Abstract
Neutrophils are innate immune cells involved in the elimination of pathogens and can also induce adaptive immune responses. Nα and Nβ neutrophils have been described with distinct in vitro capacity to generate antigen-specific CD8 T-cell responses. However, how these cell types exert their role in vivo and how manipulation of Nβ/Nα ratio influences vaccine-mediated immune responses are not known. In this study, we find that these neutrophil subtypes show distinct migratory and motility patterns and different ability to interact with CD8 T cells in the spleen following vaccinia virus (VACV) infection. Moreover, after analysis of adhesion, inflammatory, and migration markers, we observe that Nβ neutrophils overexpress the α4β1 integrin compared to Nα. Finally, by inhibiting α4β1 integrin, we increase the Nβ/Nα ratio and enhance CD8 T-cell responses to HIV VACV-delivered antigens. These findings provide significant advancements in the comprehension of neutrophil-based control of adaptive immune system and their relevance in vaccine design.
Subject terms: Adaptive immunity, Innate immunity, Vaccines, Vaccines, Virology
Introduction
Neutrophils are innate immune effector cells that form the first line of defense against microbial pathogens1. They immediately migrate to the inflammation site and neutralize the insult in a process termed microbial sterilization2, which is characterized by the engulfing of pathogens, infected and apoptotic cells3,4, the disarming of microorganisms through neutrophil extracellular traps5, the generation of reactive oxygen intermediates6, and the release of lytic enzymes from their granules2. Neutrophils can also induce antigen-specific T-cell responses by migrating to the lymph nodes (LN) and bone marrow7,8 where they efficiently cross-prime naive T cells9.
Neutrophils polarize to distinct phenotypes in response to environmental signals10, and in the presence of granulocyte macrophage-colony stimulating factor (GM-CSF) and interferon (IFN)-γ, neutrophils can acquire antigen-presenting cell (APC) functions and trigger CD8 T-cell activation11. Recent studies characterized distinct functional neutrophil subsets in cancer and infection contexts12–14. The cytokine/chemokine environment in tumor tissue polarizes tumor-associated neutrophils (TAN) to control tumor angiogenesis and growth15. In the presence of transforming growth factor (TGF)-β, TAN polarize from the anti-tumoral N1 to the pro-tumoral N2 phenotype, characterized by impaired ability to activate CD8 T cells16.
During virus infection, pro-inflammatory milieu can induce specific neutrophil subtypes13. By manipulating the vaccinia virus (VACV) vector, robust NFκB activation generates and alters neutrophil recruitment to the infection site in mice14,17. Distinct neutrophil subtypes (classified as Nα and Nβ) are recruited as a result of the cytokine/chemokine profile produced. Nβ cells overexpress costimulatory molecules CD80 and CD86 and show a greater capacity to generate VACV-specific CD8 T-cell responses than Nα cells in vitro14.
Enhanced neutrophil trafficking from the infection site to secondary lymphoid organs correlates with increased CD8 T-cell responses to HIV vector-delivered antigens14. Conversely, study of vaccine immunization showed that neutrophils generate poor antigen-specific CD4 responses, as their contacts with dendritic cells (DC) in the draining LN are very brief, and DC–T-cell interactions are improved in neutropenic mice18.
The balance between Nα and Nβ neutrophil subtypes could influence the generation of antigen-specific CD8 and CD4 T-cell immune responses and then improve the virus-based vaccine efficacy. However, the ability of neutrophil subtypes to relocate to secondary lymphoid organs, their migratory patterns, and their capacity to regulate antigen-specific CD8 T-cell responses have not been evaluated in vivo.
Attenuated VACV vectors are considered promising HIV vaccine candidates and several strategies have been developed to improve immunogenicity to HIV antigens19. Even if VAVC vector approaches gave encouraging results in primates20, the effectiveness of RV144 phase III HIV/AIDS human clinical trial was limited21. Given the importance of understanding the mechanism of neutrophil-dependent control of T-cell subset reactions to virus-delivered antigens and the interest for the generation of vaccines that preferentially induce HIV-specific CD8 T-cell responses, in this work we studied those processes in secondary lymphoid organs.
Here, we characterized Nα and Nβ splenic migratory features by multiphoton intravital microscopy (MP-IVM) and we detailed the transcriptomic profiles of these distinct neutrophil populations by RNA-sequencing and flow cytometry. Finally, we found an alternative approach to increase the antigen-specific CD8 T-cell responses. Indeed, since Nβ neutrophils compared to Nα overexpressed α4β1 integrin, also called very-late-antigen (VLA-4), administration of low dose of anti-VLA-4 increased the Nβ/Nα ratio in the peritoneal cavity (PEC) and spleen and in turn augmented the HIV-specific CD8 T-cell responses. These findings provide important insights into the neutrophil-dependent mechanism of the poxvirus-induced immune response and offer alternative strategies for HIV vaccine vector design.
Results
HIV-specific T-cell responses are generated in the spleen after neutrophil accumulation
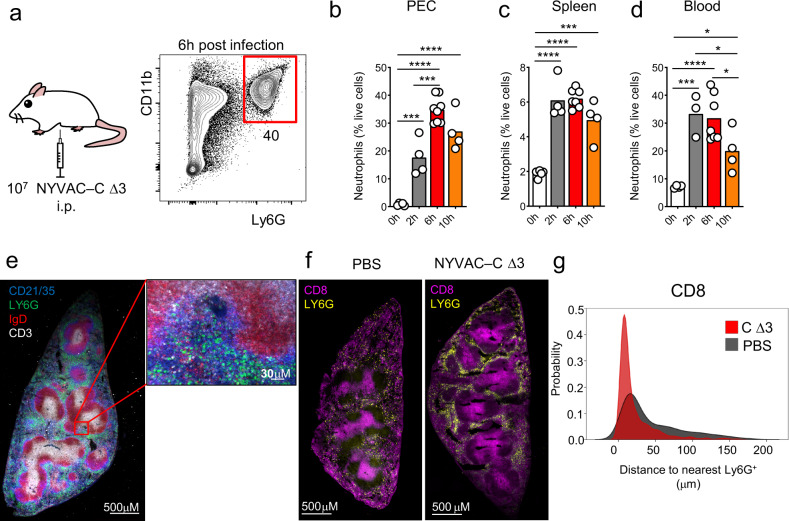
To define the dynamic recruitment of neutrophil from the blood to the PEC and to the spleen, BALB/c mice were infected by intraperitoneal (i.p.) injection of 107 plaque-forming units (PFU) of VACV strain NYVAC-C ΔA52RΔB15RΔK7R deletion mutant (also termed triple deletion mutant, NYVAC-C Δ3 or C Δ3) and we identified by flow cytometry Ly6G+CD11b+ neutrophils (Fig. 1a). To determine how neutrophil trafficking occurs from the primary site of infection to the spleen, we quantified neutrophils in PEC, spleen, and blood at 2, 6, and 10 h post infection (p.i.) (Fig. 1b–d). Neutrophils started to accumulate in PEC, spleen and blood after 2 h p.i. compared to PBS-treated mice and they decreased at 10 h p.i. in all cases (Fig. 1b–d). After infection, in the spleen, Ly6G+ neutrophils were located close to the CD21/35+ areas in the marginal zone and in the proximity of the CD3+ T-cell zone (Fig. 1e). NYVAC-C Δ3-infected mice specifically showed neutrophil accumulation surrounding the CD8+ areas (Fig. 1f). Indeed, the probability of CD8+ cells to be located close to neutrophils was significantly higher in the infected mice compared to those injected with PBS (Fig. 1g).
Fig. 1. In vivo time-course of neutrophil accumulation after poxvirus infection.
