Abstract
Accurate segmentation of the optic disc (OD) depicted on color fundus images may aid in the early detection and quantitative diagnosis of retinal diseases, such as glaucoma and optic atrophy. In this study, we proposed a coarse-to-fine deep learning framework on the basis of a classical convolutional neural network (CNN), known as the U-net model, to accurately identify the optic disc. This network was trained separately on color fundus images and their grayscale vessel density maps, leading to two different segmentation results from the entire image. We combined the results using an overlap strategy to identify a local image patch (disc candidate region), which was then fed into the U-net model for further segmentation. Our experiments demonstrated that the developed framework achieved an average intersection over union (IoU) and a dice similarity coefficient (DSC) of 89.1% and 93.9%, respectively, based on 2,978 test images from our collected dataset and six public datasets, as compared to 87.4% and 92.5% obtained by only using the sole U-net model. The comparison with available approaches demonstrated a reliable and relatively high performance of the proposed deep learning framework in automated OD segmentation.
Keywords: Image segmentation, Optic disc, Convolutional neural networks, U-net model, Color fundus images
1. Introduction
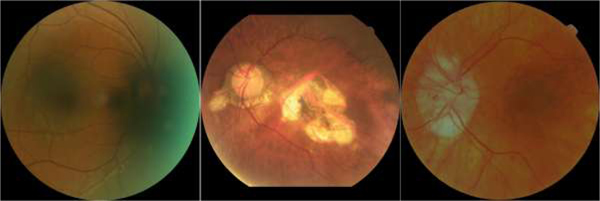
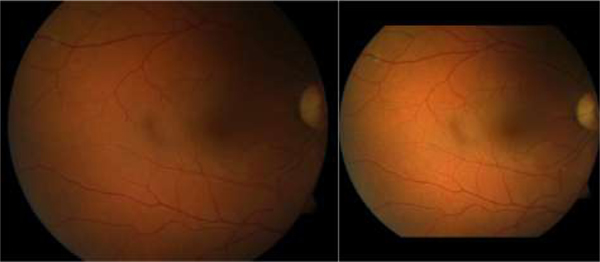
As one of the fundamental anatomic structures of the retina, the optic disc (OD) [1–3] appears as a bright circular region on color fundus images. The OD is the location of the optic nerve and the entry point for major blood vessels [4–5]. This unique anatomy makes the OD a landmark for disease assessment [6–8]. Although there is relatively high contrast between the OD and the surrounding regions on color fundus images, identifying the OD is often a challenging task [9–10], because the image quality can be confounded by a number of factors, such as light exposure, camera focus, motion artifact, and existing diseases. These factors may lead to inhomogeneous image quality [11–12], as illustrated by the examples in Fig. 1, where the OD locations and their boundaries are not well depicted.
Fig. 1.
Illustration of three fundus images with severe inhomogeneity and anomalous tissue variability
In the past, extensive efforts [13–18] have been made to develop computerized methods for identifying the OD depicted on fundus images. Most of these methods [19–20] leveraged the anatomical characteristics of the OD and its relationship with other structures or anatomical landmarks, such as the elliptical / circular shape of the OD [21–23] and the parabola shape of the blood vessels [24–25], to initially locate the OD. On the basis of the OD location, additional procedures were used to identify the OD’s boundaries [7]. The experiments showed that these methods had limited segmentation accuracy when facing fundus images with weak contrast and / or severe inhomogeneity. In this study, we proposed to approach this segmentation issue using deep convolutional neural network (CNN) technology [26–28]. The most distinctive advantage of this technology [29,31,32] is the ability to precisely learn and capture a large number of underlying image features with varying hierarchies and locations, and to optimally integrate these features to obtain a desirable result. Considering the unique characteristics of retinal images, we developed a novel coarse-to-fine deep learning framework on the basis of the U-net model [29]. Specifically, we firstly extracted retinal blood vessels from color fundus images and then constructed a vessel density map [30]. The fundus images and their vessel density maps were separately fed into the given U-net model to obtain two different segmentation results, which were combined further to obtain the possible location of the OD. Segmentation of the OD was finally performed using the same U-net model. A detailed description of this method and its performance validation follows.
