Summary Paragraph
Advances in genetics and sequencing reveal a plethora of disease-associated and disease-causing genetic alterations. Resolving causality between genetics and disease requires generating accurate models for molecular dissection; however, the rapid expansion of single-cell landscapes presents a major challenge to accurate comparisons between mutants and their wild-type equivalents. Here, we generated mouse models of human severe congenital neutropenia (SCN) using patient-derived mutations in the Growth factor independent-1 (GFI1) transcription factor. To delineate the impact of SCN mutations, we generated single-cell references for granulopoietic genomic states with linked epitopes1, aligned mutant cells to their wild-type equivalent and identified differentially expressed genes and epigenetic loci. We find that Gfi1-target genes are altered sequentially, as cells traverse successive states during differentiation. These cell-state-specific insights facilitated genetic rescue of granulocytic specification but not post-commitment defects in innate-immune effector function; underscoring the importance of evaluating the impact of mutations and therapy within each relevant cell state.
Introduction
In severe congenital neutropenia (SCN) patients, inherited and de novo mutations lead to a profound block in neutrophil granulopoiesis. We introduced SCN-patient mutations in the Growth factor independent-1 (GFI1) transcription factor into the murine Gfi1 allele, which resulted in steady-state murine dysgranulopoiesis and broad in vivo susceptibility to neutrophil-dependent pathogens. Notably, SCN mutations divorce neutropenia from Gfi1−/− defects in hematopoietic stem cell and lymphocyte numbers. To delineate mechanism, we mapped the successive genomic states encompassing neutrophil granulocyte specification (capacity to form neutrophils) and commitment (cell-fate restriction)2, including a new transitional intermediate (see online viewer3), then confirmed the trajectory. Using comparative genomics4,5 we assigned Gfi1-mutant cells to wild-type cell states to determine differential single-cell gene expression and chromatin. Few Gfi1-target genes are deregulated across granulopoiesis, but are instead deregulated in successive granulopoietic cell states; associated with failure of Gfi1-bound chromatin to close after specification. However, genetically rescuing specification did not resolve post-commitment defects in immune defense in mice or in G-CSF-rescued human SCN patient neutrophils. These experiments underscore the dominance of cell state in integrating the impact of both mutations and therapy, and illustrate a workflow that can be broadly applied to molecularly dissect translationally-relevant models of disease.
Patient mutations induce neutropenia
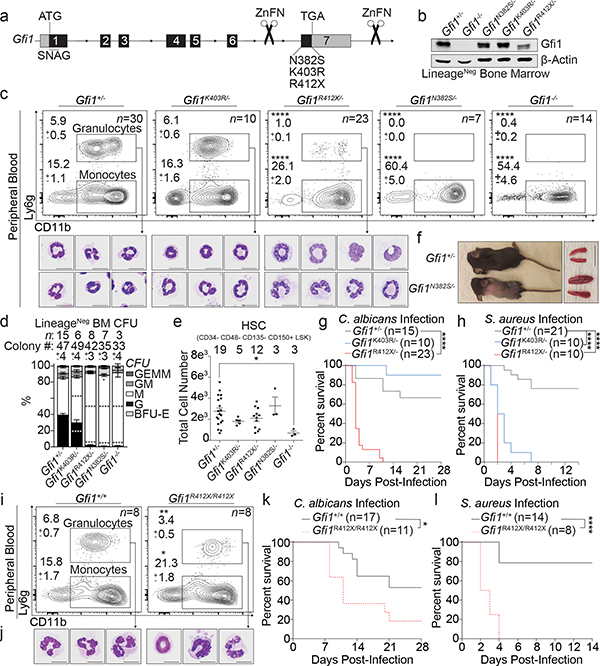
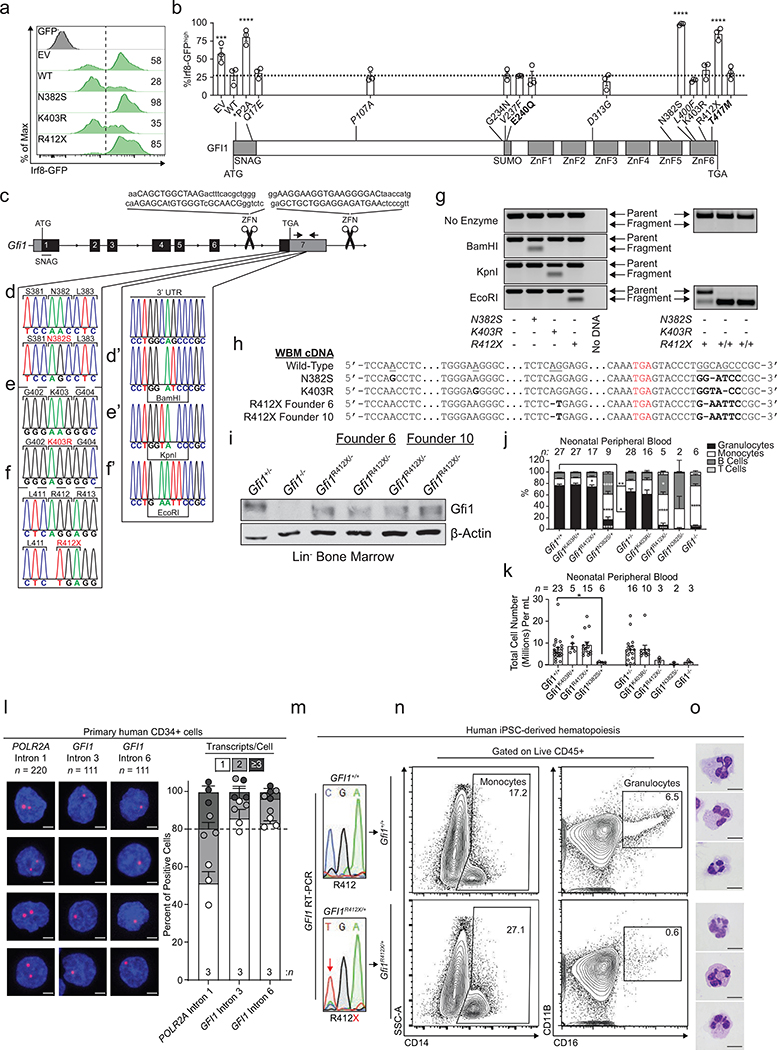
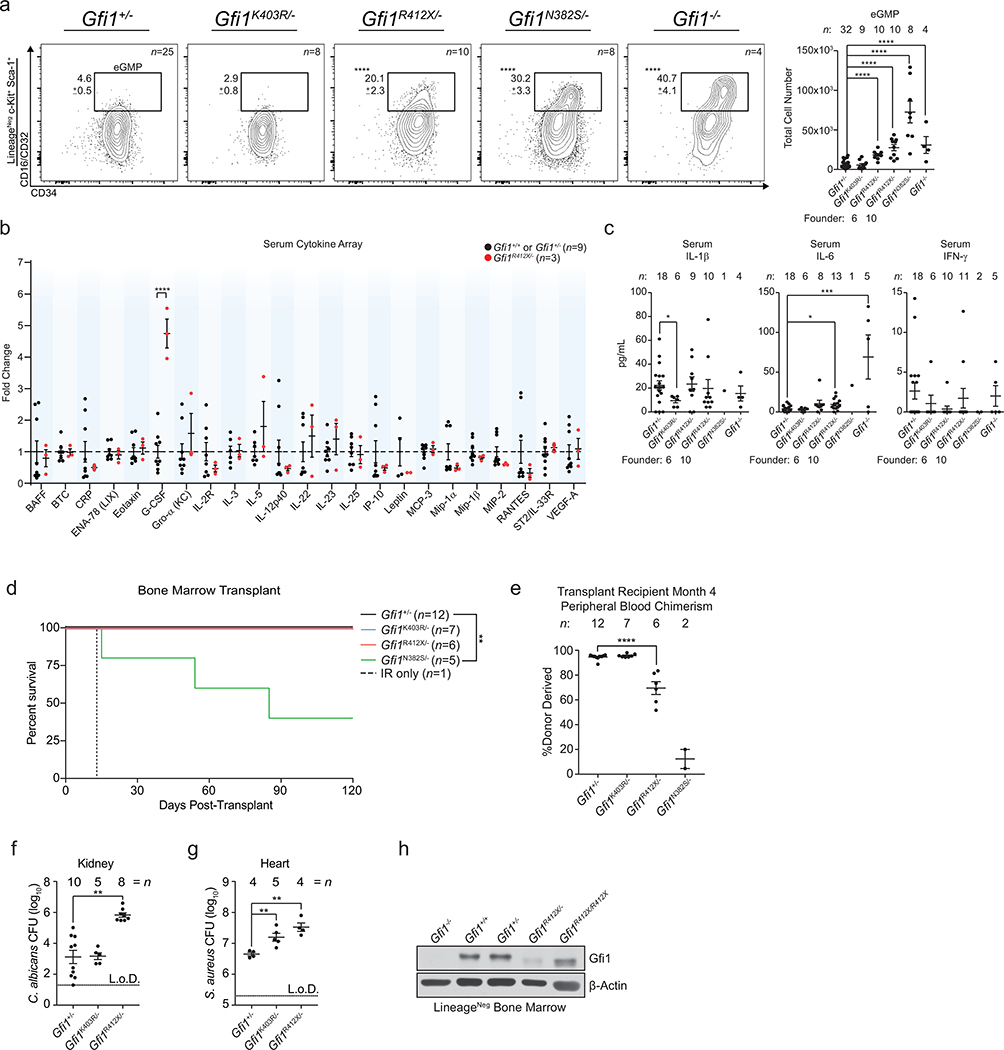
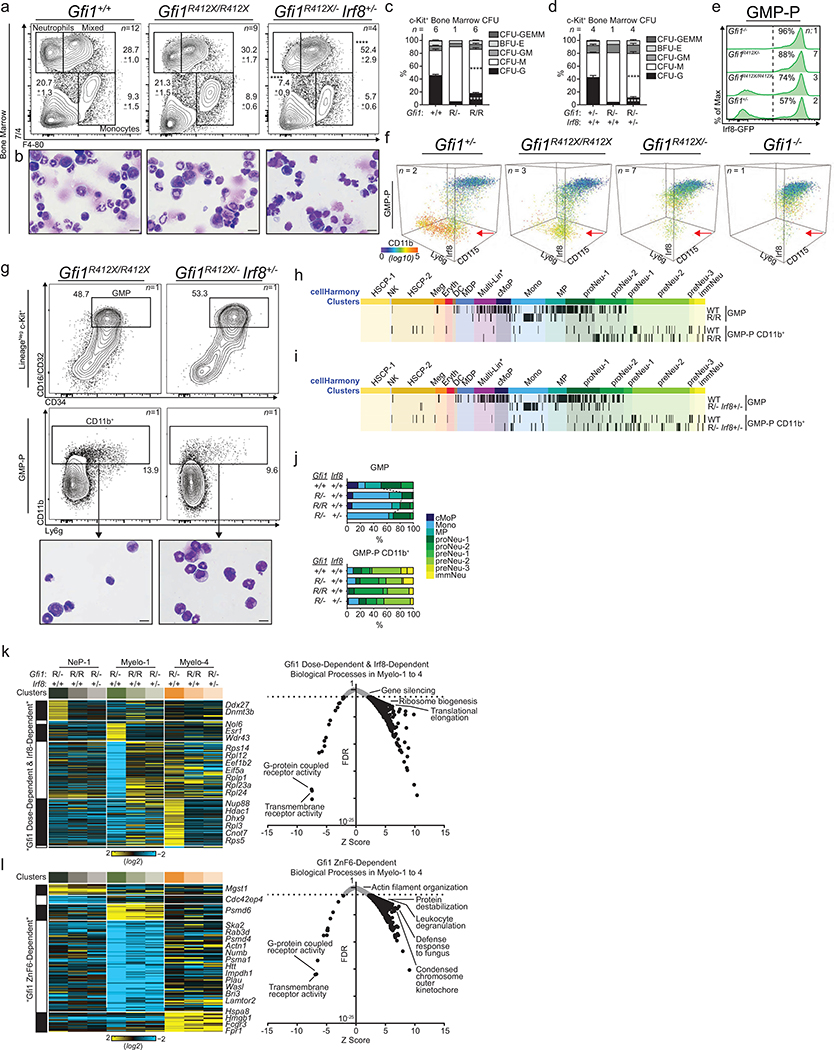
Clinical resequencing of 225 neutropenia-patients identified previously-reported6,7 and novel GFI1 sequence polymorphisms of unknown significance (Supplementary Information), which we screened for defects in repressing an Irf8-eGFP-reporter allele8 (Extended Data Fig. 1a, b). Next, we edited endogenous murine Gfi1 alleles to introduce N382S, K403R, or R412X mutations (Fig. 1a, Extended Data Fig. 1c–h, phenotypic summary: Supplementary Information). Mutant Gfi1 proteins accumulate normally, but R412X mutation results in a smaller, less abundant protein (Fig. 1b, Extended Data Fig. 1i). On postnatal day 1, only Gfi1N382S/+ mice exhibit a blunted response to neonatal gut-bacteria-driven cues9, comparable to the exacerbated clinical presentation of SCN in neonatal humans (Extended Data Fig. 1j, k). Human GFI1 is predicted to be imprinted.10 RNAScope analysis with GFI1 intronic probes (Extended Data Fig. 1l) revealed a pattern consistent with monoallelic expression.11 Human induced pluripotent stem cells (iPSC) with one GFI1R412X allele only express the mutant GFI1 allele (Extended Data Fig. 1m), and are blocked in granulopoiesis (Extended Data Fig. 1n, o). Therefore, to mimic predicted human gene regulation we genetically silenced one Gfi1 allele12 to find Gfi1R412X/-, Gfi1N382S/-, and Gfi1−/− neonatal mice profoundly neutropenic (Extended Data Fig. 1j, k). Peripheral blood revealed a range of neutropenia (Fig. 1c, Extended Data Fig. 2a), and Gfi1R412X/- and Gfi1N382S/- mice exhibited monocytosis as a result of cell-intrinsic lineage skewing (Fig. 1c, d, Extended Data Fig. 2a–c), supported by a lack of mature neutrophils in the bone marrow (Extended Data Fig. 2d–f). Gfi1R412X/- neutrophils exhibited abnormal or immature morphologies (Fig. 1c), suggesting a Gfi1 function beyond neutrophil specification. Notably, Gfi1−/−, Gfi1N382S/-, Gfi1R412X/- mice display steady-state Sca-1+ emergency granulocyte-monocyte-progenitor gate cells (eGMP)13,14 (Extended Data Fig. 3a). However, like SCN-patient serum15, extensive profiling of Gfi1R412X/- serum revealed elevated G-CSF, but not eGMP-associated inflammatory cytokines (Extended Data Fig. 3b–c, Supplementary Information).
