Abstract
Host cell DNA methylation analysis in urine provides promising triage markers for women diagnosed with a high-risk (HR) human papillomavirus (HPV) infection. In this study, we have investigated a panel of six host cell methylation markers (GHSR, SST, ZIC1, ASCL1, LHX8, ST6GALNAC5) in cervicovaginal secretions collected within the first part of the urine void (FVU) from a referral population. Cytology, histology, and HPV DNA genotyping results on paired FVU and cervical samples were available. Urinary median methylation levels from HR-HPV (n = 93) positive women were found to increase for all markers with severity of underlying disease. Significantly elevated levels were observed for GHSR and LHX8 in relation to high-grade cervical intraepithelial neoplasia (CIN2 +; n = 33), with area under de curve values of 0.80 (95% Confidence Interval (CI) 0.59–0.92) and 0.76 (95% CI 0.58–0.89), respectively. These findings are the first to support the assertion that methylation analysis of host cell genes is feasible in FVU and holds promise as molecular, triage strategy to discern low- from high-grade cervical disease in HR-HPV positive women. Molecular testing on FVU may serve to increase cervical cancer screening attendance in hard-to-reach populations whilst reducing loss to follow-up and await further optimization and validation studies.
Subject terms: Biomarkers, Epigenetics, Cancer
Introduction
Cervical cancer screening based on cytology and/or (primary) detection of high-risk human papillomavirus (HR-HPV) DNA—the main etiological agent of cervical cancer—has successfully reduced cervical cancer incidence and mortality, but is hampered by suboptimal screening coverage. Though population based approaches such as sending personal invitation letters and reminders for a scheduled appointment have proven to be effective in increasing participation rates, barriers for attending cervical cancer screening have persevered1. These barriers can be diverse, including practical, emotional, and cognitive barriers2. Self-sampling could overcome part of these issues3 and HPV testing of self-collected cervicovaginal samples (SS) has shown similar accuracy compared to clinician-collected cervical samples (CS) using a validated PCR-based method3,4. Notwithstanding that SS are well-accepted by women, highest preference is given to urine self-sampling5–10. In recent trials, good HR-HPV DNA agreement and clinical sensitivity has been reported in first-void urine (FVU) compared to CS6–8,11–15. FVU allows for self-collection of cervicovaginal secretions that accumulate between the small labia and around the urethra opening, and are captured within the first part of the urine void. It is often mistaken for the first urine of the day (morning urine), which—in contrast to FVU—does not improve urinary HPV detection6,7,16.
Based on the same concept behind identifying more human and HPV DNA in FVU than in subsequent fractions17–19, FVU may also harbour other biomarkers. The use of urine to detect biomarkers for cervical screening has been receiving close appraisal in the last decade (previously reviewed20). Similar to the limitations of SS, FVU will likely not fulfil the high-quality cellularity standards required for morphological biomarkers such as cytology. Molecular biomarkers on the other hand have the potential to overcome this issue and are likely to yield high-throughput, objective, and reproducible results. Host cell methylation markers have shown promise as they are able to distinguish low-grade cervical intraepithelial neoplasia (CIN) with productive HR-HPV infections from high-grade CIN with transforming HR-HPV infections, especially those with a high short-term risk of progression to cancer, and detect all carcinomas21,22. Their clinical value to discern HR-HPV infected women with clinically relevant disease has been validated in CS23–26 and SS27–30. More recently, methylation of host cell genes in urine has shown promise as biomarker as well31–35. Combining primary HPV detection, and in case HR-HPV positive methylation marker triage on the same FVU sample could pose a non-invasive strategy to identify HR-HPV women with clinically relevant cervical disease in need of referral. Such strategy may be especially interesting in hard-to-reach populations where an approach based on clinician-collected samples is not effective, as well as in light of pandemics such as the current SARS-CoV-2 pandemic where access to healthcare is hampered and home-based self-sampling could aid prevention initiatives.
Therefore, the primary objective of this study was to investigate the potential of methylation analysis of six host cell genes (GHSR, SST, ZIC1, ASCL1, LHX8, and ST6GALNAC526,29) in FVU as biomarker for cervical cancer prevention. These markers were previously identified by unbiased genome-wide methylation analysis in HR-HPV transformed cell lines/tissue biopsies (GHSR, SST, ZIC1)26 and SS (ASCL1, LHX8, and ST6GALNAC5)29 and validated as triage markers for HR-HPV positive women. All markers revealed a very good diagnostic performance for the detection of CIN3 + in CS and/or SS26,29. This study more specifically aimed to study feasibility of testing for these six methylation markers in FVU to discern underlying high-grade cervical disease from normal tissue and low-grade lesions in women diagnosed with HR-HPV.
Materials and methods
Study population
This cohort included 119 women (aged 25–64) referred to the Antwerp University Hospital (UZA, Belgium) colposcopy clinic due to abnormal cytology and/or infection with one or multiple HPV genotypes (January-November 2016) as described previously11. For methylation marker analysis, only women with a positive HR-HPV test result were included. In this subset of HR-HPV positive women, performance of each individual methylation marker was evaluated using either (i) cytology or (ii) histology outcomes as reference. The study protocol was registered on clinicialtrials.gov (NCT02714127) and approved by the central ethics committee of the University of Antwerp and UZA (B300201525585; B300201734143). All included participants signed informed consent prior to participating in study-related procedures which were in accordance with the Declaration of Helsinki (1964) and its later amendments as well as the ethical standards of the institutional review board.
