Abstract
Lung cancer is the leading cause of cancer-associated death worldwide and exhibits intrinsic and acquired therapeutic resistance to cisplatin (CIS). The present study investigated the role of mTOR signaling and other signaling pathways after metformin (MET) treatment in control and cisplatin-resistant A549 cells, mapping pathways and possible targets involved in CIS sensitivity. MTT, flow cytometry, clonogenic assay, western blotting, proteomic analysis using the Stable Isotope Labeling by Amino acids in Cell culture (SILAC) approach and reverse transcription-quantitative PCR were performed. The results revealed that CIS treatment induced mTOR signaling pathway overactivation, and the mTOR status was restored by MET. MET and the mTOR inhibitor rapamycin (RAPA) decreased the viability in control and resistant cells, and decreased the cell size increase induced by CIS. In control cells, MET and RAPA decreased colony formation after 72 h and decreased IC50 values, potentiating the effects of CIS. Proteomics analysis revealed important pathways regulated by MET, including transcription, RNA processing and IL-12-mediated signaling. In CIS-resistant cells, MET regulated the apoptotic process, oxidative stress and G2/M transition. Annexin 4 (ANXA4) and superoxide dismutase 2 (SOD2), involved in apoptosis and oxidative stress, respectively, were chosen to validate the SILAC analysis and may represent potential therapeutic targets for lung cancer treatment. In conclusion, the chemosensitizing and antiproliferative effects of MET were associated with mTOR signaling and with potential novel targets, such as ANXA4 and SOD2, in human lung cancer cells.
Keywords: lung cancer, cisplatin resistance, metformin, mTOR, proteomics
Introduction
Cancer is a relevant global public health problem, considered as the most important barrier to increasing global life expectancy in the 21st century (1). Each year, ~88% of people diagnosed with lung cancer have a death outcome, accounting for 2,093,876 new cases and 1,761,007 deaths worldwide in 2018 (1). Despite the development of early diagnosis methods and new treatment modalities, the 5-year survival rate of patients with lung cancer remains poor, increasing only by 3.7% between 1985 and 2004 (2). In advanced stages, when surgical resection is not possible, chemotherapy using platinum drugs, including cisplatin, is the current standard treatment for non-small cell lung cancer (NSCLC) (3,4), although it is strongly associated with intrinsic and acquired resistance (5).
The common strategy used to improve the sensitivity to platinum compounds and overcome their resistance is the combination with radiotherapy, antibodies, selective inhibitors or already prescribed drugs (3). Studies exploring the molecular profile characterization of lung cancer have allowed the development of targeted therapies, such as monoclonal antibodies against vascular endothelial growth factor, epidermal growth factor receptor (6), programmed cell death 1 ligand 1 (7), anaplastic lymphoma kinase (8), proto-oncogene tyrosine-protein kinase ROS1 (9) and serine/threonine-protein kinase B-raf (10) inhibitors, replacing or enhancing basic cytotoxic therapies (11).
Repurposing drugs already approved by the Food and Drug Administration can speed up therapeutic management due to overcoming the steps of new drug development. Metformin, a well-known oral antidiabetic drug, is widely associated with a decreased cancer risk (12) and increases the chemotherapeutic effects for different types of cancer, including endometrial cancer (13,14), osteosarcoma (15), hepatocarcinoma (16), non-small cell lung cancer (17) and gastric cancer (18). Metformin decreases the proliferation of lung cancer cells treated with cisplatin compared with cisplatin treatment alone (19) and sensitizes cells to tyrosine kinase inhibitors (20) and crizotinib treatment through the insulin growth factor 1 (IGF1) signaling pathway (21). The mechanisms by which metformin exerts antineoplastic effects remain unclear, but the AMPK-driven inhibition of mTOR seems to be required for its antimitotic activity (22).
The serine/threonine kinase mTOR is a widely evolutionarily conserved protein essential for cellular metabolism, acting as a sensor for the availability of nutrients and growth factors (23). Overactivation of the mTOR signaling pathway contributes to several disorders (24) and is associated with a poor cancer prognosis (25,26). Most cases of lung cancer have a mutation in liver kinase B1 (LKB1), such as A549 cells (27), which leads to the partial impairment of AMPK and overactivation of mTOR signaling (28). Studies have revealed that cisplatin sensitivity is associated with mTOR inhibition, especially in LKB1- and KRAS-mutant cancer (29-31). Despite the overactivation of mTOR signaling, the lack of LKB1 can also sensitize to metformin due to the inability to restore energy homeostasis (32,33). This makes mTOR signaling an important regulatory mechanism in lung cancer progression and metformin treatment.
The present study aimed to investigate the effects of metformin in cisplatin resistance and regulation of metabolic cancer pathways, including mTOR signaling, in lung cancer. The proteomes upon metformin treatment in the context of resistance and sensitivity to cisplatin in A549 cells were compared, revealing new possible molecular strategies and targets for NSCLC treatment to overcome cisplatin resistance.
Materials and methods
Cell culture
A549 cells were maintained in HAM-F12 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), penicillin (100 U/ml) and streptomycin (100 µg/ml). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Cells were treated at 37°C with 10 µM cisplatin (Sigma-Aldrich; Merck KGaA) for 72 h to generate the resistant population [CIS vs. control (C)] and were exposed to 10 mM metformin (Sigma-Aldrich; Merck KGaA) (MET or CISMET) or 100 nM rapamycin at 37°C (Sigma-Aldrich; Merck KGaA) (RAPA or CISRAPA) for 24, 48 or 72 h according to different assays.
MTT viability assay
A549 cells were seeded at a density of 3×104 cells/well for the C group (sensitive cells) and 9×104 cells/well for the CIS population (resistant cells) in 96-well plates and treated with cisplatin for 72 h and metformin or rapamycin for another 72 h. After incubation, 12 mM MTT solution was added to each well for 2 h at 37°C. The culture medium was aspirated and the formazan crystals were solubilized with a solution of 1M HCl:isopropanol (1:25) for 15 min at 37°C, and the absorbance of each sample was measured at 570 nm.
Clonogenic assay and microscopy
A549 cells were seeded at low density in 6-well plates (3×102 cells/well), treated with 10 mM metformin or 100 nM rapamycin for 72 h and incubated for 7 days at 37°C. After incubation, cells were washed with PBS and stained with 3 ml methylene blue dye (0.3% in 50% ethanol) for 30 min at room temperature. The plates were washed with deionized water and staining was measured at an absorbance of 590 nm by eluting with 10% acetic acid. The size of the colonies was calculated using ImageJ software v1.53 (National Institutes of Health). Colonies >0.01 pixel2 were considered as hits by the ImageJ software and were then manually divided into size ranges using Excel (Microsoft Corporation). The size ranges were hits <1 pixel2, between 1 and 25 pixel2 and >25 pixel2. Images of the cells for morphology analysis were captured using a light microscope coupled to a camera (magnification, ×40; Leica Microsystems, Inc.), using scale bars of 100 µm.
Cell cycle and size analysis
A549 cells were seeded in 6-well plates at a density of 3×105 cells/well and treated with 10 µM cisplatin for 72 h and 10 mM metformin or 100 nM rapamycin for 24 h for cell size analysis or 72 h for cell cycle analysis. After treatment, cells were washed with 1X PBS (0.137 M NaCl and 0.05 M NaH2PO4, pH 7.4) and resuspended in 500 µl HAM-F12 medium. Cells were centrifuged at 300 × g for 5 min at room temperature and resuspended in 100 µl propidium iodide (PI) solution (0.1% Triton X-100, 20 µg/ml PI and 10 µg/ml RNAse in PBS) for cell cycle analysis or 100 µl PBS for cell size analysis. The PI fluorescence and cell profile were determined using a BD Accuri™ C6 flow cytometer and analyzed using the BD Accuri™ software v1.0.264.21 (BD Biosciences).
Western blotting
Protein extracts obtained by lysing cells (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, protease inhibitor cocktail and phosphatase inhibitor cocktail) were quantified using a bicinchoninic acid assay and samples containing 20-40 µg of protein were separated by SDS-PAGE (8 and 10% gels). The gels were electrotransferred to 0.45-µm nitrocellulose membranes (Bio-Rad Laboratories, Inc.) and the membranes were incubated for 2 h at room temperature with 5% non-fat powdered milk dissolved in TBS-Tween (TBST; 50 mM Tris-Cl, pH 7.5; 150 mM NaCl; 0.1% Tween-20) to saturate unspecific binding sites, followed by an overnight incubation at 4°C with primary antibodies. Membranes were washed 3 times with TBST and then incubated with HRP-conjugated goat anti-mouse IgG (Sigma-Aldrich; Merck KGaA; cat. no. AP308P; 1:2,000) and goat anti-rabbit IgG (Sigma-Aldrich; Merck KGaA; cat. no. AP307P; 1:5,000) secondary antibodies for 1 h at room temperature. Protein bands were visualized using Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.) and densitometry was performed using ImageJ software v1.53. Primary antibodies (all 1:2,000) against mTOR (cat. no. 2972), phospho-mTOR (Ser2448; cat. no. 2971), AMPKα (cat. no. 5831), phospho-AMPKα (Thr172; cat. no. 50081), p70-S6K1 (cat. no. 2708), phospho-p70-S6K1 (Thr389; cat. no. 9234), S6 (cat. no. 2317), phospho-S6 (Ser240/244; cat. no. 2215) and GAPDH (cat. no. 2118) were purchased from Cell Signaling Technology, Inc.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from A549 cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). The cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit with 2,000 ng of total RNA according to the manufacturer's protocol (Thermo Fisher Scientific, Inc.). The qPCR reaction was performed with SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was performed with an initial denaturation step at 95°C for 10 min and then 40 cycles of 95°C for 15 sec and 60°C for 1 min. A melting curve was performed after the PCR from 60°C to 95°C. The 2−ΔΔCq method was used for quantification (34). β-actin was used as the normalizing gene. Samples for this reaction were added in triplicates in a 96-well plate (MicroAmp; Applied Biosystems; Thermo Fisher Scientific, Inc.) for amplification and reading in the Step One Plus Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers used were designed using Primer-BLAST (35) (National Institutes of Health): Superoxide dismutase 2 (SOD2) forward, 5′-AAG GAA CGG GGA CAC TTA CAA A-3′ and reverse, 5′-AGC AGT GGA ATA AGG CCT GTT G-3′; Annexin 4 (ANXA4) forward, 5′-CAG AGG AAC AAC CAG GAA CTT G-3′ and reverse, 5′-CAA GCA GAA GTT CTT CGA GGC-3′; and β-actin forward, 5′-GCC GCC AGC TCA CCA T-3′ and reverse, 5′-CCA CGA TGG AGG GGA AGA C-3′.
