Abstract
Neuropeptide S modulates important neurobiological functions including locomotion, anxiety, and drug abuse through interaction with its G protein-coupled receptor known as neuropeptide S receptor (NPSR). NPSR antagonists are potentially useful for the treatment of substance abuse disorders against which there is an urgent need for new effective therapeutic approaches. Potent NPSR antagonists in vitro have been discovered which, however, require further optimization of their in vivo pharmacological profile. This work describes a new series of NPSR antagonists of the oxazolo[3,4-a]pyrazine class. The guanidine derivative 16 exhibited nanomolar activity in vitro and 5-fold improved potency in vivo compared to SHA-68, a reference pharmacological tool in this field. Compound 16 can be considered a new tool for research studies on the translational potential of the NPSergic system. An in-depth molecular modeling investigation was also performed to gain new insights into the observed structure–activity relationships and provide an updated model of ligand/NPSR interactions.
Introduction
Neuropeptide S (NPS), identified in 2002 by a reverse pharmacology approach,1 is the endogenous ligand of a previous orphan G protein-coupled receptor (GPCR), now named neuropeptide S receptor (NPSR). NPS is a 20 amino acid neuropeptide (primary sequence in humans: SFRNGVGTGMKKTSFQRAKS) highly conserved among different species, and it owes its name to the serine residue at the 1-position of the peptide sequence. NPSR shows a moderate homology with the other members of the GPCR family. The in vitro pharmacology of the human and mouse NPSR showed that NPS increases both intracellular calcium levels and cAMP accumulation with EC50 values in the low nanomolar range. This indicates that NPSR can signal via both Gq and Gs pathways to increase cellular excitability.2,3 In the rodent brain, NPS is expressed only in few neurons in the peri-locus coeruleus region. On the contrary, NPSR is widely expressed in several brain regions (i.e., hypothalamus, endopiriform nucleus, amygdala, subiculum, cortex, and nuclei of the thalamic midline).4,5In vivo, NPS has been shown to control several biological functions in rodents including stress, anxiety, social behavior, locomotor activity, wakefulness, food intake and gastrointestinal functions, memory processes, pain, and drug abuse.6,7 As far as the therapeutic potential of selective NPSR ligands is concerned, NPSR agonists may be useful as innovative anxiolytics devoid of sedative effects, analgesics, and nootropics. On the other hand, NPSR antagonists may be useful to treat substance abuse disorders against which there is an urgent need for the exploration of novel potential drug targets and for developing innovative therapeutic approaches.8
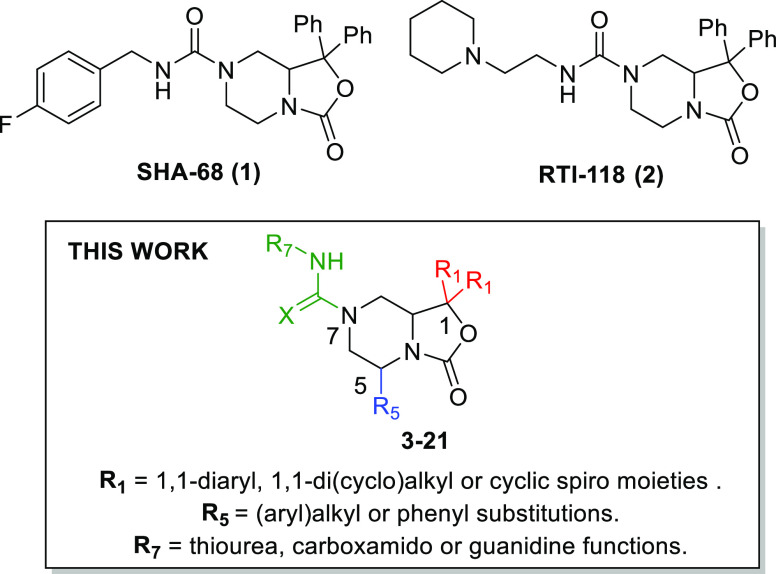
NPSR antagonists with potent in vitro activity have been developed in the last few years and a few compounds are currently in use as pharmacological tools.7 Among these, oxazolo[3,4-a]pyrazine derivatives have been first reported in 2005 by Takeda Pharmaceuticals,9 and SHA-68 (1, Figure 1) is the most representative member of this class.10 Compound 1 was shown to display nanomolar antagonist potency values (pA2/pKB) ranging from 7.28 to 8.16 toward the hNPSR-Asn107 variant and from 7.55 to 8.03 toward the hNPSR-Ile107 variant. Also, compound 1 exhibited high affinity for the hNPSR in radioligand-binding experiments (pKi = 7.32) and high selectivity over several unrelated GPCRs.7
Figure 1.
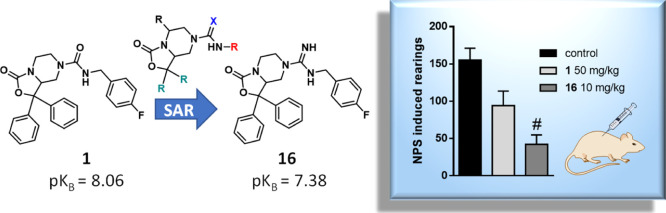
SAR extension performed in this work around the oxazolo[3,4-a]pyrazine nucleus of known NPSR antagonists 1 and 2.
The in vivo pharmacological profile of 1 has been explored in various animal models in which considerably variable effectiveness was observed according to different assays, which has been interpreted as due to suboptimal pharmacokinetic properties of the molecule.10−15 As a first attempt to overcome these limits, Hassler et al. developed the piperidine derivative RTI-118 (2, Figure 1) that exhibited lower potency (hNPSR-Asn107 pA2 = 6.31; hNPSR-Ile107 Ca2+ pA2 = 6.96) in vitro(16) but a slightly improved in vivo effectiveness in reducing cocaine self-administration and seeking behavior in rats; these results were ascribed to the higher water solubility of the molecule.17 Nonetheless, there is a generally recognized need for further optimizing the pharmacological profile and, above all, the drug-likeness properties of oxazolo[3,4-a]pyrazine ligands to obtain even more potent NPSR antagonist tools to be employed in vivo in preclinical studies. These ligands could be extremely useful for understanding the real therapeutic potential of the NPSergic system. This prompted us to extend the structure–activity relationship studies in this field investigating new and unexplored modifications of the bicyclic piperazine nucleus of compounds 1 and 2. Thus, in this work, we describe the synthesis and the in vitro and in vivo biological evaluation of oxazolo[3,4-a]pyrazine derivatives resulting from a series of substitutions at the 1-, 5-, and 7-positions, as summarized in Figure 1. Moreover, molecular modeling studies were performed to gain new insights into the structure–activity relationships observed for the newly discovered NPSR ligands and provide an updated atomistic model of ligand/NPSR interactions.
Results and Discussion
Chemistry
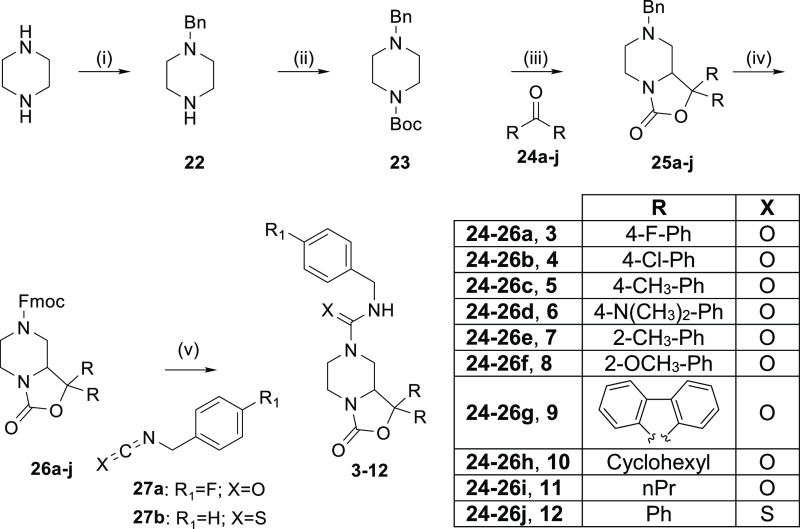
As depicted in Scheme 1, N-Fmoc-oxazolo-piperazines with a general structure 26 were employed as synthetic precursors to obtain the final compounds 3–12 in analogy with the approach previously applied for the synthesis of 1 by Okamura et al.10 Specifically, intermediates 26 were obtained starting from unsubstituted piperazine that was first monoalkylated with benzyl-bromide and next Boc-protected on the second piperazine nitrogen to give compound 23. Subsequently, the desired N-benzyl-protected oxazolo[3,4-a]pyrazines 25a–j were obtained through an ortho-lithiation reaction, in the presence of sec-butyllithium (sec-BuLi) as the base and various symmetric aromatic/aliphatic ketones (24a–j) as electrophiles. The benzyl function was next replaced with an Fmoc-group by treatment with FmocCl and finally, in order to achieve compounds 3–11, 26a–i were reacted with 4-F-benzyl-isocyanate, while compound 12 was obtained from 26j by treatment with benzyl isothiocyanate.
Scheme 1. Synthesis of Final Compounds 3–12.
Reagents and conditions: (i) BnBr, EtOH, 75 °C, 5 h; (ii) Boc2O, DMAP, tetraethylammonium, CH2Cl2, rt, 0.5 h; (iii) sec-BuLi, TMEDA, THF, −78 °C, 6 h; (iv) FmocCl, MeCN, 90 °C, 5 h; (v) DBU, THF, rt, 2 h.
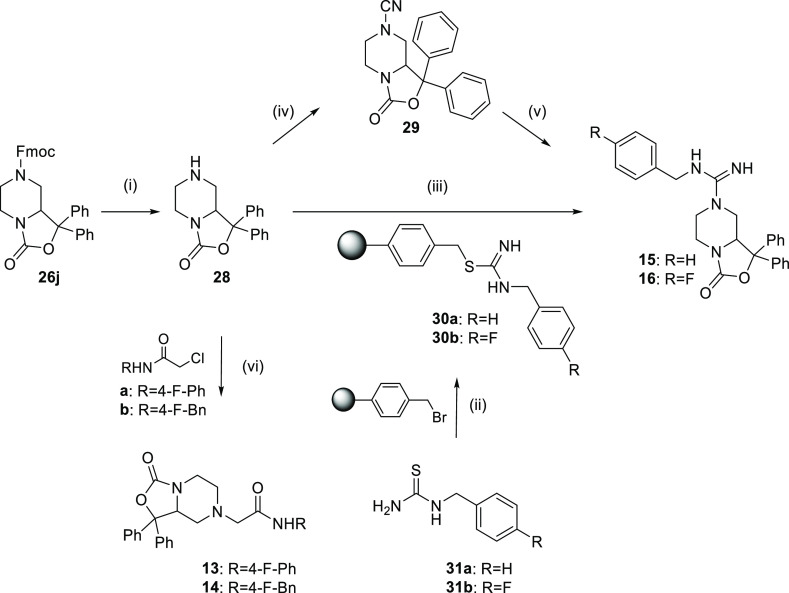
The amide derivatives 13–14 and the guanidine analogues 15–16 were synthesized according to Scheme 2 starting from 26j that was first deprotected by treatment with 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU). The N-alkylation of 28 with 2-chloro-N-(4-fluorophenyl)acetamide or 2-chloro-N-(4-fluorobenzyl)acetamide produced the final compounds 13 and 14, respectively. In order to obtain the guanidine derivatives 15 and 16, we explored two different synthetic strategies. The first approach, developed in the liquid phase, involved the reaction of 28 with cyanogen bromide, giving the key intermediate 29. Then, the addition of benzylamine or 4-fluorobenzylamine in the presence of p-toluenesulfonic acid (p-TsOH) provided the desired final products.
Scheme 2. Synthesis of Final Compounds 13–16.
Reagents and conditions: (i) DBU, THF, rt, 2 h; (ii) CH2Cl2–DMF (2:1), 50 °C, 4 h; (iii) HgCl2, CH3CN, 90 °C, 24 h; (iv) BrCN,CH2Cl2, NaHCO3, H2O, 30 min at 0 °C, 24 h rt; (v) benzyl amine or 4-fluoro benzylamine, p-TsOH, DMSO, 60 °C, 18 h; (vi) K2CO3, CH3CN, 90 °C, 4 h.
The second pathway resulted from the optimization of a known solid-phase approach.18 In this case, a bromomethyl polymeric resin was functionalized with 1-(4-fluorobenzyl)thiourea or 1-(benzyl)thiourea affording 30a–b. Subsequently, the loaded resin was reacted with 28 in the presence of HgCl2 to produce the desired guanidine derivatives in good yields.
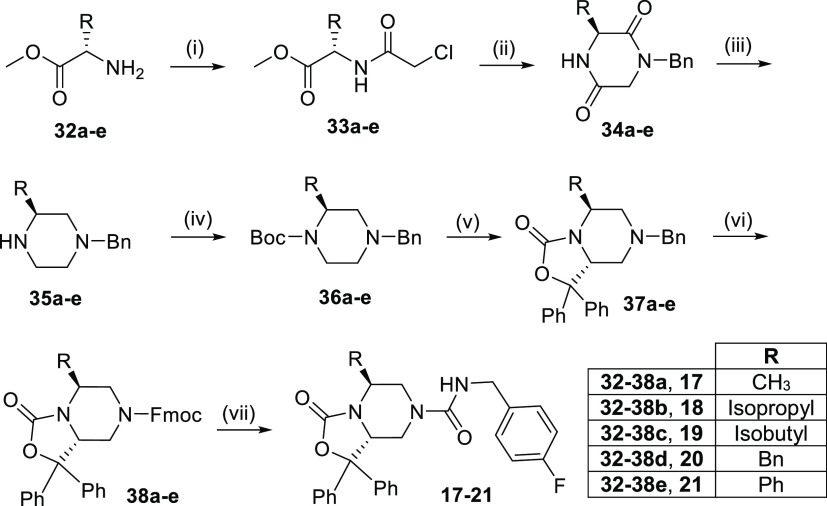
Finally, compounds 17–21 were prepared as depicted in Scheme 3 starting from five different commercially available l-amino acid methyl esters (32a–e). Specifically, the piperazine-diones 34a–e were obtained in two simple steps involving a first amino acid acylation with chloroacetyl chloride, followed by cyclization with benzylamine.19 The subsequent reduction with LiAlH4 gave 35a–e that were then protected with di-tert-butyl dicarbonate (Boc2O).20 The resulting orthogonally protected piperazines 36a–e were employed in the ortho-lithiation reaction to give the benzyl derivatives 37a–e, followed by treatment with FmocCl and the final addition reaction with 4-fluorobenzyl isocyanate as described above.
Scheme 3. Synthesis of Final Compounds 17–21.
Reagents and conditions: (i) NaHCO3, chloroacetyl chloride, toluene, 0 °C to rt, overnight; (ii) Et3N, benzylamine, dioxane, reflux, 20 h; (iii) LiAlH4, THF, reflux, 3 h; (iv) Boc2O, THF, 0 °C to rt, 1 h; (v) benzophenone, sec-BuLi, TMEDA, THF, −78 °C, 3 h; (vi) FmocCl, MeCN, 90 °C, 5 h and then rt, 18 h; (vii) 4-fluorobenzyl isocyanate, DBU, THF, rt, 2 h.
The ortho-lithiation step is of key importance for the stereochemical course of the synthetic approach leading to the final compounds 17–21. This reaction takes advantage of the defined stereochemistry at C-2 of intermediates 36 that is imposed by the choice of the starting amino acid as demonstrated in the literature for analogous piperazine systems obtained through the same strategy.21 The spatial orientation of the substituents around the asymmetric C-8a, generated in the bicyclic derivatives 37 during the ortho-lithiation reaction, was driven by the absolute configuration previously introduced at C-2. A single diastereoisomer was isolated in all cases in which an antirelative stereochemistry between the substituent at the C-5 and the oxazole ring fused at C-8a was expected according to previous studies21 and as confirmed by NOE spectroscopy performed on the reference compound 17 (Figures S1 and S2). In this experiment, the irradiation of the methyl protons at the 5-position produced an important enhancement of the signal of the proton at the C-8a position which is in accordance with a syn relationship. Thus, the absolute (5S,8aR)-configuration was assigned to 17 and to the final compounds 18–21, the latter obtained from α-amino acid with even more hindered side chains. The maintenance of a significant in vitro activity of these derivatives would indirectly confirm that C-8a would assume the absolute configuration of the eutomer of 1 that has been previously identified following the separation of its enantiomers.22 According to this study, the interaction of 1 with the human NPSR would be markedly enantioselective with the R-isomer showing a pKB value of 8.28 (hNPSR-N107) in calcium mobilization experiments, while the S-enantiomer would display a considerably reduced potency (pKB < 6).
In Vitro Structure–Activity Relationships
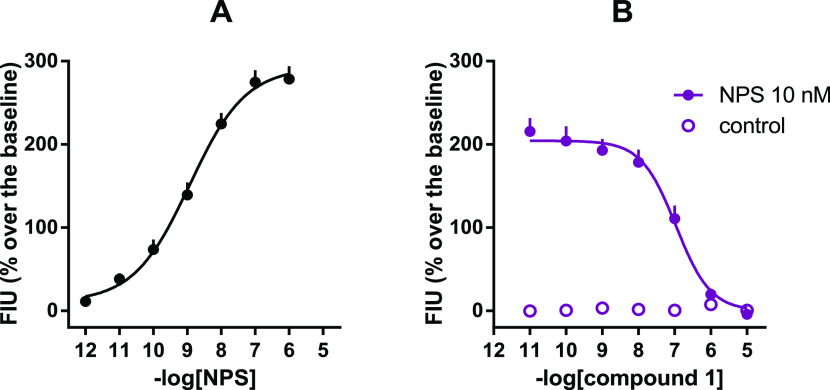
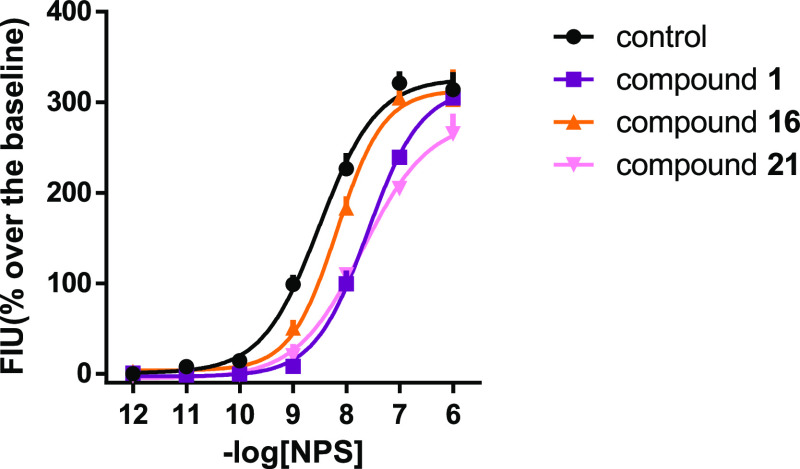
In the calcium mobilization assay, NPS increased intracellular calcium levels in a concentration-dependent manner with pEC50 and Emax values of 8.95 and 287 ± 26% over the basal values, respectively. Inhibition response curves to 1 (0.1 nM to 10 μM), used as an internal reference, were performed against the stimulatory effect of 10 nM NPS, approximately corresponding to NPS EC80. As shown in Figure 2, compound 1 concentration-dependently inhibited 10 nM NPS stimulatory effects with a pKB value of 8.12. These results agree with previously reported data.11 The pharmacological activity of compounds 3–21 was evaluated under the same experimental conditions, and the corresponding results are reported in Table 1.
