Abstract
Despite considerable research efforts, pancreatic cancer is associated with a dire prognosis and a 5-year survival rate of only 10%. Early symptoms of the disease are mostly nonspecific. The premise of improved survival through early detection is that more individuals will benefit from potentially curative treatment. Artificial intelligence (AI) methodology has emerged as a successful tool for risk stratification and identification in general health care. In response to the maturity of AI, Kenner Family Research Fund conducted the 2020 AI and Early Detection of Pancreatic Cancer Virtual Summit (www.pdac-virtualsummit.org) in conjunction with the American Pancreatic Association, with a focus on the potential of AI to advance early detection efforts in this disease. This comprehensive presummit article was prepared based on information provided by each of the interdisciplinary participants on one of the 5 following topics: Progress, Problems, and Prospects for Early Detection; AI and Machine Learning; AI and Pancreatic Cancer—Current Efforts; Collaborative Opportunities; and Moving Forward—Reflections from Government, Industry, and Advocacy. The outcome from the robust Summit conversations, to be presented in a future white paper, indicate that significant progress must be the result of strategic collaboration among investigators and institutions from multidisciplinary backgrounds, supported by committed funders.
Key Words: artificial intelligence, machine learning, pancreatic cancer, early detection
Pancreatic cancer (pancreatic ductal adenocarcinoma [PDAC]) is associated with a dire prognosis and a 5-year survival rate of only 10%.1 This statistic is somewhat misleading given that 52% of the patients will develop metastatic disease, with a resulting 2.9%, 5-year relative survival rate. However, for those patients with localized cancer where the tumor is confined to the primary site, the 5-year relative survival rate is 39.4%. It is estimated that in 2020, there will be 57,600 new cases of PDAC and an estimated 47,050 will die of this disease.1
Early symptoms of PDAC are mostly nonspecific, with both intrinsic and extrinsic risk factors believed to be involved.2 The premise of improved survival through early detection is that more individuals will benefit from potentially curative treatment. Because symptoms typically occur late in the course of the disease, detection of early/resectable pancreatic cancer will possibly require screening asymptomatic subjects. Although it remains cost-prohibitive and challenging with current technology to screen the general population for PDAC, the ability to define high-risk groups with an increased likelihood of harboring such lesions may lead to earlier interception and improved survival.
To address this need, Kenner Family Research Fund conducted the 2014 Early Detection of Sporadic Pancreatic Cancer Summit (www.kennerfamilyresearchfund.org/early-detection-sporadic-pancreatic-cancer-summit-conference/). This seminal meeting convened international representatives from science, practice, clinical research, and government and was presented in conjunction with the 45th Anniversary Joint Meeting of the American Pancreatic Association and Japan Pancreas Society. Four distinct panels of experts prepared presummit analyses in a foundational article on Case for Early Detection: Definitions, Detection, Survival, and Challenges; Biomarkers for Early Detection; Imaging; and Collaborative Studies.3 Familial PDAC emerged as a separate theme.4 Substantial material was provided via this in-depth review of the state of the science to inform each invited expert as he/she planned for involvement in the Summit's interdisciplinary conversations.
The subsequent Summit debate and vigorous discussions resulted in a shared vision for the future of early detection of pancreatic cancer and defined parameters for a new pathway.5 The Strategic Map for Innovation illustrated the pathway and included the primary factors necessary for successful innovation (Fig. 1).
FIGURE 1.
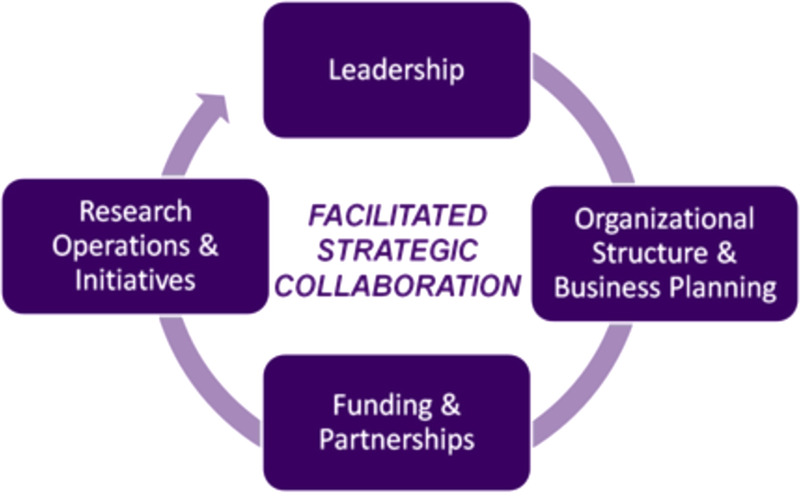
Strategic map for innovation (© Kenner Family Research Fund, 2015).
Four congruent priorities were indicated in the integrated model: leadership, organizational structure and business planning, funding and partnerships, and research operations and initiatives. The core of the model is Facilitated Strategic Collaboration to drive an accelerated pace of entrepreneurial organizational development, idea generation, significant research findings, and translation into clinical practice.
Several forums were subsequently presented by Kenner Family Research Fund, including the 2015 Early Detection of Pancreatic Cancer: Lessons Learned from Other Cancers6 and the 2016 The Role of Industry in the Development of Biomarkers.7 In 2018, the Role of Depression and Anxiety as a Precursor for Disease8,9 was the core presentation in a symposium at the American Pancreatic Association. After these initiatives and the publication of several articles for dissemination to the broader community, significant additional funding was invested in early detection research.
Despite considerable effort across the field over the past 6 years, the 5-year survival rate for PDAC remains exceedingly low. In early 2020, Kenner Family Research Fund sought an outside-the-box approach to identify additional high-risk groups for surveillance. Artificial intelligence (AI) methodology had emerged during this period as a tool for risk stratification and identification in general health care; hence, plans were initiated for the 2020 AI and Early Detection of Pancreatic Cancer Virtual Summit (www.pdac-virtualsummit.org). Participants were selected given their areas of expertise, knowledge, and/or commitment to the development of an early detection protocol for pancreatic cancer.
Against the background of a global pandemic, this comprehensive presummit article was prepared based on a synthesis of information provided by each participant. The focus was the potential of AI and how it could effectively be used to advance early detection efforts in PDAC. Each participant was asked to contribute in 1 of the following 5 topics, which organized both this article and the Summit design:
Section A: Progress, Problems, and Prospects for Early Detection
Section B: AI and Machine Learning
Section C: AI and Pancreatic Cancer—Current Efforts
Section D: Organizational Structures and Collaborative Opportunities
Section E: Moving Forward: Reflections from Government, Industry, and Advocacy
The extensive information provided via this updated comprehensive document served to prepare the participants to actively engage in strategic interdisciplinary conversations during the Summit. Expectations for the presummit article exceeded in both the scope of the information provided and in the generosity of the contributors. We are appreciative of every effort in developing this article and are looking forward to building upon this foundation.
More specifically, Progress, Problems, and Prospects for Early Detection presents the rationale for early detection along with a description of the genomic features of pancreatic cancer, the role and challenges of identifying biomarkers, use of endoscopic screening, and the importance of risk stratification to early detection.
Artificial Intelligence and Machine Learning explores the complexities of this technology, including its role in risk assessment, and understanding of human biology and disease continuum. The emergence of model-based deep learning is considered, as well as how AI has the potential to transform the practice of medicine.
Artificial Intelligence and Pancreatic Cancer—Current Efforts provides an extensive and comprehensive global overview of planned, current, or completed research initiatives that use this technology. Other critical topics covered in this section include funding support, strengths and challenges in using AI for risk stratification, data requirements, the importance of developing uniform standard operating procedures, and opportunities for near-term progress toward early detection.
Organizational Structures and Collaborative Opportunities builds on the information introduced in the previous sections, distinguishing the multiple types of data than can be used to develop and validate models to identify individuals at high risk for PDAC. It also introduces an organizational structure and conceptual approach using contributions of multidisciplinary teams and AI methodologies to provide a progressive and sustainable reduction in pancreatic cancer mortality.
Moving Forward includes reflections from government, industry, and advocacy on the need for collaborative efforts in the use of AI in early detection. It is acknowledged that additional research is being conducted in all areas and this document should not be considered an exhaustive review of all possible approaches for early detection.
Ideas generated from the presummit article and the resulting discussions will create a renewed shared vision for early detection. This article was provided to all participants before the Summit to encourage rich and productive dialogue during the October 19 and 20 meetings. The goals of the conversations include agreement upon a conceptual framework using AI and machine learning as tools for risk stratification in early detection of pancreatic cancer, establishment of communication channels to share information over time, collaboration between participants in this fast evolving area of study, and formation of strategic relationships with key stakeholders to facilitate systemic change and future action.
Innovation in research and subsequent translation to clinical practice is essential for significant advances in early detection of PDAC. Artificial intelligence and machine learning as tools for risk stratification have the potential to change the detection landscape. However, it is clear that significant progress will be the result of strategically designed collaboration among investigators and institutions from multidisciplinary backgrounds and support by committed funders.
SECTION A: PROGRESS, PROBLEMS, AND PROSPECTS OF EARLY DETECTION OF PANCREATIC CANCER
Co-leads: Suresh T. Chari, MD, and David S. Klimstra, MD
Group members: Marcia Irene Canto, MD, MHS; Christine Iacobuzio-Donahue, MD, PhD; Anirban Maitra, MBBS; and Lynn M. Matrisian, PhD, MBA
What Is “Early Pancreatic Cancer” and the Rationale for Early Detection
Pancreatic ductal adenocarcinoma (PDAC) is on track to become the number 2 cancer killer in the United States within the next decade, unless there is a major improvement in outcomes.10 There is little doubt that the relatively advanced stage at which most pancreatic cancers are diagnosed contributes to the poor survival that characterizes this disease. Breast, prostate, and colorectal cancers have all enjoyed a decrease in cancer deaths in recent years, with advances in early detection a major contributor to the decline.11 In fact, the World Health Organization assumes that earlier detection would lead to as much as a 30% greater cure rate for most cancer types (www.who.int/cancer/en/index.html). Thus, early detection holds significant promise for improving outcomes in pancreatic cancer, as seen in other major cancer types.
Pancreatic ductal adenocarcinoma has the poorest overall survival of all the major cancer types, with a 5-year relative survival rate that just reached 10%.12 This is due in part to the late stage at presentation, so that 49.6% of cases of newly diagnosed PDAC present with distant metastases, 29.1% present with regional lymph node involvement, and only 10.8% have tumors that are localized solely within the pancreas (Surveillance, Epidemiology, and End Results [SEER]-21, 2008–2017, accessed June 10, 2020). Survival rates are stage dependent: 39.4% for localized disease, 13.3% regional, and 2.9% metastatic, for an overall 5-year relative survival rate of 10.0% (SEER-18, 2010–2016, accessed June 10, 2020). If the stage distribution could be reversed, to 50% localized and 10% metastatic, survival would be more than doubled without any additional improvements in therapy.3
Surgical resection remains the only reasonable hope for “cure” from PDAC.13,14 In fact, over time, the proportion of patients with very early-stage PDAC (stages IA and IB) has increased and the survival of these patients has improved,15 such that the 5-year survival for stage IA (node-negative PDAC measuring <2 cm) is in excess of 80%.16 Note, however, that the survival for other resectable PDAC stages (stages IIA and IIB) has improved only marginally, emphasizing the need to diagnose PDAC very early; only 1.8% of the patients in this SEER analysis were diagnosed at stage I (Fig. 2).16
FIGURE 2.
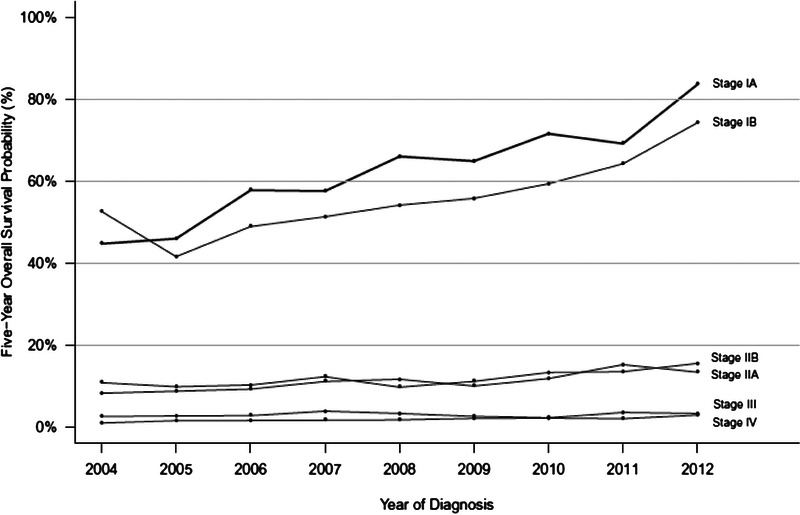
Survival rates by stage.16
Recent changes to the staging system for PDAC allow for a more precise delineation of early-stage disease (American Joint Committee on Cancer, eighth edition). Within the node-negative group undergoing surgical resection, the size of the tumor strongly correlates with outcome.17 The earliest stage (pT1N0M0) is now subdivided based on size into pT1a (<0.5 cm), pT1b (0.5–1.0 cm), and pT1c (1.0–2.0 cm) to allow for a better understanding of the size at which surgical resection has the best chance of cure (Fig. 3).
FIGURE 3.
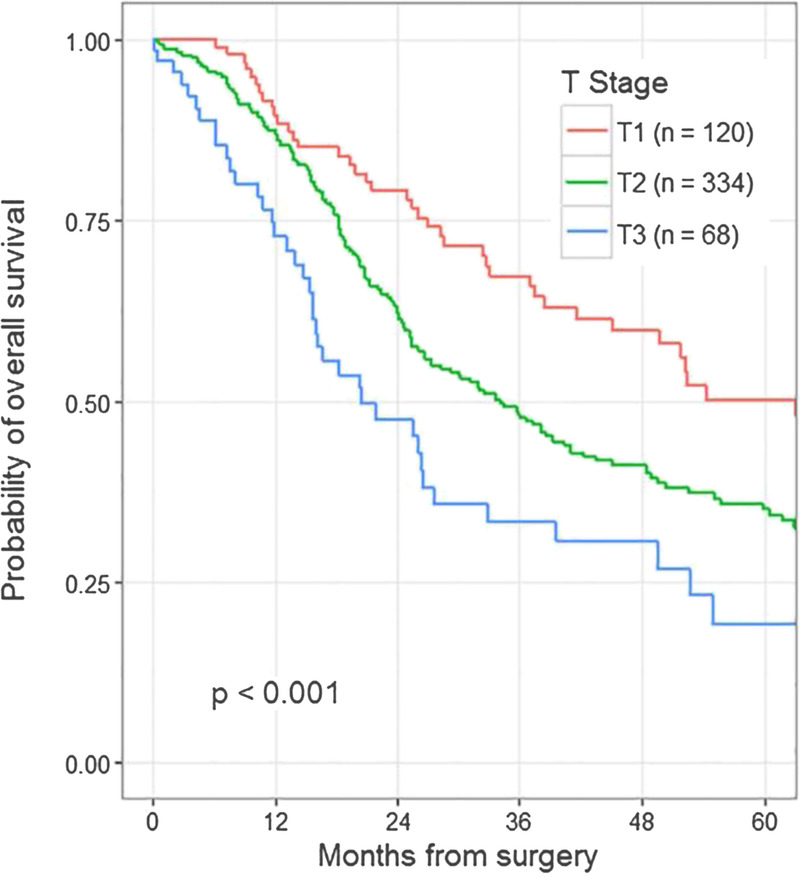
Probability of overall survival following surgery.17
Attempts to detect pancreatic cancer at an early stage that would enable surgical cure have been thwarted by the difficulty of imaging early pancreatic cancer, the lack of circulating biomarkers of early disease, the inaccessibility of the pancreas to biopsy, and the relative inability to define sufficiently high-risk populations that could benefit from screening. Defining the stage of early pancreatic cancer at which intervention would be both effective and warranted has also been challenging, and in fact, there is no accepted definition of “early pancreatic cancer.” As noted previously, small, node-negative carcinomas can be cured surgically, but many of the cases detected at this stage arise in association with a macroscopic precursor lesion such as an intraductal papillary mucinous neoplasm (IPMN),18 which has come to clinical attention due to the precursor, rather than the cancer. Precursor lesions to invasive pancreatic cancer are now well defined,19 and detection at the stage preceding the development of invasive carcinoma allows for the best opportunity for cure. Thus, understanding the phenotypes of the immediate precursors of invasive carcinoma can potentially reveal attractive targets for screening.
Most pancreatic cancer arises from a microscopic intraductal precursor lesion known as pancreatic intraepithelial neoplasia (PanIN).20 Originally graded in 3 tiers as PanIN1, PanIN2, and PanIN3, with each tier reflecting a greater degree of dysplasia, PanINs are now dichotomously graded as high grade and low grade,19 with the previous PanIN2 group now considered to be low grade. Low-grade PanINs have mucinous cells replacing the normal cuboidal ductal epithelium and demonstrate only mild to moderate abnormalities in the nuclei and architecture. High-grade PanINs show more marked architectural complexity and nuclear abnormalities. Although the terminology may suggest a sharp transition from low to high grade, PanINs often show a spectrum of cytoarchitectural atypia, which argues for a gradual transformation from a minimally dysplastic, low-risk lesion to a highly dysplasia precursor with nearly all of the features of carcinoma but invasion. Low-grade PanIN is very common. It is estimated that 40% to 75% of adults harbor low-grade PanIN,21 which is strong evidence that this lesion has a very low risk of progression to invasive carcinoma. High-grade PanIN is rarely detected in the absence of invasive carcinoma, and all types of PanINs are microscopic lesions without well-defined radiographic findings or clinical symptoms.22 The time required for progression from low-grade to high-grade PanIN is not known, and even the progression from high-grade PanIN to invasive carcinoma is rarely documented clinically and has a vague timeline. The features suggest that low-grade PanIN is not of sufficient risk to justify surgical intervention. High-grade PanIN may be the ideal stage for intervention, but it is very difficult to detect.
The other precursors to invasive carcinoma are macroscopic and cystic and can be diagnosed radiographically: IPMN and mucinous cystic neoplasm.19 Both also show a spectrum of dysplasia similar to PanINs, although there are some key genetic differences. Recognizing these neoplasms at the stage of high-grade dysplasia, before invasive carcinoma has developed, allows for timely surgical resection and may be a model for biomarker development to enable high-grade PanIN detection.
The Genomic Features of Pancreatic Cancer and Its Precursor Lesions
The genomic features of PDAC have been described over the past 2 decades.23–27 Initially, gene-focused studies identified the common driver genes of this disease and its major hereditary components.28 Subsequent large-scale sequencing studies have revealed the recurrent genomic features of this disease that target a defined number of core cellular pathways and confer genome instability.23–26
The genes somatically altered at high frequency in PDAC are KRAS, CDKN2A, TP53, and SMAD4, signifying the cellular pathways that most often are targeted during pancreatic carcinogenesis.27KRAS activation is among the earliest events known in PDAC where it signifies the transition from a normal centroacinar or ductal cell to an initiated cell.29KRAS is a 21-kDa small GTPase that activates MAPK/ERK signaling, thus controlling cellular processes relating to proliferation, differentiation, migration, and survival.30KRAS mutations are the most common oncogenic alteration in PDAC, occurring in ~90% of cases, indicating that hyperactivity of MAPK/ERK signaling is a requisite to maintain survival of the cell. Virtually all KRAS mutations in PDAC are single-nucleotide variants occurring in codons 12 (~91%), 13 (~2%), and 61 (~7%).23–26 Interestingly, the remaining 10% of PDACs are KRAS wild type; these tumors often have alternative mechanisms of activating Mapk/Erk signaling such as via mutations or fusions of oncogenes such as BRAF, GNAS, or EGFR, among others.26
Inactivation of the tumor suppressor gene CDKN2A is found in 90% of PDACs where it leads to loss of cell cycle regulation.27 In PDACs where CDKN2A is not inactivated, RB1 inactivation or hypermethylation has been identified, indicating phenotypic convergence on loss of the G1/S checkpoint. TP53 is also a tumor suppressor gene whose protein product serves as a major guardian of genome integrity. Alterations of TP53 in cancer occur in 80% of PDAC and are mainly found in DNA-binding domains, leading to gains of function via altered DNA binding and interactions with other transcription factors. Consequences of these GOF mutations include cell cycle activation and loss of apoptosis regulation. Most TP53 somatic alterations are missense mutations that confer gains of oncogenic function, although a subset of PDACs exhibit loss of TP53 expression via truncating mutations or homozygous deletion. Although TP53 plays central roles in several biochemical and/or carcinogenesis pathway including transcription, DNA repair, genomic stability, cell cycle control, and apoptosis, many of the molecular mechanisms underlying TP53's tumor suppressor function remain unclear despite more than 30 years of investigation.27 SMAD4, also a tumor suppressor, is a mediator of the canonical transforming growth factor β (TGF-β) signaling pathway that controls tissue homeostasis within the pancreas and other tissue types. Inactivation of SMAD4 occurs in just over 50% of PDACs by homozygous deletion or somatic alteration with loss of the wild-type allele. Loss of SMAD4 leads to loss of TGF-β signaling and thus facilitates epithelial-mesenchymal transition and TGF-β–dependent growth in invasive PDAC.27
Inactivating mutations in chromatin modifier genes are present in up to one-third of PDACs; however, mutations in any one gene are typically mutually exclusive of each other, indicating convergence for loss of epigenetic regulation.31 Independently, each gene is affected in <10% of PDACs.23,26 ARID1A is a member of the ATP-dependent chromatin remodeling complex SWI/SNF, which is thought to regulate transcription of genes by reconstructing chromatin and breaking its structural constraints around those genes. ARID1A itself has a DNA-binding domain that can specifically bind an AT-rich DNA recognized by a SWI/SNF complex. In cancer, ARID1A is thought to be a tumor suppressor gene and loss of ARID1A function alters genome-wide chromatin structure and regulation of transcriptions of target genes. Other chromatin modifiers such as KMT2C, KMT2D, KDM6A, ARID2, SMARCA4, or PBRM1 play important roles for genesis and progress of pancreatic cancer.
