Abstract
Cytokinins and ethylene control plant development via sensors from the histidine kinase (HK) family. However, downstream signaling pathways for the key phytohormones are distinct. Here we report not only cytokinin but also ethylene is able to control root apical meristem (RAM) size through activation of the multistep phosphorelay (MSP) pathway. We find both cytokinin and ethylene-dependent RAM shortening requires ethylene binding to ETR1 and its HK activity. The receiver domain of ETR1 interacts with MSP signaling intermediates acting downstream of cytokinin receptors, further substantiating the role of ETR1 in MSP signaling. Our studies find both cytokinin and ethylene induce the MSP in similar and distinct cell types with ETR1-mediated ethylene signaling controlling MSP output specifically in the root transition zone. We identified members of the MSP pathway specific and common to both hormones and show that ETR1-regulated ARR3 controls RAM size. ETR1-mediated MSP spatially differs from canonical CTR1/EIN2/EIN3 ethylene signaling and is independent of EIN2, indicating that both pathways can be spatially and functionally separated. Furthermore, we demonstrate that canonical ethylene signaling controls MSP responsiveness to cytokinin specifically in the root transition zone, presumably via regulation of ARR10, one of the positive regulators of MSP signaling in Arabidopsis.
Keywords: cytokinin, ethylene, crosstalk, signaling, root development
Short Summary:
Here we show that ethylene regulates root growth via the multistep phosphorelay (MSP) pathway, typically controlled by cytokinins. We find the histidine kinase activity of ethylene sensor ETR1 necessary for ethylene-mediated regulation of the MSP pathway and identify specific downstream targets involved in the control of root growth, some common, and some specific to each class of hormone.
Introduction
In plants the cytokinin hormones are known plant growth regulators that in cooperation with other hormones control a number of fundamental developmental and physiological processes, including shoot initiation, leaf senescence, apical dominance, nutrient uptake, sink and source of metabolites and stress responses among other known functions [for review see (Kamínek, 2015; Kieber and Schaller, 2014)]. In the root, cytokinin controls the equilibrium between cell division and differentiation, a fundamental process during the development of multicellular organisms (Dello Ioio et al., 2008).
Cytokinin signaling is transmitted via a multistep phosphorelay (MSP), a signaling scheme with evolutionary origins in bacterial signaling systems [for a recent review see (Hwang et al., 2012)]. The current model for MSP requires cytokinin binding to the CHASE domain of an ARABIDOPSIS HISTIDINE KINASE (AHK) receptor resulting in autophosphorylation on a conserved histidine (His) residue and subsequent intramolecular transfer to a conserved aspartate (Asp) residue within the receptor receiver domain (RD) to begin the phosphorelay. The phosphate is next transferred from the AHK RD to an AHP protein (AHP1-5), which are small cytosolic proteins required to transmit the cytokinin signal (phosphate) from membrane-located AHKs into the nucleus (Hwang and Sheen, 2001; Punwani et al., 2010; Yamada et al., 2004). In the final step of MSP, the phosphate is transferred from the AHP His residue to the RD Asp of a type-B ARABIDOPSIS RESPONSE REGULATOR (ARRs-B), a Myb family transcription factor with a GARP DNA binding domain. Phosphorylation of the ARR-B RD results in DNA binding and transcriptional regulation of MSP target genes. Among the immediate-early (Brenner et al., 2005) target genes of ARRs-B are the type-A ARRs (ARRs-A), being upregulated by cytokinin even in the absence of translation and thus considered as cytokinin primary response genes (Brandstatter and Kieber, 1998; D'Agostino et al., 2000). ARRs-A contain a RD but lack the GARP DNA binding domain found in the ARRs-B and are believed to act as negative regulators of phosphotransfer, possibly by competition with ARRs-B for the AHP phosphate transfer (Hwang and Sheen, 2001; To et al., 2004). Due to their direct regulation by ARRs-B, ARRs-A gene expression tightly reflects the activity of MSP in cells and their quantification has been used as a proxy for the status of MSP signaling (Brenner and Schmulling, 2015a; Gordon et al., 2009; Mason et al., 2005; Pernisova et al., 2009).
Crosstalk between the action of cytokinin and the gaseous plant hormone ethylene has been known for decades [reviewed in (Zdarska et al., 2015)]. Cytokinin has been shown to upregulate biosynthesis of 1-aminocyclopropane-1-carboxylic acid (ACC), the rate-limiting precursor in the ethylene biosynthetic pathway, via stabilization of ACC SYNTHASE 5 (ACS5) and ACS9 (Hansen et al., 2009; Chae et al., 2003). Proteomic studies revealed a non-transcriptional cytokinin-mediated upregulation of METHIONINE SYNTHASE 1, METHIONINEADENOSYL TRANSFERASE 3 and ACC OXIDASE 2. These three enzymes catalyze the final steps in the ethylene metabolic pathway, resulting in a prompt and robust increase in ethylene production specifically in the root that impacts ethylene-mediated root growth (Zd'arska et al., 2013). The HK sensors ETR1 and ETHYLENE RESPONSE SENSOR 1 (ERS1) recognize ethylene as do the HK-like sensor Ser/Thr kinases ETR2, ERS2 and ETHYLENE INSENSITIVE 4 (EIN4). However, in contrast to MSP-mediated cytokinin signaling, both HK sensor and HK-like Ser/Thr mediated ethylene signal transduction is thought primarily to utilize the CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1)/EIN2/EIN3 pathway [for detailed review see (Cho and Yoo, 2015)]. Plant histidine kinase receptor cross-talk between MSP and CTR1/EIN2/EIN3 signaling has been proposed at both the receptor level as well as the downstream components of MSP signal transfer i.e. via AHPs or ARRs (Schaller et al., 2011; Street et al., 2015; Zdarska et al., 2015). ETR1 is the only ethylene-perceptive hybrid HK in Arabidopsis (Gamble et al., 1998; Chang et al., 1993; Moussatche and Klee, 2004), making it a good candidate for mediating potential cytokinin/ethylene MSP crosstalk. Nonetheless, in spite of extensive evidence demonstrating the involvement of HK activity contributing to ETR1 function in plant development (Street et al., 2015), the role of ethylene in the control of MSP activity remains largely unknown.
Herein we demonstrate cytokinin and ethylene signaling can be integrated into the MSP response. This requires the HK activity of ETR1, in addition to the HK activity of the AHK receptor, which we find indispensable for the early root developmental control of RAM size in response to both hormones. We further document that cytokinin- and ethylene-activated MSP as well as the canonical ethylene-signaling pathway are functionally and spatially separated and we identified their common and specific regulatory targets from the ARRs-A family. In addition, we show that ETR1-mediated upregulation of ARR3 in the root TZ contributes to RAM shortening by ethylene. Finally, we show that EIN2 controls MSP responsiveness to cytokinins and demonstrate ethylene-mediated upregulation of ARR10, a member of ARRs-B, as another mechanism allowing EIN2-mediated control over MSP signaling.
Results
Histidine kinase activity of ETR1 contributes to both cytokinin and ethylene induced root apical meristem regulation
RAM size is determined by the equilibrium between cell division and cell differentiation (Beemster and Baskin, 1998; Dello Ioio et al., 2007; Dolan et al., 1993). The cytokinin-activated MSP was shown to reduce RAM size by promoting cell differentiation in the transition zone (TZ) (Dello Ioio et al., 2007). However, contrasting data on the involvement of ethylene in RAM control has been reported (Ruzicka et al., 2007; Street et al., 2015).
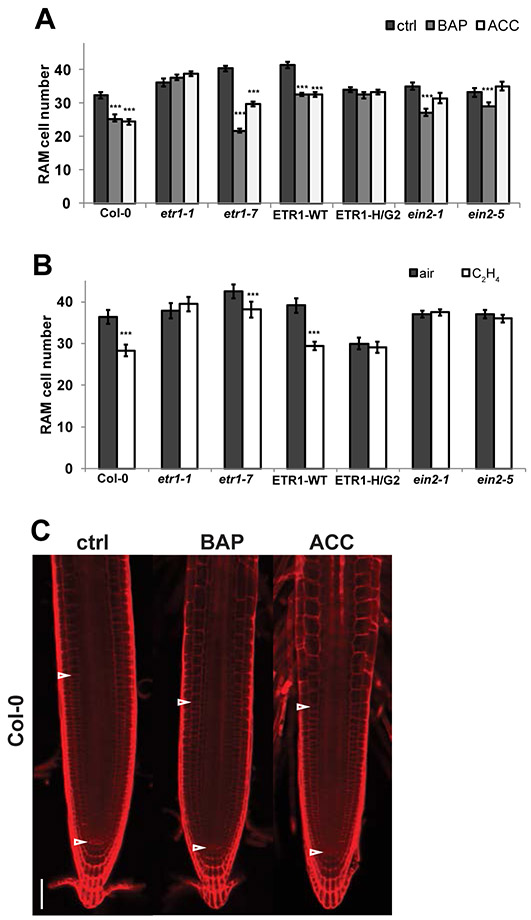
In order to uncover the individual contributions of each hormone and the potential role of the HK activity of ETR1 in the regulation of RAM size, we examined early RAM response to cytokinin [5 μM 6-benzylaminopurine (BAP)] and ethylene [5 μM ACC] in WT (Col-0) and several ethylene-signaling mutants (etr1-1, etr1-7, ETR1-WT, ETR1-HG2, ein2-1, and ein2-5). In agreement with previous reports (Street et al., 2015), our analyses show that both cytokinin and ethylene reduce RAM size in Col-0 (Figure 1A, B). In our assay (RAM size scored after 24 h of hormonal treatment), the ethylene insensitive etr1-1 mutant exhibits complete resistance to either BAP or ACC. In contrast, mutants for the loss-of-function etr1-7 allele, which is hypersensitive to ethylene in a hypocotyl elongation assay (Cancel and Larsen, 2002), has a similar ACC response as observed in Col-0 but shows a higher sensitivity to BAP-mediated RAM shortening when compared with Col-0. The double knock-out mutant etr1-9 ers1-3 complemented by a wild type ETR1 (ETR1-WT) (Hall et al., 2012a) behaves like WT in terms of a reduction in RAM size in response to both hormone treatments, while the HK-inactive ETR1-HG2 in the same genetic background shows reduced RAM size, even in untreated controls. In comparison to ETR1-WT, ETR1-HG2 does not further respond to either cytokinin or ACC application. Finally, both ein2-1 and ein2-5 deficient in canonical ethylene signaling show complete resistance to either ACC or ethylene, while still being sensitive to cytokinin (Figures 1A, B).
