Abstract
Background
Glioblastomas (GBMs) are the main primary brain tumors in adults with almost 100% recurrence rate. Patients with lateral ventricle proximal GBMs (LV-GBMs) exhibit worse survival compared to distal locations for unknown reasons. One hypothesis is the proximity of these tumors to the cerebrospinal fluid (CSF) and its chemical cues that can regulate cellular phenotype. We therefore investigated the role of CSF on GBM gene expression and the role of a CSF-induced gene, SERPINA3, in GBM malignancy in vitro and in vivo.
Methods
We utilized human CSF and GBM brain tumor-initiating cells (BTICs). We determined the impact of SERPINA3 expression in glioma patients using The Cancer Genome Atlas (TCGA) database. SERPINA3 expression changes were evaluated at mRNA and protein levels. The effects of knockdown (KD) and overexpression (OE) of SERPINA3 on cell migration, viability and cell proliferation were evaluated. Stem cell characteristics on KD cells were evaluated by differentiation and colony formation experiments. Tumor growth was studied by intracranial and flank injections.
Results
GBM-CSF increased BTIC migration accompanied by upregulation of the SERPINA3 gene. In patient samples and TCGA data, we observed SERPINA3 to correlate directly with brain tumor grade and indirectly with GBM patient survival. SERPINA3 KD induced a decrease in cell proliferation, migration, invasion, and stem cell characteristics, while SERPINA3 OE increased cell migration. In vivo, SERPINA3 KD BTICs showed increased survival in a murine model.
Conclusions
SERPINA3 plays a key role in GBM malignancy and its inhibition results in a better outcome using GBM preclinical models.
Keywords: alpha 1-antichymotrypsin, brain tumor, cerebrospinal fluid, glioblastoma, SERPINA3
Key Points
1. Cancer CSF alters the transcriptome of GBM cells and increases SERPINA3 expression.
2. SERPINA3 is overexpressed in high-grade gliomas and is linked to poor survival.
3. SERPINA3 expression in GBM contributes to tumor malignancy and decreased survival in vivo.
Importance of the Study.
SERPINA3 encodes the antiprotease α1-antichymotrypsin (ACT). α1-Antichymotrypsin is produced in the brain and other tissues and is overexpressed in multiple cancers, including glioma. We hypothesize that the overexpression (OE) of ACT is a contributing factor to GBM malignancy. We tested this hypothesis in vitro and in vivo by investigating the effects of SERPINA3 KD and OE. Our data showed that cellular migration, invasion, proliferation, and stem cell characteristics are affected after SERPINA3 KD. In contrast, SERPINA3 OE increased GBM cells migration and invasion. We also detected a decreased activation of MAPK, and a decreased matrix metalloproteinase (MMP) activity with compensatory effects of tissue inhibitor metalloproteinases (TIMPS). Mice injected with SERPINA3 KD cells showed a prolonged overall survival. In summary, this study highlights the importance of SERPINA3 and ACT for GBM aggressiveness. It shows that SERPINA3 represents a potential new therapeutic target for GBM.
Glioblastoma (GBM) is the most frequent and aggressive primary brain tumor in adults.1 In the United States, 13 000 new cases are detected annually.2 Current GBM standard of care includes surgical resection, chemotherapy, and radiotherapy.3,4 However, tumor recurrence is inevitable and the overall survival is still unacceptable.5
GBMs proximal to the lateral ventricles (LV-GBMs) have a decreased median overall survival by more than 20% (compared to noncontacting GBMs).6-8 Genetic analyses of bulk GBM samples have not identified a distinct molecular signature on LV-GBMs and the reason for the worse outcome of these tumors has not been defined.9 However, the interaction of GBM cells with components of the subventricular zone (SVZ) neurogenic niche, including the cerebrospinal fluid (CSF), may contribute to this event. The SERPINA3 gene encodes for the serine protease inhibitor α1-antichymotrypsin (ACT), which inhibits chymotrypsin and cathepsin G among other proteases.10 This gene and its protein expression correlate with tumor progression in gliomas and other cancers.11-15 However, the impact of SERPINA3 expression on glioma malignancy is unknown. This study aims to elucidate the contribution of SERPINA3 knockdown (KD) and overexpression (OE) on GBM aggressiveness. We observed that CSF induces an increase in migration of human GBM-derived brain tumor-initiating cells (BTICs); moreover, CSF induces increased expression of SERPINA3. We identified SERPINA3 as one of the genes responsible for an increase in malignancy (as defined by increased migration, invasion, and proliferation) of GBM-BTICs.
Materials and Methods
Human Glioma Tissue and GBM Cell Lines
All human BTIC primary cultures were derived from intraoperative tissue samples from newly diagnosed GBM patients without prior treatment. Human glioma tissues were obtained upon patient informed consent at the Johns Hopkins Hospital and Mayo Clinic Jacksonville under the approval of the Institutional Review Board of each institution, respectively. Written informed consent was obtained from all patients’ previous samples collection. Primary GBM cell line 1A was obtained from Galli et al,16 and cell lines 612 and 965 were used and analyzed by our group previously.17,18 BTICs were cultured without serum and in the presence of growth factors (epidermal growth factor and fibroblast growth factor) to promote their undifferentiated status, as described previously.17 Clinical data of the cell lines is included in Supplementary Figure 1.
Microarray
BTICs at 70% confluency were treated with GBM and noncancer CSF for 24 hours. RNA was extracted and submitted (2 µg of RNA) for RNA expression microarray using the Ilumina Human HT- 12 v4 chip. The log2-transformed array probe-level data across samples were normalized using robust multiarray average (RMA).19 Only probes with detection P values <.05 in at least 2 samples were considered “present” and kept for further analysis. SERPINA3 was identified by analysis of variance (ANOVA) and Bonferroni multiple test correction.
Lentivirus Construction for Knockdown, Overexpression, and Genetic Rescue
SERPINA3 ORF tagged with TurboGFP at the C-terminal end was amplified by PCR from the RG200509 ORF clone. The resulting product was cloned in a lentiviral backbone under the CMV promoter along with a polyadenylation signal to increase the translation of the mRNA. For SERPINA3 KD, 2 different shRNA sequences were used; KD1 = shRNA1 and KD2 = shRNA2. An empty vector (EV) was used as a negative control of SERPINA3 OE and KD (see Supplementary Table 1). A third shRNA sequence against SERPINA3 (Sigma, TRCN0000006658) targeting the 3′UTR region in the mRNA was used. Lentiviral particles production, titration, and cellular transduction methods were included in Supplementary Material.