Sample collection and storage
Women collected a FVU sample with a first-void urination device (20 ml collector vial, Colli-Pee, Novosanis, Belgium) at the hospital prior to their visit with the gynaecologist for a CS and colposcopy examination. Women were requested beforehand to not extensively wash their genitals before the visit at the clinic and to not urinate at least one hour prior to this visit. Upon addition of Urine Conservation Medium (UCM, UAntwerpen18) in a 1:2 UCM:FVU ratio, whole UCM-buffered FVU samples were preserved on dry ice in individual aliquots within a median time span of 12 min (interquartile range (IQR): 11–16 min) after sample collection. FVU samples were subjected to batches in random order for DNA extraction, HPV DNA genotyping, and methylation of host cell genes.
Data from CS (HPV DNA genotyping, liquid based cytology (LBC)) and colposcopy (with an optional biopsy) were retrieved from the women’s medical records. CS (Cervex-Brush, Rovers Medical Devices, The Netherlands) were transferred in 20 ml collection medium (PreservCyt Solution, Hologic Europe, Belgium), analysed at UZA laboratory with the ThinPrep Pap Test (Hologic Europe), and graded according to the Bethesda classification. When indicated (according to the guidelines of the European Federation of Colposcopy), a biopsy for histological confirmation was taken during colposcopy by the clinician and graded at the UZA pathology lab using the CIN classification system. Women graded with different colposcopy and histological outcomes were classified according to the most severe stage.
DNA extraction of first-void urine samples
DNA extraction was performed per in-house protocol developed by Vorsters A et al.18 as previously described11,18. In brief, 4 ml of UCM-buffered whole FVU was transferred to an Amicon Ultra-4 50K filter (Merck Millipore, Belgium), centrifuged (20 min; 3820 × g; 20 °C), and incubated (10 min, room temperature) after addition of 2 ml NucliSENS Lysis Buffer (BioMérieux, Benelux) to the retentate on the filter. After NucliSENS easyMag (BioMérieux, Benelux) DNA extraction, 35 of the 55 µl of DNA eluate was diluted with elution buffer (BioMérieux, Benelux) to a final volume of 75 µl used for HPV DNA genotyping. The remaining 20 µl DNA eluate was used for measuring methylation of host cell genes.
HPV DNA genotyping using quantitative PCR
HPV DNA genotyping data for FVU and CS were generated by Riatol quantitative PCR HPV genotyping assay (qPCR) as previously described11. Briefly, this assay quantifies 13 HR-HPV genotypes: IARC Group 1 (HPV16/18/31/33/35/39/45/51/52/56/58/59) and 2A (HPV68); three possibly HR-HPV genotypes: IARC Group 2B (HPV53/66/67); and two low-risk (LR) HPV genotypes: IARC Group 3 (HPV6/11)36,37. β-globin was amplified to assess the DNA quality and to determine the concentration of human (h)DNA present in the sample. The 75 µl FVU DNA extracts were directly pipetted into the 96-well plate, followed by qPCR. For HPV DNA genotyping in CS, 400–800 µl of the remaining LBC specimens were subjected to automatic nucleic acid preparation, followed by qPCR.
Methylation analysis of GHSR, SST, ZIC1, ASCL1, LHX8, and ST6GALNAC5 using quantitative methylation-specific PCR
Methylation analysis of a marker panel consisting of six host cell genes (GHSR, SST, ZIC1, ASCL1, LHX8, and ST6GALNAC5) was performed as described before26,29,38,39. In brief, 250 ng of isolated FVU DNA (or 20 µl when concentration was < 12.5 ng/µl) was subjected to bisulphite treatment using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA). For multiplex qMSP, 2.5 µl of bisulphite converted DNA (≤ 50 ng) was added to 10 µl amplification mix26,29,38,39. The housekeeping gene ACTB was used to assess DNA quality and successful bisulphite conversion. Cycle threshold (CT)-values of methylation markers were normalised to the reference gene ACTB using the comparative method (2−ΔCT × 100)40, obtaining methylation marker ratios. As a threshold for sample validity, a CT-value below or equal to 32 for ACTB was required for each sample.
Statistical analysis
The Cohen’s Kappa (κ) was calculated to assess the HPV genotype agreement between paired samples and was judged as follows: κ ≤ 0.20, poor; 0.21 ≤ κ ≤ 0.40, fair; 0.41 ≤ κ ≤ 0.60, moderate; 0.61 ≤ κ ≤ 0.80, good; and κ ≥ 0.81, very good agreement41. Based on a good κ-agreement for HR-HPV DNA in paired FVU and CS, women with a positive HR-HPV DNA test result in FVU and/or CS were included for methylation marker analysis. Square root transformed methylation marker ratios (√(CT ratio)) were visualized according to cytology and histology classification via scatter plots with overlying box plots, indicating median methylation levels and according interquartile ranges (25th and 75th percentile). For each methylated gene, differences in marker ratios according to disease outcome (HSIL +, CIN2 +, or CIN3) were tested using the Mann Whitney U-test. The performance of each host cell methylation gene was visualized by a receiver operating characteristic (ROC)-curve, and evaluated by the area under the curve (AUC). The 95% CI for AUC were derived by using 5,000 bootstrap samples. Statistical analyses were performed at a significance level of 5% using the statistical software JMP Pro 13.
Results
Study population
The median participant age in our referral population (n = 119, Fig. 1) was 36 years (IQR: 29–44 years old). Samples from 119 (FVU) and 114 (CS) women were available for HPV DNA genotyping. All samples were valid, indicated by measurable amounts of β-globin, using qPCR36,42. From the 114 women with HPV genotyping results available for both FVU and CS, a good Cohen’s Kappa (κ) agreement was observed for HR-HPV (κ: 0.647; 95% confidence interval (CI): 0.494–0.800), whereas this agreement was found to be excellent for HPV16/18 (κ: 0.905; 95% CI: 0.814–0.996). Samples from 93 women with a positive test result in FVU and/or CS for HR-HPV (71 FVU + /CS +; 15 FVU + /CS−; 7 FVU−/CS +) were selected for methylation marker analysis. Performance of each methylation marker was evaluated using either (i) cytology or (ii) histology outcomes as reference, available for 89 and 33 out of the 93 HR-HPV positive samples, respectively. Demographics and HPV genotype/viral load data are detailed in Supplementary Table S1 and Van Keer et al.11.