Figure 1. ETR1 function potentiates cytokinin-mediated root apical meristem (RAM) size reduction.
Root tips of six-day-old seedlings followed 24h of hormonal treatment (5 μM BAP, 5 μM ACC and 0.1% DMSO as a control supplemented in liquid media) (A,C) and in the presence of 10 ppm ethylene gas and air as a control (B) were stained with propidium iodide (PI) to visualize and calculate the root cells. The RAM size was determined as a number of cortex cells from the quiescent center (QC) to the first elongated cell (mean +/− SE, n=15). Analyzed mutants are in Col-0 background except for ETR1-WT and ETR1-H/G2 mutants that are in etr1-9 ers1-3 background. The statistical significance of differences between control and hormonal treatments (t test) at alpha < 0.001 is denoted by asterisks (***). (C) Representative images of RAM size measurements. Arrowheads determine the position of the QC (bottom) and the first elongated cortex cell (up) on the Col-0 example. ctrl, mock-treated control; BAP, benzyladenine; ACC, 1-aminocyclopropane-1-carboxylic acid. Scale bar represents 50 μm.
These data show that both cytokinin and ethylene control RAM size and reveal that the early RAM response to cytokinin is dependent on sensitivity to ethylene. The HK activity of ETR1 is contributing to the regulation of RAM size and mediates the response to exogenous cytokinin- and ethylene-treatment. This points to a functional role for ETR1 in the MSP-mediated regulation of RAM size.
Identification of spatially specific effects on MSP activation in the root tip in response to cytokinin and ethylene treatments
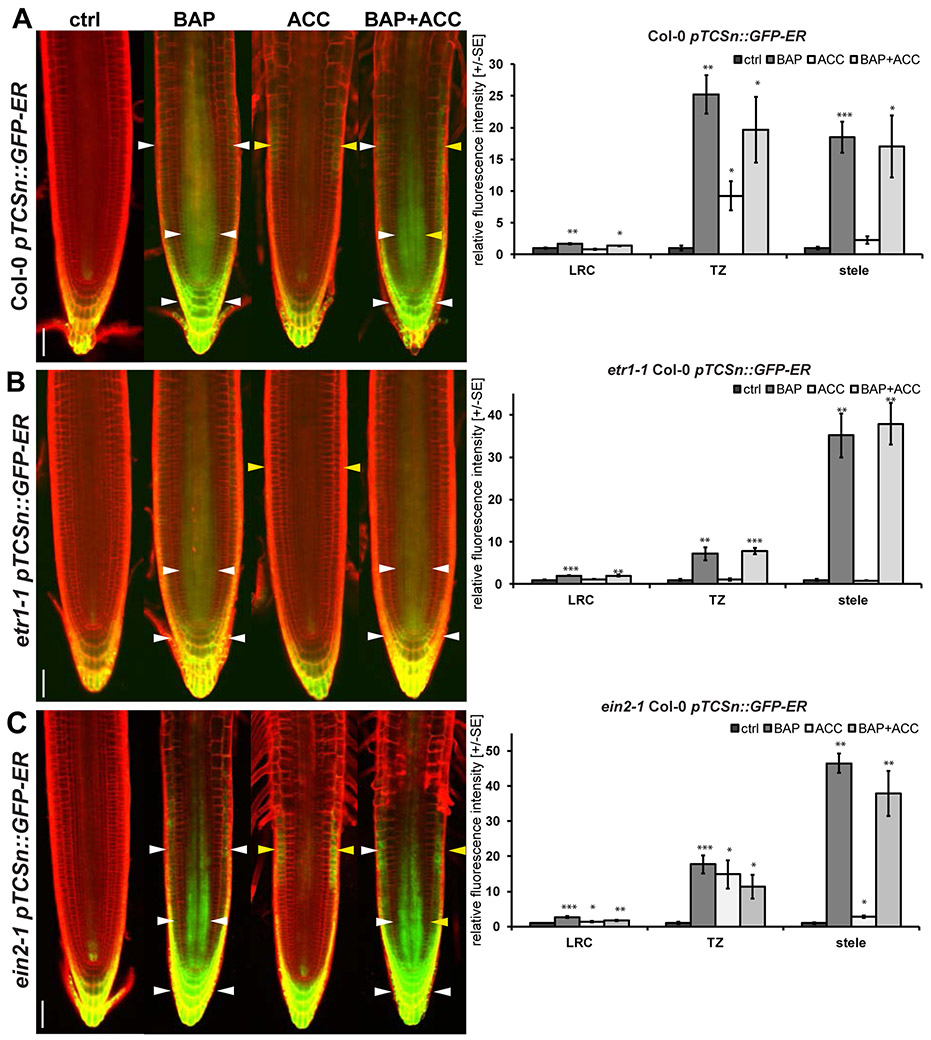
Two-Component Sensor (TCS) was previously shown to represent a sensitive and highly dynamic readout of transcriptional activity of phosphorelay signaling networks in plants (Zurcher et al., 2013). To understand any spatial and cellular specificity in cytokinin- and/or ethylene-mediated MSP response in the root, we first analyzed the response of the cytokinin-responsive pTCSn::GFP to exogenous treatments of ethylene and/or cytokinin. In the presence of BAP, pTCSn::GFP shows pronounced MSP response in the columella and lateral root cap (LRC), weaker activation was observable in epidermal cells of the TZ and stele (Figures 2A, S1A) (2013). In contrast to BAP-treated roots, the ACC-mediated induction of pTCSn::GFP signal occurs predominately in the epidermal cells of the TZ (Figures 2A, S1A). This ethylene-specific pattern is clearly recognizable in roots treated with increasing concentrations of ACC, either alone or in a combination with different BAP amounts (Figure S2). This suggests that the broader pTCSn::GFP domain observed after ACC treatment in the presence of BAP (Figures 2A, C) is a sum of partially overlapping but still spatial-specific activation of MSP signaling by each of the two regulators and cannot be explained by a higher ethylene dose or response during combined treatments. We also find 10 ppm of ethylene gives similar responses in the TZ as ACC treatment (Figure S1B, S1C), confirming that the ACC effect is evoked by ethylene production. The ethylene-mediated pTCSn::GFP activation in the root TZ is abolished in the etr1-1 mutant but still could be observed in the ein2-1 line (Figures 2B, C). Also, both etr1-1 and ein2-1 show decreased responsiveness of pTCS::GFP towards BAP specifically in the root TZ, with the effect being stronger in etr1-1 (Figures 2B, C).
Figure 2. Ethylene activates ETR1-dependent but EIN2-independent MSP signaling in the root TZ.
Six-day-old seedlings following 24h of hormonal treatment (5 μM BAP, 5 μM ACC, 5 μM BAP + 5 μM ACC and 0.1% DMSO as a control) were used for imaging the expression pattern of individual reporter lines in the root apical meristem zone. Representative figures of ER-localized pTCSn-driven GFP signal in Col-0 (A), etr1-1 (B) and ein2-1 (C). The relative fluorescence intensity quantification data are presented in adjacent charts. The fluorescence intensity was quantified separately in three regions of interest (ROIs) - lateral root cap and columella (LRC), transition zone (TZ) and stele as a fold change relative to DMSO control +/− SE n=5). The statistical significance of differences between control and hormonal treatments (t test) at alpha < 0.05, 0.01 and 0.001 is depicted by asterisks (*, ** and ***, respectively). For quantification details see Materials and Methods and Figure S1. The membrane signal from PI staining is shown in red; GFP in green. ctrl, mock-treated control; BAP, benzyladenine; ACC, 1-aminocyclopropane-1-carboxylic acid. The white arrowheads point to the specific localization of the BAP-induced signal while yellow ones to ethylene-induced responses. Scale bars represent 50 μm.
These data demonstrate the ability of ethylene signaling to activate MSP through ETR1 independently of the canonical EIN2 pathway. While BAP MSP response is broadly distributed across several domains of the root, ethylene activation of MSP is spatially restricted to the TZ. Importantly, the sensitivity to ethylene seems to be necessary for the ability of cytokinins to upregulate MSP response in the root TZ.
Cytokinin and ethylene control a subset of ARRs-A with spatial specificity
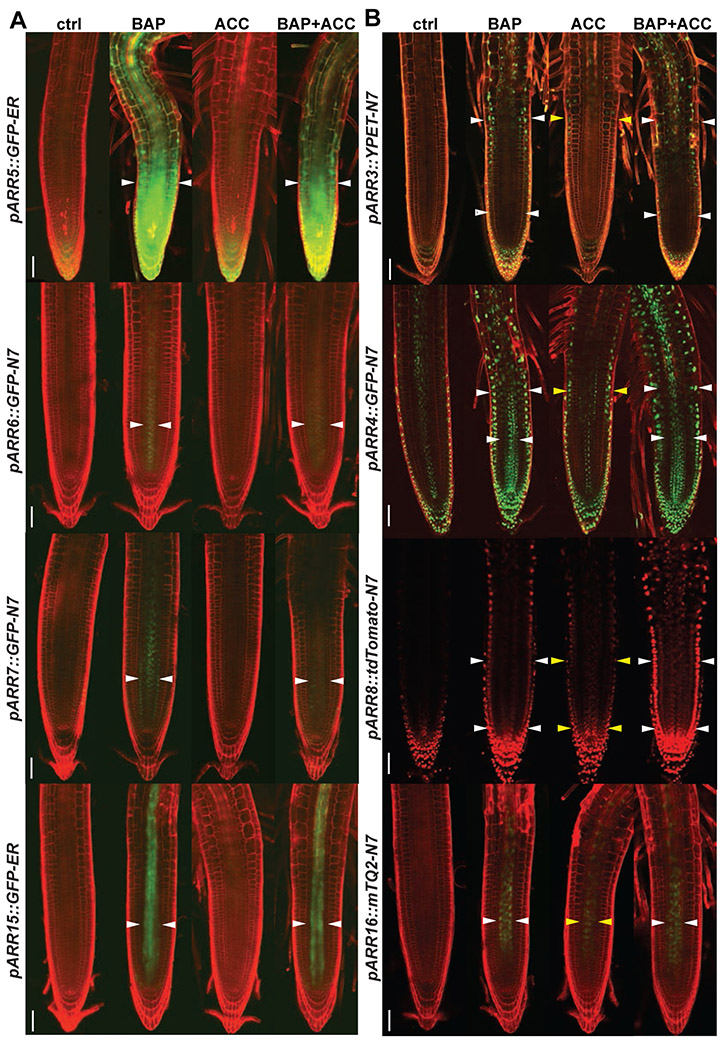
ARR-A genes are primary response targets of cytokinin signaling (D'Agostino et al., 2000). Their expression has been used as a proxy reflecting the location and strength of MSP-mediated cytokinin signaling response (Brenner and Schmulling, 2015b; Gordon et al., 2009; Mason et al., 2005; Pernisova et al., 2009). Consistent with the pTCSn::GFP expression analysis we find cytokinin- and ethylene-specific patterns of induction for individual ARR-A genes. Utilizing transcriptional reporter transgenes for the ARR-A family we observe that cytokinin treatment induces pARR6::GFP-N7, pARR7::GFP-N7 and pARR15::GFP (Figure 3A) in the stele. Contrary to this cytokinin-mediated induction, none of the aforementioned ARRs-A is upregulated by ethylene alone. However, co-treatment with cytokinin and ethylene results in an altered expression pattern for ARR6 and ARR7. In comparison to treatment with BAP alone, combined treatment with BAP and ACC results in an enlarged ARR6 expression domain while ARR7 expression appears to be reduced in the stele (Figure 3A). The pARR5::GFP line reveals the strongest cytokinin-mediated upregulation in the proximal meristem, where the signal is activated in the majority of the cell files, but ethylene does not seem to contribute further to this ARR5 response (Figure 3A).
