Abstract
Background
Medulloblastoma (MB) is a malignant brain tumor in childhood. It comprises 4 subgroups with different clinical behaviors. The aim of this study was to characterize the transcriptomic landscape of MB, both at the level of individual tumors as well as in large patient cohorts.
Methods
We used a combination of single-cell transcriptomics, cell culture models and biophysical methods such as nanoparticle tracking analysis and electron microscopy to investigate intercellular communication in the MB tumor niche.
Results
Tumor cells of the sonic hedgehog (SHH)–MB subgroup show a differentiation blockade. These cells undergo extensive metabolic reprogramming. The gene expression profiles of individual tumor cells show a partial convergence with those of tumor-associated glial and immune cells. One possible cause is the transfer of extracellular vesicles (EVs) between cells in the tumor niche. We were able to detect EVs in co-culture models of MB tumor cells and oligodendrocytes. We also identified a gene expression signature, EVS, which shows overlap with the proteome profile of large oncosomes from prostate cancer cells. This signature is also present in MB patient samples. A high EVS expression is one common characteristic of tumors that occur in high-risk patients from different MB subgroups or subtypes.
Conclusions
With EVS, our study uncovered a novel gene expression signature that has a high prognostic significance across MB subgroups.
Keywords: extracellular vesicle, gene expression signature, medulloblastoma, single-cell RNA sequencing, tumor microenvironment
Key Points.
1. Single-cell transcriptomics reveal metabolic reshaping of SHH-MB tumor cells.
2. EVS is identified as an EV-related gene expression signature in MB tumor cells.
3. High EVS expression earmarks high-risk patients in all 4 MB subgroups.
Importance of the Study.
High-risk tumors can occur in MB subgroups of different cellular origins and with divergent epigenetic profiles. With EVS, our study provides a molecular tool that can identify such high-risk tumors beyond the boundaries of the existing MB classification. EVS-high constitutes a novel, independent risk factor that has a particularly good prognostic power for Group 3 and Group 4 MB, where risk stratification poses a particular challenge.
Medulloblastoma (MB) is the most frequent pediatric malignancy of the central nervous system and demonstrates a wide spectrum of clinicopathological features. The current World Health Organization classification integrates the 4 consensus molecular subgroups of MB—wingless (WNT), sonic hedgehog (SHH), Group 3, and Group 4—with previous histological variants.1,2 Several large-scale studies revealed additional layers of heterogeneity within these subgroups and proposed additional MB subtypes.3–5 However, this progressive division of MB does not provide an explanation for the occurrence of high-risk (HR) tumors in different subgroups or subtypes that show little to no overlap in their (epi)genotypes.3,5,6 While DNA methylome and transcriptome patterns separate SHH and WNT from Group 3 and Group 4 tumors, proteome profiles of SHH and Group 3 tumor subsets are clustered together and converge into a MYC-like protein signaling pathway for which MYC amplification is dispensable.7–9
More recent single-cell studies have focused on the cellular origins, developmental programs, and intratumoral diversity at the cellular level in MB.10–14 The tumor microenvironment (TME) is of major importance for tumor progression and therapeutic response in different cancer types. Cellular communication in the TME is multidimensional. One main route involves the exchange of extracellular vesicles (EVs), which transfer bioactive cargo molecules to recipient cells and can induce epigenetic alterations. Tumor-derived EVs include small EVs (sEV)/exosomes, microvesicles (MV), large vesicles (LV), and so-called large oncosomes (LO).15 Here we used single-cell RNA sequencing (scRNA-seq) to characterize the cellular TME in murine SHH-MB. We identified a transcription signature called EVS, which indicates EV-based communication of cells in the MB tumor niche. This signature is found in gene expression data from 2 independent, large MB patient cohorts. EVS can be used as a prognostic tool to identify HR cases, especially in Group 3 and Group 4, and EVS-high represents a new, independent risk factor in MB.
Materials and Methods
Animal Model
All animal work was performed in accordance with National Research Council recommendations and guidelines provided by the local regulatory authorities (City of Hamburg; 113/16). To generate the Atoh1-cre:SmoM2 tumor model,16Atoh1-cre mice were crossed with SmoM2flox/flox mice.
Sample Processing
Tumors from 4 Atoh1-cre:SmoM2 littermates were collected independently of sex at postnatal day 20. Single cell suspensions of vital tumor cells were further processed as described.17
Cells and Cell Culture
The human MB cell lines Daoy and D341 were purchased from American Type Culture Collection via LGC Standards GmBH. Oli-neu cells were a kind gift of Tanja Kuhlmann (Institute of Neuropathology, University Münster). MB1 cells were established from tumors of Atoh1-cre:SmoM2 mice at P12 as described.18 Cells were maintained at 37°C and 5% CO2 in DMEM/F-12 medium containing B-27 and N-2 supplements (Gibco, Thermo Fisher Scientific), 20 ng/mL of recombinant murine epidermal growth factor and basic fibroblast growth factor (PeproTech Germany), and 1% penicillin/streptomycin (Gibco). For co-culture experiments, 30 mm Millicell cell culture inserts with hydrophilic polytetrafluoroethylene membranes (0.4 µm pore size) were used (Millipore, Merck).
Single-Cell RNA Sequencing
Approximately 10 000 single cells per individual tumor were used as input for scRNA-seq. Single cell capture, barcoding, cDNA amplification, and cleanup were done using Chromium technology (10x Genomics) as described.17 Libraries were generated using the Library Bead Kit and i7 Multiplex Kit, and quality controls were done with a Tapestation 2000 instrument (Agilent Technologies). Sequencing was performed on an Illumina NextSeq 500 instrument using High Output Kit v2 with 75 cycles at the Core Facility Genomics, University Hospital Münster.
Bulk mRNA Sequencing
Total RNA was isolated using RNeasy Mini Kit (Qiagen). Extraction of mRNA was done using the NEBNext Poly(A) mRNA Magnetic Isolation module; libraries were generated with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs), and sequencing was done using Illumina’s High Output Kit v2.5 with 75 cycles on an NextSeq500 instrument at the Institute of Human Genetics, University Hospital Münster.
Isolation and Analysis of Extracellular Vesicles
EVs were isolated from cell culture supernatants by differential ultracentrifugation. Supernatants were centrifuged at 500 g (5 min) to pellet residual cells, followed by a 15 min spin at 1500 g for LV, a 30 min spin at 17 000 g for MV and a 90 min spin at 140 000 g for sEV, respectively. Size and concentration of EVs were determined by nanoparticle tracking analysis using a Zetaview PMX-120 instrument (Particle Metrix). For western blot analysis, primary antibodies against the following proteins were used: ALIX (#sc-53540), alpha-actinin-4 (#sc-390205), Rgap1 (#sc-271110), CK18 (#sc-6259), Tsg101 (#sc-7964, all from Santa Cruz Biotechnology), Syntenin (#ab133267, Abcam), and GM130 (#12480, Cell Signaling). Signals were detected using the ECL ChemoStar Imager HR 9.0 (Intas).
