Abstract
Cadmium exposure is ubiquitous and has been linked to diseases including cancers and reproductive defects. Since cadmium is nonmutagenic, it is thought to exert its gene dysregulatory effects through epigenetic reprogramming. Several studies have implicated germline exposure to cadmium in developmental reprogramming. However, most of these studies have focused on maternal exposure, while the impact on sperm fertility and disease susceptibility has received less attention. In this study, we used reduced representation bisulfite sequencing to comprehensively investigate the impact of chronic cadmium exposure on mouse spermatozoa DNA methylation. Adult male C57BL/J6 mice were provided water with or without cadmium chloride for 9 weeks. Sperm, testes, liver, and kidney tissues were collected at the end of the treatment period. Cadmium exposure was confirmed through gene expression analysis of metallothionein-1 and 2, 2 well-known cadmium-induced genes. Analysis of sperm DNA methylation changes revealed 1788 differentially methylated sites present at regulatory regions in sperm of mice exposed to cadmium compared with vehicle (control) mice. Furthermore, most of these differential methylation changes positively correlated with changes in gene expression at both the transcription initiation stage as well as the splicing levels. Interestingly, the genes targeted by cadmium exposure are involved in several critical developmental processes. Our results present a comprehensive analysis of the sperm methylome in response to chronic cadmium exposure. These data, therefore, highlight a foundational framework to study gene expression patterns that may affect fertility in the exposed individual as well as their offspring, through paternal inheritance.
Keywords: cadmium, DNA methylation, spermatogenesis, epigenetics
Through occupational and nonoccupational means, millions of people worldwide are chronically exposed to cadmium, a ubiquitous heavy metal found in the earth’s crust (Dharmadasa et al., 2017). In occupational exposure, individuals are exposed to high levels of cadmium, which has been implicated in many diseases including lung and prostate cancers, impaired renal function, and bone disease (Kolonel and Winkelstein, 1977; Liu et al., 2020; Stayner et al., 1992). However, numerous epidemiological studies provide evidence of significant risks from low-level chronic cadmium exposure with the consequences of cancers, osteoporosis, and cardiovascular diseases (Akesson et al., 2008; Eriksen et al., 2014; Julin et al., 2012a,b; Satarug, 2012). Chronic, low-dose exposure occurs mainly through tobacco smoke and ingestion of contaminated food and water (Mannino et al., 2004; National Toxicology Program, 2016). Cadmium has a low elimination rate (Dharmadasa et al., 2017). Thus, upon absorption, high concentrations of cadmium can persist in the body, mainly in the liver and kidney, setting up potential health risks over time (Peng et al., 2018). A clear mechanistic understanding of how cadmium causes disease pathogenesis and changes in gene expression remains elusive.
Cadmium is not a mutagen yet causes changes in gene expression. One proposed mechanism by which it mediates these changes is through dysregulation of the epigenome. Epigenetic regulation refers to changes in gene expression not caused by alterations to DNA sequence, but rather by changes in accessibility to the chromatin structure. Epigenetic regulatory factors implicated in cadmium-induced diseases include changes to DNA methylation, histone posttranslational modifications, histone variants, noncoding RNAs, and circRNAs (reviewed in Arita and Costa, 2009; Genchi et al., 2020). The dynamic nature of these epigenetic regulators, therefore, provides phenotypic responses to environmental cues. DNA methylation, the most stable of these regulators, is important in cellular memory, which maintains cell identity and is the most studied in relation to cadmium exposure (Bird, 2002; Kim and Costello, 2017).
Cadmium-induced DNA methylation alterations have been found in a variety of tissue types, including rat liver cells, cancer cell models, maternal, and newborn blood samples, as well as in mouse testes (Mohanty et al., 2015; Peng et al., 2018; Sanders et al., 2014; Takiguchi et al., 2003). On the other hand, studies on how cadmium dysregulates DNA methylation in the germ cells, with consequences in fertility and transgenerational inheritance, are limited. Furthermore, these studies have focused mainly on the impact of cadmium-induced DNA methylation changes with regards to maternal exposure, while the potential impact of cadmium-mediated changes on the sperm methylome has received little attention (Cowley et al., 2018; Jimenez-Chillaron et al., 2009; Mohanty et al., 2015; Zeng et al., 2019). One possible explanation for this discrepancy is the long-held belief that DNA methylation is erased before and during fertilization (Davis et al., 2000). However, recent evidence now suggests that at least some methylation marks in the sperm, especially at imprinted genes, are retained and can be passed on to the next generation (Carone et al., 2014; Hammoud et al., 2009; Samans et al., 2014).
Recent studies provide evidence linking paternal lifestyle with offspring epigenome. For instance, paternal obesity has been associated with hypomethylation at key imprinted genes in offspring. Interestingly, this hypomethylation was independent of maternal obesity (Soubry et al., 2013, 2015). In addition, epigenetic profiling of offspring livers showed that food availability and paternal diet increased the risk of cardiovascular diseases in the offspring (Carone et al., 2010). Another study observed that a paternal high fat diet reprogrammed the spermatozoan epigenome to drive metabolic dysregulation that persists in the F2 generation (de Castro Barbosa et al., 2016). These studies implicate a major role of paternal inheritance.
In vitro studies investigating cadmium exposure on spermatozoa reported a dose dependent decrease in sperm motility (Wang et al., 2017; Zhao et al., 2017). Similarly, in vivo studies of cadmium exposure have shown reduction in sperm quality, integrity, motility, progressive motility, and viability in both rats and mice (Adamkovicova et al., 2016; Ige et al., 2012; Oliveira et al., 2009; Wang et al., 2017). These effects were mostly seen at higher doses of cadmium. Although high doses affect sperm parameters, the effects of chronic low-dose cadmium exposure on the sperm epigenome are currently unknown.
Several studies have shown dysregulation of the sperm methylome in response to specific environmental factors. These include exposure to perfluoroalkyl and polyfluoroalkyl substances (Leter et al., 2014), radiation (Kumar et al., 2013), alcohol consumption (Ouko et al., 2009), nicotine/alcohol (Zhang et al., 2019), cigarette smoke (Jenkins et al., 2017), and vinclozolin (Skinner et al., 2019). These studies show dysregulated methylation at both imprinted and nonimprinted levels, correlating with processes that might affect sperm quality and fertilization, as well as progeny. How cadmium impacts the sperm methylome and what transgenerational impacts cadmium exposure causes are unknown. In this study, we investigated the impact of chronic cadmium exposure on DNA methylation patterns in mice spermatozoa. We hypothesized that chronic exposure to cadmium modifies DNA methylation levels in spermatozoa, targeting specific genes leading to aberrant gene expression patterns. To that end, we exposed mice to cadmium and assessed testicular morphology, measured global and locus-specific DNA methylation changes and gene expression patterns. We provide, for the first time, a comprehensive analysis of the sperm methylome and its associated gene expression patterns in response to cadmium exposure. Our data provide further evidence of gene expression patterns that may affect offspring in a paternal inheritance context.
MATERIALS AND METHODS
Mice Treatment and Spermatozoa Collection
The study was carried out at the University of Kentucky according to an approved Institutional Animal Care and Use Committee protocol and in accordance with the National Institutes of Health standards for care and use of experimental animals, as stated in the Guide for the Care and Use of Laboratory Animals, 8th edition.
Thirty C57BL/6J male mice were purchased from The Jackson Laboratory (Bar Harbor, Maine) at 7 weeks of age. They were housed singly in an environmentally controlled vivarium between 68°F and 72°F under a controlled photoperiod (14/10 light dark cycle) and acclimated for a week during which they were fed 10% Kcal from fat diet (Product D12450J Research Diets; New Brunswick, New Jersey). Mice were fed the same 10% Kcal diet for the duration of the study to ensure purity of ingredients and matched nutritional exposure (and to decrease the risk of any cadmium being present in the food).
After acclimation, the mice were randomized by body weight and divided into 2 groups. The vehicle (control) group was given reverse osmosis water. The cadmium-exposed group was given reverse osmosis water containing 0.9 PPM of cadmium with doses ranging from 0.56 to 1.00 mg of cadmium/kg of body weight and an average of 0.69 mg of cadmium/kg of body weight for 9 weeks (prepared from cadmium chloride, Product No. 202908-10 G Sigma Aldrich; St. Louis, Missouri). This concentration of cadmium was chosen as it falls below the EPA regulatory level where cadmium is considered hazardous waste (EPA, 2020). Mice were allowed ad libitum access to their respective water formulations over the course of the study. Weekly body weights, food, and water intake were measured throughout the study. Water intake was determined by measuring the difference between pre-weighed water bottles and the weight of remaining bottles at the end of each week. Since the density of water is nearly 1 g/ml, weight differences were converted directly from grams to milliliter for the water intake analysis. Cadmium dose was calculated as (cadmium concentration × water intake)/body weight. After 9 weeks of exposure, estimates of total fat tissue, lean tissue (muscle), and water were assessed in liver in conscious mice by nuclear magnetic resonance (EchoMRI; EchoMedical Systems; Houston, Texas).
Glucose tolerance test
Mice were fasted for 3 h and given 2 g/kg of dextrose orally (Bimeda Inc, Le Sueur, Minnesota). Blood glucose concentrations were quantified just prior to glucose administration (time zero) and at 15, 30, 60, and 120 min with a handheld glucometer (AlphaTRAK 2 Blood Glucose Monitoring from Zoetis Inc. Kalamazoo, Michigan).
Tissue collection
Nine weeks after the exposure, mice were fasted for 3 h and euthanized with CO2. Both cauda epididymides were isolated and rinsed in a petri dish containing PBS (Product 10010023 ThermoFisher Scientific, Waltham, Massachusetts). Mature sperm were collected from the cauda epididymis and allowed to swim in PBS solution with 1% bovine serum albumin (BSA) (Product SC 2323, Santa Cruz Biotechnology INC, Dallas, Texas) in an Eppendorf tube. The supernatant was collected and centrifuged at 4000 rpm for 10 min at 4°C to pellet sperm. The sperm were resuspended in pure sperm wash (Product PSW-100/Spectrum Technologies, Healdsburg, California) and the motility and viability were determined. Testis was fixed in 10% formalin (Product SF100-4, ThermoFisher Scientific) for histology. Formalin fixed paraffin embedded tissue slices were then stained with hematoxylin and eosin (H&E). Other tissues were snap frozen in liquid nitrogen.