BALB/c mice were infected by intraperitoneal (i.p.) injection of 107 PFU of NYVAC-C Δ3 virus and Ly6G+ CD11b+ neutrophils were measured at 6 h post infection (a). Percentages of neutrophils in peritoneal cavity (PEC) (b), spleen (c), and blood (d) at 0, 2, 6, and 10 h post infection from NYVAC-C Δ3 injected mice. Spleen tissue section and its magnification from NYVAC-C Δ3-infected mice. Ly6G+ neutrophils in green, CD3+ T cells in white, IgD+ B cells in red, and CD21/35+ cells in blue (e). Spleen tissue section from PBS and NYVAC-C Δ3-injected mice. Ly6G+ neutrophils in yellow, CD8+ T cells in purple (f). Distance of the nearest Ly6G+ cell from CD8+ cells. The nearest neighbor euclidean distance to Ly6G+ is expressed as probability density for each cell belonging to CD8+ clusters (g). Graphs show mean; each point represents an individual mouse. Data are representative of two independent experiments. *P < 0.05, ***P < 0.001, ****P < 0.0001.
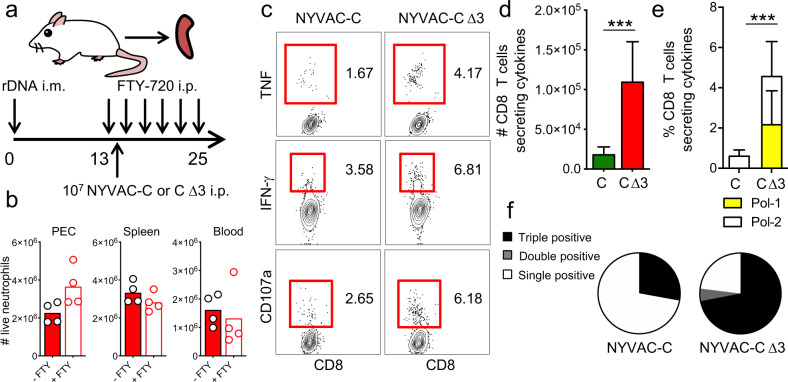
We previously demonstrated that neutrophils are involved in the enhanced HIV-specific T-cell responses of NYVAC-C Δ3-infected mice compared to those infected with parental NYVAC-C14. To study the ability of NYVAC-C Δ3 to induce an antigen T-cell response specifically in the spleen, we used a DNA intramuscular prime/poxvirus i.p. boost approach and we concurrently blocked lymphocyte egress from secondary lymphoid organs using the functional S1P receptor antagonist FTY720 (Fig. 2a) without interfering with neutrophil accumulation in the organ of interest (Fig. 2b). This heterologous immunization protocol is more immunogenic than a homologous combination in generating T-cell responses against HIV antigens22. To study the HIV-specific T-cell response, splenocytes from infected mice were stimulated with HIV-1 Pol-1, or Pol-2 peptides, which are the most immunogenic MHC class I-restricted cytotoxic T lymphocytes (CTLs) peptides of HIV clade C in BALB/c mice23. To determine the functional profile of CD8 T cells, we measured the levels of IFN-γ, TNF, and lysosomal-associated membrane protein-1 (LAMP-1 or CD107a) as a surrogate marker for induction of killing (Fig. 2c).
Fig. 2. Splenic HIV-specific T-cell responses is increased after NYVAC-C Δ3 infection.
Vaccine-induced HIV-1-specific CD8 T-cell responses from spleen of mice (n = 4 per group) primed intramuscularly with recombinant DNA, infected with 107 PFUs of NYVAC-C or NYVAC-C Δ3 and injected with FTY720 every other day starting from the day before infection (a). Neutrophil total numbers in peritoneal cavity (PEC), spleen, and blood of NYVAC-C Δ3-injected and FTY720- or PBS-injected mice (b). The response was measured 11 days after the last immunization, after splenocyte stimulation with HIV-1 peptides. Contour plots of CD8 T cells that produce IFN-γ, TNF, and CD107a after NYVAC-C or NYVAC-C Δ3 infection (c). Total numbers (d) or percentages of Pol-1-, Pol-2-specific (e) CD8 T cells that express IFN-γ and/or TNF and/or CD107a. Nonspecific responses of mice infected with control NYVAC-WT were subtracted from the total value. Graphs show mean ± confidence interval (CI). Pie chart colors indicate the relative percentage of cells producing three (black), two (gray), or one (white) activation markers (f). Data are representative of two independent experiments. ***P < 0.001.
We quantified the magnitude of the T-cell response as the number and percentage of T cells that expressed IFN-γ and/or TNF and/or CD107a and the polyfunctionality of the response as the capacity to express more than one of these activation markers. We showed that, compared to the parental strain, the triple deletion mutant induced a robust increase of the quality of HIV-specific CD8 T-cell responses since NYVAC-C Δ3 injected-mice had higher number and percentage and even more polyfunctional HIV-specific CD8 T cells (Fig. 2d–f).
These findings show that antigen-specific T-cell response was induced and increased in the spleen after NYVAC-C Δ3 infection and correlated with a robust neutrophil infiltration in this lymphoid organ.
Neutrophil subtypes display distinct dynamic behaviors in the spleen
After NYVAC-C Δ3 i.p. infection, PEC of C57BL/6 J mice showed two neutrophil populations (Nα and Nβ) that differed in Ly6G/CD11b expression, size, and complexity, and expressed Ly6B.2 similarly at high levels (Supplementary Fig. 1a). Compared to neutrophil precursors (preNeu), immature and mature neutrophils from bone marrow (Supplementary Fig. 1b), peritoneal Nα and Nβ show levels of CXCR4 and CXCR2 similar to mature neutrophils (Supplementary Fig. 1c). Bulk RNA sequencing analysis reveals that Nα neutrophils are quite dissimilar from Nβ (Supplementary Fig. 1d). Indeed, these two subsets showed vast transcriptomic differences and had 2655 differentially expressed genes (Supplementary Fig. 1e). Furthermore, analysis of genes differentially expressed by tumor-associated neutrophils and mostly overexpressed by N1 compared to N224 shows that Nβ downregulated many of those genes in comparison to Nα (Supplementary Fig. 1f).
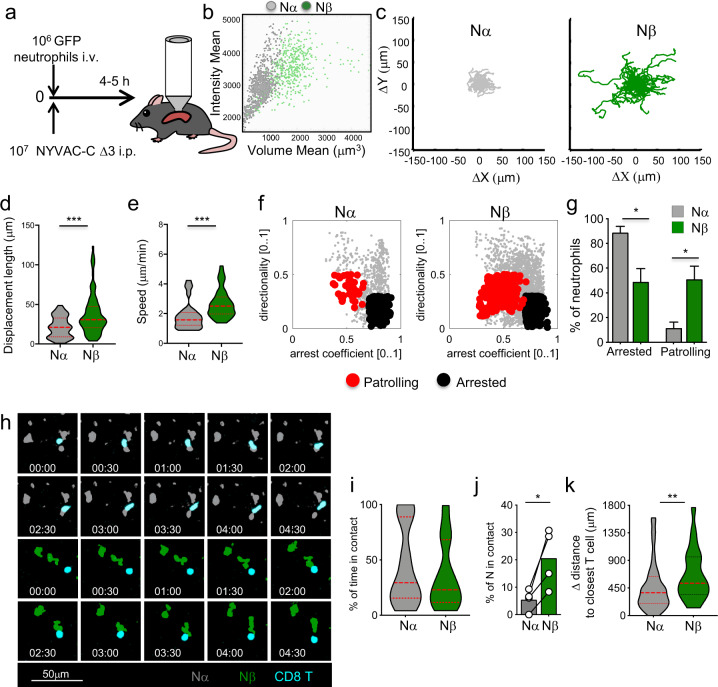
Compared to 2 h p.i., the ratio Nβ/Nα significantly increased in the PEC, spleen, and blood at 6 h and quickly decreased at 10 h after infection (Supplementary Fig. 1g–i), suggesting that the time window between 2 and 10 h represents an opportunity for Nβ to be involved in processes of immune regulation. We tested whether Nα and Nβ showed distinct dynamic behaviors in vivo in the spleen within this range of time. GFP+ neutrophils isolated from UBC (ubiquitin C)-GFP mice were intravenously (i.v.) injected in NYVAC-C Δ3-infected mice and analyzed by splenic MP-IVM from 4 to 5 h after injection (Fig. 3a). By the tracked mean volume of GFP+ cells, we could distinguish two neutrophil populations that differed in volume (Fig. 3b) and showed comparable sizes to the Nα and Nβ populations previously described14. The bigger population in green (Nβ) was not found in the PBS-injected mice (data not shown) as we previously showed14.
