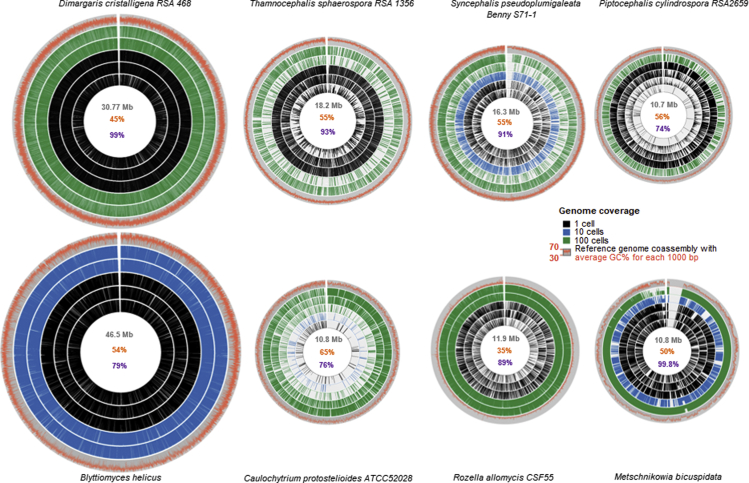
Figure 5.
Intra- and interspecific genome coverage variability
Each species Circos map is scaled relative to the largest genome (Blyttiomyces helicus) true to size. Reference genome (coassembly) is shown as the outer gray circle. Coassembly GC% plotted as a red line over the gray circle. 1,000-bp bins were plotted against reference co-assembly genome and scaled proportionally to the co-assembly genome size. Five representative libraries from single-cell (black); 10-cell, 30-cell, or 50-cell depending on the species (blue); and 100-cell sorts were chosen for each species. For each cell sort category one worst case, one average case, and one best case were picked when available. In the middle of the plot numbers are coassembly genome in gray, GC% in red, and CEGMA completeness in purple. See also Figures S6 and S8.