Abstract
Purpose
The present scoping review aims to assess the non-inferiority of distributed learning over centrally and locally trained machine learning (ML) models in medical applications.
Methods
We performed a literature search using the term “distributed learning” OR “federated learning” in the PubMed/MEDLINE and EMBASE databases. No start date limit was used, and the search was extended until July 21, 2020. We excluded articles outside the field of interest; guidelines or expert opinion, review articles and meta-analyses, editorials, letters or commentaries, and conference abstracts; articles not in the English language; and studies not using medical data. Selected studies were classified and analysed according to their aim(s).
Results
We included 26 papers aimed at predicting one or more outcomes: namely risk, diagnosis, prognosis, and treatment side effect/adverse drug reaction. Distributed learning was compared to centralized or localized training in 21/26 and 14/26 selected papers, respectively. Regardless of the aim, the type of input, the method, and the classifier, distributed learning performed close to centralized training, but two experiments focused on diagnosis. In all but 2 cases, distributed learning outperformed locally trained models.
Conclusion
Distributed learning resulted in a reliable strategy for model development; indeed, it performed equally to models trained on centralized datasets. Sensitive data can get preserved since they are not shared for model development. Distributed learning constitutes a promising solution for ML-based research and practice since large, diverse datasets are crucial for success.
Keywords: Machine learning, Clinical trial, Privacy, Ethics, Distributed learning, Federated learning
Introduction
Artificial intelligence has gained significant attention because of the achievements of machine learning (ML) and deep learning algorithms that rapidly accelerate research and transform practices in multiple fields, including medicine. However, data-driven learning necessitates “big data”-sets not to suffer from overfitting and underspecification. Indeed, studies on a small sample size using ML are affected by an inherent methodological bias that might undermine their validity. The adequate sample size is thus crucial for ML as for classical statistics [1–3]. An adequate sample is a hurdle in rare diseases (e.g. thymic malignancies, sarcomas) or low-prevalence conditions (e.g. refractory lymphoma, iodine-negative thyroid cancer) [4]. The small sample size is acknowledged as a limitation in almost every research study on image mining [1].
Other commonly recognized weaknesses in image mining studies are the monocentric retrospective design (i.e. localized learning) and the lack of independent validation. Collectively, all these aspects negatively affect the reproducibility and the generalizability of the results [1, 5]. These limitations might be overcome by multicenter or benchmarking trials. However, benchmarking trials require considerable infrastructural efforts to develop data repository platforms, while traditional multicenter studies (i.e. centralized learning) are affected by many logistical difficulties mainly related to sharing of clinical and imaging data. Data transfer is indeed burdened by legal, ethical, and privacy issues [5, 6].
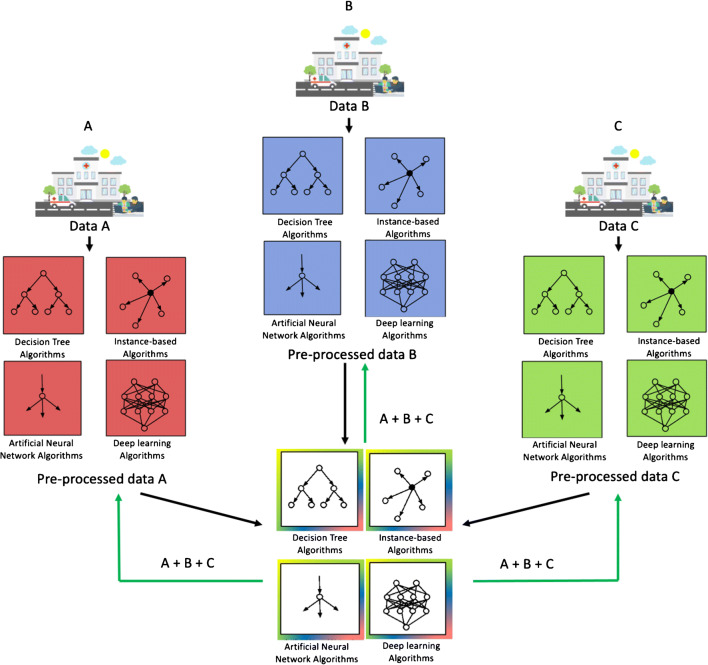
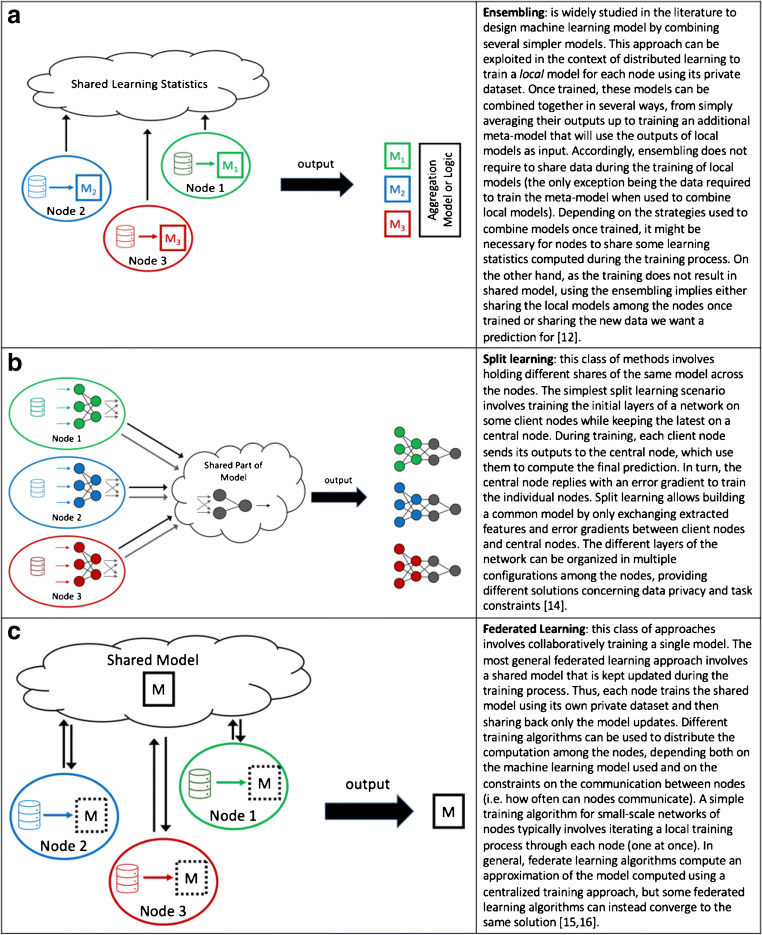
Given these constraints, distributed learning has emerged as a strategy for effective collaboration between centres while preserving governance and regulatory aspects [7]. Distributed learning aims to train one or more machine learning models within a network of nodes, each one owning a local dataset. Individual institutions do not share patients’ data externally. Just post-processed data in the form of model updates (e.g. coefficients and weight parameters) are shared among centres to build the final model [8, 9] (Fig. 1). Distributed learning methods may be distinguished according to computational principles (e.g. data parallelism and communication topology [10]) (Fig. 2) [11–13]. However, some general principles, (i) how the model parameters are displaced over the network of nodes, (ii) how nodes interact and which type of data they exchange, (iii) limitations on the kind of disclosed data, (iv) technical and technological constraints related to the task, are relevant to design the best-distributed learning model. Accordingly, distributed learning methods include different approaches, namely ensemble [14, 15], split [16], and federated learning [17, 18] described in details in Fig. 3.