2. Methods and Materials
2.1. Scheme overview
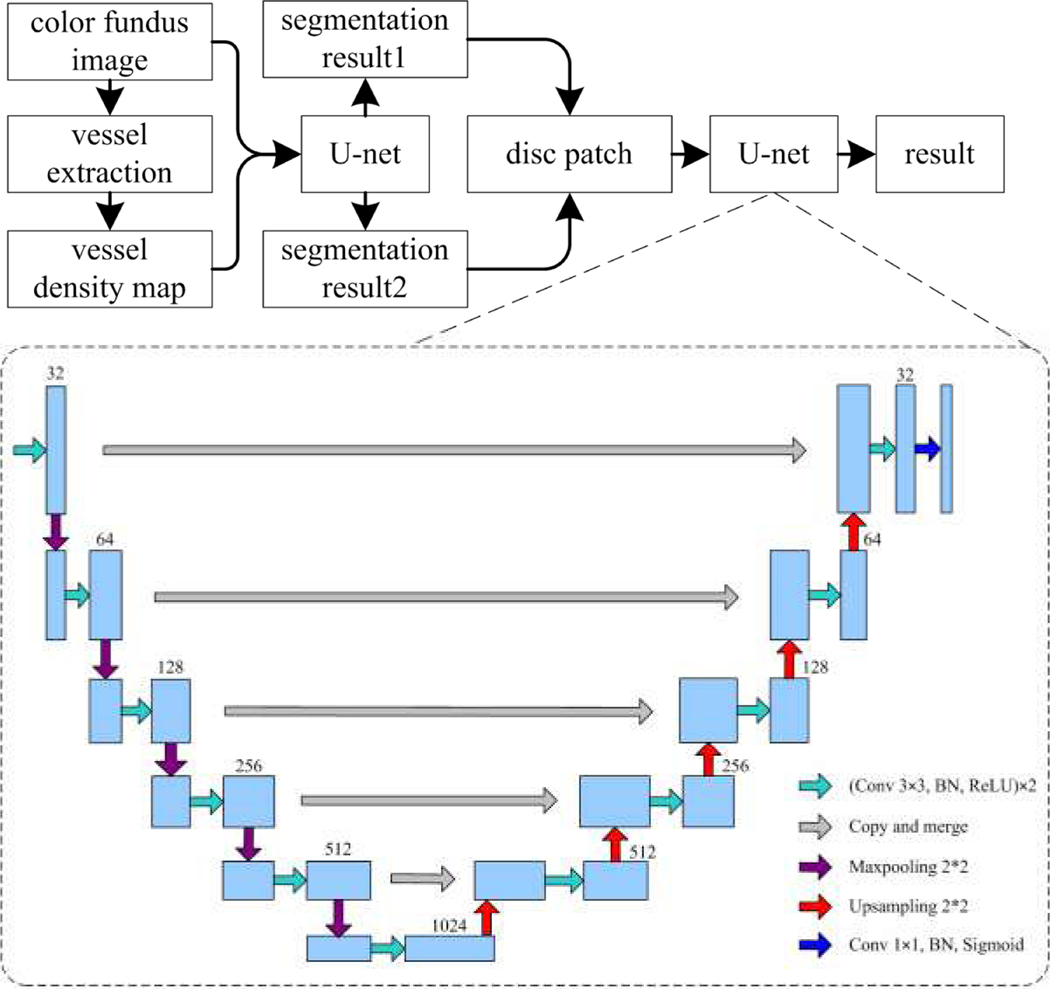
The architecture of the developed method for segmenting the optic disc in a coarse-to-fine way consists of several components, as shown in Fig. 2. We derived the vessel density map from the retinal vessels to highlight the location of the optic disc with respect to the vessels. We used the U-net model to segment the OD depicted on color fundus images and their vessel density map, respectively. This resulted in two different segmentations that often contain multiple OD candidate regions. Thereafter, we used an overlapping strategy to exclude the false segmentation and identify the candidate region with the highest probability of being the OD. Applying the U-net model to the final candidate regions led to the identification of the optimal OD region.
Fig. 2.
The flowchart of the proposed method for segmenting the optic disc depicted on color fundus images.
2.2. Vessel density map
The OD is the entry point for the retinal blood vessels. From the branching patterns of the retinal vessels, it is relatively straightforward for our human eyes, but not for the computer, to estimate the location of the OD. In this study, we proposed to use the vessel density map [30] to characterize the underlying property of the retinal vessels and their relationship with the OD. First, we used the method presented in [33] to extract the retinal vessels, which was represented as a grayscale image. On the retinal vessel images, each pixel denotes the contrast between the vessels and the background, and has large intensity for major vessels near the disc region. Then, we computed the vessel density map using the formulation:
| (1) |
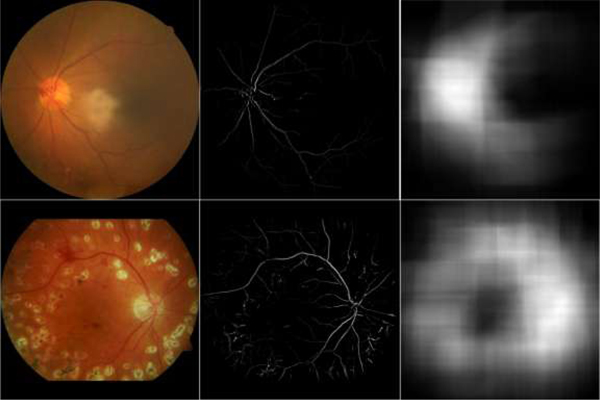
where V(·) and D(·) denote and the pixel value of the vessel image and its density map at a given position, respectively. is a given local region centered at x, with a radius of R. D(·) is the sum of the intensities of pixels in vessel feature image and actually denotes the density of the vessels within the region Ωx. This strategy is different from the map proposed in paper [30], which counted the number of vessel pixels in local regions. In contrast, our strategy considered the density values of the vessel pixels. The difference enables the proposed density to emphasize the disc region because the retinal vessels near the disc region have relatively larger densities than other regions. The examples in Fig. 3 were used to demonstrate the advantages of the proposed density maps generated from the given fundus images. It can be seen from the vessel density maps that (1) the vessels around the disc regions typically have high intensity / contrast, (2) the location of the OD is obviously associated with high density values, and (3) existing diseases / abnormalities have very limited impact on the characterization of the OD on the vessel density maps.
Fig. 3.
Illustration of the original fundus images and their vessel density maps. The first column corresponds to the original images, the last two columns to the vessel features and their density maps, respectively.
2.3. The U-net architecture
With the color fundus images and their vessel density maps, we used a relatively deep U-net convolutional neural network to perform a coarse-to-fine segmentation of the optic disc, as illustrated in Fig. 2. This network learned and captured image features by stacking two 3×3 convolutional kernels in each convolutional block, followed by a batch normalization (BN) [34] and an element-wise rectified linear unit (ReLU) activation operation [28]. The learned convolution features were down-sampled by leveraging a 2×2 Max-pooling layer with a stride of 2 to reduce the amount of image information. After alternating the convolutional block and Max-pooling layer, the input images were encoded as a large number of high-dimensional features. These features were used to locate the target objects; however, they might fail to precisely specify the boundaries of objects. To obtain both location and boundary information, the high-dimensional features were then decoded by stacking several 2×2 up-sampling layers and convolution blocks mentioned above. During the decoding procedure, a skip connection was employed to add pixel position information and guide the image segmentation. The final decoded results were fed into a 1×1 convolution layer with a sigmoid activation to finalize the target regions.
