Abstract
Axonal injury is a major contributor to the clinical symptomatology in patients with traumatic brain injury. Conventional neuroradiological tools, such as CT and MRI, are insensitive to diffuse axonal injury (DAI) caused by trauma. Diffusion tensor MRI parameters may change in DAI lesions; however, the nature of these changes is inconsistent. Multidimensional MRI is an emerging approach that combines T1, T2, and diffusion, and replaces voxel-averaged values with distributions, which allows selective isolation of specific potential abnormal components. By performing a combined post-mortem multidimensional MRI and histopathology study, we aimed to investigate T1-T2-diffusion changes linked to DAI and to define their histopathological correlates. Corpora callosa derived from eight subjects who had sustained traumatic brain injury, and three control brain donors underwent post-mortem ex vivo MRI at 7 T. Multidimensional, diffusion tensor, and quantitative T1 and T2 MRI data were acquired and processed. Following MRI acquisition, slices from the same tissue were tested for amyloid precursor protein (APP) immunoreactivity to define DAI severity. A robust image co-registration method was applied to accurately match MRI-derived parameters and histopathology, after which 12 regions of interest per tissue block were selected based on APP density, but blind to MRI. We identified abnormal multidimensional T1-T2, diffusion-T2, and diffusion-T1 components that are strongly associated with DAI and used them to generate axonal injury images. We found that compared to control white matter, mild and severe DAI lesions contained significantly larger abnormal T1-T2 component (P = 0.005 and P < 0.001, respectively), and significantly larger abnormal diffusion-T2 component (P = 0.005 and P < 0.001, respectively). Furthermore, within patients with traumatic brain injury the multidimensional MRI biomarkers differentiated normal-appearing white matter from mild and severe DAI lesions, with significantly larger abnormal T1-T2 and diffusion-T2 components (P = 0.003 and P < 0.001, respectively, for T1-T2; P = 0.022 and P < 0.001, respectively, for diffusion-T2). Conversely, none of the conventional quantitative MRI parameters were able to differentiate lesions and normal-appearing white matter. Lastly, we found that the abnormal T1-T2, diffusion-T1, and diffusion-T2 components and their axonal damage images were strongly correlated with quantitative APP staining (r = 0.876, P < 0.001; r = 0.727, P < 0.001; and r = 0.743, P < 0.001, respectively), while producing negligible intensities in grey matter and in normal-appearing white matter. These results suggest that multidimensional MRI may provide non-invasive biomarkers for detection of DAI, which is the pathological substrate for neurological disorders ranging from concussion to severe traumatic brain injury.
Keywords: traumatic, brain injury, diffuse axonal injury, traumatic axonal injury, radiological-pathological correlations, multidimensional MRI
Can microscopic traumatic axonal injury be imaged non-invasively? Benjamini et al. show that multidimensional MRI, which combines T1, T2 and diffusion, can reveal damage that is invisible using quantitative MRI modalities, and provide an injury-only image that visualizes microscopic lesions in the brain.
Introduction
Traumatic brain injury (TBI) presents major medical, social and economic challenges world-wide and is associated with high mortality, co-morbidities, and long-term disabilities (Faul and Coronado, 2015; Sharp et al., 2016). Among the various pathological brain lesions produced by impact (e.g. intracerebral bleedings, necrotic-ischaemic lesions, tissue avulsion), diffuse axonal injury (DAI) is caused by mechanical damage to axons multifocally throughout the white matter (Adams et al., 1982).
From severe closed-head injuries to concussions, the degree of injury and the duration of loss of consciousness and coma directly correlate with the extent of axonal pathology (Johnson et al., 2013; Smith and Stewart, 2020). Axons become stiff and brittle following rapid deformation caused by rotation acceleration of the brain, which leads to immediate mechanical damage to the axonal cytoskeleton. As the axonal transport is disrupted, pathological accumulation of different axonal proteins causes axonal swelling, which leads to axonal degeneration (Johnson et al., 2013). Amyloid precursor protein (APP) is one of those accumulated proteins, and it can be detected immunohistochemically within a few hours post injury (Gentleman et al., 1993, 1995; Sherriff et al., 1994). Because of that, and because of its high sensitivity and robustness, APP remains the gold standard for the identification of axonal pathology (Johnson et al., 2013).
Following mechanical impact, DAI is frequently neuroanatomically localized along the different portions of the corpus callosum, particularly at the level of its more anterior portion (Johnson et al., 2013). While most of the brain appears structurally normal and only a minority of axons are affected even in severe TBI, DAI causes collective impairment that can lead to disruption of signal transfer and can have significant implications for cognitive function (McInnes et al., 2017). Although the development of non-invasive biomarkers for DAI has become an important topic within the neuroradiological community, there are few radiological-pathological studies that correlate histopathological findings, including DAI lesions, and MRI data following acute and chronic TBI exposure.
The microscopic nature of the axonal swelling makes DAI nearly invisible to conventional neuroimaging approaches, such as CT and conventional MRI. Their low spatial resolution, usually above 400 µm, is orders of magnitude larger than the structures of interest. Moreover, mild TBI cases present sparse clusters of affected axons with low density that may reside in different parts of the brain. Showing more promise, diffusion MRI is a technique that provides higher effective spatial sensitivity to changes that are in the order of the tissue microenvironment (below 100 µm) by using water mobility as a probe of tissue microstructure (Le Bihan et al., 1986). The most established diffusion MRI framework is diffusion tensor imaging (DTI) (Basser et al., 1994). Most notably, DTI provides the macroscopic orientationally averaged diffusivity of water in tissue, termed mean diffusivity (MD), and the diffusion anisotropy, termed fractional anisotropy (FA).
Since its first use to investigate DAI following TBI (Arfanakis et al., 2002), DTI has been established as a sensitive tool for depicting axonal integrity in regions with TBI-associated lesions (Bazarian et al., 2007; Budde and Frank, 2010; Haacke et al., 2010; Shenton et al., 2012; Hulkower et al., 2013; Skinner et al., 2017). In particular, FA had stood out as being a sensitive biomarker for TBI. Unfortunately, despite DTI’s high sensitivity, conflicting findings of either decreases (Arfanakis et al., 2002; Inglese et al., 2005; Mac Donald et al., 2007; Miles et al., 2008; Kinnunen et al., 2011; Matsushita et al., 2011) or increases (Bazarian et al., 2007, 2012; Mayer et al., 2010, 2012; Henry et al., 2011; Borich et al., 2013) of the FA in TBI groups compared with control subjects have been reported. Importantly, these studies reported contradictory results with respect to FA regardless of the time from injury.
Predominantly, an additional and more fundamental obstacle for using conventional MRI in detecting tissue alterations following mild TBI is that it provides information averaged over the entire voxel. Voxel-averaged scalar images can only provide macroscopic information with respect to the voxel size (typically on the order of 1 mm3), thus limited by MRI’s relatively low image resolution. In a mammalian brain, an individual voxel contains multiple chemical and physical microenvironments such as axons, neurons, glia, myelin, blood vessels, and CSF. It should be expected then, that DAI would affect a very small volume fraction of any given voxel and would most likely remain undetectable using voxel-averaged MRI methods. As a result, the inability to separate normal and pathological tissue within a voxel is the most likely cause of the non-specificity of conventional MRI methods, including DTI, in detecting and diagnosing DAI lesions.
Following recent technological breakthroughs (Benjamini and Basser, 2016; Kim et al., 2017; Topgaard, 2017; Hutter et al., 2018), multidimensional correlation MRI is currently emerging as an attractive MRI modality to study heterogeneous biological media (Benjamini, 2020; Benjamini and Basser, 2020). Overcoming the aforementioned voxel-averaging (i.e. partial volume) limitation, this approach jointly encodes several magnetic resonance contrasts (such as T1, T2, and diffusion) and yields a multidimensional distribution of those parameters within each voxel, thus accomplishing two main goals: (i) providing a distribution instead of a mean, thus allowing the identification of multiple components within a voxel (Does and Gore, 2002; Ronen et al., 2006); and (ii) adding more magnetic resonance dimensions to help separate different biological components (Celik et al., 2013; Benjamini and Basser, 2017; Slator et al., 2019; Benjamini et al., 2020; Kim et al., 2020; Pas et al., 2020).
To provide a basis for future clinical translation, we aimed here to define the ways in which DAI pathology affects MRI biomarkers, and to establish whether multidimensional MRI can be used to obtain axonal injury images. To this end, we performed comprehensive APP histopathological assessments on the same specimens that were previously scanned using multidimensional and conventional MRI. We compared sections from control corpus callosum, normal-appearing corpus callosum in TBI cases, and DAI lesions in the corpus callosum, to enable the identification of subvoxel tissue components that are linked to axonal injury. Because our study focuses on TBI patients, we henceforth use the term traumatic axonal injury (TAI) to refer to DAI caused by a trauma (Sharp and Ham, 2011; Hill et al., 2016). Our hope is that non-invasive imaging and prediction of pathological APP accumulation would provide an important biomarker for DAI and means to detect it clinically.
Materials and methods
Donor specimens
We evaluated autopsy-derived brain autopsy specimens from two different human brain collections. Formalin-fixed portions of approximately 20 × 20 × 10 mm3 of the corpus callosum were obtained from four civilian subjects enrolled in the Transforming Research and Clinical Knowledge in Traumatic Brain Injury study (TRACK-TBI; https://tracktbi.ucsf.edu/transforming-research-and-clinical-knowledge-tbi) (Cases 1, 2, 4, and 5), and seven military subjects from the Center for Neuroscience and Regenerative Medicine Brain Tissue Repository (https://www.researchbraininjury.org, Uniformed Services University of the Health Sciences, Bethesda, MD; Cases 3 and 6–11). For each case, a next-of-kin or legal representative provided a written consent for donation of the brain for use in research. The brain tissues used have undergone procedures for donation of the tissue, its storage, and use of available clinical information that have been approved by the USU Institutional Review Board (IRB) prior to the initiation of the study. All experiments were performed in accordance with current federal, state, Department of Defense, and NIH guidelines and regulations for post-mortem analysis. A detailed description of demographics for the subjects from whom brain tissue samples were obtained is listed in Table 1 and Supplementary Table 1.
Table 1.
