Abstract
Background
Acquired chemoresistance is a major challenge in the clinical treatment of glioblastoma (GBM). Circular RNAs have been verified to play a role in tumor chemoresistance. However, the underlying mechanisms remain unclear. The aim of this study was to elucidate the potential role and molecular mechanism of circular (circ)RNA ADP-ribosylation factor GTPase activating proteins with Src homology 3 domain, ankyrin repeat and Pleckstrin homology domain 1 (circASAP1) in temozolomide (TMZ) resistance of GBM.
Methods
We analyzed circRNA alterations in recurrent GBM tissues relative to primary GBM through RNA sequencing. Real-time quantitative reverse transcription PCR verified the expression of circASAP1 in tissues and cells. Knockdown and overexpressed plasmids were used to evaluate the effect of circASAP1 on GBM cell proliferation and TMZ-induced apoptosis. Mechanistically, fluorescent in situ hybridization, dual-luciferase reporter, and RNA immunoprecipitation assays were performed to confirm the regulatory network of circASAP1/miR-502-5p/neuroblastoma Ras (NRAS). An intracranial tumor model was used to verify our findings in vivo.
Results
CircASAP1 expression was significantly upregulated in recurrent GBM tissues and TMZ-resistant cell lines. CircASAP1 overexpression enhanced GBM cell proliferation and TMZ resistance, which could be reduced by circASAP1 knockdown. Further experiments revealed that circASAP1 increased the expression of NRAS via sponging miR-502-5p. Moreover, circASAP1 depletion effectively restored the sensitivity of TMZ-resistant xenografts to TMZ treatment in vivo.
Conclusions
Our data demonstrate that circASAP1 exerts regulatory functions in GBM and that competing endogenous (ce)RNA-mediated microRNA sequestration might be a potential therapeutic strategy for GBM treatment.
Keywords: circASAP1, glioblastoma, miR-502-5p, NRAS, temozolomide resistance
Key Points.
1. CircASAP1 was highly elevated in recurrent GBM patients.
2. Eukaryotic translation initiation factor 4A3 (EIF4A3) bound to a circASAP1 flanking sequence upregulated the expression of circASAP1.
3. CircASAP1 contributed to GBM progression and TMZ resistance via sponging miR-502-5p and activating NRAS/mitogen-activated protein kinase kinase 1/extracellular signal-regulated kinase 1 and 2 signaling.
Importance of the Study.
Glioblastoma is the most malignant tumor of the central nervous system and has a high incidence of TMZ resistance. Here, we identified an abnormally upregulated circular RNA. CircASAp1 in glioblastoma recurred after treatment with TMZ. Upregulation of circASAP1 promoted GBM proliferation and TMZ resistance. The high expression of circASAP1 is due to the combination of EIF4A3 with flank sequence of circASAP1, which induces an increase in backsplicing. CircASAP1 sponges miR-502-5p to regulate NRAS expression through a ceRNA mechanism, thus promoting tumor growth and TMZ resistance in vitro and in vivo. Targeting circASAP1/miR-502-5p/NRAS signaling appears to be a promising treatment for GBM.
Gliomas are the most common primary brain tumors, more than half of which are glioblastomas (GBMs), the most malignant tumor of the central nervous system. Even with maximum feasible surgical resection, patients treated with radiotherapy and chemotherapy have a median survival time of less than 15 months.1,2 Temozolomide (TMZ), a second-generation oral alkylating agent, can readily pass through the blood–brain barrier and is currently used as a first-line therapy for GBM treatment.3,4 However, major impediments to effective treatment are postoperative tumor recurrence and acquired resistance to TMZ.5
Noncoding (nc)RNAs, including micro (mi)RNAs, long noncoding (lnc)RNAs, and circular (circ)RNAs, play crucial roles in cell development and diseases.6–9 CircRNAs are a class of widely existing ncRNAs produced by RNA backsplicing and are characterized by covalent closed-loop structures.10,11 Due to their unique structure, circRNAs are highly stable, predominantly distributed in the cytoplasm, and conserved across species.12 Many circRNAs have been detected in a variety of eukaryotes and found to be involved in the pathology of diseases.13–15 Three important functions of circRNAs have been identified: (i) acting as miRNA sponges, (ii) binding to proteins, and (iii) undergoing translation.16,17 CircRNAs can communicate with miRNAs, and they regulate each other by competitively binding to miRNA response elements to further regulate miRNA-targeted gene expression.18 Some studies raised the hypothesis that backsplicing could be mediated by the dimerization of RNA-binding proteins (such as protein quaking [HQK; encoded by QKI] or the RNA binding protein Fused in Sarcoma (FUS) that binds to specific motifs in flanking introns.19,20 Recently, EIF4A3 has been reported to bind RNA and form exon junction complexes, which participate in exon splicing.21 One study showed that EIF4A3 combined with the flanking sequence of circular matrix metalloproteinase 9 (circMMP9), promoting its backsplicing.22
Methods
More method details are in Supplementary Methods.
Ethics Approval and Consent to Participate
The study was approved by the ethics committee of Nanjing Medical University (code: 2019-SR-479), and written informed consent was obtained from all patients.
Clinical Samples
The 35 primary GBM specimens and 15 recurrent GBM specimens used in this study were obtained by surgical resection from patients undergoing TMZ chemotherapy at the Department of Neurosurgery, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China, with written informed consent provided by the patients. The diagnosis of glioma was confirmed by pathologists. Detailed patient information is presented in Supplementary Table 1.
RNA Sequencing Analysis
Total RNA was isolated, and RNA quantification and quality assessment were performed. RNA integrity and genomic (g)DNA contamination were tested by denaturing agarose gel electrophoresis. RNA from each sample was removed by ribosomal RNA prior to RNA-seq library construction. The sequencing library was evaluated and adjusted to 10 nM before cluster generation. The cDNA library was then sequenced and 100 base pairs were subjected to a paired-end run.
Public Data Collection
Microarray datasets and their associated clinical information were downloaded from the Chinese Glioma Genome Atlas (CGGA).
Cell Lines and Cell Culture
Four drug-related cell lines (N3S, N3T3rd, U251, and U251T3rd) were described in our previous report.23 All cells were cultured in high-glucose DMEM supplemented with 10% fetal bovine serum at 37°C with 5% CO2.
RNA Extraction, Treatment with RNase R, and Quantitative Real-Time PCR Assays
Total RNA was extracted from tissue samples and cell lines. Two milligrams total RNA was incubated with or without RNase R (Epicentre Technologies). Nuclear and cytoplasmic fractions were separated with the PARIS Kit (Invitrogen). Quantitative real-time (qRT) PCR analyses were performed with SYBR Green Premix Ex Taq (Takara). Primer sequences are shown in Supplementary Table 2. The relative quantitative value for each gene was determined by using the 2−∆∆CT method.