Figure 2.
Calcium mobilization assay performed on HEK293mNPSR cells. Concentration–response curve to NPS [panel (A)] and inhibition–response curves to 1 against the stimulatory effect of 10 nM NPS [panel (B)]. Data are mean ± SEM of at least five separate experiments made in duplicate.
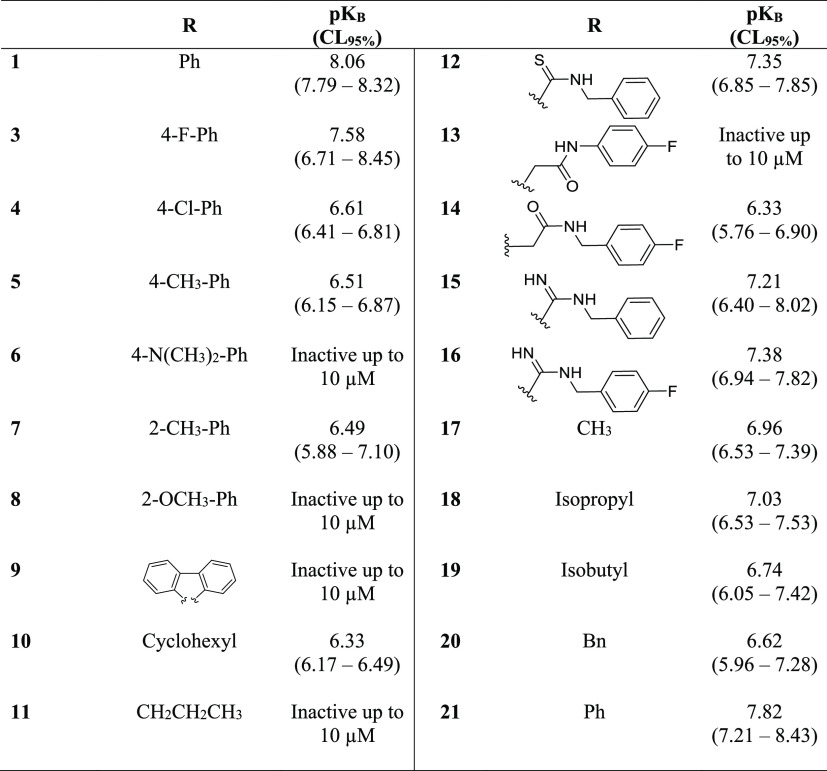
Table 1. In Vitro Pharmacological Activity of Compounds 1, 3–21 as NPSR Antagonistsa.
Calcium mobilization assay performed in HEK293mNPSR cells. Data are the mean of five separate experiments made in duplicate.
None of the novel compounds stimulated calcium mobilization up to 10 μM. On the other hand, the substitution or the replacement of the 1,1-diphenyl moiety of 1, such as in compounds 3–11, significantly affected the antagonist potency although to a different extent. In particular, the para-substitution of both the aromatic rings at the 1-position was slightly tolerated only in the case of the 4-fluoro derivative 3 which was only 3-fold less potent than 1, while a progressive reduction of potency was observed by increasing the steric hindrance of the para-substituents. For example, the bulky dimethylamino groups of compound 6 resulted in a complete loss of activity (KB > 10 μM). These data suggest that the phenyl rings at the 1-position could occupy the NPSR binding pocket in a region with highly stringent steric requirements.
Our results indicated that also a proper spatial orientation of the geminal phenyl groups relative to the oxazolidinone ring seems to be of particular importance to promote activity. Indeed, the ortho-substitution of the 1,1-diaryl moiety such as in compounds 7 and 8 should induce a conformational distortion with respect to the nonsubstituted 1, determining a marked or total loss of potency. This observation was further confirmed by the inactivity of compound 9 in which the 1,1-phenyl rings were forced into a coplanar arrangement due to their inclusion in the spiro-fluorene fusion.
The aromaticity of 1,1-substituents seems to be important as well since the 1,1-dicyclohexyl derivative 10 was more than 50-fold less potent than 1. Even more unfavorable was the replacement of the diaryl template with linear propyl chains (compound 11, KB > 10 μM).
In compounds 12–16, we explored the effect of a few modifications at the 7-position of the oxazolo[3,4-a]pyrazine core that has not been explored before.23 In particular, we introduced side chains containing thiourea (12), N-substituted acetamide (13, 14) and guanidine (15, 16) functions. In this subset of molecules, compounds 13–16 have been specifically designed to modulate the hydrophilic/lipophilic balance of 1, which might be important for its in vivo effectiveness as suggested in different studies.10,11 In particular, it has been demonstrated that 1, at the high dose of 50 mg/kg, can only partially counteract NPS effects, with different levels of efficacy, depending on the assay used.10−15 These findings have been hypothetically attributed to suboptimal physicochemical properties of the compound, in particular, its high lipophilicity.7 Thus, in a first attempt to overcome these limits, the acetamide derivatives 13 and 14 have been synthesized as possible bioisosteres of 1 in which a methylene spacer was interposed between the piperazine nitrogen and the carbonyl function of the 7-side chain. The modification was conceived to increase the basicity of the piperazine nitrogen thus opening the possibility to obtain hydrochloride salts with improved water solubility. Nonetheless, the compounds were shown to display very low (14, pKB = 6.33) or null activity (13, KB > 10 μM) in the calcium mobilization assay. However, the partial recovery of activity of compound 14, in which the 4-F phenyl moiety is not directly linked to the amide nitrogen, suggested that also the conformational freedom of this pharmacophoric portion may be important for the interaction with NPSR.
In compounds 15 and 16, we replaced the urea moiety of 1 with a guanidine function as an alternative strategy to obtain NPSR antagonists with improved hydrophilicity. The fluorinated derivative 16 was conceived as a close analogue of 1 bearing an NH-group in place of the urea oxygen atom. This modification does not interfere with the ability of the ligand to establish polar interactions with the receptor. Of note, compounds incorporating a guanidine moiety have aroused an increasing interest for their potential in the development of novel drugs due to the ability of the guanidinium group to form strong noncovalent interactions and to provide obvious advantages in terms of hydrophilicity.24−26 Compound 16 displayed low nanomolar potency in antagonizing the stimulatory activity of NPS with a pKB value of 7.38. The nonfluorinated guanidine derivative 15 was slightly less potent (pKB = 7.21) indicating some importance of the fluorine atom at the para position of the terminal benzyl moiety.
Finally, we introduced different substitutions at the 5-position of the oxazolo[3,4-a]pyrazine core whose effect on NPSR modulation has not been explored before. To this aim, we developed a highly accessible diastereoselective synthesis that provided compounds 17–21 in which the 5-position was functionalized with the side chains of a series of l-amino acids employed as the starting material. The introduction of a −CH3 (compound 17) or an isopropyl chain (compound 18) determined about a 10-fold reduction of potency if compared to 1. Even more detrimental was the introduction of a bulkier branched alkyl moiety (19) or a benzyl group (20). In contrast, the l-phenylglycine derivative 21 showed a recovery in activity becoming the most active compound of the newly reported series.
These data indicated that the 5-position tolerates substitutions with hydrophobic chemical groups of different size generating derivatives with 2 (21) to 30 (compound 20)—fold reduction of potency. Intriguingly, in the latter compounds, a subtle chemical modification such as the introduction of a methylene spacer between C5 and the phenyl ring promoted a consistent reduction of bioactivity. Given these data, we cannot exclude that the C5 phenyl ring of 21 might be recognized by a previously unexplored region of the NPSR binding pocket.
To confirm and better define the antagonist properties of compounds 16 and 21, the concentration–response curve of NPS has been reassessed in the absence and presence of 100 nM of 1, 16, and 21 (Figure 3). 1, 16, and 21 shifted the concentration–response curve of NPS to the right without changing its maximal effects. The following pA2 values have been derived from these experiments: 7.82 (7.40–8.24) for 1; 7.10 (6.65–7.55) for compound 16; and 7.59 (7.08–8.10) for compound 21. Thus, the rank order of potency of these NPSR antagonists is 1 > 21 > 16. These results are superimposable to those obtained in inhibition experiments.
Figure 3.
Calcium mobilization assay performed on HEK293mNPSR cells. Concentration–response curve to NPS in the absence and in the presence of 100 nM of 1, 16, and 21. Data are mean ± SEM of at least five separate experiments made in duplicate.
Molecular Modeling Studies
To gain major insights into the reasons for the structure–activity relationships (SARs) observed for the newly discovered NPSR ligands, molecular modeling studies were attempted. So far, the three-dimensional (3D) structure of the target receptor has not been determined so that homology-modeling techniques had to be employed to first construct a viable model of NPSR in its inactive state. The available SAR data were obtained from human HEK293 cells expressing the mouse NPSR (mNPSR). On the other hand, the sequence alignment between the human (hNPSR) and mNPSR revealed that the two proteins share 89.22% sequence identity with all the differences residing in the N-terminal region distant from the putative ligand-binding site. Thus, considering the high structural homology of NPSR across the two species, we decided to model the pharmacologically relevant hNPSR in its I107 variant in the present work. In 2010, Dal Ben et al.27 published the first model of the two NPSR variants, namely, NPSR-N107 and -I107. In this seminal work, the authors modeled the NPSR receptors starting from the X-ray crystal structure of bovine rhodopsin.28 The choice of using this latter structure as a template was dictated by preliminary modeling studies indicating that the NPSR extracellular loop 2 (ECL2) had a propensity to adopt a β-sheet conformation which was partially present in the bovine rhodopsin structure rather than in the structures of the human β1, β2 adrenergic and adenosine A2A receptors that were available at that time. Since then, more than 340 structures of GPCR have been deposited in the protein data bank (PDB) thereby allowing for a re-evaluation of the optimal template to employ in the in silico construction of the NPSR receptor variants. Thus, the hNPSR sequence (Uniprot entry code Q6W5P4) was used to interrogate the PDB and select the solved X-ray structures sharing the highest homology with the target structures. In this analysis, we decided to retain all the structures that shared with NPSR a sequence identity higher than 20%, a sequence coverage higher than 70%, and that were crystallized in their inactive states (i.e., bound to an antagonist ligand). These criteria allowed selecting 7 human GPCR structures in which, interestingly, 6 of them turned out to be receptors for endogenous peptides and 5 of these feature a twisted β-hairpin in the ECL2 region (Table S1). Indeed, the β-hairpin motif is usually found in the ECL2 of peptide-activated GPCRs such as Neuropeptide Y Y1 receptor,29 orexin receptor type 1 (OX1R),30 chemokine receptor type 4 (CXCR4),31 delta,32 and kappa33 opioid receptors, protease-activated receptor 1 (PAR1),34 neurotensin receptor 1 (NTSR1),35 endothelin ETB receptor,36 and angiotensin receptors AT137 and AT2.38 Unfortunately, for one of the selected 7 templates (Table S1), the human CC chemokine receptor type 9 (CCR9), ECL2 was unresolved; thus, this template structure was not considered further. Subsequently, the primary sequences of the remaining six GPCRs were all pairwise aligned to the one of the hNPSR-I107 variant, and the phylogenetic tree was calculated (see Figures S3–S9). Then, these templates were all used to construct six models of hNPSR, one for each template, using the Prime software within the Schrodinger’s Maestro suite. The constructed models were all used to perform docking calculations of all the newly identified analogues employing the Glide program. The results of these simulations were then analyzed in light of the available SAR data. In this step, we first verified whether Glide was able to find a viable binding pose for each active compound reported in Table 1 (namely, 1, 3–5, 7, 10, 12, and 14–21) in each of the NPSR models constructed employing the aforementioned 6 template structures. This first analysis was instrumental for the selection of the best model structure that was able to host all the newly discovered NPSR ligands. In particular, the NPSR model constructed starting from the human neuropeptide Y Y1 receptor (hNPY1R, PDB code 5ZBH)29 was the only one able to fulfill the above-mentioned selection criteria. Subsequently, we decided to analyze the docking results achieved for the most potent antagonists 16 and 21 as well as the control compound 1. Interestingly, for all three ligands, Glide was able to suggest two possible binding poses (i.e. featuring comparable docking scores) in which the ligand pendant benzyl substituent is alternatively pointing downward [inside the transmembrane (TM) bundle] or upward (toward the NPSR extracellular region). In this work, the two alternative docked positions will be referred to as binding mode 1 (BM1) and 2 (BM2), respectively (Figure 4).
Figure 4.
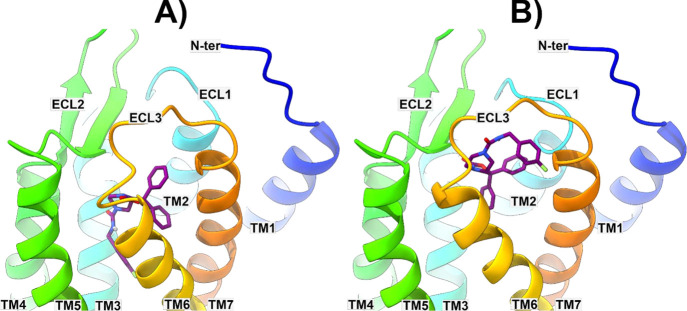
Energy-minimized docked poses of compound 1 in BM1 and BM2 (panels A and B, respectively) in the model of NPSR constructed starting from the human neuropeptide Y Y1 receptor (hNPY1R, PDB code 5ZBH).291 and the protein are represented as violet sticks and multicolored ribbons, respectively.
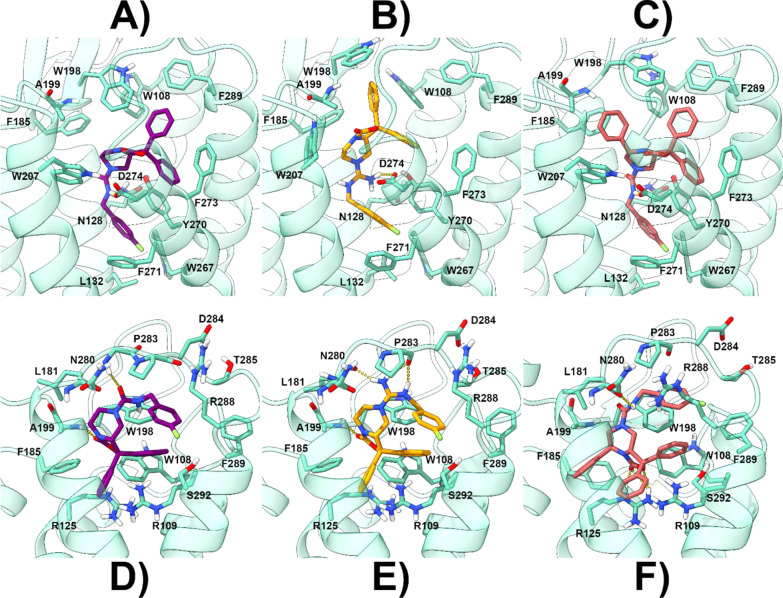
In BM1, the 1,1-diphenyl moiety of 1 is pointing toward TM7 and TM2 and establishes several π–π interactions with aromatic residues present in the outer region of NPSR (W108, W198, F273, and F289) (Figures 4A and 5A). The limited extension of the cleft lodging this moiety should explain why incrementing its steric hindrance has a detrimental effect on the antagonist potency of its analogues (compounds 3–8). Moreover, the nature of the established ligand–protein interactions (charge transfer contacts) as well as the relative position of the two phenyl rings (not coplanar) explains why compounds 9–11 are less active or devoid of an evident antagonist potency. The 1 bicyclic piperazine core orients its pendant benzylamide chain inside the TM bundle between TM3 and TM6 where its NH forms a charged-reinforced H-bond with D274 and the terminal fluorophenyl ring is lodged in a well-defined lipophilic gorge establishing π–π interactions, reinforced by the electron-withdrawing effect of the fluorine atom, with W267, Y270, F271, and van der Waal contacts with L132 (Figure 5A). The tight interactions established by the benzylamide chain in this receptor region would explain why modifications in this position result in a reduction (compound 14) or abrogation (compound 13) of the antagonist potency thereby indicating a limited tolerance for 7-position substituents.23
Figure 5.
Energy-minimized docked poses of compound 1 [panel (A) for BM1 and (D) for BM2], 16 [panel (B) for BM1 and (E) for BM2], and 21 [panel (C) for BM1 and (F) for BM2] in the model of NPSR constructed starting from the human neuropeptide Y Y1 receptor (hNPY1R, PDB code 5ZBH).291, 16, and 21 are represented as violet, orange, and red sticks, respectively. The protein is represented as cyan ribbons and sticks. H-bonds are represented as dashed yellow lines.
Almost the same binding orientation was also found for 16 with all the aforementioned interactions being maintained (Figure 5B). Here, the protonated guanidine group is now engaged in an ionic interaction with the D274 side chain. 21 is also predicted to adopt BM1 where the presence of the additional phenyl ring on the core structure allows to form supplementary charge-transfer and lipophilic interactions with F185, W198, A199, and W207 (Figure 5C). These contacts should explain why the substitution of the phenyl ring with alkyl or benzyl chains (compounds 17–20) still allows observing an antagonist activity at NPSR, although resulting in less potent antagonists if compared to 21.
