Significance
Our understanding of fundamental biophysical mechanisms that underlie temperature-dependent gating remains limited despite the elucidation of structures of thermosensitive ion channels. According to current thinking, thermosensitive channels are expected to undergo a significant change in solvation upon activation by temperature, but direct evidence of such a change is lacking. Here, we utilize NMR spectroscopy and atomistic simulations to probe the solvation dynamics of an engineered temperature-sensitive variant of Shaker potassium channel at two different temperatures. Our studies reveal that this temperature-sensitive variant undergoes a dramatic shift in hydration at higher temperatures in contrast to the wild-type channels. Our findings illustrate how point mutations can cause dramatic differences in overall solvation dynamics and thereby alter temperature-dependent function.
Keywords: ion channels, thermosensing NMR, MD, voltage sensor domain
Abstract
Temperature-dependent regulation of ion channel activity is critical for a variety of physiological processes ranging from immune response to perception of noxious stimuli. Our understanding of the structural mechanisms that underlie temperature sensing remains limited, in part due to the difficulty of combining high-resolution structural analysis with temperature stimulus. Here, we use NMR to compare the temperature-dependent behavior of Shaker potassium channel voltage sensor domain (WT-VSD) to its engineered temperature sensitive (TS-VSD) variant. Further insight into the molecular basis for temperature-dependent behavior is obtained by analyzing the experimental results together with molecular dynamics simulations. Our studies reveal that the overall secondary structure of the engineered TS-VSD is identical to the wild-type channels except for local changes in backbone torsion angles near the site of substitution (V369S and F370S). Remarkably however, these structural differences result in increased hydration of the voltage-sensing arginines and the S4–S5 linker helix in the TS-VSD at higher temperatures, in contrast to the WT-VSD. These findings highlight how subtle differences in the primary structure can result in large-scale changes in solvation and thereby confer increased temperature-dependent activity beyond that predicted by linear summation of solvation energies of individual substituents.
The human body is finely tuned to respond to subtle changes in temperature, and much of this response is mediated by specialized ion channels expressed in peripheral nerves innervating the skin and tongue. The founding members of this class of ion channels are the TRPV1 and TRPM8 channels, both of which have also been widely studied (1–5). Structures (6, 7) of the isolated TRPV1 channel and in complex with peptide toxin, DKTx, led to the identification of a hydrophobic cluster behind the selectivity filter of the channel. Mutagenesis studies of residues within this cluster (8–11) had been previously shown to alter temperature sensitivity without affecting activation by capsaicin, suggesting that this region may act an allosteric temperature-activation pathway. Most strikingly, swapping the pore region of the TRPV1 channel on to the Shaker potassium channel makes the activation of the chimeric channel temperature sensitive (12).
Despite the wealth of evidence that the hydrophobic stretch in the selectivity filter of TRPV1 channel is critical for temperature sensing, the mechanisms of thermosensing remain far from fully understood for multiple reasons. First, even in the case of TRPV1 channels, mutagenesis studies have shown that the other regions of the channel also alter the ability to sense temperature (9, 10, 13–21). It remains unclear whether these other sites are part of the sensing element or they constitute the transduction machinery coupling the sensors to pore gates. Second, we have not been able to directly measure thermodynamic parameters associated with temperature-dependent gating. These measurements are necessary to determine the contributions of putative sensors to temperature-sensitivity. Finally, we have very little firsthand information about the structural changes that are associated with temperature in these channels. Recently, Sobolevsky and colleagues used cryo-EM to solve the structures of thermosensitive TRPV3 channels after preincubation at two different temperatures (22). Their studies show that the pore lining S6 helices undergo a rotation upon opening which changes the solvent exposure of ∼8 residues. It remains unclear, however, whether the pore-lining helices are the primary temperature sensors in TRPV3 channels.
Given these uncertainties about the identity and conformation of the temperature-sensing motif in thermosensitive TRP channels, we sought to characterize temperature-induced structural changes in a more well-defined system. Previously, Chowdhury et al. (23) were able to engineer temperature sensitivity into the prototypical Shaker potassium channel, which is naturally temperature insensitive, by systematically introducing mutations in the voltage-sensing domain (VSD) (23). In contrast to the naturally occurring temperature-sensitive ion channels, the primary advantage of this designed system is that we know precisely the location and identity of the changes that cause increased temperature dependence. In one of the temperature-sensitive variants (S2bot), two hydrophobic residues between the third and fourth positive charge in the voltage-sensing helix were replaced by serines (Fig. 1A). Remarkably, these two substitutions dramatically increase the temperature dependence of channel opening wherein at 0 mV, most of the channels are open at 28 °C but not at 8 °C [see figure 3E in Chowdhury et al. (23)].
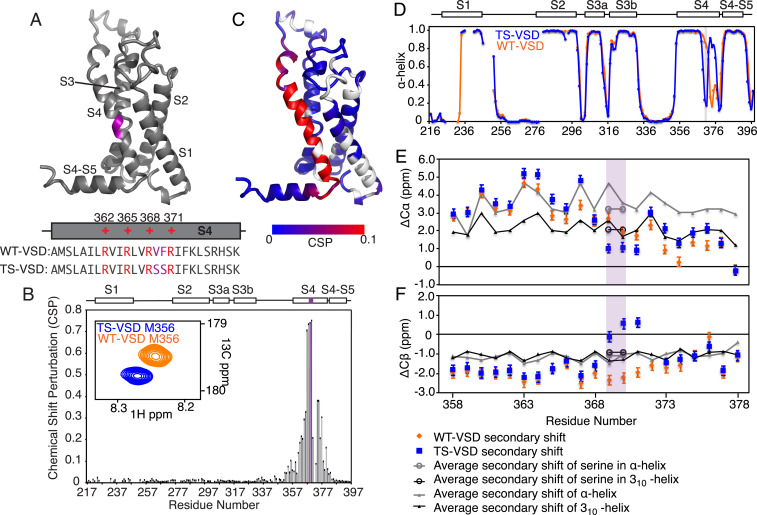
Fig. 1.
Serine mutations in S2bot alter the structure of Shaker-VSD. (A) The two-amino-acid S2bot mutation (V369F370 to S369S370) is highlighted in magenta on the homology model of isolated Shaker-VSD and in the S4 helix sequence (Lower Inset, voltage-sensing arginines in red). (B) CSP between WT-VSD and TS-VSD was calculated as described in Materials and Methods using the well-resolved 3D-HNCO spectra at 45 °C. The graph is numbered according to the full-length Shaker sequence, with the S2bot mutation (magenta) and transmembrane helices S1 to S4 indicated at the top. An overlay of the WT-VSD and TS-VSD peaks (A, Inset) corresponding to M356 in the 3D-HNCO illustrates the significance of a CSP value of 0.1. (C) The CSP data are plotted on the homology model of the isolated Shaker-VSD (blue to red color scale as shown, unassigned residues in white). Significant CSP ≥ 0.1 (red in B) is observed at the site of mutation and extending along the S4 transmembrane helix and into the adjoining loops. Similar results are observed at 28 °C (SI Appendix, Fig. S2). (D) The α-helical propensity of WT-VSD (orange) and TS-VSD (blue) was calculated using TALOS+ (25) analysis of the backbone NMR chemical shifts (HN, N, Co, C-α, and C-β). The scale extends from 0 (no helical propensity) to 1.0 (α-helix). The gaps in data reflect residues without backbone assignments. TS-VSD is generally similar to WT-VSD except for a loss of α-helical propensity at the S2bot mutation site (purple shading) and extending toward the C-terminal end of S4. Almost identical predictions are obtained with PECAN (26) (SI Appendix, Fig. S2). The raw data underlying the secondary structure analysis include the C-α (E) and C-β (F) secondary chemical shifts, which are shown for the S4 transmembrane helix of WT-VS (orange) and TS-VSD (blue) plotted against the average secondary chemical shifts for the corresponding amino acid sequence in a fully α-helix (gray) or 310-helix (black) conformation as extracted from the PACSY database (27).
