Abstract
Long noncoding RNAs (lncRNAs) are key mediators of biological regulation with diagnostic value as disease biomarkers. We explored serum lncRNA levels in early pregnancy as potential biomarkers of pregnancy-induced hypertension (PIH), including gestational hypertension (GH) and preeclampsia (PE). We performed a two-phase nested case-control study in pregnant women before 20 weeks’ gestation (before clinical diagnosis). The screening phase assessed lncRNA expression profiles with a human lncRNA microarray in 5 pairs of serum samples (5 PE patients and 5 matched controls). The second phase validated levels of 8 candidate lncRNAs selected via the random walk method by quantitative real-time polymerase chain reaction (qRT-PCR). Serum levels of the 8 lncRNAs were markedly increased in women with PIH compared with matched normotensive pregnant (NP) women (p < 0.001), consistent with the microarray results. In addition, 7 candidate lncRNAs were correlated with PIH severity. Logistic regression analysis revealed that serum levels of ENST00000527727 (odds ratio [OR], 1.113; 95% confidence interval [CI], 1.024–1.209; p = 0.0113) and ENST00000415029 (OR, 1.126; 95% CI, 1.000–1.267; p = 0.0496) were associated with adverse pregnancy outcomes, such as fetal growth restriction (FGR) and placenta accreta of PIH. Nine pathways associated with the candidate lncRNAs had confirmed associations with PIH.
Keywords: lncRNA, biomarker, pregnancy-induced hypertension, gestational hypertension, preeclampsia
Graphical abstract
This study indicates a macro predictive and functional pathophysiological correlation of circulating lncRNAs during the premature phase of the disease. Moreover, placing more emphasis on the detection of lncRNA expression in early pregnancy might reduce some adverse maternal and fetal outcomes, such as placenta accreta and FGR.
Introduction
Pregnancy-induced hypertension (PIH), a pregnancy-specific disorder that includes both gestational hypertension (GH) and preeclampsia (PE), is one of the primary causes of maternal and perinatal mortality and morbidity worldwide and complicates 4.1%–19.4% of all pregnancies.1
Women with PIH have new onset of hypertension after the first 20 weeks of pregnancy (GH) or new hypertension with features of multi-organ involvement (PE) and are not only at a greater risk of adverse pregnancy outcomes2,3 but also at increased risk (approximately 1.5–3 times) of developing cardiovascular disease later in life.4,5 Although the clinical symptoms of PIH present after 20 weeks of gestation, the molecular events resulting in its onset have been suggested to occur early in pregnancy.6 To date, there is neither proper treatment to cure PIH nor an ideal early clinical biomarker for the prediction of PIH.7 The prediction of PIH, including PE, is one of the meaningful studies in maternal-fetal medicine due to the lack of an effective therapy for this specific disorder, which affects the mother and the fetus, frequently requiring a premature delivery. Without appropriate and timely prevention, PIH may progress to a more serious stage known as eclampsia, which is combined with seizures and other severe complications. Therefore, it is of great significance for this disease to discover non-invasive and reliable biomarkers in the early stage of pregnancy for the prediction of PIH. Although many factors such as soluble fms-like tyrosine kinase-1 (sFlt-1), placental growth factor (PLGF) and ELABELA (ELA) have been used as biomarkers for predicting PIH,8, 9, 10, 11 these biomarkers for predicting or diagnosing PIH are subject to nonspecific prediction and poor accuracy during the early stage of gestation.12, 13, 14, 15
Long noncoding RNA (lncRNA), defined as transcripts longer than 200 nucleotides, has been increasingly thought to be correlated with various physiological and pathological processes of diseases.16,17 In addition, previous studies on placental lncRNAs in PIH, including GH and PE, have indicated the involvement of lncRNAs in the pathophysiology of PIH. He et al.18 first conducted a lncRNA microarray study in 2013 and compared placentas from PE and healthy pregnancies and found 738 differentially expressed lncRNAs. Jin et al.19 revealed that the overexpression of the lncRNA uc.187 could induce PE-like symptoms in a pregnant rat model by affecting the distribution of β-catenin in the nucleus and cytoplasm.
Circulating lncRNAs have emerged as novel potential biomarkers for a large number of diseases due to their stability and presence in the circulation.20 A recent study showed that the circulating lncRNA BC030099 can be used as a diagnostic marker for PE.21 However, these reports were conducted using serum/plasma/placenta samples from a more advanced stage of gestation (third trimester), and the expression and potential role of circulating lncRNAs in the early stage of pregnancy in women who later develop PIH have yet to be clarified.
In this study, to identify potential lncRNAs as early circulating biomarkers for PIH, including PE, we sought to determine the lncRNA expression profiles in the serum of patients with PE before the 20th week of gestation and evaluated the efficacy of lncRNAs for diagnosing PIH before the onset of clinical symptoms. Moreover, we ran univariate and multivariate logistic regression analyses for predicting the pregnancy outcomes of PIH to further assess the effects of lncRNAs, gaining new insight into the role of lncRNAs in the pathogenesis of PIH.
Results
Participant characteristics
The demographic characteristics of the screening phase participants are presented in Table S1. The clinical characteristics of the validation phase participants are shown in Table 1. There were no significant differences between the study groups in terms of maternal age, height, rate of smoking, rate of alcohol consumption, gestational age at sampling, fetal distress, or fetal heart rate during pregnancy.
Table1.