In BM2, the 1,1-diphenyl moiety of 1 is predicted to point toward the inner part of NPSR making contact with TM2, TM6, and TM7 (Figure 4B). In this position, this moiety established π–π and cation−π contacts with W108 and R109, respectively (Figure 5D). The carbonyl oxygen of the bicyclic piperazine scaffold accepts an H-bond from the A199 backbone NH while the pendant fluorobenzylamide chain laced beneath ECL3 (Figure 4B) in a cleft lined by D284, T285, R288, F289, and S292. Almost the same interaction pattern is also predicted for 16 in BM2 (Figure 5E), and the presence of the protonated guanidine group allows the ligands to establish charge-reinforced H-bonds with N280 and P283 backbone COs as well as a cation−π interaction with W198. 21 recapitulates the same interactions established by 1 in BM2 (Figure 5F) and takes advantage of its additional phenyl ring in position 7 of the core scaffold to establish a π–π interaction with F185. Also in this case, BM2 would match the SAR data acquired in this manuscript as well as the ones already present in the literature.23
The binary complexes calculated using the docking program for 1, 16, and 21 in both BM1 and BM2 were then subjected to 100 ns molecular dynamics (MD) simulations with Desmond39 to refine the predicted binding geometries. Most importantly, given the dichotomy of binding modes predicted for these ligands, results of MD simulations could suggest a preferential binding orientation for each ligand. Analysis of the 6 MD simulations was attained through the Desmond SID tool which allowed us to analyze the ligand–receptor interactions during the MD trajectory. Attention was given to the ligand root-mean-square fluctuation (RMSF) (Figures S10–S15) and the stability of the ligand–residue interactions (Figures S16–S27).
The ligand RMSF was useful for characterizing changes in the ligand atom positions during the MD. Analysis of this parameter demonstrated that in BM1 and BM2, the three ligands display a different degree of flexibility. In particular, regardless of the adopted binding mode, the pendant fluorobenzylamide chain is the most flexible part of the molecule with BM1 being more stable than BM2 (see Figures S10–S15). In the latter binding position, the fluorobenzylamide moiety rapidly loses its interactions with the residues belonging to ECL3 to point toward the external part of the receptor without taking stable contacts with any NPSR residue. This is further outlined by plotting the most frequent (>30%) ligand–protein contacts (Figures S16–S27) showing that for the three ligands in BM2, the benzyl chain is always solvent exposed. On the contrary, in BM1, the same portion remains anchored to the receptor although experiencing a partial relocation during the MD. All in all, MD results would suggest that only in BM1, the fluorobenzylamide chain would play a role in the ligand–receptor recognition as underscored by the experimental SAR data23 available for this ligand moiety, thereby suggesting that 1, 16, and 21 should adopt this binding orientation. Plotting of the ligand–receptor interactions also shows that while binding of 1 is mainly governed by hydrophobic and charge-transfer contacts with the receptor, 16 is stably anchored to NPSR also through the polar interaction with D274 while 21 finds additional contacts with W207 and F185.
In Vivo Characterization in the Mouse Locomotor Activity Test
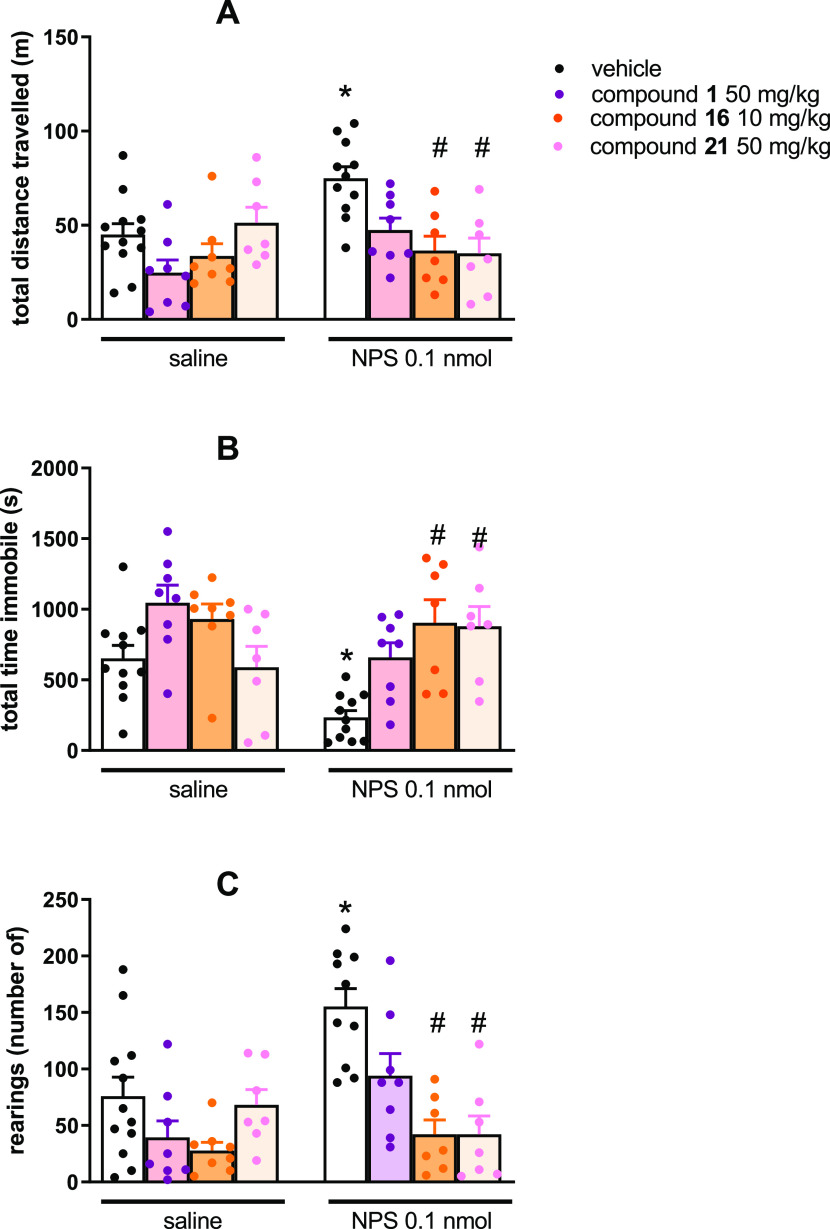
Among the synthesized molecules, 16 and 21 have been selected to be tested in vivo in the mouse locomotor activity assay. As shown in Figure 6 and in line with previous findings,11,40−42 NPS injected by the i.c.v. route at 0.1 nmol concentration was able to stimulate mouse locomotor activity by increasing the distance traveled (panel A) and the number of rearings (panel C) and reducing the immobility time (panel B) with statistically significant effects. 1 (50 mg/kg) did not significantly modify mice locomotor activity and only partially counteracted NPS-induced stimulant effects, confirming previous studies.10,11 All mice treated with 50 mg/kg of 16 displayed an important reduction of locomotor activity (data not shown); thus, in the present study, the 10 mg/kg dose was used for this compound. The doses of 10 mg/kg for 16 and 50 mg/kg for 21 did not elicit by themselves statistically significant effects on mouse locomotor activity (Figure 6).
Figure 6.
Effects of NPS, 1, 16, and 21 on mouse locomotor activity. The cumulative effects exerted on distance traveled are shown in panel (A), while the total time immobile and number of rearings over the 30 min observation period are shown in panels (B,C), respectively. Data are mean ± SEM of four separate experiments (vehicle + saline, 12 mice; 1 + saline, 8 mice; 16 + saline, 8 mice; 21 + saline, 7 mice; vehicle + NPS, 11 mice, 1 + NPS, 8 mice; 16 + NPS, 7 mice; and 21 + NPS, 7 mice). The two-way ANOVA NPS × antagonist revealed for the total distance traveled, an effect of NPS F(1,59) = 6.63 and of the interaction NPS × antagonist F(3,59) = 4.48; for the immobility time, an effect of NPS F(1,59) = 8.09 and of the interaction NPS × antagonist F(3,59) = 4.26; for the number of rearings, an effect of NPS F(1,59) = 7.35, of antagonist F(3,59) = 10.65, and of the interaction NPS × antagonist F(3,59) = 4.32. *p < 0.05 vs saline, #p < 0.05 vs vehicle according to Bonferroni’s test for multiple comparisons.
This result is in line with previous studies performed with different NPSR antagonists10,11,41 and with the lack of locomotor phenotype of NPSR knockout mice,43−45 collectively suggesting that the endogenous NPS does not control mouse locomotor activity in the open field. When administered 30 min before NPS, both 16 and 21 were able to completely block the stimulant effects of the peptide. Compound 21 was in vivo slightly more effective than 1 when injected intraperitoneally (ip) at the same dose (50 mg/kg). Compound 16, despite its lower potency in vitro, was at least 5-fold more potent than 1in vivo. It must be remarked that compound 2 (see the Introduction section) was shown to exert a significant blockade of NPS-induced locomotor activity at an i.p. dose of 50 mg/kg, thus being equipotent to the parental compound 1.16 Of note, the ability of both 16 and 21 to completely block the stimulant NPS effects confirms previous studies demonstrating that the stimulant NPS effects are selectively due to the activation of the NPSR receptor (see Tables 1 and 2 of the recent review by Ruzza et al.).7 It is worth noting that at the higher doses tested, compound 1 was able to only partially block NPS effects. This may somewhat limit the usefulness of compound 1 as a research tool to explore the biological actions modulated by the endogenous NPS. Both compounds 16 and 21 seem to overcome this limitation; in fact, at doses statistically inactive, they were able to fully block NPS stimulant effects. This feature, that is, complete NPSR occupation and blockage, makes these compounds essential pharmacological tools for investigating those conditions in which the endogenous NPS/NPSR signaling is activated, for example, cocaine/alcohol seeking and relapse.46−48 On the other hand, in proposing compound 16 as an innovative research tool, we should also underline that selectivity concerns must be taken into account. In fact, at the high dose of 50 mg/kg, 16 strongly reduces mouse locomotor activity. This is most probably an off-target effect since NPSR selective peptide antagonists did not modify mouse locomotion at doses able to completely block NPS stimulatory effects,41 and NPSR(−/−) mice did not display a locomotor phenotype in this assay.43 Thus, the high potency and antagonist effectiveness of compound 16 is associated with somewhat limited selectivity of action. Further studies, for example, CEREP receptogram, are needed to eventually identify the mechanisms involved in the putative off-target effects of compound 16 at high doses. Based on these considerations, we certainly recommend the use of compound 16 but with special caution in selecting the range of doses to be used for NPSR physiopathological investigations. The in vivo action of 21 reflects the in vitro potency of the compound that was very similar to that of the reference tool 1. On the other hand, the improved in vivo antagonist effectiveness of 16, deriving from the bioisosteric replacement of the urea function of 1 with a guanidine moiety, may be attributed to its relatively higher hydrophilic properties that could compensate for the slight loss of in vitro potency. Lipophilicity is indeed considered one of the most important physicochemical properties to be addressed for drug design purposes. Typically, highly hydrophilic compounds suffer from poor membrane permeability and faster renal clearance. On the other hand, water solubility and metabolism are more likely to be compromised at high lipophilicity values.49 Noteworthily, the optimum region of lipophilicity for candidate drug molecules has been generally suggested to lie within a narrow range of log D7.4 that has been approximately determined between 1 and 3.49 Thus, the improved in vivo potency of 16 (Clog P = 3.43 ± 0.89, ACDLabs; Clog D7.4 = 1.87, ChemAxon predictor) with respect to the known NPSR antagonist 1 (Clog P = 4.32 ± 0.86, ACDLabs; Clog D7.4 = 4.05, ChemAxon predictor) can be reasonably justified in light of the optimization of lipophilicity parameters that are closer to the recommended values. This could also account for the lower in vivo potency of compound 21 (Clog P = 6.18 ± 0.87, ACDLabs; Clog D7.4 = 5.83, ChemAxon predictor) despite its higher in vitro activity. However, we would like to underline the fact that these hypotheses are only supported by theoretical calculations; firm conclusions on this issue can be drawn only after performing experiments investigating the pharmacokinetic profile of compound 16 eventually in comparison with compounds 1 and 21.
Conclusions
The therapeutic potential of selective NPSR ligands in psychiatric disorders is supported by a series of preclinical studies.7 In particular, NPSR antagonists were shown to reduce cocaine/alcohol seeking and relapse in animal models, and this makes them potentially useful for treating drug addiction. Patients suffering from such a condition typically show resistance to the very few treatments currently available. Thus, the identification of innovative drugs able to ameliorate these conditions, which are essentially untreated, is still urgent. Additionally, it can be speculated that NPSR antagonists may be useful also for the treatment of other types of drug abuse, although no preclinical data are currently available. NPSR antagonists may open new perspectives for addressing this unmet clinical need. Moreover, NPSR antagonists still represent important research tools to investigate the neurobiology of the NPS/NPSR system and to study those biological functions for which the NPSergic tone is important. However, even though in vitro potent NPSR antagonists have been reported, NPSR antagonists with a good in vivo pharmacological profile are still missing. This is probably due to pharmacokinetic issues and represents an undeniable limit for preclinical studies on the NPSergic system aimed at translating results from basic pharmacology into clinical utility. In the present study, the in vitro pharmacological activity of a new series of oxazolo[3,4-a]pyrazines as NPSR antagonists was investigated and a molecular-modeling study helped to rationalize the resulting SARs. The most promising compounds (16 and 21) in terms of in vitro potency and/or drug-likeness properties have been also evaluated in vivo for their capability to counteract NPS-induced stimulant effects on mouse locomotor activity. Our findings demonstrated that these compounds behave in vitro as pure NPSR antagonists with nanomolar potency in inhibiting the NPS stimulatory effects in the calcium mobilization assay (pKB values of 7.38 and 7.82 for 16 and 21, respectively). Importantly, the guanidine derivative 16 exhibited a significantly (5-fold) improved potency and increased antagonist effectiveness in vivo when compared to the reference compound 1, although this is associated with somewhat reduced selectivity of action. Collectively, our efforts can be considered an important advancement in this research field culminating in the identification of a new pharmacological tool that combines in vitro and in vivo potency in blocking NPSR. This could be useful to investigate possible pharmacological treatments in all the pathological conditions in which the endogenous NPSergic system is activated.
Experimental Section
Chemistry
Materials and Methods
The chemicals, including the 2-(4-bromomethyl-phenoxy)ethyl polystyrene HL resin for solid-phase synthesis, were purchased from Fluorochem, Novabiochem Iris Biotech GmbH, or Sigma-Aldrich. Reaction progress and product mixtures were monitored by thin-layer chromatography (TLC) on silica gel (precoated F254 Macherey-Nagel plates) and visualized with a UV lamp (254 nm light source). Compounds were purified through silica gel flash chromatography (silica gel 60, 40–63 μm) using appropriate eluent mixtures or on a reverse-phase Waters Prep 600 HPLC system equipped with a Jupiter column C18 (250 × 30 mm, 300 Å, 15 μm spherical particle size). Reverse-phase purification of crude compounds was carried out using a gradient of CH3CN/H2O [with 0.1% trifluoroacetyl (TFA)] programed time by time, with a flow rate of 20 mL/min and a UV detector with a wavelength of 220 nm. Analytical HPLC analyses were performed on a Beckman 116 liquid chromatograph equipped with a Beckman 166 diode array detector. Analytical purity of the final compounds were assessed using a XBridge C18 column (4.6 × 150 mm, 5 μm particle size) at a flow rate of 0.7 mL/min with a linear gradient from 100% of solvent A (H2O + 0.1% TFA) to 100% of solvent B (CH3CN + 0.1% TFA) over 25 min. Analytical determinations were reported as column retention time (TR) in minutes, and the purity of final compounds was >95% as determined by HPLC analysis carried out at a wavelength of 220 nm. Mass spectra were recorded with a Waters ESI Micromass ZQ dissolving the samples in a solution of H2O/CH3CN/TFA (40:60:0.1). Melting points for purified products 3–21 were determined using glass capillaries on a Stuart Scientific electrothermal apparatus SMP3 and are uncorrected. NMR analyses were performed in CDCl3 or DMSO-d6 at ambient temperature using a Varian 200 or 400 MHz spectrometer. Chemical shifts (δ) are reported in parts per million (ppm) using the peak of tetramethylsilane as an internal standard in deuterated solvents, and coupling constants (J) are reported in Hertz. Splitting patterns are designed as s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; and b, broad. Optical rotations were measured on a Jasco P-2000 polarimeter dissolving the samples in methanol, with a path length of 1 dm, using sodium D line, 589 nm.
General Procedure for the Synthesis of 25a–j
Tetramethylethylenediamine (TMEDA, 2.7 mmol) was added under an argon atmosphere at room temperature to a stirring solution of 23(10) (1 mmol) in freshly distilled tetrahydrofuran (THF) (5 mL). After cooling at −78 °C, sec-BuLi (2.7 mmol) was added and the reaction was allowed to reach −30 °C over 2 h. A solution of appropriate benzophenone 24a–j (2 mmol) in THF (5 mL) was added and the reaction solution was left stirring at −30 °C for 30 min, then it was slowly warmed to rt and stirred for 16 h. The reaction was quenched with a saturated solution of NH4Cl (15 mL), and the solvents were concentrated under vacuum to half volume and the aqueous phase was extracted with EtOAc (3 × 15 mL). The organic layers were combined, dried over Na2SO4, and the solvent was removed under vacuum. The resulting crude product was purified by flash column chromatography on silica gel using a mixture of EtOAc/PEt 1:2 as the eluent.
7-Benzyl-1,1-bis(4-fluorophenyl)tetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25a)
White solid (227 mg, 54% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.70–7.59 (m, 2H); 7.40–7.16 (m, 11H); 4.66 (dd, J = 8.0, 4.0 Hz, 1H); 3.62–3.49 (m, 2H); 3.38–3.21 (m, 1H); 3.16–2.98 (m, 1H); 2.63–2.58 (m, 2H); 1.88–1.69 (m, 1H); 1.52–1.38 (m, 1H). MS (ESI): m/z calcd for C25H23F2N2O2 [M + H]+, 421.47; found, 421.03.