Stable isotope labeling by amino acids in cell culture (SILAC)
For heavy (H)- or light (L)-lysine labeling experiments, A549 cells were maintained in a T25 flask with SILAC™ HAMF12 medium (Thermo Fisher Scientific, Inc.) supplemented with 10% dialyzed FBS without lysine and arginine. To obtain H and L conditions, SILAC™ HAMF12 medium was supplemented with 13C6 L-lysine-2HCl (H) or 12C6 L-lysine-2HCl (L) and 12C6 L-arginine-2HCl (H and L). The final concentration of amino acids were 0.46 mM (lysine) and 0.47 mM (arginine). After 5 passages, C and CIS cells were treated in triplicate with 10 mM metformin for 72 h (CxMET representing the Ctrl population; CISxCISMET representing the CIS population), washed two times with PBS in a 6-well plate and collected using 100 µl lysis buffer as aforementioned.
The mixed H-/L-labeled protein sample (27.5 µg of total protein per group) was separated by electrophoresis in reducing 10% SDS-PAGE, heated for 10 sec in a microwave in Coomassie Brilliant Blue stain (Sigma-Aldrich; Merck KGaA; 2% Coomassie in 50% ethanol and 2.5% acetic acid) and incubated for 1 h at room temperature. After overnight incubation of the gel for background de-staining, each gel lane was sliced into 10 pieces. The excised sections were unstained twice with a de-staining solution (50% ethanol and 2.5% acetic acid in purified water) in 1.5-ml microtubes to start the gel bands digestion protocol using trypsin. The percentage of isotopic label incorporation was tested according to the following equation: (Ratio H/L x 100)/(Ratio H/L + 1) (36).
Trypsin digestion, mass spectrometry (MS) and data analysis
The protein bands were reduced, alkylated and digested with trypsin overnight at 37°C. An aliquot of the peptide mixture was separated using a 2-40% acetonitrile gradient in 0.1% formic acid using an analytical PicoFrit Column (20 cm x ID75 µm; 5-µm particle size; New Objective, Inc.), at a flow rate of 300 nl/min over 35 min. Peptides were analysed using the EASY-nLC II (Proxeon Biosystems) coupled to LTQ Orbitrap Velos (Thermo Fisher Scientific, Inc.), with electrospray ionization in positive mode and set up in the data-dependent acquisition mode. Full scan MS spectra (m/z 300-1,600) were acquired in the Orbitrap analyzer after accumulation to a target value of 1×106. Resolution in the Orbitrap was set to r=60,000, and the 20 most intense peptide ions with charge states ≥2 were sequentially isolated to a target value of 5,000 and fragmented in the linear ion trap by low-energy collision-induced dissociation (normalized collision energy of 35%). Three independent experiments were performed.
Bioinformatics analysis
For SILAC data analysis, the raw files were processed using MaxQuant 2012 version 1.3.0.5 (https://www.maxquant.org) and the MS/MS spectra were searched using the Andromeda search engine against the Uniprot Human Protein Database (37) (release 17 February 2016; 91,974 sequences; 36,693,332 amino acid residues). The initial maximal allowed mass tolerance was set to 20 ppm for precursor, 6 ppm in the main search afterward and then to 0.5 Da for fragment ions. Enzyme specificity was set to trypsin with a maximum of two missed cleavages. Carbamidomethylation of cysteine (57.021464 Da) was set as a fixed modification, and oxidation of methionine (15.994915 Da) and protein N-terminal acetylation (42.010565 Da) were selected as variable modifications. The minimum peptide length was set to 6 amino acids, including heavy label Lys6. For protein quantification, a minimum of two ratio counts was set and the 'requantify' and 'match between runs' functions were enabled. The false discovery rates (FDRs) of peptide and protein were both set to 0.01. Data processing was performed using Perseus v.1.2.7.4 available in the MaxQuant database (https://www.maxquant.org/). First, reverse and contaminant entries were excluded from further analysis. A protein ratio intensity between H and L was used to compare differential protein expression in the total extract from C and MET cells, and CIS and CISMET treated cells. The protein ratios were calculated from the median of all normalized peptide ratios using unique peptides or peptides assigned to the protein group with the highest number of peptides (razor peptides).
For statistical analysis of differentially expressed proteins, the ratios were converted into log2 and Student's unpaired t-test was applied on the C and CMET treated groups and CIS and CISMET treated groups. All MS raw files and search parameter settings associated with the present study are available for download via the PRIDE data repository at https://www.ebi.ac.uk/pride/archive/ (accession no. PXD017645). After the t-test, volcano plots were constructed to each replicate separately using the Volcano Plot Plugin of the OriginLab 2019b software (v9.65; OriginLab) considering P<0.05[Log10(0.05)=1.3010] and log2 ratio >±1.0 (fold change of CxMET and CISxCISMET ratio H/L normalized) as exclusion parameters. Heatmaps were generated on Morpheus online (https://software.broadinstitute.org/morpheus) considering common proteins in both groups with P<0.05. The heatmap represents the standardized values to robust z-score clustered by Euclidian distance using the average linkage method. Proteome networks were generated using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) v11 (https://string-db.org/), using the significant proteins list after t-test analysis and setting a High Confidence value of 0.700 as a parameter. FDRs of Gene Ontology (GO) classification were calculated using STRING v11 for biological processes and performed as previously described by Szklarczyk et al (38). Biological processes were arbitrarily chosen so as not to overlap redundant classifications and obtain the largest number of classified targets. The FDR of GO and the P-value of the protein-protein interaction (PPI) enrichment were automatically calculated by STRING and the detailed description of the enrichment algorithm has been previously described (39).
Survival rates of patients with lung adenocarcinoma and lung squamous cell carcinoma were assessed using The Cancer Genome Atlas Pan-Lung Cancer database (40) (dbGaP Study Accession: phs000488.v1.p1) for ANXA4 and SOD2 in the cBioPortal software v3 5.0 (https://www.cbioportal.org/) (41). Each sample was defined as altered or unaltered for each gene based on the Onco Query Language, which considers non-synonymous mutations, fusions, amplifications and deep deletions (https://www.cbioportal.org/oql).
Statistical analysis
Statistical analyses were assessed using GraphPad Prism 8.01 software (GraphPad Software, Inc.) applying Student's unpaired t-test or one-way ANOVA followed by Tukey's post-hoc test when n sample was homogeneous between groups and Bonferroni's test when not meeting that condition. P<0.05 was considered to indicate a statistically significant difference. For proteomics analysis, Student's t-tests were performed using Perseus.
Results
Metformin (MET) decreases cell viability and size without changes in the cell cycle
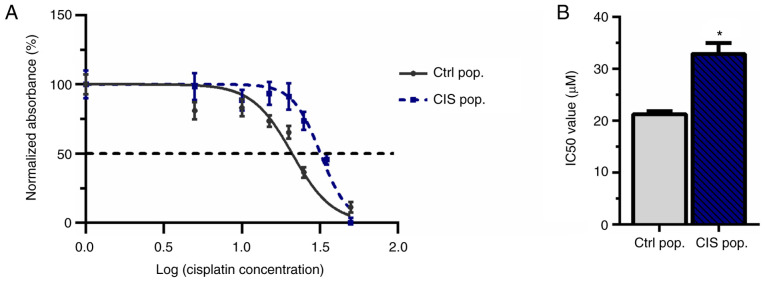
To demonstrate that 10 µM cisplatin was able to create resistance in A549 cells, an MTT assay was performed to calculate the IC50. This cisplatin concentration was adopted since it had been previously reported to confer resistance to new cisplatin exposure in melanoma (42). A549 cells were subjected to 10 µM cisplatin treatment for 72 h (called CIS population or CIS pop) and subsequent doses of cisplatin (5, 10, 15, 20, 25, 35 and 50 µM) for another 72 h. The CIS pop presented a significant increase in the IC50 compared with the untreated control population (Ctrl pop), suggesting the acquisition of resistance (21.24 µM for Ctrl pop vs. 32.86 µM for CIS pop; Fig. 1A and B).
Figure 1.
CIS (10 µM) generates resistance, increasing the IC50 in A549 lung cancer cells. A549 cells exposed to 10 µM CIS (CIS pop) or not (Ctrl pop) for 72 h were subsequently subjected to different concentrations of CIS and MTT assay was performed. (A) Data was expressed in a non-fit curve of log of cisplatin concentration versus normalized percentage of viability. (B) Bar graph shows the mean with SEM of 3 independent experiments. *P<0.05. CIS, cisplatin; Ctrl, control; pop, population.
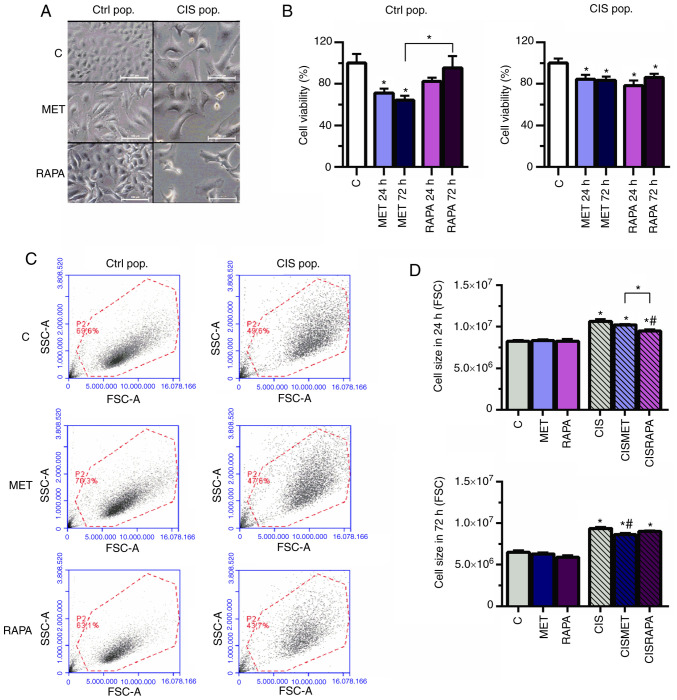
To investigate the effects of MET in lung cancer cells, MTT assay was performed (Fig. 2B) in the CIS pop compared with the Ctrl pop. MET decreased cell viability more extensively in the Ctrl pop (71.2% for 24 h and 64.2% for 72 h exposure) compared with in the CIS pop (84.5% for 24 h and 83.5% for 72 h) (Fig. 2B). Additionally, the viability of cells treated with MET was significantly decreased compared with that of cells treated with rapamycin (RAPA) in the Ctrl pop after 72 h (64.2 vs. 95.5%; Fig. 2B).
Figure 2.