Figure 3. Cytokinin and ethylene treatment reveal spatially specific effects on MSP activation in the root tip.
Six-day-old seedlings following 24h of hormonal treatment (5 μM BAP, 5μM ACC, 5 μM BAP + 5 μM ACC and 0.1% DMSO as a control) were used for imaging the expression pattern of individual reporter lines in the root apical meristem zone and representative images are shown. (A) ER-localized GFP showing spatial distribution of promoter activity of ARR5 (pARR5::GFP-ER) and nuclear-localized (N7) GFP, driven by the promoters and 3’UTRs for ARR6 (pARR6::GFP-N7), pARR7::GFP-N7 and pARR15::GFP-ER (B) pARR3::YPET-N7, pARR4::GFP-N7, pARR8::tdTomato-N7, pARR16::mTurquoise2-N7 (mTQ2). The membrane signal from PI staining and tdTomato signal (ARR8 only) is shown in red; GFP, YPET and mTQ2 signals are in green. ctrl, mock-treated control; BAP, benzyladenine; ACC, 1-aminocyclopropane-1-carboxylic acid. The white arrowheads point to the specific localization of the BAP-mediated signal while yellow ones to ethylene-mediated responses. Scale bars represent 50 μm.
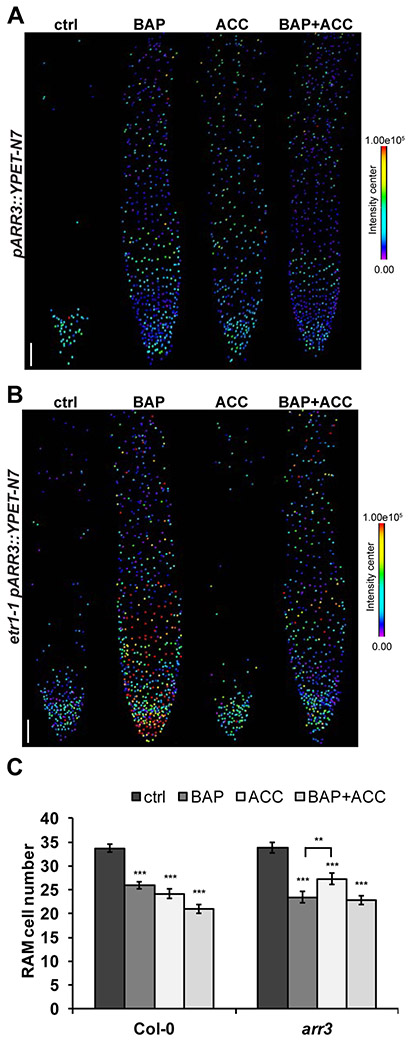
Expression analyses for pARR3::YPET-N7, pARR4:: GFP-N7, pARR8::tdTomato-N7 and pARR16::mTurquoise2-N7 (mTQ2) reveal these ARR-A family members are activated by each of the hormones (Figure 3B). The pARR3::YPET-N7 signal is induced by cytokinin in LRC, columella, and elongating epidermal cells in and proximal to the TZ. The activation of ARR3 by ACC seemed to occur in the same cell type where TCSn is activated solely by ACC and ethylene, i.e. in the epidermis of the root TZ (Figures 3B, S3). However, a detailed quantification of pARR3::YPET-N7 positive nuclei) shows the induction of ARR3 promoter activity by either cytokinin or ACC or both is spatially less restricted, being detectable in the epidermis and stele of the root tip (Figure 4A). Furthermore, cotreatment with cytokinin and ethylene results in additive regulation of ARR3 expression as both the number and intensity of pARR3::YPET-N7 positive nuclei is increased relative to the individual treatments (Figure S3B). Similarly to pTCSn::GFP, induction of ARR3 by ethylene in the root TZ is impaired in the etr1-1 background (Figures 4B, S3A), pointing to its dependence on ETR1-mediated ethylene signaling. Cytokinin upregulates ARR4 in the stele, TZ, and in the elongating cells located proximally to the TZ (Figure 3B; see also Figure S3C showing GFP signal without PI counterstaining). In the epidermis of the TZ and in the root region proximal to that, ACC also induces pARR4::GFP-N7 signal. ARR8 is induced by either cytokinin or ethylene in the LRC, columella, epidermis (including TZ), and stele (Fig. 3B). However, when both cytokinin and ACC are applied, the pARR8::tdTomato-N7 signal is markedly amplified, suggesting an additive effect of the two hormones in the control of ARR8 expression. pARR16::mTQ2-N7 is activated by cytokinin or ethylene treatment. However, while cytokinin upregulates pARR16::mTQ2-N7 throughout the stele, ACC activates pARR16::mTQ2-N7 only in a subset of central stele cells. Simultaneous application of both cytokinin and ethylene confers a pARR16::mTQ2-N7 response very similar to the cytokinin-specific expression (Fig. 3B).
Figure 4. ARR3 contributes to ethylene-induced RAM shortening in an ETR1-dependent fashion.
Six-day-old seedlings following 24h of hormonal treatment (5 μM BAP, 5 μM ACC, 5 μM BAP + 5 μM ACC and 0.1% DMSO as a control supplemented in liquid media were analyzed. (A, B) Heat maps of each quantified pARR3::YPET-N7 expressing nuclei showing the spatial induction of ARR3 promoter activity in the root of Col-0 (A) and etr1-1 (B) background. The images show results of signal intensities measured in single root per each treatment as a representative example (n=5). Scale bars represent 50 μm. (C) RAM size of Col-0 and the arr3 mutant was determined as a number of cortex cells per file from the quiescent center (QC) to the first elongated cell (mean +/− SE, n=15). The mean numbers of RAM cells are shown at the bottom of the corresponding bar. The statistical significance (t test) of differences between control and hormonal treatments or ACC and BAP treated roots (bracket) at alpha < 0.01 and < 0.001 are denoted by asterisks (** and ***, respectively). ctrl, control; BAP, benzyladenine; ACC, 1-aminocyclopropane-1-carboxylic acid.
The ethylene-mediated, ETR1-dependent upregulation of ARR3 in the TZ area suggests a possible role in mediating the RAM response to ethylene. Accordingly, we observed reduced sensitivity of RAM shortening in an arr3 loss of function mutant when treated by ACC. The effect was similar to that in Col-0 when the plant was treated with BAP or BAP and ACC together (Figure 4C).
Taken together, these data reveal spatially distinct MSP-induced target genes in the cells that comprise the root tip following cytokinin or ethylene treatment or a combination of both. Of particular note is the ETR1-dependent activation of ARR3 that contributed to RAM shortening, suggesting that ARR3 is a possible connection for the integration of both hormonal signals.
ETR1 interacts with all AHPs except AHP4
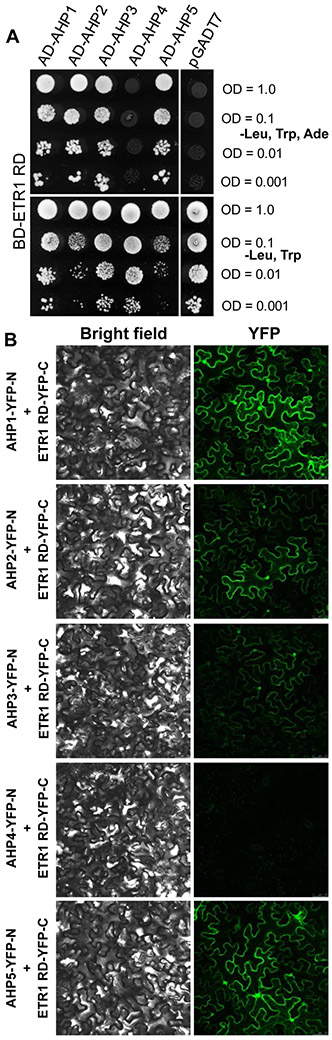
ETR1 is the only Arabidopsis ethylene receptor with HK activity possessing a receiver domain, which is necessary for the phosphorelay activity in hybrid HKs (Horak et al., 2011; Schaller et al., 2011). The RD of hybrid HKs AHK2, 3, 4, CKI1, and AHK5 was shown to mediate interaction with the downstream AHP members of the MSP in Arabidopsis (Bauer et al., 2013; Dortay et al., 2008; Dortay et al., 2006; Pekarova et al., 2011). Thus, to examine a possible molecular mechanism mediating the contribution of ETR1 to MSP signaling, we inspected the interaction of the receiver domain of ETR1 (ETR1RD) with those Arabidopsis AHP proteins previously shown to be positive regulators of cytokinin signaling (AHP1-5) (Hutchison et al., 2006). Using a yeast two-hybrid assay and confirmed by in planta bimolecular fluorescence complementation (BiFC) assays (Walter et al., 2004), we find ETR1RD interacts with AHP1, AHP2, AHP3, and AHP5 (Figure 5; for the immunoblots confirming the expression of all interacting partners see Figure S4).
