labeled hydrolysis probe. Thermal cycling is performed on a QuantStudio3 (ThermoFisher Scientific) real-time quantitative PCR (qPCR) instrument. Viral RNA cp/reaction is interpolated using quantification cycle data. Raw data is quality-controlled, positive and negative controls are checked, and the mean viral RNA cp/mL is calculated. Over the course of this study, the sensitivity limits for accurate vRNA quantification using 0.5 ml of NHP plasma improved from 250 RNA cp/ml to 62 RNA cp/mL. We chose a conservative threshold of 100 RNA cp/mL for a limit of detection and 250 RNA cp/mL for the limit of quantification.
Viral sequencing, pixel plots and LASSIE analysis.
Single genome sequencing of SHIV 3’ half genomes was performed as previously described (1, 9). Geneious R7 was used for alignments and sequence analysis and sequences were visualized using the LANL Highlighter and Pixel tools https://www.hiv.lanl.gov/content/sequence/pixel/pixel.html https://www.hiv.lanl.gov/content/sequence/HIV/HIVTools.html (44). The specific implementation of this software for this project is described in the figure legends.
IgG isolation from plasma.
Total polyclonal IgG was isolated from rhesus plasma using the Protein A/Protein G GraviTrap kit (GE Healthcare). Plasma was heat-inactivated (1 hour at 57C), clarified by centrifugation at 21,000g for 4 min, and applied to the Protein A/G column. The sample was washed and eluted per the manufacturer’s instructions, and then buffer-exchanged with phosphate buffered saline (PBS). The concentration of purified IgG sample was quantified using the Pierce BCA Protein Assay Kit (ThermoScientific).
Neutralizing antibody assay.
Assays for neutralizing antibodies were performed using TZM-bl indicator cells, as previously described (9, 18). This assay is essentially identical to that employed by Montefiori, Seaman and colleagues (85) https://www.hiv.lanl.gov/content/nab-reference-strains/html/home.htm, the only difference being that in our assay we plate virus and test plasma onto adherent TZM-bl cells and hold the concentration of test plasma constant (5% vol/vol) constant across all wells, which contain 10% heat-inactivated fetal bovine serum in the complete RPMI1640 culture medium. Target viruses express HIV-1 Envs whose complete designations and subtype classifications are included in Fig. S22 and Table S3 and reported elsewhere (26, 27, 86). In Fig. 1A, we calculated neutralization breadth (ID50 ≤ 0.05) across all time points for each animal from titers displayed in Table S2 and plotted in Fig. 1A. We calculated the maximum geometric mean titer (GMT) of neutralization at any one time point from values with ID50 ≤ 0.05 as displayed in Table S2 and plotted in Fig. 1A.
Binding antibody assays.
HIV-1 Env binding by recombinant mAbs, and plasma or sera, were tested in ELISA as previously described (50, 87). In brief, recombinant Envs were coated directly to Nunc-absorb (ThermoFisher) plates overnight at 4°C or captured using AbC- mAb (AVIDITY, Colorado, USA) that was directly coated to Nunc-absorb plates overnight at 4°C. Antibody binding was detected with goat anti-human or goat anti-rhesus HRP-labeled anti-IgG Fc antibodies (Jackson ImmunoResearch Laboratories), and HRP detection was subsequently quantified with 3,3′,5,5′-tetramethylbenzidine (TMB). Competitive ELISA to assess cross-blocking of recombinant mAbs or plasma antibodies were previously described (7, 88). We biotinylated the antibodies using the following product: BIOTIN-X-NHS, Cayman Chemicals, CAT# 13316. Competitive inhibition of biotiniylated-mAbs was measured as a percent of binding in the presence of a competing non-biotinylated mAb relative to binding in the absence of this competing mAb. MAbs were also tested for binding HIV-1 Envs using Biolayer interferometry (BLI) as described (89). Here, antibody binding was measured using mAb-captured sensors that were placed into solutions of CH505 gp120 or SOSIP trimers at 50 μg/ml for 1000s. MAbs were captured using anti-human IgG Fc sensors, and non-specific or background binding was subtracted using binding levels by anti-influenza HA mAb (CH65).
Rhesus B cell staining and sorting of strain-specific mAbs.
CH505 gp120 T/F CD4bs-specific antibodies were isolated from memory B cells in PBMCs, lymph node or bone marrow collected at weeks 20, 24, 32 and 52 using two approaches: direct single-cell sorting into PCR plates (weeks 20, 32 and 52) (23, 90), and memory B cell cultures (week 24) (91, 92). For direct sorting, we performed single-cell isolation of memory B cells decorated with AlexaFluor® 647 (AF647) or Brilliant Violet 421 (BV421)-tagged HIV-1 CH505 TF gp120 using a BD FACSAria™ or a BD FACSAria™ II (BD Biosciences, San Jose, CA), as previously described (23). The flow cytometry data were analyzed using FlowJo (Treestar, Ashland, OR) (90, 93, 94). For our sort strategy, we isolated antigen-specific IgD-negative, CD27-All memory B cells that bound BV421-tagged CH505 T/F gp120, but not AF647-tagged CH505 T/F gp120 Δ371 mutant protein; antibodies isolated from these B cells were referred to as CH505 differential binders or CD4BS antibodies (23). For memory B cell cultures, from 8 million PBMCs, we sorted 30,792 CH505 T/F gp120-specific B cells, defined as CD3-negative, CD14-negative, CD16-negative, IgD-negative, CD27-All, CD20-positive, AF647-tagged CH505 T/F gp120-positive and BV421-tagged CH505 T/F gp120-positive. As previously described (92), cells were flow sorted in bulk into wells containing 5,000 MS40L feeder cells, RPMI-1640 supplemented with 15% FBS, 1 mM sodium pyruvate, 1% non-essential amino acids, 25 mM HEPES buffer, 2.5 μg/ML ODN2006 (Invivogen, TLRL-2006-5), 5 μM CHK2-inhibitor (Calbiochem, 220486), 100 ng/mL recombinant human interleukin (IL)-21 (Peprotech, Cat. no. 2001-21), 10 ng/mL recombinant Human BAFF (Peprotech, Cat. no. 310-13), 200 U/ml IL-2 (from the myeloma IL-2 producing cell line IL2-t6, kindly provided by Dr. Antonio Lanzavecchia, IRB, Bellinzona, Switzerland), and 100 μL supernatant of the Herpesvirus papio (HVP)-infected Baboon cell line S594 (NHP Reagent Resource). The concentration of each supplement was previously determined to achieve optimal in vitro stimulation. Following overnight incubation at 37 °C in 5% CO2, memory B cells were transferred at limiting dilution into 96-well round bottom tissue culture plates containing 5,000 MS40L feeder cells. Culture medium was refreshed 7 days after plating and harvested 2 weeks after plating to test for binding to CH505 T/F gp120, CH505 T/F gp120 Δ371, RSC3 (95) and RSC3 Δ371 P363N, as well as for neutralization of pseudotyped the CH505 T/F HIV-1 strain in the TZM-bl-based neutralization assay using a single dilution of supernatant (91, 96). CD4bs DH650 lineage autologous tier 2 NAbs were isolated from CH505 differential-binding memory B cells in PBMCs from week 20, 24 and 32 post SHIV CH505 infection of RM6072. In capturing maximum numbers of B cells bearing candidate CD4bs antibodies, we used a less stringent gating strategy for capturing CH505 T/F gp120 (+) and CH505 T/F gp120 Δ371 (−) B cells. In so doing, we also captured nonCD4-binding site autologous tier 2 NAbs, including DH647 and DH648 that were isolated from memory B cells in PBMCs at week 20 post SHIV infection of RM6072.
Rhesus B cell staining, culture and microneutralization screening for V2 apex bNAbs.
Cryopreserved PBMCs from RM5695 at week 65 post-SHIV infection were thawed and stained with LIVE/DEAD Fixable Aqua Dead Cell Stain (Life Technologies), as previously described (97, 98). Cells were washed and stained with an antibody cocktail of CD3 (clone SP34-2, BD Biosciences), CD4 (clone OKT4, BioLegend), CD8 (clone RPA-T8, BioLegend), CD14 (clone M5E2, BioLegend), CD20 (clone 2H7, BioLegend), IgG (clone G18-145, BD Biosciences), IgD (polyclonal, Dako) and IgM (clone G20-127, BD Biosciences) at room temperature in the dark for 20 mins. The stained cells were washed 3 times with PBS, re-suspended in 1 ml of PBS and passed through a 70 μm cell mesh (BD Biosciences). Total memory B cells (CD3-CD4-CD8-CD14-CD20+IgD-IgM-IgG+) were sorted with a modified 3-laser FACSAria cell sorter using the FACSDiva software (BD Biosciences) and flow cytometric data was subsequently analyzed using FlowJo (v9.7.5). B cells were sorted at 1 cell per well of a 384-well plate containing B cell culture media based on a human B cell culture protocol (99) that was optimized for the expansion of rhesus B cells. Briefly, sorted B cells were expanded for 14 days in B cell culture medium consisting of Iscove’s modified Dulbecco’s medium (IMDM) with GlutaMAX™ supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1X MycoZap Plus-PR, 100 U/ml IL-2, 0.05 ug/ml IL-4, 0.05 ug/ml IL-21, 0.05 ug/ml BAFF, 2 ug/ml CpG ODN2006, and 3T3-msCD40L feeder cells at a density of 5,000 cells per well. Supernatants from ~14,000 individual wells were evaluated for neutralization of HIV-1 Q23.17 and T250-4 Env-pseudotyped viruses using the high throughput NVITAL automated microneutralization assay, as previously described (100). Wells were selected for RT-PCR based on 50% reduction in infectivity of at least one virus. Out of nearly 14,000 wells tested, five met these neutralization criteria. Paired heavy and light Ig chains were successfully amplified from four of these wells. These amplicons were sequenced, cloned and expressed as IgG1 (RHA1.V2.01-04), and all four mAbs exhibited broad and potent neutralization.
Rhesus B cell cloning and expression.
Heavy (IGHV) and light (IGKV, IGLV) chain genes were isolated via single cell PCR approaches (90, 101), and the gene sequences were computationally analyzed using rhesus Cloanalyst program (102-104). Antibody immunogenetics were reported as gene families and segments, mutation frequencies, and CDR3 lengths using the rhesus Cloanalyst database of reference genes (21). Using the rhesus cloanalyst program, we identified B cell clonal lineages for antibodies with the same inferred IGHV VDJ rearrangement and CDR3 length, and paired with the same light chain (Ig VJ segments). For DH650 lineage, the unmutated common ancestor (UCA) and intermediate (IA) genes were inferred computationally using the Cloanalyst program. The automated inference of antibody clonality was followed up by visual inspection of the DNA sequence alignments for confirmation. The heavy and light chain gene sequences from the sorted B cells or inferred UCA and IAs were commercially generated, and used to express purified recombinant mAbs as described (90). Heavy and light chain immunoglobulin repertoire next generation sequencing of monkey RM6072 was performed with the Illumina MiSeq platform utilizing primers targeting the VH1 and Vk2 families to identify DH650 clonal members using a previously described protocol (23, 105). For mAb isolation from RM5695, bulk cDNA was synthesized from the five neutralization-positive B cell culture wells using random hexamers as previously described (106). Subsequently, immunoglobulin heavy chain (IgG) and light chain (IgK and IgL) variable regions were separately amplified by nested PCR cycles using pools of rhesus macaque primers as previously described (107). Sequences were analyzed using Cloanalyst (Kepler et al, Front Imunol 2014) to infer putative variable region germline genes and identify clonal lineages. The heavy and lambda chain variable regions of the RM5695 lineage were codon optimized, synthesized with a murine immunoglobulin signal peptide (tripeptide sequence VHS) immediately preceding the 5’ end of FRW1, and cloned into rhesus IgG1 (RhCMV-H) and IgL (RhCMV-L) expression vectors directly upstream of the respective constant regions using AgeI/NheI and AgeI/ScaI restriction sites, respectively (107). Recombinant mAbs were expressed by cotransfection of paired heavy and lambda chain plasmids into 293Freestyle cells as previously described (95), purified from cell supernatant using the Protein A/Protein G GraviTrap kit (GE Healthcare), and buffer-exchanged into PBS.
Next generation sequencing of naïve B cells and IgDiscover analysis
PBMCs isolated from RM5695 plasma obtained at week 48 post-infection were stained with LIVE/DEAD Aqua, CD3-PerCP-Cy55, CD4-BV785, CD8-BV711, CD14-PE-Cy7, CD20-BV605, IgD-FITC, IgG-Ax680, and IgM-BV650. Approximately 300,000 naive B cells (CD20+, IgG-, IgD+, IgM+) were bulk sorted into RPMI with 10% FBS and 1% Pen-Strep using a BD FACSAria II. Total RNA was extracted using RNAzol RT per the manufacturer’s guidelines (Molecular Research Center, Inc). Reverse transcription of mRNA transcripts, IgM and IgL variable region library preparation, and next generation sequencing were performed as previously described (108), as these methods can be efficiently used for both humans and rhesus macaques. Both heavy and lambda immunogloblulin libraries were sequenced on Illumina Miseq with 2x300 bp runs using the MiSeq Reagent V3 kit (600-cycle). Filtered, quality-controled IgM and IgL sequences were analyzed using IgDiscover (57) to curate a personalized immunogloblin repertoire library for RM5695. A naïve IgK variable region library was not prepared since the RM5695 lineage utilizes a lambda chain. IgDiscover can identify functional VH, VL, VK, JH, JL and JK genes, and denotes any novel genes and/or alleles with respect to the provided reference database. For this analysis, a recently published rhesus macaque database was used as the template (21). The resulting personalized database was then used in SONAR (109) to accurately assign germline immunoglobulin genes and precisely determine somatic hypermutation frequencies for the RHA1.V2 lineage.
Auto/Polyreactivity analysis.
RHA1.V2.01 mAb reactivity to nine autoantigens was measured using the AtheNA Multi-Lyte ANA kit (Zeus scientific, Inc, #A21101). Antibodies were 2-fold serially diluted starting at 50 μg/mL and kit methods were followed per manufacturer’s instructions. Samples were analyzed using AtheNA software. Indirect immunofluorescence binding of RHA1.V2.01 mAb to human epithelial (HEp-2) cells (Zeus Scientific, Somerville, NJ) was also performed per manufacturer’s instructions. Briefly, 20μL of diluted antibody (50μg/ml) was added to antinuclear antibody (ANA) test slides. Slides were incubated for 20 minutes at room temperature in a humid chamber, and then washed with 1X PBS. Goat-anti-rhesus Ig-FITC (Southern Biotech, Birmingam, AL) secondary antibody was added at a concentration of 30μg/ml to each well. The slides were incubated for 20 minutes, washed twice, dried, fixed with 33% glycerol and cover-slipped. Slides were imaged using an Olympus AX70 microscope with a SpotFlex FX1520 camera. Images were acquired on a 40X objective using the FITC fluorescence channel. Positivity was determined by comparison with positive and negative control non-human primate mAbs DH1037 and DH570.30, respectively. Staining patterns were identified using the Zeus Scientific pattern guide.
Hierarchical clustering of RHA1.V2.01 and other V2 apex bNAb profiles.
We used the Heatmap webtool at the Los Alamos HIV database https://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html (95, 108, 109). Input data were Log10 transformed IC50 titers for RHA1.V2.01 and other V2 apex bNAbs for the 208 virus panel. For clustering, Euclidean distances and the Ward algorithm (110) were used. Bootstraps were calculated using 1000 iterations.
