Abstract
Gas vesicles (GVs) are cylindrical or spindle‐shaped protein nanostructures filled with air and used for flotation by various cyanobacteria, heterotrophic bacteria, and Archaea. Recently, GVs have gained interest in biotechnology applications due to their ability to serve as imaging agents and actuators for ultrasound, magnetic resonance and several optical techniques. The diameter of GVs is a crucial parameter contributing to their mechanical stability, buoyancy function and evolution in host cells, as well as their properties in imaging applications. Despite its importance, reported diameters for the same types of GV differ depending on the method used for its assessment. Here, we provide an explanation for these discrepancies and utilize electron microscopy (EM) techniques to accurately estimate the diameter of the most commonly studied types of GVs. We show that during air drying on the EM grid, GVs flatten, leading to a ~1.5‐fold increase in their apparent diameter. We demonstrate that GVs' diameter can be accurately determined by direct measurements from cryo‐EM samples or alternatively indirectly derived from widths of flat collapsed and negatively stained GVs. Our findings help explain the inconsistency in previously reported data and provide accurate methods to measure GVs dimensions.
Keywords: critical collapse pressure, cryo‐electron microscopy, diameter, electron microscopy, gas vesicles, negative staining
1. INTRODUCTION
Gas vesicles (GVs) are hollow, gas‐filled protein nanostructures natively expressed in certain types of cyanobacteria, heterotrophic bacteria, and Archaea as a buoyancy aid. 1 Recently, it was discovered that the unique physical properties of GVs enable them to serve as genetically encodable contrast agents for ultrasound and other imaging methods, allowing deep tissue imaging of cellular function. 2 , 3 , 4 , 5 , 6 , 7 , 8 In addition, GVs are being applied to acoustic manipulation and therapeutic uses of engineered cells. 9 , 10
Fully formed GVs adopt two predominant shapes—cylinders with conical ends or spindle‐like. The GVs may be 0.1–2 μm in length, or even longer when heterologously expressed in more spacious mammalian cells. 6 The mean diameter of GVs isolated from different species widely varies, but is relatively constant for the same type of GV. There is an inverse correlation between diameter and critical collapse pressure. 11 This correlation has important evolutionary consequences. While wider GVs can provide buoyancy at a lower energetic cost, they collapse at lower pressure. This is perhaps best reflected by analyzing the widths and collapse pressure of GVs isolated from Planktothrix spp. from Nordic lakes of different depths. 12 , 13 Three types of GVs isolated from Planktothrix spp. had widths of ~51, 58, and 67 nm with respective collapse pressures of 1.1, 0.9, and 0.7 MPa, allowing them to adapt to the hydrostatic pressure in different lakes. 12 , 14
Despite the importance of GVs' diameter for their biophysical properties, there are significant discrepancies in values reported in the literature. For example, the width of GVs from Anabaena flos‐aquae (Ana) measured inside cells by thin‐section electron microscopy (EM) was ~70 nm, 15 which is considerably smaller than the value obtained by negative stain EM (ns‐EM) for isolated GVs—136 nm. 16 Similar discrepancies can be observed for GVs from Halobacterium salinarum (Halo), whose reported values range from 45 to 250 nm. 16 , 17 , 18 , 19 To some extent, these discrepancies could be explained by natural variability in diameter. However, analysis of width distributions for GVs from several species of cyanobacteria 11 or Bacillus megaterium (Mega) 20 shows a narrow range. This inconsistency in diameter measurement was investigated almost 50 years ago by Walsby. 15 He observed that Ana GVs have a constant width of 70 nm when measured inside cells by thin‐section EM, which was close to the value measured for the purified sample imaged using a freeze‐etching technique (75 nm). In contrast, estimations by ns‐EM ranged from 70 to 114 nm. 15 He suggested that the stain used in EM leads to swelling of GVs, which increases their diameter but has little effect on the length. As an alternative approach for assessing GV diameter, Walsby proposed indirect measurement based on the widths of flat collapsed GVs. The diameter of Ana GVs measured using this strategy was ~85 nm. 21 Archer and King gave another potential explanation for discrepancies in GV measurements. They proposed that the isolation process leads to deformations, increasing the width of GVs. 22 Regardless of these concerns, the diameter of GVs has been routinely assessed for isolated specimens by ns‐EM.