Western Blot Assay
Samples were lysed for total protein extraction in radioimmunoprecipitation buffer (Solarbio). Protein concentrations were determined by bicinchoninic acid protein assay (Thermo Fisher Scientific). Equivalent amounts of protein (50 μg) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Merck KGaA). The membranes were incubated with primary antibodies, followed by incubation with a secondary antibody. Proteins were detected by using an enhanced chemiluminescence substrate kit (Thermo Fisher Scientific) and ImageJ software. The antibodies used are listed in Supplementary Table 3.
Plasmid Construction and Cell Transfection
Lipofectamine 2000 (Invitrogen) was used to transfect small interfering (si)RNAs, miRNA mimics, and plasmids into cells. All siRNA and short hairpin (sh)RNA sequences are listed in Supplementary Table 4. We synthesized full-length complementary (c)DNAs of human circASAP1 and EIF4A3 and cloned these cDNAs into the expression vector pLCDH-ciR (Geneseed). CircASAP1-specific shRNAs and a negative control RNA (sh-Ctrl) were designed and synthesized by GeneChem. Stable cell lines were selected with puromycin beginning at 48 h after injection. EIF4A3-specific siRNA, NRAS-specific siRNA, miR-502-5p mimics, and a miR-502-5p inhibitor were purchased from GeneChem.
RNA Fluorescence In Situ Hybridization
A cyanine 3–labeled specific probe for circASAP1 was designed and synthesized by RiboBio, and signals were detected with a fluorescence in situ hybridization (FISH) kit (RiboBio) according to the manufacturer’s instructions. Confocal images were captured using Zeiss AIM software and a Zeiss LSM 700 confocal microscope system.
Flow Cytometry Analysis of Apoptosis
For apoptosis analysis, cells were cultured with TMZ and stained with propidium iodide and annexin V‒fluorescein isothiocyanate according to the manufacturer’s instructions (Roche).
Terminal Deoxynucleotidyl Transferase Deoxyuridine Triphosphate Nick End Labeling Assay
GBM cells were fixed in 4% paraformaldehyde. The cells were then stained with the In Situ Cell Death Detection Kit, POD (peroxidase; Roche) according to the manufacturer’s protocol. Images were acquired with a Nikon ECLIPSE E800 fluorescence microscope.
CCK-8 and Colony Formation Assays
GBM cells were seeded in 96-well plates, and cell viability was evaluated with Cell Counting Kit 8 (CCK-8; Dojindo). For a colony formation assay, cells were seeded in 6-well plates and cultured. The resulting colonies were stained with 0.1% crystal violet.
RNA Pull-Down Assay
Biotinylated sense and antisense circASAP1 flanking RNA sequences were obtained with a MEGAscript kit (Ambion) and the Pierce RNA 3′ Desthiobiotinylation Kit (Thermo Fisher Scientific). Biotinylated flanking sequences were incubated with cell lysates and streptavidin magnetic beads with rotation. The eluted proteins were validated by silver staining and western blotting.
Luciferase Reporter Assays
Cells were seeded in a 24-well plate 24 h before transfection. The cells were cotransfected with a mixture of pmirGLO vectors containing circASAP1-miR-502-5p binding sequences or mutant sequences and miRNA mimics (50 nM). After 24 h, luciferase activity was measured using a dual-luciferase reporter assay system (Promega) according to the manufacturer’s protocol.
Biotinylated MiRNA Capture
A miRNA pull-down assay was performed using a biotinylated miR-502-5p mimic or control RNA (RiboBio) transfected into human embryonic kidney (HEK) 293T cells. Biotin-coupled RNA complexes were pulled down by incubating cell lysates with streptavidin magnetic beads (Thermo Fisher Scientific).
RNA Immunoprecipitation
RIP experiments were performed with the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s protocol.
In Vivo Xenograft Model
To establish an intracranial tumor model, 2.5 × 105 N3T3rd cells were separately implanted stereotactically into the nude mouse brain. After surgery, the mice were treated with or without TMZ. Bioluminescence imaging (IVIS Spectrum, PerkinElmer) was used to confirm intracranial tumor formation, and tumors were measured each week. The procedures used for animal treatments and experiments conformed with the Guide for the Care and Use of Laboratory Animals, and this study was approved by the Nanjing Medical University Animal Experimental Ethics Committee.
Immunohistochemistry
The mouse brain was fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 3.5–4 µm thick sections. The sections were stained with an antibody against NRAS or Ki-67.
Statistical Analysis
All statistical analyses were performed with GraphPad software v7.0 or IBM SPSS Statistics v23.0 software. The significance of intergroup differences was estimated with Student’s t-test, the chi-square test, or one-way ANOVA as appropriate. The Kaplan–Meier method with the log-rank test was used to calculate overall survival (OS) for comparisons between different groups. The correlations between variables were analyzed with the Pearson correlation coefficient. All results are shown as the mean ± standard error of the mean (SEM) of 3 independent experiments. Statistical significance was considered to be indicated by a value of P < 0.05.
Results
CircASAP1 Is Highly Expressed in TMZ-Resistant GBM Cells and Tissues After TMZ Treatment
We collected 3 paired primary and recurrent GBM tissue samples from 3 GBM patients after surgery and standardized treatment with TMZ. RNA-seq of ribosomal RNA-depleted total RNA was performed. The expression of circRNA transcripts varied between recurrent GBM tissues and matched primary GBM tissues (Figure 1A). Among the 56 differentially expressed circRNAs (log2 fold change ≥1, P < 0.05), 33 were upregulated and 23 were downregulated in recurrent GBM tissues relative to primary GBM tissues (Figure 1B). In addition, we cultured 2 TMZ-resistant cell lines: N3T3rd and U251T3rd. These cells exhibited a poor response to TMZ (Supplementary Figure 1A, B).