Neutralization Signature Analyses.
These analyses were performed using the webtool GenSig (https://www.hiv.lanl.gov/content/sequence/GENETICSIGNATURES/gs.html) (59) as described (60). Briefly, IC50 titers for RM5695 RHA1.V2.01 and other V2 apex bNAbs, together with Env sequences from the 208 strain virus panel, were used as inputs. Two statistical strategies were used: i) Fisher’s test with a binary phenotype, IC50 titer above or below the threshold (resistant and sensitive, respectively), and ii) Wilcoxon test that compares distribution of IC50 titers with and without a feature. The latter analyses were conducted two ways, including and excluding resistant viruses. In both strategies, every amino acid and glycan at each site were tested for associations, with and without a phylogenetic correction. A relaxed multiple test false discovery rate (FDR) (q-value < 0.2) was used to first filter all possible hypothetical associations. At this high threshold, several phylogenetically uncorrected signatures were found, raising the suspicion that phylogenetic artifacts could be at play. However, some of these signatures could be relevant and underlie the clade-specific sensitivity/resistance of the bNAb in question. Thus, to down-select the most statistically robust and relevant signature sites, each selected site was required to meet at least two of the following three criteria: i) contact site (as defined below), ii) at least one phylogenetically corrected signature, and iii) at least one strong signature at q < 0.1. All associations with q < 0.2 at selected sites were used to identify sequence features associated with sensitivity or resistance to the bNAb. The Wilcoxon signatures did not identify any new sites or associations. Env contact sites were defined using the cryo-EM structure of PGT145 in complex with Env trimer (PDB:5V8L, Lee2017 PMID:28423342) and a cutoff of 8.5Å from any antibody heavy atom (HXB2 positions: 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 184, 191, 192, 200, 201, 305, 306, 307, 308, 309, 312, 313, 314, 315, 321).
Soluble HIV-1 envelope trimer generation.
HIV Env trimer BG505 DS-SOSIP.664 was produced in transiently transfected 293F cells as previously described (111, 112). Briefly, the plasmid encoding the SOSIP trimer and a plasmid encoding furin were mixed at 4:1 ratio and transfected into 293F cells at 0.75 mg plasmid / 1 L cells (1.5 x 10E6 /ml) using 293Fectin (Thermo Scientific) or Turbo293 transfection reagent (Speed BioSystems). Cells were then incubated in a shaker at 120 rpm, 37 °C, 9% CO2. The following day, 80 ml HyClone SFM4HEK293 medium and 20 ml FreeStyle™ 293 Expression Medium were added to each liter of cells. Env trimer protein was purified from the day-7 supernatant with VRC01 affinity chromatography, followed by gel filtration on a Sephadex200 16/60HL column and V3-exposed trimers were removed with negative selection on a 447-52D affinity column. The antigenicity of the trimers was confirmed with a panel of antibodies in the Meso Scale Discovery (MSD) platform.
Antibody Fab preparation.
Variable regions of the RHA1.V2.01 antibody heavy and light chain genes were synthesized (Genscript) and subcloned into the pVRC8400 vector, in which a HRV3C cleavage site was inserted in the heavy-chain hinge region. The heavy and light chain pair was co-transfected in Expi293F cells (Thermo Fisher) using Turbo293 transfection reagent (Speed BioSystems) as described previously (113). The culture supernatant was harvested 6 days post transfection and loaded on a protein A column, the column was washed with PBS, and IgG proteins were eluted with a low pH buffer. The eluted IgG proteins were cleaved by HRV3C, and the cleavage mixture was passed through a protein A column.
Cryo-EM data collection and processing.
Antibody Fab fragments of RHA1.V2.01 were incubated with BG505 DS-SOSIP with the Fab in molar excess. 2.3 μl of the complex at 1 mg/ml concentration was deposited on a C-flat grid (protochip.com). An FEI Vitrobot Mark IV was used to vitrify the grid with a wait time of 30 seconds, blot time of 3 seconds and blot force of 1. Automated data collection was performed with Leginon (114) on a Titan Krios electron microscope equipped with a Gatan K2 Summit direct detection device. Exposures were taken in movie mode for 10 s with the total dose of 71.06 e–/Å2 fractionated over 50 raw frames. Images were pre-processed through Appion (115, 116); MotionCor2 (117) was used for frame alignment and dose-weighting. The CTF was determined using CTFFind4 (118, 119). Initial particle picking was done with DoG Picker (116, 120). RELION (121) was then used for particle extraction. CryoSPARC 2.12 (122) was used for 2D classifications, ab initio 3D reconstruction, homogeneous refinement, and nonuniform 3D refinement. Initial 3D reconstruction was performed using C1 symmetry, confirming 1 Fab per trimer and C1 symmetry was applied for the final reconstruction and refinement. Coordinates from PDB ID 6NNF (123) and 6CA6 (58) were used for initial fit to the reconstructed map. Simulated annealing was performed on the first refinement followed by iterative manual fitting and real space refinement in Phenix (124) and Coot (125). Geometry and map fitting were evaluated throughout the process using Molprobity (126) and EMRinger (127). PyMOL (www.pymol.org) was used to generate figures.
Expression and crystallization of CH505 gp120:DH650 Fab complex.
CH505 gp120 core (residues 44-492, ΔV1-V2 and ΔV3) (128) was expressed in HEK293S GnTI− cells and purified by affinity chromatography on Galanthus nivelis lectin (Vector Laboratories) followed by gel filtration on Superdex 200 column (GE). Deglycosylation was carried out with endoglycosidase H (New England biolabs) in deglycosylation buffer (50 mM sodium acetate, pH 6, 5 mM EDTA, 500 mM NaCl, 10uL endoH, 1 μg/μl leupeptin, 1 μg/μl aprotonin) at 37° overnight, followed by buffer exchange to 40 mM Tris HCl, pH 7.4, 1 M NaCl, 2 mM MnCl2, 2 mM CaCl2 and passage through a concanavalin A column (Sigma) to remove any gp120 that had not been fully deglycoylated by the endoH treatment. The eluate was buffer exchanged by passage over a Superdex200 column (GE) into 2.5mM Tris-HCl pH7.5, 350mM NaCl, followed by concentration to 4mg/ml. The Fab fragment of mAb DH650 was expressed in HEK293T cells, as described (128). DH650-gp120 core complex was formed by incubating gp120 core with DH650 in 1:1.3 molar ratio followed by gel filtration on Superdex 200 (GE) column in buffer 2.5mM Tris-HCl pH 7.5, 350mM NaCl. The complex was concentrated to 8.5 mg/ml and crystallized by hanging-drop vapor diffusion in 20% PEG 8K, 100mM Tris pH 8, 500mM NaCl.
Determination of CH505 gp120:DH650 Fab crystal structure.
X-ray diffraction data to 2.8 Å resolution were collected at the Advanced Photon Source (Argonne National Laboratories) on NE-CAT beamline 24-ID-C. Intensity data were integrated, scaled and merged with XDS and XSCALE (129) (see Table S4). An initial model, obtained by molecular replacement with Phaser (130) using the ZM176.6 gp120 core (PDB ID:4LST), was rebuilt and refined with COOT (131) and Buster (Global Phasing, Ltd., Cambridge, UK), respectively. The final model (PDB ID: 6XCJ) includes residues 52-128, 207-299, 327-396 and 409-488 of CH505 gp120, and heavy chain residues 1-223 and light chain residues 1-219 of Fab DH650.
Negative-stain electron microscopy of CH505 SOSIP:DH650 Fab complex.
CH505 DS-SOSIP, prepared as described (7), was mixed with a DH650 Fab in a 1:4 molar ratio. After incubation for 1 hour, the complex was loaded on a Superose 6 column (GE). The SOSIP-Fab complex was diluted to 0.1mg/ml and applied to freshly glow-discharged, carbon-coated EM grids and negatively stained with 1% uranyl formate. Images were recorded on an FEI Tecnai T12 electron microscope, operated at 120 kV and equipped with a ccd detector. Particles were selected and class averages computed with EMAN2 (132).
Neutralization fingerprint.
The neutralization fingerprint of a monoclonal antibody is defined as the potency pattern with which the antibody neutralizes a set of diverse viral strains. The neutralization fingerprints of V2 targeting broadly neutralizing antibodies, including VRC26, PGT145, PG9, and VRC38 classes, as well as a set of other HIV-1 broadly neutralizing antibodies were compared and clustered according to fingerprint similarity, as described previously (59), using a panel of 208 HIV-1 viral strains.
Statistical analyses.
Statistical tests were calculated by using GraphPad Prism 7 software. The Mann-Whitney test was used to determine whether the viral loads (peak and setpoint) of anti-CD8 treated animals were significantly different from untreated animals and whether viral loads (peak and setpoint) were greater in animals that developed bNAbs compared with those that did not. We chose a nonparametric rank-based test because both peak and setpoint viral loads of the untreated group and the bNAb/non-bNAb groups failed the D’Agostino & Pearson normality test (P-values < 0.05). The Spearman’s rank correlation test was used to determine if there is a significant correlation between setpoint viral loads and autologous tier 2 NAb titers. The geometric means were calculated using the Column statistics function of GraphPad Prism. The Chi-squared test was used on 2x2 contingency tables of shared and non-shared mutations within or between groups of humans and animals infected by viruses bearing the same (homologous) or different (heterologous) Envs to determine if Env residue mutations demonstrated significant strain-specificity. Separate tests were run to analyze total shared and non-shared mutations for the groups overall and for any discordant CH505, CH848 or CAP256SU group pairing.
Data and software availability.
GenBank accession numbers for all HIV-1 env sequences analyzed in this study are as follows: MN471655-MN472016, MT484339-MT487491, MT580365-MT580445, MT509359, GQ999989, HQ625604, KC863461-KC863464, KF241776, KF996577-KF996601, KF996604, KF996606, KF996610-KF996630, KF996632-KF996662, KF996664-KF996678, KF996680, KF996682-KF996683, KF996685-KF996716, KT698223-KT698227, MF572809-MF572829, EF203980-EF203981, MK205498-MK205507. HIV-1 Env sequences from the human subject CAP256 were previously published (3, 5, 133). GenBank accession numbers for immunoglobulin genes are: MT581213-MT581268, MT610888-MT610895 and MT656172-MT656253. Cryo-EM maps and fitted coordinates have been deposited with database codes EMDB-22295 and PDB ID 6XRT, respectively. DH650 bound to the gp120 core was deposited with database code PDB ID 6XCJ.
Supplementary Material
Fig. S1. Design and characterization of SHIV.CAP256SU. (A) SHIV.CAP256SU was constructed using a design scheme previously reported (9) and represents a chimera of HIV-1 CAP256SU (blue), HIV-1 191859 (red) and SIVmac766 (grey) sequences (GenBank accession #MT509359). Env375-Ser (S) is the wildtype allele found in HIV-1 CAP256SU and in most M group viruses. This residue was replaced with each of five aliphatic, basic or aromatic residues (M, Y, H, W, F). (B) SHIVs expressing each of the six Env375 alleles were tested for neutralization sensitivity to a panel of prototypic human mAbs that targeted canonical bNAb epitopes, a linear V3 epitope (447-52D) and a CD4-induced bridging sheet epitope (17b). All six Env375 alleles retained native antigenicity and tier 2 neutralization profiles. (C) Replication of SHIV.CAP256SU Env375 variants in primary, activated rhesus and human CD4 T cells is depicted. All six allelic variants replicated efficiently in human cells but only the W, F, Y and H variants replicated in rhesus cells. This result highlights the ability of a single amino acid substitution at position 375 in Env to confer dramatic differences in replication efficiency in primary rhesus CD4 T cells.
Fig. S2. Virus load and autologous neutralizing antibody kinetics in the plasma of rhesus macaques and humans. Human and rhesus infections by CH505 (left panels), CH848 (middle panels) and CAP256SU (right panels) viruses are depicted. Animals pretreated with anti-CD8 mAbs are shown in red, untreated animals in blue, and humans in open black boxes. Crosses indicate animals that developed AIDS-defining events requiring euthanasia. Dashed lines demarcate the limits of quantification of the plasma virus load assay (250 vRNA molecules/ml).
Fig. S3. Glycan shield evolution in the human subject CH505 and SHIV.CH505 infected RMs. (A) Longitudinal glycan frequencies at selected positions show similar patterns between human (red) and RMs (different shades of blue and green). N332 is gained in all hosts with similar kinetics. N234 is gained in human and most RMs. Glycan 130 is lost over time in the human and in all RMs except RM5695 where it was absent from the outset of infection. (B) Longitudinal glycan shield evolution in the CH505 trimer (side view) over time. Blue patches indicate the location of time point consensus PNGs and green their predicted glycan coverage. Magenta and pink regions indicate glycan holes at regions that are glycan shielded in >80% or 50-80% of M-group sequences, respectively. The light grey area in the central region of the trimer is the inter-protomer surface that forms a cleft with low glycan coverage (45).
Fig. S4. Conserved Env evolutionary patterns between the HIV-1 infected human subject CH848 and SHIV.CH848 infected rhesus macaques. A description of the PIXEL plots (left panels) and the LASSIE analysis (right panels) is provided in the legend to Fig. 2. Sixteen of 29 residues that mutated to >75% fixation in the human subject CH848 also mutated in one or more monkey and these are highlighted by green flags in the PIXEL plots and by green lettering in the LASSIE plot. Deletions in V1 associated with the development of neutralization breadth in the human subject CH848 and in RMs 6163 and 6167 are indicated by a yellow flag.
Fig. S5. Glycan shield evolution in the human subject CH848 and SHIV.CH848 infected RMs. (A) Common patterns of glycan shield evolution in the CH848 trimer between human and RM6167 and RM6163. Longitudinal evolution of the viral quasispecies in CH848 human resulted in two glycan changes by week 75. The loss of N413 glycan does not impact the glycan shield but could alter sensitivity to autologous NAbs. The loss of N289 creates a glycan hole. Glycan shield evolution in RM6167 at week 28 and in RM6163 at week 80 shows similar changes. (B) Longitudinal glycan frequencies over time in human and RMs for selected glycans that show similar patterns across human (red) and RMs (different shades of blue and green). N413 is lost in all hosts with similar kinetics. N289 is transiently or permanently lost in human and most RMs. (C) Longitudinal glycan shield evolution over time in each host. Glycan shield calculation and coloring is the same as in Fig. S3B.