Testis histopathological evaluation
H&E-stained testis sections from 6 mice per group were scanned at 200× magnification using a Zeiss Axio Scan Z1 digital scanner (Zeiss, Oberkochen, Germany). The acquired whole slide images were evaluated with Zen Software (Zeiss). Per recommendations issued by the Society of Toxicologic Pathology (Lanning et al., 2002), a detailed qualitative examination of transverse sections of the testes was performed, taking into account the tubular stages of the spermatogenic cycle in order to identify treatment related effects such as missing germ cell layers/types, spermatid retention, sloughing of spermatogenic cells into the lumen and damaging of Sertoli and Leydig cells. Although the fixation of testes in formalin instead of Davidson’s fluid might have negatively impacted histomorphological clarity, no subtle morphological alterations of spermatogenesis were observed in either the early (I–VII) or late (VIII–XII) stages of spermatogenic cycle. Likewise, other potentially at-risk cell populations, such as Leydig cells and Sertoli cells, appeared to be unaffected.
RNA extraction
Total RNA was extracted from liver tissue using Invitrogen Pure Link RNA Mini Kit following the manufacturer’s protocol (Product 12183025, ThermoFisher Scientific). RNA concentration was measured using Nanodrop 2000 UV-Vis Spectrophotometer (Product ND-2000, ThermoFisher Scientific). Samples were diluted to 100 ng/μl and cDNA was synthetized from 0.4 μg RNA with qScript cDNA Supermix (Product 101414-098, Quanta Biosciences, Beverly, Massachusetts) at 25°C for 5 min, 42°C for 30 min, and 85°C for 5 min in a thermal cycler (Product 185-1148, Bio-Rad Laboratories Inc., Hercules, California).
RNA quantification with real-time polymerase chain reaction
Reverse transcriptase followed by quantitative real-time polymerase chain reaction was run in Step One Plus Applied Biosystems (Product 4376600, ThermoFisher Scientific) on the RNA extracted from liver tissues. The diluted cDNA was run along with TaqMan Fast Advanced Master Mix (Product 4444964, Applied Biosystems, Waltham, Massachusetts) and TaqMan probes. The probes used are as follows: glucuronidase beta (Gusb, product Mm00151108_cn) (housekeeping gene), Metallothionein 1 (Mt1) (Mm00496660_g1), and Mt2 (Mm00809556_s1). Quantification was reported via value (calculated by methods described in Schmittgen and Livak, 2008).
Cadmium analysis
Cadmium in whole blood was measured by inductively coupled plasma mass spectrometry (ICP-MS) according to previously validated methods (Palmer et al., 2006). Whole blood was collected in trace-metal free microtainer collection tubes with K2EDTA anti-coagulant (BD, Franklin Lakes, New Jersey). Homogenized whole blood was digested in trace-metal free polypropylene centrifuge tubes (VWR) after combining with ultra-pure concentrated HNO3 (Aristar Ultra, VWR) using a microwave digestion system (CEM Mathews, North Carolina). Reagent blanks and standard reference materials (National Institute of Standards and Technology SRM 955c, toxic elements in caprine blood, Gaithersburg, Maryland) were processed with the samples. Samples were diluted to 50X the original blood volume and analyzed by ICP-MS (Agilent 7900, Santa Clarita, California) using indium as an internal standard.
DNA extraction from spermatozoa
Genomic DNA was extracted from 1 × 106 cells. Briefly, samples were pelleted at 5000 rpm at 4°C for 5 min and re-suspended in 500 µl of digestion buffer (83% buffer RLT, 15% 1 M dithiothreitol (DTT), and 2% 20 mg/ml Proteinase K). The mixture was pulse vortexed for 5 min then incubated at 56°C for 2 h. Subsequently, DNA isolation was performed using Qiagen DNeasy Tissue Kit (Cat No.: 69504) following the manufacturer’s protocol. Sample was eluted in 0.25× TE and the concentrations and quality were determined by Thermo Scientific NanoDrop One Microvolume UV-Vis Spectrophotometer.
Quantification of the 5-mC level in DNA
Total 5-mC was determined using the 5-mC DNA ELISA Kit (Zymo Research, Cat Nos: D5325 and D5326) per the manufacturer’s protocol. Briefly, DNA was denatured and then incubated with a diluted antibody mix containing an anti-5-mC monoclonal antibody that is both sensitive and specific to 5-mC antibody, a secondary antibody and 5-mC ELISA buffer. Finally, horseradish peroxidase developer was to detect 5-mC. Values are expressed as percent 5-mC in a DNA sample calculated through a standard curve generated with specially designed control that is included in the kit. Each ELISA was performed with 6 biological replicates and students t test (p < .05) determined statistical significance.
Reduced representation bisulfite sequencing
Reduced representation bisulfite sequencing was performed on samples representing 15 control and 15 cadmium-treated mice by Diagenode, Inc. as previously described (Veillard et al., 2016). Sequencing was carried out on an Illumina HiSeq 3000/4000 in single-end mode, generating 50 base reads (SE50). Quality control of sequencing reads was performed using FastQC (Andrews, 2010) version 0.11.8. Adapter removal was performed using Trim Galore (Krueger, 2012) version 0.4.1. Reads were then aligned to the murine reference genome mm10/GRCm38 using bismark version 0.20.0 (Krueger and Andrews, 2011), followed by in silico bisulfite conversion and methylation calling using the corresponding bismark functionality. Reported cytosines were filtered to get only the cytosines followed by guanine residues (CpGs) covered in each sample. The spike-in control sequences were used at this step to check the bisulfite conversion rates and to validate the efficiency of the bisulfite treatment. Methylkit version 1.7.0 (Akalin et al., 2012), a R/Bioconductor package, was used to perform the differential methylation analysis between control and cadmium-treated samples. The CpG data set was filtered for low coverage (CpGs with coverage <10× in all samples per comparative group were discarded) and for extremely high coverage to discard reads with possible PCR bias (CpGs with coverage higher than the 99.9th percentile were discarded). Differentially methylated CpGs, as well as DMRs were identified. DMRs were determined using 1 kb sliding window. A window was considered a DMR if the global methylation level in one condition was significantly different from the global methylation of the same window in the other condition, considering a Benjamini-Hochberg adjusted p-value threshold of .01 and regardless of the number of CpGs included in that window. The DMRs therefore varied in the number of CpGs they contain from 0, when the CpGs was not detected in one sample (this can happen as CpGs present in at least 80% of the samples were considered), to around 15 CpGs. p-Values were normalized to q-value using the sliding window model for multiple comparison tests. Statistically significant DMCs and DMRs were identified with a q-value cutoff < 0.01 and a methylation difference higher than 25% in at least 80% of the samples. We used permutation tests to determine the enrichment of DMCs at various genic regions. We carried out 1000 permutations to generate a distribution count and determine a p-value.
Bisulfite conversion and pyrosequencing validation
Isolated genomic DNA was digested with HindIII. Digested DNA (500 ng) was used in bisulfite conversion per the manufacturer’s protocol using the EZ DNA Methylation-Lightning Kit–Zymo Research. Converted DNA was eluted in 12 μl of provided elution buffer, and 1 μl was used in PCR reactions. Primers were designed using Zymo Research Bisulfite Primer Seeker (http://www.zymoresearch.com/tools/bisulfite-primer-seeker; last accessed December 1, 2019) around the CpG sites identified by reduced representation bisulfite sequencing as being differentially methylated. PCR conditions for Bisulfite using ZymoTaq PreMix (Cat No.: 76211-542) were as follows: (1) 95°C 10 min; (2) 95°C 30 s; (3) 50°C–58°C (dependent on primer pair) 40 s; (4) 72°C 45 s; (5) repeat steps 2–4 for 40 cycles; and (6) 72°C 7 min. After amplification, PCR products were separated on a 1% agarose gel, and appropriate bands were cut and purified. Purified PCR product was then blunt-end modified (End-It DNA End-Repair Kit–Epicentre—Cat No.: ER81050), and subcloned into pUC19 vector predigested with HincII. Eight to ten clones from each product were sequenced (University of Chicago Comprehensive Cancer Center DNA Sequencing and Genotyping Facility). Sequences were aligned with the genomic sequence using BiQ Analyzer. This program aligns the sequences, determines the converted cytosine residues and calculates percent methylation at specific CpG sites in the sequence.
PCR Primers Used
Spata13: NC_000080.6: 60732486-60733285
Forward: 5′-ATTAATAGTTTTGGGAAAGTGTTTTGTAGGG-3′
Reverse: 5′-AACACACACAACAAATTAAAAATATAACCAAAAAAACC-3′
Lefty2: NC_000067.6: 180893511-180894310
Forward: 5′-TGGGGTGATTATTTAGGATTATTTGGGATAG-3′
Reverse: 5′-ACAAAACCRTTACAATTCTATACCTTTTTAAAATC-3′
nCounter expression analysis (NanoString Technologies)
Sperm cell lysates containing 60 × 103 cells (sperm) were prepared according to NanoString recommendations and ran on a Gene Expression Panel designed for the top differentially methylated genes from reduced representation bisulfite sequencing. nSolver Analysis Software (NanoString Technologies, Seattle, Waltham) was used to normalize counts by lane using the positive control samples. Data were then normalized to the reference genes Gapdh, Eif3f, B2m, Actb, Rplpo, and Rpl19. Genes that passed a nominal significance cutoff between control and cadmium-treated samples were considered for further validation (p < .05, n = 10 per group).
GO analysis
GO for biological processes was performed using the ConsensusPath Database (ConsensusPathDB; Herwig et al., 2016; Kamburov et al., 2013). Data on transcripts involved in biological processes related to male fertility were retrieved by systematically searching for key GO terms as follows: acrosome (GO term 0001669), acrosome reaction (0007340), sperm binding to zona pellucida (0007339), copulation (0007620), embryo implantation (0007566), embryonic development (0009790), female pregnancy (0007565), fertilization (0009566), male gamete generation (0048416), genitalia development (0048806), germ cell development (0007281), gonad development (0008406), insemination (0007320), mating (0007618), placenta development (0001890), reproduction (0000003), reproductive process (0022414), sexual reproduction (0019953), acrosomal vesicle (0001669) sperm motility (0030317), spermatid development (0007286), spermatid nucleus differentiation (0007289), spermatogenesis exchange of chromosomal proteins (0035093), and spermatogenesis (0007283).