Transmission Electron Microscopy
For ultrastructural analyses by transmission electron microscopy (TEM) MB cells cells were grown on poly-L-lysin coated aclar film. All methodology and instrumentation were done as previously described in detail.19
Data Analyses
Bioinformatics data analyses were carried out in R version 3.5.1 and Bioconductor version 3.7. The Seurat package20 was used for quality control, filtering, dimensionality reduction, clustering, and differential expression analysis. Trajectory inference was performed using Monocle 2.21 Other analyses and packages are listed in the Supplementary Material. The scRNA-seq data of murine SHH-MB tumors have been deposited in Gene Expression Omnibus (GEO) under the accession number GSE127997, and bulk mRNA-seq data of co-cultured MB1, D341, and Oli-neu cells have been deposited in GEO under accession number GSE159763.
Results
Transcriptomic Landscape of Murine Sonic Hedgehog Medulloblastoma
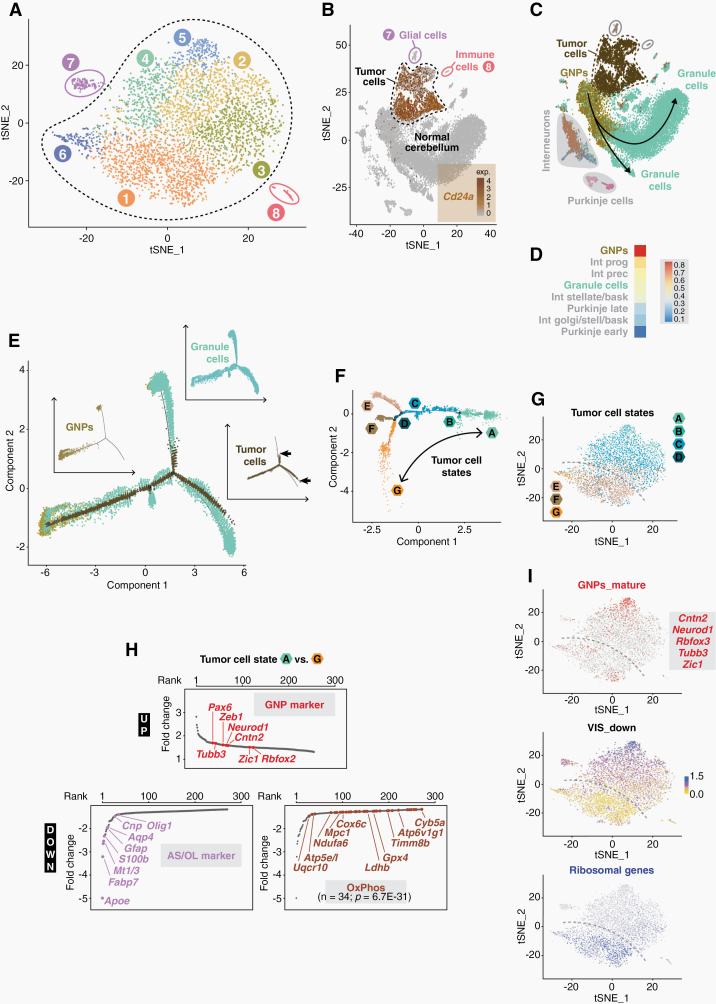
Human MB can be modeled in mice by introducing mutations in the transmembrane protein Smoothened (Smo). We used Atoh1-cre:SmoM2 mice, which develop SHH-MB from granule neuron precursors (GNPs) in the cerebellum (CB),16 to determine the transcriptional landscape of this tumor. ScRNA-seq provided a record of almost 4800 individual transcriptomes from cells of 4 tumor-bearing mice at postnatal day 20 (P20). Nearest-neighbor clustering and visualization revealed 8 distinct cell clusters, which were subsequently characterized as tumor cells (clusters 1–6) and associated nontumor cells, namely glial cells (cluster 7) and immune cells (cluster 8) (Fig. 1A; Supplementary Table 1). The CD24 glycoprotein has been established as a specific SHH-MB tumor cell marker.22 Embedding SHH-MB profiles into a single-cell transcriptome map of the developing mouse cerebellum23 revealed that high Cd24a expression was confined to cells of clusters 1–6, but largely absent in cluster 7/8 cells or normal CB cells (Fig. 1B). The tumor cells showed the closest positioning (ie, relationship) to GNPs in relation to all cerebellar cell types (Fig. 1C, D). Lineage trajectory analysis was performed to link tumor cells with developmental stages by projecting their transcriptomes onto a pseudotime-ordered axis of granule cell development (Supplementary Figure 1). While proliferating GNPs from normal tissue split into 2 branches of mature granule cell populations, SHH-MB tumor cells showed a developmental block and did not reach a fully differentiated stage (Fig. 1E).
Fig. 1.
The transcriptome landscape of murine SHH medulloblastoma. (A) t-distributed stochastic neighbor embedding (t-SNE) analysis of cell clusters in SHH-MB tumors at P20. (B) t-SNE plot of Cd24a expression in SHH-MB cells and in cerebellar cells from healthy mice.23 (C) Relation of SHH-MB cells to different cell populations of the cerebellum. (D) Similarity of SHH-MB cells and different cerebellar cell types as calculated by a logistic regression-based model. (E) Pseudo-temporal lineage trajectory of granule cells in which SHH-MB tumor cells are embedded. The insets show the individual trajectories of the 3 cell populations separately, with arrows indicating the termination points of the tumor cell trajectory. (F) Intratumoral heterogeneity as determined by trajectory inference of pseudotime-ordered SHH-MB tumor cells. (G) t-SNE plot of tumor cell states. The dashed line illustrates the bisection into states ABCD and EFG. (H) Ranked list of differentially expressed genes between tumor cells states A and G. Various marker genes of granule neuron precursor (GNP) cells, astrocytes/oligodendrocytes (AS/OL) and oxidative phosphorylation (OxPhos) are highlighted. (I) t-SNE plots of markers for mature GNPs,13 of ribosomal gene expression, or of VIS_down, a gene expression signature found in SHH-MB tumor cells which are strongly depleted by vismodegib.14
We also used trajectory inference to illuminate transcriptomic heterogeneity within Cd24a-positive tumor cells. Two major subpopulations, consisting of states ABCD and EFG, were resolved (Fig. 1F, G). The former is characterized by increased expression of postmitotic granular cell markers (eg, “GNP_mature” signature from10), the latter by increased expression of glial, oxidative phosphorylation (OxPhos) and ribosomal genes (Fig. 1H, I). This bisection is relevant at the therapeutic level. It is reflected by the VIS_down gene signature, which contains 65 differentially expressed genes from those SHH-MB tumor cells that respond most sensitively to treatment with the Smo inhibitor vismodegib14 (Fig. 1I; see Supplementary Figure 2 for details on deriving VIS_down).