Transwell With and Without Matrigel (2D/3D)
BTICs were resuspended in 200 µL of GBM media with 0.5% of fetal bovine serum (FBS) onto the top chamber of noncoated or matrigel-coated transwell migration chambers. The bottom well was filled with 500 µL of GBM media plus 2% FBS. For specific experimental conditions, recombinant ACT protein at 4 µM or CSF at 25 µg/mL was added to the bottom chamber. After 24 hours cells were fixed, stained with DAPI and counted.
Real-time PCR
Total RNA was extracted using RNeasy Mini Kit. Quantitative real-time PCR (RT-qPCR) was performed using Power SYBR Green PCR Master Mix. Primer sequences are listed in Supplementary Table 1.
Cell Cycle Analysis and Cellular Viability
Cell cycle analysis was performed using EdU and cellular viability was measured using alamarBlue. Manufacturer’s instructions were followed for both methods.
Fluorescence-activated Cell Sorting
Cell sorting for CD133-positive cells was performed using the BD FACS ARIA II Flow Cytometer. Unfixed cells were stained with CD133/2(293C3)-PE antibody following manufacturer’s protocol.
Colony Formation Assay
We cultured 1 × 103 cells per well in 6-well plates. After 14 days, colonies were fixed in methanol, stained with 0.05% crystal violet, and counted using ImageJ software. A positive colony was considered >500 µm in diameter or at least 50 cells.21
Limiting Dilution Assay In Vitro and In Vivo
We seeded 1, 5, 25, and 50 cells per well in a 96-well plate with 12 wells per dilution. The number of tumor spheres (tight, spherical, nonadherent masses >50 µm in diameter)20 that subsequently formed in each well were quantified after 14 days. For extreme limiting dilution assay (ELDA) in vivo, 2000, 20 000, and 200 000 cells were injected in the left flank of 7 mice per group. The number of tumors (tight, spherical, masses >80 µm in diameter) that subsequently formed in each animal were measured in length and width with a caliper after 10 weeks; these tumors were also weighed with a balance after extraction. Limiting dilution plots and stem cell frequencies for both experiments were calculated with the Walter and Eliza Hall Institute of Medical Research (http://bioinf.wehi.edu.au/software/elda/index.html).
Cellular Differentiation
We seeded 3.5 × 105 cells per well on 6-well plates. After 24 hours, media containing 10% FBS was added to the cells for 7 days to induce differentiation.
Immunoblotting and Immunohistochemistry
Fifty micrograms of whole cell lysate proteins were resolved using the NuPAGE gel and transferred to PVDF membranes. Membranes were blocked at room temperature and primary antibody was incubated overnight at 4°C. After washing, the appropriate secondary antibody was added for 1 hour (see Supplementary Table 2). For immunohistochemistry analysis, paraffin-embedded mouse brain sections were deparaffinized and rehydrated, followed by antigen recovery. Endogenous peroxidase activity was inhibited, after which slides were washed in phosphate-buffered saline and stained using the appropriate antibodies (see Supplementary Table 2).
Matrix Metalloproteinase Antibody Array
Human matrix metalloproteinase (MMP) antibody array membrane was used following manufacturer’s instructions with 250 µg of cell lysate.
Tumor Implantation
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Mayo Clinic (protocol A00002260-16). Intracranial tumor implantation was performed as previously described.22 3.5 × 105 luciferase-bearing 1A-green fluorescent protein (GFP) cells were injected in (15 mice per group) 2 µL of GBM base media at coordinates X = 1.5 mm, Y = 1.35 mm, and Z = 3.5 mm relative to bregma. For survival analysis, 10 mice per group were used. For tumor volume assessment, 5 mice per group were sacrificed 4 weeks after injection.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 8. Data distribution was determined by D’Agostino-Pearson test. Statistical significance for 2 group experiments was determined by T test and for multiple groups by a 1-way ANOVA, followed by Tukey or Kruskal-Wallis analysis based on data distribution. Overall survival data were analyzed by Log-rank test (Mantel-Cox). Results represent the mean ± SEM. Statistical significance is represented by **P <.01, *P <.05.
Results
GBM-CSF Increases Migration and SERPINA3 Expression in BTICs
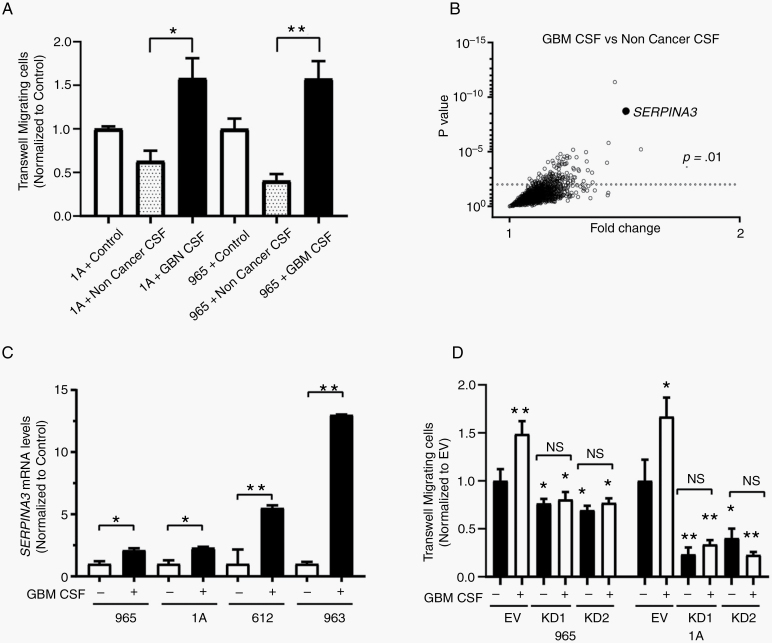
To understand how CSF contributes to the aggressiveness of GBM cells, we treated BTICs with either noncancer or GBM-CSF for 24 hours. The stimulation of BTICs with GBM-CSF induced an increase in cell migration in comparison to noncancer CSF (965 by 57.3% and in 1A by 57.8%) (P < .01) (Figure 1A). We also determined the transcriptome response of BTICs to GBM and noncancer CSF using an RNA expression array. We identified SERPINA3 as one of the genes most overexpressed in BTICs when exposed to GBM-CSF in comparison to noncancer CSF (Figure 1B). This observation was confirmed by RT-qPCR in 4 different BTICs (Figure 1C). Remarkably, we detected levels of ACT protein in CSF to be higher in high-grade glioma patients than in controls (P < .05) (Supplementary Figure 2). Upon SERPINA3 KD with 2 different short hairpin RNAs (Supplementary Figure 3), we observed a reduction in the migratory response of GBM-BTICs to CSF in the evaluated lines (P < .05) (Figure 1D). These results indicate that cancer CSF increases the migration of BTICs and that expression of SERPINA3 is necessary for this response.