MET decreases cell viability and size in Ctrl and CIS-treated A549 cells. A549 cells exposed to 10 µM CIS (CIS pop) or not (Ctrl pop) for 72 h were subsequently exposed to 10 mM MET or 100 nM RAPA for 24 and 72 h. (A) Cell size increased in CIS-treated A549 cells and decreased in cells treated with 10 mM MET or 100 nM RAPA after 72 h, as seen by microscopy. Scale bar, 100 µm. (B) MET and RAPA significantly decreased cell viability in the Ctrl and CIS pop after 72 h. (C and D) CIS altered cell size after 24 and 72 h, as analyzed by flow cytometry, and was then restored by MET after 72 h and by RAPA after 24 h. *P<0.05 vs. C; #P<0.05 vs. CIS. CIS, cisplatin; Ctrl/C, control; pop, population; MET, metformin; RAPA, rapamycin; FSC, forward scatter; SSC, side scatter.
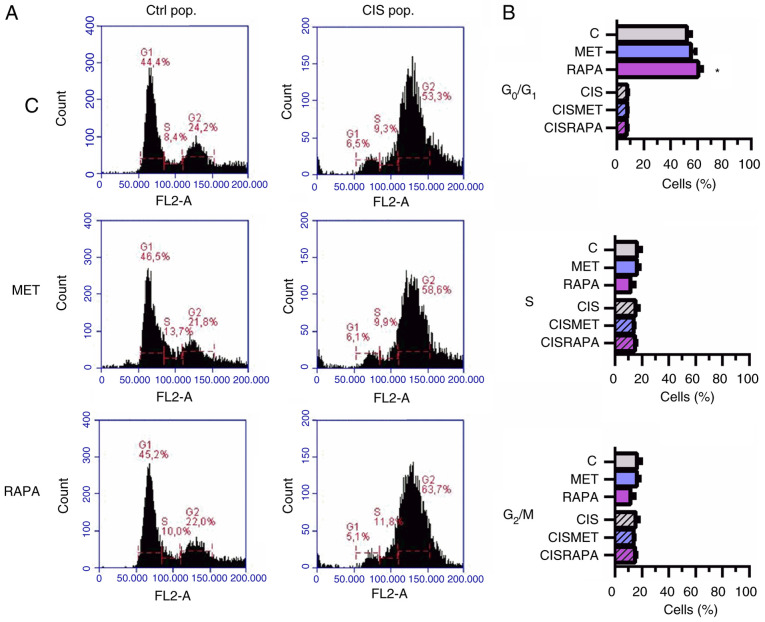
Cisplatin led to increased cell size and granularity, as seen by light microscopy and forward scatter and side scatter flow cytometry parameters (Fig. 2A and C, respectively), conferring more heterogeneity to the CIS pop compared with the Ctrl pop. In the CIS pop, flow cytometry data revealed that RAPA significantly decreased cell size after 24 h, while MET significantly decreased cell size after 72 h treatment, both compared with cisplatin alone (Fig. 2D). However, only RAPA led to G0/G1 cell cycle arrest after 72 h treatment (Fig. 3A and B). The present data suggested that 72 h of MET treatment decreased cell viability and size in cisplatin-sensitive and -resistant cells without cell cycle impairment. In summary, MET decreased lung cancer cell viability and size without significantly changing the cell cycle.
Figure 3.
MET does not change the cell cycle in Ctrl and CIS-treated A549 cells. A549 cells exposed to 10 µM CIS (CIS pop) or not (Ctrl pop) for 72 h were subsequently exposed to 10 mM MET or 100 nM RAPA for 24 and 72 h. (A and B) Cell cycle was analyzed by flow cytometry in the Ctrl and CIS pop treated with MET and RAPA for 72 h. *P<0.05 vs. C. CIS, cisplatin; Ctrl/C, control; pop, population; MET, metformin; RAPA, rapamycin.
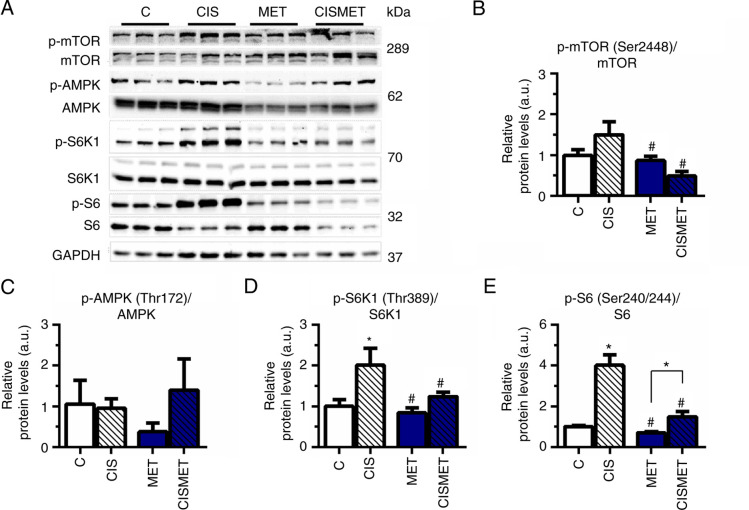
MET reverts mTOR activation induced by cisplatin in A549 cells
The mTOR signaling pathway status was evaluated by western blotting after MET treatment in Ctrl and CIS pops. The analysis revealed that the mTOR signaling pathway was overactivated after cisplatin treatment, with a significant increase in S6K1 and S6 phosphorylation (Fig. 4A, D and E) compared with the control group. Subsequent MET treatment significantly decreased mTOR, S6K1 and S6 activation compared with CIS treatment, corroborating the aforementioned decreases in cell viability and size. A non-significant difference was observed in AMPK phosphorylation after MET treatment in A549 cells (Fig. 4C). The present results indicated robust activation of the mTOR signaling pathway after cisplatin treatment in A549 cells.
Figure 4.
MET reverts mTOR activation induced by cisplatin in A549 lung cancer cells. (A) Analysis of protein content of the mTOR signaling pathway in A549 lung cancer cells after treatment with 10 µM CIS for 72 h and 10 mM MET for 72 h analyzed via western blotting. Band densitometry performed using ImageJ of the protein expression levels of (B) p-mTOR/mTOR (n=2), (C) p-AMPK/AMPK (n=2), (D) p-S6K1/S6K1 (n=3) and (E) p-S6/S6 (n=3). *P<0.05 vs. C; #P<0.05 vs. CIS. CIS, cisplatin; C, control; MET, metformin; p, phosphorylated; a.u., arbitrary units.
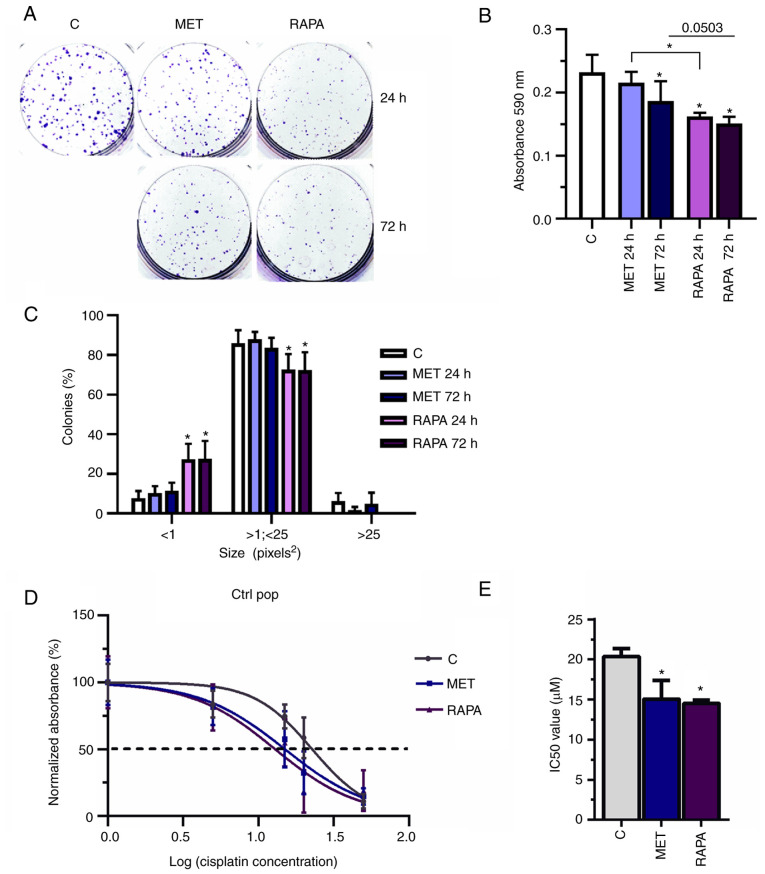
MET and RAPA decrease colony formation and sensitize A549 cells to cisplatin
To determine the colony formation potential of MET- and RAPA-treated cells, A549 cells exposed to 10 mM MET or 100 nM RAPA for 24 or 72 h were evaluated in a clonogenic cell assay (Fig. 5A). Compared with the control group, MET and RAPA treatments significantly decreased the absorbance of colonies formed (MET at 72 h and RAPA at 24 and 72 h) compared with the control group (Fig. 5B), but only RAPA decreased the size of colonies compared with the control group (Fig. 5C). Both MET and RAPA significantly decreased the IC50 in Ctrl pop cells (IC50=15.4 and 14.5 µM, respectively, vs. 20.4 µM for Ctrl; Fig. 5D and E). Thus, the decrease of IC50 indicated that previous treatment with MET or RAPA may potentiate and sensitize cells to cisplatin treatment.
Figure 5.
MET and RAPA decrease colony formation and IC50 in A549 lung cancer cells. (A) Ctrl pop cells were treated with 10 mM MET or 100 nM RAPA for 24 and 72 h, and the clonogenic assay was performed. (B) Graph data presents the absorbance of crystal violet and (C) the size of colonies was measured using ImageJ. (D) Ctrl pop was treated with MET and RAPA, and subsequently subjected to CIS at different concentrations. (E) MET and RAPA pre-treatment in Ctrl pop followed by CIS treatment significantly decreased the IC50 values of CIS. *P<0.05 vs. C. CIS, cisplatin; Ctrl/C, control; pop, population; MET, metformin; RAPA, rapamycin.