Fig. S6. Selected sites identified by LASSIE in human subjects CH505 and CH848 are rarely mutated in RMs infected by SHIVs bearing heterologous Envs. The uppermost LOGO plots at the top of this illustration depict HIV-1 Env residues in the human subjects CH505 (panel A) and CH848 (panel B) that were identified by LASSIE analysis to be under positive selection (Figs. 2 and S4). The position of each of these residues (HXB2 numbering) is shown at the bottom of each panel. Only four residues were identified to be under positive selection in both human subjects, and these positions were 130, 300, 413 and 620 (indicated by the vertical red lines). The lower LOGO plots residues in the heterologous T/F Env (CH848 - panel A; CH505 - panel B), and these were tracked over time in the SHIV infected monkeys for comparison to the matched analyses presented in Figs. 2 (CH505) and S4 (CH848). Over each animal’s LOGO display, there are two numbers in green at the upper right. The first digit shows the total number of sites under positive selection in the Env mismatched human and monkey, including sites highlighted in red. The second digit shows the number of sites under positive selection in the Env mismatched human and monkey after excluding the shared sites. Excluding the shared sites, we compared the patterns of selection in the different groups of animals. We found four of six animals infected with CH848 had no mutations in selected sites from the human subject CH505, RM6700 had 1 shared site, and RM6167 had 2 (panel A). In a comparison with Fig. 2 from the main text, 3 of the six animals shared 9 sites, two shared 7 sites, and one shared 6 sites. A Wilcoxon test showed that the distribution of CH505 selected sites in SHIV.CH505 macaques (9, 9, 9, 7, 7, 6), versus six SHIV.CH848 macaques (2, 1, 0, 0, 0, 0), was highly significant (p = 0.002). We also compared the human CH848 selected sites shared among the six SHIV.CH505 macaques shown in part B (3, 2, 1, 0, 3, 7 shared sites) with the six SHIV.CH848 infected macaques (Fig. S4) (11, 10, 6, 9, 4), and again found the distributions to be significantly different (p = 0.006). Note that because the human CAP256 was infected with two divergent HIV-1 strains, this control analysis was not applicable for this SHIV.
Fig. S7. Conserved Env evolutionary patterns between the HIV-1 infected human subject CAP256 and SHIV.CAP256SU infected rhesus macaques. (A) PIXEL plot highlighting amino acid mutations between the SHIV.CAP256SU infecting strain and subsequent samples in each RM. Red pixels indicate a change in amino acid relative to the infecting strain, and black indicates an Indel, as in Figs. 2 and S3. Green tags indicate residue positions that are selected in multiple RMs. Black numbers indicate HXB2 numbering and white numbers approximate HXB2 numbering in hypervariable regions. Yellow tags indicate identical Indels. (B) LASSIE analysis of the same longitudinal Env RM sequences, summarizing mutations under positive selection in any of the four RMs with a threshold of 75% replacement of the infecting CAP256SU strain shown at the top. Green numbers represent mutations observed in more than one animal. (C) Recombination map of CAP256SU and CAP256PI. The primary infection (PI) variants are shown as solid blue lines at week 6 and the superinfection (SU) variants as red lines at week 15. In subsequent time points, amino acids where the two forms match are grey. Places where they differ are indicated by blue if they match the PI, red if they match SU, and black if the differ from both. Extensive recombination is seen as early as week 23. (D) The mutations under selection in the RMs shown in panel B are tracked in the human relative to the CAP256SU form that dominates at w015. Positions under selection in the RMs that differ in the SU and PI forms are evident as the w006 forms are different than SU reference amino acids along the top. The mutations that arose de novo in the macaques were also often favored in the human and were most often explained by recombination of the PI forms into the SU backbone (e.g., E87K and T525A), although sometimes the mutation selected in RMs arose independently in the human and was not evident in either the PI or SU forms (e.g., R46K and K340E). Mutations at residue 169 in the human and in RMs 40591 and 42056 conferred escape from V2 apex targeted bNAbs (Fig. 3B,C).
Fig. S8. Glycan shield evolution in the human subject CAP256 and SHIV.CAP256SU infected RMs. (A) CAP256SU has a glycan hole due to absence of N339. During longitudinal evolution in the human and RM40591, N396 is gained over time. (B) Longitudinal glycan frequencies over time in human and RMs for N396 that shows similar patterns across human (red) and RMs (different shades of blue and green). (C) Longitudinal glycan shield evolution over time in each host. Glycan shield calculation and coloring same as Fig. S3B.
Fig. S9. Summary of Indels in the CH505 human and SHIV.CH505 infected RMs. The HXB2 reference and CH505T/F sequences are indicated at the top for orientation. Hypervariable regions within the loops are shown in red. Each Indel is aligned independently to the T/F as multiple alignments obscure the precise nature of the Indel. Precise Indel boundaries are indicated and unique indels are assigned a color. Only Indels that were found in the human or repeated in multiple individuals are shown. With the exception of a single codon deletion (in red), all V1 Indels were insertions. The T/F is shown at the top and the Indel beneath it. The translated sequences are depicted on the right. Insertions are generally direct repeats, and the section that is repeated is highlighted (e.g. the 9 bases in green in V1) and then shown repeated in the inserted sequence and translated on the right. The V1 insertions almost always included the addition of at least one N-linked glycosylation site. The V5 deletions always changed the relative location of a glycan within the V5 loop. The distinct hosts that carried an identical Indel are indicated by the colored boxes on the right. The vast majority of V1 Indels were insertions in the seven CH505 Env infections, and the histogram on the right shows that insertions in V1 restore it to a more typical length. The distribution represents V1 loop lengths based on the global Env M group alignment from the Los Alamos HIV database (www.hiv.lanl.gov). CH505 T/F was unusually short in V1 (20 amino acids long, magenta column), while the median V1 length was 30 amino acids (red column). Similarly, the deletions found in the long CH505 T/F in V4 bring the viral quasispecies closer to the median length of 31, and the repeated insertions in V5 bring the short CH505 TF V5 closer to a typical length. Only four time points collected during the first year of infection are shown here for simplicity; after the first year of infection, Indels on top of Indels begin to accrue and the evolutionary history becomes unresolvable.
Fig. S10. Summary of Indels in the CH848 human and SHIV.CH848 infected RMs. The figure organization is the same as in Fig. S9. Unlike CH505, the V1 length in CH848 T/F was atypically long and all Indels were comprised of deletions bringing the V1 loop length closer to the global median. Ten Indels in V1, V4 or V5 were precisely replicated between the human subject CH848 and one or more macaques. Additional Indels were replicated among macaques only.
Fig. S11. Epitope mapping of autologous neutralizing antibody responses in the SHIV.CH505 infected RM6072. (A) Mutations at residues N130, T234, N279, V281, K302, Y330, N334, H417 and K460 were strongly selected in human CH505 and in RM6072 (see Fig. 2A). Here, the left panel depicts fold changes in NAb titers of week 10 and 20 plasma against these mutant versus wildtype CH505 T/F Env pseudotyped viruses. The right panel shows neutralization curves of RM6072 wk20 plasma against CH505 T/F and single and combination mutants that evolve in human and rhesus. MLV is a murine leukemia virus control for non-specific inhibition of virus entry. (B) Left, neutralization of wildtype CH505 T/F and observed in vivo mutants by autologous monoclonal antibodies (mAbs) DH647 and DH648 isolated from RM6072. Neutralization potency is expressed as IC50 values (μg/ml). Right, neutralization curves of mAbs against CH505 T/F and single and combination mutants. (C) Neutralization of wildtype CH505 T/F and observed in vivo mutants by mAb DH650 isolated from RM6072 and its inferred germline unmutated common ancestor. mAbs isolated from human subject CH505 (6) are included as controls. Neutralization titers are shown as IC50 values (μg/ml).
Fig. S12. Epitope mapping of heterologous neutralizing activity in plasma from RMs infected by SHIVs CH505 (RM5695 and RM6070) or CAP256SU (RM40591 and RM42056). (A) RM5695, B) RM6070, C) RM40591, D) RM42056. Neutralization was tested in the TZM-bl assay using plasma or isolated plasma IgG against viruses expressing wildtype Envs or Envs with mutations at residues 156, 166, 169 or 171. In a diverse set of heterologous Env backbones (Q23.17, T250, BG505, 246F3, MT145K, CE0217 and ZM233), bNAb activity was found to be dependent on basic residues (R or K) at positions 166 and 169, and to a lesser extent, on a PNG at position 156 and a basic residue at position 171. These results, together with in vivo escape mutations at residues 166, 169 and 171 (Fig. 3B), map the bNAb activity in these plasma specimens to the V2 apex.
Fig. S13. Variable effects of N160K substitution on heterologous neutralization in RMs infected by SHIVs CH505 (RM5695 and RM6070) or CAP256SU (RM40591 and RM42056). (A) RM5695, (B) RM6070, (C) RM40591, (D) RM42056. Neutralization was tested in the TZM-bl assay using plasma or isolated plasma IgG against viruses expressing wildtype Envs or Envs with N160K substitutions in a diverse set of heterologous Env backbones (Q23.17, T250, BG505, 246F3, MT145K, and ZM233). Plasma from every animal potently neutralized viruses expressing heterologous wildtype Envs. However, the effects of N160K substitution ranged from elimination of neutralization activity (e.g, RM5695 plasma against HIV-1 BG505) to enhancement of neutralization (RM5695 plasma against HIV-1 Q23.17) (panel A). In some animals, the enhancement of neutralization against heterologous N160K containing viruses was extraordinary [e.g., RM6070 (panel B) and RM42056 (panel D)]. N160K dependent neutralization was strictly dependent on antibody binding to basic C-strand residues as shown by N160K.K169E double mutants (panels B and D). The most likely explanation for these results is that SHIV-infected animals contain three specificities of V2 apex targeted C-strand dependent neutralizing antibodies. The first are the typical V2 apex bNAbs that depend on interactions with C-strand residues and N160 glycans. The second C-strand targeted bNAbs that do not require binding to N160 and instead are enhanced in their potency by the absence of N160 glycans. The third are “2909-like” antibodies (83, 84, 135) that are strain-specfic and entirely lacking in neutralization breadth, but whose potency against the homologous virus strain is enhanced by the absence of N160 glycans. We have evidence for the existence of all three types of V2 apex NAbs in SHIV-infected RMs (see Supplement Extended Discussion).
Fig. S14. Epitope mapping of autologous neutralizing antibody responses in the SHIV.CH848 infected RM6163 and RM6167. (A) At weeks 10 and 36 post SHIV infection of animal RM6167, neutralizing antibody titers (reciprocal ID50) against the T/F virus reached 1:220 and 1:435, respectively. A strongly selected early substitution at Env residue R336 occurred in RM6167 and in the other SHIV.CH848 infected RMs and the human subject CH848. The R336G substitution in RM6167 resulted in a 67% reduction in neutralization potency of week 10 plasma. Similarly, a naturally occurring 10 amino acid deletion in V1 resulted in a 41% reduction in neutralizing antibody titer in the week 10 plasma but enhanced neutralization titers in concurrent wk 36 plasma. This V1 deletion, together with week 36 mutations at positions 169, 240, 290, 336, 339, 413 and 65 (denoted V1Δ10aa+Mut), led to complete escape from neutralizing antibodies in week 10 and 36 plasma, indicating again that selected mutations in Env arise as a consequence of neutralizing antibody pressure. (B) A six member autologous strain-specific neutralizing Ab lineage isolated from RM6163 (136) potently neutralized CH848 T/F and was dependent on N332 for activity. The naturally occurring V1 deletion in CH848 T/F (panel A) reduced neutralization titers by five-fold, indicating modulation by V1 of binding by this Nab lineage.
Fig. S15. Characterization of CD4bs antibodies in RM6072. (A) Plasma antibody binding to wildtype and CD4-binding site mutant (Δ371I) variants of CH505 TF gp120 and resurfaced core 3, RSC3 (95). Bottom panels depict differential binding ratios. ELISA results are from a representative experiment. (B) Plasma antibody blocking of Env binding by CD4bs bnAbs CH235 and CH106 (from the CH103 lineage) and by soluble CD4 (sCD4) as determined by competitive ELISA in a representative experiment. (C) Phylogram of memory B cell-derived antibodies designated the “DH650 lineage” that were isolated from blood (black) and bone marrow aspirate (purple) cells by an antigen-specific B cell sort approach or from blood by memory B cell culture (blue). The unmutated common ancestor (UCA) was computational inferred using the Cloanalyst software of macaque immunoglobulin reference genes (21). (D) Immunogenetics of the DH650 heavy and light chain genes was inferred by the Cloanalyst software. (E) Binding of DH650 mAbs to CH505 T/F gp120 was measured by ELISA and titers reported as EC50 (μg/ml). (F) DH650 mAbs were tested by ELISA for their ability to block Env binding by CH235, CH106 and sCD4. Inhibition is reported as percent blocking in a representative experiment.
Fig. S16. Binding and neutralization profiles of DH650 lineage antibodies. (A) Binding of DH650 mAbs to CH505 T/F gp120 and CH505 T/F SOSIPS was assessed by biolayer interferometry (BLI). mAb-captured sensors were placed into solutions of Envs at 50 μg/ml to measure binding depicted as response units over time. (B) Neutralization by DH650 lineage mAbs in TZM-bl cells against wildtype CH505TF and mutant CH505TF depleted of four glycans surrounding the CD4bs (137). IA refers to computationally inferred intermediate antibodies. Murine leukemia virus (MuLV) was used as a negative virus control. (C) Neutralization by TZM-bl assay of DH650 lineage mAbs against heterologous HIV-1 strains previously used to track neutralization breadth in the plasma of the HIV-1 infected CH505 human subject (2). Neutralization results were confirmed and extended in a test of 118 heterologous virus strains (Table S3).
Fig. S17. Genetic and biological characterization of DH650 lineage. (A) The HCDR2 region of the VH gene of CH235 and 8ANC131 plays a critical role in engaging the CD4 binding loop of HIV-1 Env. Asterisks identify CH235VH Env contact residues. Dots indicate identical residues at each position and dashes indicate deletions. A residue 57 serine (S) to arginine (R) mutation in the CH235 UCA was found to be critical for binding of CH235 lineage bNAbs to HIV-1 Envs (6). We found a similar mutation at residue 57 asparagine (N) to R in the DH650 lineage NAbs. (B) Binding by ELISA of DH650 mAb to CH505 T/F Env monomer and SOSIP depends on R57 in CDRH2. Controls included CH235, 2G12 and influenza-reactive mAb CH65. (C) Complete VH sequences of DH650 lineage antibodies isolated from single cell sorts of blood derived B cells 20, 24 and 32 weeks post SHIV infection (DH650-DH650.13) and sequences generated by next generation sequencing from blood mononuclear cells 52 weeks post SHIV infection (DH650.15-34_NGS). (D) DH650.6, which contained VH residues S35 and P62, failed to bind CH505 T/F Env (Figs. S15E, 16A) or neutralize CH505 T/F or T/F-gly4 virus (Figs. S16B,C). Substitution of either S35 to N35 or P62 to Q62 restored functionality as measured by ELISA binding to CH505 T/F gp120 (Fig. S17D). However, for DH650.15_NGS and DH650.16_NGS heavy chains paired with the DH650 VL chain, N35 and Q62 were not sufficient to support Env binding, suggesting that authentic paired VL chains are essential for DH650.15-16 functionality. (E) VL chain sequences of DH650 lineage antibodies compared with CH235 UCA and mature bNAbs. DH650 lineage antibodies contained an exceptionally long LCDR1 of 17 amino acids compared with 11 amino acids in CH235 antibodies.
Fig. S18. Heavy and light chain sequences of RHA1 bNAbs from RM5695. (A) Nucleotide and amino acid alignments of rhesus mAb heavy chains RAH1.V2.01-04 compared with RM5695 germline gene IGHV4-ABB-5*01_S8200. (B) Nucleotide and amino acid alignments of rhesus mAb light chains RAH1.V2.01-04 compared with RM5695 germline gene IGLV1-ACN*02.