Figure 1.
GBM-CSF effects on SERPINA3 expression and migratory capacity in BTICs. (A) GBM-CSF increases the migratory response of 1A and 965 BTICs compared to noncancer CSF. (B) RNA expression microarray showed that SERPINA3 was one of the genes upregulated on BTICs after conditioning of GBM-CSF. (C) Confirmation of higher levels of SERPINA3 mRNA by RT-qPCR following GBM-CSF stimulus in different BTICs. (D) SERPINA3 KD impairs the migration response of 965 and 1A BTICs to CSF stimulation. *P < .05, **P < .01. BTIC, brain tumor-initiating cell; GBM-CSF, glioblastoma-cerebrospinal fluid; KD, knockdown.
SERPINA3 Expression Correlates With Glioma Grade and Indirectly With Patient Survival in GBM Tissues and Cells
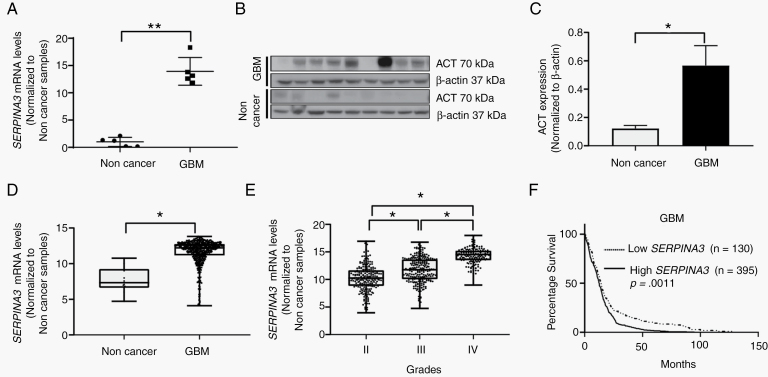
We evaluated the expression of the SERPINA3 gene and its protein ACT in human brain. SERPINA3 is overexpressed in gliomas and other human cancers when compared to noncancer tissue.11,12,23,24 We observed that GBM brain samples had a higher expression of the SERPINA3 transcript (fold change of 13.1) (P = .0075) (Figure 2A) and ACT protein versus noncancer tissue (P = .0315) (Figure 2B and C). To determine the impact of LV proximity with the expression of SERPINA3, we identified that 33% of the tumors were contacting only the SVZ, while 66% were contacting both the SVZ and the cortex. There were no samples contacting only the cortex (Supplementary Figure 1B). We then explored The Cancer Genome Atlas (TCGA) dataset and observed a higher expression of SERPINA3 mRNA in GBM samples when compared to noncancer tissue (median expression levels of 7.3 in noncancer vs 12.2 in GBM samples) (P < .05) (Figure 2D) and lower-grade gliomas (mean difference: grade II vs III: −1.4 ± 0.19; grade II vs IV: −4.0 ± 0.22; and grade III vs IV: −2.5 ± 0.2) (P < .05) (Figure 2E). Moreover, we observed that high levels of SERPINA3 correlate with lower patient survival (median survival: low SERPINA3 14.10 mo vs high SERPINA3 12.50 mo) (P = .0011) (Figure 2F). After stratifying by molecular subtype, a significant survival effect of SERPINA3 expression was evident in the proneural subtype (median survival: low SERPINA3 16.45 mo vs high SERPINA3 11.80 mo) (P = .0011) (Supplementary Figure 4).
Figure 2.
SERPINA3 and ACT expression in cancer samples and relation to patient survival. (A) Higher expression of SERPINA3 in GBM tissues compared to noncancer as measured by RT-qPCR in samples from the Mayo Clinic Biobank. (B, C) Immunoblot quantification of multiple GBM tissues expressing higher levels of ACT than control samples from Mayo Clinic Biobank. (D) TCGA dataset depicting higher levels of SERPINA3 mRNA in GBM samples. (E) Correlation between increased SERPINA3 mRNA expression and glioma grade (TCGA data). (F) Survival analysis of TCGA dataset on glioma patients reveals that patients with high SERPINA3 expression have significantly worse outcome. *P < .05, **P < .01. ACT, α1-antichymotrypsin; GBM, glioblastoma; TCGA, The Cancer Genome Atlas.
SERPINA3 Silencing Impairs Proliferation and Stem Cell Characteristics of BTICs
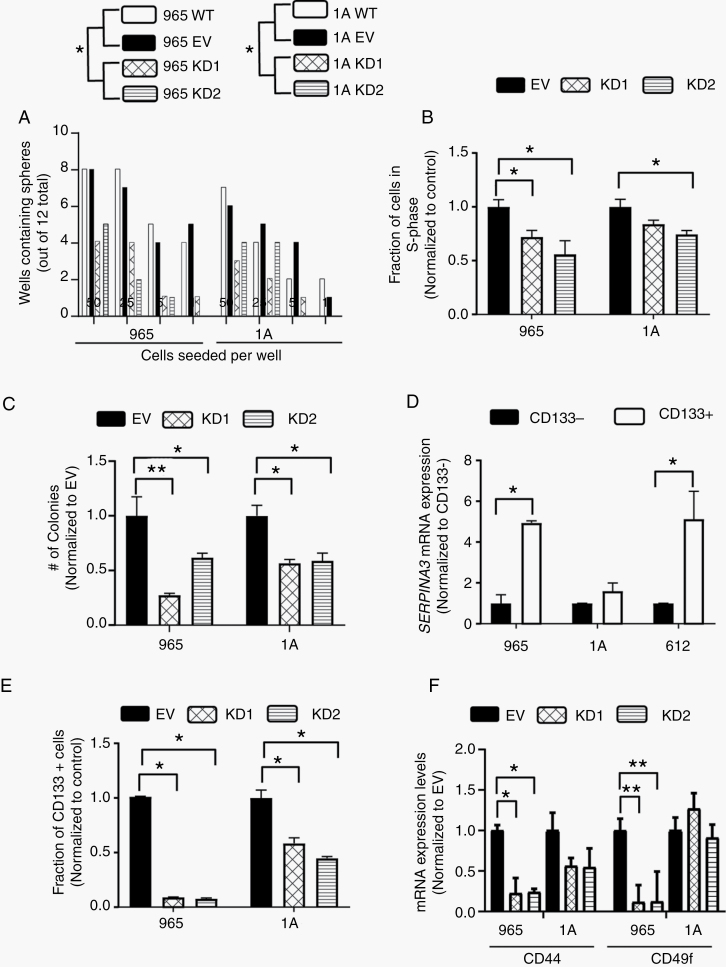
Next, we evaluated the contribution of SERPINA3 expression to GBM malignancy by knocking down SERPINA3 in BTICs using 2 shRNA constructs. We first measured the effects of SERPINA3 KD on cancer stem cell characteristics of BTICs. Using an ELDA,20 we observed a decrease in the clonal capacity of BTICs 965 and 1A upon SERPINA3 KD (cells seeded/percentage of colonies formed: WT: 50/62%, 25/50%, 5/29%, 1/25%; EV: 50/58%, 25/50%, 5/33%, 1/25%; KD1: 50/29%, 25/25%, 5/8.3%,1/4.1%; KD2: 50/37%, 25/25%, 5/4%, 1/0%) (P < .05) (Figure 3A) and (statistics and ELDA plots in Supplementary Figure 