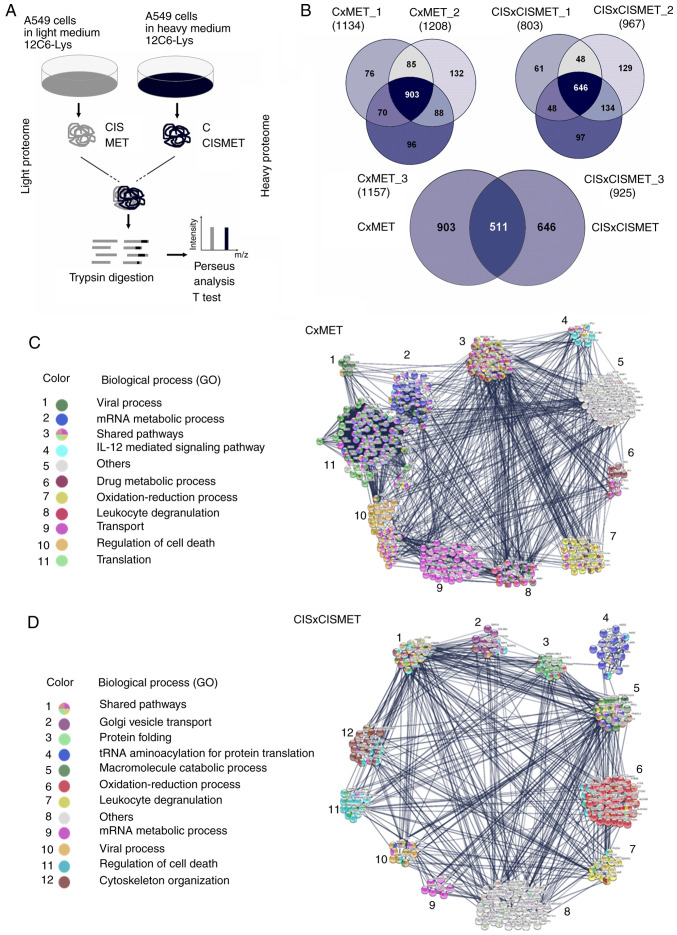
Proteomics analysis reveals MET treatment profile in Ctrl and CIS populations
In addition to the mTOR signaling pathway, MET exerts important changes in crucial pathways involved in cancer (22). To investigate other possible molecular mechanisms involved in cisplatin sensitivity induced by MET, a proteomics analysis of A549 cells after MET treatment was performed, and the proteomes in control and cisplatin-resistant cells were compared using the SILAC approach (CxMET and CISxCISMET; Fig. 6A). A total of 903 proteins were quantified in CxMET and 646 in CISxCISMET, with 511 common proteins (Fig. 6B). The Student's t-test analysis indicated 361 differentially expressed and statistically significant proteins (P<0.05) for CxMET ratios and 254 for CISxCISMET ratios (Tables SI and SII). These proteins were classified and grouped based on their involvement in biological processes (Fig. 6C and D). GO terms and FDRs generated by STRING analysis are presented in Table I. PPI enrichment P-values were <1.00−16 for CxMET and CISxCISMET networks (data not shown).
Figure 6.
Proteomics analysis reveals modulated pathways after MET treatment in control and CIS-resistant A549 cells. (A) Experimental design of proteomics analysis using the Stable Isotope Labeling by Amino acids in Cell culture approach. (B) Venn diagrams presenting 903 proteins detected in control (CxMET) and 646 proteins in CIS-resistant cells (CISxCISMET), as well as 511 common proteins in both groups. Network analysis performed using the Search Tool for the Retrieval of Interacting Genes/Proteins revealing (C) 361 proteins in CxMET and (D) 254 proteins in CISxCISMET separated according to GO biological processes. CIS, cisplatin; MET, metformin; GO, Gene Ontology.
Table I.
Biological processes, GO numbers and FDRs associated with each proteomic network.
| Biological processes | GO no. | CxMET | CISxCISMET | Upregulated | Downregulated |
|---|---|---|---|---|---|
| Viral process | 0016032 | 8.62×1011 | 6.97×1007 | - | - |
| Regulation of mRNA metabolic process | 1903311 | - | - | - | 0.0283 |
| mRNA metabolic process | 0016071 | 3.57×1035 | 0.0076 | - | - |
| IL-12 mediated signaling pathway | 0035722 | 1.94×1007 | - | - | 0.0019 |
| Translation | 0006412 | 2.30×1042 | 2.83×1007 | 6.65×1005 | - |
| Regulation of cell death | 0010941 | 3.78×1007 | 3.02×1009 | - | - |
| Transport | 0006810 | 8.36×1027 | - | - | 0.0290 |
| Leukocyte degranulation | 0043299 | 4.18×1006 | 5.24×1010 | - | - |
| Oxidation-reduction process | 0055114 | 4.55×1015 | 1.76×1020 | 9.19×1006 | 0.0021 |
| Drug metabolic process | 0017144 | 3.11×1013 | - | - | - |
| Golgi vesicle transport | 0048193 | - | 6.64×1005 | - | - |
| Protein folding | 0006457 | - | 3.41×1013 | 0.00014 | - |
| Macromolecule catabolic process | 0009057 | - | 0.00032 | - | - |
| Cytoskeleton organization | 0007010 | - | 0.00055 | - | - |
| Negative regulation of apoptotic process | 0043066 | - | - | - | 0.0019 |
| Regulated exocitosis | 0045055 | - | - | 4.45×1006 | - |
| Regulation of apoptotic process/apoptosis | 0042981 | - | - | 0.0048 | - |
GO, Gene Ontology; FDR, false discovery rate; CIS, cisplatin; MET, metformin.
In the CxMET group, MET regulated 'viral process', 'mRNA metabolic process', 'IL-12 mediated signaling pathway', 'drug metabolic process', 'oxidation-reduction process', 'leukocyte degranulation', 'transport', 'regulation of cell death' and 'translation' (Fig. 6C). In the CISxCISMET group, MET regulated 'Golgi vesicle transport', 'protein folding', 'tRNA aminoacylation for protein translation', 'macromolecule catabolic process', 'oxidation-reduction process', 'leucocyte degranulation', 'mRNA metabolic process', 'viral process', 'regulation of cell death' and 'cytoskeleton organization' (Fig. 6D). The data revealed five shared pathways between groups ('mRNA metabolic process', 'oxidation-reduction process', 'leukocyte degranulation', 'viral process' and 'regulation of cell death'). The raw list of proteins identified by Perseus in both groups is presented in Tables SI and SII. Therefore, these molecular pathways may be potentially important signatures of the mechanisms of action of MET in lung cancer.
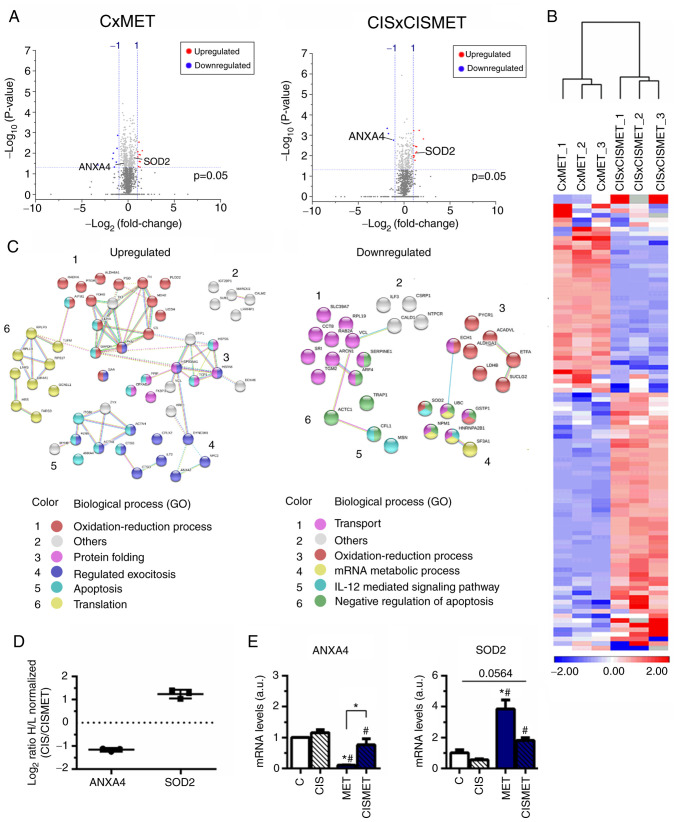
MET alters translation, oxidative stress, apoptosis and metabolic pathways in control and cisplatin-resistant A549 cells
Volcano plots displayed the 1,448 proteins in CxMET and 1,158 in CISxCISMET (Fig. 7A), both separated by magnitude of evidence (P-value) and change (fold change of log2 ratio values) [cut-off values, log10(0.05)=1.3010; log2 ratio=1.00]. A total of 186 and 184 proteins were significantly downregulated and upregulated in MET compared with C, respectively, and 102 and 167 proteins were significantly downregulated and upregulated in CISMET compared with CIS, respectively. Applying the cut-off P-value, in CxMET the volcano plot expressed 3 significantly downregulated proteins, including ANXA4, and 12 significantly upregulated proteins, including SOD2. In CISxCISMET, the volcano plot expressed 7 significantly down-regulated proteins and 12 significantly upregulated proteins. Subsequently, 99 common significant proteins between CxMET and CISxCISMET analysis (ACADVL, ACTC1, ACTN1, ACTN4, AIFM1, AK3, AKR1C1, ALDH1A1, ALDH6A1, ANXA1, ANXA2, ANXA4, ARCN1, ARF3/ARF1, ARF4, C1orf57/NTPCR, CALD1, CALM2/CALM1, CANX, CCT8, CFL1, CPLX2, CRYAB, CS, CSRP1, CTSB, CTSD, DDX46, DYNC1H1, DYNC1LI2, ECH1, EIF4A1, EPRS, ETFA, ETFB, FARSB, FH, FKBP3, FLNA, G6PD, GAA, GAPDH, GCN1L1, GRPEL1, GSTP1, HADHA, HMGA1, HNRNPA2B1, HNRNPL, HSP90AA1, HSPA8, HSPD1, IARS, IGF2BP1, ILF2, ILF3, ITGB1, LARS, LDHA, LDHB, LRRFIP1, MARCKS, MDH2, MSN, MYH9, NCL, NPC2, NPM1, PDHB, PGD, PKM2, PLOD2, PPIF, PTBP1, PTGR1, PYCR1, RAB2A/RAB2B, RPL19, RPL3, RPLP0, RPS17, SERPINE1, SF3A1, SLC39A7, SOD2, SRI, STIP1, SUB1, SUCLG2, TCP1, TGM2, TKT, TPM4, TRAP1, TUFM, UBC, UGDH, VCL and ZYX), altered after MET treatment and expressed by standardized values, were grouped in clusters by Euclidian distance using the average linkage in a heatmap (Fig. 7B). These proteins were then classified into two different networks, according to their upregulation or downregulation in resistant cells compared with control cells (Fig. 7C and Table I). Cisplatin resistance decreased proteins associated with 'transport' and 'mRNA metabolic process', while it upregulated proteins involved in 'translation' (Fig. 7C), corroborating the aforementioned activation of the mTOR signaling pathway. Regulation of 'oxidation-reduction process' and apoptotic processes were common biological processes found to be upregulated or downregulated by cisplatin treatment (Fig. 7C).
Figure 7.