2.4. Coarse-to-fine segmentation strategy
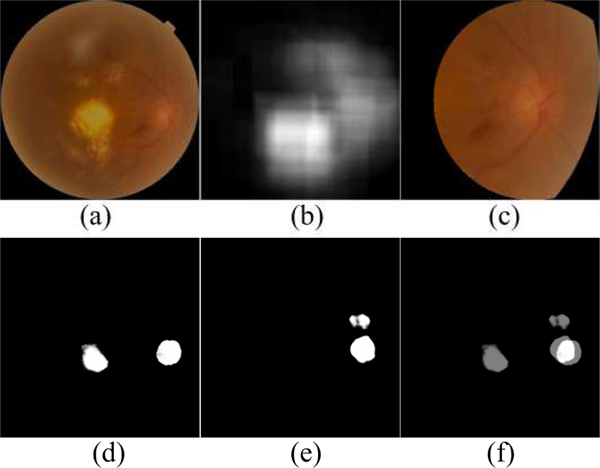
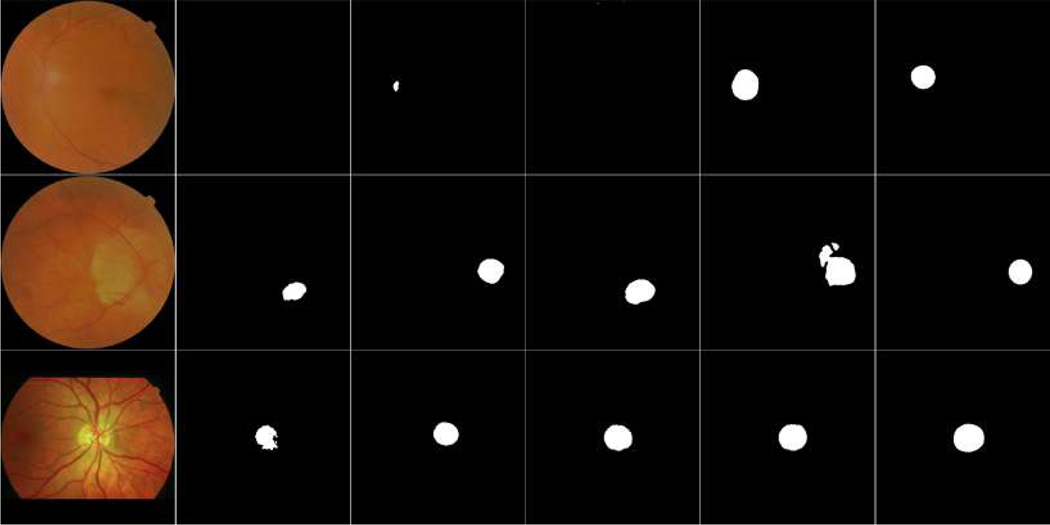
In implementation, the U-net model was firstly trained on the fundus images with three color channels (i.e., RGB), and their vessel density maps with one grayscale channel, respectively, to generate two different segmentation results. The results were susceptible to relatively large segmentation errors, since the entire images were used and their resolutions were greatly reduced. In addition, the segmentation results often contained false positive regions due to the image quality and / or existing abnormalities. To reduce false positive detection and obtain optimal segmentation results, we combined the above two segmentation results by identifying their overlapped regions and selected the largest one as the most reliable OD candidate region. The selection was based on the fact that most of false positive regions, similar to the OD, were typically caused by some kinds of pathological disease in the original images, and it is very seldom to have these false positive regions on both color fundus images and the vessel density maps. Based on the selection criterion, an image patch centered at the overlapped region was cropped from the original fundus images. The size of the image patch was approximately three times the diameter of an OD to fully enclose the OD and the surrounding regions. Since the OD appears as a circular region, we proposed to use a circular image patch in this study, as shown in Fig. 4. This overlapping strategy can largely exclude the false positive regions, and meanwhile the obtained circular image patch can characterize the OD regions using a relatively high image resolution. Most important, these local image patches improved the contrast between the OD and its surrounding regions as compared to their contrast in the original fundus image. The identified circular image patches with three color channels were then fed into the U-net model again to perform a refined segmentation.
Fig. 4.
Example of location of local disc patches by combining two coarse segmentation results. (a) is the original fundus image, (b) is vessel density map of (a), (c) is local disc patch of (a), (d) and (e) are the segmentation results of (a) and (b), respectively, and (f) is the overlapped map of (d) and (e).
2.5. Training the U-net models
To train the involved models, we collected a total of 7,614 color fundus images (CFI) acquired on different subjects in the Shaanxi Provincial People’s Hospital, among which there were about 1810 images with weak contrast or severe intensity inhomogeneity. These images have a resolution ranging from 1,924×1,556 pixels to 2,336×2,336 pixels. Before the training, we excluded the dark background and resized the field of view from the original fundus images to a dimension of 512×512 pixels. We also normalized the images in order to reduce the impact caused by different illumination conditions, as shown in Fig. 5. These images were manually processed by two experienced ophthalmologists to annotate the disc regions with ImageJ software (https://imagej.net/) using an ellipse. The annotated elliptic regions were used as the ground truths in this study. These ground truths, together with the normalized fundus images, were randomly divided into three independent parts for training, validation, and testing purposes at a ratio of 0.75, 0.15 and 0.1, respectively, and fed into the above proposed methods for segmenting the optic disc. The three independent datasets contained 1377, 276 and 157 images with weak contrast and severe inhomogeneity, respectively.