Statistical analysis
Results are presented as absolute values and expressed as mean ± SEM. The effects of cadmium on final body weight, fat accumulation, cadmium blood levels, and 5-mC methylation were analyzed by a Mann-Whitney U test (data were not normally distributed). Reduced representation bisulfite sequencing data were analyzed as described in the Materials and Methods section. Statistical analyses were performed using GraphPad Prism version 8 (La Jolla, California).
RESULTS
Chronic Exposure to Cadmium Did Not Have Any Adverse Effects on Body Weight, Fat Accumulation, or Structure of Mice Testes
Due to a renewed interest in paternal effects on inheritance of epigenetic marks, we asked whether cadmium impacts methylation patterns in mice spermatozoa. We treated mice with water lacking cadmium (vehicle) and water containing cadmium for 9 weeks (see Materials and Methods section). During this exposure, we measured body weight and fat accumulation in cadmium-exposed and vehicle mice since these parameters have been implicated in spermatogenesis and infertility (Oliveira et al., 2017). We observed no significant changes in body weight or fat accumulation (Figs. 1A and 1B).
Figure 1.
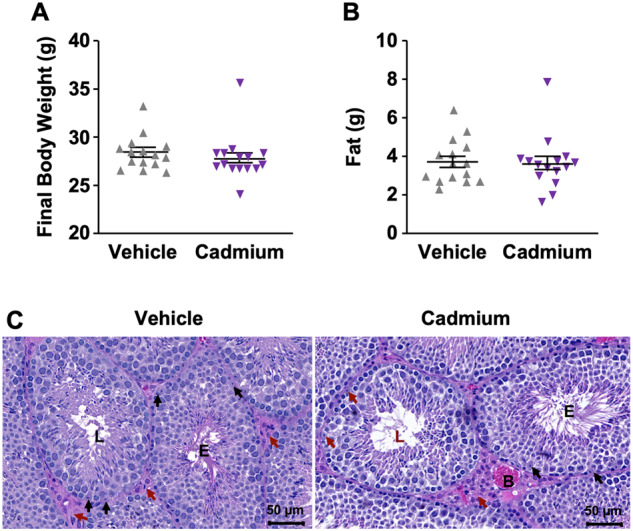
Assessment of mice shows no impact of cadmium exposure to mice body weight and fat accumulation. After 9-week exposure of mice to cadmium, we measured and compared (A) final body weight and (B) fat accumulation between vehicle and cadmium-exposed mice. Data are shown as mean ± SEM (n = 15). A p < .05 determines significance as assessed by a Mann Whitney test. Data show no statistical difference between the 2 groups. C, Histological examination of testes with an awareness of the stages of the spermatogenic cycle did not show any morphological difference between vehicle and cadmium-exposed mice. E, tubule in early stages of spermatogenesis; L, tubule in late stages of spermatogenesis; red arrow, Leydig cells; black arrows, Sertoli cells; B, blood vessel, normal (hematoxylin & eosin stain).
Cadmium is also known to specifically target Sertoli cells, affecting the blood-testis barrier and leading to disruption in spermatogenesis (Wong et al., 2004). Indeed, several studies have reported that acute cadmium exposure not only disrupts the blood-testis barrier, but causes testicular edema, hemorrhage and necrosis in various mammalian species including rodents and rabbits (Lui et al., 2003; Siu et al., 2009; Terada et al., 2005; Wong and Cheng, 2005; Wong et al., 2004). We therefore asked whether morphological changes of the testes occurred in our chronic cadmium exposure experiment by evaluating formalin-fixed paraffin-embedded sections of the testes stained with H&E. The histopathological examination of transverse sections of seminiferous tubules with awareness of the stages of the spermatogenic cycle (stages I–XII; Creasy, 1997; Lanning et al., 2002) did not show any morphological difference between testes from vehicle and cadmium-exposed animals, including germ cell death and depletion, spermatid retention, Sertoli cell and Leydig cell damage (Figure 1C).
These results show that this chronic exposure to cadmium in drinking water did not cause any adverse effects on growth, fat accumulation, or normal testicular features of exposed mice. Additionally, perhaps because cadmium largely partitions to the liver and kidneys, cadmium concentrations in the blood did not differ between groups (Supplementary Figure 1). Yet, we confirmed that these mice were exposed to cadmium through gene expression analysis of metallothionein-1 and 2 (Mt1 and Mt2), which are known to be inducible by cadmium (Kimura et al., 2019; Nordberg, 2009; Nordberg et al., 2009). As expected, liver Mt1 and Mt2 RNA levels were significantly higher in the cadmium-treated group (Supplementary Figure 2), suggesting that these mice received cadmium levels that caused differential gene expression.
Cadmium Induces Global DNA Methylation Alterations in Mice Spermatozoa
After establishing no dramatic phenotypic adverse effects on body weight, fat accumulation or testicular features of cadmium-exposed mice, we next investigated possible cadmium-induced changes to the sperm methylome globally. Purified DNA from sperm of vehicle and cadmium-exposed mice was used in a methylcytosine antibody-based analysis for global DNA methylation (5-methylcytosine [5-mC] ELISA) assay using the Zymo 5-mC DNA ELISA kit (Eckstein et al., 2017; Rea et al., 2017). Quantification of the 5-mC levels, showed a 10% global reduction of methylation level in the cadmium-exposed group compared with vehicle. The difference however was not statistically significant, most likely due to the low sample size used and high variability (Figure 2).
Figure 2.
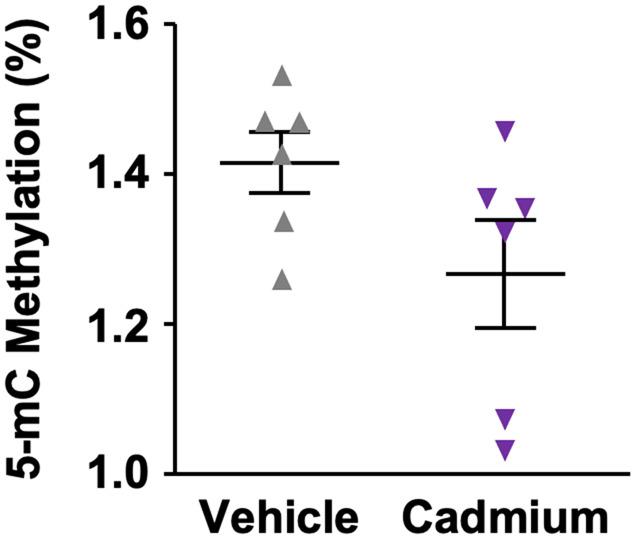
Global DNA methylation levels in vehicle compared with cadmium-exposed mice. Methylation levels were measured using a 5-methylcytosine -specific antibody ELISA assay on purified DNA from sperm of vehicle and cadmium-exposed mice (n = 6). Data are shown as mean ± SEM (n = 3). A p < .05 determines significance as assessed by a Mann Whitney test. We found exposure to cadmium in drinking water was (nonsignificantly) associated with a 10% reduction of global DNA methylation (p = .09).
The 5-mC ELISA is a low-resolution technique providing global methylation levels but not base-pair resolution of methylation. To achieve nucleotide-level resolution of DNA methylation, we performed reduced representation bisulfite sequencing with genomic DNA isolated from spermatozoa of each mice. We used pairwise comparison of DNA methylation levels in sperm DNA from vehicle and cadmium-exposed mice to quantify the differential methylation changes due to cadmium-exposure. DNA methylation changes were deemed significant if a CpG was present in all of the samples, with at least 25% difference between the groups, and at a 10× read coverage with a q ≤ 0.01. Using this stringent cutoff, only 320 000 CpGs were found, of which only 69 differentially methylated (Table 1).
Table 1.
Data Filtering of the CpG Coverage in Sperm of Vehicle Versus Cadmium-Exposed Mice
| CpG Covered in ≥80% of Samples |
CpG Covered in All Samples |
|||
|---|---|---|---|---|
| Covered >10× (1065 495 CpGs) |
Covered >10× (320 000 CpGs) |
|||
| Methylation Difference Threshold | Methylation Difference Threshold | |||
| q-value | 25% | 10% | 25% | 10% |
| 0.01 | 1788 | 52 302 | 69 | 9429 |
| 0.05 | 1788 | 52 302 | 69 | 9429 |
We compared different sequence depth (10×) in 80% of the samples versus 100% of the samples. Due to the high variability in animals, CpGs covered in at least 80% of the samples were used for further analysis.
Due to high variability in animal samples, we then relaxed the stringency to CpGs found in 80% of the samples, with at least 25% difference between groups, and a 10X read coverage with a q ≤ 0.01. This stringency of 25% is still higher than the 10% used in other studies (Cuna et al., 2015; Hahn et al., 2018; Murphy et al., 2018). This relaxed approach, covered 1 065 495 CpGs, including those identified in our first stringent analysis (see Table 1). Significant methylation changes were observed in 1788 CpGs, of which 58.78% (1051) were hypermethylated and 41.22% (737) hypomethylated (Figs. 3A and 3B). These results therefore suggest that chronic exposure to cadmium induces global reprogramming of the sperm methylome.
Figure 3.
