Abstract
Viruses and transposable elements are major drivers of evolution and make up over half the sequences in the human genome. In some cases, these elements are co-opted to perform biological functions for the host. Recent studies made the surprising observation that the neuronal gene Arc forms virus-like protein capsids that can transfer RNA between neurons to mediate a novel intercellular communication pathway. Phylogenetic analyses showed that mammalian Arc is derived from an ancient retrotransposon of the Ty3/gypsy family and contains homology to the retroviral Gag polyproteins. The Drosophila Arc homologues, which are independently derived from the same family of retrotransposons, also mediate cell-to-cell signaling of RNA at the neuromuscular junction; a striking example of convergent evolution. Here we propose an Arc “life cycle”, based on what is known about retroviral Gag, and discuss how elucidating these biological processes may lead to novel insights into brain plasticity and memory.
Keywords: Endogenous retrovirus, Arc, extracellular vesicle, synaptic plasticity, neuron, neurodegeneration, intercellular communication, RNA, retrotransposon
Retroviral Origins of Synaptic Plasticity and Memory
Brains encode and store information from the outside world but how these processes evolved remains unclear. Sequencing of the human genome revealed that as much as 50% of the non-protein coding sequences have viral or transposable element (TE) (see glossary) origins [1]. In some cases, these random sequence insertions led to the generation of new genes with important functions [2, 3]. While characterizing the biochemistry of the mammalian immediate early gene Arc, a key mediator of synaptic plasticity and cognition [4], we unexpectedly found that Arc protein forms virus-like protein capsids (Figure 1A,B) that can transfer RNA between neurons in a previously unknown intercellular communication pathway mediated by specialized extracellular vesicles (EVs) [5]. Phylogenetic analyses by a number of research groups showed that Arc is derived from an ancient retrotransposon of the Ty3/gypsy family (Figure 1C) [5–7]. The Gag polyprotein of HIV and other retroviruses are also related to Ty3/gypsy retrotransposons and share structural homology with Arc [6, 8, 9]. Moreover, a number of genes in animals are predicted to have similar homology to Gag [8] and may have retained similar biology as Arc, suggesting that virus-like intercellular communication may be used in a variety of cell types and physiological processes. These novel cell-to-cell signaling pathways may reveal new mechanisms of information transfer between cells, but much of the cell biology of these processes is unknown.
Figure 1. Rat and Drosophila Arc proteins form virus-like capsids via a conserved retroviral Gag capsid region.
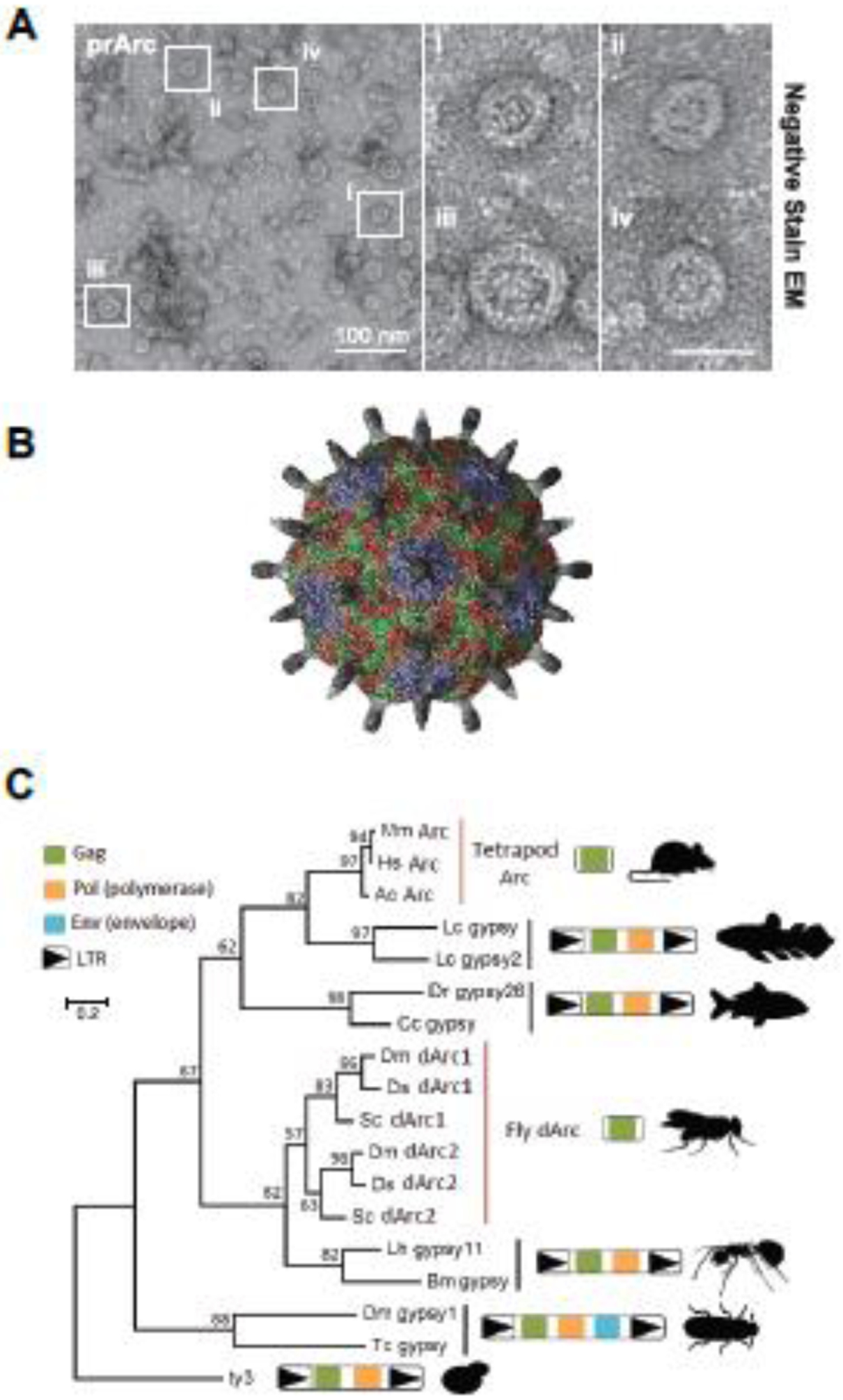
A. Representative negative stain EM images of purified rat Arc protein (prArc). Inset shows magnified Arc capsids (from [5]). B. Atomic resolution model of a dArc1 capsid derived from cryo-EM data as reported in [17] (image courtesy of Simon Erlendsson). C. Phylogeny based on an amino acid alignment of tetrapod Arc, fly dArc1, and Gag sequences from related Ty3/gypsy retrotransposons. In lineages without Arc genes, the most closely related sequences to Arc are Gag-pol polyproteins flanked by long terminal repeats (LTRs) as expected in bona fide Ty3/gypsy retrotransposons (from [5]).
In the brain, Arc intercellular signaling may mediate a non-canonical pathway required for information encoding and storage. We propose that aspects of long-term synaptic plasticity and memory may share similarities with the cell biological processes used by retroviruses. Here, we discuss the “life cycle” of Arc (Figure 2), comparing it to retrovirus virion assembly and trafficking. We speculate how neurons may use Arc’s virus-like properties to regulate memory.
Figure 2. Proposed model of the Arc “life cycle”.