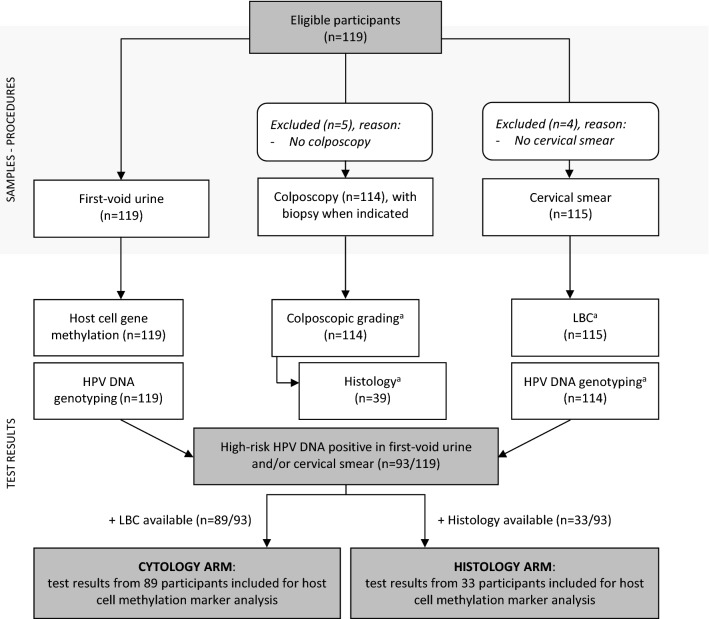
Figure 1.
Flow diagram for the inclusion of study participants, samples, and medical records. Results from 119 participants were used for HPV DNA genotyping in first-void urine and cervical samples. Test results from high-risk HPV DNA positive women (in first-void urine and/or cervical smear, n = 93) were used to examine methylation marker performance; using either cytology results (CYTOLOGY ARM), or histology outcomes (HISTOLOGY ARM) as reference. aWhen unavailable at D0 (day of study visit, i.e. first-void urine collection), the HPV DNA genotyping, liquid based cytology (LBC), colposcopy, and/or histology results from D0 ± 3 months were included for data analysis instead. Thus, three additional HPV DNA genotyping results were included, as well as four LBC and colposcopy, and six histology results.
Methylation marker levels in first-void urine according to disease outcome
Cytology endpoints
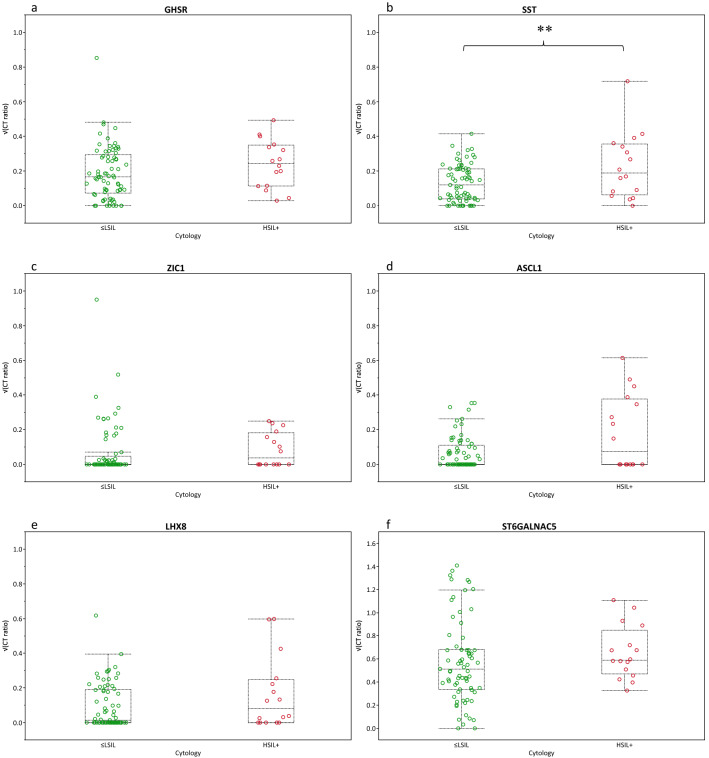
Firstly, we analysed the methylation levels in relation to cytology using the complete series of HR-HPV positive samples. A set of 89 out of 93 HR-HPV positive samples with cytology endpoint available were selected (Fig. 1), resulting in 73 ≤ LSIL (NILM, ASC-US, LSIL) and 16 HSIL + (HSIL, ASC-H) cases. Increased median methylation levels were observed for all markers in HSIL + as to ≤ LSIL, with a significant increase observed for SST (Fig. 2). All tested FVU samples were valid for the qMSP (CT-value ACTB ≤ 32).
Figure 2.
Difference in methylation ratios (square root transformed; y-axis) for normal/low-grade and high-grade cervical disease according to cytology outcome (HSIL +; x-axis); ≤ LSIL (green circles, n = 73) and HSIL + (red circles, n = 16). Box-plots indicate median methylation levels of (a) GHSR, (b) SST, (c) ZIC1, (d) ASCL1, (e) LHX8, and (f) ST6GALNAC5 and according interquartile ranges (25th and 75th percentile). P-values (Mann Whitney U-test) indicated by a double asterisk mean that the null hypothesis (H0) is rejected (p ≤ 0.05) and that the median square root transformed methylation marker ratios between normal/low-grade and high-grade cervical disease are not equal. No trends were observed (0.05 < p ≤ 0.10).