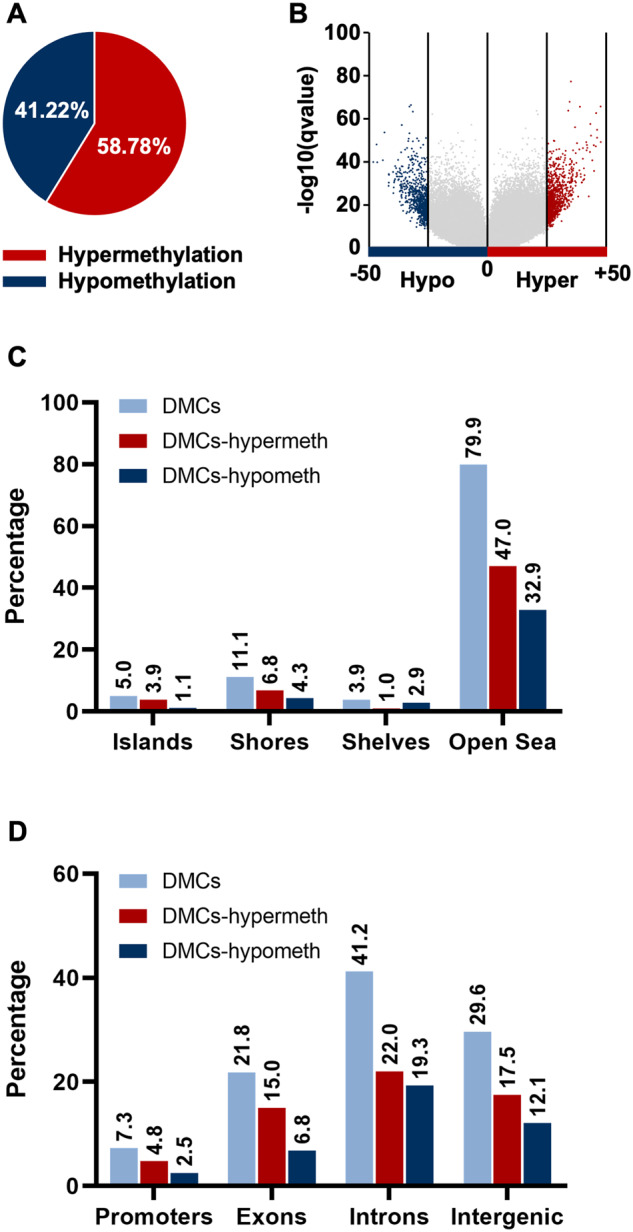
Cadmium exposure alters site-specific differential DNA methylation patterns across genomic locations and gene regulatory regions in mice sperm cells. A, Pie chart of the relative percentage of differential hyper-or hypo differentially methylated cytosines (DMCs) in spermatozoa of vehicle versus cadmium-exposed mice. B, Volcano plot of comparisons of all DMCs assessed in reduced representation bisulfite sequencing analysis of spermatozoa of vehicle versus cadmium-exposed mice. The distribution of 1788 DMCs including 1051 hypermethylated and 737 hypomethylated DMCs across (C) genomic locations in relation to CGIs, and (D) different genic features.
DNA Methylation Patterns at Specific Genomic Loci Are Altered during Chronic Cadmium Exposure
Chromosomal DNA methylation patterns
To assess the specific targets of cadmium in mice spermatozoa, we grouped differentially methylated cytosines (DMCs) according to their associated chromosomes. We then assessed whether cadmium exposure caused a chromosome-specific methylation change. Our analyses showed that the distribution of DMCs was similar across all chromosomes except at chromosome 6, which exhibited significant hypomethylation (Supplementary Figure 3). Although these results were perplexing at first, a number of genes mapped on chromosome 6 have been implicated in spermatogenesis. For example, the activator of cAMP-responsive element modulator in testis (ACT) gene is found on chromosome 6q16.1–16.3 and is critical for the differentiation of spermatids into mature spermatozoa (Palermo et al., 2001).
Average DNA methylation changes at specific gene regulatory regions
Looking at chromosomes does not show specific targeted regulatory regions. To determine the impact of cadmium exposure in mice sperm, we next analyzed the distribution of cadmium-induced DMCs across specific gene regulatory regions. DNA methylation is thought to occur mainly at CpG islands (CGIs), about 1000 base pairs in length and found at gene promoters (Deaton and Bird, 2011; Eckstein et al., 2017; Saxonov et al., 2006). However, DNA methylation also occurs at other regulatory sites scattered throughout the genome. To gain an understanding of the different regions where cadmium impacts the sperm methylome, we divided the genome into genomic locations: CGIs, shores (regions up to 2 kb from CGIs), shelves (regions from 2 to 4 kb from CGIs), and open sea (the rest of the genome; Irizarry et al., 2009; Sandoval et al., 2011). Comparative analysis of cadmium-induced methylation changes revealed that 79.92% of the DMCs were found in open sea, followed by 11.13% at shores, 5.03% at CGIs and 3.91% at shelves (Figure 3C). Although the majority of DMCs was mapped to open sea, we observed a significant difference in the distribution of DMCs with respect to hyper- and hypomethylated CpGs. Most of the DMCs (approximately 78%) in CGIs were hypermethylated in contrast to hypomethylated, followed by approximately 61.3% of the DMCs in shores and approximately 58.8% in open sea (see Figure 3C). A greater proportion (approximately 74.4%) of the DMCs in shelves were hypomethylated (see Figure 3C).
Methylation patterns at different genic features
To further investigate the impact of cadmium-mediated DNA methylation changes at the gene level, we divided the genome into 4 genic features (promoters, exon, intron, and intergenic) and then categorized CpG islands by these genic features. We found that DMCs were highest in gene bodies (63.01%), of which 41.24% were intronic and 21.77% were exonic (Permutation test; p < 0.01). The intergenic regions contained 29.64% of the DMCs, while promoters had 7.35% of the DMCs (Figure 3D). Despite the majority of DMCs mapping to gene bodies, we also observed that the highest proportion (65.8%) of hypermethylated DMCs were found in promoters when considering the number of DMCs in each genic feature (see Figure 3D). These methylation distributions are in line with previous studies (Eckstein et al., 2017; Rea et al., 2017) demonstrating that the process of DNA methylation modifications is nonuniform and intricate. It also raises the level of complexity in understanding how these different factors may alter gene expression.
It is possible that methylation at individual CpGs regulates gene expression (Chen, 1983; Venza et al., 2012). However, methylation levels at neighboring CpGs are often correlated and provide regulatory mechanisms associated with genomic regions. We therefore performed similar analysis on differentially methylated regions (DMRs) with a sliding window of 1 kb (see Materials and Methods section). We found 284 DMRs, of which 148 (52.11%) were hypermethylated and 136 (47.89%) hypomethylated (Supplementary Figs. 4A and 4B). The number of CpGs found in the DMRs of vehicle and cadmium-treated mice varied slightly with an average of 3.26 CpGs in the hypermethylated DMRs and 3.09 in the hypomethylated DMRs. Comparative analysis of cadmium-induced methylation changes revealed that 84.7% of the DMRs mapped to open sea, followed by 7.6% at shores, 6% at shelves and 1.7% at promoters (Supplementary Figure 4C). When these analyses were done at genic regions, most of the DMRs (61.41%) were found in gene bodies with 18.48% exonic and 42.93% intronic. The intergenic regions contained 29.62% of the DMRs followed by promoters with 8.97% of the DMRs (Supplementary Figure 4D). Similar to our observation when comparing the genomic locations/genic features of hypo- and hypermethylated DMCs, we observed a significant difference in the distribution of DMRs. Notably, a greater proportion (approximately 76.4%) of the DMRs in CpG islands were hypermethylated in contrast to hypomethylated, with the latter found at a greater proportion (approximately 78.3%) of the DMRs in shelves (see Supplementary Figure 4C). We also observed that a greater proportion (approximately 72.2%) of the DMRs in promoters were hypermethylated while the other genic features exhibited similar hypo/hyper methylation (see Supplementary Figure 4D).
Gene ontology associated with differentially methylated genes
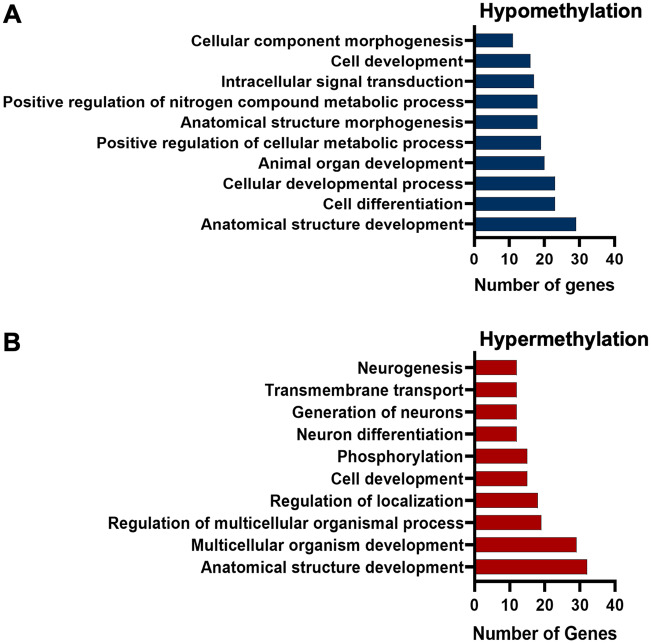
To further understand the biological significance of cadmium-induced DNA methylation in mice spermatozoa, we performed gene ontology (GO) analysis using ConsensusPathDB (Herwig et al., 2016; Kamburov et al., 2013). For these analyses, we used only genes with DMCs at both promoter and gene body regions (introns and exons) so we could experimentally link the differential methylation changes to gene expression. We used 483 unique genes with hypomethylated DMCs and 284 unique genes with hypermethylated DMCs (Supplementary Table 1). The associated general biological processes of the differentially methylated genes (hyper- and hypomethylated) were broadly classified according to their GO annotations (see Supplementary Table 1). We show that the top 10 GO terms for the hypomethylated genes include anatomical structure development, cell differentiation, cellular metabolic processes, and anatomical structure morphogenesis (Figure 4A). Similarly, among the top 10 GO terms of the hypermethylated genes were anatomical structure development, multicellular organism development, regulation of localization, and cell development (Figure 4B). Both hypo- and hypermethylated genes are involved in relatively similar pathways, which supports the idea that these pathways could be targeted by cadmium. Since we are interested in differential methylation in spermatozoa, it is logical to ask whether cadmium alters genes associated either directly or indirectly with pathways involving sperm function, differentiation, and maturation. Deeper targeted analysis of these pathways revealed genes targeted by cadmium exposure, which play roles in spermatogenesis, germ cell differentiation and maturation. These genes are listed in Supplementary Tables 2 and 3.
Figure 4.
Biological processes of genes with differentially methylated cytosines in mice sperm in response to cadmium exposure. (A) Gene ontology of promoters of hypomethylated genes and (B) hypermethylated genes.