Here, we use NMR spectroscopy to characterize the temperature-induced conformational changes and water accessibility in the voltage-sensing domain of the wild-type Shaker VSD (WT-VSD) and the temperature-sensitive S2bot mutant (TS-VSD). We have recently shown that the isolated WT-VSD is structurally stable in 1-palmitoyl-2-hydroxy-3-sn-phosphatidylglycerol (LPPG) micelles and suitable for NMR studies (24). Here, we find that the two serine substitutions introduced in the S4 helix of the Shaker potassium channel not only alters the distribution between the resting and activated Shaker-VSD upon thermal perturbation in the isolated TS-VSD but also modifies the structure such that the water-filled crevices are much more hydrated. Molecular dynamics (MD) simulations of analogous mutations in a homologous channel reveal a similar increase in hydration. Together, these findings provide insight into the role of solvation in temperature-dependent gating of ion channels.
Results
Serine Mutations in S2bot Alter the Structure and Dynamics of Shaker-VSD.
To assess the impact of the S2bot mutations on the structure and hydration of the Shaker-VSD by NMR, we first need to assign the backbone resonances for the mutant VSD. Using the previously described methodology for backbone assignments of the WT-VSD (24), we assigned 85% of the TS-VSD mutant in LPPG micelles at 45 °C (SI Appendix, Fig. S1). We evaluated the chemical shift perturbation (CSP) between WT-VSD and TS-VSD using TROSY-HNCO spectra collected at 45 °C because of the improved resolution of the three-dimensional (3D) experiments at this temperature enable quantitative analysis of more residues. Plotting the CSP data onto the homology model of the WT-VSD structure shows that the largest differences are at the site of the mutations as expected, but significant perturbations extend along the whole length of the S4 transmembrane helix (Fig. 1 B and C). To compare the structural differences at two temperatures, the CSP analysis was repeated with TROSY-HNCO spectra of WT-VSD and TS-VSD collected at 28 °C, with the resonance assignments transferred by temperature titrations. The data were very similar at both 28 and 45 °C, indicating that there are no significant structural changes in the isolated VSDs in this temperature range (SI Appendix, Fig. S2 A and B) as expected. Unfortunately, due to the slow tumbling time of the micelle-solubilized protein and consequent low sensitivity, we were not able to extend this analysis to 8 °C.
Backbone chemical shifts (HN, 15N, 13CO, 13C-α, and 13C-β) are also highly sensitive to the structure of the backbone and can be used to determine the secondary structure and backbone torsion angle. We calculated the residue-specific α-helical propensity of TS-VSD using TALOS+ (25) for comparison with WT-VSD (Fig. 1D). Overall, the α-helical propensity is very similar between WT-VSD and TS-VSD. However, there are major differences in the S4 transmembrane helix, with a significant decrease in the α-helical propensity at the site of the mutations and an increase in helical propensity in the C-terminal portion of S4 in the TS-VSD. Similar results are obtained using a different program, PECAN (26), to predict torsion angle and secondary structure based on backbone chemical shifts (SI Appendix, Fig. S2C). These results strongly indicate that the S2bot mutations alter the secondary structure of the S4 transmembrane helix near the site of substitutions.
Next, we examined the secondary chemical shifts of C-α and C-β for the S4 transmembrane helix (Fig. 1 E and F). Also shown as reference are the average C-α and C-β secondary chemical shifts derived from the Protein structure and Chemical Shift NMR spectroscopy database for various positions in the S4 helix if it were either purely α-helical or 310-helical structure (27). For WT-VSD, the C-α and C-β secondary chemical shifts at the C terminus of the S4 transmembrane helix agree well with 310-helix (24). However, the secondary chemical shifts of TS-VSD deviate from helical values near the site of mutation and are closer to the random coil values. In addition, the model-free order parameters (S2) predicted from the backbone chemical shifts (28) suggest increased flexibility at the site of the mutations (SI Appendix, Fig. S3A). Together, these results indicate that the S4 helix is much shorter in the TS-VSD compared to the WT-VSD.
We previously observed peak doubling in the S4 transmembrane helix of WT-VSD (24), including the voltage-sensing arginines, indicating two conformational states. Interestingly, we only observe a single peak for the residues in S4 in TS-VSD (SI Appendix, Fig. S3B). Either the S2bot mutation has stabilized a single conformational state or it has increased the rate of exchange between conformational states so that it is fast on the NMR timescale and only a single, average chemical shift is observed. Further experiments will be required to distinguish these two possibilities. Overall, these observations indicate that the serine mutations in S2bot perturb the structure of the S4 helix in the TS-VSD.
Shaker TS-VSD Mutant Has Enhanced Temperature Sensitivity Compared to WT-VSD.
Solvation of hydrophobic surfaces is an important factor in the heat capacity hypothesis used to design the temperature-sensitive S2bot mutant. We therefore assessed the temperature-dependent water accessibility of both WT-VSD and TS-VSD using NMR spectroscopy. An important consideration in choosing the S2bot mutant for this work is the observation that temperature alone can shift the S2bot channel from the resting state to the activated state at 0 mV (23), where our NMR experiments are performed. The WT-VSD and TS-VSD samples were purified and prepared according to our previously published protocols (24). In this experiment, we focus on the four arginines (R362, R365, R368, and R371) in the S4 helix which are primarily involved in voltage sensing and are expected to undergo a change in environment upon channel activation (29–31). The S4 helix is also the region with the largest backbone chemical shift differences between WT-VSD and TS-VSD, making this region most likely to have different hydration in the temperature-sensitive mutant. In addition, the increased motion of the arginine side chains relative to the backbone makes them more sensitive probes for NMR detection, particularly at low temperatures where the slow tumbling of the Shaker-VSD/micelle complex results in extreme line broadening of backbone resonances. Thus, the voltage-sensing arginines are ideal residues to monitor changes in water accessibility within the WT-VSD and TS-VSD at 28 and 8 °C for direct comparison with the electrophysiology measurements.