7-Benzyl-1,1-bis(4-chlorophenyl)tetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25b)
White solid (200 mg, 44% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.44–7.18 (m, 13H); 4.63–4.20 (m, 1H); 3.82 (dd, J = 8.0, 4.0 Hz, 1H); 3.44–3.04 (m, 2H); 2.76 (d, J = 10.6 Hz, 1H); 2.53 (d, J = 11.1 Hz, 1H); 2.01–1.88 (m, 1H); 1.60 (t, J = 7.4 Hz, 2H). MS (ESI): m/z calcd for C25H23Cl2N2O2 [M + H]+, 454.37; found, 454.26.
7-Benzyl-1,1-di-p-tolyltetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25c)
Pale yellow solid (260 mg, 63% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.44 (d, J = 8.4 Hz, 2H); 7.31–7.12 (m, 11H); 4.60 (dd, J = 11.1, 3.7 Hz, 1H); 3.61–3.46 (m, 2H); 3.34 (s, 1H); 3.16–2.96 (m, 1H); 2.68–2.52 (m, 2H); 2.26 (d, J = 3.4 Hz, 6H); 1.84–1.65 (m, 1H); 1.58–1.39 (m, 1H). MS (ESI): m/z calcd for C27H29N2O2 [M + H]+, 413.54; found, 413.46.
7-Benzyl-1,1-bis(4-(dimethylamino)phenyl)tetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25d)
Dark blue solid (169 mg, 36% yield). 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.31–7.20 (m, 7H); 6.99 (d, J = 8.6 Hz, 2H); 6.66 (t, J = 8.2 Hz, 4H); 4.45 (d, J = 8 Hz, 1H); 3.53 (t, J = 4 Hz, 2H); 3.42–3.31 (m, 1H); 3.01 (d, J = 12 Hz, 1H); 2.87 (s, 12H); 2.63–2.41 (m, 2H); 1.78 (t, J = 12 Hz, 1H); 1.48 (t, J = 12 Hz, 1H). MS (ESI): m/z calcd for C29H35N4O2 [M + H]+, 471.63; found, 471.44.
7-Benzyl-1,1-di-o-tolyltetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25e)
White solid (82 mg, 28% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.89–7.79 (m, 1H); 7.67–7.55 (m, 1H); 7.40–6.96 (m, 11H); 4.88 (d, J = 10.5 Hz, 1H); 3.86 (d, J = 11.2 Hz, 2H); 3.52 (d, J = 12.1 Hz, 1H); 3.32–3.10 (m, 2H); 2.79–2.58 (m, 1H); 2.11–1.95 (m, 2H); 1.79 (d, J = 9.8 Hz, 6H). MS (ESI): m/z calcd for C27H29N2O2 [M + H]+, 413.54; found, 413.44.
7-Benzyl-1,1-bis(2-methoxyphenyl)tetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25f)
White solid (169 mg, 38% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.49–7.21 (m, 8H); 6.96–6.75 (m, 5H); 4.54 (d, J = 8 Hz, 1H); 3.90 (dd, J = 8.0, 4.0 Hz, 1H); 3.51 (s, 6H); 3.40–3.05 (m, 2H); 2.81–2.52 (m, 2H); 2.20–2.01 (m, 1H); 1.75–1.56 (m, 2H). MS (ESI): m/z calcd for C27H29N2O4 [M + H]+, 445.54; found, 445.30.
7′-Benzyl-6′,7′,8′,8a′-tetrahydrospiro[fluorene-9,1′-oxazolo[3,4-a]pyrazin]-3′(5′H)-one (25g)
Yellow solid (279 mg, 73% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.85–7.79 (m, 4H); 7.61–7.09 (m, 9H); 4.20 (dd, J = 10.6, 3.2 Hz, 1H); 3.72 (d, J = 11.1 Hz, 1H); 3.43–3.36 (m, 2H); 3.20–3.05 (m, 1H); 2.80 (d, J = 8.2 Hz, 1H); 2.31–1.85 (m, 3H). MS (ESI): m/z calcd for C25H23N2O2 [M + H]+, 383.47; found, 383.29.
7-Benzyl-1,1-dicyclohexyltetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25h)
White solid (162 mg, 41% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.40–7.22 (m, 4H); 7.02 (s, 1H); 3.78–3.64 (m, 2H); 3.58–3.42 (m, 2H); 3.34 (s, 1H); 2.90–2.61 (m, 3H) 2.24 (t, J = 12 Hz, 1H); 1.97–1.41 (m, 13H); 1.28–0.98 (m, 9H). MS (ESI): m/z calcd for C25H37N2O2 [M + H]+, 397.58; found, 397.31.
7-Benzyl-1,1-dipropyltetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25i)
White solid (136 mg, 43% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.76 (d, J = 6.8 Hz, 1H); 7.44–7.25 (m, 4H); 4.80–4.44 (m, 2H); 4.30–4.00 (m, 2H); 3.67 (d, J = 12.0 Hz, 1H); 3.29–3.15 (m, 1H); 3.00–2.56 (m, 3H); 1.61–1.00 (m, 8H); 0.94–0.84 (m, 6H). MS (ESI): m/z calcd for C19H29N2O2 [M + H]+, 317.45; found, 317.46.
7-Benzyl-1,1-diphenyltetrahydro-1H-oxazolo[3,4-a]pyrazin-3(5H)-one (25j)
White solid (277 mg, 72% yield). 1H NMR (200 MHz, CDCl3): δ (ppm): 7.60–7.49 (m, 1H); 7.38–7.16 (m, 14H); 4.52 (dd, J = 8.0, 4.0 Hz, 1H); 3.72–3.59 (m, 2H); 3.48–3.21 (m, 1H); 3.16–2.98 (m, 1H); 2.63–2.58 (m, 2H); 1.88–1.69 (m, 1H); 1.52–1.38 (m, 1H). MS (ESI): m/z calcd for C25H25N2O2 [M + H]+, 385.49; found, 385.28.
General Procedure for the Synthesis of 26a–j
9-Fluorenylmethoxycarbonyl chloride (FmocCl, 1.1 mmol) was added to a solution of 25a–j (1 mmol) in CH3CN (5 mL). The reaction solution was heated at 90 °C for 5 h and then stirred at room temperature for 18 h. The solvent was evaporated giving a residue that was dissolved in EtOAc (30 mL), and the resulting organic phase was washed with water (3 × 15 mL), dried over anhydrous Na2SO4, and concentrated to dryness. All crude residues were finally purified via flash column chromatography on a silica gel using the appropriate mixture of petroleum ether and EtOAc as an eluent (see below).
(9H-Fluoren-9-yl)methyl 1,1-Bis(4-fluorophenyl)-3-oxotetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26a)
White solid (448 mg, 81% yield). Eluent for chromatography purification: EtOAc/PEt 1:1; 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.94–7.84 (m, 2H); 7.62–7.18 (m, 14H); 4.65–4.39 (m, 3H); 4.25 (t, J = 5.4 Hz, 1H); 3.82 (d, J = 10.5 Hz, 1H); 3.56 (dd, J = 10.6, 2.2 Hz, 1H); 3.05–2.85 (m, 1H); 2.83–2.64 (m, 2H); 2.31–2.16 (m, 1H). MS (ESI): m/z calcd for C33H27F2N2O4 [M + H]+, 553.59; found, 553.57.
(9H-Fluoren-9-yl)methyl 1,1-Bis(4-chlorophenyl)-3-oxotetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26b)
White solid (298 mg, 51% yield). Eluent for chromatography purification: EtOAc/PEt 1:2; 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.90 (t, J = 6.8 Hz, 2H); 7.59–7.28 (m, 14H); 4.43–4.39 (m, 2H); 4.24 (t, J = 5.6 Hz, 2H); 3.82 (d, J = 11.2 Hz, 1H); 3.56 (d, J = 10.6 Hz, 1H); 3.19 (s, 1H); 3.07–2.64 (m, 2H); 2.24 (t, J = 12.4 Hz, 1H). MS (ESI): m/z calcd for C33H27Cl2N2O4 [M + H]+, 586.49; found, 586.63.
(9H-Fluoren-9-yl)methyl 3-Oxo-1,1-di-p-tolyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26c)
White solid (327 mg, 60% yield). Eluent for chromatography purification: EtOAc/PEt 1:4; 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.90 (t, J = 7.4 Hz, 2H); 7.59 (d, J = 7.0 Hz, 2H); 7.44–7.16 (m, 10H); 6.99 (s, 2H); 4.77–4.59 (m, 1H); 4.50–4.35 (m, 1H); 4.26 (s, 1H); 4.16–4.09 (m, 1H); 3.80 (d, J = 10.6 Hz, 1H); 3.53 (dd, J = 11.2, 1.8 Hz, 1H); 3.22–3.05 (m, 1H); 2.95–2.58 (m, 2H); 2.30 (d, J = 4.6 Hz, 6H); 2.20–2.08 (m, 1H). MS (ESI): m/z calcd for C35H33N2O4 [M + H]+, 545.66; found, 545.59.
(9H-Fluoren-9-yl)methyl 1,1-Bis(4-(dimethylamino)phenyl)-3-oxotetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26d)
Pink solid (380 mg, 63% yield). Eluent for chromatography purification: EtOAc/PEt 1:4; 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.88 (t, J = 6.6 Hz, 2H); 7.59 (d, J = 7.2 Hz, 2H); 7.44–7.32 (m, 5H); 7.11 (s, 2H); 6.75–6.63 (m, 5H); 4.79–4.61 (m, 1H); 4.52–4.33 (m, 1H); 4.26 (t, J = 5.6 Hz, 1H); 4.01–3.90 (m, 1H); 3.82–3.60 (m, 1H); 3.53 (d, J = 10.6 Hz, 1H); 3.22–3.05 (m, 1H); 3.11–2.98 (m, 1H); 2.90 (d, J = 4.8 Hz, 12H); 2.83–2.77 (m, 1H); 2.22–2.06 (m, 1H). MS (ESI): m/z calcd for C37H39N4O4 [M + H]+, 603.74; found, 603.51.
(9H-Fluoren-9-yl)methyl 3-Oxo-1,1-di-o-tolyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26e)
White solid (408 mg, 75% yield). Eluent for chromatography purification: EtOAc/PEt 3:7; 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.89–7.79 (m, 2H); 7.67–7.55 (m, 2H); 7.40–6.96 (m, 12H); 2.51–4.07 (m, 1H); 4.88 (d, J = 10.5 Hz, 1H); 3.86 (d, J = 11.2 Hz, 2H); 3.52 (d, J = 12.1 Hz, 1H); 3.32–3.10 (m, 2H); 2.79–2.58 (m, 1H); 2.11–1.95 (m, 2H); 1.79 (d, J = 9.8 Hz, 6H). MS (ESI): m/z calcd for C35H33N2O4 [M + H]+, 545.66; found, 545.25.
(9H-Fluoren-9-yl)methyl 1,1-Bis(2-methoxyphenyl)-3-oxotetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26f)
White solid (363 mg, 63% yield). Eluent for chromatography purification: EtOAc/PEt 1:1; 1H NMR (200 MHz, CDCl3): δ (ppm): 7.72 (d, J = 7.2 Hz, 2H); 7.48–7.29 (m, 10H); 7.01–6.88 (m, 4H); 4.58–4.35 (s, 2H); 4.24–4.06 (m, 4H); 4.01–3.36 (m, 7H); 2.89–2.69 (m, 2H); 2.64–2.42 (m, 1H). MS (ESI): m/z calcd for C35H33N2O6 [M + H]+, 577.66; found, 577.65.
(9H-Fluoren-9-yl)methyl 3′-Oxo-5′,6′,8′,8a′-tetrahydro-3′H,7′H-spiro[fluorene-9,1′-oxazolo[3,4-a]pyrazine]-7′-carboxylate (26g)
White solid (283 mg, 55% yield). Eluent for chromatography purification: EtOAc/PEt 1:4; 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.95 (s, 2H); 7.90–7.02 (m, 14H); 4.56–4.25 (m, 2H); 4.19–4.05 (s, 1H); 3.86 (d, J = 12.40 Hz, 2H); 3.69 (d, J = 12.90 Hz, 2H); 2.98–2.64 (m, 3H). MS (ESI): m/z calcd for C33H27N2O4 [M + H]+, 515.59; found, 515.50.
(9H-Fluoren-9-yl)methyl 1,1-Dicyclohexyl-3-oxotetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26h)
White solid (434 mg, 82% yield). Eluent for chromatography purification: EtOAc/PEt 1:4; 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.88 (d, J = 7.5 Hz, 2H); 7.65 (t, J = 7.0 Hz, 2H); 7.42 (t, J = 7.4 Hz, 2H); 7.33 (dd, J = 13.8, 7.1 Hz, 2H); 4.63–4.42 (m, 2H); 4.28 (t, J = 5.6 Hz, 1H); 3.94–3.58 (m, 2H); 3.50–3.18 (m, 3H); 2.98–2.63 (m, 2H); 1.78–1.03 (m, 22H). MS (ESI): m/z calcd for C33H41N2O4 [M + H]+, 529.70; found, 529.66.
(9H-Fluoren-9-yl)methyl 3-Oxo-1,1-dipropyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26i)
White solid (166 mg, 37% yield). Eluent for chromatography purification: EtOAc/PEt 3:7; 1H NMR (200 MHz, CDCl3): δ (ppm) 7.76 (d, J = 6.8 Hz, 2H); 7.55 (d, J = 7.2 Hz, 2H); 7.44–7.25 (m, 4H); 4.80–4.44 (m, 2H); 4.30–4.00 (m, 3H); 3.67 (d, J = 12.0 Hz, 1H); 3.29–3.15 (m, 1H); 3.00–2.56 (m, 3H); 1.61–1.00 (m, 8H); 0.94–0.84 (m, 6H). MS (ESI): m/z calcd for C27H33N2O4 [M + H]+, 449.57; found, 449.55.
(9H-Fluoren-9-yl)methyl 3-Oxo-1,1-diphenyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (26j)
White solid (305 mg, 59% yield). Eluent for chromatography purification: EtOAc/PEt 1:1; 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.89 (t, J = 7.4 Hz, 2H); 7.59 (d, J = 7.4 Hz, 2H); 7.54–7.03 (m, 14H); 4.71–4.38 (m, 2H); 4.26 (t, J = 5.4 Hz, 2H); 3.79 (d, J = 10.6 Hz, 1H); 3.56 (dd, J = 11.4, 1.8 Hz, 2H); 3.00–2.84 (m, 2H); 2.28–2.09 (m, 1H). MS (ESI): m/z calcd for C33H29N2O4 [M + H]+, 517.61; found, 517.29.
General Procedure for the Synthesis of Final Compounds 3–12
The appropriate iso(thio)cyanate 27a–b (2 mmol) and DBU (1.2 mmol) were sequentially added at room temperature to a stirring solution of 26a–j (1 mmol) in anhydrous THF (5 mL). The reaction solution was continued stirring for 2 h after which it was quenched with a saturated solution of NH4Cl (10 mL). The mixture was concentrated under vacuum to half volume, and the aqueous phase was extracted with EtOAc (3 × 15 mL). The organic layers were combined, dried over Na2SO4, and concentrated under vacuum. The crude products were purified by preparative RP-HPLC.
N-(4-Fluorobenzyl)-1,1-bis(4-fluorophenyl)-3-oxotetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (3)
White solid (289 mg, 60% yield); mp 104–107 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.65–7.54 (m, 2H); 7.40–7.19 (m, 9H); 7.18–7.05 (m, 2H); 4.51 (dd, J = 11.2, 3.6 Hz, 1H); 4.21 (d, J = 5.5 Hz, 2H); 3.93 (d, J = 10.5 Hz, 1H); 3.82 (dd, J = 13.1, 2.9 Hz, 1H); 3.59 (dd, J = 13.2, 2.6 Hz, 1H); 3.05 (td, J = 12.8, 3.6 Hz, 1H); 2.68 (td, J = 12.9, 3.4 Hz, 1H); 2.17–2.00 (m, 1H). 13C NMR (DMSO-d6): δ 162.94, 162.70, 162.12, 160.49, 160.27, 159.71, 157.05, 154.80, 138.44, 136.75, 134.48, 128.89, 128.09, 127.78, 115.45, 115.24, 114.70, 84.07, 59.51, 45.51, 42.79, 42.52, 41.09. MS (ESI): m/z calcd for C26H23F3N3O3 [M + H]+, 482.48; found, 482.21. TR = 24.77 min.
1,1-Bis(4-chlorophenyl)-N-(4-fluorobenzyl)-3-oxotetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (4)
White solid (412 mg, 80% yield); mp 77–79 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.63–7.55 (m, 2H); 7.53–7.44 (m, 4H); 7.37–7.22 (m, 5H); 7.15–7.07 (m, 2H); 4.52 (dd, J = 11.2, 3.6 Hz, 1H); 4.21 (d, J = 5.2 Hz, 2H); 3.93 (d, J = 11.6 Hz, 1H); 3.83 (dd, J = 12.9, 2.9 Hz, 1H); 3.59 (dd, J = 13.1, 2.7 Hz, 1H); 3.05 (td, J = 12.7, 3.6 Hz, 1H); 2.68 (td, J = 13.0, 3.4 Hz, 1H); 2.11 (dd, J = 12.8, 11.4 Hz, 1H). 13C NMR (DMSO-d6): δ 162.14, 159.73, 157.07, 154.68, 140.92, 136.96, 136.73, 133.18, 132.84, 128.92, 128.55, 127.77, 127.47, 114.71, 83.94, 59.33, 45.45, 42.81, 42.50, 41.14. MS (ESI): m/z calcd for C26H23Cl2FN3O3 [M + H]+, 515.39; found, 515.45. TR = 22.16 min.
N-(4-Fluorobenzyl)-3-oxo-1,1-di-p-tolyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (5)
White solid (312 mg, 66% yield); mp 89–90 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm): 7.41–7.39 (m, 3H); 7.30–7.25 (m, 3H); 7.21–7.09 (m, 7H); 4.43 (dd, J = 11.1, 3.7 Hz, 1H); 4.20 (d, J = 5.3 Hz, 2H); 3.92 (d, J = 11.6 Hz, 1H); 3.84 (dd, J = 12.9, 2.9 Hz, 1H); 3.59 (dd, J = 13.3, 2.9 Hz, 1H); 3.02 (td, J = 12.7, 3.6 Hz, 1H); 2.64 (td, J = 12.9, 3.4 Hz, 1H); 2.29 (s, 3H); 2.27 (s, 3H), 2.08–2.02 (m, 1H). 13C NMR (DMSO-d6): δ 162.66, 160.26, 157.63, 155.68, 140.15, 138.08, 137.59, 137.33, 136.28, 129.46, 129.38, 126.18, 125.93, 115.36, 115.15, 85.31, 60.08, 46.32, 43.33, 43.12, 41.52, 21.01. MS (ESI): m/z calcd for C28H29FN3O3 [M + H]+, 474.56; found, 474.43. TR = 18.53 min.