Fig. 3. Distinct dynamic behaviors from different neutrophil subtypes.
106 GFP+ neutrophils isolated from UBC-GFP mice were intravenously (i.v.) injected in C57BL6/J mice that were i.p. infected with 107 PFU of NYVAC-C Δ3 virus and the spleen was imaged from 4 to 5 h post infection (a). Multiphoton intravital microscopy dot plot determined by intensity and volume mean of GFP+ neutrophils from spleen of NYVAC-C Δ3-infected mice (b). Cell track plot of Nα and Nβ neutrophil from spleen of NYVAC-C Δ3 infected mice from 4 to 5 h post infection (c). Violin plots of displacement length (μm) (d) and speed (μm/min) (e) of Nα and Nβ neutrophils from 4 to 5 h post infection. Dot plots based on directionality and arrest coefficient of neutrophil subset activities (arrested: black; patrolling: red) (f). Percentages of arrested and patrolling Nα and Nβ from 4 to 5 h post infection (g). Nα (gray), Nβ (green) neutrophil and cognate CD8 T cells (cyan) from spleen of NYVAC-C Δ3-infected mice from 4 to 5 h post infection (h). Violin plots of percentage of time in contact of Nα and Nβ (i), graph bars of percentage of Nα and Nβ in contact with CD8 T cells (j), and violin plots of differential distance of Nα and Nβ to closest T cell (k). Graphs show mean ± SEM; each point represents an individual mouse. Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
We analyzed these neutrophil subtypes and found that Nβ cell tracks (in green) showed an increased migratory pattern compared to Nα cell tracks (in gray) (Fig. 3c and Supplementary Videos 1 and 2). Moreover, the displacement length as well as the speed of Nβ were significantly higher than Nα between 4 and 5 h p.i. (Fig. 3d, e). To understand if these different neutrophil subsets have distinct biological functions, we analyzed their migratory patterns based on their directionality and arrest coefficient25. Indeed, we found that Nβ neutrophils had distinct migratory pattern compared to Nα subset between 4 and 5 h p.i. (Fig. 3f). Nβ were significantly more patrolling cells than Nα, which conversely showed a significantly more arrested behavior between 4 and 5 h p.i. (Fig. 3g). Accordingly, compared to Nα, Nβ upregulated several G protein-coupled receptors (GPCR) genes associated with cell migration, and specifically those belonging to the C and adhesion classes (Supplementary Fig. 2).
To test whether these distinct motility and migratory patterns correlate with dissimilar neutrophil ability to interact with cognate CD8 T cells, labeled CD8 T cells from poxvirus-infected mice were adoptively transferred together with GFP neutrophils in recipient mice that were NYVAC-C Δ3-infected. Neutrophil and CD8 T-cell interactions were analyzed and quantified by splenic MP-IVM between 4 and 5 h p.i. (Fig. 3h and Supplementary Videos 3 and 4). Both subtypes showed interactions with a similar duration (less than 5 mins) and spent comparable percentage of imaged time in contact with CD8 T cells (Fig. 3h, i). Nevertheless, Nβ showed an increased ability to interact and find CD8 T cells compared to Nα as evidenced by a higher percentage of Nβ that established CD8 T-cell contacts (Fig. 3j) as well as their increased capacity to travel longer distances to get closer to CD8 T cells (Fig. 3k and Supplementary Videos 3 and 4).
These results show that Nβ neutrophils were more motile and prone to establish CD8 T-cell contacts than Nα in secondary lymphoid organs after NYVAC-C Δ3 infection.
Neutrophil subtypes show a different expression of α4β1 integrin
To understand better the different dynamics of recruitment of both neutrophil subsets from the primary site of infection to secondary lymphoid organs, we analyzed the expression of the surface markers involved in neutrophil migration and adhesion, such as IL-1 receptor (IL-1R), CXCR2, and very-late-antigen (VLA-4) α4β1. IL-1R has a clear role in neutrophil recruitment to the peritoneal cavity26 and migration to secondary lymphoid organs25. CXCR2 regulates neutrophil release from bone marrow27 and neutrophil migration in tumor microenvironment and during severe sepsis28,29; it has been shown that failure of CXCR2 downregulation controls neutrophil mobilization during inflammation in the peritoneum30. VLA-4 α4β1 integrin is involved in leukocyte transendothelial migration31 and neutrophils use this integrin in the rolling phase during the inflammatory response32.
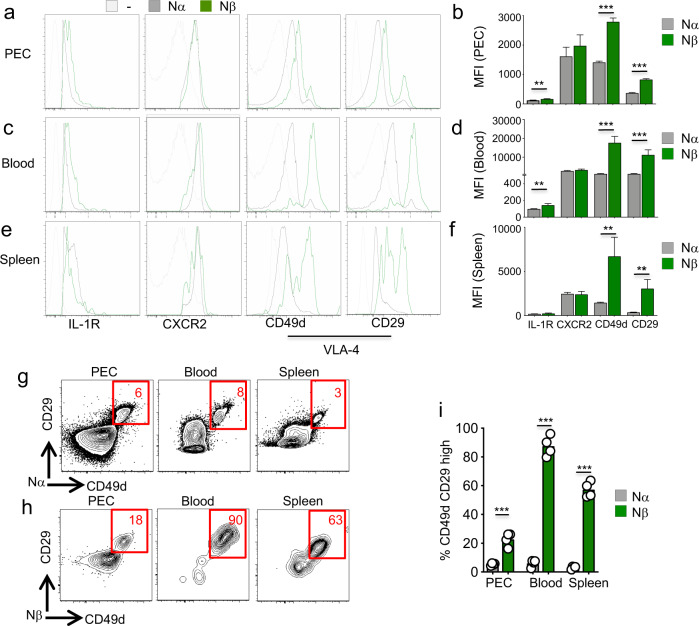
Nβ in the peritoneal cavity of NYVAC-C Δ3-infected C57BL/6 J mice had similar levels of CXCR2 compared to Nα (Fig. 4a) but they started to overexpress IL-1R and CD49d (α4) CD29 (β1) integrins. Indeed, the levels of these last surface markers were significantly higher in Nβ than in Nα neutrophils (Fig. 4b). In the blood, we detected no difference in CXCR2 levels between these populations; however, Nβ upregulated CD49d and CD29 compared to surface levels of these cells in the PEC (Fig. 4c). By analyzing the surface expression of these markers, we found that Nβ expressed more IL-1R and VLA-4 than Nα did (Fig. 4d). Finally, in the spleen, Nβ showed the same levels of IL-1R and CXCR2 as Nα (Fig. 4e); however, they still significantly overexpressed VLA-4 compared to Nα, even if VLA-4 expression levels were lower than those in the blood (Fig. 4e, f).
Fig. 4. Different expression of VLA-4 integrin by distinct neutrophil subsets.
Layouts and median fluorescence intensity (MFI) of IL-1R, CXCR2, CD49d, and CD29 in Nα (gray) and Nβ (green) neutrophils at 6 h post infection in PEC (a–b), blood (c–d), and spleen (e–f) of NYVAC-C Δ3-injected mice. Isotype control in gray. Columns show mean ± SEM of four mice. Contour plots of CD29 and CD49d markers of Nα (g) and Nβ (h) neutrophils from PEC, blood, and spleen of NYVAC-C Δ3-injected mice at 6 h post infection. Percentages of CD49d and CD29 high Nα (gray) and Nβ (green) neutrophils from PEC, blood, and spleen of NYVAC-C Δ3-injected mice (i). Graphs show mean; each point represents an individual mouse. Data are representative of three independent experiments. **P < 0.01, ***P < 0.001.