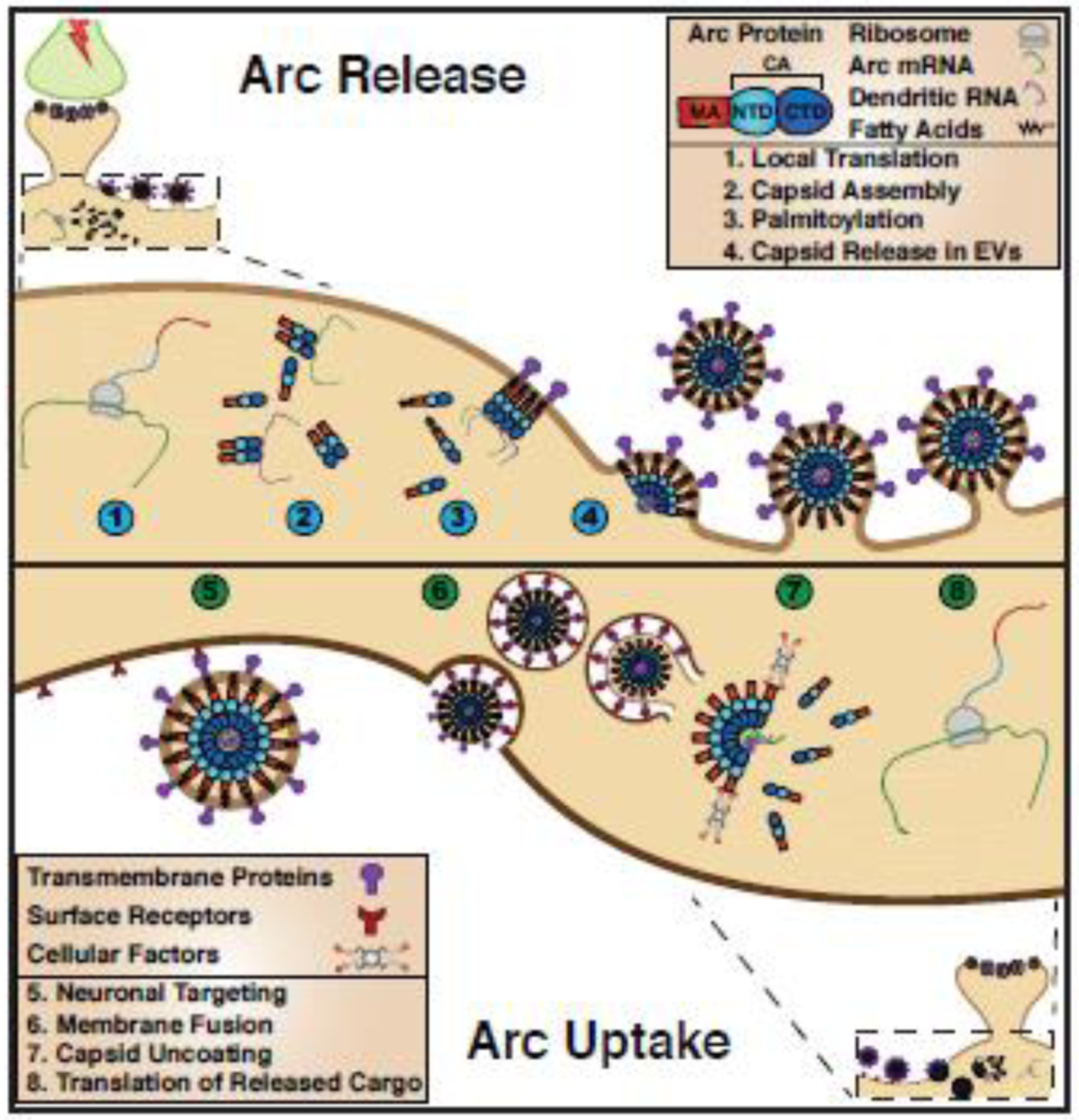
Arc protein release (steps 1–4) 1. Neuronal activity induces transcription and trafficking of Arc mRNA to active dendrites where it is locally translated. 2. Arc protein oligomerizes and assembles into capsids, which encapsulate RNA. 3. Post translational modifications, such as palmitoylation, promote Arc interactions with the plasma membrane. 4. Arc capsids are released within extracellular vesicles (EVs) directly at the cell surface. Arc EV uptake (steps 5–8) 5. Cell surface receptors specify Arc EV uptake. 6. Arc EVs are endocytosed. 7. Arc capsid crosses the endosomal membrane and is disassembled. 8. Delivered Arc mRNA is translated into protein.
The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein
Viruses have influenced the evolution of animal genes through several mechanisms, including evolution of specific immune defenses to combat viruses and direct gene transfer that can integrate into a host’s genome [10]. The latter is particularly evident for retrotransposons and retroviruses, which if inserted into the host’s germline, can lead to permanent changes in the infected organism’s progeny [11]. The Ty3/gypsy retrotransposons are members of ancient RNA-based self-replicating TEs that are present in animal, plant, and fungal kingdoms. Retrotransposons are considered either ancestral to, or derived from, modern retroviruses [11], but the line that distinguishes them is blurry. Retrotransposons share sequence and structural similarities to bonafide retroviruses, and many contain virus-like genes [1]. The main distinction lies in the ability of retroviruses to spread from organism-to-organism, whereas retrotransposons are usually only transmitted through the germline.
Over half the human genome is composed of sequences derived from viruses or TEs [1]. Some of these sequences encode genes that enable virus-like propagation and spread, termed endogenous retroviruses (ERVs) [11]. The genomes of virtually all animals are laden with ERVs and TEs. Over time, ERV genomes are often mutated and become inert. These remnants may no longer be capable of transposition and eventually encode genes that become beneficial for the organism. For example, multiple retroviral envelope genes were co-opted during mammalian evolution to promote the cell-cell fusion events essential for placental development [12]. The molecular functions of repurposed genes often relate to the original role of the viral gene in the virus replication cycle [13].
Retroviral Gag proteins contain three major elements that perform specific functions during viral assembly: MA (matrix) mediates membrane interactions, CA (capsid) mediates virion assembly, and NC (nucleocapsid) binds viral RNA. Arc has both sequence [8] and structural similarity to the retroviral CA element [6, 9]. Sequence analyses suggests that Arc may also contain an MA element, but lacks a canonical NC element for specific viral RNA interactions. However, the HIV MA element also interacts with viral RNA [14] and other retroviruses, such as foamy viruses, have evolved various RNA binding motifs [15]. Bioinformatic analysis revealed that the Drosophila Arc homologues (dArc1 and dArc2) also have a conserved CA element [7], which is corroborated by recent structure studies [16, 17]. Interestingly, tetrapod Arc genes cluster with Ty3/gypsy retrotransposons from fish, whereas the fly Arc homologs clusters with a separate lineage of Ty3/gypsy retrotransposons from insects (Figure 1C). This suggests that the tetrapod and fly Arc genes originated independently from distinct lineages yet share significant homology in their Gag-like regions. dArc1 protein is packaged within EVs and transfers RNA from the pre-synaptic neuromuscular junction synapse into post-synaptic muscle cells [18]. While mammalian Arc is localized predominately post-synaptic in the brain, it is unclear whether dArc is expressed pre- or post-synaptic in the fly brain. Purified dArc proteins form structures that are similar to those formed by mammalian Arc [5], indicating that the ability to form capsids is evolutionarily conserved. The atomic resolution model of fly Arc capsids (Figure 1B) show that these capsids are structurally similar to known retrovirus capsids [17]. Thus, the fly and tetrapod Arc genes provide a striking example of convergent evolution to regulate intercellular communication in neurons.