Main demographic and histopathological findings in patients with history of TBI and healthy controls
| Case | Age | Sex | Cause of death | PMI, h | Survival time | Relevant medical history | TBI clinical outcome | DAI grade b | Microscopically confirmed TAI in CC, APP-positive |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Extent | Severity, left/right | |||||||||
| 1 | 70 | Male | MVA resulting in SAH and SDH | <12 | 3 days | Alcohol abuse, depression, diabetes | Fatal | II | Bilateral | +++/+ |
| 2 | 60 | Male | MVA resulting in IPH | <12 | 26 days | NA | Fatal | II | Bilateral | +/+ |
| 3 | 33 | Male | MVA resulting in IPH | 30 | NA | Cholecystectomy, anxiety | Fatal | I | Bilateral | +/+ |
| 4 | 49 | Male | Fall resulting in IPH and SAH | <12 | 16 h | NA | Fatal | III | Bilateral | +/+ |
| 5 | 23 | Female | MVA resulting in SDH | <12 | 5 days | NA | Fatal | II | Unilateral (right) | 0/+ |
| 6 | 36 | Male | Suicide resulting in IPH | <12 | >1 year | Blast-induced TBI, anxiety, paranoia, depression, psychosis | Non-fatala | I | Unilateral (right) | NA/++ |
| 7 | 58 | Male | Hepatic cirrhosis with ascites | <12 | >1 year | Impact-induced TBI, alcohol abuse, drug abuse | Non-fatal | II | Unilateral (left) | +/0 |
| 8 | 32 | Male | Hypoxemia and cerebral anoxia | 12 | >1 year | Impact-induced TBI, severe headache, hypertension. | Non-fatal | I | Unilateral (right) | 0/+ |
| 9 | 52 | Male | Sudden cardio-respiratory arrest | 19 | NA | NA | Control | NA | Absent | 0/NA |
| 10 | 44 | Male | Sudden cardio-respiratory arrest | 22 | NA | NA | Control | NA | Absent | 0/0 |
| 11 | 54 | Male | Sudden cardio-respiratory arrest | 29 | NA | NA | Control | NA | Absent | 0/0 |
+/++/+++ = semi-quantitative severity assessment based on APP staining; IPH = intraparenchymal hemorrhage; MVA = motor vehicle accident; NA = not available; PMI = post-mortem interval; SAH = subarachnoid haemorrhage; SDH = subdural haemorrhage.
Non-fatal refers to the documented blast-induced TBI and not the immediate cause of death.
Based on Adams et al. (1989).
MRI acquisition
Prior to MRI scanning, each formalin-fixed brain specimen was transferred to a phosphate-buffered saline (PBS) filled container for 12 days to ensure that any residual fixative was removed from the tissue. The specimen was then placed in a 25 mm tube, and immersed in perfluoropolyether (Fomblin LC/8, Solvay Solexis), a proton-free fluid void of a proton-MRI signal. Specimens were imaged using a 7 T Bruker vertical bore MRI scanner equipped with a microimaging probe and a 25 mm quadrupole RF coil.
Multidimensional data were acquired using a 3D inversion recovery diffusion-weighted sequence with a repetition time of 1000 ms, and an isotropic voxel dimension of 300 µm. To encode the multidimensional magnetic resonance space spanned by T1 and T2 (i.e. T1-T2), by T1 and diffusion (i.e. MD-T1), and by T2 and diffusion (i.e. MD-T2), 56, 223, and 223 images were acquired, respectively, according to a previously published sampling scheme (Pas et al., 2020), which resulted in respective acquisition times of 4.5, 17.8, and 17.8 h. Additional parameters of the MRI pulse sequence can be found in the Supplementary material.
A standard DTI imaging protocol was applied with the same imaging parameters as the multidimensional data and using 21 diffusion gradient directions and four b-values ranging from 0 to 1400 s/mm2, which resulted in an acquisition time of 4.5 h.
Lastly, a high-resolution MRI scan with an isotropic voxel dimension of 100 µm was acquired using a fast low angle shot (FLASH) sequence (Matthaei et al., 1985) with a flip angle of 49.6° to serve as a high resolution reference image and facilitate co-registration of histopathological and magnetic resonance images. The acquisition time was approximately 2 h.
Histology and immunohistochemistry
After MRI scanning, each corpus callosum tissue block was transferred for histopathological processing. Tissue blocks from each brain specimen were processed using an automated tissue processor (ASP 6025, Leica Biosystems). After tissue processing, each tissue block was embedded in paraffin and cut in a series of 5-µm thick consecutive sections. The first two sections were selected for haematoxylin and eosin, and Luxol fast blue (LFB) stains, while the remaining sections were selected for immunohistochemistry procedures using a Leica Bond III automated immunostainer with a diaminobenzidine chromogen detection system (DS9800, Leica Biosystems). Immunohistochemistry was performed for anti-APP for the detection of TAI, anti-ionized calcium-binding adapter molecule 1 (Iba-1) to assess microglia reactivity, glial fibrillary acidic protein (GFAP), to evaluate presence of astrogliosis and myelin basic protein (MBP) to evaluate possible myelin loss. Two sections per antibody were stained at 300-µm apart from each other, in accordance with the MRI slice thickness. Staining procedures details can be found in the Supplementary material.
All stained sections were digitally scanned using an Aperio scanner system (Aperio AT2 - High Volume, Digital whole slide scanning scanner, Leica Biosystems, Inc.) and stored in Biolucida, a hub for 2D and 3D image data (MBF Bioscience) for further assessment and analyses. A Zeiss Imager A2 (ImagerA2 microscope, Zeiss) bright-field microscope with ×40 and ×63 magnification lenses was used to identify and photograph histological and pathological details, as needed.
Grading of the injury
It is well recognized that both clinical and pathological aspects of TBI (i.e. closed head injury) are heterogeneous (Thompson et al., 2020). We therefore used three separate classifications and gradings to evaluate the cases examined in this study. First, the clinical outcomes in each case where a distinction was made based on whether the TBI was fatal or non-fatal. Second, the DAI grading was based on the distribution of axonal injury in the brain as proposed by Adams et al. (1989). Lastly, we used a semi-quantitative grading based on APP immunostaining, i.e. APP-positive DAI lesions (Blumbergs et al., 1995). The extent of the DAI lesions was classified as unilateral or bilateral according to their presence in one or both hemispheres and their level of severity was semi-quantitatively assessed based on APP estimated loads as occupying <50% (+), ∼50% (++) or >50% (+++) of the microscopically-examined cross-sectional section surface.
Quantification of axonal damage
Images of APP stained sections were digitized using an Aperio whole slide scanning scanner system (Leica Biosystems) at ×20 magnification. The following steps, all implemented using MATLAB (The Mathworks, Natick, MA), were taken to allow for a quantitative analysis of the APP images. First, the images were transformed into a common, normalized space to enable improved quantitative analysis (Macenko et al., 2009). Then, the normalized images were deconvolved to unmix the primary (APP) and secondary (haematoxylin and eosin) stains, and background to three separate channels (Ruifrok and Johnston, 2001). Once an APP-only image has been obtained, a final thresholding step was taken to exclude non-specific staining and to allow for a subsequent per cent area calculation. These steps are illustrated in Fig. 1F, M and T.
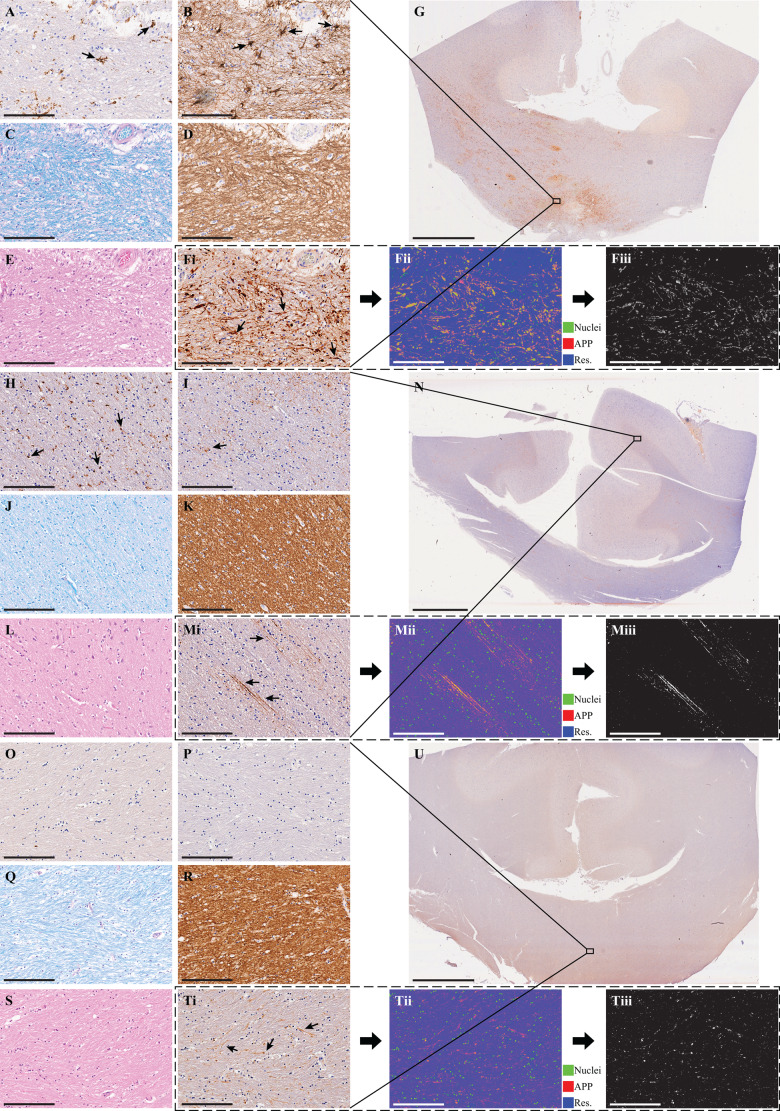
Figure 1.
Histological findings from three representative cases. Case 1 (A–F), Case 2 (H–M), and Case 4 (O–T). Micrographs from approximately the same regions of (A, H and O) Iba1 (microglia), (B, I and P) GFAP (astrocytes), (C, J and Q) Luxol fast blue (myelin), (D, K and R) MBP (myelin), (E, L and S) haematoxylin and eosin. [F(i), M(i) and T(i)] Original APP images, [F(ii), M(ii) and T(ii)], which are deconvolved to obtain the APP density, in F(iii), M(iii) and T(iii). Scale bar = 200 µm in all micrographs and 5 mm in the whole-mount corpus collosum APP images in G, N and U. Case 1 survived 3 days post TBI and presented reactive microglia (A), astrogliosis (B), no apparent myelin loss (C and D), and TAI APP lesions (F). Case 2 survived 26 days post TBI and presented reactive microglia (H), very sparse astrogliosis (I), no apparent myelin loss (J and K), and TAI APP lesions (M). Case 4 survived 16 h post TBI and did not present any glial reactivity (O and P), no apparent myelin loss (Q and R), but did present TAI APP lesions (T).