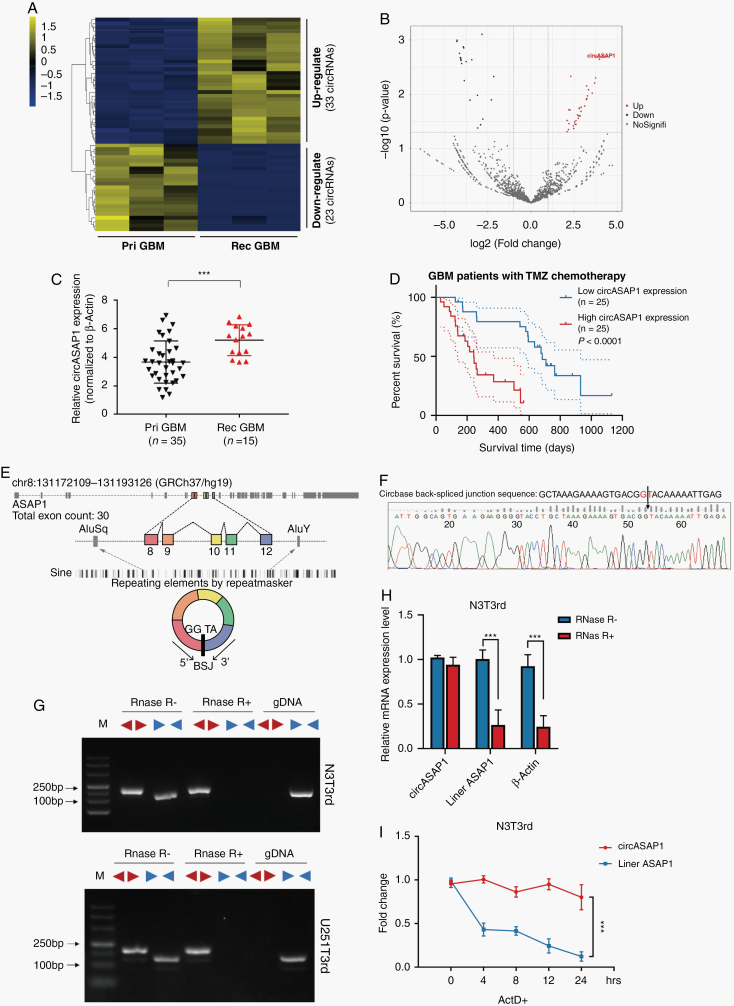
Fig. 1.
CircASAP1 expression is upregulated in TMZ-resistant GBM tissue samples and cell lines. (A) Hierarchical clustering of 56 differentially expressed circRNAs in 3 primary and 3 corresponding recurrent TMZ-refractory GBM tumor samples. (B) Volcano plot of differentially expressed circRNAs. (C) Relative expression of circASAP1 in primary and recurrent GBM tissue samples, normalized to β-actin. (D) Kaplan–Meier survival analysis of the OS of GBM patients treated with TMZ chemotherapy (n = 50, P < 0.0001). (E) Schematic diagram of the genomic location and splicing pattern of hsa_circ_0002330 (480 bp). (F) The arrow represents the “head to tail” splicing sites of circASAP1 identified by Sanger sequencing. (G) The expression levels of the backspliced and canonical forms of ASAP1 in cDNA and gDNA isolated from GBM cells. Red arrows represent divergent primers; blue arrows represent convergent primers. (H) Relative expression of circASAP1 and linear ASAP1 and β-actin in the presence or absence of RNase R supplementation. (I) The abundances of circASAP1 and linear ASAP1 in N3T3rd cells treated with actinomycin D at the indicated time points. Data are presented as the mean ± SEM of 3 independent experiments. Significant results are presented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We focused on the most highly upregulated and downregulated circRNAs and matched them with data in CircBase (http://www.circbase.org/). Hsa_circ_0002330 (termed circASAP1 in the remainder of the article), which was the most highly upregulated circRNA, attracted our attention. CircASAP1 was significantly upregulated in recurrent GBM tissues compared with primary GBM tissues (Figure 1C). Furthermore, circASAP1 was markedly increased in GBM tissues compared with normal tissues (Supplementary Figure 1C). Similar results were observed by FISH (Supplementary Figure 1D). CircASAP1 expression was significantly higher in N3T3rd and U251T3rd than in TMZ-sensitive cell lines (N3s and U251s) (Supplementary Figure 1E). However, the expression of linear ASAP1 mRNA showed no significant difference between cell lines and GBM tissues (Supplementary Figure 1F, G). Then, Kaplan–Meier analysis was used to determine whether the expression level of circASAP1 in GBM tissue was related to the clinical response to TMZ treatment. Results of our cohort showed that patients with low circASAP1 expression displayed better OS after TMZ treatment than patients with high circASAP1 expression (Figure 1D). Moreover, the level of linear ASAP1 mRNA had no effect on OS in GBM patients according to the CGGA database (Supplementary Figure 1H).
Through the University of California Santa Cruz Genomics Institute Bioinformatics site (http://genome.ucsc.edu/), we identified that circASAP1 was transcribed from the ASAP1 gene locus and formed by backsplicing between the splicing acceptor of exon 8 and the splicing donor of exon 12. CircASAP1 contains 5 exons that ultimately create a transcript of 480 nucleotides via backsplicing (Figure 1E). The backsplicing sequence, as reported in the CircBase database, was confirmed by Sanger sequencing (Figure 1F).
In order to eliminate the possibility of genomic rearrangements and trans-splicing, PCR and agarose gel electrophoresis assays were performed to detect the expression level of backspliced or canonical forms of ASAP1 in cDNA and gDNA. Divergent primers detected circASAP1 in cDNA even with RNase R treatment, but no products were detected in gDNA, which indicated that circASAP1 was truly circular and could not be amplified from gDNA. The linear form of ASAP1 products were amplified from cDNA and gDNA by convergent primers, but these products could not endure RNase R treatment (Figure 1G). Moreover, qRT-PCR results also showed that circASAP1 could resist RNase R–mediated digestion (Figure 1H and Supplementary Figure 1I). Then, the stability of circASAP1 was investigated by transcription inhibitor actinomycin D treatment. The results exhibited that circRNA isoform was highly stable, with a transcriptional half-life of more than 24 h, while the associated linear transcript exhibited a half-life of <4 h (Figure 1I and Supplementary Figure 1J).
Taken together, these results suggest that circASAP1 expression is upregulated in TMZ-resistant GBM cell lines and tissues, pointing to a possible relationship between circASAP1 and acquired TMZ resistance.