5). Through EdU incorporation, we observed that SERPINA3 KD decreased the DNA synthesis phase of BTICs cell cycle (between 17% and 45%) resulting in an overall decrease in cell proliferation of 965 and 1A lines (P < .05) (Figure 3B). Cell viability was also decreased after SERPINA3 KD in all evaluated primary cultures (965: 71%–86%, 1A 56%–75%, and 612 62%–82% vs control) (P < .05) (Supplementary Figure 6). SERPINA3 KD also impaired the colony formation capacity of BTICs (965 KD1 27.5% decrease and KD2 62% decrease compared to EV, 1A KD1 56.8% decrease and KD2 58.8% decrease compared to EV) (P < .05) (Figure 3C). These results support the hypothesis that SERPINA3 expression is required for the self-renewal and proliferation of BTICs.
Figure 3.
SERPINA3 effect on proliferation and cancer stem cell characteristics. (A) BTICs with SERPINA3 KD exhibit lower sphere formation capacity than controls (1A and 965 cells). (B) S-phase of cell cycle and (C) colony formation capacity are decreased after SERPINA3 KD in BTICs 965 and 1A. (D) The CD133+ population of BTICs has increased expression of SERPINA3 mRNA by RT-qPCR (965, 1A, and 612 cells). (E) SERPINA3 KD decreases CD133+, CD44, and CD49f expression in BTICs (965 and 1A cells) (F). *P < 0.05, **P < .01. BTIC, brain tumor-initiating cell; KD, knockdown.
We then observed that cellular differentiation of 965 and 1A BTICs (corroborated by a decrease in stem cell markers SOX2 and NESTIN, and an increase in GFAP) induced a decrease in SERPINA3 expression (P < .05) (Supplementary Figure 7). Furthermore, expression of CD133+, a frequently used marker of BTICs,25 was also affected by SERPINA3 expression. We determined that the CD133-positive fraction of BTICs showed higher expression of SERPINA3 in our evaluated lines compared to the CD133-negative portion (in 965 by 390%, in 1A by 40%, and in 612 by 410%) (P < .05) (Figure 3D). Moreover, the silencing of SERPINA3 induced a decrease in the percentage of CD133-positive cells (in 965 KD1 by 90%–93% and in 1A KD1 by 42%–56% compared to EV) (P < .05) (Figure 3E). We also identified that SERPINA3 KD decreases the expression of stem cell markers CD44 in our 2 cell lines and of CD49f in line 965 (P < .05) (Figure 3F). These results indicate SERPINA3 expression is higher in undifferentiated BTICs and that the silencing of this gene induces a more differentiated phenotype in human GBM-BTICs.
ACT Increases the Migratory and Invasive Capabilities of BTICs
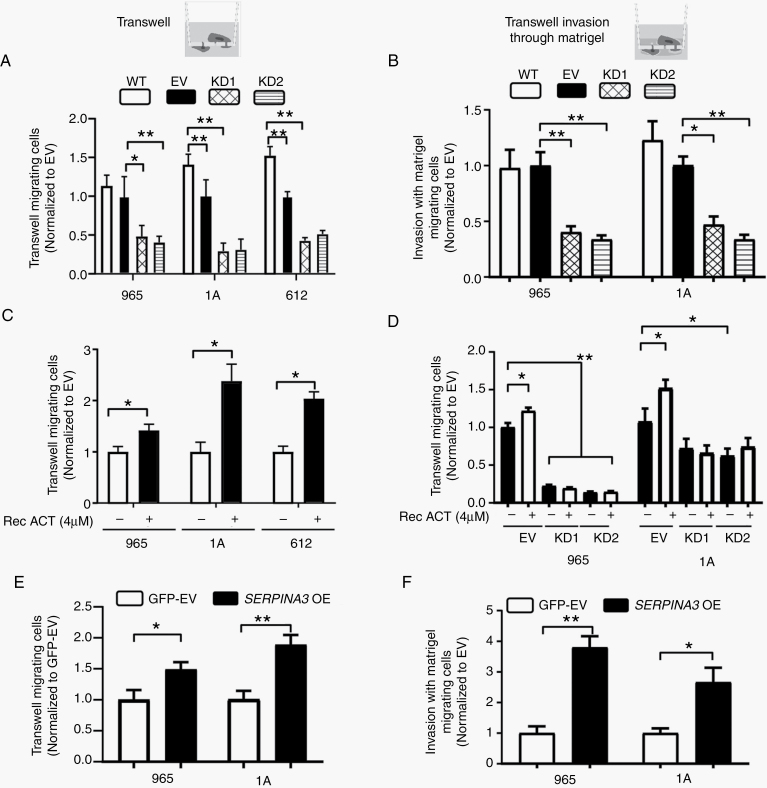
To evaluate whether the expression of ACT correlates with BTICs migration capacity, we subjected them to transwell migration assays. Interestingly, we observed a correlation between ACT expression and migratory capabilities of different BTICs at baseline (P < .05) (Supplementary Figure 8). We then evaluated the effects of SERPINA3 KD and recombinant ACT on the migratory and invasive capacity of BTICs. We observed that silencing of SERPINA3 induced a decrease in cell migration and invasion of GBM-BTICs when compared to EV. In all the lines tested, SERPINA3 KD induced a decrease in the number of migrating cells between 13% and 55% compared to EV-transduced cells (P < .05) (Figure 4A). The invasion capacity of GBM-BTICs was similarly impaired upon SERPINA3 silencing in all lines tested with 54%–67% less invasive cells when compared to EV-transduced cells (P < .05) (Figure 4B). Conversely, when BTICs were treated with recombinant ACT, we observed an increase in their migration capacity (in 965 by 37%, in 1A by 128%, and in 612 by 104% compared to nontreated) (P < .05) (Figure 4C). Interestingly, treatment with recombinant ACT did not recover the migratory capacity of SERPINA3 KD cells (Figure 4D).