Fig. 5.
Example of the original fundus image (left) and its normalized version (right).
The proposed network was implemented using the Keras library (https://keras.io/) and trained on a PC with a 3.50GHz Intel(R) Xeon(R) E5–1620 CPU, 8GB RAM and NVIDIA Quadro P5000. During the training, we resized all involved images to a dimension of 256×256 pixels and used a batch size of 16 samples, considering the computer capability. We used the intersection over union (IoU) [35] as the loss function, which can be given by:
| (2) |
where N(·) indicates the number of pixels enclosed set. A and B denote the segmentation results obtained by a specified algorithm and manual annotation, respectively. ∪ and ∩ denote the intersection and union operators. This function was maximized using the stochastic gradient descent (SGD) with a momentum of 0.95 and an initial learning rate of 0.001. The rate was decreased by a factor of 10 for every 30 epochs. The number of epochs was set at 300 in this study. An early stopping scheme was applied to reduce unnecessary training time if the values of IoU were not improved after 35 epochs. A division point of 0.5 was used to threshold the training results and to decide whether a pixel belongs to the optic disc or not. To prevent overfitting, the fundus images were randomly flipped along each axis, and then rotated [36] from 0 to 360 degrees. These data augmentation techniques increased the diversity of the image samples and were able to significantly improve the generalization and reliability of the deep learning network for these samples.
2.6. Performance evaluation
We validated the developed method on the testing part of our collected CFI images and six publicly available fundus image datasets [1, 3, 22, 33], including DIARETDB0, DIARETDB1, DRIONS-DB, DRIVE, MESSIDOR and ORGIA. These datasets have been widely used for detecting and diagnosing ocular diseases, such as diabetic retinopathy, glaucoma, and age-related macular degeneration. Along with these images, the disc boundaries manually delineated by clinical experts were also available. DIARETDB0 consisted of 130 images, of which 110 had diabetic retinopathy and 20 were normal images. DIARETDB1 contained a total of 89 images, where 84 had mild and non-proliferative diabetic retinopathy. DRIONS-DB was comprised of 110 images, most of which had moderate peripapillary atrophy (PPA) and fuzzy rim. DRIVE contained 40 images, where 33 were normal images and 7 diagnosed with mild diabetic retinopathy. In the MESSIDOR and ORGIA datasets, there were 1200 and 650 fundus images acquired to facilitate the detection of diabetic retinopathy and glaucoma, respectively. In three of these datasets, namely DIARETDB0, DIARETDB1 and MESSIDOR, the images with ocular diseases were annotated. As a result, we setup a normal group consisting of 845 images and an abnormal group consisting of 571 images to evaluate the performance of the developed approach in the presence of disease.
With these publicly available datasets, we compared the developed method with available OD segmentation methods using different performance metrics. These metrics included the IoU, the dice similarity coefficient (DSC), and balanced accuracy (Acc) [3], which are defined, respectively, as:
| (3) |
| (4) |
| (5) |
| (6) |
where Tp, Tn, Fp and Fn are the number of true positives and true negatives, false positives and false negatives, respectively. Se and Sp [18] denote the sensitivity and specificity of a given segmentation method, the former is used to measure the desired pixels with positive values while the latter measures the undesired pixels with negative values. These metrics take values in the range from 0 to 1, and a higher value means better segmentation accuracy. In addition, we used the disc location accuracy (DLA) and the mean distance to closest point (MDCP) [7] for performance comparison. The DLA was defined as the ratio of the number of correctly estimated disc centers and the total number of tested images. Similar to [25], we considered the estimated OD center correct if it was within 60 pixels of the manually identified center. MDCP was used to characterize the degree of mismatch between the segmented contour and the ground truth contour. Analysis of variance (ANOVA) was performed to assess the statistical differences among these involved methods, where we considered a p-value less than 0.05 statistically significant.
3. Results
Table 1 and 2 show the segmentation results of the collected CFI images and six public datasets using the U-net model trained on color fundus images, vessel density maps, 4-channel composite images (comprised of the original RGB image and its vessel density map), and the local disc image patches, respectively. The values of IoU, DSC, Acc and Se obtained by our method in Table 1 were, on average, higher by 1.73%, 1.40%, 0.59% and 1.51%, respectively, than the counterparts of the U-net model trained on the same set of original fundus images. The p-values for these four metrics were less than 0.001. Our method also had larger average values of IoU and DSC than those of the U-net model trained on 4-channel composite images along with the p-value of 0.001. This suggested that there were statistically significant differences between the proposed method and the U-net method trained separately on the original image and 4-channel composite images. The sole U-net model had the lowest segmentation accuracy when trained on the grayscale vessel density maps, as compared to the color fundus images, suggesting that the developed method was superior to the U-net model trained merely on the original fundus images or their vessel density maps. In particular, our method in Table 2 achieved the average IOU and DSC of 0.894 and 0.941 for the abnormal group, and 0.904 and 0.947 for the normal group, respectively. In contrast, the counterparts of the U-net model were 0.875 and 0.927 for the abnormal group, 0.896 and 0.942 for the normal group.
Table 1.