Participant characteristics
| Characteristics | NP (n = 97) | PIH (n = 97) | Z/t/χ2 | p value |
|---|---|---|---|---|
| Maternal age, years | 29 (27–31) | 30 (28–32) | −1.8949 | 0.0581 |
| Height, cm | 164.55 ± 4.94 | 162.7 ± 5.25 | 1.8723 | 0.0637 |
| Smoking, n (%) | 1 (1.03) | 2 (2.06) | 0.0000 | 1.0000 |
| Alcohol consumption, n (%) | 1 (1.03) | 4 (4.12) | 0.8212 | 0.3648 |
| Primipara, n (%) | 94 (96.91) | 81 (83.51) | 9.8605 | 0.0017 |
| At sampling gestational age <20 weeks | ||||
| Gestational age, weeks | 16 (16–17) | 16 (16–17) | 1.6325 | 0.1026 |
| Weight, kg | 58 (54–64) | 65 (57–77) | −4.2557 | <0.0001 |
| BMI, Kg/m2 | 21.68 (20.55–23.63) | 24.09 (22.1–28.73) | −4.0428 | 0.0001 |
| At delivery | ||||
| Gestational age at delivery, weeks | 39.71 (38.86–40.29) | 38.57 (37.43–39.57) | 5.5139 | <0.0001 |
| Weight at delivery, kg | 72.16 ± 8.98 | 81.67 ± 12.6 | −5.7267 | <0.0001 |
| BMI at delivery, Kg/m2 | 26.62 (24.8–28.81) | 30.11 (27.66-33.44) | −5.2219 | <0.0001 |
| ΔBMI | 12.5 (10–16) | 14 (11–19) | −2.1087 | 0.0350 |
| Systolic blood pressure, mm Hg | 117 (109–122) | 150 (142–159) | −11.7098 | <0.0001 |
| Diastolic blood pressure, mm Hg | 76 (69–80) | 96 (90–104) | −11.3651 | <0.0001 |
| CHOL, mmol/L | 6.29 (5.39–7.16) | 6.14 (5.42–7.09) | 0.4824 | 0.6295 |
| TG, mmol/L | 2.75 (2.32–3.41) | 3.61 (2.62–4.55) | −4.2394 | 0.0001 |
| Cesarean delivery, n (%) | 43 (44.79) | 76 (78.35) | 22.9845 | <0.0001 |
| Preterm birth, n (%) | 2 (2.06) | 20 (20.62) | 15.8167 | <0.0001 |
| Fetal growth restriction, n (%) | 1 (1.03) | 11 (11.34) | 8.8828 | 0.0029 |
| Fetal distress, n (%) | 4 (4.12) | 8 (8.25) | 1.4212 | 0.2332 |
| Fetal heart rate, bpm | 145 (140–149) | 145 (140.5–148) | 0.3507 | 0.7258 |
| Infant | ||||
| Birth length, cm | 50 (50–51) | 50 (48–50) | −3.9184 | 0.0001 |
| Birth wight, g | 3,345 (3085–3545) | 3145 (2712.5–3600) | −2.1226 | 0.0338 |
| Sex, male, n (%) | 43 (48.31) | 52 (54.17) | 0.6331 | 0.4262 |
BMI, body mass index; ΔBMI, BMI at delivery – BMI at sampling; SBP, systolic blood pressure; DBP, diastolic blood pressure; CHOL, total cholesterol; TG, triglyceride; FGR, fetal growth restriction; NP, normotensive pregnancy; PIH, pregnancy-induced hypertension.
Compared with patients in the NP group, patients in the PIH group had a lower gestational age at delivery, delivered infants with a significantly lower birth weight, had higher rates of primipara, caesarean delivery, preterm birth, fetal growth restriction (FGR), and fetal distress, higher blood pressures (BPs; both systolic [SBP] and diastolic [DBP]) and triglyceride (TG) levels, a higher weight and body mass index (BMI) at both sampling and delivery, and a higher ΔBMI between sampling and delivery.
Differential expression analysis in the PIH dataset
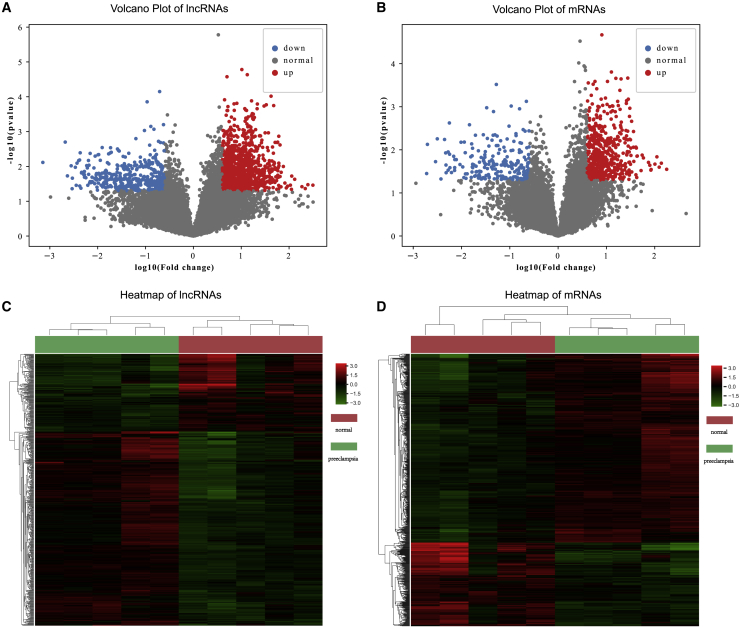
In the screening phase, to assess the differential expression of lncRNAs and mRNAs between the 5 samples from women before the 20th week of gestation who later developed PE and 5 paired NP control samples, a human lncRNA microarray was utilized. We used unpaired t test and the FC method to obtain differentially expressed lncRNAs and mRNAs. Finally, there were 417 upregulated and 167 downregulated lncRNAs and 510 upregulated and 226 downregulated mRNAs (Figures 1A and 1B), scatter points marked with red and blue are up- and downregulated lncRNAs/mRNAs). By applying the hierarchical clustering method, both the disease and normal samples can be clustered together using the differentially expressed lncRNAs/mRNAs we detected (Figures 1C and 1D). These significantly dysregulated lncRNAs/mRNAs may play a role in PIH formation or treatment. Besides, we observed that lncRNAs have better performance to separate disease and normal samples, which indicates the important role of lncRNAs in PIH. In the following study, we focused on these identified differentially expressed lncRNAs/mRNAs.
Figure 1.
Differential expression analysis in PIH
In total, 417 upregulated and 167 downregulated lncRNAs and 510 upregulated and 226 downregulated mRNAs were identified; the scatter points marked in red and blue correspond to up- and downregulated lncRNAs/mRNAs. (A) Volcano plot of lncRNAs. (B) Volcano plot of mRNAs. (C) Cluster heatmap of differentially expressed lncRNAs. (D) Cluster heatmap of differentially expressed mRNAs. (Statistics: unpaired t test, p < 0.05).
Detection of PIH-related lncRNA biomarkers based on network analysis
First, we performed Pearson correlation tests between each pair of differentially expressed lncRNAs and mRNAs and selected significantly correlated lncRNA-mRNA pairs (p < 0.001), which constituted a large network. This network comprised 1,311 nodes and 65,114 edges (Data S1). Next, we obtained experiment-supported biomarkers of PIH in the DISEASE database22 and intersected these markers with the differentially expressed lncRNAs/mRNAs we identified. In total, there were 31 mRNAs and 1 lncRNA in both sets (Table S2). We defined them as seed nodes in the network and applied a random walk algorithm to identify candidate lncRNA biomarkers. The 8 top-ranked lncRNAs, NR_002187, ENST00000415029, ENST00000398554, ENST00000586560, TCONS_00008014, ENST00000546789, ENST00000610270, and ENST00000527727, were used as potential biomarkers and were retained for subsequent analysis. The ranked candidate lncRNAs are presented in Table S3.