Figure 5. ETR1RD interacts with a specific subset of AHP proteins in yeast two-hybrid assays and in planta.
(A) Growth of the yeast clones expressing BD-ETR1RD and the indicated AD-AHP protein was documented after incubation for 4 days on either interaction-selective media lacking leucine (Leu), tryptophan (Trp) and adenine (Ade) or vector-selective media lacking Leu and Trp. Each test was performed with yeast suspension of OD600 = 0.001, 0.01, 0.1 and 1.0. The empty pGADT7 vector was used as a negative control. (B) Confocal images of abaxial tobacco leaf cells co-expressing the indicated AHP-YFP-N fusion proteins with ETR1 RD-YFP-C. YFP fluorescence of the reconstituted fluorophore as a result of ETR1RD interaction with a given AHP is documented in the right-hand column. The left-hand column shows the corresponding bright-field images.
These results imply that crosstalk in the MSP may occur between cytokinin- and ethylene-signaling pathways via direct interaction of ethylene receptor ETR1 with AHP proteins.
ETR1 controls early MSP response to ethylene in the root
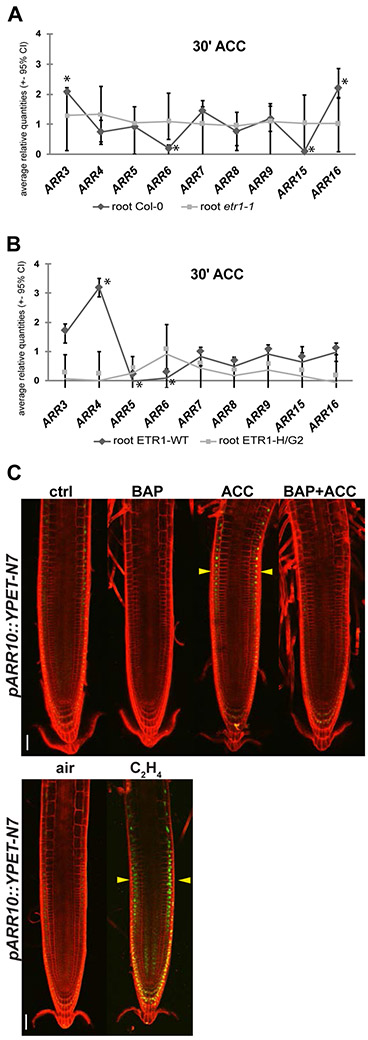
ARRs-A were shown to be upregulated by cytokinin very quickly with most of them showing peak at RNA level between 30-40 min. after exogenous cytokinin application (D'Agostino et al., 2000). However, the need for fluorescent protein maturation does not allow assessment of prompt activation of MSP in ARRs-A reporter lines. To quantify the contribution of both hormones in the early MSP response (30 min. after hormone treatment), we utilized quantitative real-time PCR (qRT-PCR) to determine the expression of 9 out of the 10 ARRs-A in roots of six-day-old seedlings (6 DAG, Col-0). 6-DAG roots were treated with exogenous BAP and ACC for 30 minutes and individual ARR-A gene expression was assessed after RNA purification and evaluated by qRT-PCR as a fold-change difference between mock-treated controls and cytokinin- and/or ACC-treated samples.
In accordance with the results obtained by imaging of fluorescent reporter transgenes and previous reports (D'Agostino et al., 2000), we detected an upregulation in the expression of all analyzed ARRs-A following 30 minutes of BAP application in Col-0 (Figure S5A). We also confirmed that ACC initiates early MSP response, similarly to BAP, although the ACC-mediated MSP response is significantly weaker than a BAP-mediated response (Figures 6A, S5B). Consistent with our live-imaging analysis of reporter transgenes, we find that, in general, cytokinin upregulates the majority of ARRs-A (Figure S5A) while ethylene treatment activates a specific subset, ARR3 and ARR16, and downregulates ARR6 and ARR15 expression in 6-DAG roots (Figures 6A, S5B). In etr1-1 roots, ACC treatment had no effect on ARR-A expression. Similarly, no observable change in ARR-A levels was detected in RNA isolated from roots of HK-inactive ETR1-H/G2 (Figures 6B, S5C).
Figure 6. Ethylene controls endogenous ARRs-A expression and upregulates ARR10 in the root TZ.
(A, B) Six-day-old whole roots followed 30 min of hormonal treatment (5 μM ACC and 0.1% DMSO as a control) were analyzed via qRT-PCR. Relative gene expression of ARRs-A in the presence of ACC double normalized to the internal control (UBQ10) and the ARRs-A transcription level in the mock-treated plants is presented. Fold-change of ARRs-A transcript levels between hormonal- and mock-treated roots is shown for ACC-induced response of Col-0 and etr1-1 (A) and ETR1-WT and ETR1-HG2 in etr1-9 ers1-3 background (B). Statistical significance of differences observed between mock- and ACC-treated roots (Mann-Whitney) at alpha < 0.05 is depicted by asterisks (*, qbase+, Biogazelle). The line connecting individual ARRs highlights the pattern of ARRs-A expression in the given genotype. (C) Six-day-old seedlings following 24h of hormonal treatment (5 μM BAP, 5 μM ACC, 5 μM BAP + 5 μM ACC and 0.1% DMSO as a control; in the presence of 10 ppm ethylene gas and air as a control) were used for imaging the expression pattern of individual reporter lines in the root apical meristem zone. Representative figures of nuclear-localized pARR10::YPET-N7 signal in Col-0 background are shown. The membrane signal from PI staining is shown in red; YPET in green. ctrl, mock-treated control; BAP, benzyladenine; ACC, 1-aminocyclopropane-1-carboxylic acid. The yellow arrowheads point to the ethylene-induced responses. Scale bars represent 50 μm.
Taken together, our qRT-PCR results confirm that, similarly to cytokinin, ethylene can regulate the MSP response. Furthermore, we demonstrate that both the ability of ETR1 to bind ethylene (impaired in etr1-1) and its HK activity contributes to the early MSP response to ethylene.
EIN2 controls MSP responsiveness to cytokinins and mediates ethylene-induced upregulation of ARR10
Our results on the quantification of TCSn:GFP in an ein2-1 mutant background suggest the role of EIN2 in controlling the responsiveness of MSP signaling to cytokinins, but not to ethylene (Figure 2C). To demonstrate the response on the level of endogenous gene regulation, we assayed the expression of ARRs-A upon cytokinin (BAP), ACC, and BAP in the presence of 2-aminoethoxyvinylglycine (AVG), an inhibitor of ethylene biosynthesis (Yang and Hoffman, 1984). Considering the weaker phenotype in cytokinin-regulated TCSn:GFP observed in ein2-1 when compared to etr1-1 (Figure 2), we isolated RNA specifically from the root tips of treated seedlings and used it for the qRT PCR-based quantification of ARRs-A (Figure S6) to avoid dilution of rare transcripts by redundant mRNAs and to be able to detect even tiny regulatory events. In line with the aforementioned results suggesting the role of EIN2 in the control of MSP responsiveness to cytokinins, we observed changes in BAP-induced ARRs-A expression (in response both to BAP and BAP+AVG, Figure S6). The mostly pronounced changes were observed in case of ARR3, ARR6-8, ARR15 and ARR16. In our fluorescent reporter experiments, the majority of these ARRs-A was shown either to respond to both cytokinin and ethylene (ARR3, ARR8 and ARR16) or were a target of combined BAP and ACC treatment (ARR6, ARR7).
To elucidate a possible mechanism of ethylene–regulated responsiveness of MSP signaling to cytokinins, we assayed the transcriptional regulation of ARRs-B by both hormones. Out of three ARRs-B tested (ARR1, ARR10 and ARR14), we observed the ability of ethylene, but not of cytokinin, to upregulate pARR10::YPET-N7 (Figure 6C). Similarly to TCSn:GFP, both ACC and ethylene upregulated ARR10 predominantly in the root TZ.
Altogether, our findings suggest that canonical ethylene signaling controls MSP response via regulation of ARR10.
Discussion
Ethylene contributes to the MSP through ethylene-mediated activation of ETR1 histidine kinase activity
In the past decades, the role of ETR1 in MSP signaling has been repeatedly assayed. The HK activity of ETR1 was shown to be dispensable for the ability of ETR1 to act via canonical CTR1/EIN2/EIN3 signaling (Gamble et al., 2002; Hall et al., 2012a; Wang et al., 2003). On the other hand, published results imply that the HK activity might at least partially be necessary for the ETR1-mediated ethylene response (Binder et al., 2018; Qu and Schaller, 2004; Street et al., 2015). Nonetheless, direct evidence for the role of ethylene in the control of MSP in planta was lacking. Our results provide genetic and molecular evidence that ethylene, in addition to cytokinin, controls the activity of the MSP pathway. We show that while overlapping in some contexts, both hormones signal through the MSP pathways in characteristic temporal and spatial patterns. These responses can be distinguished at the level of ARRs-A gene regulation and final MSP output through the TCSn reporter.
It has been shown that ACC might act independently of ethylene (Xu et al., 2008; Yoon and Kieber, 2013). Our combined analysis of RNA levels by qRT-PCR and in vivo promoter activity by the pTCSn::GFP and pARR3::YPET-N7 transgenes indicate ACC can regulate the MSP in an ETR1-dependent way. This would suggest that ACC controls the MSP through ethylene production, as ethylene, not ACC, is known to bind ETR1 (Schaller and Bleecker, 1995). Additionally, we demonstrate that ethylene at a concentration of 10 ppm is able to upregulate pTCSn::GFP in the epidermis of the root TZ, mimicking the effect of ACC (Figure S1B).
