Abstract
Background:
Humans and environmental organisms are constantly exposed to complex mixtures of chemicals. Extending our knowledge about the combined effects of chemicals is thus essential for assessing the potential consequences of these exposures. In this context, comprehensive molecular readouts as retrieved by omics techniques are advancing our understanding of the diversity of effects upon chemical exposure. This is especially true for effects induced by chemical concentrations that do not instantaneously lead to mortality, as is commonly the case for environmental exposures. However, omics profiles induced by chemical exposures have rarely been systematically considered in mixture contexts.
Objectives:
In this study, we aimed to investigate the predictability of chemical mixture effects on the whole-transcriptome scale.
Methods:
We predicted and measured the toxicogenomic effects of a synthetic mixture on zebrafish embryos. The mixture contained the compounds diuron, diclofenac, and naproxen. To predict concentration- and time-resolved whole-transcriptome responses to the mixture exposure, we adopted the mixture concept of concentration addition. Predictions were based on the transcriptome profiles obtained for the individual mixture components in a previous study. Finally, concentration- and time-resolved mixture exposures and subsequent toxicogenomic measurements were performed and the results were compared with the predictions.
Results:
This comparison of the predictions with the observations showed that the concept of concentration addition provided reasonable estimates for the effects induced by the mixture exposure on the whole transcriptome. Although nonadditive effects were observed only occasionally, combined, that is, multicomponent-driven, effects were found for mixture components with anticipated similar, as well as dissimilar, modes of action.
Discussion:
Overall, this study demonstrates that using a concentration- and time-resolved approach, the occurrence and size of combined effects of chemicals may be predicted at the whole-transcriptome scale. This allows improving effect assessment of mixture exposures on the molecular scale that might not only be of relevance in terms of risk assessment but also for pharmacological applications. https://doi.org/10.1289/EHP7773
Introduction
Understanding the combined effects of substances that occur due to mixture exposures is of long-standing interest in the biosciences. While physiologists and pharmacologists search to optimize intended drug activity by combination therapy (Kuhn-Nentwig et al. 2019; Zimmer et al. 2016), toxicologists seek to safeguard against the unintended combined effects concerning adverse outcomes in humans (Goodson et al. 2015; Jiang et al. 2018) and environmental organisms (Cedergreen 2014) resulting from mixture exposure. Society demands the inclusion of mixtures in chemical hazard and risk assessment (EC 2019). The European Commission in its European Green Deal, for example, calls for a regulatory framework that can reflect risks posed by the combined effects of multiple chemicals (EC 2019). The consideration of combined effects can also be found on the agenda of international policy advisory (Meek et al. 2011) and regulatory institutions (Rotter et al. 2018; Wegner et al. 2016).
Environmental chemists have argued that multiple substances can occur in large varieties (Escher et al. 2020) and that resulting environmental exposures may be complex in composition and dynamic over time for individuals (Jiang et al. 2018). Thus, the exposure of humans and environmental organisms to mixtures of chemicals instead of to single compounds solely is rather the rule than the exception. Universal experimental evaluation of the effects resulting from each potentially occurring mixture exposure is not feasible. Thus, reasonable predictions of the effects of mixtures should be based on models that can use knowledge of the bioactivity of the compounds to estimate their combined effects in a mixture (Altenburger et al. 2013).
Most existing models for combined effects are based on simplistic toxicodynamic assumptions of either simple similar or independent action (Altenburger et al. 2013). Among those, the concept of concentration addition (CA) has been shown to be rather useful to quantitatively predict the combined effects in short-term in vivo studies and the observations of gross changes, that is, apical effects such as death, growth impairment, or developmental dysfunction (reviewed by Kortenkamp et al. 2009). The situation is less clear, however, for sublethal, long-term effects and low-dose exposures. Here, the evidence is limited regarding the predictivity for outcomes. Moreover, qualitative interaction, that is, the emergence of novel outcomes not seen for the individual compounds is suggested to occur for mixture exposures in some studies (Rodea-Palomares et al. 2016; Zimmer et al. 2016).
The standard in long-term toxicity assessment, for example, for prospective risk assessment, is based on in vivo assays under chronic chemical exposure. Such assays are, however, highly time and cost demanding and are, therefore, limited in throughput. As an alternative to in vivo testing, high-content in vitro assays are being conducted, and researchers have found that molecular outcomes, such as those identified using toxicogenomic measurements, may be indicative of long-term effects, for example, carcinogenicity (Li et al. 2019). Thus, it has been anticipated that toxicogenomic methods may also help in advancing insight into questions of mixture toxicology for long-term and multivariate effect detection (Altenburger et al. 2012). Evaluating mixture effects using toxicogenomics would support the establishment of this method to obtain comprehensive information for chemical hazard assessment (EFSA et al. 2018) and environmental monitoring (Bahamonde et al. 2016) in an untargeted manner.
Despite these ambitious expectations, mixture effects are still perceived as the elephant in the room in the field of toxicogenomics (Schroeder et al. 2016) and there is no consensus on terminology, approaches, and assessment. A major obstacle in present toxicogenomic mixture studies results from the use of experimental designs whereby the doses used to study the compounds individually are simply put together in a mixture (Altenburger et al. 2012). The observed effect of the mixture is then compared against the effects of the single compounds. Acknowledging that toxicogenomic responses are known to be concentration dependent (Kopec et al. 2010; Smetanová et al. 2015), this assessment strategy is of limited value because any changes in the mixture response have to be judged as interactive given that reasonable expectations for the combined effect cannot be obtained (Altenburger et al. 2012; Berenbaum 1981). The challenge for mixture studies using toxicogenomic methods, therefore, is to show the validity, limits, and usefulness of established mixture methodology (Altenburger et al. 2012), which we tackle in this study.
The basis for mixture prediction, when applying a concept such as CA, therefore, requires the explicit description of single-substance effects resolved at least for concentration. Regression models for concentration-dependent toxicogenomic responses have been described before (Smetanová et al. 2015; Thomas et al. 2007) and have recently been extended for the time domain in a concentration- and time-dependent response model (CTR-model) (Schüttler et al. 2019). The CTR-model describes the logarithmic fold-change () during chemical exposure. It follows a sigmoidal shape for the concentration dependence and a biphasic shape for the time dependence of the response. Among the model parameters, represents the maximum sensitivity, S. The sensitivity is defined as the reciprocal of the minimum of over time, that is, the minimum concentration over time leading to a half-maximum effect. Thus, indicates the concentration dependence of the response. The parameter stands for the point in time when is reached, thus indicating the time dependence of response. The CTR-model has been applied to describe the toxicogenomic responses of single substances (Schüttler et al. 2019). In the present study, it provides the basis for calculating mixture expectations and for describing the observed mixture responses.
To reduce the dimensionality of toxicogenomic responses, reduce noise, and improve the robustness of the CTR-model fits, the analyses were not performed on individual genes but, rather, on groups of coexpressed genes. Based on a compiled data set of many toxicogenomic zebrafish studies, similar responding genes had been clustered in 3,600 nodes on a two-dimensional grid (a toxicogenomic universe) in a previous study (Schüttler et al. 2019). This was achieved with the help of the self-organizing map (SOM) algorithm (Kohonen 1982). The approach is based on the assumption that genes, which are functionally related or affected by the same regulators, are coexpressed (Nikkilä et al. 2002). Thus, different areas of the toxicogenomic universe can be assigned to specific functions or anatomical regions (Schüttler et al. 2019). The mapping of toxicogenomic measurements on this toxicogenomic universe results in toxicogenomic fingerprints that depict the effects of an investigated compound on the whole transcriptome for each concentration and time point (Schüttler et al. 2019). A subsequent CTR-model fit for each of the 3,600 nodes results in model parameter values that can as well be mapped on the toxicogenomic universe. These dynamic toxicogenomic fingerprints allow direct comparisons between different compounds and simplify the formulation of hypotheses regarding the affected physiological or molecular functions.
In the present study, we performed a case study and evaluated the effects of a three-component synthetic mixture on the transcriptome of the zebrafish embryo (ZFE). We calculated explicit time-resolved expectations for the toxicogenomic effects of the mixture applying the concept of CA. Subsequently, we compared these predictions with experimental observations using a fixed mixture ratio (Altenburger et al. 2012), or so-called diagonal design (Berenbaum 1981), that also allows comparisons with alternative mixture concepts.
In this study, we investigated the mixture of the three compounds—diuron, diclofenac, and naproxen—for which we had earlier obtained toxicogenomic fingerprints (Schüttler et al. 2019). The compounds are known to co-occur as freshwater contaminants (Bradley et al. 2017; Busch et al. 2016). Moreover, because they have been developed for specific pharmacological or toxicological applications, their molecular targets and intended action in organisms are well characterized (as summarized by Schüttler et al. 2019). We intentionally mixed two compounds with the same known molecular target and pharmacological mode of action [cyclooxygenase (COX) inhibition: diclofenac and naproxen] and one compound with a target and mode of action assumed to be dissimilar to the above (the herbicide diuron, which has no specific mode of action in fish). In our previous investigation of the components, we indeed found similar toxicogenomic responses for the two COX inhibitors, such as an alteration of transcripts related to the arachidonic acid metabolism and the activation of cyp2k18 (Schüttler et al. 2019). However, distinct differences between the effects of the two COX inhibitors and commonalities to the third compound diuron also became apparent. For example, we found genes related to pancreas development that were affected by naproxen and diuron but not by diclofenac (Schüttler et al. 2019). The same was found for the induction of cyp1a genes. We also showed that the curves for the uptake of the individual compounds into ZFE over time are very different for the three compounds (Schüttler et al. 2019). The observed induction of genes coding for respective metabolizing enzymes, such as Cyp2k18 (a zebrafish orthologue of CYP2C9) due to diclofenac and naproxen exposure, as well as the induction of Cyp1a caused with diuron and naproxen but not with diclofenac, indicates differences between the compounds and underlines the metabolic activity of ZFE, which seems to be qualitatively similar to that of mammals.
In the present study, we predicted and investigated how these observations on gene expression, made under single-substance exposure, translate to effects under mixture exposure. Our objectives were to evaluate whether we can quantitatively and qualitatively predict the effects of a mixture from the effects of its components on the whole-transcriptome scale. Moreover, we tackled the question of whether we can trace the toxicogenomic effects of the components under a mixture exposure.
Materials and Methods
In this study, we used the previously published time-resolved toxicogenomic fingerprints obtained with the ZFE after exposure to diuron [Chemical Abstracts Service Registry Number (CAS RN): 330-54-1], diclofenac sodium salt (CAS RN: 15307-79-6), and naproxen sodium salt (CAS RN: 26159-34-2) (Schüttler et al. 2019) to predict the effects and component contributions in quality and quantity for a mixture of these compounds. The CTR-model parameters for all nodes of the previously published toxicogenomic fingerprints can be found in Excel Table S1.
Furthermore, we experimentally retrieved a concentration- and time-resolved toxicogenomic fingerprint of the mixture of these three compounds. All data analyses were performed in R (version 3.4.3; R Core Development Team). Functions used for analysis were compiled in the custom-built R package omixR (Schüttler 2020). All scripts and code used for the data analysis can be found in the Supplemental Material, “SupplementaryInformation.html.”
Zebrafish Husbandry
Wild-type adult zebrafish, originally received from OBI pet shop (Leipzig, Germany) were kept in fish tanks containing carbon-filtered tap water at 26°C under a 10 h:14 h dark:light cycle (30 fish per tank, male:female ). Eggs were collected approximately 1 h after light onset and inspected using a light microscope (Olympus SZx7-ILLT). Fertilized eggs were incubated at 26°C, whereas unfertilized eggs were discarded.
Mixture Preparation
The mixture contained three substances: diuron (CAS RN: 330-54-1; purity: 99.6%; batch: #SZBB265XV; Fluka), diclofenac sodium salt (CAS RN: 15307-79-6; purity: not available; batch: #BCBP9916V; Sigma), and naproxen sodium salt (CAS RN: 26159-34-2; purity: 98-102%; batch: #MKBV4690V; Sigma). The ratios of the three substances in the mixture were diuron 11%, diclofenac 2.6%, and naproxen 86.4% (the process of mixture design is described in more detail below in section “Mixture Design”). One day before the experiment, diclofenac and naproxen stock solutions were prepared in oxygenated ISO water (ISO 7346-3: calcium chloride dihydrate, magnesium sulfate heptahydrate, sodium bicarbonate, potassium chloride; pH 7.4, oxygenized) and methanol [CAS RN: 67-56-1; purity: high-performance liquid chromatography (LC) grade, J.T. Baker] and was applied as the solubilizing agent for the diuron stock solution preparation. On the day of exposure, mixture exposure solutions were prepared by diluting the respective stock solutions of mixture constituents to the desired concentration. Therefore, a specific amount of concentrated single-substance stock solution (Excel Table S2) was transferred into a volumetric flask. Subsequently, a specific amount of methanol was added to guarantee an equal fraction of solvent (0.1%) throughout all exposure solutions and, finally, the flask was filled with oxygenated ISO water up to its benchmark. Mixture exposure solutions were stirred, pH and oxygen content checked, and stored at room temperature until usage. pH (measured with pH-Meter 765 Calimatic; Knick) was adjusted to pH and remained at or above pH 6 throughout the test. Oxygen saturation (measured with Oxi 340 with probe CellOx 325; WTW) was maintained at a level between 80% (beginning of the test) and 60% (end of the test).