1,1-Bis(4-(dimethylamino)phenyl)-N-(4-fluorobenzyl)-3-oxotetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (6)
White solid (500 mg, 94% yield); mp 110–113 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.33–7.19 (m, 5H); 7.17–7.06 (m, 2H); 7.01–6.98 (m, 2H); 6.70–6.67 (m, 4H); 4.29 (dd, J = 11.2, 3.5 Hz, 1H); 4.22–4.17 (m, 2H); 3.93 (d, J = 11.2 Hz, 1H); 3.79 (dd, J = 13.0, 2.7 Hz, 1H); 3.59 (dd, J = 12.9, 2.7 Hz, 1H); 3.01 (td, J = 12.8, 3.4 Hz, 1H); 2.90–2.86 (m, 12H); 2.63 (td, J = 13.2, 3.5 Hz, 1H); 2.06 (dd, J = 13.0, 11.4 Hz, 1H). 13C NMR (DMSO-d6): δ 162.11, 157.10, 155.50, 149.86, 149.44, 136.81, 129.87, 128.93, 128.85, 126.74, 126.41, 114.82, 114.61, 111.65, 111.54, 85.23, 59.63, 46.00, 42.77, 42.65, 40.84. MS (ESI): m/z calcd for C30H35FN5O3 [M + H]+, 532.64; found, 532.41. TR = 11.44 min.
N-(4-Fluorobenzyl)-3-oxo-1,1-di-o-tolyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (7)
White solid (260 mg,55% yield); mp 217–219 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.92–7.81 (m, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.35–7.19 (m, 7H); 7.18–7.05 (m, 4H); 4.95 (dd, J = 11.1, 3.7 Hz, 1H); 4.24 (dd, J = 15.3, 5.7 Hz, 1H); 4.13 (dd, J = 15.1, 5.3 Hz, 1H); 3.88 (d, J = 11.4 Hz, 1H); 3.61 (dd, J = 13.3, 2.9 Hz, 1H); 3.38–3.22 (m, 2H); 2.81 (td, J = 13.3, 3.4 Hz, 1H); 2.19 (dd, J = 12.9, 11.2 Hz, 1H); 1.76 (s, 3H); 1.74 (s, 3H). 13C NMR (DMSO-d6): δ 162.12, 159.73, 157.08, 154.97, 137.30, 137.00, 136.80, 136.21, 133.44, 132.71, 131.69, 128.98, 128.90, 128.69, 128.14, 126.74, 126.55, 125.54, 114.80, 114.60, 85.57, 56.28, 45.03, 43.11, 42.81, 41.26, 21.01, 20.45. MS (ESI): m/z calcd for C28H29FN3O3 [M + H]+, 474.56; found, 474.34. TR = 19.20 min.
N-(4-Fluorobenzyl)-1,1-bis(2-methoxyphenyl)-3-oxotetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (8)
White solid (435 mg, 86% yield); mp 172–173 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.45–7.42 (m, 1H); 7.39–7.22 (m, 4H); 7.20–7.15 (m, 1H); 7.14–6.89 (m, 7H); 4.37 (d, J = 8.1 Hz, 1H); 4.27–4.22 (m, 1H); 4.16–4.10 (m, 1H); 3.99–3.86 (m, 2H); 3.64 (s, 3H); 3.62 (s, 4H); 3.06 (td, J = 12.9, 3.6 Hz, 1H); 2.73 (td, J = 13.2, 3.6 Hz, 1H); 2.36–2.27 (m, 1H). 13C NMR (DMSO-d6): δ 162.09, 159.68, 156.90, 156.78, 155.95, 154.76, 136.84, 129.64, 129.56, 128.90, 128.82, 128.20, 127.00, 125.98, 120.18, 119.93, 114.76, 114.55, 112.54, 111.90, 83.94, 59.04, 55.35, 55.29, 44.76, 42.78, 42.62, 40.57. MS (ESI): m/z calcd for C28H29FN3O5 [M + H]+, 506.55; found, 506.52. TR = 18.71 min.
N-(4-Fluorobenzyl)-3′-oxo-5′,6′,8′,8a′-tetrahydrospiro[fluorene-9,1′-oxazolo[3,4-a]pyrazine]-7′(3′H)-carboxamide (9)
White solid (421 mg, 95% yield); mp 120–123 °C; 1H NMR (400 MHz, DMSO): δ (ppm) 7.91–7.78 (m, 3H); 7.67 (d, J = 7.5 Hz, 1H); 7.56–7.47 (m, 2H); 7.42–7.36 (m, 2H); 7.20–7.15 (m, 3H); 7.11–6.95 (m, 2H); 4.19–4.00 (m, 4H); 3.74 (dd, J = 12.4, 2.4 Hz, 1H); 3.52 (dd, J = 13.0, 2.6 Hz, 1H); 3.08 (td, J = 12.3, 3.5 Hz, 1H); 2.88 (td, J = 13.1, 3.4 Hz, 1H); 2.70 (dd, J = 12.8, 11.4 Hz, 1H). 13C NMR (DMSO-d6): δ 162.08, 156.81, 156.01, 142.93, 140.31, 139.82, 139.63, 136.66, 130.62, 130.52, 128.84, 128.77, 128.61, 128.00, 125.84, 124.86, 120.94, 120.45, 114.77, 114.56, 86.35, 58.93, 44.40, 42.66, 42.21, 41.00. MS (ESI): m/z calcd for C26H23FN3O3 [M + H]+, 444.49; found, 444.26. TR = 18.91 min.
1,1-Dicyclohexyl-N-(4-fluorobenzyl)-3-oxotetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (10)
White solid (105 mg, 23% yield); mp 94–95 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.37–7.24 (m, 3H); 7.18–7.05 (m, 2H); 4.31–4.16 (m, 2H); 4.06–3.93 (m, 2H); 3.55–3.42 (m, 2H); 2.98–2.72 (m, 3H); 2.00–1.40 (m, 12H); 1.38–0.85 (m, 10H). 13C NMR (DMSO-d6): δ 162.14, 159.74, 157.13, 155.32, 136.82, 128.96, 128.89, 114.82, 114.61, 85.71, 55.26, 43.74, 42.82, 42.18, 40.90, 27.61, 26.54, 26.20, 25.84, 25.77, 25.53, 25.38. MS (ESI): m/z calcd for C26H37FN3O3 [M + H]+, 458.60; found, 458.47. TR = 21.11 min.
N-(4-Fluorobenzyl)-3-oxo-1,1-dipropyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (11)
White solid (189 mg, 50% yield); mp 74–77 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.34–7.21 (m, 3H); 7.15–7.10 (m, 2H); 4.23 (d, J = 5.5 Hz, 2H); 4.00 (t, J = 14.1 Hz, 2H); 3.52 (dd, J = 12.7, 2.6 Hz, 1H); 3.42–3.30 (m, 1H); 2.95–2.63 (m, 3H); 1.72–1.43 (m, 4H); 1.42–1.15 (m, 4H); 0.92–0.88 (m, 6H). 13C NMR (DMSO-d6): δ 162.14, 159.73, 156.99, 155.22, 136.86, 128.92, 128.84, 114.84, 114.63, 83.14, 59.10, 44.12, 42.77, 42.58, 33.80, 16.11, 15.88, 14.26, 14.13. MS (ESI): m/z calcd for C20H29FN3O3 [M + H]+, 378.47; found, 378.38. TR = 13.08 min.
N-Benzyl-3-oxo-1,1-diphenyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carbothioamide (12)
White solid (306 mg, 69% yield); mp 184–186 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.50 (s, 1H); 7.73–7.00 (m, 15H); 4.85–4.76 (m, 3H); 4.59–4.54 (m, 2H); 3.64 (d, J = 12.6 Hz, 1H); 3.15–2.98 (m, 2H); 2.40 (t, J = 12.2 Hz, 1H). 13C NMR (DMSO-d6): δ 182.31, 155.12, 142.22, 139.22, 138.34, 128.54, 128.45, 128.36, 128.07, 127.99, 126.99, 126.58, 125.70, 125.42, 84.66, 59.42, 50.63, 48.53, 46.14, 40.82. MS (ESI): m/z calcd for C26H26N3O2S [M + H]+, 444.57; found, 444.23. TR = 15.84 min.
1,1-Diphenylhexahydro-3H-oxazolo[3,4-a]pyrazin-3-one (28)
DBU (1.2 mmol) was added to a stirring solution of 26j (1 mmol) in anhydrous THF (10 mL). The reaction solution was allowed stirring at room temperature for 18 h and monitored by TLC. The solvent was removed under vacuum, and the residue was dissolved in EtOAc (20 mL) and washed with water (20 mL). The organic phase was separated, dried over Na2SO4, and the solvent was evaporated to give a residue that was purified via flash column chromatography on silica gel using a 1:3 mixture of petroleum ether and EtOAc as the eluent.
Off-white solid (256 mg, 87% yield). 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.52 (d, J = 7.2 Hz, 2H); 7.41–7.24 (m, 8H); 4.32 (d, J = 10.1 Hz, 1H); 4.10–3.71 (m, 2H); 3.16 (t, J = 5.6 Hz, 1H); 2.97–2.76 (m, 1H); 2.28 (td, J = 11.6, 3.4 Hz, 1H); 1.83–1.75 (m, 1H), 1.21 (br s, 1H). MS (ESI): m/z calcd for C18H19N2O2 [M + H]+, 295.36; found, 295.38.
General Procedure for the Synthesis of Final Compounds 13 and 14
2-Chloro-N-(4-fluorobenzyl)acetamide or 2-chloro-N-(4-fluorophenyl)acetamide (1 mmol) was added to a mixture of compound 28 (1 mmol) and K2CO3 (1.5 mmol) in CH3CN (15 mL). The reaction mixture was heated at 90 °C for 4 h after which the solvent was removed under vacuum, and the residue was partitioned between water (15 mL) and CH2Cl2 (15 mL). The aqueous phase was further extracted with CH2Cl2 (2 × 15 mL), and the combined organic layers were washed with brine (10 mL) and dried over Na2SO4. After evaporation, the residue was purified by flash column chromatography on silica gel using a 1:1 mixture of EtOAc/PEt as the eluent.
N-(4-Fluorophenyl)-2-(3-oxo-1,1-diphenyltetrahydro-3H-oxazolo[3,4-a]pyrazin-7(1H)-yl)acetamide (13)
White solid (196 mg, 44% yield); mp 205–208 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 9.76 (s, 1H); 7.62–7.60 (m, 4H); 7.44–7.29 (m, 8H); 7.17–7.13 (m, 2H); 4.77 (dd, J = 11.2, 3.6 Hz, 1H); 3.59 (dd, J = 13.1, 2.9 Hz, 1H); 3.23–3.13 (m, 3H); 2.74 (d, J = 10.5 Hz, 1H); 2.61 (dd, J = 13.2, 2.6 Hz, 1H); 2.19 (td, J = 12.9, 3.4 Hz, 1H); 1.78 (t, J = 12.2 Hz, 1H). 13C NMR (DMSO-d6): δ 167.92, 159.26, 156.88, 155.21, 142.80, 138.58, 134.74, 128.51, 128.36, 128.19, 127.72, 125.53, 125.18, 121.46, 115.24, 115.02, 84.54, 60.85, 60.11, 54.19, 50.66, 41.24. MS (ESI): m/z calcd for C26H25FN3O3 [M + H]+, 446.50; found, 446.69. TR = 18.30.
N-(4-Fluorobenzyl)-2-(3-oxo-1,1-diphenyltetrahydro-3H-oxazolo[3,4-a]pyrazin-7(1H)-yl)acetamide (14)
White solid (170 mg, 37% yield); mp 137–140 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.41–8.37 (m, 1H); 7.56–7.54 (m, 2H); 7.43–7.39 (m, 2H); 7.37–7.25 (m, 8H); 7.17–7.13 (m, 2H); 4.76 (dd, J = 10.9, 3.4 Hz, 1H); 4.30–4.22 (m, 2H); 3.62–3.56 (m, 1H); 3.21–3.08 (m, 1H); 3.04 (d, J = 13.2 Hz, 1H); 2.88 (d, J = 12.9 Hz, 1H); 2.67–2.63 (m, 1H); 2.49–2.44 (m, 1H); 2.29–2.05 (m, 1H); 1.60 (t, J = 12.2 Hz, 1H). 13C NMR (DMSO-d6): δ 168.94, 162.24, 155.12, 142.77, 138.49, 135.83, 129.10, 128.51, 128.28, 128.22, 127.72, 125.46, 125.10, 115.38, 101.57, 84.43, 60.51, 59.96, 54.32, 50.97, 41.14. MS (ESI): m/z calcd for C27H27FN3O3 [M + H]+, 460.53; found, 460.20. TR = 18.16.
Method A for the Synthesis of Guanidine Derivatives 15 and 16
A solution of NaHCO3 (1.7 mmol) in H2O (1 mL) was added at 0 °C to a stirring solution of 28 (1 mmol) in CH2Cl2 (5 mL). At the same temperature, a solution of cyanogen bromide (1.2 mmol) in CH2Cl2 (5 mL) was added. The heterogeneous mixture was vigorously stirred at 0 °C for 30 min, then warmed to room temperature, and stirred for further 24 h. After this time, the layers were separated and the organic phase was washed with a saturated solution of NaHCO3 (2 × 10 mL), dried with anhydrous Na2SO4, and concentrated under vacuum to give a residue from which compound 29 was purified via flash column chromatography on silica gel using a 1:1 mixture of petroleum ether and EtOAc as an eluent.
White solid (160 mg, 50% yield). 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.52–7.25 (m, 10H), 4.60 (dd, J = 11.2, 3.6 Hz, 1H), 3.98–3.88 (m, 1H), 3.37–3.22 (m, 2H), 3.10–3.02 (m, 2H), 1.27–1.18 (m, 1H); MS (ESI): m/z calcd for C19H18N3O2 [M + H]+, 320.37; found, 320.40.
Benzylamine or 4-fluorobenzylamine (3 mmol) was added to a stirring solution of 29 (1 mmol) in DMSO (3 mL) in the presence of a catalytic amount of p-toluenesulfonic acid. After 18 h of stirring at 60 °C, the reaction solution was diluted with water (10 mL) and extracted with EtOAc (3 × 15 mL). The organic layers were dried over Na2SO4 and concentrated in vacuum after which crude products were purified by flash column chromatography on silica gel using a 4:1 mixture of CH2Cl2 and MeOH.
Method B for the Synthesis of Guanidine Derivatives 15 and 16
Compounds 15 and 16 were alternatively synthetized according to a manual solid-phase synthesis approach described previously.18 Briefly, compounds 31a or 31b (0.62 mmol) was added to a suspension of 2-(4-bromomethyl-phenoxy)ethyl polystyrene HL resin (substitution: 1.23 mmol/g, 0.62 mmol) in a 2:1 mixture of CH2Cl2/DMF (3 mL). The mixture was heated at 50 °C until starting material consumption was observed (4 h). After that, each of the two differently functionalized resins was filtered, washed with DMF (2 × 5 mL) and CH2Cl2 (2 × 5 mL), and dried. Subsequently, the respective functionalized resin (0.62 mmol) was suspended in CH3CN (2 mL) before adding compound 28 (1.55 mmol) and HgCl2 (0.93 mmol). After heating at 90 °C for 24 h, a simple filtration was performed and the filtrates were purified by flash column chromatography on silica gel using a 4:1 mixture of CH2Cl2 and MeOH.
N-Benzyl-3-oxo-1,1-diphenyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboximidamide (15)
White solid (method A: 56 mg, 21% yield; method B: 103 mg, 39% yield); mp 68–69 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.38 (m, 1H), 7.90 (s, 2H), 7.61–7.54 (m, 2H), 7.46–7.25 (m, 14H), 4.77 (dd, J = 11.3, 3.5 Hz, 1H), 4.43 (d, J = 5.7 Hz, 2H), 3.78–3.58 (m, 2H), 3.23–3.17 (m, 1H), 3.08–3.03 (m, 1H), 2.64–2.57 (m, 1H). 13C NMR (DMSO-d6): δ 156.79, 155.36, 142.90, 138.39, 137.37, 129.02, 128.56, 128.00, 127.57, 126.10, 85.18, 59.76, 49.02, 45.63, 45.56, 41.20. MS (ESI): m/z calcd for C26H27N4O2 [M + H]+, 427.53; found, 427.44. TR = 17.68.
N-(4-Fluorobenzyl)-3-oxo-1,1-diphenyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboximidamide (16)
White solid (method A: 28 mg, 10% yield; method B: 85 mg, 31% yield); mp 82–85 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.43 (br s, 1H), 7.95 (s, 2H), 7.60 (d, J = 7.4 Hz, 2H), 7.52–7.30 (m, 9H), 7.29–7.09 (m, 3H), 4.84–4.74 (m, 1H), 4.42 (d, J = 5.5 Hz, 2H), 3.85–3.53 (m, 3H), 3.23–3.12 (m, 1H), 2.71–2.57 (m, 1H). 13C NMR (DMSO-d6): δ 162.69, 156.14, 154.85, 142.41, 137.86, 133.08, 129.26, 129.18, 128.48, 128.03, 125.54, 115.24, 84.65, 59.26, 54.27, 48.49, 45.03, 44.37, 40.66. MS (ESI): m/z calcd for C26H26FN4O2 [M + H]+, 445.52; found, 445.39. TR = 18.09.