Similar mutations occur at varying frequencies in the precursors to PDAC, and although the exact sequence of mutations is not always predictable, there is an accumulation of mutations through the morphological spectrum from low to high grade. Telomere shortening and KRAS mutations usually occur in low-grade PanINs. Inactivation of p16/CDKN2A is later, and alterations in TP53, SMAD4, and BRCA2 occur in high-grade PanIN. However, studies to define the genomic make-up of high-grade PanIN have been hampered by the rarity with which high-grade PanIN is detected in the absence of an invasive carcinoma component. Because invasive carcinoma has a propensity to colonize the pancreatic ducts and the resulting morphology closely resembles that of high-grade PanIN, it is difficult to interpret genomic studies of high-grade PanIN that relied on samples also containing an invasive carcinoma. The macroscopic precursors to PDAC also accumulate mutations in similar genes but additionally have mutations in GNAS and/or RNF43, which are rarely involved in conventional PDAC development.
Detection of Biomarkers for Pancreatic Cancer Diagnosis
Despite the plethora of published research studies on PDAC biomarkers, carbohydrate antigen (CA) 19-9, first identified in 1979, remains the only US Food and Drug Administration (FDA)–approved biomarker for diagnosis and monitoring of this disease. Unfortunately, CA 19-9 carries an overall sensitivity in the range of 25% to 50% in early-stage disease, and conversely, the levels of CA 19-9 can be elevated in nonneoplastic conditions, such as benign biliary obstruction.32,33 In addition, 5% to 10% of the population lack the genes encoding the Lewis blood group antigen, which then undergoes modification into CA 19-9. Thus, identifying credentialed biomarkers for early detection of PDAC remains an area of great unmet need. At the same time, in 2019, the US Prevention and Screening Task Force explicitly recommended against screening for PDAC in the general population.34 This recommendation is based on the relative infrequency of PDAC in the general population (~13 cases per 100,000) and the potential for identifying a large number of false-positives, even with a relatively “perfect” biomarker, let alone CA 19-9. In light of this, the US Prevention and Screening Task Force instead recommends that any screening efforts be focused on well-defined high-risk cohorts, such as patients with germline mutation in PDAC predisposition genes. Other recognized high-risk PDAC cohorts include patients with mucinous pancreatic cysts and adults with new-onset diabetes (NOD).35 Prospective biomarker validation efforts should ideally be focused in such high-risk cohorts, before extrapolating to the general population-at-large.
There are 5 well-recognized phases of cancer biomarker discovery elaborated by Pepe and colleagues,36 with the overwhelming majority of published data at phase 1 (biomarker discovery) and phase 2 (validation in symptomatic disease), typically using “convenience” blood samples collected from patients who present with symptomatic disease to a medical center (Fig. 4).
FIGURE 4.
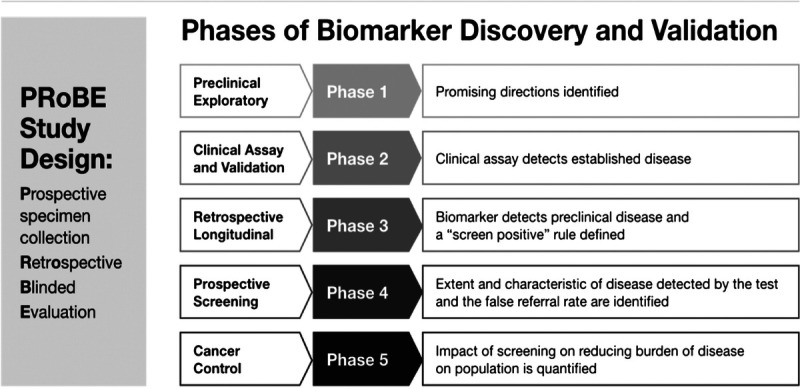
Phases of biomarker discovery and validation.7
In contrast, biomarker performance in presymptomatic samples obtained from a cohort of individuals before subsequent cancer diagnosis (ie, a phase 3 study) is not commonly evaluated. Such retrospective sample cohorts are challenging to obtain, although repositories such as the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial,37 Women's Health Initiative,38 and the UK Biobank39 represent invaluable resources for these samples. Phase 4 studies involve demonstration of stage shift or survival benefit in a statistically meaningful prospective cohort where the validated biomarker (or panel) is used as a screening tool for clinical decision making. Given this expensive and tortuous road to cancer biomarker approval, the paucity of markers beyond CA 19-9 is not surprising, although the landscape is gradually evolving with the advent of newer platforms and private-public partnerships.
Although a comprehensive review of circulating biomarkers that have been evaluated in early detection of PDAC is beyond the scope of this article, we will highlight some of the most promising results that have emerged over the past few years. Pancreatic ductal adenocarcinoma cells secrete a large number of aberrant proteins, and these can be identified in the circulation in quantities that are significantly higher than levels observed in otherwise healthy controls, or patients with nonneoplastic pancreatic diseases (eg, chronic pancreatitis). Such biomarker “panels” often build upon CA 19-9 and demonstrate improved performance in early-stage disease compared with CA 19-9 alone.40–43 In contrast to pan-cancer genomic markers (see discussion hereinafter), protein biomarkers have the potential to be cancer-specific and, when used in conjunction with genomic assays, provide both greater sensitivity and putative “organ-of-origin” information.44 Another class of markers is autoantibodies, either free in circulation or complexed with proteins, which can be detected using antigen arrays.45 Of interest, these PDAC autoantibodies are often targeted against exosomal surface proteins, and exosomes might represent a “decoy” function by binding to these autoantibodies and diminishing the humoral immune response against cancer cells.46
Cell-free DNA, which includes the entire compendium of circulating DNA, including shed DNA from nonneoplastic cells (mostly from the bone marrow compartment) and tumor-derived circulating tumor DNA (ctDNA), has emerged as an important tool in the cancer early-diagnosis armamentarium.47 In the context of early detection, detection of somatic mutations in ctDNA by next-generation sequencing has demonstrated exceptionally high specificity but lacks sensitivity in early-stage disease.48,49 This limit of detection is not a technical flaw, but simply represents the absence of sufficient shed ctDNA in many early-stage PDAC cases. Thus, some investigators have taken the approach of combining next-generation sequencing on ctDNA with protein-based markers, marrying the sensitivity of latter with the specificity of the former.50 Although detection of somatic mutations in ctDNA has been most commonly studied in PDAC, other cell-free approaches, including the detection of circulating methylated DNA and nucleosomal fragments, have all shown preclinical utility. Nucleosomal fragment size differs between healthy controls and patients with cancer and provides a mutation-free approach to cancer detection.50 Of note, in addition to cell-free DNA, recent studies have also shown that detection of nucleic acid cargo within circulating extracellular microvesicles (including exosomes) provides an alternative avenue for assessing the genomic landscape of PDAC.51,52 The challenge remains in translating these encouraging preclinical findings (mostly in aforementioned phases 1 and 2) into the next phases of cancer biomarker discovery and eventual regulatory approval.
In passing, it also needs to be stated that, although blood remains the most commonly studied biospecimen for early detection, many of the analytes are also present in other proximate samples, including pancreatic juice,53,54 stool,55 urine,56,57 saliva,58 and pancreatic cyst fluid.59 In the context of mucinous pancreatic cysts, cyst fluid has been demonstrated to be an attractive substrate for demonstrating aberrant somatic mutations, microRNAs or proteins, which are then correlated with either the presence of mucinous epithelium per se or the progression to high-grade dysplasia and cancer.60,61
The Case for Risk Stratification of Pancreatic Cancer
It is recognized that approximately 10% of PDAC cases have a familial predisposition. Of these, one-fourth are due to germline mutations in a known pancreatic cancer gene (Table 1); the remainder have familial clustering without a known genetic basis. Many of the pancreatic cancer risk genes have low penetrance. Patients with hereditary PDAC average 5 to 8 years younger at diagnosis than those without a family history. Histologically, most hereditary PDACs are conventional, and both PanIN and IPMNs are found to be associated with PDACs (Table 1).
TABLE 1.
Risk of Pancreatic Cancer in Hereditary Syndromes
| % of Families | Increased Risk | Age 50 y, % | Age 70 y, % | |
|---|---|---|---|---|
| No history | — | 1 | 0.05% | 0.5 |
| Hereditary nonpolyposis colorectal cancer | ? | 8 | 1 | 3.7 |
| BRCA2 (breast-ovarian) | 6–12 | 3.5–10 | 0.5–2 | 5 |
| PALB2 | 3 | ? | ? | ? |
| Familial atypical multiple mole melanoma (p16) | 1–3 | 20–34 | 1 | 10–17 |
| Familial pancreatitis (PRSS1) | <1 | 50–80 | 2.5 | 25–40 |
| Peutz-Jeghers (STK11/LKB1) | <1 | 132 | 6.6 | 30–60 |
| ATM | <2 | ? | ? | ? |
Data extracted from https://seer.cancer.gov/statfacts/html/pancreas.html.
Familial PDAC cases without a known genetic basis are variably defined as having 2 or 3 first-degree relatives (FDRs) with PDAC, which confers a 6× to 30× increased risk of the disease. Studies are ongoing to define the basis for their inheritance. Comprehensive genomic analysis of the germline from PDAC patients is also revealing unexpected germline mutations in known cancer predisposition genes, especially in patients with a family history of other cancers. Although individuals in families with hereditary PDAC represent an attractive population to screen for early pancreatic cancer, these families are relatively uncommon and account for a rather small proportion of PDACs overall.
It is accepted that PDAC's low prevalence makes population screening unrealistic. Through risk stratification, one can enrich the target population for PDAC. Here we define various levels of risk and the associated prevalence of PDAC in that population.
Baseline Risk
Calculated from SEER data (https://seer.cancer.gov/statfacts/html/pancreas.html), the age-adjusted incidence of PDAC in US subjects 50 years or older is 37/100,000 per year, or 0.037%. Assuming a biomarker/imaging study could identify PDAC up to 3 years before its diagnosis, the 3-year incidence or number of potentially identifiable PDAC in 100,000 subjects (prevalence) over 3 years is 111/100,000, or 0.11%. Thus, 0.11% is the baseline 3-year risk of having PDAC in subjects older than 50 years (Fig. 5).
FIGURE 5.
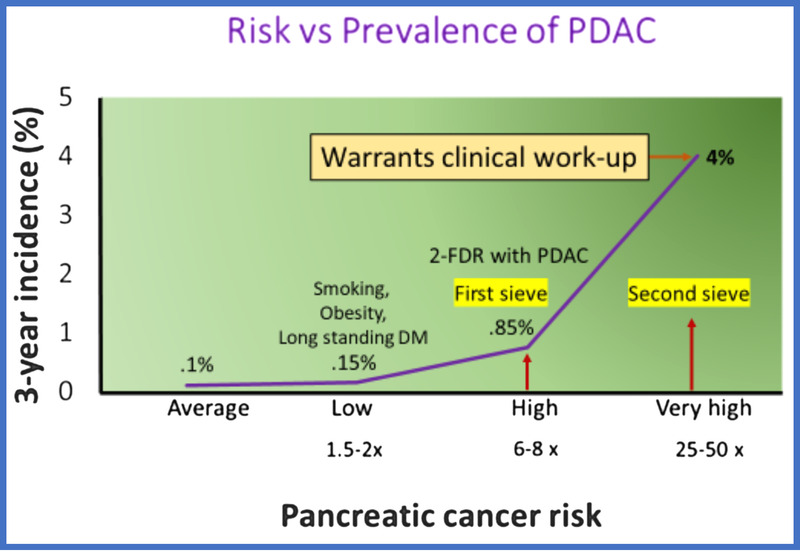
Risk versus prevalence of PDAC. Courtesy of Suresh Chari.
Low-Risk Group
These cohorts have a risk slightly higher than baseline, that is, 1.5 to 3× higher than baseline (0.2%–0.3% 3-year risk). Examples include long-standing diabetes, smoking, and obesity.62 Some studies on NOD using emergency medical response diagnosis codes to identify NOD have found NOD to have a low risk.63
Modest Risk Groups
These cohorts have 3 to 6× higher risk (0.35%–66%). Depending on how NOD is studied or defined, some studies have reported risk in this range in NOD.63
High-Risk Group
These groups have 6 to 10× higher risk (0.67%–1.0% absolute risk). The risk of PDAC in this group is very comparable to the risk of lung cancer in subjects with a 20-pack-year history of smoking,64 colon cancer in subjects older than 50 years,65 or mammography in subjects older than 50 year.66 Studies have shown this group to be cost-effective to screen. Subjects with 2 FDRs with PDAC have a lifetime risk that falls in this category. New-onset diabetes defined by glycemic criteria also has this risk.67
Intermediate High-Risk Group
These groups have 10 to 25× higher risk. Subjects with NOD and Enriching New-onset Diabetes for Pancreatic Cancer (ENDPAC) score of >0 fall in this category.68 Currently, they are the targets for screening in the Early Detection Initiative.
Very High-Risk Group
With a risk of PDAC of 3% to 4%, this group's risk is high enough for triggering a clinical work-up for PDAC. The risk in this cohort compared with baseline is at least 25 to 50× higher. New-onset diabetes cohort with an ENDPAC score of ≥3 falls in this category.68
The DEF Approach to PDAC Screening
Because the baseline risk of PDAC even in subjects older than 50 years is very low, a 3-step (DEF) approach to its early detection has been suggested6: (1) Define a high-risk group for pancreatic cancer, (2) Enrich the high-risk group further to define a very high-risk group for pancreatic cancer, and (3) Find the lesion in the very high-risk cohort (Fig. 6).
FIGURE 6.
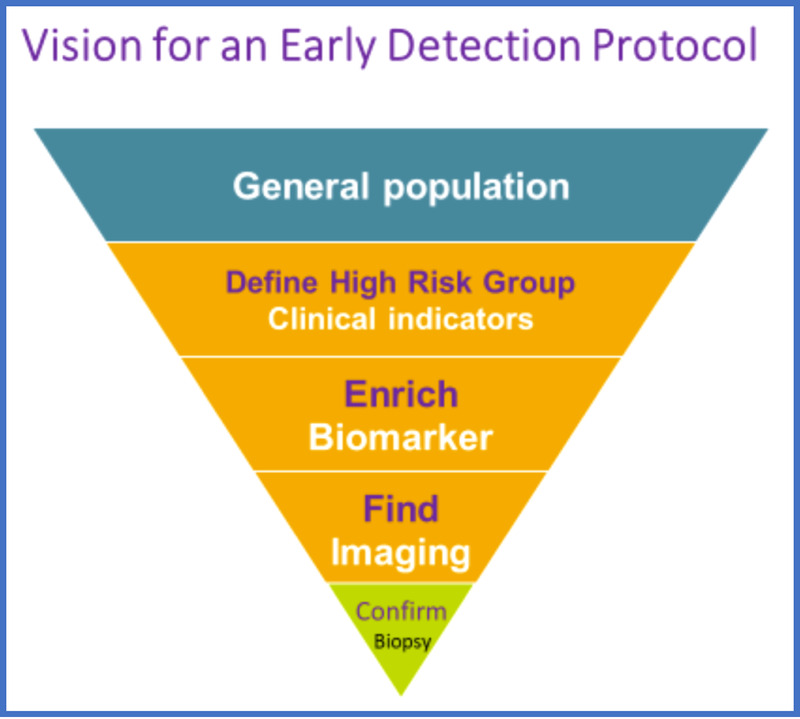
Vision for an early detection protocol. Modified from Kenner et al.6
Approach to Defining High-Risk Groups
Defining high-risk group (HRG) and very high-risk group (vHRG) using clinical indicators: in the familial cancer setting, this has been done by counting the number of FDRs affected by PDAC, with 2 affected FDRs defining an HRG and 3 FDRs defining a vHRG.69 Another approach has been to consider mutation carriers in genetic syndromes known to cause PDAC as an HRG.70 For sporadic pancreatic cancer, there is currently only one HRG and that is glycemically defined NOD.68,71 As noted previously, other methods of ascertaining NOD have a significantly lower risk of PDAC. The ENDPAC score further risk-stratifies NOD into below-average-risk, modest-risk group, and vHRG based on age, rapidity of rise of glucose, and change in weight in the 12 months before NOD date.68
Using serum biomarkers to define HRG and vHRG: because NOD occurs only in 20% of PDAC and can be difficult to ascertain, there has been considerable interest in defining an HRG and a vHRG using serum biomarkers. This heavily depends on their sensitivity in the prediagnostic stage of PDAC, a yet unknown performance characteristic. Biomarker performance fades rapidly as we go farther away from clinical diagnosis (say >12 months of lead time). Hence, the biomarker performance will depend on lead time distribution of the prediagnostic samples being analyzed and a lead time–adjusted performance needs to be calculated. Based on sensitivity and specificity, one could develop either a single highly specific (99%), modestly sensitive (40%–50%) biomarker to define a vHRG or tandem biomarkers that could define an HRG and a vHRG in sequential testing.
Imaging to “Find” the Lesion
The success of the DEF approach to screening will depend on how early imaging can identify PDAC. In a recent study, Singh et al72 reconstructed the timeline of progression of computed tomography (CT) changes in prediagnostic PDAC. They showed that, on average, CT changes started around 12 months before diagnosis, with pancreatic duct cutoff without a mass being the earliest sign. A mass appeared, on average, 9 months prior, with peripancreatic involvement at 6 months, vascular involvement at 3 months, and metastases only at diagnosis (realistically in the last 3 months). The sensitivity of CT scan findings suspicious for PDAC at −18 months was only 15%. Even 6 months before diagnosis, it was only ~50%. At 3 months, it was 85%.
The study highlighted some important lessons for early detection:
a) Changes seen on prediagnostic CT were overlooked in real time (human error): a robotic reading of every abdominal CT done could identify changes in the pancreas that should be further reviewed by the radiologist, thus avoiding human error in reading the scans.
b) CT scans have a limited role in finding PDAC beyond 12 months from diagnosis (true false-negative). The role of artificial intelligence (radiomics) to discern changes not visible to the human eye would be crucial when CT scans are “normal” to the human eye.
c) A repeat imaging study would be helpful in high-risk patients. However, the timing of the second CT will have to be tailored to the changes in clinical parameters rather than be at a fixed time after the first one. The study also highlights the importance of the need to study other modalities to identify cancers >6 months before clinical diagnosis (Fig. 7).
FIGURE 7.
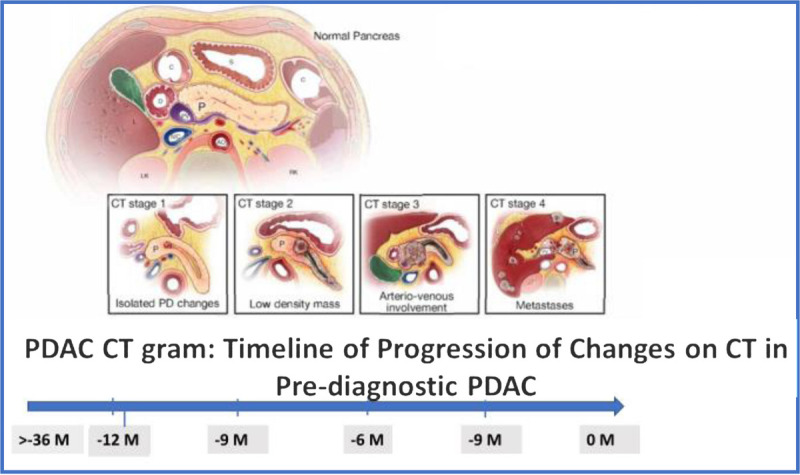
Timeline of changes on CT scan in prediagnostic PDAC.72
Role and Challenges of Endoscopic Screening
The potential for early detection of asymptomatic pancreatic neoplasms in high-risk individuals (HRI) using an endoscopic approach was first reported in 1999 when the group at the University of Washington (Seattle, Wash) first reported the detection of “dysplasia” in 3 unusual familial pancreatic cancer (FPC) kindreds with multiple affected members. Using endoscopic ultrasonography (EUS) and endoscopic retrograde cholangiopancreatography, along with CT and serum carcinoembryonic antigen (CEA) and CA 19-9, Brentnall et al73 performed prophylactic pancreatectomy in the relatives of PDAC patients with abnormal EUS and endoscopic retrograde cholangiopancreatography. In this early period of PDAC screening, the endoscopic findings largely influenced the decision to perform surgery.
However, in the next 2 decades, multiple centers of excellence around the world initiated pancreatic cancer surveillance programs and research on early detection in a well-defined expanding subset of HRI—the intermediate high-risk and very high-risk groups defined previously. These have consisted of FPC relatives with at least a pair of affected relatives but no known germline mutation, and germline mutation carriers (BRCA1, BRCA2, PALB2, ATM, HNPCC, with at least 1 affected blood relative, or patients with Peutz-Jeghers syndrome or familial atypical mole melanoma [FAMMM] syndrome).
