Abstract
A faculty member’s ability to develop meaningful research-oriented laboratories in neurobiology is often hampered by the rapid pace of new technologies and the increasing cost of equipment. To help undergraduate neuroscience faculty meet these challenges, we introduce two important neuroscience research tools we designed and built. The first is a precision micromanipulator for neurophysiology applications costing less than $40 USD. We compare data generated using the DIY manipulator with commercial micromanipulators costing over $1000. The second tool is our newly designed 3D printed epi-fluorescence microscope. Commercial fluorescence imaging devices often cost over $20,000, but our 3D printed version is constructed for less than $1200. This epi-fluorescence microscope uses interchangeable LED light sources and filter sets to image static fluorescence in prepared slides and calcium imaging of neuronal activity in living Drosophila brains. This later technique uses transgenic flies with a genetically encoded calcium indicator, GCaMP, linked to green fluorescent protein (GFP). During an action potential, calcium ions (Ca2+) enter neurons and are observed as an increase in fluorescence intensity from a series of video images. These neuronal firing patterns can be assessed qualitatively and quantitatively to understand neural circuits leading to specific behaviors. We plan to develop curricula around the use of the epi-fluorescence microscope for calcium imaging in the next year, and to provide detailed parts sources and construction guides for the student and faculty DIY experience.
Keywords: micromanipulator, neurophysiology, epsp, epi-fluorescence, GFP, mCherry, GCaMP, Calcium imaging
This report is based on our presentation at the 2020 FUN Virtual Meeting where we introduced two new teaching and research tools for undergraduate neuroscience faculty. The rapid pace of new technologies and the increasing cost of equipment can restrict the development of meaningful research-oriented laboratories in neuroscience. However, the present availability of inexpensive industrial parts, efficient supply chains for parts distribution, 3-D printing and open source software development can facilitate faculty development of do-it-yourself (DIY) laboratory teaching and research tools to overcome curricular and research financial limitations.
Previously the Cornell Hoy Neurobiology group and collaborators have designed a variety of inexpensive or free tools for neuroscience teaching and research laboratories. These include an extracellular amplifier (Land et al., 2001), suction electrodes (Land et al., 2001; Johnson et al., 2007), a temperature control device (Krans and Hoy, 2005), micromanipulators (Krans et al., 2006), physiological stimulators (Land et al., 2004), LED control boxes for optogenetic stimulation (Pulver et al., 2010; Vilinsky et al., 2018), and electrophysiological data acquisition and analysis software (Lott et al., 2009). Here we describe our recent progress developing an inexpensive micromanipulator for student neurobiology teaching labs and research, and an inexpensive fluorescence imaging device for dynamic and static fluorescence visualization.
Precision micromanipulators from commercial sources cost ~$500 to over $1000 USD. The one described below can be constructed by faculty or students for about $40.00 in parts. Fluorescence microscopes with imaging cameras are expensive, often costing over $20,000 each. This cost often puts fluorescence microscopes out of reach for many undergraduate neuroscience courses and faculty/student research. Our imaging microscope can be constructed using about $1200 in parts. A major goal for our DIY work is to increase the toolbox for neuroscience teaching, learning and research for students and faculty, especially at national and foreign institutions with limited financial resources.