Diuron, naproxen, and diclofenac were quantified in the exposure media by LC coupled to high-resolution mass spectrometry (LC-HRMS) using an LTQ Orbitrap XL instrument (Thermo) with positive-mode electrospray ionization (ESI). The separation was carried out using a Thermo Ultimate 3000 LC system consisting of degasser, ternary pump, autosampler, and column oven. We used a reversed-phase gradient separation with water (Eluent A) and methanol (Eluent B), both containing 0.1% formic acid on a Kinetex C18 column (, particle size; Phenomenex) at a flow rate of . The gradient started at 20% of Eluent B, was held for 0.5 min, then increased to 100% of Eluent B in 5.5 min, and subsequently held at 100% Eluent B for 8 min before reequilibration to the initial conditions. The injection volume was and the column oven was kept at 40°C. The ESI voltage was set to , the heater temperature to 250°C, the sheath gas flow rate to 20 a.u., and the auxiliary gas flow rate to 5 a.u. The LTQ Orbitrap was operated in full-scan mode (m/z 80–600) at a nominal resolving power of 30,000 (referenced to m/z 400). Calibration standards were prepared matrix-matched in ISO water (calibration range ) and the samples were diluted in ISO water to match this range. Quantification was done against the internal standards isoproturon-d6 (for diuron; obtained from CDN Isotopes) and diclofenac-d4 (for diclofenac and naproxen; obtained from CDN Isotopes) using the QuanBrowser of the Xcalibur software (Thermo). The internal standards were added as a mixture in methanol:water 50:50 to the samples to obtain a final concentration of .
Exposure of Zebrafish Embryos
Our earlier findings (Schüttler et al. 2019) revealed that compound-specific, mode-of-action–related effects occur together with unspecific, development-related effects. Both depend on the embryonic stage because molecular targets or respective cell types need to be present to be affected. Particularly, in a developing system, such as the ZFE, toxicity is dependent not only on exposure duration but also on the exposure start point, age, and developmental stage of the embryo [addressed in more detail by Jakobs et al. (2020)]. Therefore, we started exposures of ZFEs to the above-described mixture solution on purpose at 24 h postfertilization (hpf) to avoid many unspecific effects that can be expected when disturbing the first hours of development (Kimmel et al. 1995). The test used in our experiments can be considered to be a ZFE toxicity test because the zebrafish larvae we used were never older than 96 hpf. This test is considered to be a nonanimal test alternative to the adult acute fish toxicity test (Scholz et al. 2008) and the fish early life stage test (Scholz et al. 2018).
Determination of Lethality
Exposure concentrations for the transcriptome analyses were anchored to lethal effects obtained after 72 h postexposure (hpe). To determine lethal effects, six technical replicates were used for controls and three for mixture treatments. Each replicate contained three embryos that were exposed to of control or diluted mixture exposure solution, respectively, and incubated for the desired exposure time in gas chromatography (GC) vials (VWR International), closed with an aluminum lid and an aluminum-coated septum (Supelco Analytical). Effects were observed with a light microscope (Olympus SZx7-ILLT). Three independent experiments were performed with a broad range of dilutions to get complete dose–response curves with 0–100% lethality.
Lethal effects were modeled similar to those proposed by Scholze et al. (2001) with two different regression models (logit and weibull; Excel Table S3) using a maximum-likelihood approach [R package bbme (Bolker and R Development Core Team 2017)]. The best-fitting model was selected based on the Akaike information criterion (AIC). From these curves, the and were determined, and a dilution factor was calculated to determine the five concentrations applied for the microarray analysis according to Equations 1 and 2. More details on this can be found in the paper by Schüttler et al. (2019).
| (1) |
| (2) |
Transcriptome Experiment
For the transcriptome experiment, 20 embryos per replicate were transferred to two GC vials at 24 hpf. A volume of of exposure medium was added to each vial, and the vials were sealed and incubated in a shaking climate chamber (climate chamber: Vötsch 1514, Vötsch Industrietechnik GmbH (Balingen-Frommern); Edmund Bühler SM-30, setting: 26°C, 75 rpm, 12 h:12 h light:dark) until sampling. For our experiment, we chose a dense sampling design probing increasing concentrations and consecutive sampling times. Such a design allows decreasing the number of replicates (Sefer et al. 2016). Embryos were exposed to a range of five different concentrations of the mixture (, , , , and ), and RNA was extracted at 3, 6, 12, 24, 48, and 72 hpe.
RNA Extraction and Microarray Analysis
Two vials of ZFE (20 ZFE in total) were harvested per replicate. We took two replicates for treatments and three for controls at each observation time point. ZFE were transferred into Eppendorf tubes and stored at after the addition of Trizol and homogenization using a T10 basic Ultra-Turrax (IKA, Werke GmbH & Co.) for 20 s at maximum speed. RNA isolation was performed using a pipetting robot (Microlab Star, Hamilton Life Science Robotics) following the manual provided for Total RNA Extraction Kit MagMAX 96 for microarrays and conducted in a 96-well plate. The quality of isolated RNA was assessed using a Bioanalyzer (Agilent 2100 Technologies) and the Agilent RNA 6000 Nano Kit. RNA samples were used for further processing if RIN values derived from ribosomal RNA absorption adopted values and calculated concentrations exceeded . We selected two replicates for all controls, and for the highest exposure concentration () at all time points, as well as for the longest exposure duration (72 h) and all concentrations, respectively. For all other time points and concentrations, the microarray was measured for one replicate. All RNA samples were diluted to a concentration level of ( in total) by the addition of RNAse free water. ( RNA) were used as the starting amount of RNA for the spike mix preparation.
Transcript abundance was measured with microarray analysis using Oaklabs ArrayXS Zebrafish microarray slides (XS-200,104, Oaklabs; National Center for Biotechnology Information Gene Expression Omnibus platform accession: GPL19785). Microarray experiments were performed using the Agilent Low Input Quick Amp WT Labeling Kit according to the Agilent One-Color Microarray-Based Exon Analysis Protocol (version 2.0; August 2015). This protocol included the introduction of spike-in RNA, RNA transcription, and amplification into complementary DNA (cDNA), and cDNA transcription and amplification into cRNA with simultaneous incorporation of Cy3 (fluorescently labeled cytidine nucleotide). The cRNA was fragmented and hybridized to Oaklabs ArrayXS Zebrafish microarray slides using the Agilent hybridization kit and protocol as well as Agilent hybridization oven and chambers. Subsequently, microarray slides were washed and scanned with the Agilent High-Resolution Microarray Scanner according to the Agilent protocol. Intensity values were extracted from captured images using Agilent Feature Extraction software (version 11.5.1.1).
Dynamic Toxicogenomic Fingerprints of the Components and the Mixture
Dynamic toxicogenomic fingerprints from the compounds diuron, diclofenac, and naproxen were taken from Schüttler et al. (2019) (Excel Table S1). Measured transcriptome data from the mixture exposure experiment were mapped on the toxicogenomic universe. This included quality control, normalization using the cyclic loess method (Bolstad et al. 2003), , and normalization of expression level against the control of the respective time point controls. Finally, all transcripts were clustered into 3,600 nodes of a previously trained SOM. Details of this process are described by Schüttler et al. (2019). Next, the CTR-model was fitted to the obtained responses under mixture exposure to retrieve model parameter values and a dynamic toxicogenomic fingerprint of the mixture as follows:
| (3) |
where corresponds to the maximum fold-change of the respective node across all conditions, is the maximum sensitivity () of the node, is the point in time with maximum sensitivity, and represents a measure of the duration of the sensitivity interval. For parameter estimation, the R implementation of the algorithm shuffled complex evolution (described by Duan et al. 1993) in the R package hydromad (Andrews et al. 2011) was applied. Further details regarding parameter estimation can be found in the paper by Schüttler et al. (2019). To check for the quality of model fits the AIC-weights were calculated in comparison with a null-model and a spline (see also the “Results” section and Figure S9).
Determination of Significantly Affected Nodes
Significantly affected nodes were determined by comparing the 95% confidence interval (CI) for the regression model fits with the 2.5% and 97.5% quantiles of control measurements. Nodes showing a sum of differences between these confidence bands above or below zero were identified as significantly affected (Schüttler et al. 2019).
Mixture Effect Prediction—Concentration Addition
Mixture effects were predicted for each node in the toxicogenomic universe applying the mixture concept of CA. The maximum s per node obtained in the experiments with the individual chemicals (Excel Table S1) were summed to determine the expected direction of regulation under mixture exposure for each node. A positive sum was taken as an indication for up-regulation, a negative sum for down-regulation. The highest (for up-regulation) or lowest (for down-regulation) measured in the single-substance exposures was set as maximum for the CTR-model. Subsequently, the CTR-model parameters were retrieved for each node and compound for the expected mixture direction. If an effect induced by a single substance was of the maximum effect or an AIC-weight for the fitted model was , we assumed no effect for this substance on the respective node. For mixture prediction, a bootstrap approach was employed and errors were sampled from an error distribution fitted for the single substances (). For each sampled error combination, the CA was calculated based on the formula for multicomponent mixtures as described by Faust et al. (2001):
| (4) |
where represents the effect concentration for effect of the mixture, the effect concentration of substance inducing the same effect as the mixture on its own, and the proportion of substance in the mixture. After calculating the concentration–response for the complete range of effect concentrations for 100 bootstrap samples, the CTR-model was fitted to the sampled predictions. By defining a threshold for a significant effect based on the control variation, we could derive qualitative statements about significantly affected nodes (see also section “Prediction of Significantly Affected Nodes” for more details).
Mixture Effect Prediction—Independent Action
Given that the prediction of independent action (IA) generally allows incorporating up- and down-regulation, model parameters for each node for the best-fitting direction were retrieved from the data of single-compound exposures (Excel Table S4). As for CA, IA mixture predictions were calculated using a bootstrap approach by sampling errors from the substance error distributions (). If an effect was of the maximum effect or an AIC-weight for the fitted model was , we assumed no effect for this substance on the respective node. Mixture expectations were calculated for up- and down-regulation separately, using the basic version of IA found in the paper by Bliss (1939), afterward, the effects for up- and down-regulation were summed, resulting in the following equation:
| (5) |
where is the effect at a given mixture concentration, is the induced by compound , is the maximum up-regulation across all conditions, and is the maximum down-regulation across all conditions. Finally, the CTR-model was fitted to the sampled predictions.
Mixture Effect Prediction—Effect Addition
The prediction applying the concept of effect addition (EA), which is based on the assumption of linear dependence of effect size on concentration (Equation 6), was conducted as for IA, only that effects were added instead of multiplying fractional effects:
| (6) |
where is the induced by the mixture, is the concentration of the mixture, and the proportion of component in the mixture.
Mixture Effect Prediction—Boolean Assumption
A concept that assumes that each node that is affected by one of the components (in any concentration or at any point in time), will be affected in the mixture, we call the Boolean assumption (BA) concept. It is based on a qualitative prediction on which nodes in the toxicogenomic universe will be affected by the mixture exposure according to the following equation:
| (7) |
where is a Boolean variable describing whether a node is affected by the mixture exposure or not; is a Boolean variable describing whether a node is affected by the nth mixture component (irrespective of exposure conditions).
In addition, in this study, dose-scaling of the Boolean assumption was possible. For this, we extrapolated the single-substance effects to the concentrations and time points applied in the mixture experiment using the CTR-model. This allowed us to formulate dose-scaled Boolean assumptions for the experiment performed in this study.
Prediction of Significantly Affected Nodes
To arrive at qualitative expectations about which and how many nodes are significantly affected based on our predictions, we identified the 2.5%/97.5% quantile of control measurements from the individual compound experiments for each node and point in time. We compared these with the respective 95% CI of CA, IA, or EA predictions retrieved from a bootstrapping approach and summed the differences between the confidence bands for each concentration, calling the result sum(CI) (Figure S1). If the sum(CI) was different than 0, we defined this node as being predicted to be significantly affected under the respective mixture exposure.
Evaluation of Mixture Predictions
Mixture predictions were evaluated using different metrics. First, the prediction deviation ratios (PDR) of the fitted parameter values for and were analyzed. The PDR was retrieved according to the following equation:
| (8) |
CTR-model fits were, furthermore, visually inspected for all nodes either predicted (with CA) or observed as significantly affected with the mixture exposure. Based on this, we defined 10 categories for reasons for deviations between measurements and predictions (plus one category for matching mixture predictions; Excel Table S5) and assigned each node to these categories.
In addition, the areas under the curve (AUC) of the predicted and observed sensitivity functions S(t) (cf. Equation 1), were retrieved. The overlap between the predicted and observed AUCs served as a measure for predictivity (see also the “Results” section and Figure S17A).