Arc – A Key Mediator of Long-term Synaptic Plasticity and Memory
Arc is a key component of the gene program induced by learning that is required for long-term synaptic plasticity and memory consolidation [4, 19, 20]. Specific memories are thought to be encoded by distinct neurons that are active during learning that then ultimately form a unique circuit through synaptic plasticity mechanisms to create the memory engram [21]. Consolidation and storage of memory requires de novo protein synthesis of a unique gene expression program in these neurons [22]. Current dogma suggests that synapse-specific plasticity is essential for creating memory circuits, but how the gene program contributes to memory storage and consolidation remains unclear. Arc expression is highly dynamic: transcription in neuronal circuits is tightly coupled to learning in vivo [23] and Arc mRNA is transported to dendrites where it is locally translated in response to neuronal activity [24]. One mechanism of Arc function is to regulate AMPA-type glutamate receptor (AMPAR) trafficking via direct interactions with the endocytic machinery that includes AP-2, dynamin and endophilin [25, 26]. Arc-dependent AMPAR trafficking is critical for maintaining optimal synaptic strength through homeostatic synaptic scaling in response to chronic changes in neuronal activity [27]. This may prevent saturation of synaptic strength, for example due to “runaway” long-term potentiation (LTP), allowing a single neuron to encode multiple memories through potentiation of specific synapses without dramatically changing the overall excitability and output. In addition, Arc knock-out (KO) mice are unable to learn from visual experience [28–30] and show various cognitive impairments [31]. This implies that Arc, which is regulated downstream of many activity-dependent signaling pathways, is one of the main effector proteins required for transforming experiences into long-lasting changes in the brain.
The observation that Arc protein forms virus-like capsids suggested that Arc mediates a previously unknown form of intercellular communication [5]. Purified Arc capsids encapsidate RNA, including Arc mRNA. Endogenous Arc from brain associates with its own mRNA and is released in EVs from primary rat and mouse cortical neurons. Recombinant purified Arc capsids and endogenous Arc-containing EVs are able to transfer rat Arc mRNA into Arc-KO mouse neurons. Thus, mammalian Arc has retained biochemical oligomerization and transport functions of the ancestral Gag protein [5]. Drosophila dArc1 transfers RNA in EVs from the neuromuscular synapse to muscle cells [18]. Thus, evolution has repurposed Gag retrotransposons at least twice to regulate synaptic function. The biology of these Arc-dependent intercellular pathways remains to be uncovered, including the functional cargo and the “message” that is conveyed. In contrast, much is known about viral Gag protein function in retrovirus biology. Here we use this known retroviral Gag biology as a template to speculate on the Arc “life cycle” (Figure 2) and how this may lead to new hypotheses on the neurobiological basis of memory formation.
A proposed model for the Arc “Life cycle”
Capsid Assembly and Structure
Gag is the central structural polyprotein of retrotransposons and retroviruses [14]. The Arc genes have strong structural homology with Gag CA [6], which is necessary and sufficient to form capsids that encapsidate RNAs. dArc capsids are 37nm in diameter, composed of 240 monomers that oligomerize to form a protein shell with icosahedral structure [17] (Figure 1B). Amphipathic helices on the N-terminus of these capsids project at regular intervals in “spikes”, suggesting a mechanism for membrane interaction. Some N-termini are buried within the capsid core and may interact with encapsidated RNAs. Mammalian Arc capsids resemble dArc under negative stain EM, but an atomic resolution structure remains to be determined. It is remarkable that the biochemical and structural features of Gag CA have been maintained in these genes; ~400 million years for mammalian Arc.
Where capsid assembly and release occurs in situ will determine the cargo and membrane composition of released vesicles. This information is critical for understanding their targeting and signaling roles. It is likely that the oligomeric state of Arc is tightly controlled. One point of regulation may be the local concentration of Arc monomers in the cell. Viral Gag capsid formation requires local concentrations that are “seeded” by protein-membrane or protein-nucleic acid interactions [32]. Synaptic activity promotes transcription of Arc mRNA, which is trafficked to active synapses where it undergoes activity-dependent translation creating a highly concentrated pool of Arc protein [4]. This rapid and focal translation may lead to conditions favoring capsid formation near neuronal synapses. Interactions with other “host” proteins may also regulate Arc capsid formation. For example, interaction with the NMDA-type glutamate receptor inhibits oligomerization of Arc protein through direct binding of the Arc capsid domain [33]. Capsid formation could also be regulated by post-translational modifications. A prime candidate is Arc palmitoylation [34], which could seed the formation of Arc capsids at membranes in a similar manner to the myristoylation of HIV MA [35]. Indeed, the N-terminal MA-like region of Arc has recently been shown to be important for oligomerization [36]. In neurons, palmitoylation is dynamic [37], suggesting that assembly could be controlled by neuronal activity. Capsid assembly may also be controlled by phosphorylation, as Calcium-calmodulin kinase II phosphorylation of Arc prevents oligomerization, and knock-in mice with mutations that prevent phosphorylation show deficits in synaptic plasticity and memory [38].
Capsid Encapsidation of RNA
Purified Arc capsids contain RNA, including Arc mRNA. Retrovirus Gags evolved to preferentially encapsidate viral RNA, using a specific “packaging” signal called the psi domain that binds to zinc finger motifs in NC [39]. However, RNA interactions can also occur through nonspecific electrostatic interactions through the basic MA element [14]. Do Arc capsids contain specific RNA cargo? Mammalian Arc contains a highly basic N-terminal region but does not contain homology to NC [40]. This suggests there may be nonspecific RNA interactions and hints that RNA packaging into Arc capsids may not be sequence specific. However, we also observe that Arc mRNA is consistently encapsidated or associated with Arc protein across bacterial expression systems, cultured cells, and mouse cortical tissue [5].
There are several mechanisms by which Arc mRNAs could be selectively packaged into Arc capsids. For example, dArc1 requires the 3’UTR to package dArc1 mRNA [18] and dArc1 also contains a conserved NC [17]. Interestingly, dArc2 lacks this motif [17] indicating differences in cargo selectivity. Packaging specificity may also arise from Arc-RNA interactions that facilitate Arc oligomerization and capsid formation. Arc does not form capsids when the purified protein is stripped of nucleic acids via polyethylenimine and ammonium sulfate precipitation [5], suggesting that RNA may nucleate Arc capsid formation. The trafficking and stability of Arc mRNA is complex, suggesting that there may be distinct pools of mRNA dedicated to specific functions [41]. Some proportion of Arc mRNA is rapidly transported to active dendrites in RNA granules where translation is repressed, whereas some mRNA is immediately translated in the soma. Arc mRNA has also been shown to undergo nonsense mediated decay after translation [42], which could potentially tune how much mRNA is encapsulated. Thus, RNA packaging may be dictated by specific RNA pools present at subcellular locations of Arc translation (Figure 3). How these distinct pools relate to capsid assembly and mRNA encapsulation remain to be determined.