As GVs have attracted more attention in biotechnology applications, accurate estimates of their diameter have become a critical input into GV engineering. For that reason, we investigated the discrepancies in reported GV diameters using modern microscopy tools. Using these updated techniques, we provide measurements for the most commonly studied GVs: Ana, Mega, and Halo. For Halo, we analyzed two different GV types, which are products of the independent gene clusters p‐vac and c‐vac.
2. RESULTS AND DISCUSSION
To more closely evaluate the behavior of stained and air‐dried GVs on the EM grid, we collected projection images for different types of GVs at 0° and 50° tilt and analyzed their morphology (Figure 1a,b). Although we predicted some degree of distortions to the cylindrical shape of GVs, the observed differences were unexpectedly large. For Ana GVs, there was an average of ~55 nm width difference between measurements at these two angles. The pattern was similar for both Mega and Halo GVs, although to a different degree. This data indicates that all types of GVs flatten during the staining procedure, adopting an elliptic cylinder shape.
FIGURE 1.
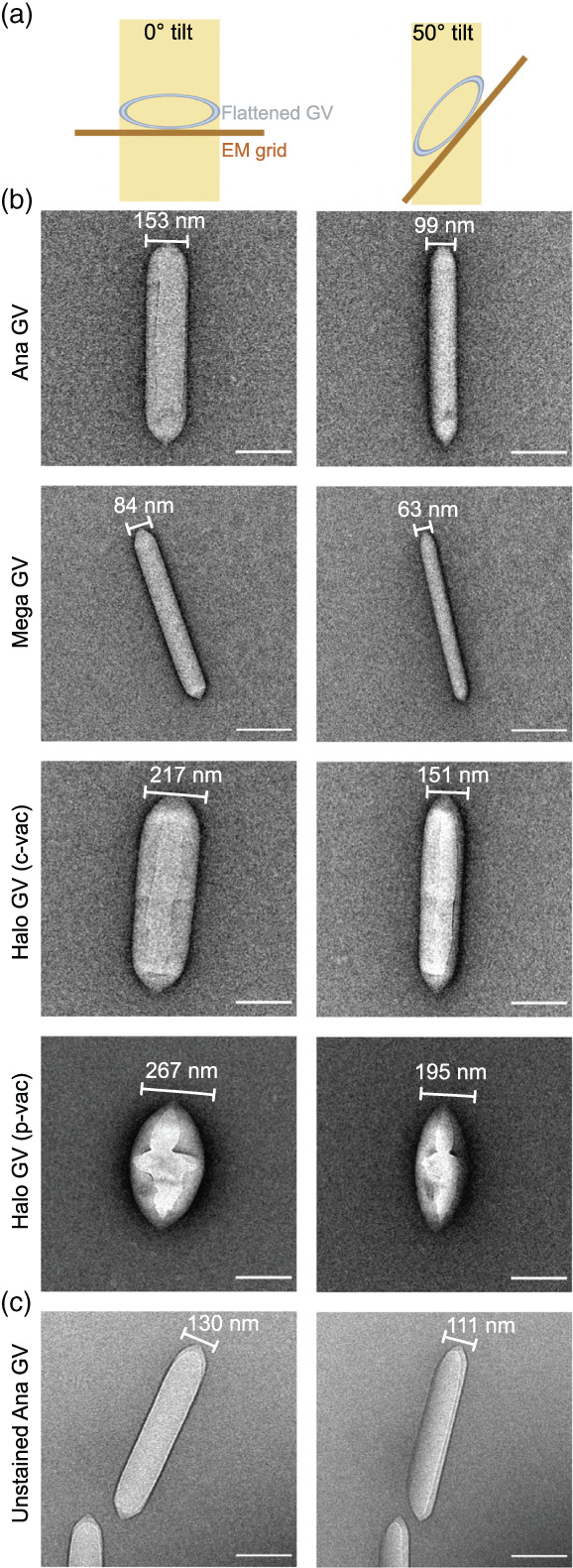
Gas vesicles (GVs) flattening on the electron microscopy (EM) grid. (a) Schematic showing cross‐section of the flattened GV at 0° and 50° tilt. (b, c) Representative projection images at 0° and 50° tilt for (b) negatively stained and air‐dried Ana, Mega, and Halo GV; and (c) unstained, air‐dried Ana GV. Scale bar, 200 nm
Certain limitations of the ns‐EM technology, such as specimen flattening or stain thickness irreproducibility, were previously described. 23 However, the observed deformation of the GV protein shell is not like the typical flattening reported before, where sample was mainly affected in z‐direction with little to no effect on the x,y‐dimensions. 23 Since GVs produce strong contrast on EM even without staining, we decided to take advantage of this unique property and evaluate the effect of the stain. Analysis of unstained, air‐dried Ana GVs samples at 0° and 50° tilts show on average ~20 nm difference in diameter (Figure 1c), which is significantly less than the stained sample, but not negligible.