DIY EQUIPMENT
Precision Micromanipulator Design
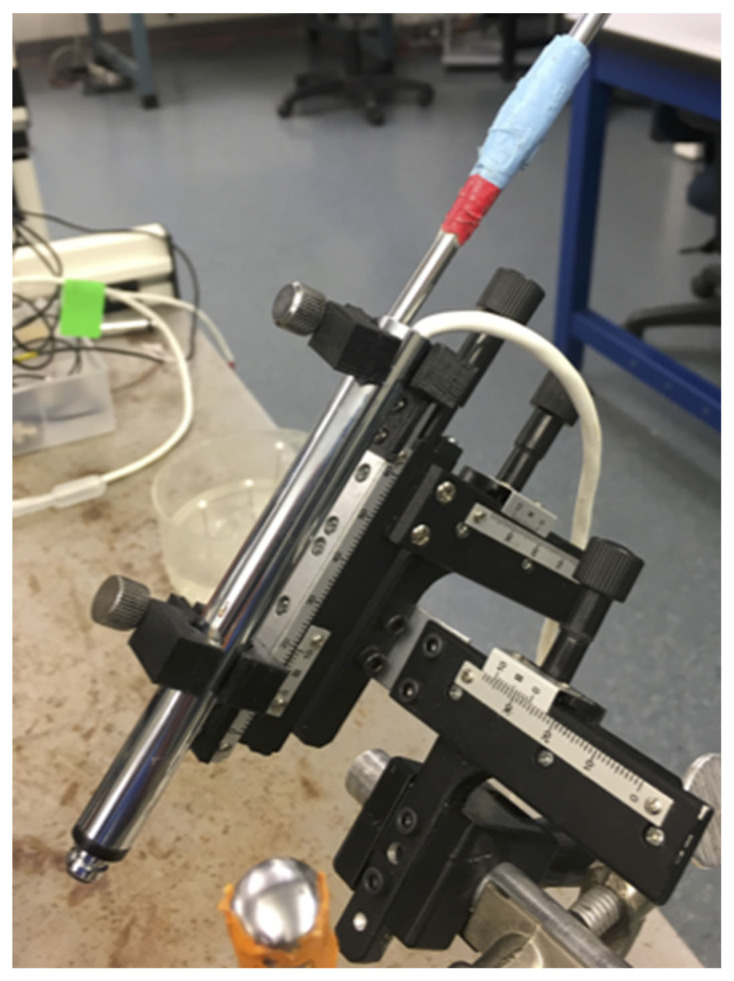
We designed an inexpensive micromanipulator that student lab teams could assemble from a kit and use for any laboratory exercise or research that requires fine probe placement. The precision micromanipulator can be built in approximately one hour. It consists of two modified x-y microscope stages, several 3D printed parts, and various nuts and bolts (Figure 1). The two x-y mechanical stages can be purchased online (Amazon, eBay) for under $20USD each. These stages have movement range of 60mm × 30mm and an accuracy of 0.1mm. The manipulator is constructed by first removing the slide holders from both x-y stages and then removing the y-axis from the x-axis on one of the two stages. The single x-axis is joined to the other x-y stage using an aluminum L-bracket with M3 screws and nuts. Finally, the completed x-y-z stage is epoxied and screwed into the 3D printed manipulator clamp. The 3D printed parts include a manipulator clamp, a thumb screw housing, and a set of 4 probe holder clamps (Figure 2). These parts are printed from .stl files using PLA, ABS, or similar filaments at 80% infill for added strength. The .stl files can be sent to a local 3D printer or uploaded to an online printing service such as CraftCloud. The 3D printed thumb screw is both threaded into the M6 hex cap and epoxied in place. The completed thumb screw is then threaded into a nut housed inside the 3D printed manipulator clamp. We are preparing parts lists, construction manuals and videos for future dissemination.
Figure 1.
The components for assembling the DIY precision micromanipulator. There are 2 x-y mechanical stages, an aluminum rod and L-bracket, a set of 3D printed parts, and miscellaneous nuts and bolts for assembly.
Figure 2.
The fully assembled micromanipulator with intracellular amplifier head-stage clamped in place.
DIY Micromanipulator Performance
We compared the stability and fine movement accuracy of our DIY manipulator with a much more expensive (~$1300) Narishige M333 micromanipulator that is normally used for student electrophysiological recordings in the “Principles of Neurophysiology” laboratory class at Cornell University (BioNB4910). A conventional electrophysiological rig (Wyttenbach et al., 2018) with 2 A-M Systems Model 1600 intracellular amplifiers, an ADInstruments Power Lab for data acquisition with LabChart software, and glass intracellular microelectrodes filled with 3M KCl (resistances 15–20 MOhm) were used to record and display the membrane potentials from crayfish muscle fibers. We recorded intracellular excitatory synaptic potentials (EPSPs) from two different crayfish superficial flexor (SF) muscle fibers with electrodes and amplifier headstages clamped to our DIY and the Narishige manipulators. Details of the saline composition, dissection and recording protocols for crayfish muscle are found in the Crawdad lab manual (Wyttenbach et al., 2014)
Figure 3 shows EPSPs recorded simultaneously from different muscle fibers using the two manipulators. Both muscle fibers were easily penetrated, and both recordings were stable and of similar fidelity. Thus, the DIY manipulator has the precision and stability to record intracellular potentials. The EPSP amplitudes recorded with the Narishige M333 micromanipulator (blue trace) have higher amplitudes than those recorded with our DIY manipulator (red trace), probably because the electrode of the blue recording was closer to a synaptic site. These muscles do not fire action potentials, and EPSP amplitude decays with distance from the synaptic site (Hoyle, 1983). The red and blue recordings in Figure 3 are not the same EPSPs because the SF muscles are poly-neuronally and multi-terminally innervated by different, spontaneously active motor neurons (Atwood, 2008).