Fig. S19. Structural, immunogenetic and neutralization properties of RHA1 bNAbs. (A) The two residue insertion in the HCDR1 region of RHA1 bNAbs results in interactions with N160 (yellow shading) and electrostatic contact between D29 and Env residue K171. (B) IGHV4-ABB-5*01_S8200 (blue), DH3-9 (red) and JH2-P (teal) plus six non-templated amino acids (black) contribute to the RHA1 HCDR3. Somatic hypermutation resulting in amino acid substitutions in D and J genes of the mature bNAb mAbs are shown. The amino-terminal Cys and carboxy-terminal Trp (underlined) are not part of the HCDR3 (IMGT). Dots indicate amino acid identities. (C) RHA1 mAbs are aligned to PCT64-35S and PGT145. RHA1 and PCT64-35S are similar in length (24 vs 25 amino acids, respectively). Seven of 24 RHA1 HCDR3 amino acids (yellow shading) are identical to those in PCT64-35S, including a critical DDY motif that is tyrosine-sulfated (red asterisks) in both antibody lineages and in PGT145. Blue shading highlights nonidentical residues that share chemical properties such as aromaticity or charge. Tyrosine sulfation plays a key role in binding HIV-1 Env by all three antibodies. (D) The neutralization fingerprint for RHA1.V2.01 illustrated in Fig. 6E showed it to cluster within the PGT145 class of bNAbs, which clustered significantly with all prototypic V2 apex bNAbs analyzed. Here, we show the additional 21 non-V2 apex targeted bNAb antibodies that were part of the same neutralization fingerprint analysis. (E) Venn diagram of the number of neutralization sensitive HIV-1 strains for RHA1.V2.01, CAP256-VRC26.25 and PGT145. Of the 101 viruses neutralized by RHA1.V2.01, 96 were neutralized by PGT145 and 84 were neutralized by CAP256-VRC26.25, supporting the neutralization fingerprint dendogram shown in Fig. 6E.
Fig. S20. RHA1.V2.01 lacks autoreactivity. (A) Indirect immunofluorescence showing lack of anti-nuclear antibody (ANA) reactivity by mAb RHA1.V2.01 in HEp-2 cells. The positive control mAb is DH1037 and negative control mAb is DH570.30 (50 μg/mL for each). The secondary antibody was a goat anti-rhesus Ig(H+L) FITC. (B) RHA1.V2.01, along with positive (4E10) and negative (CH65) control mAbs, were tested for reactivity to nine autoantigens in the ZEUS AtheNA Multi-Lyte ANA-II Plus Test System, according to manufacturer’s recommendations. All antibodies were 2-fold serially diluted starting at 50 μg/mL.
Fig. S21. Heterologous neutralization by RHA1 mAbs. Rhesus mAbs were tested for neutralization against the autologous CH505 TF virus and the same global panel of heterologous viruses used to test all macaque plasma in this study (Figs. 1). Neutralization titers are reported as IC50 (μg/ml) values. This same panel of viruses was also tested for neutralization by the RM5695 plasma from wk56 post infection, expressed as reciprocal dilution ID50 (μg/ml) values. The breadth of the RHA1 mAbs recapitulated the breadth observed in the polyclonal plasma.
Fig. S22. RHA1.V2.01 neutralization of a global panel of 208 heterologous HIV-1 viruses. Neutralization titers are reported as IC50 (μg/ml) values. This data is reflected in the dendrogram in Fig. 6B. 49% of viruses were neutralized at a concentration <50 μg/ml.
Fig. S23. Heterologous neutralization of RHA1 lineage bNAbs maps to the V2 apex. In multiple heterologous Env backbones, neutralization maps to N160, R166 and C-strand residues R/K169.
Fig. S24. Neutralization profiles of RHA1.V2.01 and other V2 apex bNAbs. (A) IC50 titers for RHA1.V2.01 and prototypic human V2 apex bNAbs are shown as a heatmap. Rows indicate viruses from the 208 strain panel (Fig. S22) and columns the bNAbs tested. IC50 titers are color-coded according to potency (red high, yellow low, blue >50μg/ml). Viruses and bNAbs are ordered using a hierarchical clustering on the Heatmap webtool (https://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html) (60, 109-113) using the Ward algorithm on Euclidean distances of Log10 transformed IC50 titers. A hierarchical clustering tree shows relatedness between IC50 profiles of different V2 apex bNAbs. Bootstrap confidence (% of 1000 bootstraps) is indicated on the splits. (B) Contingency tables highlight the overlap between sensitive and resistant viruses between RHA1.V2.01 and other V2 apex bNAbs. Resistant viruses had IC50 titers above the threshold. Below each table are listed Fisher’s exact test p-values, odds ratios (OR) and accuracy estimates (Acc), which measure the fraction of concordant viruses (i.e. sensitive or resistant to both bNAbs). (C) Scatterplots of IC50 titers with RHA1.V2.01 titers on the y-axis and other V2 apex bNAb titers on the x-axis. Viruses resistant to one but not the other bNAb are shown above or to the right of the dotted lines. The number in the top right indicates the number of viruses resistant to both. Pearson R2 and p-values are listed in the bottom right of each panel and were calculated using viruses that were sensitive to both bNAbs. These analyses together show that RHA1.V2.01 is most similar to PCT64-35M.
Fig. S25. Neutralization profiles of RHA1.V2.01 and other V2 apex bNAbs across HIV-1 group M subtypes. Neutralization IC50 titers are shown for the 208 virus global panel, broken down according to subtypes. IC50 titers are color-coded from red to green indicating potent to less potent IC50 titers, with black cells indicating IC50 titers greater than maximum concentration tested (50μg/ml). V2 apex bNAbs are grouped according to similarities in their subtype-specific neutralization profiles into three sub-groups: a) PCT64-35M and CAP256 bNAbs; b) CH01; and c) PG9, PGT145 and PGDM1400. The rhesus bNAb RHA1.V2.01 is most similar to the PCT64-35M subgroup.
Fig. S26. Signature analyses for RHA1.V2.01 and other V2 apex bNAbs. (A) Signature analyses for RHA1.V2.01 were performed as described (60). Significant signatures had two or more of the following: i) phylogenetic correction at false-discovery rate (FDR, q) < 0.2; ii) contact site; and iii) at least one strong signature (q < 0.1). This analysis identified sites 130, 160, 166, 167 and 169 as signature sites for RHA1.V2.01. The variability at these sites in M-group is shown using sequence logo in the left panel. On the right, variability in RM5695 longitudinal sequences is shown. Height of the letters indicate the frequency, and O is a potential N-linked glycan site (NXT/S). Blue letters are associated with sensitivity, red resistance and grey neutral. (B) Comparison of RHA1.V2.01 signatures to those of other V2 apex bNAbs. The results at signature sites are shown for each bNAb on a single row, using amino acids colored according to sensitivity or resistance and with the height of letters indicating the frequency in M-group sequences (www.hiv.lanl.gov). Notably, only PCT64-35M shared all signature sites for RHA1.V2.01.
Figure S27. Cryo-EM Details of RHA1.V2.01 in complex with HIV-1 Env BG505 DS-SOSIP. (A) Representative micrograph and contrast transfer function of the micrograph are shown. (B) Representative 2D class averages are shown. (C) The orientations of all particles used in the final refinement are shown as a heatmap. (D) The gold-standard fourier shell correlation resulted in a resolution of 3.90 Å using non-uniform refinement with C1 symmetry. We note that local refinement (beta) of the same particles resulted in a resolution of 3.8 Å. (E) The local resolution of the full map is shown generated through cryoSPARC using a Fourier shell correlation (FSC) cutoff of 0.5. Two contour levels are shown. (F) Representative density is shown for the Fab (top left) with a magnified region of the inserted sulfated tyrosine, Tys (top right), and for N160 of each protomer labeled P1, P2 and P3 (bottom panels).
ACKNOWLEDGMENTS
We thank A. Abuahmad, H. Chen, A. Eaton, A. Foulger, M. Gladden, T. Gurley, G. Hernandez, A. Huang, C. Jones, J. Kim, J. Meyer, A. Monroe, R. Parks, A. Ransier, J. Rathmann, P. Rawls, A. Sanzone, J. Sprenz, K. Tilahun, R. Verardi, T. Von Holle, A. Wang and R. Zhang for technical assistance and the flow cytometry core staff at the Duke Human Vaccine Institute and NIAID/NIH Vaccine Research Center. We thank T. Denny, T. Demarco and N. DeNaeyer and members of the Nonhuman Primate Virology Core Laboratory at the Duke Human Vaccine Institute for SHIV plasma vRNA measurements, and J. Baalwa, D. Ellenberger, F. Gao, K. Hong, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson and L. Zhang for contributing HIV-1 Env plasmids. We thank K. McKee, C. Moore, S. O’Dell, G. Padilla, S.D. Schmidt, C. Whittaker, A.B. McDermott and M. Seaman for assistance with neutralization assays, D. Fera for helpful advice and discussion, and the staff at Bioqual for exceptional care and assistance with nonhuman primates. Cryo-EM was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center and was supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences GM103310. X-ray diffraction data were collected on the Northeastern Collaborative Access Team beamline 24 ID-C (AdvancedPhoton Source) funded by NIH Grant P41 GM103403. This work was funded by the Bill & Melinda Gates Foundation (OPP1145046, OPP1206647/INV-007939); by the Intramural Research Program of the NIAID/NIH Vaccine Research Center; and by the Division of AIDS (NIAID/NIH) through support of the Duke Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (UM1 AI100645), the Duke Consortium for HIV/AIDS Vaccine Development (UM1 AI144371), the Penn Center for AIDS Research (P30 AI045008), and grants AI131251, AI131331, AI128832, AI050529, AI150590 and AI140897. R. Roark was supported by an NIH training grant in HIV Pathogenesis (T32-AI007632).
REFERENCES
- 1.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM, Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105, 7552–7557 (2008). 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, N. C. S. Program, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF, Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013). 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, N. C. S. Program, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR, Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62 (2014). 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, Lynch RM, Alam SM, Moody MA, Ferrari G, Berrong M, Kelsoe G, Shaw GM, Hahn BH, Montefiori DC, Kamanga G, Cohen MS, Hraber P, Kwong PD, Korber BT, Mascola JR, Kepler TB, Haynes BF, Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 158, 481–491 (2014). 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza T, Kitchin D, Botha G, Gorman J, Garrett NJ, Abdool Karim SS, Shapiro L, Williamson C, Kwong PD, Mascola JR, Morris L, Moore PL, Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med 21, 1332–1336 (2015). 10.1038/nm.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, Alam SM, Bradley T, Gladden MA, Hwang KK, Iyengar S, Kumar A, Lu X, Luo K, Mangiapani MC, Parks RJ, Song H, Acharya P, Bailer RT, Cao A, Druz A, Georgiev IS, Kwon YD, Louder MK, Zhang B, Zheng A, Hill BJ, Kong R, Soto C, N. C. S. Program, Mullikin JC, Douek DC, Montefiori DC, Moody MA, Shaw GM, Hahn BH, Kelsoe G, Hraber PT, Korber BT, Boyd SD, Fire AZ, Kepler TB, Shapiro L, Ward AB, Mascola JR, Liao HX, Kwong PD, Haynes BF, Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165, 449–463 (2016). 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonsignori M, Kreider EF, Fera D, Meyerhoff RR, Bradley T, Wiehe K, Alam SM, Aussedat B, Walkowicz WE, Hwang KK, Saunders KO, Zhang R, Gladden MA, Monroe A, Kumar A, Xia SM, Cooper M, Louder MK, McKee K, Bailer RT, Pier BW, Jette CA, Kelsoe G, Williams WB, Morris L, Kappes J, Wagh K, Kamanga G, Cohen MS, Hraber PT, Montefiori DC, Trama A, Liao HX, Kepler TB, Moody MA, Gao F, Danishefsky SJ, Mascola JR, Shaw GM, Hahn BH, Harrison SC, Korber BT, Haynes BF, Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci Transl Med 9, (2017). 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landais E, Murrell B, Briney B, Murrell S, Rantalainen K, Berndsen ZT, Ramos A, Wickramasinghe L, Smith ML, Eren K, de Val N, Wu M, Cappelletti A, Umotoy J, Lie Y, Wrin T, Algate P, Chan-Hui PY, Karita E, I. P. C. Investigators, I. A. H. R. Network, Ward AB, Wilson IA, Burton DR, Smith D, Pond SLK, Poignard P, HIV envelope glycoform heterogeneity and localized diversity govern the initiation and maturation of a V2 apex broadly neutralizing antibody lineage. Immunity 47, 990–1003 e1009 (2017). 10.1016/j.immuni.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, Brocca-Cofano E, Deleage C, Hao X, Chuang GY, Gorman J, Gardner M, Lewis MG, Hatziioannou T, Santra S, Apetrei C, Pandrea I, Alam SM, Liao HX, Shen X, Tomaras GD, Farzan M, Chertova E, Keele BF, Estes JD, Lifson JD, Doms RW, Montefiori DC, Haynes BF, Sodroski JG, Kwong PD, Hahn BH, Shaw GM, Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113, E3413–3422 (2016). 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong PD, Mascola JR, HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871 (2018). 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 11.Sok D, Burton DR, Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol 19, 1179–1188 (2018). 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM, Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308, 1906–1908 (2005). 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 13.Chuang GY, Zhou J, Acharya P, Rawi R, Shen CH, Sheng Z, Zhang B, Zhou T, Bailer RT, Dandey VP, Doria-Rose NA, Louder MK, McKee K, Mascola JR, Shapiro L, Kwong PD, Structural survey of broadly neutralizing antibodies targeting the HIV-1 env trimer delineates epitope categories and characteristics of recognition. Structure 27, 196–206 e196 (2019). 10.1016/j.str.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes EC, Error thresholds and the constraints to RNA virus evolution. Trends Microbiol 11, 543–546 (2003). 10.1016/j.tim.2003.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo J, Robertson DL, Lovell SC, Constraints on HIV-1 diversity from protein structure. J Virol 84, 12995–13003 (2010). 10.1128/JVI.00702-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haddox HK, Dingens AS, Bloom JD, Experimental estimation of the effects of all amino-acid mutations to HIV's envelope protein on viral replication in cell culture. PLoS Pathog 12, e1006114 (2016). 10.1371/journal.ppat.1006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG, The antigenic structure of the HIV GP120 envelope glycoprotein. Nature 393, 705–711 (1998). 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 18.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM, Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003). 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 19.Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Josephs SF, Gallo RC, Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45, 637–648 (1986). 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 20.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Calken I, Fung M, Katinger H, Parren PWLH, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J, HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 402, 678–682 (2002). 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 21.Ramesh A, Darko S, Hua A, Overman G, Ransier A, Francica JR, Trama A, Tomaras GD, Haynes BF, Douek DC, Kepler TB, Structure and diversity of the rhesus macaque immunoglobulin loci through multiple de novo genome assemblies. Front Immunol 8, 1407 (2017). 10.3389/fimmu.2017.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore PL, Sheward D, Nonyane M, Ranchobe N, Hermanus T, Gray ES, Abdool Karim SS, Williamson C, Morris L, Multiple pathways of escape from HIV broadly cross-neutralizing V2-dependent antibodies. J Virol 87, 4882–4894 (2013). 10.1128/JVI.03424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams WB, Zhang J, Jiang C, Nicely NI, Fera D, Luo K, Moody MA, Liao HX, Alam SM, Kepler TB, Ramesh A, Wiehe K, Holland JA, Bradley T, Vandergrift N, Saunders KO, Parks R, Foulger A, Xia SM, Bonsignori M, Montefiori DC, Louder M, Eaton A, Santra S, Scearce R, Sutherland L, Newman A, Bouton-Verville H, Bowman C, Bomze H, Gao F, Marshall DJ, Whitesides JF, Nie X, Kelsoe G, Reed SG, Fox CB, Clary K, Koutsoukos M, Franco D, Mascola JR, Harrison SC, Haynes BF, Verkoczy L, Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Nat Commun 8, 1732 (2017). 10.1038/s41467-017-01336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ndhlovu ZM, Kamya P, Mewalal N, Kloverpris HN, Nkosi T, Pretorius K, Laher F, Ogunshola F, Chopera D, Shekhar K, Ghebremichael M, Ismail N, Moodley A, Malik A, Leslie A, Goulder PJ, Buus S, Chakraborty A, Dong K, Ndung'u T, Walker BD, Magnitude and kinetics of CD8+ T Cell activation during hyperacute HIV infection impact viral set point. Immunity 43, 591–604 (2015). 10.1016/j.immuni.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robb ML, Eller LA, Kibuuka H, Rono K, Maganga L, Nitayaphan S, Kroon E, Sawe FK, Sinei S, Sriplienchan S, Jagodzinski LL, Malia J, Manak M, de Souza MS, Tovanabutra S, Sanders-Buell E, Rolland M, Dorsey-Spitz J, Eller MA, Milazzo M, Li Q, Lewandowski A, Wu H, Swann E, O'Connell RJ, Peel S, Dawson P, Kim JH, Michael NL, Team RVS, Prospective study of acute HIV-1 infection in adults in east Africa and Thailand. N Engl J Med 374, 2120–2130 (2016). 10.1056/NEJMoa1508952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR, Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84, 1439–1452 (2010). 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, Greene K, Gao H, Daniell X, Sarzotti-Kelsoe M, Gorny MK, Zolla-Pazner S, LaBranche CC, Mascola JR, Korber BT, Montefiori DC, Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 88, 2489–2507 (2014). 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT, Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169 (2014). 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.HIV-1 strains 25710 and Q23.17 have been variably reported as tier 1b or tier 2 in neutralization sensitivity to different panels of heterologous HIV-1 positive plasma. In all studies, these two virus strains have been shown to be much closer to tier 2 viruses than to lab-adapted tier 1A viruses in their neutralization sensitivity patterns. See refs. 29-31.