From each tissue section, based on APP staining, corpus callosum TAI lesions were identified by an experienced neuropathologist (D.I.) as white matter areas with swollen axonal varicosities, axonal bulbs, or distorted axons. Accordingly, regions of interest of normal-appearing white matter and TAI lesions were manually defined. Regions of interest of normal white matter from control cases were defined as well. Additionally, grey matter regions of interest from adjacent cingulate cortex were defined in all sections. Twelve regions of interest, covering together on average 81 mm2 of tissue, were identified per tissue section. Four cases and their regions of interest are shown in Supplementary Fig. 5. After extracting the regions of interest, APP density was expressed as the percentage of total area within the region of interest in the binary deconvolved APP image. In total, 132 regions of interest from 11 cases were included in this study.
MRI analysis
Diffusion tensor MRI processing
Diffusion tensor imaging parameters (Basser et al., 1994) were calculated using in-house MATLAB (The Mathworks, Natick, MA) code based on previous work (Barmpoutis and Vemuri, 2010).
Voxel-averaged T1 and T2 map processing
Conventional quantitative relaxation maps were first computed by fitting the signal decay to monoexponential functions. The T1 value was computed by fitting a subset of the multidimensional data that included 20 images with inversion times in the range of 12 ms and 980 ms. The T2 value was computed by fitting a subset of the multidimensional data that included 20 images with echo times in the range of 10.5 ms and 125 ms.
Multidimensional MRI processing
Here we implemented a marginally-constrained, l2-regularized, non-negative least square optimization to compute the multidimensional distribution in each voxel, as previously described (Benjamini and Basser, 2017; Pas et al., 2020). It is a well-tested approach that had been proved robust and reliable (Provencher, 1982; Kroeker and Henkelman, 1986; Song et al., 2002; Hürlimann et al., 2003; Mitchell et al., 2012; Celik et al., 2013; Benjamini and Basser, 2018), which in this study had resulted in three types of distributions in each voxel: T1-T2, MD-T2, and MD-T1.
If one considers the multidimensional distributions as spectra, it is possible to use them to generate images of specific spectral components by means of integration over a predefined parameter range (e.g. the rectangle T1-T2 range highlighted in Fig. 2). The integral value is a number between 0 and 1, representing a certain spectral component (SC) in a given multidimensional distribution, which can be computed in each voxel to generate an image of that specific SC (Labadie et al., 1994).
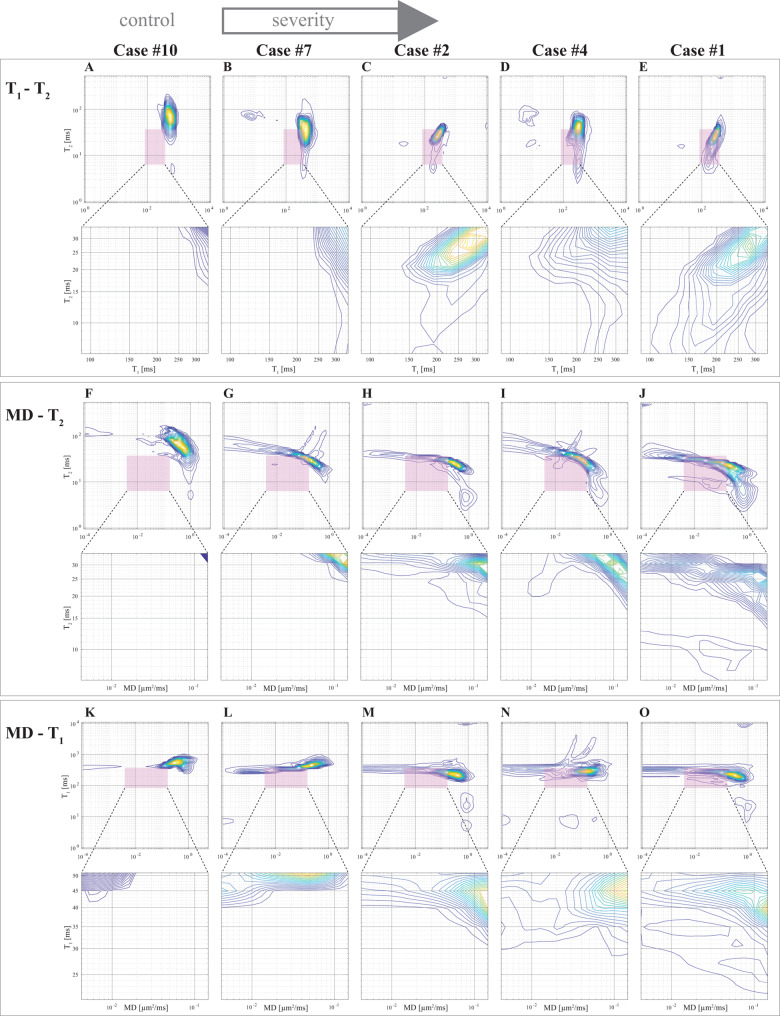
Figure 2.
Multidimensional spectra of the TAI lesions in representative cases with increasing degree of severity (left to right). (A–E) T1-T2, (F–J) MD-T2, and (K–O) MD-T1. The preselected T1-T2-MD range, T1 = [91.03, 339.32] ms, T2 = [6.70, 34.85] ms, and MD = [0.004, 0.146] µm2/ms is highlighted as pink rectangles. Below each distribution, a magnification of the highlighted spectral region of interest is shown. The progressive shift towards shorter T1, shorter T2, and slower diffusivity as the severity of the injury increases is evident.
While the T1-T2-MD spectra contain rich information from multiple tissue components such as myelin (Laule et al., 2008), we focused here on discovering spectral substrates of axonal injury. Two factors guided us in the search: (i) we know the injury is primarily a white matter injury; and (ii) we know the T1-T2-MD range of what is deemed as ‘normal’ white matter from previous multidimensional MRI studies (and current results) (Benjamini and Basser, 2017; Pas et al., 2020). Our driving hypothesis was therefore that microscopic white matter injury would affect the normal white matter spectral component, and that the injury-associated spectral information would be found in the vicinity of the normal white matter component. Based on this notion, we have identified a T1-T2-MD range, T1 = [91.03, 339.32] ms, T2 = [6.70, 34.85] ms, and MD = [0.004, 0.146] µm2/ms, in which we observed the presence of injury-associated spectral information. We developed a spectral thresholding algorithm to account for variability between different corpus callosum samples, and to extracts the injury information in a more accurate and unsupervised way. We used an adaptive threshold in the spectral domain to detect the edge that distinguishes between the normal white matter and the axonal injury spectral components. Additional information regarding multidimensional MRI processing can be found in the Supplementary material and in Supplementary Fig. 1.
Repeatability and reproducibility of the T1-T2, MD-T1, and MD-T2 TAI SC images were evaluated by repeating the multidimensional MRI acquisitions for Cases 1 and 2. For each of these subjects, the duplicate datasets were processed using the same pipeline and then compared. Bland-Altman plots were used to quantify the agreement between all the non-zero voxels in the injury SC images from the first and second acquisitions (Bland and Altman, 1986). The standard deviation of the measurement error (in units ×100 SC) in Case 1 was 5.3 for T1-T2 injury SC, 5.5 for MD-T1 injury SC, and 5.3 for MD-T2 injury SC (Supplementary Fig. 2), and in Case 2 it was 4.8 for T1-T2 injury SC, 4.3 for MD-T1 injury SC, and 6.5 for MD-T2 injury SC (Supplementary Fig. 3).
Histology MRI co-registration
The high-resolution magnetic resonance images were used as anatomical references to which the histological images were registered to. Areas in the histological images that grossly diverged from the wet tissue state (i.e. the magnetic resonance images) due to deformation were manually removed, while maintaining the image aspect ratio. Following convergence of 2D affine co-registration of histology and magnetic resonance images (Image Processing Toolbox, MATLAB, The Mathworks, Natick, MA), we performed a 2D diffeomorphic registration refinement between the APP image slices and MRI volumes. This was done to recover true in-plane tissue shape and bridge over residual differences between the modalities. The diffeomorphic registration procedure in this study was performed using an efficient implementation of the greedy diffeomorphic algorithm (Joshi et al., 2004), provided as an open-source software package (https://github.com/pyushkevich/greedy). The greedy software was initialized and used as previously described (Adler et al., 2018). The transformed histology images were overlaid on magnetic resonance images to assess the quality of the co-registration.
Statistical analysis
Demographic variables and post-mortem interval (i.e. time between death and start of autopsy) were compared between controls, non-fatal and fatal TBI patients (per clinical outcome) using Mann-Whitney U-test or unpaired two-tailed Student’s t-test.
Linear mixed-effects models were used to compare MRI-derived and histopathological measures. We fitted a linear mixed-effects model using three predictors: region of interest (control corpus callosum, normal-appearing white matter in mild TAI corpus callosum, normal-appearing white matter in severe TAI corpus callosum, mild TAI lesions, and severe TAI lesions), clinical outcome (control, fatal and non-fatal TBI), and subject (i.e. case number). To allow unbiased statistical comparisons, the voxel-averaged MRI parameters were adjusted across all tissue samples by dividing them by the mean of each parameter in each specific sample to correct for possible between-sample differences arising from post-mortem effects. We fitted the model for each of the measured quantities (magnetic resonance variables and APP density), with fixed-effects for region of interest, and potentially correlated random-effects for intercept and region of interest grouped by the clinical outcome, and for intercept and region of interest grouped by the case number. The random-effects were selected to account for MRI differences that might exist due to clinical outcome, patient-specific variations, and possible within-subject correlation among measures. False discovery rate (FDR) correction was carried out to take the overall number of pairwise contrasts into account.
The potential correlation between all conventional voxel-averaged and multidimensional MRI measures and APP density in the different tissue types was investigated using linear regression, with Pearson’s correlation coefficient, r, reported.
A P-value of 0.05 was considered statistically significant. MATLAB was used for the computation.