Figure 3.
Simultaneous intracellular recordings of excitatory postsynaptic potential (EPSPs) from 2 different crayfish muscle fibers. Top red trace DIY manipulator; resting potential, −84 mV; calibration bars, 1 mV. Bottom blue trace, Narishige M3330 manipulator; resting potential, −75 mV; calibration bar, 2 mV. Note higher gain in red trace. Horizontal calibration bar, 500 ms.
Epi-fluorescence Microscope Design
The goal here was to design an affordable epi-fluorescence microscope that small student teams could also assemble from a kit and use to bring cutting edge research into the undergraduate neuroscience curriculum. To meet this goal, we designed and built an epifluorescence microscope for approximately $1200 USD. The 3D printed epi-fluorescence microscope described here is suitable for many neuroscience applications including calcium imaging in living neurons.
The scope consists of several off the shelf components coupled with a set of 3D printed parts for the optical housing (Figure 4). The base of the scope is an aluminum optical breadboard drilled for M6 metric threads. Attached to this base is a 2×2 inch extruded aluminum column with t-slots which carries the 3D printed optical components and a precision lab jack for raising and lowering the specimen stage.
Figure 4.
The epi-fluorescence microscope with LED controller.
Attached to the top of the precision lab jack is a clear plexiglass plate (130 × 190 mm) that holds the specimen slide or dish. Positioning of the specimen in the x-y planes is achieved by a standard x-y mechanical microscope stage mounted to the upper surface of the plexiglass stage. The lab jack allows the entire stage assembly to be lowered easily to replace specimens or add fluids.
On the vertical column, the microscope uses a linear stage with micrometer adjustment for precision focusing the objective lens. Attached to the linear stage is the 3D printed optical housing. The main housing contains the fluorescence filters and has a male c-mount on top for attaching the CMOS camera and a female c-mount on the bottom for the 20x long working distance objective lens.
The monochrome global shutter CMOS camera (~$180 from Basler, model da1280-54um/uc) is capable of acquiring images in low light at up to 54 frames per second. This camera has a resolution of 1280×960 pixels and uses a 1/3″ Aptina AR0134 sensor. However, any camera with a c-mount can be attached to this port. Inside the main housing is a double filter slider that accepts two sets of fluorescence filters. The excitation and emission filters are 25mm diameter and the dichroic mirror (beamsplitter) is 25.5×36mm. The calcium imaging described below used a single band filter set tailored for green fluorescent protein (GFP) from Iridian Spectral Technologies: 469-35nm excitation, 497nm dichroic mirror, and 525-39nm emission filter ($450 USD). A second filter set for mCherry or Alexa Fluor 595 can be added to the filter slider if desired. In addition, these filter sliders are very inexpensive to 3D print, allowing the user to have several sliders and filter combinations ready to swap out in seconds.
The light source for the scope is an interchangeable high-power LED with heat sink connected to an LED controller box powered by either AC or DC and capable of being driven by a computer stimulus generator via a BNC cable (Figure 4). The wiring diagram for the LED controller has been described previously (Pulver et al., 2011 Vilinsky et al., 2018). For GFP a Cree XLamp XP-E2 high power blue (470nm peak, $4USD, LED Supply.com) LED is coupled to a BuckPuck DC LED driver ($20USD, LEDSupply.com). The LED and a 20mm narrow spot LED lens are housed in a 1 inch diameter aluminum heat sink ($15USD, LEDSupply.com). The LED and its heat sink housing are inserted into the 3D printed LED port. This design allows rapid exchange of different colored LEDs.