Proteomics analysis revealing upregulated and downregulated targets after MET treatment in control and CIS-resistant A549 cells. (A) Volcano plots of the proteins significantly downregulated and upregulated with a cut-off of P<0.05, represented by Log10 (0.05) value of threshold (1.301), and normalized H/L ratio >±1 for both groups. (B) Heatmap of the common significant proteins with P<0.05. (C) Upregulated and downregulated proteins in CISxCISMET compared with CxMET were classified using the Search Tool for the Retrieval of Interacting Genes/Proteins according to GO biological processes. (D) Normalized H/L ratios of two selected proteins from the proteomics analysis, SOD2 and ANXA4. (E) Reverse transcription-quantitative PCR reported altered expression levels of ANXA4 and SOD2 after MET treatment. *P<0.05 vs. C; #P<0.05 vs. CIS. CIS, cisplatin; C, control; MET, metformin; GO, Gene Ontology; ANXA4, annexin 4; SOD2, superoxide dismutase 2; H/L, heavy to light ratio; a.u., arbitrary units.
Two proteins reported by the proteomics analysis were chosen for further validation: ANXA4, involved in apoptosis (43), and SOD2, involved in oxidative stress pathways (44), which presented decreased and increased expression levels in the CIS versus CISMET analysis, respectively (Fig. 7D). The mRNA expression levels of these targets were further evaluated by RT-qPCR, revealing that MET significantly decreased ANXA4 expression compared with control, CIS and CISMET groups (Fig. 7E). Additionally, MET significantly increased SOD2 expression compared with control and CIS groups (Fig. 7E). Furthermore, treatment with MET in the Cis pop (CISMET group) significantly decreased ANXA4 expression and increased SOD2 expression compared with the CIS group (Fig. 7E). RT-qPCR data corroborated the proteomics analysis presented in the heatmap. A complementary analysis of the survival rates of patients with lung cancer using the cBioPortal revealed that alterations in the ANXA4 gene decreased the median survival time after initial treatment from 43.9 months to 19.5 months (Fig. S1). The altered group for SOD2 is represented by 10 patients (plus 2 deceased), with 5 patients with deep deletions of the gene (homodeleted) and 5 patients with missense mutations (G141C, L176R, R123H, G126A and S127F) (data not shown). The altered group for ANXA4 is represented by 12 patients (plus 5 deceased), with 8 patients with gene amplification and 4 patients with missense mutations (W188L, G33C, L316V and Q52E) (data not shown). Overall, MET decreased ANXA4 expression and increased SOD2 expression in A549 cells, validating the presented proteomics data.
Discussion
Over the last years, MET has been widely used as an antidiabetic agent and has been characterized to present antitumor properties and several advantages in cancer therapy (45). The mechanisms by which MET decreases tumor progression and presents chemosensitivity abilities are not fully described, but it seems that AMPK-driven inhibition of mTOR is an important regulating axis in the tumorigenic process (22). The mTOR signaling pathway is also described as a pathway with implications in tumor development and progression, metastasis and chemoresistance (46). The present study indicated that MET may potentially affect lung cancer progression and cisplatin chemosensitivity, and may be associated with mTOR signaling and other pathways, such as translation, oxidative stress and apoptosis.
In the present study, MET decreased the viability in Ctrl and cisplatin-treated cells, as described in breast (47,48), ovarian (49) and lung cancer cell lines (50). Stronger effects of MET on viability may be possible due to its extensive and embracing mechanisms of action compared with the point target effects of RAPA, as previously reported in pancreatic cancer cells (51). Although other studies have reported cell cycle arrest after MET treatment in different cell lines (52-54), no changes were observed in the present study in the A549 cell cycle. However, it was confirmed that cisplatin led to cell cycle arrest in the G2/M phase, which is a characteristic of platinum drugs (55), and that RAPA led to G0/G1 arrest, which has been also previously reported (56-58). Additionally, the current study indicated that MET reverted the increase in cell size induced by cisplatin after 72 h, which was consistent with decreased mTOR signaling. Wang et al (59) reported that cell cycle arrest induced by MET in myeloma is dependent on the mTOR signaling pathway, possibly via intact LKB1-AMPK axis also observed in A549 cells (60). mTOR inhibition seems to be an important strategy to improve cisplatin sensitivity, mediating the chemotherapy resistance in KRAS-mutant lung cancer (31). Several studies have demonstrated that cisplatin resistance induces activation of the mTOR/Akt signaling pathway and decreases apoptosis, whereas inhibitors of the mTOR/Akt signaling pathway sensitize and enhance the effects of cisplatin in different cancer cell lines, including lung cancer (61) and hepatocarcinoma cells (62). In esophageal squamous cell carcinoma (ESCC) xenografts, a small interfering RNA against mTOR significantly increased apoptosis when combined with cisplatin (63). Moreover, patients with endometrial cancer treated with MET presented decreased levels of plasma IGF-1 and PI3K, phospho-Akt, phospho-S6K1 and phospho-4EBP1 in biopsy specimens, reinforcing its antiproliferative potential (64). In accordance with the aforementioned studies, the present study confirmed in vitro that MET decreases cell viability and clonogenic potential, sensitizes cells to cisplatin by decreasing the IC50 and is associated with decreased mTOR signaling, indicating that MET may be a potential coadjuvant in NSCLC therapy.
Galluzzi et al (65,66) defined mechanisms of cisplatin resistance and its associated targets in different signaling pathways. Cisplatin resistance is generally multifactorial, characterized by successive molecular alterations, including the binding of cisplatin to its targets, increased repair mechanisms, decreased apoptosis and stimulation of pro-survival mechanisms (67,68). Considering cisplatin resistance and tumor progression, it is important to target more than one molecular mechanism to efficiently circumvent cisplatin resistance (65,66).
Understanding the MET-induced proteomic changes in sensitive and resistant contexts provides important information about specific modifications acquired by cells (67). Regarding the multifactorial resistance profile, the present study aimed to investigate other potential signaling pathways involved in cisplatin sensitivity induced by MET in addition to mTOR signaling. In both sensitive and resistant cells, MET altered transcriptional processes, regulated apoptosis, oxidation-reduction processes and proteins associated with leukocyte degranulation, with 99 common significantly altered targets, of which some have been previously described: AK3 (68), ALDH1A1 (69), ANXA2 and 4 (70,71), CFL1 (72), CRYAB (73), filamin A (FLNA) (74), GSTP1 (75), HMGA1 and G6PD (76), hnRNPA2B1 (77), HSP90 (78), IGF2BP1 (79), integrin b1 (ITGB1) (80), MYH9 (81), PKM2 (82-84), STIP1 (85), TGM2 (86), TKT (87) and TRAP1 (88).
In the present study, comparing sensitive and resistant contexts, MET decreased several oncogenes, such as CD29, FLNA, CTSD, MSN and ANXA4. Despite IL-12-mediated signaling having been associated with antitumor effects (89), the pathway component MSN has been largely associated with tumor progression (90-92). The present proteomic analysis revealed that MET increased apoptosis in cisplatin-resistant cells, as previously reported (33,53,93), by decreasing anti-apoptotic proteins such as TRAP1, CFL1 and SOD2. On the other hand, potential tumor progressors, such as CD29, cathepsin D (CTSD), FLNA and ANXA4, were upregulated. CD29, also known as ITGB1, is a transmembrane cell surface receptor that has been associated with metastasis, tumor migration and drug resistance (94,95). ITGB1 is associated with resistance to gefitinib in NSCLC (96) and its knockdown overcomes erlotinib resistance in lung cancer cells and decreases the activation of Akt after erlotinib treatment (97). FLNA acts as a scaffold for cancer-associated signaling pathways and is associated with the aggressive pattern and poor survival outcomes in patients with NSCLC treated with platinum-based drugs, such as cisplatin (98). Furthermore, FLNA may interact with other oncogenes, such as Akt, K-RAS, TRAF2, NIK and 14-3-3σ (99-102). CTSD is an intracellular aspartic protease of the pepsin superfamily associated with inhibition of SERPINE1 and is a tumor marker for invasion and metastasis (103,104). Overexpression of CTSD promotes breast cancer cell migration, invasion and metastasis through intercellular cell adhesion molecule-1 both in vitro and in vivo (105).
For the validation of the proteomics analysis, two targets were chosen, SOD2 (upregulated) and ANXA4 (downregulated), whose roles in cisplatin resistance are already known and well-documented. SOD2, a superoxide scavenger, may be directly involved in carcinogenesis by protecting cells against increased levels of reactive oxygen species (ROS) (106). It has been previously described that SOD2 can protect against DNA damage-inducing agents, especially in radiation (107-109). ROS act as mediators of DNA damage and SOD2, an endogenous antioxidant, protects the cells from DNA damage by scavenging reactive molecules, such as superoxide (110). One of the mechanisms described for the action of cisplatin is the depletion of antioxidant molecules to tilt the redox balance towards oxidative stress, which facilitates DNA damage (65). Additionally, cisplatin treatment increases ROS content in human lung cancer cells, including A549 cells (111). On the other hand, the overexpression of SOD2 in mitochondria enhance the survival of HeLa cells and contribute to cisplatin resistance in human ESCC and oral squamous cell carcinoma cell lines (110,112).
ANXA4 is largely involved in the proliferation, platinum resistance and migration in different types of cancer cells, such as ovarian (43,113) and endometrial cancer cells (114). ANXA4 overexpression is associated with tumor cell invasion and poor prognosis in patients with gallbladder cancer (115). Furthermore, overexpression of ANXA4 confers carboplatin resistance in ovarian carcinoma cells (114). ANXA4-knockdown increases sensitivity to platinum-based drugs both in vitro and in vivo (70,116) and the gain of its expression can restore cisplatin resistance in mesothelioma cells (116), which reinforces the beneficial effects of MET in decreasing ANXA4 expression. According to the present survival analysis, alterations in ANXA4 may decrease patient survival. Since treatment with MET significantly decreased ANXA4 expression, ANXA4 may be explored as a potential marker for survival and cisplatin responsiveness in lung cancer.
In conclusion, the present study demonstrated that MET sensitized cells to cisplatin treatment and decreased clonogenic survival and viability in A549 lung cancer cells. The mTOR signaling pathway was overactivated after cisplatin treatment, which was restored by MET regardless of the LKB1-AMPK axis. Therefore, MET may be able to improve the chemotherapeutic effects of cisplatin in A549 cells by decreasing mTOR signaling and modulating apoptosis, translation-associated processes and oxidative pathways, thus providing potential new therapeutic targets to circumvent cisplatin resistance in NSCLC.
Supplementary Data
Acknowledgments
The authors would like to acknowledge the Mass Spectrometry Laboratory at Brazilian Biosciences National Laboratory (Campinas, Brazil) for their support with mass spectrometry analysis.
Funding Statement
The present study was funded by the São Paulo Research Foundation (FAPESP; grant nos. 2012/13558-7, 2018/14818-9, 2016/06457-0 and 2015/22814-5; fellowship nos. 2016/02483-7, 2017/04269-5, 2019/00607-9, 2015/003111 and 2015/16601-9) and by the National Council for Scientific and Technological Development (grant no. 447553/2014-3).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Additionally, the proteomic datasets generated and/or analyzed during the current study are available in the PRIDE repository (https://www.ebi.ac.uk/pride/). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (117) with the dataset identifier PXD017645 (https://www.ebi.ac.uk/pride/archive/projectPXD017645).