Fig. 1.
Distributed learning framework
Fig. 2.
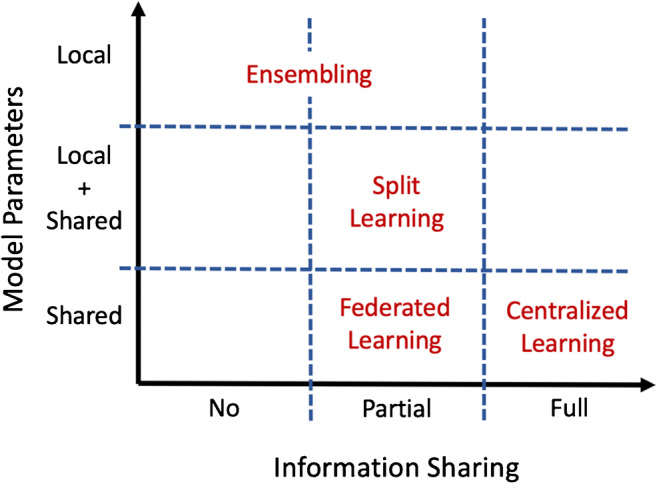
Schematic overview of the most popular distributed learning methods. Differences among methods are summarized according to two general design principles: (i) how the model parameters are displaced over the network of nodes, local, local + shared, and shared (y-axis), and (ii) how nodes interact and which type of data they exchange, no, partial, and complete exchange (x-axis)
Fig. 3.
Distributed learning methods
Therefore, distributed learning dealing with a network of nodes, each one owning a local dataset, has been proposed as a method to cross the hurdles related to patient data sharing [7, 9]. However, there are some uncertainties related to distributed learning performance compared to centrally trained models. Therefore, the present scoping review, providing an overview of distributed learning in medical research, aimed at assessing the non-inferiority of distributed learning over centrally and locally trained models. The non-inferiority of distributed learning over centrally and locally trained models is an essential requirement to appoint distributed learning as a suitable approach for data sharing within multicenter collaborations.
Materials and methods
Literature search and selection
We performed an extensive literature search using the term “distributed learning” OR “federated learning” in the PubMed/MEDLINE and EMBASE databases. No start date limit was used, and the search was extended until July 21, 2020. Two authors (MS and GN) independently searched the literature and performed an initial screening of identified titles and abstracts. The following exclusion criteria were applied: (a) articles outside the field of interest (i.e. distributed learning); (b) guidelines or expert opinion, review articles and meta-analyses, editorials, letters or commentaries, and short abstracts presented at conferences and scientific meetings; (c) articles not in the English language; and (d) studies not testing distributed approaches using medical data (i.e. electronic health records, genomic data, signals, or images). Selected papers were retrieved in full text, and reference lists were screened in order to identify additional records. Screened papers were included in the scoping review when considered eligible by both reviewers. In case of discrepancies, papers were reviewed by a third researcher, blinded to previous assessments. The majority vote was used to include/exclude a paper finally.
Analysis
Each selected article was tabulated in an Excel ®2017 (Microsoft®, Redmond, WA) file. The following information was collected: (i) clinical setting; (ii) type of data (clinical data, images, genomic data); (iii) source of data (open-access or local dataset); (iv) type of distributed network (simulated versus real); (v) machine learning algorithms’ architecture; (vi) distributed learning approach (ensembling, split, or federated learning); (vii) performance metrics; (viii) comparative analysis (distributed versus centralized, distributed versus local); (ix) analysis on data distribution among nodes; (x) other results. Later on, selected studies were classified according to their aim, namely (a) risk prediction; (b) diagnosis; (c) prognosis; and (d) treatment side effect/adverse drug reaction prediction. Papers having more than one aim were analysed as separate works.
Results
Study selection
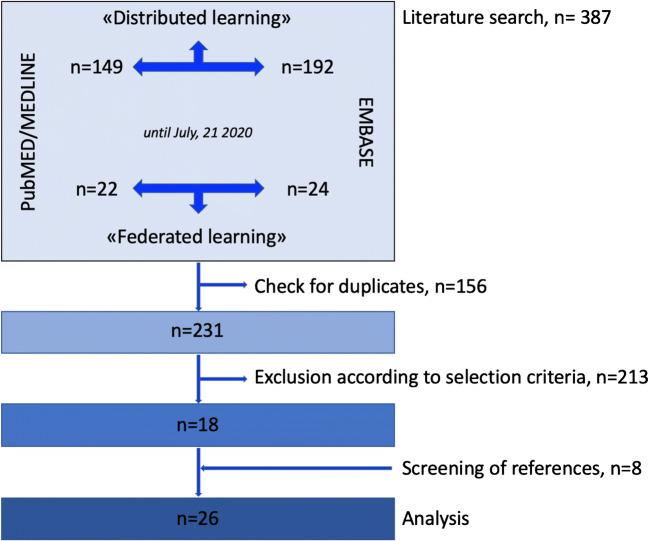
Overall, the search criteria resulted in 387 articles. After removing duplicates and initial screening of titles and abstracts, 347 papers were excluded, and the remaining 40 were retrieved in full text. Subsequent analysis of full-text articles excluded 22 papers (two using non-medical data, 13 outside the field of interest, five review articles). Eight additional research studies of interest were identified as screening reference lists. A total of 26 studies were eventually selected. Figure 4 summarizes the process of study selection.
Fig. 4.