CircASAP1 Promotes Proliferation and TMZ Resistance In Vitro
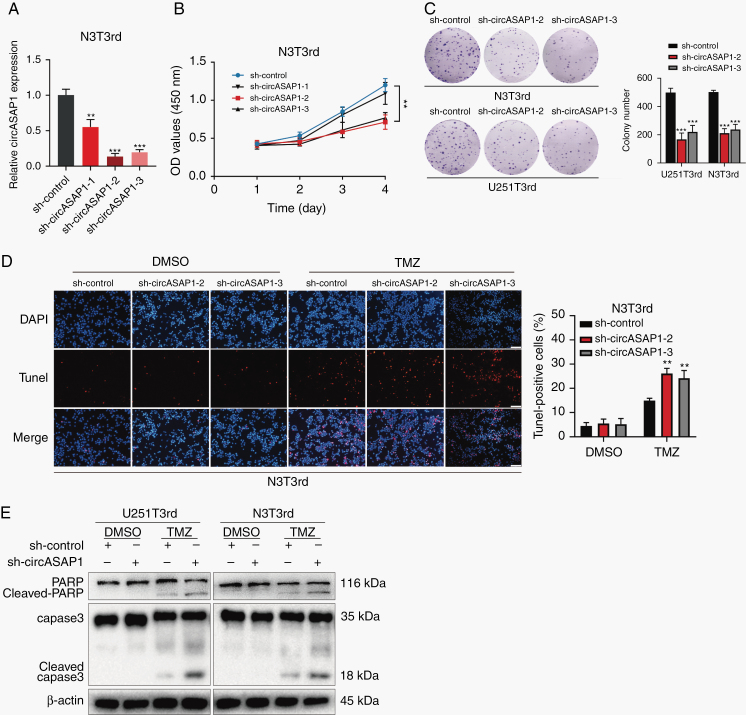
To explore the role of circASAP1 in GBM tumorigenesis and acquired TMZ resistance, 3 independent shRNAs specific for circASAP1, which had no effect on linear ASAP1 (Supplementary Figure 3A, B), were designed and transfected into TMZ-resistant cells. CircASAP1 expression level was detected by qRT-PCR (Figure 2A and Supplementary Figure 2A). Sh-circASAP1 was specific to circASAP1. Knockdown of circASAP1 significantly inhibited cell proliferation (Figure 2B and Supplementary Figure 2B), and such inhibition depends on the effectiveness of shRNA. Thus, sh-circASA1–2 and sh-circASAP1–3 were chosen for subsequent experiments. The colony formation assays showed that sh-circASAP1 reduced colony numbers compared with sh-Ctrl (Figure 2C). Moreover, downregulation of circASAP1 markedly enhanced the level of cell apoptosis (Figure 2D and Supplementary Figure 2C, D) and activated the cleavage of caspase-3 and its substrate poly(ADP-ribose) polymerase (PARP) (Figure 2E). Likewise, we got the similar results in sh-circASAP1 transducted TMZ-sensitive cells (Supplementary Figure 3C, D). Moreover, when overexpressing circASAP1, cell proliferation was promoted, and cell apoptosis induced by TMZ was reduced (Supplementary Figures 4A–F).
Fig. 2.
Knocking down circASAP1 expression inhibits proliferation and restores TMZ sensitivity in TMZ-resistant GBM cells. (A) CircASAP1 expression in N3T3rd ells. (B) CCK-8 assay of N3T3rd cells. (C) Colony formation assay of TMZ-resistant cells transducted by sh-circASAP1. (D) TUNEL analysis of circASAP1 knockdown cells or vehicle control-treated cells with or without TMZ treatment (200 μM, 48 h). Scale bar = 100 μm. (E) Western blot analysis of caspase-3 and PARP in TMZ-resistant cells treated with the vehicle control or TMZ (200 μM) for 48 h. β-actin was used as the loading control. Data are presented as the mean ± SEM of 3 independent experiments. Significant results are presented as *P < 0.05, **P < 0.01, and ***P < 0.001.
Collectively, these results showed that knocking down circASAP1 inhibits proliferation and restores TMZ sensitivity in TMZ-resistant cells.
RNA Binding Protein EIF4A3 Regulates the Expression of CircASAP1
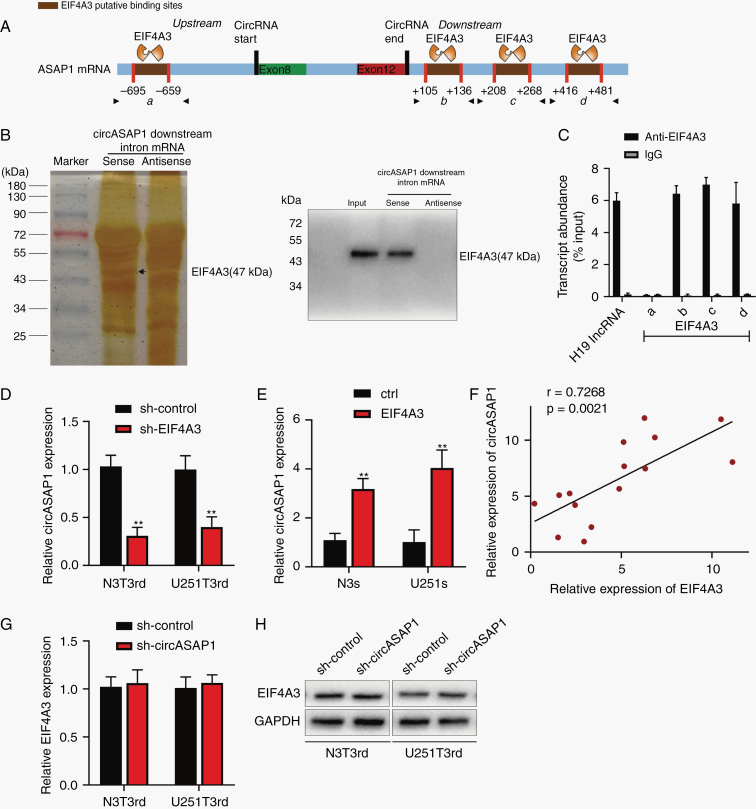
Studies have shown that some RNA-binding proteins can bind to circRNAs flanking intron sequences, playing an important role in the generation of circRNAs, such as FUS.19 Therefore, to explore how circASAP1 differentially expressed in GBM, we used CircInteractome (https://circinteractome.nia.nih.gov/) to predict RNA-binding protein sites matching the flanking regions of circASAP1, with EIF4A3 having the most binding sites (Figure 3A). EIF4A3 was reported to promote the expression of circMMP9.22 EIF4A3 expression level increased with the grade of glioma and was higher in recurrent GBM than in primary GBM (Supplementary Figure 5A–C). Similarly, the expression of EIF4A3 was higher in TMZ-resistant cells than in normal human astrocyte (Supplementary Figure 5D). Although expression of EIF4A3 is not prognostic of glioma cases in the database of The Cancer Genome Atlas (TCGA) (Supplementary Figure 5E), patients with high EIF4A3 expression had shorter survival time than those with low expression in the CGGA database (Supplementary Figure 5F, G).
Fig. 3.
The RNA binding protein EIF4A3 regulates the expression of circASAP1. (A) The binding sites for EIF4A3 in the flanking sequences of the ASAP1 mRNA transcript were predicted using CircInteractome. (B) Pull-down silver staining and western blotting were used to demonstrate the interaction between EIF4A3 and the circASAP1 downstream region. (C) RIP assay to verify EIF4A3 binding at the putative sites. H19 lncRNA was used as the positive control. (D) N3T3rd and U251T3rd cells were transfected with a control or sh-EIF4A3 and circASAP1 expression was detected by qRT-PCR. (E) Expression of circASAP1 in sh-control or sh-EIF4A3 tranducted TMZ-resistant cells. (F) The correlation between EIF4A3 and circASAP1 (n = 15). (G–H) Both the mRNA and protein levels of EIF4A3 were evaluated in circASAP1-downregulated cells. Data are presented as the mean ± SEM of 3 independent experiments. Significant results are presented as *P < 0.05 and **P < 0.01.