Histology endpoints
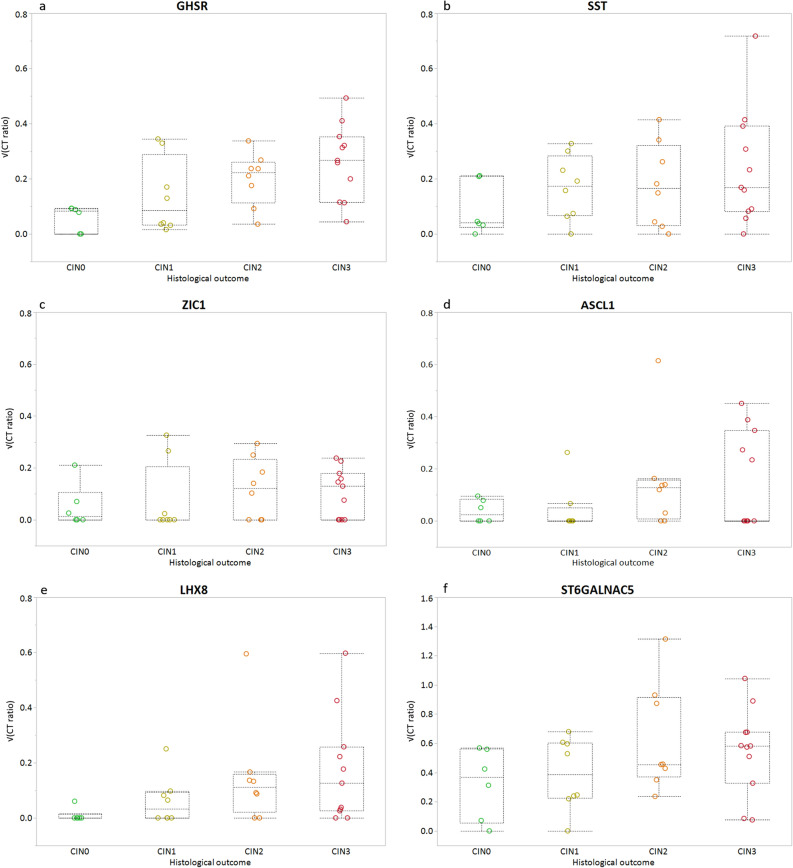
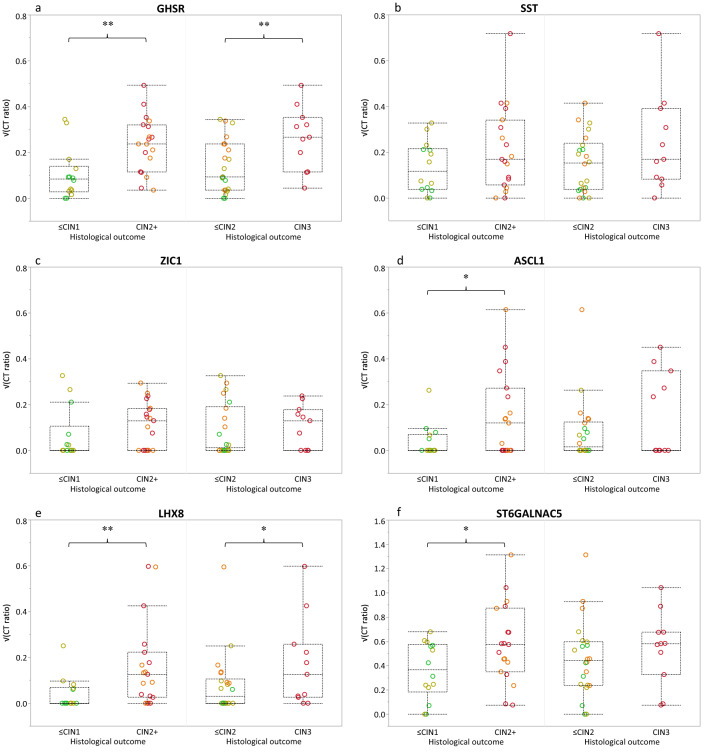
Secondly, we assessed the presence of the methylation marker levels in FVU of HR-HPV positive women with known histological outcome. This resulted in a set of 33 samples; 6 normal/CIN0, 8 CIN1, 8 CIN2, and 11 CIN3 (n = 33; Fig. 1). A gradual increase in methylation level was observed for GHSR, SST, ASCL1, and LHX8, but not for ZIC1 and ST6GALNAC5 (Fig. 3) with increasing severity of underlying disease. Comparison of ≤CIN1 to CIN2 + revealed increased median marker levels for all six markers in FVU, significantly elevated (p ≤ 0.05) for GHSR and LHX8. A similar trend (0.05 < p ≤ 0.10) was observed for ASCL1 and ST6GALNAC5 (0.05 < p ≤ 0.10) (Fig. 4 and Table 1). Comparison of ≤CIN2 to CIN3 revealed significantly elevated median methylation levels in FVU for GHSR (p ≤ 0.05), and a similar trend for LHX8 (0.05 < p ≤ 0.10).
Figure 3.
Host cell DNA methylation levels (square root transformed; y-axis) for (a) GHSR, (b) SST, (c) ZIC1, (d) ASCL1, (e) LHX8, and (f) ST6GALNAC5 according to histological outcome (x-axis); normal/CIN0 (green circles, n = 6), CIN1 (yellow circles, n = 8), CIN2 (orange circles, n = 8), and CIN3 (red circles, n = 11). Box-plots indicate median methylation levels and according interquartile ranges (25th and 75th percentile).