Figure 1. Neutropenia-patient-derived mutations induce steady-state murine dysgranulopoiesis and broad immune defects.
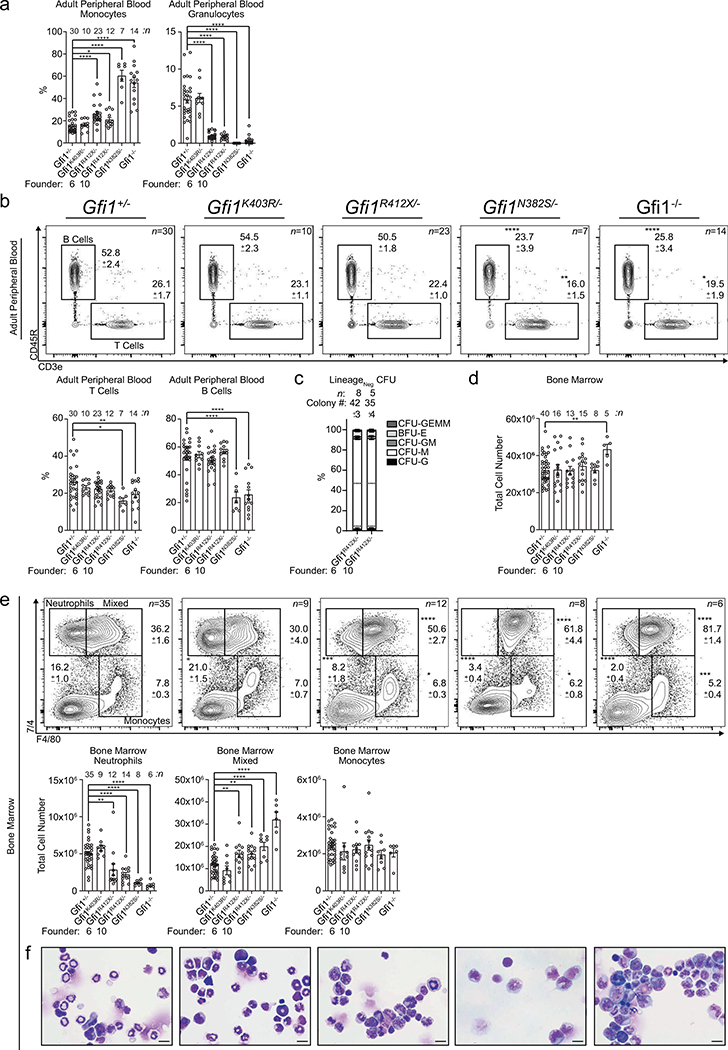
a, Schematic of Gfi1 locus; locations of mutations introduced by genome editing. b, Immunoblot of adult murine LineageNeg bone marrow. c, FACS plots with percentages of total adult peripheral blood, and cytospins of FACS-sorted CD11b+ Ly6g+ cells. d, Colony-forming-unit (CFU) assays performed on LineageNeg bone marrow cells. G; granulocyte, M; monocyte, GM; granulocyte-monocyte, BFU-E; burst-forming-unit erythroid, GEMM; granulocyte, erythrocyte, monocyte, megakaryocyte. e, FACS analysis of adult murine bone marrow. HSC, hematopoietic stem cell; LSK, LineageNeg Sca-1+ c-Kit+. f, Representative images of adult mice and spleens. g-h, Survival of mice infected with g, an LD50 dose of Candida albicans or h, 5 × 107 CFU of Staphylococcus aureus. Mouse numbers in parentheses. i, FACS plots and j, cytospins from R412X homozygotes. k-l, Survival of mice infected with k, an LD50 dose of C. albicans or l, 5 × 107 CFU of S. aureus. Mouse numbers in parentheses. Data are displayed as mean ± s.e.m. from independent biological replicates in c-e, and i. Data in b represents 3 independent experiments. Images in f are representative of 2 littermate mice for each genotype. * p < 0.05, ** p < 0.01, and **** p < 0.0001 as determined by two-tailed t-test in c-e, and i or by two-sided Mantel-Cox test in g, h, k, and i. The scale bars represent 10 μm in c and j and 1 cm in f.
Unlike Gfi1−/− mice, numbers of phenotypic hematopoietic stem cells (HSC) were normal in mutants (Fig. 1e, but Gfi1N382S/- HSC fitness was defective after transplantation. In contrast, Gfi1R412X/- HSC rescue lethal irradiation, but display a modest defect in chimerism during competitive transplant (Extended Data Fig. 3d, e). Similar to Gfi1−/− mice12,16–18, Gfi1N382S/- mutants displayed several defects, including runting, lymphopenia, splenomegaly (Fig. 1f, Extended Data Fig. 2b), and rarely survive to adulthood. However, neither Gfi1K403R/- nor Gfi1R412X/- mutants recapitulate these Gfi1−/− defects (Extended Data Fig. 2b, 3d, e), so we focused on Gfi1K403R/- and Gfi1R412X/- mice to isolate Gfi1 control of granulopoiesis.
Gfi1R412X/- mice were susceptible to Candida albicans with reduced survival and increased pathogen burden (Fig. 1g, Extended Data Fig. 3f). By contrast, Gfi1R412X/- and Gfi1K403R/- mice showed increased susceptibility to Staphylococcus aureus (Fig. 1h, Extended Data Fig. 3g). Impaired host defense in Gfi1R412X/- mice likely does not reflect reduced Gfi1 protein levels, since Gfi1R412X/R412X partially restored Gfi1 protein levels (Extended Data Fig. 3h), and neutrophil numbers (Fig. 1i) albeit with abnormal morphology (Fig. 1j) but failed to rescue susceptibility to C. albicans and S. aureus (Fig. 1k, l). Thus, Gfi1 control of granulocyte production is functionally dissociated from innate host defense against invasive microbial infection.
Neutrophil specification and commitment
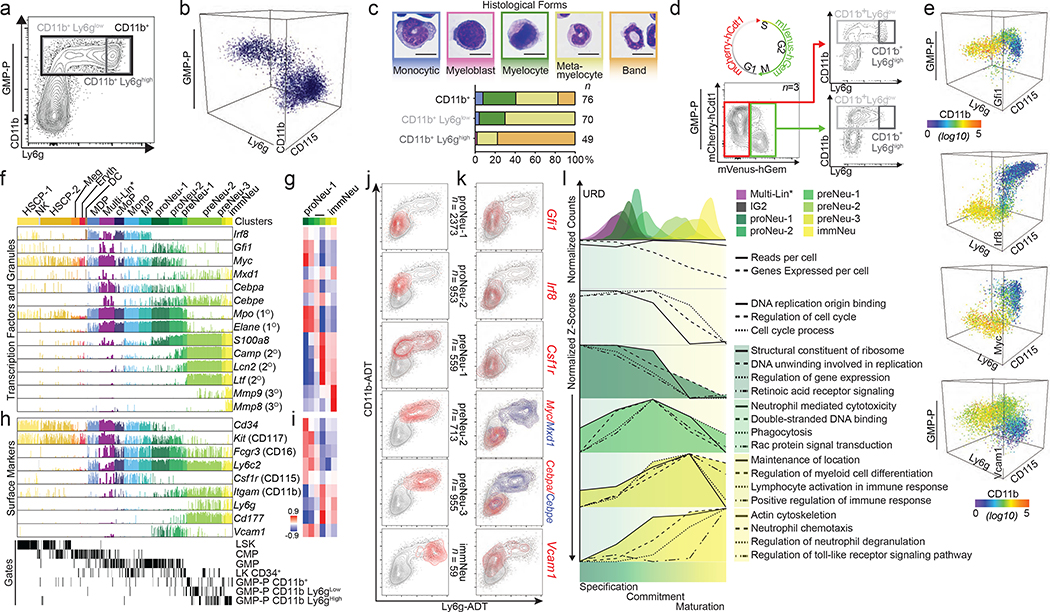
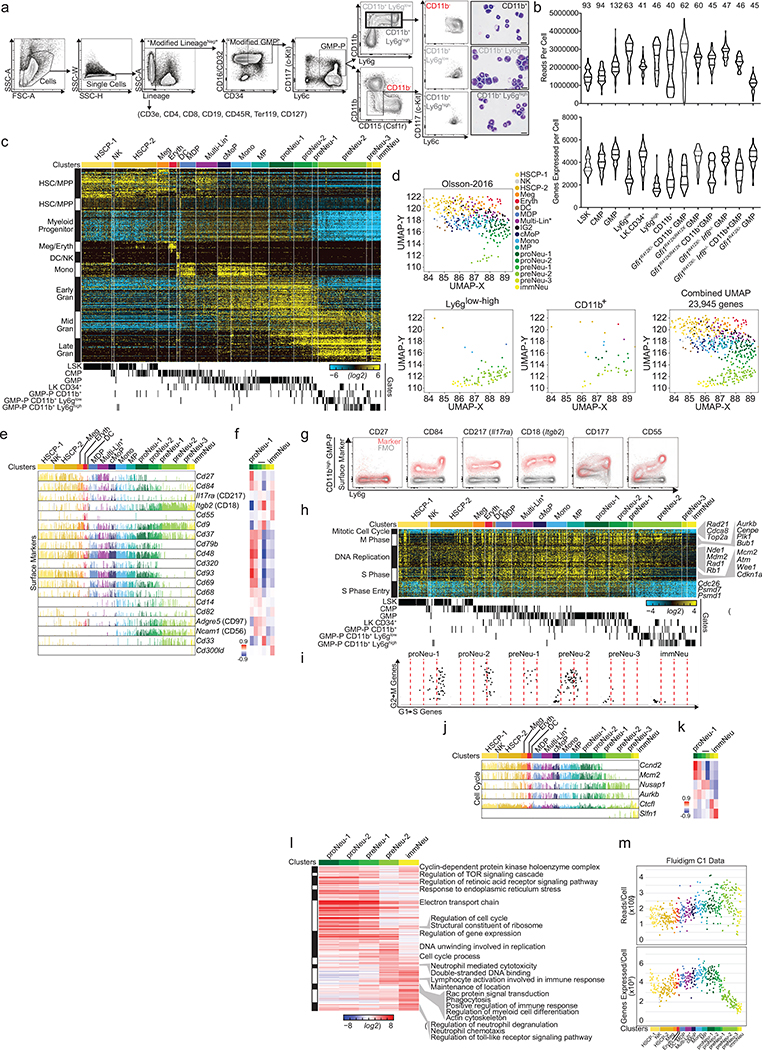
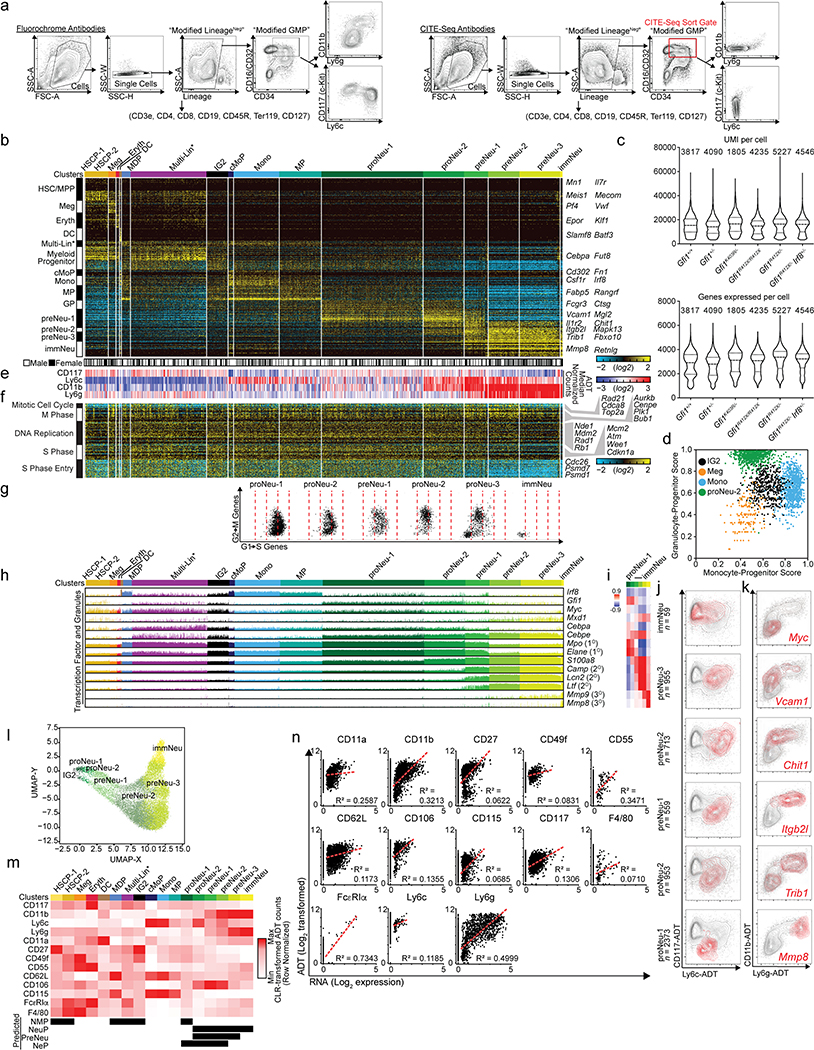
First, we established the genomic states encountered during neutrophil granulopoiesis using a novel flow gate (“GMP-P”) (Extended Data Fig. 4a), with reciprocal expression of Ly6g (a granulocytic marker) and CD115 (Csf1r, a monocytic marker) based on CD11b (Fig. 2a, b, Extended Data Fig. 4a). Histological forms, a transgenic cell cycle reporter (Fucci2)19, and transgenic reporters for Gfi1, Irf8 and Myc validated cells undergoing terminal granulopoiesis (Fig. 2c–e and Extended Data Fig. 4a). Notably, Gfi1 expression is highest in CD11bintLy6gneg cells (Fig. 2e). These analyses suggest that CD11bintLy6gneg cells are specifying, while CD11bhighLy6gneg-high cells are committing to neutrophil granulopoiesis. We captured these cells ( Extended Data Fig. 4b, c) and incorporated them into prior scRNA-Seq data20,21 (Extended Data Fig. 4d and Supplementary Information). An unsupervised discovery workflow (ICGS)20 (Extended Data Fig. 4c) revealed a dramatic shift in gene expression between early (specifying proNeu) and late (committed preNeu) granulocytic clusters, as well as a transitional population (preNeu-1) expressing both programs (proNeu, preNeu, immNeu names adapted from a review22). To validate the cluster order we identified known reciprocal patterns of transcription factor genes (Irf8/Gfi1, Myc/Mxd1 and Cebpa/Cebpe), as well as primary (Elane, Mpo), secondary (Camp, Lcn2, Ltf), and tertiary (Mmp8, Mmp9) granule-protein genes (Fig. 2f) whose expression was significantly correlated to individual clusters (Fig. 2g). We delineated surface markers that are statistically correlated to ICGS clusters (Fig. 2h, i and Extended Data Fig. 4e–g) to find those that are extinguished (Cd34, Csf1r, Kit) or induced (Itgam, Ly6g, Cd177). Surprisingly, Vcam1 is uniquely expressed in granulocyte-specifying cells, as confirmed by Vcam1/CD106-flow-cytometric analysis and 3D visualization of GMP-P gate cells (Fig. 2e) similar to the Gfi1 reporter (Fig. 2e). Cell-cycle gene expression terminated in the last cluster (Extended Data Fig. 4h–l), along with an overall decrease in expressed genes at the preNeu-1 stage independent of the number of reads per cell (Extended Data Fig. 4m, suggesting that global transcriptional silencing is concomitant with granulocytic commitment.