A deep analysis of VLA-4 expression in those organs showed that Nα almost uniformly expressed this integrin at low levels, while a fraction of Nβ was highly positive for this marker (Fig. 4g, h). Indeed, Nβ were around 20% highly positive cells in the PEC, they were almost homogenously highly VLA-4 expressing cells in the blood, and still 60% highly positive cells in the spleen (Fig. 4h). Overall, the percentages of highly positive VLA-4 Nβ cells were significantly higher than Nα in all the tested organs (Fig. 4i).
These data indicate that IL-1R was upregulated in PEC and blood, while VLA-4, which is involved in transendothelial migration, was predominantly overexpressed in blood and spleen. In addition, VLA-4 was ubiquitously at higher levels in Nβ neutrophils compared to Nα to possibly promote their trafficking from and towards secondary lymphoid organs.
α4β1 integrin inhibitor enhances CD8 T-cell responses by increasing Nβ/Nα ratio
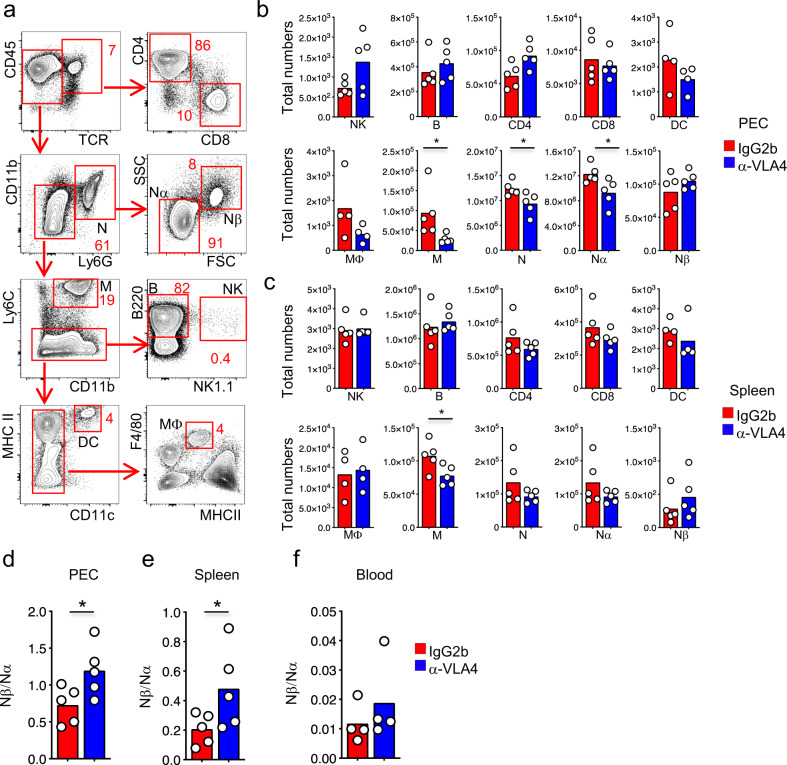
To determine whether the different expression of α4β1 VLA-4 integrin by distinct neutrophil subtypes results in the possibility to manipulate their infiltration in the primary site of infection and the secondary lymphoid organ, we injected mice with a low dose (250 μg) of α4β1 antagonist Natalizumab (monoclonal anti-VLA-4) that controls the migration of immune cells33. We broadly characterized the major immune subsets that could express α4β1 integrin (Fig. 5a). We quantified the abundance of these immune subsets in the PEC and in the spleen of mice that were treated with IgG2b isotype antibody or anti-VLA4 antibody and infected with 107 PFU of NYVAC-C Δ3. With the exception of monocytes (M) and neutrophils (specifically Nα), the total numbers of CD4, CD8, NK, B, DC, and macrophages (Mϕ) were similar in the PEC and spleen of IgG2b- or anti-VLA4 antibody-treated mice at 6 h p.i. (Fig. 5b, c). Conversely, there were no differences in Nβ neutrophil total numbers (Fig. 5b, c). As a result of this distinct neutrophil accumulation, the ratio Nβ/Nα was significantly increased in the PEC and spleen of those mice treated with α4β1 inhibitor, and a similar trend was observed in the blood (Fig. 5d–f).
Fig. 5. Inhibition of VLA-4 increases Nβ/Nα ratio.
Contour plot composition of peritoneal immune infiltrate after NYVAC-C Δ3 infection. Proportion of TCR+CD4+ and TCR+CD8+ T cells, CD11b+ Ly6G+ neutrophils divided by SSC and FSC in Nα and Nβ, CD11b+ Ly6G− Ly6C+ monocytes (M), B220+ NK1.1− B cells, NK1.1+ NK cells, MHCII+ CD11c+ dendritic cells (DC) and F4/80+ MHCII+ macrophages (MΦ) were measured at 6 h post infection (a). Total numbers of NK, B, CD4, CD8, DC, MΦ, M, N, Nα, and Nβ from PEC (b) and spleen (c) of NYVAC-C Δ3 infected and IgG2b or anti-VLA4 antibody-treated mice. Mice were injected with IgG2b isotype or anti-VLA4 antibody 16 h before- and 1 h post infection. Nβ/Nα ratio from PEC (d), spleen (e), and blood (f) of infected and IgG2b or anti-VLA4 antibody-treated mice. Graphs show mean; each point represents an individual mouse. Data are representative of two independent experiments. *P < 0.05.
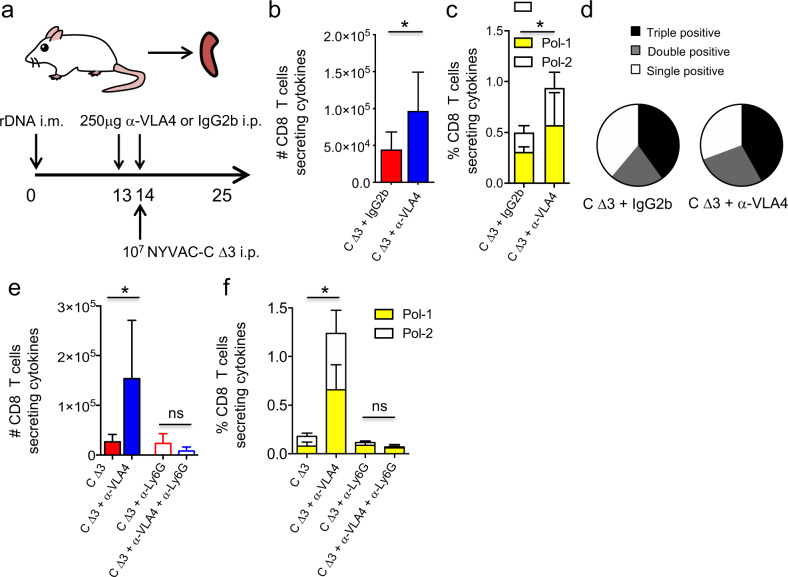
To test whether the increase of Nβ/Nα ratio in anti-VLA4 treated mice could influence the CD8 adaptive immune responses, BALB/c mice were immunized following the previously described heterologous immunization protocol (DNA prime/ NYVAC-C Δ3 boost) and treated with IgG2b isotype or anti-VLA4 antibody. CD8 T-cell responses to HIV Pol-1/Pol-2 antigens were analyzed from spleen (Fig. 6a). We quantified the magnitude of the T-cell responses as the number and percentage of T cells secreting IFN-γ and/or TNF and/or CD107a and the polyfunctionality as the capacity to express more than one of these markers. We observed in NYVAC-C Δ3-infected mice that, compared to IgG2b-treated mice, the inhibition of α4β1 integrin and infection with NYVAC-C Δ3 induced a significant increase of the magnitude of the HIV-specific CD8 T-cell responses (Fig. 6b, c), while the polyfunctionality was similar (Fig. 6d).
Fig. 6. Inhibition of VLA-4 increases HIV-specific T-cell responses.