- 30.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM, Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201, 1407–1419 (2005). 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis KL, Bibollet-Ruche F, Li H, Decker JM, Kutsch O, Morris L, Salomon A, Pinter A, Hoxie JA, Hahn BH, Kwong PD, Shaw GM, Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J Virol 83, 1240–1259 (2009). 10.1128/JVI.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B, HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science 349, 191–195 (2015). 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, Ramos A, Bian CB, Simek M, Allen S, Karita E, Kilembe W, Lakhi S, Inambao M, Kamali A, Sanders EJ, Anzala O, Edward V, Bekker LG, Tang J, Gilmour J, Kosakovsky-Pond SL, Phung P, Wrin T, Crotty S, Godzik A, Poignard P, Broadly neutralizing antibody responses in a large longitudinal sub-Saharan HIV primary infection cohort. PLoS Pathog 12, e1005369 (2016). 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landais E, Moore PL, Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers. Retrovirology 15, 61 (2018). 10.1186/s12977-018-0443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbian HJ, Decker JM, Bibollet-Ruche F, Galimidi RP, West AP Jr., Learn GH, Parrish NF, Iyer SS, Li Y, Pace CS, Song R, Huang Y, Denny TN, Mouquet H, Martin L, Acharya P, Zhang B, Kwong PD, Mascola JR, Verrips CT, Strokappe NM, Rutten L, McCoy LE, Weiss RA, Brown CS, Jackson R, Silvestri G, Connors M, Burton DR, Shaw GM, Nussenzweig MC, Bjorkman PJ, Ho DD, Farzan M, Hahn BH, Neutralization properties of simian immunodeficiency viruses infecting chimpanzees and gorillas. mBio 6, (2015). 10.1128/mBio.00296-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez-Merino V, Fabra-Garcia A, Gonzalez N, Nicolas D, Merino-Mansilla A, Manzardo C, Ambrosioni J, Schultz A, Meyerhans A, Mascola JR, Gatell JM, Alcami J, Miro JM, Yuste E, Detection of broadly neutralizing activity within the first months of HIV-1 infection. J Virol 90, 5231–5245 (2016). 10.1128/JVI.00049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, B CCC, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ, The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 206, 1253–1272 (2009). 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM, Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206, 1273–1289 (2009). 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, Hraber P, Fischer W, Li H, Wang S, Sterrett S, Keele BF, Ganusov VV, Perelson AS, Korber BT, Georgiev I, McLellan JS, Pavlicek JW, Gao F, Haynes BF, Hahn BH, Kwong PD, Shaw GM, Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog 8, e1002721 (2012). 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markowitz M, Louie M, Hurley A, Sun E, Di Mascio M, Perelson AS, Ho DD, A novel antiviral intervention results in more accurate assessment of human immunodeficiency virus type 1 replication dynamics and T-cell decay in vivo. J Virol 77, 5037–5038 (2003). 10.1128/jvi.77.8.5037-5038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M, Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126 (1995). 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 42.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. , Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373, 117–122 (1995). 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 43.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM, Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med 3, 205–211 (1997). 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 44.Hraber P, Korber B, Wagh K, Giorgi EE, Bhattacharya T, Gnanakaran S, Lapedes AS, Learn GH, Kreider EF, Li Y, Shaw GM, Hahn BH, Montefiori DC, Alam SM, Bonsignori M, Moody MA, Liao HX, Gao F, Haynes BF, Longitudinal antigenic sequences and sites from intra-host evolution (LASSIE) identifies immune-selected HIV variants. Viruses 7, 5443–5475 (2015). 10.3390/v7102881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagh K, Kreider EF, Li Y, Barbian HJ, Learn GH, Giorgi E, Hraber PT, Decker TG, Smith AG, Gondim MV, Gillis L, Wandzilak J, Chuang GY, Rawi R, Cai F, Pellegrino P, Williams I, Overbaugh J, Gao F, Kwong PD, Haynes BF, Shaw GM, Borrow P, Seaman MS, Hahn BH, Korber B, Completeness of HIV-1 envelope glycan shield at transmission determines neutralization breadth. Cell Rep 25, 893–908 e897 (2018). 10.1016/j.celrep.2018.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coffin JM, Structure, replication, and recombination of retrovirus genomes- some unifying hypotheses. J Gen Virol 42, (1979). 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 47.Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, Daniels M, Ferrari G, Haynes BF, McMichael A, Shaw GM, Hahn BH, Korber B, Seoighe C, HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog 5, e1000414 (2009). 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mabvakure BM, Scheepers C, Garrett N, Abdool Karim S, Williamson C, Morris L, Moore PL, Positive selection at key residues in the HIV envelope distinguishes broad and strain-specific plasma neutralizing antibodies. J Virol 93, (2019). 10.1128/JVI.01685-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, Pancera M, Cale EM, Ernandes MJ, Louder MK, Asokan M, Bailer RT, Druz A, Fraschilla IR, Garrett NJ, Jarosinski M, Lynch RM, McKee K, O'Dell S, Pegu A, Schmidt SD, Staupe RP, Sutton MS, Wang K, Wibmer CK, Haynes BF, Abdool-Karim S, Shapiro L, Kwong PD, Moore PL, Morris L, Mascola JR, New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol 90, 76–91 (2016). 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saunders KO, Wiehe K, Tian M, Acharya P, Bradley T, Alam SM, Go EP, Scearce R, Sutherland L, Henderson R, Hsu AL, Borgnia MJ, Chen H, Lu X, Wu NR, Watts B, Jiang C, Easterhoff D, Cheng HL, McGovern K, Waddicor P, Chapdelaine-Williams A, Eaton A, Zhang J, Rountree W, Verkoczy L, Tomai M, Lewis MG, Desaire HR, Edwards RJ, Cain DW, Bonsignori M, Montefiori D, Alt FW, Haynes BF, Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science 366, (2019). 10.1126/science.aay7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrabi R, Voss JE, Liang CH, Briney B, McCoy LE, Wu CY, Wong CH, Poignard P, Burton DR, Identification of common features in prototype broadly neutralizing antibodies to HIV envelope V2 apex to facilitate vaccine design. Immunity 43, 959–973 (2015). 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorman J, Soto C, Yang MM, Davenport TM, Guttman M, Bailer RT, Chambers M, Chuang GY, DeKosky BJ, Doria-Rose NA, Druz A, Ernandes MJ, Georgiev IS, Jarosinski MC, Joyce MG, Lemmin TM, Leung S, Louder MK, McDaniel JR, Narpala S, Pancera M, Stuckey J, Wu X, Yang Y, Zhang B, Zhou T, N. C. S. Program, Mullikin JC, Baxa U, Georgiou G, McDermott AB, Bonsignori M, Haynes BF, Moore PL, Morris L, Lee KK, Shapiro L, Mascola JR, Kwong PD, Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol 23, 81–90 (2016). 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorman J, Chuang GY, Lai YT, Shen CH, Boyington JC, Druz A, Geng H, Louder MK, McKee K, Rawi R, Verardi R, Yang Y, Zhang B, Doria-Rose NA, Lin B, Moore PL, Morris L, Shapiro L, Mascola JR, Kwong PD, Structure of super-potent antibody CAP256-VRC26.25 in complex with HIV-1 envelope reveals a combined mode of trimer-apex recognition. Cell Rep 31, 107488 (2020). 10.1016/j.celrep.2020.03.052. [DOI] [PubMed] [Google Scholar]
- 54.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC, Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011). 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiehe K, Bradley T, Meyerhoff RR, Hart C, Williams WB, Easterhoff D, Faison WJ, Kepler TB, Saunders KO, Alam SM, Bonsignori M, Haynes BF, Functional relevance of improbable antibody mutations for HIV broadly neutralizing antibody development. Cell Host Microbe 23, 759–765 e756 (2018). 10.1016/j.chom.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, Kwon YD, Longo NS, Louder MK, Luongo T, McKee K, Schramm CA, Skinner J, Yang Y, Yang Z, Zhang Z, Zheng A, Bonsignori M, Haynes BF, Scheid JF, Nussenzweig MC, Simek M, Burton DR, Koff WC, N. C. S. Program, Mullikin JC, Connors M, Shapiro L, Nabel GJ, Mascola JR, Kwong PD, Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258 (2013). 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corcoran MM, Phad GE, Vazquez Bernat N, Stahl-Hennig C, Sumida N, Persson MA, Martin M, Karlsson Hedestam GB, Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat Commun 7, 13642 (2016). 10.1038/ncomms13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rantalainen K, Berndsen ZT, Murrell S, Cao L, Omorodion O, Torres JL, Wu M, Umotoy J, Copps J, Poignard P, Landais E, Paulson JC, Wilson IA, Ward AB, Co-evolution of HIV envelope and apex-targeting neutralizing antibody lineage provides benchmarks for vaccine design. Cell Rep 23, 3249–3261 (2018). 10.1016/j.celrep.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O'Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD, Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756 (2013). 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- 60.Bricault CA, Yusim K, Seaman MS, Yoon H, Theiler J, Giorgi EE, Wagh K, Theiler M, Hraber P, Macke JP, Kreider EF, Learn GH, Hahn BH, Scheid JF, Kovacs JM, Shields JL, Lavine CL, Ghantous F, Rist M, Bayne MG, Neubauer GH, McMahan K, Peng H, Cheneau C, Jones JJ, Zeng J, Ochsenbauer C, Nkolola JP, Stephenson KE, Chen B, Gnanakaran S, Bonsignori M, Williams LD, Haynes BF, Doria-Rose N, Mascola JR, Montefiori DC, Barouch DH, Korber B, HIV-1 neutralizing antibody signatures and application to epitope-targeted vaccine design. Cell Host Microbe 25, 59–72 e58 (2019). 10.1016/j.chom.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker LM, Sok D, Nishimura Y, Donau O, Sadjadpour R, Gautam R, Shingai M, Pejchal R, Ramos A, Simek MD, Geng Y, Wilson IA, Poignard P, Martin MA, Burton DR, Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc Natl Acad Sci U S A 108, 20125–20129 (2011). 10.1073/pnas.1117531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jia M, Lu H, Markowitz M, Cheng-Mayer C, Wu X, Development of broadly neutralizing antibodies and their mapping by monomeric gp120 in human immunodeficiency virus Type 1-infected humans and simian-human immunodeficiency virus SHIVSF162P3N-infected macaques. J Virol 90, 4017–4031 (2016). 10.1128/JVI.02898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao N, Wang W, Wang C, Gu T, Guo R, Yu B, Kong W, Qin C, Giorgi EE, Chen Z, Townsley S, Hu SL, Yu X, Gao F, Development of broad neutralization activity in simian/human immunodeficiency virus-infected rhesus macaques after long-term infection. AIDS 32, 555–563 (2018). 10.1097/QAD.0000000000001724. [DOI] [PubMed] [Google Scholar]
- 64.Gao N, Gai Y, Meng L, Wang C, Zhang X, Wang W, Qin C, Yu X, Gao F, Development of antibodies with broad neutralization specificities against HIV-1 after long term SHIV infection in macaques. Viruses 12, (2020). 10.3390/v12020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao HX, Etemad-Moghadam B, Montefiori DC, Sun Y, Sodroski J, Scearce RM, Doms RW, Thomasch JR, Robinson S, Letvin NL, Haynes BF, Induction of antibodies in guinea pigs and rhesus monkeys against the human immunodeficiency virus type 1 envelope: neutralization of nonpathogenic and pathogenic primary isolate simian/human immunodeficiency virus strains. J Virol 74, 254–263 (2000). 10.1128/jvi.74.1.254-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haynes BF, Ma B, Montefiori DC, Wrin T, Petropoulos CJ, Sutherland LL, Scearce RM, Denton C, Xia SM, Korber BT, Liao HX, Analysis of HIV-1 subtype B third variable region peptide motifs for induction of neutralizing antibodies against HIV-1 primary isolates. Virology 345, 44–55 (2006). 10.1016/j.virol.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 67.Wang S, Pal R, Mascola JR, Chou TH, Mboudjeka I, Shen S, Liu Q, Whitney S, Keen T, Nair BC, Kalyanaraman VS, Markham P, Lu S, Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology 350, 34–47 (2006). 10.1016/j.virol.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 68.Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, Eroshkin AM, Guenaga J, Kaushik K, Kulp DW, Liu J, McCoy LE, Oom AL, Ozorowski G, Post KW, Sharma SK, Steichen JM, de Taeye SW, Tokatlian T, Torrents de la Pena A, Butera ST, LaBranche CC, Montefiori DC, Silvestri G, Wilson IA, Irvine DJ, Sanders RW, Schief WR, Ward AB, Wyatt RT, Barouch DH, Crotty S, Burton DR, Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46, 1073–1088 e1076 (2017). 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saunders KO, Verkoczy LK, Jiang C, Zhang J, Parks R, Chen H, Housman M, Bouton-Verville H, Shen X, Trama AM, Scearce R, Sutherland L, Santra S, Newman A, Eaton A, Xu K, Georgiev IS, Joyce MG, Tomaras GD, Bonsignori M, Reed SG, Salazar A, Mascola JR, Moody MA, Cain DW, Centlivre M, Zurawski S, Zurawski G, Erickson HP, Kwong PD, Alam SM, Levy Y, Montefiori DC, Haynes BF, Vaccine induction of heterologous tier 2 HIV-1 neutralizing antibodies in animal models. Cell Rep 21, 3681–3690 (2017). 10.1016/j.celrep.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torrents de la Pena A, de Taeye SW, Sliepen K, LaBranche CC, Burger JA, Schermer EE, Montefiori DC, Moore JP, Klasse PJ, Sanders RW, Immunogenicity in rabbits of HIV-1 SOSIP trimers from clades A, B, and C, given individually, sequentially, or in combination. J Virol 92, (2018). 10.1128/JVI.01957-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kong R, Duan H, Sheng Z, Xu K, Acharya P, Chen X, Cheng C, Dingens AS, Gorman J, Sastry M, Shen CH, Zhang B, Zhou T, Chuang GY, Chao CW, Gu Y, Jafari AJ, Louder MK, O'Dell S, Rowshan AP, Viox EG, Wang Y, Choi CW, Corcoran MM, Corrigan AR, Dandey VP, Eng ET, Geng H, Foulds KE, Guo Y, Kwon YD, Lin B, Liu K, Mason RD, Nason MC, Ohr TY, Ou L, Rawi R, Sarfo EK, Schon A, Todd JP, Wang S, Wei H, Wu W, N. C. S. Program, Mullikin JC, Bailer RT, Doria-Rose NA, Karlsson Hedestam GB, Scorpio DG, Overbaugh J, Bloom JD, Carragher B, Potter CS, Shapiro L, Kwong PD, Mascola JR, Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell 178, 567–584 e519 (2019). 10.1016/j.cell.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klasse PJ, Ozorowski G, Sanders RW, Moore JP, Env exceptionalism: Why are HIV-1 env glycoproteins atypical immunogens? Cell Host Microbe 27, 507–518 (2020). 10.1016/j.chom.2020.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Barnes CO, Gautam R, Lorenzi JCC, Mayer CT, Oliveira TY, Ramos V, Cipolla M, Gordon KM, Gristick HB, West AP, Nishimura Y, Raina H, Seaman MS, Gazumyan A, Martin MA, Bjorkman PJ, Escolano A, Nussenzweig MC, A broadly neutralizing macaque monoclonal antibody against the HIV-1 V3-glycan patch. bioRxiv (2020). 10.1101/2020.08.10.244582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haynes BF, Burton DR, Mascola JR, Multiple roles for HIV broadly neutralizing antibodies. Sci Transl Med 11, (2019). 10.1126/scitranslmed.aaz2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stephenson KE, Wagh K, Korber B, Barouch DH, Vaccines and broadly neutralizing antibodies for HIV-1 prevention. Annual Revew of Immunology 39, 673–703 (2020). 10.1146/annurev-immunol-080219-023629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Escolano A, Gristick HB, Abernathy ME, Merkenschlager J, Gautam R, Oliveira TY, Pai J, West AP Jr., Barnes CO, Cohen AA, Wang H, Golijanin J, Yost D, Keeffe JR, Wang Z, Zhao P, Yao KH, Bauer J, Nogueira L, Gao H, Voll AV, Montefiori DC, Seaman MS, Gazumyan A, Silva M, McGuire AT, Stamatatos L, Irvine DJ, Wells L, Martin MA, Bjorkman PJ, Nussenzweig MC, Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 570, 468–473 (2019). 10.1038/s41586-019-1250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steichen JM, Lin YC, Havenar-Daughton C, Pecetta S, Ozorowski G, Willis JR, Toy L, Sok D, Liguori A, Kratochvil S, Torres JL, Kalyuzhniy O, Melzi E, Kulp DW, Raemisch S, Hu X, Bernard SM, Georgeson E, Phelps N, Adachi Y, Kubitz M, Landais E, Umotoy J, Robinson A, Briney B, Wilson IA, Burton DR, Ward AB, Crotty S, Batista FD, Schief WR, A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 366, (2019). 10.1126/science.aax4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA, A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011). 