When to screen? The age for initiating pancreatic surveillance remains controversial. Experts have recommended starting at age 50 or 55 years in FPC, with the latter age recommended because most PDACs are diagnosed in HRI ≥60 years old, unless there is an affected blood relative with young-onset PDAC <50 years old. In this case, surveillance should start earlier. Furthermore, in genetic mutation carriers, surveillance is recommended to start much earlier, with age 40 years for CKDN2A mutation carriers (FAMMM syndrome) and ages 30 to 40 years for those with Peutz-Jeghers syndrome.
How to screen? The approach for early detection has been EUS and/or magnetic resonance imaging (MRI), with most surveillance programs currently using both in varying degrees. Pancreatic protocol CT has not been the mainstay for surveillance because of the concern for repeated exposure to radiation over time. The question of what imaging modality to use was addressed in part by a comparative study by the Rotterdam group showing that MRI was better for pancreatic cyst detection, and EUS was better for solid lesions.74 A 3-way blinded comparison of EUS, MRI, and CT also showed that EUS and MRI had higher comparable diagnostic yield.75 Hence, most surveillance programs now use a combination of these 2 imaging modalities.
The diagnostic yield for screen-detected lesions varies among the published studies, but the largest series report a high prevalence of pancreatic abnormalities in HRI. The American Cancer of the Pancreas Screening consortium found a solid pancreatic mass or cyst at baseline screening in 42% at baseline screening, most were cysts, frequently multiple, with 10 times the prevalence of the asymptomatic cysts detected by MRI in the general population. Many of these cysts are branch-duct IPMNs, which are considered low-risk lesions for PDAC. However, based on pathological correlation with imaging in operated HRI with detected lesions, many are also incipient (or small <10 mm) branch-duct IPMNs or larger duct precursors (PanINs).76
One of the challenges for EUS-based screening is the need for sedation, which has minimal risks but is more invasive than abdominal imaging. The frequency of EUS performed over a lifetime of surveillance of an HRI can be offset by alternating with MRI. There is no consensus of the optimal surveillance schedule, with most groups performing annual imaging detecting no pancreatic lesions, often with alternating EUS and MRI. Other groups use EUS less frequently (ie, every third year)77 or only if there is a detected pancreatic lesion on MRI. The frequency of imaging should be adjusted (3–6 months) depending on the lesion(s) under surveillance if surgery is not planned. The risk category of the HRI should also be considered: germline mutation carriers have double the risk of neoplastic progression compared with FPC relatives without a mutation.78
Another challenge of EUS is that it is operator dependent, and access to EUS expertise is not routinely available to HRI. Furthermore, in mutation carriers with an increased risk of other cancers (such as patients with Peutz-Jeghers syndrome, BRCA mutation carriers, patients with Lynch syndrome), EUS cannot visualize extrapancreatic organs.
When should screening end? Another challenge of surveillance of HRI in general is when to stop. There is no agreement on the age to stop screening, but using a cost-effectiveness perspective, no screening is favored once patients reach the age of 75 years.79 From a practical viewpoint, discontinuing surveillance makes sense when the HRI is not a surgical candidate and/or the competing risk of death from non–pancreas-related causes exceeds that of the risk of developing and dying of PDAC.
The big question is, “Is pancreatic surveillance worthwhile in HRI?” If cost-effectiveness of surveillance of HRI is considered, abdominal imaging followed by pancreatectomy of screen-detected lesions is cost-effective as a preventive measure to prevent PDAC.79 Stratification of risk as detailed previously is ideal in selecting EUS over MRI. In a Markov model–based cost-effectiveness analysis comparing no screening, EUS, and MRI for screening of HRI, analysis of a cohort with a 5-fold relative risk of PDAC higher than the general US population (“moderate risk group” defined above), MRI was the most cost-effective strategy. However, in HRIs with >20-fold relative risk (the “intermediate high risk” and “very high risk” group), EUS became the dominant strategy.79 Furthermore, the cost of MRI and EUS can vary, and if MRI is more costly than US $1600, EUS becomes more cost-effective.
In a highly selected cohort of 354 HRIs undergoing long-term surveillance (>16 years) at one institution, EUS and MRI surveillance can lead to detection of early PDAC and high-grade precursor IPMNs and PanINs, with most (90%) resected screen-detected PDACs found to be resectable, at an earlier stage than symptomatic PDAC.76 Importantly, the 3-year survival of the former was 85%, as compared with that for HRI who developed symptomatic unresectable PDAC outside surveillance (25%). The median time to progression from baseline was 4.8 years. The impact of pancreatic surveillance on survival needs to be validated in larger studies, but nonetheless provides hope for HRI facing the prospect of incurable PDAC. For now, individualized decision making within a center of excellence with regard to the risk and benefit for initiating screening, undergoing surgery, or stopping surveillance is in line with the precision medicine initiative.
SECTION B: AI AND MACHINE LEARNING
Co-leads: Adam Yala, PhD Candidate, and James A. Taylor, MD
Group members: Søren Brunak, PhD; Yonina C. Eldar, PhD; and Chris Sander, PhD
Introduction: What Is Machine Learning?
Advances in artificial intelligence (AI) are driving a revolution across science, enabling exciting new results across chemistry,80–82 biology,83,84 and medicine.85–88 Common to these diverse successes are a specific approach to building AI systems, namely, machine learning (ML). In this article, we aim to introduce the reader to ML, to showcase a few of the many research areas that are being revolutionized by ML and to outline the ongoing challenges in translating the promise of ML technologies into real improvements in clinical care.
What Is ML?
Machine learning refers to the study of algorithms that learn their behavior from data. To see why such algorithms are important, consider the following basic task, building a program to predict if an image contains a dog or a cat. Although it is exceedingly difficult for us to manually specify the exact rules to determine that a dog is a dog, it is comparatively straightforward to prepare a reference set of images and labels (ie, dogs or cat). This setting, where knowledge is more easily encoded in data rather than as a descriptive set of rules, is the focus of ML algorithms.
Given a reference set, that is, training data, and a performance metric to optimize, that is, a model objective, ML algorithms typically begin with a random guess. For instance, consider the following simplified model in Figure 8. A random guess could correspond to Figure 8A, where many mistakes are made. Given this initial guess, an ML algorithm will then measure its performance and then iteratively refine its guess to maximize its performance (Fig. 8B). Although this general framework has been explored since the 1960s89 in the context of simple linear models such as the perceptron or logistic regression, there has recently been explosive progress in speech recognition,90 natural language processing,91 and computer vision92 because of a resurgence of deep neural networks,93 a special class of ML models that are able to learn complex hierarchical representations. The study of deep neural networks is commonly referred to as deep learning.93 We note that this conceptual framework extends far beyond tasks that humans can easily perform, such as distinguishing between dogs and cats; for instance, the reference set we wish to learn from could contain mammograms and whether or not a patient developed cancer within 5 years. In this way, ML offers powerful tools to discover signals, difficult for humans to deduce or describe, directly from data, and to expand the frontiers of our scientific capabilities.
FIGURE 8.
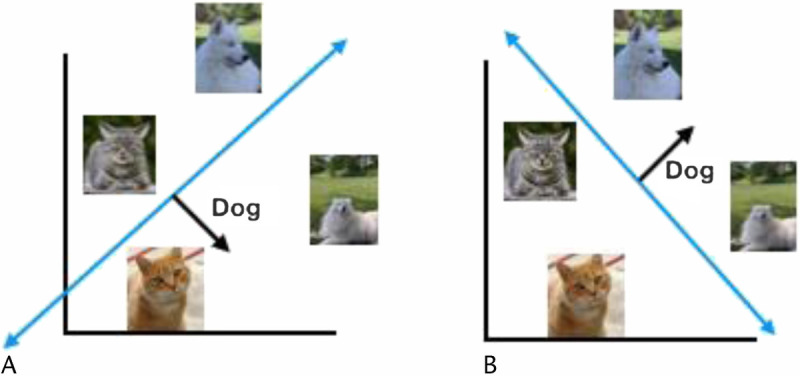
Learning to classify images of dogs versus cats. A, Initial model guess. B, The refined estimate after measuring its performance on the reference set. Courtesy of Adam Yala.
For a more in-depth introduction to deep learning models, we refer the reader to LeCun et al,93 and for a simple programming hands-on tutorial for deep learning applied to images, natural language and graphs, we refer the reader to ajfisch/deeplearning_bootcamp_2020 on GitHub (Fig. 8).94
AI and Risk Assessment
The goal of cancer screening programs95–98 is to enable earlier cancer diagnosis while minimizing screening harms. To achieve this goal, all screening programs rely on cancer risk models, which predict who is likely to develop cancer at a future point in time. Some screening programs, such as lung cancer95 and breast cancer screening,98 use risk factors as simple as age and smoking history, whereas others, such as guidelines for screening breast MRI,99 use more sophisticated statistical risk models100,101 that combine age, family history, and other clinical factors. For pancreatic cancer, sufficient risk models are still needed to enable effective screening programs. The key to developing more effective screening guidelines, which enable both earlier detection and less overtreatment, is to develop more accurate risk models.
Deep learning applied to medical imaging has the potential to transform risk modeling and screening guidelines. The use of imaging to inform cancer risk has long been explored in the context of breast cancer.101,102 As early as 1976,103 mammographic breast density, which measures the amount of fibroglandular tissue in the breast, has been associated with an increased risk of breast cancer. Breast density is widely adopted; for instance, it is a part of US federal reporting requirements with screening mammography104 and a component of major risk models.100 Despite its widespread adoption, the benefit of using mammographic breast density in risk models is relatively small. For instance, Brentnall et al101 incorporated mammographic breast density into the Gail risk model and Tyrer-Cuzick model and found that their areas under the receiver operating characteristic curve (AUCs) improved from 0.55 and 0.57 to 0.59 and 0.61, respectively. We note that AUCs are a commonly used accuracy metric in risk modeling, where a score of 0.50 corresponds to random chance and 1.0 corresponds to perfect prediction. The central limitation of breast density is that it is a poor proxy of the rich information contained within a mammogram; breast density summarizes the millions of pixels captured in digital mammography into a coarse human-designed biomarker. Deep learning image–based models for breast cancer risk87,105,106 offer a promising alternative paradigm. Instead of manually selecting which mammography patterns may be informative for breast cancer risk, these models instead rely directly on the data to deduce these patterns. By training these models to directly predict future cancer risk from a mammography, these models learn to identify which mammographic cues are most predictive of future cancer. In recent work, Yala et al106 showed that their breast cancer risk model could obtain an AUC of 0.78 compared with an AUC of 0.62 by the Tyrer-Cuzick model, the current clinical standard. In practical terms, the authors showed their risk model identified 42% of patients with future cancer as high risk, compared with 23% by the Tyrer-Cuzick model. Although the current performance of these models is promising, we note that can still be improved through better algorithm design and by leveraging richer sources of information, such as tomosynthesis or sequences of mammograms. In other words, we are still only scratching the surface of what is possible in image-based risk modeling both in breast cancer and in other cancers.
In developing the next generation of risk models, we have the responsibility to make our models equitable. We note that the issue of bias in risk modeling has existed long before deep learning. Most existing traditional risk models were developed on predominantly White populations101,107 and have known limitations in predicting risk for other racial groups.108–111 For instance, although Yala et al87 found that their deep learning model obtained AUCs of 0.71 and 0.71 on African American and White patients, respectively, they showed that the Tyrer-Cuzick model obtained AUCs of 0.45 and 0.62 for African American and White patients, respectively. One of the common causes of bias in ML systems is “distribution shift.” Deep learning models are typically trained with the assumption that both the training and testing sets are collected from the same distribution of patients; thus, if models are developed in one relatively homogenous population, it may not generalize to the diverse patient populations or clinical environments in the real world. Moreover, this form of bias does not only take shape in terms of patient demographics but can even surface itself in details, such as which machine the medical image was captured on. The first step in tackling these challenges is testing for bias and measuring model performance on diverse population groups. Exploring how model performance varies by demographics should be a common evaluation standard required for publishing new works in risk modeling and for their clinical implementation. This issue is especially important given recent research demonstrating that a model for patient stratification in use across several hospitals was racially biased.112 Once a source of bias is identified, we can work to remove it. For instance, Yala et al106 leveraged a conditional adversarial training scheme to remove imaging machine–related bias. Moreover, there is a rich and rapidly evolving body of work113–116 in computer science for devising algorithmic remedies to bias and creating equitable ML models, which in turn offers a promising path for more equitable health care.
Although our discussion focused on breast cancer, the potential of deep learning applied to medical imaging to produce both more accurate and equitable risk models spans across disease systems. We are still only scratching the surface.
AI and Biology
Early detection of pancreatic cancer requires basic research, identification of populations at risk, technological development of assays and protocols, clinical trials, professional and legal approval, cost-benefit analyses, and economic implementation—before any real-life impact.
It is of some interest to review the use of computational techniques, in particular statistical learning and ML (AI) in biology, for 2 reasons. Understanding of human biology at the level of cell, organs, and the organism is useful in any prevention or early detection program. In addition, computational methods used in basic biological research may be transferable or directly useful in the pancreatic cancer program.
In this context, basic research has 2 main dimensions. One is accumulating basic knowledge at the level of molecules, cells, and organisms. Another is basic research at the population level, focusing on risk factors, disease states, progression of disease, and impact of therapies.
Artificial intelligence methods make contributions to both of these dimensions. The boundary between physics-inspired computational methods, statistical approaches, and AI or ML methods is not sharp. It is useful to consider the entire spectrum of computational biology methods with emphasis on the extraordinary recent progress and near-term promise of the latest ML technologies. Hereinafter is a brief first set of examples, 1 or 2 at each of the 3 scales. Some examples are given in each of these categories and extrapolate to the most important near-term challenges and opportunities in moving the early detection of pancreatic cancer forward.
Molecular Biology
Computational molecular biology is a highly developed field with major advances as the result of sequencing technology for the last 20 or so years. For example, massive sequencing data permitted the construction of statistical models that link genetic and epigenetic patterns to the expression of genes and gene products, and computing the intricate and highly specific 3-dimensional shapes of protein molecules became a realistic option using evolutionary information from sequences and adapting methods from statistical physics, with further recent improvements using ML83,117 and large US national projects, in particular. The Cancer Genome Atlas (TCGA), provided large data sets of genetic and molecular profiles of cancer samples, associated with bioinformatics analyses.118,119 The computation of the full spectrum of interactions between macromolecules and with small molecules remains a major challenge. Machine learning has opened new doors to the engineering of biomolecules, especially for diagnostic and therapeutic purposes.120
Cell Biology
Knowledge of cell biological processes typically has relied on accumulation of information in publications over several decades. However, most of this knowledge is not computational. Attempts to capture cell biological processes in information systems, such as the Reactome121,122 or Pathway Commons123,124 knowledge bases, have been very useful and do have a formal computational basis (BioPax ontology125). However, these knowledge systems do not nearly capture all available knowledge and generally do not contain executable models that can be used for making nontrivial quantitative predictions. Although there are serious efforts to develop whole cell computational systems, projects such as the Japanese E-cell (e-cell.org), the UConn Health VCell (vcell.org) projects, CellML.org, BioModels.org, and others, we are still a long way from having reliable models of human cell biology, which permit the mechanistic simulation of cellular processes or the quantitative prediction of the results of unseen and potentially therapeutically useful perturbations.
One impending opportunity in the next few years is to use high-throughput technologies to relate perturbations of appropriate biological systems to massive readout of the response of the systems, which can then be the basis of ML processes that derive much more comprehensive and genuinely predictive computational models.126 If the technology of data acquisition in such systems can be scaled up to achieve reliable data sets, then there is a major opportunity for ML to suggest preventive or therapeutic interventions for desirable outcomes.
An additional major opportunity arises from technologies of single-cell observations, starting from single-cell sequencing of mRNAs. Single-cell measurements of metabolites and proteins are also on the horizon. Given such data sets, ML may be able to derive 2 kinds of predictive models. One branch has ML models that in an end-to-end fashion compute the result of perturbations in terms of desired outcomes. A second type of models relies on the combination of mechanistic and ML models, such that the ML process provides parameters for explicit simulations of interpretable mechanistic models.127
For the early detection of pancreatic cancer, a better quantitative description of cell biological processes and the power of dynamic simulations could make valuable contributions. Focus on particular cell types and on metabolic processes and aiming for biomarker discovery would be good starting points.
Organismic Biology
In terms of clinical and disease relevance, computational physiology and computational pathology are classic domains of quantitative science,128,129 with some interesting work related to healthy and diseased pancreas. Systems biology has the declared ambition to capture the connections between molecular, cell biological, and organ level biology, but the comprehensive quantitative description at this level of detail is prohibitively complicated.
A major potential near-term opportunity for early detection and for the identification of predictive biomarkers is plausibly the large-scale analysis of abdominal images, such as CT scans, and their interpretation in terms of physiological processes related to precancerous states and metabolic states that predispose to pancreatic cancer.130 Making focused progress in this direction requires interdisciplinary collaboration between experts in ML, disease physiology, and molecular cell biology.
Another very promising area of applying ML to the problem of early detection of pancreatic cancer is the analysis of real-world clinical records using ML methods.131 Analysis of such records can include a wide variety of factors, not limited to disease codes, but also including environmental factors, personal history, and the effects of therapeutic intervention. This work is in progress and described elsewhere in this collection.
AI and Disease Trajectories
A lesson learned from the human genome project has been that the number of protein coding genes is much lower than originally anticipated. This likely means that pathways overlap extensively and that the molecular etiology of diseases overlaps correspondingly. This in turn gives rise to disease co-occurrences linked to these overlaps, in addition to longitudinal disease correlations where diseases provoke each other over time.132–134 Some of the time-correlated disease patterns are obviously treatment provoked and would not arise if one treatment strategy is chosen over another, for example, in the case of chemotherapy versus surgery in the pancreatic cancer domain.135
In this context, the big data field provides a conceptual framework for analysis across the full spectrum of disease that may better capture patient subcategories, in particular when considering longitudinal disease development in a lifelong perspective. Here, variation in “healthy” diagnosis-free routes toward disease and later differences in disease comorbidities are currently of high interest. Using health care sector, socioeconomic, and consumer data, the precision medicine field works increasingly toward such a disease spectrum-wide approach. Ideally, this involves data describing healthy individuals, many of whom will later become sick—to have long-range correlations that relate to outcomes available for analysis. This notion extends the traditional disease trajectory concept into healthy life-course periods potentially enabling stratification of patient cohorts by systematically observed differences present before the onset and diagnosis of disease (Fig. 9).
FIGURE 9.
Disease progression over the life course. The figure illustrates how diseases follow each other and how different patients develop different complications to the same disease over time. Events in early disease-free periods will in many cases influence disease risks and disease severity later in life. Big data disease analytics aim at finding such early events and symptoms with predictive value in relation to early diagnosis of for example pancreatic cancer. Courtesy of Jessica Xin Hjaltelin, University of Copenhagen.
Today, industrialized, high-throughput technologies create realms of patient-specific data, begging for novel computational strategies to make better diagnoses and prognoses and to create improved understanding of disease development. Many of these techniques, such as genomic sequencing or proteomic profiling, produce biomarker data that potentially link to more than one disease.136 This development at the molecular level is synergistic with the idea of analyzing the disease spectrum in a more holistic manner.
Patients with pancreatic cancer are among the most difficult groups to stratify.135 Although pancreatic cancer can be classified into a few discreet subtypes based on symptomology, in reality it represents a disease continuum. This complex disorder is associated with several comorbid conditions and complications, one of the most common being diabetes. Many other diseases, such as dyslipidemia, hypertension, liver disease, obesity, and other cancers, may influence the risk profiles in unknown ways, and the influence of shared genetic loci, prior treatment, and other exposures is far from being understood. Given the increasing levels of pancreatic cancer incidence worldwide, a major challenge is to understand the transition from the healthy state or other diseases to, for example, prediabetes, diabetes, and further on to pancreatic cancer. Disease trajectories not including diabetes are of course also of similar high interest in the context of early detection of pancreatic cancer.
Most health data–driven projects work from data obtained from the life span with diagnoses, or at least periods with higher diagnosis density that manifest closer to an index disease time point.137 However, the initiation of large precision medicine initiatives, such as those spearheaded by the United States and the United Kingdom, data on seemingly healthy individuals are growing dramatically. In the United States, the renaming of the national precision medicine repository as the “All-of-Us” database is one sign of this development. Other types of projects also focus directly on data from healthy live course periods; this is, for example, the case for blood donor studies that are studying an extreme phenotype that is healthier than that of the average population. One of the largest examples worldwide is the Danish Blood Donor Study (www.danishhealthdata.com/find-health-data/Det-Danske-Bloddonorstudie) initiated in 2010, where blood samples are obtained longitudinally, and as the blood donors eventually get sick and retire as blood donors, these can be used to characterize molecular-level trajectories of initially healthy participants transitioning into a given disease, for example, pancreatic cancer. The Danish Blood Donor Study has now genotyped around ~150,000 of the participants included so far. In Scandinavia, one can combine such data with near population-wide data from the socioeconomic, health registry, and complete electronic patient record domains, with the aim of modeling life-course disease development for specific disease groups or just millions of patients. In the case of Denmark, one of the most digital countries in the world, we can exploit data for population-wide analyses over at least 40 years for around 10 million individuals with reasonably well-known family relations.