Data availability
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Results
Traumatic axonal injury pathology in the corpus callosum
Table 1 and Supplementary Table 1 summarize the main demographic data for each examined brain and histopathological findings observed in each studied corpus callosum. No significant differences were found for age and post-mortem interval between fatal and non-fatal TBI, and controls. No significant differences were found for APP-based TAI grade, APP-based TAI extent and APP-based severity between the fatal and non-fatal TBI cases (Supplementary Table 1).
The post-injury survival time in all fatal TBI cases was sufficiently long to allow for pathological APP accumulation (Sherriff et al., 1994). Except for Case 6, subjects who died of causes unrelated to TBI had documented head trauma. The subject described in Case 6 sustained a fatal head trauma that resulted in immediate death, which would likely have prevented significant pathological accumulation of APP. Considering that APP is the histopathological marker we evaluate in this study, and because of their documented blast-induced TBI, we classified Case 6 as non-fatal TBI. It is noteworthy that classification as fatal or non-fatal TBI is important for understanding the clinical context; however, it does not affect the MRI findings and determinations in this study, which are made regardless of the cause of death.
Figure 1 shows simultaneous views from three representative TBI cases immunostained for markers of microglia (Iba-1), astrocytes (GFAP), myelin (MBP), and TAI lesions (APP), histology stains for myelin (LFB) and general histological assessment (haematoxylin and eosin), and the deconvolved APP images along with the obtained APP densities.
Levels of myelin density as assessed by both MBP immunohistochemistry and LFB stain did not show any marked loss across all considered regions (e.g. Fig. 1C, D, J, K, Q and R). By contrast, microglia activation and reactivity varied across all cases (e.g. Fig. 1A, H and O). Likewise, astroglial reactivity was variable as well (e.g. Fig. 1B, I and P). Astroglial and microglial cells reactivities depend on various biological timing factors, most notably, time interval differences between the TBI exposure and autopsy (Ramlackhansingh et al., 2011), which, in our study, varied greatly between the fatal and non-fatal TBI groups.
Additionally, apart from Case 1, haematoxylin and eosin-based assessment of each corpus callosum did not reveal evidence of ischaemic-necrotic lesions, presence of vascular lesions, microhaemorrhages, or tissue rarefaction.
One hundred and thirty-two tissue regions of interest were selected based on the APP density images and blind to MRI, consisting of four APP-positive regions from each TAI case (total of 32), four normal-appearing white matter regions from each TAI case (total of 32), eight white matter regions from the control cases (total of 24), and four cortical grey matter regions from all cases (total of 44). In total, 132 regions of interest from 11 cases were included in this study.
Transverse and longitudinal relaxations shorten with traumatic axonal injury severity
We observed a consistent and gradual change in the way in which T1, T2, and MD were distributed as the APP-based severity of the TAI was increasing. Figure 2 shows five cases with increased injury severity and their respective T1-T2 (Fig. 2A–E), MD-T2 (Fig. 2F–J), and MD-T1 (Fig. 2K–O) distributions averaged from 20 voxels within control white matter and TAI lesions. The shift in the distributions can be illustrated by examining the most affected T1-T2-MD ranges, which are highlighted as pink rectangles. A magnification of these ranges in the spectra are shown below each distribution. Qualitatively, the magnified regions show a gradual shortening of T1 and T2 as the severity of the injury is increased (Fig. 2, left to right). A shift towards lower MD values is also noticeable, although not as consistent as changes in the relaxation distributions.
Multidimensional and voxel-averaged MRIs of traumatic axonal injury
Based on the control and TAI cases we examined here, we have identified a T1-T2-MD range, (T1 = [91.03, 339.32] ms, T2 = [6.70, 34.85] ms, and MD = [0.004, 0.146] µm2/ms) in which we assessed most of the spectral information regarding the injury resides. Integration according to the spectral signature within this range, as detailed in the ‘Materials and methods’ section, was performed, resulting in MD-T1, MD-T2, and T1-T2 injury SC images.
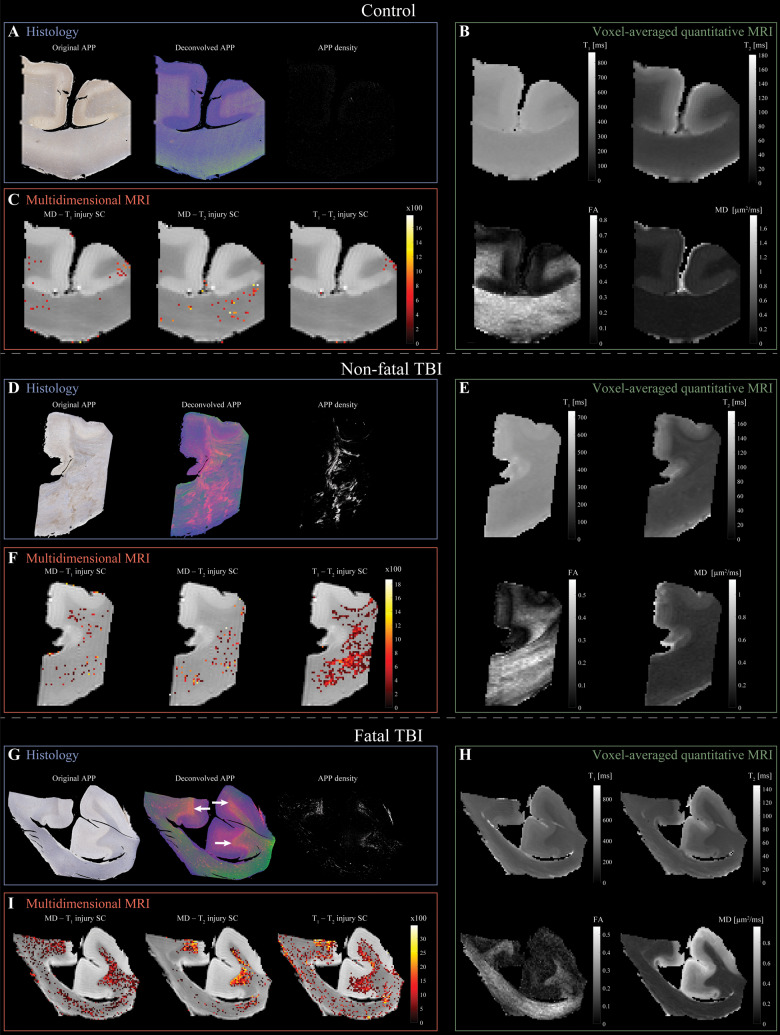
Figure 3 shows conventional and multidimensional magnetic resonance and histological images of three representative cases: control, non-fatal and fatal TBI (Cases 10, 6, and 2, respectively). Histological images of the control case show negative APP staining (Fig. 3A), compared with positive APP staining in the injured samples (Fig. 3D and G). Conventional, voxel-averaged relaxation and diffusion images of the control, non-fatal and fatal cases, are shown in Fig. 3B, E and H, respectively. Lastly, the multidimensional injury images of the control, non-fatal and fatal cases, are shown in Fig. 3C, F and I, respectively. Note that to facilitate visualization, the injury SC images were thresholded at 10% of the maximal intensity and overlaid on grayscale proton density images. More multidimensional injury images are shown in Supplementary Fig. 4.
Figure 3.
Multidimensional and voxel-averaged MRIs of TAI. (A–C) Control brain (Case 10). (A) APP histological images co-registered to MRIs. Deconvolved histological image: red = APP stain. Negligible APP density was detected. (B) Conventional MRI maps of T1, T2, FA and MD do not show visible abnormalities. (C) Multidimensional injury maps overlaid onto proton density images show absent of significant injury. (D–F) Non-fatal TBI brain (Case 6). (D) APP histological images co-registered to MRIs show visible TAI lesions in the corpus callosum. (E) Conventional MRI maps of T1, T2, FA and MD do not show visible abnormalities in the corpus callosum. (F) Multidimensional injury maps overlaid onto proton density images show significant injury in white matter (in particular, the T1-T2 injury SC). (G–I) Fatal TBI brain (Case 2). (G) APP histological images co-registered to MRIs show TAI lesions in regions of white matter/grey matter interface (white arrows). (H) Conventional MRI maps of T1, T2, FA and MD do not show visible abnormalities in white matter/grey matter interface. (I) Multidimensional injury maps overlaid onto proton density images show substantial injury along the white matter/grey matter interface.
Histopathological, multidimensional and conventional MRI findings
We grouped the 132 tissue regions into controls, normal-appearing white matter in TBI patients, and TAI lesions in TBI patients. We further partitioned the TBI patients into mild and severe TAI according to the APP staining-based severity in Table 1 to assess whether the examined MRI metrics are able to identify TAI with different degrees of APP-based severity (Cases 1–4 and 6 were grouped as severe TAI and Cases 5, 7 and 8 as mild TAI). We also applied a commonly used strategy (Holleran et al., 2017; Cooper et al., 2019) to correct for possible between-subject differences arising from post-mortem effects; we adjusted each voxel-averaged MRI parameter by dividing them by the mean for that parameter across all white matter (in control subjects) and normal-appearing white matter (in subjects with TBI) voxels in each brain sample.
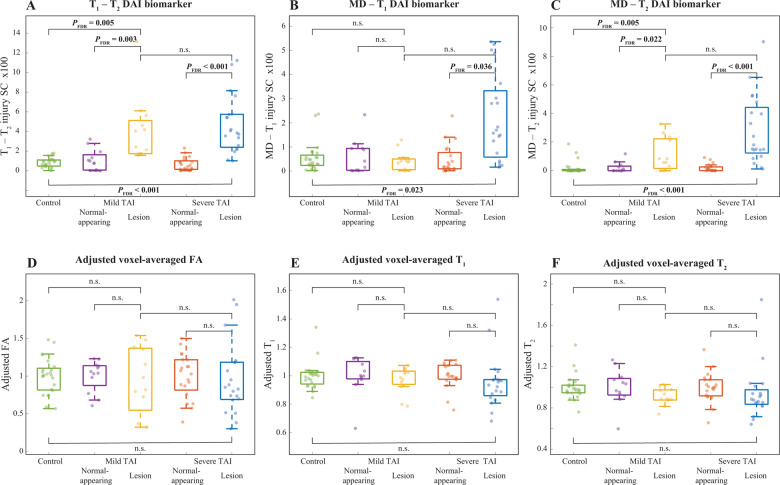
Compared to controls’ white matter, mild TAI lesions showed increased T1-T2 injury SC (FDR-corrected P-value, PFDR = 0.005), increased MD-T2 injury SC (PFDR = 0.005), and increased per cent area APP (PFDR < 0.001) (Fig. 4 and Supplementary Table 2).