Authors' contributions
APM, ICBP and FRS performed the cell experiments and revised the manuscript. APM, ICBP, AFPL, DCG, BAP and RRD performed mass spectrometry experiments and proteomics data analysis. GFP performed bioinformatics analysis. APM wrote the manuscript and headed the execution of all experiments. APM, FMS, RMNB, TCT, LPDM, DCG, AFPL and RC designed the experiments and revised the manuscript. APM and FMS confirmed the authenticity of the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ridge CA, McErlean AM, Ginsberg MS. Epidemiology of lung cancer. Semin Intervent Radiol. 2013;30:93–98. doi: 10.1055/s-0033-1342949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Wu YL, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 4.Talebian Yazdi M, Schinkelshoek MS, Loof NM, Taube C, Hiemstra PS, Welters MJ, van der Burg SH. Standard radiotherapy but not chemotherapy impairs systemic immunity in non-small cell lung cancer. Oncoimmunology. 2016;5:e1255393. doi: 10.1080/2162402X.2016.1255393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarin N, Engel F, Kalayda GV, Mannewitz M, Cinatl J, Jr, Rothweiler F, Michaelis M, Saafan H, Ritter CA, Jaehde U, Frötschl R. Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS One. 2017;12:e0181081. doi: 10.1371/journal.pone.0181081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Hackshaw A, Feng Q, Fu X, Zhang Y, Mao C, Tang J. Comparison of gefitinib, erlotinib and afatinib in non-small cell lung cancer: A meta-analysis. Int J Cancer. 2017;140:2805–2819. doi: 10.1002/ijc.30691. [DOI] [PubMed] [Google Scholar]
- 7.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–228. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 8.Uchibori K, Inase N, Araki M, Kamada M, Sato S, Okuno Y, Fujita N, Katayama R. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat Commun. 2017;8:14768. doi: 10.1038/ncomms14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Facchinetti F, Rossi G, Bria E, Soria JC, Besse B, Minari R, Friboulet L, Tiseo M. Oncogene addiction in non-small cell lung cancer: Focus on ROS1 inhibition. Cancer Treat Rev. 2017;55:83–95. doi: 10.1016/j.ctrv.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Planchard D, Kim TM, Mazieres J, Quoix E, Riely G, Barlesi F, Souquet PJ, Smit EF, Groen HJ, Kelly RJ, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: A single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:642–650. doi: 10.1016/S1470-2045(16)00077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 12.Decensi A, Puntoni M, Goodwin P, Cazzaniga M, Gennari A, Bonanni B, Gandini S. Metformin and cancer risk in diabetic patients: A systematic review and meta-analysis. Cancer Prev Res (Phila) 2010;3:1451–1461. doi: 10.1158/1940-6207.CAPR-10-0157. [DOI] [PubMed] [Google Scholar]
- 13.Cantrell LA, Zhou C, Mendivil A, Malloy KM, Gehrig PA, Bae-Jump VL. Metformin is a potent inhibitor of endometrial cancer cell proliferation-implications for a novel treatment strategy. Gynecol Oncol. 2010;116:92–98. doi: 10.1016/j.ygyno.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong L, Zhou Q, Zhang Z, Zhu Y, Duan T, Feng Y. Metformin sensitizes endometrial cancer cells to chemotherapy by repressing glyoxalase I expression. J Obstet Gynaecol Res. 2012;38:1077–1085. doi: 10.1111/j.1447-0756.2011.01839.x. [DOI] [PubMed] [Google Scholar]
- 15.Duo J, Ma Y, Wang G, Han X, Zhang C. Metformin synergistically enhances antitumor activity of histone deacetylase inhibitor trichostatin a against osteosarcoma cell line. DNA Cell Biol. 2013;32:156–164. doi: 10.1089/dna.2012.1926. [DOI] [PubMed] [Google Scholar]
- 16.Fujita H, Hirose K, Sato M, Fujioka I, Fujita T, Aoki M, Takai Y. Metformin attenuates hypoxia-induced resistance to cisplatin in the HepG2 cell line. Oncol Lett. 2018;17:2431–2440. doi: 10.3892/ol.2018.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin CC, Yeh HH, Huang WL, Yan JJ, Lai WW, Su WP, Chen HH, Su WC. Metformin enhances cisplatin cytotoxicity by suppressing signal transducer and activator of transcription-3 activity independently of the liver kinase B1-AMP-activated protein kinase pathway. Am J Respir Cell Mol Biol. 2013;49:241–250. doi: 10.1165/rcmb.2012-0244OC. [DOI] [PubMed] [Google Scholar]
- 18.Yu G, Fang W, Xia T, Chen Y, Gao Y, Jiao X, Huang S, Wang J, Li Z, Xie K. Metformin potentiates rapamycin and cisplatin in gastric cancer in mice. Oncotarget. 2015;6:12748–1262. doi: 10.18632/oncotarget.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teixeira SF. Metformin synergistically enhances antiproliferative effects of cisplatin and etoposide in NCI-H460 human lung cancer cells. J Bras Pneumol. 2013;39:644–649. doi: 10.1590/S1806-37132013000600002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Han R, Xiao H, Lin C, Wang Y, Liu H, Li K, Chen H, Sun F, Yang Z, Jiang J, He Y. Metformin sensitizes EGFR-TKI-resistant human lung cancer cells in vitro and in vivo through inhibition of IL-6 signaling and EMT reversal. Clin Cancer Res. 2014;20:2714–2726. doi: 10.1158/1078-0432.CCR-13-2613. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Wang Y, Peng T, Zhang K, Lin C, Han R, Lu C, He Y. Metformin restores crizotinib sensitivity in crizotinib-resistant human lung cancer cells through inhibition of IGF1-R signaling pathway. Oncotarget. 2016;7:34442–34452. doi: 10.18632/oncotarget.9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pernicova I, Korbonits M. Metformin-mode of action and clinical implications for diabetes and cancer. Nat Rev Endocrinol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- 23.Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavares MR, Pavan IC, Amaral CL, Meneguello L, Luchessi AD, Simabuco FM. The S6K protein family in health and disease. Life Sci. 2015;131:1–10. doi: 10.1016/j.lfs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 26.Amaral CL, Freitas LB, Tamura RE, Tavares MR, Pavan IC, Bajgelman MC, Simabuco FM. S6Ks isoforms contribute to viability, migration, docetaxel resistance and tumor formation of prostate cancer cells. BMC Cancer. 2016;16:602. doi: 10.1186/s12885-016-2629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong D, Guo L, de Aguirre I, Liu X, Lamb N, Sun SY, Gal AA, Vertino PM, Zhou W. LKB1 mutation in large cell carcinoma of the lung. Lung Cancer. 2006;53:285–294. doi: 10.1016/j.lungcan.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Kullmann L, Krahn MP. Controlling the master-upstream regulation of the tumor suppressor LKB1. Oncogene. 2018;37:3045–3057. doi: 10.1038/s41388-018-0145-z. [DOI] [PubMed] [Google Scholar]
- 29.Jiang G, Liu CT. Knockdown of SALL4 overcomes cisplatin-resistance through AKT/mTOR signaling in lung cancer cells. Int J Clin Exp Pathol. 2018;11:634–641. [PMC free article] [PubMed] [Google Scholar]
- 30.Teng X, Fan XF, Li Q, Liu S, Wu DY, Wang SY, Shi Y, Dong M. XPC inhibition rescues cisplatin resistance via the Akt/mTOR signaling pathway in A549/DDP lung adenocarcinoma cells. Oncol Rep. 2019;41:1875–1882. doi: 10.3892/or.2019.6959. [DOI] [PubMed] [Google Scholar]
- 31.Liang SQ, Bührer ED, Berezowska S, Marti TM, Xu D, Froment L, Yang H, Hall SRR, Vassella E, Yang Z, et al. mTOR mediates a mechanism of resistance to chemotherapy and defines a rational combination strategy to treat KRAS-mutant lung cancer. Oncogene. 2019;38:622–636. doi: 10.1038/s41388-018-0479-6. [DOI] [PubMed] [Google Scholar]
- 32.Algire C, Amrein L, Bazile M, David S, Zakikhani M, Pollak M. Diet and tumor LKB1 expression interact to determine sensitivity to anti-neoplastic effects of metformin in vivo. Oncogene. 2011;30:1174–1182. doi: 10.1038/onc.2010.483. [DOI] [PubMed] [Google Scholar]
- 33.Moro M, Caiola E, Ganzinelli M, Zulato E, Rulli E, Marabese M, Centonze G, Busico A, Pastorino U, de Braud FG, et al. Metformin enhances cisplatin-induced apoptosis and prevents resistance to cisplatin in Co-mutated KRAS/LKB1 NSCLC. J Thorac Oncol. 2018;13:1692–1704. doi: 10.1016/j.jtho.2018.07.102. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bremang M, Cuomo A, Agresta AM, Stugiewicz M, Spadotto V, Bonaldi T. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol Biosyst. 2013;9:2231–2247. doi: 10.1039/c3mb00009e. [DOI] [PubMed] [Google Scholar]
- 37.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 38.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database Issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database Issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Körbel C, Laschke MW, Gimotty PA, Philipp SE, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1Bhigh cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mogami T, Yokota N, Asai-Sato M, Yamada R, Koizume S, Sakuma Y, Yoshihara M, Nakamura Y, Takano Y, Hirahara F, et al. Annexin A4 is involved in proliferation, chemo-resistance and migration and invasion in ovarian clear cell adenocarcinoma cells. PLoS One. 2013;8:e80359. doi: 10.1371/journal.pone.0080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N, Huang HQ, Zhang GS. Association between SOD2 C47T polymorphism and lung cancer susceptibility: A meta-analysis. Tumor Biol. 2014;35:955–959. doi: 10.1007/s13277-013-1127-y. [DOI] [PubMed] [Google Scholar]
- 45.Morales DR, Morris AD. Metformin in cancer treatment and prevention. Annu Rev Med. 2015;66:17–29. doi: 10.1146/annurev-med-062613-093128. [DOI] [PubMed] [Google Scholar]