Validation of lncRNAs by quantitative real-time PCR
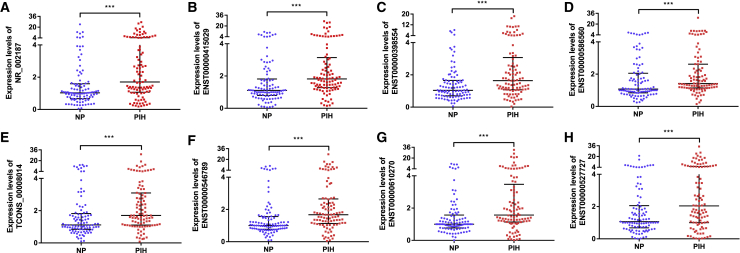
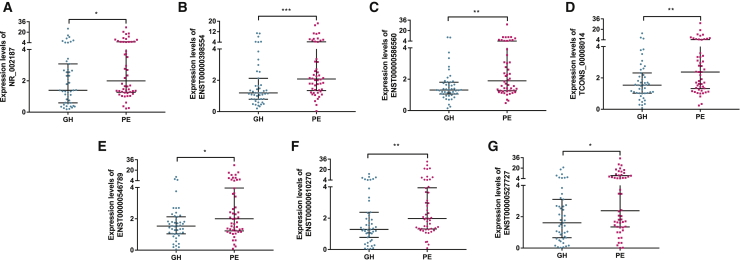
In the validation phase, quantitative real-time PCR was performed to confirm the expression of the 8 lncRNAs in serum samples obtained from women before the 20th week of gestation who later developed PIH (n = 97) and NP women (n = 97). As shown in Figures 2A–2H, the 8 lncRNAs were markedly increased in women with PIH compared with matched NP women (p < 0.001), and the results were highly consistent with the data derived from the microarray. This finding demonstrated that the 8 lncRNAs play a potential role in disease progression, so we further divided the PIH patients into the GH group (n = 47) and PE group (n = 50) and then analyzed the lncRNA expression characteristics between the two groups. Of the 8 lncRNAs, 7 lncRNAs (NR_002187, ENST00000398554, ENST00000586560, TCONS_00008014, ENST00000546789, ENST00000610270, and ENST00000527727) had elevated expression levels in the PE group compared to the GH group (p < 0.05); the exception was ENST00000415029 (p = 0.0770). These results emphasized that these 7 lncRNAs are underlying markers of disease development (Figures 3A–3G).
Figure 2.
Expression of serum lncRNAs in the validation phase
(A–H) Circulating expression levels of lncRNAs were determined by quantitative real-time PCR with serum samples from pregnant women at an early stage of gestation who later developed PIH (n = 97) compared to women with normotensive pregnancy (n = 97). (A) NR_002187 (p = 4.60E-05). (B) ENST00000415029 (p = 4.87E-06). (C) ENST00000398554 (p = 1.52E-04). (D) ENST00000586560 (p = 9.79E-04). (E) TCONS_00008014 (p = 9.79E-04). (F) ENST00000546789 (p = 8.74E-06). (G) ENST00000610270 (p = 5.90E-05). (H) ENST00000527727 (p = 6.28E-04). The expression of the 8 lncRNAs was significantly higher in pregnant women who later developed PE than in NP women (statistics: Wilcoxon rank sum test, ∗∗∗p < 0.001).
Figure 3.
Expression of serum lncRNAs in PIH patients
(A–E) 7 of the 8 lncRNAs were associated with the severity of PIH and showed higher expression levels in the PE group (n = 50) than in the GH group (n = 47), excluding ENST00000415029 (p = 0.0770). (A) NR_002187 (p = 0.0414). (B) ENST00000398554 (p = 0.0004). (C) ENST00000586560 (p = 0.0015). (D) TCONS_00008014 (p = 0.0060). (E) ENST00000546789 (p = 0.0262). (F) ENST00000610270 (p = 0.0093). (G) ENST00000527727 (p = 0.0162). (Statistics: Wilcoxon rank sum test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
lncRNAs can serve as biomarkers for PIH and risk factors for pregnancy outcomes of PIH
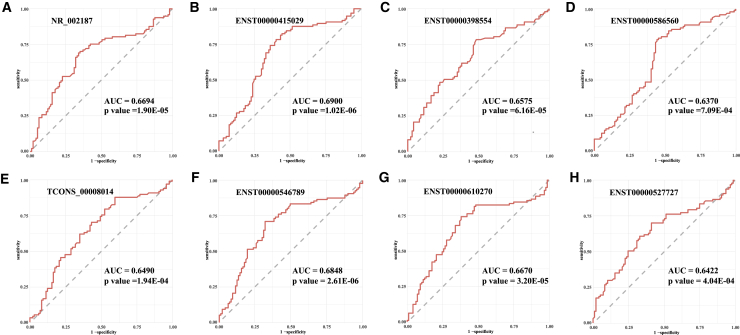
First, receiver operating characteristic (ROC) curves and the areas under the ROC curves (AUCs) were used to confirm the relationship between the lncRNAs and PIH (Figures 4A–4H). The AUCs of the 8 lncRNAs (NR_002187, ENST00000415029, ENST00000398554, ENST00000586560, TCONS_00008014, ENST00000546789, ENST00000610270, and ENST00000527727) were 0.6694 (95% confidence interval [CI] = 0.5918–0.7471), 0.6900 (95% CI = 0.6138–0.7662), 0.6575 (95% CI = 0.5804–0.7345), 0.6370 (95% CI = 0.5577–0.7164), 0.6490 (95% CI = 0.5706–0.7273), 0.6848 (95% CI = 0.6077–0.7619), 0.6670 (95% CI = 0.5883–0.7457), and 0.6422 (95% CI = 0.5634–0.7209).These results indicated that the 8 serum lncRNAs could be used as potential early biomarkers for predicting PIH.
Figure 4.
ROC analysis of early pregnancy serum lncRNAs for predicting PIH
(A–H) NR_002187, AUC 0.6694 (95% CI = 0.5918–0.7471) (A); ENST00000415029, AUC 0.6900 (95% CI = 0.6138–0.7662) (B); ENST00000398554, AUC 0.6575 (95% CI = 0.5804–0.7345) (C); ENST00000586560, AUC 0.6370 (95% CI = 0.5577–0.7164) (D); TCONS_00008014, AUC 0.6490 (95% CI = 0.5706–0.7273) (E); ENST00000546789, AUC 0.6848 (95% CI = 0.6077–0.7619) (F); ENST00000610270, AUC 0.6670 (95% CI = 0.5883–0.7457) (G); and ENST00000527727, AUC 0.6422 (95% CI = 0.5634–0.7209) (H).