The 3D printed parts were designed using the free, online CAD program TinkerCad (AutoDesk). TinkerCad allows you to build complex parts from a library of basic shapes or to import and modify existing .stl files. Finished parts are exported as .stl files for 3D printing (Figure 5). The .stl files can be printed on any 3D printer. However, printing these parts using an online print company such as CraftCloud allow greater choice of print materials. The optical housing described here was printed using MJF Nylon or resin. These materials are stronger and provide higher detail than that of PLA or ABS plastics.
Figure 5.
3D printable microscope parts shown in TinkerCad prior to export as .stl files. A) Main optical housing, B) Double filter slider, and C) the filter holder components.
Finally, the microscope is modular in design. Removing the fluorescence filters and adding an LED light source from below the specimen converts the scope into a conventional light microscope. Screw a post and micromanipulator into the base and it becomes a neurophysiology rig.
Fly Stocks for Imaging Preps
Fluorescent Drosophila lines were raised on standard cornmeal-molasses food. The stocks used were UAS GCaMP7b; P{GMR94G06-GAL4}attP2 for calcium imaging from the Bloomington Stock Center (#80907and #40701) and jus-GAL4 UAS-CD8-GFP (available from D. Deitcher) for static fluorescent imaging of membrane-bound GFP.
Larval and CNS Preparation
Wandering third instar larvae were selected with forceps, placed in a Sylgard petri dish and rinsed with water, then pinned at the mouth hooks and posterior end with fine insect pins with the dorsal (trachea) side oriented up. Room temperature saline (135 mM NaCl, 5 mM KCl, 2 mM CaCl2, 4 mM MgCl2, 5 mM BES, pH 7.5) for calcium imaging was added to the preparation. A superficial incision is made from the tail to the mouth hook with micro-scissors and the “guts” are carefully removed to reveal the CNS composed of brain lobes and ventral ganglion. Additional pins are placed to flatten the cuticle flaps into a “filet” (Pulver et al., 2011).
For static fluorescent imaging, the Sylgard disc was adhered to a glass slide with double-sided tape and then imaged using appropriate camera settings. For dynamic calcium imaging, the CNS was excised from the larval filet and gently placed on a Sylgard-coated glass depression slide and covered with saline. The slide was placed on the microscope stage. Initially the camera was set to autoexposure to locate the preparation, then manually adjusted to reveal the GCaMP7b labeled motoneurons.
Flourescent Imaging
To test the epifluorescence microscope, we first attempted to image neurons expressing GFP in the ventral ganglion of the larva. Figure 6 shows static GFP imaging of a ventral ganglion of a third instar larva expressing the membrane-bound GFP fusion, CD8-GFP in selected neurons driven by the GAL4/UAS binary expression system. CD8-GFP is a useful label to highlight the morphology of neurons.
Figure 6.
Expression of membrane-bound GFP in ventral ganglion neurons that express the neural gene, julius seizure (jus).
Next, to show the utility of the microscope we imaged a ventral ganglion expressing the calcium-sensitive protein GCaMP7b in a pair of motor neurons in each segment. Images were sent at 10 fps for 300 seconds from the camera to a lap top computer, using camera supplied software, and converted into an AVI format with Fiji (Image J) software. Figure 7 shows two frames of dynamic imaging of actively firing motor neurons in the abdominal ganglia of a fruit fly larvae from the video link (https://youtu.be/zB55QNQ_IrM). For a short while after the larval fly’s CNS is removed, CNS motor networks will produce spontaneous neural activity that is correlated with larval crawling and turning (fictive locomotion, Pulver et al., 2015). Action potential-induced depolarization results in calcium entry which is detected by the genetically encoded calcium indicator, GCaMP that is expressed in selective motoneurons. Thus, calcium-induced fluorescence during neural activity is a proxy for identifying actively firing neurons that normally drive muscles to execute larval locomotion. These neuronal firing patterns can be assessed qualitatively and quantitatively to understand neural circuits leading to specific behaviors.
Figure 7.