Figure 4.
Between-group comparisons of voxel-averaged and multidimensional biomarkers and histopathological measures. Box plots showing between-group differences among control corpus callosum, normal-appearing white matter in mild TAI corpus callosum, mild TAI lesions, normal-appearing white matter in severe TAI corpus callosum, and severe TAI lesions. (A) T1-T2 injury SC; (B) MD-T1 injury SC; (C) MD-T2 injury SC; (D) adjusted voxel-averaged FA; (E) adjusted voxel-averaged T1; and (F) adjusted voxel-averaged T2. All parameters are averaged across their corresponding spatial regions of interest.
Compared to controls white matter, severe TAI lesions showed increased T1-T2 injury SC (PFDR < 0.001), increased MD-T1 injury SC (PFDR = 0.023), increased MD-T2 injury SC (PFDR < 0.001), and increased per cent area APP (PFDR < 0.001) (Fig. 4 and Supplementary Table 2).
Compared to normal-appearing white matter tissue regions in TBI patients with mild TAI, mild TAI lesions showed increased T1-T2 injury SC (PFDR = 0.003), increased MD-T2 injury SC (PFDR = 0.022), and increased per cent area APP (PFDR < 0.001) (Table 2 and Fig. 4).
Table 2.
Comparisons of MRI parameters and pathological measures among mild TAI, severe TAI, NAWM in mild and severe TAI tissue regions
| Variable |
NAWM in mTAI
n = 12 |
mTAI lesions
n = 12 |
NAWM in sTAI
n = 20 |
sTAI lesions
n = 20 |
mTAI lesions versus NAWM in mTAI |
sTAI lesions versus NAWM in sTAI |
sTAI lesions versus mTAI lesions |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean (SE) | Mean (SE) | Mean (SE) | Mean (SE) | Estimated mean difference (95% CI) | P/PFDR |
Estimated mean difference
(95% CI) |
P/PFDR |
Estimated mean difference
(95% CI) |
P/PFDR | |
| T1-T2 SC, ×100 | 1.09 (0.39) | 4.09 (0.97) | 0.67 (0.29) | 4.70 (1.03) | 3.00 (0.95, 5.05) | <0.001/0.003 | 4.03 (1.93, 6.13) | <0.001/<0.001 | 0.74 (1.93, 3.41) | 0.667/0.943 |
| MD-T1 SC, ×100 | 0.56 (0.29) | 0.41 (0.20) | 0.46 (0.19) | 2.20 (0.64) | −0.15 (−0.84, 0.54) | 0.707/0.793 | 1.74 (0.43, 3.05) | 0.018/0.036 | 1.57 (0.24, 2.90) | 0.008/0.060 |
| MD-T2 SC, ×100 | 0.22 (0.24) | 1.14 (0.24) | 0.19 (0.19) | 2.84 (0.65) | 0.92 (0.25, 1.58) | 0.008/0.022 | 2.65 (1.32, 3.98) | <0.001/<0.001 | 1.72 (0.36, 3.08) | 0.015/0.060 |
| Adjusted FA | 1.00 (0.07) | 0.94 (0.23) | 1.00 (0.05) | 0.96 (0.15) | −0.06 (−0.54, 0.41) | 0.793/0.793 | −0.04 (−0.36, 0.28) | 0.790/0.790 | 0.02 (−0.53, 0.57) | 0.943/0.943 |
| Adjusted MD | 1.00 (0.06) | 1.04 (0.06) | 1.00 (0.05) | 1.13 (0.11) | 0.04 (−0.13, 0.21) | 0.675/0.793 | 0.13 (−0.09, 0.36) | 0.261/0.418 | −0.09 (−0.14, 0.33) | 0.452/0.903 |
| Adjusted T1 | 1.00 (0.03) | 0.97 (0.03) | 1.00 (0.02) | 0.95 (0.06) | −0.03 (−0.11, 0.05) | 0.504/0.793 | −0.04 (−0.17, 0.09) | 0.523/0.597 | −0.01 (−0.15 0.12) | 0.847/0.943 |
| Adjusted T2 | 1.00 (0.04) | 0.91 (0.04) | 1.00 (0.03) | 0.94 (0.08) | −0.09 (−0.20, 0.02) | 0.126/0.251 | −0.06 (−0.23, 0.11) | 0.497/0.597 | 0.03 (−0.15, 0.21) | 0.730/0.943 |
| % area APP | 0.40 (1.99) | 20.67 (3.71) | 0.97 (1.54) | 26.65 (6.35) | 20.27 (12.02, 28.52) | <0.001/<0.001 | 25.68 (12.87, 38.49) | <0.001/<0.001 | 5.96 (−8.45, 20.37) | 0.419/0.903 |
Values in bold indicate statistical significance. CI = confidence interval; mTAI = mild TAI; NAWM = normal-appearing white matter; SE = standard error; sTAI = severe TAI.
Compared to normal-appearing white matter tissue regions in TBI patients with severe TAI, severe TAI lesions showed increased T1-T2 injury SC (PFDR < 0.001), increased MD-T1 injury SC (PFDR = 0.036), increased MD-T2 injury SC (PFDR < 0.001), and increased per cent area APP (PFDR < 0.001) (Table 2 and Fig. 4).
The MD-T1 and MD-T2 injury biomarkers were the only ones that were able to distinguish between mild TAI and severe TAI lesions (P = 0.008 and P = 0.015, respectively); however, they did not survived correction for multiple comparisons.
None of the other MRI metrics showed significant between-group differences (Table 2, Fig. 4 and Supplementary Table 2).
Correlation between MRI measures and APP density
To determine whether MRI could potentially detect TAI in TBI patients, we performed radiological–pathological correlation analyses with histological APP density and all the investigated MRI parameters.
Figure 5 summarizes the association between the investigated magnetic resonance metrics—conventional voxel-averaged and multidimensional images—and the pathological findings in normal white matter, cortical grey matter, and TAI tissue blocks.
Figure 5.
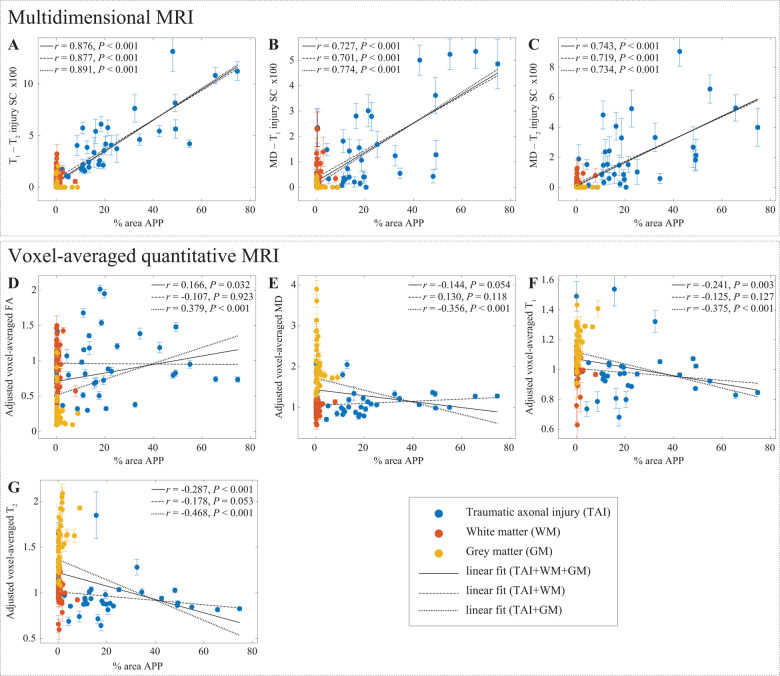
APP density. APP density (% area) from 132 tissue regions, consisting of 4 APP-positive regions from each TAI case (total of 32, blue dots), four to six normal-appearing white matter regions from all cases (total of 56, red dots), and four cortical grey matter regions from all cases (total of 44, yellow dots), and the corresponding magnetic resonance parameter correlations. Individual data-points represent the mean region of interest value from each post-mortem tissue sample. Scatterplots of the mean (with 95% confidence interval error bars) % area APP and (A) T1-T2, (B) MD-T1, and (C) MD-T2 injury SCs show positive and significant correlation with APP density. Both T1-T2 and MD-T2 injury SCs demonstrated specificity exclusively towards TAI (all grey matter and normal-appearing white matter had negligible intensities). The conventional MRI metrics (D–G) did not result in strong and significant correlations with % area APP.
To assess the relationship of the MRI parameters with the degree of injury, all tissue regions of interest were grouped together and correlated with the APP density (Fig. 5, solid lines). We found that APP density was strongly and significantly correlated with the T1-T2 injury SC (r = 0.876, P < 0.001), the MD-T1 injury SC (r = 0.727, P < 0.001), and the MD-T2 injury SC (r = 0.743, P < 0.001). Importantly, these results indicated that higher intensity of the multidimensional injury biomarkers is associated with increased DAI severity, regardless of the tissue type studied. From the conventional voxel-averaged images, we found that APP density had very weak yet significant correlations with adjusted FA (r = 0.166, P = 0.032), with adjusted T1 (r = −0.241, P = 0.003) and with adjusted T2 (r = −0.287, P < 0.001).
To assess the capability of the MRI parameters to differentiate between normal and injured white matter with respect to APP density, normal white matter and TAI regions of interest were grouped together (Fig. 5, dashed lines). We found that the correlations of multidimensional injury biomarker images with APP density have barely changed by excluding the cortical grey matter regions of interest. Conversely, this grouping resulted in insignificant voxel-averaged MRI metrics correlations with respect to APP density, showing that none of these variables can be used to distinguish lesion and non-lesion white matter tissue.
To assess the capability of the MRI parameters to differentiate between grey matter and injured white matter with respect to APP density, grey matter and TAI regions of interest were grouped together (Fig. 5, dotted lines). Here too, we found that the correlations of multidimensional injury biomarker images with APP density have barely changed by excluding the normal white matter regions of interest. All of the voxel-averaged MRI parameters were significantly correlated with APP density (adjusted FA: r = 0.379, P < 0.001; adjusted MD: r = −0.356, P < 0.001; adjusted T1: r = −0.375, P < 0.001; adjusted T2: r = −0.468, P < 0.001), showing that they are able to distinguish between grey matter and axonal injury. However, given their inability to differentiate normal white matter and axonal injury, these findings merely show that voxel-averaged parameters can distinguish grey matter and white matter.