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sok D, Pauthner M, Briney B, Lee JH, Saye-Francisco KL, Hsueh J, Ramos A, Le KM, Jones M, Jardine JG, Bastidas R, Sarkar A, Liang CH, Shivatare SS, Wu CY, Schief WR, Wong CH, Wilson IA, Ward AB, Zhu J, Poignard P, Burton DR, A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity 45, 31–45 (2016). 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA, Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9, e1003342 (2013). 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haynes BF, Kelsoe G, Harrison SC, Kepler TB, B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 30, 423–433 (2012). 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moore PL, The neutralizing antibody response to the HIV-1 env protein. Curr HIV Res 16, 21–28 (2018). 10.2174/1570162X15666171124122044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S, Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol 79, 5232–5237 (2005). 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Honnen WJ, Krachmarov C, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A, Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J Virol 81, 1424–1432 (2007). 10.1128/JVI.02054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC, Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods 409, 131–146 (2014). 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han Q, Jones JA, Nicely NI, Reed RK, Shen X, Mansouri K, Louder M, Trama AM, Alam SM, Edwards RJ, Bonsignori M, Tomaras GD, Korber B, Montefiori DC, Mascola JR, Seaman MS, Haynes BF, Saunders KO, Difficult-to-neutralize global HIV-1 isolates are neutralized by antibodies targeting open envelope conformations. Nat Commun 10, 2898 (2019). 10.1038/s41467-019-10899-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Williams LD, Ofek G, Schatzle S, McDaniel JR, Lu X, Nicely NI, Wu L, Lougheed CS, Bradley T, Louder MK, McKee K, Bailer RT, O'Dell S, Georgiev IS, Seaman MS, Parks RJ, Marshall DJ, Anasti K, Yang G, Nie X, Tumba NL, Wiehe K, Wagh K, Korber B, Kepler TB, Munir Alam S, Morris L, Kamanga G, Cohen MS, Bonsignori M, Xia SM, Montefiori DC, Kelsoe G, Gao F, Mascola JR, Moody MA, Saunders KO, Liao HX, Tomaras GD, Georgiou G, Haynes BF, Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci Immunol 2, (2017). 10.1126/sciimmunol.aal2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alam SM, Liao HX, Tomaras GD, Bonsignori M, Tsao CY, Hwang KK, Chen H, Lloyd KE, Bowman C, Sutherland L, Jeffries TL Jr., Kozink DM, Stewart S, Anasti K, Jaeger FH, Parks R, Yates NL, Overman RG, Sinangil F, Berman PW, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Karasavva N, Rerks-Ngarm S, Kim JH, Michael NL, Zolla-Pazner S, Santra S, Letvin NL, Harrison SC, Haynes BF, Antigenicity and immunogenicity of RV144 vaccine AIDSVAX clade E envelope immunogen is enhanced by a gp120 N-terminal deletion. J Virol 87, 1554–1568 (2013). 10.1128/JVI.00718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alam SM, Aussedat B, Vohra Y, Meyerhoff RR, Cale EM, Walkowicz WE, Radakovich NA, Anasti K, Armand L, Parks R, Sutherland L, Scearce R, Joyce MG, Pancera M, Druz A, Georgiev IS, Von Holle T, Eaton A, Fox C, Reed SG, Louder M, Bailer RT, Morris L, Abdool-Karim SS, Cohen M, Liao HX, Montefiori DC, Park PK, Fernandez-Tejada A, Wiehe K, Santra S, Kepler TB, Saunders KO, Sodroski J, Kwong PD, Mascola JR, Bonsignori M, Moody MA, Danishefsky S, Haynes BF, Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide. Sci Transl Med 9, (2017). 10.1126/scitranslmed.aai7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, Yang Y, Chen X, Gao F, Munshaw S, Kepler TB, Denny T, Moody MA, Haynes BF, High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods 158, 171–179 (2009). 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O'Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao HX, Haynes BF, Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 85, 9998–10009 (2011). 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blasi M, Negri D, LaBranche C, Alam SM, Baker EJ, Brunner EC, Gladden MA, Michelini Z, Vandergrift NA, Wiehe KJ, Parks R, Shen X, Bonsignori M, Tomaras GD, Ferrari G, Montefiori DC, Santra S, Haynes BF, Moody MA, Cara A, Klotman ME, IDLV-HIV-1 Env vaccination in non-human primates induces affinity maturation of antigen-specific memory B cells. Commun Biol 1, 134 (2018). 10.1038/s42003-018-0131-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moody MA, Yates NL, Amos JD, Drinker MS, Eudailey JA, Gurley TC, Marshall DJ, Whitesides JF, Chen X, Foulger A, Yu JS, Zhang R, Meyerhoff RR, Parks R, Scull JC, Wang L, Vandergrift NA, Pickeral J, Pollara J, Kelsoe G, Alam SM, Ferrari G, Montefiori DC, Voss G, Liao HX, Tomaras GD, Haynes BF, HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. J Virol 86, 7496–7507 (2012). 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiehe K, Easterhoff D, Luo K, Nicely NI, Bradley T, Jaeger FH, Dennison SM, Zhang R, Lloyd KE, Stolarchuk C, Parks R, Sutherland LL, Scearce RM, Morris L, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Sinangil F, Phogat S, Michael NL, Kim JH, Kelsoe G, Montefiori DC, Tomaras GD, Bonsignori M, Santra S, Kepler TB, Alam SM, Moody MA, Liao HX, Haynes BF, Antibody light-chain-restricted recognition of the site of immune pressure in the RV144 HIV-1 vaccine trial is phylogenetically conserved. Immunity 41, 909–918 (2014). 10.1016/j.immuni.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR, Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Montefiori DC, Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter 12, Unit 12 11 (2005). 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 97.Perfetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, Roederer M, Amine-reactive dyes for dead cell discrimination in fixed samples. Curr Protoc Cytom Chapter 9, Unit 9 34 (2010). 10.1002/0471142956.cy0934s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Donaldson MM, Kao SF, Eslamizar L, Gee C, Koopman G, Lifton M, Schmitz JE, Sylwester AW, Wilson A, Hawkins N, Self SG, Roederer M, Foulds KE, Optimization and qualification of an 8-color intracellular cytokine staining assay for quantifying T cell responses in rhesus macaques for pre-clinical vaccine studies. J Immunol Methods 386, 10–21 (2012). 10.1016/j.jim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang J, Doria-Rose NA, Longo NS, Laub L, Lin CL, Turk E, Kang BH, Migueles SA, Bailer RT, Mascola JR, Connors M, Isolation of human monoclonal antibodies from peripheral blood B cells. Nat Protoc 8, 1907–1915 (2013). 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Doria-Rose N, Doria-Rose N, Bailer R, Louder M, Lin C-L, Turk E, Laub L, Longo N, Connors M, Mascola J, High throughput HIV-1 microneutralization assay. Protocol Exchange, (2013). 10.1038/protex.2013.069. [DOI] [Google Scholar]
- 101.Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, Wilson PC, Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 4, 372–384 (2009). 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ewing B, Hillier L, Wendl MC, Green P, Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8, 175–185 (1998). 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 103.Volpe JM, Cowell LG, Kepler TB, SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics 22, 438–444 (2006). 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
- 104.Kepler TB, Munshaw S, Wiehe K, Zhang R, Yu JS, Woods CW, Denny TN, Tomaras GD, Alam SM, Moody MA, Kelsoe G, Liao HX, Haynes BF, Reconstructing a B-cell clonal lineage. II. mutation, selection, and affinity maturation. Front Immunol 5, 170 (2014). 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang R, Verkoczy L, Wiehe K, Munir Alam S, Nicely NI, Santra S, Bradley T, Pemble C. W. t., Zhang J, Gao F, Montefiori DC, Bouton-Verville H, Kelsoe G, Larimore K, Greenberg PD, Parks R, Foulger A, Peel JN, Luo K, Lu X, Trama AM, Vandergrift N, Tomaras GD, Kepler TB, Moody MA, Liao HX, Haynes BF, Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci Transl Med 8, 336ra362 (2016). 10.1126/scitranslmed.aaf0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H, Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329, 112–124 (2008). 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mason RD, Welles HC, Adams C, Chakrabarti BK, Gorman J, Zhou T, Nguyen R, O'Dell S, Lusvarghi S, Bewley CA, Li H, Shaw GM, Sheng Z, Shapiro L, Wyatt R, Kwong PD, Mascola JR, Roederer M, Correction: Targeted isolation of antibodies directed against major sites of SIV Env vulnerability. PLoS Pathog 12, e1005674 (2016). 10.1371/journal.ppat.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krebs SJ, Kwon YD, Schramm CA, Law WH, Donofrio G, Zhou KH, Gift S, Dussupt V, Georgiev IS, Schatzle S, McDaniel JR, Lai YT, Sastry M, Zhang B, Jarosinski MC, Ransier A, Chenine AL, Asokan M, Bailer RT, Bose M, Cagigi A, Cale EM, Chuang GY, Darko S, Driscoll JI, Druz A, Gorman J, Laboune F, Louder MK, McKee K, Mendez L, Moody MA, O'Sullivan AM, Owen C, Peng D, Rawi R, Sanders-Buell E, Shen CH, Shiakolas AR, Stephens T, Tsybovsky Y, Tucker C, Verardi R, Wang K, Zhou J, Zhou T, Georgiou G, Alam SM, Haynes BF, Rolland M, Matyas GR, Polonis VR, McDermott AB, Douek DC, Shapiro L, Tovanabutra S, Michael NL, Mascola JR, Robb ML, Kwong PD, Doria-Rose NA, Longitudinal analysis reveals early development of three MPER-directed neutralizing antibody lineages from an HIV-1-Infected individual. Immunity 50, 677–691 e613 (2019). 10.1016/j.immuni.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schramm CA, Sheng Z, Zhang Z, Mascola JR, Kwong PD, Shapiro L, SONAR: A high-throughput pipeline for inferring antibody ontogenies from longitudinal sequencing of B cell transcripts. Front Immunol 7, 372 (2016). 10.3389/fimmu.2016.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ward JH, Hierarchiacal grouping to optimze an objectivefunction. Journal of the American Statistical Association 58, 236–244 (1963). 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 111.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP, A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9, e1003618 (2013). 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD, Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014). 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kwon YD, Chuang GY, Zhang B, Bailer RT, Doria-Rose NA, Gindin TS, Lin B, Louder MK, McKee K, O'Dell S, Pegu A, Schmidt SD, Asokan M, Chen X, Choe M, Georgiev IS, Jin V, Pancera M, Rawi R, Wang K, Chaudhuri R, Kueltzo LA, Manceva SD, Todd JP, Scorpio DG, Kim M, Reinherz EL, Wagh K, Korber BM, Connors M, Shapiro L, Mascola JR, Kwong PD, Surface-matrix screening identifies semi-specific interactions that improve potency of a near pan-reactive HIV-1-neutralizing antibody. Cell Rep 22, 1798–1809 (2018). 10.1016/j.celrep.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B, Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 115.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B, DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. Journal of Structural Biology 166, 205–213 (2009). 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B, Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166, 95–102 (2009). 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA, MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang K, Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B, DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166, 205–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 123.Liu Q, Lai YT, Zhang P, Louder MK, Pegu A, Rawi R, Asokan M, Chen X, Shen CH, Chuang GY, Yang ES, Miao H, Wang Y, Fauci AS, Kwong PD, Mascola JR, Lusso P, Improvement of antibody functionality by structure-guided paratope engraftment. Nat Commun 10, 721 (2019). 10.1038/s41467-019-08658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat 11, 53–55 (2004). 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 125.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 126.Davis IW, Murray LW, Richardson JS, Richardson DC, MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32, W615–619 (2004). 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, Fraser JS, EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946 (2015). 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fera D, Schmidt AG, Haynes BF, Gao F, Liao HX, Kepler TB, Harrison SC, Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proc Natl Acad Sci U S A 111, 10275–10280 (2014). 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kabsch W, XDS. Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010). 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ, EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46 (2007). 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 133.Treurnicht FK, Seoighe C, Martin DP, Wood N, Abrahams MR, Rosa Dde A, Bredell H, Woodman Z, Hide W, Mlisana K, Karim SA, Gray CM, Williamson C, Adaptive changes in HIV-1 subtype C proteins during early infection are driven by changes in HLA-associated immune pressure. Virology 396, 213–225 (2010). 10.1016/j.virol.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Andrabi R, Pallesen J, Allen JD, Song G, Zhang J, de Val N, Gegg G, Porter K, Su CY, Pauthner M, Newman A, Bouton-Verville H, Garces F, Wilson IA, Crispin M, Hahn BH, Haynes BF, Verkoczy L, Ward AB, Burton DR, The chimpanzee SIV envelope trimer: structure and deployment as an HIV vaccine template. Cell Rep 27, 2426–2441 e2426 (2019). 10.1016/j.celrep.2019.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Spurrier B, Sampson JM, Totrov M, Li H, O'Neal T, Williams C, Robinson J, Gorny MK, Zolla-Pazner S, Kong XP, Structural analysis of human and macaque mAbs 2909 and 2.5B: implications for the configuration of the quaternary neutralizing epitope of HIV-1 gp120. Structure 19, 691–699 (2011). 10.1016/j.str.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Williams WB, Meyerhoff RR, Edwards RJ, Li H, Nicely NI, Henderson R, Zhou Y, Janowska K, Mansouri K, Manne K, Stalls V, Hsu AL, Borgnia MJ, Stewart-Jones G, Lee MS, Bronkema N, Perfect J, Moody MA, Wiehe K, Bradley T, Kepler TB, Alam SM, Parks RJ, Foulger A, Bonsignori M, LaBranche CC, Montefiori DC, Seaman M, Santra S, Francica JR, Lynn GM, Aussedat B, Walkowicz WE, Laga R, Kelsoe G, Saunders KO, Fera D, Kwong PD, Seder R, Bartesaghi A, Shaw GM, Acharya P, Haynes BF, Fab-dimerized glycan-reactive antibodies neutralize HIV and are prevalent in humans and rhesus macaque. bioRxiv (2020). 10.1101/2020.06.30.178921. [DOI] [Google Scholar]
- 137.Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, O'Dell S, Sastry M, Schmidt SD, Xu K, Chen L, Chen RE, Louder MK, Pancera M, Wanninger TG, Zhang B, Zheng A, Farney SK, Foulds KE, Georgiev IS, Joyce MG, Lemmin T, Narpala S, Rawi R, Soto C, Todd JP, Shen CH, Tsybovsky Y, Yang Y, Zhao P, Haynes BF, Stamatatos L, Tiemeyer M, Wells L, Scorpio DG, Shapiro L, McDermott AB, Mascola JR, Kwong PD, Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation. Cell Rep 19, 719–732 (2017). 10.1016/j.celrep.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Design and characterization of SHIV.CAP256SU. (A) SHIV.CAP256SU was constructed using a design scheme previously reported (9) and represents a chimera of HIV-1 CAP256SU (blue), HIV-1 191859 (red) and SIVmac766 (grey) sequences (GenBank accession #MT509359). Env375-Ser (S) is the wildtype allele found in HIV-1 CAP256SU and in most M group viruses. This residue was replaced with each of five aliphatic, basic or aromatic residues (M, Y, H, W, F). (B) SHIVs expressing each of the six Env375 alleles were tested for neutralization sensitivity to a panel of prototypic human mAbs that targeted canonical bNAb epitopes, a linear V3 epitope (447-52D) and a CD4-induced bridging sheet epitope (17b). All six Env375 alleles retained native antigenicity and tier 2 neutralization profiles. (C) Replication of SHIV.CAP256SU Env375 variants in primary, activated rhesus and human CD4 T cells is depicted. All six allelic variants replicated efficiently in human cells but only the W, F, Y and H variants replicated in rhesus cells. This result highlights the ability of a single amino acid substitution at position 375 in Env to confer dramatic differences in replication efficiency in primary rhesus CD4 T cells.