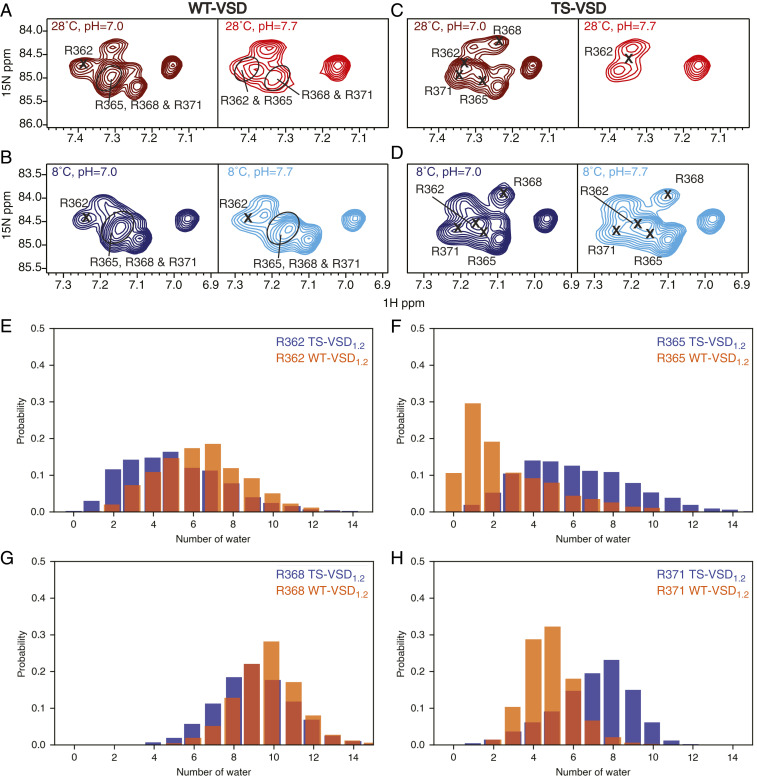
We use established NMR methods for specific detection of the -NεHε resonance of the arginine side chains (32), which fall in a unique region of 1H-15N TROSY-HSQC spectra (Fig. 2). There is significant overlap of the arginine side-chain resonances in the two-dimensional (2D) 1H-15N TROSY-HSQC spectra, so each of the four voltage-sensing arginines were assigned by mutagenesis (R to K) in both WT-VSD and TS-VSD under each set of conditions to account for peak shifts as a function of temperature and pH (SI Appendix, Fig. S4). Hydrogen/deuterium (H/D) exchange is traditionally performed by rapidly transferring the protein from water to D2O, a more challenging experiment for detergent-solubilized membrane proteins since detergent is often lost during these procedures. Spectral overlap complicates the use of quantitative methods to monitor H/D exchange that require precise measurement of peak intensity. Thus, we turned to pH perturbation since hydrogen exchange with water is pH dependent and exchange rates increase at pH values above neutral (33, 34). This increased exchange leads to enhanced line broadening of water-exposed -NεHε resonances of lysine and arginine side chains at high pH (35, 36). We therefore compared arginine -NεHε spectra of WT-VSD and TS-VSD at neutral pH (pH 7.0) and slightly basic pH (pH 7.7) to qualitatively assess water accessibility of the voltage-sensing arginines at 28 and 8 °C (Fig. 2).
Fig. 2.
Temperature dependence of solvent accessibility for the voltage-sensing arginines in WT-VSD and TS-VSD. 1H-15N TROSY-HSQC spectra of the arginine sidechain NεHε group for both WT-VSD (A and B) and TS-VSD (C and D) are shown at 28 °C (A and C) and 8 °C (B and D). For each condition, spectra at pH = 7.0 and 7.7 are compared to assess susceptibility to base-catalyzed H/D exchange as a measure of water accessibility. Peaks corresponding to arginine sidechains that are well protected from water will be present in the spectra at both pH values, while those that are exposed to water will disappear at the higher pH. Peaks corresponding to the sidechains of the voltage-sensing arginines (R362, R365, R368, and R371) were assigned by Arg to Lys mutagenesis at each position (SI Appendix, Fig. S4). The voltage-sensing arginine sidechains are observable at both pH values for WT-VSD at 8 and 18 °C and for TS-VSD at 8 °C, indicating good protection from water under these conditions. In contrast, for TS-VSD at 28 °C, the peaks corresponding to R365, R368, and R371 in TS-VSD at 28 °C disappear at pH = 7.7, indicating that these residues are no longer protected from H/D exchange in the TS-VSD at this temperature. (E–H) Histograms of the number of water molecules within 6 Å of Hε in the voltage-sensing arginines based on 1 μs of conventional MD simulations (histograms calculated over entire trajectory) also show a redistribution of water around the voltage-sensing arginines in TS-VSD1.2. A slightly larger amount of water is observed around R362 (E) and R368 (G) in the WT-VSD1.2, while the TS-VSD1.2 features more water around R365 (F) and R371 (H).
For WT-VSD (Fig. 2 A and B), no significant peak loss was observed in the arginine TROSY-HSQC between pH 7.0 and 7.7 at 8 °C. Some peak broadening is observed at 28 °C when the pH is raised. This is expected for arginines on the surface of the VSD in the loop or tail regions due to the intrinsic temperature-dependent increase in the rate of hydrogen exchange with water. However, the -NεHε resonances of R362, R365, R368, and R371 in S4 remain visible and are not broadened at pH 7.7, indicating that these important residues remain relatively well-protected from water at 28 °C. We note that we explicitly assigned these four individual -NεHε resonances at every condition, and the method we used (SI Appendix, Fig. S4) will detect whether the resonance for a particular side chain is present or exchange broadened at each pH, even when they are not well-resolved in the 2D spectrum. Thus, the four voltage-sensing arginines in S4 are protected from water and this protection is insensitive to temperature, consistent with our expectation from the electrophysiology data showing WT Shaker channel is temperature insensitive from 8 to 28 °C.
The NMR spectra for TS-VSD at 8 °C (Fig. 2D) show similar results to WT-VSD, with all four voltage-sensing arginines again protected from water. For TS-VSD at 28 °C (Fig. 2C), many arginine side-chain resonances again disappear when the pH is increased, and in contrast to WT-VSD, the resonance that disappear in TS-VSD include the side chains for R365, R368, and R371. Only the side chain of R362 at the top of the S4 helix is relatively protected from water in TS-VSD at 28 °C (for further explanation see SI Appendix, Fig. S4). This demonstrates a clear temperature-dependent change in water-accessibility for voltage-sensing arginines in TS-VSD compared to the WT-VSD at 0 mV as temperature is increased from 8 to 28 °C.
The exact pattern of the temperature-dependent differences between WT-VSD and TS-VSD does not match our original expectation. In the electrophysiology experiments, at 0 mV the S2bot channel is closed at 8 °C and open at 28 °C, while the WT Shaker channel is open at both temperatures. Thus, our initial assumption was that the difference in hydration between TS-VSD and WT-VSD would be detected at 8 °C, not at 28 °C where we experimentally observe it by NMR. This prompted us to perform further NMR experiments and MD simulations.
Increased Hydration within the Water Crevices of TS-VSD.
To further probe the differences in solvent accessibility between the WT-VSD and TS-VSD, we performed 15N-edited NOESY-TROSY experiments. NOESY experiments provide a sensitive measurement of intramolecular distances for protons within ∼5 Å and are commonly used for structure determination. Intermolecular nuclear Overhauser effects (NOEs) are also observable, and NOESY experiments were first used decades ago to monitor protein hydration (37, 38). Although concerns have been raised regarding potential artifacts due to the influence of water dynamics on the NOE, spin diffusion, and potential long-range nature of NOEs (39–41), recent comparison of MD simulations and NOESY spectra indicates that NOEs to water correlate remarkably well with close proximity to surface water in the simulations, and this experiment is therefore a useful monitor of protein hydration (42).
Therefore, 15N-edited NOESY-TROSY experiments spectra of WT-VSD and TS-VSD were collected at 45 °C using identical parameters for the comparison of solvent accessible surface at high temperature (Fig. 3). We do not attempt to distinguish NOESY cross-peaks due to H/D exchange from true NOEs since both reflect accessibility to water. These experiments were performed at 45 °C to maximize spectral resolution and take advantage of the more complete backbone assignments at this temperature. Our NMR data (Fig. 1 B and C and SI Appendix, Fig. S2) indicate that there is no significant structural change in either WT-VSD or TS-VSD between 28 and 45 °C, and both electrophysiology studies (23) and the NMR data (Fig. 2 A–D) show that the structural and functional transition occurs between 8 and 28 °C. Thus, the hydrated surface of WT-VSD and TS-VSD observed at 45 °C should be very similar to the hydrated surface at 28 °C.