Distortions to the GV shape are the effect of the unique mechanical properties of GVs' protein shell. In ns‐EM, the sample lies on a carbon support; thus, we suspect that GVs are compressed by the surface tension of evaporating water. Notably, the degree of deformation appears to be correlated with critical collapse pressure. Halo GVs, which experience the most flattening, are also the least robust among investigated GVs, with collapse pressure of 0.1 Mpa. 16 In contrast, Mega GVs, which have a much higher collapse pressure of ~0.7 Mpa, 16 flatten the least.
To obtain more accurate measurements of GV diameter, we used two complementary methods. First, we imaged the GVs with cryo‐EM, which preserves GVs' cylindrical shape. Unfortunately, cryo‐EM is a more demanding technique, requiring time‐consuming sample optimization, larger sample quantities, and access to a more sophisticated instrument. Alternatively, we inferred GV diameter from the widths of flat collapsed GVs with negative staining, as measured by Walsby and Bleything. 21 This method, which equates the collapsed GV width with half of the intact cylindrical circumference, should allow for a faster and more accessible estimation of GV dimensions. We decided to analyze diameter distribution for Mega, Ana, and Halo GVs using both strategies.
Cryo‐EM of intact GVs and collapsed ns‐GV imaging resulted in similar values for each analyzed GV type (Figure 2, Table 1), with differences within statistical error. Mega and Ana GVs appear to have a uniform diameter, varying within a narrow range (Figure 2c, Table 1). In contrast, Halo GV diameters varied. Halo is capable of producing two types of GVs. Spindle‐shaped GVs are encoded by the p‐vac gene cluster located on an endogenous plasmid, while the c‐vac cluster located on a mini‐chromosome generates cylindrical GVs. 19 According to our measurements, the diameter of both types of Halo GVs varies (Figure 2c). However, some of this variability may be due to imperfect classification. All GV types begin their assembly as bicones, which look like smaller spindle‐shape p‐vac Halo GVs. 19 Thus, some c‐vac GVs, in their bicone phase, could have been classified as p‐vac GVs. This misclassification could have made a minor contribution to the overall diameter distribution. Overall, the range of diameter values for different GV types suggest that Ana and Mega GVs have tighter regulation over diameter compared to Halo GVs. However, it is not yet known what the physiological consequences of this regulation are or how exactly the diameter is adjusted in growing GVs.
FIGURE 2.