Study selection
Overall results
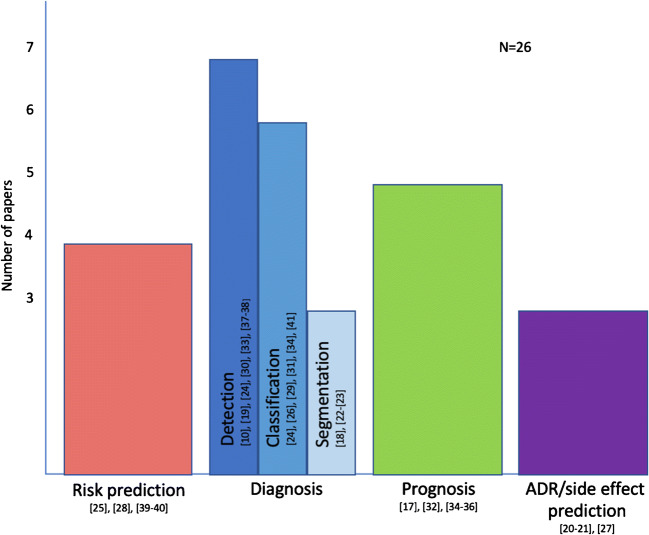
Distributed learning was based on federated learning with [12, 19–21] or without [22–43] other methods in the great majority of the selected studies (4/26 and 22/26, respectively); in few articles (3/26), the ensembling approach was preferred [38, 40, 42]. Distributed training was compared to centralized and/or localized training in 21/26 [12, 19, 20, 22–24, 26, 28–31, 34–43] and 14/26 [12, 21, 22, 25, 27, 28, 30, 32, 33, 36, 37, 39, 40, 42] selected papers, respectively. Area under the curve (AUC) [19, 22–25, 27–29, 36, 43] and accuracy [12, 21, 27, 34, 35, 37, 39–41] were the most commonly used metrics to assess models’ performance in the selected papers (10/26 and 7/26). Other metrics included mean squared error [26, 31, 39, 41], dice score [20, 32, 33], F1 score [27, 42], precision [27, 37], normalized mutual information and signal-to-noise ratio [39], recall [37], sensitivity and positive predictive value [27], hazard ratio [38], relative bias, error ratio, and odd ratio [30]. Figure 5 summarizes the studies by the aim.
Fig. 5.
Summary of the topics covered by selected studies. (ADR, adverse drug reaction)
Risk prediction
Three out of the 26 included papers developed a distributed learning framework for risk prediction (Table 1). Brisimi et al. [29] and Wang et al. [38] used clinical data to predict hospitalization risks for cardiac events and re-hospitalization for heart failure, respectively. Duan et al. [30] used distributed clinical data to build a risk model for foetal loss in pregnant women. Wu et al. [35] instead performed a federated genome-wide association studies analysis to predict the risk of developing ankylosing spondylitis in healthy individuals. In the four studies, distributed and centralized learning model performance was compared. In all cases, the two approaches showed substantial equivalence.
Table 1.
Studies using distributed learning models for risk prediction
| Reference | Data, type | Aim | Distributed network | Machine learning model | Distributed learning method | Distributed vs centralized learning | Distributed vs localized learning |
|---|---|---|---|---|---|---|---|
| [29] | Clinical data | Prediction of risk of hospitalisations for cardiac events | Simulated distribution among 5 and 10 nodes | SVM | Federated learning | Federated learning performs close to centralized learning | NA |
| [30] | Clinical data | Prediction of foetal loss risk | Simulated distribution among 10 nodes | Logistic regression | Federated learning | Federated learning performs close to centralized learning | Federated learning outperforms localized learning models |
| [35] | Genomic data | Prediction of risk of developing ankylosing spondylitis | Real distribution among 3 nodes | PCA | Federated learning | Federated learning performs close to centralized learning | NA |
| [38] | Clinical data | Prediction of readmission for heart failure within 30 days | Simulated distribution among 50 and 200 nodes | Cox proportional hazards model | Ensembling | Ensembling performs close to centralized learning | NA |
NA, not assessed; SVM, support vector machine
Diagnosis
Out of the 26 papers on distributed learning, 15 focused on diagnosis (Table 2). Among these, three works [12, 34, 42] used different datasets as input for their experiments. Specifically, Chang et al. [12] tested distributed learning to analyse retinal fundus images and mammograms, Balachandar et al. [34] to analyse retinal fundus images and chest x-ray, and Tuladhar et al. [42] to analyse digital fine-needle aspiration (FNA) images and clinical data.
Table 2.
Studies using distributed learning models for diagnosis
| Reference | Data, type | Aim | Distributed network | Machine learning model | Distributed learning method | Distributed vs. centralized learning | Distributed vs. localized learning |
|---|---|---|---|---|---|---|---|
| [42] | Images (digital FNA images) | Detection of breast cancer | Simulated distribution among many different numbers of nodes | ANN, SVM, RF | Ensembling | Ensembling performs close to centralized learning | Ensembling outperforms localized learning models |
| [42] | Clinical data | Detection of MCI | Real distribution among 43 nodes | ANN, SVM, RF | Ensembling | Centralized learning outperforms ensembling | Ensembling outperforms localized learning models |
| [42] | Clinical data | Detection of diabetes | Simulated distribution among many different numbers of nodes | ANN, SVM, RF | Ensembling | Ensembling performs close to centralized learning | Ensembling outperforms localized learning models |
| [42] | Clinical data | Detection of heart disease | Simulated distribution among many different numbers of nodes | ANN, SVM, RF | Ensembling | Ensembling performs close to centralized learning | Ensembling outperforms localized learning models |
| [43] | Image-derived data (radiomic features from head and neck CT scans) | Prediction of HPV status in head and neck cancer | Real distribution among 6 nodes | Logistic regression | Federated learning | Federated learning performs close to Centralized Learning | NA |
| [12] | (Retinal fundus images) | Diabetic retinopathy detection | Simulated distribution among 4 nodes | CNN | Federated learning + ensembling | Federated learning performs close to centralized Learning | NA |
| [12] | Images (mammograms) | Breast cancer detection | Simulated distribution among 4 nodes | CNN | Federated learning + ensembling | Federated learning performs close to centralized learning | NA |
| [27] | Clinical data (ICU data) | Disease detection | Distribution among 2 nodes based on the data provisioning system | Autoencoder | Federated learning | NA | Federated learning outperforms localized learning models |
| [28] | Clinical data (ICU data) | Disease detection | Simulated distribution among 2 and 3 nodes | Hashing | Federated learning | Federated learning performs close to centralized learning | Federated learning outperforms localized learning models |
| [20] | Images (brain MRI) | Brain tumour segmentation | Simulated distribution among 4, 8,16, and 32 nodes | CNN | Federated learning, CIIL, IIL | Federated learning performs close to centralized learning | NA |
| [31] | Images (brain MRI) | Classification of neurodegenerative diseases | Real distribution among 4 nodes | PCA | Federated learning | Federated learning performs close to centralized learning | NA |
| [32] | Images (head CT scans) | Haemorrhage segmentation | Real distribution among 4 nodes (only 2 nodes used for training) | CNN | Federated learning | NA | Federated learning outperforms localized learning models |
| [33] | Images (head CT scans) | Haemorrhage segmentation | Real distribution among 2 nodes | CNN | Federated learning | NA | Federated learning outperforms localized learning models |
| [34] | Images (retinal fundus images) | Diabetic retinopathy detection | Simulated distribution among 4 nodes | CNN | Federated learning | Federated learning performs close to centralized learning | NA |
| [34] | Images (chest X-rays) | Thoracic disease classification | Simulated distribution among 4 nodes | CNN | Federated learning | Federated learning performs close to centralized learning | NA |
| [21] | Images (brain fMRI) | Autism spectrum disorder detection | Real distribution among 4 nodes | ANN | Federated learning + ensembling | NA | Federated learning outperforms localized learning models |
| [36] | Images (chest CT scans) | Pneumonia classification | Real distribution among 4 nodes | CNN | Federated learning | Federated learning performs close to centralized learning (except on viral pneumonia other than COVID-19) | Federated learning outperforms localized learning models |
| [39] | Images (digital FNA images) | Breast lesion classification | Simulated distribution among 5 nodes | Semi-supervised ANN + ELM | Federated learning | Centralized learning outperforms federated learning | Federated learning outperforms localized learning models |
| [40] | Images (brain MRI) | Schizophrenia detection | Real distribution among 4 nodes | SVM | Ensembling | Ensembling performs close to centralized learning | Ensembling outperforms localized learning models |
| [41] | Images (digital FNA images) | Breast lesion classification | Simulated distribution among 10 nodes | ANN | Federated learning | Federated learning performs close to centralized learning | NA |
ANN, artificial neural network; CIIL, cyclic institutional incremental learning; CNN, convolutional neural network; CT, computed tomography; ELM, extreme learning machine; FNA, fine-needle aspiration; HPV, human papilloma virus; ICU, intensive care unit; IIL, institutional incremental learning; MCI, mild cognitive impairment; MRI, magnetic resonance imaging; NA, not assessed; PCA, principal component analysis; RF, random forest; SVM, support vector machine
In the group of 26 papers, seven articles aimed at using distributed clinical [27, 28, 42, 43] or imaging [12, 21, 34, 40, 42] data to detect several diseases. Included diseases were diabetic retinopathy, breast cancer, cardiac diseases, diabetes, and neuropsychiatric disorders.