RNA pull-down assay was used to verify the ability of EIF4A3 to bind to circASAP1 flanking sequences. Results showed that EIF4A3 could bind to the downstream flanking sequence of circASAP1 (Figure 3B), but not upstream sequences (Supplementary Figure 5H). Besides, mass spectrometry results showed that EIF4A3 was the most abundant protein bound to circASAP1 flanking sequences (Supplementary Figure 5I). Figure 3C indicates that EIF4A3 could combine with flanking sequences through 3 downstream putative binding sites (named b, c, and d), but not the upstream site (named a). Downregulation of EIF4A3 leads to a reduction in circASAP1 expression (Figure 3D), while overexpression of EIF4A3 increased circASAP1 expression (Figure 3E). In addition, there was a positive expression correlation between circASAP1 and EIF4A3 in 15 clinical samples (Figure 3F). In contrast, silencing circASAP1 did not change EIF4A3 mRNA or protein levels (Figure 3G, H). In conclusion, EIF4A3 increased expression of circASAP1 by combining with the flanking sequences.
CircASAP1 Serves as a Sponge of MiR-502-5p
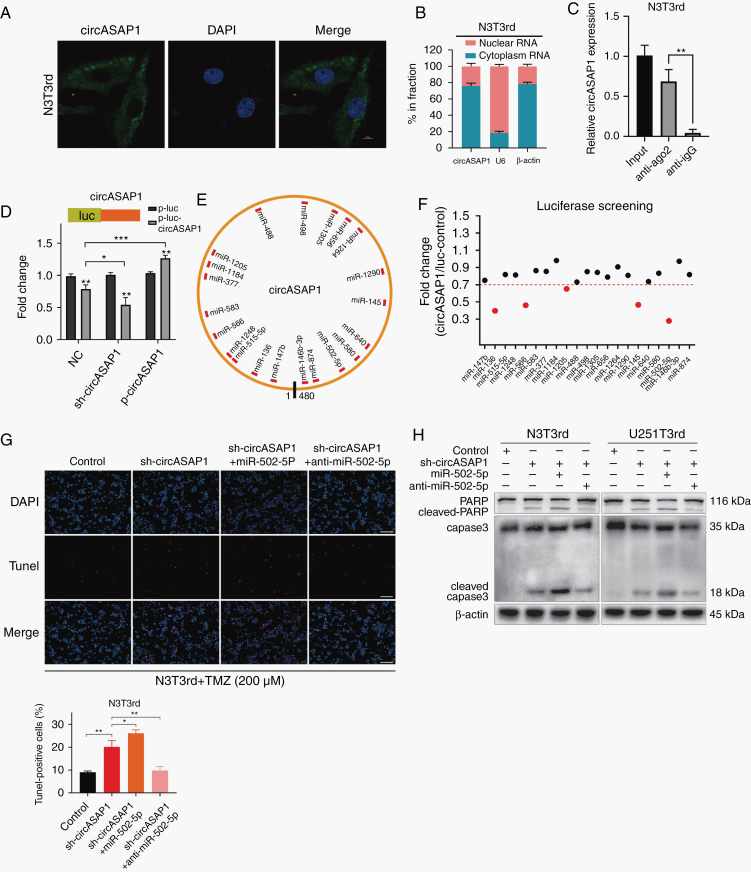
Because circASAP1 has been shown to play important roles in glioblastoma tumorigenesis and TMZ resistance, we next investigated the mechanisms underlying circASAP1 functions. CircRNAs have been reported to regulate target gene expression by acting as ceRNAs for miRNAs in cytoplasm.24 We first analyzed the circASAP1 distribution using a nuclear mass separation assay and FISH analysis. CircASAP1 was mainly distributed in the cytoplasm (Figure 4A, B and Supplementary Figure 6A). CircASAP1 could directly combine with Argonaute-2 (Ago2), which is a core component of the RNA-induced silencing complex (Figure 4C and Supplementary Figure 6B, C). A luciferase reporter gene with a circASAP1 fragment was constructed and inserted. Subsequently, knocking down endogenous circASAP1 expression further decreased luciferase activity, and vice versa (Figure 4D). These results suggested that circASAP1 might serve as a binding platform for Ago2 and miRNAs.
Fig. 4.
CircASAP1 serves as a sponge for miR-502-5p. (A) FISH analysis indicated the subcellular location of circASAP1 in N3T3rd cells (green). Nuclei were stained with DAPI (blue). Scale bar = 10 μm. (B) Relative circASAP1 expression levels in nuclear and cytosolic fractions of N3T3rd cells are shown. U6 was used as the nuclear control. β-actin was used as the cytosolic control. (C) RIP experiments were performed using an anti-Ago2 antibody, and specific primers were used to detect the enrichment of circASAP1 in N3T3rd cells by qRT-PCR. (D) A luciferase reporter assay was conducted to measure the luciferase activity of p-luc-circASAP1 in HEK-293T cells cotransfected with the sh-circASAP1 or pLCDH-circASAP1 vector. (E) A schematic drawing shows the putative binding sites for miRNAs associated with circASAP1. (F) A luciferase reporter assay was used to evaluate the luciferase activity of p-luc-circASAP1 in HEK-293T cells transfected with a library of 21 miRNA mimics to identify miRNAs that were able to bind to the circASAP1 sequence. The 5 miRNAs that inhibited luciferase activity by 30% are indicated by red dots. (G) TUNEL staining was used to analyze the effect of a miR-502-5p mimic or inhibitor on N3T3rd cells with circASAP1 knockdown after treatment with 200 μM TMZ for 48 h. Scale bar = 100 μm. (H) Cleaved caspase-3 and PARP in N3T3rd and U251T3rd cells after 200 μM TMZ treatment for 48 h. Data are presented as the mean ± SEM of 3 independent experiments. Significant results are presented as *P < 0.05, **P < 0.01, and ***P < 0.001.
Twenty-one candidate miRNAs were predicted to have binding sites along the circASAP1 sequence in the CircInteractome database (Figure 4E). To identify the target miRNAs, we performed luciferase screening of a miRNA library. Five miRNAs (miR-136, miR-566, miR-1205, miR-145, and miR-502-5p) out of the 21 miRNAs were able to reduce luciferase reporter activity by at least 30% (Figure 4F). These miRNAs did not decrease circASAP1 and linear ASAP1 expression (Supplementary Figure 6D, E), suggesting that circASAP1 may not be digested by these miRNAs. To determine which miRNA plays a role in tumor inhibition, individual miRNA mimics were transfected into TMZ-resistant cells. A cell proliferation assay revealed that miR-502-5p significantly inhibited cell growth (Supplementary Figure 6F). In addition, a specific biotinylated miR-502-5p probe successfully captured circASAP1 (Supplementary Figure 6G). Next, we found that miR-502-5p–mediated luciferase activity suppression was restored by mutation of circASAP1 within the predicted binding site (Supplementary Figure 6H). Therefore, the direct interaction between circASAP1 and miR-502-5p was confirmed.