Experimental results on the collected CFI images and six public datasets using the U-net model trained separately on the fundus images, vessel density maps, composite images and local disc patches.
| Datasets | No. of images | Color fundus images | Vessel density maps | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IoU | DSC | Acc | Se | IoU | DSC | Acc | Se | ||
| CFI | 762 | 0.873 | 0.923 | 0.960 | 0.921 | 0.687 | 0.799 | 0.903 | 0.809 |
| DIARETDB0 | 128 | 0.837 | 0.898 | 0.937 | 0.874 | 0.684 | 0.800 | 0.937 | 0.878 |
| DIARETDB1 | 88 | 0.833 | 0.904 | 0.934 | 0.869 | 0.686 | 0.807 | 0.932 | 0.869 |
| DRIONS-DB | 110 | 0.818 | 0.885 | 0.930 | 0.862 | 0.657 | 0.786 | 0.859 | 0.719 |
| DRIVE | 40 | 0.778 | 0.863 | 0.895 | 0.790 | 0.688 | 0.802 | 0.874 | 0.750 |
| MESSIDOR | 1200 | 0.894 | 0.940 | 0.970 | 0.942 | 0.739 | 0.845 | 0.935 | 0.874 |
| ORIGA | 650 | 0.864 | 0.920 | 0.976 | 0.954 | 0.722 | 0.831 | 0.938 | 0.880 |
| Overall | 2978 | 0.874 | 0.925 | 0.964 | 0.930 | 0.714 | 0.824 | 0.924 | 0.851 |
| Datasets | No. of images | 4-channel composite images | Local disc patches | ||||||
| IoU | DSC | Acc | Se | IoU | DSC | Acc | Se | ||
| CFI | 762 | 0.880 | 0.928 | 0.967 | 0.935 | 0.886 | 0.935 | 0.965 | 0.935 |
| DIARETDB0 | 128 | 0.856 | 0.916 | 0.947 | 0.895 | 0.848 | 0.909 | 0.935 | 0.872 |
| DIARETDB1 | 88 | 0.841 | 0.905 | 0.936 | 0.873 | 0.849 | 0.916 | 0.935 | 0.873 |
| DRIONS-DB | 110 | 0.870 | 0.923 | 0.954 | 0.909 | 0.885 | 0.936 | 0.960 | 0.924 |
| DRIVE | 40 | 0.766 | 0.854 | 0.889 | 0.778 | 0.783 | 0.864 | 0.896 | 0.794 |
| MESSIDOR | 1200 | 0.900 | 0.943 | 0.970 | 0.940 | 0.907 | 0.949 | 0.972 | 0.948 |
| ORIGA | 650 | 0.873 | 0.930 | 0.989 | 0.979 | 0.889 | 0.940 | 0.989 | 0.984 |
| Overall | 2978 | 0.882 | 0.931 | 0.969 | 0.939 | 0.891 | 0.939 | 0.970 | 0.944 |
Table 2.
Experimental results on the normal group and the abnormal group from DIARETDB0, DIARETDB1, and MESSIDOR using the U-net model trained separately on the fundus images, vessel density maps, composite images and local disc patches.
| Datasets | No. of images | Color fundus images | Vessel density maps | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IoU | DSC | Acc | Se | IoU | DSC | Acc | Se | ||
| Normal | 571 | 0.875 | 0.927 | 0.959 | 0.920 | 0.740 | 0.845 | 0.936 | 0.876 |
| Abnormal | 845 | 0.896 | 0.942 | 0.970 | 0.940 | 0.725 | 0.834 | 0.935 | 0.873 |
| Overall | 1416 | 0.885 | 0.934 | 0.965 | 0.931 | 0.731 | 0.838 | 0.935 | 0.874 |
| Datasets | No. of images | 4-channel composite images | Local disc patches | ||||||
| IoU | DSC | Acc | Se | IoU | DSC | Acc | Se | ||
| Normal | 571 | 0.899 | 0.943 | 0.968 | 0.938 | 0.904 | 0.947 | 0.970 | 0.944 |
| Abnormal | 845 | 0.887 | 0.935 | 0.964 | 0.928 | 0.894 | 0.941 | 0.964 | 0.932 |
| Overall | 1416 | 0.892 | 0.939 | 0.966 | 0.932 | 0.898 | 0.944 | 0.967 | 0.937 |
Some examples in Fig. 6 demonstrate the segmentation results by these methods. The OD regions were not effectively identified from the original images using the U-net model when these images had severe intensity inhomogeneity and / or abnormalities, while the OD regions were successfully located from the local disc patches. The proposed method took an average of 0.04 seconds to segment the OD for a single image.
Fig. 6.
Illustration of segmentation results of the U-net model on different images. The first column showed three original fundus images, the last five columns from left to right were results of these fundus images, vessel density maps, 4-channel composite images, local disc patches, and their ground truths, respectively.
We also trained the U-net model on 4-channel disc patches (comprised of local disc regions and their local vessel density maps) in the fine segmentation step, and achieved the average IoU, DSC and Acc of 0.890, 0.939, and 0.966, respectively, on the same testing datasets. The results were slightly inferior to those (i.e., 0.891, 0.939 and 0.970) of our method trained on local disc patches with RGB channel, but there were no significant differences between the two results.
Table 3 summarizes a performance comparison of the proposed method with several available OD segmentation methods [37–42] based on six public datasets. As demonstrated by the results, the proposed method achieved, on average, 99.82% disc location accuracy and 2.47 pixel distance to closest contour point in ground truths, demonstrating a better performance than available approaches.
Table 3.