Figure 3. Putative role of Arc-dependent intercellular RNA trafficking.
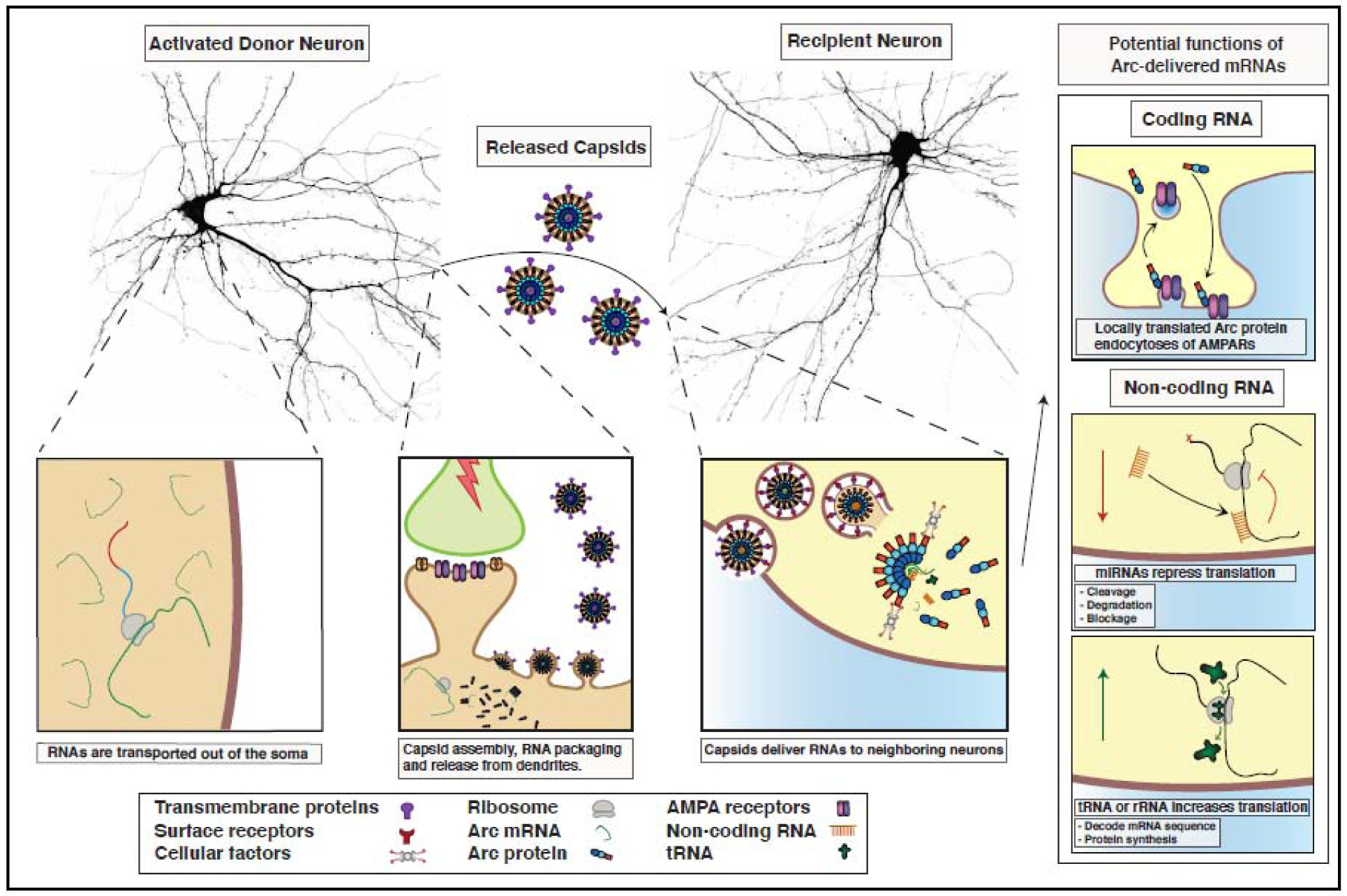
Arc mRNA traffics to active dendrites where translation and capsid assembly takes place. Extracellular vesicles containing Arc are released and deliver Arc mRNA and/or non-coding RNA cargo to neighboring cells. In recipient cells, translation of transferred Arc mRNA in dendrites could lead to the endocytosis of AMPA-type glutamate receptors at synapses and regulate circuit-level form of heterosynaptic plasticity. Alternatively, delivery of non-coding RNAs such as miRNAs or tRNAs/rRNAs could regulate protein expression in recipient cells. (Neuron images obtained from cultured rat hippocampal neurons transfected with a cell filler, mCherry, courtesy of Andrew Taibi).
Surprisingly, half of the RNA encapsidated by nascent retroviruses are not viral genomes, but instead host-derived RNAs [43]. The majority of these host-derived RNAs are non-coding RNAs (ncRNAs) less than 300 nucleotides in length, including tRNAs, rRNAs, and small nuclear RNAs. [43]. Ectopically expressed mRNAs can be packaged and transferred in Arc capsids [5], and it is likely that other endogenous RNAs are transferred between cells by Arc. Along with RNAs, retroviruses incorporate host proteins during virion biogenesis [44]. Arc associates with multiple proteins that regulate synaptic activity and neuronal function, including PSD95, TARPγ2, and CaMKII [6, 45], but it is unclear whether these proteins associate with Arc capsids or incorporate into Arc EVs. Given that Arc is present in the nucleus, soma and dendrites of neurons, the extreme spatial-temporal expression of Arc could result in a variety of cargo.
Release from Cells in Extracellular Vesicles
Retroviral capsids take advantage of host cell biology, such as EVs, to exit the cell. We propose that a major function of Gag-like genes is to produce a unique set of EVs that carry specific cargo involved in intercellular communication. These endogenous EVs use similar mechanisms of virion biogenesis, through the formation of capsids. How Arc capsids become enveloped in EVs is an important mechanistic question, with critical implications for how they recognize target cells and transfer information. In principle, Arc capsids may become enveloped by any of the different cellular pathways that create EVs. For example, exosomes first bud into endosomal compartments or multivesicular bodies (MVBs), which are then released in bulk when the MVB fuses with the plasma membrane. Little is known about exosome release from neurons, although MVBs are prevalent in synapses and dendrites [46–48]. Thus, Arc EVs may be released directly from the cell surface (like in HIV budding) or through an exosome pathway.
HIV capsids use the Endosomal Sorting Complex Required for Transport (ESCRT) proteins for release [49]. EVs can be released using a variety of mechanisms that include ESCRT activity and ESCRT-independent ceramide-mediated budding from sphingolipid and sterol-rich membrane microdomains [50]. Arc does not contain a classic retrovirus ESCRT interaction domain and may be released by non-ESCRT mechanisms. Another EV release pathway involves autophagy. Conventional autophagy pathways degrade proteins and organelles through autophagosomes, which fuse with lysosomes. However, recent observations have shown that autophagosomes can be targeted to the cell membrane, releasing materials in extracellular vesicles [51–53]. This pathway uses components of the autophagy machinery to selectively package RNA and proteins, although the function of this pathway is largely unknown [53]. Interestingly, rats show increased expression of both Arc and autophagy proteins following a learning task [54]. Additionally, blocking the autophagy pathway leads to deficits in long-term memory and improves when mice are perfused with the activating peptide Beclin1 [55]. When Arc is downregulated the autophagy pathway is no longer recruited after learning, suggesting an interaction between Arc and the autophagy pathway. These observations suggest that Arc capsids might be released through a non-canonical autophagy pathway to regulate long-term memory formation.