Figure 4.
Difference in methylation ratios (square root transformed; y-axis) for normal/low-grade and high-grade cervical disease according to histological outcome (CIN2 + and CIN3; x-axis); normal/CIN0 (green circles, n = 6), CIN1 (yellow circles, n = 8), CIN2 (orange circles, n = 8), and CIN3 (red circles, n = 11). Box-plots indicate median methylation levels of (a) GHSR, (b) SST, (c) ZIC1, (d) ASCL1, (e) LHX8, and (f) ST6GALNAC5 and according interquartile ranges (25th and 75th percentile). P-values (Mann Whitney U-test) indicated by a double asterisk mean that the null hypothesis (H0) is rejected (p ≤ 0.05) and that the median square root transformed methylation marker ratios between normal/low-grade and high-grade cervical disease are not equal. P-values indicated by a single asterisk indicate a trend (0.05 < p ≤ 0.10).
Table 1.
Performance of individual host cell methylation markers to discern high-grade cervical disease.
| Marker | P-value MWUa | AUC (95% CI)b | ||||
|---|---|---|---|---|---|---|
| Cytology | Histology | Cytology | Histology | |||
| HSIL + | CIN2 + | CIN3 | HSIL + | CIN2 + | CIN3 | |
| GHSR | 0.128 | 0.004** | 0.014** | 0.622 (0.488–0.757) | 0.801 (0.591–0.919) | 0.769 (0.561–0.897) |
| SST | 0.049** | 0.251 | 0.221 | 0.658 (0.498–0.798) | 0.620 (0.464–0.789) | 0.634 (0.473–0.809) |
| ZIC1 | 0.345 | 0.228 | 0.588 | 0.568 (0.435–0.707) | 0.620 (0.489–0.788) | 0.558 (0.400–0.742) |
| ASCL1 | 0.125 | 0.068* | 0.524 | 0.614 (0.451–0.778) | 0.677 (0.497–0.819) | 0.566 (0.473–0.849) |
| LHX8 | 0.198 | 0.009** | 0.087* | 0.598 (0.480–0.741) | 0.763 (0.575–0.890) | 0.682 (0.465–0.842) |
| ST6GALNAC5 | 0.177 | 0.061* | 0.244 | 0.609 (0.465–0.725) | 0.695 (0.501–0.846) | 0.628 (0.453–0.804) |
Performance of each marker was assessed using cytology (HSIL + , n = 89) and histology (CIN2 + and CIN3, n = 33) as reference.
aP-values (Mann Whitney U-test (MWU)) indicated by a double asterisk indicate that the null hypothesis (H0) is rejected (p ≤ 0.05) and that the median square root transformed methylation marker ratios between normal/low-grade and high-grade cervical disease are not equal. P-values values indicated by a single asterisk indicate a trend (0.05 < p ≤ 0.10).
bAn estimation of the performance of each marker was evaluated by the area under the curve (AUC) and accompanying 95% confidence interval (CI).
Triage marker performance of GHSR, SST, ZIC1, ASCL1, LHX8, and ST6GALNAC5 in first-void urine to discern high-grade disease
We have furthermore investigated the ability of these methylation markers to discriminate HR-HPV positive women with (i) cytology and (ii) biopsy confirmed high-grade disease from normal/low-grade disease. To do so, an estimation of the clinical performance of each methylation marker in FVU was evaluated by the AUC, and visualized by ROC-curves (Supplementary Fig. S1). When categorizing according to CIN2 +, AUC’s closest to 1 were observed for GHSR (0.80; 95% CI 0.59–0.92) and LHX8 (0.76; 95% CI 0.58–0.89), followed by ST6GALNAC5, ASCL1, and SST/ZIC1, in the respective order (Supplementary Fig. S1 and Table S1). Discriminating CIN3 from ≤ CIN2 lesions yields slightly lower AUC’s, with values closest to 1 again for GHSR (0.77; 95% CI 0.56–0.90) and LHX8 (0.68; 95% CI 0.47–0.84). Stratifying according to cytology (HSIL +) results in overall lower AUC’s, ranging between 0.66 (SST; 95% CI 0.50–0.80) and 0.57 (ZIC1; 95% CI 0.44–0.71), using HSIL + as endpoint (Supplementary Fig. S1 and Table S1).
Discussion
To the best of our knowledge, this study is the first to report on the potential of methylation analysis of this panel of six host cell genes (GHSR, SST, ZIC1, ASCL1, LHX8, and ST6GALNAC526,29), previously identified in HPV transformed cell lines, tissue biopsies, and cervicovaginal self-samples, in FVU. All six methylation markers showed an increase in median methylation levels in FVU for underlying disease (HSIL + and CIN2 +). Four out of six markers showed a gradual increase in median methylation levels with increasing lesion severity (normal/CIN0 through CIN3). The rise in methylation levels with increasing lesion severity was predominantly pronounced for GHSR, LHX8 and SST, where significant increases were observed using CIN2 + /CIN3, CIN2 +, and HSIL + as endpoint, respectively.