To estimate the accuracy of the qualitative mixture predictions obtained with the different mixture concepts, we calculated the -score, which is the harmonic mean of precision and recall of a model (Zhang and Zhang 2009). The equation for calculating the -score is summarized below:
| (9) |
Calculation of Components Contributions
The expected contributions of the individual mixture components to the effect of the mixture can be derived based on the known effects of the individual substances. The expected component contributions depend on the applied mixture concept and were calculated for each node of the toxicogenomic universe. For the concept of CA, we calculated effect units (EUs) analog to toxic units (Sprague 1970). Because component contributions are dependent on the effect concentration and the point in time in our study, we took the integral over time of the EUs for . This is analogous to the integral of the (scaled) sensitivity over time:
| (10) |
where represents the relative EU for component , the sensitivity function for component , and the expected sensitivity function for the mixture based on CA. The corresponding equations for the IA concept (Equation 11) and EA concept (Equation 12) are shown below.
| (11) |
| (12) |
Mixture Design
The general setup of our study should have worked with any selected mixture proportion. Therefore, we only briefly elaborate on the mixture design and the chosen mixture proportions as this is not trivial and not an essential key point for this study. The three-component mixture in this study was designed in a way that combined effects were expected. This means that more than one compound should contribute to the response of a node under the mixture exposure for as many nodes as possible. To determine the ratio at which we could expect distinct combined effects, we first predicted the expected effects for an array of different mixture ratios. For this, we set up a matrix of different mixture proportions (Excel Table S6), roughly aligned along the concentration ranges between and of each component to ensure contributions of each compound to the mixture exposure. Then, for each proportion in the matrix, we predicted mixture effects for lethality. This was based on the observed lethal effects for the single compounds (Excel Tables S7 and S8) and a CA-assumption (Equation 4). Based on the predicted lethal mixture effects, we derived design concentrations for the hypothetical mixture exposure. This included five dilutions from to according to Equations 1 and 2. For these hypothetical concentrations and the exposure durations 3, 6, 12, 24, 48, and 72 h, we predicted the toxicogenomic effects across the whole toxicogenomic universe (using the CA concept; Equation 4). Next, for each node, we calculated and summed the difference between the effect predicted with CA and the effect of the most effective mixture component for each concentration and point in time. Each node with a sum larger than 1.5 was counted as a node showing a combined effect. This information helped us to select a mixture ratio with a high expected combined effect (cf. Excel Table S6 and Figure S2).
Finally, the mixture composition selected for this study was diuron 11%, diclofenac 2.6%, and naproxen 86.4%. For measuring the toxicogenomic fingerprint of the mixture, we chose a diagonal mixture design (Berenbaum 1981), that is, a mixture of constant ratio was applied in several dilutions. For this mixture, we determined the concentration–response relationship for lethality and derived exposure concentrations for the microarray experiment from to as described above and by Schüttler et al. (2019). The final mixture concentrations were 43.1, 61.01, 86.49, 102.93, and for C1 () to C5 (), respectively.
Results
Applying the CA concept, we predicted the concentration- and time-resolved effect of a mixture based on the previously published fingerprints of the three mixture components: diuron, diclofenac, and naproxen (Schüttler et al. 2019). This was performed for all nodes in the toxicogenomic universe, which contains all genes of the zebrafish transcriptome (Schüttler et al. 2019). We experimentally measured the effects of the mixture and compared expected and observed mixture responses and evaluated hypotheses on similar and dissimilar joint actions of the mixture components.
Using the CA concept requires extrapolation and scaling of the concentration–effect relationships for responses obtained with individual compound exposures (Berenbaum 1985). It allows predictions for a multitude of potential mixture exposure scenarios regarding compound concentrations and proportions in the mixture. The mixture proportions selected for this study were optimized in a way that effects were expected for all three substances in the mixture and a high total combined effect in terms of gene expression (Excel Table S6 and Figure S2). This means that more than one compound should contribute to the response of a node under the mixture exposure. Furthermore, the expected effect should be substantially stronger than the effect induced by the most effective single compound (Figure S2A). Only effects on gene expression were considered for optimizing the mixture proportions; lethal effects were not taken into account. This led to the design of the mixture containing 11% diuron, 2.6% diclofenac, and 86.4% naproxen. For exposure experiments, we applied dilutions of this mixture while keeping the mixture proportions constant [diagonal mixture design, cf. Berenbaum (1981)]. To define the dilution range for exposure in the toxicogenomic experiments, lethal concentrations of the designed mixture were determined (Figure S3). The toxicity of the mixture increased with exposure duration and the determined after 72 hpe was (measured compound concentrations in the mixture exposure solution are provided in Excel Table S2). The mixture was then investigated in five dilutions between and (Equations 1 and 2) in the toxicogenomic experiment. This design allowed us to compare the CA predictions to corresponding measurements for the whole ZFE transcriptome.
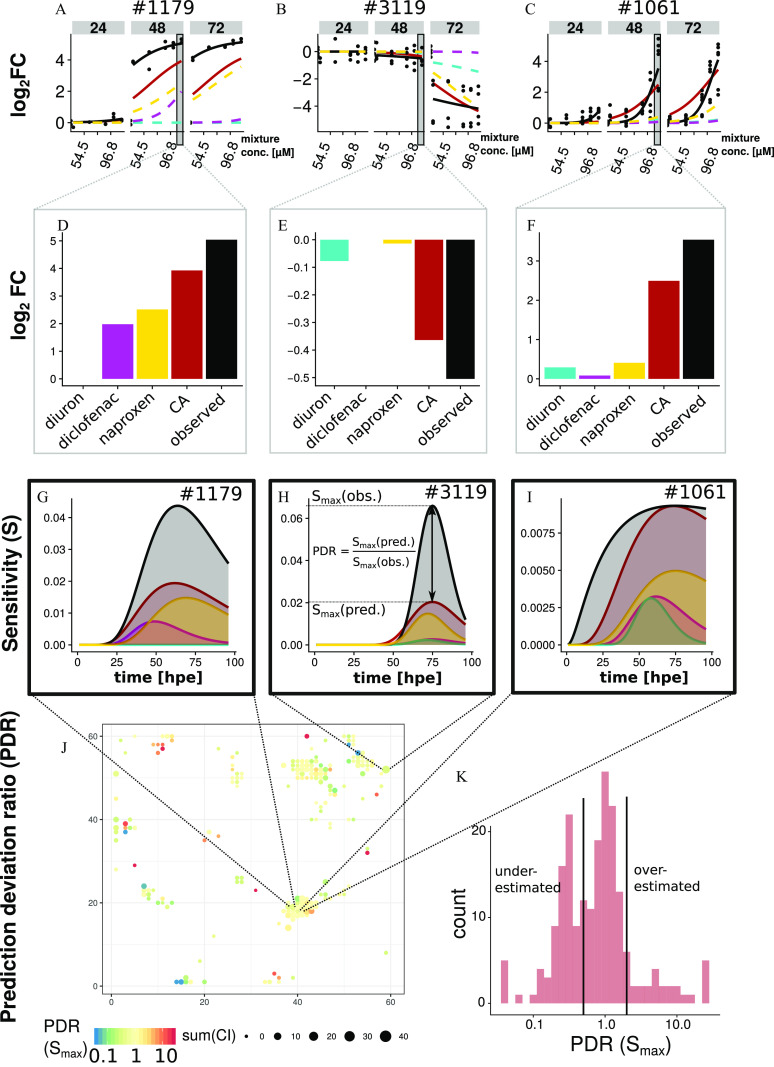
The result of calculating mixture predictions with the CA concept for the nodes in the toxicogenomic universe is exemplarily shown for Node #1061 (Figure 1). The figure is structured in a way that it also illustrates our approach. The example node contains four genes coding for small proteins related to guanosine triphosphate (GTP)-binding or GTPase activity. The logarithmic fold-change, relative to control conditions () after single-compound exposure, was described by a CTR-model (Figure 1A and Equation 3) (Schüttler et al. 2019). The CTR-model provided compound-specific sensitivity curves (Figure 1B) with sensitivity denominating the inverse value of a time-dependent value, that is, the concentration at half-maximum . The node-specific maximum was approximated by the maximum across all concentrations and durations of the single-substance exposures. The sensitivity curves for each of the three substances were scaled (i.e., multiplied) by the compound proportion in the mixture (Figure 1C) and then added up to arrive at the mixture effect prediction (Figure 1E). Subsequently, the CTR-model was fitted to the mixture predictions (of the full range of effects concentrations) to retrieve predicted parameter values for each node (Figure 1E). While provides information about the concentration dependence of the node response, provides information about time dependence by indicating the point in time when the sensitivity reaches . For the example node, we predicted an value of 0.009 and a value of 73.8 hpe for the mixture response. The calculations, illustrated here for Node #1061, were performed individually for all nodes in the toxicogenomic universe. Eventually, the fitted values for those nodes, which were predicted to be significantly affected (, see also Figure S1), were mapped on the toxicogenomic universe (Figure 1D and Excel Table S9).
Figure 1.
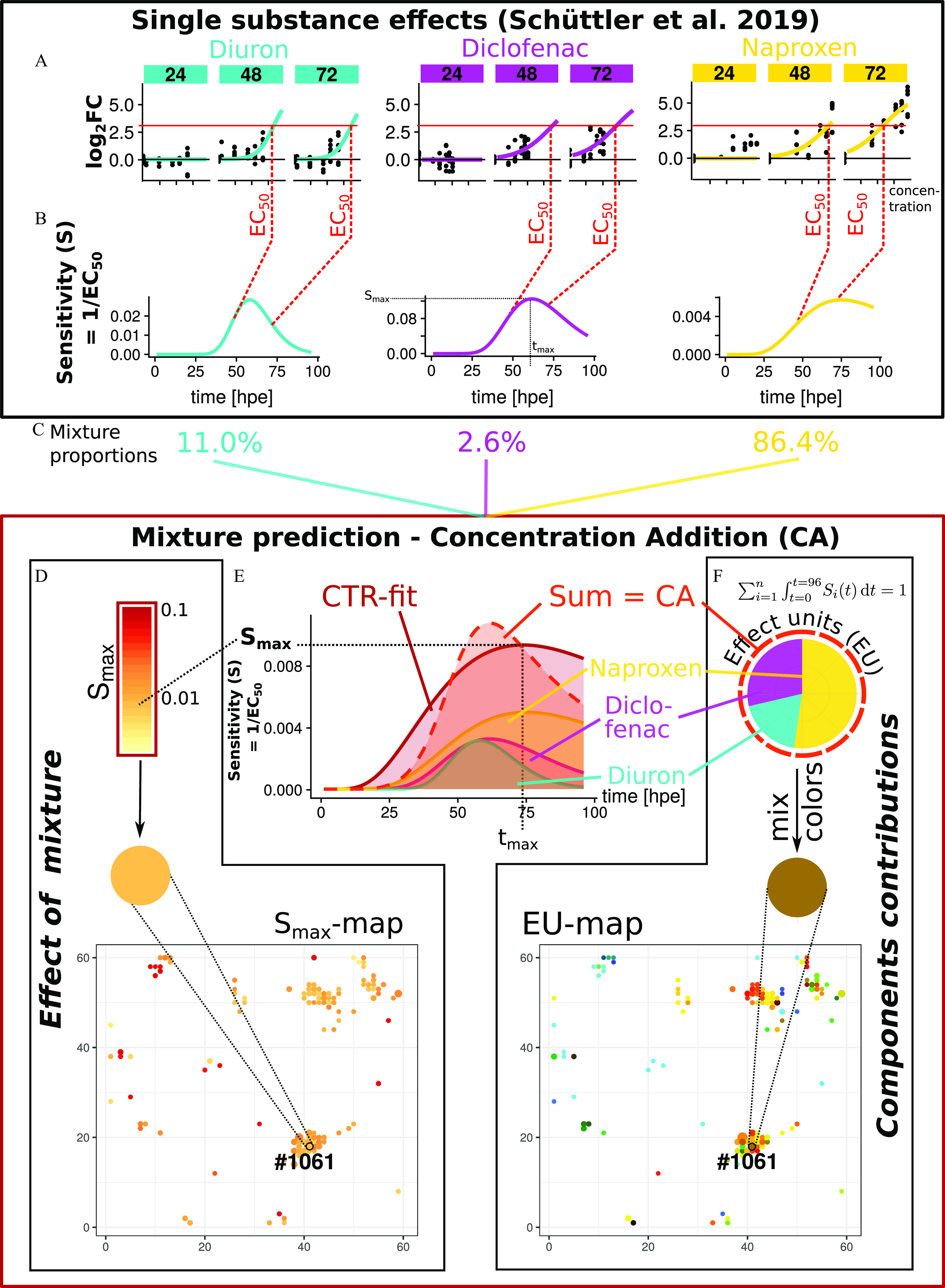
Mixture effect prediction (CA concept and results for Node #1061). (A) Effect concentrations for the half-maximum response () were derived from the changes () compared with the controls for each node and time point (exemplarily shown for three of six time points) based on previous data from individual compound exposures; (B) sensitivity curves over time derived from values; (C) sensitivities were scaled according to mixture proportions; and (E) predicted sensitivity (S) for the mixture exposure resulted from the sum of scaled sensitivities obtained for the individual mixture components according to the CA concept. The maximum sensitivity value () and the time point [; hour postexposure (hpe)], where is reached, were derived by fitting a concentration- and time-dependent response model (CTR-model) on the mixture prediction data. (D) The map shows the predicted values for all nodes predicted to be significantly affected under mixture exposure. (D) Effect of mixture and (F) Component contributions to the predicted total effect [relative effect units (EUs)] were derived from the ratios of the areas under the sensitivity curves. EU values are presented in CMYK-colors and subtractive color theory (e.g., the sum of yellow and magenta gives red, the sum of yellow and cyan gives green). The EU-map contains EU values for all nodes predicted to be significantly affected under mixture exposure. Axis numbers in (D) and (F) give the x and y coordinates of nodes in the toxicogenomic universe. All data are provided in Excel Table S9. Note: CA, concentration addition; CMYK, cyan, magenta, yellow, and black; CTR, concentration- and time-dependent response.