It is clear that EVs can be released from multiple internal membrane compartments. Retroviruses also show a variety of release mechanisms, assemble capsids in different cellular compartments and interact with many different host proteins [56]. Thus, we envisage that Gag-like genes have evolved unique mechanisms of cell release that are integral to the cellular processes governed by these proteins. Differences in assembly and release would ultimately lead to key differences in EV contents, providing an essential cargo sorting mechanism.
Capsid Delivery and Uptake
Enveloped viruses enter cells using specific virus-encoded fusogenic proteins that confer cell-type specificity and mediate virus-cell fusion to deliver the capsid into the cytoplasm. For example, the HIV envelope glycoprotein binds cellular CD4, which are expressed on the surfaces of target T-cells [57]. Arc EVs can deliver Arc mRNA into recipient cell cytosols that can then be translated [5, 18]. Arc mRNA transfer is much more efficient in neurons than in HEK293 cells [5], indicating some specificity of uptake or transfer. Surprisingly, purified Arc capsids that lack membranes can deliver cargo into neurons. This is a regulated process that is endocytosis dependent, which suggests a mechanism that may require a surface receptor interaction. It remains to be determined how Arc capsids “escape” the endosome prior to degradation. Intriguingly, dArc capsids contain amphipathic regions that extend from the capsid surface in “spikes” [17], reminiscent of viral amphipathic membrane-penetrating proteins [58]. The mammalian Arc N-terminus changes structure upon exposure to low pH conditions [40]. Thus, it is possible that Arc utilizes membrane-penetrating mechanisms, rather than membrane fusion, to deliver its contents into the cell.
Capsid Disassembly and Cargo Transfer
Virus capsid disassembly is a complicated process that requires host factors. In the case of retroviruses, once delivered into the cytoplasm, viral RNAs must be reverse transcribed into DNA and trafficked to the nucleus. HIV capsids interact with microtubule machinery to facilitate post-entry trafficking and disassemble either during trafficking, or upon docking at the nuclear pore complex [59]. Capsid disassembly allows reverse-transcribed viral DNA to be released and enter the nucleus through the nuclear pore complex, which has to be tightly coordinated in time. Once delivered, HIV DNAs embed into the cell genome and begin expressing several viral proteins that are not on the Gag polyprotein to subvert cellular immune responses and hijack host translational machinery [60, 61]. In doing so, delivered DNAs begin the process of producing progeny viruses in the newly infected cell. While we do not predict that Arc can highjack cellular translation machinery, Arc may contain a putative internal ribosome entry site (IRES), which could allow preferential non cap-dependent translation [62].
The fate of Arc capsids after cytoplasmic delivery is unclear. Arc protein has been observed in the nucleus of neurons [63] and it is possible that Arc capsids traffic to the nucleus. However, since the main cargo seems to be RNA there would be little need to localize to the nucleus. While the premature disassembly of the HIV capsid by TRIM5α in the cytoplasm blocks virus infection [64], Arc-delivered RNAs may need to disassemble in the cytoplasm for cargo to be translated. Thus, TRIM5α and other proteins known to interact with retrovirus capsids may also be involved in regulating Gag-derived capsid disassembly.
Concluding Remarks
Viruses have been intricately intwined with the evolution of life. While the viral or TE origins of some genes are clear, whether the proteins encoded by these genes retain their ancestral functions has remained unknown. The ability of the Arc genes to encode proteins that retain the ancestral capsid function, suggests a new mechanism for the biogenesis of a subset of EVs through virus-like capsids. EV signaling has emerged as a critical component of a diverse array of biological processes; from plant defenses against microbes [65] to the spread of pathology in neurodegenerative disorders [66]. However, the mechanisms of EV biogenesis and signaling are poorly understood [67], especially in the brain. In particular, it remains unclear how EVs sort and enrich for specific cargo. Similar to retroviruses, these endogenous capsids can enrich for specific cargo and dictate release/uptake mechanisms. The Arc genes represent the first known examples of repurposed TEs that encode for endogenous proteins that can form capsids, but this may be the tip of the iceberg; there are ~100 Gag-like genes [8] and over 1500 putative Gag open reading frames contained in ERVs in the human genome [68]. Few of these genes have been extensively studied and it remains to be determined whether they have also retained virus-like properties (see Table 1). Some of these genes are expressed in the brain, such as RTL4/Sirh11, which regulates noradrenaline levels in the prefrontal cortex and plays a role in spatial working memory [69]. The Gag-like gene RTL2/Peg10 is involved in promoting cell proliferation and cell-to-cell RNA trafficking [70]. The paraneoplastic Ma antigens (PNMAs) are another family of genes found in the CNS [71, 72] that encode proteins that have homology to Gag [8]. An exciting direction of research will be to determine whether the functions of Gag-like proteins also involve capsid biology. It is intriguing to speculate that multiple capsid proteins facilitate distinct forms of intercellular communication in specific cell types and in various organs.
Table 1.
The known biological activities of Gag-like proteins.
| Gag-like genea | Species | Tissue expressionb | Biological activities | Virus-like properties | References |
|---|---|---|---|---|---|
| Arc | Mammals and some insects | Brain, testes, and dendritic cells | Regulates endocytosis at synapses to tune synaptic strength. Essential for long-term memory. | Forms virus-like particles and transfers RNA between cells. | [4, 5, 18] |
| PNMA4/MOAP1 | Mammals | Ubiquitous | Promotes apoptosis | Unknown | [91] |
| RTL1/Peg11 | Eutherian mammals | Adrenal gland, brain, and placenta | Essential to maintain placental capillaries. | Unknown | [92] |
| RTL2/Peg10 | Eutherian mammals | Placenta, adrenal gland, pituitary gland, and brain | Promotes cell proliferation, inhibits apoptosis, and blocks TGF-beta signaling. Contributes to tumorigenesis. | Transfers RNA between cells, unknown if can form capsids. | [70, 93, 94] |
| RTL3/Sirh9 | Eutherian mammals | Ubiquitous | Controls collagen release through transcriptional regulation. | Unknown | [95] |
| RTL4/Sirh11 | Eutherian mammals | Brain, testes, ovaries, kidneys, and liver | Regulates noradrenaline levels and influences spatial working memory. | Unknown | [69] |
| RTL7/Sirh7/Ldoc1 | Eutherian mammals | Ubiquitous | Controls placental development and suppresses immune signaling. | Unknown | [96, 97] |
As determined by sequence homology
As found in the Human Protein Atlas (https://www.proteinatlas.org/)
Since mammalian Arc seems to function in multiple aspects of synaptic plasticity, it will be critical to tease out the specific functions of capsid vs. monomeric Arc. Arc capsids are released in an activity-dependent manner and can be taken up by other neurons [5]. Does Arc release regulate cells in the local environment or act as a long-range signal? The function of intercellular Arc signaling will be determined by the functional cargo being transferred (Figure 3). Delivered Arc mRNA can be translated, and thus the downstream consequence of capsid uptake at neighboring synapses could result in Arc-mediated AMPAR endocytosis and synapse weakening. Arc has already been implicated in the removal of AMPA receptors from weak synapses on the same dendrite in a form of heterosynaptic depression [73, 74]. However, Arc mRNA transfer between synapses could mediate heterosynaptic plasticity at the circuit level, facilitating the stabilization of memory circuits by weakening connections with surrounding neurons that were not active during learning. Alternatively, Arc capsids could deliver non-coding RNAs that could alter protein expression in recipient cells. For example, neurons express a variety of micro-RNAs in an activity-dependent manner and some micro-RNAs are expressed in dendrites [75, 76]. Arc may package select micro-RNAs depending on the prior activity history of a particular neuron. We speculate that Arc EVs could thus contain distinct cargo and “read-out” the activity status of neurons at the circuit level. Given the relatively small capacity of Arc capsids for nucleic acids, small RNAs could be packaged at high enough concentrations to significantly alter cellular processes in recipient cells.