Discussion
This study crucially identified potential imaging biomarkers of DAI pathology from the joint analysis of multidimensional MRI and histopathological data in the corpus callosum. The ability to selectively extract a specific T1-T2-MD spectral range, or ‘spectral signature’, and provide its corresponding image, allows multidimensional MRI to achieve good separation between subjects and microscopic lesion and non-lesion regions. We found that the multidimensional MRI axonal injury images are significantly and strongly correlated with histological evidence of DAI. Furthermore, these non-invasive DAI biomarkers produce negligible intensities in grey matter and in normal-appearing white matter.
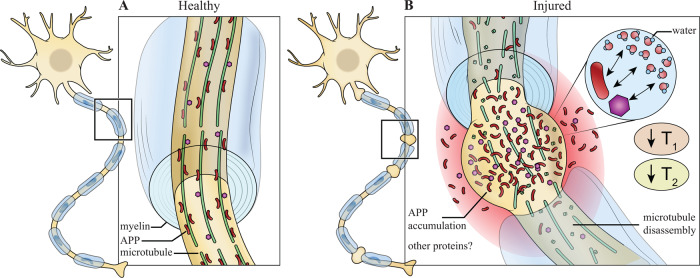
Multidimensional MRI allows us to capture the strong association of T1 and T2 shortening and the severity of the microscopic axonal injury. Proteins, which have very short T2 values, interact with free and partially bound water, reducing both T1 and T2 (Edzes and Samulski, 1977; Zhang et al., 2007; Filo et al., 2019). Axonal injury causes microtubule disruption that induces long-term pathological co-accumulation of APP and other intra-axonal proteins (Chen et al., 2004), and to the formation of axonal varicosities (Fig. 6). While non-invasive and direct detection of proteins may not be feasible with adequate spatial resolution, we suggest that these protein aggregates could act as endogenous relaxation enhancers of the surrounding water, which can be captured by observed shortened T1-T2 (Fig. 6). However, quantitative biophysical modelling and further detailed experiments will be required to confirm or refute this hypothesis.
Figure 6.
Proposed explanation of T1 and T2 shortening in TAI lesions. Schematic representation of (A) healthy neuron and (B) traumatized neuron. In healthy neurons, APP and other proteins transported through axons via microtubules by fast axonal transport without reaching detectable levels. In traumatized neurons, microtubules undergo mechanical failure resulting in transport disruption that leads to accumulation of APP and possibility other proteins, which causes local axonal swelling. As known T1 and T2 relaxation enhancers, proteins accumulate and affect the surrounding water by shortening their observed T1 and T2 values.
Robust quantitative radiological-pathological correlations allowed us to visualize, within each TBI patient, regions with substantial TAI lesions and regions that were not affected by trauma. We showed that voxel-averaged MRI approaches were unable to detect any differences between controls, normal-appearing white matter in TBI subjects, and TAI lesions (Fig. 4, Table 2 and Supplementary Table 2). The main reason for this insensitivity is that using these approaches one cannot selectively extract the injury-affected spectral range per subject because these images contain only scalar averages, not distributions. Conversely, multidimensional MRI parameters, primarily T1-T2 and MD-T2, were able to distinguish between lesions and normal-appearing white matter regions within TBI patients (Table 2 and Supplementary Table 2). Replacing the conventionally obtained voxel-averaged scalar value with a distribution increases the sensitivity to detect subtle changes, that would have been otherwise averaged-out.
Consistent with previous literature (Johnson et al., 2013), we found that the corpus callosum was affected by DAI following trauma. Apart from Case 1, none of the corpus callosa we evaluated showed macroscopic damage, cytotoxic oedema, or large (>1 mm) lesions. No myelin loss was observed in all the examined cases, and glial cells reactivity varied greatly between cases, most likely due to variability in the survival time. These histopathological findings suggest that the damage from TAI in the cases we examined was microscopically limited, and in most cases did not result in observable macroscopic loss.
Remarkably, there have been only a few quantitative MRI studies of TBI that combined relaxometry and diffusion with histological correlation. Our findings of decreased T1 and T2 are in contrast with previous animal models studies that reported an increase in T2 following TBI. The increase in T2 in those studies was associated with oedema (Ducreux et al., 2005; Obenaus et al., 2007; Immonen et al., 2009; Li et al., 2016), necrosis (Mac Donald et al., 2007), and haemorrhage (Long et al., 2015). In animal models of TBI, where moderate to severe injury is prevalent, macroscopic damage that causes oedema and haemorrhage may dominate the MRI signal. Most of the cases in our study had a survival time longer than 14 days, which is typically the time it takes oedema to resolve. Of the three acute cases with survival time under 7 days (Cases 1, 4, and 5), oedema and loss of tissue were observed only in Case 1 (care was taken such that none of the analysed tissue blocks were selected from that area).
Although DTI metrics are considered to be sensitive to microstructural alterations due to TBI (Mac Donald et al., 2007; Holleran et al., 2017), it is becoming increasingly evident that they are inconsistent in their observed response (Budde and Annese, 2013; Kamnaksh et al., 2015; Dennis et al., 2018). Reflecting this heterogeneity, voxel-averaged FA and MD in our study presented a large variability in all groups that had led to insignificant changes due to injury.
Common to all ex vivo human MRI studies, our data include the effects of post-mortem degeneration, fixation and resulting dehydration. First, the fixation process by itself changes the tissue properties, such that a direct comparison with in vivo data is not possible. In the case of human tissue, delayed fixation (i.e. post-mortem interval) affects the measured MRI parameters as well (Shepherd et al., 2009; Roebroeck et al., 2019), which further complicates comparison with in vivo human results. The post-mortem interval in this study was reasonably homogeneous, with 7 of 11 cases ≤12 h, and the rest ≤30 h. Moreover, histopathology results indicate preservation of APP and other immunostains in all samples, regardless of post-mortem interval. Although this study was designed to include both fatal and non-fatal TBI outcomes, this design results in inherently heterogeneous survival time, which presents a limitation. Glial reactivity, which is highly dependent on the time from injury (Ramlackhansingh et al., 2011), cannot be controlled for in our study, and cannot be ruled out as a contributing factor to our MRI findings. Future studies, in which the whole brain is imaged and processed, may yield additional information regarding TAI localization and severity in the brain following different types of trauma.
Here, we would like to emphasize the T1-T2 MRI sequence results because of its potential ease of migration towards clinical applications. Although technological progress is being made (Wu et al., 2016; Hutter et al., 2018), substantial gaps remain between preclinical MRI, like the one used here, and clinical MRI systems, and achieving satisfactory diffusion encoding in the latter is a particularly challenging task (Mukherjee et al., 2008; Jones et al., 2018). Conversely, T1-T2-weighted acquisition is more clinically feasible (Deoni et al., 2008; Shepherd et al., 2020), especially considering the relatively small number of T1-T2-weighted images that were required in our study, i.e. 56. Here we used roughly half of the amount of data that has been previously used in the human brain in vivo (Kim et al., 2020), which would have reduced the acquisition time of a single-slice dataset from 20 to 10 min. Because T1, T2, and diffusion dynamics are different in fixed tissue compared with living systems, further research will be needed to establish whether and how the T1-T2-MD range of multidimensional magnetic resonance parameters is altered in vivo.
A non-invasive method to detect DAI pathology with high specificity and in vivo would be a major medical contribution. Here, we have shown that DAI due to trauma has a multidimensional spectral signature, which can be used to generate injury biomarker images that closely follow APP histopathology: axonal injury image intensity scales with pathological APP accumulation, and importantly, lack of APP corresponds to negligible intensity. Our results indicate that the T1-T2-MD range we identified is directly linked to microscopic tissue alterations due to DAI. We believe that the improved sensitivity of these novel injury biomarkers towards DAI is advancing the neuroimaging field closer towards non-invasive quantitative ‘histology’. This neuroimaging tool provides an injury-only image that may help clinicians and clinical investigators to detect and visualize microscopic DAI lesions in the brain. As evidence that indicates DAI is a likely pathological substrate for concussion is mounting (Smith and Stewart, 2020), the importance of developing a means to detect it clinically becomes clear.
Supplementary Material
Acknowledgements
We thank the subjects’ families that consented for brain donations for the better understanding of TBI consequences. The authors thank Dr Jessica Ettedgui for fruitful discussions, and Dr Thaddeus Haight for insights into the statistical analysis. We also thank Mrs Patricia Lee, Mrs Nichelle Gray and Mr Paul Gegbeh for their valuable technical work. We are grateful to Mrs Stacey Gentile, Mrs Deona Cooper and Mr Harold Kramer Anderson for their administrative assistance. We thank the TRACK-TBI Investigators: Opeolu Adeoye, University of Cincinnati; Neeraj Badjatia, University of Maryland; Kim Boase, University of Washington; Jason Barber, MS, University of Washington; Yelena Bodien, Massachusetts General Hospital; M. Ross Bullock, University of Miami; Randall Chesnut, University of Washington; John D. Corrigan, ABPP, Ohio State University; Karen Crawford, University of Southern California; Ramon Diaz-Arrastia, University of Pennsylvania; Sureyya Dikmen, University of Washington; Ann-Christine Duhaime, MassGeneral Hospital for Children; Richard Ellenbogen, University of Washington; V Ramana Feeser, Virginia Commonwealth University; Adam R. Ferguson, University of California, San Francisco; Brandon Foreman, University of Cincinnati; Raquel Gardner, University of California, San Francisco; Etienne Gaudette, University of Southern California; Joseph Giacino, Spaulding Rehabilitation Hospital; Dana Goldman, University of Southern California; Luis Gonzalez, TIRR Memorial Hermann; Shankar Gopinath, Baylor College of Medicine; Rao Gullapalli, University of Maryland; J. Claude Hemphill, University of California, San Francisco; Gillian Hotz, University of Miami; Sonia Jain, University of California, San Diego; C. Dirk Keene, University of Washington; Frederick K. Korley, University of Michigan; Joel Kramer, PsyD, University of California, San Francisco; Natalie Kreitzer, University of Cincinnati; Harvey Levin, Baylor College of Medicine; Chris Lindsell, Vanderbilt University; Joan Machamer, MA, University of Washington; Christopher Madden, UT Southwestern; Geoffrey T Manley, University of California, San Francisco; Alastair Martin, University of California, San Francisco; Thomas McAllister, Indiana University; Michael McCrea, Medical College of Wisconsin; Randall Merchant, Virginia Commonwealth University; Pratik Mukherjee, University of California, San Francisco; Lindsay Nelson, Medical College of Wisconsin; Laura B. Ngwenya, University of Cincinnati; Florence Noel, Baylor College of Medicine; Amber Nolan, University of California, San Francisco; David Okonkwo, University of Pittsburgh; Eva Palacios, University of California, San Francisco; Daniel Perl, Uniformed Services University; Ava Puccio, University of Pittsburgh; Miri Rabinowitz, University of Pittsburgh; Claudia Robertson, Baylor College of Medicine; Jonathan Rosand, MSc, Massachusetts General Hospital; Angelle Sander, Baylor College of Medicine; Gabriella Satris, University of California, San Francisco; David Schnyer, UT Austin; Seth Seabury, University of Southern California; Mark Sherer, TIRR Memorial Hermann; Murray Stein MPH, University of California, San Diego; Sabrina Taylor, University of California, San Francisco; Nancy Temkin, University of Washington; Arthur Toga, University of Southern California; Alex Valadka, Virginia Commonwealth University; Mary Vassar, RN MS, University of California, San Francisco; Paul Vespa, University of California, Los Angeles; Kevin Wang, University of Florida; John K. Yue, University of California, San Francisco; Esther Yuh, University of California, San Francisco; Ross Zafonte, Harvard Medical School.