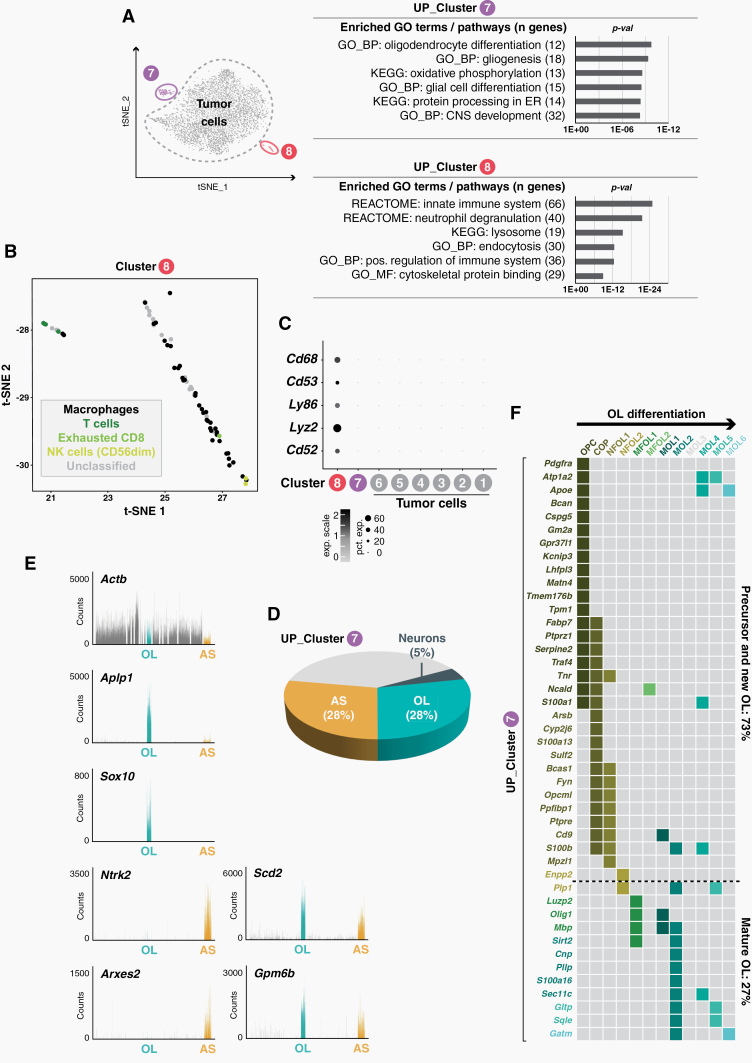
Gene functional classification identified the nontumor cells of clusters 7 and 8 as glial cells and immune cells, respectively (Fig. 2A). Based on the enrichment of typical marker genes, macrophages were revealed as the most prevalent cell type of cluster 8 (Fig. 2B, C). This is consistent with a macrophage-dominated immunological landscape in SHH-MB.24–26 Immune cells accounted for only 1.4% of all cells within the SHH-MB tumors, which is in line with similarly low numbers of tumor-infiltrating immune cells in murine SHH-MB10 and human SHH-MB.27
Fig. 2.
Tumor-associated glial and immune cells. (A) Gene ontology (GO) analysis of upregulated genes of clusters 7 and 8. (B) t-SNE plot showing immune cell phenotypes of cluster 8 cells based on the expression of cell type-specific marker genes.39 (C) Dot plot of immune cell marker genes in clusters 1–8. (D) Overlap of upregulated cluster 7 genes with astrocyte (AS), oligodendrocyte (OL) and neuron marker genes.28 (E) Upregulated cluster 7 genes are highly expressed in AS and OL cells as revealed by interrogation of a molecular atlas of cell types in the mouse brain vasculature.30 (F) A matrix showing stage-specific cluster 7 gene expression during OL differentiation.29
Cluster 7 represents a mixture of astrocytes (AS) and oligodendrocytes (OL), as shown by comparison to AS and OL profiles from several independent datasets28–30 (Fig. 2D, E). OL cells of cluster 7 were most similar to oligodendrocyte progenitor cells (OPCs) (Fig. 2F), which is in agreement with SHH-MB scRNA-seq data from the Gershon group, which showed that in oligodendrocytes in murine, SHH-MB tumors predominantly show OPC characteristics.14
Transcriptome Reprogramming in SHH-MB Tumor Cells
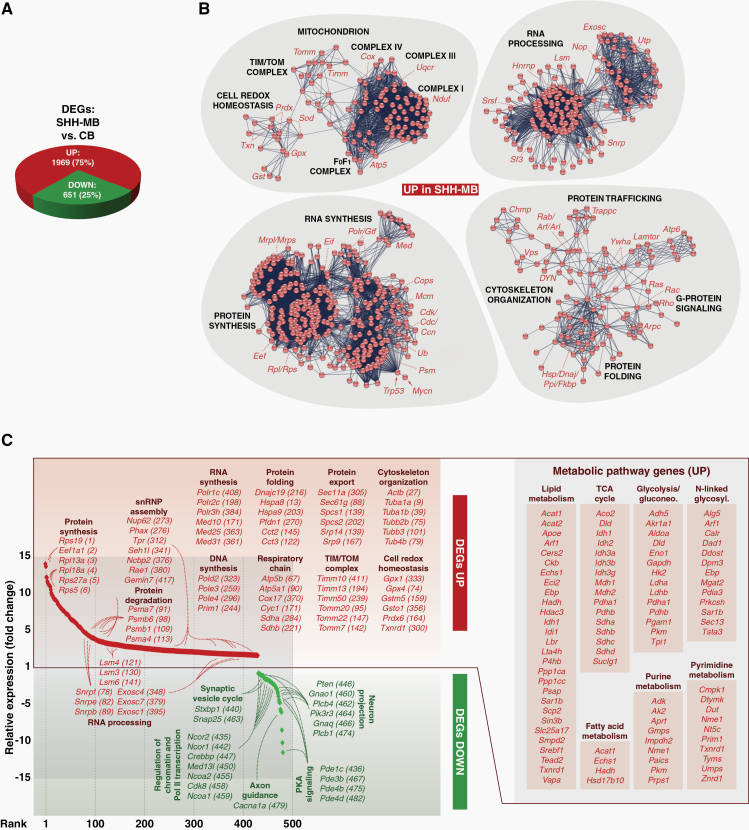
To identify the molecular framework underlying the transformation of SHH-MB tumor cells, we compared their transcriptomes with those of non-transformed CB cells. In total 2620 differentially expressed genes (DEGs) were found, of which 75% are upregulated in tumor cells (Fig. 3A; Supplementary Table 2). Network analyses of this gene set showed the concerted activation of functionally linked gene-regulatory circuits. Genes involved play important roles in the life cycle of RNA and proteins (ie, their synthesis, maturation, modification, transport, localization, and degradation), in cytoskeletal organization, or in G-protein signaling (Fig. 3B). In addition, the cellular metabolism is extensively reshaped in tumor cells, including lipid, fatty acid. and nucleic acid metabolism or major energy pathways (Fig. 3C; Supplementary Figure 3; Supplementary Table 3).