- 46.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Sharma A, Bandyopadhayaya S, Chowdhury K, Sharma T, Maheshwari R, Das A, Chakrabarti G, Kumar V, Mandal CC. Metformin exhibited anticancer activity by lowering cellular cholesterol content in breast cancer cells. PLoS One. 2019;14:e0209435. doi: 10.1371/journal.pone.0209435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JO, Kang MJ, Byun WS, Kim SA, Seo IH, Han JA, Moon JW, Kim JH, Kim SJ, Lee EJ, et al. Metformin overcomes resistance to cisplatin in triple-negative breast cancer (TNBC) cells by targeting RAD51. Breast Cancer Res. 2019;21:115. doi: 10.1186/s13058-019-1204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dang JH, Jin ZJ, Liu XJ, Hu D, Wang J, Luo Y, Li LL. Metformin in combination with cisplatin inhibits cell viability and induces apoptosis of human ovarian cancer cells by inactivating ERK 1/2. Oncol Lett. 2017;14:7557–7564. doi: 10.3892/ol.2017.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riaz MA, Sak A, Erol YB, Groneberg M, Thomale J, Stuschke M. Metformin enhances the radiosensitizing effect of cisplatin in non-small cell lung cancer cell lines with different cisplatin sensitivities. Sci Rep. 2019;9:1282. doi: 10.1038/s41598-018-38004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang JW, Zhao F, Sun Q. Metformin synergizes with rapamycin to inhibit the growth of pancreatic cancer in vitro and in vitro. Oncol Lett. 2018;15:1811–1816. doi: 10.3892/ol.2017.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin DH, Kim Y, Lee B, Han J, Kim HK, Shim YM, Kim DH. Metformin induces cell cycle arrest at the G1 phase through E2F8 suppression in lung cancer cells. Oncotarget. 2017;8:101509–101519. doi: 10.18632/oncotarget.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Queiroz EAIF, Puukila S, Eichler R, Sampaio SC, Forsyth HL, Lees SJ, Barbosa AM, Dekker RF, Fortes ZB, Khaper N. Metformin induces apoptosis and cell cycle arrest mediated by oxidative stress, AMPK and FOXO3a in MCF-7 breast cancer cells. PLoS One. 2014;9:e98207. doi: 10.1371/journal.pone.0098207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie W, Wang L, Sheng H, Qiu J, Zhang D, Zhang L, Yang F, Tang D, Zhang K. Metformin induces growth inhibition and cell cycle arrest by upregulating MicroRNA34a in renal cancer cells. Med Sci Monit. 2017;23:29–37. doi: 10.12659/MSM.898710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lundholm L, Hååg P, Zong D, Juntti T, Mörk B, Lewensohn R, Viktorsson K. Resistance to DNA-damaging treatment in non-small cell lung cancer tumor-initiating cells involves reduced DNA-PK/ATM activation and diminished cell cycle arrest. Cell Death Dis. 2013;4:e478. doi: 10.1038/cddis.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatterjee A, Mukhopadhyay S, Tung K, Patel D, Foster DA. Rapamycin-induced G1 cell cycle arrest employs both TGF-β and Rb pathways. Cancer Lett. 2015;360:134–140. doi: 10.1016/j.canlet.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Z, Peng K, Wang N, Liu HM, Hou G. Downregulation of p70S6K enhances cell sensitivity to rapamycin in esophageal squamous cell carcinoma. J Immunol Res. 2016;2016:7828916. doi: 10.1155/2016/7828916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song J, Wang X, Zhu J, Liu J. Rapamycin causes growth arrest and inhibition of invasion in human chondrosarcoma cells. J BUON. 2016;21:244–251. [PubMed] [Google Scholar]
- 59.Wang Y, Xu W, Yan Z, Zhao W, Mi J, Li J, Yan H. Metformin induces autophagy and G0/G1 phase cell cycle arrest in myeloma by targeting the AMPK/mTORC1 and mTORC2 pathways. J Exp Clin Cancer Res. 2018;37:63. doi: 10.1186/s13046-018-0731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong DS, Sun LL, Dong LX. Molecular mechanisms of LKB1 induced cell cycle arrest. Thorac Cancer. 2013;4:229–233. doi: 10.1111/1759-7714.12003. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Bao C, Mu Q, Chen J, Wang J, Mi Y, Sayari AJ, Chen Y, Guo M. Reversal of cisplatin resistance by inhibiting PI3K/Akt signal pathway in human lung cancer cells. Neoplasma. 2016;63:362–370. doi: 10.4149/304_150806N433. [DOI] [PubMed] [Google Scholar]
- 62.Sheng J, Shen L, Sun L, Zhang X, Cui R, Wang L. Inhibition of PI3K/mTOR increased the sensitivity of hepatocellular carcinoma cells to cisplatin via interference with mitochondrial-lysosomal crosstalk. Cell Prolif. 2019;52:e12609. doi: 10.1111/cpr.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hou G, Yang S, Zhou Y, Wang C, Zhao W, Lu Z. Targeted inhibition of mTOR signaling improves sensitivity of esophageal squamous cell carcinoma cells to cisplatin. J Immunol Res. 2014;2014:845763. doi: 10.1155/2014/845763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Y, Sun H, Feng M, Zhao J, Zhao X, Wan Q, Cai D. Metformin is associated with reduced cell proliferation in human endometrial cancer by inbibiting PI3K/AKT/mTOR signaling. Gynecol Endocrinol. 2018;34:428–432. doi: 10.1080/09513590.2017.1409714. [DOI] [PubMed] [Google Scholar]
- 65.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 66.Galluzzi L, Vitale I, Michels J, Brenner C, Szabadkai G, Harel-Bellan A, Castedo M, Kroemer G. Systems biology of cisplatin resistance: Past, present and future. Cell Death Dis. 2014;5:e1257. doi: 10.1038/cddis.2013.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 68.Chang X, Ravi R, Pham V, Bedi A, Chatterjee A, Sidransky D. Adenylate kinase 3 sensitizes cells to cigarette smoke condensate vapor induced cisplatin resistance. PLoS One. 2011;6:e20806. doi: 10.1371/journal.pone.0020806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei Y, Wu S, Xu W, Liang Y, Li Y, Zhao W, Wu J. Depleted aldehyde dehydrogenase 1A1 (ALDH1A1) reverses cisplatin resistance of human lung adenocarcinoma cell A549/DDP. Thorac Cancer. 2017;8:26–32. doi: 10.1111/1759-7714.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morimoto A, Serada S, Enomoto T, Kim A, Matsuzaki S, Takahashi T, Ueda Y, Yoshino K, Fujita M, Fujimoto M, et al. Annexin A4 induces platinum resistance in a chloride-and calcium-dependent manner. Oncotarget. 2014;5:7776–7787. doi: 10.18632/oncotarget.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng X, Liu H, Zhang Z, Gu Y, Qiu H, He Z. Annexin A2 contributes to cisplatin resistance by activation of JNK-p53 pathway in non-small cell lung cancer cells. J Exp Clin Cancer Res. 2017;36:123. doi: 10.1186/s13046-017-0594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Becker M, De Bastiani MA, Müller CB, Markoski MM, Castro MA, Klamt F. High cofilin-1 levels correlate with cisplatin resistance in lung adenocarcinomas. Tumor Biology. 2014;35:1233–1238. doi: 10.1007/s13277-013-1164-6. [DOI] [PubMed] [Google Scholar]
- 73.Wittig R, Nessling M, Will RD, Mollenhauer J, Salowsky R, Münstermann E, Schick M, Helmbach H, Gschwendt B, Korn B, et al. Candidate genes for cross-resistance against DNA-damaging drugs. Cancer Res. 2002;62:6698–6705. [PubMed] [Google Scholar]
- 74.Zeller C, Dai W, Steele NL, Siddiq A, Walley AJ, Wilhelm-Benartzi CS, Rizzo S, van der Zee A, Plumb JA, Brown R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene. 2012;31:4567–4576. doi: 10.1038/onc.2011.611. [DOI] [PubMed] [Google Scholar]
- 75.Yang M, Li Y, Shen X, Ruan Y, Lu Y, Jin X, Song P, Guo Y, Zhang X, Qu H, et al. CLDN6 promotes chemoresistance through GSTP1 in human breast cancer. J Exp Clin Cancer Res. 2017;36:157. doi: 10.1186/s13046-017-0627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang R, Tao F, Ruan S, Hu M, Hu Y, Fang Z, Mei L, Gong C. The TGFβ1-FOXM1-HMGA1-TGFβ1 positive feedback loop increases the cisplatin resistance of non-small cell lung cancer by inducing G6PD expression. Am J Transl Res. 2019;11:6860–6876. [PMC free article] [PubMed] [Google Scholar]
- 77.Wang JM, Liu BQ, Zhang Q, Hao L, Li C, Yan J, Zhao FY, Qiao HY, Jiang JY, Wang HQ. ISG15 suppresses translation of ABCC2 via ISGylation of hnRNPA2B1 and enhances drug sensitivity in cisplatin resistant ovarian cancer cells. Biochim Biophys Acta Mol Cell Res. 2020;1867:118647. doi: 10.1016/j.bbamcr.2020.118647. [DOI] [PubMed] [Google Scholar]
- 78.Di Martino S, Amoreo CA, Nuvoli B, Galati R, Strano S, Facciolo F, Alessandrini G, Pass HI, Ciliberto G, Blandino G, et al. HSP90 inhibition alters the chemotherapy-driven rearrangement of the oncogenic secretome. Oncogene. 2018;37:1369–1385. doi: 10.1038/s41388-017-0044-8. [DOI] [PubMed] [Google Scholar]
- 79.Qin X, Sun L, Wang J. Restoration of microRNA-708 sensitizes ovarian cancer cells to cisplatin via IGF2BP1/Akt pathway. Cell Biol Int. 2017;41:1110–1118. doi: 10.1002/cbin.10819. [DOI] [PubMed] [Google Scholar]
- 80.Xu Z, Zou L, Ma G, Wu X, Huang F, Feng T, Li S, Lin Q, He X, Liu Z, Cao X. Integrin β1 is a critical effector in promoting metastasis and chemo-resistance of esophageal squamous cell carcinoma. Am J Cancer Res. 2017;7:531–542. [PMC free article] [PubMed] [Google Scholar]
- 81.Li Y, Liu X, Lin X, Zhao M, Xiao Y, Liu C, Liang Z, Lin Z, Yi R, Tang Z, et al. Chemical compound cinobufotalin potently induces FOXO1-stimulated cisplatin sensitivity by antagonizing its binding partner MYH9. Signal Transduct Target Ther. 2019;4:48. doi: 10.1038/s41392-019-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Depeng S, Wu J, Guo L, Xu Y, Liu L, Lu J. Metformin increases sensitivity of osteosarcoma stem cells to cisplatin by inhibiting expression of PKM2. Int J Oncol. 2017;50:1848–1856. doi: 10.3892/ijo.2017.3950. [DOI] [PubMed] [Google Scholar]