Such longitudinal data analysis may be essential to relate patient trajectories covering many subsequent illnesses to biomarker data from the omics domain. There is likely a strong potential in deconvoluting disease progression scenarios and disease co-occurrence patterns influenced by shared, pleiotropic causes and those that represent consequent development in which one disease state leads to others. Exposure or shared genetic links may often be involved in the former category of co-occurrence, whereas the latter category comprises a large number of chronic disease progression modes.
It is increasingly acknowledged that not only gene loci but also proteins and pathways can possess multiple context- and time-dependent roles. Such multifunctionalities can be involved in pleiotropy (the effect of a genetic locus on more than one trait), resulting in comorbidities where 2 diseases coexist in the same individual. In this complex background of disease interaction, predictive approaches, such as data-driven ML models, can predict patient survival scores or rank treatment options, taking both molecular and clinical data into account. They can also take advantage of the interactions between these data types.137 It was recently shown that preadmission disease history alone may outcompete intensive care data obtained during the first 24 hours after admission for the task of predicting intensive care patient survival. The optimal strategy is of course to combine these types of data and obtain an even better prognostic value for the patient by reusing data generated in routine care.138–140
In summary, we would like to highlight the importance of the previous disease history as a basis for predicting the outcome of pancreatic cancer patients and for understanding the molecular level etiology. We suggest that the temporal disease history should be an additional aspect of personalized medicine, as opposed to a snapshot view of the current condition. Another option is that the temporal disease history may systematically be implemented as a stratification parameter in clinical trials.
AI and Medical Imaging Workflows
One of the most promising areas of innovation in medical imaging in the past decade has been the application of deep learning. Deep learning has the potential to impact the entire medical imaging workflow from image acquisition, image registration, to interpretation.141,142
Traditional image processing is dominated by algorithms that are based on statistical models. These statistical model-based processing algorithms carry out inference based on a complete knowledge of the underlying statistical model relating the observations at hand and the desired information, and do not require data to learn their mapping. In practice, accurate knowledge of the statistical model relating the observations and the desired information is typically unavailable. The past decade has witnessed a deep learning revolution. Deep learning methods have surpassed the state of the art for many problems in signal processing, imaging, and vision with unprecedented performance gains. However, most deep learning approaches are purely data-driven, and the underlying structures are difficult to interpret. In addition, their practical success is often overly dependent on the quantity and quality of training data. They also do not always generalize well to unknown settings. This is particularly important in medical imaging where interpretability and generalization are crucial.
In the past few years, a new paradigm within deep learning has emerged, which we refer to as model-based deep learning. This framework attempts to combine models within deep networks in several interesting ways. One approach is based on the seminal work of Gregor and LeCun,143 which introduced a promising technique called algorithm unrolling. This approach helps connect iterative model-based algorithms to neural networks. The past few years have seen a surge of efforts that unroll iterative algorithms for many significant problems in signal and image processing.144 Examples in ultrasound imaging are reviewed by Solomon et al145 and van Sloun et al.146 An unrolled network may be trained using back-propagation, and the trained network can be naturally interpreted as a parameter optimized algorithm. An additional benefit is that prior knowledge inherent in traditional iterative algorithms may be transferred. Furthermore, networks constructed in this fashion usually achieve a more favorable trade-off between increase in parameters and improvement in performance and are readily interpretable.
A second approach is based on hybrid methods, where an underlying well-established method is used in combination with a deep network. In this setting, the network is used not in order to learn an end-to-end task, but rather in place of a specific block that relies on unknown parameters or models. This approach has been used extensively (eg, in communications)147 but has the potential to be used in many medical imaging problems as well.
Besides using a model-based deep network for image recovery and processing tasks, we can also consider designing the acquisition side as well. Given that the recovery is now going to be performed using deep networks, it makes sense to also learn how best to acquire the signal. For example, we may wish to learn the optimal sequences in an MRI machine, or to learn the optimal angles in a CT scanner. Learning how best to distribute the elements within an ultrasound probe and which samples to take in time and space can also lead to more efficient designs of medical imaging systems.
Today, interpreting medical imaging (x-rays, CT, MRI, ultrasound) scans is a highly skilled, manual job requiring many years of training. Model-based deep learning methods can improve medical imaging capabilities in an interpretable manner without relying on huge databases to learn and train. They can pave the way to faster, more accurate diagnoses. For example, they can lead to clean and sharper images, super resolution, separation between different features in an image, and efficient segmentation, and they can aid in image interpretation. The higher efficiency provided by deep learning in medical imaging will allow radiologists to perform higher-value tasks such as medical judgment, communication of diagnosis, engaging multidisciplinary teams, interventional procedures, and more.
Bringing AI to the Clinic
Ultimately, it is likely that AI will transform much of the practice of medicine. AI will be used to interpret radiographs, ultrasounds, CT, and MRI, either as an adjunct to the clinician's interpretation or as the standalone reading.88 Health care organizations will use AI systems to extract and analyze electronic health record (EHR) data to better allocate staff and other resources, identify patients at risk for acute decompensation, and prevent medication errors.148 Using sensors on commodity devices such as smartphones, wearables, smart speakers, laptops, and tablets, individuals will be able to share health data during their daily lives and help generate a longitudinal personal health record, with pertinent information incorporated into their EHR. By extracting information from the EHR and incorporating data during an encounter with a patient, clinicians can be provided with a differential diagnosis in real-time with probabilities included.
Despite its potential, the use of AI in clinical medicine is in its infancy with little widespread adoption.149 Not surprisingly, the preponderance of AI technologies that have been cleared by the FDA for clinical use to date is based on image classification, a task well suited for deep learning methodologies.150,151 In a 2019 review, Topol identified 11 AI technologies focusing on interpretation of imaging studies that had been cleared by the FDA for use.88 Artificial intelligence interpretation of photographic images of the retinal fundus is used for an FDA-cleared device, IDx-DR, to diagnose diabetic retinopathy.88 Probably the most widely used medical device that incorporates AI is atrial fibrillation detection on the Apple Watch. However, there are limited data on its clinical impact post-FDA clearance and some questions about its accuracy in detecting dysrhythmias.152
There are numerous reasons for the wide gap between promising study results and FDA-cleared, AI-based medical devices. Chief among these is the “AI Chasm,” a term highlighting the fact that accuracy, typically using the metric of AUC, demonstrated in a research study, does not necessarily translate to clinical utility.149,150 It is intuitive that an AI-based clinical tool will generate the most accurate classifications when presented with data similar to that used for its training. However, it may be impossible to have enough training data to adequately account for all the clinical settings in which the system might be used. Although an algorithm might generate highly accurate results in almost every setting, even a small number of outlier results with large errors from “edge cases” may have catastrophic consequences. There is also the well-documented problem of “algorithm bias,” in which the accuracy of specific algorithms has been found to be lower in individuals from underrepresented groups.88,149,153 Implementation of these algorithms into clinical practice tends to increase the disparity in health outcomes in these groups rather than leading to a more equitable health care system, which should be an overriding goal of all AI-based medical devices.88
Because of the “black box” quality of many deep learning algorithms, clinicians and patients may be hesitant to depend on AI-based solutions. This fear is not unfounded. For example, it was discovered that an algorithm evaluating data from images of skin lesions was more likely to classify the lesion as malignant if a ruler was included in the photograph.149 The reticence by clinicians to embrace AI-based medical devices may also be explained by the paucity of peer-reviewed prospective studies assessing the efficacy of these systems.88,149 Finally, regulatory assessment of the effectiveness and safety of AI-based products is different from that of traditional medical devices.150 Regulatory agencies are working to find the best processes for determining whether an AI medical device should be cleared for clinical use.
The speed at which these obstacles are overcome will determine how quickly AI-based solutions will be incorporated into routine clinical care. There is a tremendous amount of digital health data that many of us now create, computing power is adequate for developing high performing algorithms, and the value of AI for use in medicine is apparent. However, there is a potential paradox; large incumbents may focus on using AI for incremental advances, whereas new, smaller entrants who are pushing frontiers may lack the resources to take an idea from development to FDA-approved device.154,155 The incorporation of AI-based solutions into clinical practice will be accelerated by companies with the necessary expertise and funding who adopt the approach used by the pharmaceutical companies where there is the expectation of multiple expensive failures yielding a few, extremely successful products. Clinicians would be more likely to use and promote AI solutions if more prospective studies, conducted in typical clinical settings on diverse populations, were published in peer-reviewed medical journals. It is crucial that the performance metrics reported in these studies be clinically relevant. To minimize the potential for a drop-off in performance from the initial validation of a new algorithm to its implementation in “real-world” clinical settings, neural network techniques specifically designed to minimize this effect have been developed. Cognizant of the fundamental difference between AI solutions and traditional medical devices, FDA's clearance processes for AI products have evolved and are evolving; however, there are a series of challenges that need to be fully addressed to facilitate the approval and clearance processes for AI-based devices in a manner that maximizes the potential of the new technologies while providing the appropriate regulatory oversight to ensure safety and effectiveness on an ongoing basis.156,157 Finally, to actually “transform medicine,” it is crucial that the implementation of AI-based medical devices be equitable so that the benefits are realized by all people and not just those with the most social advantage.
SECTION C: AI AND EARLY DETECTION OF PANCREATIC CANCER—CURRENT EFFORTS
Co-leads: David Kelsen, MD, and Michael Rosenthal, MD, PhD
Group members: David Bernstein, PhD; Elliot K. Fishman, MD; Sung Poblete, RN, PhD; Uri Shalit, PhD; and Brian Wolpin, MD, MPH
The ability to reliably detect very early-stage PDAC in asymptomatic patients should result in a major improvement in survival. This hypothesis is based on the observation that the prognosis for PDAC is clearly related to the pathological stage of the tumor at the time of diagnosis. Using the SEER database, Ansari et al reported that 5-year survival for patients with lymph node–negative primary PDAC less than 1-cm cancers is ~60%; with primary tumors of 2 cm or larger even without lymph node metastasis, survival was less than 20%. However, less than 1% of patients are found with primary PDAC less than 1 centimeter in size. Pancreatic ductal adenocarcinoma is diagnosed in the large majority of even stage IA patients because of symptoms, not as a result of an early detection program. The hypothesis that the earlier the stage of a PDAC, the better the outcome, is in concert with data from many other solid tumors, including breast, non–small cell lung, colorectal, prostate, and gastric cancers.12
Although the velocity of growth of PDAC metastases can be very rapid, more recent data suggest that the time to the development of incurable metastatic PDAC, measured from the first genetic event resulting in a primary cancer, may be much longer than previously thought. Yachida et al158 reported the results of a detailed genomic analysis performed on tumor specimens obtained at autopsy soon after death in 7 PDAC patients. They studied the clonal relationships between the primary tumor and metastatic foci. Cell lines and xenografts were developed. Next-generation sequencing was performed; somatic mutations at different metastatic sites were compared with the primary tumor and metastases at other sites. A quantitative analysis of the genetic evolution of the metastatic clones in comparison to the primary cancer was performed. The analysis indicated that there is a prolonged period of approximately 10 years from the first genetic mutation in a normal pancreas cell to the development of a clearly malignant PDAC cell. They further estimated that, on average, another 5 years is required after the cell becomes malignant before additional mutations confer the ability for malignant PDAC cells to metastasize.
These data suggest that the commonly observed rapid progression of established PDAC in the individual patient is a late development. Yachida et al158 suggest that there should be a substantial window of opportunity, measured in at least several years, in which to detect very early-stage PDAC in asymptomatic patients. The net effect of these data (higher cure rates for earlier pathology stage cancers and a substantial window of opportunity to detect early-stage tumors) is that early detection of small asymptomatic PDAC should markedly increase survival. This will change the current clinical paradigm of waiting until symptoms develop to diagnose PDAC to the routine use of effective surveillance and screening programs.
The working group AI and Pancreatic Cancer—Current Efforts has addressed the potential power of AI as an aid to developing effective early detection methodologies in PDAC. Outlined hereinafter are approaches in early detection of PDAC in which AI may be a critical methodology, and currently underway or planned efforts. We sought to identify AI in early detection of PDAC projects by contacting individual principal investigators (PIs) of early detection programs, individuals involved in funding of early detection efforts, and databases of clinical trials. We included areas of investigation for early detection of PDAC involving the following modalities and approaches:
Imaging
Blood based assays
Microbiome (including bacterial colonization of the pancreas as an inciting factor to PDAC)
-
Patient characteristics including but not limited to the following:
∘ Analysis of electronic medical record for changes in weight and laboratory tests
∘ Lifestyle
∘ Social media
∘ Pharmacy records
∘ Insurance claim records
-
Integrative approaches
∘ Combine all of the above
∘ Plus genomic alterations
We attempted to identify planned or ongoing efforts in which AI (including ML and deep learning) was being used as an aid to early detection in PDAC underway in North America, Europe, and Israel. We were not able to identify PIs who were including AI efforts in PDAC early detection in Asia. We also considered active AI efforts in the early detection of PDAC versus projects that are clearly adjacent to AI space, but may benefit from AI or use components of AI.
As noted previously, we primarily used personal contact with individuals who we thought would be informed regarding these efforts; one of our suggestions noted hereinafter is that there should be a centralized, ongoing effort to identify planned/on-going AI in PDAC projects. A Web-based site should be established so that investigators could identify AI efforts in pancreatic cancer both for early detection and for other areas of investigation. We believe this will considerably ease the ability to develop collaborations on a national and International basis.
We identified the following planned, current, or completed efforts in AI and early detection of PDAC:
Imaging With and Without Patient Characteristics (Using Electronic Medical Records)
Project Felix is a Lustgarten Foundation initiative led by Elliott Fishman at Johns Hopkins University to develop deep learning tools that can detect pancreatic tumors when they are smaller and with greater reliability than human readers alone. This effort has involved meticulous manual segmentation of thousands of abdominal CT scans to serve as a training and testing cohort, which represents the largest effort in this domain in the world. In collaboration with the computer scientist Alan Yuille. Project Felix has produced at least 17 articles on techniques to automatically detect and characterize lesions within the pancreas (https://www.ctisus.com/responsive/deep-learning/felix.asp).
Wansu Chen and Bechien U. Wu of the Kaiser Permanente Southern California have previously reported work using natural language processing to identify individuals at risk for pancreatic cancer based on radiology reports.159 Their group is in the early stages of participating in the recently National Institutes of Health (NIH)/National Cancer Institute (NCI)–funded NOD cohort (https://prevention.cancer.gov/news-and-events/blog/new-onset-diabetes-cohort) and with the NIH-NCI Early Detection Research Network (EDRN) effort (described hereinafter). Although the details of the research plan have not yet been published, their work will apply AI techniques to risk estimation in this cohort.
The Pancreatic Surgery Consortium assembled a cohort of 1073 patients with resected IPMN's to assess for the risk of recurrence of high-grade dysplasia or invasive cancer. Their logistic regression model to assess the risk of high-grade dysplasia or invasive carcinoma was based on patient characteristics and IPMN imaging features and showed significant stratification between low-risk and high-risk groups.160 No specific AI-based analysis was described in this work.
-
The Pancreatic Cancer Collective, an initiative of the Lustgarten Foundation and Stand Up To Cancer to improve pancreatic cancer patient outcomes, has funded 2 efforts to use AI to screen for pancreatic cancer. The first team is using clinical records and images to identify individuals at high risk for future pancreatic cancer, and the second team is using genomic and immune factors to identify at-risk individuals. The term of these projects is May 2019 to April 2021.
The records-based team is led by Chris Sander (Dana-Farber Cancer Institute [DFCI]) and Regina Barzilay (Massachusetts Institute of Technology). Their effort includes 4 major components: assembly of cohorts of >4 million patient records at 3 study sites that include both future pancreatic cancer cases and asymptomatic controls, implementation of a common data model to which all local site data can be mapped to allow site-agnostic analysis and generalizability, development of AI models that can identify signs and symptoms (so-called intermediate phenotypes) of known relevance using medical records and images, and development of AI models that can integrate structured clinical data, images, and AI-based intermediate phenotypes into overall individual risk scores. The long-term goal of this work is to be able to deploy AI models into health care systems that can automatically identify individuals who require evaluation and/or surveillance for pancreatic cancer (https://standuptocancer.org/research/research-portfolio/research-teams/pcc-ai-identifying-individuals/).
Eugene Koay from The University of Texas MD Anderson Cancer Center (MDACC) has previously characterized subtypes of PDAC on CT scans, whereby conspicuous (high delta) PDAC tumors are more likely to have aggressive biology, a higher rate of common pathway mutations, and poorer clinical outcomes compared with inconspicuous (low delta) tumors.161 His group has recently completed an analysis, currently under review, that shows that high-delta tumors demonstrate higher growth rates and shorter initiation times than their low-delta counterparts in the prediagnostic period. Although not strictly an AI initiative, his work serves as a rich foundation for future AI initiatives in this space. Drs Koay and Anirban Maitra at the MDACC are leading the NCI-sponsored EDRN initiative to assemble a prediagnosis pancreatic cancer cohort that could facilitate AI research into screening and early detection.
Blood-Based Assays
CancerSeek Biomarker Assay (Bert Vogelstein, PI; Cristian Tomasetti): ML methods were used in the development of the CancerSEEK assay,44 for example, logistic regression for combining the mutation and protein scores and random forest for tissue localization. The current evolution of CancerSEEK and associated algorithms also uses ML methods.
Mayo Clinic (Gloria Petersen and Shounak Majumdar): using Mayo Clinic's large database of pancreatic cancer patients from whom the PIs and their group over the past several years collected biospecimens in combination with imaging, clinical, and genetic data, they are exploring AI tools for early detection. They are also using ML approaches to molecular and imaging biomarker discovery and validation and exploring the role of natural language processing in identifying HRIs. The high-risk pancreas clinic serves as the translational hub for these activities; they are in early stages of establishing a prospective high-risk patient registry that will facilitate sequential biospecimen collection and data archiving for the study of early detection using AI tools in the years to follow.
-
Planned efforts
∘ Memorial Sloan Kettering (MSK) in collaboration with Weill Cornell, Weizmann Institute, Sheba Medical Center, and Shaare Zedek Medical Center, and Cold Spring Harbor Lab are planning to use AI as an aid to analyzing blood based biomarkers in sporadic and high-risk populations (D. Kelsen, PI). Collection of the deeply annotated biospecimens from the high-risk population (including BRCA mutation carriers with PDAC and controls) started in 2014; there are more than 400 participants in the BRCAmut Registry. Collection of annotated blood and tissue specimens from sporadic PDAC patients and controls, including benign pancreatic diseases such as pancreatic cysts, IPMN, and pancreatitis, and normal controls, began in 2017.
∘ DFCI (Brian Wolpin, PI): funded by a U01 and a Lustgarten Foundation grant, Dr Wolpin leads a multicenter project for early detection of PDAC, with the goal of developing a blood-based biomarker assay.162,163 The project includes high-risk populations and control populations. Analysis is planning to use AI as an aid in analyzing the biomarker data.
∘ New York University has an extensive registry and annotated biospecimen collection for early detection of PDAC (Diane Simeone, PI). Dr Simeone is in the planning stage for including AI in analysis.
-
Earlier efforts
∘ Earlier efforts in developing blood-based biomarkers for early detection of PDAC largely used limited component panels (eg, CA 19-9, CEA, thrombospondin2). A very limited number of clinical variables were considered; standard biostatistical approaches to analyze the data were used. However, current efforts involve the use of multiple assay approaches involving much larger numbers of data points, including but not limited to plasma exosomes protein cargo, ctDNA, proteomic spectrum (serum), and methylated DNA, resulting in much more complex data sets. For example, in a collaboration between MSK, Weill Cornell, the Weizmann Institute, and Cold Spring Harbor Laboratory, exosome protein cargo are being studied in the Lyden Lab at Weill Cornell. More than 1400 individual proteins in exosomes are isolated from plasma of patients with PDAC and controls.164 Using serum from the same blood specimens, serum proteomic spectrum involving hundreds of individual proteins, plasma ctDNA, and more conventional blood-based biomarkers such as CA 19-9 are annotated by patient characteristics involving scores of factors (including sex, age, presence or absence of diabetes with details regarding duration of diabetes, and agents used to treat diabetes, body mass index, ethnicity, comorbidities, etc). Artificial intelligence approaches to analyzing these data sets may be more effective than standard bio statistical approaches.
Early Detection of PDAC by Analyzing Microbiome
Gregory Poore and Robert Knight reported their reanalysis of TCGA data for a variety of cancers, in which they studied whole-genome and whole transcriptome data for microbial reads. They used ML to identify microbial signatures that discriminate among different types of cancer and compared their performance.165
Patient Characteristics
We are aware of few formal efforts to use patient characteristics from general medical records to identify individuals who are at increased risk for pancreatic cancer. The Pancreatic Cancer Collective has funded one such effort, described previously. The team led by Søren Brunak from Denmark has also published on the analysis of temporal sequences of International Classification of Diseases codes to predict cancer risk before their involvement with The Pancreatic Cancer Collective.
-
The Pancreatic Cancer Collective has funded 2 efforts to use AI to screen for pancreatic cancer. The first team is using clinical records and images to identify individuals at high risk for future pancreatic cancer, and the second team is using genomic and immune factors to identify at-risk individuals. The term of these projects is May 2019 to April 2021.