Studies have confirmed miR-502-5p as a tumor suppressor in a variety of cancers.25–27 To investigate whether miR-502-5p participates in the circASAP1-mediated mechanisms involved in tumor growth and TMZ resistance, we knocked down or overexpressed miR-502-5p in circASAP1-depleted TMZ-resistant cells. Colony formation and CCK-8 assays showed that miR-502-5p overexpression markedly suppressed cell growth, while knocking down miR-502-5p promoted cell proliferation (Supplementary Figure 7A, B). Moreover, miR-502-5p restored TMZ resistance in circASAP1-depleted TMZ-resistant cells, while miR-502-5p inhibitor reversed such effect (Supplementary Figure 7C). Similarly, assay by terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL) and western blot analysis showed that miR-502-5p overexpression markedly increased the apoptosis ratio and activation of caspase-3 (Figure 4G, H and Supplementary Figure 7D). Likewise, when circASAP1 was overexpressed in TMZ-sensitive cells, cell proliferation was promoted and miR-502-5p inhibitor could enhance this effect, but miR-502-5p mimics could inhibit this effect (Supplementary Figure 8A). In addition, pLCDH-circASAP1 plasmid decreased the apoptosis rates of TMZ-sensitive cells, but it was blocked by miR-502-5p mimic (Supplementary Figure 8B). Finally, we discovered that the copy numbers of miR-502-5p almost equaled the copy numbers of circASAP1 in TMZ-resistant cells, but were more than circASAP1 in TMZ-sensitive cells by absolute quantification PCR (Supplementary Figure 8C).
In summary, circASAP1 acts as a molecular sponge of miR-502-5p, both of which are involved in the molecular mechanism underlying acquired TMZ resistance in GBM.
NRAS Is an Endogenous Target of MiR-502-5p
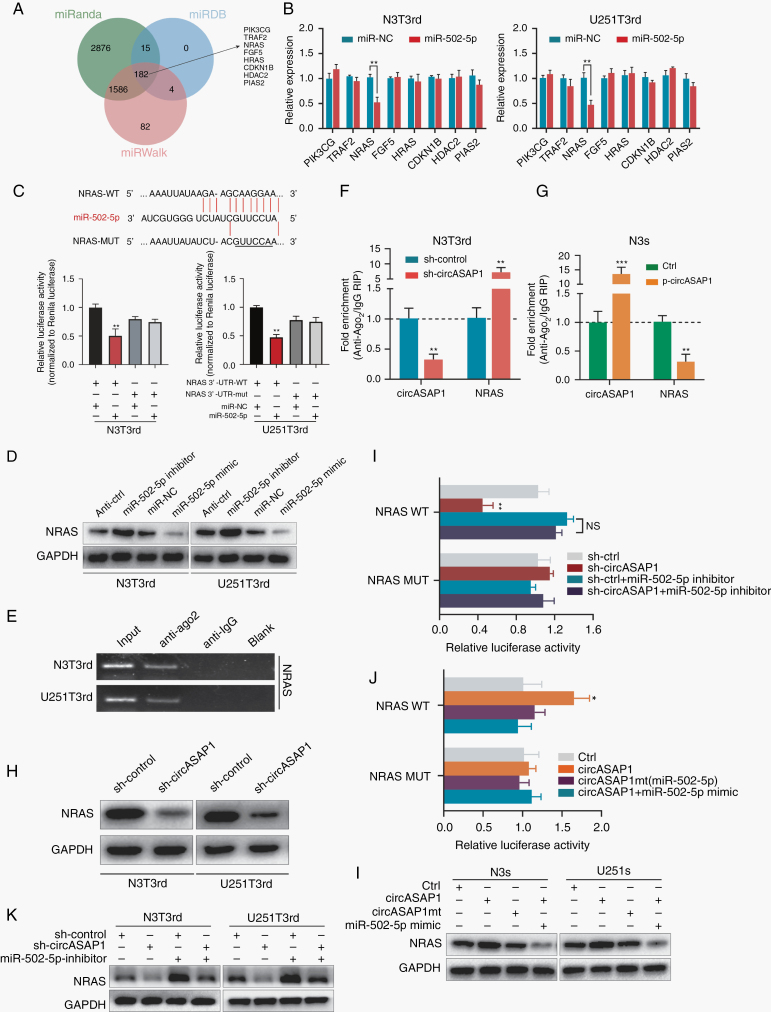
We predicted miR-502-5p targets by analyzing 3 databases together: miRanda, miRDB, and miRWalk; 182 putative target genes were identified. Among these targets, we selected 8 genes related to tumor growth in the Kyoto Encyclopedia of Genes and Genomes pathway analysis as potential components of the circASAP1–miR-502-5p ceRNA network (Figure 5A). Upregulation of miR-502-5p decreased NRAS expression, while anti–miR-502-5p exhibited the opposite effect (Figure 5B and Supplementary Figure 9A). NRAS expression was higher in TMZ-resistant cells than in TMZ-sensitive cells (Supplementary Figure 9B). CircASAP1 was positively correlated with NRAS expression (Supplementary Figure 9C). Furthermore, miR-502-5p directly bound to the NRAS 3′ untranslated region (UTR) (Figure 5C). NRAS protein levels were increased and decreased by miR-502-5p inhibitor and mimic, respectively (Figure 5D). RIP assays revealed that NRAS could bind to Ago2 (Figure 5E). CircASAP1 knockdown in TMZ-resistant cells elicited a marked increase in the recruitment of Ago2 to NRAS transcripts (Figure 5F and Supplementary Figure 9D). Overexpression of circASAP1 in TMZ-sensitive cells led to increased enrichment of Ago2 but decreased enrichment of Ago2 bound to NRAS transcripts (Figure 5G and Supplementary Figure 9E). In addition, knocking down circASAP1 expression remarkably reduced NRAS mRNA and protein levels (Figure 5H and Supplementary Figure 9F).
Fig. 5.