Figure 2. Cell states traversed during commitment to terminal granulopoiesis include a rare transitional state (bridging specification to commitment).
a, FACS plot with annotated gates used to sort cells for scRNA-Seq (see Extended Data Fig. 4a; granulocyte-monocyte-progenitor and precursor “GMP-P” gate). b, 3D FACS plot of GMP-P gate (see Extended Data Fig. 4a). c, Quantified histological forms of FACS-sorted cells (from gates in a). d, Schematic of fluorescent cell cycle reporter and representative FACS plots of GMP-P gate from Fucci2 transgenic mice. e, 3D FACS plots of bone marrow isolated from adult mice bearing Gfi1, Irf8 or Myc fluorescent-protein reporters, or Vcam1 expression. Events are pseudocolored for CD11b expression. f, Bar chart displaying incidence and amplitude for selected genes (Fluidigm C1; see Extended Data Fig. 4c)(n = 509 cells). ICGS clusters are annotated (top) FACS gates are annotated (bottom). g, Heatmap of correlation between gene expression and each displayed cluster (MarkerFinder). h, Bar chart displaying selected genes (same cells as in f). i, Heatmap of correlation between gene expression and each displayed cluster (MarkerFinder). j-k, Plots of ADT UMIs (10X 3’: see Extended Data Fig. 5b, e) where grey indicates all captured cells: j, red identifies cells within ICGS clusters (right). k, red or blue indicates the top 1% of cells expressing the indicated gene(s). l, URD pseudotime analysis (of data in Extended Data Fig. 5b), and transcriptional features, or normalized Z-scores of enriched biological processes (of data in Extended Data Fig. 4c). Data are representative of three independent biological replicates in a, b, d, and e. Data in c represents cumulative total cell numbers from one experiment using cells pooled from 3 male mice. Data in g and i display Pearson correlation values. LSK, LineageNeg c-Kit+ Sca-1+; CMP, common myeloid progenitor; LK CD34+, LineageNeg c-Kit+ Sca-1Neg CD34+.
To directly link GMP-P flow gates (Fig. 2a, Extended Data Fig. 4a) to cell clusters (Extended Data Fig. 4c), we performed cellular indexing of transcriptomes and epitopes by sequencing (CITE-Seq)1 (Fig. 2j, Extended Data Fig. 5a). We supervised analysis of the CITE-Seq data using centroids of ICGS clusters (Extended Data Fig. 4c and 20) to classify the newly captured cells (Extended Data Fig. 5b–e), which replicated mixed-lineage and specifying cell states (including Multi-Lin*20, IG220 and preNeu-1 transitional populations)(Extended Data Fig. 5b, d) with no apparent sex-based bias in any cluster (Extended Data Fig. 5b, bottom). Myeloid-associated, cell cycle, and surface-marker gene expression in the CITE-Seq data displayed similar trends (compare Fig. 2f, g and Extended Data Fig. 4h–k to Extended Data Fig. 5f–i). Transformed antibody-dependent-tag (ADT) counts revealed patterns closely resembling FACS data (compare Extended Data Fig. 5a with Extended Data Fig. 5e). Surprisingly, Ly6g surface expression is discordant with the underlying processes; creating overlap between preNeu and immNeu clusters. Plotting ICGS-clusters or individual genes (significantly correlated to clusters) illustrates a similar progression through the CD11b versus Ly6g plots (Fig. 2j, k, Extended Data Fig. 5j, k). Plotting the top 1% of cells expressing Gfi1, Irf8, Csf1r, Myc/Mxd1, Cebpa/Cebpe, and Vcam1 (Fig. 2k) confirmed the trajectory, which was further evidenced using pseudotemporal ordering with URD23 (Fig. 2l, top). Clusters were confirmed in a published data set24 (Extended Data Fig. 5l), and reconciled to reported neutrophil progenitor subsets (NMP,25 NeuP,26 preNeu,27 and NeP24) using 13 of the surface markers used to identify them. Notably, NMP, NeuP, preNeu, and NeP span multiple ICGS clusters (Extended Data Fig. 5m, n). Next, we identified key pathways and biological processes that are dynamically regulated during terminal granulopoiesis (Fig. 2l and Extended Data Fig. 4l). Specification and commitment are accompanied by distinct biological processes inducing withdrawal from the cell-cycle, a general collapse of gene expression concurrent with the sequential induction of innate immune effector gene expression (Fig. 2l).
Mutation impacts cell states differently
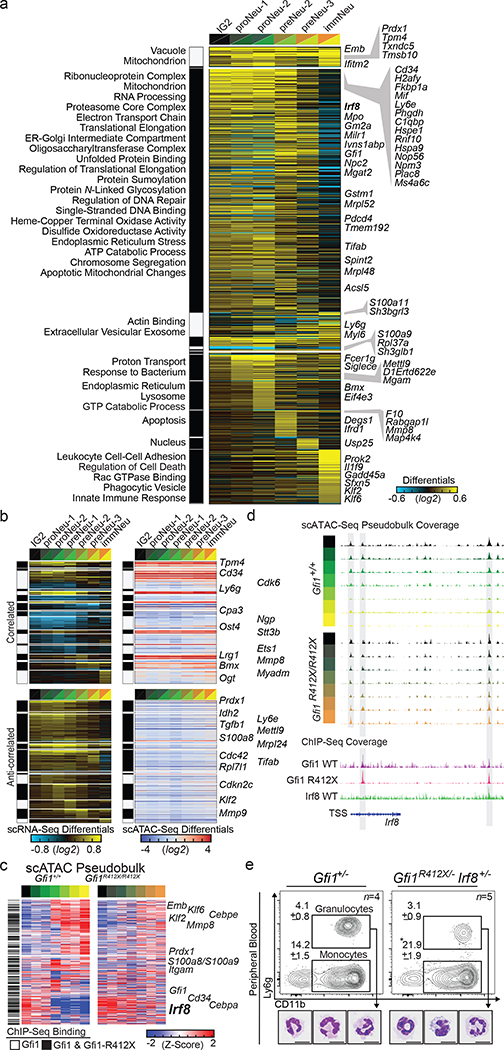
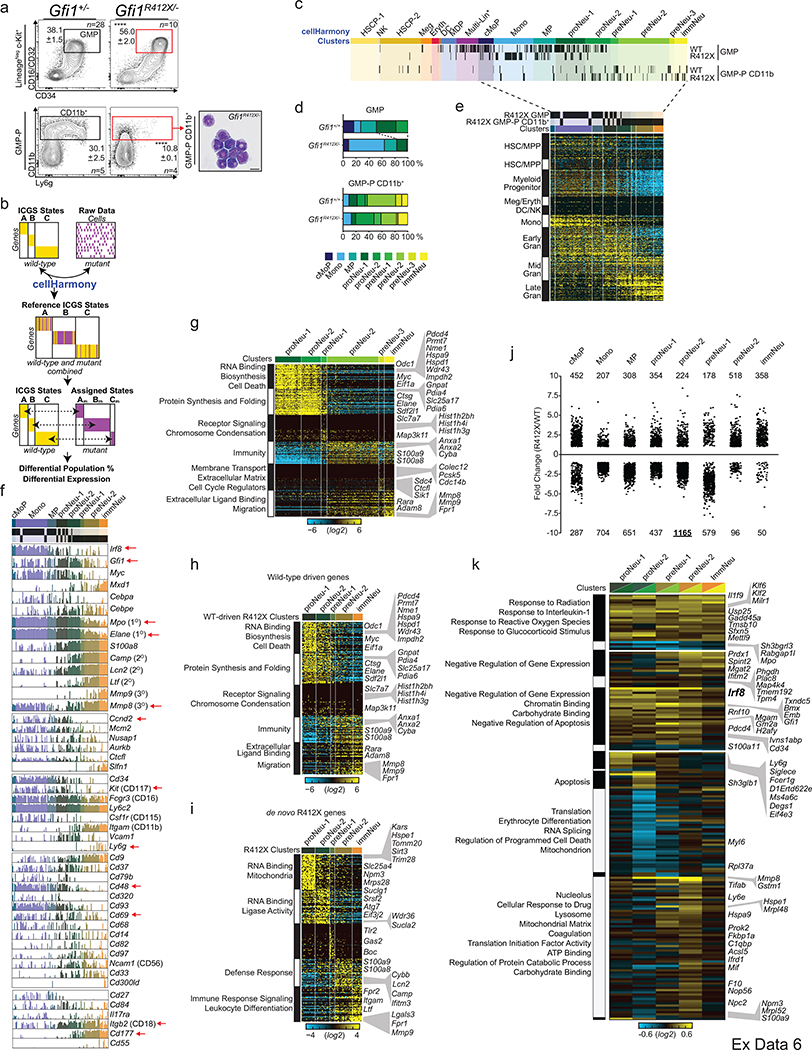
Given references for granulopoietic genomic states, we could now determine the transcriptional deregulation underlying the Gfi1R412X/- defects in neutrophil production and function. We extended CITE-Seq and complementary Fluidigm captures to Gfi1+/− and Gfi1R412X/- GMP (Extended Data Fig. 4b, see online viewer3, Extended Data Fig. 6a–k, 7a–d, and 20). Next, we applied a computational workflow (cellHarmony4 Extended Data Fig. 6b) to assign Gfi1-mutant cells to ICGS clusters and determine differential gene expression. Most Gfi1R412X/- GMP are monocytic progenitors (51% by CITE-Seq, 55% by Fluidigm), a ~3 fold increase relative to Gfi1+/− or Gfi1+/+. Few cells were assigned beyond proNeu-1 (6.5 fold decrease)(Extended Data Fig. 6c, d, 7b, c). However, capturing rare Gfi1R412X/- CD11bhigh GMP-P (Extended Data Fig. 6a) revealed cells within each of the granulocytic clusters (Extended Data Fig. 6c–e, 7b, c) resembled that of the wild-type (Extended Data Fig. 4c), including known myeloid, cell cycle, and surface marker genes (Extended Data Fig. 6f). Strikingly, of Gfi1 targets defined by ChIP-Seq20 (57% of differentially expressed genes), only a minority are disrupted across all clusters (see Gfi1, Mpo, Irf8 and Mmp8 expression in Extended Data Fig. 6f). Instead, using either 10x or Fluidigm technology most deregulated expression occurs during specification or in specific clusters (Fig. 3a, Extended Data Fig. 6k).