Validation of the reduced representation bisulfite sequencing DNA methylation changes
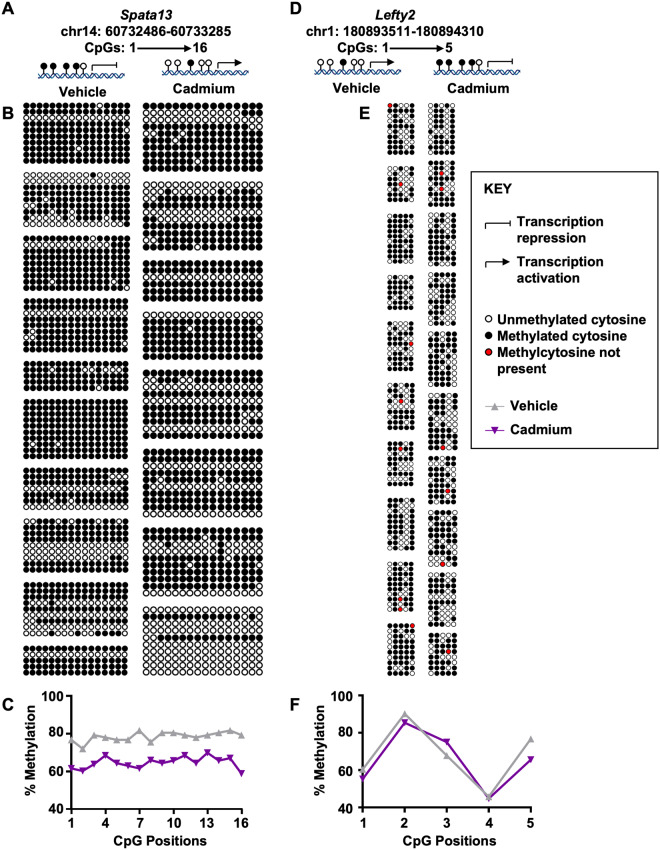
We next validated the reduced representation bisulfite sequencing methylation data using bisulfite conversion followed by pyrosequencing. Due to the expense and number of samples, we chose 2 genes to validate on 10 vehicle and 10 cadmium-exposed mice. Spata13, was hypomethylated at the CpG site—chr14: 60732886 in our reduced representation bisulfite sequencing analyses and the protein has been implicated in cell migration (Sagara et al., 2009). Upon bisulfite conversion, cloning, and pyrosequencing, we observed hypomethylation at both the specific locus and neighboring CpG sites in the Spata13 promoter (Figs. 5A–C and Supplementary Figure 5A). The second gene, Lefty2, was hypermethylated in our reduced representation bisulfite sequencing analysis at the CpG site—chr1: 180893911 and postulated to be important in determination of left-right asymmetry in organ systems during development (Puri et al., 2014). In the case of Lefty2, we show that CpG site 3 was hypermethylated (Figs. 5C–E and Supplementary Figure 5B).
Figure 5.
Validation by pyrosequencing of specific gene promoter loci of the reduced representation bisulfite sequencing data. A, Cartoon representation of chromosomal and CpG positions for Spata13 (chr14: 60732886) and possible effect of methylation status on transcriptional activity. B, Lollipop representation of bisulfite converted DNA (vehicle vs cadmium, n = 8–10 mice). C, Percent methylation at each of the CpG positions at the Spata13 promoter (top) and the Lefty2 promoter (bottom). D, Cartoon representation of chromosomal and CpG positions for Lefty2 (chr1: 180893911) and possible effect of methylation status on transcriptional activity. E, Lollipop representation of bisulfite converted DNA (vehicle vs cadmium, n = 8–10 mice). Each group (box) depicts a mouse. CpG locations are depicted from left to right and the number of clones after bisulfite pyrosequencing from top to bottom. Erroneous sequences and those with low conversation rate were removed, thus the differences in the sizes of each group (box).
Coupling of differential methylation with gene expression
Investigating methylation changes at specific genomic loci provides important insights into their biological functions, yet the fate and impact of these differential methylation changes across the genome are still not fully understood. However, methylation changes at the promoters and gene body are associated with gene expression. For instance, promoter hypermethylation correlates with gene repression, whereas hypomethylation correlates with gene activation (Jones, 1999; Kuroda et al., 2009).
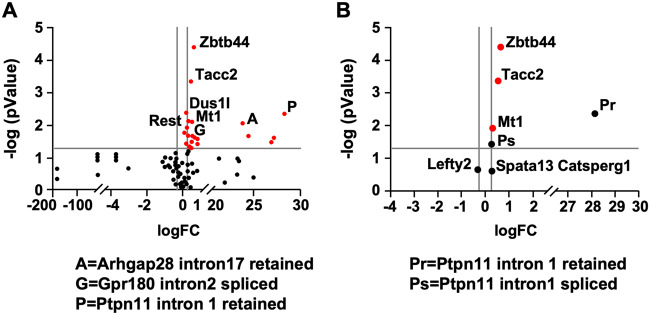
The consequences of DNA methylation within gene bodies are not as clear as they are at promoters. Gene body methylation has been linked with increased gene expression, allowing the fast movement of RNA polymerase II through the gene body (Jjingo et al., 2012). Thus, the effect of DNA methylation on RNA polymerase elongation rate will not only impact the level of transcripts made but also the types of transcriptionally spliced transcripts (Lev Maor et al., 2015; Maunakea et al., 2013; Shukla et al., 2011). We therefore asked whether cadmium-induced DNA methylation changes are linked to gene expression at the transcript initiation and splicing levels. For this, we performed gene expression analysis using direct digital detection through NanoString nCounter Analysis System (NanoString Technologies). This method provides analysis of the level of mRNA in cell lysates without the need to purify the low levels of mRNA present in sperm. Additionally, the optical molecular barcode labeling provided by this technology allowed for simultaneous quantification of up to 800 unique mRNA transcripts. We chose the top 117 genes that were differentially methylated at either promoter or gene body regions (Figure 6A). We normalized their expression to 5 housekeeping genes and expression was considered significant if log fold change (logFc) ≥ 0.263 and p < .05.
Figure 6.
Cadmium-induced methylation patterns correlate to changes in gene expression patterns at both the transcript and alternative splicing levels in mice sperm. NanoString Direct Digital Detection through NanoString nCounter Analysis System was used to determine gene expression changes. Volcano plots of differential gene expression in sperm of vehicle versus cadmium-exposed mice are shown. A, Volcano plots showing top 177 genes. Genes with significant differential expression are shown in red while genes with no significant differential expression are shown in black. B, Volcano plots of selected genes of interest. Red dots depict the genes with significant differential expression; black dots depict genes hypothesized to play a role in spermatogenesis. Gray vertical lines depict the cutoffs for significance (logFc ≥ 0.263), while horizontal gray line depicts the threshold for significance (p < .05).
Our analysis showed that Mt1 expression increased in response to cadmium, which is consistent with our finding in the liver (see Supplementary Figure 2) and previous studies by others showing that Mt1 is inducible by cadmium (Nordberg, 2009; Nordberg et al., 2009). We next examined Spata13, which is involved in cell migration (Sagara et al., 2009) and hypomethylated at the promoter region according to our reduced representation bisulfite sequencing data. NanoString comparative analysis at the transcript level revealed a trend toward increase in gene expression, which correlates with the observed hypomethylation at the promoter region. It is important to note, however, the fold increase was not significant (p > .05; Figure 6B). This could partially be due to the fact that not all individual CpGs regulate gene expression and the level of methylated marks is critical.
Another gene we analyzed was Ptpn11, which is essential for stem cell transition to progenitor spermatogonia (Puri et al., 2014). Excision of exon 4 of Ptpn11 is linked to decreased expression and infertility in mice (Hu et al., 2015). In our reduced representation bisulfite sequencing data, we observed hypomethylation at a CpG site on intron 1. We hypothesized that methylation causes a change in the spliced isoforms that are critical in spermatogenesis. To test this, we designed 2 probes, one that maps intron 1 retention and one targeting and mapping intron 1 excision. Our analyses showed that Ptpn11 expression was significantly increased when intron 1 was retained and its expression level was reduced upon excision of intron 1 (see Figure 6B). These results are in line with the finding that intron hypomethylation is associated with increased gene expression (Anastasiadi et al., 2018). These results could also suggest that the methylation status of intron 1 regulates differential splicing, leading to exon 4 retention, needed for normal Ptpn11 expression. This is consistent with the observed ablation of Ptpn11 expression upon exon 4 excision (Hu et al., 2015).
We next examined the transcript levels of Lefty2, which is involved in the determination of left-right asymmetry in organ systems during development (Li et al., 2017) and is hypermethylated at its promoter. Our data showed that hypermethylation led to decreased expression. Similar to Spata13, the change was not statistically significant (p > .05), although log fold change (logFc) was > 0.263 (see Figure 6B). Finally, we analyzed Catsperg1, a member of the Catsper complex important for sperm hyperactivated motility and male fertility (Wang et al., 2009). Though its promoter was hypermethylated, we were not able to show any expected significant decrease in gene expression (see Figure 6B). The fact that some methylation changes did not lead to significant gene expression does not refute an epigenetic role, but rather supports the complexity of such process and a potential role of other factors such as other epigenetic marks in dynamically regulating this process.
DISCUSSION
Cadmium is a nonmutagen that has been associated with increased risks of various diseases (Akesson et al., 2008; Eriksen et al., 2014; Julin et al., 2012a,b; Kolonel and Winkelstein, 1977; Liu et al., 2020; Satarug, 2012; Stayner et al., 1992). One mechanism by which cadmium has been hypothesized to cause diseases is via dysregulation of the epigenome, causing differential methylation at target genes. Although this mechanism has been shown in different cell types, a thorough analysis of cadmium-induced gene specific DNA methylation dysregulation in the sperm is missing.
We carried out a comprehensive analysis to determine how cadmium impacts DNA methylation in mice spermatozoa. We used Methyl-Seq which provided a population-based, unbiased, global high-resolution view of cadmium-induced differential DNA methylation in sperm. At a global scale, generally no chromosome specific-methylation changes were observed. However, the methylation pattern on chromosome 6 seems to be impacted more than that of the other chromosomes (see Supplementary Figure 3). This finding is interesting as balanced translocation of chromosome 6 is associated with reproductive failure (Mierla et al., 2014; Vozdova et al., 2013; Yang et al., 2018; Zhang et al., 2015). Additionally, several genes critical for sperm development and function are located on human chromosome 6. These include: (1) The ACT gene (mentioned earlier), (2) The sperm acrosomal membrane-associated protein 32 gene (SAMP32), which encodes a testis-specific protein involved in binding sperm to the oocyte complex (Hao et al., 2002); and (3) The human leukocyte antigen, Class II, DR alpha whose variants have been identified as risk factors for nonobstructive azoospermia (Zhao et al., 2012). These findings suggest that changes in the methylation pattern of chromosome 6 could impact the expression of genes that are important for sperm function. However, future studies will be needed to test this directly.