General Procedure for the Synthesis of 36a–e
Enantiomerically pure monosubstituted piperazines 35a–e were prepared starting from commercial chiral amino-esters following previously reported procedures, and the analytical data for intermediates 33–35a–e are in agreement with data from the literature.19,50 A solution of di-tert-butyl dicarbonate (1.1 mmol) in anhydrous THF was added at 0 °C to a solution of compounds 35a–e (1 mmol) in dry THF (10 mL). The reaction mixture was warmed at room temperature and further stirred for 1 h. The solvent was removed in vacuo, and the residue was dissolved in CH2Cl2 (15 mL) and washed with water (3 × 10 mL) and brine (1 × 10 mL). The organic phase was dried over anhydrous Na2SO4 and concentrated to dryness. Flash column chromatography on silica gel using a 0.5:9.5 mixture of EtOAc/PEt as the eluent provided the desired compounds with good yields.
(S)-tert-Butyl 4-Benzyl-2-methylpiperazine-1-carboxylate (36a)
Colorless oil (192 mg, 66% yield). 1H NMR (200 MHz, CDCl3-d): δ (ppm) 7.41–7.20 (m, 5H), 4.28–4.10 (m, 1H), 3.89–3.73 (m, 1H), 3.63–3.33 (m, 2H), 3.23–3.00 (m, 1H), 2.87–2.67 (m, 1H), 2.67–2.50 (m, 1H), 2.23–1.90 (m, 2H), 1.45 (s, 9H), 1.24 (d, J = 6.6 Hz, 3H). MS (ESI): m/z calcd for C17H27N2O2 [M + H]+, 291.42; found, 291.04.
(S)-tert-Butyl 4-Benzyl-2-isopropylpiperazine-1-carboxylate (36b)
Colorless oil (223 mg, 70% yield). 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.37–7.17 (m, 5H), 3.88–3.73 (m, 1H), 3.60–3.43 (m, 2H), 3.28 (s, 1H), 3.00–2.70 (m, 3H), 2.40–2.20 (m, 1H), 2.00–1.77 (m, 2H), 1.37 (s, 9H), 0.77–0.70 (m, 6H). MS (ESI): m/z calcd for C19H31N2O2 [M + H]+, 319.47; found, 319.46.
(S)-tert-Butyl 4-Benzyl-2-isobutylpiperazine-1-carboxylate (36c)
Colorless oil (239 mg, 72% yield). 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.39–7.17 (m, 5H), 4.00 (br s, 1H), 3.83–3.70 (m, 1H), 3.60–3.47 (m, 1H), 3.31 (s, 1H), 3.03–2.85 (m, 1H), 2.80–2.70 (m, 1H), 2.60–2.50 (m, 1H), 2.00–1.82 (m, 2H), 1.60–1.47 (m, 2H), 1.38 (s, 9H), 1.30–1.20 (m, 1H), 0.88–0.80 (m, 6H). MS (ESI): m/z calcd for C20H33N2O2 [M + H]+, 333.50; found, 333.07.
(S)-tert-Butyl 2,4-Dibenzylpiperazine-1-carboxylate (36d)
Colorless oil (136 mg, 37% yield). 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.42–7.22, (m, 5H), 7.22–7.10 (m, 3H), 7.03–6.95 (m, 2H), 4.13–4.00 (m, 1H), 3.87–3.72 (m, 1H), 3.63–3.52 (m, 1H), 3.27–3.16 (m, 1H), 3.00–2.73 (m, 4H), 2.60 (m, 1H), 2.10–1.83 (m, 2H), 1.26 (s, 9H). MS (ESI): m/z calcd for C23H31N2O2: [M + H]+, 367.51; found, 367.02.
(S)-tert-Butyl 4-Benzyl-2-phenylpiperazine-1-carboxylate (36e)
Colorless oil (229 mg, 65% yield). 1H NMR (200 MHz, DMSO-d6): δ (ppm) 7.34–7.18 (m, 10 H), 5.17–5.06 (m, 1H), 3.90–3.73 (m, 1H), 3.60–3.44 (m, 2H), 3.30–3.20 (m, 1H), 3.00–2.83 (m, 1H), 2.83–2.70 (m, 1H), 2.37–2.25 (m, 1H), 2.13–1.93 (m, 1H), 1.39 (s, 9H). MS (ESI): m/z calcd for C22H29N2O2 [M + H]+, 353.49; found, 353.45.
General Procedure for the Synthesis of 37a–e
TMEDA (2.7 mmol) was added under an argon atmosphere at room temperature to a stirring solution of 36a-e (1 mmol) in freshly distilled THF (5 mL). After cooling at −78 °C, sec-BuLi (2.7 mmol) was added, and the reaction solution was allowed to reach −30 °C over 2 h. A solution of benzophenone (2 mmol) in THF (5 mL) was added, and the reaction mixture was left stirring at −30 °C for 30 min, then slowly warmed to room temperature, and stirred for 18 h. The reaction was quenched with a saturated solution of NH4Cl (15 mL), and the solvents were concentrated under vacuum to half volume giving a residue, which was extracted with EtOAc (3 × 15 mL). The organic layers were combined, dried over Na2SO4, and the solvent was removed under vacuum. The resulting crude product was purified by flash column chromatography on silica gel using a 1:9 mixture of EtOAc/PEt as the eluent.
(5S,8aR)-7-Benzyl-5-methyl-1,1-diphenylhexahydro-3H-oxazolo[3,4-a]pyrazin-3-one (37a)
Yellow oil (100 mg, 25% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.56–7.44 (m, 2H), 7.44–7.20 (m, 13H), 4.70 (d, J = 9.1 Hz, 1H), 4.15–4.01 (m, 1H), 3.60–3.45 (m, 1H), 3.33–3.22 (m, 1H), 2.63–2.47 (m, 2H), 2.11–1.97 (m, 1H), 1.62–1.50 (m, 2H), 1.39–1.29 (m, 2H). MS (ESI): m/z calcd for C26H27N2O2 [M + H]+, 399.51; found, 399.44.
(5S,8aR)-7-Benzyl-5-isopropyl-1,1-diphenylhexahydro-3H-oxazolo[3,4-a]pyrazin-3-one (37b)
Colorless oil (77 mg, 18% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.70–7.60 (m, 2H), 7.60–7.14 (m, 13H), 4.73–4.40 (m, 1H), 3.70–3.03 (m, 3H), 2.91–2.70 (m, 1H), 2.61–2.47 (m, 1H), 2.34–2.18 (m, 1H), 1.94–1.91 (m, 1H), 1.27–1.22 (m, 1H), 0.79 (s, 3H), 0.78 (s, 3H). MS (ESI): m/z calcd for C28H31N2O2 [M + H]+, 427.57; found, 427.48.
(5S,8aR)-7-Benzyl-5-isobutyl-1,1-diphenylhexahydro-3H-oxazolo[3,4-a]pyrazin-3-one (37c)
Yellow oil (79 mg, 18% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.57–7.47 (m, 2H), 7.43–7.29 (m, 13H), 4.76–4.64 (m, 1H), 4.06–3.86 (m, 1H), 3.56–3.47 (m, 1H), 3.27–3.16 (m, 1H), 2.69–2.25 (m, 2H), 2.11–1.98 (m, 1H), 1.65–1.63 (m, 2H), 1.03–0.89 (m, 2H), 0.86 (dd, J = 6.5, 3.7 Hz, 6H). MS (ESI): m/z calcd for C29H33N2O2 [M + H]+, 441.60; found, 441.46.
(5S,8aR)-5,7-Dibenzyl-1,1-diphenylhexahydro-3H-oxazolo[3,4-a]pyrazin-3-one (37d)
Yellow oil (247 mg, 52% yield). 1H NMR (200 MHz, CDCl3): δ (ppm) 7.46–7.44 (m, 2H), 7.39–7.27 (m, 13H), 7.15–7.08 (m, 3H), 7.01–6.94 (m, 2H), 4.77 (dd, J = 11.1, 3.3 Hz, 1H), 4.19–4.07 (m, 1H), 3.54 (d, J = 12.9 Hz, 1H), 3.23 (d, J = 12.9 Hz, 1H), 3.12–2.98 (m, 2H), 2.63 (d, J = 11.5 Hz, 2H), 1.95 (dd, J = 11.7, 3.9 Hz, 1H), 1.73–1.60 (m, 1H). MS (ESI): m/z calcd for C32H31N2O2 [M + H]+, 475.61; found, 475.35.
(5S,8aR)-7-Benzyl-1,1,5-triphenylhexahydro-3H-oxazolo[3,4-a]pyrazin-3-one (37e)
C31H28N2O2 (88 mg, 19% yield); 1H NMR (400 MHz, CDCl3): δ (ppm) 7.61–7.58 (m, 2H), 7.37–7.28 (m, 6H), 7.24–6.88 (m, 11H), 3.84–3.75 (m, 1H), 3.55–3.38 (m, 2H), 3.32–3.29 (m, 2H), 2.96–2.80 (m, 1H), 2.64–2.47 (m, 1H), 2.35–2.20 (m, 1H), 1.36–1.22 (m, 1H). MS (ESI): m/z calcd for C31H29N2O2 [M + H]+, 461.59; found, 461.49.
General Procedure for the Synthesis of 38a–e
9-Fluorenylmethoxycarbonyl chloride (FmocCl, 1.1 mmol) was added to a solution of 37a–e (1 mmol) in CH3CN (5 mL). The reaction solution was heated at 90 °C for 5 h and then stirred at room temperature for 18 h. The solvent was evaporated giving a residue that was dissolved in EtOAc (15 mL), and the resulting organic phase was washed with water (3 × 10 mL), dried over anhydrous Na2SO4, and concentrated to dryness. All crude residues were finally purified via flash column chromatography on silica gel using a 1:4 mixture of EtOAc/PEt as the eluent.
(9H-Fluoren-9-yl)methyl (5S,8aR)-5-Methyl-3-oxo-1,1-diphenyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (38a)
White solid (440 mg, 83% yield); 1H NMR (400 MHz, DMSO): δ (ppm) 7.97–7.84 (m, 3H), 7.47–7.22 (m, 13H), 7.06–6.93 (m, 2H), 5.02–4.91 (m, 1H), 4.51–4.33 (m, 2H) 4.33–4.21 (m, 1H), 4.13–4.05 (m, 1H), 3.11–3.00 (m, 1H), 3.00–2.87 (m, 1H), 2.87–2.75 (m, 1H), 2.33–2.11 (m, 1H), 0.87–0.79 (d, J = 7.2 Hz, 3H); 13C NMR (400 MHz, DMSO): δ 154.61, 144.05, 141.94, 140.67, 137.74, 128.64, 128.46, 127.86, 127.60, 127.54, 127.07, 127.00, 125.43, 124.95, 120.02, 84.15, 64.96, 55.01, 47.27, 46.36, 45.96, 45.02, 24.02. MS (ESI): m/z calcd for C34H31N2O4 [M + H]+, 531.63; found, 531.52.
(9H-Fluoren-9-yl)methyl (5S,8aR)-5-Isopropyl-3-oxo-1,1-diphenyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (38b)
White solid (318 mg, 57% yield); 1H NMR (400 MHz, CDCl3): δ (ppm): 7.87–7.73 (m, 2H), 7.64–7.16 (m, 14H), 7.07–6.96 (m, 2H), 5.29–5.20 (dd, 1H), 4.56–4.40 (m, 1H), 4.24–4.11 (m, 2H), 3.40–3.16 (m, 1H), 2.80–2.64 (m, 1H), 2.26–2.01 (m, 1H), 1.81–1.56 (m, 2H), 1.16–1.04 (m, 1H), 0.94 (d, J = 6.4 Hz, 3H), 0.62 (d, J = 6.8 Hz, 3H); 13C NMR (400 MHz, CDCl3): δ 155.96, 144.29, 143.23, 142.11, 141.41, 138.79, 138.02, 128.61, 127.96, 127.85, 127.18, 125.64, 125.34, 124.56, 120.00, 119.63, 84.87, 64.89, 57.60, 56.77, 48.27, 46.09, 43.89, 25.64, 19.98, 19.47. MS (ESI): m/z calcd for C36H35N2O4 [M + H]+, 559.69; found, 559.57.
(9H-Fluoren-9-yl)methyl (5S,8aR)-5-Isobutyl-3-oxo-1,1-diphenyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (38c)
White solid (309 mg, 54% yield). 1H NMR (400 MHz, CDCl3): δ (ppm): 7.78–7.75 (m, 2H), 7.64–7.16 (m, 14H), 7.07–6.96 (m, 2H), 5.29–5.20 (m, 1H), 4.56–4.40 (m, 1H), 4.24–4.11 (m, 2H), 3.40–3.16 (m, 1H), 2.80–2.64 (m, 1H), 2.26–2.01 (m, 1H), 1.81–1.56 (m, 2H), 1.40–1.23 (m, 2H), 1.16–1.04 (m, 1H), 0.98–0.91 (d, J = 6.4 Hz, 3H), 0.67–0.60 (d, J = 6.8 Hz, 3H). MS (ESI): m/z calcd for C37H37N2O4 [M + H]+, 573.71; found, 573.73.
(9H-Fluoren-9-yl)methyl (5S,8aR)-5-Benzyl-3-oxo-1,1-diphenyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (38d)
White solid (376 mg, 62% yield). 1H NMR (400 MHz, CDCl3): δ (ppm) 7.84–7.71 (m, 2H), 7.71–7.01 (m, 21H), 5.22–5.11 (m, 1H), 4.63–4.47 (m, 2H), 4.26–4.15 (m, 1H), 4.13–3.89 (m, 2H), 3.42–3.31 (m, 1H), 2.89–2.82 (m, 1H), 2.82–2.61 (m, 1H), 2.61–2.44 (m, 1H), 2.27–2.00 (m, 1H). MS (ESI): m/z calcd for C40H35N2O4 [M + H]+, 607.73; found, 607.49.
(9H-Fluoren-9-yl)methyl (5S,8aR)-3-Oxo-1,1,5-triphenyltetrahydro-3H-oxazolo[3,4-a]pyrazine-7(1H)-carboxylate (38e)
White solid (350 mg, 59% yield); 1H NMR (400 MHz, CDCl3): δ (ppm) 7.95–6.88 (m, 23H), 5.10–4.98 (m, 1H), 4.68–4.52 (m, 1H), 4.40–4.30 (m, 1H), 4.30–4.18 (m, 1H), 4.18–4.09 (m, 1H), 3.83–3.78 (m, 1H), 3.78–3.67 (m, 1H), 2.93–2.73 (m, 2H). MS (ESI): m/z calcd for C39H33N2O4 [M + H]+, 593.70; found, 593.50.
General Procedure for the Synthesis of Final Compounds 17–21
4-Fluorobenzyl isocyanate (2 mmol) and DBU (1.2 mmol) were sequentially added at room temperature to a stirring solution of 38a–e (1 mmol) in anhydrous THF (5 mL). The reaction solution was stirred for 2 h and then it was quenched with a saturated solution of NH4Cl (10 mL). The mixture was concentrated under vacuum to half volume and the aqueous phase was extracted with EtOAc (3 × 15 mL). The organic layers were combined, dried over Na2SO4, and concentrated in vacuum. The desired products were purified by preparative RP-HPLC.
(5S,8aR)-N-(4-Fluorobenzyl)-5-methyl-3-oxo-1,1-diphenyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (17)
White solid (225 mg, 49% yield); mp 153–156 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.57–7.55 (m, 2H), 7.44–7.25 (m, 11H), 7.14–7.09 (m, 2H), 4.70 (dd, J = 11.3, 3.8 Hz, 1H), 4.28–4.14 (m, 2H), 3.98–3.84 (m, 2H), 3.79 (d, J = 13.7 Hz, 1H), 2.87 (dd, J = 13.7, 3.9 Hz, 1H), 2.15–2.01 (m, 1H), 1.15 (d, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6): δ 162.12, 157.39, 154.69, 142.36, 138.46, 136.92, 128.83, 128.75, 128.55, 128.40, 128.24, 127.84, 125.61, 125.35, 114.83, 114.62, 84.82, 55.76, 46.83, 46.11, 45.45, 42.80, 15.33. MS (ESI): m/z calcd for C26H27FN3O3 [M + H]+, 460.53; found, 460.49. TR = 23.10. [α]D22 = +1059.13 (c 0.023).
(5S,8aR)-N-(4-Fluorobenzyl)-5-isopropyl-3-oxo-1,1-diphenyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (18)
White solid (229 mg, 47% yield); mp 186–187 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.65 (d, J = 7.5 Hz, 2H); 7.43–7.38 (m, 6H); 7.35–7.28 (m, 2H); 7.28–7.20 (m, 3H); 7.13–7.08 (m, 2H); 4.67 (dd, J = 11.2, 3.9 Hz, 1H); 4.19 (dd, J = 15.1, 5.4 Hz, 2H); 4.01 (d, J = 14.2 Hz, 1H); 3.71 (dd, J = 12.8, 3.6 Hz, 1H); 3.26 (dd, J = 12.3, 3.8 Hz, 1H); 2.88 (dd, J = 13.8, 3.9 Hz, 1H); 2.36–2.21 (m, 1H); 1.93–1.76 (m, 1H); 0.92 (d, J = 6.6 Hz, 3H); 0.60 (d, J = 6.6 Hz, 3H). 13C NMR (DMSO-d6): δ 162.11, 159.71, 157.18, 155.27, 142.90, 138.34, 136.94, 128.82, 128.75, 128.50, 128.43, 128.15, 127.79, 125.25, 125.17, 114.80, 114.59, 84.77, 57.01, 56.58, 45.15, 43.05, 42.82, 26.10, 19.47, 18.98. MS (ESI): m/z calcd for C29H31FN3O3 [M + H]+, 488.58; found, 488.54. TR = 26.07. [α]D22 = +1058.5 (c 0.012).
(5S,8aR)-N-(4-Fluorobenzyl)-5-isobutyl-3-oxo-1,1-diphenyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (19)
White solid (100 mg, 20% yield); mp 216–219 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.61–7.55 (m, 2H), 7.42–7.07 (m, 13H), 4.69 (dd, J = 11.1, 3.6 Hz, 1H), 4.29–4.13 (m, 3H), 3.85–3.75 (m, 2H), 2.91 (dd, J = 13.6, 3.9, 1H), 2.17 (m, 1H), 1.63–1.52 (m, 1H), 1.40–1.23 (m, 2H), 0.80 (d, J = 6.4, 3H), 0.76 (d, J = 6.8, 3H). 13C NMR (DMSO-d6): δ 162.12, 157.29, 155.77, 155.01, 142.58, 138.36, 136.94, 128.96, 128.85, 128.78, 128.44, 128.19, 127.79, 125.42, 125.28, 115.13, 114.96, 114.75, 114.57, 84.79, 55.89, 48.69, 45.99, 45.16, 42.75, 37.37, 24.13, 22.64, 21.65. MS (ESI): m/z calcd for C30H33FN3O3 [M + H]+, 502.61; found, 502.40. TR = 26.28. [α]D22 = + 794.29 (c 0.022).