Vaccine-induced HIV-1–specific CD8 T-cell response from spleen of mice (n = 4 per group) primed intramuscularly with recombinant DNA, infected with 107 PFUs of NYVAC-C Δ3 and injected with IgG2b isotype or anti-VLA4 antibody 16 h before and 1 h post infection (a). The response was measured 11 days after the last immunization, after splenocyte stimulation with HIV-1 Pol-1/Pol-2 peptides. Total numbers (b) or percentages of Pol-1-, Pol-2-specific (c) CD8 T cells that express IFN-γ and/or TNF and/or CD107a. Nonspecific responses of mice infected with control NYVAC-WT were subtracted from the total value. Graphs show mean ± SD. Pie chart colors indicate the relative percentage of cells producing three (black), two (gray), or one (white) activation markers (d). Total numbers (e) or percentages of Pol-1-, Pol-2-specific (f) CD8 T cells that express IFN-γ and/or TNF and/or CD107a in mice infected with 107 PFUs of NYVAC-C Δ3, injected with IgG2b isotype or anti-VLA4 and treated or untreated with anti-Ly6G. *P < 0.05.
To demonstrate whether the increase of the magnitude of T-cell responses in mice treated with anti-VLA4 antibody was neutrophil-dependent, we depleted these cells in vivo using anti-Ly6G mAb (1A8), administered 24 h before virus boost. Compared to control, mice treated with anti-VLA4 showed significant increase in total numbers and percentage of the Pol-1–, Pol-2–specific CD8 T-cell responses only when neutrophils were present (Fig. 6e, f). Indeed, mice injected with anti-Ly6G show low and similar CD8 T-cell response if were treated with or without anti-VLA4 (Fig. 6e, f).
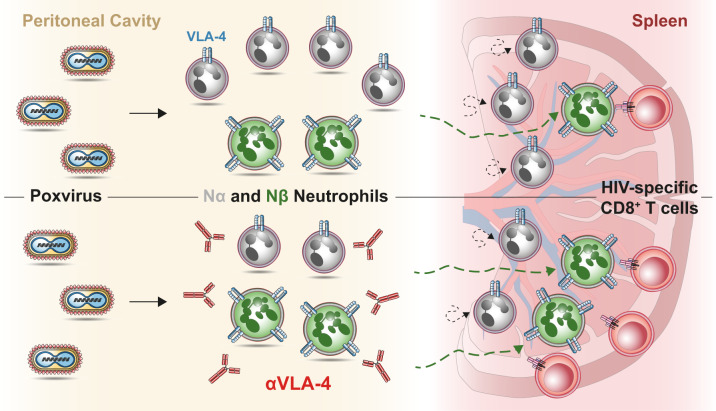
These findings show that HIV-specific CD8 T-cell response was enhanced whenever we increased Nβ/Nα ratio. Hence, systemic inhibition of α4β1 integrin, by shaping neutrophil subsets accumulation in primary site of infection and secondary lymphoid organs and by increasing antigen-specific CD8 T-cell responses (Fig. 7), provides new opportunities for vaccine design.
Fig. 7. Model of action.
After poxvirus infection in the peritoneal cavity, two distinct subtypes of neutrophil (Nα and Nβ) are generated. Nβ are characterized by overexpression of the trans-endothelial migration marker VLA-4 and an increased ability to interact with antigen-specific CD8+ T cells in the spleen (top panel). Poxvirus infection in combination with VLA-4 inhibition increases Nβ/Nα ratio in peritoneal cavity and spleen leading to augmented HIV-specific CD8 T-cell responses (bottom panel).
Discussion
In this study we described the in vivo accumulation of distinct neutrophil subsets in the primary site of VACV infection, their capacity to relocate to secondary lymphoid organs where CD8 T cells adaptive and memory responses take place. Furthermore, we characterized several markers associated with their ability to migrate to different tissues and defined, by shaping their ratio, a new strategy to increase HIV-specific CD8 responses that could be successfully applied in the design of new poxvirus-based vaccines.
Attenuated poxvirus vectors are one of the best vaccine candidates and, specifically, MVA and NYVAC poxvirus strains are used against cancer and infectious diseases in humans34. With respect to VACV vectors towards Acquired Immune Deficiency Syndrome (AIDS), several approaches have been developed to improve their immunogenicity to HIV antigens19. Among those, innate immune stimulation showed a clear therapeutic effect in monkeys infected with simian immunodeficiency virus35. So far, among all the clinical trials carried out with distinct AIDS vaccine candidates, only the RV144 phase III showed modest protection21. Nonetheless, new MVA/NYVAC-based clinical trials have been recently performed with promising results36–38. Based on these studies, there is major interest in understanding the VACV vectors-based mechanisms of induction of T-cell responses and, in turn, in increasing the effectiveness of those vaccines.
In this regard, NYVAC-C ΔA52RΔB15RΔK7R deletion mutant showed to be able to enhance HIV antigen-specific CD8 T cellular and humoral immune responses compared to many other NYVAC-C viruses. The mechanism of action is related with the targeting of the NFκB central host-cell signaling pathway and with the increasing of neutrophil migration in the peritoneal cavity and spleen of infected mice14,17. Upon infection, neutrophil characterization revealed two distinct cell size subsets that showed different APC properties and capacities to generate VACV-specific CD8 T-cell responses in vitro14.
Here, in order to extend those previous findings, we better characterized these neutrophil subsets by defining their transcriptional profile. Bulk RNA-sequencing analysis showed clearly two distinct neutrophil subpopulations: Nα upregulated more than 2000 genes compared to Nβ, and among those several N1 genes. A previous study39 defined N1 and N2 markers but a more recent RNA-sequencing analysis24 highlighted 138 genes with the strongest alteration between N1 and N2. Among those, 136 genes were downregulated by N2 neutrophils and approximately 100 of those genes were downregulated by Nβ cells in our study. Together, these data indicate a transcriptomic similarity between N2 and Nβ and between N1 and Nα neutrophils. Further studies, where neutrophil subsets isolated from two different environments (tumor and infection) are contemporaneously RNA-sequenced, have to demonstrate how similar these neutrophil subpopulations are.
Additionally, we defined how both neutrophil subtypes behave in the spleen and showed that Nβ moved faster, migrated more, and spend more time patrolling than Nα. Nβ higher speed and patrolling ability could facilitate the interactions of this neutrophil subset with HIV-specific T-cell clones. Indeed, a recent study also distinguished two subsets of splenic Ly6G+ cells, immature and mature neutrophils, that moved differently in the spleen and contributed differentially to the eradication of Streptococcus pneumoniae40. In this direction, our in vivo movies show how Nβ move towards CD8+ T cells and they establish short interactions more frequently than Nα neutrophils. Further studies will have to specifically address how these Nβ–T-cell interactions and alternative mechanisms, such as release of factors from Nβ in the T-cell proximity, might be regulating T-cell activation/proliferation.
The higher expression levels of VLA-4 by Nβ cells and their larger percentages of highly positive cells compared to Nα, indicate that Nβ have advantage in cell recruitment and displacement. Indeed, the role of VLA-4 has been previously shown in neutrophil recruitment32, and by blocking VLA-4 it is possible to reduce neutrophil infiltration and limit the neuronal injury from ischemic stroke41. Along the same lines, here, a low dose of VLA-4 inhibitor decreased the amount of VLA-4low Nα cells that infiltrated the peritoneal cavity and the spleen, without limiting the recruitment of VLA-4high Nβ neutrophils. As a consequence, in the presence of the VLA-4 inhibitor, Nβ/Nα ratio was increased in both tissues. High doses of VLA-4 inhibitor have been shown to reduce the amounts of lymphocytes42. However, in our study, the mice that received a low dose of antibody had comparable CD4 and CD8 T cell total numbers to those mice that did not receive the inhibitor.
The increase of Nβ/Nα ratio correlated with a higher HIV-1 Pol-1/Pol-2-specific CD8 T-cell responses in mice that received the VLA-4 α4β1 integrin inhibitor. Pol is one of the best conserved HIV-1 proteins43 and increased number of CD8 response from variable Env epitopes to Pol epitopes is associated with control of viremia in the first years of HIV-1 infection44. Pol-specific CD8 T-cell response in NYVAC-C Δ3-infected and anti-VLA-4 treated mice is increased and is still mainly polyfunctional compared to those mice only infected with NYVAC-C Δ3. Considering that human HIV non progressors preferentially maintain highly functional HIV-specific CD8 T cells45, the NYVAC-C Δ3 vaccinia vector in combination with α4β1 integrin inhibitor might result in a promising approach for HIV prophylaxis and AIDS treatment.