The genomics and immune factor team is led by Raul Rabadan (Columbia University, New York, NY) and Núria Malats (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain). Their work will combine several large multinational genomic data sets with clinical and tumor microenvironmental factors to produce an integrated estimate of pancreatic cancer risk (https://standuptocancer.org/research/research-portfolio/research-teams/pcc-ai-genomic-immune-factors/).
Integrative
Many of the efforts described previously incorporate components of structured patient characteristics into their analysis plan. The Pancreatic Cancer Collective records-based team is specifically working to integrate structured data, natural language processing of medical notes, and neural network analysis of medical image data into a combined risk score.
Anirban Maitra's team at the MDACC was recently funded under the NCI–MCL (Molecular and Cellular Characterization of Screen-Detected Lesions; see government activity hereinafter) to develop a framework that integrates imaging data with host immune responses and circulating biomarkers in patients with pancreatic cystic neoplasms.
Who Has Funded AI in Early Detection of PDAC?
-
Government and industry activity and support
US Federal support: there is ongoing discussion in the NCI in developing AI tools for improving detection of precancer lesions, early-stage cancer, and stratification of indolent and cancer based on preclinical and clinical images along with “Omic” data.
-
The NIH-NCI sponsors the Alliance of Pancreatic Cancer Consortia, which includes 4 pancreatic cancer consortia: Pancreatic Cancer Detection Consortium; Chronic Pancreatitis, Diabetes, and Pancreatic Cancer; Early Detection Research Network; and Molecular and Cellular Characterization of Screen-Detected Lesions. Several of these groups are conducting work that either explicitly includes or is relevant to AI in pancreatic cancer. Matthew Young at the NCI coordinates the meetings. The members of the group are as follows:
∘ Pancreatic Cancer Detection Consortium (https://prevention.cancer.gov/major-programs/pancreatic-cancer-detection-consortium) develops and tests new molecular and imaging biomarkers to detect early-stage PDAC and its precursor lesions. This consortium currently includes 8 distinct projects. None of the projects are specifically focused on AI.
∘ Chronic Pancreatitis, Diabetes, and Pancreatic Cancer (https://cpdpc.mdanderson.org/) seeks to understand the clinical, epidemiological, and biological characteristics of patients with chronic pancreatitis and NOD, including the subsequent risk of pancreatic cancer.166 This group has established several prospective cohorts, including the NOD cohort described previously, that will likely be pivotal for future AI work on risk prediction.167
∘ Early Detection Research Network (https://edrn.nci.nih.gov/) is funded by the NCI to accelerate biomarker development to improve early detection of cancer. Eugene Koay's group is leading a project within the NCI-sponsored EDRN initiative to assemble a prediagnosis pancreatic cancer image and clinical data cohort that could facilitate AI research into screening and early detection.
∘ Molecular and Cellular Characterization of Screen-Detected Lesions (https://mcl.nci.nih.gov/) is a large NCI-sponsored effort to perform molecular characterization of early cancers. There are currently 4 funded subprojects related to the pancreas. None specifically mentions AI.
∘ The US Department of Defense recently published an “Idea Development Award” as part of its Pancreatic Cancer Research Program that included as a focus area “Integration of biologic and imaging biomarkers to drive more precise and earlier detection.” The Funding Opportunity, number W81XWH-20-PCARP-IDA, closed to preapplications on August 25, 2020, and will undergo programmatic review in March 2021. The program details are available at: https://cdmrp.army.mil/funding/pcarp.
-
Philanthropy
The Stand Up To Cancer Foundation supports numerous research programs in pancreatic cancer. At least 2 of which are specific to AI research (funded by the Pancreatic Cancer Collective collaboration with the Lustgarten Foundation). These 2 programs were discussed in the preceding Current Research section. The full Stand Up To Cancer research portfolio is available here: https://standuptocancer.org/research/research-portfolio/
The Lustgarten Foundation supports numerous research programs in pancreatic cancer. The largest AI-specific program in their portfolio is the FELIX Project at Johns Hopkins University, as discussed previously. The Lustgarten research portfolio and funding opportunities are available here: https://lustgarten.org/research/funded-researchers-programs-topic/
The Pancreatic Cancer Action Network is supporting the Early Detection Initiative to provide imaging to individuals with NOD that are further stratified by the ENDPAC clinical model.68 Preliminary work using AI techniques to identify clinical indicators of pancreatic cancer in the UK Biobank data set was supported but not used for the study. Blood-based biospecimens will become part of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)/NCI-supported NOD cohort study for biomarker validation studies. Computed tomography images from ENDPAC-high individuals will be made publicly available at an appropriate time for AI-based analysis.168 Additional biospecimen collection and analysis is anticipated. The study is institutional review board approved, and enrollment is expected to start early 2021 (ClinicalTrials.Gov: NCT04662879).
Strengths and Challenges in Using AI for Risk Stratification
Artificial intelligence encompasses a large family of techniques to distill complex data into simplified representations that can be used for classification or decision making. To date, most of the AI efforts in early detection of PDAC that have been identified involve ML. In the last 15 years, AI techniques have been developed to interpret complex sets of image, text, categorical, and time series data. Modern neural networks may contain tens of millions of parameters and have the capacity to model exceptionally complex interactions among their input fields. These methods have also evolved from requiring explicit human design of every step of the analysis process to enabling fully autonomous unsupervised learning in which the systems can identify salient features on their own.
The strengths of using AI for risk stratification include the capacity to integrate data from large, diverse feature sets; the ability to process irregular time-series data; and simultaneous estimation of multiple risk types and landmarks.
The challenges of AI include the requirements for very large, high-quality training data sets; the difficulty of assuring generalizability across sites and cohorts; and the difficulty of understanding what features an AI system is relying upon when producing a result (also known as interpretability). There are public policy concerns related to the explicit sharing of large volumes of detailed patient data. There are relative few standard operating procedures that govern data collection and encoding in this space, leading to challenges with data sharing and interoperability (see discussion hereinafter). There are also terminology and networking challenges in connecting disparate fields like data science, cancer biology, oncology, and epidemiology communities.
Accessing and Aggregating Data to Enable Early Detection Research
As noted previously, AI projects require relatively large, diverse data sets to successfully train systems that can generalize broadly. The data requirements of AI can be in conflict with privacy requirements like Health Insurance Portability and Accountability Act rules, and there have been relatively few efforts to systematically address data sharing within the pancreatic cancer domain.
There are 2 major competing strategies for accessing large data sets for AI training: centralization and federation. Centralized databases bring data from multiple sources together into a shared repository, which greatly simplifies model training but can be limited by privacy concerns, institutional data sharing restrictions, and maintenance costs. Federated methods retain data at local repositories, distribute the computational work to local resources, and then return the model training results to a central system for integration.
The NIH-NCI–sponsored EDRN effort exemplifies the centralization approach. This effort is focused on imaging studies and limited correlative clinical data. The Stand Up To Cancer–sponsored pancreatic cancer risk study on medical records has a federated component to facilitate multicenter research.
We have identified the following public and private databases as potential sources for future PDAC AI research:
NIH-NCI EDRN (Anirban Maitra and Eugene Koay, MDACC): centralized repository for early-stage and prediagnosis imaging in PDAC. Maintained and access controlled under NIH mechanism.
The Pancreatic Cancer Collective screening cohort (Chris Sander, DFCI, and Regina Barzilay, Massachusetts Institute of Technology): 1.5-million-person cohort from general hospital population developed to support PDAC risk detection. Private hospital holding constructed under federated model with Observational Medical Outcomes Partnership common data model.
UK Biobank: database of 500,000 volunteers with a wide variety of health data. It is maintained and access controlled by the UK government (https://www.ukbiobank.ac.uk/).
Danish National Medical Record: Denmark maintains a comprehensive family of databases of medical care for its population. Søren Brunak, in conjunction with the Stand Up To Cancer–funded medical records team, is active in AI research for early PDAC detection using these data (https://econ.au.dk/the-national-centre-for-register-based-research/danish-registers/).
Blood-based biomarker registries is listed previously, including MSK-CSHL-Weizmann, Mayo Clinic, New York University, and DFCI.
We have identified the following resources for federated learning:
Tensorflow federated: Google has extended their tensorflow platform to include tools for federated learning. Their code and documentation are available here: https://www.tensorflow.org/federated.
NVidia Clara: The NVidia corporation has created a set of tools for image-based AI that includes a federated learning component. The effort is described on their Web site here: https://blogs.nvidia.com/blog/2019/12/01/clara-federated-learning/.
Developing Uniform Standard Operating Procedures
To maximize the ability of AI as an aid to early detection of pancreatic cancer, we feel that it is important to develop standard operating procedures for the collection both of biological materials and the demographic, clinical, pathological, imaging, and genomic data that annotate these biospecimens. Uniform standard operating procedures would improve the ability to ensure high-quality data and compatibility of data extraction across sites. Some examples are the following:
-
It is likely that effective early detection programs will include assays of a body substance for a biomarker associated with a developing pancreatic cancer in an asymptomatic person. Although most current efforts involve a study of a blood component (eg, plasma or serum), urine, the stool microbiome (or saliva), or exhaled respiratory air may also provide the substrate for an early detection biomarker. Standard operating procedures for collection of biological material may be crucial for the development of the assays that will eventually be used in the standard of care clinical setting.169 For example, assays for blood-based biomarkers may be influenced by preparation and storage of the appropriate blood component. Although the goal is the development of an assay that can be performed in a CLIA-approved laboratory, so that preparation and storage should be achievable in a community setting, particularly during the early stages of assay development, adopting uniform SOP for preparation, storage, and performance of the assay allowing for multi-institutional validation studies is important. Uniform SOP applies not only to blood and tissue but also to imaging and data collection, and storage (as discussed previously). Examples include the following:
∘ The Observational Health Data Sciences and Informatics group is an organization dedicated to enabling observational research through standardization of data formats, methods, and tools. They developed and maintain the Observational Medical Outcomes Partnership Common Data Model and associated tools. The Common Data Model defines a set of standard terminology and formats that facilitate data interchange across institutions in a site-agnostic manner. Details of this effort are available here: https://www.ohdsi.org/data-standardization/the-common-data-model/.
∘ The NIH-NCI EDRN project has adopted a standardized data dictionary, initially developed at the DFCI, for data collection in their multisite study. A standardized RedCAP database model is used to implement many of the data dictionary standards.
∘ The Digital Imaging and Communications in Medicine standard is used to allow for interoperability across imaging vendors, archiving and storage systems, and image analysis platforms. Compliance with the Digital Imaging and Communications in Medicine standard allows image data and annotation objects to seamlessly move across storage platforms and provides access to a large body of image analysis tools (https://www.dicomstandard.org/).
Opportunities and Needs for Near-Term Progress Toward Early Detection
We have identified both opportunities and needs in each of the areas discussed in this section. The most promising and highest yields of those are as follows:
Current Research
Most of the identified efforts are using established epidemiological or ML techniques rather than true deep learning techniques.
Integration of disparate data sources like imaging, genetics, -omics, patient characteristics, and microbiome data remains limited. Although a few active projects have been supported by NCI, Lustgarten, and Stand Up To Cancer, there remains relatively little activity in this domain.
No ongoing PDAC-specific microbiome early detection research was identified (although several projects are under discussion), so to date, we have not been able to identify an opportunity to integrate these data into risk models using AI techniques. We anticipate, however, that microbiome/bacterial analysis PDAC early detection studies will be or are starting.
Data science techniques for natural language processing, time series analysis, and integrative risk analysis remain poorly represented in active PDAC research and in the PDAC literature.
-
The absence of a public or semipublic data set for PDAC risk may be a barrier to recruiting nonmedical PDAC researchers to this important cause.
∘ Training and testing data are needed across the spectrum of imaging (including CT, MRI, and ultrasound), genomics, proteomics, immune factors, and metabolomics.
Data Accessibility for Pancreatic Cancer AI Research
There are few centralized, anonymized data sets that can be semipublicly accessed by researchers for PDAC research. The NIH-NCI–sponsored EDRN effort is currently the only major public effort that we are aware of in this space.
There has been no peer-reviewed demonstration of a federated learning system in this space. Although Stand Up To Cancer has funded a project with this as a core goal, these techniques will remain inaccessible to most research groups until proven systems are established and made available.
Multidisciplinary Collaboration
-
There is a need for a mechanism for sharing knowledge of PDAC AI projects and for increasing outreach and involvement of non-PDAC AI researchers.
∘ As noted previously, we suggest that a Web-based site that would allow posting in the public domain of both planned and ongoing AI efforts in early detection (and other aspects of pancreatic cancer research including but not limited to diagnosis, staging, and treatment), and including contact information to the principal investigators, funding sources, and scope of the program, as well as aggregation of information of available cohort sets, will considerably improve national and international efforts in AI and pancreatic cancer research.
The development of international efforts for COVID-19 pandemic may serve as a model to develop AI in early detection of pancreatic cancer collaborations.
SECTION D: ORGANIZATIONAL STRUCTURES AND COLLABORATIVE OPPORTUNITIES FOR PANCREATIC CANCER EARLY DETECTION
Co-leads: Stephen J. Pandol, MD, and Anil K. Rustgi, MD
Group members: Noura Abul-Husn, MD, PhD; Debiao Li, PhD; and Lawrence H. Schwartz, MD
Overview
Several studies have shown that early detection of PDAC improves outcome. This result is based on the findings that surgery is the only therapy to date associated with a durable long-term outcome. Furthermore, the size of the lesion at the time of surgery is a predictor of long-term survival.3,166,167 Thus, using methods to analyze large and diverse data sets of risk factors to develop prediction models for identifying individuals with increased probability of developing PDAC is essential for monitoring at risk populations with biomarker tests and imaging methods tests for early detection when the pancreatic lesion is small. This strategy should lead to improved outcome for patients with this disease. However, there are many challenges to increasing the number of patients identified with early lesions including the fact that there are no specific symptoms in patients with early disease. This issue is confounded by the fact that the disease is relatively rare so that screening tests without nearly 100% specificity and high sensitivity will lead to a large number of false-positive tests requiring follow-through with imaging and invasive biopsy tests that are associated with risk to the patient.
Finally, standard abdominal CTs for the general population are fraught with missed diagnosis with standard approaches to interpretation,170 and the currently used blood-based biomarker, CA 19-9, accurately predicts only 65% of those with early disease when surgical treatment is a viable option.3,167 This article presents an overview of the challenges that need to be traversed to markedly improve outcome in patients with PDAC. We propose a collaborative enterprise bringing together a multidisciplinary group of experts using advanced analytic methods of AI and ML to identify and validate predictive factors in specific data sets as well as across data sets to establish a comprehensive approach to risk prediction, testing and diagnosing PDAC at its very earliest stages.
Identifying High-Risk Populations for Screening and Monitoring
To address the challenges outlined previously, screening for early PDAC requires identification of groups of patients with increased risk to maximize the identification of cases with the disease and decrease the number of false-positives that lead to invasive procedures for definitive diagnosis resulting in risks to the patient. Epidemiological studies indicate that susceptibility to PDAC is a complex interplay of modifiable risk factors and genetics that must be considered in developing models for risk prediction.
In a major effort to developing tests (liquid biopsy and imaging), the US Consortium on Chronic Pancreatitis, Diabetes, and Pancreatic Cancer has initiated a longitudinal cohort study, the New-Onset Hyperglycemia and Diabetes Cohort, to discover and validate high-sensitive and specificity biomarkers to identify patients with a high probability of having early PDAC.167 This study enrolls patients with the onset of hyperglycemia or diabetes after age 50 years. Of note, epidemiological studies support that this group of patients has about 1% probability of diagnosis for 3 years after the onset of diabetes. The study will recruit 10,000 patients, which will yield approximately 100 cases of pancreatic cancer. Developing liquid biopsy and imaging tests that distinguish between those who develop PDAC and those who do not with high sensitivity and specificity is the key goal of this cohort study. Of note, several studies show that the probability PDAC development can be further increased by including additional factors such as weight change, ethnicity, and lifestyle factors that are commonly contained in electronic medical records.171–178
Other well-known high-risk populations must be considered in identifying patients who need screening with imaging and liquid biopsy approaches. For example, chronic pancreatitis especially those who develop diabetes with chronic pancreatitis have the highest proportional risk of developing PDAC.178,179 Another risk population are those patients who are increasingly identified with pancreatic cysts.178 Germline pathogenic (ie, disease-causing) variants in a number of genes associated with hereditary cancer syndromes or hereditary pancreatitis are known to increase the risk of pancreatic cancer. Recent studies estimate that genomic risk contributes to up to 8% of PDAC cases,180–184 and that 10% of patients with PDAC harbor germline pathogenic variants.185 These include monogenic (single gene) variants in double-strand DNA damage repair genes associated with hereditary cancer syndromes, such as BRCA1 and BRCA2 (hereditary breast and ovarian cancer syndrome), ATM (ataxia telangiectasia syndrome), and PALB2. Other genes associated with an increased lifetime risk of PDAC are as follows: MLH1, MSH2, MSH6, PMS2, and EPCAM (associated with Lynch syndrome), APC (familial adenomatous polyposis), CDKN2A (familial atypical multiple mole/melanoma syndrome), TP53 (Li-Fraumeni syndrome), and STK11 (Peutz-Jegher syndrome).186 Germline variants in BRCA1 and BRCA2 are especially common in the general population187 and have a predictive value in terms of treatment response and survival.185,188
Medical history–based features to identify individuals with germline pathogenic variants linked to pancreatic cancer include a young age at PDAC diagnosis and a family history of pancreatic or other cancers. However, these classic features have poor sensitivity in identifying individuals at high genomic risk for pancreatic cancer.182–185 Recently published recommendations outline the need for genetic evaluation in all patients diagnosed with pancreatic cancer, regardless of age at diagnosis or family cancer history.189 Screening of unaffected individuals who have a significant family history suggestive of FPC is also recommended.189 An emerging strategy is population genomic screening to uncover the genomic risk of certain cancers in asymptomatic or presymptomatic individuals.187 The rationale is that identifying germline pathogenic variants can facilitate cancer prevention and increase early detection through enhanced surveillance and risk-reducing interventions in individuals harboring such variants.190 The suitability of this genomic screening approach at a population level will depend on pilot studies demonstrating downstream patient benefit and clinical utility.191
Common genetic variants identified through genome-wide association studies of pancreatic cancer can also inform risk stratification.192,193 Genome-wide association studies have shown that the genetic underpinning of most common diseases, including pancreatic cancer, is highly polygenic, comprising hundreds to thousands of variants that each have a small effect on disease risk. Polygenic risk scores are constructed by aggregating weighted genotypes for risk alleles into a single, integrated measure of risk.194 Polygenic scores promise to revolutionize genomic screening for common diseases by identifying individuals with disease risk that is equivalent to monogenic risk.195 However, there is still considerable debate around the clinical utility of polygenic scores to determine cancer risk.194
As genomic testing costs continue to rapidly decrease, and clinical utility of genomic screening and polygenic scores is better understood, it is reasonable to consider that genomic applications will become a routine part of clinical care. To maximize the potential predictive value of genomics in pancreatic cancer, it will be critical to evaluate the integration of germline monogenic risk, polygenic risk, and family history information with other clinical factors, to generate a comprehensive genomic risk assessment. Individuals identified as having the highest genomic risk of pancreatic cancer can then be offered genomic-driven surveillance and risk-reducing interventions to mitigate that risk.
Another consideration for identifying patients at risk for PDAC include use of social media and Internet-based commercial data.178 The enormous expansion of use of the Internet for communication and purchasing creates an interesting and powerful mechanism to identify early signs of PDAC outside the walls of health care facilities and constraints of research studies. The contribution of mining Internet interactions may identify behavioral changes in early PDAC patients that are not discerned by the medical system as early symptoms are vague and not easily recognized by medical professions as PDAC. The science of social netnography (a type of ethnography) that analyzes perceptions and behaviors of individuals online178 may be able to discern online behavioral patterns that occur that are associated with PDAC at its earliest stages. As an example of how this would work, identifying individuals with a self-reported PDAC diagnosis online can provide the ability to collect their previous deidentified and publicly available online posts and purchases that occurred before their cancer diagnosis. By using these data, researchers can identify online behavioral signals and develop a behavioral phenotype that can then be validated in a prospective manner again using online users. This approach was recently described in more detail.178 Of note, if needed, this approach can apply to specific geographic regions or ethnic groups to bring emphasis to address diversities.
In sum, this section shows that there are multiple types of data that can be used to develop and validate risk models to identify individuals at high risk that should be monitored using more specific diagnostic tools. Because the risk models require integrating and analyzing large data sets coming from different sources and disciplines, advanced data management method and AI-based analysis will be necessary to provide robust models to apply to the population. Of note, it is possible that adjustments for geographic, ethnic, and possible other biologic differences will be needed. Notably, the risk models will also likely yield opportunities for prevention of PDAC in specific patient subgroups at high risk where lifestyle alterations or prevention therapeutics can be applied.