Five studies instead tested distributed learning using diagnostic images for various classification tasks, namely differentiation of neurodegenerative diseases [31], thoracic pathologies [34], types of pneumonia [36], benign vs malignant breast lesions [39, 41], and HPV+ vs HPV- head and neck cancer [43].
Finally, three studies [20, 31, 32] utilized distributed head computed tomography (CT) and brain magnetic resonance imaging (MRI) data to perform segmentation of intraparenchymal haemorrhages and brain tumours.
In 13/15 papers focusing on the diagnosis, the authors compared distributed and centralized approaches for model training. The performance achieved using a model trained on distributed data was always comparable to the one obtained using a centralized approach, except for a few experiments [36, 42]. Specifically, in the experiment by Tuladhar et al. [42] on real data distribution, the skewness of the training dataset towards patient examples affected the ensembling approach’s performance in predicting mild cognitive impairment (MCI), but not those of the centralized learning. However, this experiment was characterized not only by imbalanced classes (189 MCI patients versus 94 healthy subjects) but also by a limited number of observations (average of 4 patients and two healthy cases) for each node [42]. Finally, centralized learning outperformed distributed learning in classifying non-COVID-19 viral pneumonia [36]. Xu et al. [34] developed a distributed CNN model [36](Xu et al. 2020) [26–28, 34]to classify CT scan as normal or with pneumonia, distinguishing among COVID-19 or other viral pneumonia and bacterial pneumonia. The federated model performed close to the centralized one in all cases, except non-COVID-19 viral pneumonia, which was scarcely represented in the whole dataset (76 cases) [36].
Prognosis
Five out of the 26 analysed articles on distributed learning were aimed at prognostication (Table 3).
Table 3.
Studies using distributed learning models for prognosis
| Reference | Data, type | Aim | Distributed network | Machine learning model | Distributed learning method | Distributed vs. centralized learning | Distributed vs. localized learning |
|---|---|---|---|---|---|---|---|
| [24] | Clinical data | Post-treatment 2-year survival prediction | Real distribution among 3 nodes | SVM with ADMM | Federated learning | Federated learning performs close to centralized learning | NA |
| [25] | Clinical data | Post-treatment 2-year survival prediction | Real distribution among 8 nodes | Bayesian network | Federated learning | NA | Localized learning models sometimes outperform federated learning |
| [43] | Image-derived data (radiomic features from head and neck CT scans) | Post-treatment 2-year survival prediction | Real distribution among 6 nodes | Logistic regression | Federated learning | Federated learning performs close to centralized learning | NA |
| [26] | Clinical data | Prediction of hospital LoS | Simulated distribution among 3, 6, and 12 nodes | Linear regression | Federated learning | Federated learning performs close to centralized learning | NA |
| [19] | Clinical data | Prediction of mortality | Real distribution among 50 nodes | Autoencoder, K-means, and nearest neighbour | Federated learning + ensembling | Federated learning performs close to centralized learning | NA |
| [19] | Clinical data | Prediction of ICU LoS | Real distribution among 50 nodes | Autoencoder, K-means, and nearest neighbour | Federated learning + ensembling | Federated learning performs close to centralized learning | NA |
ADMM, alternative direction method of multipliers; CT, computed tomography; ICU, intensive care unit; LoS, length of stay; NA, not assessed; SVM, support vector machine
Jochems et al. [24] set up a network of systematic clinical data collection and sharing through distributed learning to predict post-treatment 2-year survival in 894 non-small cell lung cancer (NSCLC) patients. The same group [25] confirmed the previous results on a larger cohort, including more than 20,000 patients in eight centres across different countries.
Bogowicz et al. [43] used a distributed approach to train a radiomic model to predict 2-year survival in a cohort of 1174 patients with head and neck cancer.
Dankar et al. [26] and Huang et al. [19] used a distributed algorithm to estimate the patient length of stay in hospital and ICU, respectively. Moreover, Huang et al. [19] used the same approach to predict inpatient mortality.
In four out of these five prognostic studies, predictive models trained using distributed and centralized data were compared (Table 3). In all cases, models did not show significant differences in terms of performance.
Adverse drug reaction/side effect prediction
Three out of the 26 studies used a distributed approach to predict either adverse drug reaction (ADR) or treatment side effect (Table 4).
Table 4.