Fig. 3.
Metabolic remodeling of SHH-MB tumor cells. (A) Differentially expressed genes (DEGs) between SHH-MB and cells from normal cerebellum (CB). (B) High-confidence interaction networks of upregulated DEGs in tumor cells. Major cellular functions and involved gene sets/families are indicated. (C) Overlap of tumor cell DEGs with the cancer cell metabolism gene database (https://bioinfo.uth.edu/ccmGDB). Shared genes are shown in ranked order from maximally upregulated to maximally downregulated, and exemplary metabolic pathway gene sets are highlighted.
Selective Gene Expression Changes in the Tumor Niche
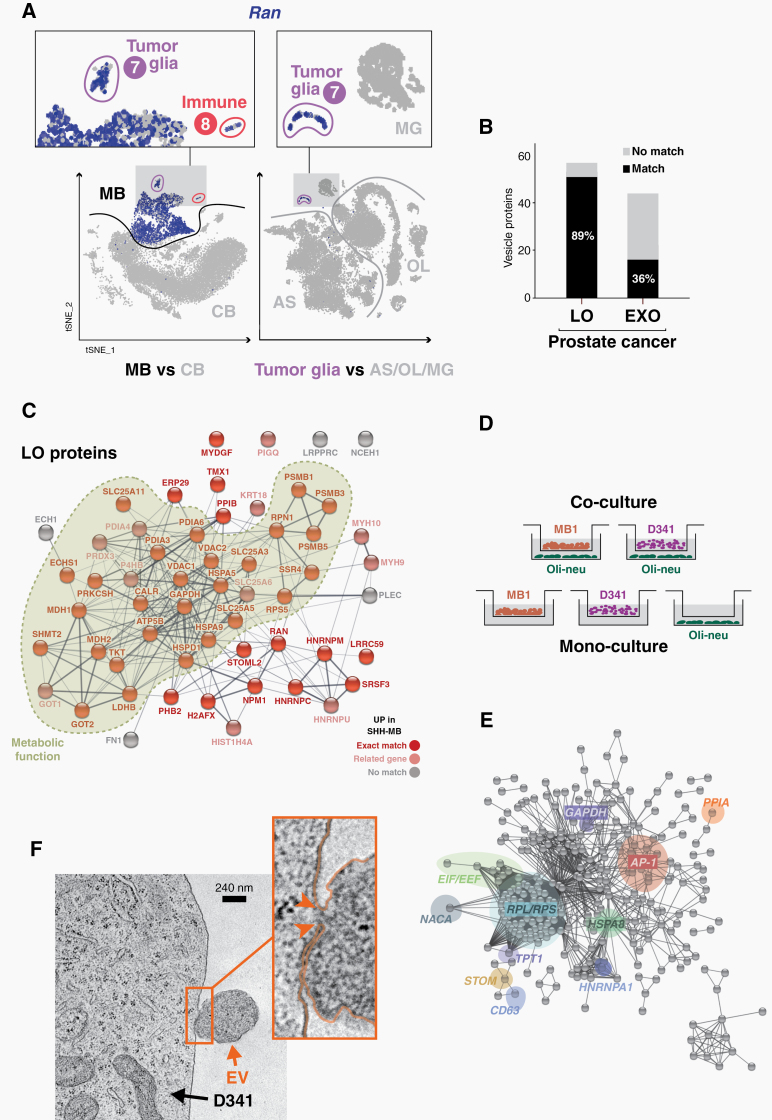
The comparison of SHH-MB cells with a scRNA-seq atlas of the developing, healthy mouse brain23 revealed unexpected gene expression changes in the TME. For example, cell type–specific genes such as Pdgfra (an OPC marker), Apoe (an AS marker), and Lgmn (an immune cell marker) are strongly expressed in their correct cell type, but also in SHH-MB tumor cells (Supplementary Figure 4A–C). Conversely, many upregulated DEGs from tumor cells become activated in tumor glia (cluster 7) and immune cells (cluster 8), but not in the corresponding cell types from healthy tissue. These include genes involved in protein biosynthesis (eg, Eef1a1, Rpl13a), in protein folding and trafficking (eg, Dnaja1, Hspa8, Ppia; Supplementary Figure 4E), or in mitochondrial electron transport (eg, Cox6c, Ndufa4). Interestingly, we also found genes that are linked to extracellular vesicles—eg, the tetraspanin genes Cd9, Cd81, and Cd63 (Supplementary Figure 4D), which encode EV membrane proteins—or genes like Atp5b, Npm1, or Ran (Fig. 4A), whose products are enriched as cargo of large oncosome EVs.31 This affects 71 genes that are upregulated in tumor and glia cells (including 13 OxPhos genes; P = 7.6E-15), and 104 genes that are upregulated in tumor and immune cells (Supplementary Figure 5; Supplementary Table 4).
Fig. 4.