We further analyzed DNA methylation alterations to gene regulatory regions (see Figure 3) to correlate these methylation changes to measurable gene expression patterns (see Figure 6). We observed DMRs and DMCs at genes such as aquaporin 2, SRY-related HMG-box 5, bone morphogenic 7 (Bmp7), growth differentiation factor 3 and 5 doublesex and mab-3-related transcription factor 1-like family C1, and membrane progesterone receptor alpha. These genes have been suggested to play multiple roles in regulating spermatogenesis and sperm function. Changes in methylation patterns were also seen at genes related to germ cell proliferation and maturation such as retinoic acid receptors, and steroid hormone synthesis-related genes. Steroid hormones coordinate the maturation of spermatogonial stem cell renewal to sperm (Schulz et al., 2010). Cadmium induced differential methylation changes were observed at gene regulatory regions of the steroid metabolism-related gene, cytochrome P450 26 (Cyp26). Cyp26 binds to its receptor, to regulate genes that are critical in initiation of meiosis during germ cell proliferation, maturation and spermatogenesis (Hogarth et al., 2015; Wang et al., 2018). In addition, the insulin-like growth factor 1 receptor (Igf1r) and matrix metalloproteinase-15 were also differentially methylated. These genes are involved in initiation of spermatogenesis. Specifically, Igf1r levels are associated with sperm count (Cannarella et al., 2019) and sperm capacitation (Nakayama et al., 1999). Hypomethylation of the H19 promoter (the negative regulator of Igf1r) drives increased H19 transcription with consequences in decreased Igf2 and Igf1r transcription (Giacone et al., 2019). Other genes of interest with cadmium-induced differential methylation patterns were those of the Kinesin family (Kif9, Kif13b, Kif2b, Kifc3, and Kif13a), which regulate acrosomal biogenesis (Khawar et al., 2019; Ma et al., 2017). Kinesins participate in the dynamic cytoskeleton rearrangements of male germ cells and essential organelle transportation during male germ cell proliferation, sperm maturation, and fertilization (Ma et al., 2017). Interestingly, we also observed differential methylation of the Bmp7 promoter which regulates spermatogonia and Sertoli cell proliferation (Puglisi et al., 2004; Xu et al., 2018). A comprehensive list of the genes with differential cadmium-induced DNA methylation patterns is found in Supplementary Table 3. In changing the methylation status/expression of these genes, cadmium could be impacting the differentiation, anatomical structure and localization of key components critical for spermatozoa development and function.
Due to the previous understanding that global DNA methylation occurs during fertilization and preimplantation, studies of the environmental impact on the dynamic changes on the sperm methylome are limiting. However, growing literature now suggests that methylation marks at imprinted and nonimprinted genes escape DNA methylation (Borgel et al., 2010; Daxinger and Whitelaw, 2012). Thus, identifying chemicals and the mechanism by which they cause male reproductive effects is critical to developing approaches that mitigate the risks of lifestyle choices. Furthermore, exposures occurring in adult life may induce DNA methylation changes in sperm, which may later be transmitted across several generations with phenotypic consequences (Dias and Ressler, 2014; Zhu et al., 2020). Transgenetional DNA methylation patterns in the sperm have been observed in exposures to cadmium (Cribiu et al., 2020), vinclozolin (Guerrero-Bosagna et al., 2010), dioxin (2,3,7,8-Tetrachlorodibenzodioxin or TCDD; Manikkam et al., 2012), N, N-diethyl-meta-toluamide (Manikkam et al., 2013), benzopyrene (Knecht et al., 2017), bisphenol A (Manikkam et al., 2013), ethanol exposure (Stouder et al., 2011), alcohol (Liang et al., 2015; Luján et al., 2019), and cigarette smoke (Jenkins et al., 2017). Interestingly, each toxicant seems to target specific DNA methylation genes/regions, thus the hypothesis of exposure-specific DMRs (Nilsson et al., 2018). Although the results presented here do not directly measure a cadmium-specific DMR, they present a platform for analysis of a cadmium-induced specific sperm methylome that can be used in further studies.
Taken together, our results present the first comprehensive, genome-wide DNA methylation profiling of mice spermatozoa in response to chronic exposure to cadmium. These results provide a strong foundation for future studies on the transgenerational effect of chronic cadmium exposure through parent-of-origin analyses and in particular, the transgenerational effect of cadmium-induced epigenetic effects. Such studies will help in the discovery of biomarkers to cadmium exposure, and also in development of potential therapeutic targets for developmental and birth defects.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
AVAILABILITY OF DATA AND MATERIALS
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this article may be requested from the authors. Additionally, the data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus with the GEO accession number: GSE158455.
AUTHOR CONTRIBUTIONS
W.S.N., J.P.D., K.J.P., and Y.F.M. conceived and designed the experiments. W.S.N., S.Y.N.T., J.D.P., J.M.U., and E.D. performed the experiments. W.S.N., S.Y.N.T., J.D.P., E.D., R.d.C., J.E.D., J.M.U., K.J.P., and Y.F.M. analyzed the data. R.d.C., K.J.P., and Y.F.M. contributed reagents/materials. W.S.N., S.Y.N.T., J.D.P., E.D., R.d.C., J.E.D., J.M.U., K.J.P., and Y.F.M. wrote the article. All authors read and approved the final article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr Rouchka and Dr Andreeva, University of Louisville for some of the statistical analyses. We appreciate the animal work contributions by Claire Crosby and Stevi Howard. This work used the Diagenode Genomics Sequencing Laboratory for reduced representation bisulfite sequencing. We also acknowledge the Markey Cancer Center’s Research Communications Office for manuscript editing and assistance with graphic design.
FUNDING
This research was supported by National Institute of Environmental Sciences (NIEHS) grants (R01 ES024478 and 1R01 ES031846 to Y.F.M.) and NIEHS grant (P30 ES026529; “UK-CARES”). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS. The authors E.D. and R.d.C. are supported by the intramural research program of National Institute on Aging, the National Institutes of Health.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Adamkovicova M., Toman R., Martiniakova M., Omelka R., Babosova R., Krajcovicova V., Grosskopf B., Massanyi P. (2016). Sperm motility and morphology changes in rats exposed to cadmium and diazinon. Reprod. Biol. Endocrinol. 14, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A., Kormaksson M., Li S., Garrett-Bakelman F. E., Figueroa M. E., Melnick A., Mason C. E. (2012). Methylkit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akesson A., Julin B., Wolk A. (2008). Long-term dietary cadmium intake and postmenopausal endometrial cancer incidence: A population-based prospective cohort study. Cancer Res. 68, 6435–6441. [DOI] [PubMed] [Google Scholar]
- Anastasiadi D., Esteve-Codina A., Piferrer F. (2018). Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin 11, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed January 21, 2021.
- Arita A., Costa M. (2009). Epigenetics in metal carcinogenesis: Nickel, arsenic, chromium and cadmium. Metallomics 1, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. [DOI] [PubMed] [Google Scholar]
- Borgel J., Guibert S., Li Y., Chiba H., Schübeler D., Sasaki H., Forné T., Weber M. (2010). Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42, 1093–1100. [DOI] [PubMed] [Google Scholar]
- Cannarella R., Liuzzo C., Mongioi L. M., Condorelli R. A., La Vignera S., Bellanca S., Calogero A. E. (2019). Decreased total sperm counts in habitants of highly polluted areas of Eastern Sicily, Italy. Environ. Sci. Pollut. Res. Int. 26, 31368–31373. [DOI] [PubMed] [Google Scholar]
- Carone B. R., Fauquier L., Habib N., Shea J. M., Hart C. E., Li R., Bock C., Li C., Gu H., Zamore P. D., et al. (2010). Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143, 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone B. R., Hung J. H., Hainer S. J., Chou M. T., Carone D. M., Weng Z., Fazzio T. G., Rando O. J. (2014). High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev. Cell 30, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. F. (1983). Endocrinologic and immunologic function in uremic patients with asthenia of the kidney. Zhong Xi Yi Jie He Za Zhi 3, 328–330. [PubMed] [Google Scholar]
- Cowley M., Skaar D. A., Jima D. D., Maguire R. L., Hudson K. M., Park S. S., Sorrow P., Hoyo C. (2018). Effects of cadmium exposure on DNA methylation at imprinting control regions and genome-wide in mothers and newborn children. Environ. Health Perspect. 126, 037003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasy D. M. (1997). Evaluation of testicular toxicity in safety evaluation studies: The appropriate use of spermatogenic staging. Toxicol. Pathol. 25, 119–131. [DOI] [PubMed] [Google Scholar]
- Cribiu P., Devaux A., Garnero L., Abbaci K., Bastide T., Delorme N., Queau H., Degli Esposti D., Ravanat J. L., Geffard O., et al. (2020). A “population dynamics” perspective on the delayed life-history effects of environmental contaminations: An illustration with a preliminary study of cadmium transgenerational effects over three generations in the Crustacean Gammarus. Int. J. Mol. Sci. 21, 4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuna A., Halloran B., Faye-Petersen O., Kelly D., Crossman D. K., Cui X., Pandit K., Kaminski N., Bhattacharya S., Ahmad A., et al. (2015). Alterations in gene expression and DNA methylation during murine and human lung alveolar septation. Am. J. Respir. Cell Mol. Biol. 53, 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. L., Yang G. J., McCarrey J. R., Bartolomei M. S. (2000). The h19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 9, 2885–2894. [DOI] [PubMed] [Google Scholar]
- Daxinger L., Whitelaw E. (2012). Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 13, 153–162. [DOI] [PubMed] [Google Scholar]
- de Castro Barbosa T., Ingerslev L. R., Alm P. S., Versteyhe S., Massart J., Rasmussen M., Donkin I., Sjogren R., Mudry J. M., Vetterli L., et al. (2016). High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol. Metab. 5, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton A. M., Bird A. (2011). CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmadasa P., Kim N., Thunders M. (2017). Maternal cadmium exposure and impact on foetal gene expression through methylation changes. Food Chem. Toxicol. 109, 714–720. [DOI] [PubMed] [Google Scholar]
- Dias B. G., Ressler K. J. (2014). Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 17, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein M., Rea M., Fondufe-Mittendorf Y. N. (2017). Transient and permanent changes in DNA methylation patterns in inorganic arsenic-mediated epithelial-to-mesenchymal transition. Toxicol. Appl. Pharmacol. 331, 6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA. (2020). Test methods for evaluating solid waste: Physical/chemical methods (Revision 5).