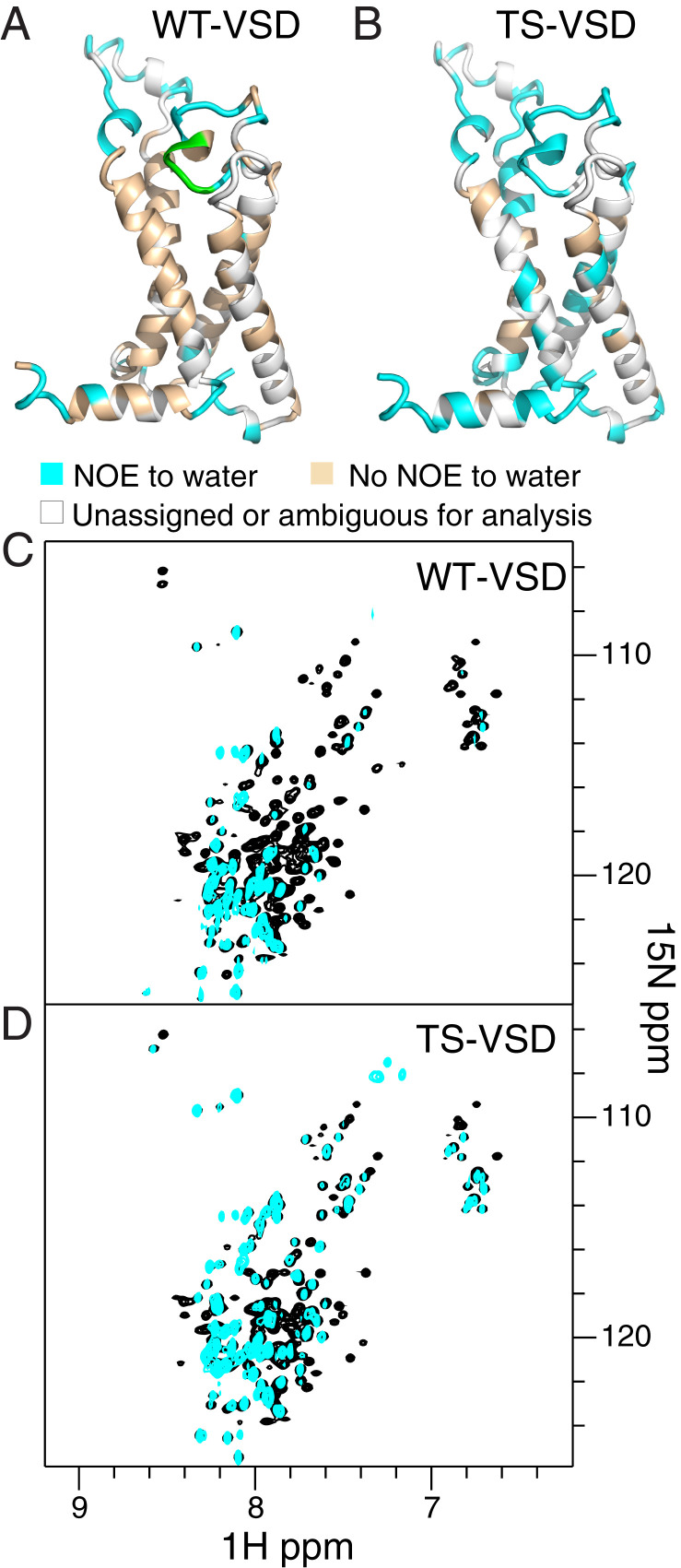
Fig. 3.
NMR reveals increased hydration within the water crevices of TS-VSD. Residues within WT-VSD (A) and TS-VSD (B), with visible crosspeaks to water in an 15N-edited NOESY-TROSY experiment are colored cyan on the homology model of the Shaker-VSD. Residues without crosspeaks to water resonances are colored wheat, and residues without assignments or peak overlap are colored gray. The raw data for WT-VSD (C) and TS-VSD (D) are shown below. The overlay of the 15N-1H TROSY-HSQC spectrum (black) and the water plane (∼4.58 ppm at 45 °C) from the 15N-edited NOESY-TROSY spectrum (cyan) clearly shows which backbone amide resonances have crosspeaks to water due to close proximity of water or H/D exchange. For WT-VSD (A and C), crosspeaks to water are not observed for the majority of residues on the transmembrane helices, while many more residues have crosspeaks to water in TS-VSD (B and D). This indicates increased water occupancy around the water crevices within the VSD. WT-VSD and TS-VSD are both stably embedded in the same way in the LPPG micelle (SI Appendix, Fig. S5), so the increased hydration of the water crevices in TS-VSD is not due to mutation-induced disruption of the protein/micelle complex. In addition, T265 to E268 in the S1–S2 linker of WT is colored green in A. Doubled resonances are observed for this region in the 15N-1H TROSY HSQC spectrum, but NOESY crosspeaks to water are only observed for one resonance, indicating that the two conformations have different water accessibility.
Since much of the Shaker-VSD is embedded in the micelle, we expect the majority of the resonances will not have any cross-peaks to water in an 15N-edited NOESY-TROSY experiment. Thus, this provides an internal control to check VSD folding and micelle insertion. This is exactly what we observe for WT-VSD (Fig. 3 A and C), with NOE cross-peaks to water resonances from amides in the loop regions but not from amides in the transmembrane helices. In contrast (Fig. 3 B and D), in TS-VSD many more cross-peaks to water are observed, including residues along the potential water crevice within the VSD helical bundle. To confirm that TS-VSD remains stably embedded within the LPPG micelles, we examined the NOE cross-peaks to the methyl group of LPPG, as a reporter of regions of the VSD covered by LPPG (SI Appendix, Fig. S5). NOE cross-peaks to the methyl groups of LPPG are observed for almost all of the assigned residues within the transmembrane helices, and the pattern is nearly identical between WT-VSD and TS-VSD (SI Appendix, Fig. S5), indicating both of them are well embedded within the micelle. Thus, the increased hydration within the TS-VSD reflects increased hydration within the water crevices of TS-VSD and not expulsion of the VSD from the micelle. The broader assessment of hydration provided by this NOESY data are consistent with the earlier analysis of voltage-sensing arginine side chains (Fig. 2 A–D), with both data sets indicating increased hydration within the TS-VSD compared to WT-VSD.
We also observe differences in hydration for WT- and TS-VSD outside the transmembrane helices. We previously noted that our NMR data reveals two conformations for the large S1–S2 loop in WT-VSD, with two distinct resonances for residues in this region (24). Interestingly, we observe NOE crosspeaks to water for only one of the two HN resonances in WT-VSD for residues T265 to E268 (Fig. 3A, highlighted in green), consistent with differential water accessibility of the two loop conformations. However, in TS-VSD (Fig. 3B), we observe NOE cross-peaks to water for both HN resonances throughout this region. We also observe hydration differences between WT- and TS-VSD in the amphipathic S4–S5 linker helix that would connect the VSD to the pore domain in the full channel (Fig. 3 A and B). Once again, increased hydration in this region is observed in TS-VSD compared to WT-VSD. Altogether, the NMR data clearly demonstrates differences in hydration between WT- and TS-VSD along the voltage-sensing S4 helix as well as in adjacent regions.
A similar approach was recently used to assess temperature-dependent hydration in the isolated human TRPV1 sensor domain, and increased hydration as a function of temperature was observed in the C-terminal region of S4 helix and in several loops (43). This pattern is similar but not as widespread as the increased hydration we observe between WT-VSD and TS-VSD.