Figure 4.
SERPINA3 effects on cellular migration and invasion. (A) SERPINA3 KD decreased migration (965, 1A, and 612 cells) and (B) invasion (965 and 1A) capacity of BTICs. (C) Recombinant ACT (4 µM) increased cellular migration in WT cells (965, 1A, and 612 cells), (D) but does not rescue SERPINA3 KD cells migration capacity (965 and 1A cells). (E, F) SERPINA3 OE increases 965 and 1A BTICs migration and invasion. *P < .05, **P < .01. ACT, α1-antichymotrypsin; BTIC, brain tumor-initiating cell; KD, knockdown; OE, overexpression; WT, wild type.
Due to the lack of rescue in cancer cells migration by recombinant ACT and to discard any off-target effect of the SERPINA3 shRNA, we performed a genetic rescue experiment. We utilized a combination of an overexpression vector that was not targeted by the shRNA utilized. Genetic re-expression of SERPINA3 rescued the migratory (50% in 965 and 96% in 1A cells) and proliferative capacity of the SERPINA3 KD cells (P < .05) (Supplementary Figure 9). Additionally, to determine whether exogenous ACT is able to enter the cell, we performed an experiment utilizing fluorescently labeled recombinant ACT (4 µM). Upon 24 hours of incubation, we observed no labeled protein inside the 1A cells (Supplementary Figure 10).
The overexpression of SERPINA3 increased the migration (between 48% and 88% vs EV) and invasion (between 165% and 279% vs EV) of GBM-BTICs (P < .05) (Figure 4E and F). However, overexpression of SERPINA3 did not affect the proliferation capacity of BTICs compared to control cells (Supplementary Figure 11). These results indicate that the expression of SERPINA3 and ACT induce a more aggressive migratory behavior in GBM cells that appears to be dependent on the endogenous expression of SERPINA3.
Silencing of SERPINA3 Suppresses BTICs-Derived Tumor Growth In Vivo
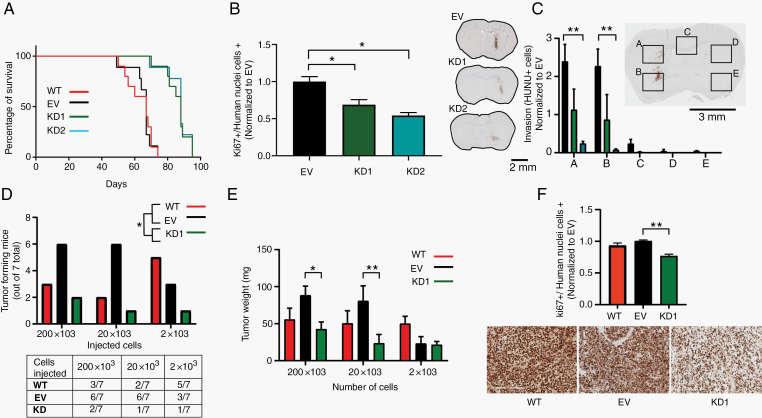
We examined the effects of SERPINA3 expression on the growth of intracranial tumors using human GBM-derived BTICs injected orthotopically in immunosuppressed mice (NuNu). Green fluorescent protein wild type (WT), EV, KD1, and KD2 transduced 1A cells were utilized (n = 15 per group). Five animals per group were euthanized 4 weeks after injection to evaluate tumor volume and levels of proliferation in vivo, and the remaining 10 mice per group were followed for survival. Silencing of SERPINA3 in GBM cells results in increased survival of mice from a median of 67 days in control groups (WT and EV) to a median of 86 days in the KD groups (P < .05) (Figure 5A). Immunohistochemistry analysis at 4 weeks postinjection demonstrated lower percentages of proliferating GBM cells (Ki67-positive cells in KD1 decreased by 32% and KD2 by 46% when normalized to the EV group) (P < .05) (Figure 5B). We also studied cell invasion by quantifying the number of human nuclei (HuNu) positive cells that migrated to the contralateral hemisphere by crossing the corpus callosum. We detected decreased invasive capacity in the KD groups (decreased in KD1 by 50%–60% and KD2 by 80%–90% compared to EV in quadrants A and B) (P < .01) (Figure 5C). Tumor histology of the groups (WT, EV, KD1, and KD2) was performed at survival endpoint, and was included in Supplementary Figure 12A. Tumor size is not different among controls and KDs since at the time of death (45 d after injection for controls vs 80 d after injection for the KDs), the tumor in the animals had reached the same dimensions. Neuropathological analysis revealed higher number of mitotic spindles and cellularity in the EV groups, and higher numbers of thin blood vessels, hemorrhage, and necrosis in the KD groups (Supplementary Figure 12B and C). These results indicate that the impact of SERPINA3 expression on GBM malignancy is maintained in the brain microenvironment in vivo.
Figure 5.
Effects of SERPINA3 KD in vivo. (A) Mice injected intracranially with 1A SERPINA3 KD cells survived longer than the control groups. (B) SERPINA3 KD groups showed decreased Ki67 proliferation index and (C) invasive capacity than the controls. (D) In vivo ELDA analysis showing lower tumor formation capacity in SERPINA3 KD BTICs than the controls. (E, F) ELDA in vivo tumors in the SERPINA3 KD group weighed less and had a decreased Ki67 proliferation index than the controls. *P < .05, **P < .01. Scale 2 mm and 200 µm. BTIC, brain tumor-initiating cell; ELDA, extreme limiting dilution assay; KD, knockdown.
We also performed a flank in vivo limiting dilution assay with 1A WT, EV, and SERPINA3 KD cells. We observed that 1A SERPINA3 KD cells showed decreased tumor formation ability when compared to the control groups (group/dilution: percentage of tumors formed WT/200 × 103: 42%, WT/20 × 103: 28%, WT/2 × 103: 71%, EV/200 × 103: 85%, EV/20 × 103: 85%, EV/2 × 103: 42%, KD1/200 × 103: 28%, KD1/20 × 103: 14%, and KD1/2 × 103: 14%) (P < .05) (Figure 5D). After tumor extraction at 10 weeks, we observed a decreased weight in the KD tumors by 52.2% in the 200 × 103 dilution, 71% in 20 × 103, and 8.6% in 2 × 103 when compared with EV (P < .05). Ki67 index of these tumors revealed decreased proliferation in the KD1 group by 20% (P < .01) (Figure 5E and F). This experiment further corroborates the role of SERPINA3 on maintaining stem cell characteristics in vivo.