Both ETR1 and ERS1 contain a functional HK domain (Moussatche and Klee, 2004). Our data show that both binding of ethylene to ETR1 (compromised in etr1-1 (Schaller and Bleecker, 1995)) and functional ETR1 HK activity (inactive in ETR1-H/G2 (Hall et al., 2012a)) are required for ethylene-mediated MSP (Figures 1 and 6). The requirement for ETR1 HK activity to regulate ARRs-A expression and the ability of the ETR1RD to interact with AHPs imply a possible mechanistic route for ETR1-triggered phosphorelay through MSP (Figures 5, S4). This hypothesis is supported by ample published evidence. ETR1 was found to autophosphorylate on the conserved His residue (Gamble et al., 1998; Moussatche and Klee, 2004) and the role of the conserved His and Asp residues was shown to be important for ETR1-mediated activation of the ARR6 promoter in Arabidopsis protoplasts (Cho and Yoo, 2007). In a cell-free phosphorelay system derived from Arabidopsis mesophyll protoplasts, ARR2, a type-B ARR, can be phosphorylated in an ETR1-dependent manner (Hass et al., 2004). However, there is also evidence speaking against the role of ETR1 in directly mediating transphosphorylation through the MSP pathway. In comparison to bacterial and other Arabidopsis RDs, ETR1RD shows important structural differences (Hung et al., 2016; Pekarova et al., 2016), raising questions about its ability to mediate an ETR1-triggered phosphorelay. Accordingly, the tobacco homologue of ETR1, NtHK2, is able to phosphorylate myelin basic protein, but not its own RD domain (Zhang et al., 2004). Alternative scenarios involving ETR1 interference with MSP signaling via ETR1 interaction with AHPs (Scharein and Groth, 2011), direct phosphorylation of ARRs (e.g. ARR2) from the ETR1 HK domain (Hass et al., 2004), and ethylene-regulated phosphatase activity of ETR1 or the ability of ethylene receptors to control the MSP via formation of heterodimers with other histidine kinases (Grefen et al., 2008) cannot be excluded. Hence, the detailed nature of the molecular mechanism allowing ETR1-dependent control over the MSP pathway remains to be identified.
Canonical ethylene signaling controls MSP responsiveness
Besides the role of HK activity of the ethylene sensor ETR1 in the control of MSP signaling, our data revealed another layer of complexity in the crosstalk between cytokinin and ethylene signaling via ethylene-mediated upregulation of ARR10. ARR10 together with ARR1 and ARR12 is one of the key positive regulators mediating the majority of cytokinin responses via MSP signaling (Argyros et al., 2008; Ishida et al., 2008; Mason et al., 2005). Genome-wide studies on the binding of ARR10 to gene regulatory elements imply its role in the control of biosynthesis, transport or signaling of various hormones like auxin, brassinosteroids, cytokinins, ethylene, gibberellin, jasmonic and salicylic acids. ARR10 also seems to mediate response to biotic and abiotic stimuli (Xie et al., 2018; Zubo et al., 2017). In line with that, ARR10 (again together with ARR1 and ARR12) was shown to mediate crosstalk of auxin with ethylene and cytokinin in the control of cell division and elongation in the Arabidopsis root (Street et al., 2015; Street et al., 2016). Compared to other B-ARRs, ARR10 seems to be more stable and this could have possible functional consequence in MSP signaling (Hill et al., 2013).
Our data suggest that both MSP and EIN2-mediated canonical ethylene signaling are necessary for the control of RAM size by ethylene. We observed that etr1-1 and both ein2 lines (ein2-1 and ein2-5) are resistant to the ethylene-mediated RAM shortening. On top of that, both etr1-1 and ein2-1 show almost complete and partial decrease, respectively, in TCSn:GFP responsiveness to cytokinin specifically in the root TZ. A possible explanation could be that, in contrast to ein2-1, etr1-1 is affected in both ethylene-regulated MSP and canonical ethylene signaling (Chang et al., 1993). Thus, in etr1-1 cytokinin (as well as ethylene) is unable to activate MSP and initiate cell differentiation in the root TZ because of low level of ARR10 that could be phosphorylated upon activation of the phosphorelay via cytokinin or ethylene HK sensors. Compared to that, ETR1-mediated MSP is still active in ein2 mutants and the ETR1-dependent ethylene activation of MSP might be sufficient to activate the synthetic TCSn promoter, a sensitive sensor for MSP activity (Zurcher et al., 2013). This could explain the WT-like TCSn:GFP activation by ethylene in ein2-1 roots. Meanwhile this same level of MSP activity may not be sufficient to activate the endogenous promoters of genes responsible for the MSP-mediated cell differentiation in the root TZ, which may be less sensitive then the synthetic TCSn promoter. The ETR1-induced MSP activity could also contribute to the overall MSP activation by cytokinin, thus leading to the observed partial sensitivity of ein2 lines to exogenous cytokinin. The need of ETR1 HK activity for mediating the sensitivity to cytokinins is supported by the cytokinin insensitivity to RAM shortening observed in ETR1-H/G2 (Figure 1). The ethylene-mediated upregulation of ARR10 seems to be effected via the canonical ethylene (CTR1/EIN2/EIN3) pathway. This is supported by the weak downregulation of ARR10 by exogenous cytokinin treatment (Figure S7) and consistent with the previously reported downregulation of endogenous ARR10 transcripts assayed via NanoString nCounter (Street et al., 2016). In accordance with our data on ethylene-mediated upregulation of pARR10::YPET-N7 dominantly in the TZ, we observed activation of EIN3-mediated signaling in the root TZ by both ACC and ethylene (Figure S8), thus explaining the spatial specificity of observed defects in responsiveness of TCSn:GFP to cytokinins to the root TZ. Alternatively, besides ethylene-mediated upregulation of ARR10, genes targeted by canonical ethylene signaling could be contributing to the initiation of cell differentiation in parallel to MSP signaling.
Cytokinin and ethylene show spatial specificity in the control of the MSP
A detailed analysis of cytokinin/ethylene crosstalk in the root tip allowed us to identify common and specific targets of both hormones in MSP signaling. The spatial specificity is apparent not only for cytokinin- and ethylene-controlled MSP, but also in case of canonical ethylene signaling. As shown using pEBS::GUS, the EIN3-specific reporter (Stepanova et al., 2007), cytokinin upregulates EIN3-dependent ethylene signaling particularly in the vascular tissue of the more mature part (differentiation zone) of the root (Zdarska et al., 2013). We found that in the root tip, BAP is able to activate pEBS::GUS specifically in the columella cells (Figure S7A). However, the ACC- and particularly ethylene-induced pEBS::GUS expression in that tissue is distinctly stronger (Figures S7A, B) when compared to induction by BAP. In addition, BAP-induced activation is inhibited by the presence of the ethylene biosynthesis inhibitor AVG, suggesting that the BAP effect on pEBS::GUS is mediated by cytokinin-induced ethylene production. While the spatial specificity of ethylene-activated MSP (as evidenced by activation of the pTCSn, pARR3, pARR4 and pARR8 reporters) overlaps with the expression domain for ETR1 in the root TZ, the ethylene-dependent upregulation of pEBS::GUS in the columella seems largely to correspond with the expression profile for ERS1 in the root vascular tissue and the columella (Grefen et al., 2008). Thus, it seems ETR1 controls MSP signaling while ERS1 might be active in regulating the canonical ethylene-signaling pathway in the root tip. However, activation of pEBS::GUS in the TZ in the presence of both ACC and ethylene (Figure S7) might suggest a possible role of ETR1 and/or other ethylene sensors also in the control of the CTR1/EIN2/EIN3 pathway in the TZ. This is in agreement with a recent study suggesting the role of the canonical ethylene signaling pathway in RAM size (Street et al., 2015). On the other hand, an ACC-dependent contribution of ERS1 to MSP signaling also cannot be excluded, as implied by slightly changed ARRs-A response in ETR1-WT (etr1-9 ers1-3 background (Hall et al., 2012b)) when compared to Col-0 (Figures 6A, B). The ethylene-induced activation of MSP signaling via ERS1, e.g. via formation of heterodimer with ERS2 or EIN4 (Grefen et al., 2008), both of which are Ser/Thr kinases but possess potentially functional RD domains, might also explain the ethylene sensitivity of the loss-of-function mutant etr1-7.
Taken together, the spatial specificity observed for cytokinin and ethylene control of the MSP pathway suggests existence of mechanisms distinguishing the origin of the signal triggering the MSP response to both hormones in the root. The spatially specific cellular expression pattern for ETR1 (Grefen et al., 2008), the cytokinin receptors AHK2, 3, and4 (Nishimura et al., 2004; Pernisova et al., 2016), and/or each of AHPs (Yuan et al., 2016) might be one way to achieve such a mechanism. Similarly, the expression specificity of ERS1 (Grefen et al., 2008) and/or its downstream targets including EIN3 seem to be responsible for the spatial-specific dissection of the ethylene canonical signaling pathway. The restriction of the ethylene-controlled MSP to a few cell types in a complex tissue might be the reason for only recent attention to the role of ETR1 and its HK activity in the control of cell differentiation in the RAM (this work and (Street et al., 2015)). However, the detailed mechanism mediating the expression of individual ARRs-A in response to cytokinin and/or ethylene remains to be identified and will likely be tissue and cell type specific. ARR3 and its closest Arabidopsis paralogue ARR4 were found to control circadian period independently of cytokinin (Salome et al., 2006). Whether ethylene-regulated ARR3 and ARR4 in the TZ represent readout of another environmental input translated to the hormonal signal in the control of the root growth remains to be seen.
Model for the cytokinin/ethylene crosstalk in the control of RAM size
Based on our findings we propose a model of cytokinin/ethylene crosstalk mediated by ETR1 and controlling cell differentiation in the root TZ (Figure 7). In the model, both ETR1-controlled MSP as well as canonical ethylene signaling are necessary for the cytokinin- and ethylene-mediated RAM shortening. Cytokinin activates MSP signaling primarily in the root stele, LRC and columella (this work and (Zurcher et al., 2013)) and via non-transcriptional regulation of ethylene biosynthesis to increase ethylene production specifically in the root (Zdarska et al., 2013). Ethylene (cytokinin-induced or produced upon other environmental and/or intrinsic stimuli) is recognized by ETR1 dominantly in the root TZ, where it contributes to the ARRs-B phosphorylation (Hass et al., 2004). The canonical ethylene signaling pathway acting via EIN2 upregulates the level of phosphorylatable ARRs-B via transcriptional upregulation of ARR10. Cytokinin- and ethylene- induced phosphorylation of ARRs-B allows specific control of target genes including subset of ARRs-A (ARR3, ARR4, ARR6, ARR7, ARR8 and ARR16) and activates cell differentiation in the root TZ. Thus, cytokinin-induced MSP output integrates both cytokinin and ethylene signals. In the absence of exogenous ethylene, the HK activity of ETR1 contributes to the maintenance of RAM size, possibly via upregulation of mitotic activity of meristematic cells and/or control over the balance between cell division/differentiation in the root TZ. However, the contribution of EIN2-mediated canonical ethylene signaling in the direct control of the genes involved in the root cell differentiation also cannot be excluded.