Studies using distributed learning models for adverse drug reaction/side effect prediction
| Reference | Data, type | Aim | Distributed network | Machine learning model | Distributed learning method | Distributed vs. centralized learning | Distributed vs. localized learning |
|---|---|---|---|---|---|---|---|
| [22] | Clinical data | Prediction of post-radiotherapy dyspnoea | Real distribution among 5 nodes | Bayesian network | Federated learning | Federated learning performs close to centralized learning | Federated learning performs close to localized learning |
| [23] | Clinical data | Prediction of post-radiotherapy dyspnoea | Real distribution among 5 nodes | SVM with ADMM | Federated learning | Federated learning performs close to centralized learning | NA |
| [37] | Clinical data | Prediction of opioid chronic use | Simulated distribution among 10 nodes | SVM, single-layer perceptron, logistic regression (all of them trained with SGD) | Federated learning | Federated learning performs close to centralized learning | Federated learning outperforms localized learning models |
| [37] | Clinical data | Prediction of antipsychotics side effects | Simulated distribution among 10 nodes | SVM, single-layer perceptron, logistic regression (all of them trained with SGD) | Federated learning | Federated learning performs close to centralized learning | Federated learning outperforms localized learning models |
ADMM, alternative direction method of multipliers; NA, not assessed; SGD, stochastic gradient descent; SVM, support vector machine
Among these, Choudhury et al. [37] performed two distributed learning experiments and built models to predict opioid chronic usage and antipsychotic side effects using clinical data.
Jochems et al. [22] tested the distributed approach using clinical parameters and built a Bayesian network model to predict dyspnoea in NSCLC patients treated with radiotherapy. The same group [23] confirmed the distributed approach’s potential in a second study using a different machine learning algorithm to predict the same outcome.
In the three studies, authors compared the distributed and centralized algorithms’ performance and showed substantial equivalence between the two approaches.
Discussion
The present scoping review aimed to gather and assess the research evidence to answer whether multi-institutional collaboration in machine learning research and practice can rely on distributed learning as on a conventional centralized approach. Our analysis confirmed that distributed learning is an effective strategy for multi-institutional collaboration with the potential advantage of being privacy-preserving. Although distributed learning has been proposed to share data guaranteeing privacy-preserving issues, it does not per se guarantee security and privacy by design. Indeed, it might be possible to retrieve estimations of the original data through a reverse engineering approach from the shared weights. Nonetheless, distributed learning should be considered as the prerequisite infrastructure to address governance and regulatory compliance. Indeed, a distributed network may be easily empowered by specific privacy preservation methods (e.g. differential privacy and cryptographic techniques) [7, 9]. To propose an effective privacy-preserving methodology in multicenter collaboration, the assessment of distributed learning performance compared to a centralized approach is the first-step requirement. Evaluation of the effectiveness of privacy-preserving methods was out of the present work’s scope and should be evaluated in future investigations.
Regardless of the aim (risk prediction, diagnosis, prognosis, or treatment side effect/adverse drug reaction prediction), the type of input (clinical data, images, or genetic data), the method (federated or ensembling), and the classifier (e.g. artificial neural networks, support vector machine, random forest), distributed training performed close to centralized training in almost all the experiments [12, 19, 20, 22–24, 26, 28–31, 34–43] (Tables 1-4). Centralized learning showed better performances than the distributed approach in two experiments among those reported in the studies by Xu et al. [36] and Tuladhar et al. [42]. However, both these experiments were burdened by a limited number of observations, probably responsible for the overfitting-related failure. Particularly, Xu et al. [36] built a model using a dataset consisting mainly of patients with SARS-CoV-2 or bacterial pneumonia (34% and 27%, respectively) and healthy subjects (33%). Other viral non-COVID pneumonia, which per se included several entities characterized by specific features (e.g. respiratory syncytial virus, cytomegalovirus, influenza A), represented only 6% of the entire dataset [36]. Similarly, the ensembling approach tested by Tuladhar et al. [42] dealt with unbalanced groups (67% MCI patients versus 33% healthy subjects), but the experiment also suffered from a limited number of observations for each node (average of 6 cases in the 43 nodes). Notably, the other experiments performed by Tuladhar et al. [42] dealing with simulated distributed data demonstrated that the dataset’s skewness did not affect the final model’s performance. This finding indicated that the imbalance of groups determined the model’s poor generalizability when the number of nodes and observations in each node is not sufficiently heterogenous to be representative for the target population. Indeed, when a sufficient number of “nodes” and of “observations for each node” is included in the model, distributed learning is not prone to the inherent limitations of locally trained models (e.g. imbalanced groups) and the overfitting related to the peculiarity of each site resulting in a competitive generalisable model. Distributed models outperformed locally trained models [21, 27, 28, 30, 32, 33, 36, 37, 39, 40, 42] with rare exceptions [22, 25]. Specifically, the distributed model developed by Jochems et al. [22] was as efficient as localized learning models in predicting post-radiotherapy dyspnoea. Diversely, Deist et al. [25] found that localized learning models sometimes outperform federated learning in 2-year survival prediction, suggesting that unobserved confounding factors or diverse outcome collection standards may affect the model’s performance. Preprocessing data harmonization – if possible – (e.g. image resampling, a clear methodology and uniform criteria for data collection) could positively impact data integration, simplifying multi-institutional collaboration for large-scale analytics [25, 44].
This scoping review aimed to produce evidence on the efficiency of the distributed learning regardless of the type of input data. We included papers dealing with clinical data (n = 12), images (hand-extracted features = 1, images = 11), both clinical data and images (n = 1), and genetic data (n = 1). This variability demonstrates that distributed learning may be implemented within any multicenter collaboration. Indeed, in precision medicine, an increasing number and kind of variables (lifestyle habits, constitutional factors, clinical, pathological, imaging, and “omics” data including treatment-related information) need to be considered to build predictive models. Medicine is evolving from a “disease-based” vision towards a “patient-based” approach based on multidimensional data that describe physiological and pathological conditions [6, 45, 46]. Distributed learning may represent a reliable framework for future multi-institutional and multidisciplinary research and clinical practice.
On the other hand, the “digital twin” approach has been proposed to respond to the challenge of the integration and analysing of large amounts of data within a dynamic framework. This technology is based on the construction of a digital model of an individual patient (depicting the molecular profile, the physiological status, and the life style habits) to virtually test a multitude of treatments to choose the optimal one [47, 48]. Therefore, both the medical and technological conceptual views that aim to provide a detailed and extensive description of an individual’s multidimensionality may benefit from a distributed learning framework.
Differences in terms of aims, input data, and methods (i.e. algorithms, data distribution among nodes, number of nodes, and metrics to assess model performance) made selected papers hardly comparable. Consequently, a preferred distributed learning method tailored for each specific task cannot be recommended. Nonetheless, ensembling emerged to be incredibly convenient when input data are heavily heterogeneous; it can be applied to any machine learning algorithm and uses different learning models for each node [14, 15]. Split learning, owing to its layered architecture, is fit for deep neural networks [16]. Finally, federated learning, which parallelly trains local models and aggregates their updates into a “central” node, can be efficiently utilized with many different machine learning algorithms [17]. Therefore, when setting up a distributed learning network, artificial data and simulated networks may be developed according to the model’s particular setting and objective to get preliminary performance results and compare distributed vs non-distributed approaches.