Performance comparison of different disc segmentation methods on six public datasets using the metrics of IoU, Se, DLA, and MDCP (pixels), respectively. The dash (“−”) denotes unavailable measures in related literature.
| DIARETDB0 | IoU | Se | DLA | MDCP |
| Kumar et al [7] | 89.6% | 93.3% | - | 8.62 |
| Zhang et al [5] | - | - | 95.5% | - |
| Sinha et al [10] | - | - | 96.9% | - |
| Our method | 84.8% | 87.2% | 98.4% | 4.97 |
| DIARETDB1 | ||||
| Welfer et al [8] | 43.7% | - | 97.8% | 8.31 |
| Abdullah et al [22] | 85.1% | 85.1% | 100.0% | - |
| Walter et al [13] | 37.3% | - | - | 15.52 |
| Seo et al [38] | 35.3% | - | - | 9.74 |
| Lupascu et al [39] | 31.0% | - | - | 13.81 |
| Kande et al [14] | 33.2% | - | - | 8.35 |
| Morales et al [16] | 81.7% | 92.2% | 99.6% | 2.88 |
| Our method | 84.9% | 87.3% | 100.0% | 2.75 |
| DRIONS-DB | ||||
| Morales et al [16] | 84.2% | 92.8% | 99.34% | 2.49 |
| Maninis et al [27] | 88.0% | - | - | - |
| Kumar et al [17] | 89.0% | - | - | - |
| Abdullah et al [22] | 85.1% | 85.1% | 99.09% | - |
| Our method | 88.5% | 92.4% | 100.0% | 2.59 |
| DRIVE | ||||
| Walter et al [13] | 29.3% | 49.9% | - | 12.39 |
| Seo et al [38] | 31.1% | 50.3% | - | 11.19 |
| Abdullah et al [22] | 78.6% | 81.9% | 100.0% | - |
| Morales et al [16] | 71.6% | 85.4% | 99.0% | 5.85 |
| Lalonde et al [40] | 80.0% | - | - | - |
| Our method | 78.3% | 79.4% | 97.5% | 9.57 |
| MESSIDOR | ||||
| Aquino et al [9] | 86.0% | - | 99.0% | - |
| Lu et al [36] | - | - | 99.8% | - |
| Kumar et al [7] | 91.6% | - | - | - |
| Abdullah et al [22] | 87.9% | 89.5% | 99.3% | - |
| Morales et al [16] | 82.3% | 93.0% | 99.5% | 4.08 |
| Our method | 90.7% | 94.8% | 99.9% | 2.10 |
| ORIGA | ||||
| Yin et al [4] | 88.7% | - | - | - |
| Cheng et al [41] | 90.0% | - | - | - |
| Cheng et al [42] | 90.5% | - | - | - |
| Fu et al [3] | 92.9% | - | - | - |
| Our method | 88.9% | 98.4% | 100.0% | 2.17 |
4. Discussion
We developed a coarse-to-fine deep learning framework on the basis of the U-net model to segment the OD depicted on color fundus images. The most unique characteristic of the proposed method was the integration of two types of image information (i.e., pixel intensities and vessel density) to locate and segment the OD. In particular, we developed a vessel density map to characterize the spatial relationship between the OD and retinal vessels, to help detect the OD and to exclude many false positive objects in the entire image region. This density map was especially useful for the images where the contrast between the OD and the surrounding region was not good but the vessels were well depicted on the images.
We quantitatively assessed the segmentation performance of the proposed method using a total of 2,978 images from the collected CFI dataset and six public datasets. The extensive validation on multiple datasets demonstrates the effectiveness and robustness of the proposed method against different fundus cameras, imaging conditions, image resolutions, and abnormalities. In contrast, the existing methods were typically assessed using a small number of test images. For example, Kumar et al. [17] only used 112 images, of 130 images, from the DIARETDB0 dataset and 40 images, of 1200 images, from the MESSIDOR dataset. Yin et al. [4] only tested 325 images, of 650 images, from the ORIGA dataset. When using the same testing images as those in paper [4], the proposed method achieved the IoU and DSC of 0.885 and 0.938, respectively, which were similar to those of the method [4] (i.e., 0.887 and 0.941). In addition, tests on the normal and abnormal groups suggested that the developed method was not very sensitive to the presence of ocular diseases (Table 2). It is notable that due to the hardware (GPU memory) limitation, our method was trained on images with a dimension of 256×256 pixels. In contrast, most of available deep learning methods [3, 27, 28] were trained on images with relatively large dimensions. For example, the images were resized to 400×400 pixels and 565×584 pixels, respectively, for training purpose in [3] and [27]. Hence, given the size and diversity of our testing dataset, our method demonstrated a relatively reliable and reasonable performance.
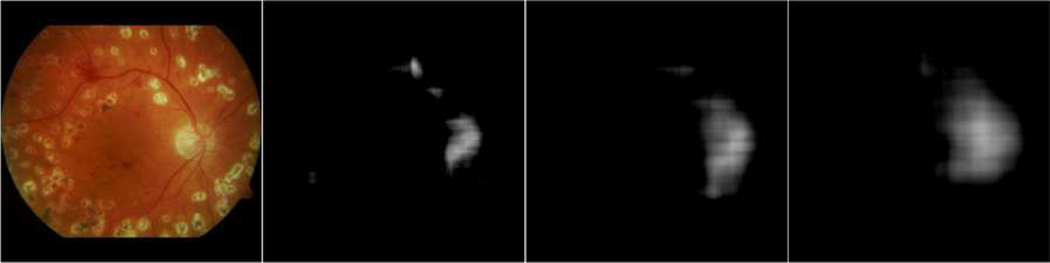
The proposed method involved several important parameters, among which R and the local patch size were important. R determines the size of the disc region as depicted on the density map, while the local image patch size may affect segmentation performance. In this study, the R value was approximately equal to the radius of the optic disc. When R was too small, the vessel density map only highlighted the vessels and only a small part of the optic disc, as demonstrated by the example in Fig. 7, where three different vessel density maps were calculated using the R values of 20, 40 and 60 pixels, respectively. When R was too large, the OD boundaries were not well characterized on the density map. The local patch size was set at a value approximately equal to three times the diameter of the optic disc, aiming to fully enclose the OD regions and provide a relatively high resolution of the OD region, and meanwhile alleviate a high requirement on computer hardware (e.g., the GPU memory).