Moreover, multivariate logistic regression analysis revealed that ENST00000527727 (odds ratio [OR], 1.113; 95% CI, 1.024–1.209; p = 0.0113) and DBP (OR, 1.095; 95% CI, 1.028–1.167; p = 0.0051) were independent risk factors for FGR of PIH. Moreover, ENST00000415029 (OR, 1.126; 95% CI, 1.000–1.267; p = 0.0496) and SBP (OR, 1.059; 95% CI, 1.013–1.108; p = 0.0119) were the independent risk factors for placenta accreta of PIH (Table 2).
Table 2.
Univariate and multivariate analysis of maternal outcomes for FGR and placenta accreta of patients with PIH
| Variable | Fetal growth restriction |
Placenta accreta |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate |
Multivariate |
Univariate |
Multivariate |
|||||||||
| OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p | |
| ENST00000415029 | 1.0386 | 0.9401–1.1475 | 0.4561 | – | – | – | 1.1236 | 1.014–1.245 | 0.0261 | 1.126 | 1.000–1.267 | 0.0496 |
| NR_002187 | 0.9023 | 0.7311–1.1135 | 0.3378 | – | – | – | 0.8142 | 0.5065–1.3087 | 0.3959 | – | – | – |
| ENST00000398554 | 1.014 | 0.8451–1.2167 | 0.8811 | – | – | – | 1.0473 | 0.8325–1.3177 | 0.6931 | – | – | – |
| ENST00000586560 | 1.0378 | 0.9024–1.1934 | 0.6031 | – | – | – | 1.0632 | 0.9061–1.2476 | 0.4523 | – | – | – |
| TCONS_00008014 | 1.0958 | 0.9789–1.2267 | 0.112 | – | – | – | 1.0174 | 0.836–1.2381 | 0.8636 | – | – | – |
| ENST00000546789 | 1.043 | 0.9185–1.1845 | 0.5161 | – | – | – | 0.6812 | 0.2873–1.615 | 0.3834 | – | – | – |
| ENST00000610270 | 1.0051 | 0.9113–1.1086 | 0.9184 | – | – | – | 0.7534 | 0.3855–1.4724 | 0.4075 | – | – | – |
| ENST00000527727 | 1.0963 | 1.0161–1.1829 | 0.0176 | 1.113 | 1.024–1.209 | 0.0113 | 0.8622 | 0.6099-1.219 | 0.4014 | – | – | – |
| Age | 1.0155 | 0.8062–1.2793 | 0.8958 | -– | -– | -– | 1.5576 | 0.9384–2.5854 | 0.0865 | – | – | – |
| SBP | 1.0357 | 1.0012–1.0713 | 0.0421 | – | – | – | 1.0625 | 1.0166–1.1105 | 0.0071 | 1.059 | 1.013–1.108 | 0.0119 |
| DBP | 1.0806 | 1.0203–1.1444 | 0.0081 | 1.095 | 1.028–1.167 | 0.0051 | 1.0554 | 0.9812–1.1352 | 0.1471 | – | – | – |
| GDM (0 as ref) | 4.5152 | 1.3698–14.8827 | 0.0132 | – | – | – | 0.7896 | 0.0896–6.9585 | 0.8315 | – | – | – |
GDM, gestational diabetes mellitus.
Functional annotation results show lncRNA biomarkers are associated with hypertension in pregnancy
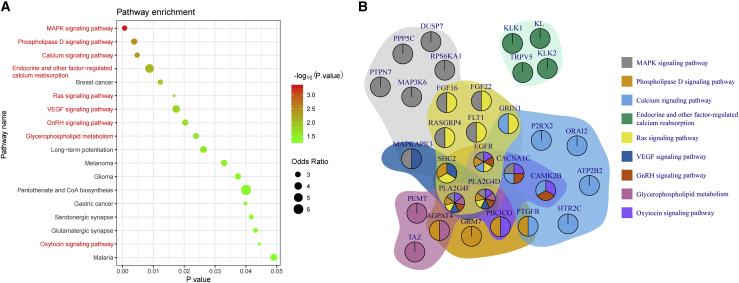
To explore the biological functions of the identified candidate lncRNAs, we extracted the neighbors of these lncRNAs in the network. In this step, we obtained 335 nonredundant mRNAs and used these mRNAs to perform functional annotation in the Enrichr web server. Of the 335 mRNAs, 40 could map into Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. We identified 18 significant pathways. Among them, 9 had been suggested to be associated with PIH or PE (marked as red text in Figure 5A). Of these pathway-related genes, only 15 genes participate in one pathway, and most of the genes are involved in at least two pathways (Figure 5B). In particular, EGFR, PLA2G4F, and PLA2G4D are multifunctional genes that participate in 6 or 7 pathways.
Figure 5.
Functional annotation of the lncRNA biomarkers of hypertension in pregnancy
(A) Bubble chart of significantly enriched KEGG pathways; the bubble size indicates the odds ratio, and the color represents the significance level. (B) The relationship of genes and pathways. The colors of each gene represent its participating pathways, and the background of each gene set indicates the corresponding pathway. (Statistics: Fisher’s exact test, p < 0.05.)
Discussion
In this nested case-control study, we obtained four novel findings. First, the serum lncRNAs of early pregnancy can serve as novel potential biomarkers for PIH, including PE. Second, we found that the severity of PIH increased as the differences in serum expression levels of 7 of the candidate lncRNAs were more pronounced, suggesting that changes in lncRNA expression can effectively reflect the extent and intensity of damage to patients owing to PIH. Third, we assessed the association between serum lncRNA levels and the individual risks of adverse pregnancy outcomes. Finally, functional enrichment analysis was performed to evaluate the possible biological effects of the candidate lncRNAs in PIH.
This present study, for the first time, demonstrated the lncRNA expression profiles of serum before the 20th week of gestation in women with PIH, including GH and PE. Our data have revealed novel dysregulated lncRNAs at the early stages of gestation (before 20 weeks) in the serum of pregnant woman who developed PIH in the third trimester of pregnancy by using a lncRNA microarray. All 8 candidate lncRNAs identified in our study show an upregulated state, which may be because the majority (23/32, 72%) of seeds used in random rank are upregulated in our PIH samples. We expect more downregulated disease markers to be validated, which could enable the identification of lncRNAs that potentially have the opposite effect in PIH. In particular, we selected 8 candidate upregulated lncRNAs based on network analysis for validation by quantitative real-time PCR, which showed that the expression levels of NR_002187, ENST00000415029, ENST00000398554, ENST00000586560, TCONS_00008014, ENST00000546789, ENST00000610270, and ENST00000527727 were increased significantly in women with PIH compared with NP women. Moreover, the AUCs for these 8 lncRNAs were more than 0.6, indicating that they are suitable as potential early biomarkers for predicting PIH. Interestingly, for the PIH group, we further found that 7 of the 8 lncRNAs, NR_002187, ENST00000398554, ENST00000586560, TCONS_00008014, ENST00000546789, ENST00000610270, and ENST00000527727, showed higher levels in the serum of pregnant women who later developed PE than in those with GH, suggesting that these 7 lncRNAs could also be used to stratify the severity of PIH.