The less pronounced difference between median methylation levels between normal/low-grade and high-grade cervical disease in FVU compared to CS/SS26,29 can be explained by several factors. Firstly, the investigated host cell genes, GHSR, SST, ZIC1, ASCL1, LHX8, and ST6GALNAC5 were identified and validated in clinician and self-collected cervicovaginal samples. As the clinical value of methylation markers is not necessarily analogous between sample types43, a similar comprehensive approach including genome-wide DNA methylation profiling of FVU samples from HR-HPV positive women with normal to high-grade cervical disease and (invasive) cancer might allow us to identify methylated genes that predict high-grade cervical disease more accurately in FVU. Nevertheless, as we are using the first part of the urine void as liquid biopsy to capture cervicovaginal secretions, similarities in biomarker profile are expected. Secondly, CIN2 and CIN3 are a heterogeneous group of disease of which only a subset has a high risk of progression to cancer. Previous research has indicated that high methylation levels are associated with an advanced stage of disease and presumable high cancer progression risk. On the other hand, CIN2 and CIN3 with low methylation levels are suggested to have a low cancer progression risk21,26. We have no data on the duration and cancer risk of the CIN2 and CIN3 included in present study.
Others have investigated difference in host cell methylation levels in urine between cancers and controls, for which larger differentiations between groups are anticipated. One study reported increasing number of hyper methylation-positive urine samples (for ≥ 1/4 genes: DAPK1, RARB, TWIST, CDH13) according to lesion severity (4% CIN0/1, 28% CIN2/3/carcinoma in situ, and 62% invasive cervix carcinoma)32. Significant different methylation levels between cancers and controls in (first-void) urine have also been described for ASCL1, FAM19A4, GHSR, LHX8, PHACTR3, PRDM14, SFRP4, SST, and ZIC131,34,35. An estimate of the diagnostic performance of the markers in our study showed similar AUC’s as those reported by van den Helder and colleagues (2020)35, between 0.62–0.80 and 0.56–0.77 to discriminate between CIN0/1 and CIN2 + and CIN0-2 and CIN3, respectively. Highest AUC’s were observed when comparing cancers versus controls. It is therefore of interest to investigate the discriminatory power of our methylation marker panel in FVU from cancer patients in future studies.
Furthermore, the controls used in this study to assess biomarker performance were all HR-HPV positive (w/o low-grade underlying cervical disease), which is different from other studies reporting on triage marker performance between (pre)cancers and controls with partial HR-HPV positivity31 or without any disease34,35. Smaller differences in methylation levels between cases and controls can thus be expected from our data set. However, as methylation marker(s) (panels) are proposed as a triage strategy to discern high grade disease in women with a positive HR-HPV test, we believe that the controls used in this study align well with the intended clinical use, empowering the results.
Limitations of our study need to be acknowledged, and firstly involve the relative small sample size, potentially contributing to the insignificant differences observed, and heterogeneous character of our study population. This study was designed to identify promising triage markers for HR-HPV positive women. Hereto, a referral population of women was targeted attending colposcopy, either because of first abnormal screen result, or for follow-up of a persistent infection and/or associated lesion. Histology outcomes from biopsies (only taken when indicated) and cervical conization were only available for one third of the study population. To overcome this, performance of the markers was also assessed based on cytology, notwithstanding that histology is the golden standard reference test. Including both endpoints was reinforced by good κ-agreements observed between cytology (HSIL +) and histology CIN2 + (κ: 0.60 (95% CI 0.35–0.86)) and CIN3 (κ: 0.68 (95% CI 0.42–0.94)) endpoints. The difference between methylation markers showing significance in cytology (i.e. SST) versus histology endpoints (i.e. GHSR and LHX8) is likely a reflection of the suboptimal performance of cytology to predict underlying disease and the relatively small sample size. For this reason and given the nature of the study being a feasibility study we did not investigate diagnostic performance of methylation marker combinations. Secondly, at the time of study initiation limited optimization experiments had been performed identifying the optimal FVU collection and pre-analytical processing method preceding methylation analysis by qMSP. Optimizations might potentially increase analytic sensitivity, and consequently distinctions between cases and controls and AUC’s. Yet, it is expected that this will not change the general conclusions from present study, showing that increases in methylation levels of host cell genes can be observed in FVU with increasing severity of underlying disease.
In summary, the data from this study propose that methylation analysis of host cell genes in FVU has the diagnostic potential to distinguish normal/low-grade from high-grade underlying cervical disease in HR-HPV infected women. Concomitantly, it is a promising liquid biopsy to offer a fully molecular screen and triage strategy based on primary HPV testing and methylation marker detection in the same sample. Its non-invasive nature, being well-accepted, and ability to be self-collected at home furthermore fortifies its use to potentially increase screening-attendance in hard-to-reach populations whilst reducing loss to follow-up. Further studies including a larger sample series with histological confirmation of underlying disease are ongoing to define the diagnostic accuracy of methylation marker testing in FVU and to extract the most discriminative methylation markers or marker combinations.
Supplementary Information
Acknowledgements
We would like to express our gratitude to all women who volunteered to participate in this study, and thank Dr. L. Ameryckx for her assistance in recruiting participants. We are grateful for the excellent laboratory and logistic assistance of Ms. S. Biesmans and furthermore thank Prof. N. Hens, PhD for statistical support. This work was supported by the Industrial Research Fund of the University of Antwerp, Belgium under grant number PS ID 32387. S. Van Keer is supported by a junior postdoctoral fellowship of the Research Foundation—Flanders (FWO), Belgium under grant number 1240220N. J. Pattyn is supported by the Royal Belgian Academy of Medicine.
Author contributions
S.V.K., A.V., P.V.D., W.A.A.T., X.V.O., and M.I. were involved in conceptualization of the study and funding acquisition. S.V.K., A.V., W.A.A.T., and J.P. were involved in sample and data collection. A.D.S. and A.P.v.S. were involved in laboratory analysis of the biological samples. S.V.K., A.V., and R.D.M.S. were involved in methodology, resources, and data interpretation. S.V.K. was involved in formal analysis and drafting the original and final version of the manuscript. S.A.H. was involved in statistical review of the formal analysis. All authors critically reviewed the manuscript and approved the final version.