Developing Tests Needed for Diagnosis in High-Risk Populations
Liquid biomarkers and imaging biomarkers will play a central role in the pathway to early diagnosis of PDAC.178 There are significant challenges in identifying liquid biomarkers for screening HRIs, a topic that has recently been reviewed. In brief, there have been thousands of publications showing biomarkers in PDAC. However, no single candidate biomarker has been translated into clinical practice. The low incidence of PDAC and the fact that early PDAC cases have minimal symptoms and signs of the disease are major obstacles to moving promising candidates through the validation process needed for approval for use in practice. Recently, large and international studies have been launched to obtain samples and monitor patients with a high risk of PDAC to identify early biomarkers.178 The example described previously is New-Onset Hyperglycemia and Diabetes Cohort study recruiting 10,000 subjects.167 Estimates are that approximately 100 individuals among the 10,000 recruits over a 3-year period will be diagnosed with PDAC. The samples collected in this cohort study will be used for discovery followed by validation of promising biomarkers. Of note, the literature addresses the fact that disease controls such as chronic pancreatitis are needed in studies as there are several examples of biomarkers present in both chronic pancreatitis and PDAC.178
Abdominal pain is the single most common reason that Americans visit the emergency department, accounting for 7 million visits per year, where an abdominal CT scan is usually performed. Although most scans do not show any signs of cancer visible to radiologists, some subjects eventually develop PDAC in the next few years.170 These prediagnostic CT images provide critical morphological information associated with biological changes at the pre-cancer or early cancer stage, which can be extracted using AI and ML methods to predict PDAC.178 In addition, nonimaging variables such as demographic, epidemiological risk factors, anthropometry, clinical comorbidities, and laboratory tests can be combined with prediagnostic imaging feature for a more accurate prediction model. Preexisting conditions such as acute pancreatitis, chronic pancreatitis, and pancreatic cysts have been associated with the future development of PDAC179,196 so separate prediction models can be developed for each of these abnormalities by exploring imaging features for each of the conditions.
For prospective imaging studies to detect early PDAC, MRI offers a nonionizing radiation alternative. Noncontrast, quantitative MRI such as T1/T2 mapping is an effective tool to characterize tissue properties such as fibrosis and inflammation to provide early indicators of biological changes. Advanced free-breathing MR techniques are available for rapid image access and enhanced resolution can be used to reveal features that could further advance detection.197–199 Alternatively, breath-hold MRI techniques may provide additional information in select patients.200 Other advances in MR techniques including chemical exchange saturation transfer imaging to evaluate extracellular pH level imaging and low-dose dynamic contrast-enhanced MRI to assess tissue vascularity can further advance detection. As with liquid biomarker discovery and validation, disease controls such as chronic pancreatitis should be included. Advanced MR techniques including chemical exchange saturation transfer imaging to evaluate extracellular pH level and low-dose dynamic contrast-enhanced MRI to assess tissue vascularity. Overall, MRI may provide information related to the pancreatic parenchyma, and the pancreatic “environment” and other MRI sequence may visualize lesions when they are small.
Artificial intelligence-based automated segmentation of the pancreas and its subregions (head, body, tail) is a prerequisite for extraction of CT or MR image features for PDAC prediction or early detection. In addition, AI may help to detect and characterize lesions of the pancreas identified on imaging studies. Finally, serial changes and further characterization of pancreatic lesions can be best assessed with automated quantitative approaches, as is done in lung cancer screening of pulmonary nodules.201
As a point of emphasis, it is important to recognize that the performance of the testing methodology is highly dependent on the risk predictive models developed by AI and ML. That is, because PDAC is a relatively rare occurrence and because any tests developed will likely not have 100% specificity, a nonselective application of testing to the general population will lead to many false-positive tests potentially leading to greater harm than benefit.
Organizational Structures and Functions Needed for Early Diagnosis
As outlined previously, 2 overall strategies are needed for developing and implementing an early diagnosis program. One is to develop a risk model including multiple inputs from genetics, medical and lifestyle data from epidemiological studies and from electronic medical record systems, and social and commercial data mined from Internet sources. Although each of these inputs requires data collection and analysis by researchers with the knowledge and skills to identify indicators that have value in risk prediction, a combination of these inputs from these disparate approaches is likely to provide a set of indicators with greater sensitivity and specificity than the ones from any individual approach. This approach will require both a multidisciplinary approach and development of methods that are able to analyze data coming from these different methodologies to create a prediction model with a combination of indicators from these dissimilar data sets. Thus, experts in multiple methodologies starting with management of large sets of data from the different sources and the skill to perform analysis of the data to meet the expected outcomes are needed. The process would start with a discovery phase to create a prediction model followed by validation. Most importantly, application of a validated prediction model to populations should be demonstrated to result in a measurable increase in the proportion of patients with PDAC who are identified at an early stage associated with improved outcomes. Of note, the prediction model(s) tested and validated when combined with the liquid biopsy and noninvasive imaging tests should provide enough sensitivity to identify a large proportion of early cases without a significant number of false-positive tests that result in unacceptable rates of complications from the diagnostic procedures such as endoscopic ultrasound and biopsy. Obviously, important discussions about benefit, risk, and ethics are needed to set the thresholds for sensitivity and specificity of the models.
With populations of patients with identified increased risk by prediction models, the second step includes liquid biopsy and imaging tests that can identify which subjects have a very high likelihood of PDAC and who should undergo a biopsy procedure for definitive diagnosis versus those who should continue to be monitored by liquid biopsy and imaging. Like the risk prediction modeling, it is possible that a combination of results from these 2 types of tests may perform better than each individually to reveal which patients should be monitored and which patients should undergo invasive biopsy procedures for diagnosis.
Considering the multiple inputs and analysis required to optimize and deliver an early diagnosis method that improves outcome in patients with PDAC, we propose an organization structure titled “Early Detection Strategy” (Fig. 10).
FIGURE 10.
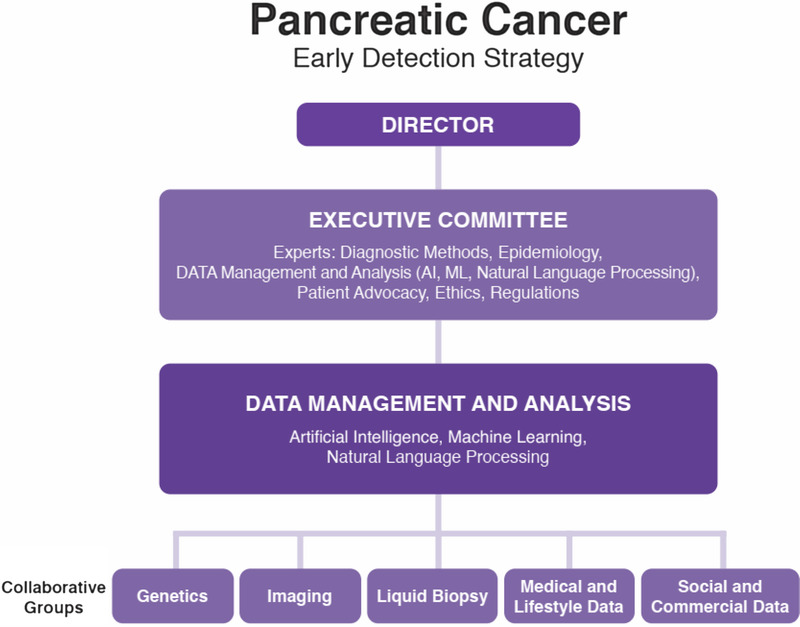
Early detection strategy. Courtesy of participants from Collaborative Opportunities and Kenner Family Research Fund.
As is evident from the draft of the Strategy in the figure, inputs from the methodologies required are organized around Collaborative Groups of the technology. In addition, to take advantage of the potential additive effects of combining different types of data, we recognize that there needs to be a centralized Data Management and Analysis Group that uses analytic tools of statistics, AI, ML, and natural language processing to produce robust risk prediction and diagnostic testing needed.
The results from the Collaborative Groups and the Data Management and Analysis Group need to be considered in the context of regulatory and ethics issues with significant input from patient advocacy representatives. An Executive Committee composed of the leaders of the Enterprise with experts in regulation, ethics, and patient advocacy would be charged with developing strategy, workflow, and milestones for the Enterprise. The Enterprise would require a director and support staff to operate the Enterprise.
Early Detection Strategy: From Concept to Reality
The initiation of the Early Detection Strategy requires selection of leadership including the Director and members of the Executive Committee. These individuals will be charged with developing a charter for the operation of the Strategy. The charter needs to include milestones and timelines to achieve an overall goal of progressively decreasing the mortality of PDAC. The charter should recognize that barriers to obtaining data needed for building the models need to be traversed to benefit society and all of its members. The charter should also develop methods to prioritize areas that analysis show have the greatest benefit in reaching to goals and at the same time remove those that show limited value from the data analysis. The charter should recognize that as more data are collected, the predictive and testing models and methods will continue to improve, which should result in continuing improvement in outcome. Finally, the Strategy will need buy-in and support from society and funding from multiple sources. Thus, the charter needs to address how budgets will be developed and how to access the resources needed.
Summary
In this dissertation focused on decreasing mortality from PDAC, we detail key challenges in the field of early diagnosis (and potentially prevention with lifestyle modification and prevention agents in high-risk patients). We provide an approach to traverse the obstacles using contributions of multidisciplinary teams with a considerable reliance on AI methodologies to provide a progressive and sustainable reduction in PDAC mortality.
SECTION E: MOVING FORWARD: REFLECTIONS FROM GOVERNMENT, INDUSTRY, AND ADVOCACY
Group members: Dana K. Andersen, MD, FACS; Jane M. Holt, BA; Graham Lidgard, PhD; and Sudhir Srivastava, PhD, MPH, MS
A Personal Viewpoint
Sudhir Srivastava
Disclaimer: The opinions expressed by the author are their own and this material should not be interpreted as representing the official viewpoint of the US Department of Health and Human Services, the NIH, or the NCI.
The ability to effectively identify earlier-stage PDAC and its aggressive precursors may represent a critical first step toward improving the survival rates. Multiple studies have shown that asymptomatic PDAC is associated with better outcomes than symptomatic disease.202,203 Early-stage detection is rare due, in part, to the highly aggressive nature of the disease, the absence of early symptoms, and the subtle imaging features. More accurate minimally invasive tests used for evaluation of early-stage, asymptomatic PDAC and its precursors are needed. Even a small increase in the sensitivity has the potential to reduce the number of misses (false negatives) and thus improve patient outcomes. Existing tests have limited sensitivity for detection of early-stage disease to ensure that few true cases are missed (low false-negative error rate). The most widely used blood-based biomarker, CA 19-9, accurately predicts only 65% of resectable pancreatic tumors. Standard abdominal CT scans (“the pancreas protocol”) have sensitivity ranging between 76% and 92% for diagnosing pancreatic cancer even in late stages of the disease.204,205 The problem is exacerbated by issues related to interobserver and intraobserver variability of CT image interpretations rendered by radiologists, which can be as high as 37%.206
Although screening for PDAC in the general population is not feasible, it may be beneficial for individuals at high risk for developing the disease. Recent discoveries have linked 35% to 50% of PDAC cases to more than 40 potential risk factors and medical conditions, such as familial risk, germline mutations, NOD,68 and pancreatic cystic lesions. Panels of noninvasive biomarkers, such as CA 19-9 and CEA, look promising, but none has been clinically validated for screening so far. Presently, the utility of screening in these high-risk groups is not well established, and the groups themselves are not well defined.206 Imaging-based screening is often recommended for people with genetic predisposition or suspicious lesions. Still, more evidence is needed on issues such as when to start screening, the interval of follow-up surveillance, and preferred modalities.204,205
The development of risk-based screening protocols for PDAC will require more accurate risk prediction models and risk scores for different high-risk groups. For example, even experienced radiologists cannot accurately separate benign and low-grade lesions from high-grade precursor lesions based on morphological features, without supporting histological evidence obtained by surgical biopsy. Typically, morphological features with or without cyst fluid analysis are used to guide the physician's choice between surveillance, surgery, and expectant management. Approximately half of pancreatic cysts detectable by CT are IPMNs, which have significant malignant potential (from 2% to 8% for branch duct IPMNs and 35% to 68% for main duct IPMNs). The current approach to preoperative identification of potentially malignant IPMNs (Fukuoka criteria) incorrectly directs benign lesions to surgery one-third of the time (a false-positive rate of 36%).207
The sharp rise in the use of abdominal CT imaging over the past decade has brought a “man-made epidemic” of pancreatic cysts.207,208 Annually, more than 50 million such scans are performed for reasons unrelated to the pancreas, and more than 6 million asymptomatic pancreatic cysts are discovered based on these scans. Most of these “incidental” cysts are benign, but some harbor aggressive precursor lesions that can rapidly give rise to invasive PDAC either directly or indirectly through field cancerization and pro-inflammatory signaling. The diagnostic uncertainty generates overtreatment as well as considerable anxiety among the affected individuals due to the concern for possible malignant transformation to pancreatic cancer. The “cyst epidemic” also causes a considerable increase in interpretation workload for radiologists, further highlighting the importance of improving existing imaging tests as well as protocols for risk assessment of cysts.208
There is a strong interest in the research community to use data science methods, such as AI, to assist radiologists in detecting visual abnormalities while minimizing both false-positives and false-negatives. Specifically, AI systems could improve early detection of PDAC and management of asymptomatic high-risk groups, including those with premalignant pancreatic lesions and FPC. Most recently, deep learning, a subset of AI, has emerged as a powerful approach for information extraction from CT volumes. It is well suited for modeling the intricate relationships between images and their subsequent interpretation. Traditional ML approaches like SVM or Random Forest are better suited for analysis of relatively modest-sized data sets with a large number of variables. For this reason, the applicability of omics-based deep learning algorithms to early detection of cancer is still limited because of the challenges associated with obtaining adequate samples from thousands of patients who have not yet developed cancer.209
The View From the NIDDK
Dana K. Andersen
The mission and scope of the NIDDK includes several diseases and conditions that are risk factors for and therefore potential harbingers of pancreatic cancer. Diabetes, obesity, and pancreatitis are all major research interests of the NIDDK, and each has been shown to increase the risk of PDAC. Diabetes is a particular interest as it is both a risk factor for the development of PDAC, with a roughly 2-fold increased incidence of PDAC in long-standing, largely type 2 diabetes mellitus (T2DM), as well as being a consequence of PDAC.210 The prevalence of diabetes among patients diagnosed with PDAC is remarkably higher than for any other solid tumor,211 and more than 50% of PDAC is accompanied by diabetes at the time of diagnosis.212 Roughly half of the diabetes associated with PDAC is of recent onset, having occurred within 24 to 36 months of PDAC diagnosis. Furthermore, more than half of the NOD resolves after resection of the tumor (and half of the pancreas) and is therefore considered a paraneoplastic process induced by the tumor.213 The form of diabetes caused by exocrine pancreatic disease including PDAC is referred to as pancreatogenic or type 3c diabetes mellitus (T3cDM).210 The mechanism(s) whereby PDAC causes T3cDM is not clearly understood, but roughly 1% to 2% of NOD in persons older 50 years is associated with PDAC.71 This indicates that the detection of T3cDM, and distinguishing it from the vastly more common T2DM, may lead to the early detection of PDAC, as many of these cases of PDAC-T3cDM develop up to a year or 2 before the appearance and diagnosis of PDAC.
The identification of T3cDM separate from T2DM is a current research focus of the NIDDK-NCI–sponsored Consortium for the Study of Chronic Pancreatitis, Diabetes, and Pancreatic Cancer. The Chronic Pancreatitis, Diabetes, and Pancreatic Cancer study titled “Evaluation of a Mixed Meal Test for Diagnosis and Characterization of Pancreatogenic Diabetes secondary to Pancreatic Cancer and Chronic Pancreatitis,” or the DETECT study, seeks to evaluate several potential biomarkers of T3cDM that have shown promise in small cohort studies.214 A total of 452 subjects with and without diabetes associated with chronic pancreatitis, pancreatic cancer, and no pancreatic exocrine disease are being recruited. A preliminary interim analysis is underway.
Artificial intelligence methods have been applied to imaging studies of the pancreas, as reviewed in the NIDDK workshop “Precision Medicine in Pancreatic Disease” held in July 2019215,216 and at the NIDDK workshop “Imaging the Pancreas in Diabetes and Benign and Malignant Pancreatic Exocrine Disease” held in January 2020. Dr. Ronald Summers and colleagues in the Advanced Imaging Center of the NIH Clinical Center have explored AI applications in pancreatic imaging to enhance the detection of early-stage PDAC.217,218 This represents a major advance in the field because the interpretation of CT radiography has been marred by a significance rate of failure to depict or detect the early-stage PDAC. Artificial intelligence applications have the potential of greatly enhancing the early detection of small, localized PDAC lesions that are amenable to surgical cure. Beyond imaging applications, applying AI to identify those asymptomatic persons who may harbor an occult PDAC and are therefore candidates for newer imaging methods is the next goal of the use of deep learning to assess large data sets to identify early-stage PDAC.
An Industry Perspective
Graham Lidgard
“You can't really know where you are going until you know where you have been.” (Maya Angelou)
And a word of caution for those with the arrogance to predict:
“There is no reason anyone would want a computer in their home.” (Ken Olsen, founder of Digital Equipment Corporation, 1977)
The United States will spend more than $4 trillion on health care in 2020, almost 20% of the gross domestic product.219 According to an NCI report, approximately 5% (>$200 billion) is spent on cancer care, and health care reports estimate another 1% is spent on cancer preventative services such as breast, colon, and cervical screening. Government through its institutions (eg, NIH, NCI, NHLBI (National Heart, Lung, and Blood Institute), NASA (National Aeronautics and Space Administration), DARPA (Defense Advanced Research Projects Agency); see GRANTS.gov) combined with state and nonprofit organizations will spend an estimated $200 billion in medical and health care related research. By contrast, the whole of the medical device/in vitro diagnostics/laboratory industry is less than $125 billion in US revenues.
Over the last half century, the medical devices and diagnostic industry has been in a close but somewhat unacknowledged “partnership” with the government. Most of the seminal discoveries and innovations have come from federal- or government-funded research around the world. These discoveries find their way into industry through formal licensing, entrepreneurial spin-out, or adoption of non-IP protected work. Industry exploits, improves, and commercializes these technologies for the benefit of all. Through this process, massive progress has been made in the field of imaging technologies, CT, MRI, ultrasound, and endoscopy. Similarly, we have seen the incredible development of in vitro diagnostics with advanced immunoassay technology, molecular technology, and automation that has fueled multiple important diagnostic tests in all areas of medicine including cancer. Biomarkers in the field of endocrinology; insulin, gastrin, growth hormone, and prolactin; and the early solid tumor associated markers such as CEA, CA 19-9, CA 125, PSA, and fecal hemoglobin have had an impact on the cancer field. With these single or small multiplexed set of analytes, simple cutoff or logistic algorithms have been sufficient to define response parameters that allow for assays that discriminate disease from nondisease, and consequently, the need for AI/ML has been limited.
However, although our knowledge and understanding of cancer are exponentially greater than it was 50 years ago, we have not made the same progress in curing, improving survival, or early detection.220 With an aging population, we are seeing increases in the total number of cancer cases and, for certain cancers, an overall increase in incidence. Decreases in deaths or increased survival in certain cancers such as cervical, breast, and colon cancers have been associated with screening, whereas a decrease in lung cancer is associated with a reduction in smoking. The decline of stomach cancer in the United States is still not understood, but it is not considered because of medical intervention.
As we moved into the era of “Big Data,” we see the collection and assembly of large public data bases such as TCGA, with more than 20,000 primary cancer and 33 cancer types, the COSMIC catalog of somatic mutations in cancer; MethylCancer, database of DNA methylation in cancer; and many, many more. The ability to access and extract useful information requires extensive computing power, and AI/ML is finding utility. However, the metadata associated with these databases are not necessarily helpful in building predictive tools—patient data may include age, sex, race but not the germline genome, family history, or medical records.
Insight may be attained from other classic disease studies. The impact the Framingham Heart Study221 has had on our understanding of cardiovascular disease and the improvement in survival and reduction of cardiovascular events is an example of what might be necessary in the future to influence cancer prevention and survival. The study is currently in its 71st year and has followed generations of subjects in Framingham, Mass. The study was enacted into law in the same legislation that founded the National Heart Institute, now the National Heart, Lung and Blood Institute. The study revolutionized our understanding of the etiology of cardiovascular disease, the simple monitoring of blood pressure, and the role of lipids: cholesterol, high-density lipoprotein, and the low-density lipoprotein fraction and their management in preventing adverse outcomes.
Do we need a similar study to follow a population and to collect data for better understanding of the overall cycle of cancer? With second- and third-generation molecular technologies entering the diagnostic field, we see large data becoming more important: whole genome, exome, targeted methylated, and more, producing terabytes of data. But just as important is all of the patient data that put the information into context: Access to patient medical records to build the longitudinal medical information that might inform the tumor data is needed.
Numerous companies, academic centers, and government agencies are attempting to build better informed databases to use data for risk prediction, identify patients for clinical trials, or develop drugs. However, the size and scope of what is needed may be beyond the funding capabilities of these organizations, and there is resistance to pooling the information and standardization of data formats. This resistance emanates from multiple sources. As examples, there can be numerous institutional review boards involved in a clinical study controlling how the collected data can be used, the extent of informed consent required, and how deidentifying must be conducted or data excluded to meet the Health Insurance Portability and Accountability Act requirements. Companies or scientists sponsoring and managing the studies may not want to enable competitors by giving others access, and both international and national groups are wary on how their populations genome data may be used, and consequently may try to limit uncontrolled future access.