In pregnant women, a higher BP level is associated with multiple adverse maternal and perinatal outcomes. Recent studies have shown a continuous relationship between BP level and adverse maternal outcomes,23, 24, 25 as we demonstrated in our analyses here. In our study, univariate and multivariate regression analyses indicated that patients with PIH with higher DBP and elevated expression of ENST00000527727 were at higher risk of combined FGR. With the rise in research on lncRNA function, lncRNAs have been confirmed to be associated with some diseases and developmental processes, including PIH and FGR. Koukoura et al.26 found increased H19 expression in placentas from pregnancies complicated with FGR, consistent with the results of our study. Moreover, increased expression of ENST00000415029 and increased SBP are independent risk factors for placenta accreta in patients with PIH. This is the first time that lncRNA in PIH patients has been demonstrated to be associated with placenta accreta. In brief, these results underpin antenatal care, in which a key component is related to the measurement of BP and detection of hypertension. In addition, detection of lncRNAs in early pregnancy may be a new method to prevent adverse pregnancy outcomes.
Based on our microarray data, we finally obtained 18 significant pathways by KEGG that may be associated with the biological functions of the 8 candidate lncRNAs. 9 of the 18 pathways had been confirmed to be relevant to PIH.27, 28, 29, 30, 31, 32, 33, 34, 35 Among the 9 pathways, the MAPK pathway was the most significant. Moreover, there were 3 upregulated mRNAs (PTPN7, DUSP7, and PPP5C) that encode inhibitors (PTP, MKP, and PP5) of the MAPK pathway. Some previous studies have verified that lncRNAs participate in the progression and development of PIH by modulating the MAPK signaling pathway.36,37 Therefore, we further speculate that patients with PIH were in a compensatory phase in early pregnancy and that the MAPK pathway was in an inhibited state. With the progression of the disease, the MAPK pathway was decompensated under the regulation of lncRNAs, and pregnant women subsequently developed PIH after the 20th week of gestation. In the future, we will carry out experiments to confirm these hypotheses.
In summary, the utilization of a non-invasive, efficient screening procedure to identify women at risk of PIH would be beneficial for early goal-directed preventive interventions. Using lncRNA microarray and quantitative real-time PCR analyses, our data demonstrate that the maternal circulating lncRNA profiles in serum samples from early PIH pregnancies are significantly different in early gestation compared with that from normotensive pregnancies. This finding indicates a macro predictive and functional pathophysiological correlation of circulating lncRNAs during the premature phase of the disease. Moreover, placing more emphasis on the detection of lncRNA expression in early pregnancy might reduce some adverse maternal and fetal outcomes, such as placenta accreta and FGR.
Materials and methods
Study design and patient characteristics
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University. All participating subjects were informed of the purpose of the study and signed an informed consent form. The investigation conformed to the principles outlined in the Declaration of Helsinki.
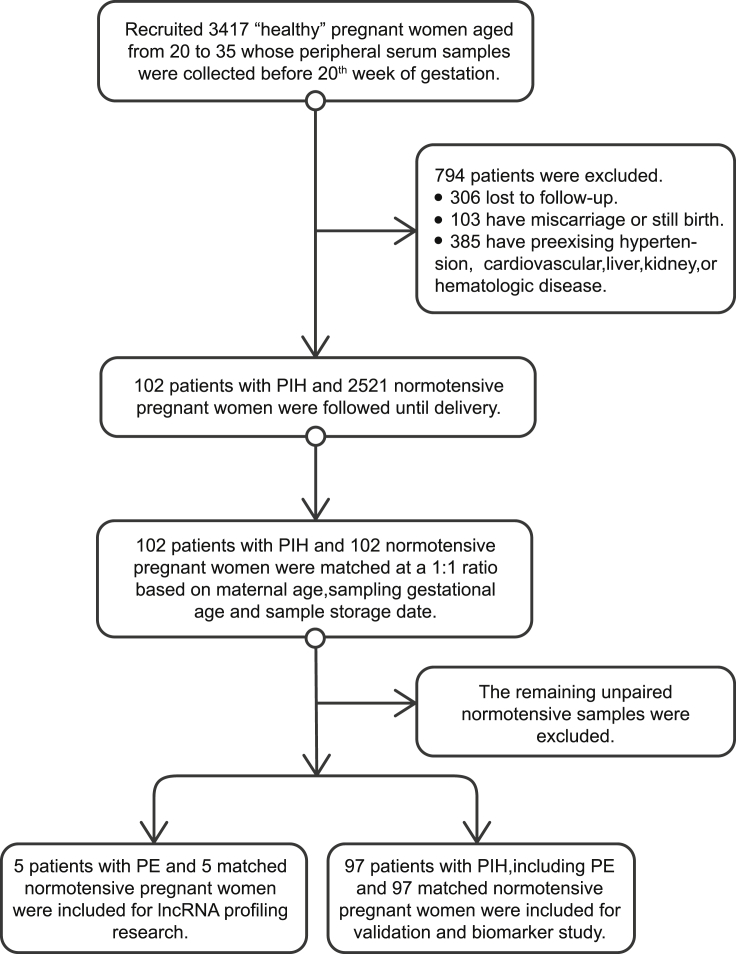
This is a nested case-control study, and we recruited a total of 3,417 “healthy” pregnant women who underwent prenatal examination and subsequently delivered at the First Affiliated Hospital of Harbin Medical University, China, between 2015 and 2019. Serum from maternal whole peripheral blood was collected before the 20th week of gestation and stored at −80°C until use.
The diagnoses of GH and PE were recognized by an evaluation of the medical records of all included women.38 GH is defined as SBP ≥ 140 mm Hg or DBP ≥ 90 mm Hg or more or both, measured twice at least 4 h apart after 20 weeks of gestation, in a woman with an antecedent normal BP. Like GH, PE is a pregnancy-specific disorder, often with new-onset proteinuria (>300 mg/d or at least +1 on sterile urine dipstick) or other maternal symptoms (cardiovascular, liver, kidney, or hematologic dysfunctions), according to the American College of Obstetricians and Gynecology criteria.2
After excluding women with pre-existing hypertension and cardiovascular, liver, kidney, or hematologic disease, patients with PIH (n = 102), which encompassed both GH (n = 47) and PE (n = 55), were included in the study, and normotensive pregnant (NP) women (n = 102) were matched at a 1:1 ratio based on maternal age, sampling gestational age and sample storage date with the PIH group.