Fig. S2. Virus load and autologous neutralizing antibody kinetics in the plasma of rhesus macaques and humans. Human and rhesus infections by CH505 (left panels), CH848 (middle panels) and CAP256SU (right panels) viruses are depicted. Animals pretreated with anti-CD8 mAbs are shown in red, untreated animals in blue, and humans in open black boxes. Crosses indicate animals that developed AIDS-defining events requiring euthanasia. Dashed lines demarcate the limits of quantification of the plasma virus load assay (250 vRNA molecules/ml).
Fig. S3. Glycan shield evolution in the human subject CH505 and SHIV.CH505 infected RMs. (A) Longitudinal glycan frequencies at selected positions show similar patterns between human (red) and RMs (different shades of blue and green). N332 is gained in all hosts with similar kinetics. N234 is gained in human and most RMs. Glycan 130 is lost over time in the human and in all RMs except RM5695 where it was absent from the outset of infection. (B) Longitudinal glycan shield evolution in the CH505 trimer (side view) over time. Blue patches indicate the location of time point consensus PNGs and green their predicted glycan coverage. Magenta and pink regions indicate glycan holes at regions that are glycan shielded in >80% or 50-80% of M-group sequences, respectively. The light grey area in the central region of the trimer is the inter-protomer surface that forms a cleft with low glycan coverage (45).
Fig. S4. Conserved Env evolutionary patterns between the HIV-1 infected human subject CH848 and SHIV.CH848 infected rhesus macaques. A description of the PIXEL plots (left panels) and the LASSIE analysis (right panels) is provided in the legend to Fig. 2. Sixteen of 29 residues that mutated to >75% fixation in the human subject CH848 also mutated in one or more monkey and these are highlighted by green flags in the PIXEL plots and by green lettering in the LASSIE plot. Deletions in V1 associated with the development of neutralization breadth in the human subject CH848 and in RMs 6163 and 6167 are indicated by a yellow flag.
Fig. S5. Glycan shield evolution in the human subject CH848 and SHIV.CH848 infected RMs. (A) Common patterns of glycan shield evolution in the CH848 trimer between human and RM6167 and RM6163. Longitudinal evolution of the viral quasispecies in CH848 human resulted in two glycan changes by week 75. The loss of N413 glycan does not impact the glycan shield but could alter sensitivity to autologous NAbs. The loss of N289 creates a glycan hole. Glycan shield evolution in RM6167 at week 28 and in RM6163 at week 80 shows similar changes. (B) Longitudinal glycan frequencies over time in human and RMs for selected glycans that show similar patterns across human (red) and RMs (different shades of blue and green). N413 is lost in all hosts with similar kinetics. N289 is transiently or permanently lost in human and most RMs. (C) Longitudinal glycan shield evolution over time in each host. Glycan shield calculation and coloring is the same as in Fig. S3B.
Fig. S6. Selected sites identified by LASSIE in human subjects CH505 and CH848 are rarely mutated in RMs infected by SHIVs bearing heterologous Envs. The uppermost LOGO plots at the top of this illustration depict HIV-1 Env residues in the human subjects CH505 (panel A) and CH848 (panel B) that were identified by LASSIE analysis to be under positive selection (Figs. 2 and S4). The position of each of these residues (HXB2 numbering) is shown at the bottom of each panel. Only four residues were identified to be under positive selection in both human subjects, and these positions were 130, 300, 413 and 620 (indicated by the vertical red lines). The lower LOGO plots residues in the heterologous T/F Env (CH848 - panel A; CH505 - panel B), and these were tracked over time in the SHIV infected monkeys for comparison to the matched analyses presented in Figs. 2 (CH505) and S4 (CH848). Over each animal’s LOGO display, there are two numbers in green at the upper right. The first digit shows the total number of sites under positive selection in the Env mismatched human and monkey, including sites highlighted in red. The second digit shows the number of sites under positive selection in the Env mismatched human and monkey after excluding the shared sites. Excluding the shared sites, we compared the patterns of selection in the different groups of animals. We found four of six animals infected with CH848 had no mutations in selected sites from the human subject CH505, RM6700 had 1 shared site, and RM6167 had 2 (panel A). In a comparison with Fig. 2 from the main text, 3 of the six animals shared 9 sites, two shared 7 sites, and one shared 6 sites. A Wilcoxon test showed that the distribution of CH505 selected sites in SHIV.CH505 macaques (9, 9, 9, 7, 7, 6), versus six SHIV.CH848 macaques (2, 1, 0, 0, 0, 0), was highly significant (p = 0.002). We also compared the human CH848 selected sites shared among the six SHIV.CH505 macaques shown in part B (3, 2, 1, 0, 3, 7 shared sites) with the six SHIV.CH848 infected macaques (Fig. S4) (11, 10, 6, 9, 4), and again found the distributions to be significantly different (p = 0.006). Note that because the human CAP256 was infected with two divergent HIV-1 strains, this control analysis was not applicable for this SHIV.
Fig. S7. Conserved Env evolutionary patterns between the HIV-1 infected human subject CAP256 and SHIV.CAP256SU infected rhesus macaques. (A) PIXEL plot highlighting amino acid mutations between the SHIV.CAP256SU infecting strain and subsequent samples in each RM. Red pixels indicate a change in amino acid relative to the infecting strain, and black indicates an Indel, as in Figs. 2 and S3. Green tags indicate residue positions that are selected in multiple RMs. Black numbers indicate HXB2 numbering and white numbers approximate HXB2 numbering in hypervariable regions. Yellow tags indicate identical Indels. (B) LASSIE analysis of the same longitudinal Env RM sequences, summarizing mutations under positive selection in any of the four RMs with a threshold of 75% replacement of the infecting CAP256SU strain shown at the top. Green numbers represent mutations observed in more than one animal. (C) Recombination map of CAP256SU and CAP256PI. The primary infection (PI) variants are shown as solid blue lines at week 6 and the superinfection (SU) variants as red lines at week 15. In subsequent time points, amino acids where the two forms match are grey. Places where they differ are indicated by blue if they match the PI, red if they match SU, and black if the differ from both. Extensive recombination is seen as early as week 23. (D) The mutations under selection in the RMs shown in panel B are tracked in the human relative to the CAP256SU form that dominates at w015. Positions under selection in the RMs that differ in the SU and PI forms are evident as the w006 forms are different than SU reference amino acids along the top. The mutations that arose de novo in the macaques were also often favored in the human and were most often explained by recombination of the PI forms into the SU backbone (e.g., E87K and T525A), although sometimes the mutation selected in RMs arose independently in the human and was not evident in either the PI or SU forms (e.g., R46K and K340E). Mutations at residue 169 in the human and in RMs 40591 and 42056 conferred escape from V2 apex targeted bNAbs (Fig. 3B,C).
Fig. S8. Glycan shield evolution in the human subject CAP256 and SHIV.CAP256SU infected RMs. (A) CAP256SU has a glycan hole due to absence of N339. During longitudinal evolution in the human and RM40591, N396 is gained over time. (B) Longitudinal glycan frequencies over time in human and RMs for N396 that shows similar patterns across human (red) and RMs (different shades of blue and green). (C) Longitudinal glycan shield evolution over time in each host. Glycan shield calculation and coloring same as Fig. S3B.
Fig. S9. Summary of Indels in the CH505 human and SHIV.CH505 infected RMs. The HXB2 reference and CH505T/F sequences are indicated at the top for orientation. Hypervariable regions within the loops are shown in red. Each Indel is aligned independently to the T/F as multiple alignments obscure the precise nature of the Indel. Precise Indel boundaries are indicated and unique indels are assigned a color. Only Indels that were found in the human or repeated in multiple individuals are shown. With the exception of a single codon deletion (in red), all V1 Indels were insertions. The T/F is shown at the top and the Indel beneath it. The translated sequences are depicted on the right. Insertions are generally direct repeats, and the section that is repeated is highlighted (e.g. the 9 bases in green in V1) and then shown repeated in the inserted sequence and translated on the right. The V1 insertions almost always included the addition of at least one N-linked glycosylation site. The V5 deletions always changed the relative location of a glycan within the V5 loop. The distinct hosts that carried an identical Indel are indicated by the colored boxes on the right. The vast majority of V1 Indels were insertions in the seven CH505 Env infections, and the histogram on the right shows that insertions in V1 restore it to a more typical length. The distribution represents V1 loop lengths based on the global Env M group alignment from the Los Alamos HIV database (www.hiv.lanl.gov). CH505 T/F was unusually short in V1 (20 amino acids long, magenta column), while the median V1 length was 30 amino acids (red column). Similarly, the deletions found in the long CH505 T/F in V4 bring the viral quasispecies closer to the median length of 31, and the repeated insertions in V5 bring the short CH505 TF V5 closer to a typical length. Only four time points collected during the first year of infection are shown here for simplicity; after the first year of infection, Indels on top of Indels begin to accrue and the evolutionary history becomes unresolvable.
Fig. S10. Summary of Indels in the CH848 human and SHIV.CH848 infected RMs. The figure organization is the same as in Fig. S9. Unlike CH505, the V1 length in CH848 T/F was atypically long and all Indels were comprised of deletions bringing the V1 loop length closer to the global median. Ten Indels in V1, V4 or V5 were precisely replicated between the human subject CH848 and one or more macaques. Additional Indels were replicated among macaques only.
Fig. S11. Epitope mapping of autologous neutralizing antibody responses in the SHIV.CH505 infected RM6072. (A) Mutations at residues N130, T234, N279, V281, K302, Y330, N334, H417 and K460 were strongly selected in human CH505 and in RM6072 (see Fig. 2A). Here, the left panel depicts fold changes in NAb titers of week 10 and 20 plasma against these mutant versus wildtype CH505 T/F Env pseudotyped viruses. The right panel shows neutralization curves of RM6072 wk20 plasma against CH505 T/F and single and combination mutants that evolve in human and rhesus. MLV is a murine leukemia virus control for non-specific inhibition of virus entry. (B) Left, neutralization of wildtype CH505 T/F and observed in vivo mutants by autologous monoclonal antibodies (mAbs) DH647 and DH648 isolated from RM6072. Neutralization potency is expressed as IC50 values (μg/ml). Right, neutralization curves of mAbs against CH505 T/F and single and combination mutants. (C) Neutralization of wildtype CH505 T/F and observed in vivo mutants by mAb DH650 isolated from RM6072 and its inferred germline unmutated common ancestor. mAbs isolated from human subject CH505 (6) are included as controls. Neutralization titers are shown as IC50 values (μg/ml).
Fig. S12. Epitope mapping of heterologous neutralizing activity in plasma from RMs infected by SHIVs CH505 (RM5695 and RM6070) or CAP256SU (RM40591 and RM42056). (A) RM5695, B) RM6070, C) RM40591, D) RM42056. Neutralization was tested in the TZM-bl assay using plasma or isolated plasma IgG against viruses expressing wildtype Envs or Envs with mutations at residues 156, 166, 169 or 171. In a diverse set of heterologous Env backbones (Q23.17, T250, BG505, 246F3, MT145K, CE0217 and ZM233), bNAb activity was found to be dependent on basic residues (R or K) at positions 166 and 169, and to a lesser extent, on a PNG at position 156 and a basic residue at position 171. These results, together with in vivo escape mutations at residues 166, 169 and 171 (Fig. 3B), map the bNAb activity in these plasma specimens to the V2 apex.