MD Simulations Support Enhanced Hydration Level of the Crevices and Structural Plasticity of the S4 Helix.
To supplement the NMR studies and gain further insights into the response of VSD to the S2bot mutations, we carried out μs-scale conventional MD simulations as well as metadynamics simulations for the WT-VSD1.2 and TS-VSD1.2 at 25 C°. Note that these simulations used the Kv1.2 channel VSD structural model (denoted VSD1.2 to distinguish from the Shaker VSD), which was validated for the WT protein in our recent joint MD–NMR study (24). Motivated by the NMR results presented above, we focus MD analysis on the hydration levels of the voltage-sensing arginine side chains, polar/charged residues in the water crevices, and the structural plasticity of the S4 helix.
Conventional MD simulations reveal that the hydration level of polar/charged residues in the water crevices generally increases in TS-VSD1.2 relative to WT-VSD1.2. In addition to the expected level of enhanced hydration near the mutation sites (SI Appendix, Figs. S6A and S7A), the hydration level of the key arginine residues increases in the TS-VSD1.2. As illustrated by the histograms in Fig. 2 E–H and radial distribution functions in SI Appendix, Fig. S8, elevated hydration is most visible for the second and fourth arginine sidechain in the S4 helix (R365 and R371 with the Shaker sequence), while the change is minimal for the first and the third arginines. The R371 sidechain is close to the base of the water crevice, and analysis of interhelical distances (SI Appendix, Fig. S11) indicates that this region expands in TS-VSD1.2, which explains a higher level of hydration for R371. Therefore, the MD simulations qualitatively captured the hydration level change observed by NMR. We note, however, that the NMR data reports increased hydration for the second, third, and fourth arginine (R365, R368, and R371 with the Shaker sequence). In addition, there is evidence for a redistribution of water around the S4–S5 linker helix (SI Appendix, Fig. S6 B and C), although it is not as uniformly an increase in hydration or proximity of water molecules in the MD simulations. The quantitative difference between MD and the NMR results might be due in part to the different time scales (see SI Appendix, Fig. S7B for the time dependence of hydration levels of the arginine sidechains) and in part to the difference between the Shaker and Kv1.2 channels used for NMR and MD, respectively.
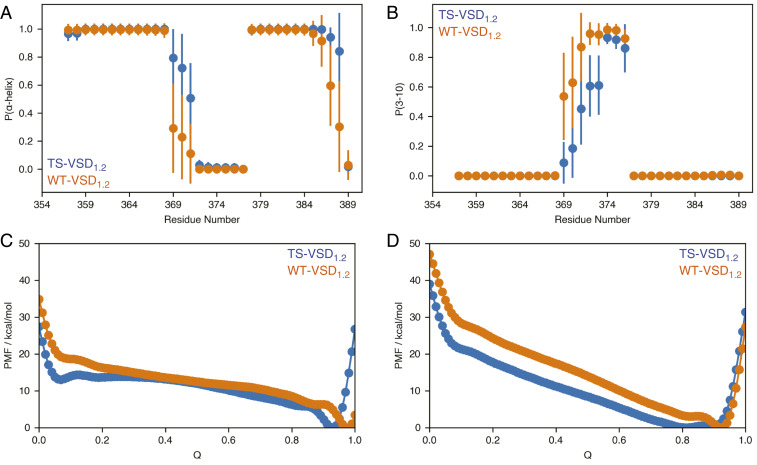
In the conventional MD simulations, S4 residues near the mutation site are observed to exhibit a higher propensity for 310- than α-helix (Fig. 4 A and B), while the reverse is true for the mutation sites (S369 and S370) and the following residues in the TS-VSD1.2. Therefore, the S4 helix remains structured in the TS-VSD1.2 during conventional MD simulations at the μs time scale, as also illustrated by the backbone ψ/ϕ-angles, secondary-structure analysis, and amide order parameters shown in SI Appendix, Fig. S9. While the high 310-helical propensity for the WT-VSD1.2 is qualitatively consistent with the NMR analysis discussed above, the conventional MD simulations do not observe any significant reduction in helicity in S4 in the TS-VSD1.2.
Fig. 4.
MD simulations capture changes in secondary structure propensity and flexibility near the site of mutation in TS-VSD. In conventional MD simulations, the secondary structures around the mutation sites are ordered but show a slight shift in α-helix (A) versus 310-helix (B) propensity in the TS-VSD. The timescale of the MD simulations may not be long enough to fully capture the loss of helical structure observed by NMR. Multiple-walker well-tempered metadynamics simulations reveal that TS-VSD features a higher level of structural plasticity near the mutation sites compared to the WT-VSD, consistent with the NMR data. The potential of mean force (PMF) along the collective variable plotted against the fraction of native contacts (Q) for α-helix (C) and 310-helix (D) from metadynamics simulations reveals an energy minimum at lower fraction of native contacts and lower energy barrier to access low-Q states for the TS-VSD. Convergence behavior of the computed PMFs is shown in SI Appendix, Fig. S10.
On one hand, since the conventional MD simulations captured the enhanced hydration level in the water crevices of TS-VSD1.2, the lack of structural disorder in S4 in these simulations implies that the change in hydration level in TS-VSD is not a consequence of the locally disordered conformations near the mutation site (i.e., the increased polarity due to mutations in a membrane-contact region of the S4 helix is sufficient to induce notably enhanced hydration within the water crevices). On the other hand, transition to locally disordered conformations induced by the S2bot mutations might occur at a time scale much longer than μs. To test the latter hypothesis, we have conducted metadynamics simulations using the fraction of local contact (44) (Q) as a collective variable. As shown in Fig. 4 C and D (for convergence behavior of the metadynamics simulations, see SI Appendix, Fig. S10), the free energy profile along Q (the fraction of native contacts) for the TS- VSD1.2 is consistently lower than WT-VSD1.2, especially in the low-Q region with few native contacts, which corresponds to locally disordered conformations. Moreover, the free energy minimum in the high-Q region is observed at a lower Q value in TS-VSD1.2 than in the WT-VSD1.2. These observations support the experimental NMR observation that the S2bot mutations modify not only the hydration level of the water crevices but also the structural ensemble of the S4 helix relative to the WT-VSD.
Discussion
According to the heat capacity hypothesis proposed by Miller and Clapham (45), temperature-dependent gating of TRP channels is associated with large changes in molar heat capacity. Thus, despite the same overall architecture between a typical voltage-gated potassium channel and a thermosensing TRP channel, the source of temperature sensitivity is entirely due to the high heat capacity of TRP channels. Since the primary origin of high heat capacity is the solvation of hydrophobic residues upon channel gating, their theory predicts that a targeted substitution of about 25 to 30 amino acids per subunit will be sufficient to endow high temperature sensitivity to any channel. Using this overarching thermodynamic framework, Chowdhury et al. (23) found that even substitution of just eight amino substitution (two per subunit) is sufficient to confer high-temperature dependence, suggesting that there may be other processes at play. Our NMR studies here on one such double mutant show that these substitutions alter the temperature-dependent solvation of a broader region, providing a possible explanation for the unexpected nonlinear effects of the two substitutions.
Since the full-length channel is too large for NMR studies, we focused on the isolated voltage-sensing domain where both the mutations in the S2bot channel are localized. Previous studies have shown that the isolated VSD, like the WT channel, is able to transport gating charge in response to depolarization, demonstrating that this domain folds correctly in the membrane (46). However, unlike the VSD in the WT channel, the isolated VSD conducts protons through the gating pore at hyperpolarized potentials. This observation suggests that the integrity of the gating septum is compromised when a strong force is applied in absence of stabilization by the pore domain. Our measurements were carried at 0 mV, and under those conditions the isolated VSD behaves similar to the VSD in the full-length channel.