SERPINA3 Expression Induces Changes in the MAPK Signaling Pathway and MMP Expression
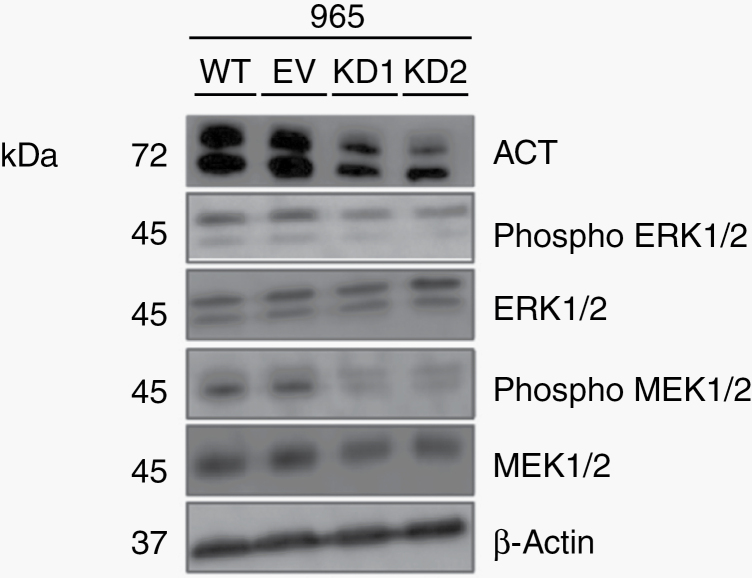
To elucidate the intracellular mechanism by which SERPINA3 mediates the different effects on BTICs, we evaluated the phosphorylation status of MAPK signaling pathway members MEK1/2 and ERK1/2, which are known to play important roles in cell cycle, migration, and apoptosis.26,27 SERPINA3 KD cells showed a decreased expression of phosphorylated MEK1/2 and ERK1/2 than controls by western blot (Figure 6).
Figure 6.
Effect of SERPINA3 expression on ERK and MEK kinases. The active forms of the mitogen activated protein kinases ERK1/2 and MEK1/2 are decreased upon KD of SERPINA3 (965 cells). KD, knockdown.
Lastly, MMPs and tissue inhibitor of metalloproteinases (TIMPs) are key regulators in extracellular matrix remodeling. Matrix metalloproteinases and TIMPs have a crucial role in cancer progression, specifically in tumor invasion into distal parenchyma.28,29,30 In SERPINA3 KD BTICs, we observed decreased protein expression of MMP 1, 2, 3, 8, 9, and 13 by 20%–80% when compared with EV (P < .05), with compensatory changes in TIMP-1 and TIMP-2 (P < .05). Interestingly, treatment of 1A WT BTICs with recombinant ACT induced an increase in MMP2 expression by 625% (P < .05), but did not affect TIMPs expression (Supplementary Figure 13). These results suggest that the activities of MAPK and MMP proteins are related to the effects of SERPINA3 expression and the malignancy of GBM cells.
Discussion
In this study, we showed that human GBM-CSF induces changes in BTICs at the cell migration and transcriptional level. Multiple studies have demonstrated that LV-GBMs show higher malignancy when compared to LV-distal tumors.6,7 Specifically, these tumors show higher distal recurrence6 and lower survival.31 The wall of the LV contains the SVZ, the largest neurogenic niches in adults.32,33,34 In rodents, neuroblast migration from the SVZ to the olfactory bulb is greatly regulated by the flow of CSF inside the LV and its contained chemokines.35 Our results indicate that GBM cells respond to CSF by increasing their migration and their expression of the SERPINA3 gene. Although we saw different levels of SERPINA3 expression in response to GBM-CSF among BTICs, this is likely due to the intrinsic cellular heterogeneity between cell lines. We also saw decreased migration of BTICs in SERPINA3 KD lines. Interestingly, the migration phenotype of BTICs is not rescued when cells are stimulated with GBM-CSF, reinforcing the role of SERPINA3 expression in this response. Our results indicate that SERPINA3 expression plays a critical role in the ability of GBM cells to respond to CSF exposure, suggesting that SERPINA3 expression is increased in LV-GBM. An increased expression of SERPINA3 in serum from patients with LV-GBMs has been previously reported.9 A recent study evaluated gene expression differences in LV-GBMs versus LV-distal GBMs and found no significant molecular difference.36 The use of bulk tumor samples along with the heterogeneity of GBM might be reasons for these findings. A study using directed biopsies and single cell analysis would be necessary to further investigate this hypothesis. The collection of such samples by neurosurgeons offers some technical challenges (ie, need of navigation, satisfy pathology tissue requirements for diagnosis, brain shift during surgery, among others) that need to be addressed.