EVS, an extracellular vesicle-related gene signature in medulloblastoma. (A) t-SNE plots showing expression of EV marker gene Ran in SHH-MB cells (MB), and in cerebellum (CB) cells (left panels) or glial cells (right panels) of the developing mouse brain.23 (B) Overlap of upregulated genes in SHH-MB tumor cells with enriched proteins of prostate cancer-derived large oncosomes (LO) and exosomes (EXO). (C) Protein interaction network of large oncosome-enriched proteins. Nodes colored in bright red indicate exact matches to upregulated DEGs of SHH-MB tumor cells, and nodes colored in light red match to closely related genes. Proteins/genes with metabolic function are highlighted. (D) Scheme illustrating the transwell co-culture of Oli-neu cells with D341 and MB1 cells. As a control, cells were also kept in mono-culture conditions. (E) A high-confidence interaction network of the top 500 most significantly upregulated DEGs in D341 cells when co-cultured with Oli-neu cells. Prominent subnetworks/nodes related to murine SHH-MB (cf. Figure 3B), metabolism (eg, GADH), EVS (eg, HSPA8, NACA) or the AP-1 network (JUN, JUND, FOS) are highlighted. (F) Electron micrograph of a D341 tumor cell and an EV detaching from its outer cell membrane. The connecting bridge between plasma and vesicle is highlighted by arrowheads in the enlarged section. Scale bar: 240 nm.
Extracellular Vesicles and a Related Gene Expression Signature in MB
The mutual activation of EV-related genes could reflect an EV-based transfer of information between cells in the SHH-MB tumor niche. EVs have been recognized as important mediators of metabolic adaptations in cancer cells. The oncogene MYC plays an important role in the metabolic reprogramming of cancer cells, and activation of MYC in stromal cells is transmitted by tumor cell–derived large oncosomes.32 Upregulated DEGs from murine SHH-MB show overlaps with EV proteomes (eg, the top 100 EV proteins from Vesiclepedia [http://microvesicles.org]), as well as with a MYC-like protein-signaling signature, which is associated with rapid death for subtypes of SHH and Group 3 MB9 (Supplementary Figure 6). Strikingly, upregulated DEGs in SHH-MB show almost 90% overlap with the proteome profile of large oncosomes from human prostate cancer cells32 (Fig. 4B; Supplementary Table 5). Many of these overlapping gene/protein pairs are involved in metabolic processes, for example Gapdh/GAPDH, Ldhb/LDHB, Mdh1/MDH1, and Tkt/TKT (Fig. 4C).
To test the hypothesis that gene expression changes in the TME of MB tumors can result from an EV-mediated process, we used a co-culture system of Oli-neu, a murine oligodendrocytic cell line, and 2 MB tumor cell lines. These were D341, a human cell line representing Group 3 MB, and MB1, one of several murine SHH-MB short-term cell cultures that we established from tumors of Atoh1-cre:SmoM2 mice at postnatal day P12. All cells were incubated in a transwell system in serum-free conditions that allows the exchange of EVs (Fig. 4D). As a control, each cell line was kept as a monoculture in the same system. After an incubation period of 4 days, total RNA was isolated from mono- and co-cultivated cells and analyzed by sequencing bulk mRNAs (mRNA-seq) in order to determine changes in the respective gene expression patterns. In the D341/Oli-neu co-culture 3570 DEGs were identified in the former, and 457 DEGs in the latter cells, while in the MB1/Oli-neu co-culture 705 and 159 DEGs were identified in the former and latter cells, respectively (Supplementary Table 6). Furthermore, we were able to identify 79 genes that behave identically in Oli-neu cells when co-cultured with D341 and MB1 cells and form a tight network that is centered around the transcription factor AP-1 (Supplementary Figure 7). AP-1 is an ubiquitous transcription factor encoded by Jun/Fos and plays an important role in glial cell proliferation and cell survival of oligodendrocytes.33,34 D341 cells showed almost 2000 upregulated DEGs in response to Oli-neu co-culture (Supplementary Table 6). Further analysis of the affected gene networks revealed a striking correspondence with the metabolic reprogramming of SHH-MB cells in our mouse model (Fig. 4E; cf. Fig. 3B). A Gene Ontology analysis of the 500 most significant upregulated genes in the co-culture revealed the following terms, among others: “eukaryotic translation elongation” (REACTOME; 67 genes; P = 5.102E-80), “ribosome” (KEGG; 63 genes; P = 2.805E-77), “metabolism of proteins” (REACTOME; 133 gene; P = 3.825E-29), “glucose metabolism” (REACTOME; 16 genes; P = 1.968E-9), or “unfolded protein binding” (GO_MF; 19 genes; P = 1.002E-9). As an example of a mutually activated gene network in D341 and Oli-neu cells, the “ATF-2 transcription factor network” (BIOCARTA; 12 genes; P = 8.885E-8), which contains FOS, JUN and DUSP1, was also found among these top 500 DEGs in D341 cells (cf. Supplementary Figure 7).
To directly prove EV release in these cells, we collected cell culture supernatants and purified EVs by differential ultracentrifugation. Size and concentration of EVs were determined for 3 subfractions, namely LV, MV, and sEV, by nanoparticle tracking analysis (Supplementary Figure 8A). While vesicle size was similar in the 3 cell lines, in relative terms about 3.3 times more LV were found in MB1 and about 1.5 times more sEV in Oli-neu cells. To confirm the nature of these EVs, we performed western blot analysis in whole cell lysates and purified EV subfractions of D341 and MB1 cells (Supplementary Figure 8B). Specific marker proteins such as Rgap1, alpha-actinin-4 (LV), ALIX and Syntenin (sEV), or Tsg101 (LV and sEV) were enriched in the corresponding subfractions; yet there are pronounced differences in the proteomic profiles of distinct EV populations in the 2 MB cell lines. To validate the release of EVs by MB tumor cells, we performed transmission electron microscopy (TEM). Ultra-thin sections of cells grown on poly-L-lysin coated Aclar films showed LV/MV-type vesicles released by D341 cells (Fig. 4F; Supplementary Figure 9A), MB1 cells (not shown), or by Daoy, a human SHH-MB cell line (Supplementary Figure 9B).