The other issue to consider is the apparent inconsistency between the electrophysiology data of the S2bot mutant channel (23) and NMR data obtained from the isolated VSD. Our NMR data shows that the solvent accessibility of the TS-VSD is similar to WT-VSD at low temperature whereas at high temperature, the gating crevice in the TS-VSD is significantly more hydrated than either the WT-VSD or TS-VSD at low temperature. If one assumes a simple model that these mutations only affect temperature dependence of voltage-sensor movement, then one would expect that the low-temperature structures are going to be different. One possible and simple explanation is that the temperature dependence of the isolated TS-VSD is flipped, which would happen if the mutation also makes the allosteric coupling temperature dependent. Multiple studies have established the crucial role of V369—one of the S2bot sites—in voltage-sensor pore coupling and Shaker channel cooperativity (47–50). Loss of voltage-sensor pore coupling in V369I mutation causes opposite shifts on the Po-V and Q-V curves (47). Furthermore, Hill slope (51) of S2bot dramatically increases at higher temperatures, indicating that the cooperativity associated with channel activation is highly temperature dependent.
The MD simulations, in addition to recapitulating the observed increase in hydration at the gating crevices in mutant VSD, also provide further insight into the mechanism. The MD simulations suggest that water penetration to solvate the polar serine sidechains in the TS-VSD1.2 occurs first, followed by structural changes that occur on a longer timescale. The additional structural change may enhance the temperature sensitivity introduced by this particular mutation by causing a greater difference in solvation between resting and activated states of the channel, although further structural studies will be needed to experimentally determine the precise details of the conformational differences between WT-VSD and TS-VSD in order to test this hypothesis. Interestingly, differences in hydration also propagate along the S4–S5 linker, suggesting that there could be secondary effects of the two serine mutations that could further influence the temperature-dependent activity of the channel.
Previous studies have shown that introducing serine in a transmembrane helix embedded in a hydrophobic lipid bilayer is able to cause a water defect (52, 53), and serine within transmembrane helices is associated with helical bending and recruitment of water molecules to functionally important regions (54–56). Thus, our observation of increased hydration and structural changes in the C-terminal half of S4 in TS-VSD is consistent with the known properties of serine. Recently, a combination of MD simulations and experimental electrophysiology experiments with hyperpolarization-activated cyclic nucleotide-gated (HCN)-channel chimeras and mutants demonstrated that a break in the S4 helix mediated by a serine residue was critical for determining channel-gating behavior, and this helix break resulted in large differences in hydration of the lower crevice within the VSD between the resting and activated state (57, 58). Amino acid substitutions that substantially alter the hydrophobicity and helical stability in the lower portion of S4 may thus have a profound impact on the S4 conformational transition between the resting and activated states with consequent effects on channel gating as a function of both voltage and temperature. These results suggest that exact amino acid sequence, particularly the presence and location of polar residues in the C-terminal region of S4, may be important for determining the regulatory mechanism and setpoint for channel gating by sensor domains.
Materials and Methods
Sample Preparation.
WT-VSD was expressed and purified as described previously (24) and the TS-VSD was expressed and purified using the same protocol, as described below. The S2bot mutation and Arg to Lys mutations used for side-chain assignment were introduced into the pET15b VSD expression vector using standard quick-change mutagenesis protocols. All protein samples were overexpressed in Escherichia coli BL21 (DE3) pRARE in M9 minimal media and induced at optical density (OD) 0.8 to 1.0 with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 25 °C overnight (18 to 20 h) and purified into LPPG micelles using immobilized metal ion affinity chromatography and size-exclusion chromatography following the protocol described previously (24). For isotopic labeling with 13C, 15N, and/or 2H, the standard media components were substituted with 15NH4Cl, 13C-glucose, and/or D2O, respectively, plus 0.5 g/L of the corresponding isotopically labeled Isogro (Sigma-Isotec). Cell lysis was performed by sonication in buffer A (50 mM Tris pH 8.0, 100 mM KCl, 1 μg/mL pepstatin, 10 μM leupeptin, and 100 μM phenylmethylsulfonyl fluoride [PMSF]) supplemented with 1 mM ethylenediaminetetraacetic acid (EDTA), 5 mM β-me, 250 mM sucrose, and 1 mg/mL lysozyme. The cell debris and inclusion bodies were removed using a low-speed spin (20 min at 7,000 x g) and the membrane fraction collected with a high-speed spin (1 h at 150,000 x g). Membranes were solubilized with 1.5% wt/vol Foscholine-12 (Anatrace) in buffer A with 15 mM tris(2-carboxyethyl)phosphine (TCEP) for 2 h with rotation at room temperature. The insoluble fraction was removed by centrifugation at 30,000 x g for 30 min. Solubilized protein in foscholine-12 was purified using Ni-NTA His-bind beads (Protino) prewashed with buffer B (50 mM Tris pH 8.0, 100 mM KCl, and 15 mM TCEP) supplemented with 0.15% wt/vol Foscholine-12 and allowed to bind for 30 min at room temperature. The beads were washed with 10 bed volumes of buffer B, followed by 10 bed volumes of buffer C (buffer B plus 20 mM imidazole and 0.15% wt/vol Foscholine-12). The protein was then exchanged into the lysolipid LPPG on column by washing the beads with 15 bed volumes of buffer B supplemented with 0.1% wt/vol LPPG. Purified protein in LPPG was eluted with 5 bed volumes of buffer B supplemented with 400 mM imidazole and 0.2% wt/vol LPPG and dialyzed against TEV cleavage buffer (20 mM Tris pH 8.0, 50 mM KCl, 5 mM β-me) containing 0.0001% wt/vol LPPG. The His-tag was cleaved using TEV protease at 1:1 protein:TEV mole ratio overnight at room temperature, with cleavage verified by SDS-PAGE. Cleaved tag and uncleaved protein were removed using Ni-NTA His-bind resin, and the flow-through was concentrated to 0.5 mL and loaded onto a Superdex 200 column pre-equilibrated in NMR buffer (100 mM 3-morpholinopropane-1-sulfonic acid [MOPS] pH 7.0, 50 mM KCl, and 4 mM TCEP) with 0.2% wt/vol LPPG. Peak fractions were pooled and concentrated to 300 μL for NMR.
For experiments at different pH, samples were prepared in the same way except that the final Superdex 200 column buffer was 50 mM MOPS, 50 mM Bicine pH 7.0, 50 mM KCl, and 4 mM TCEP with 0.2% wt/vol LPPG instead. The final NMR sample pH was adjusted at the temperature of the NMR experiment (8 or 28 °C) using a micro pH electrode calibrated at that temperature.
NMR Experiments and Data Analysis.
NMR samples contained 0.2 to 0.5 mM protein in LPPG micelles with 10% D2O and 0.05% NaN3. The 1H-15N TROSY-HSQC, TROSY HNCO, TROSY HN(ca)CO, TROSY HNCA, TROSY HN(co)CA, TROSY HNCACB, and 15N-edited NOESY-TROSY spectra were recorded on a 750 MHz Bruker spectrometer with cryoprobe at the National Magnetic Resonance Facility at Madison (NMRFAM). Spectra were processed using nmrPipe (59) and analyzed with CcpNmr Analysis (60). Backbone walk experiments for assignment were supplemented with residue-specific labeling using 1–13C′-Val and Phe (61).