Whether intercellular Arc plays a direct role in memory or synaptic plasticity remains to be tested. In addition to functions in the brain, Arc transcripts are also found in blood serum [77] and expressed in dendritic immune cells, which are important for aspects of the immune response [78]. A recent study found that Arc is released from peripheral dorsal root ganglion neurons, which may be important for controlling neuroinflammation in the skin [79]. Intriguingly, preliminary studies also suggest that an active Gypsy retrotransposon in C.elegans can mediate the transfer of avoidance memory through the formation of virus-like capsids [80].
While the function of the Drosophila Arc proteins have not been as extensively studied as in mammals, there are some striking similarities between them and mammalian Arc that could shed light on the function of intercellular Arc signaling. dArc1 and dArc2 expression is upregulated by neuronal activity [81–83], dArc1 is involved in synaptic plasticity [18] and dArc2 has been implicated in memory [81]. Additionally, neurons expressing dArc regulate the metabolic state and responses to starvation in flies [82, 83]. The physiological role of the dArc capsids and intercellular communication in these phenotypes remains to be determined. However, the conservation of the Gag capsid domain and the ability to form retrovirus-like capsids suggest strong evolutionary pressure to maintain capsid biology.
Accumulating evidence suggests that ERVs may become expressed during aging and contribute to neurological diseases such as Amyotrophic Lateral Sclerosis, Alzheimer’s disease, multiple sclerosis, and Aicardi-Goutieres syndrome [84, 85]. EVs have been proposed as one mechanism by which pathology is spread over time [86]. Arc may also be involved in the spread of pathology in neurodegenerative diseases; evidence is accumulating that this spread occurs via synaptic connections, and is modulated by neuronal activity [87]. Arc itself interacts with the amyloid precursor protein and facilitates its processing into pathogenic β-amyloid [88], which may include packaging into EVs. Pathogenic misfolded proteins like tau can also induce ERV expression [89], and the expression of ERVs in the mouse hippocampus coincides with memory decline [90]. In addition to Arc misregulation, it is possible that Gag proteins expressed by ERVs could contribute to spread of pathology through abnormal intercellular communication or interfere with the signaling of endogenous Gag proteins.
The recent progress in gene editing tools, such as CRISPR, promises to significantly impact genetic diseases. However, an intractable obstacle in the translation to gene therapy remains: how to safely and efficiently deliver nucleic acids. Established gene delivery methods rely on modified viruses, and are limited by host immune reactions, inadequate target specificity and small cargo loads. The ability to deliver packaged RNAs into target cells through endogenous capsids make these proteins intriguing candidates for gene delivery. Arc capsids encapsidate and deliver not only Arc mRNA, but also ectopically expressed RNAs [5, 18]. Arc protein is also expressed in the periphery, suggesting that Arc capsids may not be immunogenic [77, 78], and may even have anti-inflammatory activities [79]. Given their likely physical and immunogenic properties, endogenous Gag-like capsids may be promising delivery vehicles for gene therapy.
Like with most new observations, there are now more questions than answers about the viral nature of Arc and its implications on brain function. Future studies on intercellular Arc could impact our understanding of memory and neurodegenerative disease. The investigation of other Gag-like genes may lead to the discovery of various new EV-dependent pathways in different physiological contexts. It is fascinating to envision that key brain functions and many other cellular processes may have evolved from ancient viral infections!
Outstanding Questions.
What are the cargoes and “signals” transported by Arc capsids?
Do Arc capsids transfer cargoes locally or long-distance?
Is there cell-type specificity of Arc intercellular signaling?
Is Arc intercellular signaling required for memory consolidation?
Do Arc extracellular vesicles propagate pathology in neurodegenerative diseases?
Do other Gag-like proteins form capsids and mediate intercellular communication?
Can endogenous capsids be used for gene delivery?
Highlights.
Ancient viruses and mobile transposable genetic elements have left permanent marks on animal genomes. Evolution has repurposed some of these elements to mediate important biological processes.
A gene critical for memory consolidation and synaptic plasticity, Arc, encodes a repurposed retrotransposon Gag protein that forms virus-like protein capsids. Arc is released from neurons in specialized extracellular vesicles that are able to transfer RNA between cells; a previously unknown type of intercellular communication. Here, we propose an Arc “life-cycle” based on known functions of retrovirus Gag.
Arc-dependent intercellular communication may alter cellular processes through the transfer of various RNAs. This pathway may be important for synaptic plasticity, memory and the spread of pathology during neurodegeneration.
The human genome contains ~100 genes that contain homology to Gag and over 1500 Gag open reading frames encoded as endogenous retroviruses, suggesting that virus-like intercellular communication may regulate many biological processes. We posit that there may be a repertoire of endogenous genes that code for proteins capable of assembling into capsids, which generate specialized extracellular vesicles with different cargoes.
Acknowledgements
We thank Simon Erlendsson for the fly Arc capsid model in Figure 1B and Andrew Taibi for the neuronal images in figure 3. We thank the Shepherd lab for helpful discussions and feedback. This work was supported by a National Institutes of Health Director’s Transformative Research Award (R01 NS115716) and a Chan Zuckerberg Initiative Ben Barres Early Career Acceleration Award (J.D.S).
Glossary
- Transposable element
a DNA sequence capable of moving between different locations in the genome.
- Transposition
the movement of DNA from one location of the genome to another, either by excision and reinsertion, or by making a copy of itself.
- Retrotransposon
a transposable element that moves through the genome via an RNA intermediate. Retrotransposon RNAs undergo reverse transcription to generate DNA copies that are reinserted into the genome.
- Gag
group specific antigen, protein domain that forms capsids in HIV
- Retrovirus
a virus that reverse transcribes its genomic RNA into DNA to embed itself in the infected host genome.
- Endogenous retrovirus
a genetic remnant of a retrovirus infection or germline retrotransposon embedded in the genome.
- Repurposed gene
a gene likely derived from a transposon or viral sequence that is incapable of infection or transposition.
- Memory engram
the physical trace of a memory encoded in a neural circuit.
- Extracellular vesicles (EVs)
small membranous vesicles released from virtually all cells that transfer cytoplasmic and membrane-bound cargos between cells.
- Internal ribosome entry site (IRES)
RNA elements that act in cis to recruit ribosomes and facilitate cap-independent translation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
J.D.S is a scientific co-founder and consultant to VNV, with a patent application related to this work.