In summary, here we describe how neutrophils, after poxvirus infection, accumulate in the primary site of infection and close to CD8 T cells on secondary lymphoid organs. Two distinct neutrophils subsets display different migratory patterns that correlate with distinct levels of markers associated with their recruitment. Moreover, by inhibiting α4β1 integrin and manipulating the migration of these neutrophil subsets in the peritoneal cavity and in the spleen, we increased the vaccine efficacy of NYVAC-C vector in terms of CD8 T-cell adaptive immune responses specific for HIV heterologous antigens. Our findings could be relevant to develop a valid prototype for future HIV vaccine and provide novel basis for virus vaccine vector design.
Methods
Mice and injections
BALB/c and C57/BL6J mice (6–8 weeks old) were purchased from Harlan and Janvier labs. UBC (ubiquitin C)-GFP mice with a C56BL/6 background46 were bred in-house. NYVAC-C vector was provided by Sanofi Pasteur and NYVAC-C Δ3 was previously described14. For the DNA prime immunization protocol to assay virus immunogenicity, mice received 100 μg of DNA-C (50 μg pcDNA-CN54GP120 + 50 μg pcDNA-CN54GagPolNef) or 100 μg DNA-ϕ (pcDNA) by the intramuscular route22. Plasmids were purified using Endofree Plasmid Mega Kit (Qiagen). After two weeks, mice were immunized i.p. with 107 PFU of NYVAC-WT, NYVAC-C, or NYVAC-C Δ3. Mice immunized with sham DNA-ϕ prime and NYVAC-WT boost were used as controls. FTY720 (1 mg kg−1 bodyweight) in 150 μl H2O was injected intraperitoneally every other day until the end of the experiment. Anti-VLA-4 antibody (PS/2, BioXcell) or IgG2b, κ isotype control antibody (LTF-2, BioXcell) were injected intraperitoneally (250 μg per mouse) on day 13 (16 h before infection) and on day 14 (1 h after infection). Animal studies were approved by the Ethical Committee of Animal Experimentation (CEEA-CNB) of the Centro Nacional de Biotecnologia (CNB-CSIC, Madrid, Spain) and by Institutional Committee of the Cantonal Veterinary (Commissione cantonale per gli esperimenti sugli animali, Ticino), in accordance with national and international guidelines and the Royal Decree (RD 1201/2005) (permit n° 12013, 13013) and with the Swiss Federal Veterinary Office guidelines (permit n° TI28/17, TI02/14, and TI07/13).
Cell and viruses
African green monkey kidney cells (BSC-40; American Type Culture Collection) and primary chicken embryo fibroblast (CEF) cells from fertilized eggs (MSD Animal Health, Salamanca, Spain) were grown in DMEM supplemented with 10% (vol/vol) FCS. All viruses were grown in primary CEF cells, purified through two 36% (w/v) sucrose cushions, and titrated by immunostaining plaque assay in BSC-40 cells as described47.
Peptides
The HIV-1 peptides Pol-1 (LVGPTPVNI) and Pol-2 (YYDPSKDLI) are H-2d-restricted CTL epitopes23 and were provided by the CNB-CSIC Proteomics Service.
Flow cytometry
Peritoneal exudate cells (PEC), peripheral blood, and spleen (SP) obtained from infected and PBS-injected mice were stained with anti-Ly6B.2 (7/4) (abcam), anti-Ly6G (1A8), -CD11b (M1/70), -CD49d (R1-2), -CD29 (HMβ1-1), -CD182 (CXCR2; SA044G4), -CD184 (CXCR4; L276F12), -CD121 (IL-1R; JAMA147), -CD45 (30-F11), -TCRβ (H57-597), -CD4 (GK1.5), -CD8 (53-6.7), -Ly6C (HK1.4), -B220 (RA3-6B2), and NK1.1 (PK136) (all from Biolegend). For intracellular cytokine staining (ICS), erythrocyte-depleted splenocytes were resuspended in RPMI 1640 with 10% FCS and 1 μg/ml Golgiplug (BD), monensin (eBioscience), and anti-CD107a (1D4B) (BD). After re-stimulation with HIV-1 peptides (6 h, 37°C, 5% CO2), splenocytes were stained for surface markers with anti-CD3 (145-2C11), -CD4 (GK1.5), and -CD8 (53-6.7) (all from BD), fixed, permeabilized (Cytofix/Cytoperm kit; BD), and stained intracellularly with anti-IFN-γ (XMG 1.2) and -TNF (MP6-X722) (all from BD). Cells were acquired using a GALLIOS (Beckman Coulter) or LSRII (BD) flow cytometer and data were analyzed with FlowJo software v.9.9.5 and 10.5.3.
Immunofluorescence
Mice were anesthetized with a mixture of ketamine (100 mg/kg bodyweight, Parke Davis) and xylazine (10 mg/kg bodyweight, Bayer) and perfused with a fixative solution made of 10 mL of 0.05 M phosphate buffer containing 0.1 M L-lysine, 4% paraformaldehyde, and 2 mg/mL NaIO4 at pH 7.4 (PLP). After collection, spleens were incubated in 30% sucrose phosphate-buffered solution overnight. Tissues were snap frozen in O.C.T. compound (VWR Chemicals Leuven). 30 mm sections were cut with a microtome and blocked with proper sera. Sections were stained with anti-CD21/CD35 (CR2/CR1) (7E9), -IgD antibody (11-26 c.2a), -Ly-6G antibody (1A8), -CD8a (53-6.7), and -CD3ε antibody (145-2C11) (all from Biolegend). Immunofluorescence was performed using a Leica TCS SP5 confocal microscope (Leica Microsystems) using sequential acquisition to limit the signal crosstalk. Images were analyzed using Imaris software (Bitplane AG).
CD8+ cells were segmented with Imaris surface segmentation to determine their positions. CD8+ cell density was calculated with the kernel density estimate function of “scikit-learn” python package. The follicles were identified by thresholding the density function to exclude isolated CD8+ cells. The nearest neighbor distance of the aggregated CD8+ cells to the Ly6G+ neutrophils was obtained with a custom made R script. The p-value between the distributions was calculated with “ttest_ind” function of SciPy package.
Multi-photon intravital microscopy
Intravital microscopy of GFP+ neutrophils and CellTrace™ Violet-labeled cognate CD8+ T cells in the spleen was performed in C57/BL6J mice from 4 to 5 h after i.p vaccination. 5 × 106 of isolated GFP+ neutrophils were i.v. injected at the time of the vaccination whereas 5 × 106 of labeled cognate CD8+ T cells were i.v injected 16 h before vaccination. Mice were anesthetized with a mixture of ketamine (100 mg/kg bodyweight, Parke Davis) and xylazine (10 mg/kg bodyweight, Bayer). Once anesthetized, mice were kept on a specific customized surgical board over a heated plate or a heated surgical bench set at 37 °C for all the time of the surgery. Hair from the left dorsal side of the animal was removed using an electric razor and a depilatory cream. Spleen was exposed through a small incision in the skin and musculature at the left dorsal side of the animal. Once out, two drops of special surgical glue were placed in the extremes of the longitudinal axis of the spleen to fix the organ on the surface of the coverslip. Finally, the organ was covered with a few drops of pre-heated PBS and petroleum jelly to guarantee proper hydration to the organ.
Deep tissue imaging was performed on a customized inverted two-photon platform (TrimScope, LaVision BioTec). Two-photon probe excitation and tissue second-harmonic generation (SHG) were obtained with a set of two tunable Ti:sapphire lasers (Chamaleon Ultra I, Chamaleon Ultra II, Coherent) and an optical parametric oscillator that emits in the range of 1010–1340 nm (Chamaleon Compact OPO, Coherent), with output wavelength in the range of 690–1080 nm. During the acquisition a temperature-controlled incubation chamber, and a 25X/NA 1.1 water immersion objective was used. The photomultipliers (PMT) used for image acquisition were either hybrid detectors or high sensitivity GaAsP. For the in vivo analysis of cell movement, two-photon micrographs were acquired in full Z stacks of 40 nm every 30 s.
