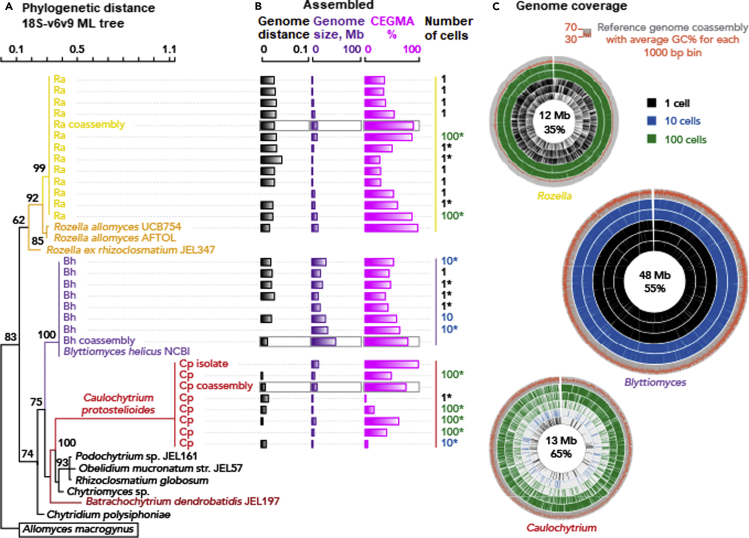
Figure 4.
Intra- and interspecific variabilities
(A) Cryptomycota and Chytridiomycota 18S rDNA (region v6 to v9) ML tree based on the HKY85 nucleotide substitution model with bootstrap values shown above 60%.
(B) Assembled: Genome distance was calculated using GGDC formula 2, designed for incomplete isolated genomes (Auch et al., 2010; Meier-Kolthoff et al., 2013); the genome size shows the degree of variation in genome recovery between single-cell and multiple-cell sorts, and the core eukaryotic gene mapping approach (CEGMA) value reflects genome completeness. ∗Assemblies used for the genome coverage Circos plots. For all the other species, see Figure 5.
(C) Genome coverage shows mapping in 1,000-bp bins from individual select single-cell or multiple-cell libraries to the reference coassembled species genome.
See also Figure S6.