Finally, pancreatic cancer detection might present a solution to the prevalence problem. If we look at the cancers that have no screening modality, can we use molecular technology (both nucleic acid and protein) to identify cancer earlier than would be detected in the routine work-up of symptoms and at what cost? Companies like Grail, Thrive, and Exact Sciences are working on technological solutions to detect the cancers earlier. The work of Ahlquist222 shows that grouping the prevalence of cancers together could justify this approach if high specificity is achieved. Artificial intelligence and ML are used to search gigabytes of sequencing data from liquid biopsies across multiple patients to examine features that correlate with disease and can identify cancer with the high specificity. The AI/ML algorithms can process large amount of data that would not be possible with the simpler logistic methods.156 To date, the limitation has been on early cancer sensitivity, but as we add in other marker data, we will see the lower boundaries expanding and detection of early stage improving.
Summary
Industry and government have a long history of success working together.
Historically, small number of targets and limited access to data are not conducive to AI/ML.
Image data have been available to use for AI/ML for many years and had some success.
New technology with multitarget and extensive deep molecular sequencing data as well as access to extensive patient metadata makes AI/ML more interesting.
The FDA has written a discussion article for the use of AI/ML in diagnostic applications.156
Challenges for early detection include identifying the population to be tested and sensitivity/specificity of a test.
For PDAC, the asymptomatic population is too large and specificity challenge is too high to make widespread screening safe and cost-effective.
Do we need an AI/ML that covers a significant fraction of the disease to identify higher-risk population: >50% with prevalence of >2%?
Can pancreatic cancer help with prevalence and specificity with enough sensitivity?
After all of this, can we make a difference in survival or cure?
Reflections From Patient Advocacy
Jane M. Holt
Early detection of cancer enhances the chances for successful treatment. This is proven by the increasing survival rates of any cancer with an early detection method. There are several critical components to early detection: first, education to promote early diagnosis and screening, and, second, an awareness of warning signs of cancer and steps for prompt action.
Patient advocacy groups can play a major role in supporting the quest for early detection of pancreatic cancer. As an example, The National Pancreas Foundation is building an awareness campaign around early detection of pancreatic cancer by focusing on high-risk groups such as those with a history of pancreatic cancer in their family, those who have hereditary pancreatitis, or individuals with NOD. By creating a high-impact, multimedia campaign to raise awareness of pancreatic cancer, pancreatic cancer patient descendants will be encouraged to seek genetic testing. This campaign has the potential to save lives and improve patient outcomes. The work of the Rolfe Pancreatic Cancer Foundation is another example of how advocacy interacts with research. They have developed a family history tool to “Know Your Risk” for pancreatic cancer. Other patient advocacy groups, including Pancreatic Cancer Action Network, Lustgarten Foundation, Project Purple, Griffith Family Foundation, Hirshberg Foundation, and the Ron Foley Pancreatic Cancer Foundation, provide critical funding for early detection initiatives. The AI and Early Detection of Pancreatic Cancer Summit presented by Kenner Family Research Fund is an additional illustration of how patient advocacy supports progress in this area.
Regrettably, the rate of pancreatic cancer patients enrolling in clinical trials is very low. However, through grass root programs, patient advocacy groups play a critical function in educating patients, encouraging participation of underserved communities, and providing guidance to researchers on how to make the clinical trial process easier for patients. Several nonprofit groups also assist with financial support for patients who would not otherwise be able to participate in trials. In addition, a program supported by the Pancreatic Cancer Action Network successfully pairs patients with appropriate clinical trials. Similarly, the National Pancreas Foundation has developed an animation module about clinical trials to inform patients about the process for enrollment. Registries developed by these 2 groups survey patients, track genetic testing results, gather demographic information, provide important data for researchers, and inform patients about clinical trials.
Patient advocacy groups have been very successful in raising critical research funds for early detection, whether it be from outreach to government, industry, or private individuals. These funds also support a younger generation of doctors and researchers interested in pancreatic cancer by providing grants to those with promising work.
The World Pancreatic Cancer Coalition (http://www.worldpancreaticcancercoalition.org) was founded in 2016 and consists of more than 90 patient advocacy groups from more than 30 countries on 6 continents. The Coalition members collaborate to raise awareness about the symptoms and risk factors of pancreatic cancer. The voice from this group strengthens each year, and through collective impact, members are focused on driving transformational change that will improve earlier detection and ultimately increase survival of pancreatic cancer.
ACKNOWLEDGMENTS
Group section leads for the AI and Early Detection of Pancreatic Cancer presummit article were Suresh Chari, MD, of MD Anderson Cancer Center; David Kelsen, MD, of Memorial Sloan Kettering Cancer Center; David Klimstra, MD, of Memorial Sloan Kettering Cancer Center; Stephen Pandol, MD, of Cedars-Sinai Medical Center; Michael Rosenthal, MD, PhD, of Dana-Farber Cancer Institute; Anil Rustgi, MD, of NewYork-Presbyterian Hospital/Columbia University Irving Medical Center, James Taylor, MD, of Google Health; and Adam Yala, PhD Candidate, of MIT. Summit Planning Committee Team members were Anil Rustgi, MD (President of the American Pancreatic Association); Vay Liang Go, MD; Suresh Chari, MD; David Klimstra, MD; Bruce Field; and William Hoos, MBA (Kenner Family Research Fund scientific board members); Laura Rothschild, MBA; Ann Goldberg, BA; and Barbara Kenner, PhD (Kenner Family Research Fund board members); and Susan Randel, BA (advisor).
Footnotes
S.T.C., D.K., D.S.K., S.P., M.R., A.K.R., J.A.T., and A.Y. are section leads. Other authors are listed in alphabetical order.
This article was prepared for the AI and Early Detection of Pancreatic Cancer 2020 Virtual Summit through the support of Kenner Family Research Fund.
The authors declare no conflict of interest. S.T.C., B. F., V.L.W.G., W.H., and D.S.K. are Kenner Family Research Fund scientific advisors. A.G., B.K., and L.R. are Kenner Family Research Fund board members.
Contributor Information
Barbara Kenner, Email: drbken50@gmail.com.
Suresh T. Chari, Email: stchari@mdanderson.org.
David Kelsen, Email: kelsend@mskcc.org.
David S. Klimstra, Email: klimstrd@mskcc.org.
Stephen J. Pandol, Email: stephen.pandol@cshs.org.
Michael Rosenthal, Email: Michael_Rosenthal@dfci.harvard.edu.
Anil K. Rustgi, Email: akr2164@cumc.columbia.edu.
James A. Taylor, Email: uncjat@google.com;uncjat@uw.edu.
Adam Yala, Email: adamyala@csail.mit.edu.
Noura Abul-Husn, Email: noura.abul-husn@mssm.edu.
Dana K. Andersen, Email: dana.andersen@nih.gov.
David Bernstein, Email: dbernstein@su2c.org.
Søren Brunak, Email: soren.brunak@cpr.ku.dk.
Marcia Irene Canto, Email: mcanto@jhmi.edu.
Yonina C. Eldar, Email: yonina.eldar@weizmann.ac.il.
Elliot K. Fishman, Email: efishman@jhmi.edu.
Julie Fleshman, Email: JFleshman@pancan.org.
Vay Liang W. Go, Email: vlwgo@mednet.ucla.edu.
Jane M. Holt, Email: jholt203@gmail.com.
Bruce Field, Email: bfield@minn.net.
Ann Goldberg, Email: anngoldberg21@gmail.com.
William Hoos, Email: williamhoos@gmail.com.
Christine Iacobuzio-Donahue, Email: iacobuzc@mskcc.org.
Debiao Li, Email: debiao.li@cshs.org.
Graham Lidgard, Email: glidgard@exactsciences.com.
Anirban Maitra, Email: amaitra@mdanderson.org.
Lynn M. Matrisian, Email: lmatrisian@pancan.org.
Sung Poblete, Email: spoblete@su2c.org.
Laura Rothschild, Email: ljrchild@gmail.com.
Chris Sander, Email: chris@sanderlab.org.
Lawrence H. Schwartz, Email: lhs2120@mail.cumc.columbia.edu.
Uri Shalit, Email: urishalit@technion.ac.il.
Sudhir Srivastava, Email: srivasts@mail.nih.gov.
Brian Wolpin, Email: bwolpin@partners.org.
REFERENCES
- 1.Cancer Stat Facts: Pancreatic Cancer. Available at: https://seer.cancer.gov/statfacts/html/pancreas.html. Accessed October 10, 2020.
- 2.Gheorghe G Bungau S Ilie M, et al. Early diagnosis of pancreatic cancer: the key for survival. Diagnostics. 2020;10:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chari ST Kelly K Hollingsworth MA, et al. Early detection of sporadic pancreatic cancer: summative review. Pancreas. 2015;44:693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canto M, Brentnall TA. Screening familial pancreatic cancer (FPC). Supplemental Digital Content 1. In: Chari ST, Kelly K, Hollingsworth MA, et al. Early detection of sporadic pancreatic cancer: summative review. Pancreas. 2015;44:693–712. Available at: http://links.lww.com/MPA/A372. Accessed August 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenner BJ Chari ST Cleeter DF, et al. Early detection of sporadic pancreatic cancer: strategic map for innovation—a white paper. Pancreas. 2015;44:686–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenner BJ Chari ST Maitra A, et al. Early detection of pancreatic cancer—a defined future using lessons from other cancers: a white paper. Pancreas. 2016;45:1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenner BJ Go VLW Chari ST, et al. Early detection of pancreatic cancer: the role of industry in the development of biomarkers. Pancreas. 2017;46:1238–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenner BJ. Early detection of pancreatic cancer: the role of depression and anxiety as a precursor for disease. Pancreas. 2018;47:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seoud T Syed A Carleton N, et al. Depression before and after a diagnosis of pancreatic cancer: results from a national, population-based study. Pancreas. 2020;49:1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahib L Smith BD Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. [DOI] [PubMed] [Google Scholar]
- 11.Fedewa SA Sauer AG Siegel RL, et al. Prevalence of major risk factors and use of screening tests for cancer in the United States. Cancer Epidemiol Biomarkers Prev. 2015;24:637–652. [DOI] [PubMed] [Google Scholar]
- 12.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 13.Chakraborty S, Singh S. Surgical resection improves survival in pancreatic cancer patients without vascular invasion—a population based study. Ann Gastroenterol. 2013;26:346–352. [PMC free article] [PubMed] [Google Scholar]
- 14.Katz MH Hu CY Fleming JB, et al. Clinical calculator of conditional survival estimates for resected and unresected survivors of pancreatic cancer. Arch Surg. 2012;147:513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pongprasobchai S Pannala R Smyrk TC, et al. Long-term survival and prognostic indicators in small (≤2 cm) pancreatic cancer. Pancreatology. 2008;8:587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackford AL Canto MI Klein AP, et al. Recent trends in the incidence and survival of stage 1A pancreatic cancer: a surveillance, epidemiology, and end results analysis. J Natl Cancer Inst. 2020;112:1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen PJ Kuk D Castillo CF, et al. Multi-institutional validation study of the American Joint Commission on Cancer (8th Edition) changes for T and N staging in patients with pancreatic adenocarcinoma. Ann Surg. 2017;265:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter JM Jiang W Basturk O, et al. Recurrence and survival after resection of small intraductal papillary mucinous neoplasm-associated carcinomas (≤20-mm invasive component): a multi-institutional analysis. Ann Surg. 2016;263:793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basturk O Hong SM Wood LD, et al. A revised classification system and recommendations from the Baltimore Consensus Meeting for neoplastic precursor lesions in the pancreas. AmJ Surg Pathol. 2015;39:1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hruban RH Adsay NV Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda Y Furukawa T Yachida S, et al. The prevalence and clinicopathological characteristics of high-grade pancreatic intraepithelial neoplasia: autopsy study evaluating the entire pancreatic parenchyma. Pancreas. 2017;46:658–664. [DOI] [PubMed] [Google Scholar]
- 22.Hruban RH Takaori K Klimstra DS, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–987. [DOI] [PubMed] [Google Scholar]
- 23.Waddell N Pajic M Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones S Zhang X Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biankin AV Waddell N Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network . Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer. 2016;16:553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansel DE, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Annu Rev Genomics Hum Genet. 2003;4:237–256. [DOI] [PubMed] [Google Scholar]
- 29.Storz P, Crawford HC. Carcinogenesis of pancreatic ductal adenocarcinoma. Gastroenterology. 2020;158:2072–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17:153–168. [DOI] [PubMed] [Google Scholar]
- 31.Shain AH Giacomini CP Matsukuma K, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A. 2012;109:E252–E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinberg W. The clinical utility of the CA 19-9 tumor-associated antigen. Am J Gastroenterol. 1990;85:350–355. [PubMed] [Google Scholar]
- 33.Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33:266–270. [DOI] [PubMed] [Google Scholar]
- 34.US Preventive Services Task Force, Owens DK Davidson KW, et al. Screening for pancreatic cancer: US Preventive Services Task Force reaffirmation recommendation statement. JAMA. 2019;322:438–444. [DOI] [PubMed] [Google Scholar]
- 35.Singhi AD Koay EJ Chari ST, et al. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology. 2019;156:2024–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pepe MS Etzioni R Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. [DOI] [PubMed] [Google Scholar]
- 37.Carrick DM Black A Gohagan JK, et al. The PLCO Biorepository: creating, maintaining, and administering a unique biospecimen resource. Rev Recent Clin Trials. 2015;10:212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prentice RL, Anderson GL. The women's health initiative: lessons learned. Annu Rev Public Health. 2008;29:131–150. [DOI] [PubMed] [Google Scholar]
- 39.Peila R, Rohan TE. Diabetes, glycated hemoglobin, and risk of cancer in the UK Biobank study. Cancer Epidemiol Biomarkers Prev. 2020;29:1107–1119. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y Kaur S Huang Y, et al. Biomarkers and strategy to detect pre-invasive and early pancreatic cancer: state of the field and the impact of the EDRN. Cancer Epidemiol Biomarkers Prev. 2020;29:2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fahrmann JF Bantis LE Capello M, et al. A plasma-derived protein-metabolite multiplexed panel for early-stage pancreatic cancer. J Natl Cancer Inst. 2019;111:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staal B Liu Y Barnett D, et al. The sTRA plasma biomarker: blinded validation of improved accuracy over CA19-9 in pancreatic cancer diagnosis. Clin Cancer Res. 2019;25:2745–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J Bamlet WR Oberg AL, et al. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci Transl Med. 2017;9:eaah5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen JD Li L Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumstrei K, Chen H, Brenner H. A systematic review of serum autoantibodies as biomarkers for pancreatic cancer detection. Oncotarget. 2016;7:11151–11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Capello M Vykoukal JV Katayama H, et al. Exosomes harbor B cell targets in pancreatic adenocarcinoma and exert decoy function against complement-mediated cytotoxicity. Nat Commun. 2019;10:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattox AK Bettegowda C Zhou S, et al. Applications of liquid biopsies for cancer. Sci Transl Med. 2019;11:eaay1984. [DOI] [PubMed] [Google Scholar]
- 48.Bettegowda C Sausen M Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sausen M Phallen J Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cristiano S Leal A Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allenson K Castillo J San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melo SA Luecke LB Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu J Sadakari Y Shindo K, et al. Digital next-generation sequencing identifies low-abundance mutations in pancreatic juice samples collected from the duodenum of patients with pancreatic cancer and intraductal papillary mucinous neoplasms. Gut. 2017;66:1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suenaga M Yu J Shindo K, et al. Pancreatic juice mutation concentrations can help predict the grade of dysplasia in patients undergoing pancreatic surveillance. Clin Cancer Res. 2018;24:2963–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kisiel JB Yab TC Taylor WR, et al. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer. 2012;118:2623–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arasaradnam RP Wicaksono A O'Brien H, et al. Noninvasive diagnosis of pancreatic cancer through detection of volatile organic compounds in urine. Gastroenterology. 2018;154:485–487.e1. [DOI] [PubMed] [Google Scholar]
- 57.Blyuss O Zaikin A Cherepanova V, et al. Development of PancRISK, a urine biomarker-based risk score for stratified screening of pancreatic cancer patients. Br J Cancer. 2020;122:692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lau C Kim Y Chia D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288:26888–26897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singhi AD McGrath K Brand RE, et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut. 2018;67:2131–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singhi AD Nikiforova MN Fasanella KE, et al. Preoperative GNAS and KRAS testing in the diagnosis of pancreatic mucinous cysts. Clin Cancer Res. 2014;20:4381–4389. [DOI] [PubMed] [Google Scholar]
- 61.Springer S Wang Y Dal Molin M, et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology. 2015;149:1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowenfels AB, Maisonneuve P. Epidemiology and risk factors for pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:197–209. [DOI] [PubMed] [Google Scholar]
- 63.Ben Q Xu M Ning X, et al. Diabetes mellitus and risk of pancreatic cancer: a meta-analysis of cohort studies. Eur J Cancer. 2011;47:1928–1937. [DOI] [PubMed] [Google Scholar]
- 64.Aberle DR Adams AM Berg CD, et al. National Lung Screening Trial Research Team . Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Imperiale TF Ransohoff DF Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. [DOI] [PubMed] [Google Scholar]
- 66.Pisano ED Hendrick RE Yaffe MJ, et al. Diagnostic accuracy of digital versus film mammography: exploratory analysis of selected population subgroups in DMIST. Radiology. 2008;246:376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bruenderman E, Martin RC, 2nd. A cost analysis of a pancreatic cancer screening protocol in high-risk populations. Am J Surg. 2015;210:409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma A Kandlakunta H Nagpal SJS, et al. Model to determine risk of pancreatic cancer in patients with new-onset diabetes. Gastroenterology. 2018;155:730–739.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klein AP Brune KA Petersen GM, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. [DOI] [PubMed] [Google Scholar]
- 70.Goggins M Overbeek KA Brand R, et al. Management of patients with increased risk for familial pancreatic cancer: updated recommendations from the International Cancer of the Pancreas Screening (CAPS) consortium. Gut. 2020;69:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chari ST Leibson CL Rabe KG, et al. Probability of pancreatic cancer following diabetes: a population-based study. Gastroenterology. 2005;129:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh DP Sheedy S Goenka AH, et al. Computerized tomography scan in pre-diagnostic pancreatic ductal adenocarcinoma: stages of progression and potential benefits of early intervention: a retrospective study. Pancreatology. 2020;20:1495–1501. [DOI] [PubMed] [Google Scholar]
- 73.Brentnall TA Bronner MP Byrd DR, et al. Early diagnosis and treatment of pancreatic dysplasia in patients with a family history of pancreatic cancer. Ann Intern Med. 1999;131:247–255. [DOI] [PubMed] [Google Scholar]
- 74.Harinck F Konings IC Kluijt I, et al. A multicentre comparative prospective blinded analysis of EUS and MRI for screening of pancreatic cancer in high-risk individuals. Gut. 2016;65:1505–1513. [DOI] [PubMed] [Google Scholar]
- 75.Canto MI Hruban RH Fishman EK, et al. Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology. 2012;142:796–804; quiz e14-e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Canto MI Almario JA Schulick RD, et al. Risk of neoplastic progression in individuals at high risk for pancreatic cancer undergoing long-term surveillance. Gastroenterology. 2018;155:740–751.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartsch DK Slater EP Carrato A, et al. Refinement of screening for familial pancreatic cancer. Gut. 2016;65:1314–1321. [DOI] [PubMed] [Google Scholar]
- 78.Abe T Blackford AL Tamura K, et al. Deleterious germline mutations are a risk factor for neoplastic progression among high-risk individuals undergoing pancreatic surveillance. J Clin Oncol. 2019;37:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corral JE Das A Bruno MJ, et al. Cost-effectiveness of pancreatic cancer surveillance in high-risk individuals: an economic analysis. Pancreas. 2019;48:526–536. [DOI] [PubMed] [Google Scholar]
- 80.Stokes JM Yang K Swanson K, et al. A deep learning approach to antibiotic discovery. Cell. 2020;180:688–702.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elton DC Boukouvalas Z Fuge MD, et al. Deep learning for molecular design—a review of the state of the art. Mol Syst Design Eng. 2019;4:828–849. [Google Scholar]
- 82.Ma J Sheridan RP Liaw A, et al. Deep neural nets as a method for quantitative structure-activity relationships. J Chem Inf Model. 2015;55:263–274. [DOI] [PubMed] [Google Scholar]
- 83.Senior AW Evans R Jumper J, et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. [DOI] [PubMed] [Google Scholar]
- 84.Leung MK Xiong HY Lee LJ, et al. Deep learning of the tissue-regulated splicing code. Bioinformatics. 2014;30:i121–i129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rajkomar A Oren E Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digital Med. 2018;1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campanella G Hanna MG Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yala A Lehman C Schuster T, et al. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology. 2019;292:60–66. [DOI] [PubMed] [Google Scholar]
- 88.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. [DOI] [PubMed] [Google Scholar]
- 89.Minsky M, Papert S. Perceptrons: An Introduction to Computational Geometry. Cambridge, MA: The MIT Press; 1969. [Google Scholar]
- 90.Hinton G Deng L Yu D, et al. Deep neural networks for acoustic modeling in speech recognition. IEEE Signal Proc Mag. 2012;29:82–97. [Google Scholar]
- 91.Sutskever I, Vinyals O, Le QV. Sequence to sequence learning with neural networks. Adv Neural Inform Proc Syst. 2014;27:3104–3112. [Google Scholar]
- 92.Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. Adv Neural Inform Proc Syst. 2012;25:1090–1098. [Google Scholar]
- 93.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 94. mit_deeplearning_bootcamp. Available at: https://github.com/ajfisch/deeplearning_bootcamp_2020. Accessed August 14, 2020.