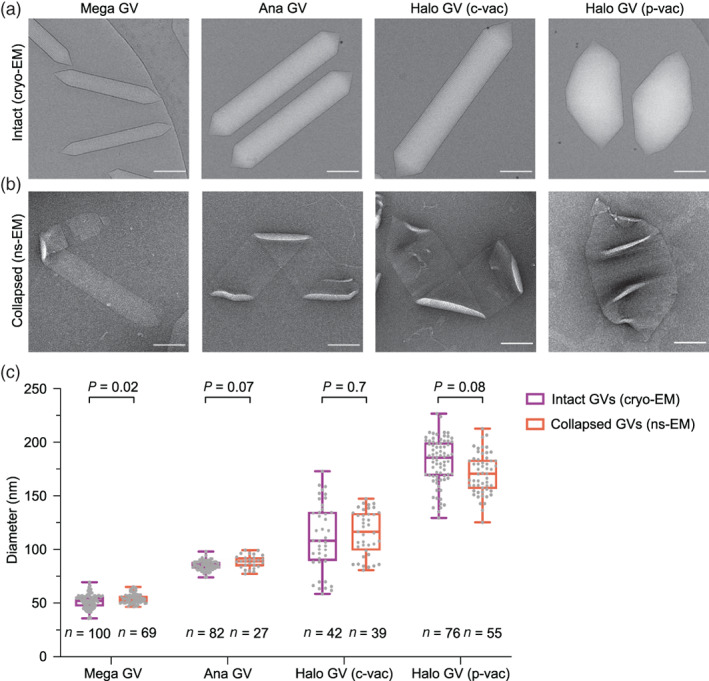
Diameter determination for Mega, Ana, and Halo gas vesicles (GVs). (a) Representative cryo‐electron microscopy (EM) of intact GVs used for direct diameter measurement. (b) Representative ns‐EM images of collapsed GVs used for indirect diameter assessment based on widths of flat collapsed regions. Scale bar, 100 nm. (c) Diameter distribution for Mega, Ana, and Halo GVs measured by cryo‐EM and collapsed ns‐EM. Center line indicates median, the box limits denote the interquartile range and the whiskers absolute range. Each dot represents an individual measurement. Paired t test was performed between directly measured (cryo‐EM) and calculated (ns‐EM) diameters for each GV type
TABLE 1.
GV diameters (mean ± SD) obtained by three EM‐based methods
| GV type | Intact GVs (ns‐EM) a (nm) | Intact GVs (cryo‐EM) (nm) | Collapsed GVs (ns‐EM) (nm) |
|---|---|---|---|
| Mega | 73 ± 14 | 52 ± 6 | 54 ± 5 |
| Ana | 136 ± 21 | 85 ± 4 | 89 ± 6 |
| Halo (c‐vac) | 251 ± 51 | 111 ± 32 | 116 ± 21 |
| Halo (p‐vac) | 182 ± 22 | 171 ± 19 |
Abbreviations: EM, electron microscopy; GVs, gas vesicles.
Previously reported by Lakshmanan et al. 16
Taken together, our findings provide an explanation for discrepancies in previous GV diameter measurements reported in the literature. Although ns‐EM is routinely used to evaluate the morphology and dimensions of intact GVs, 16 , 20 , 24 , 25 , 26 our data show that this method causes GV flattening and inaccurate apparent diameter. Instead, cryo‐EM of intact GVs and ns‐EM of flat collapsed GVs provide correct dimensions that are mutually consistent between the two methods, as shown here for three commonly studied GVs variants.
3. MATERIALS AND METHODS
3.1. GV preparation
GVs were either isolated from native sources (Ana and Halo) or expressed heterologously in Rosetta 2(DE3) pLysS Escherichia coli cells (Mega) as previously described. 16 In the final two or three rounds of buoyancy purification, sample buffer was exchanged to 10 mM HEPES, pH 7.5. Concentrations were measured by optical density (OD) at 500 nm using a spectrophotometer (NanoDrop ND‐1000, Thermo Scientific). To prepare collapsed GV samples, diluted samples were pressurized in a sealed syringe until the solution turned transparent.
3.2. Negative stain EM
For imaging of intact GVs, the purified sample was diluted to OD500 ~ 0.5 for Ana and Halo, and OD500 ~ 0.2 for Mega. Three microliters of the target sample was applied to a freshly glow‐discharged (Pelco EasiGlow, 15 mA, 1 min) Formvar/carbon‐coated, 200 mesh copper grid (Ted Pella) for 1 min before blotting. Afterward, the sample was incubated for 1 min with a 0.75% uranyl formate solution before blotting and air‐dried. Image acquisition was performed using a Tecnai T12 (FEI, now Thermo Fisher Scientific) EM at 120 kV, equipped with a Gatan Ultrascan 2 k × 2 k CCD.