Extracellular Vesicle Signatures in MB Patients
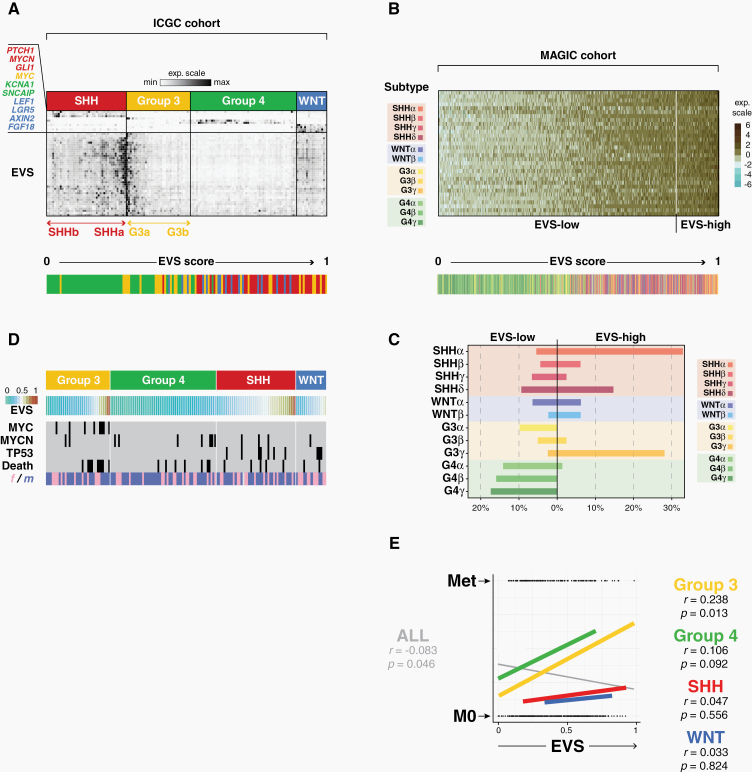
Based on the above observations, we developed a gene expression signature—termed EVS, or Extracellular Vesicle Signature—to examine human patient data. EVS contains 38 genes, the selection criteria of which are explained in details in the Supplementary Methods section. In summary, EVS signature genes were selected according to the following criteria: (i) they were found in the top 20th percentile of the most significantly activated genes in SHH-MB cells compared with healthy cerebellar cells; (ii) they constituted exact matches or were functionally closely related to the proteome signature of large oncosome proteins of prostate cancer cells; (iii) they could be linked to other EVS members through stringent functional association network analyzes (eg, Naca; see Fig. 4E) and/or had been previously linked to tumorigenic processes (eg, Tpt1, tumor protein, translationally controlled 1; Fig. 4E). It shows overlaps with different gene sets from the Molecular Signatures Database MsigDB, such as metabolic process, secretome, MYC targets, stem cell annotations and a number of cancer signatures (Supplementary Figure 10A). EVS and a derived EVS score (0 to 1, or lowest to highest) was applied to bulk mRNA expression data from the International Cancer Genome Consortium (ICGC) cohort (148 patients) and MAGIC cohort (763 patients).3 These analyses revealed a pronounced intertumoral heterogeneity of EVS, both across all 4 and within individual MB subgroups. Patients with SHH-MB tumors showed the highest and patients with Group 4 tumors the lowest average EVS score, while EVS ranged from extremely low to high scores in Group 3 patients (Supplementary Figure 10B). Large differences in EVS expression levels were also observed within individual subgroups. In particular we found cases with significantly increased EVS expression in the SHH and Group 3 subgroups of the ICGC cohort (Fig. 5A). We next expanded and refined this analysis to the 12 subtypes of the MAGIC cohort (Fig. 5B). Here the EVS score was highest in subtypes SHH-alpha and G3-gamma (Fig. 5C). This pronounced intertumoral heterogeneity of EVS led us to investigate whether heterogeneity also exists at the cellular level within single MB tumors. The analysis of human scRNA-seq data10 indeed revealed a comparable range of intertumoral EVS heterogeneity as found in the large MB cohorts (Supplementary Figure 11). More importantly, it also showed that even within individual tumors there can be considerable differences in EVS levels between tumor cell subpopulations. We confirmed intratumoral heterogeneity of EVS in published scRNA-seq data of the SmoM2 MB mouse model14 and in our own scRNA-seq data (Supplementary Figure 12). Interestingly, a clear anticorrelation between expression of EVS, the maturation status and vismodegib-sensitivity of tumor cells is observed (Supplementary Figures 2, 12).
Fig. 5.
EVS in 2 medulloblastoma patients cohorts. (A) Heatmap of relative expression levels of EVS and subgroup-specific marker genes in the ICGC cohort. Color-coding indicates subgroup affiliation of patients, and the bar graph below shows the distribution when patient samples are ordered by EVS score. (B) As in (A), showing EVS across 12 MB subtypes of the MAGIC cohort. (C) Distribution (in percentage) of samples with low or high EVS score, grouped by tumor subtype (MAGIC cohort). (D) Patient characteristics in the ICGC cohort, including focal amplifications of MYC or MYCN, TP53 mutations, survival/death or gender. (E) Overlaid correlation plots of EVS score vs metastatic status in the 4 subgroups or for the whole cohort. Pearson correlation coefficients (r) and t-test p-values (p) are reported.
EVS as a Prognostic Tool in MB
We next investigated whether high EVS expression levels are a risk factor for the survival of MB patients, and compared EVS with known other risk factors. Indeed, there was a positive correlation between MYC amplification with a high EVS score and increased mortality of the patients (ICGC cohort: Fig. 5D; MAGIC cohort: Supplementary Figure 13A). In addition, EVS-high correlated with a high incidence of metastases, especially in Group 3 and Group 4 tumors (Fig. 5E). In contrast, there were no clear links between EVS and other known risk factors, including chromosomal abnormalities such as isochromosome 17q, in the different MB subgroups/subtypes represented in the MAGIC cohort (Supplementary Figure 13A).
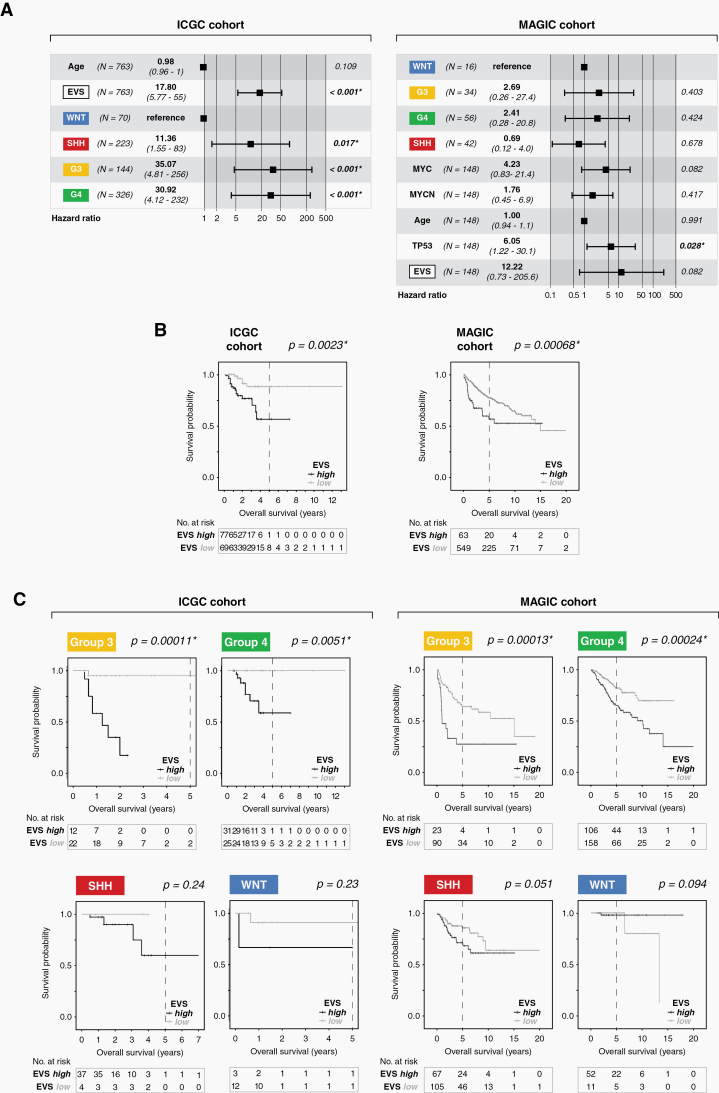
Finally, by correlating EVS with clinical data of both cohorts we were able to demonstrate its suitability as a prognostic tool in MB. In both cohorts EVS represents an independent, strong risk factor (hazard ratio 12.2 in ICGC, and 17.8 in MAGIC, respectively; Fig. 6A). Kaplan–Meier survival curves clearly illustrate the significant difference in clinical outcome of EVS-high versus EVS-low patients. This was true when considering all 4 subgroups (Fig. 6B), but especially pronounced for Group 3 and Group 4 patients (Fig. 6C).