(5S,8aR)-N-(4-Fluorobenzyl)-5-benzyl-3-oxo-1,1-diphenyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (20)
White solid (134 mg, 25% yield); mp 95–98 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.51–7.48 (m, 2H); 7.43–7.25 (m, 11H); 7.16–7.04 (m, 7H); 4.87 (dd, J = 11.2, 3.7 Hz, 1H); 4.29–4.18 (m, 2H); 4.03–3.99 (m, 1H); 3.94–3.88 (m, 1H); 3.82 (dd, J = 13.1, 3.2 Hz, 1H); 2.99–2.79 (m, 3H); 2.26–2.14 (m, 1H). 13C NMR (DMSO-d6): δ 162.12, 159.73, 157.43, 154.95, 142.44, 138.22, 137.71, 136.86, 128.96, 128.85, 128.37, 128.29, 128.05, 127.93, 127.79, 126.05, 125.57, 125.33, 114.82, 114.61, 84.76, 56.42, 52.19, 45.66, 45.35, 42.87, 34.85. MS (ESI): m/z calcd for C33H31FN3O3 [M + H]+, 536.63; found, 536.38. TR = 26.10. [α]D23 = +539.8 (c 0.024).
(5S,8aR)-N-(4-Fluorobenzyl)-3-oxo-1,1,5-triphenyltetrahydro-1H-oxazolo[3,4-a]pyrazine-7(3H)-carboxamide (21)
White solid (83 mg, 16% yield); mp 105–107 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 7.63 (d, J = 7.9 Hz, 2H); 7.50 (t, J = 7.7 Hz, 2H); 7.39 (t, J = 7.1 Hz, 1H); 7.24 (t, J = 6.9 Hz, 2H); 7.15–6.99 (m, 11H); 6.97–6.88 (m, 2H); 5.03 (d, J = 13.9 Hz, 1H); 4.17–4.06 (m, 2H); 3.75–3.62 (m, 2H); 2.85–2.80 (m, 1H), 2.71–2.55 (m, 2H). 13C NMR (DMSO-d6): δ 162.04, 159.64, 155.95, 139.25, 138.42, 136.73, 134.11, 128.60, 128.01, 127.42, 125.92, 125.61, 114.69, 114.47, 87.97, 71.18, 47.37, 42.64, 42.11. MS (ESI): m/z calcd for C32H29FN3O3 [M + H]+, 522.60; found, 522.46. TR = 26.72. [α]D23 = +8.8 (c 0.025).
Pharmacology
Calcium Mobilization Assay
HEK293mNPSR cells were generated as previously described3 and maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and hygromycin B (100 mg/L) and cultured at 37 °C in 5% CO2 humidified air. HEK293mNPSR cells were seeded at a density of 50,000 cells/well into poly-d-lysine coated 96-well black, clear-bottom plates. The following day, the cells were incubated with medium supplemented with 2.5 mM probenecid, 3 μM of the calcium sensitive fluorescent dye Fluo-4 AM, and 0.01% pluronic acid, for 30 min at 37 °C. After that time, the loading solution was aspirated and 100 μL/well of assay buffer (Hanks’ balanced salt solution; HBSS) supplemented with 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 2.5 mM probenecid, and 500 μM Brilliant Black (Sigma-Aldrich) was added. Concentrated solution (1 mM) of NPS was made in bidistilled water and kept at −20 °C. Concentrated solutions (10 mM) of NPSR antagonists were made in DMSO and kept at −20 °C. Serial dilutions were carried out in HBSS/HEPES (20 mM) buffer (containing 0.02% bovine serum albumin fraction V). After placing both plates (cell culture and master plate) into the fluorometric imaging plate reader FlexStation II (Molecular Devices, Sunnyvale, CA), fluorescence changes were measured. On-line additions were carried out in a volume of 50 μL/well. To facilitate drug diffusion into the wells in antagonist type experiments, the present studies were performed at 37 °C, and three cycles of mixing (25 μL from each well moved up and down 3 times) were performed immediately after antagonist injection to the wells. Inhibition response curves were determined against the stimulatory effect of 10 nM NPS. Additionally, the concentration–response curve to NPS has been tested in the absence and in the presence of 100 nM of 1, compound 16, and compound 21 (Figure 3). NPSR antagonists were injected into the wells 24 min before adding NPS.
Mouse Locomotor Activity Test
All animal care and experimental procedures conformed with the European Communities Council Directives (2010/63/EU) and national regulations (D.L. 26/2014). Studies involving animals are reported in accordance with the ARRIVE guidelines.51 This study was approved by the Italian Ministry of Health (authorization number 120/2014-PR). The experiments were performed with CD-1 mice (2–4 month old, from the Laboratory for Preclinical Research (LARP) of the University of Ferrara, Italy). Mice were housed under standard conditions (22 °C, 55% humidity, 12 h light/dark cycle, light on at 7:00 am), with free access to food and water. Appropriate environmental enrichment was present in each cage. Mice were killed with CO2 overdose. Each animal was used only once. Experiments were performed during the light cycle (between 09.00 and 13.00) according to Guerrini et al. (2009).42 For in vivo studies, 1, 16, and 21 were solubilized in water containing 1% DMSO and 10% Cremophor EL (Sigma-Aldrich). NPS was solubilized in saline solution. Vehicle, 1, 16, and 21 were injected by the ip route 30 min before saline or NPS injection. NPS or saline were given by the i.c.v. route 15 min before the beginning of the test and the locomotor activity was recorded for 30 min. The i.c.v. injections (2 μL/mouse) were given under light (just enough to produce loss of the righting reflex) isofluorane anesthesia into the left ventricle according to the procedure described by Laursen and Belknap (1986)52 and routinely adopted in our laboratory. For these experiments, the ANY-maze video tracking system was used (Ugo Basile, application version 4.52c Beta). Mice were positioned in a square plastic cage (40 × 40 cm), one mouse per cage. Four mice were monitored in parallel. Mouse horizontal activity was monitored by a camera while vertical activity was measured by an infrared beam array. The parameters measured were cumulative distance traveled (total distance in m that the animal traveled during the test), immobility time (the animal is considered immobile when 90% of it remains in the same place for a minimum of 2.5 s), and the number of rearings (the number of beam breaks due to the vertical movements; this input is triggered only when the beam is interrupted for at least 200 ms). Previous studies performed under the present experimental conditions demonstrated that NPS stimulated locomotor activity in a dose-dependent way;42 from these studies, the dose of 0.1 nmol was selected as the lower dose inducing statistically significant effects.
Data Analysis and Terminology
Data are expressed as means ± standard error of the mean (SEM) of n experiments. Nonlinear regression analysis using GraphPad Prism software (v.4.0) allowed logistic iterative fitting of the resultant responses and the calculation of agonist potencies and maximal effects. Agonist potency was expressed as pEC50, which is the negative logarithm to base 10 of the agonist molar concentration that produces 50% of the maximal possible effect of that agonist. In inhibition response experiments (i.e., increasing concentrations of antagonist vs a fixed concentration of agonist), the antagonist potency was expressed as pKB, derived from the following equation
where IC50 is the concentration of antagonist that produces 50% inhibition of the agonist response, [A] is the concentration of agonist, EC50 is the concentration of agonist producing a 50% maximal response, and n is the Hill coefficient of the concentration–response curve to the agonist. When the concentration–response curve to NPS has been tested in the absence and in the presence of antagonists, the antagonist potency was expressed as pA2, derived from the following equation
where CR means the ratio between agonist potency in the presence and absence of antagonist and [A] the molar concentration of the antagonist.
In vivo data are expressed as mean ± SEM of n animals. Data were analyzed using two-way ANOVA, followed by Bonferroni’s post-hoc test. Differences were considered statistically significant when p < 0.05.
Molecular Modeling
hNPSR Model Construction
The query sequence of the hNPSR in its I107 variant (Q6W5P4, FASTA format) was downloaded from the Universal Protein Resource.53 BLAST (Basic Local Alignment Search Tool) was used to search the homologous sequences to be used as template structures. The sequences of templates were obtained from the Uniprot web server. The human C5a anaphylatoxin chemotactic receptor 1 (P21730, PDB 6C1R),54 the human κ opioid receptor (P41145, PDB 4DJH),33 the human M2 muscarinic receptor (P08172, PDB 5ZKC),55 the human neuropeptide Y Y1 Receptor (Q15761, PDB 5ZBH),29 the human orexin-1 receptor (O43613, PDB 6TOD),56 and the human type-2 angiotensin receptor (P50052, PDB 4ZUD)38 were chosen as the templates. The sequence identity and coverage between hNPSR and the six templates are reported in Table S1 along with the phylogenetic tree (Figure S3) and pairwise sequence alignments (Figures S4–S9). The choice of templates was dictated by the resolution and sufficient similarity sequence coverage with the target. The alignments reported in Figures S4–S9 were used to build six hNPSR models using the Prime module within Schrodinger (Prime; Schrödinger, LLC, version 2020-1). The X-ray structures of the templates were obtained from the Protein Data Bank. Homology built models were obtained using the knowledge-based method. Validation of the model was carried by generating the Ramachandran plots for each model. This analysis allowed supporting the viability of the constructed models that all had >90% of the residues in the allowed regions of the plot.
Docking Calculations
Docking calculations were attained employing the Glide tool implemented in Maestro 12.4.57 The 3D structures of 1, 3–5, 7, 10, 12, and 14–21 were generated with the Maestro fragment Build tool and then energetically minimized with Macromodel.58 The 6 models were all prepared through the Protein Preparation Wizard of the Maestro 12.4 graphical user interface, which assigns bond orders, adds hydrogen atoms, and generates appropriate protonation states.
The docking grid box was centered on the residues lining the putative binding orthosteric binding, with a grid box dimension equal to 31 Å × 31 Å × 31 Å. Finally, docking runs were carried out using the standard precision method. Pictures were rendered employing UCSF Chimera software.59
MD Simulation System Setup
The complexes obtained from docking experiments of 1, 16, and 21 in both BM1 and BM2 employing the hNPSR model built using the human neuropeptide Y Y1 receptor (hNPY1R, PDB code 5ZBH) structure as a template were used to build an MD simulation system. The complex was embedded in a membrane of phosphatidylcholine lipids60 using Maestro’s system builder and was located in the membrane using the default parameters. Next, the system was solvated in an orthorhombic water box with a buffer distance of 10 Å.61 The system was neutralized with 9 Cl– ions for the complexes with 1 and 21 while 10 Cl– ions were included in the 16/hNPSR complexes. The salt concentration was set to 0.15 M NaCl. The OPLS3 force field was used for the constructed receptor/ligand/membrane system.62
MD Simulation Protocols
The Desmond module within the Schrödinger suite was used for MD simulations.63−65 First, the system was relaxed using the following relaxation protocol for membrane proteins:
-
1.
minimization with restraints on all solute-heavy atoms;
-
2
unrestrained minimization;
-
3
Brownian Dynamics in NVT ensemble (constant number of particles, constant volume, constant temperature of 300 K) using small timesteps and restraints on solute heavy atoms (50 kcal/mol) at 10 K for 50 ps;
-
4
NPT ensemble simulation (constant number of particles, constant pressure of 1 bar, and constant temperature of 100 K) Gaussian barrier potential on water molecules, membrane restrained in z, protein restrained (20 kcal/mol) for 20 ps;
-
5
NPγT ensemble simulation (constant number of particles, constant pressure of 1 bar, constant temperature of 100 K, and lateral surface tension of membranes) Gaussian Barrier potential on water molecules, membrane restrained in z, protein restrained (10 kcal/mol) for 100 ps;
-
6
NPγT ensemble simulation with increasing temperature from 100 to 300 K, Gaussian Barrier potential on water molecules, and gradual release of restraints for 150 ps.
-
7
NVT production run (constant number of particles, constant volume, and constant temperature of 300 K) without any restraint for 100 ps.
After the relaxation, 100 ns production runs were conducted under the NPγT ensemble for each of the six systems using the default protocol. In detail, the temperature was controlled using the Nosé–Hoover thermostat66,67 with a coupling constant of 1.0 ps. The pressure was controlled using the Martyna–Tuckerman–Klein barostat68,69 with a coupling constant of 2.0 ps. The cutoff distance for short-range nonbonded interactions was 9 Å, and the long-range van der Waals interactions were based on a uniform density approximation. To minimize the computation time, nonbonded forces were calculated using an r-RESPA integrator70 where the short-range forces were updated every two steps and the long-range forces were updated every six steps. The trajectories were saved at 100.0 ps. The Desmond SID tool was used to analyze the receptor–ligand interactions during the MD trajectory. Particular attention was given to ligand–residue interactions and ligand RMSF.
Acknowledgments
D.P., C.R., G.C., and R.G. were supported by the FAR (Fondo di Ateneo per la Ricerca Scientifica) grants from the University of Ferrara; C.R. was supported by a FIR (Fondo per l’Incentivazione alla Ricerca) grant from the University of Ferrara. For the determination of optical rotations, the authors thank Dr. Julie Oble, Sorbonne Université, Faculté des Sciences et Ingénierie, Campus Pierre et Marie Curie, Institut Parisien de Chimie Moléculaire.
Glossary
Abbreviations
- BLAST
Basic Local Alignment Search Tool
- BM
binding mode
- Boc2O
di-tert-butyl dicarbonate
- CL95%
95% confidence limit
- CCR9
CC chemokine receptor type 9
- CXCR4
chemokine receptor type 4
- DMEM
Dulbecco’s modified Eagle’s medium
- ECL
extracellular loop
- EtOAc
ethyl acetate
- FmocCl
Fmoc chloride
- HBSS
Hanks’ balanced salt solution
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- hNPY1R
human neuropeptide Y Y1 receptor
- NPS
neuropeptide S
- NPSR
neuropeptide S receptor
- NTSR1
neurotensin receptor 1
- OX1R
orexin receptor type 1
- PAR1
protease-activated receptor 1
- PEt
petroleum ether
- p-TsOH
p-toluenesulfonic acid
- RMSF
root-mean-square fluctuation
- sec-BuLi
sec-butyllithium
- SEM
standard error of the mean
- TM
transmembrane
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c02223.
1H NMR and NOE NMR analyses of compound 17; sequence identity and coverage between the hNPSR and selected hGPCRs; phylogenetic tree of the hNPSR and the six selected template structures; pairwise sequence alignments of the hNPSR and the six selected template structures; ligand-root-mean-square fluctuation of compounds 1, 16, and 21; and ligand–NPSR interactions for compounds 1, 16, and 21 (PDF)
Molecular formula strings (CSV)
3D coordinates of the energy-minimized docked pose of 1 in the constructed model of NPSR during MD simulation in BM1 (PDB)
3D coordinates of the energy-minimized docked pose of 1 in the constructed model of NPSR during MD simulation in BM2 (PDB)
3D coordinates of the energy-minimized docked pose of 16 in the constructed model of NPSR during MD simulation in BM1 (PDB)
3D coordinates of the energy-minimized docked pose of 16 in the constructed model of NPSR during MD simulation in BM2 (PDB)
3D coordinates of the energy-minimized docked pose of 21 in the constructed model of NPSR during MD simulation in BM1 (PDB)
3D coordinates of the energy-minimized docked pose of 21 in the constructed model of NPSR during MD simulation in BM2 (PDB)
Author Contributions
∇ V.A and C.R. contributed equally to the work. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Syuji S.; Yasushi S.; Nobuyuki M.; Koji Y.. cDNA and protein sequence of a novel human and animal G protein-coupled receptors and their uses in drug screening, diagnosis, and therapeutics. WO2002031145A1, April 18, 2002.
- Xu Y.-L.; Reinscheid R. K.; Huitron-Resendiz S.; Clark S. D.; Wang Z.; Lin S. H.; Brucher F. A.; Zeng J.; Ly N. K.; Henriksen S. J.; de Lecea L.; Civelli O. Neuropeptide S: a neuropeptide promoting arousal and anxiolytic-like effects. Neuron 2004, 43, 487–497. 10.1016/j.neuron.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Reinscheid R. K.; Xu Y.-L.; Okamura N.; Zeng J.; Chung S.; Pai R.; Wang Z.; Civelli O. Pharmacological characterization of human and murine neuropeptide S receptor variants. J. Pharmacol. Exp. Ther. 2005, 315, 1338–1345. 10.1124/jpet.105.093427. [DOI] [PubMed] [Google Scholar]
- Xu Y.-L.; Gall C. M.; Jackson V. R.; Civelli O.; Reinscheid R. K. Distribution of neuropeptide S receptor mRNA and neurochemical characteristics of neuropeptide S-expressing neurons in the rat brain. J. Comp. Neurol. 2007, 500, 84–102. 10.1002/cne.21159. [DOI] [PubMed] [Google Scholar]
- Clark S. D.; Duangdao D. M.; Schulz S.; Zhang L.; Liu X.; Xu Y.-L.; Reinscheid R. K. Anatomical characterization of the neuropeptide S system in the mouse brain by in situ hybridization and immunohistochemistry. J. Comp. Neurol. 2011, 519, 1867–1893. 10.1002/cne.22606. [DOI] [PubMed] [Google Scholar]
- Guerrini R.; Salvadori S.; Rizzi A.; Regoli D.; Calo’ G. Neurobiology, pharmacology, and medicinal chemistry of neuropeptide S and its receptor. Med. Res. Rev. 2010, 30, 751–777. 10.1002/med.20180. [DOI] [PubMed] [Google Scholar]
- Ruzza C.; Calò G.; Di Maro S.; Pacifico S.; Trapella C.; Salvadori S.; Preti D.; Guerrini R. Neuropeptide S receptor ligands: a patent review (2005-2016). Expert Opin. Ther. Pat. 2017, 27, 347–362. 10.1080/13543776.2017.1254195. [DOI] [PubMed] [Google Scholar]
- Blough B.; Namjoshi O. Small molecule neuropeptide S and melanocortin 4 receptor ligands as potential treatments for substance use disorders. Handb. Exp. Pharmacol. 2020, 258, 61–87. 10.1007/164_2019_313. [DOI] [PubMed] [Google Scholar]
- Kohji F.; Yutaka N.; Tarui N.; Masaaki M.; Hirokazu M.; Osamu K.; Hiroshi B.. Bicyclic piperazine compound and use thereof. WO2005021555A1, March 10, 2005.