Data availability
All genotyping data generated and analysed during this study are included in this published article (and its Supplementary Information files; Supplementary Tables S1, S2 and S3). The methylation marker dataset generated and analysed during the current study is available from the corresponding author on reasonable request.
Competing interests
P. Van Damme and A. Vorsters are co-founders of Novosanis (Belgium), a spin-off company of the University of Antwerp, and were minority shareholder until January 2019. R. D. M. Steenbergen has a minority stake in Self-screen B.V. (The Netherlands), a spin-off company of VU University Medical Center Amsterdam, which owns patents related to this work. The other authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87329-1.
References
- 1.Everett T, et al. Interventions targeted at women to encourage the uptake of cervical screening. Cochrane Database Syst. Rev. 2011 doi: 10.1002/14651858.CD002834.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waller J, Bartoszek M, Marlow L, Wardle J. Barriers to cervical cancer screening attendance in England: A population-based survey. J. Med. Screen. 2009;16:199–204. doi: 10.1258/jms.2009.009073. [DOI] [PubMed] [Google Scholar]
- 3.Arbyn M, Smith SB, Temin S, Sultana F, Castle P. Detecting cervical precancer and reaching underscreened women by using HPV testing on self samples: Updated meta-analyses. BMJ. 2018;363:k4823. doi: 10.1136/bmj.k4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polman NJ, et al. Performance of human papillomavirus testing on self-collected versus clinician-collected samples for the detection of cervical intraepithelial neoplasia of grade 2 or worse: A randomised, paired screen-positive, non-inferiority trial. Lancet Oncol. 2019 doi: 10.1016/S1470-2045(18)30763-0. [DOI] [PubMed] [Google Scholar]
- 5.Sellors JW, et al. Comparison of self-collected vaginal, vulvar and urine samples with physician-collected cervical samples for human papillomavirus testing to detect high-grade squamous intraepithelial lesions. CMAJ. 2000;163:513–518. [PMC free article] [PubMed] [Google Scholar]
- 6.Senkomago V, et al. Comparison of urine specimen collection times and testing fractions for the detection of high-risk human papillomavirus and high-grade cervical precancer. J. Clin. Virol. 2016;74:26–31. doi: 10.1016/j.jcv.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Leeman A, et al. HPV testing in first-void urine provides sensitivity for CIN2+ detection comparable to a physician-taken smear or brush-based self-sample: Cross-sectional data from a triage population. BJOG. 2017 doi: 10.1111/1471-0528.14682. [DOI] [PubMed] [Google Scholar]
- 8.Rohner E, et al. Racial and ethnic differences in acceptability of urine and cervico-vaginal sample self-collection for HPV-based cervical cancer screening. J. Womens Health (Larchmt) 2020;29:971–979. doi: 10.1089/jwh.2019.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sargent A, Fletcher S, Bray K, Kitchener HC, Crosbie EJ. Cross-sectional study of HPV testing in self-sampled urine and comparison with matched vaginal and cervical samples in women attending colposcopy for the management of abnormal cervical screening. BMJ Open. 2019;9:e025388. doi: 10.1136/bmjopen-2018-025388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tranberg M, Jensen JS, Bech BH, Andersen B. Urine collection in cervical cancer screening: Analytical comparison of two HPV DNA assays. BMC Infect. Dis. 2020;20:926. doi: 10.1186/s12879-020-05663-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Keer S, et al. Human papillomavirus genotype and viral load agreement between paired first-void urine and clinician-collected cervical samples. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:859–869. doi: 10.1007/s10096-017-3179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tshomo U, et al. Evaluation of the performance of Human Papillomavirus testing in paired urine and clinician-collected cervical samples among women aged over 30 years in Bhutan. Virol. J. 2017;14:74. doi: 10.1186/s12985-017-0744-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Combita AL, et al. Comparison between urine and cervical samples for HPV DNA detection and typing in young women in Colombia. Cancer Prev. Res. 2016;9:766–771. doi: 10.1158/1940-6207.CAPR-16-0038. [DOI] [PubMed] [Google Scholar]
- 14.Vorsters A, et al. Long-term follow-up of HPV infection using urine and cervical quantitative HPV DNA Testing. Int. J. Mol. Sci. 2016;17:15. doi: 10.3390/ijms17050750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchegger K, et al. Detection and genotyping of human papillomavirus virus (HPV): A comparative analysis of clinical performance in cervical and urine samples in Chilean women. Int. J. Clin. Exp. Pathol. 2018;11:5413–5421. [PMC free article] [PubMed] [Google Scholar]
- 16.Pattyn J, et al. Human papillomavirus detection in urine: Effect of a first-void urine collection device and timing of collection. J. Virol. Methods. 2019;264:23–30. doi: 10.1016/j.jviromet.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Pathak N, Dodds J, Zamora J, Khan K. Accuracy of urinary human papillomavirus testing for presence of cervical HPV: Systematic review and meta-analysis. BMJ. 2014;349:g5264. doi: 10.1136/bmj.g5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vorsters A, et al. Optimization of HPV DNA detection in urine by improving collection, storage, and extraction. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:2005–2014. doi: 10.1007/s10096-014-2147-2. [DOI] [PubMed] [Google Scholar]