Fig. 6.
Prognostic value of EVS. (A) Forest plots showing hazard ratios and 95% confidence intervals associated with the indicated variables for the ICGC cohort (left) and MAGIC cohort (right). The hazard ratio HR was calculated using the EVS score as a continuous variable in a multivariate Cox regression model of overall survival. (B) Kaplan–Meier curves showing the overall survival probabilities of patients in EVS-high versus EVS-low for the entire cohort, or (C) for the individual MB subgroups separately. The P-values were determined using a log-rank test, where P < 0.05 was considered significant (*).
Discussion
The results of our study are conceptually relevant in 2 ways: on the one hand with regard to general aspects of tumor biology and intercellular communication within the tumor niche, and on the other hand with regard to prognostics, and potentially diagnostics, of MB.
One lead observation is the marked similarity between the protein signature of large oncosomes generated by human prostate cancer cells and the EVS signature found in a murine CNS tumor. This implies a broader relevance of this shared signature, and one can easily anticipate its existence in additional, unrelated tumor entities. Indeed, preliminary data from our group indicate that there are other, unrelated tumors in which EVS expression is linked to patient survival (data not shown).
Tumor-secreted EVs are known to be important mediators of intercellular communication in both local and distant tumor microenvironments.35 A novel aspect of our study is to uncover an epigenetically stable anchoring of “foreign” gene expression patterns in both cancer and noncancer cells within the tumor niche. This holds true for several genes of the tumor-derived EV signature, which are upregulated in tumor-associated glial and immune cells, as well as for canonical glial and immune cell–specific genes, which are activated in the tumor cells. There are several possible scenarios that can explain these mutual adaptations of gene expression programs. On the one hand, these could occur as a result of EV-mediated communication. However, it remains largely speculative how such changes would be stably fixed in the recipient cells. Another explanation is provided by an elegant study that was published only recently, while our paper was being prepared. According to this study, so-called TuAstrocytes, or tumor-associated astrocytes, arise through trans-differentiation of transformed GNPs in SHH-activated MB.36
Our study establishes EVS as a prognostic tool in MB with high predictive power, which achieves high levels of significance especially for tumors in Group 3 and Group 4. This is all the more important, since for the latter subgroup there are hardly any robust prognostic tools so far.37 This becomes clear in the analysis of the MAGIC cohort, in which the authors used their 12-subtype spectrum to determine the likelihood of survival of the affected patients.3 There was no statistically significant difference in overall survival (OS) between the Group 4 subtypes G4 alpha, beta, and gamma (P-value 0.13). In striking contrast, the retrospective classification of the same patients using EVS revealed a highly significant difference in OS between EVS-high and EVS-low groups (P-value 0.00024; Fig. 6C). Similarly, the classification of Group 3 patients according to subtypes G3 alpha, beta, and gamma achieved a much lower significance level (P-value 0.0362) than the stratification according to EVS-high versus EVS-low (P-value 0.00013; Fig. 6C). The prognosis for Group 3 patients belonging to the EVS-high group is even worse than that of patients in the same subgroup with focal MYC amplification (P-value 0.0264).3 In this context it is interesting that key genes involved in glycolysis are upregulated in Group 3 tumors; of these genes, LDHA (encoding subunit A of lactate dehydrogenase/LDH) expression positively correlated with MYC and is associated with poor prognosis of Group 3 patients.38 Our study established Ldhb (encoding LDH subunit B) as one of the 38 EVS signature genes (Fig. 5C), which suggests that upregulation of LDH is common in Group 3 and SHH high-risk tumor subtypes (Fig. 5D).7 Cell culture models and a limited number of preclinical studies show that various chemical compounds have the potential to block or at least partially limit the formation and release of EVs. However, the specificity/selectivity of many of these compounds is questionable, which is not surprising in view of the complex biogenesis pathways of different EV classes. More extensive studies are now required to examine the activities of these drugs, also in combination with other inhibitor classes, in a broader range of in vitro and in vivo models. In this context, our study represents a promising starting point for such studies in medulloblastoma.
Taken together, the above results underline that patient classification according to EVS enables high-risk patients to be identified in each of the 4 molecular subgroups of MB. The identification of EVS as a novel clinicobiological trait broadens our understanding of MB and may be applicable to future molecular diagnostics, clinical risk stratification, and translational research.
Supplementary Material
Acknowledgments
We thank Buket Celik (Department of Medicine A, Hematology, Oncology, and Pneumology, UKM), Ilka Wolff and Kerstin Seiling (Department of Anatomy and Molecular Neurobiology, WWU Münster) for technical assistance during NTA and Western blot analyes or electron microscopy, respectively, Prof Tanja Kuhlmann and Dr Stefanie Albrecht (Institute of Neuropathology, University Münster) for the kind gift of Oli-neu cells and technical advice, and Carolin Ruckert and Dr Judit Horvath (Institute of Human Genetics, WWU Münster) for bulk mRNA-sequencing.