The relative contributions of the individual substances to the effect of the mixture (i.e., the EU) were derived from the area under the scaled sensitivity curves for each component (Equation 10). They were mapped on the toxicogenomic universe for all nodes predicted to be affected under mixture exposure with CA (Figure 1E,F). For the example Node #1061, we expected that all three compounds would contribute to the effect (, , ). Here, we therefore expected a combined effect due to the mixture exposure.
Similar to Figure 1 and the CA concept, the results for the predictions based on the other concepts are provided in the Supplemental Material [number of nodes predicted to be significantly affected due to the mixture exposure: IA concept: 20 nodes (Figure S4); EA concept: 13 nodes (Figure S5); BA concept: 432 nodes (Figure S6); all data in Excel Table S9].
Finally, we experimentally measured the concentration- and time-resolved toxicogenomic fingerprint of the mixture exposure, for which the effects were predicted before. We mapped the responses on the toxicogenomic universe and fitted the CTR-model (Equation 3) to each node as described by Schüttler et al. (2019). The results are again exemplarily shown for Node #1061 (Figure S7A,B). Model parameters, such as , were calculated and mapped for all significantly affected nodes (; Excel Table S9) on the toxicogenomic universe (Figure S7C,D). For the example node, we received an value of 0.009 and a value of 75 hpe. The whole toxicogenomic fingerprint of the mixture can be accessed using our previously published toxicogenomic fingerprint browser (http://webapp.ufz.de/itox/tfpbrowser). The results of the measurement and the different predictions are provided in Excel Table S9.
Prediction of Whole-Transcriptome Mixture Response Using the CA Concept
Based on the results from the CA predictions and measurements, we compared the expected and observed mixture effects. The projections of predicted (Figure 2A,C) and observed (Figure 2B,D) parameter values for and on the toxicogenomic universe demonstrate a striking similarity, though some differences also become apparent. The distributions of and values as well as results on the model fits are shown in Figures S8 and S9, respectively.
Figure 2.
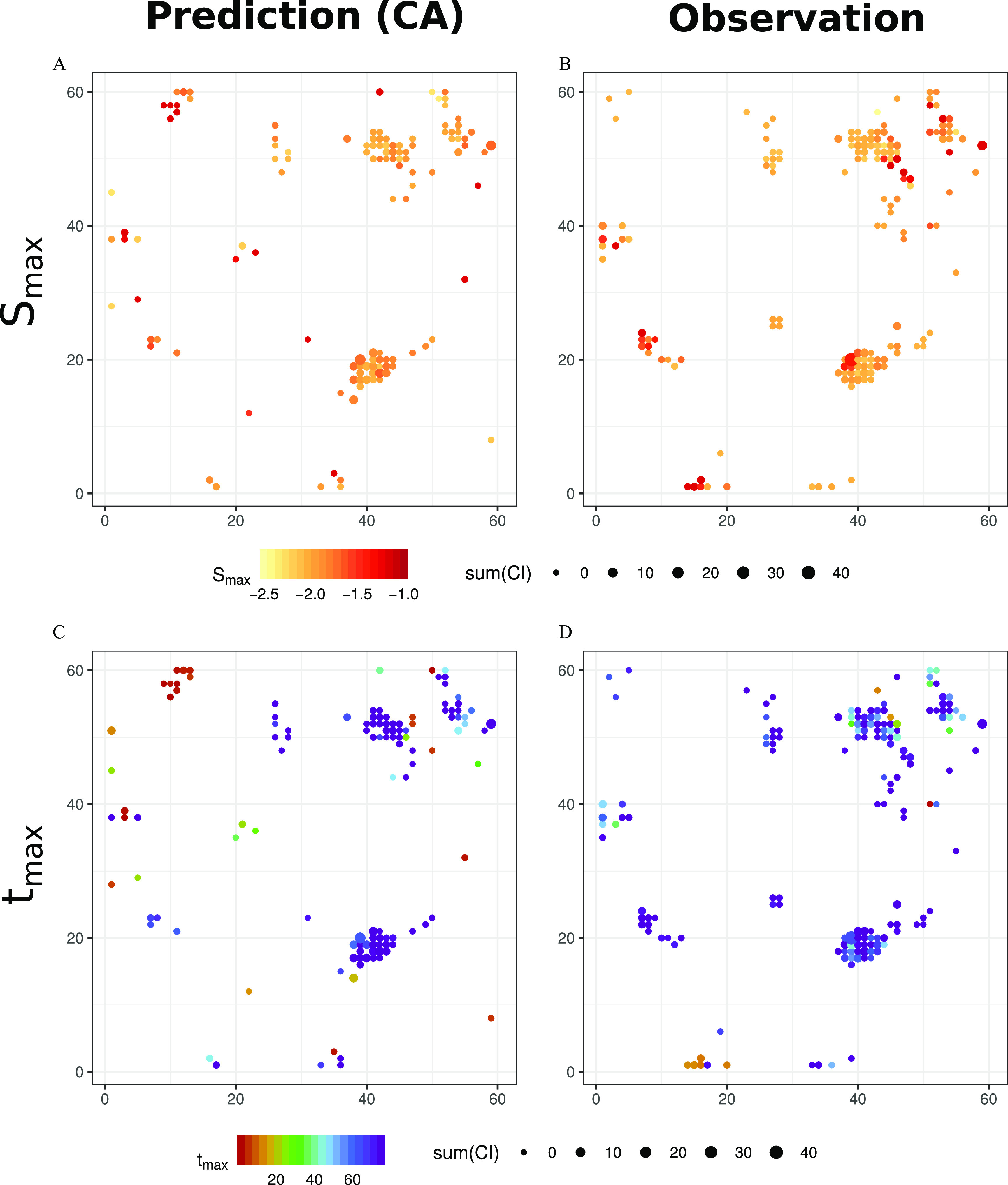
Predicted and observed toxicogenomic fingerprints. (A,C) Predicted with the concept of concentration addition (CA), and (B,D) observed values for maximum sensitivity () and time point of maximum sensitivity () (both parameters of the CTR-model), respectively, mapped for mixture-affected significant nodes on the toxicogenomic universe. Dot size represents significant effect size given by the sum of confidence interval differences between treatment and control [sum(CI)]. Data are available in Excel Table S9. Note: CTR, concentration- and time-dependent response.
Having applied the CA concept allowed us to analyze these similarities and differences in detail from different perspectives. In the following, we summarize which nodes in the toxicogenomic universe were affected. With this, we took a qualitative perspective by looking at the lists of nodes that were either predicted or observed to be significantly affected due to the mixture exposure. Next, we will report how the affected nodes responded in a concentration- and time-dependent manner, that is, we took a quantitative perspective on deviations between measured and predicted effects.
Qualitative perspective.
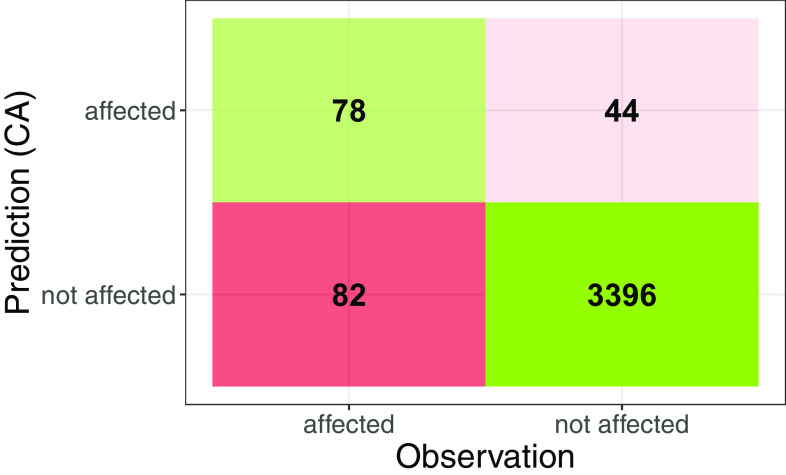
The common perspective on toxicogenomic effects is of a qualitative nature, for example, by providing and interpreting lists of differentially expressed genes. Here we compared lists of nodes that were predicted and measured to be significantly affected under the mixture exposure with CA. Applying a bootstrapping approach on the CA predictions, we expected 122 nodes to be significantly affected under the mixture exposure, and we observed 160 nodes to be significantly affected in the experiment (Excel Table S9). The overlap between expected and observed nodes was 78 (true-positives) (Figure 3). Eighty-two nodes were deemed false-negatives, meaning that they were not predicted to be affected due to the mixture exposure (according to the CA concept) but were observed experimentally to be significantly affected. Forty-four nodes were deemed to be false-positives, meaning that they were predicted to be affected according to the CA concept but not judged as significantly affected in the experiment. In total, 204 nodes were either observed or predicted to be affected due to the mixture exposure, whereas 3,396 nodes, the majority of the transcriptome, were not affected by the mixture as predicted (true-negatives). We further looked into quantitative comparisons and deviations from predictions for these 204 nodes.
Figure 3.
Qualitative comparison of prediction and observation: Confusion matrix for qualitative expectation regarding affected nodes in the zebrafish embryo toxicogenomic universe due to exposure to a mixture of diuron, diclofenac, and naproxen between 24 and 96 h postfertilization. Predictions were based on the concept of concentration addition. Significantly affected nodes were determined by comparing the 95% CI for the regression model fits with the 2.5% and 97.5% quantiles of control measurements. Nodes showing a sum of differences between these curves above or below zero were identified as significantly affected. For the prediction of affected nodes due to the mixture exposure the 2.5–97.5% quantile of control measurements from the individual compound experiments was compared with the 95% CI of CA predictions (see also the “Materials and Methods” section and Excel Table S9). Note: CI, confidence interval; CA, concentration addition.
Quantitative perspective.
Detailed quantitative analysis and comparison of predicted and measured effects were possible due to the applied concentration- and time-resolved approach and the CTR-regression model that describes the respective responses using uniform parameters. We exemplarily show the results of predicted and observed effects for the top three affected nodes [according to sum(CI), Excel Table S9] under the mixture exposure determined in the experiment (Figure 4A–I).
Figure 4.
Quantitative comparison of prediction and observation. (A–C) Observed (black) and predicted mixture effects [ (CA)], and component contributions (, , ) as for three of six observed time points for the three nodes with largest difference to control [largest sum(CI)] in the mixture exposure experiment; (D–F) individual values obtained and predicted for 48 h postexposure (hpe) with highest concentration (); (G–I) sensitivity curves; (J) prediction deviation ratios (PDRs) for mapped on the toxicogenomic universe for all 204 nodes either predicted or observed to be affected; (K) distribution of PDRs for . As discussed in more detail below, under- and overestimation of cannot be treated as equivalent to observed synergisms or antagonisms because such assessments would require distinct criteria and discrimination from technical reasons for variation. Data are available in Excel Table S9.
The observed (Figure 4A, black dots, with the black line as model fit) were underestimated for Node #1179 by the CA prediction (Figure 4A, red line) whereas measurements were closer to the predictions in the other two nodes (#3119, #1061; Figure 4B,C). For all three nodes individual , for example, at 48 hpe and the highest exposure concentration of (Figure 4D–F), indicate a slight underestimation of the combined effect predicted using CA. Concentration- and time-dependent deviations from predictions become obvious when comparing the sensitivity curves (Figure 4G–I). To quantify this systematically, first, we calculated the percent overlap between areas under the predicted and observed sensitivity curves (Figure S8 A,B). These were 47%, 40%, and 78% for the top three Nodes #1179, #3119, #1061, respectively (Figure 4 and Excel Table S9). Second, we considered the distance between the observed and predicted model parameter values and (PDR) (Figure 4H). The PDRs for the model parameters for these three nodes were 0.44, 0.31, and 1.0 for and 0.97, 1.0, and 0.98 for , respectively. The PDR values of all 204 nodes (Excel Table S9), were mapped on the toxicogenomic universe (Figure 4J). The distribution of these values shows two peaks (Figure 4K). One peak represents a group of 104 nodes for which the prediction matched the observation within the range of . A second peak indicates a group of 69 nodes for which sensitivity was underestimated within the range of . The sensitivity was underestimated with a for 6 nodes, whereas it was overestimated for 25 nodes with . This analysis was also performed for the values that were well predicted for 162 of 204 nodes showing a PDR between 0.5 and 2 (cf. Figure S10B,D).