GCaMP7b fluorescence in select motoneurons of the ventral ganglion. Two single frames taken from series illustrate waves of neural activity during fictive locomotion. A. Earlier fame shows cell at red arrow with low light intensity. B. Later frame shows higher intensity light emission from the same neuron, indicating greater electrical excitability. See short video link (https://youtu.be/zB55QNQ_IrM) for entire series of images.
DISCUSSION
We present two new teaching and research tools as “proof of concept” of our recent DIY efforts to increase the faculty toolbox for neuroscience teaching, learning and research. Our DIY manipulator performed well compared to the commercial Narishige M333 when recording synaptic potentials. The DIY manipulator could be used for any student lab exercise or research that requires fine probe placement, such as for cell injection of dyes or genetic material. Although students and faculty have not yet formally tested and assessed it, initial and limited feedback for manipulator assembly and use was positive with constructive criticisms from faculty participating in the January 2020 CrawFly workshop in San Antonio. A manipulator made by one of the 2020 CrawFly participants was used to record miniature end-plate potentials from fruit fly larval muscle during the workshop (data not saved). The inexpensive manipulator could expand the range of electrophysiological exercises possible in a student/faculty lab. For example, faculty could add conventional intracellular and whole cell patch recording to student electrophysiology exercises. Our students will construct and use the DIY manipulator for electrophysiological student lab exercises. Learning outcomes, specific student skill objectives, and data analysis suggestions for our electrophysiology exercises are found in the Crawdad lab manual (Wyttenbach et al., 2014).
Our initial results of imaging static GFP fluorescence in fixed tissue and dynamic GFP fluorescence from living neurons during fictive larval locomotion (Pulver et al., 2015) show that our fluorescence microscope is operationally ready to be brought into the student laboratory classroom and into faculty research labs. Any type of fluorescence tissue could be imaged with the appropriate excitation and emission filters. Our fluorescence microscope facilitates unique and high powered research experiences for students and faculty with limited financial resources. We are not the first to propose inexpensive fluorescence imaging for the student neuroscience lab or for research. Our imaging microscope design was inspired by previous efforts including from the “FlyPi” group (Chagas et al., 2017), the miniscope produced by Zhang et al. (2019) and an imaging system recently adapted to a classroom compound microscope (Sane et al., 2020- this JUNE issue). For an imaging microscope design that fit our needs, we searched for the least expensive, yet high quality options for the microscope imaging components, light filters, camera, microscope and mounting stage parts, and JR designed the 3-D printed optical housing, filter holders, and slider.
Our student learning objectives and skills for the imaging microscope and dynamic imaging project will promote interdisciplinary thinking and skills in our neuroscience students. For example, they will study the excitable properties of neurons though calcium imaging as a proxy for signal transmission by action potentials, and the advantages of imaging for exploring neuronal circuit activity (Yang and Yuste, 2017). Students will examine the properties of light and optics, and understand principles of fluorescence imaging (Cox, 2012) as they build and use their imaging microscopes to measure fluorescence in living preparations expressing fictive locomotion. They will learn introductory coding to control camera software and analyze images, and practice engineering design and construction by building the microscope. This includes 3D printing, soldering, fine scale assembly, and troubleshooting. Finally, students will perfect their dissection and dexterity skills by preparing Drosophila larval nervous systems for imaging.
We are still preparing imaging analysis protocols appropriate for our undergraduate student use. μManager software can be used to gather the imaging movies from the camera, and image J to process the images and select “regions of interest”. Data analysis programs such as Dataview (Heitler, 2009; https://www.standrews.ac.uk/~wjh/dataview/) can be used to visualize and measure fluorescent amplitude and activity patterns (see Sane et al., 2020, this JUNE issue). Our imaging software protocols, in prep, will be designed for student interaction to modify Python code at various phases of image acquisition, analysis and visualization.