The secondary structure of WT-VSD and TS-VSD are predicted using TALOS+ (25) and PECAN (26), which yield similar predictions (Fig. 1D and SI Appendix, Fig. S2C). The secondary chemical shifts were calculated using NMRFAM-SPARKY (62). CSPs were determined using HNCO spectra of WT-VSD and TS-VSD according to the following equation:
MD Simulations.
The voltage-sensing domain in the crystal structure of the Shaker Potassium Channel Kv1.2 (Protein Data Bank, 3LUT) serves as the starting structure of the WT-VSD1.2 in the activated state. For the TS-VSD1.2, V154 and F155 are mutated to serines.
The systems are assembled using the CHARMM-GUI (63, 64) membrane builder. For each system, the starting structure is first embedded in a pre-equilibrated 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine lipid bilayer with a side length of 80 Å. Rectangular TIP3P water boxes with a dimension of 15 Å are then added to the top and the bottom of the lipid bilayer with periodic boundary condition, respectively. A total of 150 mM KCl ions are randomly placed in water to neutralize the system and mimic the physiological condition.
Simulations are performed with the OpenMM (65) package with graphics processing unit (GPU) acceleration using the CHARMM36m force field (66). Particle-mesh Ewald with an Ewald error tolerance of 0.0005 is used to calculate electrostatic interactions. Van der Waals interactions are treated with a nonbonded cutoff of 12 Å and a switch distance of 10 Å.
Each system is first minimized for 5,000 steps with the Limited-Memory Broyden–Fletcher–Goldfarb–Shanno algorithm. Then, a six-step equilibration in the constant particle number, volume, and temperature (NVT) ensemble is carried out with decreasing restraint forces on the backbone carbons, side chain heavy atoms, and the lipid bilayer (with both positional and dihedral restraints). Production simulations are carried out in the constant particle number, pressure, and temperature (NPT) ensemble for 1,000 ns at 298 K. The Langevin Integrator is used with a 1 fs time step and a collision frequency of 1 ps−1. MonteCarloMembraneBarostat is applied at 1 bar with an update frequency of every 100 steps. The x- and y-axes are treated isotropically, while the z-axis is updated independently. In equilibration and production runs, all water molecules are rigid, and all bonds involving hydrogen atoms are constrained using HBonds constraints in OpenMM. Overall translations and rotations are removed, and all frames are aligned against the crystal structure before any ensemble analysis.
Metadynamics Simulations.
The multiple-walkers well-tempered (MWWT) metadynamics method with a temperature bias factor of 10 is utilized to extensively sample the conformations near the mutation site. The collective variable is chosen as the fraction of native contact (44) (Q) between the backbone O of the i-th residue (i = 154 and 155) and the backbone HN of the (i + 4)th residue in one set of simulations, and the backbone HN of the (i + 3)th residue in another set of simulations. Accordingly, Q = 1 represents an ideal α-helix and an ideal 310-helix, respectively, in the two sets of MWWT simulations; Q = 0 corresponds to a random coil state.
where is the distance between atoms i and j, is the corresponding value in the reference conformation, S is the number of atom pairs; β is chosen to be 2 Å−1, and λ is set to be 1.8. All MWWT metadynamics simulations are performed using OpenMM 7.3.1 interfaced with PLUMED 2.5.3. A new Gaussian biasing potential is added every 1 ps with an initial height of 1 kJ/mol and a width of 0.05. A total of eight walkers are used for the WT-VSD1.2 and the TS-VSD1.2, respectively. Each walker is simulated for 90 ns in parallel while sharing hill history with other seven walkers every 1 ps, leading to 720 ns sampling for each system; to illustrate convergence, potentials of mean force computed with different amount of data are compared in SI Appendix, Fig. S10.
Supplementary Material
Acknowledgments
We thank Scott Wildman for generating the homology model of Shaker-VSD, Marco Tonelli and Claudia Cornilescu for technical assistance with NMR experiments, and Woonghee Lee for help with secondary chemical shift analysis. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH Grant P41GM103399 (NIGMS) (old number: P41RR002301). Equipment was purchased with funds from the University of Wisconsin–Madison, the NIH (P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, and S10RR029220), the NSF (DMB-8415048, OIA-9977486, and BIR-9214394), and the US Department of Agriculture. Computational resources from the Extreme Science and Engineering Discovery Environment, which is supported by NSF Grant Number OCI-1053575, are greatly appreciated; part of the computational work was performed on the Shared Computing Cluster, which is administered by Boston University’s Research Computing Services (http://www.bu.edu/tech/support/research/). Grant support was from the NIH R01NS081293 (B.C.) and R35NS116850 (B.C.) for the NMR studies and the NSF CHE-1829555 (Q.C.) for the molecular dynamics simulations.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017280118/-/DCSupplemental.
Data Availability
The backbone assignments of WT-VSD were previously deposited in the Biological Magnetic Resonance Data Bank (BMRB) (http://www.bmrb.wisc.edu/) as BMRB: 27866. The backbone assignments of TS-VSD have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) as BMRB: 28036. All study data are included in the article and/or SI Appendix.
References
- 1.Cynx J., Williams H., Nottebohm F., Hemispheric differences in avian song discrimination. Proc. Natl. Acad. Sci. U.S.A. 89, 1372–1375 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caterina M. J., Rosen T. A., Tominaga M., Brake A. J., Julius D., A capsaicin-receptor homologue with a high threshold for noxious heat. Nature 398, 436–441 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Bandell M., Macpherson L. J., Patapoutian A., From chills to chilis: Mechanisms for thermosensation and chemesthesis via thermoTRPs. Curr. Opin. Neurobiol. 17, 490–497 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Julius D., TRP channels and pain. Annu. Rev. Cell Dev. Biol. 29, 355–384 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Huffer K. E., Aleksandrova A. A., Jara-Oseguera A., Forrest L. R., Swartz K. J., Global alignment and assessment of TRP channel transmembrane domain structures to explore functional mechanisms. eLife 9, e58660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao E., Liao M., Cheng Y., Julius D., TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao M., Cao E., Julius D., Cheng Y., Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grandl J., et al., Pore region of TRPV3 ion channel is specifically required for heat activation. Nat. Neurosci. 11, 1007–1013 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers B. R., Bohlen C. J., Julius D., A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron 58, 362–373 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandl J., et al., Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat. Neurosci. 13, 708–714 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F., Swartz K. J., Jara-Oseguera A., Conserved allosteric pathways for activation of TRPV3 revealed through engineering vanilloid-sensitivity. eLife 8, e42756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F., et al., Heat activation is intrinsic to the pore domain of TRPV1. Proc. Natl. Acad. Sci. U.S.A. 115, E317–E324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brauchi S., Orta G., Salazar M., Rosenmann E., Latorre R., A hot-sensing cold receptor: C-terminal domain determines thermosensation in transient receptor potential channels. J. Neurosci. 26, 4835–4840 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordero-Morales J. F., Gracheva E. O., Julius D., Cytoplasmic ankyrin repeats of transient receptor potential A1 (TRPA1) dictate sensitivity to thermal and chemical stimuli. Proc. Natl. Acad. Sci. U.S.A. 108, E1184–E1191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B., Qin F., Single-residue molecular switch for high-temperature dependence of vanilloid receptor TRPV3. Proc. Natl. Acad. Sci. U.S.A. 114, 1589–1594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao J., Liu B., Qin F., Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc. Natl. Acad. Sci. U.S.A. 108, 11109–11114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlachová V., et al., Functional role of C-terminal cytoplasmic tail of rat vanilloid receptor 1. J. Neurosci. 