Deviations from predictions may have different reasons. To systematically consider these reasons, we classified the 204 nodes into different deviation categories (Excel Tables S5 and S10). Of the 100 nodes that showed a PDR for outside the range of , we could identify potential technical reasons for the deviations. These comprise, for example, that nodes were affected in two different directions, for example, over time or for different compounds (40 nodes; Excel Table S10, exemplarily shown for Node #876 in Figure S11). Deviations from predictions also occurred when the CTR-model could not correctly fit the calculated mixture expectation data (25 nodes). This was the case, for example, if a node showed sensitivity peaks at more than one point in time (15 nodes). Deviations from qualitative predictions occurred, for example, because of undue control variation in single-substance exposures (exemplarily shown for Node #34 in Figure S12) or when individual genes within a node were not coexpressed or showed very different effect amplitudes (18 nodes, exemplarily shown for Node #818 in Figure S13). Next to these deviations, explainable by technical limitations of the approach, we also found effects of the mixture for 18 of the 204 analyzed nodes that clearly did not resemble the effect that was previously observed for the single substances (Excel Table S10, exemplarily shown for Node #20 in Figure S14).
Evaluation of Other Mixture Concepts
Qualitative perspective.
A comparison of the list of the 160 observed nodes, affected due to the mixture exposure in our experiment, with those predicted with the other three above-described concepts (IA, EA, and BA) showed, that the Boolean assumption led to a large number of false-positive predictions (294 nodes, Figure S15A). In contrast, the concepts of effect addition and IA resulted in large numbers of falsely negative predicted nodes (148 and 147, respectively) because the overall numbers of predicted affected nodes were quite small with these two concepts, with 13 and 20 nodes, respectively (Figure S15B,C). Similar qualitative comparisons between numbers of observed and expected nodes could be performed for each time point and mixture concentration separately, based on the Boolean assumption, adapted for each concentration and time point tested. These comparisons showed differing qualities of predictions dependent on the concentration and time point (Figure S16A). We calculated the -score, an indicator balancing false-positive and false-negative predictions, for all concepts. Comparing these results revealed that overall the CA concept showed the highest -score (Figures S15D and S16B).
Quantitative perspective.
CTR-model parameters, such as and , could not only be calculated for the CA concept but also for the EA and IA concepts, respectively. The distributions of concept-dependent PDRs for and are shown in Figure S10C and S10D, respectively. All concepts showed similar distributions and high prediction accuracy for with PDRs of (Figure S10D). In comparison with the CA concept, for which we found 104 nodes with -value PDRs between 0.5 and 2, we found 97 nodes with the EA concept and 95 nodes with the IA concept with (Figure S10C).
Contributions of the Three Mixture Components to the Effects of the Mixture on the Transcriptome
Exposures to chemical mixtures can cause combined effects when more than one of the mixture components contribute to the overall effect. This is the case in the three examples in Figure 4 where naproxen (yellow) and diclofenac (magenta) were predicted to contribute to an effect under mixture exposure in Node #1179 (Figure 4G), and all three compounds were expected to contribute in Nodes #3119 (Figure 4H) and #1061 (Figure 4I), for example. In all of these cases, the predicted effect of the mixture in terms of or sensitivity was higher or longer than would have been expected by looking at any of the mixture components alone.
In some cases, combined effects may lead to situations where there is no measurable effect observed or expected for the single-compound exposure, but a significant effect is observed under mixture exposure. This was the case for some selected concentrations and points in time in our study. For example, based on the small or not observed effects for any single-compound exposure, no strong effect would have been expected at 48 hpe and the highest mixture exposure concentration for the Nodes #3119 (Figure 4E) and #1061 (Figure 4F), whereas a substantial effect was predicted with CA, which was even stronger in the experiment. The occurrence of such combined effects is called the something-from-nothing effect, referring to an effect under mixture exposure when the mixture components alone do not show a measurable effect (Silva et al. 2002; Walter et al. 2002).
To quantify to what extent each compound was expected to contribute to the effect of the mixture, relative EUs were derived from the areas under the sensitivity curves for each node (cf. Figure 1F). The EUs were color coded with the help of the cyan, magenta, yellow, and black (CMYK)–color model and subtractive color theory (e.g., the sum of yellow and magenta gives red, the sum of yellow and cyan gives green), and all nodes predicted to be affected sorted in a triangle plot according to the expected component contribution (Figure 5A). For some nodes, we predicted only one of the three mixture components to contribute to the effect of the mixture (single-origin effect). In this case, the relative EU () was larger than 0.9 for one of the components and the node was plotted in one of the three corners of the triangle. This was the case for 17 nodes driven by naproxen, 15 nodes driven by diuron, and no node driven by diclofenac. The other 90 nodes were predicted to be jointly affected by two or all three components ( for each component). Thus, we predicted the effect of the mixture on the whole transcriptome to emerge as a blend of single-origin and combined-affected nodes, with the majority of nodes (90 of 122) expected to show combined effects (Figure 5A).
Figure 5.
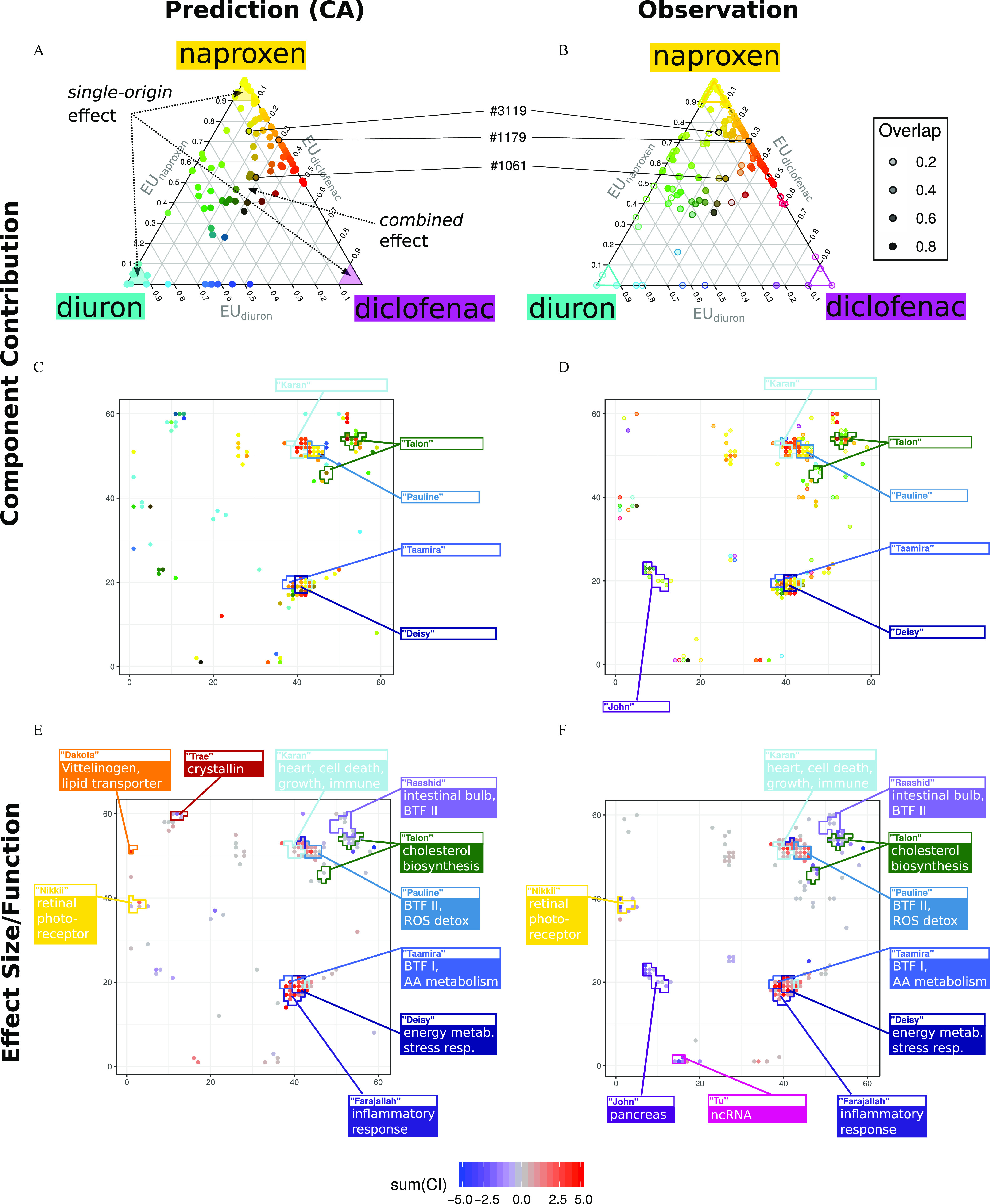
Component contributions and functional comparisons: Relative effect units (EUs) as representations for component contributions to the effect of the mixture for each of the (A,C) 122 CA-predicted nodes, and (B,D) 160 observed nodes; all nodes are colored according to the CMYK subtractive color code (e.g., the sum of yellow and magenta gives red, the sum of yellow and cyan gives green); opacity indicates prediction accuracy according to the overlap between predicted and observed sensitivity curves in (B) and (D). Summed effect size for (E) 122 predicted nodes, and (F) 160 observed nodes with the top 10 affected clusters and their summarized function, , (values clipped above 5 and below ). Data on EUs are provided in Excel Table S9. Cluster names and functional enrichment of clusters are according to Schüttler et al. 2019. Note: AA, arachidonic acid; BTF I/II, biotransformation phase I/II; CA, concentration addition; CMYK, cyan, magenta, yellow, and black; energy metab., energy metabolism; ncRNA, noncoding RNA; ROS detox, detoxification of reactive oxygen species; stress resp., stress response.
The prediction of compound contributions could not be easily experimentally validated, given that the observed effects under mixture exposure did not come with a name tag. However, we could plot the experimentally observed mixture-affected nodes on the triangle too and color them according to the compound contributions calculated with CA, whereas the overall prediction accuracy for each node (i.e., the overlap between prediction and observation, cf. Figure S17A) is indicated by the opacity (Figure 5B). We found, for example, that experimentally determined effects on some nodes are potentially driven by diclofenac but were not predicted by CA. In addition, we analyzed whether the expected fraction and type of compound contribution influenced the quality of predictions, but we did not find a clear association or correlation between both (Figure S17C). The calculated EUs for all 204 nodes predicted or observed to be affected are provided in Excel Table S9.
By mapping calculated compound contributions on the toxicogenomic universe, it can be illustrated how the previously obtained single-compound fingerprints are expected to reoccur under mixture exposure (cf. Figures 1F and 5C), and how the observed effects resemble these expectations (Figure 5D). For example, certain nodes predicted to be affected under mixture exposure mainly due to naproxen (Figure 5C, top middle, yellow nodes) reoccurred in Figure 5D, whereas other nodes, predicted to be affected by the mixture but solely driven by diuron (Figure 5C, top left area, cyan nodes) did not reoccur in Figure 5D, that is, these nodes were not significantly affected in the mixture experiment. Furthermore, evaluating the effects of the mixture can provide information on the potential individual contributions of each of the individual chemicals to unexpected mixture effects. For example, there are four nodes in the middle of the fingerprint that were observed but not predicted to be affected by the mixture (Figure 5C,D). These nodes might potentially be affected by the different mixture components respectively, as indicated by the different colors (Figure 5D).
Analysis of Joint Action for Compounds with Similar and Dissimilar Modes of Action
The 3,600 nodes of the ZFE toxicogenomic universe were assigned to 118 clusters (Schüttler et al. 2019). Each cluster was assigned to a random name and analyzed for enriched functional terms [contained in the databases GO, Reactome, ZFIN, and Interpro (Schüttler et al. 2019), cf. also Excel Table S11] in our previous study to support the functional interpretation of obtained toxicogenomic fingerprints due to chemical exposures. Here, we compared the predicted and observed effects of the mixture exposure based on the predefined clusters in the toxicogenomic universe (Figure 5E,F). Almost all clusters expected to be most prominently affected (based on the highest proportion of nodes predicted to be significantly affected with CA) could be found among those top clusters where the observed affected nodes were assigned to (Figure 5E,F and Excel Table S11). For example, 12 nodes that were predicted to show an effect due to the mixture exposure are assigned to cluster Pauline. This cluster contained 13 nodes in total, comprising 56 genes, and was functionally enriched among others for Phase II–compound conjugation, glutathione, and detoxification of reactive oxygen species (Excel Table S11) (Schüttler et al. 2019). All 13 nodes were observed as significantly affected in our mixture experiment. Other clusters with strong overlap between prediction and observation were Deisy, Farajallah, Karan, Taamira, Talon, Nikkii, and Raashid. These clusters were functionally enriched for different terms related to immune and stress reactions (Deisy, Farajallah, and Karan), different metabolic processes (Taamira, Talon, and Raashid), and the retina (Nikkii) (Figure 5E,F and Excel Table S11). Differences among the lists of predicted and observed nodes were found for the clusters Trae and Dakota (enriched for crystallin and vitellogenin genes, respectively), which were expected but not observed to be significantly affected. Meanwhile, the cluster Tu was affected by the mixture exposure but was not predicted to be. It contains a set of noncoding RNAs with mostly unknown functions. Furthermore, cluster John, functionally enriched for pancreas genes, was much more affected than expected (Figure 5E,F and Excel Table S11).