We hope that our goals for the imaging microscope project in particular will be broadly applicable at other institutions. The National Research Council (2003) emphasizes that future science research progress will require greater STEM interdisciplinary training. We plan to create an interdisciplinary classroom where students from different STEM studies work together to develop the confidence to build and understand equipment and solve biological problems. Interdisciplinary science education is considered critical for the future recruitment of talented scientists (Kazar and Elrod, 2012). Our imaging project promotes interdisciplinary learning with engineering design, physics (optics and light principles), biology (genetics and neuroscience), and computer science (introductory coding to control camera software and analyze images). Team-based learning skills required for research (Bennett and Gadlin, 2012) will be fostered as student teams build, test, and trouble shoot their own instrumentation, and gather and analyze data to answer biological questions. Too often our students treat instrumentation as a “black box”, with little understanding of how scientific equipment works. We are in the process of developing written and video guides for construction and use of the manipulator and the imaging microscope, along with laboratory modules to guide inquiry driven research experiences for students (Fromherz et al., 2018).
Our plans for a Spring 2020 rollout of student team construction and use of the manipulator and imaging microscope in the laboratory classroom were derailed by the Covid-19 pandemic, but we will continue our curriculum development for both Cornell and Hobart and William Smith students this Spring 2021. We will follow inclusive classroom practices of clearly structured assignments and expectations, and emphasize that all ideas are welcome, respected and open for discussion (Penner, 2018). We will assess student interdisciplinary awareness (Basu et al., 2017; Crisp and Muir, 2012) before and after imaging microscope use, and student perceived experience and confidence in building and using the imaging microscope (Glover and Luzon, 2018). Our student learning outcomes will be examined by grading student mastery of microscope construction, designing and testing hypotheses of neuronal network activity through data gathering and interpretation, student written journal-like articles and oral presentations on their work. Our project results will be disseminated through additional journal articles and faculty workshops such as CrawFly (https://www.adinstruments.com/training/education/application-workshops?field_event_type_tid=2977).
Footnotes
This work was funded by a grant from the Cornell Center for Teaching Innovation (DD and BRJ), and support from Hobart-William Smith Colleges (JR). We thank Dr. Stefan Pulver of the University of St. Andrews in Scotland for DIY inspiration and advice.
REFERENCES
- Atwood HL. Parallel ‘phasic’ and ‘tonic’ motor systems of the crayfish abdomen. J Exp Biol. 2008;211:2193–2195. doi: 10.1242/jeb.010868. [DOI] [PubMed] [Google Scholar]
- Basu A, Mondoux MA, Whitt JL, Isaacs AK, Narita T. Integrative approach to STEM concepts in an introductory neuroscience course: Gains in interdisciplinary awareness. J Undergrad Neurosci Educ. 2017;16(1):A102–A111. [PMC free article] [PubMed] [Google Scholar]
- Bennett ML, Gadlin H. Collaboration and team science: From theory to practice. JIM. 2012;60:768–775. doi: 10.231/JIM.0b013e318250871d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagas AM, Prieto-Godino LL, Arrenberg AB, Baden B. The €100 lab: A 3D- printable open-source platform for fluorescence microscopy, optogenetics, and accurate temperature control during behaviour of zebrafish, Drosophila, and Caenorhabditis elegans. PLoS Biol. 2017;15(7):e2002702. doi: 10.1371/journal.pbio.2002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. Optical imaging techniques in cell biology. Boca Raton, FL: CRC Press LLC; 2012. Fluorescence and fluorescence microscopy; pp. 35–48. [Google Scholar]
- Crisp KM, Muir GM. Assessing development of an interdisciplinary perspective in an undergraduate neuroscience course. J Undergrad Neurosci Educ. 2012;10(2):A88–95. [PMC free article] [PubMed] [Google Scholar]