Cell detection, tracking, and volumetric reconstruction were performed using Imaris (Oxford Instruments, v7.7.2). Raw data were processed and analyzed with a custom Matlab script. Distinct neutrophil subsets were differentiated based on Volume Mean (Nα < 1000 μm3 and Nβ > 1500 μm3). Cell tracks were generated semi-automatically. Tracks with duration less than 300 s were excluded from the analysis.
Measurements of cell motility were computed using the Imaris software (Oxford Instruments, v9.2) and a custom Matlab script: track duration (time interval between the first and the last time points in which a cell is tracked), track length (total length of the cell trajectory), track speed mean (track length/track duration), track displacement (length of the vector from the first to the latest centroid position of the cell), directionality (straightness of a cell track, ranging from 0 to 1: from poorly directional to highly directional), arrest coefficient (the percentage of time in which a cell moves below a threshold of 2 μm/min ranging from 0 to 1: from always moving to never moving), and differential distance (the distance variation of each neutrophil towards the closest CD8+ T cell). The association of the motility patterns of each neutrophil to the cell actions “patrolling” and “arrested” were determined as previously described25.
Cell isolation
GFP+ neutrophils were purified from bone marrow of UBC (ubiquitin C)-GFP mice using a three-layer Percoll gradient of 78, 69, and 52% as previously described48. Neutrophils were collected at the 69–78% interface and with purity >90% injected. Cognate CD8+ T cells were obtained from spleens of WT mice that were DNA intramuscularly primed and NYVAC-C Δ3 i.p. boosted two weeks before. Cognate CD8+ T cells were purified by negative selection using the The EasySep™ Mouse CD8+ T Cell Isolation Kit (STEMCELL Technologies) and labeled with 5 μM of CellTrace™ Violet dye (Thermofisher).
Low-input RNA-sequencing
Nα and Nβ neutrophils from peritoneal washes of infected mice were FACS sorted based on their expression of CD11b and Ly6G and FSC/SSC characteristics (Supplementary Fig 1a). Up to 1000 cells of each subtype were directly sorted in a 96-well plate containing Lysis Buffer and RNase Inhibitor. One sample was excluded for technical reason. cDNA amplification was performed directly from lysed cells, with no previous RNA isolation, using the SMART-Seq v4 Ultra Low Input RNA Kit (Takara). Amplified cDNA was used for preparing index tagged sequencing libraries using the Nextera XT kit (Illumina). Average library size and size distribution were determined using the 2100 Bioanalyzer High Sensitivity DNA chip (Agilent). Libraries were quantified with the Qubit fluorometer using the dsDNA HS assay (ThermoFisher Scientific). Libraries were normalized and sequenced as a pool in one lane of a Rapid flow cell in the HiSeq2500 sequencer (Illumina) to generate 61 bases single-end reads. FastQ files were obtained using the bcl2fastq v2.20.0.422 software (Illumina).
RNA sequencing data analysis
Read quality was determined with the application FastQC v0.11.5. (‘https://www.bioinformatics.babraham.ac.uk/projects/fastqc/‘). For data analysis, sequencing adaptor contaminations were removed from reads using Cutadapt v1.7.1 and the resulting reads were mapped on the transcriptome (GRCm38 Ensembl gene-build 84) and quantified using RSEM v1.2.2049. Only genes with at least 1 count per million in at least 6 of the initial samples were considered for statistical analysis. The R package limma v3.46.050 was used to normalize the estimated counts from RSEM and to test differential expression using the moderated t statistics51. Raw and Benjamini–Hochberg adjusted p-values (FDR) were calculated for each of the tests. An adjusted p value of less than 0.05 was considered statistically significant.
Statistical analysis
A two-tailed Student’s t-test was used for comparisons between two groups, and a one-way ANOVA with Tukey post hoc test (single time points) was used for comparisons across multiple groups. For statistical analysis of CD8 T-cell response to HIV-1 Pol antigens, we utilized an approach that corrects measurements for medium response and calculates confidence intervals and p values of hypothesis tests52. Background levels (splenocytes in RPMI) were subtracted from all values used.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We would like to thank Elena Prieto and David Jarrossay for FACS technical support. This study was funded by FEBS Short-Term Fellowship and a Marie Curie Global Fellowship (750973) (M.D.P.), FEBS Long-Term Fellowship (M.P.-S.), the Swiss National Foundation (SNF) grants, R’equipt (145038) Ambizione (148183) and the European Commission Marie Curie Reintegration Grant (612742) (S.F.G.), Formación del Profesorado Universitario (E.M.P.), EMBO Short-Term Fellowship (P.P.), SystemsX.ch for a grant 2013/124 (D.U.P.), Ministerio de Ciencia e Innovación Fellowships BES-2016-076635 (A.R.-P.) RTI2018- 095497-B-I00 (A.H.) and Spanish SAF-2017-88089-R-Mineco-FEDER, AIDS Research Network RD16/0025/0014-ISCIII-FEDER, and the PTVDC Program with support from the Bill & Melinda Gates Foundation (M.E.).
Author contributions
M.D.P. conceived the study, initiated, designed, performed and analyzed the experiments, and wrote the manuscript. M.P.-S. initiated the project, designed, and performed experiments. E.M.-P. designed, performed, and analyzed the experiments. P.P., C.E.G., I.B., A.D., and A.B. performed experiments. A.R.-P., R.K.R., R.D.A., D.U.P., and C.O.S.S. analyzed the data. A.H. helped to develop protocols and contributed with essential reagents. S.F.G. and M.E. supervised the work and conceived the study.
Data availability
All datasets generated during the current study are available from the corresponding authors upon reasonable request. Neutrophils bulk RNAseq data are available as a super-series in GEO under the accession number GSE165227.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mauro Di Pilato, Miguel Palomino-Segura.
These authors contributed equally: Mariano Esteban, Santiago F. Gonzalez.
Contributor Information
Mauro Di Pilato, Email: mdi@mdanderson.org.
Mariano Esteban, Email: mesteban@cnb.csic.es.
Santiago F. Gonzalez, Email: santiago.gonzalez@irb.usi.ch
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-021-00314-7.