Figure 3. A single mutation in a lineage-determining transcription factor differentially impacts target gene expression in distinct cell states.
a, cellHarmony differential expression heatmap. scRNA-Seq libraries from Gfi1R412X/- and Gfi1+/− mice were analyzed (10X 3’ mod-GMP gate and Ly6ghigh GMP-P gate), then restricted to Gfi1 ChIP-Seq targets (841 out of 1462 total), and genes commonly deregulated across 10x and Fluidigm platforms are displayed (right)(see Extended Data Fig. 6k). Each row represents a single gene, each column the fold difference, and enriched Gene Ontology terms (left). b, Differential cicero gene activity scores correlated (top) or anticorrelated (bottom) with scRNA-Seq differentially expressed genes (from panel a). c, Heatmaps of scATAC-Seq pseudo-bulk data from Gfi1 and Gfi1-R412X bound loci (black bars on left, and see Extended Data Fig. 8e). ICGS clusters (top). Each row is a gene locus (right). d, Schematic of the Irf8 locus displaying scATAC-Seq pseudobulk accessibility with regions bound by Gfi1 shaded (top). ChIP-Seq reads (bottom). ICGS clusters (left). e, FACS plots and representative cytospins of FACS-sorted CD11b+ Ly6g+ cells from adult murine peripheral blood. In e, data are displayed as mean ± s.e.m. from independent biological replicates; * p < 0.05 as determined by two-tailed t-test, and scale bars represent 10 μm.
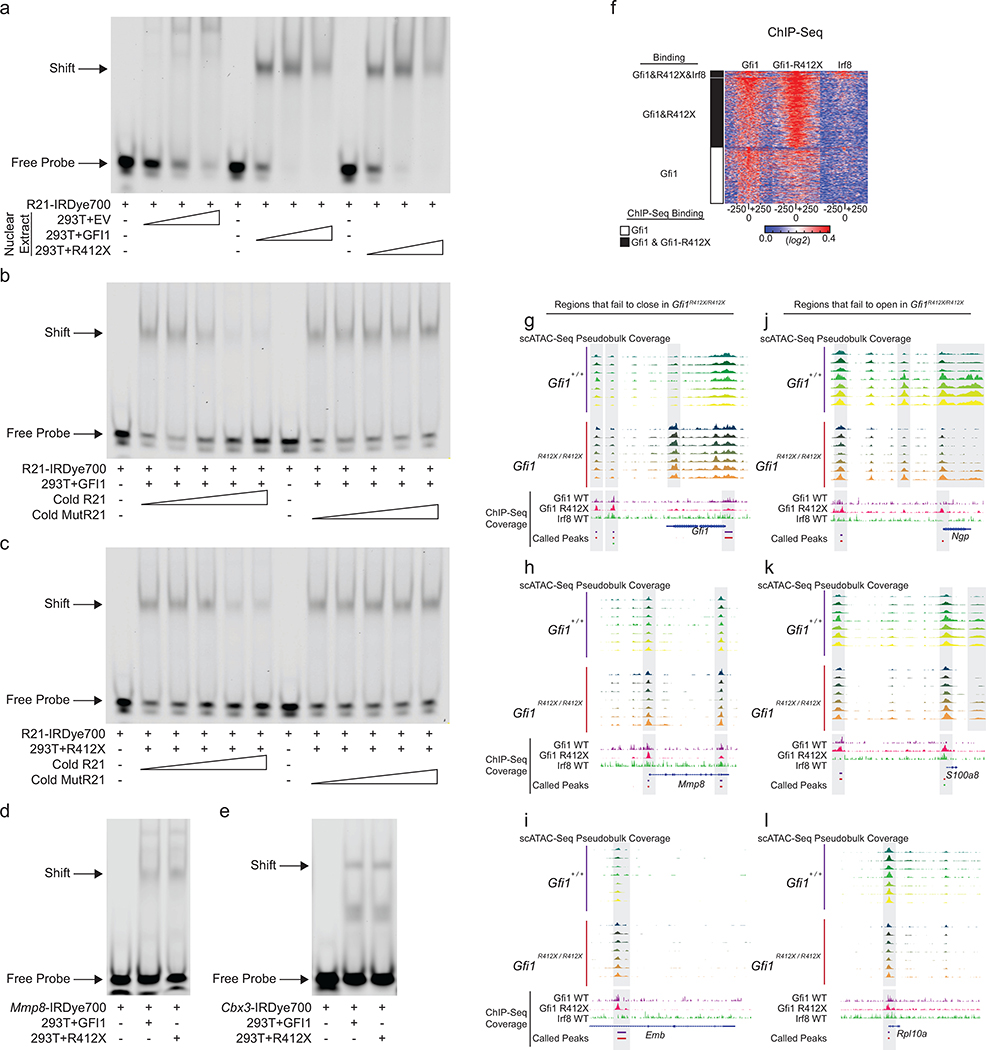
To understand the underlying gene regulatory mechanisms, we performed scATAC-Seq on Gfi1+/ + and Gfi1R412X/ R412X Modified-GMP-gate cells (Extended Data Fig. 7e–j). The scATAC-Seq clusters were aligned to CITE-Seq cell populations (Extended Data Fig. 7e, f) using label transfer in Seurat35 (consistent with SNAP-ATAC28; see Extended Data Fig. 7i), and cluster-specific ATAC-peaks were associated with lineage-specific genes and sequential neutrophil programming (Extended Data Fig. 7k, Supplementary Information: UCSC Genome Browser Session to Visualize ChIP-Seq and scATAC-Seq Data). We computed differentially accessible loci (cicero gene-activity scores), identified regions correlated or anticorrelated with differential gene expression (Fig. 3b), and inferred dynamic transcription factor activity changes (Extended Data Fig. 7l). Accessible DNA in Gfi1R412X/R412X clusters were differentially enriched for Gfi1 and Irf8 motifs; suggesting the failure of Gfi1-R412X to silence them. Notably, zinc-finger 6 is dispensable for DNA binding in vitro29, which we confirmed for Gfi1-R412X (Extended Data Fig. 8a–e). Moreover, Gfi1R412X/R412X ChIP-Seq on sorted GMP revealed most loci normally bound by Gfi1 (Extended Data Fig. 8f). However, while Gfi1-target loci demonstrate dynamic chromatin accessibility in wild-type cells (Fig. 3c, left), these patterns are disrupted in Gfi1R412X/R412X cells, including Irf8 (Fig. 3c, right, Fig. 3d, Extended Data Fig. 8g–l). Thus, Gfi1-R412X occupancy is not associated with normal dynamic regulation of chromatin accessibility.
We noted Gfi1-R412X deregulation of both Irf8 expression and Gfi1-bound chromatin accessibility (Fig. 3a, c, d, Extended Data Fig. 6f, k), so we limited Irf8 alleles. Similar to Gfi1R412X/R412X which repressed Irf8-GFP expression and partially rescued granulopoietic specification (Fig. 1i, Extended Data Fig. 9a–f), Gfi1R412X/-Irf8+/− induces a partial rescue of neutrophil production (Fig. 3e, Extended Data Fig. 9a–d). However, Gfi1R412X/R412X and Gfi1R412X/-Irf8+/− neutrophils still exhibit abnormal morphologies (Fig. 1c, j, Fig. 3e).
Rescuing neutropenia but not function
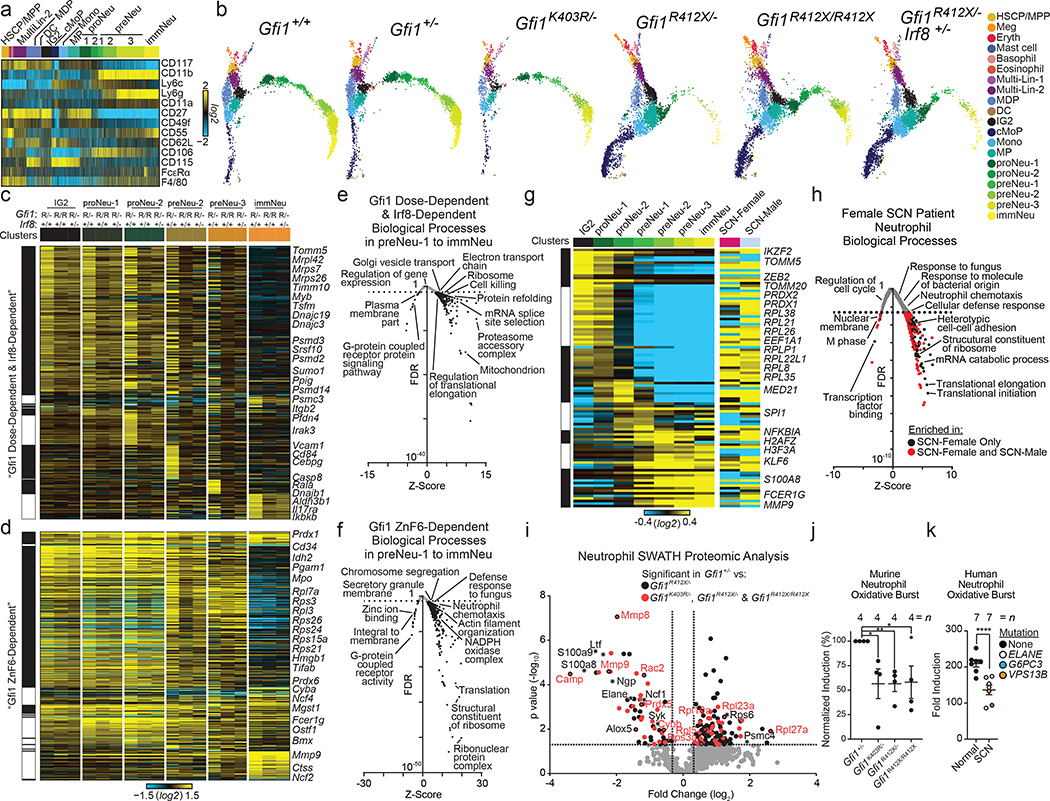
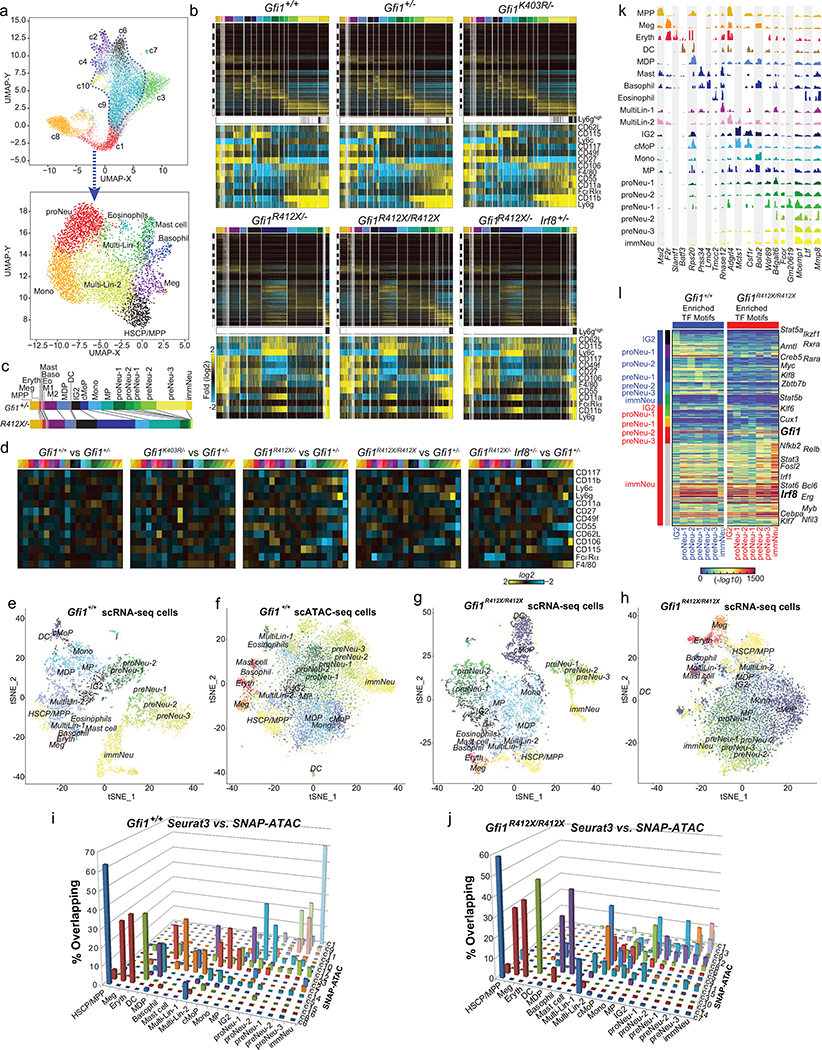
To determine the genes underlying genetic rescue, we extended Fluidigm captures to genetically-rescued GMP-gate cells (Extended Data Fig. 4b, 9g–l) and CITE-Seq of Modified-GMP-gate cells to all mice (Fig. 4a–f, Extended Data Fig. 5c, 7b, d). Joint embedding of CITE-Seq datasets into a single Uniform Manifold Approximation and Projection (UMAP) reveals similar embedding of cell populations in Gfi1+/+, Gfi1+/−, and Gfi1K403R/- mice. All Gfi1-R412X mutants display a dramatically altered trajectory (Fig. 4b), but genetic rescue increased the fraction of granulocytic specifying cells (Fig. 4b, Extended Data Fig. 7b, 9j). Similarly, ADT expression was only subtly different between Gfi1+/+, Gfi1+/−, and Gfi1K403R/-; however, Gfi1R412X/- and genetic rescues showed modest but consistent ADT perturbation in Ly6g, CD55, CD106, CD115 and F4/80 (Fig. 4a, Extended Data Fig. 7b, d).