Fig. S13. Variable effects of N160K substitution on heterologous neutralization in RMs infected by SHIVs CH505 (RM5695 and RM6070) or CAP256SU (RM40591 and RM42056). (A) RM5695, (B) RM6070, (C) RM40591, (D) RM42056. Neutralization was tested in the TZM-bl assay using plasma or isolated plasma IgG against viruses expressing wildtype Envs or Envs with N160K substitutions in a diverse set of heterologous Env backbones (Q23.17, T250, BG505, 246F3, MT145K, and ZM233). Plasma from every animal potently neutralized viruses expressing heterologous wildtype Envs. However, the effects of N160K substitution ranged from elimination of neutralization activity (e.g, RM5695 plasma against HIV-1 BG505) to enhancement of neutralization (RM5695 plasma against HIV-1 Q23.17) (panel A). In some animals, the enhancement of neutralization against heterologous N160K containing viruses was extraordinary [e.g., RM6070 (panel B) and RM42056 (panel D)]. N160K dependent neutralization was strictly dependent on antibody binding to basic C-strand residues as shown by N160K.K169E double mutants (panels B and D). The most likely explanation for these results is that SHIV-infected animals contain three specificities of V2 apex targeted C-strand dependent neutralizing antibodies. The first are the typical V2 apex bNAbs that depend on interactions with C-strand residues and N160 glycans. The second C-strand targeted bNAbs that do not require binding to N160 and instead are enhanced in their potency by the absence of N160 glycans. The third are “2909-like” antibodies (83, 84, 135) that are strain-specfic and entirely lacking in neutralization breadth, but whose potency against the homologous virus strain is enhanced by the absence of N160 glycans. We have evidence for the existence of all three types of V2 apex NAbs in SHIV-infected RMs (see Supplement Extended Discussion).
Fig. S14. Epitope mapping of autologous neutralizing antibody responses in the SHIV.CH848 infected RM6163 and RM6167. (A) At weeks 10 and 36 post SHIV infection of animal RM6167, neutralizing antibody titers (reciprocal ID50) against the T/F virus reached 1:220 and 1:435, respectively. A strongly selected early substitution at Env residue R336 occurred in RM6167 and in the other SHIV.CH848 infected RMs and the human subject CH848. The R336G substitution in RM6167 resulted in a 67% reduction in neutralization potency of week 10 plasma. Similarly, a naturally occurring 10 amino acid deletion in V1 resulted in a 41% reduction in neutralizing antibody titer in the week 10 plasma but enhanced neutralization titers in concurrent wk 36 plasma. This V1 deletion, together with week 36 mutations at positions 169, 240, 290, 336, 339, 413 and 65 (denoted V1Δ10aa+Mut), led to complete escape from neutralizing antibodies in week 10 and 36 plasma, indicating again that selected mutations in Env arise as a consequence of neutralizing antibody pressure. (B) A six member autologous strain-specific neutralizing Ab lineage isolated from RM6163 (136) potently neutralized CH848 T/F and was dependent on N332 for activity. The naturally occurring V1 deletion in CH848 T/F (panel A) reduced neutralization titers by five-fold, indicating modulation by V1 of binding by this Nab lineage.
Fig. S15. Characterization of CD4bs antibodies in RM6072. (A) Plasma antibody binding to wildtype and CD4-binding site mutant (Δ371I) variants of CH505 TF gp120 and resurfaced core 3, RSC3 (95). Bottom panels depict differential binding ratios. ELISA results are from a representative experiment. (B) Plasma antibody blocking of Env binding by CD4bs bnAbs CH235 and CH106 (from the CH103 lineage) and by soluble CD4 (sCD4) as determined by competitive ELISA in a representative experiment. (C) Phylogram of memory B cell-derived antibodies designated the “DH650 lineage” that were isolated from blood (black) and bone marrow aspirate (purple) cells by an antigen-specific B cell sort approach or from blood by memory B cell culture (blue). The unmutated common ancestor (UCA) was computational inferred using the Cloanalyst software of macaque immunoglobulin reference genes (21). (D) Immunogenetics of the DH650 heavy and light chain genes was inferred by the Cloanalyst software. (E) Binding of DH650 mAbs to CH505 T/F gp120 was measured by ELISA and titers reported as EC50 (μg/ml). (F) DH650 mAbs were tested by ELISA for their ability to block Env binding by CH235, CH106 and sCD4. Inhibition is reported as percent blocking in a representative experiment.
Fig. S16. Binding and neutralization profiles of DH650 lineage antibodies. (A) Binding of DH650 mAbs to CH505 T/F gp120 and CH505 T/F SOSIPS was assessed by biolayer interferometry (BLI). mAb-captured sensors were placed into solutions of Envs at 50 μg/ml to measure binding depicted as response units over time. (B) Neutralization by DH650 lineage mAbs in TZM-bl cells against wildtype CH505TF and mutant CH505TF depleted of four glycans surrounding the CD4bs (137). IA refers to computationally inferred intermediate antibodies. Murine leukemia virus (MuLV) was used as a negative virus control. (C) Neutralization by TZM-bl assay of DH650 lineage mAbs against heterologous HIV-1 strains previously used to track neutralization breadth in the plasma of the HIV-1 infected CH505 human subject (2). Neutralization results were confirmed and extended in a test of 118 heterologous virus strains (Table S3).
Fig. S17. Genetic and biological characterization of DH650 lineage. (A) The HCDR2 region of the VH gene of CH235 and 8ANC131 plays a critical role in engaging the CD4 binding loop of HIV-1 Env. Asterisks identify CH235VH Env contact residues. Dots indicate identical residues at each position and dashes indicate deletions. A residue 57 serine (S) to arginine (R) mutation in the CH235 UCA was found to be critical for binding of CH235 lineage bNAbs to HIV-1 Envs (6). We found a similar mutation at residue 57 asparagine (N) to R in the DH650 lineage NAbs. (B) Binding by ELISA of DH650 mAb to CH505 T/F Env monomer and SOSIP depends on R57 in CDRH2. Controls included CH235, 2G12 and influenza-reactive mAb CH65. (C) Complete VH sequences of DH650 lineage antibodies isolated from single cell sorts of blood derived B cells 20, 24 and 32 weeks post SHIV infection (DH650-DH650.13) and sequences generated by next generation sequencing from blood mononuclear cells 52 weeks post SHIV infection (DH650.15-34_NGS). (D) DH650.6, which contained VH residues S35 and P62, failed to bind CH505 T/F Env (Figs. S15E, 16A) or neutralize CH505 T/F or T/F-gly4 virus (Figs. S16B,C). Substitution of either S35 to N35 or P62 to Q62 restored functionality as measured by ELISA binding to CH505 T/F gp120 (Fig. S17D). However, for DH650.15_NGS and DH650.16_NGS heavy chains paired with the DH650 VL chain, N35 and Q62 were not sufficient to support Env binding, suggesting that authentic paired VL chains are essential for DH650.15-16 functionality. (E) VL chain sequences of DH650 lineage antibodies compared with CH235 UCA and mature bNAbs. DH650 lineage antibodies contained an exceptionally long LCDR1 of 17 amino acids compared with 11 amino acids in CH235 antibodies.
Fig. S18. Heavy and light chain sequences of RHA1 bNAbs from RM5695. (A) Nucleotide and amino acid alignments of rhesus mAb heavy chains RAH1.V2.01-04 compared with RM5695 germline gene IGHV4-ABB-5*01_S8200. (B) Nucleotide and amino acid alignments of rhesus mAb light chains RAH1.V2.01-04 compared with RM5695 germline gene IGLV1-ACN*02.
Fig. S19. Structural, immunogenetic and neutralization properties of RHA1 bNAbs. (A) The two residue insertion in the HCDR1 region of RHA1 bNAbs results in interactions with N160 (yellow shading) and electrostatic contact between D29 and Env residue K171. (B) IGHV4-ABB-5*01_S8200 (blue), DH3-9 (red) and JH2-P (teal) plus six non-templated amino acids (black) contribute to the RHA1 HCDR3. Somatic hypermutation resulting in amino acid substitutions in D and J genes of the mature bNAb mAbs are shown. The amino-terminal Cys and carboxy-terminal Trp (underlined) are not part of the HCDR3 (IMGT). Dots indicate amino acid identities. (C) RHA1 mAbs are aligned to PCT64-35S and PGT145. RHA1 and PCT64-35S are similar in length (24 vs 25 amino acids, respectively). Seven of 24 RHA1 HCDR3 amino acids (yellow shading) are identical to those in PCT64-35S, including a critical DDY motif that is tyrosine-sulfated (red asterisks) in both antibody lineages and in PGT145. Blue shading highlights nonidentical residues that share chemical properties such as aromaticity or charge. Tyrosine sulfation plays a key role in binding HIV-1 Env by all three antibodies. (D) The neutralization fingerprint for RHA1.V2.01 illustrated in Fig. 6E showed it to cluster within the PGT145 class of bNAbs, which clustered significantly with all prototypic V2 apex bNAbs analyzed. Here, we show the additional 21 non-V2 apex targeted bNAb antibodies that were part of the same neutralization fingerprint analysis. (E) Venn diagram of the number of neutralization sensitive HIV-1 strains for RHA1.V2.01, CAP256-VRC26.25 and PGT145. Of the 101 viruses neutralized by RHA1.V2.01, 96 were neutralized by PGT145 and 84 were neutralized by CAP256-VRC26.25, supporting the neutralization fingerprint dendogram shown in Fig. 6E.
Fig. S20. RHA1.V2.01 lacks autoreactivity. (A) Indirect immunofluorescence showing lack of anti-nuclear antibody (ANA) reactivity by mAb RHA1.V2.01 in HEp-2 cells. The positive control mAb is DH1037 and negative control mAb is DH570.30 (50 μg/mL for each). The secondary antibody was a goat anti-rhesus Ig(H+L) FITC. (B) RHA1.V2.01, along with positive (4E10) and negative (CH65) control mAbs, were tested for reactivity to nine autoantigens in the ZEUS AtheNA Multi-Lyte ANA-II Plus Test System, according to manufacturer’s recommendations. All antibodies were 2-fold serially diluted starting at 50 μg/mL.
Fig. S21. Heterologous neutralization by RHA1 mAbs. Rhesus mAbs were tested for neutralization against the autologous CH505 TF virus and the same global panel of heterologous viruses used to test all macaque plasma in this study (Figs. 1). Neutralization titers are reported as IC50 (μg/ml) values. This same panel of viruses was also tested for neutralization by the RM5695 plasma from wk56 post infection, expressed as reciprocal dilution ID50 (μg/ml) values. The breadth of the RHA1 mAbs recapitulated the breadth observed in the polyclonal plasma.
Fig. S22. RHA1.V2.01 neutralization of a global panel of 208 heterologous HIV-1 viruses. Neutralization titers are reported as IC50 (μg/ml) values. This data is reflected in the dendrogram in Fig. 6B. 49% of viruses were neutralized at a concentration <50 μg/ml.
Fig. S23. Heterologous neutralization of RHA1 lineage bNAbs maps to the V2 apex. In multiple heterologous Env backbones, neutralization maps to N160, R166 and C-strand residues R/K169.
Fig. S24. Neutralization profiles of RHA1.V2.01 and other V2 apex bNAbs. (A) IC50 titers for RHA1.V2.01 and prototypic human V2 apex bNAbs are shown as a heatmap. Rows indicate viruses from the 208 strain panel (Fig. S22) and columns the bNAbs tested. IC50 titers are color-coded according to potency (red high, yellow low, blue >50μg/ml). Viruses and bNAbs are ordered using a hierarchical clustering on the Heatmap webtool (https://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html) (60, 109-113) using the Ward algorithm on Euclidean distances of Log10 transformed IC50 titers. A hierarchical clustering tree shows relatedness between IC50 profiles of different V2 apex bNAbs. Bootstrap confidence (% of 1000 bootstraps) is indicated on the splits. (B) Contingency tables highlight the overlap between sensitive and resistant viruses between RHA1.V2.01 and other V2 apex bNAbs. Resistant viruses had IC50 titers above the threshold. Below each table are listed Fisher’s exact test p-values, odds ratios (OR) and accuracy estimates (Acc), which measure the fraction of concordant viruses (i.e. sensitive or resistant to both bNAbs). (C) Scatterplots of IC50 titers with RHA1.V2.01 titers on the y-axis and other V2 apex bNAb titers on the x-axis. Viruses resistant to one but not the other bNAb are shown above or to the right of the dotted lines. The number in the top right indicates the number of viruses resistant to both. Pearson R2 and p-values are listed in the bottom right of each panel and were calculated using viruses that were sensitive to both bNAbs. These analyses together show that RHA1.V2.01 is most similar to PCT64-35M.
Fig. S25. Neutralization profiles of RHA1.V2.01 and other V2 apex bNAbs across HIV-1 group M subtypes. Neutralization IC50 titers are shown for the 208 virus global panel, broken down according to subtypes. IC50 titers are color-coded from red to green indicating potent to less potent IC50 titers, with black cells indicating IC50 titers greater than maximum concentration tested (50μg/ml). V2 apex bNAbs are grouped according to similarities in their subtype-specific neutralization profiles into three sub-groups: a) PCT64-35M and CAP256 bNAbs; b) CH01; and c) PG9, PGT145 and PGDM1400. The rhesus bNAb RHA1.V2.01 is most similar to the PCT64-35M subgroup.
Fig. S26. Signature analyses for RHA1.V2.01 and other V2 apex bNAbs. (A) Signature analyses for RHA1.V2.01 were performed as described (60). Significant signatures had two or more of the following: i) phylogenetic correction at false-discovery rate (FDR, q) < 0.2; ii) contact site; and iii) at least one strong signature (q < 0.1). This analysis identified sites 130, 160, 166, 167 and 169 as signature sites for RHA1.V2.01. The variability at these sites in M-group is shown using sequence logo in the left panel. On the right, variability in RM5695 longitudinal sequences is shown. Height of the letters indicate the frequency, and O is a potential N-linked glycan site (NXT/S). Blue letters are associated with sensitivity, red resistance and grey neutral. (B) Comparison of RHA1.V2.01 signatures to those of other V2 apex bNAbs. The results at signature sites are shown for each bNAb on a single row, using amino acids colored according to sensitivity or resistance and with the height of letters indicating the frequency in M-group sequences (www.hiv.lanl.gov). Notably, only PCT64-35M shared all signature sites for RHA1.V2.01.
Figure S27. Cryo-EM Details of RHA1.V2.01 in complex with HIV-1 Env BG505 DS-SOSIP. (A) Representative micrograph and contrast transfer function of the micrograph are shown. (B) Representative 2D class averages are shown. (C) The orientations of all particles used in the final refinement are shown as a heatmap. (D) The gold-standard fourier shell correlation resulted in a resolution of 3.90 Å using non-uniform refinement with C1 symmetry. We note that local refinement (beta) of the same particles resulted in a resolution of 3.8 Å. (E) The local resolution of the full map is shown generated through cryoSPARC using a Fourier shell correlation (FSC) cutoff of 0.5. Two contour levels are shown. (F) Representative density is shown for the Fab (top left) with a magnified region of the inserted sulfated tyrosine, Tys (top right), and for N160 of each protomer labeled P1, P2 and P3 (bottom panels).