References
- 1.Lander ES et al. (2001) Initial sequencing and analysis of the human genome. Nature 409 (6822), 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Kaneko-Ishino T and Ishino F (2012) The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front Microbiol 3, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuong EB et al. (2017) Regulatory activities of transposable elements: from conflicts to benefits. Nature Reviews Genetics 18 (2), 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shepherd JD and Bear MF (2011) New views of Arc, a master regulator of synaptic plasticity. Nature Neuroscience 14 (3), 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastuzyn ED et al. (2018) The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 172 (1–2), 275–288.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W et al. (2015) Structural Basis of Arc Binding to Synaptic Proteins: Implications for Cognitive Disease. Neuron 86 (2), 490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrusan G et al. (2013) Turning gold into ‘junk’: transposable elements utilize central proteins of cellular networks. Nucleic Acids Res 41 (5), 3190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campillos M et al. (2006) Computational characterization of multiple Gag-like human proteins. Trends in Genetics 22 (11), 585–589. [DOI] [PubMed] [Google Scholar]
- 9.Taylor WR et al. (2017) A comparative analysis of the foamy and ortho virus capsid structures reveals an ancient domain duplication. BMC Structural Biology 17 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koonin EV and Dolja VV (2013) A virocentric perspective on the evolution of life. Curr Opin Virol 3 (5), 546–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson WE (2019) Origins and evolutionary consequences of ancient endogenous retroviruses. Nature Reviews Microbiology 17 (6), 355–370. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis G et al. (2015) Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci U S A 112 (5), E487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields BN et al. (2013) Fields virology, Wolters Kluwer Health/Lippincott Williams & Wilkins.
- 14.Lee EG and Linial ML (2004) Basic Residues of the Retroviral Nucleocapsid Play Different Roles in Gag-Gag and Gag- RNA Interactions. Journal of Virology 78 (16), 8486–8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stenbak CR and Linial ML (2004) Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. Journal of virology 78 (17), 9423–9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cottee MA et al. (2020) Structure of Drosophila melanogaster ARC1 reveals a repurposed molecule with characteristics of retroviral Gag. Sci Adv 6 (1), eaay6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erlendsson S et al. (2020) Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins. Nat Neurosci 23 (2), 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashley J et al. (2018) Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell 172 (1–2), 262–274.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolaienko O et al. (2018) Arc protein: a flexible hub for synaptic plasticity and cognition. Semin Cell Dev Biol 77, 33–42. [DOI] [PubMed] [Google Scholar]
- 20.Plath N et al. (2006) Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52 (3), 437–44. [DOI] [PubMed] [Google Scholar]
- 21.Josselyn SA and Tonegawa S (2020) Memory engrams: Recalling the past and imagining the future. Science 367 (6473). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap EL and Greenberg ME (2018) Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 100 (2), 330–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzowski JF et al. (1999) Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci 2 (12), 1120–4. [DOI] [PubMed] [Google Scholar]
- 24.Steward O et al. (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21 (4), 741–51. [DOI] [PubMed] [Google Scholar]
- 25.Chowdhury S et al. (2006) Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52 (3), 445–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall MJ and Correa SAL (2018) The mechanistic link between Arc/Arg3.1 expression and AMPA receptor endocytosis. Semin Cell Dev Biol 77, 17–24. [DOI] [PubMed] [Google Scholar]
- 27.Shepherd JD et al. (2006) Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52 (3), 475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCurry CL et al. (2010) Loss of Arc renders the visual cortex impervious to the effects of sensory experience or deprivation. Nat Neurosci 13 (4), 450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenks KR and Shepherd JD (2020) Experience-Dependent Development and Maintenance of Binocular Neurons in the Mouse Visual Cortex. Cell Rep 30 (6), 1982–1994 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenks KR et al. (2017) Arc restores juvenile plasticity in adult mouse visual cortex. Proc Natl Acad Sci U S A 114 (34), 9182–9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manago F et al. (2016) Genetic Disruption of Arc/Arg3.1 in Mice Causes Alterations in Dopamine and Neurobehavioral Phenotypes Related to Schizophrenia. Cell Rep 16 (8), 2116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lingappa JR et al. (2014) How HIV-1 Gag assembles in cells: Putting together pieces of the puzzle. Virus Research 193, 89–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen LD et al. (2019) The Capsid Domain of Arc Changes Its Oligomerization Propensity through Direct Interaction with the NMDA Receptor. Structure 27 (7), 1071–1081 e5. [DOI] [PubMed] [Google Scholar]
- 34.Barylko B et al. (2018) Palmitoylation and Membrane Binding of Arc/Arg3.1: A Potential Role in Synaptic Depression. Biochemistry 57 (5), 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Provitera P et al. (2006) The effect of HIV-1 Gag myristoylation on membrane binding. Biophysical Chemistry 119 (1), 23–32. [DOI] [PubMed] [Google Scholar]
- 36.Eriksen MS et al. (2020) Arc self-association and formation of virus-like capsids are mediated by an N-terminal helical coil motif. FEBS J. DOI: 10.1111/febs.15618. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi T et al. (2005) Differential Regulation of AMPA Receptor Subunit Trafficking by Palmitoylation of Two Distinct Sites. Neuron 47 (5), 709–723. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W et al. (2019) Arc Oligomerization Is Regulated by CaMKII Phosphorylation of the GAG Domain: An Essential Mechanism for Plasticity and Memory Formation. Molecular Cell 75 (1), 13–25.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comas-Garcia M et al. (2017) Dissection of specific binding of HIV-1 Gag to the ‘packaging signal’ in viral RNA. eLife 6, e27055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallin EI et al. (2018) Structure of monomeric full-length ARC sheds light on molecular flexibility, protein interactions, and functional modalities. Journal of Neurochemistry 147 (3), 323–343. [DOI] [PubMed] [Google Scholar]
- 41.Steward O et al. (2015) Localization and local translation of Arc/Arg3.1 mRNA at synapses: some observations and paradoxes. Frontiers in molecular neuroscience 7, 101–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giorgi C et al. (2007) The EJC Factor eIF4AIII Modulates Synaptic Strength and Neuronal Protein Expression. Cell 130 (1), 179–191. [DOI] [PubMed] [Google Scholar]
- 43.Eckwahl MJ et al. (2016) Host RNA Packaging by Retroviruses: A Newly Synthesized Story. mBio 7 (1), e02025–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott DE (2008) Cellular proteins detected in HIV-1. Rev Med Virol 18 (3), 159–75. [DOI] [PubMed] [Google Scholar]
- 45.Fernández E et al. (2017) Arc Requires PSD95 for Assembly into Postsynaptic Complexes Involved with Neural Dysfunction and Intelligence. Cell Rep 21 (3), 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Von Bartheld CS and Altick AL (2011) Multivesicular bodies in neurons: distribution, protein content, and trafficking functions. Prog Neurobiol 93 (3), 313–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooney JR et al. (2002) Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J Neurosci 22 (6), 2215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rind HB et al. (2005) Synaptic targeting of retrogradely transported trophic factors in motoneurons: comparison of glial cell line-derived neurotrophic factor, brain-derived neurotrophic factor, and cardiotrophin-1 with tetanus toxin. J Neurosci 25 (3), 539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrus JE et al. (2001) Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding. Cell 107 (1), 55–65. [DOI] [PubMed] [Google Scholar]