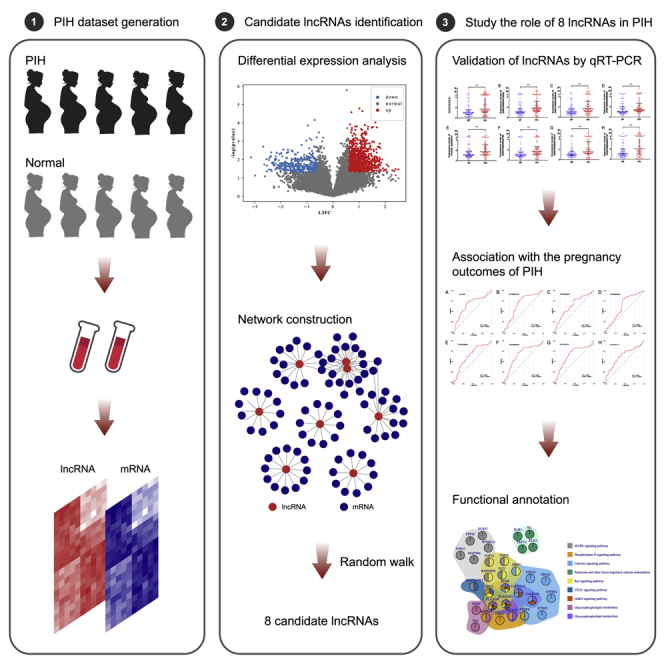
This study contained two phases: the screening phase (lncRNA profiling research) and the validation phase (biomarker study). In the screening phase, we finally included patients with PE (n = 5) and matched NP women (n = 5). The validation phase was the prospective development study of NR_002187, ENST00000415029, ENST00000398554, ENST00000586560, TCONS_00008014, ENST00000546789, ENST00000610270, and ENST00000527727 in maternal serum to predict PIH before 20 weeks’ gestation. The remaining participants, including patients with PIH (n = 97; including GH [n = 47] and PE [n = 50]) and matched normotensive women (n = 97), were included in the validation phase study. The study design is shown in Figure 6.
Figure 6.
Study design
Sample collection and RNA extraction
A total of 3∼5 mL of fasting peripheral venous blood samples was centrifuged (3,000 RCF(Reactive Centrifugal Force) for 10 min at 4°C), and the serum was separated into a 2-mL RNase-free centrifuge tube (Axygen, Union City, CA, USA) and immediately stored at −80°C until assayed. Total RNA was extracted using the RNA Isolation Kit (Axygen, Union City, CA, USA) according to the manufacturer’s instructions.
Human lncRNA and mRNA microarray
Human LncRNA Microarray V4.0 (Arraystar, USA) procedures and data analysis were performed by Kangchen Bio-tech (Shanghai, China) to detect the relative levels of lncRNA and mRNA expression. Approximately 40,173 lncRNAs and 20,730 coding transcripts could be detected. Total RNA was extracted from 5 pairs of serum samples using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA), and microarray hybridization was performed based on the manufacturer’s instructions.
Identification of differentially expressed lncRNAs and mRNAs
First, we performed log2 transformation and quantile normalization for the raw intensity of lncRNAs and mRNAs separately. During this process, we also did normalization of lncRNAs and mRNAs together and compared the results derived from two methods. Results show they have highly consistent value (Figure S1), demonstrating the expression value used in our study is robust. Next, we applied the unpaired t test and the fold change (FC) method to obtain differentially expressed lncRNAs and mRNAs. In this process, those lncRNAs/mRNAs with a p value < 0.05 and absolute FC statistic >1.5 were selected as differentially expressed lncRNAs/mRNAs. The samples were clustered using the differentially expressed lncRNAs or mRNAs by applying hierarchical clustering method based on euclidean distance. (GEO: GSE150317)
lncRNA-mRNA interaction network construction
Only those lncRNAs/mRNAs that passed the differential expression test were used for network construction. We aimed to identify significantly correlated lncRNA-mRNA pairs, which includes both the positive and negative pairs. We performed the Pearson correlation test for each lncRNA-mRNA pair and retained the significantly correlated pairs (p < 0.001). The retained lncRNA-mRNA pairs formed the final interaction network.
PIH-related lncRNA biomarker discovery
We downloaded PIH-related markers from the DISEASES website (https://diseases.jensenlab.org/Search).22 These markers included genes, miRNAs, circRNAs, and lncRNAs. We retained only genes and lncRNAs and intersected them with the differentially expressed lncRNAs/mRNAs in the microarray dataset, and the lncRNAs/mRNAs in both sets were defined as seed markers. Next, by using the seed markers and the global network, we applied the random walk method to discover potential PIH-associated lncRNAs. Each lncRNA was assigned a score, and a higher score indicated that the corresponding lncRNA was more important.
Functional enrichment analysis
We explored the biological functions of the identified candidate lncRNAs by investigating the functions of the correlated mRNAs. Specifically, we extracted protein-coding genes that interact with the 8 lncRNAs in the network and used the Enrichr web server39,40 to perform functional enrichment analysis. We selected significantly enriched KEGG pathways (p < 0.05).
Quantitative real-time RT-PCR
cDNA was synthesized from total RNA with the TOYOBO kit (Osaka, Japan). Subsequently, 40 qRT-PCR cycles are preprogrammed to perform the qRT-PCR procedure, and qRT-PCR was detected on an Applied Biosystems 7500 instrument by FastStart Universal SYBR Green Master (ROX; Roche, USA). For each 20 μL reaction, the details of PCR master mix are demonstrated in Table S4. All reactions were analyzed by using the 2–ΔΔCT method after normalization for the expression of controls, and the level of GAPDH served as an endogenous control. The primer sequences of the lncRNAs and GAPDH are shown in Table S5. The efficiency of the primers and PCR products are shown in Figures S2 and S3.
Statistical analysis
Normally distributed data are presented as the mean ± standard deviation, and nonnormally distributed data are presented as the median (interquartile range). The differences between two groups were assessed using a two-tailed unpaired t test or the Wilcoxon rank-sum test. Categorical data were compared by the chi-square test and chi-square test of continuity correction. Paired data were analyzed using paired t test, Wilcoxon matched-pairs signed-ranks test, and McNemar’s test. ROC curve analysis was performed to calculate the AUC and then predict PIH. The potential effects of the clinicopathological variables on clinical outcome were first examined using univariate analysis. Multivariate logistic regression analysis was used to further assess the effects of these variables. All statistical analyses were performed using R 3.6.2 software, and a two-sided p value of less than 0.05 was considered statistically significant.