- Okamura N.; Habay S. A.; Zeng J.; Chamberlin A. R.; Reinscheid R. K. Synthesis and pharmacological in vitro and in vivo profile of 3-oxo-1,1-diphenyl-tetrahydro-oxazolo[3,4-a]pyrazine-7-carboxylic acid 4-fluoro-benzylamide (SHA 68), a selective antagonist of the neuropeptide S receptor. J. Pharmacol. Exp. Ther. 2008, 325, 893–901. 10.1124/jpet.107.135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzza C.; Rizzi A.; Trapella C.; Pela’ M.; Camarda V.; Ruggieri V.; Filaferro M.; Cifani C.; Reinscheid R. K.; Vitale G.; Ciccocioppo R.; Salvadori S.; Guerrini R.; Calo’ G. Further studies on the pharmacological profile of the neuropeptide S receptor antagonist SHA 68. Peptides 2010, 31, 915–925. 10.1016/j.peptides.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Li M. S.; Peng Y. L.; Jiang J. H.; Xue H. X.; Wang P.; Zhang P. J.; Han R. W.; Chang M.; Wang R. Neuropeptide S increases locomotion activity through corticotropin-releasing factor receptor 1 in substantia nigra of mice. Peptides 2015, 71, 196–201. 10.1016/j.peptides.2015.07.024. [DOI] [PubMed] [Google Scholar]
- Han R.-W.; Zhang R.-S.; Xu H.-J.; Chang M.; Peng Y.-L.; Wang R. Neuropeptide S enhances memory and mitigates memory impairment induced by MK801, scopolamine or Aβ1-42 in mice novel object and object location recognition tasks. Neuropharmacology 2013, 70, 261–267. 10.1016/j.neuropharm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Ruzza C.; Asth L.; Guerrini R.; Trapella C.; Gavioli E. C. Neuropeptide S reduces mouse aggressiveness in the resident/intruder test through selective activation of the neuropeptide S receptor. Neuropharmacology 2015, 97, 1–6. 10.1016/j.neuropharm.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Okamura N.; Garau C.; Duangdao D. M.; Clark S. D.; Jüngling K.; Pape H.-C.; Reinscheid R. K. Neuropeptide S enhances memory during the consolidation phase and interacts with noradrenergic systems in the brain. Neuropsychopharmacology 2011, 36, 744–752. 10.1038/npp.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler C.; Zhang Y.; Gilmour B.; Graf T.; Fennell T.; Snyder R.; Deschamps J. R.; Reinscheid R. K.; Garau C.; Runyon S. P. Identification of neuropeptide S antagonists: structure-activity relationship studies, X-ray crystallography, and in vivo evaluation. ACS Chem. Neurosci. 2014, 5, 731–744. 10.1021/cn500113c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoutz C. D.; Zhang Y.; Runyon S. P.; Goeders N. E. Antagonism of the neuropeptide S receptor with RTI-118 decreases cocaine self-administration and cocaine-seeking behavior in rats. Pharmacol. Biochem. Behav. 2012, 103, 332–337. 10.1016/j.pbb.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Sauer D. R.; Djuric S. W. A facile and practical one-pot ’catch and release’ synthesis of substituted guanidines. Tetrahedron Lett. 2009, 50, 5145–5148. 10.1016/j.tetlet.2009.06.113. [DOI] [Google Scholar]
- Maity P.; König B. Synthesis and structure of 1,4-dipiperazino benzenes: Chiral terphenyl-type peptide helix mimetics. Org. Lett. 2008, 10, 1473–1476. 10.1021/ol8002749. [DOI] [PubMed] [Google Scholar]
- Liu B.; Xu G. Y.; Yang C. H.; Wu X. H.; Xie Y. Y. A novel and facile method to synthesize (R)- and (S)-2-methylpiperazine. Synth. Commun. 2004, 34, 4111–4118. 10.1081/scc-200036590. [DOI] [Google Scholar]
- Reginato G.; Di Credico B.; Andreotti D.; Mingardi A.; Paio A.; Donati D. A new versatile and diastereoselective synthesis of polysubstituted 2-oxopiperazines from naturally occurring amino acids. Tetrahedron: Asymmetry 2007, 18, 2680–2688. 10.1016/j.tetasy.2007.10.027. [DOI] [Google Scholar]
- Trapella C.; Pela M.; Del Zoppo L.; Calo G.; Camarda V.; Ruzza C.; Cavazzini A.; Costa V.; Bertolasi V.; Reinscheid R. K.; Salvadori S.; Guerrini R. Synthesis and separation of the enantiomers of the neuropeptide S receptor antagonist (9R/S)-3-oxo-1,1-diphenyl-tetrahydro-oxazolo[3,4-a]pyrazine-7-carboxylic acid 4-fluoro-benzylamide (SHA 68). J. Med. Chem. 2011, 54, 2738–2744. 10.1021/jm200138r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Gilmour B. P.; Navarro H. A.; Runyon S. P. Identifying structural features on 1,1-diphenyl-hexahydro-oxazolo[3,4-a]pyrazin-3-ones critical for Neuropeptide S antagonist activity. Bioorg. Med. Chem. Lett. 2008, 18, 4064–4067. 10.1016/j.bmcl.2008.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Xu Z.; Zhang Z.; Yang Z.; Liu Y.; Wang J.; Cai T.; Li S.; Chen K.; Shi J.; Zhu W. Like-charge guanidinium pairing between ligand and receptor: an unusual interaction for drug discovery and design?. J. Phys. Chem. B 2015, 119, 11988–11997. 10.1021/acs.jpcb.5b04130. [DOI] [PubMed] [Google Scholar]
- Saczewski F.; Balewski Ł. Biological activities of guanidine compounds. Expert Opin. Ther. Pat. 2009, 19, 1417–1448. 10.1517/13543770903216675. [DOI] [PubMed] [Google Scholar]
- Saczewski F.; Balewski L. Biological activities of guanidine compounds, 2008-2012 update. Expert Opin. Ther. Pat. 2013, 23, 965–995. 10.1517/13543776.2013.788645. [DOI] [PubMed] [Google Scholar]
- Dal Ben D.; Antonini I.; Buccioni M.; Lambertucci C.; Marucci G.; Vittori S.; Volpini R.; Cristalli G. Molecular modeling studies on the human neuropeptide S receptor and its antagonists. ChemMedChem 2010, 5, 371–383. 10.1002/cmdc.200900467. [DOI] [PubMed] [Google Scholar]
- Okada T.; Fujiyoshi Y.; Silow M.; Navarro J.; Landau E. M.; Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 5982–5987. 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Han S.; Keller M.; Kaiser A.; Bender B. J.; Bosse M.; Burkert K.; Kögler L. M.; Wifling D.; Bernhardt G.; Plank N.; Littmann T.; Schmidt P.; Yi C.; Li B.; Ye S.; Zhang R.; Xu B.; Larhammar D.; Stevens R. C.; Huster D.; Meiler J.; Zhao Q.; Beck-Sickinger A. G.; Buschauer A.; Wu B. Structural basis of ligand binding modes at the neuropeptide Y Y1 receptor. Nature 2018, 556, 520–524. 10.1038/s41586-018-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J.; Babaoglu K.; Brautigam C. A.; Clark L.; Shao Z.; Scheuermann T. H.; Harrell C. M.; Gotter A. L.; Roecker A. J.; Winrow C. J.; Renger J. J.; Coleman P. J.; Rosenbaum D. M. Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat. Struct. Mol. Biol. 2016, 23, 293–299. 10.1038/nsmb.3183. [DOI] [PubMed] [Google Scholar]
- Wu B.; Chien E. Y. T.; Mol C. D.; Fenalti G.; Liu W.; Katritch V.; Abagyan R.; Brooun A.; Wells P.; Bi F. C.; Hamel D. J.; Kuhn P.; Handel T. M.; Cherezov V.; Stevens R. C. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 2010, 330, 1066–1071. 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenalti G.; Giguere P. M.; Katritch V.; Huang X.-P.; Thompson A. A.; Cherezov V.; Roth B. L.; Stevens R. C. Molecular control of delta-opioid receptor signalling. Nature 2014, 506, 191–196. 10.1038/nature12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Wacker D.; Mileni M.; Katritch V.; Han G. W.; Vardy E.; Liu W.; Thompson A. A.; Huang X.-P.; Carroll F. I.; Mascarella S. W.; Westkaemper R. B.; Mosier P. D.; Roth B. L.; Cherezov V.; Stevens R. C. Structure of the human kappa-opioid receptor in complex with JDTic. Nature 2012, 485, 327–332. 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Srinivasan Y.; Arlow D. H.; Fung J. J.; Palmer D.; Zheng Y.; Green H. F.; Pandey A.; Dror R. O.; Shaw D. E.; Weis W. I.; Coughlin S. R.; Kobilka B. K. High-resolution crystal structure of human protease-activated receptor 1. Nature 2012, 492, 387–392. 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. F.; Noinaj N.; Shibata Y.; Love J.; Kloss B.; Xu F.; Gvozdenovic-Jeremic J.; Shah P.; Shiloach J.; Tate C. G.; Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature 2012, 490, 508–513. 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihoya W.; Nishizawa T.; Yamashita K.; Inoue A.; Hirata K.; Kadji F. M. N.; Okuta A.; Tani K.; Aoki J.; Fujiyoshi Y.; Doi T.; Nureki O. X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog. Nat. Struct. Mol. Biol. 2017, 24, 758–764. 10.1038/nsmb.3450. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Unal H.; Gati C.; Han G. W.; Liu W.; Zatsepin N. A.; James D.; Wang D.; Nelson G.; Weierstall U.; Sawaya M. R.; Xu Q.; Messerschmidt M.; Williams G. J.; Boutet S.; Yefanov O. M.; White T. A.; Wang C.; Ishchenko A.; Tirupula K. C.; Desnoyer R.; Coe J.; Conrad C. E.; Fromme P.; Stevens R. C.; Katritch V.; Karnik S. S.; Cherezov V. Structure of the angiotensin receptor revealed by serial femtosecond crystallography. Cell 2015, 161, 833–844. 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Unal H.; Desnoyer R.; Han G. W.; Patel N.; Katritch V.; Karnik S. S.; Cherezov V.; Stevens R. C. Structural basis for ligand recognition and functional selectivity at angiotensin receptor. J. Biol. Chem. 2015, 290, 29127–29139. 10.1074/jbc.m115.689000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kalé L.; Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarda V.; Ruzza C.; Rizzi A.; Trapella C.; Guerrini R.; Reinscheid R. K.; Calo G. In vitro and in vivo pharmacological characterization of the novel neuropeptide S receptor ligands QA1 and PI1. Peptides 2013, 48, 27–35. 10.1016/j.peptides.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Ruzza C.; Rizzi A.; Camarda V.; Pulga A.; Marzola G.; Filaferro M.; Novi C.; Ruggieri V.; Marzola E.; Vitale G.; Salvadori S.; Guerrini R.; Calo’ G. [tBu-D-Gly5]NPS, a pure and potent antagonist of the neuropeptide S receptor: in vitro and in vivo studies. Peptides 2012, 34, 404–411. 10.1016/j.peptides.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Guerrini R.; Camarda V.; Trapella C.; Calò G.; Rizzi A.; Ruzza C.; Fiorini S.; Marzola E.; Reinscheid R. K.; Regoli D.; Salvadori S. Synthesis and biological activity of human neuropeptide S analogues modified in position 5: identification of potent and pure neuropeptide S receptor antagonists. J. Med. Chem. 2009, 52, 524–529. 10.1021/jm8012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzza C.; Pulga A.; Rizzi A.; Marzola G.; Guerrini R.; Calo’ G. Behavioural phenotypic characterization of CD-1 mice lacking the neuropeptide S receptor. Neuropharmacology 2012, 62, 1999–2009. 10.1016/j.neuropharm.2011.12.036. [DOI] [PubMed] [Google Scholar]
- Duangdao D. M.; Clark S. D.; Okamura N.; Reinscheid R. K. Behavioral phenotyping of neuropeptide S receptor knockout mice. Behav. Brain Res. 2009, 205, 1–9. 10.1016/j.bbr.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Mingler M. K.; McBride M. L.; Murphy A. J.; Valenzuela D. M.; Yancopoulos G. D.; Williams M. T.; Vorhees C. V.; Rothenberg M. E. Abnormal response to stress and impaired NPS-induced hyperlocomotion, anxiolytic effect and corticosterone increase in mice lacking NPSR1. Psychoneuroendocrinology 2010, 35, 1119–1132. 10.1016/j.psyneuen.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannella N.; Economidou D.; Kallupi M.; Stopponi S.; Heilig M.; Massi M.; Ciccocioppo R. Persistent increase of alcohol-seeking evoked by neuropeptide S: an effect mediated by the hypothalamic hypocretin system. Neuropsychopharmacology 2009, 34, 2125–2134. 10.1038/npp.2009.37. [DOI] [PubMed] [Google Scholar]
- Kallupi M.; Cannella N.; Economidou D.; Ubaldi M.; Ruggeri B.; Weiss F.; Massi M.; Marugan J.; Heilig M.; Bonnavion P.; de Lecea L.; Ciccocioppo R. Neuropeptide S facilitates cue-induced relapse to cocaine seeking through activation of the hypothalamic hypocretin system. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 19567–19572. 10.1073/pnas.1004100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paneda C.; Huitron-Resendiz S.; Frago L. M.; Chowen J. A.; Picetti R.; Lecea L. d.; Roberts A. J. Neuropeptide S reinstates cocaine-seeking behavior and increases locomotor activity through corticotropin-releasing factor receptor 1 in mice. J. Neurosci. 2009, 29, 4155–4161. 10.1523/jneurosci.5256-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J. Lipophilicity in drug discovery. Expet Opin. Drug Discov. 2010, 5, 235–248. 10.1517/17460441003605098. [DOI] [PubMed] [Google Scholar]
- Jida M.; Ballet S. An efficient one-pot synthesis of chiral N-protected 3-substituted (diketo)piperazines via Ugi-4CR/de-Boc/cyclization Process. ChemistrySelect 2018, 3, 1027–1031. 10.1002/slct.201702943. [DOI] [Google Scholar]
- Kilkenny C.; Browne W.; Cuthill I. C.; Emerson M.; Altman D. G.; Group N. C. R. R. G. W. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br. J. Pharmacol. 2010, 160, 1577–1579. 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen S. E.; Belknap J. K. Intracerebroventricular injections in mice. Some methodological refinements. J. Pharmacol. Methods 1986, 16, 355–357. 10.1016/0160-5402(86)90038-0. [DOI] [PubMed] [Google Scholar]
- The Universal Protein Resource (UniProt) databases. https://www.uniprot.org/ (accessed Sept 15, 2020).
- Liu H.; Kim H. R.; Deepak R. N. V. K.; Wang L.; Chung K. Y.; Fan H.; Wei Z.; Zhang C. Orthosteric and allosteric action of the C5a receptor antagonists. Nat. Struct. Mol. Biol. 2018, 25, 472–481. 10.1038/s41594-018-0067-z. [DOI] [PubMed] [Google Scholar]
- Suno R.; Lee S.; Maeda S.; Yasuda S.; Yamashita K.; Hirata K.; Horita S.; Tawaramoto M. S.; Tsujimoto H.; Murata T.; Kinoshita M.; Yamamoto M.; Kobilka B. K.; Vaidehi N.; Iwata S.; Kobayashi T. Structural insights into the subtype-selective antagonist binding to the M2 muscarinic receptor. Nat. Chem. Biol. 2018, 14, 1150–1158. 10.1038/s41589-018-0152-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappas M.; Ali A. A. E.; Bennett K. A.; Brown J. D.; Bucknell S. J.; Congreve M.; Cooke R. M.; Cseke G.; de Graaf C.; Doré A. S.; Errey J. C.; Jazayeri A.; Marshall F. H.; Mason J. S.; Mould R.; Patel J. C.; Tehan B. G.; Weir M.; Christopher J. A. Comparison of Orexin 1 and Orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J. Med. Chem. 2020, 63, 1528–1543. 10.1021/acs.jmedchem.9b01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2020-1: Maestro; Schrödinger, LLC: New York, NY, 2020.
- Schrödinger Release 2020-1: MacroModel; Schrödinger, LLC: New York, NY, 2020.
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Lyman E.; Higgs C.; Kim B.; Lupyan D.; Shelley J. C.; Farid R.; Voth G. A. A role for a specific cholesterol interaction in stabilizing the Apo configuration of the human A2A adenosine receptor. Structure 2009, 17, 1660–1668. 10.1016/j.str.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark P.; Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. 10.1021/jp003020w. [DOI] [Google Scholar]
- Harder E.; Damm W.; Maple J.; Wu C.; Reboul M.; Xiang J. Y.; Wang L.; Lupyan D.; Dahlgren M. K.; Knight J. L.; Kaus J. W.; Cerutti D. S.; Krilov G.; Jorgensen W. L.; Abel R.; Friesner R. A. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016, 12, 281–296. 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- Bowers K. J. C.; Xu H.; Dror R. O.; Eastwood M. P.; Gregersen B. A.; Klepeis J. L.; Kolossvary I.; Moraes M. A.; Sacerdoti F. D.; Salmon J. K.; Shan Y.; Shaw D. E.. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, Florida, November 11–17, 2006.
- Schrödinger Release 2020-1: Desmond Molecular Dynamics System; D. E. Shaw Research: New York, NY, 2020.
- Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, 2020.
- Nosé S. A molecular-dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. 10.1080/00268978400101201. [DOI] [Google Scholar]
- Hoover W. G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A: At., Mol., Opt. Phys. 1985, 31, 1695–1697. 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- Martyna G. J.; Tobias D. J.; Klein M. L. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. 10.1063/1.467468. [DOI] [Google Scholar]
- Martyna G. J.; Tuckerman M. E.; Tobias D. J.; Klein M. L. Explicit reversible integrators for extended systems dynamics. Mol. Phys. 1996, 87, 1117–1157. 10.1080/00268979600100761. [DOI] [Google Scholar]
- Tuckerman M.; Berne B. J.; Martyna G. J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992, 97, 1990–2001. 10.1063/1.463137. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.