Figure 4. Rescuing specification does not repair innate immune effector functions that are programmed after commitment.
a, Heatmap of median-normalized values of antibody-dependent tags (ADT) from CITE-Seq of sorted wild-type mod-GMP-gate cells. b, Joint UMAP projection of CITE-Seq from the indicated genotypes, each dot represents one cell, and cluster identity is color coded (cellHarmony) to highlight genetic repair (Gfi1R412X/R412X and Gfi1R412X/-Irf8+/−)(also see Extended Data Fig. 7b). c-d, Heatmap of gene expression that is c, genetically repaired or d, not genetically repaired in each cluster (top), with representative genes indicated (right). e-f, Enriched Gene Ontology from e, genetically repaired genes (in c), or f, genes not genetically repaired (in d). g, Heatmap of genes that are differentially expressed in SCN-patient neutrophils (right), and their dynamic expression across murine wild-type clusters is shown (left). h, Enriched biological processes (from genes in g)(Grey dots; control versus female. Red dots: changes conserved to control versus male). i, Volcano plot displaying SWATH proteomics data. (Black dots: differential proteins between Gfi1R412X/- and Gfi1+/− neutrophils. Red dots: change conserved in Gfi1R412X/-, Gfi1K403R/- and Gfi1R412X/R412X neutrophils. j-k, Oxidative burst in purified neutrophils from j, murine bone marrow or k, human peripheral blood. Data in j and k are represented as mean ± s.e.m. from independent biological replicates. * p < 0.05, ** p < 0.01, **** p < 0.0001 as determined by two-tailed t-test for j and k.
Using cluster-specific comparisons between groups in both datasets, we identified those genes transcriptionally repaired by both genetic rescues (Fig. 4c, Extended Data Fig. 9k) and those that are not repaired by either genetic rescue (Fig. 4d, Extended Data Fig. 9l). Notably, processes controlling neutrophil chemotaxis, NADPH oxidation, defense response, translation, and chromosome condensation were not repaired (Fig. 4f, Extended Data Fig. 9l). Indeed, Gfi1-R412X mutant neutrophils failed to condense their chromatin (Fig. 1c, j, Fig. 3e).
Next, we captured SCN-patient and age-matched-donor neutrophils (Fluidigm C1v4; Extended Data Fig. 10, Supplementary Information). Differentially expressed genes are dynamically expressed during normal murine granulopoiesis (Fig. 4g), and enriched for many of the same Gfi1-ZnF6-dependent biological processes (e.g., translation, neutrophil chemotaxis and defense response)(Fig. 4h).
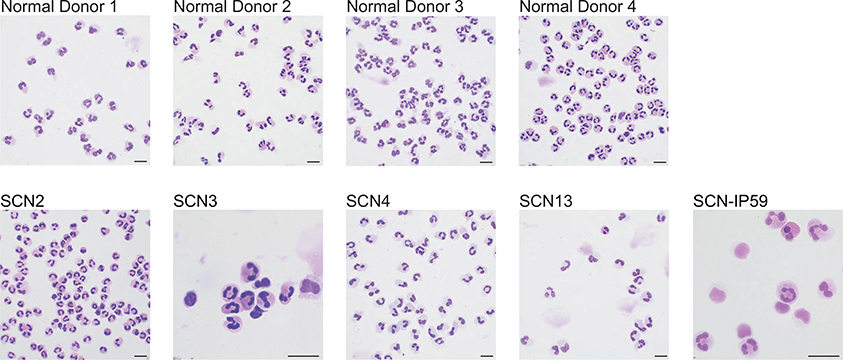
To validate defects in translation, we performed SWATH30 proteomic analysis on purified bone marrow neutrophils. Dissimilar to their elevated gene expression (Extended Data Fig. 6f), we find loss of neutrophil granule protein expression (e.g. Camp, Mmp8, and Mmp9) in all Gfi1-mutant neutrophils (Fig. 4i). In addition, NADPH oxidase complex proteins were low (Fig. 4i), which manifested as a blunted oxidative burst response in vitro (Fig. 4j). SCN patients display innate immune defects in spite of G-CSF therapy,31 and their neutrophils exhibit deregulated genes involved in innate immune functions (Fig. 4h) suggesting that their cytokine-rescued neutrophils may be functionally defective. Similar to Gfi1-mutant murine neutrophils (which are also produced under conditions of high G-CSF, Extended Data Fig. 3b), we found that SCN-patient peripheral blood neutrophils exhibited a blunted oxidative burst response (Fig. 4k). Our findings provide evidence that genetic or cytokine rescue of granulocyte specification is not sufficient to rescue post-commitment innate effector programs.
Discussion
Delineating disease-associated genetic variation is the focus of intensive research; however, determining which sequence alterations are pathogenic requires generating and analyzing genetic models. Exploiting models to resolve the pathobiological impact of mutations is limited by both technology and our understanding of normal biology. We describe a transferable workflow that can be harnessed to answer developmental questions across disciplines. Our results suggest that such analyses can reveal cell-state-specific impacts of mutations (likely due to accompanying changes in the composition of transcription factors and chromatin accessibility at their target genes), with direct consequences for attempts to repair defects. We think that this finding has significant implications for the study of congenital and acquired genetic changes; especially cancers which are multiclonal. Moreover, this approach could be extended to the analysis of the therapeutic impact of new small molecules, where bulk cell analyses or current single cell analytic pipelines might gloss over rare but important cell states.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were blinded to allocation for C. albicans and S. aureus experiments.
Human Samples.
De-identified clinical data and peripheral blood from healthy donors or neutropenic patients was obtained at Cincinnati Children’s Hospital Medical Center through informed consent under an approved institutional review board research protocol.
Mice.
Irf8eGFP 1, Gfi1Δex2−3/Δex2−3 2, Rosa26Fucci2 3, and Irf8tm1.2Hm/J 4 mice were maintained on a C57BL/6 background, and Ptprca (BoyJ) mice were purchased from Jackson Laboratories. Mice were maintained under specific pathogen-free conditions by Cincinnati Children’s Hospital Medical Center (CCHMC) Veterinary Services and all procedures were approved by the Children’s Hospital Research Foundation Institutional Animal Care and Use Committee (Protocol Number IACUC2017–0021).
To generate Gfi1N382S, Gfi1K403R, and Gfi1R412X mice, two pairs of ZFN RNAs that recognize target sites flanking Gfi1 coding exon 6 were designed and generated by Millipore-Sigma (Genome Editing division). mRNAs encoding site-specific ZFNs (50ng/μl) were introduced into 1-cell mouse oocytes by standard pronuclear injection approach, and injected oocytes were transferred to a pseudo pregnant surrogate mother. To introduce specific mutations, oocytes were co-injected with ZFN mRNAs as well as a homologous donor construct (2ng/ml) containing the R412X mutation. Positive founder pups were identified using mutation-specific primers and mated to C57BL/6 mice to generate stable heritable knock-in lines. Constructs consisted of 1.5kb 5’ and 0.8kb 3’ arms of homology flanking the mutant Gfi1 exon 6. The ZFN target sequences used were:
5’-GTCCCCTTCACCTTCCTTcccggaGCTGCTGGAGGAGATGAA-3’ and
5’-CGTTGCGACCCACATGCTCTtgctaaCAGCTGGCTAAG-3’, where the capital letters denote nucleotides actually bound by right and left ZFN proteins. Positive founders and subsequent progeny were genotyped by PCR using the primers: 5’-CAGAAAGCACACAGGCTTCA-3’ and 5’-GATGAGCTTTGCACACTGGA-3’ then subsequent restriction enzyme digestion using BamHI for Gfi1N382S, KpnI for Gfi1K403R, and EcoRI for Gfi1R412X. Expression of the mutant alleles was confirmed by TOPO cloning/TOP10 transformation (Thermo Fisher) of RT-PCR products generated using the same primers that were used for genotyping, followed by Sanger sequencing of individual clones with T3 and T7 primers and analysis using ApE (version 2.0.49.0).
Flow cytometry and cell sorting.
Mice were euthanized using carbon dioxide followed by cervical dislocation. Peripheral blood was collected into EDTA-coated tubes, and hind limb bones (femurs, tibias, and the iliac crest) were harvested immediately after euthanasia and stored in cold FACS buffer (1% FBS, 0.01% NaN3 in DPBS) under sterile conditions. Bones were flushed using a syringe for transplantation or crushed using a mortar and pestle for all other applications, then passed through a 40 μm cell strainer (Becton, Dickinson and Company) to obtain single cell suspensions for downstream applications. Prior to analytical flow cytometry, erythrocytes were lysed using ACK buffer (Gibco), then cells were washed in FACS Buffer and resuspended in FACS Buffer containing DAPI (Thermo Fisher) or 7-AAD (Becton, Dickinson, and Company). Flow cytometry analyses were conducted on a FACS LSRII or FACS LSRFortessa (Becton, Dickinson, and Company) and analyzed using FlowJo software (FlowJo, Ashland, OR, USA).
To enrich for murine stem/progenitor populations, freshly isolated bone marrow cells were incubated with CD117 Microbeads or with components of the Mouse Lineage Depletion kit and separated on an AutoMACS Pro separator (Miltenyi) according to manufacturer specifications. Alternatively, murine LSK cells were purified by first using the Sca-1 Multisort Kit (Miltenyi) for separation on an AutoMACS Pro separator then cells were subsequently incubated with CD117 Microbeads and separated again on an AutoMACS Pro separator. Murine bone marrow neutrophils were isolated using the EasySep™ Mouse Neutrophil Enrichment Kit (Stem Cell Technologies) according to manufacturer’s specifications. Human peripheral blood neutrophils were enriched from whole blood using the EasySep™ Direct Human Neutrophil Isolation Kit (Stemcell Technologies) according to manufacturer’s specifications.
Peripheral blood analysis: Peripheral blood cells were stained in FACS buffer with a mix of antibodies: CD16/CD32 (clone 2.4G2, Becton, Dickinson and Company), CD3-APC (clone 145–2C11, Becton, Dickinson and Company), CD45R-PE (clone RA3–6B2, Becton, Dickinson and Company), CD11b-PacificBlue (clone M1/70, BioLegend), and Ly6g-PerCP-Cy5.5 (clone 1A8, BioLegend). For analysis of transplant chimerism, cells were also stained with CD45.1-BV605 (clone A20, Biolegend) and CD45.2-AlexaFluor700 (clone 104, Biolegend). To quantify neonatal murine blood numbers, samples were supplemented with 5.24 μm AccuCount blank particles (Spherotech, Lake Forest, IL, USA) immediately prior to analysis. To purify peripheral blood neutrophils for cytospin analysis, cell sorting was performed on a FACSAria II (Becton, Dickenson, and Company) and cells were collected in a solution of DPBS + 50% FBS (Atlanta Biologicals).
Mature myeloid bone marrow analysis: Whole bone marrow cells were stained in FACS buffer with a mix of antibodies: CD16/CD32 (clone 2.4G2, Becton, Dickinson and Company), and Ly-6B.2-AlexaFluor647 (clone 7/4, Bio-Rad), F4–80-AlexaFluor488 (clone Cl:A3–1, Bio-Rad) for 30 minutes on ice.
Hematopoietic stem cell analysis: Whole bone marrow cells were stained for 30 minutes on ice with a cocktail of antibodies: Fc Block (Becton, Dickinson, and Company), biotin-conjugated anti-CD3e (clone 145–2C11, BioLegend), biotin-conjugated anti-CD4 (clone RM4–5, Thermo Fisher), biotin-conjugated anti-CD8 (clone 53–6.7), biotin-conjugated anti-CD11b (clone M1/70, Becton, Dickinson, and Company), biotin-conjugated anti-CD19 (clone 6.D5, BioLegend), biotin-conjugated anti-CD127 (clone B12–1, Becton, Dickinson, and Company), biotin-conjugated anti-B220 (clone RA3–6B2, BioLegend), biotin-conjugated anti-Gr1 (clone RB6–8C5, Becton, Dickinson, and Company), and biotin-conjugated anti-Ter119 (clone TER-119, BioLegend). Cells were then washed twice with FACS buffer and incubated on ice overnight with a cocktail of antibodies; streptavidin-APC-Cy7 (Becton, Dickinson, and Company), APC-conjugated anti-CD117 (clone 2B8, BioLegend), PE-Cy7-conjugated anti-Sca-1 (clone D7, Becton, Dickinson, and Company), FITC-conjugated anti-CD34 (clone RAM34, Becton, Dickinson, and Company), PacificBlue-conjugated anti-CD48 (clone HM48–1, BioLegend), PE-conjugated anti-CD135 (clone A2F10, BioLegend) and Brilliant Violet 510-conjugated anti-CD150 (clone TC15–12F12.2, BioLegend).
Myeloid progenitor analysis: Prior to FACS sorting, Miltenyi AutoMacs CD117-enriched bone marrow cells were stained for 1 hour at room temperature protected from light with a mix of antibodies: PerCP-eFluor710-conjugated anti-CD16/CD32 (clone 93, Thermo Fisher), biotin-conjugated anti-CD3e (clone 145–2C11, BioLegend), biotin-conjugated anti-CD4 (clone RM4–5, Thermo Fisher), biotin-conjugated anti-CD8 (clone 53–6.7), biotin-conjugated anti-CD11b (clone M1/70, Becton, Dickinson, and Company), biotin-conjugated anti-CD19 (clone 6.D5, BioLegend), biotin-conjugated anti-CD127 (clone B12–1, Becton, Dickinson, and Company), biotin-conjugated anti-B220 (clone RA3–6B2, BioLegend), biotin-conjugated anti-Gr1 (clone RB6–8C5, Becton, Dickinson, and Company), and biotin-conjugated anti-Ter119 (clone TER-119, BioLegend), Brilliant Violet 421-conjugated anti-CD34 (clone RAM34, Becton, Dickinson and Company), APC-conjugated anti-CD117 (clone 2B8, BioLegend), and PE-Cy7-conjugated anti-Sca-1 (clone D7, Becton, Dickinson, and Company). Cells were then washed twice with FACS buffer and incubated for 15 minutes at room temperature with streptavidin-APC-Cy7 (Becton, Dickinson, and Company). Cells were washed once in FACS Buffer, and resuspended in FACS Buffer for cell sorting. Cell sorting was performed on a FACSAria II (Becton, Dickenson, and Company) and cells were collected in a solution of DPBS + 50% FBS (Atlanta Biologicals).