- Fromherz S, Whitaker-Fornek J, Andrew A, Sharp AA. Classroom-based research experiences to support underserved STEM student success: From introductory inquiry to optogenetics in the embryonic chicken. J Undergrad Neurosci Educ. 2018;17(1):A97–A110. [PMC free article] [PubMed] [Google Scholar]
- Glover EM, Lauzon O. Using a contrast illusion to teach principles of neural processing. J Undergrad Neurosci Educ. 2018;17(1):A81–A88. [PMC free article] [PubMed] [Google Scholar]
- Heitler WJ. Practical tools for analysing rhythmic neural activity. J Neurosci Methods. 2009;185:151–64. doi: 10.1016/j.jneumeth.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Hoyle G. Muscles and their neural control. New York, NY: John Wiley and Sons; 1983. pp. 483–525. [Google Scholar]
- Johnson BR, Hauptman SA, Bonow RH. Construction of a simple suction electrode for extracellular recording and Stimulation. J Undergrad Neurosci Educ. 2007;6(1):A21–A26. [PMC free article] [PubMed] [Google Scholar]
- Kazar A, Elrod S. Facilitating interdisciplinary learning: Lessons from Project Kalioedoscope. Change. 2012;44(1):16–25. [Google Scholar]
- Krans JL, Hoy RR. Tools for physiology labs: An inexpensive means of temperature control. J Undergrad Neurosci Educ. 2005;4(1):A22–A26. [PMC free article] [PubMed] [Google Scholar]
- Krans J, Gilbert C, Hoy R. Teaching insect retinal physiology with newly designed, inexpensive micromanipulators. Adv Physiol Educ. 2006;30:254–261. doi: 10.1152/advan.00029.2006. [DOI] [PubMed] [Google Scholar]
- Land BR, Wyttenbach RA, Johnson BR. Tools for the student physiology lab: An inexpensive high-performance amplifier and suction electrode for extracellular recording. J Neurosci Meth. 2001;106:47–55. doi: 10.1016/s0165-0270(01)00328-4. [DOI] [PubMed] [Google Scholar]
- Land BR, Johnson BR, Wyttenbach RA, Hoy RR. Tools for Physiology Labs: Inexpensive equipment for physiological stimulation. J Undergrad Neurosci Educ. 2004;3:A30–A35. [PMC free article] [PubMed] [Google Scholar]
- Lott GK, III, Johnson BR, Bonow RH, Land BR, Hoy RR. g-PRIME: A free, windows based data acquisition and event analysis software package for physiology in classrooms and research labs. J Undergrad Neurosci Educ. 2009;8(1):A50–A54. [PMC free article] [PubMed] [Google Scholar]
- National Research Council. BIO2010: Transforming undergraduate education for future research biologists. 2003. Available at https://www.nap.edu/catalog/10497/bio2010-transforming-undergraduate-education-for-future-research-biologists. [PubMed]
- Penner MR. Building an inclusive classroom. J Undergrad Neurosci Educ. 2018;16(3):A268–A272. [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Hornstein NJ, Land BL, Johnson BR. Optogenetics in the teaching laboratory: Using channelrhodopsin-2 to study the neural basis of behavior and synaptic physiology in Drosophila. Adv Physiol Educ. 2011;35(1):82–91. doi: 10.1152/advan.00125.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Bayley TG, Taylor AL, Berni J, Hedwig B. Imaging fictive locomotor patterns in larval Drosophila. J Neurophysiol. 2015;114(5):2564–77. doi: 10.1152/jn.00731.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth JRH, Sane V, Gather M, Pulver SR. Inexpensive Methods for Live Imaging of Central Pattern Generator Activity in the Drosophila Larval Locomotor System. J Undergrad Neurosci Educ. 2020;19(1):A124–A132. [PMC free article] [PubMed] [Google Scholar]
- Vilinsky I, Hibbard KL, Johnson BR, Deitcher DL. Probing synaptic transmission and behavior in Drosophila with optogenetics: A laboratory exercise. J Undergrad Neurosci Educ. 2018;16(3):A289–A295. [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach RA, Johnson BR, Hoy RR. Crawdad: An Online Lab Manual for Neurophysiology. Sinauer Associates, Inc; Sunderland, MA: 2014. [Google Scholar]
- Wyttenbach RA, Johnson BR, Hoy RR. Reducing the cost of electrophysiology in the teaching laboratory. J Undergrad Neurosci Educ. 2018;16(3):A277–A281. [PMC free article] [PubMed] [Google Scholar]
- Yang W, Yuste R. In vivo imaging of neural activity. Nat Methods. 2017;14(4):349–359. doi: 10.1038/nmeth.4230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Bo Liang B, Barbera G, 1, Hawes S, Zhang Y, Stump K, Baum I, Yang Y, Li Y, Lin D-T. Miniscope GRIN lens system for calcium imaging of neuronal activity from deep brain structures in behaving animals. Curr Protocols Neurosci. 2019;86:e56. doi: 10.1002/cpns.56.. [DOI] [PMC free article] [PubMed] [Google Scholar]