The projection of the observed nodes with their respective calculated component contributions on the toxicogenomic universe showed that certain clusters were dominated by certain components (Figure 5C,D). Although a majority of effects observed in John and Talon were potentially caused by the joint action of naproxen and diuron, indicated by green-colored nodes (Figure 5C,D), the clusters Deisy, Taamira, and Karan were rather affected by the joint action of diclofenac and naproxen, indicated by orange or red nodes (Figure 5C,D). Furthermore, a remarkable number of nodes in clusters Pauline and Deisy seem to be mainly affected by naproxen alone, indicated by the yellow color. Naproxen and diclofenac are known COX inhibitors and certain functional terms of the mentioned clusters could be related to an organismic reaction to COX inhibition (cf. Schüttler et al. 2019). The affected nodes of these clusters were all up-regulated due to the mixture exposure (red in Figure 5F). Although some of those nodes were induced more strongly than predicted, the majority was observed as predicted (cf. Figure 5E,F). This supports the hypothesis of concentration additivity for toxicogenomic effects induced by compounds with a similar mode of action.
The affected nodes of the clusters John and Talon were all down-regulated due to the mixture exposure (blue in Figure 5F). Down-regulation of these clusters was previously observed with diuron and naproxen (Schüttler et al. 2019). The joint action of the compounds in the mixture and down-regulation of 11 nodes in the two clusters was, therefore, predicted with CA. Eight of those nodes were observed to be down-regulated by the mixture exposure in the experiment, most of them more strongly than predicted (PDR for for 7 of 8 nodes). Another 11 nodes were observed to be down-regulated in these clusters. Those were not predicted (Tables S9 and S11). Although no common mode of action is known for diuron and naproxen, a more-than-additive combined effect was found on the transcriptome.
These results show that combined effects in a whole organism can occur on the molecular level not only with mixtures of similar-acting compounds but also with compounds expected to have dissimilar modes of action. The results also show that effects induced by different individual compounds can combine under mixture exposure and cause a multitude of responses at the whole transcriptome that can be predicted with the CA concept.
Discussion
Understanding the effects of intended or unintended exposure of organisms to chemical mixtures is key in designing combination therapies in pharmacy (Kuhn-Nentwig et al. 2019; Menden et al. 2019) and understanding potential exposure outcomes, such as carcinogenesis (Goodson et al. 2015) and toxic mechanisms threatening environmental and human health (Cedergreen 2014). Furthermore, it can facilitate chemical risk assessment and management (Bopp et al. 2019; Rotter et al. 2018). Predictions and interpretations of mixture effects on the whole-transcriptome scale were believed to be challenging due to expected interdependences between expressed transcripts and emergent effects based on interconnected processes in gene regulatory networks (Bluhm et al. 2014; Gong et al. 2008; Zhang et al. 2017). In the present study, we showed that the prediction and interpretation of whole-transcriptome mixture responses are possible and that predictable combined effects on the molecular level are not only caused by compounds with an anticipated similar mode of action.
Quantitative Modeling to Predict Combined Effects
Interpreting observable combined effects from mixture exposures requires the identification of differences between expectations and observations. Such expectations could be of qualitative and/or quantitative nature. Quantitative predictions of effects of chemical mixtures, based on their components, require knowledge about the concentration dependence of the same effect caused by the individual mixture components (Altenburger et al. 2013). This has been established in toxicology and pharmacology for single apical effects or selected receptor responses, for example, lethality or receptor activation (Kortenkamp et al. 2009). Meanwhile, it has been an unresolved challenge when simultaneously considering multiple effects, such as changes in transcript abundance across the whole transcriptome. This is not least because the specific dynamics of each signal depend on compound concentration and exposure time (Altenburger et al. 2012; Schüttler et al. 2019). Employing quantitative information on the effect dynamics and their variability provoked by individual mixture components, aggregating signals of a transcriptome into nodes, modeling these in their concentration and time dependence, and recruiting existing mixture concepts, we were able to formulate quantitative and qualitative expectations about mixture effects.
Using the developed workflow for modeling transcriptomic mixture responses, we were able to predict the occurrence of so-called something-from-nothing effects (Silva et al. 2002) that have previously been described for apical effects in unicellular organisms and cell cultures (Faust et al. 2001; Silva et al. 2002; Walter et al. 2002). Something-from-nothing effects designate responses that are observed only under mixture exposure, but not for any of the mixture components individually. The occurrence of this type of combined effect is coherent with the dilution principle underlying the concept of CA whereby any low concentration of a mixture component contributes to a combined effect (Faust et al. 2001). If this type of combination effect occurs for multiple mixture exposures, it implies a major challenge for chemical risk assessment, which currently focuses on assessing risks compound by compound (Drakvik et al. 2020; Escher et al. 2020).
CA Compared with Other Mixture Concepts
The availability of curated chemical–gene/protein–pathway interaction data, such as through the comparative toxicogenomics database (CTD) (Davis et al. 2021), have led to the idea to derive toxicogenomic mixture response expectations from such information (Berninger et al. 2019; Bradley et al. 2017; Schroeder et al. 2016). This concept, which we call BA, assumes that all signals detected in any of the single-compound exposures—regardless of exposure concentration or duration—reoccur under mixture exposure. This concept is applied in cases where only qualitative information about the effects of mixture components is available (e.g., lists of affected genes). Applying this concept, we found, not surprisingly, the highest number of false-positive and the lowest number of false-negative responses compared with the other concept-based predictions. This stresses our hypothesis that interpreting mixture responses at the transcriptome level requires a dose-scaled point of reference.
When quantitatively evaluating toxicogenomic mixture effects, one approach could be to add up the effect sizes (e.g., ) observed for single-compound exposures to arrive at an effect expectation for the mixture exposure, which we called EA. EA builds on the assumption of a linear relationship between concentration and effects. This notion has been challenged on theoretical grounds (Berenbaum 1981). In addition, combined effects from very low-effect concentrations are not to be expected but have been demonstrated in experimental mixture studies for various compounds using cells and unicellular organisms (Faust et al. 2001; Silva et al. 2002; Walter et al. 2002) and also for higher organisms, such as rats, for example, for endocrine disruptors (Hass et al. 2007). In line with this, the results of our study also demonstrate examples of evident nonlinear transcriptomic and low-dose responses and a substantial underestimation of the number of observed mixture responses with EA (cf. Excel Table S9 and Figures S5 and S15).
From a probability perspective, one may assume that multiple transcript responses under mixture exposure follow the principle of independence, which is labeled as IA in the mixture toxicology arena (Altenburger et al. 2012). IA accommodates for nonlinear dose–response relationships. In a whole-transcriptome study, Labib et al. (2017) exposed mice lung tissues to eight different PAHs and their mixtures. For six cancer-related pathways, the concept of IA was judged to most accurately predict pathway perturbations. Earlier, Dardenne et al. (2008) reported a good predictivity from the IA concept for binary mixture effects on stress gene reporter assays. In both studies, however, the component contributions were not explicitly evaluated; therefore, the extent of actual combined effects compared with effects that could be explained by single components remains unclear. In our study, we also found quantitative reasonable predictivity, whereas the number of responding nodes was substantially underestimated, similar as with EA (cf. Figures S4 and S15). When considering the effects from mixtures at low-effect concentrations, a challenging requirement for IA lies in the need to estimate low-effect levels accurately (Kortenkamp et al. 2007).
In mixture pharmacology, the concept of CA has been formulated as yet another reasonable concept to anticipate combined effects (Altenburger et al. 2012; Greco et al. 1995). It follows the dilution rule by assuming that one compound can be replaced by another compound in proportion to its individual effect concentration to achieve the same effect. In the present study, we found the concept of CA to be the most accurate for predicting mixture effects in comparison with the aforementioned concepts. For example, for this concept we found the prediction deviation ratio to fall within a 2-fold deviation for 50% of the nodes concerning the maximum sensitivity () and for 79% for the time of maximum response (). We see these findings as a proof of concept for the usefulness of our prediction approach. A reanalysis of apical responses in aquatic organisms found a 2-fold deviation range for and values describing 88% of all observed binary mixture effects in 207 experiments (Belden et al. 2007). This proportion is higher than what we detected in our experiment, yet one would need more evidence from different mixture studies to decide on the suitability of a 2-fold deviation range as a heuristic criterion for discrimination between expected and unexpected combined effects. After all, apical and transcriptomic responses might not be directly comparable.
Using the concept of CA in comparison with the other concepts discussed above, we also achieved a larger portion of false-negative but far fewer false-positive predictions of signal occurrence. Overall, a better balance between false-negative and false-positive predictions, indicated by a higher -score (Figure S15D), was achieved. The similarity between the predicted and observed mixture effects on the whole-transcriptome scale was found to be striking, as described above (cf. Figure 2).
The predictive power of the different mixture concepts has rarely been comparatively evaluated on a whole-transcriptome basis. In this study, the highest overall predictivity scores and the quantitatively least deviations between prediction and observation were obtained for CA in comparison with EA, IA, and BA. This finding showed up, despite the hypothesis that underlying complex networks of gene regulation would lead to ill-defined interactions (Rodea-Palomares et al. 2016; Zimmer et al. 2016). Here we show that the effect induced by the mixture exposure could be quantitatively and qualitatively well predicted by describing the signals individually, without taking interactions into account for the majority of cases. Moreover, only by formulating explicit expectations about noninteractive combined effects, the identification of interactions became unambiguous.
Distinguishing Combined Effects on the Transcriptome
A notion commonly held in the regulation of chemicals presumes that chemicals with dissimilar action will not have any joint impact on an organism (EC 2012), that is, there is implicitly an expectation of no combined effect for dissimilarly acting compounds. In our study, we predicted and detected combined effects of chemicals irrespective of their individual mode of action. For example, we predicted some nodes to be jointly affected by naproxen and diuron, two chemicals known to act through different modes of action. Down-regulation of these nodes and the assigned clusters was previously observed with diuron and naproxen and interpreted to indicate an unspecific disturbance of the pancreas development in the ZFE (Schüttler et al. 2019). Indeed, all pancreas-development–related nodes, expected to be affected in the mixture, were either correctly predicted or showed a more-than-additive combined effect. No underestimation was observed for any of these nodes. This indicates that, although known modes of action may differ and thus suggest a dissimilar joint action, compounds may indeed act together and may also cause combined effects that are predictable by CA. This is a novel and exciting finding because the similarity criterion is used, for example, to identify common exposure assessment groups for mixture risk assessment (EC 2012; EFSA Scientific Committee et al. 2019).
For some nodes, observations clearly deviated from the prediction of no observable effects. If we can exclude technical reasons, these deviations may hint at interactive mechanisms. For example, there is a group of nodes that showed a clear down-regulation in the mixture fingerprint. This would not have been expected from the single-substance observations (cluster Tu; Figure 5F and Figure S10). These nodes mostly contain different noncoding RNAs (ncRNAs), with some of them connected to RNA-polymerase III, which points to a change in transcriptional activity. Given that the effect is partly concentration dependent, it is rather unlikely that we observed an artifact here. However, we did not come up with an explanation for this truly unexpected effect. Given the provided means to inspect experimental findings, the example demonstrates how to identify qualitatively unexpected mixture effects with greater confidence by separating them from noninteractive additive combined effects.
Furthermore, we identified 15 nodes where measured effects emerged stronger than predicted with the CA concept (cf. Excel Table S9). Of these, 7 nodes were down-regulated, showing a similar pattern as Node #3119 depicted in Figure 4B. For these nodes we mainly expected contributions from diuron and naproxen, neither of which is known to share the same molecular target nor to have a similar mode of action. Still, both were found to down-regulate genes related to pancreas development, probably through an unspecific mechanism of disturbance (Schüttler et al. 2019). On the other hand, we found 8 nodes that were more strongly up-regulated than predicted by CA. Three of these nodes (#34, #1179, and #2925) appeared to be specifically targeted by the two COX inhibitors in our experiment and mainly comprise genes coding for different metabolic enzymes that are presumably involved in the biotransformation of diclofenac and naproxen. The more-than-additive up-regulation is most pronounced in Node #1179 (cf. Figure 4A). This node contains the gene cyp2k18, which has previously been shown to be induced in zebrafish by COX inhibitors (Poon et al. 2017).
With the help of kinetic modeling, Fitzgerald et al. (2006) showed that such more-than-additive or, if one likes, synergistic, effects may occur due to the topology of the affected pathway. For example, they simulated that two compounds directly targeting the same receptor may, in a mixture, induce a more-than-additive effect if the compounds act in a mutually nonexclusive manner on the receptor. Furthermore, if different elements of one pathway are affected simultaneously, the resulting effect may also be more than additive. However, they could also show in their exemplary calculations that tight regulation in pathway loops, such as feedback inhibition, may nevertheless neutralize such synergisms, leading to effects as predicted by CA. These different simulation outcomes may help to explain the observation of this study finding a majority of apparently nonadditive effects, despite the notion that combined effects at a molecular level may show various deviations on theoretical grounds.