Fig. 7.
Illustration of the original image and its vessel density maps calculated with three different R values of 20, 40 and 60 pixels, respectively.
Finally, there are some limitations with this study. First, the ground truth for the optic disc may not be perfectly delineated in color fundus images due to the influence of the peripapillary atrophy [1] and subjective bias. This makes the U-net model potentially unable to locate desirable object boundaries when trained on local disc patches, since the peripapillary atrophy and fuzzy rim were enlarged and might affect the estimation of the pixels within the disc regions, leading to reduced segmentation accuracy. Second, the accuracy of the blood vessel identification may affect the results as well. When the vessels are not well depicted on the images or there are severe diseases, the vessel density maps may fail to highlight the position of the optic disc, and introduce irrelevant object regions in image segmentation. Third, we selected a low image resolution (i.e., 256×256) in this study with consideration of the limitation of the GPU hardware in memory. This resolution might limit the amount of image features in the deep learning network. Fu et al [3] and Maninis et al [27] trained different variants of the U-net model on images with a resolution of 400×400 and 584×565, respectively, which were much larger than the counterpart used in our experiment. Despite these limitations, the proposed method can automatically identify the OD regions with very promising accuracy.
5. Conclusion
In this paper, we introduced a coarse-to-fine deep learning framework based on the U-net model to accurately and automatically segment the OD depicted on color fundus images. Its novelty mainly lies in integrating different image information (i.e., image contrast and retinal vessels) and leveraging a coarse-to-fine segmentation strategy. Our extensive experiments on the collected images and six public datasets, which consisted of a total of 2,978 fundus images, demonstrated that the proposed framework could achieve a relatively reliable segmentation performance.
Acknowledgements
This work is supported by National Institutes of Health (NIH) (Grant No. R21CA197493, R01HL096613) and Jiangsu Natural Science Foundation (Grant No. BK20170391).
References
- [1].Almazroa A, Burman R, Raahemifar K, Lakshminarayanan V, Optic Disc and Optic Cup Segmentation Methodologies for Glaucoma Image Detection: A Survey, Journal of Ophthalmology, 2015 (2015) 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Singh A, Dutta M, Sarathi M, Uher V, Burget R, Image processing based automatic diagnosis of glaucoma using wavelet features of segmented optic disc from fundus image, Computer Methods and Programs in Biomedicine, 124 (2016) 108–120 [DOI] [PubMed] [Google Scholar]
- [3].Fu H, Cheng J, Xu Y, Wong D, Liu J, Cao X, Joint Optic Disc and Cup Segmentation Based on Multi-label Deep Network and Polar Transformation, IEEE Transactions on Medical Imaging, DOI 10.1109/TMI.2018.2791488, (2018) [DOI] [PubMed] [Google Scholar]
- [4].Yin F, Liu J, Ong S, et al. , Model-based optic nerve head segmentation on retinal fundus images, Conference of the IEEE Engineering in Medicine and Biology Society (EMBS), 2626–2629, (2011) [DOI] [PubMed] [Google Scholar]
- [5].Zhang D, Yi Y, Shang X, Peng Y, Optic disc localization by projection with vessel distribution and appearance characteristics, International Conference on Pattern Recognition (ICPR), 3176–3179, (2012). [Google Scholar]
- [6].Zilly J, Buhmann J, Mahapatra D, Glaucoma detection using entropy sampling and ensemble learning for automatic optic cup and disc segmentation, Computerized Medical Imaging and Graphics 55:28–41, (2017) [DOI] [PubMed] [Google Scholar]
- [7].Kumar V, Sinha N, Automatic optic disc segmentation using maximum intensity variation, Proceedings of the IEEE TENCON Spring Conference, 29–33, (2013) [Google Scholar]
- [8].Welfer D, Scharcanski J, Kitamura C, Dal Pizzol M, Ludwig L, Marinho D, Segmentation of the optic disk in color eye fundus images using an adaptive morphological approach, Computers in Biology and Medicine, 40(2):124–137, (2010) [DOI] [PubMed] [Google Scholar]
- [9].Aquino M, Gegundez E, Mar D, Detecting the optic disc boundary in digital fundus images using morphological, edge detection, and feature extraction techniques, IEEE Transactions on Medical Imaging, 29(11):1860–1869, (2010) [DOI] [PubMed] [Google Scholar]
- [10].Sinha N, Babu R, Optic disk localization using L1 minimization, Proceedings of the 19th IEEE International Conference on Image Processing (ICIP), 2829–2832, (2012) [Google Scholar]
- [11].Wang L, Chang Y, Wang H, Wu Z, Pu J, Yang X, An active contour model based on local fitted images for image segmentation, Information Sciences, 418 (2017) 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang L, Chen G, Shi D, Chang Y, Chan S, Pu J, Yang X, Active contours driven by edge entropy fitting energy for image segmentation, Signal Processing, 149 (2018) 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Walter T, Klein J, Massin P, Erginay A, A contribution of image processing to the diagnosis of diabetic retinopathy-detection of exudates in color fundus images of the human retina, Transactions on Medical Imaging, 21(10):1236–1243, (2002). [DOI] [PubMed] [Google Scholar]
- [14].Kande G, Subbaiah P, Savithri T, Segmentation of exudates and optic disc in retinal images, Sixth Indian Conference on Computer Vision, Graphics and Image Processing (ICVGIP), 535–542, (2008) [Google Scholar]