Myeloid progenitor and precursor gate (GMP-P):
Prior to FACS sorting, CD117-enriched bone marrow cells were stained for 1 hour at room temperature protected from light with a mix of antibodies: PerCP-eFluor710-conjugated anti-CD16/CD32 (clone 93, Thermo Fisher), biotin-conjugated anti-CD3e (clone 145–2C11, BioLegend), biotin-conjugated anti-CD4 (clone RM4–5, Thermo Fisher), biotin-conjugated anti-CD8 (clone 53–6.7), biotin-conjugated anti-CD19 (clone 6.D5, BioLegend), biotin-conjugated anti-CD127 (clone B12–1, Becton, Dickinson, and Company), biotin-conjugated anti-B220 (clone RA3–6B2, BioLegend), biotin-conjugated anti-Ter119 (clone TER-119, BioLegend), AlexaFluor700-conjugated anti-CD11b (clone M1/70, BioLegend), Brilliant Violet 785-conjugated anti-Ly6c (clone HK1.4, BioLegend), FITC-conjugated anti-Ly6g (clone 1A8, BioLegend), Brilliant Violet 421-conjugated anti-CD34 (clone RAM34, Becton, Dickinson and Company), APC-conjugated anti-CD117 (clone 2B8, BioLegend), and Brilliant Violet 605-conjugated anti-CD115 (clone T38–320, Becton, Dickinson, and Company). Cells were then washed twice with FACS buffer and incubated for 15 minutes at room temperature with streptavidin-APC-Cy7 (Becton, Dickinson, and Company). Cells were washed once in FACS Buffer, and resuspended in FACS Buffer for cell sorting. Cell sorting was performed on a FACSAria II (Becton, Dickenson, and Company) and cells were collected in a solution of DPBS + 50% FBS (Atlanta Biologicals).
Alternate GMP-P gate (incorporating additional markers):
Alternatively, prior to analysis, CD117-enriched bone marrow cells were stained for 1 hour at room temperature protected from light with a mix of antibodies: Brilliant UV 395-conjugated anti-CD16/CD32 (clone 93, Becton, Dickenson, and Company), biotin-conjugated anti-CD3e (clone 145–2C11, BioLegend), biotin-conjugated anti-CD4 (clone RM4–5, Thermo Fisher), biotin-conjugated anti-CD8 (clone 53–6.7), biotin-conjugated anti-CD19 (clone 6.D5, BioLegend), biotin-conjugated anti-CD127 (clone B12–1, Becton, Dickinson, and Company), biotin-conjugated anti-B220 (clone RA3–6B2, BioLegend), biotin-conjugated anti-Ter119 (clone TER-119, BioLegend), PE-Cy7-conjugated anti-CD11b (clone M1/70, BioLegend), Brilliant Violet 785-conjugated anti-Ly6c (clone HK1.4, BioLegend), PerCP-Cy5.5-conjugated anti-Ly6g (clone 1A8, BioLegend), Brilliant Violet 421-conjugated anti-CD34 (clone RAM34, Becton, Dickinson and Company), Brilliant Violet 650-conjugated anti-CD117 (clone 2B8, BioLegend), and Brilliant Violet 605-conjugated anti-CD115 (clone T38–320, Becton, Dickinson, and Company). Additional analyses were performed by also individually staining cells with the following antibodies: PE-conjugated anti-CD18 (clone M18/2, BioLegend), PE-conjugated anti-CD27 (clone LG.3A10, BioLegend), PE-conjugated anti-CD55 (clone RIKO-3, BioLegend), PE-conjugated anti-CD84 (clone mCD84.7, BioLegend), Alexa Fluor 700-conjugated anti-CD177 (clone 1171A, R&D Systems, Minneapolis, MN, USA), PE-conjugated anti-CD106 (Vcam1, clone 429, BioLegend), PE-conjugated anti-CD217 (IL-17R, clone PAJ-17R, Thermo Fisher). Cells were then washed twice with FACS buffer and incubated for 15 minutes at room temperature with streptavidin-APC-Cy7 (Becton, Dickinson, and Company).
Cell Culture and viral transduction.
Lentiviral particles were generated as described previously5. Briefly, Lenti-X 293T cells (Clontech) were seeded one day prior to transfection with individual expression vectors (pLVX-EF1α-IRES-Puro base vector, Clontech), the lentiviral packaging plasmid Δ8.9, and envelope plasmid VSV-G using Transit-LT1 (Mirus) according to manufacturer instructions. Growth medium was changed the following day. Virus-containing supernatant was collected 48 and 72 hours post-transfection, pooled and concentrated via ultracentrifugation over a 20% sucrose cushion, then aliquoted and frozen at −80°C.
Freshly isolated murine LSK bone marrow cells were cultured in IMDM (ThermoFisher Scientific) supplemented with 100U/mL Penicillin-Streptomycin (ThermoFisher Scientific), 10% BIT (Stemcell Technologies), 20 ng/mL IL-3, 20 ng/mL IL-6, and 50 ng/mL SCF (Miltenyi) with lentiviral particles overnight at 37°C, 5% CO2. Media was changed and supplemented with 1.5 μg/mL puromycin (Invivogen, San Diego, CA, USA) 48 hours after transduction, and the Irf8-GFP levels were determined by FACS 72 hours later.
RNAscope Imaging.
Human CD34+ cells were isolated from either G-CSF mobilized peripheral blood or fresh bone marrow aspirate using CliniMACS® CD34+ (Miltenyi, Cat. No. 130–017-501) or FACS sorting. Cells were fixed with 4% neutral buffered formalin at 37°C for 1 hour and cyto-centrifuged onto SuperFrost Plus® Slides (Fisher Scientific, Cat. No. 12–550-15). Samples were then processed according to the RNAscope® V2 User Manual, (ACDBio). Channel 1 of the 3-plex Negative Control Probe (ACDBio, Cat. No. 320871), probing for the bacterial gene dabB, was developed to determine background fluorescence. Negligible fluorescent signal was generated by this probe in all samples. Channel 1 of experimental probes for POLR2A intron 1 (ACDBio, Cat. No. 588751), GFI1 intron 3 (ACDBio, Cat. No. 575371), and GFI1 intron 6 (ACDBio, Cat. No. 588761) was also developed. Samples were stained using the Opal™ 570 reagent (Perkin Elmer, Cat. No. FP1488001KT) at a dilution of 1:1500. Slides were imaged with a Nikon A1 confocal on an Eclipse TiE inverted microscope with a 60X Plan Apo IR DIC-Water Immersion objective. Z-stacks were acquired with a 0.11 μm pixel size and 0.25 μm Z interval. Z-stacked data were analyzed using Imaris 9 with transcripts per cell defined using “spot” detection. Transcripts per cell were quantified using Imaris XT MatLab algorithms. Cells with no transcripts were excluded from final analysis. Images shown are maximum intensity projections of Z-stacks.
GFI1-R412X iPSC generation and differentiation.
iPSC lines were generated as described previously6. CRISPR-Cas9 was used to introduce the R412X mutation to the GFI1 locus of an iPSC line (iPSC286C11) derived from a healthy individual. A guide RNA targeting the last intron of GFI1 (5’-GTGACTCCGTTCTAATTCAG-3’) was designed according to the web tool (http://CRISPOR.org) and cloned into a modified pX458 vector (Addgene #48138) that expresses an optimized sgRNA scaffold and a high-fidelity eSpCas9(1.1)-2A-GFP. The editing activity of the plasmid was validated in HEK293T cells by T7E1 assay. The donor plasmid was constructed by inserting the DNA fragments containing desired mutations and homologous arms to a pUC57 vector carrying a loxP-flanked EF1a-GFP-2A-Puro cassette6. For gene editing, a single cell suspension of iPSC286C11 cells was prepared using accutase and 0.75×106 cells were reverse-transfected in a 12-well matrigel-coated dish with 0.6 μg of the gRNA-containing pX458 vector, 0.6 μg of donor plasmid, and 0.18 μg of pRetroSuper-p53sh (a kind gift from Dr. Jim Mulloy) using the manufacturer’s recommended protocol (TransIT-LT1; Mirus). One day post-transfection, mTeSr1 containing 1 μg/mL puromycin was added to cells for 48 hours to eliminate non-transfected cells. Five days post-transfection, a single cell suspension of surviving cells was prepared using accutase. Cells were replated at 0.25×106 per well in a matrigel-coated 6-well dish and cultured for 4 days in mTeSR1 containing 10% CloneR (StemCell Technologies), using the manufacturer’s recommended protocol. Cells were subsequently fed daily with mTeSR1 containing 1 μg/mL puromycin for an additional 1–2 weeks before colonies with stereotypical iPSC morphology were manually excised and expanded for genotyping. Correctly targeted clones were identified by PCR, enzyme digestion and Sanger Sequencing and confirmed to have normal karyotype. To remove the loxP-flanked EF1a-GFP-2a-Puro cassette, cells were incubated with TAT-CRE recombinase according to manufacturer’s recommendations (Millipore Sigma). GFP− cells were subsequently purified by FACS sorting.
Granulocytes were generated from iPSC using a three-step differentiation process. First, CD34+/CD45+ hematopoietic progenitors were derived using the STEMdiff hematopoietic kit (Stem Cell Technologies, Vancouver, BC). Floating CD34+/CD45+ cells were harvested and cultured in RPMI 1640 medium (Gibco) containing 10% FBS (Hyclone), 50 ng/mL SCF, 10 ng/mL IL-3 and 10 ng/mL GM-CSF (Peprotech) and cultured for 5 days. After 5 days, culture medium was transitioned to RPMI 1640 medium supplemented with 10% FBS and 50 ng/ml G-CSF (Peprotech) and cultured for another 5 days. Cells were stained with PE-conjugated anti-CD45 (clone HI30, Becton, Dickenson, and Company), APC-conjugated anti-CD34 (clone 581, Becton, Dickenson, and Company), APC-Cy7-conjugated anti-CD11B (clone ICRF44, Becton, Dickenson, and Company), Vioblue-conjugated anti-CD14 (clone REA599, Miltenyi), and PE-Cy7-conjugated anti-CD16 (clone REA423, Miltenyi) prior to FACS analysis. To analyze GFI1 expression, cells were lysed in TriZol (Thermo Fisher) and total RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Amplification of cDNA was performed using GoTaq Green Master Mix (Promega) using 5’-CAGGAACGGAGCTTTGACTG-3’ and 5’-GAAGGAGGAGCAACCTGGTA-3’ primers prior to sequencing using 5’-CAAGAGGTCATCCACACTGTC-3’ or 5’- TCTGGAAAGTCAGAAGGGAGT-3’ primers.
Immunoblot analysis.
Freshly isolated LineageNeg bone marrow cells were lysed in laemmli buffer, briefly sonicated, and boiled at 100°C for 5 minutes before being subjected to SDS-PAGE. The resolved proteins were transferred onto nitrocellulose membranes (GE Healthcare) that were subsequently blocked in 5% nonfat dried milk (Carnation) in TBS-T (0.1% Tween 20, Sigma Aldrich) for 1 hour at room temperature with agitation. Following washing with TBS-T, membranes were incubated overnight at 4°C with agitation in a solution of 5% BSA Fraction V/TBS-T (Fisher Scientific) containing goat anti-Gfi1 (R&D Systems) antibody diluted at 1:1,000. Following a wash with TBS-T, membranes were incubated for 2 hours at room temperature with agitation in a solution of 2% nonfat dried milk in TBS-T containing a 1:5,000 dilution of HRP-conjugated anti-goat secondary antibody (Invitrogen). Following three washes with TBS-T, chemiluminescent detection of blotted proteins was performed using ECL (Thermo Fisher) and detected using X-ray film (Lab Scientific). Next, membranes were washed thoroughly in TBS-T, then stripped in Restore Plus (Thermo Fisher) for 10 minutes at room temperature with agitation. After washing in TBS-T, membranes were blocked as before, and incubated as described above with a 1:5,000 dilution of mouse anti-β-Actin (Sigma Aldrich) and then a 1:10,000 dilution of HRP-conjugated anti-mouse secondary antibody (GE Healthcare) before chemiluminescent detection was achieved as before.
Cytospin.
Freshly prepared cells were diluted in 200 μL of FACS buffer and spun onto VWR VistaVision Histobond slides (VWR) for 3 minutes at 900 rpm using a Cytospin 4 apparatus (Thermo Scientific). Slides were then fixed and stained using the Camco stain pak (Cambridge Diagnostic Products, Inc), and once dry, sealed using Cytoseal 60 (Thermo Scientific) and microscope cover glass (Globe Scientific Inc.).
Colony forming unit assays.
Freshly isolated LineageNeg or c-Kit+ cells were plated at 1,000 cells/mL in M3434 (Stemcell Technologies) and the differential colony number was determined 7 days later.
Bone marrow transplantation.
A single cell suspension of 1×106 unfractionated bone marrow cells from 6-week old mice (CD45.2) was injected into lethally irradiated (11.75 Gy) CD45.1 congenic recipients.
Code availability.
All described code is provided in Github (https://github.com/nsalomonis/altanalyze).
Data availability.
scRNA-Seq, CITE-Seq, scATAC-Seq, and ChIP-Seq data is deposited as GEO SuperSeries GSE120409 and in Synapse (https://www.synapse.org/#!Synapse:syn16806696).
Proteomic data are available via ProteomeXchange (www.ebi.ac.uk/pride/archive/) with identifier PXD010943. Extensive processed and primary data are provided and organized in Synapse, including 10x Genomics count matrices, Fluidigm expression, single-cell populations, quality control metrics, 10x Genomics Cell Ranger outputs (summary report, loupe browser files), genomic coordinate peak files and differential expression results (cellHarmony).