The discussed examples demonstrate that our approach fosters unambiguous identification of unexpected mixture effects by separating them from simple noninteractive, yet additive, effects. Furthermore, the assessment that the occurrence of nonadditive effects was rather the exception than the rule suggests that CA can be seen as a useful reference concept for defining synergy (Greco et al. 1995) at the level of whole-transcriptome responses.
Methodological Limitations
By analyzing the nodes, for which predicted values deviated by more than a factor of two from the experimentally determined values, we found that these deviations could be explained in most cases with methodological limitations due to experimental uncertainty, the approach of data reduction via the SOM approach, as well as the CTR regression model.
The accuracy of a mixture effect prediction depends to a large extent on the accuracy of the effect estimations for the mixture components. Thus, uncertainty in single-compound CTR-model parameter values propagates to uncertainty and accuracy in the mixture predictions. In addition, it was shown previously that the Hill model (being part of the CTR regression model used in this study) might not capture low-dose effects very well, which may result in biased mixture predictions (Scholze et al. 2014). An approach that uses an array of different models and selects the one fitting best could be a way forward (e.g., (Scholze et al. 2001; Smetanová et al. 2015). In theory, it would be feasible to include different models into our fitting strategy. Yet, we assume that for toxicogenomic data, issues like high variability and nonmonotonic regulation (see below) have a more significant influence on prediction quality. In addition, undue control variation in single-substance exposures can introduce bias into the qualitative predictions. Because controls are used as references for normalization, control outliers may inadequately shift obtained . Although this would show up in a high significance threshold for effect assessment in the qualitative analysis, it could also lead to false-negative predictions for the mixture exposure (cf. Figure S12).
In our approach, we tried to increase robustness in transcript responses by using the SOM approach and performing predictions for nodes instead of individual transcripts. However, although the SOM approach provides the intended data reduction, functional grouping, and noise reduction, this clustering may in some cases lead to the masking of relevant single-gene effects by nonresponding genes aggregated into the same node. For example, in Node #818 the gene for cyp1c1 was evidently affected under mixture exposure. However, it was induced with a higher than the remaining genes in the node. This resulted in a rather high uncertainty estimate in the CTR-model fit for the observation (cf. Figure S13). Consequently, no exceedance of a significance threshold could be found and this resulted in an assessment as a deviation from the qualitative prediction. The quantitative prediction for the average sensitivity of this node, was, however, quite accurate using the CA concept (cf. Figure S13), which underlines the joint induction of expression of cyp1 genes by naproxen and diuron, but not diclofenac. This example shows the tradeoff between noise reduction and specificity.
The CTR-model is limited in capturing more complex patterns of differential node regulation. For example, if a node is affected in different directions at different points in time or shows a biphasic behavior across different concentrations, this behavior cannot be captured by the CTR-model. Advancing the CTR-model and allowing more flexibility, for example, by including the time dependence as a differential equation instead of a regression model, might further improve the predictivity for mixture effects. In addition, this would simplify the inclusion of toxicokinetic parameters into the model. It would come at the cost of extended experimental efforts. One may also think about an extended heuristic approach to capture more of the time dependence by using the information on the internal concentrations of the components.
Furthermore, it is not covered by the assumptions of the CA concept and can, hence, not be calculated correctly if a node is regulated in two directions under different substance exposures (cf. Figure S11). Such cases could in principle be captured by applying an extended IA concept that, again, would require a more flexible CTR-model. Capturing such more complex cases of responses of genes or nodes should be the topic of further research.
Mixture Assessment with Omics Technologies
Short-term molecular effects as retrieved by omics measurements have been shown to indicate long-term effects, such as carcinogenicity (Li et al. 2019) or reduced fitness and reproduction in Xenopus caused by endocrine metabolic disruption (Regnault et al. 2018). Nevertheless, appropriate study designs are required to tap the potential of omics technologies for providing proxies for sublethal and potentially long-term mixture effects. This can only be achieved with experimental designs that vary treatments systematically for concentration and time (Diaz et al. 2020; Schüttler et al. 2017) as it allows extrapolation to expected concentration–response patterns of mixtures. These extrapolations also allow predictions for mixtures that do not occur in designed ratios as in this proof-of-concept study. Mixture modeling and the identification of effect driving components, as shown for chemically characterized environmental mixtures (Altenburger et al. 2004, 2018), could thus become possible also on the level of the whole transcriptome.
Earlier, we proposed a novel analysis pipeline of response aggregation using the toxicogenomic universe and concentration and time regression modeling (Schüttler et al. 2019). We extended this pipeline now to mixture-concept-based combined effect prediction. On this route, we believe it is now possible to distinguish unexpected synergism from predictable noninteractive additive combined effects. This should be helpful to disentangle the multistep nature of long-term adverse outcomes and renders omics data applicable for establishing and validating networks of adverse outcome pathways. Respective evidence could then be based on qualitative information about potential key events jointly affected in mixtures. Jointly affected key events could in turn be identified via quantitative comparisons of mixture effects to single-compound effects. Examples for this are the joint induction of Cyp2k18 and the repression of pancreas-related nodes due to mixture exposure, as shown in the present study.
Furthermore, the results of our study show, that toxicogenomic effect patterns of single compounds, which indicate distinct biological effects, reappear in the toxicogenomic fingerprints under mixture exposure. This promises progress in using omics observations on the road to nontarget biological effect assessments of mixtures.
In the light of the innovation rate and increase in chemical uses, advancing our understanding of intended as well as unintended effects of chemical mixtures in organisms is critical for future assessment and management of chemical impacts on humans and the environment. Adapting our analytical tools for this challenge requires advancement in conceptual thinking, experimental design, and data evaluation.
Supplementary Material
Acknowledgments
The authors’ contributions were as follows: conceptualization: A.S., W.B., and R.A.; data curation: A.S.; data analysis: A.S.; funding acquisition: A.S., W.B., and R.A.; investigation: A.S., W.B., J.K., J.M.F., G.J., D.L., and M.K.; project administration: W.B.; software: A.S.; supervision: W.B.; visualization: A.S.; writing—original draft: A.S., W.B., and R.A.; writing—review and editing: A.S., W.B., J.K., J.M.F., M.K., G.J., D.L., and R.A.
We acknowledge N. Schweiger, S. Aulhorn, and S. Schmidt for their support with the experiments. We thank B. Escher, J. Hackermüller, N. Klüver, and K. Schubert as well as three unknown reviewers for the critical review of this manuscript. We also acknowledge the constructive support of the Environmental Health Perspectives editors. We acknowledge the Young Investigators Group Bioinformatics and Transcriptomics at the Helmholtz Centre for Environmental Research for support and inspiration building a reproducible data analysis workflow. We thank the administration and support staff of EVE and WOMBAT who kept everything running and supported us with our scientific computing needs: T. Schnicke, B. Langenberg, C. Krause, S. Petruschke, M. Abbrent, M. Garbe, and N. Ziegner.
This work was supported by the German Federal Environmental Foundation Scholarship Programme (DBU, AZ: 20014/350) and the European Union 7th Framework Programme project SOLUTIONS (grant agreement no. 603437).
The microarray raw data are available in the Gene Expression Omnibus repository via the accession number GSE109498. R functions used for data analysis are compiled in the custom R package omixR (Schüttler 2020).
References
- Altenburger R, Backhaus T, Boedeker W, Faust M, Scholze M. 2013. Simplifying complexity: mixture toxicity assessment in the last 20 years. Environ Toxicol Chem 32(8):1685–1687, PMID: 23843317, 10.1002/etc.2294. [DOI] [PubMed] [Google Scholar]
- Altenburger R, Scholze M, Busch W, Escher BI, Jakobs G, Krauss M, et al. 2018. Mixture effects in samples of multiple contaminants—an inter-laboratory study with manifold bioassays. Environ Int 114:95–106, PMID: 29499452, 10.1016/j.envint.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Altenburger R, Scholz S, Schmitt-Jansen M, Busch W, Escher BI. 2012. Mixture toxicity revisited from a toxicogenomic perspective. Environ Sci Technol 46(5):2508–2522, PMID: 22283441, 10.1021/es2038036. [DOI] [PubMed] [Google Scholar]
- Altenburger R, Walter H, Grote M. 2004. What contributes to the combined effect of a complex mixture? Environ Sci Technol 38(23):6353–6362, PMID: 15597892, 10.1021/es049528k. [DOI] [PubMed] [Google Scholar]
- Andrews FT, Croke BFW, Jakeman AJ. 2011. An open software environment for hydrological model assessment and development. Environ Model Softw 26(10):1171–1185, 10.1016/j.envsoft.2011.04.006. [DOI] [Google Scholar]
- Bahamonde PA, Feswick A, Isaacs MA, Munkittrick KR, Martyniuk CJ. 2016. Defining the role of omics in assessing ecosystem health: perspectives from the Canadian environmental monitoring program. Environ Toxicol Chem 35(1):20–35, PMID: 26771350, 10.1002/etc.3218. [DOI] [PubMed] [Google Scholar]
- Belden JB, Gilliom RJ, Lydy MJ. 2007. How well can we predict the toxicity of pesticide mixtures to aquatic life? Integr Environ Assess Manag 3(3):364–372, PMID: 17695109, 10.1002/ieam.5630030307. [DOI] [PubMed] [Google Scholar]
- Berenbaum MC. 1981. Criteria for analyzing interactions between biologically active agents. Adv Cancer Res 35:269–335, PMID: 7041539, 10.1016/s0065-230x(08)60912-4. [DOI] [PubMed] [Google Scholar]
- Berenbaum MC. 1985. The expected effect of a combination of agents: the general solution. J Theor Biol 114(3):413–431, PMID: 4021503, 10.1016/s0022-5193(85)80176-4. [DOI] [PubMed] [Google Scholar]
- Berninger JP, DeMarini DM, Warren SH, Simmons JE, Wilson VS, Conley JM, et al. 2019. Predictive analysis using chemical-gene interaction networks consistent with observed endocrine activity and mutagenicity of U.S. streams. Environ Sci Technol 53(15):8611–8620, PMID: 31287672, 10.1021/acs.est.9b02990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss CI. 1939. The toxicity of poisons applied jointly. Ann Appl Biol 26(3):585–615, 10.1111/j.1744-7348.1939.tb06990.x. [DOI] [Google Scholar]
- Bluhm K, Otte JC, Yang L, Zinsmeister C, Legradi J, Keiter S, et al. 2014. Impacts of different exposure scenarios on transcript abundances in Danio rerio embryos when investigating the toxicological burden of riverine sediments. PLoS One 9(9):e106523, PMID: 25187966, 10.1371/journal.pone.0106523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker B, R Development Core Team. 2017. bbmle: tools for general maximum-likelihood estimation. R package version 1.0.20. https://CRAN.R-project.org/package=bbmle [accessed 22 March 2021].
- Bolstad BM, Irizarry RA, Åstrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19(2):185–193, PMID: 12538238, 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bopp SK, Kienzler A, Richarz AN, van der Linden SC, Paini A, Parissis N, et al. 2019. Regulatory assessment and risk management of chemical mixtures: challenges and ways forward. Crit Rev Toxicol 49(2):174–189, PMID: 30931677, 10.1080/10408444.2019.1579169. [DOI] [PubMed] [Google Scholar]
- Bradley PM, Journey CA, Romanok KM, Barber LB, Buxton HT, Foreman WT, et al. 2017. Expanded target-chemical analysis reveals extensive mixed-organic-contaminant exposure in U.S. streams. Environ Sci Technol 51(9):4792–4802, PMID: 28401767, 10.1021/acs.est.7b00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Schmidt S, Kühne R, Schulze T, Krauss M, Altenburger R. 2016. Micropollutants in European rivers: a mode of action survey to support the development of effect-based tools for water monitoring. Environ Toxicol Chem 35(8):1887–1899, PMID: 27299692, 10.1002/etc.3460. [DOI] [PubMed] [Google Scholar]
- Cedergreen N. 2014. Quantifying synergy: a systematic review of mixture toxicity studies within environmental toxicology. PLoS One 9(5):e96580, PMID: 24794244, 10.1371/journal.pone.0096580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardenne F, Nobels I, De Coen W, Blust R. 2008. Mixture toxicity and gene inductions: can we predict the outcome? Environ Toxicol Chem 27(3):509–518, PMID: 17983273, 10.1897/07-303.1. [DOI] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, Wiegers TC, et al. 2021. Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res 49(D1):D1138–D1143, PMID: 33068428, 10.1093/nar/gkaa891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz JE, Ahsen ME, Schaffter T, Chen X, Realubit RB, Karan C, et al. 2020. The transcriptomic response of cells to a drug combination is more than the sum of the responses to the monotherapies. eLife 9:e52707, PMID: 32945258, 10.7554/eLife.52707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakvik E, Altenburger R, Aoki Y, Backhaus T, Bahadori T, Barouki R, et al. 2020. Statement on advancing the assessment of chemical mixtures and their risks for human health and the environment. Environ Int 134:105267, PMID: 31704565, 10.1016/j.envint.2019.105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan QY, Gupta VK, Sorooshian S. 1993. Shuffled complex evolution approach for effective and efficient global minimization. J Optim Theory Appl 76(3):501–521, 10.1007/BF00939380. [DOI] [Google Scholar]
- EC (European Commission). 2012. Toxicity and assessment of chemical mixtures. https://op.europa.eu/en/publication-detail/-/publication/ffab4074-6ce5-4f87-89b7-fbd438943b54/language-en [accessed 22 March 2021].