Figure 7. Model for ETR1-mediated integration of cytokinin and ethylene in the control of RAM size.
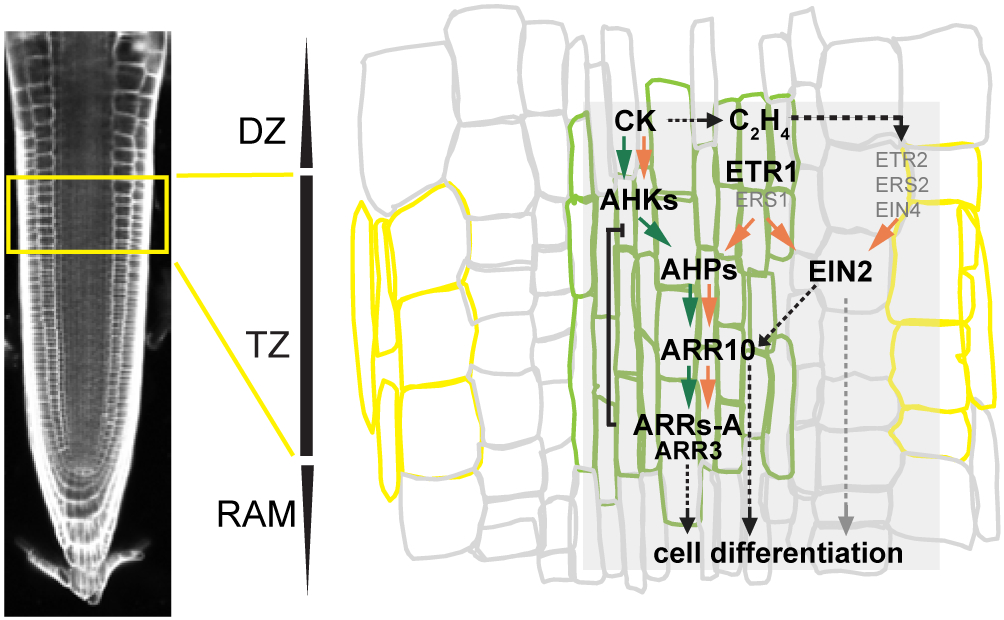
Cytokinin activates the MSP signaling pathway in the root stele and upregulates ethylene biosynthesis, creating ethylene that is recognized by ethylene receptors, preferentially by ETR1 and possible participation of ERS1. ETR1 interacts with AHP proteins and contributes to the MSP output via phosphorylation of ARRs-B including ARR10 that target individual ARRs-A, particularly ARR3. The ability of ETR1 to bind ethylene and ETR1 HK activity are necessary for the ethylene-dependent control of MSP output that integrates both cytokinin and ethylene signals in the control of cell differentiation. EIN2-mediated canonical ethylene signaling controls MSP sensitivity to cytokinins in the root TZ via upregulating the amount of phosphorylatable ARR10. However, the direct control of cell differentiation via canonical ethylene signaling (grey dashed arrows and grey characters) in parallel to MSP also cannot be excluded. Green colored rectangle corresponds to stele cells and yellow to epidermis in the root TZ. Green arrows depict cytokinin-specific regulation, the orange arrows represent ethylene-mediated control of the MSP DZ, differentiation zone; TZ, transition zone; RAM, root apical meristem.
Materials and methods
Plant material and growth conditions
The following lines of Arabidopsis thaliana were used: etr1-1 (NASC,N237), etr1-7 (Hua and Meyerowitz, 1998), ETR1-WT, ETR1-H/G2 (Hall et al., 2012b), arr3 (N25265), ein2-1 (N3071) and ein2-5 (N16707). A.thaliana Col-0 (N1092) was used as a control. pEBS::GUS line was constructed by Anna Stepanova in Joe Ecker lab. Fluorescent reporter lines previously prepared: pTCSn::GFP-ER (Zurcher et al., 2013), pARR5::GFP (Yanai et al., 2005) and pARR15::GFP (Muller and Sheen, 2008) and newly constructed: pARR3::YPET-N7, pARR4::GFP-N7, pARR6::GFP-N7, pARR7::GFP-N7, pARR8::tdTomato-N7, pARR10::YPET-N7 and pARR16::mtQ2-N7. etr1-1 and ein2-1 were crossed into the pTCSn::GFP-ER, etr1-1 was crossed into the pARR3::YPET-N7, the F3 homozygous plants were analyzed.
All plants were grown under long-day conditions 16/8 h light/dark at 21/19 °C, light period at the intensity of 150 μmol m−2 s−1 as described previously (Pernisova et al., 2009). For our experiments we employed six-day-old (6 DAG) seedlings grown on ½ strength Murashige and Skoog (MS) media containing 1% sucrose under on vertical plates in growth chambers (CLF Plant Climatics, GmbH). 6 DAG were treated with liquid MS media containing 5 μM benzylaminopurine (BAP, Duchefa), 5 μM 1-aminocyclopropane-1-carboxylic acid (ACC, Sigma), 5 μM BAP + 5 μM ACC and 0.1% DMSO as a control (mock) for 30 minutes in qRT-PCR analyses and for 24 h in spatial-specific analyses of fluorescent reporter lines, RAM measurements and GUS staining assay under the same conditions as seedling cultivation. For RAM and fluorescent reporter lines 6 DAG seedlings were also analyzed followed 24 h of 10 ppm ethylene gas treatment (gas mixture: 99% of N2 + 1% C2H4, 500mL/min and air as a control; ethylene gas growth chambers developed in collaboration with Photon Systems Instruments; http://psi.cz/).
Cloning ARRs-A reporters
The genomic region upstream and downstream of the ORF for ARR3, ARR4, ARR6, ARR7, ARR8, and ARR16 and ARR10 was PCR amplified from Col-0 genomic DNA using Pfu II Ultra (Agilent). The size of the genomic region amplified in each case depended on the proximity of genes in the genomic neighborhood. When possible 5-4.5 kilobases of the proximal promoter upstream the translation start site and 2-1.5 kb downstream the stop codon of the ARR ORF was amplified. The PCR fragments were subcloned into pBJ36 and a Gateway conversion cassette (R1/R2 reference frame B, Invitrogen) was inserted between the ARR promoter and 3’UTR region using a unique Sma1 site. The ARR-A promoter-gateway-3’UTR was subcloned into the binary vector pMOA33 using a unique Not1 site. This generated the pMOA33 pARR-A-GWrefB and pMOA34 pARR10-GWrefBI destination vector. Standard gateway recombination with LR clonase II (Invitrogen) was used to generate the reporter construct with pENTR/D 2xGFP-N7, pENTR/D 2xYPET-N7, pENTR/D 2xmTurquoise-N7, or pENTR/D tdTomato-N7 acting as the fluorescent protein donors. This generated the final pMOA33 and pMOA34 pARR::2xmTurquoise-N7, pARR::2xGFP-N7, pARR::2xYPET-N7, pARR::tdTomato-N7. The final plasmid was sequenced after recombination and the binary vector was transformed into the agrobacterium strain GV3101. Transgenic Arabidopsis (Col-0) plants were generated by the floral dip method (Clough and Bent, 1998).
Cloning primer sequences, sizes of fragments amplified, and restriction sites used:
| ARR-A | Primer sequence | Restriction site used |
Amplicon size |
|---|---|---|---|
| ARR3 Prom F | cccacgcgtgtaagatttgagttccttattatatagaaaagg | Mlu1 | 3382 bp |
| ARR3 Prom R | cccctcgagaggcctagtgagaaaagagtaggaaatgaaaagatagaa | Xho1/Stu1 | |
| ARR3 3'UTR F | cccggatccaggcctagagatccgtgtgataacgatctccg | BamH1/Stu1 | 1596 bp |
| ARR3 3'UTR | cccgctagccagagggaatagaggcaagtggaatgta | Nhe1 | |
| ARR4 Prom F | cccgtcgacgtgaatcaagtcatgggataatcttatttt | Sal1 | 4541 bp |
| ARR4 Prom R | ccccgggagacgagcttatagtaactgtgaggagagg | Sma1 | |
| ARR4 3'UTR F | ccccgggtggaactattctgggatgccgtgtaatatt | Sma1 | 1553 bp |
| ARR4 3'UTR R | cccgctagcctactatatattcaaaaccatgagttatca | Nhe1 | |
| ARR6 Prom F | cccacgcgtctgcttatagccaacaacaacgagcaagaa | Mlu1 | 2030 bp |
| ARR6 Prom R | ccccgggtgatcaacgaatgttggaggatttggaaga | Sma1 | |
| ARR6 3'UTR F | cccggatcccccggggctctccgatgcaaattccgtgactggatc | BamH1/Sma1 | 729 bp |
| ARR6 3"UTR R | cccgctagcaaggagaggtgcggttttctcaaccatgtt | Nhe1 | |
| ARR7 Prom F | cccacgcgtAAGTTCGTAAACATTGACCCTGTTACTTAT | Mlu1 | 3122 bp |
| ARR7 Prom R | ccccgggtgtcaaactcagaataaaaaagagagcaga | Sma1 | |
| ARR7 3'UTR F | ccccgggattatatgtatagaggccaagaccttagca | BamH1/Sma1 | 1018 bp |
| ARR7 3"UTR R | cccgctagcGAGGTGTTAACACGCGGGTTTCGATTTCGC | Nhe1 | |
| ARR8 Prom F | cccacgcgtcggtaaaatattctcgctaaaatacgtaag | Mlu1 | 4079 bp |
| ARR8 Prom R | ccctcgagcccgggtgtagatattcaatcgaaaaaaactaagat | Xho1/Sma1 | |
| ARR8 3'UTR F | cccggatcccccgggaggaggactaacgcaacccgagaagccact | BamH1/Sma1 | 1681 bp |
| ARR8 3'UTR R | cccgctagctgacatttccccaacgaactaata | Nhe1 | |
| ARR15 Prom F | cccgtcgacACCATTTCCTCAAACATCACAACCGCATTC | Sal1 | 2202 bp |
| ARR15 Prom R | ccccgggtgttttctctcgggaaagtaaacaacaaaacag | Sma1 | |
| ARR15 3'UTR F | cccggatcccccgggatgattgagctcaatcttttctcacgttttctc | BamH1/Sma1 | 1170 bp |
| ARR15 3'UTR R | cccgctagccaagatttagattacttgttcaataagcttg | Nhe1 | |
| ARR16 Prom F | cccacgcgtcgtattattttacagAATAAAGAAGAGTTGTC | Mlu1 | 3004 bp |
| ARR16 Prom R | ccctcgagcccggggatctcctttgttccttaaatgtcaagtaact | Xho1/Sma1 | |
| ARR16 3'UTR F | cccggatcccccgggccatgagagatttataatgttagccctaacca | BamH1/Sma1 | 1500 bp |
| ARR16 3'UTR R | Cccgctagcgaagaccaaaagtttaagaatctctttaattc | Nhe1 | |
| ARR10 Prom F | cccGTCGACCCAATCACGAGACGTCACCACATCGCCAGC | Sal1 | 3934 bp |
| ARR10 Prom R | ccccGGTACCCCCGGGcgccgtcaaagattatgaattgaagtagaa | Kpn1/Sma1 | |
| ARR10 3’UTR F | cccGGATCCCCCGGGgaaacctgttttattcagacacaatcaatt | BamH1/Sma1 | 1500 bp |
| ARR10 3’UTR R | cccGCTAGCGATGCTGTCCGAGAAACCAATGCTGCAGCC | Nhe1 |
RAM measurements and reporters image analysis
6 DAG seedlings followed 24 h of hormonal treatment were stained with propidium iodide to visualize root cells and imaged employing inverted confocal microscope system Zeiss LSM 780 or LSM 880. For RAM measurements at least 15 roots were evaluated. RAM size was determined as a number of cortex cells filed from QC to the first elongated cell. For reporters at least 10 roots were examined and representative images are shown. The variability of signal intensity is due to imaging at different time points and different laser intensity of used microscopes. The majority of images were analyzed with LSM 780. Several representative images and experiments were repeated or added using LSM 880 in order to gain better results, namely representative images of pARR3::YPET-N7 and pARR4::GFP-N7 (Fig.3); etr1-1 pARR3::YPET-N7 quantification Z-stack assay (Fig. 4B) and representative images (Fig. S3); pTCSn::GFP-ER in dosage assay (Fig. S2); pARR10::YPET-N7 in Fig.6C All experiments were performed in three independent biological repeats.