Technical constraints and artificial intelligence (AI) acceptance may be the main barriers to widespread distributed learning in healthcare. Technical challenges consist of computational burden and the communication overload (i.e. the amount of data shared among nodes) that need to meet the infrastructure constraints. In this regard, the number of nodes and kinds of data distributed among nodes is crucial hyperparameters that need to be tuned. Nonetheless, as a proof of concept, some commercial and open source solutions (e.g. DistriM from Oncoradiomics [49], Varian Learning Portal from Varian [50], Clara platform from Nvidia [51], and GRIN from Genomics Research and Innovation Network [52]) have recently become available, supporting the feasibility of implementing a distributed learning infrastructure.
Moreover, the lack of confidence and critical appraisal of the ML-based tools by healthcare personnel may limit technology implementation. Consequently, educational material and programs [6, 17, 53] involving clinicians, researchers, and regulatory officers are progressively getting available to promote the awareness on opportunities of multi-institutional trials and practices based on distributed learning towards a responsible AI. Additionally, the trustworthiness of AI-based methods is challenged by the barrier of explainability. The so-called eXplainable AI (XAI) field is growing, intending to develop responsible AI and encourage experts and professionals to embrace the new technology’s benefits to overcome the limitation related to explainability [54].
Furthermore, data have to be structured or preprocessed before their use to train a model. Distributed learning strategies were first introduced to analyse clinical data from electronic health records (EHRs). The rigid structure of EHRs has been influenced by research data collection systems and technology advances applied in clinical trials. Indeed, in clinical research, study design, data collection, analysis, and sharing have evolved over the last 20 years [55]. At present, data within clinical trials are recorded according to demanding rules aimed at making them standardized and structured or semi-structured. Overall, these approaches lead to high-quality information, adequate assessment of outcomes, endpoints, events, timely data analysis, reliable models, and efficient and comprehensive results’ publication and dissemination. The trend towards structured data is getting translated to routine medical EHRs. Generally, EHRs share common terminology, codes, and sections that could be easily collected from different centres or countries and subsequently analysed, while other domains, including medical imaging, are less structured. In the past years, several initiatives and registries containing structured data have been developed mainly as public health and descriptive epidemiological tools [55, 56]. The CancerLinQ has been the first and the foremost initiative sponsored by the American Society of Clinical Oncology and its Institute for Quality to collect “protected” EHRs data with the final goal of assessing, monitoring, and improving delivered care in cancer patients [57]. However, all these initiatives were challenged by privacy and security concerns on collecting, using, and patient information disclosure.
Additionally, concerns related to ethical and reliability aspects of AI-based algorithms may influence distributed learning technology spread. A potential drawback of AI prediction is its dependence on the data being used to train the algorithm. Training data have to represent the diseases and patient populations under evaluation and be balanced to perform when exposed to diverse patient data. Additionally, underspecification in ML pipelines may contribute to the failure of ML algorithms in a real-world deployment. Underspecification occurs when many distinct solutions can solve the problem equivalently [58]. Modelling pipelines need to explicitly account for this problem, also in the medical imaging domain.
The great potential of distributed learning has been already recognized by companies as proofed by the EHR4CR project. Within that project, pharmaceutical companies created a precompetitive data environment that relied on distributed learning methods that allowed them to safely perform clinical trials, sharing commercially confidential information [59].
This scoping review has some limitations. The high heterogeneity of included studies in terms of aims, input data, methods, and performance metrics prevented a comparison between studies, leaving no space for systematic considerations. Therefore, the present review aimed at assessing if multisite collaboration could efficiently rely on distributed learning. Therefore, our interest focused on assessing the non-inferiority of distributed learning over centrally and locally trained models instead of producing a systematic review. Nonetheless, the review gathered evidence that distributed learning is feasible, safe, and non-inferior for localized and centralized learning. Given this premise, we expect that distributed learning’s usefulness and applicability will be objects of many more investigations in different medical settings shortly.
In conclusion, distributed learning-based models showed to be reliable; indeed, they performed equally to models trained on centralized datasets. Sensitive data can get preserved by distributed learning since they are not shared for model development. Distributed learning constitutes a promising solution, especially for AI research, since large, diverse datasets are crucial for success. We foresee distributed learning being the bridge to large-scale, multi-institutional collaboration in research and medical practice.
Acknowledgements
M. Sollini is supported by the Investigator Grant 2019-23959, funded by AIRC (Italian Association for Cancer Research) won by A.C.
Availability of data and material
The manuscript represents valid work, and neither this manuscript nor one with substantially similar content under the same authorship has been published or is being considered for publication elsewhere. Arturo Chiti had full access to all the data in the study and takes responsibility for the data integrity and the accuracy of the data analysis. Raw data are available on specific request to the corresponding author.
Code availability
Not applicable.
Author contribution
DL, MK, MS, and AC conceptualized the review; MS performed the literature search; MS, MK, and GN performed the article selection; GN, EG, DL, MS, MK, and NG interpreted the article analyses; GN, EG, DL, MS, and MK, drafted the paper; MS, GN, MK, EG, and DL provided the figures; NG, EG, DL, FA, LM, PLL, MA, and AC critically commented the paper; all the authors critically revised the paper and approved the submitted version of the manuscript.
Declarations
Ethics approval
Not applicable.
Consent to participate and consent for publication
Not applicable for a scoping review.
Conflict of interest
Prof. Chiti reports a fellowship grant from Sanofi, personal fees from AAA, Blue Earth Diagnostics, and General Electric Healthcare outside the submitted work. Prof. Amigoni reports personal fees from Sandoz, outside the submitted work. The other authors do not report any conflict of interest.
Footnotes
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence).
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Margarita Kirienko, Email: margarita.kirienko@icloud.com.
Martina Sollini, Email: martina.sollini@hunimed.eu.
Gaia Ninatti, Email: gaia.ninatti@st.hunimed.eu.
Daniele Loiacono, Email: daniele.loiacono@polimi.it.
Edoardo Giacomello, Email: edoardo.giacomello@polimi.it.
Noemi Gozzi, Email: noemi.gozzi@humanitas.it.
Francesco Amigoni, Email: francesco.amigoni@polimi.it.
Luca Mainardi, Email: luca.mainardi@polimi.it.
Pier Luca Lanzi, Email: pierluca.lanzi@polimi.it.
Arturo Chiti, Email: arturo.chiti@hunimed.eu.