Acknowledgments
We gratefully acknowledge Xin Li and Lining Zhang for their assist in bioinformatics analysis. We appreciate Fan Zhang’s advice on experimental design. This work was supported by Key Program of the National Natural Science Foundation of China (81830012 and 82070336), General Program of the National Natural Science Foundation of China (81670297), Youth Program of the National Natural Science Foundation of China (81900374 and 81900302), and National Key Research and Development Program of China (2017YFC1307400).
Author contribution
C.Dai designed and performed experiments, analyzed the data, and wrote the manuscript. C.Zhao, M.Xu, and X.Sui performed experiments. L.Sun, Y. Liu, M.Su, and H.Wang designed and performed experiments. J. Shi has made an outstanding contribution to the revision of the article. Y.Yuan and S.Zhang designed experiments, analyzed data, and assisted with drafting the manuscript. J. Sun and Y. Li designed experiments and corrected the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.03.010.
Contributor Information
Jingxia Sun, Email: sjxsw2013@163.com.
Yue Li, Email: ly99ly@vip.163.com.
Supplemental information
References
- 1.Umesawa M., Kobashi G. Epidemiology of hypertensive disorders in pregnancy: prevalence, risk factors, predictors and prognosis. Hypertens. Res. 2017;40:213–220. doi: 10.1038/hr.2016.126. [DOI] [PubMed] [Google Scholar]
- 2.Espinoza J., Vidaeff A., Pettker C.M., H S Gestational hypertension and preeclampsia. Obstet. Gynecol. 2019;133:e1–e25. [Google Scholar]
- 3.Brown M.A., Magee L.A., Kenny L.C., Karumanchi S.A., McCarthy F.P., Saito S., Hall D.R., Warren C.E., Adoyi G., Ishaku S., International Society for the Study of Hypertension in Pregnancy (ISSHP) Hypertensive Disorders of Pregnancy: ISSHP Classification, Diagnosis, and Management Recommendations for International Practice. Hypertension. 2018;72:24–43. doi: 10.1161/HYPERTENSIONAHA.117.10803. [DOI] [PubMed] [Google Scholar]
- 4.Honigberg M.C., Zekavat S.M., Aragam K., Klarin D., Bhatt D.L., Scott N.S., Peloso G.M., Natarajan P. Long-Term Cardiovascular Risk in Women With Hypertension During Pregnancy. J. Am. Coll. Cardiol. 2019;74:2743–2754. doi: 10.1016/j.jacc.2019.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster K., Fishburn S., Maresh M., Findlay S.C., Chappell L.C., Guideline C., Guideline Committee Diagnosis and management of hypertension in pregnancy: summary of updated NICE guidance. BMJ. 2019;366:l5119. doi: 10.1136/bmj.l5119. [DOI] [PubMed] [Google Scholar]
- 6.Mol B.W.J., Roberts C.T., Thangaratinam S., Magee L.A., de Groot C.J.M., Hofmeyr G.J. Pre-eclampsia. Lancet. 2016;387:999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 7.Rana S., Lemoine E., Granger J.P., Karumanchi S.A. Preeclampsia: Pathophysiology, Challenges, and Perspectives. Circ. Res. 2019;124:1094–1112. doi: 10.1161/CIRCRESAHA.118.313276. [DOI] [PubMed] [Google Scholar]
- 8.Levine R.J., Lam C., Qian C., Yu K.F., Maynard S.E., Sachs B.P., Sibai B.M., Epstein F.H., Romero R., Thadhani R., Karumanchi S.A., CPEP Study Group Soluble endoglin and other circulating antiangiogenic factors in preeclampsia. N. Engl. J. Med. 2006;355:992–1005. doi: 10.1056/NEJMoa055352. [DOI] [PubMed] [Google Scholar]
- 9.Herraiz I., Simón E., Gómez-Arriaga P.I., Martínez-Moratalla J.M., García-Burguillo A., Jiménez E.A., Galindo A. Angiogenesis-Related Biomarkers (sFlt-1/PLGF) in the Prediction and Diagnosis of Placental Dysfunction: An Approach for Clinical Integration. Int. J. Mol. Sci. 2015;16:19009–19026. doi: 10.3390/ijms160819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho L., van Dijk M., Chye S.T.J., Messerschmidt D.M., Chng S.C., Ong S., Yi L.K., Boussata S., Goh G.H.Y., Afink G.B. ELABELA deficiency promotes preeclampsia and cardiovascular malformations in mice. Science. 2017;357:707–713. doi: 10.1126/science.aam6607. [DOI] [PubMed] [Google Scholar]
- 11.Panaitescu B., Romero R., Gomez-Lopez N., Pacora P., Erez O., Vadillo-Ortega F., Yeo L., Hassan S.S., Hsu C.D. ELABELA plasma concentrations are increased in women with late-onset preeclampsia. J. Matern. Fetal Neonatal Med. 2020;33:5–15. doi: 10.1080/14767058.2018.1484089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinrouweler C.E., Wiegerinck M.M., Ris-Stalpers C., Bossuyt P.M., van der Post J.A., von Dadelszen P., Mol B.W.J., Pajkrt E., EBM CONNECT Collaboration Accuracy of circulating placental growth factor, vascular endothelial growth factor, soluble fms-like tyrosine kinase 1 and soluble endoglin in the prediction of pre-eclampsia: a systematic review and meta-analysis. BJOG. 2012;119:778–787. doi: 10.1111/j.1471-0528.2012.03311.x. [DOI] [PubMed] [Google Scholar]
- 13.Hassan S.S., Gomez-Lopez N. Reducing maternal mortality: can elabela help in this fight? Lancet. 2019;394:8–9. doi: 10.1016/S0140-6736(19)30543-4. [DOI] [PubMed] [Google Scholar]
- 14.Noori M., Donald A.E., Angelakopoulou A., Hingorani A.D., Williams D.J. Prospective study of placental angiogenic factors and maternal vascular function before and after preeclampsia and gestational hypertension. Circulation. 2010;122:478–487. doi: 10.1161/CIRCULATIONAHA.109.895458. [DOI] [PubMed] [Google Scholar]
- 15.Hirashima C., Ohkuchi A., Nagayama S., Suzuki H., Takahashi K., Ogoyama M., Takahashi H., Shirasuna K., Matsubara S. Galectin-1 as a novel risk factor for both gestational hypertension and preeclampsia, specifially its expression at a low level in the second trimester and a high level after onset. Hypertens. Res. 2018;41:45–52. doi: 10.1038/hr.2017.85. [DOI] [PubMed] [Google Scholar]
- 16.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 17.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 18.He X., He Y., Xi B., Zheng J., Zeng X., Cai Q., Ouyang Y., Wang C., Zhou X., Huang H. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS ONE. 2013;8:e81437. doi: 10.1371/journal.pone.0081437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J., Qian Y., Cheng Q., Yang J., Ding H., Jia R. Over expression of long non-coding RNA uc.187 induces preeclampsia-like symptoms in pregnancy rats. Am. J. Hypertens. 2020;33:439–451. doi: 10.1093/ajh/hpaa011. [DOI] [PubMed] [Google Scholar]