The opinions expressed herein are those of the authors and are not necessarily representative of those of the Uniformed Services University of the Health Sciences (USUSH), the Department of Defense (DOD), the NIH or any other US government agency.
Glossary
- DAI
diffuse axonal injury
- DTI
diffusion tensor imaging
- FA
fractional anisotropy
- MD
mean diffusivity
- SC
spectral component
- TAI
traumatic axonal injury
- TBI
traumatic brain injury
Funding
This research was partially supported by a grant from the U.S. Department of Defense, Program Project 308430 Uniformed Services University of the Health Sciences (USUHS). Support for this work also included funding from the U.S. Department of Defense to the Brain Tissue Repository and Neuropathology Core, Center for Neuroscience and Regenerative Medicine (CNRM). D.B. and M.E.K. were supported by the CNRM Neuroradiology-Neuropathology Correlations Core. D.I., D.P.P., and D.L.B. were supported by the CNRM and USUHS. PJB was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- Adams JH, Doyle D, Ford I, Gennarelli TA, Graham DI, Mclellan DR.. Diffuse axonal injury in head injury: definition, diagnosis and grading. Histopathology 1989; 15: 49–59. [DOI] [PubMed] [Google Scholar]
- Adams JH, Graham DI, Murray LS, Scott G.. Diffuse axonal injury due to nonmissile head injury in humans: an analysis of 45 cases. Ann Neurol 1982; 12: 557–63. [DOI] [PubMed] [Google Scholar]
- Adler DH, Wisse LEM, Ittyerah R, Pluta JB, Ding S-L, Xie L, et al. Characterizing the human hippocampus in aging and Alzheimer’s disease using a computational atlas derived from ex vivo MRI and histology. Proc Natl Acad Sci USA 2018; 115: 4252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfanakis K, Haughton VM, Carew JD, Rogers BP, Dempsey RJ, Meyerand ME.. Diffusion tensor MR imaging in diffuse axonal injury. AJNR Am J Neuroradiol 2002; 23: 794–802. [PMC free article] [PubMed] [Google Scholar]
- Barmpoutis A, Vemuri BC.. A unified framework for estimating diffusion tensors of any order with symmetric positive-definite constraints. In: 2010 IEEE International Symposium on Biomedical Imaging: from Nano to Macro. IEEE; 2010. pp. 1385–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D.. MR diffusion tensor spectroscopy and imaging. Biophys J 1994; 66: 259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazarian JJ, Zhong J, Blyth B, Zhu T, Kavcic V, Peterson D.. Diffusion tensor imaging detects clinically important axonal damage after mild traumatic brain injury: a pilot study. J Neurotrauma 2007; 24: 1447–59. [DOI] [PubMed] [Google Scholar]
- Bazarian JJ, Zhu T, Blyth B, Borrino A, Zhong J.. Subject-specific changes in brain white matter on diffusion tensor imaging after sports-related concussion. Magn Reson Imaging 2012; 30: 171–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini D. Nonparametric inversion of relaxation and diffusion correlation data. In: Topgaard D, editor. Advanced diffusion encoding methods in MRI. Cambridge, UK: Royal Society of Chemistry; 2020. pp. 278–316. [Google Scholar]
- Benjamini D, Basser P.. Use of marginal distributions constrained optimization (MADCO) for accelerated 2D MRI relaxometry and diffusometry. J Magn Reson 2016; 271: 40–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini D, Basser PJ.. Magnetic resonance microdynamic imaging reveals distinct tissue microenvironments. Neuroimage 2017; 163: 183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini D, Basser PJ.. Towards clinically feasible relaxation-diffusion correlation MRI using MADCO. Microporous Mesoporous Mater 2018; 269: 93–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini D, Basser PJ.. Multidimensional correlation MRI. NMR Biomed 2020; 33: e4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini D, Hutchinson EB, Komlosh ME, Comrie CJ, Schwerin SC, Zhang G, et al. Direct and specific assessment of axonal injury and spinal cord microenvironments using diffusion correlation imaging. Neuroimage 2020; 221: 117195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M.. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology 1986; 161: 401–7. [DOI] [PubMed] [Google Scholar]
- Bland MJ, Altman DG.. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986; 327: 307–10. [PubMed] [Google Scholar]
- Blumbergs PC, Scott G, Vis JM, Wainwright H, Simpson DA, McLean AJ.. Topography of axonal injury as defined by amyloid precursor protein and the sector scoring method in mild and severe closed head injury. J Neurotrauma 1995; 12: 565–72. [DOI] [PubMed] [Google Scholar]
- Borich M, Makan N, Boyd L, Virji-Babul N.. Combining whole-brain voxel-wise analysis with in vivo tractography of diffusion behavior after sports-related concussion in adolescents: a preliminary report. J Neurotrauma 2013; 30: 1243–9. [DOI] [PubMed] [Google Scholar]
- Budde MD, Annese J.. Quantification of anisotropy and fiber orientation in human brain histological sections. Front Integr Neurosci 2013; 7: 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde MD, Frank JA.. Neurite beading is sufficient to decrease the apparent diffusion coefficient after ischemic stroke. Proc Natl Acad Sci USA 2010; 107: 14472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik H, Bouhrara M, Reiter DA, Fishbein KW, Spencer RG.. Stabilization of the inverse Laplace transform of multiexponential decay through introduction of a second dimension. J Magn Reson 2013; 236: 134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-H, Siman R, Iwata A, Meaney DF, Trojanowski JQ, Smith DH.. Long-term accumulation of amyloid-β, β-secretase, presenilin-1, and caspase-3 in damaged axons following brain trauma. Am J Pathol 2004; 165: 357–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G, Finke C, Chien C, Brandt AU, Asseyer S, Ruprecht K, et al. Standardization of T1w/T2w ratio improves detection of tissue damage in multiple sclerosis. Front Neurol 2019; 10: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EL, Wilde EA, Newsome MR, Scheibel RS, Troyanskaya M, Velez C, et al. ENIGMA military brain injury: a coordinated meta-analysis of diffusion MRI from multiple cohorts. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE; 2018. pp. 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deoni SCL, Rutt BK, Arun T, Pierpaoli C, Jones DK.. Gleaning multicomponent T 1 and T 2 information from steady-state imaging data. Magn Reson Med 2008; 60: 1372–87. [DOI] [PubMed] [Google Scholar]
- Does MD, Gore JC.. Compartmental study of T1 and T2 in rat brain and trigeminal nerve in vivo. Magn Reson Med 2002; 47: 274–83. [DOI] [PubMed] [Google Scholar]
- Mac Donald CL, Dikranian K, Bayly P, Holtzman D, Brody D.. Diffusion tensor imaging reliably detects experimental traumatic axonal injury and indicates approximate time of injury. J Neurosci 2007; 27: 11869–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducreux D, Huynh I, Fillard P, Renoux J, Petit-Lacour MC, Marsot-Dupuch K, et al. Brain MR diffusion tensor imaging and fibre tracking to differentiate between two diffuse axonal injuries. Neuroradiology 2005; 47: 604–8. [DOI] [PubMed] [Google Scholar]
- Edzes HT, Samulski ET.. Cross relaxation and spin diffusion in the proton NMR of hydrated collagen. Nature 1977; 265: 521–3. [DOI] [PubMed] [Google Scholar]
- Faul M, Coronado V.. Epidemiology of traumatic brain injury. In: Grafman J, Salazar AM, editor(s). Handbook of clinical neurology: traumatic brain injury, part I. Oxford, UK: Elsevier; 2015. pp. 3–13. [DOI] [PubMed] [Google Scholar]
- Filo S, Shtangel O, Salamon N, Kol A, Weisinger B, Shifman S, et al. Disentangling molecular alterations from water-content changes in the aging human brain using quantitative MRI. Nat Commun 2019; 10: 3403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman SM, Nash MJ, Sweeting CJ, Graham DI, Roberts GW.. β-Amyloid precursor protein (βAPP) as a marker for axonal injury after head injury. Neurosci Lett 1993; 160: 139–44. [DOI] [PubMed] [Google Scholar]
- Gentleman SM, Roberts GW, Gennarelli TA, Maxwell WL, Adams JH, Kerr S, et al. Axonal injury: a universal consequence of fatal closed head injury? Acta Neuropathol 1995; 89: 537–43. [DOI] [PubMed] [Google Scholar]
- Haacke EM, Duhaime AC, Gean AD, Riedy G, Wintermark M, Mukherjee P, et al. Common data elements in radiologic imaging of traumatic brain injury. J Magn Reson Imaging 2010; 32: 516–43. [DOI] [PubMed] [Google Scholar]
- Henry LC, Tremblay J, Tremblay S, Lee A, Brun C, Lepore N, et al. Acute and chronic changes in diffusivity measures after sports concussion. J Neurotrauma 2011; 28: 2049–59. [DOI] [PubMed] [Google Scholar]
- Hill CS, Coleman MP, Menon DK.. Traumatic axonal injury: mechanisms and translational opportunities. Trends Neurosci 2016; 39: 311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran L, Kim JH, Gangolli M, Stein T, Alvarez V, McKee A, et al. Axonal disruption in white matter underlying cortical sulcus tau pathology in chronic traumatic encephalopathy. Acta Neuropathol 2017; 133: 367–80. [DOI] [PubMed] [Google Scholar]
- Hulkower MB, Poliak DB, Rosenbaum SB, Zimmerman ME, Lipton ML.. A decade of DTI in traumatic brain injury: 10 years and 100 articles later. AJNR Am J Neuroradiol 2013; 34: 2064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hürlimann MD, Flaum M, Venkataramanan L, Flaum C, Freedman R, Hirasaki GJ.. Diffusion-relaxation distribution functions of sedimentary rocks in different saturation states. Magn Reson Imaging 2003; 21: 305–10. [DOI] [PubMed] [Google Scholar]