- 95.Moyer VA. Screening for lung cancer: US Preventive Services Task Force recommendation statement. Ann Intern Med. 2015;160:330–338. [DOI] [PubMed] [Google Scholar]
- 96.US Preventive Services Task Force, Bibbins-Domingo K Grossman DC Curry SJ, et al. Screening for colorectal cancer: US Preventive Services Task Force recommendation statement. JAMA. 2016;315:2564–2575. [DOI] [PubMed] [Google Scholar]
- 97.US Preventive Services Task Force, Curry SJ Krist AH Owens DK, et al. Screening for cervical cancer: US Preventive Services Task Force recommendation statement. JAMA. 2018;320:674–686. [DOI] [PubMed] [Google Scholar]
- 98.Siu AL, U.S. Preventive Services Task Force . Screening for breast cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2016;164:279–296. [DOI] [PubMed] [Google Scholar]
- 99.Saslow D Boetes C Burke W, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin. 2007;57:75–89. [DOI] [PubMed] [Google Scholar]
- 100.Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Stat Med. 2004;23:1111–1130. [DOI] [PubMed] [Google Scholar]
- 101.Brentnall AR Harkness EF Astley SM, et al. Mammographic density adds accuracy to both the Tyrer-Cuzick and Gail breast cancer risk models in a prospective UK screening cohort. Breast Cancer Res. 2015;17:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:1159–1169. [DOI] [PubMed] [Google Scholar]
- 103.Wolfe JN. Breast patterns as an index of risk for developing breast cancer. Am J Roentgenol. 1976;126:1130–1137. [DOI] [PubMed] [Google Scholar]
- 104.Keating NL, Pace LE. New federal requirements to inform patients about breast density: will they help patients? JAMA. 2019;321:2275–2276. [DOI] [PubMed] [Google Scholar]
- 105.Dembrower K Liu Y Azizpour H, et al. Comparison of a deep learning risk score and standard mammographic density score for breast cancer risk prediction. Radiology. 2020;294:265–272. [DOI] [PubMed] [Google Scholar]
- 106.Yala A Mikhael PG Strand F, et al. Toward robust mammography-based models for breast cancer risk. Sci Transl Med. 2021;13:eaba4373. [DOI] [PubMed] [Google Scholar]
- 107.Gail MH Brinton LA Byar DP, et al. Projecting individualized probabilities of developing breast cancer for White females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. [DOI] [PubMed] [Google Scholar]
- 108.Boggs DA Rosenberg L Adams-Campbell LL, et al. Prospective approach to breast cancer risk prediction in African American women: the Black women's health study model. J Clin Oncol. 2015;33:1038–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gail MH. Twenty-five years of breast cancer risk models and their applications. J Natl Cancer Inst. 2015;107:djv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gail MH Costantino JP Pee D, et al. Projecting individualized absolute invasive breast cancer risk in African American women. J Natl Cancer Inst. 2007;99:1782–1792. [DOI] [PubMed] [Google Scholar]
- 111.Matsuno RK Costantino JP Ziegler RG, et al. Projecting individualized absolute invasive breast cancer risk in Asian and Pacific Islander American women. J Natl Cancer Inst. 2011;103:951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Obermeyer Z Powers B Vogeli C, et al. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366:447–453. [DOI] [PubMed] [Google Scholar]
- 113.Kearns M, Roth A, Sharifi-Malvajerdi S. Average individual fairness: algorithms, generalization and experiments. arXiv preprint. 2019;1905:10607. [Google Scholar]
- 114.Agarwal A Beygelzimer A Dudik M, et al. A reductions approach to fair classification. Proceedings of the 35th International Conference on Machine Learning. PMLR. 2018;80:60–69. [Google Scholar]
- 115.Mitchell S Potash E Barocas S, et al. Prediction-based decisions and fairness: a catalogue of choices, assumptions, and definitions. Annu Rev Stat Appl. 2021;8:1. [Google Scholar]
- 116.Sorelle AF Scheidegger C Venkatasubramanian S, et al. A comparative study of fairness-enhancing interventions in machine learning. In: Proceedings of Conference on Fairness, Accountability, and Transparency (FAT* ‘19). New York, NY: ACM; 2019:329–338. Available at: 10.1145/3287560.3287589. Accessed August 14, 2020. [DOI] [Google Scholar]
- 117.Marks DS Colwell LJ Sheridan R, et al. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hutter C, Zenklusen JC. The Cancer Genome Atlas: creating lasting value beyond its data. Cell. 2018;173:283–285. [DOI] [PubMed] [Google Scholar]
- 119.Sanchez-Vega F Mina M Armenia J, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173:321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ingraham J Garg V Barzilay R, et al. Generative models for graph-based protein design. Adv Neural Inform Proc Syst. 2019;32:15820–15831. [Google Scholar]
- 121.Croft D O'Kelly G Wu G, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jassal B Matthews L Viteri G, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cerami EG Gross BE Demir E, et al. Pathway commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rodchenkov I Babur O Luna A, et al. Pathway Commons 2019 Update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 2020;48:D489–D497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Demir E Cary MP Paley S, et al. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010;28:935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Korkut A Wang W Demir E, et al. Perturbation biology nominates upstream-downstream drug combinations in RAF inhibitor resistant melanoma cells. Elife. 2015;4:e04640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yuan B Shen C Luna A, et al. CellBox: interpretable machine learning for perturbation biology with application to the design of cancer combination therapy. Cell Syst. 2021;12:128–140.e4. [DOI] [PubMed] [Google Scholar]
- 128.Bordbar A, Palsson BO. Using the reconstructed genome-scale human metabolic network to study physiology and pathology. J Intern Med. 2012;271:131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Louis DN Gerber GK Baron JM, et al. Computational pathology: an emerging definition. Arch Pathol Lab Med. 2014;138:1133–1138. [DOI] [PubMed] [Google Scholar]
- 130.Chu LC Park S Kawamoto S, et al. Application of deep learning to pancreatic cancer detection: lessons learned from our initial experience. J Am Coll Radiol. 2019;16:1338–1342. [DOI] [PubMed] [Google Scholar]
- 131.Santus E Schuster T Tahmasebi AM, et al. Exploiting rules to enhance machine learning in extracting information from multi-institutional prostate pathology reports. JCO Clin Cancer Inform. 2020;4:865–874. [DOI] [PubMed] [Google Scholar]
- 132.Hu JX, Thomas CE, Brunak S. Network biology concepts in complex disease comorbidities. Nat Rev Genet. 2016;17:615–629. [DOI] [PubMed] [Google Scholar]
- 133.Jensen AB Moseley PL Oprea TI, et al. Temporal disease trajectories condensed from population-wide registry data covering 6.2 million patients. Nat Commun. 2014;5:4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Siggaard T Reguant R Jørgensen IF, et al. Disease trajectory browser for exploring temporal, population-wide disease progression patterns in 7.2 million Danish patients. Nat Commun. 2020;11:4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu JX Helleberg M Jensen AB, et al. A large-cohort, longitudinal study determines pre-cancer disease routes across different cancer types. Cancer Res. 2019;79:864–872. [DOI] [PubMed] [Google Scholar]
- 136.Maretty L Jensen JM Petersen B, et al. Sequencing and de novo assembly of 150 genomes from Denmark as a population reference. Nature. 2017;548:87–91. [DOI] [PubMed] [Google Scholar]
- 137.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13:395–405. [DOI] [PubMed] [Google Scholar]
- 138.Thorsen-Meyer HC Nielsen AB Nielsen AP, et al. Dynamic and explainable machine learning prediction of mortality in patients in the intensive care unit: a retrospective study of high-frequency data in electronic patient records. Lancet Digit Health. 2020;2:e179–e191. [DOI] [PubMed] [Google Scholar]
- 139.Nielsen AB Thorsen-Meyer HC Belling K, et al. Survival prediction in intensive-care units based on aggregation of long-term disease history and acute physiology: a retrospective study of the Danish National Patient Registry and electronic patient records. Lancet Digit Health. 2019;1:e78–e89. [DOI] [PubMed] [Google Scholar]
- 140.Moseley PL, Brunak S. Identifying sepsis phenotypes. JAMA. 2019;322:1416–1417. [DOI] [PubMed] [Google Scholar]
- 141.Lee JG Jun S Cho YW, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hosny A Parmar C Quackenbush J, et al. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gregor K, LeCun Y. Learning fast approximations of sparse coding. In: Proceedings of the 27th International Conference on International Conference on Machine Learning. Haifa, Israel; 2010:399–406. [Google Scholar]
- 144.Monga V, Li Y, Eldar YC. Algorithm unrolling: interpretable, efficient deep learning for signal and image processing. IEEE Signal Proc Mag. 2021;38:18–44. [Google Scholar]
- 145.Solomon O Cohen R Zhang Y, et al. Deep unfolded robust PCA with application to clutter suppression in ultrasound. IEEE Trans Med Imaging. 2020;39:1051–1063. [DOI] [PubMed] [Google Scholar]
- 146.van Sloun RJG, Cohen R, Eldar YC. Deep learning in ultrasound imaging. Proc IEEE. 2020;108:11–29. [Google Scholar]
- 147.Farsad N Shlezinger N Goldsmith AJ, et al. Data-driven symbol detection via model-based machine learning. 2020. Submitted to Communications in Information and Systems, special issue in honor of Thomas Kailath's 85th birthday. arXiv:2002.07806.
- 148.Rough K Dai AM Zhang K, et al. Predicting inpatient medication orders from electronic health record data. Clin Pharmacol Ther. 2020;108:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kelly CJ Karthikesalingam A Suleyman M, et al. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Keane PA, Topol EJ. With an eye to AI and autonomous diagnosis. NPJ Digit Med. 2018;1:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Esteva A Robicquet A Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med. 2019;25:24–29. [DOI] [PubMed] [Google Scholar]
- 152.Seshadri DR Bittel B Browsky D, et al. Accuracy of the apple watch 4 to measure heart rate in patients with atrial fibrillation. IEEE J Transl Eng Health Med. 2019;8:2700204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rajkomar A Hardt M Howell MD, et al. Ensuring fairness in machine learning to advance health equity. Ann Intern Med. 2018;169:866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Farr C. Apple health team faces departures as tensions rise over differing visions for the future. CNBC.com. August 20, 2019. Available at: https://www.cnbc.com/2019/08/20/apple-health-employee-departures-show-split-over-ambitions.html. Accessed August 11, 2020.
- 155.Kimmell J. What ‘Google health care’ could look like in 5 years. Advisory Board March 13, 2019. Available at: https://www.advisory.com/daily-briefing/2019/03/13/google. Accessed August 14, 2020.
- 156.US Food & Drug Administration. 2019. Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD). Discussion paper and request for feedback. Available at: https://www.fda.gov/media/122535/download. Accessed August 14, 2020.
- 157.Gerke S Babic B Evgeniou T, et al. The need for a system view to regulate artificial intelligence/machine learning-based software as medical device. NPJ Digit Med. 2020;3:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yachida S Jones S Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen W Butler RK Zhou Y, et al. Prediction of pancreatic cancer based on imaging features in patients with duct abnormalities. Pancreas. 2020;49:413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Attiyeh MA Fernández-Del Castillo C Al Efishat M, et al. Development and validation of a multi-institutional preoperative nomogram for predicting grade of dysplasia in intraductal papillary mucinous neoplasms (IPMNs) of the pancreas: a report from the pancreatic surgery consortium. Ann Surg. 2018;267:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koay EJ Lee Y Cristini V, et al. A visually apparent and quantifiable CT imaging feature identifies biophysical subtypes of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2018;24:5883–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Aguirre AJ Nowak JA Camarda ND, et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 2018;8:1096–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mayers JR Wu C Clish CB, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014;20:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hoshino A Kim HS Bojmar L, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Poore GD Kopylova E Zhu Q, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 166.Serrano J Andersen DK Forsmark CE, et al. Consortium for the study of chronic pancreatitis, diabetes, and pancreatic cancer: from concept to reality. Pancreas. 2018;47:1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Maitra A Sharma A Brand RE, et al. A prospective study to establish a new-onset diabetes cohort: from the Consortium for the Study of Chronic Pancreatitis, Diabetes, and Pancreatic Cancer. Pancreas. 2018;47:1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Boursi B Finkelman B Giantonio BJ, et al. A clinical prediction model to assess risk for pancreatic cancer among patients with new-onset diabetes. Gastroenterology. 2017;152:840–850.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fisher WE Cruz-Monserrate Z McElhany AL, et al. Standard operating procedures for biospecimen collection, processing, and storage: from the Consortium for the Study of Chronic Pancreatitis, Diabetes, and Pancreatic Cancer. Pancreas. 2018;47:1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gangi S Fletcher JG Nathan MA, et al. Time interval between abnormalities seen on CT and the clinical diagnosis of pancreatic cancer: retrospective review of CT scans obtained before diagnosis. AJR Am J Roentgenol. 2004;182:897–903. [DOI] [PubMed] [Google Scholar]
- 171.Pannala R Basu A Petersen GM, et al. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 2009;10:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Setiawan VW Stram DO Porcel J, et al. Pancreatic cancer following incident diabetes in African Americans and Latinos: The Multiethnic Cohort. J Natl Cancer Inst. 2019;111:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huang BZ Pandol SJ Jeon CY, et al. New-onset diabetes, longitudinal trends in metabolic markers, and risk of pancreatic cancer in a heterogeneous population. Clin Gastroenterol Hepatol. 2020;18:1812–1821.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huang BZ Stram DO Le Marchand L, et al. Interethnic differences in pancreatic cancer incidence and risk factors: The Multiethnic Cohort. Cancer Med. 2019;8:3592–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu L Zhang J Deapen D, et al. Differences in pancreatic cancer incidence rates and temporal trends across Asian subpopulations in California (1988–2015). Pancreas. 2019;48:931–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Baecker A Kim S Risch HA, et al. Do changes in health reveal the possibility of undiagnosed pancreatic cancer? Development of a risk-prediction model based on healthcare claims data. PLoS One. 2019;14:e0218580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jeon CY Chen Q Yu W, et al. Identification of individuals at increased risk for pancreatic cancer in a community-based cohort of patients with suspected chronic pancreatitis. Clin Transl Gastroenterol. 2020;11:e00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pereira SP Oldfield L Ney A, et al. Early detection of pancreatic cancer. Lancet Gastroenterol Hepatol. 2020;5:698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Solomon S Das S Brand R, et al. Inherited pancreatic cancer syndromes. Cancer J. 2012;18:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Johns AL McKay SH Humphris JL, et al. Lost in translation: returning germline genetic results in genome-scale cancer research. Genome Med. 2017;9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shindo K Yu J Suenaga M, et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J Clin Oncol. 2017;35:3382–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grant RC Selander I Connor AA, et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology. 2015;148:556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Holter S Borgida A Dodd A, et al. Germline BRCA mutations in a large clinic-based cohort of patients with pancreatic adenocarcinoma. J Clin Oncol. 2015;33:3124–3129. [DOI] [PubMed] [Google Scholar]
- 185.Yurgelun MB Chittenden AB Morales-Oyarvide V, et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med. 2019;21:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Syngal S Brand RE Church JM, et al. ACG clinical guideline: genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015;110:223–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Abul-Husn NS Soper ER Odgis JA, et al. Exome sequencing reveals a high prevalence of BRCA1 and BRCA2 founder variants in a diverse population-based biobank. Genome Med. 2019;12:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lucas AL Frado LE Hwang C, et al. BRCA1 and BRCA2 germline mutations are frequently demonstrated in both high-risk pancreatic cancer screening and pancreatic cancer cohorts. Cancer. 2014;120:1960–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Stoffel EM, McKernin SE, Khorana AA. Evaluating susceptibility to pancreatic cancer: ASCO Clinical Practice Provisional Clinical Opinion Summary. J Oncol Pract. 2019;15:108–111. [DOI] [PubMed] [Google Scholar]
- 190.Yurgelun MB. Germline testing for individuals with pancreatic cancer: the benefits and challenges to casting a wider net. J Clin Oncol. 2017;35:3375–3377. [DOI] [PubMed] [Google Scholar]
- 191.Buchanan AH Lester Kirchner H Schwartz MLB, et al. Clinical outcomes of a genomic screening program for actionable genetic conditions. Genet Med. 2020;22:1874–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Klein AP Wolpin BM Risch HA, et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun. 2018;9:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nakatochi M Lin Y Ito H, et al. Prediction model for pancreatic cancer risk in the general Japanese population. PLoS One. 2018;13:e0203386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Khera AV Chaffin M Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cho J Scragg R Pandol SJ, et al. Exocrine pancreatic dysfunction increases the risk of new-onset diabetes mellitus: results of a nationwide cohort study. Clin Transl Sci. 2021;14:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Christodoulou AG Shaw JL Nguyen C, et al. Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging. Nat Biomed Eng. 2018;2:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang L Gaddam S Wang N, et al. Multiparametric mapping magnetic resonance imaging of pancreatic disease. Front Physiol. 2020;11:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang N Gaddam S Wang L, et al. Six-dimensional quantitative DCE MR multitasking of the entire abdomen: method and application to pancreatic ductal adenocarcinoma. Magn Reson Med. 2020;84:928–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lavdas E Vlychou M Arikidis N, et al. How reliable is MRCP with an SS-FSE sequence at 3.0 T: comparison between SS-FSE BH and 3D-FSE BH ASSET sequences. Clin Imaging. 2013;37:697–703. [DOI] [PubMed] [Google Scholar]
- 201.Massion PP Antic S Ather S, et al. Assessing the accuracy of a deep learning method to risk stratify indeterminate pulmonary nodules. Am J Respir Crit Care Med. 2020;202:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Takeda Y Saiura A Takahashi Y, et al. Asymptomatic pancreatic cancer: does incidental detection impact long-term outcomes? J Gastrointest Surg. 2017;21:1287–1295. [DOI] [PubMed] [Google Scholar]
- 203.Seufferlein T, Mayerle J. Pancreatic cancer in 2015: precision medicine in pancreatic cancer—fact or fiction? Nat Rev Gastroenterol Hepatol. 2016;13:74–75. [DOI] [PubMed] [Google Scholar]
- 204.Chari ST. Detecting early pancreatic cancer: problems and prospects. Semin Oncol. 2007;34:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Granata V Fusco R Catalano O, et al. Multidetector computer tomography in the pancreatic adenocarcinoma assessment: an update. Infect Agent Cancer. 2016;11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Langlotz CP Allen B Erickson BJ, et al. A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology. 2019;291:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hanania AN Bantis LE Feng Z, et al. Quantitative imaging to evaluate malignant potential of IPMNs. Oncotarget. 2016;7:85776–85784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sharib J, Kirkwood K. Early and accurate diagnosis of pancreatic cancer? Oncotarget. 2016;7:85676–85677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chu LC Park S Kawamoto S, et al. Utility of CT radiomics features in differentiation of pancreatic ductal adenocarcinoma from normal pancreatic tissue. AJR Am J Roentgenol. 2019;213:349–357. [DOI] [PubMed] [Google Scholar]
- 210.Hart PA Bellin MD Andersen DK, et al. Type 3c (pancreatogenic) diabetes mellitus secondary to chronic pancreatitis and pancreatic cancer. Lancet Gastroenterol Hepatol. 2016;1:226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Aggarwal G, Kamada P, Chari ST. Prevalence of diabetes mellitus in pancreatic cancer compared to common cancers. Pancreas. 2013;42:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Huxley R Ansary-Moghaddam A Berrington de González A, et al. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. Br J Cancer. 2005;92:2076–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Pannala R Leirness JB Bamlet WR, et al. Prevalence and clinical profile of pancreatic cancer–associated diabetes mellitus. Gastroenterology. 2008;134:981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hart PA Andersen DK Mather KJ, et al. Evaluation of a mixed meal test for diagnosis and characterization of pancreatogenic diabetes secondary to pancreatic cancer and chronic pancreatitis. Rationale and methodology for the DETECT study from the Consortium for the Study of Chronic Pancreatitis, Diabetes and Pancreatic Cancer. Pancreas. 2018;47:1239–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Roth HR Lu L Lay N, et al. Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med Image Anal. 2018;45:94–107. [DOI] [PubMed] [Google Scholar]
- 216.Lowe ME Andersen DK Caprioli RM, et al. Precision medicine in pancreatic disease – knowledge gaps and research opportunities. Summary of a National Institute of Diabetes and Digestive and Kidney Diseases workshop. Pancreas. 2019;48:1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Summers RM. Progress in fully automated abdominal CT interpretation. AJR Am J Roentgenol. 2016;207:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yan K Wang X Lu L, et al. DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J Med Imaging (Bellingham). 2018;5:036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.American Cancer Society . Cancer Facts & Figures 2020. Atlanta, GA: American Cancer Society; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Roser M, Ritchie H. Cancer. Our World in Data. Updated April 2018. Available at: https://ourworldindata.org/cancer. Accessed August 14, 2020.
- 221.Mahmood SS Levy D Vasan RS, et al. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet. 2014;383:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ahlquist DA. Universal cancer screening: revolutionary, rational, and realizable. NPJ Precis Oncol. 2018;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are few centralized, anonymized data sets that can be semipublicly accessed by researchers for PDAC research. The NIH-NCI–sponsored EDRN effort is currently the only major public effort that we are aware of in this space.
There has been no peer-reviewed demonstration of a federated learning system in this space. Although Stand Up To Cancer has funded a project with this as a core goal, these techniques will remain inaccessible to most research groups until proven systems are established and made available.