- EC. 2019. The European Green Deal. Communication from the Commission to the European Parliament, the European Council, the Council, the European Economic and Social Committee and the Committee of the Regions. COM(2019) 640 final. https://ec.europa.eu/info/sites/info/files/european-green-deal-communication_en.pdf [accessed 22 March 2021].
- EFSA (European Food Safety Authority), Aguilera J, Aguilera-Gomez M, Barrucci F, Cocconcelli PS, Davies H, et al. 2018. EFSA Scientific Colloquium 24—’omics in risk assessment: state of the art and next steps. EFSA Support Publ 15(11):1512E, 10.2903/sp.efsa.2018.EN-1512. [DOI] [Google Scholar]
- EFSA Scientific Committee, More SJ, Bampidis V, Benford D, Bennekou SH, Bragard C, et al. 2019. Guidance on harmonised methodologies for human health, animal health and ecological risk assessment of combined exposure to multiple chemicals. EFSA J 17(3):e05634, PMID: 32626259, 10.2903/j.efsa.2019.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher BI, Stapleton HM, Schymanski EL. 2020. Tracking complex mixtures of chemicals in our changing environment. Science 367(6476):388–392, PMID: 31974244, 10.1126/science.aay6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust M, Altenburger R, Backhaus T, Blanck H, Boedeker W, Gramatica P, et al. 2001. Predicting the joint algal toxicity of multi-component s-triazine mixtures at low-effect concentrations of individual toxicants. Aquat Toxicol 56(1):13–32, PMID: 11690628, 10.1016/s0166-445x(01)00187-4. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. 2006. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol 2(9):458–466, PMID: 16921358, 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- Gong P, Guan X, Inouye LS, Deng Y, Pirooznia M, Perkins EJ. 2008. Transcriptomic analysis of RDX and TNT interactive sublethal effects in the earthworm Eisenia fetida. BMC Genomics 9(suppl 1):S15, PMID: 18366604, 10.1186/1471-2164-9-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson WH III, Lowe L, Carpenter DO, Gilbertson M, Manaf Ali A, Lopez de Cerain Salsamendi A, et al. 2015. Assessing the carcinogenic potential of low-dose exposures to chemical mixtures in the environment: the challenge ahead. Carcinogenesis 36(suppl 1):S254–S296, PMID: 26106142, 10.1093/carcin/bgv039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco WR, Bravo G, Parsons JC. 1995. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev 47(2):331–385, PMID: 7568331. [PubMed] [Google Scholar]
- Hass U, Scholze M, Christiansen S, Dalgaard M, Vinggaard AM, Axelstad M, et al. 2007. Combined exposure to anti-androgens exacerbates disruption of sexual differentiation in the rat. Environ Health Perspect 115(suppl 1):122–128, PMID: 18174960, 10.1289/ehp.9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs G, Krüger J, Schüttler A, Altenburger R, Busch W. 2020. Mixture toxicity analysis in zebrafish embryo: a time and concentration resolved study on mixture effect predictivity. Environ Sci Eur 32(1):143, 10.1186/s12302-020-00409-3. [DOI] [Google Scholar]
- Jiang C, Wang X, Li X, Inlora J, Wang T, Liu Q, et al. 2018. Dynamic human environmental exposome revealed by longitudinal personal monitoring. Cell 175(1):277–291.e31, PMID: 30241608, 10.1016/j.cell.2018.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203(3):253–310, PMID: 8589427, 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kohonen T. 1982. Self-organized formation of topologically correct feature maps. Biol Cybern 43(1):59–69, 10.1007/BF00337288. [DOI] [Google Scholar]
- Kopec AK, Burgoon LD, Ibrahim-Aibo D, Burg AR, Lee AW, Tashiro C, et al. 2010. Automated dose–response analysis and comparative toxicogenomic evaluation of the hepatic effects elicited by TCDD, TCDF, and PCB126 in C57BL/6 mice. Toxicol Sci 118(1):286–297, PMID: 20702594, 10.1093/toxsci/kfq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortenkamp A, Backhaus T, Faust M. 2009. State of the Art Report on Mixture Toxicity. https://ec.europa.eu/environment/chemicals/effects/pdf/report_mixture_toxicity.pdf [accessed 22 March 2021].
- Kortenkamp A, Faust M, Scholze M, Backhaus T. 2007. Low-level exposure to multiple chemicals: reason for human health concerns? Environ Health Perspect 115(suppl 1):106–114, PMID: 18174958, 10.1289/ehp.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn-Nentwig L, Langenegger N, Heller M, Koua D, Nentwig W. 2019. The dual prey-inactivation strategy of spiders—in-depth venomic analysis of Cupiennius salei. Toxins (Basel) 11(3):167, PMID: 30893800, 10.3390/toxins11030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib S, Williams A, Kuo B, Yauk CL, White PA, Halappanavar S. 2017. A framework for the use of single-chemical transcriptomics data in predicting the hazards associated with complex mixtures of polycyclic aromatic hydrocarbons. Arch Toxicol 91(7):2599–2616, PMID: 27858113, 10.1007/s00204-016-1891-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Lu X, Natoli T, Bittker J, Sipes NS, Subramanian A, et al. 2019. The carcinogenome project: in vitro gene expression profiling of chemical perturbations to predict long-term carcinogenicity. Environ Health Perspect 127(4):47002, PMID: 30964323, 10.1289/EHP3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek MEB, Boobis AR, Crofton KM, Heinemeyer G, Van Raaij M, Vickers C. 2011. Risk assessment of combined exposure to multiple chemicals: a WHO/IPCS framework. Regul Toxicol Pharmacol 60(2):S1–S14, PMID: 21466831, 10.1016/j.yrtph.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Menden MP, Wang D, Mason MJ, Szalai B, Bulusu KC, Guan Y, et al. 2019. Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen. Nat Commun 10(1):2674, PMID: 31209238, 10.1038/s41467-019-09799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkilä J, Törönen P, Kaski S, Venna J, Castrén E, Wong G. 2002. Analysis and visualization of gene expression data using self-organizing maps. Neural Netw 15(8–9):953–966, PMID: 12416686, 10.1016/s0893-6080(02)00070-9. [DOI] [PubMed] [Google Scholar]
- Poon KL, Wang X, Lee SGP, Ng AS, Goh WH, Zhao Z, et al. 2017. Transgenic zebrafish reporter lines as alternative in vivo organ toxicity models. Toxicol Sci 156(1):133–148, PMID: 28069987, 10.1093/toxsci/kfw250. [DOI] [PubMed] [Google Scholar]
- Regnault C, Usal M, Veyrenc S, Couturier K, Batandier C, Bulteau AL, et al. 2018. Unexpected metabolic disorders induced by endocrine disruptors in Xenopus tropicalis provide new lead for understanding amphibian decline. Proc Natl Acad Sci USA 115(19):E4416–E4425, PMID: 29686083, 10.1073/pnas.1721267115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodea-Palomares I, Gonzalez-Pleiter M, Gonzalo S, Rosal R, Leganes F, Sabater S, et al. 2016. Hidden drivers of low-dose pharmaceutical pollutant mixtures revealed by the novel GSA-QHTS screening method. Sci Adv 2(9):e1601272, PMID: 27617294, 10.1126/sciadv.1601272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter S, Beronius A, Boobis AR, Hanberg A, van Klaveren J, Luijten M, et al. 2018. Overview on legislation and scientific approaches for risk assessment of combined exposure to multiple chemicals: the potential EuroMix contribution. Crit Rev Toxicol 48(9):796–814, PMID: 30632445, 10.1080/10408444.2018.1541964. [DOI] [PubMed] [Google Scholar]
- Scholz S, Fischer S, Gündel U, Küster E, Luckenbach T, Voelker D. 2008. The zebrafish embryo model in environmental risk assessment—applications beyond acute toxicity testing. Environ Sci Pollut Res Int 15(5):394–404, PMID: 18575912, 10.1007/s11356-008-0018-z. [DOI] [PubMed] [Google Scholar]
- Scholz S, Schreiber R, Armitage J, Mayer P, Escher BI, Lidzba A, et al. 2018. Meta-analysis of fish early life stage tests—association of toxic ratios and acute-to-chronic ratios with modes of action. Environ Toxicol Chem 37(4):955–969, PMID: 29350428, 10.1002/etc.4090. [DOI] [PubMed] [Google Scholar]
- Scholze M, Boedeker W, Faust M, Backhaus T, Altenburger R, Grimme LH. 2001. A general best-fit method for concentration–response curves and the estimation of low-effect concentrations. Environ Toxicol Chem 20(2):448–457, PMID: 11351447, 10.1002/etc.5620200228. [DOI] [PubMed] [Google Scholar]
- Scholze M, Silva E, Kortenkamp A. 2014. Extending the applicability of the dose addition model to the assessment of chemical mixtures of partial agonists by using a novel toxic unit extrapolation method. PLoS One 9(2):e88808, PMID: 24533151, 10.1371/journal.pone.0088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder AL, Ankley GT, Houck KA, Villeneuve DL. 2016. Environmental surveillance and monitoring—the next frontiers for high-throughput toxicology. Environ Toxicol Chem 35(3):513–525, PMID: 26923854, 10.1002/etc.3309. [DOI] [PubMed] [Google Scholar]
- Schüttler A. 2020. omixR. https://git.ufz.de/itox/omixR [accessed 22 March 2021].
- Schüttler A, Altenburger R, Ammar M, Bader-Blukott M, Jakobs G, Knapp J, et al. 2019. Map and model—moving from observation to prediction in toxicogenomics. Gigascience 8(6):giz057, PMID: 31140561, 10.1093/gigascience/giz057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüttler A, Reiche K, Altenburger R, Busch W. 2017. The transcriptome of the zebrafish embryo after chemical exposure: a meta-analysis. Toxicol Sci 157(2):291–304, PMID: 28329862, 10.1093/toxsci/kfx045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefer E, Kleyman M, Bar-Joseph Z. 2016. Tradeoffs between dense and replicate sampling strategies for high-throughput time series experiments. Cell Syst 3(1):35–42, PMID: 27453445, 10.1016/j.cels.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Rajapakse N, Kortenkamp A. 2002. Something from “nothing” − eight weak estrogenic chemicals combined at concentrations below NOECs produce significant mixture effects. Environ Sci Technol 36(8):1751–1756, PMID: 11993873, 10.1021/es0101227. [DOI] [PubMed] [Google Scholar]
- Smetanová S, Riedl J, Zitzkat D, Altenburger R, Busch W. 2015. High-throughput concentration–response analysis for omics datasets. Environ Toxicol Chem 34(9):2167–2180, PMID: 25900799, 10.1002/etc.3025. [DOI] [PubMed] [Google Scholar]
- Sprague JB. 1970. Measurement of pollutant toxicity to fish. II. Utilizing and applying bioassay results. Water Res 4(1):3–32, 10.1016/0043-1354(70)90018-7. [DOI] [Google Scholar]
- Thomas RS, Allen BC, Nong A, Yang L, Bermudez E, Clewell HJ III, et al. 2007. A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicol Sci 98(1):240–248, PMID: 17449896, 10.1093/toxsci/kfm092. [DOI] [PubMed] [Google Scholar]
- Walter H, Consolaro F, Gramatica P, Scholze M, Altenburger R. 2002. Mixture toxicity of priority pollutants at no observed effect concentrations (NOECs). Ecotoxicology 11(5):299–310, PMID: 12463676, 10.1023/a:1020592802989. [DOI] [PubMed] [Google Scholar]
- Wegner S, Heiger-Bernays W, Dix D. 2016. Spanning regulatory silos in the U.S. EPA’s endocrine disruptor screening program: letter to the editor re: Evans et al. “should the scope of human mixture risk assessment span legislative and regulatory silos for chemicals?” Sci Total Environ 553:671–672, PMID: 26972864, 10.1016/j.scitotenv.2016.02.192. [DOI] [PubMed] [Google Scholar]
- Zhang E, Zhang Y. 2009. F-measure. In: Encyclopedia of Database Systems. Liu L, Özsu MT, eds. Boston, MA: Springer US, 1147. [Google Scholar]
- Zhang J, Liu L, Ren L, Feng W, Lv P, Wu W, et al. 2017. The single and joint toxicity effects of chlorpyrifos and beta-cypermethrin in zebrafish (Danio rerio) early life stages. J Hazard Mater 334:121–131, PMID: 28407539, 10.1016/j.jhazmat.2017.03.055. [DOI] [PubMed] [Google Scholar]
- Zimmer A, Katzir I, Dekel E, Mayo AE, Alon U. 2016. Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc Natl Acad Sci USA 113(37):10442–10447, PMID: 27562164, 10.1073/pnas.1606301113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.