NRAS is an endogenous target of miR-502-5p. (A) A schematic drawing of the screening procedure for miR-502-5p candidate targets. (B) After transfection of miR-NC or miR-502-5p into N3T3rd and U251T3rd cells, the expression levels of 8 potential targets of miR-502-5p were analyzed using qRT-PCR. (C) A luciferase reporter plasmid carrying wildtype (WT) or mutant (MUT) NRAS was cotransfected into N3T3rd and U251T3rd cells with miR-502-5p mimics in parallel with an empty vector. Relative luciferase activity in the N3T3rd and U251T3rd cells was determined. (D) RIP experiments were performed using an anti-Ago2 antibody, and specific primers were used to detect the enrichment of NRAS. (E) A RIP assay was used to assess the enrichment of Ago2 on circASAP1 and NRAS transcripts relative to immunoglobulin G in N3s and U251s cells transfected with an empty vector or pLCDH-circASAP1. (F) A RIP assay was used to assess the enrichment of Ago2 on circASAP1 and NRAS transcripts relative to immunoglobulin G in N3T3rd and U251T3rd cells transfected with sh-Ctrl or sh-circASAP1. (G) The relative protein levels of NRAS in N3T3rd and U251T3rd cells transfected with miRNA-NC, a miR-502-5p inhibitor, or miR-502-5p mimics were determined. (H) NRAS protein levels were measured in N3T3rd and U251T3rd cells following knockdown of circASAP1 expression. (I, J) The luciferase activity of luciferase reporters containing the WT or MUT (miR-502-5p miRNA response elements) NRAS 3′-UTR was assessed in HEK-293T cells given the indicated treatment. (K) Relative protein levels of NRAS in N3T3rd and U251T3rd cells following knockdown of circASAP1 and/or inhibition of miR-502-5p. (L) Relative protein levels of NRAS in N3s and U251s cells transfected with an empty vector, pLCDH-circASAP1, or pLCDH-circASAP1mt along with miR-502-5p mimics. Data are presented as the mean ± SEM of 3 independent experiments. The significance of results is indicated as NS, nonsignificant; *P < 0.05 and **P < 0.01.
To further study whether the circASAP1-mediated sequestration of miR-502-5p was responsible for the upregulation of NRAS expression, NRAS wildtype and mutant reporters were transfected with miR-502-5p mimic or inhibitor. The luciferase activity of the NRAS reporters was decreased upon circASAP1 knockdown and was rescued by the miR-502-5p inhibitor (Figure 5I). Conversely, the luciferase activity of the NRAS wildtype reporters but not the mutant reporters was elevated upon circASAP1 overexpression, whereas the miR-502-5p mimic abolished this effect (Figure 5J). The RNA and protein levels of NRAS were regulated by circASAP1 and miR-502-5p (Figure 5K, L and Supplementary Figure 9G, H). Collectively, these data demonstrate that circASAP1 functions as a molecular sponge for miR-502-5p to facilitate the expression of NRAS.
CircASAP1 Promotes Tumor Growth and TMZ Resistance by Regulating NRAS/MEK1/ERK1‒2 Signaling, and CircASAP1 Inhibition Restores TMZ Sensitivity In Vivo
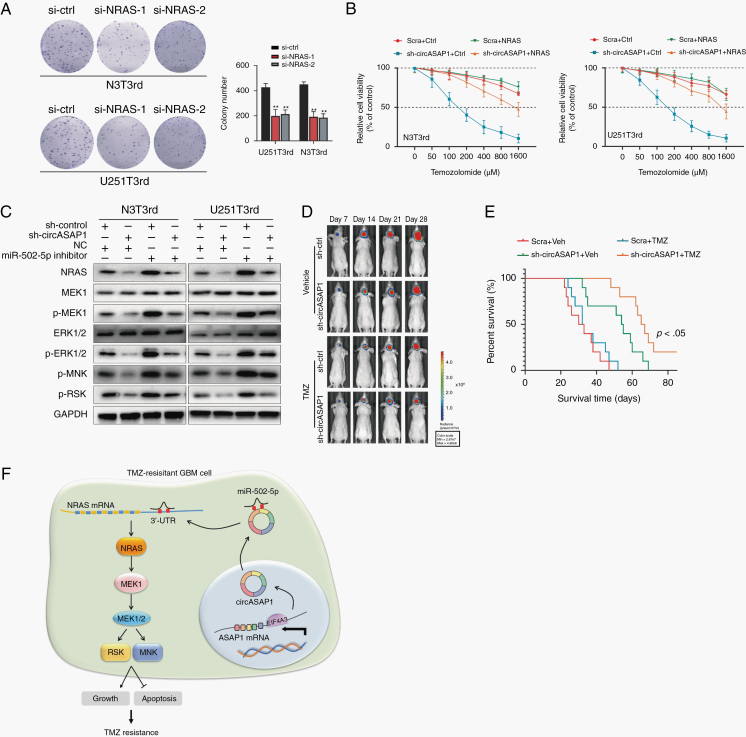
NRAS has been reported to be involved in the regulation of multiple signaling pathways.28–30 We observed that knocking down NRAS expression decreased colony formation in TMZ-resistant cells (Figure 6A). Overexpression of NRAS restored the resistance to TMZ in circASAP1 deleted TMZ-resistant cells (Figure 6B). Si-NRAS increased TMZ-induced cell apoptosis, while overexpression of circASAP1 reversed this effect (Supplementary Figure 10A). Furthermore, we found that sh-circASAP1 inhibited the activation of NRAS/MEK1/ERK1‒2 signaling, but overexpression of circASAP1 activated this signaling (Figure 6D and Supplementary Figure 10B). Thus, all these results demonstrate that NRAS exerts regulatory effects on cell growth and apoptosis inhibition in circASAP1-mediated TMZ resistance and that circASAP1 can regulate apoptosis through the NRAS/MEK1/ERK1‒2 pathway.
Fig. 6.
CircASAP1 affects tumor growth and TMZ resistance by regulating NRAS/MEK1/ERK1–2 signaling, and circASAP1 inhibition restores TMZ sensitivity in vivo. (A) Colony formation assay revealing the effect of NRAS knockdown on TMZ-resistant cells. (B) CCK-8 assay analysis of the effect of NRAS overexpression on TMZ-resistant cells after knocking down circASAP1 expression upon TMZ treatment at the indicated concentrations for 48 h. (C) Western blot analysis of the activity of NRAS/MEK1/ERK1–2 signaling after cotransfection of a miR-502-5p inhibitor and sh-circASAP1 into TMZ-resistant cells. (D) Representative bioluminescence images of intracranial xenografts composed of circASAP1-depleted or control N3T3rd cells treated with or without TMZ and imaged on the days indicated. (E) Kaplan–Meier survival curve of nude mice. (F) Schematic of the mechanisms involving circASAP1 in the regulation of GBM cell proliferation and TMZ resistance. Data are presented as the mean ± SEM of 3 independent experiments. Significant results are presented as *P < 0.05 and **P < 0.01.