Funding
KK is supported by funds from the Deutsche Krebshilfe e.V. (111537 and 111784), by Kinderkrebshilfe Münster e.V. and by the Deutsche Kinderkrebsstiftung. GMzH is supported by grants from the Deutsche Forschungsgemeinschaft (ME4050/4-1 and 8-1), and from the Ministerium für Innovation, Wissenschaft und Forschung of North-Rhine-Westfalia. CFK is supported by grants from the Deutsche Forschungsgemeinschaft (SFB1192). MM is supported by grants from the Deutsche Forschungsgemeinschaft (SFB1348 - TP A03).
Conflict of interest statement. The authors declare that there is no conflict of interest regarding the publication of this article.
Authorship statement. Conceptualization: TKA, MI, KK, US, NJ, SMP. Methodology: NM, MG, TL, GMzH, MM, CFK. Validation: NJ, MS, SMP, AB. Data analysis: TKA, MI, MS, KK, NJ, VM, KM, AR. Experiments: TKA, NM, MG, DH, MS, SK, KM, AR. Original draft: TKA, MI, KK. Supervision: MD, US, MCF, SMP, AB, MM, KK. Funding acquisition: KK, GMzH, CFK.
References
- 1. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31(6):737–754.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwalbe EC, Lindsey JC, Nakjang S, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 2017;18(7):958–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramaswamy V, Remke M, Bouffet E, et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 2016;131(6):821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Archer TC, Ehrenberger T, Mundt F, et al. Proteomics, post-translational modifications, and integrative analyses reveal molecular heterogeneity within medulloblastoma subgroups. Cancer Cell. 2018;34(3):396– 410.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rivero-Hinojosa S, Lau LS, Stampar M, et al. Proteomic analysis of medulloblastoma reveals functional biology with translational potential. Acta Neuropathol Commun. 2018;6(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zomerman WW, Plasschaert SLA, Conroy S, et al. Identification of 2 protein-signaling states delineating transcriptionally heterogeneous human medulloblastoma. Cell Rep. 2018;22(12):3206–3216. [DOI] [PubMed] [Google Scholar]
- 10. Hovestadt V, Smith KS, Bihannic L, et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature. 2019;572(7767): 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jessa S, Blanchet-Cohen A, Krug B, et al. Stalled developmental programs at the root of pediatric brain tumors. Nat Genet. 2019;51(12):1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vladoiu MC, El-Hamamy I, Donovan LK, et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature. 2019;572(7767):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang L, He X, Liu X, et al. Single-cell transcriptomics in medulloblastoma reveals tumor-initiating progenitors and oncogenic cascades during tumorigenesis and relapse. Cancer Cell. 2019;36(3):302–318.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ocasio J, Babcock B, Malawsky D, et al. scRNA-seq in medulloblastoma shows cellular heterogeneity and lineage expansion support resistance to SHH inhibitor therapy. Nat Commun. 2019;10(1):5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Menck K, Sivaloganathan S, Bleckmann A, Binder C. Microvesicles in cancer: small size, large potential. Int J Mol Sci. 2020;21(15):5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schüller U, Heine VM, Mao J, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14(2):123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Melcher V, Graf M, Interlandi M, et al. Macrophage-tumor cell interaction promotes ATRT progression and chemoresistance. Acta Neuropathol. 2020;139(5):913–936. [DOI] [PubMed] [Google Scholar]
- 18. Huang X, Ketova T, Litingtung Y, Chiang C. Isolation, enrichment, and maintenance of medulloblastoma stem cells. J Vis Exp. 2010;(43):2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Niesmann K, Breuer D, Brockhaus J, et al. Dendritic spine formation and synaptic function require neurobeachin. Nat Commun. 2011;2:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu X, Mao Q, Tang Y, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sandén E, Dyberg C, Krona C, et al. Aberrant immunostaining pattern of the CD24 glycoprotein in clinical samples and experimental models of pediatric medulloblastomas. J Neurooncol. 2015;123(1):1–13. [DOI] [PubMed] [Google Scholar]
- 23. Rosenberg AB, Roco CM, Muscat RA, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360(6385):176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee C, Lee J, Choi SA, et al. M1 macrophage recruitment correlates with worse outcome in SHH Medulloblastomas. BMC Cancer. 2018;18(1):535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Margol AS, Robison NJ, Gnanachandran J, et al. Tumor-associated macrophages in SHH subgroup of medulloblastomas. Clin Cancer Res. 2015;21(6):1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maximov V, Chen Z, Wei Y, et al. Tumour-associated macrophages exhibit anti-tumoural properties in Sonic Hedgehog medulloblastoma. Nat Commun. 2019;10(1):2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bockmayr M, Mohme M, Klauschen F, et al. Subgroup-specific immune and stromal microenvironment in medulloblastoma. Oncoimmunology. 2018;7(9):e1462430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cahoy JD, Emery B, Kaushal A, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marques S, Zeisel A, Codeluppi S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352(6291):1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vanlandewijck M, He L, Mäe MA, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554(7693):475–480. [DOI] [PubMed] [Google Scholar]
- 31. Minciacchi VR, You S, Spinelli C, et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6(13):11327–11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Minciacchi VR, Spinelli C, Reis-Sobreiro M, et al. MYC mediates large oncosome-induced fibroblast reprogramming in prostate cancer. Cancer Res. 2017;77(9):2306–2317. [DOI] [PubMed] [Google Scholar]
- 33. Barnett SC, Rosario M, Doyle A, Kilbey A, Lovatt A, Gillespie DA. Differential regulation of AP-1 and novel TRE-specific DNA-binding complexes during differentiation of oligodendrocyte-type-2-astrocyte (O-2A) progenitor cells. Development. 1995;121(12):3969–3977. [DOI] [PubMed] [Google Scholar]
- 34. Vollgraf U, Wegner M, Richter-Landsberg C. Activation of AP-1 and nuclear factor-kappaB transcription factors is involved in hydrogen peroxide-induced apoptotic cell death of oligodendrocytes. J Neurochem. 1999;73(6):2501–2509. [DOI] [PubMed] [Google Scholar]
- 35. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30(6):836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao M, Ventura PB, Jiang Y, et al. Astrocytic trans-differentiation completes a multicellular paracrine feedback loop required for medulloblastoma tumor growth. Cell. 2020;180(3):502–520.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frappaz D, Faure-Conter C, Meyronet D, et al. Are molecular subgroups of medulloblastomas really prognostic? Curr Opin Neurol. 2018;31(6):747–751. [DOI] [PubMed] [Google Scholar]
- 38. Tao R, Murad N, Xu Z, et al. MYC drives group 3 medulloblastoma through transformation of Sox2+ astrocyte progenitor cells. Cancer Res. 2019;79(8):1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Danaher P, Warren S, Dennis L, et al. Gene expression markers of tumor Infiltrating leukocytes. J Immunother Cancer. 2017;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.