- Hutter J, Slator PJ, Christiaens D, Teixeira RPAG, Roberts T, Jackson L, et al. Integrated and efficient diffusion-relaxometry using ZEBRA. Sci Rep 2018; 8: 15138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen RJ, Kharatishvili I, Gröhn H, Pitkänen A, Gröhn OHJ.. Quantitative MRI predicts long-term structural and functional outcome after experimental traumatic brain injury. Neuroimage 2009; 45: 1–9. [DOI] [PubMed] [Google Scholar]
- Inglese M, Makani S, Johnson G, Cohen BA, Silver JA, Gonen O, et al. Diffuse axonal injury in mild traumatic brain injury: a diffusion tensor imaging study. J Neurosurg 2005; 103: 298–303. [DOI] [PubMed] [Google Scholar]
- Johnson VE, Stewart W, Smith DH.. Axonal pathology in traumatic brain injury. Exp Neurol 2013; 246: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Alexander DC, Bowtell R, Cercignani M, Dell'Acqua F, McHugh DJ, et al. Microstructural imaging of the human brain with a ‘super-scanner’: 10 key advantages of ultra-strong gradients for diffusion MRI. Neuroimage 2018; 182: 8–38. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G.. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage 2004; 23: S151–S160. [DOI] [PubMed] [Google Scholar]
- Kamnaksh A, Budde MD, Kovesdi E, Long JB, Frank JA, Agoston DV.. Diffusion tensor imaging reveals acute subcortical changes after mild blast-induced traumatic brain injury. Sci Rep 2015; 4: 4809.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Doyle EK, Wisnowski JL, Kim JH, Haldar JP.. Diffusion-relaxation correlation spectroscopic imaging: a multidimensional approach for probing microstructure. Magn Reson Med 2017; 78: 2236–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Wisnowski JL, Nguyen CT, Haldar JP.. Multidimensional correlation spectroscopic imaging of exponential decays: from theoretical principles to in vivo human applications. NMR Biomed 2020; 33: e4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen KM, Greenwood R, Powell JH, Leech R, Hawkins PC, Bonnelle V, et al. White matter damage and cognitive impairment after traumatic brain injury. Brain 2011; 134: 449–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker RM, Henkelman MR.. Analysis of biological NMR relaxation data with continuous distributions of relaxation times. J Magn Reson 1986; 69: 218–35. [Google Scholar]
- Labadie C, Lee JH, Vetek G, Springer CS.. Relaxographic imaging. J Magn Reson Ser B 1994; 105: 99–112. [DOI] [PubMed] [Google Scholar]
- Laule C, Kozlowski P, Leung E, Li DKB, MacKay AL, Moore GRW.. Myelin water imaging of multiple sclerosis at 7 T: correlations with histopathology. Neuroimage 2008; 40: 1575–80. [DOI] [PubMed] [Google Scholar]
- Li W, Long JA, Watts L, Shen Q, Liu Y, Jiang Z, et al. Spatiotemporal changes in diffusion, T 2 and susceptibility of white matter following mild traumatic brain injury. NMR Biomed 2016; 29: 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Watts LT, Chemello J, Huang S, Shen Q, Duong TQ.. Multiparametric and longitudinal MRI characterization of mild traumatic brain injury in rats. J Neurotrauma 2015; 32: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Guan X, et al. A method for normalizing histology slides for quantitative analysis. In: 2009 IEEE international symposium on biomedical imaging: from nano to macro. IEEE; 2009. pp. 1107–1110. [Google Scholar]
- Matsushita M, Hosoda K, Naitoh Y, Yamashita H, Kohmura E.. Utility of diffusion tensor imaging in the acute stage of mild to moderate traumatic brain injury for detecting white matter lesions and predicting long-term cognitive function in adults. JNS 2011; 115: 130–9. [DOI] [PubMed] [Google Scholar]
- Matthaei D, Frahm J, Haase A, Hanicke W.. Regional physiological functions depicted by sequences of rapid magnetic resonance images. Lancet 1985; 326: 893.. [DOI] [PubMed] [Google Scholar]
- Mayer AR, Ling J, Mannell MV, Gasparovic C, Phillips JP, Doezema D, et al. A prospective diffusion tensor imaging study in mild traumatic brain injury. Neurology 2010; 74: 643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AR, Ling JM, Yang Z, Pena A, Yeo RA, Klimaj S.. Diffusion abnormalities in pediatric mild traumatic brain injury. J Neurosci 2012; 32: 17961–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes K, Friesen CL, MacKenzie DE, Westwood DA, Boe SG.. Mild traumatic brain injury (mTBI) and chronic cognitive impairment: a scoping review. PLoS One 2017; 12: e0174847.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles L, Grossman RI, Johnson G, Babb JS, Diller L, Inglese M.. Short-term DTI predictors of cognitive dysfunction in mild traumatic brain injury. Brain Inj 2008; 22: 115–22. [DOI] [PubMed] [Google Scholar]
- Mitchell J, Chandrasekera TC, Gladden LF.. Numerical estimation of relaxation and diffusion distributions in two dimensions. Prog Nucl Magn Reson Spectrosc 2012; 62: 34–50. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Chung SW, Berman JI, Hess CP, Henry RG.. Diffusion tensor MR imaging and fiber tractography: technical considerations. AJNR Am J Neuroradiol 2008; 29: 843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenaus A, Robbins M, Blanco G, Galloway NR, Snissarenko E, Gillard E, et al. Multi-modal magnetic resonance imaging alterations in two rat models of mild neurotrauma. J Neurotrauma 2007; 24: 1147–60. [DOI] [PubMed] [Google Scholar]
- Pas K, Komlosh ME, Perl DP, Basser PJ, Benjamini D.. Retaining information from multidimensional correlation MRI using a spectral regions of interest generator. Sci Rep 2020; 10: 3246.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher SW. A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput Phys Commun 1982; 27: 213–27. [Google Scholar]
- Ramlackhansingh AF, Brooks DJ, Greenwood RJ, Bose SK, Turkheimer FE, Kinnunen KM, et al. Inflammation after trauma: microglial activation and traumatic brain injury. Ann Neurol 2011; 70: 374–83. [DOI] [PubMed] [Google Scholar]
- Roebroeck A, Miller KL, Aggarwal M.. Ex vivo diffusion MRI of the human brain: technical challenges and recent advances. NMR Biomed 2019; 32: e3941.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen I, Moeller S, Ugurbil K, Kim D-S.. Analysis of the distribution of diffusion coefficients in cat brain at 9.4 T using the inverse Laplace transformation. Magn Reson Imaging 2006; 24: 61–8. [DOI] [PubMed] [Google Scholar]
- Ruifrok AC, Johnston DA.. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 2001; 23: 291–9. [PubMed] [Google Scholar]
- Sharp DJ, Fleminger S, Powell J, Traumatic brain injury. In: Husain M, Schott JM, editors. Oxford textbook of cognitive neurology and dementia. New York: Oxford University Press; 2016. pp. 435–452. [Google Scholar]
- Sharp DJ, Ham TE.. Investigating white matter injury after mild traumatic brain injury. Curr Opin Neurol 2011; 24: 558–63. [DOI] [PubMed] [Google Scholar]
- Shenton ME, Hamoda HM, Schneiderman JS, Bouix S, Pasternak O, Rathi Y, et al. A review of magnetic resonance imaging and diffusion tensor imaging findings in mild traumatic brain injury. Brain Imaging Behav 2012; 6: 137–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd TM, Flint JJ, Thelwall PE, Stanisz GJ, Mareci TH, Yachnis AT, et al. Postmortem interval alters the water relaxation and diffusion properties of rat nervous tissue—implications for MRI studies of human autopsy samples. Neuroimage 2009; 44: 820–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd TM, Hoch MJ, Bruno M, Faustin A, Papaioannou A, Jones SE, et al. Inner SPACE: 400-micron isotropic resolution MRI of the human brain. Front Neuroanat 2020; 14: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherriff FE, Bridges LR, Sivaloganathan S.. Early detection of axonal injury after human head trauma using immunocytochemistry for?-amyloid precursor protein. Acta Neuropathol 1994; 87: 55–62. [DOI] [PubMed] [Google Scholar]
- Skinner NP, Kurpad SN, Schmit BD, Tugan Muftuler L, Budde MD.. Rapid in vivo detection of rat spinal cord injury with double-diffusion-encoded magnetic resonance spectroscopy. Magn Reson Med 2017; 77: 1639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slator PJ, Hutter J, Palombo M, Jackson LH, Ho A, Panagiotaki E, et al. Combined diffusion‐relaxometry MRI to identify dysfunction in the human placenta. Magn Reson Med 2019; 82: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DH, Stewart W.. ‘Concussion’ is not a true diagnosis. Nat Rev Neurol 2020; 16: 457–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y-Q, Venkataramanan L, Hürlimann MD, Flaum M, Frulla P, Straley C.. T1-T2 correlation spectra obtained using a fast two-dimensional Laplace inversion. J Magn Reson 2002; 154: 261–8. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Jahanshad N, Ching CRK, Salminen LE, Thomopoulos SI, Bright J; for the ENIGMA Consortium, et al. ENIGMA and global neuroscience: a decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl Psychiatry 2020; 10: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topgaard D. Multidimensional diffusion MRI. J Magn Reson 2017; 275: 98–113. [DOI] [PubMed] [Google Scholar]
- Wu W, Poser BA, Douaud G, Frost R, In M-H, Speck O, et al. High-resolution diffusion MRI at 7T using a three-dimensional multi-slab acquisition. Neuroimage 2016; 143: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang L, Kao Y-T, Qiu W, Yang Y, Okobiah O, et al. Mapping hydration dynamics around a protein surface. Proc Natl Acad Sci USA 2007; 104: 18461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.