Quantification of reporter gene expression
Quantification of pTCSn::GFP-ER signal (38 slides of Z-stacks) was performed in Fiji (Schindelin, J.; Arganda-Carreras, I. & Frise, E. et al. (2012), "Fiji: an open-source platform for biological-image analysis", Nature Methods 9(7): 676-682, PMID 22743772). Images where processed as follows prior to GFP quantification. First the despeckle function, a median filter, was used to denoise the image followed by bilateral filtering to smooth the pixel values while preserving the edges of the image detail. After denoising and smoothing the background of the image black space was determined and the lower threshold was adjusted to this value. For the quantification of the pTCSn::GFP-ER signal three domains were selected by use of the region of interest (ROI) tool in Fiji: 1. Root cap, 2. Transition zone, and 3. Stele. ROIs were selected using the propidium iodide labeled channel to select the cells for each region. These ROIs were then translated onto the pTCSn::GFP-ER channel and the multi-measure function in the ROI manager was used for grey value quantification and ROI area quantification (see Figure S1). The sum of the grey value was used for each of these regions and the values were normalized to the total area of each ROI.
Prior to quantification of pARR3::2xYPET-N7 in Col-0 and etr1-1 background pixel intensity denoising of the image was performed in FIJI using the despeckle (median filter) function. For the quantification of ARR3 promoter activity denoised images were processed using the spot detection in IMARIS 9.0 (Bitplane, http://www.bitplane.com/imaris/imaris) with an unassigned pixel intensity threshold set at 200. The spot segmentation determined the unassigned pixel values, X position, Y position, and Z position for all nuclei segmented in each treated root image. Following segmentation the unassigned pixel values were transformed by dividing the values by (2^16)-1 allowing each nuclei to be represented on an arbitrary scale from 0 to 1. Plotting of the individual nuclei data distribution was done in R using ggplot2. Statistical analysis of nuclei pixel intensity distribution was conducted using the Kolmogorov-Smirnov test. Total cell number with active ARR3 expression, and X, Y position of nuclei as heat maps (individual nuclei data distribution) were generated by IMARIS functions. Statistical analysis of the number of ARR3 positive cells was performed using the student’s t-test.
Yeast two-hybrid assay and BiFC
DNA constructs for the expression of appropriate fusion proteins were prepared by means of Gateway cloning (Invitrogen) according to the manufacturer’s instructions. For yeast two-hybrid assay, we cloned AHP1-AHP5 cDNAs into the prey vector pGADT7-Dest (AD:AHPs) and ETR1RD in bait vector pGBKT7-Dest (BD-ETR1RD), which were derived from the Matchmaker™ system (Clontech, http://www.clontech.com) (Horak et al., 2008). BD:ETR1RD and the respective AD:AHP were co-transformed into the yeast strain J694A and cultivated on either vector selective media as a control and an interaction selective media to assess the interaction as described previously (Horak et al., 2008). Expression of all proteins in yeast was confirmed by immunoblot. For detection of of AD fusions primary rat monoclonal anti-HA antibodies clone 3F10 (Roche) and secondary anti rat IgG alkaline phosphatase-conjugated antibodies (Sigma, A8438) were used. For detection of BD fusions we used primary mouse anti c-myc polyclonal antibodies (Sigma, M4439) and secondary anti-mouse IgG alkaline phosphatase conjugated antibodies (Sigma, 3562). For BiFC, we generated YFP fusion constructs driven by 35S promoter. C-terminal YFP fragment was used for fusion with ETR1RD (ETR1RD:YFP-C) and N-terminal YFP fragment for fusion with indicated AHPs (AHP:YFP-N). The constructs were co-transformed using Agrobacterium tumefaciens into Nicotiana benthamiana leaves and transiently expressed in their mesophyll cells.
All procedures required for in vivo interaction experiments, such as tobacco infiltration, confocal laser-scanning microscopy, yeast transformation, growth assay or western blotting were performed as described previously (Horak et al., 2008; Pekarova et al., 2011).
qRT-PCR and data analysis.
After the treatment, whole roots (Fig. 6, S5) or 1 cm long root tips (Fig. S6) were detached with a scalpel, collected, immediately frozen in liquid nitrogen and analyzed separately using four independent biological replicas. Total RNA from plant tissue was isolated using the RNAqueous Small Scale Phenol-Free Total RNA Isolation Kit (Ambion) according to the manufacturer’s instructions. cDNA was prepared using RTP3 primer and Superscript III (Invitrogen) according to the manufacturer’s instructions, and qRT-PCR was performed using the FastStart SYBR Green Master Kit (Roche) according to the manufacturer’s instructions on a Rotor-Gene 6000 (CORBETT RESEARCH) instrument.
RTP3 primer: 5′ CGT TCG ACG GTA CCT ACG TTT TTTTTTTTTTTT TT 3′
For relative quantification of ARR transcripts ARR3, ARR4, ARR6, ARR8, ARR9 and ARR16 we used primers as described in (Pernisova et al., 2009), ARR5 as described in (Jeon et al, 2010), ARR7 as described in (Tran et al., 2007) and for ARR15, we used the following primers:
fARR15rt (5’-GAC GGA TTA TTC AAT GCC-3’) rARR15rt (5’-TTA ACC CCT AGA CTC TAA TTT G3’).
For individual ARR pairs of primers we used following conditions:
Q10, ARR4, ARR8, ARR9, ARR16: 95°C/7 min – 35x (95 °C/15 s + 60 °C/20 s + 72 °C/20 s) – 72 °C/1 min – melt
ARR3: 95°C/7 min – 35x (95 °C/15 s + 60 °C/30 s + 72 °C/30 s) – 72 °C/1 min – melt
ARR5: 95°C/7 min – 35x (95 °C/15 s + 55 °C/30 s + 72 °C/20 s) – 72 °C/1 min – melt
ARR6: 95°C/7 min – 35x (95 °C/20 s + 59 °C/20 s + 72 °C/45 s) – 72 °C/1 min – melt
ARR7: 95°C/7 min – 30x (95 °C/15 s + 60 °C/30 s) – 72 °C/1 min – melt
ARR15: 95°C/7 min – 35x (95 °C/15 s + 58 °C/30 s + 72 °C/30 s) – 72 °C/1 min – melt
Average relative quantities were normalized to internal controls UBIQUITIN10 (Q10) and to mock- treated controls that correspond to value 1 (qbase+, Biogazelle). For statistical analysis Mann-Whitney test was performed (qbase+, Biogazelle).
GUS staining assay
After the treatment, GUS staining with seedlings was performed as previously described (Malamy and Benfey, 1997). At least 20 seedlings were evaluated using differential interference contrast microscopy (Olympus BX61) in at least three independent experiments.
Supplementary Material
Acknowledgements
The work was supported by the Czech Science Foundation (19-23108Y) and by the Ministry of Education, Youth and Sports of the Czech Republic under the projects CEITEC 2020 (LQ1601), CZ.02.1.01/0.0/0.0/16_026/0008446. We acknowledge the core facility CELLIM of CEITEC supported by the Czech-BioImaging large RI project (LM2015062 funded by MEYS CR) for their support with obtaining scientific data presented in this paper. Plant Sciences Core Facility of CEITEC MU is acknowledged for the cultivation of experimental plants used in this paper. Work performed in Elliot Meyerowitz’s laboratory was supported by NIH NIGMS grant R01 GM104244, Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation through Grant GBMF3406.