- 83.Zhu H, Wu J, Zhang W, Luo H, Shen Z, Cheng H, Zhu X. PKM2 enhances chemosensitivity to cisplatin through interaction with the mTOR pathway in cervical cancer. Sci Rep. 2016;6:30788. doi: 10.1038/srep30788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y, Hao F, Nan Y, Qu L, Na W, Jia C, Chen X. PKM2 inhibitor shikonin overcomes the cisplatin resistance in bladder cancer by inducing necroptosis. International J Biol Sci. 2018;14:1883–1891. doi: 10.7150/ijbs.27854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krafft U, Tschirdewahn S, Hess J, Harke NN, Hadaschik BA, Nyirády P, Szendröi A, Szücs M, Módos O, Olah C, et al. STIP1 tissue expression is associated with survival in chemotherapy-treated bladder cancer patients. Pathol Oncol Res. 2020;26:1243–1249. doi: 10.1007/s12253-019-00689-y. [DOI] [PubMed] [Google Scholar]
- 86.Li C, Cai J, Ge F, Wang G. TGM2 knockdown reverses cisplatin chemoresistance in osteosarcoma. Int J Molr Med. 2018;42:1799–1808. doi: 10.3892/ijmm.2018.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang H, Wu XL, Wu KH, Zhang R, Ju LL, Ji Y, Zhang YW, Xue SL, Zhang YX, Yang YF, Yu MM. MicroRNA-497 regulates cisplatin chemosensitivity of cervical cancer by targeting transketolase. Am J Cancer Res. 2016;6:2690–2699. [PMC free article] [PubMed] [Google Scholar]
- 88.Matassa DS, Amoroso MR, Lu H, Avolio R, Arzeni D, Procaccini C, Faicchia D, Maddalena F, Simeon V, Agliarulo I, et al. Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ. 2016;23:1542–1554. doi: 10.1038/cdd.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yue T, Zheng X, Dou Y, Zheng X, Sun R, Tian Z, Wei H. Interleukin 12 shows a better curative effect on lung cancer than paclitaxel and cisplatin doublet chemotherapy. BMC Cancer. 2016;16:665. doi: 10.1186/s12885-016-2701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barros FBA, Assao A, Garcia NG, Nonogaki S, Carvalho AL, Soares FA, Kowalski LP, Oliveira DT. Moesin expression by tumor cells is an unfavorable prognostic biomarker for oral cancer. BMC Cancer. 2018;18:53. doi: 10.1186/s12885-017-3914-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alam F, Mezhal F, El Hasasna H, Nair VA, Aravind SR, Saber Ayad M, El-Serafi A, Abdel-Rahman WM. The role of p53-microRNA 200-Moesin axis in invasion and drug resistance of breast cancer cells. Tumor Bio. 2017;39:1010428317714634. doi: 10.1177/1010428317714634. [DOI] [PubMed] [Google Scholar]
- 92.Wang Q, Lu X, Zhao S, Pang M, Wu X, Wu H, Hoffman RM, Yang Z, Zhang Y. Moesin expression is associated with glioblastoma cell proliferation and invasion. Anticancer Res. 2017;37:2211–2218. doi: 10.21873/anticanres.11556. [DOI] [PubMed] [Google Scholar]
- 93.Wang J, Gao Q, Wang D, Wang Z, Hu C. Metformin inhibits growth of lung adenocarcinoma cells by inducing apoptosis via the mitochondria-mediated pathway. Oncol Lett. 2015;10:1343–1349. doi: 10.3892/ol.2015.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: Regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blandin AF, Renner G, Lehmann M, Lelong-Rebel I, Martin S, Dontenwill M. β1 integrins as therapeutic targets to disrupt hallmarks of cancer. Front Pharmacol. 2015;6:279. doi: 10.3389/fphar.2015.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ju L, Zhou C, Li W, Yan L. Integrin beta1 over-expression associates with resistance to tyrosine kinase inhibitor gefitinib in non-small cell lung cancer. J Cel Biochem. 2010;111:1565–1574. doi: 10.1002/jcb.22888. [DOI] [PubMed] [Google Scholar]
- 97.Kanda R, Kawahara A, Watari K, Murakami Y, Sonoda K, Maeda M, Fujita H, Kage M, Uramoto H, Costa C, et al. Erlotinib resistance in lung cancer cells mediated by integrin β1/Src/Akt-driven bypass signaling. Cancer Res. 2013;73:6243–6253. doi: 10.1158/0008-5472.CAN-12-4502. [DOI] [PubMed] [Google Scholar]
- 98.Gachechiladze M, Skarda J, Janikova M, Mgebrishvili G, Kharaishvili G, Kolek V, Grygarkova I, Klein J, Poprachova A, Arabuli M, Kolar Z. Overexpression of filamin-A protein is associated with aggressive phenotype and poor survival outcomes in NSCLC patients treated with platinum-based combination chemotherapy. Neoplasma. 2016;63:274–281. doi: 10.4149/214_150224N108. [DOI] [PubMed] [Google Scholar]
- 99.Ji ZM, Yang LL, Ni J, Xu SP, Yang C, Duan P, Lou LP, Ruan QR. Silencing filamin a inhibits the invasion and migration of breast cancer cells by up-regulating 14-3-3σ. Curr Med Sci. 2018;38:461–466. doi: 10.1007/s11596-018-1901-6. [DOI] [PubMed] [Google Scholar]
- 100.Li L, Lu Y, Stemmer PM, Chen F. Filamin A phosphorylation by Akt promotes cell migration in response to arsenic. Oncotarget. 2015;6:12009–12019. doi: 10.18632/oncotarget.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo Y, Li M, Bai G, Li X, Sun Z, Yang J, Wang L, Sun J. Filamin a inhibits tumor progression through regulating BRCA1 expression in human breast cancer. Oncol Lett. 2018;16:6261–6266. doi: 10.3892/ol.2018.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Donadon M, Di Tommaso L, Soldani C, Franceschini B, Terrone A, Mimmo A, Vitali E, Roncalli M, Lania A, Torzilli G. Filamin A expression predicts early recurrence of hepatocellular carcinoma after hepatectomy. Liver Int. 2018;38:303–311. doi: 10.1111/liv.13522. [DOI] [PubMed] [Google Scholar]
- 103.Maynadier M, Farnoud R, Lamy PJ, Laurent-Matha V, Garcia M, Rochefort H. Cathepsin D stimulates the activities of secreted plasminogen activators in the breast cancer acidic environment. Int J Oncol. 2013;43:1683–1690. doi: 10.3892/ijo.2013.2095. [DOI] [PubMed] [Google Scholar]
- 104.Hah YS, Noh HS, Ha JH, Ahn JS, Hahm JR, Cho HY, Kim DR. Cathepsin D inhibits oxidative stress-induced cell death via activation of autophagy in cancer cells. Cancer Lett. 2012;323:208–214. doi: 10.1016/j.canlet.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 105.Zhang C, Zhang M, Song S. Cathepsin D enhances breast cancer invasion and metastasis through promoting hepsin ubiquitin-proteasome degradation. Cancer Lett. 2018;438:105–115. doi: 10.1016/j.canlet.2018.09.021. [DOI] [PubMed] [Google Scholar]
- 106.Kang SW. Superoxide dismutase 2 gene and cancer risk: Evidence from an updated meta-analysis. Int J Clin Exp Med. 2015;8:14647–14655. [PMC free article] [PubMed] [Google Scholar]
- 107.Lee JH, Choi IY, Kil IS, Kim SY, Yang ES, Park JW. Protective role of superoxide dismutases against ionizing radiation in yeast. Biochim Biophys Acta. 2001;1526:191–198. doi: 10.1016/S0304-4165(01)00126-X. [DOI] [PubMed] [Google Scholar]
- 108.Epperly MW, Gretton JE, Sikora CA, Jefferson M, Bernarding M, Nie S, Greenberger JS. Mitochondrial localization of superoxide dismutase is required for decreasing radiation-induced cellular damage. Radiat Res. 2003;160:568–578. doi: 10.1667/RR3081. [DOI] [PubMed] [Google Scholar]
- 109.Takada Y, Hachiya M, Park SH, Osawa Y, Ozawa T, Akashi M. Role of reactive oxygen species in cells overexpressing manganese superoxide dismutase: Mechanism for induction of radioresistance. Mol Cancer Res. 2002;1:137–146. [PubMed] [Google Scholar]
- 110.Hosoki A, Yonekura S, Zhao QL, Wei ZL, Takasaki I, Tabuchi Y, Wang LL, Hasuike S, Nomura T, Tachibana A, et al. Mitochondria-targeted superoxide dismutase (SOD2) regulates radiation resistance and radiation stress response in HeLa cells. J Radiat Res. 2012;53:58–71. doi: 10.1269/jrr.11034. [DOI] [PubMed] [Google Scholar]
- 111.Cruz-Bermúdez A, Laza-Briviesca R, Vicente-Blanco RJ, García-Grande A, Coronado MJ, Laine-Menéndez S, Palacios-Zambrano S, Moreno-Villa MR, Ruiz-Valdepeñas AM, Lendinez C, et al. Cisplatin resistance involves a metabolic reprogramming through ROS and PGC-1α in NSCLC which can be overcome by OXPHOS inhibition. Free Radic Biol Med. 2019;135:167–181. doi: 10.1016/j.freeradbiomed.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 112.Zuo J, Zhao M, Liu B, Han X, Li Y, Wang W, Zhang Q, Lv P, Xing L, Shen H, Zhang X. TNF-α-mediated upregulation of SOD-2 contributes to cell proliferation and cisplatin resistance in esophageal squamous cell carcinoma. Oncol Rep. 2019;42:1497–1506. doi: 10.3892/or.2019.7252. [DOI] [PubMed] [Google Scholar]
- 113.Yan X, Pan L, Yuan Y, Lang JH, Mao N. Identification of platinum-resistance associated proteins through proteomic analysis of human ovarian cancer cells and their platinum-resistant sublines. J Proteome Res. 2007;6:772–780. doi: 10.1021/pr060402r. [DOI] [PubMed] [Google Scholar]
- 114.Matsuzaki S, Enomoto T, Serada S, Yoshino K, Nagamori S, Morimoto A, Yokoyama T, Kim A, Kimura T, Ueda Y, et al. Annexin A4-conferred platinum resistance is mediated by the copper transporter ATP7A. Int J Cancer. 2014;134:1796–1809. doi: 10.1002/ijc.28526. [DOI] [PubMed] [Google Scholar]
- 115.Yao HS, Sun C, Li XX, Wang Y, Jin KZ, Zhang XP, Hu ZQ. Annexin A4-nuclear factor-κB feedback circuit regulates cell malignant behavior and tumor growth in gallbladder cancer. Sci Rep. 2016;6:31056. doi: 10.1038/srep31056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yamashita T, Nagano K, Kanasaki S, Maeda Y, Furuya T, Inoue M, Nabeshi H, Yoshikawa T, Yoshioka Y, Itoh N, et al. Annexin A4 is a possible biomarker for cisplatin susceptibility of malignant mesothelioma cells. Biochem Biophys Res Commun. 2012;421:140–144. doi: 10.1016/j.bbrc.2012.03.144. [DOI] [PubMed] [Google Scholar]
- 117.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Additionally, the proteomic datasets generated and/or analyzed during the current study are available in the PRIDE repository (https://www.ebi.ac.uk/pride/). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (117) with the dataset identifier PXD017645 (https://www.ebi.ac.uk/pride/archive/projectPXD017645).