Extended Data
Extended Data Figure 1. Functional assessment of SCN patient-derived GFI1 variants and generation of Gfi1 ZnF-mutant mice.
a, Representative FACS plots of lentiviral transduced LSK cells isolated from adult Irf8-eGFP transgenic mice with the %Irf8-eGFPhigh indicated. b, Graphical summary of FACS analysis of lentiviral transduced LSK cells isolated from adult Irf8-eGFP transgenic mice (top) with locations of the variants mapped to the GFI1 protein (bottom). EV; empty vector, *P2A; mutation not found in patients, italics; other variants detected in the same patient, bold; also found in patients diagnosed with a malignancy, ATG; start codon, TGA; stop codon, gray blocks; characterized protein domains, SNAG; Snail/Gfi1 family domain, SUMO; sumoylation domain, ZnF; zinc-finger. c, Schematic of the Gfi1 locus annotated with relevant features. Line with small arrows; intronic regions, numbered blocks; exons, black blocks; coding regions, gray blocks; noncoding regions, ATG; start codon, TGA; stop codon, SNAG; Snail/Gfi1 family domain-encoding region, ZFN; zinc-finger nucleases, large arrows; noncoding region used for genotyping. d-f, Schematic of the nucleotide changes made to the coding region and 3’ UTR to introduce the d, d’ N382S, e, e’ K403R, and f, f’ R412X mutations. g, Representative genotyping of the ZnF-mutant mice with or without restriction enzyme digestion. h, Representation of Sanger sequencing analysis of cDNA from adult whole bone marrow. Targeted wild-type nucleotides are underlined, mutated nucleotides are in bold, and red nucleotides indicate the location of the stop codon. i, Immunoblot analysis of two R412X founder lines using adult murine LineageNeg bone marrow lysates. j, Graphical summary of FACS analysis of neonatal murine peripheral blood. k, Total cell counts per mL of neonatal peripheral blood as determined by FACS. l, Representative RNAscope images (left) and transcript quantitation (right) of the indicated transcripts in primary human CD34+ cells. The number of cells scored (left) and the number of donors tested (right) is indicated. m, Representative elecropherogram plots from Sanger sequencing of GFI1 RT-PCR products derived from iPSC. The arrow indicates the nucleotide substitution made to introduce the R412X mutation. n, Representative FACS plots of iPSC cells at the end of the 10-day hematopoietic differentiation protocol. o, Representative cytospin images of iPSC-derived neutrophils. Data in a, b, and l are representative of three biological replicates, data in j and k are representative of individual biological replicates, and is plotted as mean in a or as mean ± s.e.m in b, j, k, and l. * p<0.05, ** p<0.01, *** p < 0.001, **** p < 0.0001 as determined by two-tailed t-test. Data in i displays two biological replicates for each founder line. Data in m-o are representative of two independent experiments. Scale bars in l represent 2 μm and 10 μm in o.
Extended Data Figure 2. Characterization of Gfi1 ZnF-mutant mice at steady-state.
a, Graphical representation of FACS analysis of adult murine peripheral blood. b, FACS plots and graphical representation of FACS analysis of adult murine peripheral blood. c, Graphical summary of colony-forming-unit (CFU) assays performed on adult LineageNeg bone marrow cells from two different founder lines. G; granulocyte, M; monocyte, GM; granulocyte-monocyte, BFU-E; burst-forming-unit erythroid, GEMM; granulocyte, erythrocyte, monocyte, megakaryocyte. d, Total cell counts of adult murine whole bone marrow harvested from two femurs and tibias per mouse. e, FACS plots and quantitation of adult murine bone marrow populations. f, Representative cytospins of adult murine whole bone marrow. Data in f are representative of three biological replicates, and the scale bar represents 10 μm. Data in a-e are displayed as mean ± s.e.m. * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001 as determined by two-tailed t-test.
Extended Data Figure 3. Inflammatory-cytokine-independent emergency granulopoiesis and high G-CSF levels in Gfi1 ZnF-mutant mice at steady-state and functional analysis of Gfi1 ZnF-mutants.
a, FACS plots and quantitation of adult murine LineageNeg, c-Kit+, Sca-1+ bone marrow cells. b, Quantitation of adult peripheral blood cytokine levels at steady-state by cytokine array. c, Quantitation of individual adult peripheral blood cytokine levels at steady-state by Luminex analysis. d, Survival analysis of lethally irradiated BoyJ recipients of adult whole bone marrow from adult Gfi1-mutant donors. e, Graphical representation of FACS analysis of peripheral blood chimerism in transplant recipients from d at 4 months post-transplant. f-g, Graphical representations of the total number of colonies in murine organs harvested at the indicated time points (see arrowheads in Fig. 1g, h) after infection with f, an LD50 dose of C. albicans or g, 5 × 107 CFU of S. aureus. h, Immunoblot analysis of adult murine LineageNeg bone marrow lysates. Data in a-c and e-g are displayed as mean ± s.e.m. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 as determined by two-tailed t-test in a-c and e-g. ** p < 0.01 as determined by two-sided Mantel-Cox test in d. Data in h are representative of 3 independent experiments.
Extended Data Figure 4. FACS and Fluidigm-based analysis of steady-state terminal granulopoiesis.
a, FACS plots demonstrating the gating strategy used for sorting. The populations sorted for scRNA-Seq are indicated in shades of grey with accompanying cytospins. b, Violin plots of the gene and read-level metrics for each of the indicated libraries. Dashed lines indicate mean, lower, and upper quartiles. Sample size (n = # cells) displayed (top). c, Heatmap of gene expression defined by ICGS (Fluidigm C1, excluding cell-cycle genes) in scRNA-Seq data (n = 516 cells). Each column represents a single cell and each row represents a single gene. ICGS clusters are annotated (top). HSCP-1, haematopoietic stem cell progenitor; NK, natural killer T-cell progenitor; HSCP-2; Meg, megakaryocytic; Eryth, erythrocytic; DC, dentritic cell; MDP, monocyte dendritic cell progenitor; Multi-Lin*, multi-lineage primed; cMoP, common monocyte progenitor; Mono, monocytic; MP, monocyte-committed progenitors; proNeu-1, neutrophil progenitor, proNeu-2; preNeu-1, neutrophil precursor, preNeu-2, preNeu-3; immNeu, immature neutrophil. d, Joint UMAP plot of scRNA-Seq data from c separated according to archival (29) and new Fludigm captures. e, Bar chart of the heatmap in c displaying the incidence and amplitude of selected genes. f, Heatmap of correlation between gene expression and each displayed cluster as generated by MarkerFinder (AltAnalyze software). g, FACS plots comparing expression of Ly6g with the indicated surface marker. FMO; fluorescence minus one control. h, Heatmap of cell cycle gene expression in ICGS-defined clusters in scRNA-Seq data (n = 509 cells). Each column represents a single cell and each row represents a single gene. Gene expression clusters were generated in AltAnalyze and the ICGS clusters are annotated (top). FACS gates are annotated (bottom). LSK, LineageNeg c-Kit+ Sca-1+; CMP, common myeloid progenitor; GMP, granulocyte monocyte progenitor; LK CD34+, LineageNeg c-Kit+ Sca-1Neg CD34+; GMP-P CD11b+, granulocyte monocyte progenitor and precursor, GMP-P CD11b+ Ly6glow, GMP-P CD11b+ Ly6ghigh. Key genes are indicated (right). i, Scatter plot representation of scRNA-Seq data from h comparing the gene expression of G1 to S phase transition genes with G2 to M phase transition genes in each cell. Each point represents a single cell. j, Bar chart of the heatmap in h, displaying the incidence and amplitude of selected genes. k, Heatmap of correlation between gene expression and each displayed cluster as generated by MarkerFinder. l, Heatmap of enrichment for Gene Ontology biological processes enriched in the granulocytic clusters from the Fluidigm scRNA-Seq data from c with key processes indicated (right). m, Scatter plot representation of scRNA-Seq data from c where each point represents a single cell. Reads per cell indicate RSEM transcript aligned read counts for each cell library. Genes expressed per cell indicate the number of genes with a TPM>1 for each single-cell library. Data in a and g are representative of three biological replicates. Data in f and k display Pearson correlation values.
Extended Data Figure 5. CITE-Seq analysis of steady-state terminal granulopoiesis.
a, FACS plots demonstrating the gating strategy used for sorting. The population sorted for scRNA-Seq is indicated in red. b, Heatmap of gene expression for cellHarmony assigned cell populations from CITE-Seq 10x Genomics captures (male and female mice, n = 11,132 cells) compared to ICGS-defined clusters from the Fluidigm scRNA-Seq data. De novo marker genes (MarkerFinder) for each assigned cluster from the 10x Genomics data are shown (top). Each column represents a single cell and each row represents a single gene. HSCP-1, haematopoietic stem cell progenitor; NK, natural killer T-cell progenitor; HSCP-2; Meg, megakaryocytic; Eryth, erythrocytic; DC, dentritic cell; MDP, monocyte dendritic cell progenitor; Multi-Lin*, multi-lineage primed; IG2, Irf8-expressing GMP subpopulation 2; cMoP, common monocyte progenitor; Mono, monocytic; MP, monocyte-committed progenitors; proNeu-1, neutrophil progenitor, proNeu-2; preNeu-1, neutrophil precursor, preNeu-2, preNeu-3; immNeu, immature neutrophil. The gender of the host mouse of each cellular barcode (bottom) and example MarkerFinder genes in common between Fluidigm and 10x Genomics data are indicated (right). c, Violin plots of the gene and read-level metrics for each of the indicated libraries. Dashed lines indicate mean, lower, and upper quartiles. Sample size (n = # cells) displayed (top). d, Lineage-priming scores for monocytic and granulocytic specification. Scatter plot displaying assigned scores for cellHarmony-assigned neutrophil progenitors (proNeu-2), monocytic progenitors (Mono), bi-potential monocytic-granuloctyic intermediates (IG2) and megakaryocyte progenitors (Meg) from b (see Online Methods). Each point represents a single cell. e, Heatmap of row normalized ADT UMI counts (log2, median substracted) for each corresponding cell from b. f, Heatmap of cell cycle gene expression displaying the same cells as b and the same genes as Extended Data Fig. 4h. g, Scatter plot representation of scRNA-Seq data from f comparing the gene expression of G1 to S phase transition genes with G2 to M phase transition genes in each cell. Each point represents a single cell. h, Bar chart of the heatmap in b, displaying the incidence and amplitude of selected genes. i, Heatmap of correlation between gene expression and each displayed cluster as generated by the MarkerFinder feature of AltAnalyze. j-k, Plots of CITE-Seq ADT UMIs where grey indicates all captured cells and j, red indicates cells classified by ICGS clusters or k, the top 1% of UMIs expressing the indicated gene. l, UMAP of 15,968 cell-barcodes colored according to cellHarmony assigned cell populations (10x genomics reference) where each dot represents a single cell. Minor contaminant populations (<30 cells per cluster) were excluded. m, Heatmap of row normalized CLR-transformed ADT counts of the indicated cell surface proteins (left) displayed as an average of all cells in the indicated clusters (top). n, Correlation plots between ADT UMI counts and the expression of genes (cellular barcode normalized UMI counts) encoding the corresponding proteins detected via CITE-Seq. Each dot represents a single cell. Linear trend lines for all cells with RNA expression >0 are indicated by dotted red lines with corresponding coefficients of determination displayed. Data in i displays Pearson correlation values.
Extended Data Figure 6. Fluidigm-based transcriptional analysis of Gfi1R412X/- cells.
a, FACS plots of adult murine bone marrow (left) and representative cytospin of FACS-sorted adult murine bone marrow (right). The populations sorted for scRNA-Seq are indicated in red. b, Schematic summary of the cellHarmony algorithm. c, cellHarmony-assigned ICGS states of combined wild-type and Gfi1R412X/- Fluidigm scRNA-Seq data (n = 624 cells, excluding cell-cycle genes). Each tick mark represents data from a single cell. Gene expression clusters were generated in AltAnalyze and the ICGS clusters are annotated (top) (see Extended Data Fig. 4c) HSCP-1, haematopoietic stem cell progenitor; NK, natural killer T-cell progenitor; HSCP-2; Meg, megakaryocytic; Eryth, erythrocytic; DC, dentritic cell; MDP, monocyte dendritic cell progenitor; Multi-Lin*, multi-lineage primed; cMoP, common monocyte progenitor; Mono, monocytic; MP, monocyte-committed progenitors; proNeu-1, neutrophil progenitor, proNeu-2; preNeu-1, neutrophil precursor, preNeu-2, preNeu-3; immNeu, immature neutrophil. FACS gates are annotated (right). GMP, granulocyte monocyte progenitor; GMP-P CD11b+, granulocyte monocyte progenitor and precursor. d, Population distribution of Fluidigm scRNA-Seq data from Extended Data Fig. 6c and Extended Data Fig. 4c. e, cellHarmony heatmap of wild-type aligned Gfi1R412X/- cell gene expression as ordered in c (dotted lines). Each column represents a single cell and each row represents a single gene from Extended Data Fig. 4c. The FACS gates and ICGS clusters are annotated (top) and gene clusters are indicated (left). f, Bar chart of the heatmap in e, displaying the incidence and amplitude for selected genes. Arrows indicate differential expression of the adjacent gene, as compared to wild-type. g, Heatmap of MarkerFinder cell population-specific genes expression from the Fluidigm scRNA-Seq data (n = 191 cells) with enriched pathway associated genes (right) and statistically enriched Gene Ontology biological processes (left). h-i, Heatmap of h, wild-type-driven (same genes as in g) or i, de novo MarkerFinder genes for cellHarmony classified Gfi1R412X/- cell populations (n = 62 cells) annotated with enriched pathway associated genes (right) and statistically enriched Gene Ontology biological processes (left). j, Representation of the scRNA-Seq data from k displaying the fold change in gene expression in Gfi1R412X/- compared to wild-type cells in the indicated clusters with the number of genes up- or down-regulated displayed. Each point represents a single gene. k, Heatmap of Fluidigm differentially expressed Gfi1-target genes (excluding cell-cycle genes) where each column represents a single cell and each row represents a single gene, with key genes (right) and enriched biological processes (left) indicated. Cytospin data in a are representative of two biological replicates. Data in a are displayed as mean ± s.e.m. **** p < 0.0001 as determined by two-tailed t-test in a.