References
- 1.Sollini M, Antunovic L, Chiti A, Kirienko M. Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imaging. 2019;46:2656–2672. doi: 10.1007/s00259-019-04372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park JE, Kim D, Kim HS, Park SY, Kim JY, Cho SJ, et al. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol. 2020;30:523–536. doi: 10.1007/s00330-019-06360-z. [DOI] [PubMed] [Google Scholar]
- 3.Sollini M, Berchiolli R, Delgado Bolton RC, Rossi A, Kirienko M, Boni R, et al. The “3M” approach to cardiovascular infections: multimodality, multitracers, and multidisciplinary. Semin Nucl Med. 2018;48:199–224. doi: 10.1053/j.semnuclmed.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Kirienko M, Ninatti G, Cozzi L, Voulaz E, Gennaro N, Barajon I, et al. Computed tomography (CT)-derived radiomic features differentiate prevascular mediastinum masses as thymic neoplasms versus lymphomas. Radiol Med. 2020;125:951–960. doi: 10.1007/s11547-020-01188-w. [DOI] [PubMed] [Google Scholar]
- 5.Sollini M, Cozzi L, Ninatti G, Antunovic L, Cavinato L, Chiti A, et al. PET/CT radiomics in breast cancer: mind the step. Methods. 2020;S1046–2023:30263–30264. doi: 10.1016/j.ymeth.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Sollini M, Bartoli F, Marciano A, Zanca R, Slart RHJA, Erba PA. Artificial intelligence and hybrid imaging: the best match for personalized medicine in oncology. Eur J Hybrid Imaging. 2020;4:24. doi: 10.1186/s41824-020-00094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaissis GA, Makowski MR, Rückert D, Braren RF. Secure, privacy-preserving and federated machine learning in medical imaging. Nat Mach Intell. 2020.
- 8.Konečný J, McMahan B, Ramage D. Federated optimization:distributed optimization beyond the datacenter. 2015;
- 9.Zerka F, Barakat S, Walsh S, Bogowicz M, Leijenaar RTH, Jochems A, et al. Systematic review of privacy-preserving distributed machine learning from federated databases in health care. JCO Clin Cancer Informatics. 2020;4:184–200. doi: 10.1200/CCI.19.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verbraeken J, Wolting M, Katzy J, Kloppenburg J, Verbelen T, Rellermeyer JS. A survey on distributed machine learning. ACM Comput Surv. 2020;53:1–33. doi: 10.1145/3377454. [DOI] [Google Scholar]
- 11.Dean J, Corrado GS, Monga R, Chen K, Devin M, Le QV, et al. Large scale distributed deep networks. Adv Neural Inf Proces Syst. 2012.
- 12.Chang K, Balachandar N, Lam C, Yi D, Brown J, Beers A, et al. Distributed deep learning networks among institutions for medical imaging. J Am Med Inform Assoc. 2018;25:945–954. doi: 10.1093/jamia/ocy017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vepakomma P, Gupta O, Swedish T, Raskar R. Split learning for health: distributed deep learning without sharing raw patient data. 2018;
- 14.Polikar R. Ensemble based systems in decision making. IEEE Circuits Syst Mag. 2006;6:21–45. doi: 10.1109/MCAS.2006.1688199. [DOI] [Google Scholar]
- 15.Wolpert DH. Stacked generalization. Neural Netw. 1992;5:241–259. doi: 10.1016/S0893-6080(05)80023-1. [DOI] [Google Scholar]
- 16.Gupta O, Raskar R. Distributed learning of deep neural network over multiple agents. J Netw Comput Appl. 2018.
- 17.Rieke N, Hancox J, Li W, Milletarì F, Roth HR, Albarqouni S, et al. The future of digital health with federated learning. npj Digit Med. 2020. [DOI] [PMC free article] [PubMed]
- 18.Brendan McMahan H, Moore E, Ramage D, Hampson S, Agüera y Arcas B. Communication-efficient learning of deep networks from decentralized data. Proc 20th Int Conf Artif Intell Stat AISTATS 2017. 2017.
- 19.Huang L, Shea AL, Qian H, Masurkar A, Deng H, Liu D. Patient clustering improves efficiency of federated machine learning to predict mortality and hospital stay time using distributed electronic medical records. J Biomed Inform. 2019;99:103291. doi: 10.1016/j.jbi.2019.103291. [DOI] [PubMed] [Google Scholar]
- 20.Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. Brainlesion. 2019;11383:92–104. doi: 10.1007/978-3-030-11723-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Gu Y, Dvornek N, Staib LH, Ventola P, Duncan JS. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results. Med Image Anal. 2020;65:101765. doi: 10.1016/j.media.2020.101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jochems A, Deist TM, van Soest J, Eble M, Bulens P, Coucke P, et al. Distributed learning: developing a predictive model based on data from multiple hospitals without data leaving the hospital – a real life proof of concept. Radiother Oncol. 2016;121:459–467. doi: 10.1016/j.radonc.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Deist TM, Jochems A, van Soest J, Nalbantov G, Oberije C, Walsh S, et al. Infrastructure and distributed learning methodology for privacy-preserving multi-centric rapid learning health care: euroCAT. Clin Transl Radiat Oncol. 2017;4:24–31. doi: 10.1016/j.ctro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jochems A, Deist TM, El Naqa I, Kessler M, Mayo C, Reeves J, et al. Developing and validating a survival prediction model for NSCLC patients through distributed learning across 3 countries. Int J Radiat Oncol. 2017;99:344–352. doi: 10.1016/j.ijrobp.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deist TM, Dankers FJWM, Ojha P, Scott Marshall M, Janssen T, Faivre-Finn C, et al. Distributed learning on 20 000+ lung cancer patients – the personal health train. Radiother Oncol. 2020;144:189–200. doi: 10.1016/j.radonc.2019.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Dankar FK, Madathil N, Dankar SK, Boughorbel S. Privacy-preserving analysis of distributed biomedical data: designing efficient and secure multiparty computations using distributed statistical learning theory. JMIR Med Inf. 2019;7:e12702. doi: 10.2196/12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Roberts K, Jiang X, Long Q. Distributed learning from multiple EHR databases: contextual embedding models for medical events. J Biomed Inform. 2019. [DOI] [PMC free article] [PubMed]
- 28.Lee J, Sun J, Wang F, Wang S, Jun C-H, Jiang X. Privacy-preserving patient similarity learning in a federated environment: development and analysis. JMIR Med Inf. 2018;6:e20. doi: 10.2196/medinform.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brisimi TS, Chen R, Mela T, Olshevsky A, Paschalidis IC, Shi W. Federated learning of predictive models from federated electronic health records. Int J Med Inform. 2018;112:59–67. doi: 10.1016/j.ijmedinf.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan R, Boland MR, Liu Z, Liu Y, Chang HH, Xu H, et al. Learning from electronic health records across multiple sites: a communication-efficient and privacy-preserving distributed algorithm. J Am Med Inform Assoc. 2020;27:376–385. doi: 10.1093/jamia/ocz199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva S, Gutman BA, Romero E, Thompson PM, Altmann A, Lorenzi M. Federated learning in distributed medical databases: meta-analysis of large-scale subcortical brain data. 2019 IEEE 16th Int Symp biomed imaging (ISBI 2019). IEEE; 2019. p. 270–4.