- 50.Trajkovic K et al. (2008) Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 319 (5867), 1244–1247. [DOI] [PubMed] [Google Scholar]
- 51.Murrow L et al. (2015) ATG12–ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nature Cell Biology 17 (3), 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sirois I et al. (2012) Caspase activation regulates the extracellular export of autophagic vacuoles. Autophagy 8 (6), 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leidal AM et al. (2020) The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol 22 (2), 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pandey K et al. (2020) Autophagy coupled to translation is required for long-term memory. Autophagy. DOI: 10.1080/15548627.2020.1775393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hylin MJ et al. (2018) A role for autophagy in long-term spatial memory formation in male rodents. J Neurosci Res 96 (3), 416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caobi A et al. (2020) Extracellular Vesicles in the Pathogenesis of Viral Infections in Humans. Viruses 12 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilen CB et al. (2012) HIV: cell binding and entry. Cold Spring Harbor perspectives in medicine 2 (8), a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spriggs CC et al. (2019) How non-enveloped viruses hijack host machineries to cause infection. Advances in virus research 104, 97–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ambrose Z and Aiken C (2014) HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology 454–455, 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Montfoort N et al. (2014) Unmasking immune sensing of retroviruses: interplay between innate sensors and host effectors. Cytokine Growth Factor Rev 25 (6), 657–68. [DOI] [PubMed] [Google Scholar]
- 61.Guerrero S et al. (2015) HIV-1 replication and the cellular eukaryotic translation apparatus. Viruses 7 (1), 199–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinkstaff JK et al. (2001) Internal initiation of translation of five dendritically localized neuronal mRNAs. Proceedings of the National Academy of Sciences of the United States of America 98 (5), 2770–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bloomer WA et al. (2007) Activity-regulated cytoskeleton-associated protein Arc/Arg3.1 binds to spectrin and associates with nuclear promyelocytic leukemia (PML) bodies. Brain Res 1153, 20–33. [DOI] [PubMed] [Google Scholar]
- 64.Stremlau M et al. (2006) Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proceedings of the National Academy of Sciences of the United States of America 103 (14), 5514–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai Q et al. (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360 (6393), 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Budnik V et al. (2016) Extracellular vesicles round off communication in the nervous system. Nature Reviews Neuroscience 17 (3), 160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shurtleff MJ et al. (2018) Extracellular Vesicles and Cancer: Caveat Lector. Annual Review of Cancer Biology 2 (1), 395–411. [Google Scholar]
- 68.Nakagawa S and Takahashi MU (2016) gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database : the journal of biological databases and curation 2016, baw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Irie M et al. (2015) Cognitive Function Related to the Sirh11/Zcchc16 Gene Acquired from an LTR Retrotransposon in Eutherians. PLOS Genetics 11 (9), e1005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abed M et al. (2019) The Gag protein PEG10 binds to RNA and regulates trophoblast stem cell lineage specification. PLOS ONE 14 (4), e0214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pang SW et al. (2018) PNMA family: Protein interaction network and cell signalling pathways implicated in cancer and apoptosis. Cell Signal 45, 54–62. [DOI] [PubMed] [Google Scholar]
- 72.Schüller M et al. (2005) The human PNMA family: Novel neuronal proteins implicated in paraneoplastic neurological disease. Journal of Neuroimmunology 169 (1), 172–176. [DOI] [PubMed] [Google Scholar]
- 73.El-Boustani S et al. (2018) Locally coordinated synaptic plasticity of visual cortex neurons in vivo. Science 360 (6395), 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okuno H et al. (2012) Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIbeta. Cell 149 (4), 886–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fiore R et al. (2009) Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. Embo j 28 (6), 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schratt GM et al. (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439 (7074), 283–9. [DOI] [PubMed] [Google Scholar]
- 77.Sanders TH et al. (2019) Cognition-Enhancing Vagus Nerve Stimulation Alters the Epigenetic Landscape. The Journal of Neuroscience 39 (18), 3454–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ufer F et al. (2016) Arc/Arg3.1 governs inflammatory dendritic cell migration from the skin and thereby controls T cell activation. Science Immunology 1 (3), eaaf8665–eaaf8665. [DOI] [PubMed] [Google Scholar]
- 79.Barragan-Iglesias P et al. (2020) Intercellular Arc signaling regulates vasodilation. bioRxiv. DOI: 10.1101/2020.08.13.250209, 2020.08.13.250209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moore RS et al. (2020) Horizontal and vertical transmission of transgenerational memories via the Cer1 transposon. bioRxiv. DOI: 10.1101/2020.12.28.424563, 2020.12.28.424563. [DOI] [Google Scholar]
- 81.Awata H et al. (2019) The neural circuit linking mushroom body parallel circuits induces memory consolidation in Drosophila. Proc Natl Acad Sci U S A 116 (32), 16080–16085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mattaliano MD et al. (2007) The Drosophila ARC homolog regulates behavioral responses to starvation. Mol Cell Neurosci 36 (2), 211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mosher J et al. (2015) Coordination between Drosophila Arc1 and a specific population of brain neurons regulates organismal fat. Dev Biol 405 (2), 280–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tam OH et al. (2019) Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mobile DNA 10, 32–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Küry P et al. (2018) Human Endogenous Retroviruses in Neurological Diseases. Trends in Molecular Medicine 24 (4), 379–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coleman BM and Hill AF (2015) Extracellular vesicles--Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol 40, 89–96. [DOI] [PubMed] [Google Scholar]
- 87.Thompson AG et al. (2016) Extracellular vesicles in neurodegenerative disease - pathogenesis to biomarkers. Nat Rev Neurol 12 (6), 346–57. [DOI] [PubMed] [Google Scholar]
- 88.Wu J et al. (2011) Arc/Arg3.1 Regulates an Endosomal Pathway Essential for Activity-Dependent Beta-Amyloid Generation. Cell 147 (3), 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo C et al. (2018) Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep 23 (10), 2874–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sankowski R et al. (2019) Endogenous retroviruses are associated with hippocampus-based memory impairment. Proceedings of the National Academy of Sciences of the United States of America 116 (51), 25982–25990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan KO et al. (2001) MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J Biol Chem 276 (4), 2802–7. [DOI] [PubMed] [Google Scholar]
- 92.Sekita Y et al. (2008) Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nature Genetics 40 (2), 243–248. [DOI] [PubMed] [Google Scholar]
- 93.Yahiro Y et al. (2019) PEG10 counteracts signaling pathways of TGF-β and BMP to regulate growth, motility and invasion of SW1353 chondrosarcoma cells. J Bone Miner Metab 37 (3), 441–454. [DOI] [PubMed] [Google Scholar]
- 94.Xie T et al. (2018) PEG10 as an oncogene: expression regulatory mechanisms and role in tumor progression. Cancer Cell Int 18, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ball HC et al. (2020) A retrotransposon gag-like-3 gene RTL3 and SOX-9 co-regulate the expression of COL2A1 in chondrocytes. Connective Tissue Research. DOI: 10.1080/03008207.2020.1828380, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao S et al. (2015) LDOC1 inhibits proliferation and promotes apoptosis by repressing NF-κB activation in papillary thyroid carcinoma. Journal of Experimental & Clinical Cancer Research 34 (1), 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Naruse M et al. (2014) Sirh7/Ldoc1 knockout mice exhibit placental P4 overproduction and delayed parturition. Development 141 (24), 4763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]