3.3. Cryo‐EM
For cryo‐EM, Quantifoil R2/2 200 Mesh, extra thick carbon, copper grids (EMS) were glow discharged (Pelco EasiGlow, 10 mA, 1 min). Freshly purified Mega (OD500 ~ 1), Ana (OD500 ~ 15), and Halo (OD500 ~ 8) GVs sample was frozen using a Mark IV Vitrobot (FEI, now Thermo Fisher Scientific) (4°C, 100% humidity, blot force 3, blot time 4 s). Micrographs were collected on a 300 kV Titan Krios microscope (FEI, now Thermo Fisher Scientific) with an energy filter (Gatan) and equipped with a K3 6k × 4 k direct electron detector (Gatan). Data were collected using SerialEM software with a pixel size of either 1.4 Å (×64,000 magnification) or 2.15 Å (×42,000 magnification) and −2.5 μm defocus. 27
3.4. Diameter determination
All measurements were made using IMOD software. 28 The cylinder/spindle diameter direct measurements from cryo‐EM micrographs were performed only once for each GVs at its widest region. Indirectly diameter was calculated as 2w/π, where w is the width of the flat collapsed GV measured from the ns‐EM micrograph. Sample from at least two independent preparations were used for each measurement.
Statistical analysis was performed in GraphPad PRISM. To ensure normal distribution of the data a Shapiro–Wilk normality test, Kolmogorov–Smirnov test, and D'Agostino & Pearson test was performed. For all data sets, at least two calculated tests suggested normal distribution, thus a paired t test was employed.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Przemysław Dutka: Conceptualization; methodology; investigation; formal analysis; visualization; writing—original draft preparation; writing—review & editing. Dina Malounda: Investigation. Lauren Ann Metskas: Investigation. Songye Chen: Investigation. Robert C. Hurt: Investigation. George J. Lu: Investigation. Grant J. Jensen: Conceptualization; writing—review & editing; supervision; funding acquisition. Mikhail G. Shapiro: Conceptualization; writing—review & editing; supervision; funding acquisition.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grant R35‐GM122588 to Grant J. Jensen and R01‐EB018975 to Mikhail G. Shapiro) and the Caltech Center for Environmental Microbial Interactions (CEMI). Electron microscopy was performed in the Beckman Institute Resource Center for Transmission Electron Microscopy at Caltech. Related research in the Shapiro Laboratory is also supported by the Heritage Medical Research Institute, the Pew Scholarship in the Biomedical Sciences, and the Packard Fellowship for Science and Engineering.
Dutka P, Malounda D, Metskas LA, et al. Measuring gas vesicle dimensions by electron microscopy. Protein Science. 2021;30:1081–1086. 10.1002/pro.4056
Funding information Caltech Center for Environmental Microbial Interactions; Heritage Medical Research Institute; National Institutes of Health, Grant/Award Numbers: R01‐EB018975, R35‐GM122588; Packard Fellowship for Science and Engineering; Pew Scholarship in the Biomedical Sciences
Contributor Information
Grant J. Jensen, Email: jensen@caltech.edu.
Mikhail G. Shapiro, Email: mikhail@caltech.edu.