- 32.Remedios SW, Roy S, Bermudez C, Patel MB, Butman JA, Landman BA, et al. Distributed deep learning across multisite datasets for generalized CT hemorrhage segmentation. Med Phys. 2020;47:89–98. doi: 10.1002/mp.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remedios S, Roy S, Blaber J, Bermudez C, Nath V, Patel MB, et al. Distributed deep learning for robust multi-site segmentation of CT imaging after traumatic brain injury. In: Angelini ED, Landman BA, editors. Med Imaging 2019 Image process. SPIE; 2019. p. 9. [DOI] [PMC free article] [PubMed]
- 34.Balachandar N, Chang K, Kalpathy-Cramer J, Rubin DL. Accounting for data variability in multi-institutional distributed deep learning for medical imaging. J Am Med Inform Assoc. 2020;27:700–708. doi: 10.1093/jamia/ocaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Zheng H, Dou Z, Chen F, Deng J, Chen X, et al. A novel privacy-preserving federated genome-wide association study framework and its application in identifying potential risk variants in ankylosing spondylitis. Brief Bioinform. 2020; [DOI] [PubMed]
- 36.Xu Y, Ma L, Yang F, Chen Y, Ma K, Yang J, et al. A collaborative online AI engine for CT-based COVID-19 diagnosis. medRxiv Prepr Serv Heal Sci. 2020.
- 37.Choudhury O, Park Y, Salonidis T, Gkoulalas-Divanis A, Sylla I, Das AK. Predicting adverse drug reactions on distributed health data using federated learning. AMIA Annu Symp Proc AMIA Symp. 2019;2019:313–322. [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Hong C, Palmer N, Di Q, Schwartz J, Kohane I, et al. A fast divide-and-conquer sparse Cox regression. Biostatistics. 2019. [DOI] [PMC free article] [PubMed]
- 39.Xie J, Liu S, Dai H. Manifold regularization based distributed semi-supervised learning algorithm using extreme learning machine over time-varying network. Neurocomputing. 2019.
- 40.Dluhoš P, Schwarz D, Cahn W, van Haren N, Kahn R, Španiel F, et al. Multi-center machine learning in imaging psychiatry: a meta-model approach. Neuroimage. 2017. [DOI] [PubMed]
- 41.Scardapane S, Di Lorenzo P. A framework for parallel and distributed training of neural networks. Neural Netw. 2017. [DOI] [PubMed]
- 42.Tuladhar A, Gill S, Ismail Z, Forkert ND. Building machine learning models without sharing patient data: a simulation-based analysis of distributed learning by ensembling. J Biomed Inform. 2020. [DOI] [PubMed]
- 43.Bogowicz M, Jochems A, Deist TM, Tanadini-Lang S, Huang SH, Chan B, et al. Privacy-preserving distributed learning of radiomics to predict overall survival and HPV status in head and neck cancer. Sci Rep. 2020. [DOI] [PMC free article] [PubMed]
- 44.Sheller MJ, Edwards B, Reina GA, Martin J, Pati S, Kotrotsou A, et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep. 2020;10:12598. doi: 10.1038/s41598-020-69250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhaskar H, Hoyle DC, Singh S. Machine learning in bioinformatics: a brief survey and recommendations for practitioners. Comput Biol Med. 2006;36:1104–1125. doi: 10.1016/j.compbiomed.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Sollini M, Gelardi F, Matassa G, Delgado Bolton RC, Chiti A, Kirienko M. Interdisciplinarity: an essential requirement for translation of radiomics research into clinical practice – a systematic review focused on thoracic oncology. Rev Española med Nucl e Imagen Mol (English Ed) SEMNIM. 2020;39:146–156. doi: 10.1016/j.remn.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Bruynseels K, Santoni de Sio F, van den Hoven J. Digital twins in health care: ethical implications of an emerging engineering paradigm. Front Genet. 2018;9. [DOI] [PMC free article] [PubMed]
- 48.Björnsson B, Borrebaeck C, Elander N, Gasslander T, Gawel DR, Gustafsson M, et al. Digital twins to personalize medicine. Genome Med. 2019. [DOI] [PMC free article] [PubMed]
- 49.Oncoradiomics - Radiomics research Software - Clinical A.I platform [Internet]. [cited 2020 Nov 9]. Available from: https://www.oncoradiomics.com/.
- 50.Home [Internet]. [cited 2020 Nov 9]. Available from: https://distributedlearning.ai/.
- 51.NVIDIA Clara Platform to Usher in Next Generation of Medical Instruments | NVIDIA Blog [Internet]. [cited 2020 Nov 9]. Available from: https://blogs.nvidia.com/blog/2018/09/12/nvidia-clara-platform/.
- 52.Mandl KD, Glauser T, Krantz ID, Avillach P, Bartels A, Beggs AH, et al. The genomics research and innovation network: creating an interoperable, federated, genomics learning system. Genet Med. 2020;22:371–380. doi: 10.1038/s41436-019-0646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirienko M, Biroli M, Gelardi F, Seregni E, Chiti A, Sollini M. Deep learning in nuclear medicine—focus on CNN-based approaches for PET/CT and PET/MR: where do we stand? Clin Transl Imaging. Springer. 2021:1–19.
- 54.Barredo Arrieta A, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, et al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion. 2020;58:82–115. doi: 10.1016/j.inffus.2019.12.012. [DOI] [Google Scholar]
- 55.Weeks J, Pardee R. Learning to share health care data: a brief timeline of influential common data models and distributed health data networks in U.S. Health Care Research. eGEMs (Generating Evid Methods to Improv patient outcomes). 2019;7:4. [DOI] [PMC free article] [PubMed]
- 56.Bilimoria KY, Stewart AK, Winchester DP, Ko CY. The national cancer data base: a powerful initiative to improve cancer care in the United States. Ann Surg Oncol. 2008;15:683–690. doi: 10.1245/s10434-007-9747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shah A, Stewart AK, Kolacevski A, Michels D, Miller R. Building a rapid learning health care system for oncology: why CancerLinQ collects identifiable health information to achieve its vision. J Clin Oncol. 2016;34:756–763. doi: 10.1200/JCO.2015.65.0598. [DOI] [PubMed] [Google Scholar]
- 58.D’Amour A, Heller K, Moldovan D, Adlam B, Alipanahi B, Beutel A, et al. Underspecification presents challenges for credibility in modern machine learning. 2020;
- 59.Claerhout B, Kalra D, Mueller C, Singh G, Ammour N, Meloni L, et al. Federated electronic health records research technology to support clinical trial protocol optimization: evidence from EHR4CR and the InSite platform. J Biomed Inform. 2019;90:103090. doi: 10.1016/j.jbi.2018.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The manuscript represents valid work, and neither this manuscript nor one with substantially similar content under the same authorship has been published or is being considered for publication elsewhere. Arturo Chiti had full access to all the data in the study and takes responsibility for the data integrity and the accuracy of the data analysis. Raw data are available on specific request to the corresponding author.