Our results of higher expression levels of SERPINA3 in brain cancer tissue are in agreement with other reports12 and with the tissue microarray data published by Li et al, where SERPINA3 expression was higher in high-grade tumors.23 In addition, our analysis of TCGA and REMBRANDT databases detected a positive correlation of SERPINA3 expression with glioma tumor grade and a negative correlation with survival. SERPINA3 is linked with poor prognosis and tumor progression in several types of cancer, including glioma.11,15,23 SERPINA3 is a member of the serpin protease family that contains 36 protein-coding genes, of which ACT is one of the most abundant.10 The main function of SERPINA3 is to act as protease inhibitor. Additional functions described to other members of the same family include hormone transporters, blood pressure regulation, and apoptosis mediators.37,38 During inflammatory processes ACT release is increased by 2- to 5-fold resulting in an increased concentration in plasma.10,39 Extracellular ACT plays a role in regulating inflammation and extracellular matrix remodeling predominantly through inhibition of neutrophil cathepsin G, and mast cells chymases.10 Through loss- and gain-of-function experiments, we identified SERPINA3 to increase malignant features of GBM in vitro and in vivo. Specifically, both overexpression of SERPINA3 and the addition of recombinant ACT induced an increase in cell migration and invasion. Conversely, SERPINA3 KD caused a decrease in cell migration, proliferation, invasion, and stem cell characteristics. Our results showed a pro-tumorigenic action of SERPINA3 expression in GBM cells. Interestingly, we observed a positive feedback of SERPINA3 expression upon stimulation with its encoded protein ACT. In the brain, ACT is secreted from reactive astrocytes as a response of IL-6 and IL-1 during inflammatory events.23,40,41 Once released, ACT is able to successfully reach the tumor site.23,42 As confirmed by our data, in GBM patients, there are increased levels of SERPINA3 in CSF. Interleukin-6 also has been described in higher levels in CSF from cancer patients.43-45 Addition of recombinant ACT did not rescue the phenotypic changes induced by SERPINA3 silencing, whereas genetic re-expression of SERPINA3 did. These observations could be the result of a differential effect between intracellular and extracellular ACT and indicate that the endogenous expression of ACT is required to induce cell migration and proliferation. While specific intracellular action of ACT needs further elucidation, SERPINA3 expression has already been reported to increase cell proliferation, migration, and invasion in other cancers like melanoma, colon, and endometrial tumors.11,14,15
Our data revealed that MERK 1/2 and ERK1/2 activation decreased after SERPINA3 KD in BTICs. These findings have also been reported in endometrial cancer.46 The decreased phosphorylated levels in MEK1/2 and ERK1/2 kinases after SERPINA3 KD can be a contributing factor of the reduced cellular proliferation and migration that we observed. However, further analysis on SERPINA3 in intracellular signaling processes is needed.
Tumor growth is dependent on extracellular matrix degradation and remodeling. The role of SERPINA3 in connective tissue turnover has previously been demonstrated in colon carcinoma cells.11 After SERPINA3 KD, we observed decreased activity levels in MMP 1, 2, 3, 8, 9, and 13, as well as compensatory increased activity in inhibitors TIMPS 1 and 2. Among MMPs, gelatinases 2 and 9 (MMP2 and MMP9) are key players in tumor progression, adhesion, migration, differentiation, and evasion of the immune system.28,30 After recombinant ACT stimulus, we observed higher activity levels of MMP2 in GBM cells, suggesting that SERPINA3 is a contributing factor for tissue remodeling that increases migration in cancer cells. However, the role of SERPINA3 in immune response against GBM should be elucidated in an immunocompetent mice model.
In summary, this study provides evidence that the baseline levels and CSF-induced increase of SERPINA3 expression are strongly linked to GBM migration, invasion, proliferation, and stem cell characteristics in vitro, as well as tumor formation capability and decreased survival expectancy in vivo in mice and human patients. Our findings encourage further studies to identify therapeutic compounds against SERPINA3 and ACT activity in GBM, particularly in the context of LV-GBMs.
Supplementary Material
Acknowledgments
M.L.-V. thanks PECEM, UNAM, and CONACYT. We thank Ms. Brandy Edenfield and Dr. Laura Lewis-Tuffin for technical assistance.
Funding
M.L.-V. was supported by Consejo Nacional de Ciencia y Tecnología, Universidad Nacional Autónoma de México, and the Uihlein Professorship Research Grant. A.Q.-H. was supported by the Mayo Clinic Professorship, a Clinician Investigator grant, and the National Institutes of Health (NIH) (R43CA221490, R01CA200399, R01CA183827, R01CA195503, and R01CA216855). H.G.-C. was supported by NIH grants (R03NS109444, R21CA221490, and K01NS11093001).
Conflict of interest statement. None declared.
Authorship statement. M.L.-V., N.Z., and H.G.-C. lead the project and analyzed the data. M.L.-V., N.Z., P.S., R.A., J.P., S.J., M.E.J., and E.S.N. performed the experiments. A.C. and Y.W.A. performed genetic analysis. K.L.C. and A.Q.-H. performed CSF and tissue collection. J.S. and T.C. contributed with result analysis. J.R.-T. collected clinical data. All authors approved the final version of the manuscript.
References
- 1. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. J Am Med Assoc. 2013;310(17):1842–1850. [DOI] [PubMed] [Google Scholar]
- 2. Ostrom Q, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20(suppl. 4):iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lara-Velazquez M, Al-Kharboosh M, Jeanneret S., et al. Advances in brain tumor surgery for glioblastoma in adults. Brain Sci. 2017;7(12):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group . Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 5. Lu VM, Goyal A, Graffeo CS, et al. Survival benefit of maximal resection for glioblastoma reoperation in the temozolomide era: a meta-analysis. World Neurosurg. 2019;127:31–37. [DOI] [PubMed] [Google Scholar]
- 6. Lim DA, Cha S, Mayo MC, et al. Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro Oncol. 2007;9(4):424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaichana KL, McGirt MJ, Frazier J, Attenello F, Guerrero-Cazares H, Quinones-Hinojosa A. Relationship of glioblastoma multiforme to the lateral ventricles predicts survival following tumor resection. J Neurooncol. 2008;89(2):219–224. [DOI] [PubMed] [Google Scholar]
- 8. Chen L, Chaichana KL, Kleinberg L, Ye X, Quinones-Hinojosa A, Redmond K. Glioblastoma recurrence patterns near neural stem cell regions. Radiother Oncol. 2015;116(2):294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gollapalli K, Ghantasala S, Kumar S, et al. Subventricular zone involvement in glioblastoma—a proteomic evaluation and clinicoradiological correlation. Sci Rep. 2017;7(1):1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker C, Belbin O, Kalsheker N, Morgan K. SERPINA3 (aka alpha-1-antichymotrypsin). Front Biosci. 2007;12:2821–2835. [DOI] [PubMed] [Google Scholar]