- 19.Vorsters A, et al. Detection of human papillomavirus DNA in urine. A review of the literature. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:627–640. doi: 10.1007/s10096-011-1358-z. [DOI] [PubMed] [Google Scholar]
- 20.Van Keer S, et al. First-void urine: A potential biomarker source for triage of high-risk human papillomavirus infected women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017;216:1–11. doi: 10.1016/j.ejogrb.2017.06.036. [DOI] [PubMed] [Google Scholar]
- 21.Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat. Rev. Cancer. 2014;14:395–405. doi: 10.1038/nrc3728. [DOI] [PubMed] [Google Scholar]
- 22.Luttmer R, et al. Management of high-risk HPV-positive women for detection of cervical (pre)cancer. Expert Rev. Mol. Diagn. 2016;16:961–974. doi: 10.1080/14737159.2016.1217157. [DOI] [PubMed] [Google Scholar]
- 23.De Strooper LMA, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev. Res. 2014;7:1251–1257. doi: 10.1158/1940-6207.Capr-14-0237. [DOI] [PubMed] [Google Scholar]
- 24.Luttmer R, et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high-risk HPV-positive women of a gynecologic outpatient population (COMETH study) Int. J. Cancer. 2016;138:992–1002. doi: 10.1002/ijc.29824. [DOI] [PubMed] [Google Scholar]
- 25.De Strooper LMA, et al. Cervical cancer risk in HPV-positive women after a negative FAM19A4/mir124-2 methylation test: A post hoc analysis in the POBASCAM trial with 14 year follow-up. Int. J. Cancer. 2018;143:1541–1548. doi: 10.1002/ijc.31539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verlaat W, et al. Genome-wide DNA methylation profiling reveals methylation markers associated with 3q gain for detection of cervical precancer and cancer. Clin. Cancer Res. 2017;23:3813–3822. doi: 10.1158/1078-0432.CCR-16-2641. [DOI] [PubMed] [Google Scholar]
- 27.Luttmer R, et al. FAM19A4 methylation analysis in self-samples compared with cervical scrapes for detecting cervical (pre)cancer in HPV-positive women. Br. J. Cancer. 2016;115:579–587. doi: 10.1038/bjc.2016.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Strooper LMA, et al. Validation of the FAM19A4/mir124-2 DNA methylation test for both lavage- and brush-based self-samples to detect cervical (pre)cancer in HPV-positive women. Gynecol. Oncol. 2016;141:341–347. doi: 10.1016/j.ygyno.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verlaat W, et al. Identification and validation of a 3-gene methylation classifier for HPV-based cervical screening on self-samples. Clin. Cancer Res. 2018;24:3456–3464. doi: 10.1158/1078-0432.CCR-17-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhoef VM, et al. Triage by methylation-marker testing versus cytology in women who test HPV-positive on self-collected cervicovaginal specimens (PROHTECT-3): A randomised controlled non-inferiority trial. Lancet Oncol. 2014;15:315–322. doi: 10.1016/S1470-2045(14)70019-1. [DOI] [PubMed] [Google Scholar]
- 31.Hoffstetter R, et al. Evaluation of DNA methylation in promoter regions of SFRP4 and ZAR1 in urine and plasma of women with cervical lesions. Transl. Cancer Res. 2017;6:157–168. doi: 10.21037/tcr.2017.01.41. [DOI] [Google Scholar]
- 32.Feng QH, et al. Promoter hypermethylation of tumor suppressor genes in urine from patients with cervical neoplasia. Cancer Epidemiol. Biomark. 2007;16:1178–1184. doi: 10.1158/1055-9965.Epi-06-0694. [DOI] [PubMed] [Google Scholar]
- 33.Guerrero-Preston R, et al. Molecular triage of premalignant lesions in liquid-based cervical cytology and circulating cell free DNA from urine, using methylated viral and host genes. Cancer Prev. Res. 2016 doi: 10.1158/1940-6207.CAPR-16-0138. [DOI] [PubMed] [Google Scholar]
- 34.Snoek BC, et al. Cervical cancer detection by DNA methylation analysis in urine. Sci. Rep. 2019;9:3088. doi: 10.1038/s41598-019-39275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Helder R, et al. Methylation analysis in urine fractions for optimal CIN3 and cervical cancer detection. Papillomavir. Res. 2020;9:100193. doi: 10.1016/j.pvr.2020.100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Micalessi IM, Boulet GAV, Bogers JJ, Benoy IH, Depuydt CE. High-throughput detection, genotyping and quantification of the human papillomavirus using real-time PCR. Clin. Chem. Lab. Med. 2012;50:655–661. doi: 10.1515/Cclm.2011.835. [DOI] [PubMed] [Google Scholar]
- 37.IARC. Agents Classified by the IARC Monographs. (2016).
- 38.Snellenberg S, et al. Development of a multiplex methylation-specific PCR as candidate triage test for women with an HPV-positive cervical scrape. BMC Cancer. 2012;12:551. doi: 10.1186/1471-2407-12-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overmeer RM, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J. Pathol. 2008;215:388–397. doi: 10.1002/path.2367. [DOI] [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Altman DG. Practical Statistics for Medical Research. Boca Raton: Chapman and Hall/CRC; 1991. p. 624. [Google Scholar]
- 42.Depuydt CE, et al. Linear viral load increase of a single HPV-type in women with multiple HPV infections predicts progression to cervical cancer. Int. J. Cancer. 2016;139:2021–2032. doi: 10.1002/ijc.30238. [DOI] [PubMed] [Google Scholar]
- 43.Hesselink AT, et al. Methylation marker analysis of self-sampled cervico-vaginal lavage specimens to triage high-risk HPV-positive women for colposcopy. Int. J. Cancer. 2014;135:880–886. doi: 10.1002/ijc.28723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genotyping data generated and analysed during this study are included in this published article (and its Supplementary Information files; Supplementary Tables S1, S2 and S3). The methylation marker dataset generated and analysed during the current study is available from the corresponding author on reasonable request.