- Eriksen K. T., Halkjær J., Sørensen M., Meliker J. R., McElroy J. A., Tjønneland A., Raaschou-Nielsen O. (2014). Dietary cadmium intake and risk of breast, endometrial and ovarian cancer in Danish postmenopausal women: A prospective cohort study. PLoS One 9, e100815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genchi G., Sinicropi M. S., Lauria G., Carocci A., Catalano A. (2020). The effects of cadmium toxicity. Int. J. Environ. Res. Public Health 17, 3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacone F., Cannarella R., Mongioi L. M., Alamo A., Condorelli R. A., Calogero A. E., La Vignera S. (2019). Epigenetics of male fertility: Effects on assisted reproductive techniques. World J. Mens Health 37, 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C., Settles M., Lucker B., Skinner M. K. (2010). Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 5, e13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn O., Stubbs T. M., Reik W., Gronke S., Beyer A., Partridge L. (2018). Hepatic gene body hypermethylation is a shared epigenetic signature of murine longevity. PLoS Genet. 14, e1007766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud S. S., Nix D. A., Zhang H., Purwar J., Carrell D. T., Cairns B. R. (2009). Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z., Wolkowicz M. J., Shetty J., Klotz K., Bolling L., Sen B., Westbrook V. A., Coonrod S., Flickinger C. J., Herr J. C. (2002). Samp32, a testis-specific, isoantigenic sperm acrosomal membrane-associated protein. Biol. Reprod. 66, 735–744. [DOI] [PubMed] [Google Scholar]
- Herwig R., Hardt C., Lienhard M., Kamburov A. (2016). Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 11, 1889–1907. [DOI] [PubMed] [Google Scholar]
- Hogarth C. A., Arnold S., Kent T., Mitchell D., Isoherranen N., Griswold M. D. (2015). Processive pulses of retinoic acid propel asynchronous and continuous murine sperm production. Biol. Reprod. 92, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Tang Z., Li Y., Liu W., Zhang S., Wang B., Tian Y., Zhao Y., Ran H., Liu W., et al. (2015). Deletion of the tyrosine phosphatase Shp2 in Sertoli cells causes infertility in mice. Sci. Rep. 5, 12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ige S. F., Olaleye S. B., Akhigbe R. E., Akanbi T. A., Oyekunle O. A., Udoh U. A. (2012). Testicular toxicity and sperm quality following cadmium exposure in rats: Ameliorative potentials of Allium cepa. J. Hum. Reprod. Sci. 5, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M., et al. (2009). The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins T. G., James E. R., Alonso D. F., Hoidal J. R., Murphy P. J., Hotaling J. M., Cairns B. R., Carrell D. T., Aston K. I. (2017). Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology 5, 1089–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Chillaron J. C., Isganaitis E., Charalambous M., Gesta S., Pentinat-Pelegrin T., Faucette R. R., Otis J. P., Chow A., Diaz R., Ferguson-Smith A., et al. (2009). Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes 58, 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jjingo D., Conley A. B., Yi S. V., Lunyak V. V., Jordan I. K. (2012). On the presence and role of human gene-body DNA methylation. Oncotarget 3, 462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. (1999). The DNA methylation paradox. Trends Genet. 15, 34–37. [DOI] [PubMed] [Google Scholar]
- Julin B., Wolk A., Bergkvist L., Bottai M., Akesson A. (2012a). Dietary cadmium exposure and risk of postmenopausal breast cancer: A population-based prospective cohort study. Cancer Res. 72, 1459–1466. [DOI] [PubMed] [Google Scholar]
- Julin B., Wolk A., Johansson J. E., Andersson S. O., Andren O., Akesson A. (2012b). Dietary cadmium exposure and prostate cancer incidence: A population-based prospective cohort study. Br. J. Cancer 107, 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A., Stelzl U., Lehrach H., Herwig R. (2013). The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 41(Database issue), D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawar M. B., Gao H., Li W. (2019). Mechanism of acrosome biogenesis in mammals. Front. Cell Dev. Biol. 7, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Costello J. (2017). DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 49, e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Hosaka T., Nakanishi T., Aozasa O. (2019). Long-term cadmium exposure enhances metallothionein-1 induction after subsequent exposure to high concentrations of cadmium in p1798 mouse lymphosarcoma cells. J. Toxicol. Sci. 44, 309–316. [DOI] [PubMed] [Google Scholar]
- Knecht A. L., Truong L., Marvel S. W., Reif D. M., Garcia A., Lu C., Simonich M. T., Teeguarden J. G., Tanguay R. L. (2017). Transgenerational inheritance of neurobehavioral and physiological deficits from developmental exposure to benzo[a]pyrene in zebrafish. Toxicol. Appl. Pharmacol. 329, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolonel L., Winkelstein W. Jr. (1977). Cadmium and prostatic carcinoma. Lancet 310, 566–567. [DOI] [PubMed] [Google Scholar]
- Krueger F. (2012). Trim Galore! Available at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- Krueger F., Andrews S. R. (2011). Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Salian S. R., Kalthur G., Uppangala S., Kumari S., Challapalli S., Chandraguthi S. G., Krishnamurthy H., Jain N., Kumar P., et al. (2013). Semen abnormalities, sperm DNA damage and global hypermethylation in health workers occupationally exposed to ionizing radiation. PLoS One 8, e69927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A., Rauch T. A., Todorov I., Ku H. T., Al-Abdullah I. H., Kandeel F., Mullen Y., Pfeifer G. P., Ferreri K. (2009). Insulin gene expression is regulated by DNA methylation. PLoS One 4, e6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanning L. L., Creasy D. M., Chapin R. E., Mann P. C., Barlow N. J., Regan K. S., Goodman D. G. (2002). Recommended approaches for the evaluation of testicular and epididymal toxicity. Toxicol. Pathol. 30, 507–520. [DOI] [PubMed] [Google Scholar]
- Leter G., Consales C., Eleuteri P., Uccelli R., Specht I. O., Toft G., Moccia T., Budillon A., Jönsson B. A., Lindh C. H., et al. (2014). Exposure to perfluoroalkyl substances and sperm DNA global methylation in arctic and European populations. Environ. Mol. Mutagen. 55, 591–600. [DOI] [PubMed] [Google Scholar]
- Lev Maor G., Yearim A., Ast G. (2015). The alternative role of DNA methylation in splicing regulation. Trends Genet. 31, 274–280. [DOI] [PubMed] [Google Scholar]
- Li G., Liu X., Xing C., Zhang H., Shimeld S. M., Wang Y. (2017). Cerberus-Nodal-Lefty-Pitx signaling cascade controls left-right asymmetry in amphioxus. Proc. Natl. Acad. Sci. U.S.A. 114, 3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Diao L., Jiang N., Zhang J., Wang H. J., Zhou W. H., Huang G. Y., Ma D. (2015). Chronic exposure to ethanol in male mice may be associated with hearing loss in offspring. Asian J. Androl. 17, 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yuan Y., Xiao Y., Li Y., Yu Y., Mo T., Jiang H., Li X., Yang H., Xu C., et al. (2020). Associations of plasma metal concentrations with the decline in kidney function: A longitudinal study of Chinese adults. Ecotoxicol. Environ. Saf. 189, 110006. [DOI] [PubMed] [Google Scholar]
- Lui W. Y., Wong C. H., Mruk D. D., Cheng C. Y. (2003). TGF-beta3 regulates the blood-testis barrier dynamics via the p38 mitogen activated protein (MAP) kinase pathway: An in vivo study. Endocrinology 144, 1139–1142. [DOI] [PubMed] [Google Scholar]
- Luján S., Caroppo E., Niederberger C., Arce J. C., Sadler-Riggleman I., Beck D., Nilsson E., Skinner M. K. (2019). Sperm DNA methylation epimutation biomarkers for male infertility and FSH therapeutic responsiveness. Sci. Rep. 9, 16786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D. D., Wang D. H., Yang W. X. (2017). Kinesins in spermatogenesis. Biol. Reprod. 96, 267–276. [DOI] [PubMed] [Google Scholar]
- Manikkam M., Tracey R., Guerrero-Bosagna C., Skinner M. K. (2012). Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One 7, e46249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M., Tracey R., Guerrero-Bosagna C., Skinner M. K. (2013). Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 8, e55387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannino D. M., Holguin F., Greves H. M., Savage-Brown A., Stock A. L., Jones R. L. (2004). Urinary cadmium levels predict lower lung function in current and former smokers: Data from the third national health and nutrition examination survey. Thorax 59, 194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea A. K., Chepelev I., Cui K., Zhao K. (2013). Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 23, 1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierla D., Jardan D., Stoian V. (2014). Chromosomal abnormality in men with impaired spermatogenesis. Int. J. Fertil. Steril. 8, 35–42. [PMC free article] [PubMed] [Google Scholar]
- Mohanty A. F., Farin F. M., Bammler T. K., MacDonald J. W., Afsharinejad Z., Burbacher T. M., Siscovick D. S., Williams M. A., Enquobahrie D. A. (2015). Infant sex-specific placental cadmium and DNA methylation associations. Environ. Res. 138, 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. K., Itchon-Ramos N., Visco Z., Huang Z., Grenier C., Schrott R., Acharya K., Boudreau M. H., Price T. M., Raburn D. J., et al. (2018). Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics 13, 1208–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y., Yamamoto T., Abe S. I. (1999). IGF-I, IGF-II and insulin promote differentiation of spermatogonia to primary spermatocytes in organ culture of newt testes. Int. J. Dev. Biol. 43, 343–347. [PubMed] [Google Scholar]
- National Toxicology Program. (2016). Report on Carcinogens, 14th edn. U.S. Department of Health and Human Services, Research Triangle Park. [Google Scholar]
- Nilsson E. E., Sadler-Riggleman I., Skinner M. K. (2018). Environmentally induced epigenetic transgenerational inheritance of disease. Environ. Epigenet. 4, dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg G. F. (2009). Historical perspectives on cadmium toxicology. Toxicol. Appl. Pharmacol. 238, 192–200. [DOI] [PubMed] [Google Scholar]