- [15].Alghamdi H, Tang H, Waheeb S, Peto T, Automatic Optic Disc Abnormality Detection in Fundus Images: A Deep Learning Approach, Proceedings of the Ophthalmic Medical Image Analysis (OMIA), 17–24, (2016) [Google Scholar]
- [16].Morales S, Naranjo V, Angulo J, Alcaniz M, Automatic detection of optic disc based on PCA and mathematical morphology, IEEE Transactions on Medical Imaging 32:786–796, (2013) [DOI] [PubMed] [Google Scholar]
- [17].Kumar J, Pediredla AC Seelamantula, Active discs for automated optic disc segmentation, 2015 IEEE Global Conference on Signal and Information Processing (GlobalSIP), 225–229, (2015) [Google Scholar]
- [18].Wang L, Zhang H, He K, Chang Y, Yang X, Active contours driven by multi-feature Gaussian distribution fitting energy with application to vessel segmentation, PLoS ONE 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Giachetti A, Ballerini L, Trucco E, Accurate and reliable segmentation of the optic disc in digital fundus images, Journal of Medical Imaging, 1(2):024001, (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hu M, Zhu C, Li X, Xu Y, Optic cup segmentation from fundus images for glaucoma diagnosis, Bioengineered, 8(1):21–28, (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gopalakrishnan A, Almazroa A, Raahemifar K, Lakshminarayanan L, Optic disc segmentation using circular hough transform and curve fitting, Conference on Opto-Electronics Applied Optics, (2015) 1–4 [Google Scholar]
- [22].Abdullah M, Fraz M, Barman S, Localization and segmentation of optic disc in retinal images using circular Hough transform and grow-cut algorithm, PeerJ, (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sigut J, Nunez O, Fumero F, Gonzalez M, Arnay R, Contrast based circular approximation for accurate and robust optic disc segmentation in retinal images, PeerJ, (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hoover A, Goldbaum M, Locating the optic nerve in a retinal image using the fuzzy convergence of the blood vessels, IEEE Transactions on Medical Imaging 22(8):951–958, (2003) [DOI] [PubMed] [Google Scholar]
- [25].Youssif A, Ghalwash A, Ghoneim A, Optic Disc Detection From Normalized Digital Fundus Images by Means of a Vessels’ Direction Matched Filter, IEEE Transactions on Medical Imaging 27(1):11–18, (2008) [DOI] [PubMed] [Google Scholar]
- [26].Tan J, Acharya U, Bhandary S, Chua K, Sivaprasad S, Segmentation of optic disc, fovea and retinal vasculature using a single convolutional neural network, arXiv:1702.00509 [Google Scholar]
- [27].Maninis K, Tuset J, Arbelaez P, Gool L, Deep retinal image understanding, Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 140–148, (2016) [Google Scholar]
- [28].Litjens G, Kooi T, Bejnordi B, Setio A, Ciompi F, Ghafoorian M, Laak J, Ginneken B, Sanchez C, “A survey on deep learning in medical image analysis,” Medical Image Analysis 42 (2017) 60–68 [DOI] [PubMed] [Google Scholar]
- [29].Ronneberger O, Fischer P, Brox T, U-Net: Convolutional Networks for Biomedical Image Segmentation, arXiv:1505.04597v1, (2015) [Google Scholar]
- [30].Budai A, Hornegger J, Michelson G, Blood Vessel Density and Distribution in Fundus Images, Investigative Ophthalmology & Visual Science, 53(14):2175, (2012) [Google Scholar]
- [31].Milletari F, Navab N, Ahmadi S, V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation, arXiv:1606.04797v1, (2016) [Google Scholar]
- [32].Badrinarayanan V, Kendall A, Cipolla R, SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation, IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(12): 2481–2495, (2017) [DOI] [PubMed] [Google Scholar]
- [33].Zhen Y, Gu S, Meng X, Zhang X, Wang N, Pu J, Automated identification of retinal vessels using a multiscale directional contrast quantification (MDCQ) strategy, Med Phys, 41(9):092702, (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ioffe S, Szegedy C, Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift, arXiv preprint arXiv:1502.03167v3 (2015) [Google Scholar]
- [35].Wang L, Zhu J, Sheng M, Cribb A, Zhu S, Pu J, Simultaneous segmentation and bias field estimation using local fitted images, Pattern Recognition, 74 (2018) 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang L, Gao X, Cui X, Liang Z, 2D/3D rigid registration by integrating intensity distance, Optics and Precision Engineering, 22(10) (2014) 2815–2824 [Google Scholar]
- [37].Lu S, Accurate and efficient optic disc detection and segmentation by a circular transformation, IEEE Transactions on Medical Imaging, 30(12):2126–2133, (2011) [DOI] [PubMed] [Google Scholar]
- [38].Seo J, Kim K, Kim J, Park K, Chung H, Measurement of ocular torsion using digital fundus image, Proceedings of the 26th Annual International Conference of the IEEE EMBS, 1711–1713, (2004). [DOI] [PubMed] [Google Scholar]
- [39].Lupascu C, Tegolo D, Rosa L, Automated detection of optic disc location in retinal images, Proceedings of the 21st IEEE International Symposium on Computer-Based Medical Systems (CBMS), 17–22, (2008) [Google Scholar]
- [40].Lalonde M, Beaulieu M, Gagnon L, Fast and robust optic disc detection using pyramidal decomposition and Hausdorff-based template matching, IEEE Transactions on Medical Imaging, 2o(11):1193–1200, (2001) [DOI] [PubMed] [Google Scholar]
- [41].Cheng J, Liu J, Wong D. et al. , Automatic optic disc segmentation with peripapillary atrophy elimination, Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 6224–6227, (2011) [DOI] [PubMed] [Google Scholar]
- [42].Cheng J, Liu J, Xu Y, et al. , Superpixel Classification Based Optic Disc and Optic Cup Segmentation for Glaucoma Screening, IEEE Transactions on Medical Imaging, 32(6):1019–1032, (2013) [DOI] [PubMed] [Google Scholar]