REFERENCES
- 1. Walsby AE. Gas vesicles. Microbiol Rev. 1994;58:94–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shapiro MG, Goodwill PW, Neogy A, et al. Biogenic gas nanostructures as ultrasonic molecular reporters. Nat Nanotechnol. 2014;9:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shapiro MG, Ramirez RM, Sperling LJ, et al. Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat Chem. 2014;6:629–634. [DOI] [PubMed] [Google Scholar]
- 4. Bourdeau RW, Lee‐Gosselin A, Lakshmanan A, et al. Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature. 2018;553:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu GJ, Farhadi A, Szablowski JO, et al. Acoustically modulated magnetic resonance imaging of gas‐filled protein nanostructures. Nat Mater. 2018;17:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farhadi A, Ho GH, Sawyer DP, Bourdeau RW, Shapiro MG. Ultrasound imaging of gene expression in mammalian cells. Science. 2019;365:1469–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farhadi A, Bedrossian M, Lee J, Ho GH, Shapiro MG, Nadeau JL. Genetically encoded phase contrast agents for digital holographic microscopy. Nano Lett. 2020;20:8127–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lakshmanan A, Jin Z, Nety SP, et al. Acoustic biosensors for ultrasound imaging of enzyme activity. Nat Chem Biol. 2020;16:988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bar‐Zion A, Nourmahnad A, Mittelstein DR, et al. Acoustically detonated biomolecules for genetically encodable inertial cavitation. bioRxiv. 2019. 10.1101/620567. [DOI] [Google Scholar]
- 10. Wu D, Baresch D, Cook C, et al. Genetically encoded nanostructures enable acoustic manipulation of engineered cells. bioRxiv. 2019. 10.1101/691105. [DOI] [Google Scholar]
- 11. Hayes PK, Walsby AE. The inverse correlation between width and strength of gas vesicles in cyanobacteria. Brit Phycol J. 1986;21:191–197. [Google Scholar]
- 12. Beard SJ, Davis PA, Iglesias‐Rodríguez D, Skulberg OM, Walsby AE. Gas vesicle genes in Planktothrix spp. from Nordic lakes: Strains with weak gas vesicles possess a longer variant of gvpCThe GenBank accession numbers for the new sequences in this paper are AJ253125–253133. Microbiology. 2000;146:2009–2018. [DOI] [PubMed] [Google Scholar]
- 13. Beard SJ, Handley BA, Hayes PK, Walsby AE. The diversity of gas vesicle genes in Planktothrix rubescens from Lake Zürich. Microbiology. 1999;145(Pt 10):2757–2768. [DOI] [PubMed] [Google Scholar]
- 14. Dunton PG, Walsby AE. The diameter and critical collapse pressure of gas vesicles in Microcystis are correlated with GvpCs of different length. FEMS Microbiol Lett. 2005;247:37–43. [DOI] [PubMed] [Google Scholar]
- 15. Walsby AE, Fogg GE. The pressure relationships of gas vacuoles. Proc R Soc Lond Ser B Biol Sci. 1971;178:301–326. [Google Scholar]
- 16. Lakshmanan A, Lu GJ, Farhadi A, et al. Preparation of biogenic gas vesicle nanostructures for use as contrast agents for ultrasound and MRI. Nat Protoc. 2017;12:2050–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simon RD. Morphology and protein composition of gas vesicles from wild type and gas vacuole defective strains of Halobacterium salinarium strain 5. Microbiology. 1981;125:103–111. [Google Scholar]
- 18. Offner S, Ziese U, Wanner G, Typke D, Pfeifer F. Structural characteristics of halobacterial gas vesicles. Microbiology. 1998;144(Pt 5):1331–1342. [DOI] [PubMed] [Google Scholar]
- 19. Pfeifer F. Distribution, formation and regulation of gas vesicles. Nat Rev Microbiol. 2012;10:705–715. [DOI] [PubMed] [Google Scholar]
- 20. Farhadi A, Ho G, Kunth M, et al. Recombinantly expressed gas vesicles as nanoscale contrast agents for ultrasound and hyperpolarized MRI. AIChE J. 2018;64:2927–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walsby AE, Bleything A. The dimensions of cyanobacterial gas vesicles in relation to their efficiency in providing buoyancy and withstanding pressure. Microbiology. 1988;134:2635–2645. [Google Scholar]
- 22. Archer DB, King NR. Isolation of gas vesicles from Methanosarcina barken. Microbiology. 1984;130:167–172. [Google Scholar]
- 23. Frank J. Three‐dimensional electron microscopy of macromolecular assemblies: Visualization of biological molecules in their native state. New York: Oxford University Press, 2006. [Google Scholar]
- 24. Li N, Cannon MC. Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli . J Bacteriol. 1998;180:2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramsay JP, Williamson NR, Spring DR, Salmond GPC. A quorum‐sensing molecule acts as a morphogen controlling gas vesicle organelle biogenesis and adaptive flotation in an enterobacterium. Proc Natl Acad Sci USA. 2011;108:14932–14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu BY, Dai YN, Zhou K, et al. Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa . Acta Crystallogr D Biol Crystallogr. 2014;70:3013–3022. [DOI] [PubMed] [Google Scholar]
- 27. Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. [DOI] [PubMed] [Google Scholar]
- 28. Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three‐dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. [DOI] [PubMed] [Google Scholar]