- Nordberg G. F., Jin T., Wu X., Lu J., Chen L., Lei L., Hong F., Nordberg M. (2009). Prevalence of kidney dysfunction in humans - Relationship to cadmium dose, metallothionein, immunological and metabolic factors. Biochimie 91, 1282–1285. [DOI] [PubMed] [Google Scholar]
- Oliveira H., Spanò M., Santos C., Pereira M. d L. (2009). Adverse effects of cadmium exposure on mouse sperm. Reprod. Toxicol. 28, 550–555. [DOI] [PubMed] [Google Scholar]
- Oliveira P. F., Sousa M., Silva B. M., Monteiro M. P., Alves M. G. (2017). Obesity, energy balance and spermatogenesis. Reproduction 153, R173–R185. [DOI] [PubMed] [Google Scholar]
- Ouko L. A., Shantikumar K., Knezovich J., Haycock P., Schnugh D. J., Ramsay M. (2009). Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: Implications for fetal alcohol spectrum disorders. Alcohol Clin. Exp. Res. 33, 1615–1627. [DOI] [PubMed] [Google Scholar]
- Palermo I., Litrico L., Emmanuele G., Giuffrida V., Sassone-Corsi P., De Cesare D., Maria Fimia G., D'Agata R., Calogero A. E., Travali S. (2001). Cloning and expression of activator of CREM in testis in human testicular tissue. Biochem. Biophys. Res. Commun. 283, 406–411. [DOI] [PubMed] [Google Scholar]
- Palmer C. D., Lewis M. E., Geraghty C. M., Barbosa F., Parsons P. J. (2006). Determination of lead, cadmium and mercury in blood for assessment of environmental exposure: A comparison between inductively coupled plasma–mass spectrometry and atomic absorption spectrometry. Spectrochim. Acta B Atom. Spectrosc. 61, 980–990. [Google Scholar]
- Peng L., Huang Y. T., Zhang F., Chen J. Y., Huo X. (2018). Chronic cadmium exposure aggravates malignant phenotypes of nasopharyngeal carcinoma by activating the Wnt/beta-catenin signaling pathway via hypermethylation of the casein kinase 1alpha promoter. Cancer Manag. Res. 11, 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi R., Balduzzi D., Galli A. (2004). In vitro sperm penetration speed and its relationship with in vivo bull fertility. Reprod. Domest. Anim. 39, 424–428. [DOI] [PubMed] [Google Scholar]
- Puri P., Phillips B. T., Suzuki H., Orwig K. E., Rajkovic A., Lapinski P. E., King P. D., Feng G. S., Walker W. H. (2014). The transition from stem cell to progenitor spermatogonia and male fertility requires the shp2 protein tyrosine phosphatase. Stem Cells 32, 741–753. [DOI] [PubMed] [Google Scholar]
- Rea M., Eckstein M., Eleazer R., Smith C., Fondufe-Mittendorf Y. N. (2017). Genome-wide DNA methylation reprogramming in response to inorganic arsenic links inhibition of CTCF binding, DNMT expression and cellular transformation. Sci. Rep. 7, 41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagara M., Kawasaki Y., Iemura S. I., Natsume T., Takai Y., Akiyama T. (2009). Asef2 and Neurabin2 cooperatively regulate actin cytoskeletal organization and are involved in HGF-induced cell migration. Oncogene 28, 1357–1365. [DOI] [PubMed] [Google Scholar]
- Samans B., Yang Y., Krebs S., Sarode G. V., Blum H., Reichenbach M., Wolf E., Steger K., Dansranjavin T., Schagdarsurengin U. (2014). Uniformity of nucleosome preservation pattern in mammalian sperm and its connection to repetitive DNA elements. Dev. Cell 30, 23–35. [DOI] [PubMed] [Google Scholar]
- Sanders A., Smeester L., Rojas D., DeBussycher T., Wu M., Wright F., Zhou Y.-H., Laine J., Rager J., Swamy G., et al. (2014). Cadmium exposure and the epigenome: Exposure-associated patterns of DNA methylation in leukocytes from mother-baby pairs. Epigenetics 9, 212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M. A., Bibikova M., Esteller M. (2011). Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6, 692–702. [DOI] [PubMed] [Google Scholar]
- Satarug S. (2012). Long-term exposure to cadmium in food and cigarette smoke, liver effects and hepatocellular carcinoma. Curr. Drug Metab. 13, 257–271. [DOI] [PubMed] [Google Scholar]
- Saxonov S., Berg P., Brutlag D. L. (2006). A genome-wide analysis of CPG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U.S.A. 103, 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(t) method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schulz R. W., de França L. R., Lareyre J.-J., Le Gac F., LeGac F., Chiarini-Garcia H., Nobrega R. H., Miura T. (2010). Spermatogenesis in fish. Gen. Comp. Endocrinol. 165, 390–411. [DOI] [PubMed] [Google Scholar]
- Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. (2011). CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu E. R., Mruk D. D., Porto C. S., Cheng C. Y. (2009). Cadmium-induced testicular injury. Toxicol. Appl. Pharmacol. 238, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. K., Nilsson E., Sadler-Riggleman I., Beck D., Ben Maamar M., McCarrey J. R. (2019). Transgenerational sperm DNA methylation epimutation developmental origins following ancestral vinclozolin exposure. Epigenetics 14, 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A., Murphy S. K., Wang F., Huang Z., Vidal A. C., Fuemmeler B. F., Kurtzberg J., Murtha A., Jirtle R. L., Schildkraut J. M., et al. (2015). Newborns of obese parents have altered DNA methylation patterns at imprinted genes. Int. J. Obes. (Lond.). 39, 650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A., Schildkraut J. M., Murtha A., Wang F., Huang Z., Bernal A., Kurtzberg J., Jirtle R. L., Murphy S. K., Hoyo C. (2013). Paternal obesity is associated with IGF2 hypomethylation in newborns: Results from a newborn epigenetics study (NEST) cohort. BMC Med. 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stayner L., Smith R., Thun M., Schnorr T., Lemen R. (1992). A quantitative assessment of lung cancer risk and occupational cadmium exposure. IARC Sci. Publ. 118, 447–455. [PubMed] [Google Scholar]
- Stouder C., Somm E., Paoloni-Giacobino A. (2011). Prenatal exposure to ethanol: A specific effect on the h19 gene in sperm. Reprod. Toxicol. 31, 507–512. [DOI] [PubMed] [Google Scholar]
- Takiguchi M., Achanzar W. E., Qu W., Li G., Waalkes M. P. (2003). Effects of cadmium on DNA-(cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp. Cell Res. 286, 355–365. [DOI] [PubMed] [Google Scholar]
- Terada N., Ohno N., Yamakawa H., Ohara O., Liao X., Baba T., Ohno S. (2005). Immunohistochemical study of a membrane skeletal molecule, protein 4.1g, in mouse seminiferous tubules. Histochem. Cell Biol. 124, 303–311. [DOI] [PubMed] [Google Scholar]
- Veillard A.-C., Datlinger P., Laczik M., Squazzo S., Bock C. (2016). Diagenode® premium RRBS technology: Cost-effective DNA methylation mapping with superior coverage. Nat. Methods 13, 184. [Google Scholar]
- Venza I., Visalli M., Fortunato C., Ruggeri M., Ratone S., Caffo M., Caruso G., Alafaci C., Tomasello F., Teti D., et al. (2012). PGE2 induces interleukin-8 derepression in human astrocytoma through coordinated DNA demethylation and histone hyperacetylation. Epigenetics 7, 1315–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozdova M., Oracova E., Kasikova K., Prinosilova P., Rybar R., Horinova V., Gaillyova R., Rubes J. (2013). Balanced chromosomal translocations in men: Relationships among semen parameters, chromatin integrity, sperm meiotic segregation and aneuploidy. J. Assist. Reprod. Genet. 30, 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu J., Cho K. H., Ren D. (2009). A novel, single, transmembrane protein CATSPERG is associated with CATSPER1 channel protein. Biol. Reprod. 81, 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. F., Chang M., Peng T. T., Yang Y., Li N., Luo T., Cheng Y. M., Zhou M. Z., Zeng X. H., Zheng L. P. (2017). Exposure to cadmium impairs sperm functions by reducing CATSPER in mice. Cell Physiol. Biochem. 42, 44–54. [DOI] [PubMed] [Google Scholar]
- Wang X., Liu Q., Xu S., Xiao Y., Wang Y., Feng C., Xue R., Zhao H., Song Z., Li J. (2018). Transcriptome dynamics during turbot spermatogenesis predicting the potential key genes regulating male germ cell proliferation and maturation. Sci. Rep. 8, 15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. H., Cheng C. Y. (2005). The blood-testis barrier: Its biology, regulation, and physiological role in spermatogenesis. Curr. Top. Dev. Biol. 71, 263–296. [DOI] [PubMed] [Google Scholar]
- Wong C. H., Mruk D. D., Lui W. Y., Cheng C. Y. (2004). Regulation of blood-testis barrier dynamics: An in vivo study. J. Cell Sci. 117, 783–798. [DOI] [PubMed] [Google Scholar]
- Xu Y. R., Wang G. Y., Zhou Y. C., Yang W. X. (2018). The characterization and potential roles of bone morphogenetic protein 7 during spermatogenesis in Chinese mitten crab Eriocheir sinensis. Gene 673, 119–129. [DOI] [PubMed] [Google Scholar]
- Yang X., Zhang H., Yu Y., Zhu H., Hu X., Jiang Y., Wang R., Liu R. (2018). Clinical features of chromosome 6 translocation in male carriers: A report of 10 cases and review of the literature. Med. Sci. Monit. 24, 4162–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Huo X., Zhang Y., Hylkema M. N., Wu Y., Xu X. (2019). Differential DNA methylation in newborns with maternal exposure to heavy metals from an e-waste recycling area. Environ. Res. 171, 536–545. [DOI] [PubMed] [Google Scholar]
- Zhang M., Fan H. T., Zhang Q. S., Wang X. Y., Yang X., Tian W. J., Li R. W. (2015). Genetic screening and evaluation for chromosomal abnormalities of infertile males in Jilin province, China. Genet. Mol. Res. 14, 16178–16184. [DOI] [PubMed] [Google Scholar]
- Zhang W., Li M., Sun F., Xu X., Zhang Z., Liu J., Sun X., Zhang A., Shen Y., Xu J., et al. (2019). Association of sperm methylation at line-1, four candidate genes, and nicotine/alcohol exposure with the risk of infertility. Front. Genet. 10, 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Xu J., Zhang H., Sun J., Sun Y., Wang Z., Liu J., Ding Q., Lu S., Shi R., et al. (2012). A genome-wide association study reveals that variants within the HLA region are associated with risk for nonobstructive azoospermia. Am. J. Hum. Genet. 90, 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. L., Ru Y. F., Liu M., Tang J. N., Zheng J. F., Wu B., Gu Y. H., Shi H. J. (2017). Reproductive effects of cadmium on sperm function and early embryonic development in vitro. PLoS One 12, e0186727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Li X., Ge R. S. (2020). Toxicological effects of cadmium on mammalian testis. Front. Genet. 11, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this article may be requested from the authors. Additionally, the data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus with the GEO accession number: GSE158455.