Extended Data Figure 7. Population analysis of 10x Genomics-based scRNA-Seq data and label transfer from CITE-Seq transcriptome to scATAC-Seq cells.
a, Identification of additional cell populations from wild-type scRNA-Seq data (ICGS unsupervised analysis, male-female CITE-Seq datasets). UMAP projections of scRNA-Seq data where the cell barcodes within the outlined region (top) were analyzed for additional heterogeneity through a second ICGS analysis (bottom). b, Heatmaps of additional CITE-Seq captures with cell assignments from cellHarmony. cellHarmony classifications were derived using the refined cluster annotation assignments from Extended Data Fig. 6c and 7a. Each panel displays a scRNA-Seq heatmap of MarkerFinder genes from the cellHarmony reference (top), where each column represents a single cell and each row represents a single gene. Those cell barcodes captured from an independent FACS sort of Ly6ghigh/CD11bhigh GMP-P cells are indicated by a black bar (middle). Relative expression of median normalized ADTs (right) are shown in the bottom heatmaps. c, Assigned cell-population frequencies for Gfi1+/− and Gfi1R412X/- CITE-Seq (Modified GMP gate) cells datasets from cellHarmony. d, Heatmaps of differential ADT expression compared to Gfi1+/− for the indicated markers (right). e, tSNE plot of CITE-Seq transcriptome for the Gfi1+/+ sample. f, tSNE plot of scATAC-Seq for the Gfi1+/+ sample. g, tSNE plot of CITE-Seq transcriptome for the Gfi1R412X/R412X sample. h, tSNE plot of scATAC-Seq for the Gfi1R412X/R412X sample. i, Comparison of Seurat transferred CITE-Seq labels to unsupervised scATAC-Seq cell population prediction methods (SNAP-ATAC) for Gfi1+/+. Percentage of overlapping cells for all pairwise comparisons between SNAP-ATAC clusters to CITE-Seq clusters derived from Seurat label transfer. j, Comparison of Seurat transferred CITE-Seq labels to unsupervised scATAC-Seq cell population prediction methods (SNAP-ATAC) for Gfi1R412X/R412X. Percentage of overlapping cells for all pairwise comparisons between SNAP-ATAC clusters to CITE-Seq clusters derived from Seurat label transfer. k, Cell-cluster combined scATAC-Seq marker peaks associated with Gfi1+/+ CITE-Seq annotated cell populations (Seurat 3 label transfer using cicero gene activity scores). Each row is a cluster (left) and each column is a locus within 50 kb of a CITE-Seq marker gene for each cluster (bottom), where the colored bars represent the normalized read count coverage. l, Heatmaps of transcription factor (TF)-motif-enrichment probabilities (-log10) in accessible regions of Gfi1+/+ (blue) or Gfi1R412X/ R412X (red) cell populations (indicated left). Each row represents a TF motif from the Cisbp2 database (indicated right). Each dot in a and e-h represents a single cell barcode that is pseudo-colored for its ICGS predicted cluster.
Extended Data Figure 8. In vitro DNA binding analysis and in vivo ChIP-Seq and scATAC-Seq analysis of Gfi1-R412X.
a-e, EMSA using nuclear extracts of 293T cells transfected with a GFI1, GFI1-R412X, or empty (EV) expression vector. Arrows indicate bound (shift) and free probe. All data represent one experiment. a-c, A probe containing a high affinity GFI1 binding site (R21) was incubated with a, a titration of nuclear extracts, b-c, a titration of cold competitors with (R21) or without (MutR21) a high affinity GFI1 binding site. d-e, Nuclear extracts were incubated with probes containing a GFI1 binding site taken from the d, Mmp8 or e, Cbx3 locus. These loci contain a GFI1 ChIP-Seq peak and the corresponding genes are differentially expressed in Gfi1R412X/- Fluidigm scRNA-Seq data. f, Heatmap of ChIP-Seq read coverage at Gfi1-specific peaks where each row represents one peak that was called in the wild-type and each column represents a DNA-base position of the peak centered on each Gfi1 called peak in the indicated ChIP samples. The cluster color (left) indicates loci bound by Gfi1 alone (white) or by Gfi1 and Gfi1-R412X (black). g-l, Schematics of the indicated genomic loci displaying scATAC-Seq pseudobulk accessibility in the color-coded clusters (top) or ChIP-Seq reads and called peaks (bottom), with differentially accessible regions bound by Gfi1 shaded.
Extended Data Figure 9. Fluidigm-based transcriptional analysis of Gfi1R412X/- genetic rescues.
a, FACS plots and b, representative cytospins of adult murine bone marrow. c-d Graphical summary of colony-forming-unit (CFU) assays performed on c-Kit+ bone marrow cells. G; granulocyte, M; monocyte, GM; granulocyte-monocyte, BFU-E; burst-forming-unit erythroid, GEMM; granulocyte, erythrocyte, monocyte, megakaryocyte. e, FACS plots of murine bone marrow cells isolated from Irf8-eGFP transgenic mice with the mean %Irf8-GFPhigh indicated. f, 3D FACS plots of bone marrow isolated from adult Irf8-eGFP mice. Events are pseudocolored for CD11b expression and arrows indicate changes in the incidence of granulocytic populations. g, FACS plots and representative cytospins of adult murine bone marrow populations sorted for scRNA-Seq. h-i, cellHarmony-assigned Gfi1R412X/R412X and Gfi1R412X/-Irf8+/− cells to wild-type reference Fluidigm scRNA-Seq populations (h, n = 86 cells, i, n = 88 cells). Each tick mark represents a single cell library. ICGS clusters are annotated (top). HSCP-1, haematopoietic stem cell progenitor; NK, natural killer T-cell progenitor; HSCP-2; Meg, megakaryocytic; Eryth, erythrocytic; DC, dentritic cell; MDP, monocyte dendritic cell progenitor; Multi-Lin*, multi-lineage primed; cMoP, common monocyte progenitor; Mono, monocytic; MP, monocyte-committed progenitors; proNeu-1, neutrophil progenitor, proNeu-2; preNeu-1, neutrophil precursor, preNeu-2, preNeu-3; immNeu, immature neutrophil. FACS gates are annotated (right). GMP, granulocyte monocyte progenitor; GMP-P CD11b+; granulocyte monocyte progenitor and precursor. j, Population distribution of Fluidigm scRNA-Seq data from h, i, Extended Data Fig. 4c, and Extended Data Fig. 6c. j-k, Fluidigm scRNA-Seq heatmaps (left) of genes that are genetically repaired in each cluster, with key genes indicated. Adjacent plot (right) of enriched biological processes in the indicated clusters. Scale bars in b and g represent 10 μm. Cytospin data in b are representative of three biological replicates. Data in a, c, and d are displayed as mean ± s.e.m or mean in e. **** p < 0.0001 as determined by two-tailed t-test in a, c, and d.
Extended Data Figure 10. G-CSF pushed SCN-patient neutrophils are morphologically normal but functionally defective.
Representative cytospins of purified human peripheral blood neutrophils. The data is representative of one experiment per donor. Scale bars in represent 10 μm.
Supplementary Material
Acknowledgements
We thank Harinder Singh for Gfi1 antisera, Herbert C. Morse for supplying Irf8-GFP mice, members of the Cincinnati Children’s Hospital Medical Center (CCHMC) DNA Sequencing and Genotyping Core and the Research Flow Cytometry Core (supported in part by NIH AR-47363, NIH DK78392 and NIH DK90971) especially Alyssa Sproles for assistance with cytokine analyses. We thank Shawn Smith, Hung Chi Liang, and Kelly Rangel in the CCHMC Gene Expression Core for generating scRNA-Seq libraries. We thank Tess Newkold for excellent technical assistance. We thank Harinder Singh, Jose Cancelas, Scott Kogan, Raphael Kopan, Jim Wells and Tanja Gruber for helpful questions and discussions. This work was partly funded by support from NIH DP1AI131080 (SS.W.) T32 ES007250 (to D.E.M), S10RR027015 (to K.D.G.), and R01HL122661 (H.L.G.).
Footnotes
The authors declare the following competing interests: DEM is currently employed by Eli Lilly and Company and KLN is employed by BioLegend Inc. The remaining authors declare no competing interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Reprints and permissions information is available at www.nature.com/reprints.
Supplementary Methods. Please see the Supplementary Information for additional methods descriptions.
References
- 1.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14, 865–868, doi: 10.1038/nmeth.4380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert SF Developmental Biology, 6th edition. (Sinauer Associates, 2000). [Google Scholar]
- 3.An interactive online viewer of murine hematopoietic gene expression, <http://www.altanalyze.org/ICGS/Neutrophil/Viewer.php> (
- 4.DePasquale EAK et al. cellHarmony: cell-level matching and holistic comparison of single-cell transcriptomes. Nucleic Acids Res, doi: 10.1093/nar/gkz789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821, doi: 10.1016/j.cell.2019.05.031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia J et al. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS and G6PC3 in patients with severe congenital neutropenia. Br J Haematol 147, 535–542, doi: 10.1111/j.1365-2141.2009.07888.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Person RE et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet 34, 308–312, doi: 10.1038/ng1170 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H et al. A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J Immunol 193, 1766–1777, doi: 10.4049/jimmunol.1301939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshmukh HS et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 20, 524–530, doi: 10.1038/nm.3542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luedi PP et al. Computational and experimental identification of novel human imprinted genes. Genome Res 17, 1723–1730, doi: 10.1101/gr.6584707 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonthuis PJ et al. Noncanonical Genomic Imprinting Effects in Offspring. Cell Rep 12, 979–991, doi: 10.1016/j.celrep.2015.07.017 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Hock H et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18, 109–120 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Belyaev NN et al. Induction of an IL7-R(+)c-Kit(hi) myelolymphoid progenitor critically dependent on IFN-gamma signaling during acute malaria. Nat Immunol 11, 477–485, doi: 10.1038/ni.1869 (2010). [DOI] [PubMed] [Google Scholar]
- 14.MacNamara KC et al. Infection-induced myelopoiesis during intracellular bacterial infection is critically dependent upon IFN-gamma signaling. J Immunol 186, 1032–1043, doi: 10.4049/jimmunol.1001893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mempel K, Pietsch T, Menzel T, Zeidler C & Welte K Increased serum levels of granulocyte colony-stimulating factor in patients with severe congenital neutropenia. Blood 77, 1919–1922 (1991). [PubMed] [Google Scholar]
- 16.Hock H et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431, 1002–1007, doi: 10.1038/nature02994 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Karsunky H et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet 30, 295–300, doi: 10.1038/ng831 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Zeng H, Yucel R, Kosan C, Klein-Hitpass L & Moroy T Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. Embo j 23, 4116–4125, doi: 10.1038/sj.emboj.7600419 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe T et al. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development (Cambridge, England) 140, 237–246, doi: 10.1242/dev.084111 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Olsson A et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702, doi: 10.1038/nature19348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanez A et al. Granulocyte-Monocyte Progenitors and Monocyte-Dendritic Cell Progenitors Independently Produce Functionally Distinct Monocytes. Immunity 47, 890–902.e894, doi: 10.1016/j.immuni.2017.10.021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng LG, Ostuni R & Hidalgo A Heterogeneity of neutrophils. Nat Rev Immunol 19, 255–265, doi: 10.1038/s41577-019-0141-8 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Farrell JA et al. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science, doi: 10.1126/science.aar3131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu YP et al. Identification of an Early Unipotent Neutrophil Progenitor with Pro-tumoral Activity in Mouse and Human Bone Marrow. Cell Rep 24, 2329–2341.e2328, doi: 10.1016/j.celrep.2018.07.097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drissen R et al. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat Immunol 17, 666–676, doi: 10.1038/ni.3412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim MH et al. A late-lineage murine neutrophil precursor population exhibits dynamic changes during demand-adapted granulopoiesis. Sci Rep 7, 39804, doi: 10.1038/srep39804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evrard M et al. Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity 48, 364–379.e368, doi: 10.1016/j.immuni.2018.02.002 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Fang R et al. Fast and Accurate Clustering of Single Cell Epigenomes Reveals Cis-Regulatory Elements in Rare Cell Types. bioRxiv, 615179, doi: 10.1101/615179 (2019). [DOI] [Google Scholar]
- 29.Zweidler-Mckay PA, Grimes HL, Flubacher MM & Tsichlis PN Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol 16, 4024–4034, doi: 10.1128/mcb.16.8.4024 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillet LC et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 11, O111.016717, doi: 10.1074/mcp.O111.016717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elsner J, Roesler J, Emmendorffer A, Lohmann-Matthes ML & Welte K Abnormal regulation in the signal transduction in neutrophils from patients with severe congenital neutropenia: relation of impaired mobilization of cytosolic free calcium to altered chemotaxis, superoxide anion generation and F-actin content. Experimental hematology 21, 38–46 (1993). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-Seq, CITE-Seq, scATAC-Seq, and ChIP-Seq data is deposited as GEO SuperSeries GSE120409 and in Synapse (https://www.synapse.org/#!Synapse:syn16806696).
Proteomic data are available via ProteomeXchange (www.ebi.ac.uk/pride/archive/) with identifier PXD010943. Extensive processed and primary data are provided and organized in Synapse, including 10x Genomics count matrices, Fluidigm expression, single-cell populations, quality control metrics, 10x Genomics Cell Ranger outputs (summary report, loupe browser files), genomic coordinate peak files and differential expression results (cellHarmony).