- 20.Qi P., Zhou X.Y., Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol. Cancer. 2016;15:39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y., Hou Y., Lv N., Liu Q., Lin N., Zhao S., Chu X., Chen X., Cheng G., Li P. Circulating lncRNA BC030099 Increases in Preeclampsia Patients. Mol. Ther. Nucleic Acids. 2019;14:562–566. doi: 10.1016/j.omtn.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pletscher-Frankild S., Pallejà A., Tsafou K., Binder J.X., Jensen L.J. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015;74:83–89. doi: 10.1016/j.ymeth.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 23.von Dadelszen P., Payne B., Li J., Ansermino J.M., Broughton Pipkin F., Côté A.M., Douglas M.J., Gruslin A., Hutcheon J.A., Joseph K.S., PIERS Study Group Prediction of adverse maternal outcomes in pre-eclampsia: development and validation of the fullPIERS model. Lancet. 2011;377:219–227. doi: 10.1016/S0140-6736(10)61351-7. [DOI] [PubMed] [Google Scholar]
- 24.Payne B.A., Hutcheon J.A., Ansermino J.M., Hall D.R., Bhutta Z.A., Bhutta S.Z., Biryabarema C., Grobman W.A., Groen H., Haniff F., miniPIERS Study Working Group A risk prediction model for the assessment and triage of women with hypertensive disorders of pregnancy in low-resourced settings: the miniPIERS (Pre-eclampsia Integrated Estimate of RiSk) multi-country prospective cohort study. PLoS Med. 2014;11:e1001589. doi: 10.1371/journal.pmed.1001589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magee L.A., Singer J., Lee T., McManus R.J., Lay-Flurrie S., Rey E., Chappell L.C., Myers J., Logan A.G., von Dadelszen P. Are blood pressure level and variability related to pregnancy outcome? Analysis of control of hypertension in pregnancy study data. Pregnancy Hypertens. 2020;19:87–93. doi: 10.1016/j.preghy.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Koukoura O., Sifakis S., Zaravinos A., Apostolidou S., Jones A., Hajiioannou J., Widschwendter M., Spandidos D.A. Hypomethylation along with increased H19 expression in placentas from pregnancies complicated with fetal growth restriction. Placenta. 2011;32:51–57. doi: 10.1016/j.placenta.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 27.Irani R.A., Xia Y. The functional role of the renin-angiotensin system in pregnancy and preeclampsia. Placenta. 2008;29:763–771. doi: 10.1016/j.placenta.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deborde S., Schofield J.N., Rademacher T.W. Placental GPI-PLD is of maternal origin and its GPI substrate is absent from placentae of pregnancies associated with pre-eclampsia. J. Reprod. Immunol. 2003;59:277–294. doi: 10.1016/s0165-0378(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 29.Boeldt D.S., Krupp J., Yi F.X., Khurshid N., Shah D.M., Bird I.M. Positive versus negative effects of VEGF165 on Ca2+ signaling and NO production in human endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 2017;312:H173–H181. doi: 10.1152/ajpheart.00924.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haché S., Takser L., LeBellego F., Weiler H., Leduc L., Forest J.C., Giguère Y., Masse A., Barbeau B., Lafond J. Alteration of calcium homeostasis in primary preeclamptic syncytiotrophoblasts: effect on calcium exchange in placenta. J. Cell. Mol. Med. 2011;15:654–667. doi: 10.1111/j.1582-4934.2010.01039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taufield P.A., Ales K.L., Resnick L.M., Druzin M.L., Gertner J.M., Laragh J.H. Hypocalciuria in preeclampsia. N. Engl. J. Med. 1987;316:715–718. doi: 10.1056/NEJM198703193161204. [DOI] [PubMed] [Google Scholar]
- 32.Versmissen J., Mirabito Colafella K.M., Koolen S.L.W., Danser A.H.J. Vascular Cardio-Oncology: Vascular Endothelial Growth Factor inhibitors and hypertension. Cardiovasc. Res. 2019;115:904–914. doi: 10.1093/cvr/cvz022. [DOI] [PubMed] [Google Scholar]
- 33.Estienne A., Bongrani A., Reverchon M., Ramé C., Ducluzeau P.H., Froment P., Dupont J. Involvement of Novel Adipokines, Chemerin, Visfatin, Resistin and Apelin in Reproductive Functions in Normal and Pathological Conditions in Humans and Animal Models. Int. J. Mol. Sci. 2019;20:4431. doi: 10.3390/ijms20184431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly R.S., Croteau-Chonka D.C., Dahlin A., Mirzakhani H., Wu A.C., Wan E.S., McGeachie M.J., Qiu W., Sordillo J.E., Al-Garawi A. Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics. 2017;13:7. doi: 10.1007/s11306-016-1149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szczepanska-Sadowska E., Cudnoch-Jedrzejewska A., Wsol A. The role of oxytocin and vasopressin in the pathophysiology of heart failure in pregnancy and in fetal and neonatal life. Am. J. Physiol. Heart Circ. Physiol. 2020;318:H639–H651. doi: 10.1152/ajpheart.00484.2019. [DOI] [PubMed] [Google Scholar]
- 36.Jiao S., Wang S.Y., Huang Y. LncRNA PRNCR1 promoted the progression of eclampsia by regulating the MAPK signal pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:3635–3642. doi: 10.26355/eurrev_201806_15240. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J., Zhao Z.M. LncRNA HOXD-AS1 promotes preeclampsia progression via MAPK pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:8561–8568. doi: 10.26355/eurrev_201812_16618. [DOI] [PubMed] [Google Scholar]
- 38.Milic N.M., Codsi E., Butler Tobah Y.S., White W.M., Kattah A.G., Weissgerber T.L., Saiki M., Parashuram S., Vaughan L.E., Weaver A.L. Electronic Algorithm Is Superior to Hospital Discharge Codes for Diagnoses of Hypertensive Disorders of Pregnancy in Historical Cohorts. Mayo Clin. Proc. 2018;93:1707–1719. doi: 10.1016/j.mayocp.2018.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90-7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zelenski J.M., Whelan D.C., Nealis L.J., Besner C.M., Santoro M.S., Wynn J.E. Personality and affective forecasting: trait introverts underpredict the hedonic benefits of acting extraverted. J. Pers. Soc. Psychol. 2013;104:1092–1108. doi: 10.1037/a0032281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.