23, 1340–1350 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito S., et al., Evolution of heat sensors drove shifts in thermosensation between Xenopus species adapted to different thermal niches. J. Biol. Chem. 291, 11446–11459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph J., Wang S., Lee J., Ro J. Y., Chung M. K., Carboxyl-terminal domain of transient receptor potential vanilloid 1 contains distinct segments differentially involved in capsaicin- and heat-induced desensitization. J. Biol. Chem. 288, 35690–35702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y., et al., Selective disruption of high sensitivity heat activation but not capsaicin activation of TRPV1 channels by pore turret mutations. J. Gen. Physiol. 139, 273–283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jara-Oseguera A., Bae C., Swartz K. J., An external sodium ion binding site controls allosteric gating in TRPV1 channels. eLife 5, e13356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh A. K., et al., Structural basis of temperature sensation by the TRP channel TRPV3. Nat. Struct. Mol. Biol. 26, 994–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury S., Jarecki B. W., Chanda B., A molecular framework for temperature-dependent gating of ion channels. Cell 158, 1148–1158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H., et al., NMR structural analysis of isolated shaker voltage-sensing domain in LPPG micelles. Biophys. J. 117, 388–398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eghbalnia H. R., Wang L., Bahrami A., Assadi A., Markley J. L., Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J. Biomol. NMR 32, 71–81 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Lee W., et al., PACSY, a relational database management system for protein structure and chemical shift analysis. J. Biomol. NMR 54, 169–179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berjanskii M. V., Wishart D. S., Application of the random coil index to studying protein flexibility. J. Biomol. NMR 40, 31–48 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Jensen M. O., et al., Mechanism of voltage gating in potassium channels. Science 336, 229–233 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Broomand A., Elinder F., Large-scale movement within the voltage-sensor paddle of a potassium channel-support for a helical-screw motion. Neuron 59, 770–777 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Starace D. M., Stefani E., Bezanilla F., Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron 19, 1319–1327 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Olson A. L., Cai S., Herdendorf T. J., Miziorko H. M., Sem D. S., NMR dynamics investigation of ligand-induced changes of main and side-chain arginine N-H’s in human phosphomevalonate kinase. J. Am. Chem. Soc. 132, 2102–2103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Englander S. W., Downer N. W., Teitelbaum H., Hydrogen exchange. Annu. Rev. Biochem. 41, 903–924 (1972). [DOI] [PubMed] [Google Scholar]
- 34.Henry G. D., Sykes B. D., Determination of the rotational dynamics and pH dependence of the hydrogen exchange rates of the arginine guanidino group using NMR spectroscopy. J. Biomol. NMR 6, 59–66 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Iwahara J., Jung Y. S., Clore G. M., Heteronuclear NMR spectroscopy for lysine NH(3) groups in proteins: Unique effect of water exchange on (15)N transverse relaxation. J. Am. Chem. Soc. 129, 2971–2980 (2007). [DOI] [PubMed] [Google Scholar]
- 36.André I., Linse S., Mulder F. A., Residue-specific pKa determination of lysine and arginine side chains by indirect 15N and 13C NMR spectroscopy: Application to apo calmodulin. J. Am. Chem. Soc. 129, 15805–15813 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Otting G., Wuethrich K., Studies of protein hydration in aqueous solution by direct NMR observation of individual protein-bound water molecules. J. Am. Chem. Soc. 111, 1871–1875 (1989). [Google Scholar]
- 38.Otting G., Liepinsh E., Wüthrich K., Protein hydration in aqueous solution. Science 254, 974–980 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Nucci N. V., Pometun M. S., Wand A. J., Site-resolved measurement of water-protein interactions by solution NMR. Nature Struct. Mol. Biol. 18, 245–249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halle B., Protein hydration dynamics in solution: A critical survey. Philos. Trans. R Soc. Lond. B Biol. Sci. 359, 1207–1223, discussion 1223-1204, 1323-1208 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melacini G., Kaptein R., Boelens R., Editing of chemical exchange-relayed NOEs in NMR experiments for the observation of protein-water interactions. J. Magn. Reson. 136, 214–218 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Braun D., Schmollngruber M., Steinhauser O., Revival of the intermolecular nuclear overhauser effect for mapping local protein hydration dynamics. J. Phys. Chem. Lett. 8, 3421–3426 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Kim M., et al., Evidence that the TRPV1 S1-S4 membrane domain contributes to thermosensing. Nat. Commun. 11, 4169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Best R. B., Hummer G., Eaton W. A., Native contacts determine protein folding mechanisms in atomistic simulations. Proc. Natl. Acad. Sci. U.S.A. 110, 17874–17879 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clapham D. E., Miller C., A thermodynamic framework for understanding temperature sensing by transient receptor potential (TRP) channels. Proc. Natl. Acad. Sci. U.S.A. 108, 19492–19497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J., Blunck R., The isolated voltage sensing domain of the Shaker potassium channel forms a voltage-gated cation channel. eLife 5, e18130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledwell J. L., Aldrich R. W., Mutations in the S4 region isolate the final voltage-dependent cooperative step in potassium channel activation. J. Gen. Physiol. 113, 389–414 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith-Maxwell C. J., Ledwell J. L., Aldrich R. W., Uncharged S4 residues and cooperativity in voltage-dependent potassium channel activation. J. Gen. Physiol. 111, 421–439 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soler-Llavina G. J., Chang T. H., Swartz K. J., Functional interactions at the interface between voltage-sensing and pore domains in the Shaker K(v) channel. Neuron 52, 623–634 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Xu Y., Ramu Y., Shin H. G., Yamakaze J., Lu Z., Energetic role of the paddle motif in voltage gating of Shaker K(+) channels. Nat. Struct. Mol. Biol. 20, 574–581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yifrach O., Hill coefficient for estimating the magnitude of cooperativity in gating transitions of voltage-dependent ion channels. Biophys. J. 87, 822–830 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacCallum J. L., Bennett W. F., Tieleman D. P., Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 94, 3393–3404 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johansson A. C., Lindahl E., Amino-acid solvation structure in transmembrane helices from molecular dynamics simulations. Biophys. J. 91, 4450–4463 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ballesteros J. A., Deupi X., Olivella M., Haaksma E. E., Pardo L., Serine and threonine residues bend alpha-helices in the chi(1) = g(-) conformation. Biophys. J. 79, 2754–2760 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilman H. R., Shi J., Deane C. M., Helix kinks are equally prevalent in soluble and membrane proteins. Proteins 82, 1960–1970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miyano M., Ago H., Saino H., Hori T., Ida K., Internally bridging water molecule in transmembrane alpha-helical kink. Curr. Opin. Struct. Biol. 20, 456–463 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Kasimova M. A., et al., Helix breaking transition in the S4 of HCN channel is critical for hyperpolarization-dependent gating. eLife 8, e53400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowgill J., et al., Bipolar switching by HCN voltage sensor underlies hyperpolarization activation. Proc. Natl. Acad. Sci. U.S.A. 116, 670–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delaglio F., et al., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 60.Vranken W. F., et al., The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Takeuchi K., Ng E., Malia T. J., Wagner G., 1-13C amino acid selective labeling in a 2H15N background for NMR studies of large proteins. J. Biomol. NMR 38, 89–98 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Lee J., et al., CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eastman P., et al., OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 13, e1005659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang J., et al., CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The backbone assignments of WT-VSD were previously deposited in the Biological Magnetic Resonance Data Bank (BMRB) (http://www.bmrb.wisc.edu/) as BMRB: 27866. The backbone assignments of TS-VSD have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) as BMRB: 28036. All study data are included in the article and/or SI Appendix.