References
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, and Schaller GE (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20:2102–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Reiss K, Veerabagu M, Heunemann M, Harter K, and Stehle T (2013). Structure-Function Analysis of Arabidopsis thaliana Histidine Kinase AHK5 Bound to Its Cognate Phosphotransfer Protein AHP1. Molecular Plant 6:959–970. [DOI] [PubMed] [Google Scholar]
- Beemster GT, and Baskin TI (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116:1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Kim HJ, Mathews DE, Hutchison CE, Kieber JJ, and Schaller GE (2018). A role for two-component signaling elements in the Arabidopsis growth recovery response to ethylene. Plant Direct 2:e00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstatter I, and Kieber JJ (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10:1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Kollmer I, Burkle L, and Schmulling T (2005). Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44:314–333. [DOI] [PubMed] [Google Scholar]
- Brenner WG, and Schmulling T (2015a). Summarizing and exploring data of a decade of cytokinin-related transcriptomics. Front Plant Sci 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, and Schmulling T (2015b). Summarizing and exploring data of a decade of cytokinin-related transcriptomics. Front Plant Sci 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel JD, and Larsen PB (2002). Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol 129:1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, and Bent AF (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, and Kieber JJ (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124:1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, and Sabatini S (2008). Emerging role of cytokinin as a regulator of cellular differentiation. Curr Opin Plant Biol 11:23–27. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, and Sabatini S (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17:678–682. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, and Scheres B (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119:71–84. [DOI] [PubMed] [Google Scholar]
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmulling T, and Heyl A (2008). Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J Proteome Res 7:3649–3660. [DOI] [PubMed] [Google Scholar]
- Dortay H, Mehnert N, Burkle L, Schmulling T, and Heyl A (2006). Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 273:4631–4644. [DOI] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, and Schaller GE (1998). Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci U S A 95:7825–7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, and Schaller GE (2002). Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol 128:1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, and Meyerowitz EM (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A 106:16529–16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Stadele K, Ruzicka K, Obrdlik P, Harter K, and Horak J (2008). Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Molecular Plant 1:308–320. [DOI] [PubMed] [Google Scholar]
- Hall B, Shakeel S, Amir M, Ul Haq N, Qu X, and Schaller GE (2012a). Histidine-Kinase Activity of the Ethylene Receptor ETR1 Facilitates the Ethylene Response in Arabidopsis. Plant Physiology 159:682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BP, Shakeel SN, Amir M, Ul Haq N, Qu X, and Schaller GE (2012b). Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol 159:682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chae HS, and Kieber JJ (2009). Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J 57:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schafer E, Kudla J, et al. (2004). The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23:3290–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Mathews DE, Kim HJ, Street IH, Wildes SL, Chiang YH, Mason MG, Alonso JM, Ecker JR, Kieber JJ, et al. (2013). Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiology 162:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J, Grefen C, Berendzen KW, Hahn A, Stierhof YD, Stadelhofer B, Stahl M, Koncz C, and Harter K (2008). The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol 8:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J, Janda L, Pekarova B, and Hejatko J (2011). Molecular Mechanisms of Signalling Specificity via Phosphorelay Pathways in Arabidopsis. Curr Protein Pept Sci 12:126–136. [DOI] [PubMed] [Google Scholar]
- Hua J, and Meyerowitz EM (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94:261–271. [DOI] [PubMed] [Google Scholar]
- Hung YL, Jiang I, Lee YZ, Wen CK, and Sue SC (2016). NMR Study Reveals the Receiver Domain of Arabidopsis ETHYLENE RESPONSE1 Ethylene Receptor as an Atypical Type Response Regulator. PLoS One 11:e0160598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. (2006). The Arabidopsis Histidine Phosphotransfer Proteins Are Redundant Positive Regulators of Cytokinin Signaling. THE PLANT CELL ONLINE 18:3073–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, and Sheen J (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413:383–389. [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, and Muller B (2012). Cytokinin signaling networks. Annu Rev Plant Biol 63:353–380. [DOI] [PubMed] [Google Scholar]
- Chae HS, Faure F, and Kieber JJ (2003). The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15:545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, and Meyerowitz EM (1993). Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262:539–544. [DOI] [PubMed] [Google Scholar]
- Cho YH, and Yoo SD (2007). ETHYLENE RESPONSE 1 histidine kinase activity of Arabidopsis promotes plant growth. Plant Physiol 143:612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, and Yoo SD (2015). Novel connections and gaps in ethylene signaling from the ER membrane to the nucleus. Front Plant Sci 5:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, and Mizuno T (2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant and Cell Physiology 49:47–57. [DOI] [PubMed] [Google Scholar]
- Kamínek M (2015). Tracking the Story of Cytokinin Research. Journal of Plant Growth Regulation 34:723–739. [Google Scholar]
- Kieber JJ, and Schaller GE (2014). Cytokinins. Arabidopsis Book 12:e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, and Benfey PN (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124:33–44. [DOI] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, and Schaller GE (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17:3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussatche P, and Klee HJ (2004). Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem 279:48734–48741. [DOI] [PubMed] [Google Scholar]
- Muller B, and Sheen J (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453:1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, and Ueguchi C (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16:1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarova B, Klumpler T, Triskova O, Horak J, Jansen S, Dopitova R, Borkovcova P, Papouskova V, Nejedla E, Sklenar V, et al. (2011). Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana. Plant J 67:827–839. [DOI] [PubMed] [Google Scholar]
- Pekarova B, Szmitkowska A, Dopitova R, Degtjarik O, Zidek L, and Hejatko J (2016). Structural Aspects of Multistep Phosphorelay-Mediated Signaling in Plants. Mol Plant 9:71–85. [DOI] [PubMed] [Google Scholar]
- Pernisova M, Klima P, Horak J, Valkova M, Malbeck J, Soucek P, Reichman P, Hoyerova K, Dubova J, Friml J, et al. (2009). Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc Natl Acad Sci U S A 106:3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisova M, Prat T, Grones P, Harustiakova D, Matonohova M, Spichal L, Nodzynski T, Friml J, and Hejatko J (2016). Cytokinins influence root gravitropism via differential regulation of auxin transporter expression and localization in Arabidopsis. New Phytol 212:497–509. [DOI] [PubMed] [Google Scholar]
- Punwani JA, Hutchison CE, Eric Schaller G, and Kieber JJ (2010). The Subcellular Distribution of the Arabidopsis Histidine Phosphotransfer Proteins is Independent of Cytokinin Signaling. Plant J. [DOI] [PubMed] [Google Scholar]
- Qu X, and Schaller GE (2004). Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol 136:2961–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Ljung K, Vanneste S, Podhorska R, Beeckman T, Friml J, and Benkova E (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19:2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, To JP, Kieber JJ, and McClung CR (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18:55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, and Bleecker AB (1995). Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270:1809–1811. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Shiu SH, and Armitage JP (2011). Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol 21:R320–330. [DOI] [PubMed] [Google Scholar]
- Scharein B, and Groth G (2011). Phosphorylation alters the interaction of the Arabidopsis phosphotransfer protein AHP1 with its sensor kinase ETR1. PLoS One 6:e24173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, and Alonso JM (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19:2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street IH, Aman S, Zubo Y, Ramzan A, Wang X, Shakeel S, Kieber JJ, and Schaller GE (2015). Ethylene Inhibits Cell Proliferation of the Arabidopsis Root Meristem. Plant Physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street IH, Mathews DE, Yamburkenko MV, Sorooshzadeh A, John RT, Swarup R, Bennett MJ, Kieber JJ, and Schaller GE (2016). Cytokinin acts through the auxin influx carrier AUX1 to regulate cell elongation in the root. Development 143:3982–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, and Kieber JJ (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16:658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, and Yamaguchi-Shinozaki K (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci U S A 104:20623–20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438. [DOI] [PubMed] [Google Scholar]
- Wang W, Hall AE, O'Malley R, and Bleecker AB (2003). Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci U S A 100:352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Chen H, Huang L, O'Neil RC, Shokhirev MN, and Ecker JR (2018). A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat Commun 9:1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, and Kieber JJ (2008). Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. The Plant cell 20:3065–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Koizumi N, Nakamichi N, Kiba T, Yamashino T, and Mizuno T (2004). Rapid response of Arabidopsis T87 cultured cells to cytokinin through His-to-Asp phosphorelay signal transduction. Biosci Biotechnol Biochem 68:1966–1976. [DOI] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, and Ori N (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15:1566–1571. [DOI] [PubMed] [Google Scholar]
- Yang SF, and Hoffman NE (1984). Ethylene Biosynthesis and Its Regulation in Higher-Plants. Annual Review of Plant Physiology and Plant Molecular Biology 35:155–189. [Google Scholar]
- Yoon GM, and Kieber JJ (2013). 1-Aminocyclopropane-1-carboxylic acid as a signalling molecule in plants. AoB Plants 5. [Google Scholar]
- Yuan L, Liu Z, Song X, Johnson C, Yu X, and Sundaresan V (2016). The CKI1 Histidine Kinase Specifies the Female Gametic Precursor of the Endosperm. Dev Cell 37:34–46. [DOI] [PubMed] [Google Scholar]
- Zd'arska M, Zatloukalova P, Benitez M, Sedo O, Potesil D, Novak O, Svacinova J, Pesek B, Malbeck J, Vasickova J, et al. (2013). Proteome analysis in Arabidopsis reveals shoot- and root-specific targets of cytokinin action and differential regulation of hormonal homeostasis. Plant Physiology 161:918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdarska M, Dobisova T, Gelova Z, Pernisova M, Dabravolski S, and Hejatko J (2015). Illuminating light, cytokinin, and ethylene signalling crosstalk in plant development. J Exp Bot 66:4913–4931. [DOI] [PubMed] [Google Scholar]
- Zdarska M, Zatloukalova P, Benitez M, Sedo O, Potesil D, Novak O, Svacinova J, Pesek B, Malbeck J, Vasickova J, et al. (2013). Proteome analysis in Arabidopsis reveals shoot- and root-specific targets of cytokinin action and differential regulation of hormonal homeostasis. Plant Physiol 161:918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZG, Zhou HL, Chen T, Gong Y, Cao WH, Wang YJ, Zhang JS, and Chen SY (2004). Evidence for serine/threonine and histidine kinase activity in the tobacco ethylene receptor protein NTHK2. Plant Physiol 136:2971–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Blakley IC, Yamburenko MV, Worthen JM, Street IH, Franco-Zorrilla JM, Zhang W, Hill K, Raines T, Solano R, et al. (2017). Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc Natl Acad Sci U S A 114:E5995–E6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, and Muller B (2013). A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiology 161:1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.