- 11. Cao LL, Pei XF, Qiao X, et al. SERPINA3 silencing inhibits the migration, invasion, and liver metastasis of colon cancer cells. Dig Dis Sci. 2018;63(9):2309–2319. [DOI] [PubMed] [Google Scholar]
- 12. Luo D, Chen W, Tian Y, et al. Serpin peptidase inhibitor, clade A member 3 (SERPINA3), is overexpressed in glioma and associated with poor prognosis in glioma patients. Onco Targets Ther. 2017;10:2173–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tahara E, Ito H, Taniyama K, Yokozaki H, Hata J. Alpha 1-antitrypsin, alpha 1-antichymotrypsin, and alpha 2-macroglobulin in human gastric carcinomas: a retrospective immunohistochemical study. Hum Pathol. 1984;15(10):957–964. [DOI] [PubMed] [Google Scholar]
- 14. Wang Y, Jiang H, Dai D, et al. Alpha 1 antichymotrypsin is aberrantly expressed during melanoma progression and predicts poor survival for patients with metastatic melanoma. Pigment Cell Melanoma Res. 2010;23(4):575–578. [DOI] [PubMed] [Google Scholar]
- 15. Zhou ML, Chen FS, Mao H. Clinical significance and role of up-regulation of SERPINA3 expression in endometrial cancer. World J Clin Cases. 2019;7(15):1996–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021. [DOI] [PubMed] [Google Scholar]
- 17. Garzon-Muvdi T, Schiapparelli P, ap Rhys C, et al. Regulation of brain tumor dispersal by NKCC1 through a novel role in focal adhesion regulation. PLoS Biol. 2012;10(5):e1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang JM, Schiapparelli P, Nguyen HN, et al. Characterization of PTEN mutations in brain cancer reveals that pten mono-ubiquitination promotes protein stability and nuclear localization. Oncogene. 2017;36(26):3673–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu Z, Irizarry R, Gentleman R, et al. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99(468):909–917. [Google Scholar]
- 20. Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–2319. [DOI] [PubMed] [Google Scholar]
- 21. Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–78. [DOI] [PubMed] [Google Scholar]
- 22. Gonzalez-Perez O, Guerrero-Cazares H, Quinones-Hinojosa A. Targeting of deep brain structures with microinjections for delivery of drugs, viral vectors, or cell transplants. J Vis Exp. 2010;(46):2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rooprai HK, McCormick D. Proteases and their inhibitors in human brain tumours: a review. Anticancer Res. 1997;17(6B):4151–4162. [PubMed] [Google Scholar]
- 24. Ahmed SI, Javed G, Laghari AA, et al. CD133 expression in glioblastoma multiforme: a literature review. Cureus. 2018;10(10):e3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mawrin C, Diete S, Treuheit T, et al. Prognostic relevance of MAPK expression in glioblastoma multiforme. Int J Oncol. 2003;23(3):641–648. [PubMed] [Google Scholar]
- 27. Mehner C, Hockla A, Miller E, Ran S, Radisky DC, Radisky ES. Tumor cell-produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget. 2014;5(9):2736–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shay G, Lynch CC, Fingleton B. Moving targets: emerging roles for MMPs in cancer progression and metastasis. Matrix Biol. 2015;44–46:200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roomi MW, Monterrey JC, Kalinovsky T, Rath M, Niedzwiecki A. In vitro modulation of MMP-2 and MMP-9 in human cervical and ovarian cancer cell lines by cytokines, inducers and inhibitors. Oncol Rep. 2010;23(3):605–614. [DOI] [PubMed] [Google Scholar]
- 30. Mistry AM, Hale AT, Chambless LB, Weaver KD, Thompson RC, Ihrie RA. Influence of glioblastoma contact with the lateral ventricle on survival: a meta-analysis. J Neurooncol. 2017;131(1):125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim D, Alvarez-Buylla A. The adult ventricular-subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harb Perspect Biol. 2016;8(5):1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leong SY, Turnley AM. Regulation of adult neural precursor cell migration. Neurochem Int. 2011;59(3):382–393. [DOI] [PubMed] [Google Scholar]
- 33. Gonzalez-Perez O, Garcia-Verdugo JM, Quinones-Hinojosa A, Luquin S, Gudino-Cabrera G, Gonzalez-Castaneda RE. Neural stem cells in the adult brain: from benchside to clinic. Stem Cells Int. 2012;2012:378356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu M, Feng Y, Dangelmajer S, et al. Human cerebrospinal fluid regulates proliferation and migration of stem cells through insulin-like growth factor 1. Stem Cells Dev. 2014;24(2):160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mistry AM, Wooten DJ, Davis LT, Mobley BC, Quaranta V, Ihrie RA. Ventricular-subventricular zone contact by glioblastoma is not associated with molecular signatures in bulk tumor data. Sci Rep. 2019;9(1):1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blanco I. Serpins and serpinopathies. In: Blanco I, ed. Blanco’s Overview of Alpha-1 Antitrypsin Deficiency. Barcelona, Spain: Elsevier Inc; 2017:13–22. [Google Scholar]
- 37. Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994;269(23):15957–15960. [PubMed] [Google Scholar]
- 38. Forsyth S, Horvath A, Coughlin P. A review and comparison of the murine alpha1-antitrypsin and alpha1-antichymotrypsin multigene clusters with the human clade A serpins. Genomics. 2003;81(3):336–345. [DOI] [PubMed] [Google Scholar]
- 39. Sun YX, Wright HT, Janciauskiene S. Glioma cell activation by Alzheimer’s peptide Abeta1-42, alpha1-antichymotrypsin, and their mixture. Cell Mol Life Sci. 2002;59(10):1734–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Michelson N, Rincon-Torroella J, Quiñones-Hinojosa A, Greenfield JP. Exploring the role of inflammation in the malignant transformation of low-grade gliomas. J Neuroimmunol. 2016;297:132–140. [DOI] [PubMed] [Google Scholar]
- 41. Castell JV, Gomez-Lechon MJ, David M, et al. Interleukin-6 is the major regulator of acute phase protein synthesis in adult human hepatocytes. FEBS Lett. 1989;242:237–239. [DOI] [PubMed] [Google Scholar]
- 42. Mehta PD, Pirttila T, Patrick BA, Barshatzky M, Mehta SP. Amyloid beta protein 1-40 and 1-42 levels in matched cerebrospinal fluid and plasma from patients with Alzheimer disease. Neurosci Lett. 2001;304(1-2):102–106. [DOI] [PubMed] [Google Scholar]
- 43. Schuhmann MU, Zucht HD, Nassimi R, et al. Peptide screening of cerebrospinal fluid in patients with glioblastoma multiforme. Eur J Surg Oncol. 2010;36(2):201–207. [DOI] [PubMed] [Google Scholar]
- 44. Shen F, Zhang Y, Yao Y, et al. Proteomic analysis of cerebrospinal fluid: toward the identification of biomarkers for gliomas. Neurosurg Rev. 2014;37(3):367–380; discussion 380. [DOI] [PubMed] [Google Scholar]
- 45. Yang GD, Yang XM, Lu H, et al. SERPINA3 promotes endometrial cancer cells growth by regulating G2/M cell cycle checkpoint and apoptosis. Int J Clin Exp Pathol. 2014;7(4):1348–1358. [PMC free article] [PubMed] [Google Scholar]
- 46. Könnecke H, Bechmann I. The role of microglia and matrix metalloproteinases involvement in neuroinflammation and gliomas. Clin Dev Immunol. 2013;2013:914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.