To investigate the effect of circASAP1 on the TMZ-resistant phenotype in vivo, 2.5 × 105 transduced N3T3rd cells luciferase-labeled as sh-circASAP1 or sh-Ctrl were injected into nude mice. Tumor progression was monitored by in vivo bioluminescence imaging. Compared with control xenografts, xenografts composed of circASAP1-depleted N3T3rd cells displayed significant tumor regression. Next, tumor-bearing mice were treated with TMZ (66 mg/kg/day, 5 days per week for 3 cycles) and analyzed by bioluminescence imaging. CircASAP1 depletion effectively restored the sensitivity of TMZ-resistant xenografts to TMZ treatment, and mice receiving this combined treatment had a prolonged lifespan (Figure 6D, E). As shown by immunohistochemistry, the expression of Ki-67 and NRAS decreased after knockdown of circASAP1 (Supplementary Figure 10C).
Discussion
Glioblastoma is the most malignant tumor of the central nervous system. Most patients are prone to developing acquired TMZ resistance, which greatly increases the mortality rate of GBM patients, especially those with recurrent GBM. In recent years, the roles of ncRNAs in tumors have been widely studied. Previous studies by our group have confirmed that several miRNAs and lncRNAs are involved in the malignant process of glioma and the acquisition of TMZ resistance.23,31 However, the mechanism of TMZ resistance mediated by circRNAs has rarely been studied.
We collected 3 pairs of GBM samples for RNA sequencing analysis and found that circASAP1 was abnormally expressed in the recurrent GBM tissue samples and associated with the TMZ-resistant phenotype. CircASAP1 was shown to promote TMZ resistance by enhancing cell growth and inhibiting apoptosis. Knocking down circASAP1 expression increased the sensitivity of TMZ-resistant cells to TMZ, which indicated that circASAP1 might be a key node in treatment intervention as part of GBM therapy.
The biogenesis of circRNAs is regulated by specific cis-acting elements and trans-acting factors. It has been shown that certain RNA-binding proteins promote circRNA expression.32 Through bioinformatic analysis and experiments, we predicted and screened that EIF4A3 could bind to a flanking sequence of circASAP1. EIF4A3-mediated exon backsplicing could be a potential mechanism to induce high expression of circASAP1.
The miRNA sponge function of circRNAs, ie, the ceRNA hypothesis, has been observed empirically. In the present study, we confirmed that miR-502-5p was a potential target of circASAP1. Next, we found that NRAS was a potential target of miR-502-5p. NRAS is a member of the RAS family and acts as a GDP/GTP-regulated on/off switch. It encodes a membrane-bound protein with GTPase activity that functions as an important regulatory element in the signal transduction processes of numerous hormones, cytokines, and growth factors, regulating survival and proliferation in cancers. The Ras/Raf/ERK (mitogen-activated protein kinase/ERK) pathway has been reported to be overactivated in many cancers, thereby promoting the malignant phenotype; this pathway is, therefore, considered to be a potential drug target.33,34 We found that circASAP1 promoted TMZ resistance by competitively binding to miR-502-5p, resulting in the dysregulation of NRAS and activation of MEK1/ERK1–2 signaling (Figure 6I).
In summary, we identified a new circRNA, circASAP1. Its role in glioma was not well defined, but it was found to be abnormally expressed in recurrent glioblastoma tissue samples and TMZ-resistant cell lines. Its high expression enhanced cell tolerance to TMZ and was associated with a relatively poor prognosis in GBM patients. Although expression of EIF4A3 is not prognostic of glioma cases in the database of TCGA, it shows prognostic significance in the CGGA database. This may be due to the difference in sample size and ethnicity between the 2 datasets. TMZ resistance is multifactorial, involving not only the internal processes of cells but also factors within the GBM microenvironment.35 Conclusively, our findings indicate that circASAP1 plays an important role in TMZ resistance and it is potential target for predicting patient prognosis and performing therapeutic intervention.
Supplementary Material
Funding
This work was supported by the National Nature Science Foundation of China (nos. 81772679, 81974389, 81772951, and 81972610); Jiangsu Province’s Key Discipline (ZDXKA2016001); and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of interest statement. The authors declare that the manuscript, or any part of it, has not been previously published or submitted concurrently to any other journal.
Authorship statement. Conception and design: Y.W., C.L. Collection and assembly of data: Y.W., P.Z. Data analysis and interpretation: Y.W., Z.S. Manuscript writing: All authors. Final approval of manuscript: All authors. Accountable for all aspect of the study: All authors. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups ; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Wesolowski JR, Rajdev P, Mukherji SK. Temozolomide (Temodar). AJNR Am J Neuroradiol. 2010;31(8):1383–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stupp R, Hegi ME, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups ; National Cancer Institute of Canada Clinical Trials Group. Changing paradigms—an update on the multidisciplinary management of malignant glioma. Oncologist. 2006;11(2):165–180. [DOI] [PubMed] [Google Scholar]
- 5. Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sabin LR, Delás MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Mol Cell. 2013;49(5):783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26(9):1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- 12. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. [DOI] [PubMed] [Google Scholar]
- 13. Zhu LP, He YJ, Hou JC, et al. The role of circRNAs in cancers. Biosci Rep. 2017;37(5):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. 2017;14(8):1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang R, Zhang S, Chen X, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78(17):4812–4825. [DOI] [PubMed] [Google Scholar]
- 17. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. [DOI] [PubMed] [Google Scholar]
- 19. Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. [DOI] [PubMed] [Google Scholar]
- 21. Chan CC, Dostie J, Diem MD, et al. E IF4A3 is a novel component of the exon junction complex. RNA. 2004;10(2):200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang R, Zhang S, Chen X, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeng A, Wei Z, Yan W, et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018;436:10–21. [DOI] [PubMed] [Google Scholar]
- 24. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. [DOI] [PubMed] [Google Scholar]
- 25. Drusco A, Fadda P, Nigita G, et al. Circulating micrornas predict survival of patients with tumors of glial origin. EBioMedicine. 2018;30:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhai H, Song B, Xu X, Zhu W, Ju J. Inhibition of autophagy and tumor growth in colon cancer by miR-502. Oncogene. 2013;32(12):1570–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J, Hou L, Liang R, et al. CircDLST promotes the tumorigenesis and metastasis of gastric cancer by sponging miR-502-5p and activating the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 2019;18(1):80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jansen B, Schlagbauer-Wadl H, Eichler H-G, et al. Activated N-ras contributes to the chemoresistance of human melanoma in severe combined immunodeficiency (SCID) mice by blocking apoptosis. Cancer Res. 1997;57(3):362–365. [PubMed] [Google Scholar]
- 29. Eisfeld AK, Schwind S, Hoag KW, et al. NRAS isoforms differentially affect downstream pathways, cell growth, and cell transformation. Proc Natl Acad Sci U S A. 2014;111(11):4179–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kwong LN, Costello JC, Liu H, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med. 2012;18(10):1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer Res. 2019;38(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. [DOI] [PubMed] [Google Scholar]
- 33. Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK–RAS–RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(1):103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. BBA-Mol Cell Res 2007;1773(8):1263–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.