References
- 1.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 3.Sorensen OE, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97:3951–3959. doi: 10.1182/blood.V97.12.3951. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto Y, Moki T, Takizawa T, Shiratsuchi A, Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J. Immunol. 2007;178:2448–2457. doi: 10.4049/jimmunol.178.4.2448. [DOI] [PubMed] [Google Scholar]
- 5.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 6.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Chtanova T, et al. Dynamics of neutrophil migration in lymph nodes during infection. Immunity. 2008;29:661–661. doi: 10.1016/j.immuni.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duffy D, et al. Neutrophils transport antigen from the dermis to the bone marrow, initiating a source of memory CD8+ T cells. Immunity. 2012;37:1145–1145. doi: 10.1016/j.immuni.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Beauvillain C, et al. Neutrophils efficiently cross-prime naive T cells in vivo. Blood. 2007;110:2965–2973. doi: 10.1182/blood-2006-12-063826. [DOI] [PubMed] [Google Scholar]
- 10.Araki H, et al. Reprogramming of human postmitotic neutrophils into macrophages by growth factors. Blood. 2004;103:2973–2980. doi: 10.1182/blood-2003-08-2742. [DOI] [PubMed] [Google Scholar]
- 11.Matsushima H, et al. Neutrophil differentiation into a unique hybrid population exhibiting dual phenotype and functionality of neutrophils and dendritic cells. Blood. 2013;121:1677–1689. doi: 10.1182/blood-2012-07-445189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sagiv JY, et al. Phenotypic diversity and plasticity in circulating neutrophil subpopulations in cancer. Cell Rep. 2015;10:562–573. doi: 10.1016/j.celrep.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 13.Lerman YV, et al. Sepsis lethality via exacerbated tissue infiltration and TLR-induced cytokine production by neutrophils is integrin alpha(3)beta(1)-dependent. Blood. 2014;124:3515–3523. doi: 10.1182/blood-2014-01-552943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Pilato M, et al. NF kappa B activation by modified vaccinia virus as a novel strategy to enhance neutrophil migration and HIV-specific T-cell responses. Proc. Natl Acad. Sci. USA. 2015;112:E1333–E1342. doi: 10.1073/pnas.1424341112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jablonska J, Leschner S, Westphal K, Lienenklaus S, Weiss S. Neutrophils responsive to endogenous IFN-beta regulate tumor angiogenesis and growth in a mouse tumor model. J. Clin. Invest. 2010;120:4163–4163. doi: 10.1172/JCI37223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fridlender ZG, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Pilato M, Mejias-Perez E, Sorzano COS, Esteban M. Distinct roles of vaccinia virus NF-kappa B inhibitor proteins A52, B15, and K7 in the immune response. J. Virol. 2017;91:e00575–17. doi: 10.1128/JVI.00575-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang CW, Strong BSI, Miller MJ, Unanue ER. Neutrophils influence the level of antigen presentation during the immune response to protein antigens in adjuvants. J. Immunol. 2010;185:2927–2934. doi: 10.4049/jimmunol.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum. Vaccines Immunother. 2012;8:1192–1207. doi: 10.4161/hv.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barouch DH, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013;155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 22.Gomez CE, et al. Generation and immunogenicity of novel HIV/AIDS vaccine candidates targeting HIV-1 Env/Gag-Pol-Nef antigens of clade C. Vaccine. 2007;25:1969–1992. doi: 10.1016/j.vaccine.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 23.Wild J, et al. Preclinical evaluation of the immunogenicity of C-type HIV-1-based DNA and NYVAC vaccines in the Balb/C mouse model. Viral Immunol. 2009;22:309–319. doi: 10.1089/vim.2009.0038. [DOI] [PubMed] [Google Scholar]
- 24.Shaul ME, et al. Tumor-associated neutrophils display a distinct N1 profile following TGFbeta modulation: a transcriptomics analysis of pro- vs. antitumor TANs. Oncoimmunology. 2016;5:e1232221. doi: 10.1080/2162402X.2016.1232221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pizzagalli DU, et al. Characterization of the dynamic behavior of neutrophils following influenza vaccination. Front. Immunol. 2019;10:2621. doi: 10.3389/fimmu.2019.02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biondo C, et al. The interleukin-1 beta/CXCL1/2/neutrophil axis mediates host protection against group B streptococcal infection. Infect. Immun. 2014;82:4508–4517. doi: 10.1128/IAI.02104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eash KJ, Greenbaum AM, Gopalan PK, Link DC. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Invest. 2010;120:2423–2431. doi: 10.1172/JCI41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jablonska J, Wu CF, Andzinski L, Leschner S, Weiss S. CXCR2-mediated tumor-associated neutrophil recruitment is regulated by IFN-beta. Int. J. Cancer. 2014;134:1346–1358. doi: 10.1002/ijc.28551. [DOI] [PubMed] [Google Scholar]
- 29.Rios-Santos F, et al. Down-regulation of CXCR2 on neutrophils in severe sepsis is mediated by inducible nitric oxide synthase-derived nitric oxide. Am. J. Resp. Crit. Care. 2007;175:490–497. doi: 10.1164/rccm.200601-103OC. [DOI] [PubMed] [Google Scholar]
- 30.von Vietinghoff S, Asagiri M, Azar D, Hoffmann A, Ley K. Defective regulation of CXCR2 facilitates neutrophil release from bone marrow causing spontaneous inflammation in severely NF-kappa B-deficient mice. J. Immunol. 2010;185:670–678. doi: 10.4049/jimmunol.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller WA. Mechanisms of leukocyte transendothelial migration. Annu. Rev. Pathol. Mech. 2011;6:323–344. doi: 10.1146/annurev-pathol-011110-130224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson RB, et al. The use of lymphocyte function-associated antigen (LFA)-1-deficient mice to determine the role of LFA-1, Mac-1, and alpha 4 integrin in the inflammatory response of neutrophils. J. Exp. Med. 2001;194:219–226. doi: 10.1084/jem.194.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niino M, et al. Natalizumab effects on immune cell responses in multiple sclerosis. Ann. Neurol. 2006;59:748–754. doi: 10.1002/ana.20859. [DOI] [PubMed] [Google Scholar]
- 34.Gomez CE, Najera JL, Krupa M, Perdiguero B, Esteban M. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr. Gene Ther. 2011;11:189–217. doi: 10.2174/156652311795684731. [DOI] [PubMed] [Google Scholar]
- 35.Borducchi EN, et al. Ad26/MVA therapeutic vaccination with TLR7 stimulation in SIV-infected rhesus monkeys. Nature. 2016;540:284-+. doi: 10.1038/nature20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couto E, et al. Comparison of safety and vector-specific immune responses in healthy and HIV-infected populations vaccinated with MVA-B. Vaccines. 2019;7:178. doi: 10.3390/vaccines7040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pantaleo G, et al. Safety and immunogenicity of a multivalent HIV vaccine comprising envelope protein with either DNA or NYVAC vectors (HVTN 096): a phase 1b, double-blind, placebo-controlled trial. Lancet HIV. 2019;6:E737–E749. doi: 10.1016/S2352-3018(19)30262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baden LR, et al. First-in-human randomized, controlled trial of mosaic HIV-1 immunogens delivered via a modified vaccinia Ankara vector. J. Infect. Dis. 2018;218:633–644. doi: 10.1093/infdis/jiy212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mishalian I, et al. Tumor-associated neutrophils (TAN) develop pro-tumorigenic properties during tumor progression. Cancer Immunol. Immunother. 2013;62:1745–1756. doi: 10.1007/s00262-013-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deniset JF, Surewaard BG, Lee WY, Kubes P. Splenic Ly6G(high) mature and Ly6G(int) immature neutrophils contribute to eradication of S-pneumoniae. J. Exp. Med. 2017;214:1333–1350. doi: 10.1084/jem.20161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann J, et al. Very-late-antigen-4 (VLA-4)-mediated brain invasion by neutrophils leads to interactions with microglia, increased ischemic injury and impaired behavior in experimental stroke. Acta Neuropathol. 2015;129:259–277. doi: 10.1007/s00401-014-1355-2. [DOI] [PubMed] [Google Scholar]
- 42.Haanstra KG, et al. Antagonizing the alpha(4)beta(1) Integrin, but Not alpha(4)beta(7), inhibits leukocytic infiltration of the central nervous system in rhesus monkey experimental autoimmune encephalomyelitis. J. Immunol. 2013;190:1961–1973. doi: 10.4049/jimmunol.1202490. [DOI] [PubMed] [Google Scholar]
- 43.Goulder PJR, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang OO, Daar ES, Ng HL, Shih R, Jamieson BD. Increasing CTL targeting of conserved sequences during early HIV-1 infection is correlated to decreasing viremia. AIDS Res. Hum. Retrovir. 2011;27:391–398. doi: 10.1089/aid.2010.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/dendritic cell interactions in vivo. Cell Immunol. 2001;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- 47.Ramirez JC, Gherardi MM, Esteban M. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 2000;74:923–933. doi: 10.1128/JVI.74.2.923-933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palomino-Segura, M., Virgilio, T., Morone, D., Pizzagalli, D. U. & Gonzalez, S. F. Imaging cell interaction in tracheal mucosa during influenza virus infection using two-photon intravital microscopy. J. Vis. Exp. 10.3791/58355 (2018). [DOI] [PMC free article] [PubMed]
- 49.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol.10.2202/1544-6115.1027 (2004). [DOI] [PubMed]
- 52.Najera JL, Gomez CE, Garcia-Arriaza J, Sorzano CO, Esteban M. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS ONE. 2010;5:e11406. doi: 10.1371/journal.pone.0011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated during the current study are available from the corresponding authors upon reasonable request. Neutrophils bulk RNAseq data are available as a super-series in GEO under the accession number GSE165227.